- 1Department of Diagnostic and Biomedical Sciences, The University of Texas Health Science Center at Houston, School of Dentistry, Houston, TX, United States
- 2Department of Cancer Systems Imaging, The University of Texas MD Anderson Cancer Center, Houston, TX, United States
- 3MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences, Houston, TX, United States
- 4Department of Anthropology, Washington University in St. Louis, St. Louis, MO, United States
- 5Departments of BioSciences and Bioengineering, Rice University, Houston, TX, United States
Colorectal cancer (CRC) is the third leading cause of cancer-related deaths among both men and women in the United States. Early detection and surgical removal of high-risk lesions in the colon can prevent disease from developing and spreading. Despite implementation of programs aimed at early detection, screening colonoscopies fail to detect a fraction of potentially aggressive colorectal lesions because of their location or nonobvious morphology. Optical colonoscopies, while highly effective, rely on direct visualization to detect changes on the surface mucosa that are consistent with dysplasia. Recent advances in endoscopy techniques and molecular imaging permit microscale visualization of the colonic mucosa. These technologies can be combined with various molecular probes that recognize and target heterogenous lesion surfaces to achieve early, real-time, and potentially non-invasive, detection of pre-cancerous lesions. The primary goal of this review is to contextualize existing and emergent CRC surface biomarkers and assess each’s potential as a candidate marker for early marker-based detection of CRC lesions. CRC markers that we include were stratified by the level of support gleaned from peer-reviewed publications, abstracts, and databases of both CRC and other cancers. The selected biomarkers, accessible on the cell surface and preferably on the luminal surface of the colon tissue, are organized into three categories: (1) established biomarkers (those with considerable data and high confidence), (2) emerging biomarkers (those with increasing research interest but with less supporting data), and (3) novel candidates (those with very recent data, and/or supportive evidence from other tissue systems). We also present an overview of recent advances in imaging techniques useful for visual detection of surface biomarkers, and discuss the ease with which these methods can be combined with microscopic visualization.
Introduction
Colorectal cancer (CRC) remains the third leading cause of cancer-related deaths among both men and women in the United States, primarily because asymptomatic early disease often has progressed to an aggressive disease before it is diagnosed (1). Current American Medical Association (AMA) guidelines for early detection of CRC, adopted in 2018, recommend screening by fecal occult blood testing, sigmoidoscopy, or optical colonoscopy. In some health centers, CT colonography (“virtual colonoscopy”) also is available. Screening recommendations include starting at age 50, 45 for African Americans, and continuing until age 75 (2). As illustrated by the recent shocking death of actor Chadwick Boseman from colon cancer at age 43, early onset CRC increasingly is being diagnosed in younger patients who present with stage III/IV cancers (3). When diagnosed early, the 5-year survival rate of CRC is 90%, but the survival rate drops drastically with a later-stage diagnosis. According to the American Cancer Society (ACS), the 5-year survival rate of CRC for patients diagnosed at stage III and IV are 71% and 14%, respectively (1).
Several modalities are in current clinical practice for the screening of adenomatous polyps and CRC. Optical colonoscopy is the most widely used CRC screening technique, and the most commonly performed endoscopic procedure in the United States (4). Despite recent efforts to implement screening programs, full adherence to colonoscopy screening has proved elusive (5). It is estimated that 35% of adults in the United States who are eligible for colonoscopy screening remain unscreened (6). A survey of over 425 ethnically and racially diverse adults ≥ 50 years of age reported fear of embarrassment, fear of getting AIDS, fear of procedural pain, and older age as reasons to avoid colonoscopy (7). In addition, aversion to bowel preparation and fear of invasive procedures are among other barriers to undergoing a colonoscopy (8).
Alternative minimally invasive screening methods, such as fecal and blood tests, rely on changes in the tumor microenvironment to suggest the presence of CRC lesions, but these methods seldom detect early lesions in time to change the course of disease (9). While the new-generation immunochemical fecal occult blood tests (iFOBT) demonstrate a significantly higher diagnostic performance when compared with the earlier guaiac-based gFOBTs, a positive result still requires the use of a follow up colonoscopy (9).
CRC is a heterogeneous disease, arising from accumulations of various stochastic genetic and epigenetic alterations, which first lead to polyp growth and then, in some cases, to dysplasia and carcinogenesis. Polyps with similar morphological features typically follow different molecular paths to overgrowth. This observation is true even for lesions in hereditary forms of CRC such as those that occur in Lynch Syndrome (10). Because optical colonoscopies depend on the direct visualization of dysplastic tissue, lesions must reach a certain size and have an atypical appearance to be detected during the endoscopy procedure.
Sessile serrated adenomas/polyps (SSA/P), for instance, are usually flat, depressed, or similar in color to the surrounding healthy mucosa. These polyps can be under-recognized or mistaken for inverted colonic diverticulum (ICD) because of their shiny, smooth-surfaced appearance and a central indentation (11). It is estimated that among those who undergo optical colonoscopy, 3.5 per 1000 persons develop CRC adenomas that were missed during the baseline colonoscopy (12).
Missed adenomas during colonoscopies are relatively common: a meta-analysis of 43 publications and over 15,000 tandem colonoscopies estimated the miss rate to be 26%, 9%, 27%, and 34% for adenomas, advanced adenomas, serrated polyps, and flat adenomas, respectively (4). Although invasive carcinomas are present in only 5.4% of flat elevated lesions, 73.2% of flat depressed lesions between 10-20mm grow rapidly and become invasive carcinomas at an early stage (13). Given that the standard recommended screening interval is 10 years, a missed flat lesion could develop readily into advanced disease in the interval between serial colonoscopies (14). Given the high prevalence of CRC and the limitations of optical colonoscopy, the development of more sensitive, specific, and less invasive diagnostic techniques is warranted to identify “difficult to see” lesions and improve clinical outcomes for CRC patients.
Biomarkers in Detection of Colorectal Lesions
Recent advances in molecular technologies have led to the discovery of multiple biomarkers that might facilitate early detection of colorectal lesions (15). The screening tests based on diagnostic biomarkers may be classified broadly as eliminating metabolites and circulating. Eliminating metabolite tests offer the opportunity to detect hemoglobin (through a peroxidase reaction), DNA or RNA, proteins such as M2 pyruvate kinase (M2-PK), and microbiome such as Streptococcus bovis, Fusobacterium nucleatum and Helicobacter pylori in stool (16). The circulating tests, used primarily as patient monitoring tools, are designed to identify CRC-specific biomarkers such as Carcinoembryonic Antigen (CEA) and carbohydrate antigen (CA19-9) in blood circulation (17).
Reasons for slow adoption of biomarker-based screening tests include the need for high throughput genotyping and phenotyping techniques, and challenges in achieving regulatory requirements for routine clinical use (18). The current biomarker screening tests require laboratory testing that is time-consuming and inconvenient. As histological analysis of biopsies remains the gold standard for a definitive CRC diagnosis (19), all abnormalities identified through biomarker tests next must be verified by colonoscopy during which removal of visible lesions may occur. To minimize the number of invasive procedures, it is typical for all suspicious lesions to be removed during the colonoscopy procedure. The fixation of the biopsy specimen, histological staining, and evaluation of the results complete the screening workflow.
The ideal biomarker(s) for early detection of pre-cancerous colorectal lesions should be highly sensitive and specific, expressed reliably on pre-cancerous or early stage lesions, and allow seamless integration into existing clinical protocols to eliminate unnecessary steps in the screening workflow (20). Because most colorectal lesions originate from the epithelial cells lining the luminal surface of the colon or their stem/progenitor cell precursors, biomarkers expressed on the polyp or lesion surface present an opportunity for early, real-time, in vivo detection of pre-cancerous lesions with high contrast against nearby healthy tissue (Figure 1). In an ideal world, a single perfect biomarker would instantly identify all lesions, regardless of phenotype. Unfortunately, the high degree of heterogeneity of colorectal polyps/lesions, both in stage and mutations, makes this notion unlikely. It is possible, however, to envision a minimum suite of biomarkers that would collectively identify the majority, if not all, of the lesions with a high risk of becoming cancerous. The necessary chemistries for tethering antibodies, aptamers, or other high-affinity recognition molecules to a contrast agent would still apply, regardless of the selected biomarker(s). With this motivation, we surveyed the literature to identify new potential early surface markers that could be combined with the established surface markers to create a future “visualization panel” that could be combined with colonoscopy or emerging methodologies to revolutionize CRC screening.
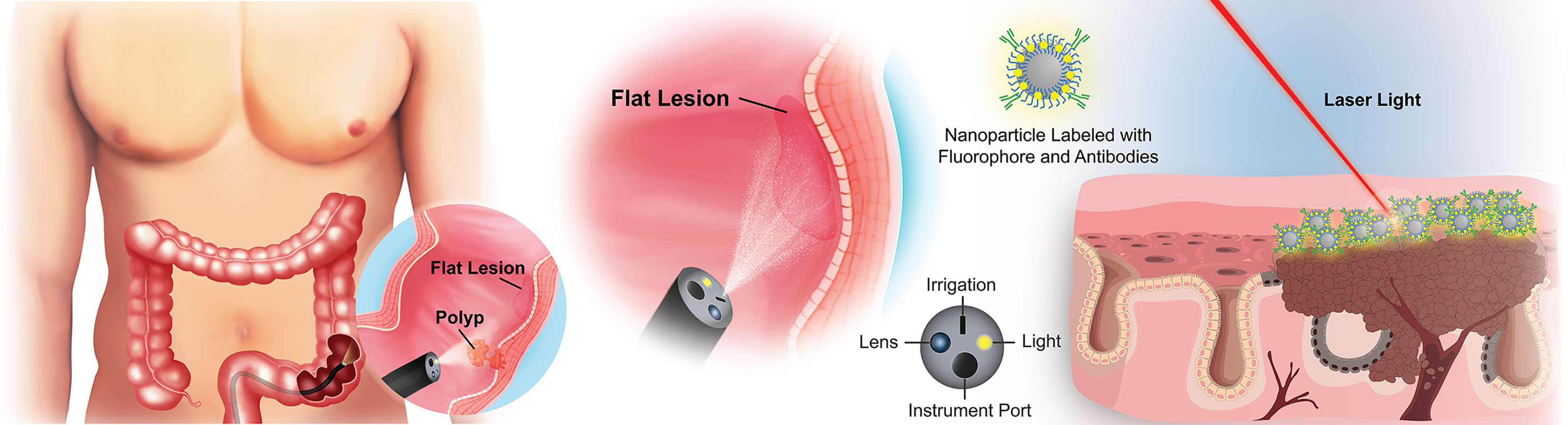
Figure 1 Overview of targeted imaging approaches to identifying flat colorectal lesions. For lesions that are not easily identified by traditional white light colonoscopy, a modified endoscope can both deliver high-contrast particles across tissue surfaces, and identify their retention through fluorescence barcoding, or via other sensitive visualization methods. Such “nanobeacons” could reveal otherwise undetectable early lesions.
Literature Review for Early Surface Biomarkers of CRC
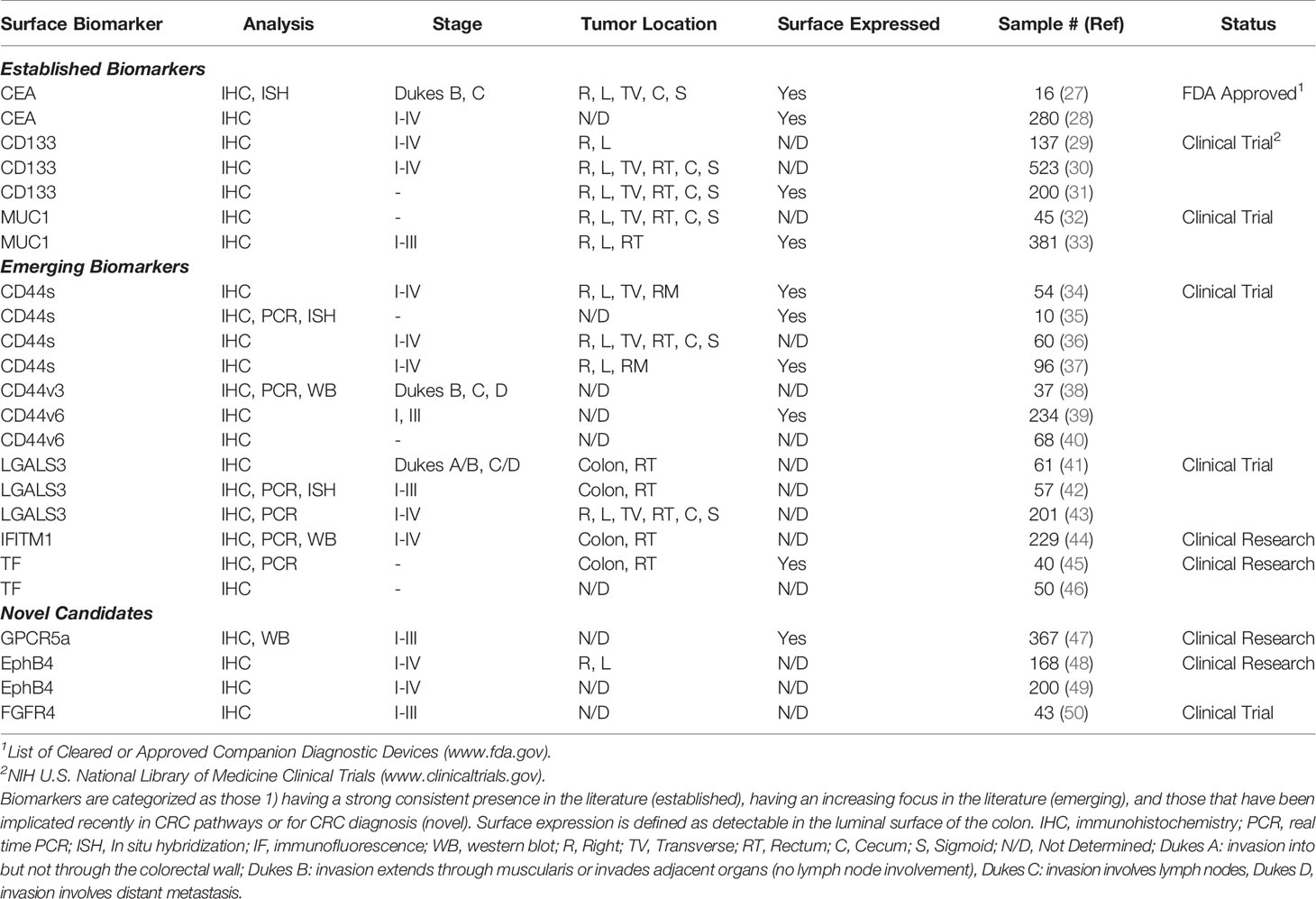
We collected 2,749 abstracts relating to proteins or other cell-surface CRC biomarkers from a variety of databases using evolving search terms and their synonyms: “colorectal cancer”, “biomarker” “cell-surface protein”, “adenocarcinoma”, “EMT”, and “cancer stem cell”. These terms were searched in AACR abstracts, ASCO Meeting Library, Cancer Epidemiology, Biomarkers, Prevention, Cochrane Library, EBSCO Databases, Embase, and PubMed (Figure 2). With an intention of focusing on relatively new discoveries, we limited results to exclude studies published before the year 2000 but did not exclude based on method of evaluating protein expression, such that a diversity of methods and results could be considered for each biomarker. Furthermore, studies that illuminated new implications in oncogenic pathways or other systems of interest were reviewed. 342 biomarkers were selected and evaluated further. From these 342 biomarkers, 44 were identified based on biomarker expression and excluded those with focus on prognosis or on other cancers. These candidate biomarkers were further refined to 10 CRC biomarkers after excluding non-surface cellular biomarkers and those with unconfirmed high expression levels or lack of diagnostic applications.
Surface Biomarkers
Recent studies on CRC biomarker discovery have provided excellent overview of diagnostic, predictive, and prognostic biomarkers identified through proteomics research (21), as well as those with therapy applications (22). Epidermal growth factor receptor (EGFR) (23), vascular endothelial growth factor (VEGF) (24), and mesenchymal-epithelial transition (c-MET or MET) (25) are among the well-established pathways that play an important role in the biology of CRC. A number of therapy agents targeting molecular biomarkers in these pathways, including cetuximab, panitumumab, and bevacizumab, have been developed and approved by the Food and Drug Administration (FDA) (26). However, these biomarkers are not currently being targeted clinically for early detection of CRC.
The selected biomarkers in this review are organized into three categories (1): established biomarkers, which have high supporting evidence in the literature and/or are currently in use in clinical practice (2), emerging biomarkers, which have had an increasing research focus and/or some evidence of utility in the pre-clinical literature, and (3) novel candidates, which have been recently implicated in CRC pathways or for CRC diagnosis, but for which there is limited pre-clinical or clinical evidence. It is important to note that the biomarkers listed in our review are not a comprehensive list but are representative of biomarkers identified by our selection criteria. We focused specifically on extracellular markers with sufficient surface accessibility in the colon to allow their identification by a functionalized probe. All biomarkers are listed in Table 1, with supporting literature evidence, and illustrated in Figure 3 as a comparison of prominent molecular features and relative size.
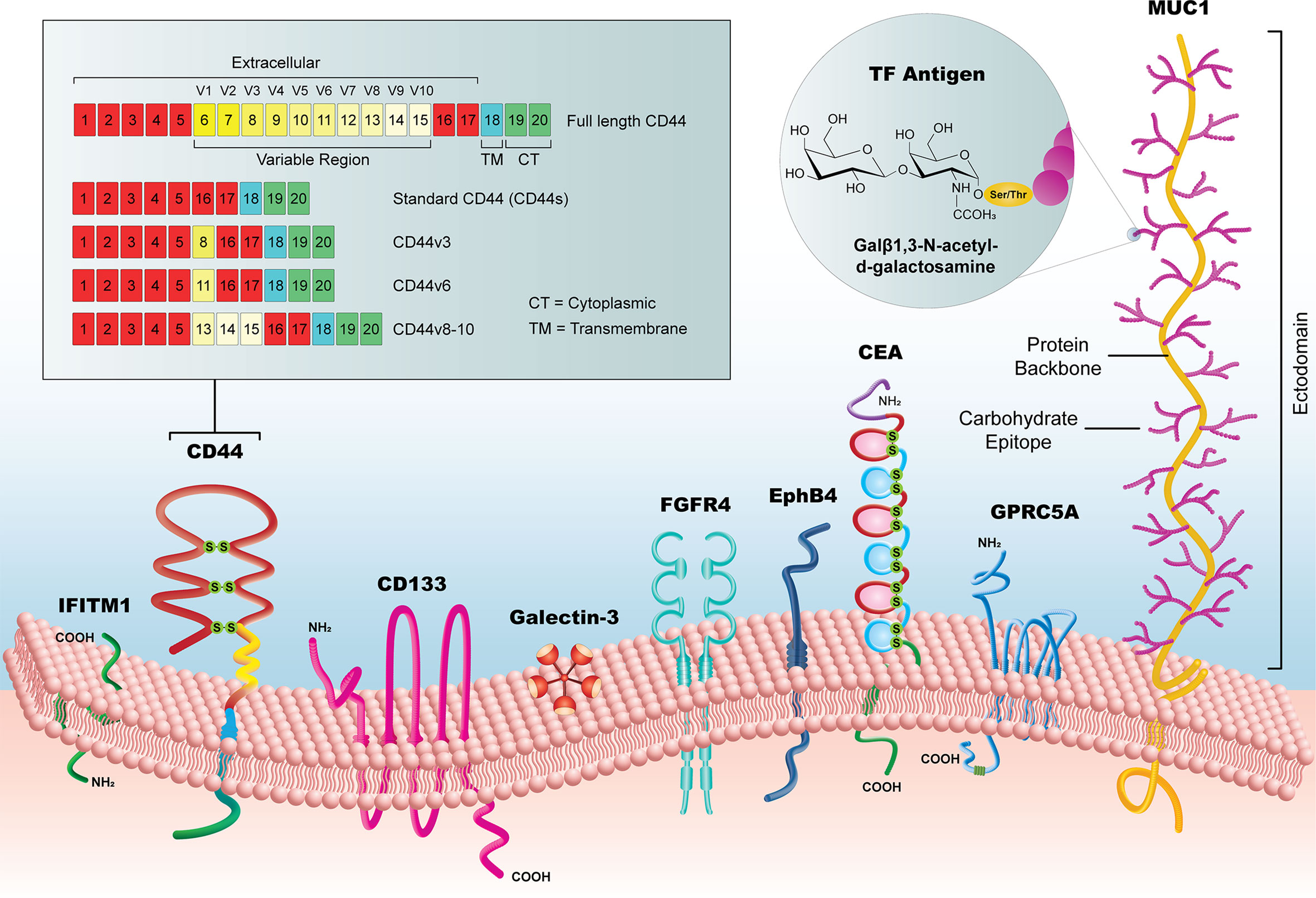
Figure 3 Summary of new candidate biomarkers, highlighted in this review. Although substantially variable in size, from the < 1kDa TF Antigen to the >1 MDa MUC1 glycoprotein, these surface biomarkers offer a potential to reveal early CRC lesion development, particularly when used as a library of markers that could identify lesions with heterogeneous subpopulations.
Established Biomarkers
CEA
Carcinoembryonic antigen (CEA) is a glycoprotein that has been implicated as a CRC biomarker since 1965. It is a member of the CEACAM family, which contains 12 independent genes. CEA, or CEACAM5, is associated with the cell membrane through a glycosylphosphatidylinositol (GPI) anchor (51). CEA is involved in mediating homophilic and heterophilic interactions, and has an understood role in cell-cell adhesion, inter and intracellular signaling, vascularization, and a variety of other physiological functions (51, 52).
CEA is expressed in healthy fetal and adult tissue in various organ systems, including the digestive tract (53). A 1993 study found that both mRNA and tissue-level expression of CEA in paired healthy and cancerous resected colon samples had a similar expression gradient in colonic crypts. The maximum amount of CEA expression was in the upper third of the colonic crypt, with a gradient of decreasing amount towards the base (27). While CEA is expressed in healthy tissue, overexpression of CEA in a variety of cancers, including breast, gastrointestinal, and respiratory, has had a role in diagnosis and prognosis since its discovery (52).
Immunohistochemical studies have highlighted the role that CEA can play as a diagnostic biomarker in CRC. Matched FFPE samples of normal colorectal mucosa and primary carcinoma stained for various biomarkers found the greatest difference in degree of expression for CEA (28). This study determined that the sensitivity and specificity of CEA used for CRC imaging would be 93.7% and 96.1% respectively. Because of this, CEA-detecting diagnostic methods have been developed and used on mouse models with human breast and pancreatic cancer and have been successful in detecting primary tumors and micrometastases (54).
The utility of serum CEA levels as a prognostic marker was demonstrated multiple times in the past 20 years (55, 56). Elevated CEA levels in serum was linked to poor prognosis in CRC patients, and was shown to independently predict higher overall mortality in both metastatic and nonmetastatic CRC patients, and across all cancer stages (57). Notably, elevated CEA is also observed in IBD and other inflammatory diseases, reducing its utility as a single marker for early CRC. Its use in combination with other candidates in this review may enhance its specificity.
CD133
CD133, also known as Prominin-1 or AC133, is a pentaspan transmembrane glycoprotein protein that often is recognized as a stem cell marker for normal and caner tissue (58). Encoded by gene PROM1, CD133 has some cell-cycle dependence, and its expression may be promoted in hypoxic environments, a key characteristic of the tumor microenvironment and of the colon (59, 60). CD133 is linked to tumor-forming and tumor-growing related processes, and is associated with an increase in tumor volume and tumorigenicity (61).
Although CD133 often is cited as a cancer stem cell marker (59), its role as a stem cell biomarker was disputed in one study. Yeung and Mortensen found that only one in 262 CD133+ cells can initiate tumors (62). Others question the efficacy of CD133 as an EMT marker as well, as it was found to localize to the nucleus during EMT, and return to the cell membrane when MET is induced (63). These findings indicate that cell-surface expression of CD133 is more likely in the early stages of CRC, which makes it a more useful tool for CRC screening.
Tissue expression of CD133 in CRC, though not without controversy, makes a strong case for its use as a CRC biomarker. Immunohistochemistry (IHC) staining of colon cancer tissue removed from a cohort of 137 patients showed high CD133 expression in 64% of stage I, 28% of stage II, and 54% of stage III tumors (29). More modest IHC results set the CD133 positive staining at 24.5% (30). However, CD133 presence in the healthy colon epithelium is controversial: Shmelkov et al. found that while CD133 is expressed ubiquitously in the majority of the human colon cancer cells, the healthy human colon also expresses CD133 robustly (58). It is worth noting that because of the glycosylation pattern of CD133, the choice of antibodies may impact the detection of this biomarker in colorectal tissue and cell line (64). Nonetheless, a metanalysis by Abbasian et al. showed higher levels of CD133 in cells derived from primary tumors than cells originated from cell lines (61).
While the relationship between CD133 expression and colorectal tumor volume or stage varies among studies (30, 31, 65), a meta-analysis of 27 tumorgenicity studies found a twofold increase in tumor volume in cells that expressed CD133 or CD44. Interestingly, co-expression of CD133 and CD44 was associated with sevenfold higher tumorgenicity (61), further implicating their combined application as surface biomarkers for in vivo screening of CRC.
MUC1
The non-neoplastic (normal) surface of the colon is lined with various types of mucins, secreted by specialized epithelial cells that protect the lining of the epithelium against invading bacteria and pathogens (66). In developing neoplasms, however, the mucin layer is decreased, presenting an opportunity for detection of surface accessible biomarkers (67). MUC1 is a large, highly accessible, transmembrane mucin that is expressed on the apical surface of many luminal epithelial cells of the respiratory, urinary, gastrointestinal, and reproductive tract (68). The mature glycosylated form of MUC1 has an estimated weight of 250-500kDa. In most epithelial cancers, including CRC, MUC1 is overexpressed and displays aberrant glycosylation (69). Thus, the glycosylated ectodomain of MUC1 can serve as a potential surface biomarker for in-vivo screening of CRC.
In one study, the immunohistochemical analysis of CRC tissues from 45 patients revealed positive expression of MUC1 in 55.6% of CRC tissue and in 0% of nontumor tissue adjacent to carcinoma (32). In another histopathological study of tissues removed from 381 CRC patients, it was discovered that MUC1 is expressed in 64% of the CRC tissue (33). The high expression of mature MUC1 mucin correlates with TNM stage, depth of invasion, lymph node metastasis, and poor overall survival (OS) outcome in CRC patients (70). These findings suggest that MUC1 is a promising prognostic and diagnostic surface biomarker of CRC.
Emerging Biomarkers
CD44 Variant Isoforms
CD44 is a 85-200 kDa transmembrane glycoprotein that has various functions in cell division, migration, adhesion, and signaling (71–75). The broad variation in molecular weights is partially attributable to multiple aberrant splice variants, which may include several exons, and multiple variations in glycosylation state (76). The standard form of CD44 (CD44s) is made of common exons 1-5 and 16-20 that when spliced together form a transcript that encodes the isoform with the smallest molecular weight (85-90 kDa). The other 10 variable exons (6–15), also known as v1-v10, are excluded either completely from CD44s or are spliced and inserted between exons 5 and 16. Both the standard form and the higher-molecular weight isoforms (CD44v) of CD44 are expressed on the cell membrane, strengthening their status as potential surface biomarkers for early visual detection of CRC.
CD44s, the most abundant form of CD44, plays an important role in cellular adhesion and three dimensional organ and tissue maintenance (77). Because of its important role in cell-to-cell adhesion, CD44s dysregulation is implicated in multiple cancers, including breast, prostate, pancreatic, and colon cancer (78). Colorectal tissue expression levels of CD44s vary notably among studies. Several studies have reported increased expression of CD44s in CRC tissue compared to adjacent normal tissue (35, 79), with CD44s expression localized at the base of colonic crypts in a healthy colon and on the luminal side of crypt in CRC tissue (34, 35). Others have shown loss or down-regulation of CD44s during transformation from normal mucosa to colon carcinoma (80). The inconsistent expression levels of CD44s in colorectal adenomas, combined with weak to moderate expression in normal colon epithelium, undermine the efficacy of CD44s as a diagnostic biomarker for CRC.
CD44v6, another splice variant of CD44 containing exon v6, has a higher affinity for hyaluronic acid than CD44s, further implicating this CD44 variant in tumor pathology (81). The immunohistochemical staining of normal (n=25) and hyperplastic colon tissue (n=45) has shown that the expression of CD44v6 in the healthy colon is rare and limited to the base of colonic crypts (39). This is consistent with the finding that CD44v6 is found in only one out of 23 benign serous effusions from adenocarcinomas (39). In normal and hyperplastic colon tissue, CD44v6 expression was absent or sporadic in the crypts or lumen epithelium, but showed as strong/diffuse staining in the dysplastic surface epithelium of tubular adenomas (86%, 49/57), tubulovillous adenomas (84%, 21/25), and villous adenomas (100%, 9/9) (39). The low expression of CD44v6 in the healthy colon and its increased expression in the dysplastic surface epithelium in adenomas makes this biomarker a strong candidate for visual CRC screening technologies.
CD44v3 is the heparan sulfate proteoglycan domain of CD44 containing the sequence encoded by variant exon 3 (82). This isoform is thought to contribute to malignant behavior in human colon through reduced binding affinity for heparan sulfate, a molecule that enhances the invasive capacity of colon cancer cells (38). Immunohistochemical staining of normal colon mucosa and primary colon adenocarcinoma samples has shown significantly higher levels of CD44v3 in the cytoplasmic membrane of cancer cells compared to normal mucosal cells (38). CD44v3,8-10, an isoform variant containing both exons v3 and v8-10, is another high molecular-weight (~260 kDa) isoform of CD44 that could be relevant for diagnosis of CRC (83). The reverse transcriptase polymerase chain reaction (RT-PCR) analysis of transcripts present in mucosal samples, removed from 53 patients, detected CD44v3,8-10 transcript in 2/23 (6%) of normal, 19/20 (95%) of adenoma, and 29/31 (93.5%) of carcinoma (84).
The limited expression of CD44v3, CD44v6, and CD44v3,v8-10 splice variants in healthy colon tissue and their apparent upregulation in CRC tissue strengthens the potential use of these biomarkers for future CRC screening.
Galectin 3
Galectin 3, or LGALS3, is a member of the galectin family, a group of carbohydrate-binding lectins characterized by their binding affinity for beta-galactosides (85). LGALS3 is expressed at the cell surface, where it interacts with the extracellular matrix, especially with glycoproteins, and has the ability to affect intracellular signaling pathways (42). LGALS3-expressing cells also possess higher ALDH1 activity, which often correlates with a dedifferentiated cancer stem cell phenotype, than do their LGALS3-negative counterparts (86).
The correlation of LGALS3 expression in CRC with clinical pathological characteristics has been explored in several immunohistochemical and RT-PCR studies. In one study, the IHC staining of CRC tissue (n=61) and normal adjacent tissue (n=23) samples showed significantly higher LGALS3 expression in cancer tissue (62.5%) versus normal cancer-adjacent tissue (13.0%) (41). In another study, 75% of CRC tissue samples stain high for LGALS3, and ten CRC cell lines were shown to have increased LGALS3 protein levels compared to HeLa cells (42).
LGALS3 expression varies according to cancer staging and the degree of differentiation of the adenocarcinoma. LGALS3 mRNA levels were higher in early stage colorectal cancers (58% in stage I) compared to advanced cancers (50% in stage IV) (43). Protein analysis found higher LGALS3 levels in primary adenocarcinomas than in metastatic adenocarcinomas, and stronger LGALS3 staining in well-differentiated tumor areas compared to poorly differentiated tumor areas (43). Conversely, colorectal adenocarcinomas may display higher levels of LGALS3 than do colorectal adenomas; one study sets the rate of colorectal adenocarcinoma expression of LGALS3 at 95% while only 73% of adenomas were positive for LGALS3 (43). The higher expression of LGALS3 in CRC tissue compared to normal adjacent tissue, and high expressions in early stage CRC, make this biomarker a potential candidate for early diagnostic applications.
IFITM1
Interferon-inducible transmembrane protein 1 (IFITM1) is a member of the IFN-inducible transmembrane protein family that mediates the antiproliferative effects of cytokines. A 2008 meta-analysis of CRC gene expression profiles identified IFITM1 as consistently upregulated in cancer conditions vs normal tissue (87), building on prior evidence from in situ hybridization studies (88). Later staining by IHC of IFITM1 protein in tissue specimens confirmed elevated IFITM1 levels in CRC, compared to paired adjacent normal tissues (44). Oligonucleotide microarray comparisons between healthy tissue and tumor tissue also showed a 1.5-fold plus increase in IFITM1 levels in CRC (89). High levels of IFITM1 are positively correlated with distant metastasis, advanced stage, and poor OS (44).
Transfection of LoVo and HT-29 colorectal cancer cells with small interfering RNA (siRNA) targeting IFITM1 reduced migration and invasion capacities of these cells in a wound-healing assay, whereas overexpression of IFITM1 enhanced these capacities (44). In a separate study, IFITM1 overexpression in SW480 cells also promoted invasiveness (90). Conversely, IFITM1 reduction by >85% using short hairpin RNA (shRNA) reduced cell proliferation by 10-35% (91). Immunoblot analysis shows that non-metastatic SW480 had low levels of IFITM1, but metastatic cell lines such as HT29, HCT116, and SW620 exhibited higher levels of IFITM1 (91). Western blot conducted by another team found similar results: high IFITM1 levels in HCT116, LoVo, and HT-29 cell lines (44).
Whether in native tissue or in cell lines, this recent data all supports the notion that high levels of IFITM1 correlates with increased migration, invasion, and behaviors associated with metastasis in CRC.
Thomsen-Friedenreich Antigen
The Thomsen-Friedenreich (TF) antigen (also known as T antigen or CD176) is a disaccharide with the structure galactose β1,3-N-acetyl-d-galactosamine (Gal-GalNAc). Over many decades, TF antigen has been appreciated increasingly for its appearance on aberrantly glycosylated proteins, particularly cell surface proteins on transformed epithelia (92). More recent literature identified TF antigen in numerous cancer types, including CRC, as well as an interesting potential interaction of this uncommon disaccharide with the galectins (93). Because this antigen also is displayed on gut bacteria, anti-TF antibodies are found in humans, and are postulated to be agents for immunosurveillance of developing tumors (94).
The immunodiagnostic potential of TF antigen in patients with CRC has been investigated in several studies using IHC. In most normal mucosa, the TF structure is not present and is instead modified (45). In two separate immunohistochemical studies, the TF antigen was undetectable in normal colon mucosal tissue (46, 95), but was reported in 60% of adenomas and adenocarcinomas (46). These findings explain the rationale for exploiting the specificity and sensitivity of using the TF antigen as a nanobeacon target in CRC imaging techniques, such as shown in Figure 1.
The TF antigen has been the subject of a study of its potential use as a molecular target of targeted nanobeacons for fluorescent light (FL) colonoscopy (96). This study showed that in a xenograft CRC mouse model the fluorescent specimens had a 35-fold stronger signal than controls, which grew to 60-fold at a later stage of tumor development. The strong signal-to-noise ratio achieved when using the TF antigen as a target in imaging techniques is congruent with protein expression levels identified in other studies.
The TF antigen can also be detected using peanut agglutinin and other lectins, which can be more cost-effective than antibodies for functionalizing micro- or nano-particles (96, 97). The TF antigen is an exciting target to consider for future CRC imaging techniques as it has already been applied in this setting with promising results. This antigen can potentially reduce the costs associated with purchasing antibodies for functionalization.
Novel Candidates
GPRC5A
GPRC5A is a retinoic acid induced G protein-coupled receptor (GPCR), class C, group 5, also known as retinoic acid-induced gene 3 (RAI3) or retinoic acid-induced gene 1 (RAIG1). The GPCRs are a broad family of transmembrane receptors, with a broad array of functions. GPRC5A is one of a group of four proteins within group 5, which is identified by the sequence similarity of its members (GPRC5A, -B, -C. and -D), and characterized by a short extracellular N-terminal domain, and its ability to be induced for transcription by retinoic acid (for all members except GPRC5D) (98). In normal tissues, GPRC5A is primarily expressed in lung, albeit followed next by the gastrointestinal system, including colon (99). Despite this, baseline GPRC5A transcripts are relatively low in normal adult colon tissue (100), and protein staining by IHC in normal colon tissue remains undetectable or limited to a few neuroendocrine cells within the colonic crypts (47).
Over the last 10-15 years, dysregulation of GPRC5A was identified in many cancers, including breast, prostate, and CRC (47, 98, 99). In one immunohistochemical study, staining of 367 CRC tumor samples displayed GPRC5A localization to the luminal membrane of 193 (62%) samples (47). In another study, the transcript levels were 2.2-fold higher in cancerous tissue than in healthy tissue of 57 paired patients (101).
The relationship between GPRC5A expression and prognosis are unclear. GPRC5A expression may be a negative prognosis factor, as an analysis of Gene Expression Omnibus databases found (101). GPRC5A expression may also be related to hypoxia, a key characteristic of many tumor microenvironments. Hypoxia-induced increases in the levels of GPRC5A was found in a stable isotope labeling by amino acids in cell culture (SILAC)-based proteomics analysis of SW640 cells grown in normoxia and hypoxia. GPRC5A deletion increased apoptosis in hypoxic conditions by 12.2-fold (102). These findings highlight the role of GPRC5A presence in CRC pathology, and strengthen its potential as a CRC biomarker.
EphB4
Ephrin type-B receptor 4 (EphB4) is a tyrosine kinase-type receptor that recognizes membrane-bound ephrin ligands on adjacent cells. One important role of ephrins and their receptors is to direct cell-cell positioning, through their bidirectional signaling and activation (103). These proteins are integral partners in the development of neural and vascular structures, and in the maintenance of tissue boundaries. Conversely, release of boundaries enables de-differentiated stem/progenitor cell phenotypes, and their dysregulation in cancer further permits the disruption of stable tissue boundaries, and ultimately favors a migratory and metastatic phenotype.
EphB4 expression in the healthy colon was reported as minimal to none (104, 105). However, EphB4 is overexpressed in multiple CRC cell lines, including SW480, LIM2405 B4, and CT26 cells, highlighting its potential as a CRC biomarker (106). Differences in IHC staining for EphB4 in tumor tissue and normal tissue was very pronounced according to one team (48). IHC staining of 50 normal colon tissue samples showed EphB4 levels high in only 8% of healthy samples (49), while others showed EphB4 expression in 73% and 85.3% of clinical CRC samples (104). EphB4 shows potential as a candidate biomarker because of the strong differences in its expression levels between healthy and cancerous tissue, and its implication in multiple processes related to cancer progression.
FGFR4
Fibroblast growth factor receptor four (FGFR4), one of the four transmembrane tyrosine kinase receptors, plays a role in cell proliferation, differentiation, and survival (107). Recent IHC studies found positive FGFR4 stains in 90.7% of biopsies removed from patients with locally advanced CRC (50). Protein levels of FGFR4 were higher in colorectal adenoma tissue as well (108).
FGFR4 levels may be related to metastasis and EMT. FGFR4 knockdown facilitated the expression of E-cadherin and decreased levels of TWIST and other EMT inducers (109). This data aligns with findings that metastatic CRC cell lines have higher FGFR4 levels than non-metastatic cell lines, but contrasts with the finding that FGFR4 is cancer-specific in early Duke’s stages (110). FGFR4 is a target of FOXC1, which when elevated is associated with worse prognosis, as is FGFR4 (111).
Although measures of FGFR4 levels in CRC through IHC remains limited, its elevation in other cancers and early evaluation of its protein levels at the cell surface suggests that FGFR4 should continue to be assessed as a CRC biomarker. The relationship between FGFR4 expression and cellular adhesion, invasion, and metastasis pathways highlights its potential as a CRC marker.
Other Biomarkers With Specificity for Flat Lesions
Non-polypoid colorectal neoplasm (NP-CRN) represent a heterogeneous group of lesions that appear to be slightly elevated, completely flat, or slightly depressed compared to normal adjacent mucosa (112). SSA/P and hyperplastic polyps, both classified as NP-CRN, share some of the same histological and molecular features. Histologically, SSA/P and hyperplastic polyps both have crypts with serrated luminal outline and epithelial cells that are rich in mucin (113). Molecularly, both polyp subtypes have the BRAF mutation, a downstream target in the EGFR signaling pathway (114). The SSA/P, differ from hyperplastic polyps in their higher degree of abnormal proliferation and potential to develop invasive adenocarcinomas (115). Because histopathological classification of polyps is critical for determining the malignant potential of colorectal lesions, surface biomarkers that can differentiate between SSA/P and hyperplastic polyps would be highly valuable in accurate characterization of lesions.
Annexin A10
Although not highlighted in our larger group of biomarkers in Figure 3, Annexin A10 was found in our broader category of 44 candidate biomarkers (Figure 2), and has particular relevance here in relation to SSA/P lesions. Annexins (ANX) are a large family of eukaryotic calcium-dependent membrane proteins that play important roles in cell life cycle, exocytosis, and apoptosis (116). Some annexins (A1, A2, A4, A10, and A11) are expressed at higher levels in CRC than in normal colon (117, 118). Several studies showed that A10 (ANXA10) is correlated with serrated pathway of colorectal carcinoma (113, 119–121). A microarray analysis of distal hyperplastic polyps (n=6) and proximal SSA/Ps (n=6) showed that ANXA10 has a 73% sensitivity and 73% specificity in the diagnosis of SSA/Ps (113). In another immunohistochemical study with larger samples (n=131), immunoreactivity for ANXA10 predicted serrated histology with sensitivity of 55% and specificity of 97% (120).
CD133
CD133 was already noted in 2.1.2. as a relevant biomarker for early CRC. While not without controversy as a diagnostic and prognostic biomarker, the stem cell biomarker CD133 has been shown additionally to be implicated in the serrated pathway. In one immunohistochemical study, CD133 was expressed more prominently in SSA/P than in hyperplastic polyps (122). This finding further strengthens the status of CD133 as a potential biomarker for early detection of CRC.
Imaging of Surface Biomarkers
The key value to identifying a battery of cell surface biomarkers is that antibodies or other targeting agents for each biomarker can be tethered to various molecular contrast agents, with the aim to improve early identification of CRC that may be difficult to visualize by other means. In addition to visual detection of surface biomarkers, contrast agents can act as carriers for selective release of drugs in cancer cells (123). Existing imaging technologies such as magnetic resonance imaging (MRI), positron emission tomography (PET), confocal laser endomicroscopy (CLE), and Raman spectroscopy offer potential improvements to conventional colonoscopy methods (Figure 4).
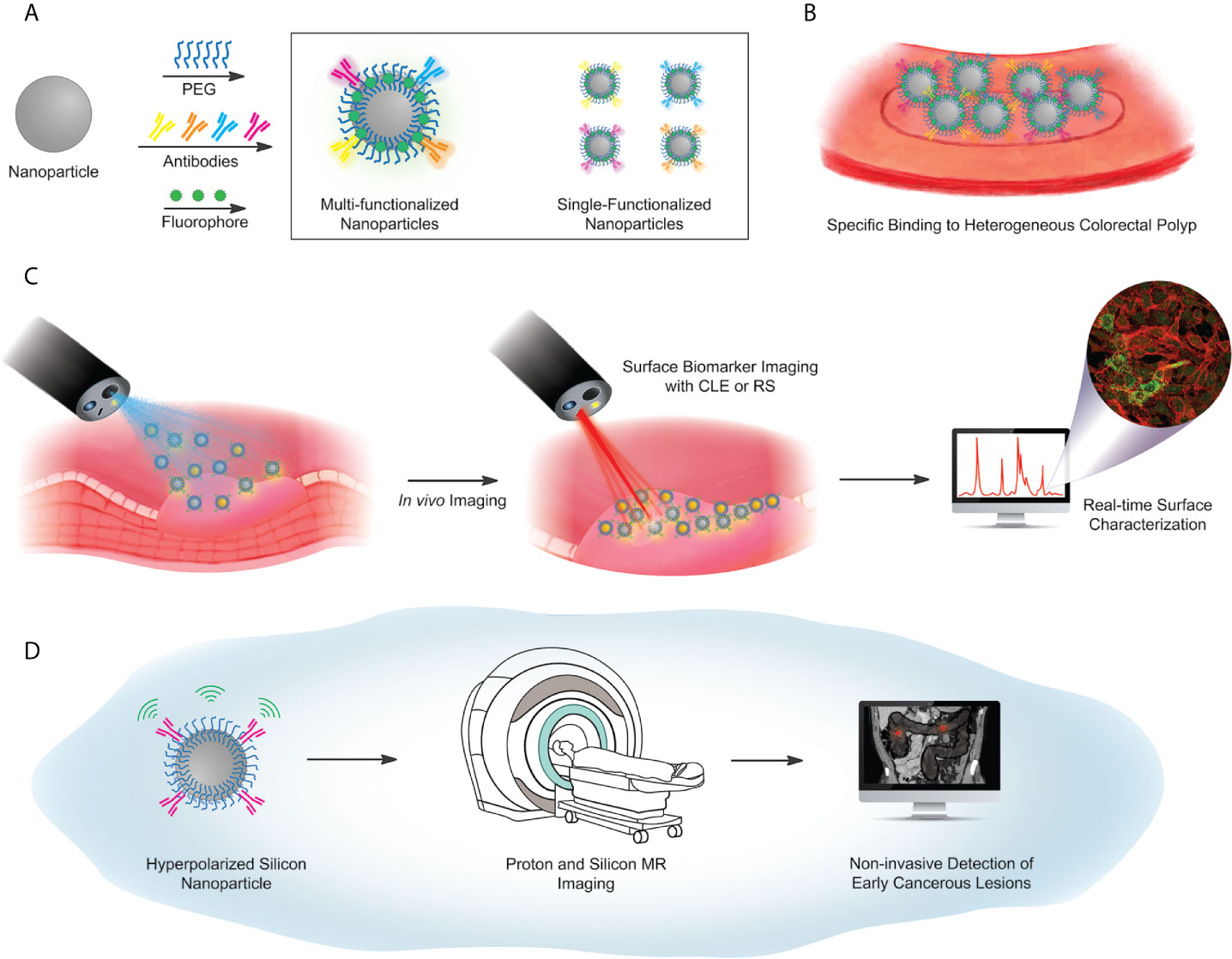
Figure 4 Proposed workflow for future concepts in particle-based targeted imaging. (A) Surface functionalization scheme, using a library of antibodies against selected surface biomarker targets. Antibodies are covalently coupled to a PEG-functionalized particle, with either a fluorescent barcode, or hyperpolarization (used in (D) below). Particle libraries may have multiple targeting antibodies on a single particle (box, left) or single-target particles, mixed to illuminate multiple biomarkers on a heterogeneous lesion (box, right). (B) Illustrated identification of lesions, via a library of particles. (C) Imaging workflow for fluorescence-based or Raman-based image capture. (D) Hyperpolarized Si particles preserve signal over a sufficient lifetime to enable magnetic resonance-based imaging. The MRI image of the abdomen adopted from the depiction of volvulus on Wikipedia under the Creative Commons license: https://en.wikipedia.org/wiki/Volvulus).
Confocal Laser Endomicroscopy
Confocal laser endomicroscopy (CLE) is an endoscopic modality that was developed for real-time histological assessment of the mucosal layer of the gastrointestinal (GI) tract (124) through the excitation and imaging of fluorescent probes that highlight specific tissue features. CLE has been used in the diagnosis and management of various diseases including squamous cell cancer of the esophagus, gastric cancer, gastritis, celiac disease, and to visualize colonic neoplasia and normal colonic mucosa (125–128). In the lower GI tract studies with CLE, systemic fluorescein combined with topical acriflavine have been used to distinguish high grade and low grade adenomas (129). These studies indicate that CLE can provide real-time histological information and aid in characterization and in vivo diagnosis of colonic lesions. Additionally, CLE can be combined with exogenous probes conjugated with fluorescein to detect tumor-specific biomarkers in CRC patients. In one study, fluorescein-conjugated peptides administered topically to dysplastic colon were detected using CLE with 81% sensitivity and 82% specificity (20).
Raman Spectroscopy and SERS
Raman spectroscopy (RS) is another modality that has shown potential for endoscopic diagnosis of diseases of the epithelium, including those of the esophagus, stomach, and colon (130–132). RS can be combined with exogenous contrast agents, such as surface-enhanced Raman-scattering (SERS) nanoparticles (NPs), for sensitive and multiplexed molecular imaging of epithelial biomarkers. Several studies have reported the detection of targeted and non-targeted SERPS NPs in small animals and on human tissue (133–136). SERS NPs can be conjugated to antibodies, peptides, lectins or other moieties that target a diverse panel of tumor-enhanced biomarkers. Although SERS NPs have low toxicity, their application in clinical studies requires further regulatory approval. Nonetheless, Zavaleta et al. have developed and tested the utility of a Raman endoscope for detection of functionalized-SERS NPs in human patients. This Raman endoscope was inserted through the accessory channel of a conventional endoscope and acquired images from the colon wall of a human patient (137).
Hyperpolarized MRI
In recent years, the implementation of virtual colonoscopy (CT and MRI) has emerged as potential alternatives to colonoscopy. However, these modalities historically suffer from significant drawbacks including poor detection accuracy for small (<10mm) lesions (138, 139), reliance on non-tumor specific contrast agents, and inaccurate diagnosis due to false-positives or false-negatives (140). One novel approach to increase the sensitivity of MRI is through dynamic nuclear polarization (DNP), whereby the nuclear spin alignment of underlying material is enhanced by 3-4 orders of magnitude, resulting in higher signal to noise ratio (SNR) (141). Hyperpolarized silicon (29Si) particles (HP SiPs), detectable via MRI, could serve as nanobeacons to discern cancerous tissue from healthy tissue in a variety of cancers including prostate, ovarian, and colorectal cancer (142–144).
Silicon particles can be functionalized with antibodies or other targeting moieties to detect specific cell surface biomarkers common to CRC, but largely absent from healthy tissue. The feasibility of imaging surface biomarkers in vivo with antibody functionalized HP SiPs has been demonstrated in a CRC mouse model expressing MUC1. 2μm HP SiPs functionalized with an IgG1 antibody to MUC1 (214D4), administered to human-MUC1 expressing mice (MUC1+) via the rectum, were detected at the location of the tumor. The 29Si MRI signal was absent in a control study where the same particles were administered to MUC1 negative mice (MUC1-), or in another control where PEGylated only particles (without the 214D4 antibody) were administered to human MUC1 expressing mice (MUC1+) (144).
PET
[18F]Fluorodeoxyglucose PET has been clinically used for the evaluation of patients with a wide variety of cancers since most malignancies, including colorectal cancer, typically show increased glucose metabolism (145, 146). However the versatility of this receptor imaging technique can be harnessed (147–150), much like hyperpolarized Silicon-based MRI to functionalized appropriate biomarker antibodies/peptides/aptamers/affibodies for colorectal cancer with a radioactive nuclei (18F, 68Ga, 124I, 89Zr) to achieve targeted molecular imaging in colorectal cancer systems.
Conclusions and the Future of Biomarkers in Cancer Screening
Although optical colonoscopy is the gold standard in CRC screening, its effectiveness in detecting early lesions critically depends upon the experience of the endoscopist and the ability to visually distinguish the lesion from normal tissue. To be visible through conventional endoscopes, colorectal lesions must reach a certain size and have an atypical appearance. This lack of specificity can lead to under-detection of potentially aggressive early lesions such as sessile serrated adenomas/polyps.
Because CRC almost always originates from the epithelial cells lining the luminal surface of the colon, surface accessible biomarkers present an opportunity for early detection of CRC. This means that instead of relying on direct visual detection of dysplastic tissue, one can use sophisticated imaging methods to search for cellular signatures that identify malignant or pre-malignant cells and growths. In this review, we surveyed the literature to identify potential surface biomarkers that might facilitate early detection of colorectal lesions. These biomarkers may be combined with colonoscopy or emerging methodologies to enhance the detection of heterogeneous tumors in diverse patient populations. The objective of future research will be to explore a cost-effective and non-invasive approach for early and real-time detection of pre-cancerous lesions with high contrast against nearby healthy tissue. Other goals are to create unique spectral signatures for different subtypes of polyps based on the binding pattern of antibodies or other high affinity recognition molecules.
Author Contributions
SR has contributed to conceptualizing, writing, organizing of figures, tables, references, and final revisions. AP has contributed to conceptualizing, literature search, and early drafting of manuscript. MF-C has contributed to conceptualizing, writing, and final revisions. PB has contributed to writing and final revisions. DH has contributed to conceptualizing, writing, final revisions, and supervision of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
SR and AP acknowledge the UTHealth Innovation for Cancer Prevention Research Training Program (Cancer Prevention and Research Institute of Texas grant #RP160015). PB acknowledges Pancreatic Cancer Action Network (PANCAN; 16-65-BHAT); Cancer Prevention and Research Institute of Texas (CPRIT; RP180873); US National Cancer Institute (NCI; U01 CA214263 and R21 CA185536), Institutional Research Grants and a Startup grant from MD Anderson Cancer Center.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the Cancer Prevention and Research Institute of Texas.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2020. CA Cancer J Clin (2020) 70(1):7–30. doi: 10.3322/caac.21590
2. Bénard F, Barkun AN, Martel M, von Renteln D. Systematic Review of Colorectal Cancer Screening Guidelines for Average-Risk Adults: Summarizing the Current Global Recommendations. World J Gastroenterol (2018) 24(1):124–38. doi: 10.3748/wjg.v24.i1.124
3. Fass OZ, Poels KE, Qian Y, Zhong H, Liang PS. Demographics Predict Stage III/IV Colorectal Cancer in Individuals Under Age 50. J Clin Gastroenterol (2020) 54(8):714–9. doi: 10.1097/MCG.0000000000001374
4. Zhao S, Wang S, Pan P, Xia T. Magnitude, Risk Factors, and Factors Associated With Adenoma Miss Rate of Tandem Colonoscopy: A Systematic Review and Meta-Analysis. Gastroenterology (2019) 156(6):1661–74. doi: 10.1053/j.gastro.2019.01.260
5. D’Andrea E, Ahnen DJ, Sussman DA, Najafzadeh M. Quantifying the Impact of Adherence to Screening Strategies on Colorectal Cancer Incidence and Mortality. Cancer Med (2020) 9(2):824–36. doi: 10.1002/cam4.2735
6. Roth JA, deVos T, Ramsey SD. Clinical and Budget Impact of Increasing Colorectal Cancer Screening by Blood- and Stool-Based Testing. Am Health Drug Benefits (2019) 12(5):256–62.
7. Bynum SA, Davis JL, Green BL, Katz RV. Unwillingness to Participate in Colorectal Cancer Screening: Examining Fears, Attitudes, and Medical Mistrust in an Ethnically Diverse Sample of Adults 50 Years and Older. Am J Health Promot AJHP (2012) 26(5):295–300. doi: 10.4278/ajhp.110113-QUAN-20
8. Young PE, Womeldorph CM. Colonoscopy for Colorectal Cancer Screening. J Cancer (2013) 4(3):217–26. doi: 10.7150/jca.5829
9. Meklin J, Syrjänen K, Eskelinen M. Fecal Occult Blood Tests in Colorectal Cancer Screening: Systematic Review and Meta-Analysis of Traditional and New-Generation Fecal Immunochemical Tests. Anticancer Res (2020) 40(7):3591–604. doi: 10.21873/anticanres.14349
10. Lynch HT, Lanspa S, Shaw T, Casey MJ, Rendell M, Stacey M, et al. Phenotypic and Genotypic Heterogeneity of Lynch Syndrome: A Complex Diagnostic Challenge. Fam Cancer (2018) 17(3):403–14. doi: 10.1007/s10689-017-0053-3
11. Lee E-J, Kim M-J, Chun S-M, Jang S-J, Kim DS, Lee DH, et al. Sessile Serrated Adenoma/Polyps With a Depressed Surface: A Rare Form of Sessile Serrated Adenoma/Polyp. Diagn Pathol (2015) 10(75):1–4. doi: 10.1186/s13000-015-0325-x
12. Pohl H, Robertson DJ. Colorectal Cancers Detected After Colonoscopy Frequently Result From Missed Lesions. Clin Gastroenterol Hepatol (2010) 8(10):858–64. doi: 10.1016/j.cgh.2010.06.028
13. Kashida H, Kudo S, Takeuchi T, Ohtsuka K. Colorectal Cancer: The Importance of Depressed Lesions in the Development of Colorectal Cancer. In: Fujita R, Jass JR, Kaminishi M, Schlemper RJ, editors. Early Cancer of the Gastrointestinal Tract: Endoscopy, Pathology, and Treatment. Tokyo: Springer Japan (2006). p. 243–7. doi: 10.1007/4-431-30173-9_21
14. Soetikno RM, Kaltenbach T, Rouse RV, Park W, Maheshwari A, Sato T, et al. Prevalence of Nonpolypoid (Flat and Depressed) Colorectal Neoplasms in Asymptomatic and Symptomatic Adults. JAMA (2008) 299(9):1027–35. doi: 10.1001/jama.299.9.1027
15. Pellino G, Gallo G, Pallante P, Capasso R, De Stefano A, Maretto I, et al. Noninvasive Biomarkers of Colorectal Cancer: Role in Diagnosis and Personalised Treatment Perspectives. Passardi A, editor. Gastroenterol Res Pract (2018) 2018:1–21. doi: 10.1155/2018/2397863
16. Toma SC, Ungureanu BS. Colorectal Cancer Biomarkers - A New Trend in Early Diagnosis. Curr Health Sci J (2018) 2:140–6. doi: 10.12865/CHSJ.44.02.08
17. Gonzalez-Pons M, Cruz-Correa M. Colorectal Cancer Biomarkers: Where Are We Now? Scartozzi M, editor. BioMed Res Int (2015) 2015:1–14. doi: 10.1155/2015/149014
18. Vacante M, Borzì AM, Basile F, Biondi A. Biomarkers in Colorectal Cancer: Current Clinical Utility and Future Perspectives. World J Clin Cases (2018) 6(15):869–81. doi: 10.12998/wjcc.v6.i15.869
19. Kiesslich R, Goetz M, Vieth M, Galle PR, Neurath MF. Technology Insight: Confocal Laser Endoscopy for In Vivo Diagnosis of Colorectal Cancer. Nat Clin Pract Oncol (2007) 4(8):480–90. doi: 10.1038/ncponc0881
20. Hsiung P, Hardy J, Friedland S, Soetikno R, Du CB, Wu APW, et al. Detection of Colonic Dysplasia In Vivo Using a Targeted Fluorescent Septapeptide and Confocal Microendoscopy. Nat Med (2008) 14(4):454–8. doi: 10.1038/nm1692
21. Alves Martins BA, de Bulhões GF, Cavalcanti IN, Martins MM, de Oliveira PG, Martins AMA. Biomarkers in Colorectal Cancer: The Role of Translational Proteomics Research. Front Oncol (2019) 9:1–11. doi: 10.3389/fonc.2019.01284
22. Sveen A, Kopetz S, Lothe RA. Biomarker-Guided Therapy for Colorectal Cancer: Strength in Complexity. Nat Rev Clin Oncol (2020) 17(1):11–32. doi: 10.1038/s41571-019-0241-1
23. de Castro-Carpeño J, Belda-Iniesta C, Sáenz EC, Agudo EH, Batlle JF, Barón MG. EGFR and Colon Cancer: A Clinical View. Clin Transl Oncol (2008) 10(1):6–13. doi: 10.1007/s12094-008-0147-3
24. Ellis LM, Takahashi Y, Liu W, Shaheen RM. Vascular Endothelial Growth Factor in Human Colon Cancer: Biology and Therapeutic Implications. Oncologist (2000) 5(S1):11–5. doi: 10.1634/theoncologist.5-suppl_1-11
25. Parizadeh SM, Jafarzadeh-Esfehani R, Fazilat-Panah D, Hassanian SM, Shahidsales S, Khazaei M, et al. The Potential Therapeutic and Prognostic Impacts of the c-MET/HGF Signaling Pathway in Colorectal Cancer. IUBMB Life (2019) 71(7):802–11. doi: 10.1002/iub.2063
26. Xie Y-H, Chen Y-X, Fang J-Y. Comprehensive Review of Targeted Therapy for Colorectal Cancer. Signal Transduct Target Ther (2020) 5(1):1–30. doi: 10.1038/s41392-020-0116-z
27. Jothy S, Yuan SY, Shirota K. Transcription of Carcinoembryonic Antigen in Normal Colon and Colon Carcinoma. In Situ Hybridization Study and Implication for a New In Vivo Functional Model. Am J Pathol (1993) 143(1):250–7.
28. Tiernan JP, Perry SL, Verghese ET, West NP, Yeluri S, Jayne DG, et al. Carcinoembryonic Antigen Is the Preferred Biomarker for In Vivo Colorectal Cancer Targeting. Br J Cancer (2013) 108(3):662–7. doi: 10.1038/bjc.2012.605
29. Coco C, Zannoni GF, Caredda E, Sioletic S, Boninsegna A, Migaldi M, et al. Increased Expression of CD133 and Reduced Dystroglycan Expression Are Strong Predictors of Poor Outcome in Colon Cancer Patients. J Exp Clin Cancer Res (2012) 31(71):1–10. doi: 10.1186/1756-9966-31-71
30. Choi D, Lee HW, Hur KY, Kim JJ, Park G-S, Jang S-H, et al. Cancer Stem Cell Markers CD133 and CD24 Correlate With Invasiveness and Differentiation in Colorectal Adenocarcinoma. World J Gastroenterol WJG (2009) 15(18):2258–64. doi: 10.3748/wjg.15.2258
31. Kazama S, Kishikawa J, Kiyomatsu T, Kawai K, Nozawa H, Ishihara S, et al. Expression of the Stem Cell Marker CD133 Is Related to Tumor Development in Colorectal Carcinogenesis. Asian J Surg (2018) 41(3):274–8. doi: 10.1016/j.asjsur.2016.12.002
32. Wang H, Wang L. The Expression and Significance of Gal-3 and MUC1 in Colorectal Cancer and Colon Cancer. OncoTargets Ther (2015) 8:1893–8. doi: 10.2147/OTT.S83502
33. Betge J, Schneider NI, Harbaum L, Pollheimer MJ, Lindtner RA, Kornprat P, et al. Muc1, MUC2, MUC5AC, and MUC6 in Colorectal Cancer: Expression Profiles and Clinical Significance. Virchows Arch Int J Pathol (2016) 469(3):255–65. doi: 10.1007/s00428-016-1970-5
34. Manhas J, Bhattacharya A, Argawal S, Gupta B. Characterization of Cancer Stem Cells From Different Grades of Human Colorectal Cancer. Tumor Biol (2016) 37(10):14069–81. doi: 10.1007/s13277-016-5232-6
35. Gorham H, Sugino T, Woodman AC, Tarin D. Cellular Distribution of CD44 Gene Transcripts in Colorectal Carcinomas and in Normal Colonic Mucosa. J Clin Pathol (1996) 49(6):482–8. doi: 10.1136/jcp.49.6.482
36. Ismaiel NEHS, Sharaf WM, Helmy DO, Zaki MM, Badawi MA, Soliman ASA. Detection of Cancer Stem Cells in Colorectal Cancer: Histopathological and Immunohistochemical Study. Open Access Maced J Med Sci (2016) 4(4):543–7. doi: 10.3889/oamjms.2016.126
37. Al-Magrabi J, Gomaa W, Abdelbaset B, Al-Qahtani M. Decreased Immunoexpression of Standard Form of CD44 Is An Independent Favourable Predictor of Nodal Metastasis in Colorectal Carcinoma. Anticancer Res (2012) 32:3455–62.
38. Kuniyasu H, Oue N, Tsutsumi M, Tahara E. Heparan Sulfate Enhances Invasion by Human Colon Carcinoma Cell Lines Through Expression of CD44 Variant Exon 3. Clin Cancer Res (2001) 7(12):4067–72.
39. Afify A, Durbin-Johnson B, Virdi A, Jess H. The Expression of CD44v6 in Colon: From Normal to Malignant. Ann Diagn Pathol (2016) 20:19–23. doi: 10.1016/j.anndiagpath.2015.10.010
40. Afify A, Pang L, Howell L. Diagnostic Utility of CD44 Standard, CD44v6, and CD44v3-10 Expression in Adenocarcinomas Presenting in Serous Fluid. Appl Immunohistochem Mol Morph (2007) 15(4):446–50. doi: 10.1097/01.pai.0000213154.49063.22
41. Tao L, Jin L, Dechun L, Hongquiang Y. Galectin-3 Expression in Colorectal Cancer and Its Correlation With Clinical Pathological Characteristics and Prognosis. Open Med Wars (2017) 12:226–30. doi: 10.1515/med-2017-0032
42. Lu W, Wang J, Yang G, Yu N, Huang Z. Posttranscriptional Regulation of Galectin-3 by miR-129 Contributes to Colorectal Cancer Progression. Oncotarget (2017) 8(9):15242–51. doi: 10.18632/oncotarget.14839
43. Gopalan V, Saremi N, Sullivan E, Sadiul K. The Expression Profiles of the Galectin Gene Family in Colorectal Adenocarcinomas. Hum Pathol (2015) 53:105–13. doi: 10.1016/j.humpath.2016.02.016
44. Yu F, Xie D, Ng S, Lum C, Cai M. IFITM1 Promotes the Metastasis of Human Colorectal Cancer Via CAV-1. Cancer Lett (2015) 368:135–43. doi: 10.1016/j.canlet.2015.07.034
45. Schneider F, Kemmer W, Haensch W, Franke G. Overexpression of Sialyltransferase Cmp-Sialic Acid:Galβ1,3galNAc-R α6-Sialyltransferase Is Related to Poor Patient Survival Inhuman Colorectal Carcinomas. Cancer Res (2001) 61(11):4605–11.
46. El-Sayed I, Lofty M, Moawad M. Immunodiagnostic Potential of Mucin (MUC2) and Thomsen-Friedenreich (TF) Antigens in Egyptian Patients With Colorectal Cancer. Eur Rev Med Pharmacol Sci (2011) 15(1):91–7.
47. Zougman A, Hutchins G, Cairns D, Verghese E. Retinoic Acid-Induced Protein 3: Identification and Characterisation of a Novel Prognostic Colon Cancer Biomarker. Eur J Cancer (2013) 49(2):531–9. doi: 10.1016/j.ejca.2012.07.031
48. Stammes M, Prevoo H, Horste M, Groot S. Evaluation of EphA2 and EphB4 as Targets for Image-Guided Colorectal Cancer Surgery. Int J Mol Sci (2017) 18(2):1–10. doi: 10.3390/ijms18020307
49. Lv J, Xia Q, Wang J, Shen Q, Zhang J, Zhou X. EphB4 Promotes the Proliferation, Invasion, and Angiogenesis of Human Colorectal Cancer. Exp Mol Pathol (2016) 100(3):402–8. doi: 10.1016/j.yexmp.2016.03.011
50. Ahmed MA, Selzer E, Dörr W, Jomrich G, Harpain F, Silberhumer GR, et al. Fibroblast Growth Factor Receptor 4 Induced Resistance to Radiation Therapy in Colorectal Cancer. Oncotarget (2016) 7(43):69976–90. doi: 10.18632/oncotarget.12099
51. Beauchemin N, Arabzadeh A. Carcinoembryonic Antigen-Related Cell Adhesion Molecules (Ceacams) in Cancer Progression and Metastasis. Cancer Metastasis Rev (2013) 32(3–4):643–71. doi: 10.1007/s10555-013-9444-6
52. Lee JH, Lee S-W. The Roles of Carcinoembryonic Antigen in Liver Metastasis and Therapeutic Approaches. Beaulieu J-F, editor. Gastroenterol Res Pract (2017) 2017:1–12. doi: 10.1155/2017/7521987
53. Nap M, Mollgard K, Burtin P, Fleuren GJ. Immunohistochemistry of Carcino-Embryonic Antigen in the Embryo, Fetus and Adult. Tumour Biol J Int Soc Oncodev Biol Med (1988) 9(2–3):145–53. doi: 10.1159/000217555
54. Ramos-Gomes F, Bode J, Sukhanova A, Bozrova SV, Saccomano M, Mitkovski M, et al. Single- and Two-Photon Imaging of Human Micrometastases and Disseminated Tumour Cells With Conjugates of Nanobodies and Quantum Dots. Sci Rep (2018) 8:4595. doi: 10.1038/s41598-018-22973-8
55. Park YJ, Park KJ, Park JG, Lee KU, Choe KJ, Kim JP. Prognostic Factors in 2230 Korean Colorectal Cancer Patients: Analysis of Consecutively Operated Cases. World J Surg (1999) 23(7):721–6. doi: 10.1007/PL00012376
56. Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, et al. ASCO 2006 Update of Recommendations for the Use of Tumor Markers in Gastrointestinal Cancer. J Clin Oncol Off J Am Soc Clin Oncol (2006) 24(33):5313–27. doi: 10.1200/JCO.2006.08.2644
57. Thirunavukarasu P, Sukumar S, Sathaiah M, Mahan M, Pragatheeshwar KD, Pingpank JF, et al. C-Stage in Colon Cancer: Implications of Carcinoembryonic Antigen Biomarker in Staging, Prognosis, and Management. J Natl Cancer Inst (2011) 103(8):689–97. doi: 10.1093/jnci/djr078
58. Shmelkov S, Butler J, Hooper A, Hormigo A. CD133 Expression Is Not Restricted to Stem Cells, and Both CD133+ and CD133- Metastatic Colon Cancer Cells Initiate Tumors. J Clin Invest (2008) 118(6):2111–20. doi: 10.1172/JCI34401
59. Glumac P, LeBeau A. The Role of CD133 in Cancer: A Concise Review. Clin Transl Med (2018) 7(18):1–14. doi: 10.1186/s40169-018-0198-1
60. Zheng L, Kelly CJ, Colgan SP. Physiologic Hypoxia and Oxygen Homeostasis in the Healthy Intestine. A Review in the Theme: Cellular Responses to Hypoxia. Am J Physiol-Cell Physiol (2015) 309(6):C350–60. doi: 10.1152/ajpcell.00191.2015
61. Abbasian M, Mousavi E, Arab-Bafrani Z, Sahekbar A. The Most Reliable Surface Marker for the Identification of Colorectal Cancer Stem-Like Cells: A Systematic Review and Meta-Analysis. J Cell Physiol (2018) 234(6):8192–202. doi: 10.1002/jcp.27619
62. Yeung T, Mortensen N. Colorectal Cancer Stem Cells. Colon Rectum (2009) 52(10):1788–96. doi: 10.1007/DCR.0b013e3181a8738c
63. Turano M, Costabille V, Cerasuolo A, Duraturo F. Characterisation of Mesenchymal Colon Tumor-Derived Cells in Tumourspheres as a Model for Colorectal Cancer Progression. Int J Oncol (2018) 53(6):2379–96. doi: 10.3892/ijo.2018.4565
64. Corbeil D, Röper K, Hellwig A, Tavian M, Miraglia S, Watt SM, et al. The Human Ac133 Hematopoietic Stem Cell Antigen Is Also Expressed in Epithelial Cells and Targeted to Plasma Membrane Protrusions. J Biol Chem (2000) 275(8):5512–20. doi: 10.1074/jbc.275.8.5512
65. Huang R, Mo D, Wu J, Ai H, Lu Y. CD133 Expression Correlates With Clinicopathologic Features and Poor Prognosis of Colorectal Cancer Patients: An Updated Meta-Analysis of 37 Studies. Med (Baltimore) (2018) 97(23):1–9. doi: 10.1097/MD.0000000000010446
66. Biemer-Hüttmann AE, Walsh MD, McGuckin MA, Ajioka Y, Watanabe H, Leggett BA, et al. Immunohistochemical Staining Patterns of MUC1, Muc2, MUC4, and MUC5AC Mucins in Hyperplastic Polyps, Serrated Adenomas, and Traditional Adenomas of the Colorectum. J Histochem Cytochem Off J Histochem Soc (1999) 47(8):1039–48. doi: 10.1177/002215549904700808
67. Sewda K, Coppola D, Enkemann S, Yue B, Kim J, Lopez AS, et al. Cell-Surface Markers for Colon Adenoma and Adenocarcinoma. Oncotarget (2016) 7(14):17773–89. doi: 10.18632/oncotarget.7402
68. Nath S, Mukherjee P. Muc1: A Multifaceted Oncoprotein With a Key Role in Cancer Progression. Trends Mol Med (2014) 20(6):332–42. doi: 10.1016/j.molmed.2014.02.007
69. Brayman M, Thathiah A, Carson DD. Muc1: A Multifunctional Cell Surface Component of Reproductive Tissue Epithelia. Reprod Biol Endocrinol RBE (2004) 2(4):1–9. doi: 10.1186/1477-7827-2-4
70. Li C, Liu T, Yin L, Zuo D, Lin Y, Wang L. Prognostic and Clinicopathological Value of MUC1 Expression in Colorectal Cancer: A Meta-Analysis. Med (Baltimore) (2019) 98(9):e14659. doi: 10.1097/MD.0000000000014659
71. Skandalis SS, Karalis TT, Chatzopoulos A, Karamanos NK. Hyaluronan-CD44 Axis Orchestrates Cancer Stem Cell Functions. Cell Signal (2019) 63:109377. doi: 10.1016/j.cellsig.2019.109377
72. Mishra MN, Chandavarkar V, Sharma R, Bhargava D. Structure, Function and Role of CD44 in Neoplasia. J Oral Maxillofac Pathol JOMFP (2019) 23(2):267–72. doi: 10.4103/jomfp.JOMFP_246_18
73. Senbanjo LT, Chellaiah MA. Cd44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front Cell Dev Biol (2017) 5:18. doi: 10.3389/fcell.2017.00018
74. Spinelli FM, Vitale DL, Sevic I, Alaniz L. Hyaluronan in the Tumor Microenvironment. Adv Exp Med Biol (2020) 1245:67–83. doi: 10.1007/978-3-030-40146-7_3
75. Liu M, Tolg C, Turley E. Dissecting the Dual Nature of Hyaluronan in the Tumor Microenvironment. Front Immunol (2019) 10:947. doi: 10.3389/fimmu.2019.00947
76. Bartolazzi A, Jackson D, Bennett K, Aruffo A, Dickinson R, Shields J, et al. Regulation of Growth and Dissemination of a Human Lymphoma by CD44 Splice Variants. J Cell Sci (1995) 108(4):1723–33.
77. Goodison S, Urquidi V, Tarin D. CD44 Cell Adhesion Molecules. Mol Pathol (1999) 52(4):189–96. doi: 10.1136/mp.52.4.189
78. Nagano O, Okazaki S, Saya H. Redox Regulation in Stem-Like Cancer Cells by CD44 Variant Isoforms. Oncogene (2013) 32(44):5191–8. doi: 10.1038/onc.2012.638
79. Liu Y-J, Yan P-S, Li J, Jia J-F. Expression and Significance of CD44s, Cd44v6, and nm23 mRNA in Human Cancer. World J Gastroenterol WJG (2005) 11(42):6601–6. doi: 10.3748/wjg.v11.i42.6601
80. Tanabe KK, Stamenkovic I, Cutler M, Takahashi K. Restoration of CD44H Expression in Colon Carcinomas Reduces Tumorigenicity. Ann Surg (1995) 222(4):493–503. doi: 10.1097/00000658-199522240-00007
81. Ma L, Dong L, Chang P. CD44v6 Engages in Colorectal Cancer Progression. Cell Death Dis (2019) 10(30):1–13. doi: 10.1038/s41419-018-1265-7
82. Bánky B, Rásó-Barnett L, Barbai T, Tímár J, Becságh P, Rásó E. Characteristics of CD44 Alternative Splice Pattern in the Course of Human Colorectal Adenocarcinoma Progression. Mol Cancer (2012) 11(83):1–15. doi: 10.1186/1476-4598-11-83
83. Bourguignon LYW, Gunja-Smith Z, Iida N, Zhu HB, Young LJT, Muller WJ, et al. Cd44v3,8–10 Is Involved in Cytoskeleton-Mediated Tumor Cell Migration and Matrix Metalloproteinase (MMP-9) Association in Metastatic Breast Cancer Cells. J Cell Physiol (1998) 176(1):206–15. doi: 10.1002/(SICI)1097-4652(199807)176:1<206::AID-JCP22>3.0.CO;2-3
84. Kopp R, Fichter M, Schalhorn G, Danescu J. Frequent Expression of the High Molecular, 673-Bp. CD44v3, v8-10 Variant in Colorectal Adenomas and Carcinomas. Int J Mol Med (2009) 24(5):677–83. doi: 10.3892/ijmm_00000279
85. Wang Y, Liu S, Wang Y, Zhang Q, Zhou X. Prognostic Role of Galectin-3 Expression in Patients With Solid Tumors: A Meta-Analysis of 36 Eligible Studies. Cancer Cell Int (2018) 18:1–15. doi: 10.1186/s12935-018-0668-y
86. Ilmer M, Mazurek N, Byrd J, Ramirez K. Cell Surface Galectin-3 Defines a Subset of Chemoresistant Gastrointestinal Tumor-Initiating Cancer Cells With Heightened Stem Cell Characteristics. Cell Death Dis (2016) 7(8):1–9. doi: 10.1038/cddis.2016.239
87. Chan K, Obi G, Tai I, Jones S. Meta-Analysis of Colorectal Cancer Gene Expression Profiling Studies Identifies Consistently Reported Candidate Biomarkers. Cancer Epidemiol Biomarkers Prev (2008) 17(3):543–52. doi: 10.1158/1055-9965.EPI-07-2615
88. Andreu P, Colnot S, Godard C, Laurent-Puig P, Lamarque D, Kahn A, et al. Identification of the IFITM Family as a New Molecular Marker in Human Colorectal Tumors. Cancer Res (2006) 66(4):1949–55. doi: 10.1158/0008-5472.CAN-05-2731
89. Kim K, Park U, Wang J, Lee J, Park S. Gene-Expression Profiling of Colonic Adenomas by Using Oligonucleotide Microarray. Surg Endosc (2008) 23(6):569–80. doi: 10.1007/s00384-008-0451-y
90. He J, Luo H, Li J, Chen L. Influences of the Interferon Induced Transmembrane Protein 1 on Proliferation, Invasion, and Metastasis of Colorectal SW480 Cell Lines. Chin Med J (2012) 125(3):517–22. doi: 10.3760/cma.j.issn.0366-6999.2012.03.02
91. Sari N, Yang Y, Phi L, Kim H. Interferon-Induced Transmembrane Protein 1 (IFITM1) Is Required for Progression of Colorectal Cancer. Oncotarget (2016) 7(52):86039–50. doi: 10.18632/oncotarget.13325
92. Kurtenkov O. Profiling of Naturally Occurring Antibodies to the Thomsen-Friedenreich Antigen in Health and Cancer: The Diversity and Clinical Potential. BioMed Res Int (2020) 2020:9747040. doi: 10.1155/2020/9747040
93. Sindrewicz P, Lian L-Y, Yu L-G. Interaction of the Oncofetal Thomsen-Friedenreich Antigen With Galectins in Cancer Progression and Metastasis. Front Oncol (2016) 6:79. doi: 10.3389/fonc.2016.00079
94. Henderson G, Ulsemer P, Schöber U, Löffler A, Alpert C-A, Zimmermann-Kordmann M, et al. Occurrence of the Human Tumor-Specific Antigen Structure Galβ1-3galnacα- (Thomsen-Friedenreich) and Related Structures on Gut Bacteria: Prevalence, Immunochemical Analysis and Structural Confirmation. Glycobiology (2011) 21(10):1277–89. doi: 10.1093/glycob/cwr058
95. Croce MV, Isla-Larrain M, Rabassa ME, Demichelis S, Colussi AG, Crespo M, et al. Lewis X Is Highly Expressed in Normal Tissues: A Comparative Immunohistochemical Study and Literature Revision. Pathol Oncol Res (2007) 13(2):130–8. doi: 10.1007/BF02893488
96. Sakuma S, Yu J, Quang T, Hiwatari K, Kumagai H. Fluorescence-Based Endocscopic Imaging of Thomsen-Friedenreich Antigen to Improve Early Detection of Colorectal Cancer. Int J Cancer (2015) 136(5):1095–103. doi: 10.1002/ijc.29093
97. Liang Y, Chen H, Zhang H-B, Jin Y-X, Guo H-Q, Chen X-G, et al. Lectin From Agrocybe Aegerita as a Glycophenotype Probe for Evaluation of Progression and Survival in Colorectal Cancer. Asian Pac J Cancer Prev APJCP (2014) 15(14):5601–5. doi: 10.7314/APJCP.2014.15.14.5601
98. Jiang X, Xu X, Wu M, Guan Z, Su X, Chen S, et al. Gprc5a: An Emerging Biomarker in Human Cancer. BioMed Res Int (2018) 2018:e1823726. doi: 10.1155/2018/1823726
99. Zhou H, Rigoutsos I. The Emerging Roles of GPRC5A in Diseases. Oncoscience (2014) 1(12):765–76. doi: 10.18632/oncoscience.104
100. Cheng Y, Lotan R. Molecular Cloning and Characterization of a Novel Retinoic Acid-Inducible Gene That Encodes a Putative G Protein-Coupled Receptor. J Biol Chem (1998) 273(52):35008–15. doi: 10.1074/jbc.273.52.35008
101. Zhang L, Li L, Gao G, Wei G, Zheng Y. Elevation of GPRC5A Expression in Colorectal Cancer Promotes Tumor Progression Through VNN-1 Induced Oxidative Stress. Int J Cancer (2017) 140(12):2734–47. doi: 10.1002/ijc.30698
102. Greenhough A, Bagley C, Heesom K, Gurevich D. Cancer Cell Adaptation to Hypoxia Involves a HIF-GPRC5A-YAP Axis. EMBO Mol Med (2018) 10(11):1–13. doi: 10.15252/emmm.201708699
103. Kania A, Klein R. Mechanisms of Ephrin–Eph Signalling in Development, Physiology and Disease. Nat Rev Mol Cell Biol (2016) 17(4):240–56. doi: 10.1038/nrm.2015.16
104. Kumar S, Schenet J, Ley E, Singh J. Preferential Induction of EphB4 Over EphB2 and Its Implication in Colorectal Cancer Progression. Cell Tumor Stem Cell Biol (2009) 69(9):3736–45. doi: 10.1158/0008-5472.CAN-08-3232
105. Herath NI, Boyd AW. The Role of Eph Receptors and Ephrin Ligands in Colorectal Cancer. Int J Cancer (2010) 126(9):2003–11. doi: 10.1002/ijc.25147
106. Kadife E, Ware T, Luwor R, Chan S. Effects of EphB4 Receptor Expression on Colorectal Cancer Cells, Tumor Growth, Vascularization and Composition. Acta Oncol (2018) 57(8):1043–56. doi: 10.1080/0284186X.2018.1429650
107. Turkington RC, Longley DB, Allen WL, Stevenson L, McLaughlin K, Dunne PD, et al. Fibroblast Growth Factor Receptor 4 (FGFR4): A Targetable Regulator of Drug Resistance in Colorectal Cancer. Cell Death Dis (2014) 5(2):e1046. doi: 10.1038/cddis.2014.10
108. Wang D, Zhang J, Li Z, Han J. Upregulation of Fibroblast Growth Factor 19 Is Associated With the Initiation of Colorectal Adenoma. Dig Disease (2018) 37(3):214–25. doi: 10.1159/000494454
109. Pelaez-Garcio A, Barderas R, Torres S, Hernandez-Varas P. Fgfr4 Role in Epithelial-Mesenchymal Transition and Its Therapeutic Value in Colorectal Cancer. PloS One (2013) 8(5):1–11. doi: 10.1371/journal.pone.0063695
110. Barderas R, Babel I, Diaz-Uriarte R, Moreno V. An Optimized Predictor Panel for Colorectal Cancer Diagnosis Based on the Combination of Tumor-Associated Antigens Obtained From Protein and Phage Microarrays. J Proteomics (2012) 75(15):4647–55. doi: 10.1016/j.jprot.2012.03.004
111. Liu J, Zhang Z, Li X, Chen J. Forkhead Box C1 Promotes Colorectal Cancer Metastasis Through Transactivating ITGA7 and FGFR4 Expression. Oncogene (2018) 37:5477–91. doi: 10.1038/s41388-018-0355-4
112. Soetikno R, Friedland S, Kaltenbach T, Chayama K, Tanaka S. Nonpolypoid (Flat and Depressed) Colorectal Neoplasms. Gastroenterology (2006) 130(2):566–76. doi: 10.1053/j.gastro.2005.12.006
113. Gonzalo DH, Lai KK, Shadrach B, Goldblum JR, Bennett AE, Downs-Kelly E, et al. Gene Expression Profiling of Serrated Polyps Identifies Annexin A10 as a Marker of a Sessile Serrated Adenoma/Polyp. J Pathol (2013) 230(4):420–9. doi: 10.1002/path.4200
114. O’Brien MJ, Yang S, Mack C, Xu H, Huang CS, Mulcahy E, et al. Comparison of Microsatellite Instability, CpG Island Methylation Phenotype, BRAF and KRAS Status in Serrated Polyps and Traditional Adenomas Indicates Separate Pathways to Distinct Colorectal Carcinoma End Points. Am J Surg Pathol (2006) 30(12):1491–501. doi: 10.1097/01.pas.0000213313.36306.85
115. Spring KJ, Zhao ZZ, Karamatic R, Walsh MD, Whitehall VLJ, Pike T, et al. High Prevalence of Sessile Serrated Adenomas With BRAF Mutations: A Prospective Study of Patients Undergoing Colonoscopy. Gastroenterology (2006) 131(5):1400–7. doi: 10.1053/j.gastro.2006.08.038
116. Mirsaeidi M, Gidfar S, Vu A, Schraufnagel D. Annexins Family: Insights Into Their Functions and Potential Role in Pathogenesis of Sarcoidosis. J Transl Med (2016) 14:89. doi: 10.1186/s12967-016-0843-7
117. Duncan R, Carpenter B, Main LC, Telfer C, Murray GI. Characterisation and Protein Expression Profiling of Annexins in Colorectal Cancer. Br J Cancer (2008) 98(2):426–33. doi: 10.1038/sj.bjc.6604128
118. Lu S-H, Yuan R-H, Chen Y-L, Hsu H-C, Jeng Y-M. Annexin A10 Is an Immunohistochemical Marker for Adenocarcinoma of the Upper Gastrointestinal Tract and Pancreatobiliary System. Histopathology (2013) 63(5):640–8. doi: 10.1111/his.12229
119. Tsai J-H, Lin Y-L, Cheng Y-C, Chen C-C, Lin L-I, Tseng L-H, et al. Aberrant Expression of Annexin A10 Is Closely Related to Gastric Phenotype in Serrated Pathway to Colorectal Carcinoma. Mod Pathol (2015) 28(2):268–78. doi: 10.1038/modpathol.2014.96
120. Sajanti SA, Väyrynen JP, Sirniö P, Klintrup K, Mäkelä J, Tuomisto A, et al. Annexin A10 Is a Marker for the Serrated Pathway of Colorectal Carcinoma. Virchows Arch (2015) 466(1):5–12. doi: 10.1007/s00428-014-1683-6
121. Kim JH, Rhee Y-Y, Kim K-J, Cho N-Y, Lee HS, Kang GH. Annexin A10 Expression Correlates With Serrated Pathway Features in Colorectal Carcinoma With Microsatellite Instability. APMIS (2014) 122(12):1187–95. doi: 10.1111/apm.12284
122. Mohammadi M, Bzorek M, Bonde JH, Nielsen HJ, Holck S. The Stem Cell Marker CD133 Is Highly Expressed in Sessile Serrated Adenoma and Its Borderline Variant Compared With Hyperplastic Polyp. J Clin Pathol (2013) 66(5):403–8. doi: 10.1136/jclinpath-2012-201192
123. Vázquez-González M, Willner I. Aptamer-Functionalized Micro- andNanocarriers for Controlled Release. ACS Appl Mater Interfaces (2021) 13(8):9520–41. doi: 10.1021/acsami.0c17121
124. Fugazza A, Gaiani F, Carra MC, Brunetti F, Lévy M, Sobhani I, et al. Confocal Laser Endomicroscopy in Gastrointestinal and Pancreatobiliary Diseases: A Systematic Review and Meta-Analysis. BioMed Res Int (2016) 2016:4638683. doi: 10.1155/2016/4638683
125. Pech O, Rabenstein T, Manner H, Petrone MC, Pohl J, Vieth M, et al. Confocal Laser Endomicroscopy for In Vivo Diagnosis of Early Squamous Cell Carcinoma in the Esophagus. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc (2008) 6(1):89–94. doi: 10.1016/j.cgh.2007.10.01
126. Li W-B, Zuo X-L, Li C-Q, Zuo F, Gu X-M, Yu T, et al. Diagnostic Value of Confocal Laser Endomicroscopy for Gastric Superficial Cancerous Lesions. Gut (2011) 60(3):299–306. doi: 10.1136/gut.2010.223586
127. Leong RWL, Nguyen NQ, Meredith CG, Al-Sohaily S, Kukic D, Delaney PM, et al. In Vivo Confocal Endomicroscopy in the Diagnosis and Evaluation of Celiac Disease. Gastroenterology (2008) 135(6):1870–6. doi: 10.1053/j.gastro.2008.08.054
128. Kiesslich R, Burg J, Vieth M, Gnaendiger J, Enders M, Delaney P, et al. Confocal Laser Endoscopy for Diagnosing Intraepithelial Neoplasias and Colorectal Cancer In Vivo. Gastroenterology (2004) 127(3):706–13. doi: 10.1053/j.gastro.2004.06.050
129. Sanduleanu S, Driessen A, Gomez-Garcia E, Hameeteman W, de Bruïne A, Masclee A. In Vivo Diagnosis and Classification of Colorectal Neoplasia by Chromoendoscopy-Guided Confocal Laser Endomicroscopy. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc (2010) 8(4):371–8. doi: 10.1016/j.cgh.2009.08.006
130. Wang J, Lin K, Zheng W, Ho KY, Teh M, Yeoh KG, et al. Fiber-Optic Raman Spectroscopy for In Vivo Diagnosis of Gastric Dysplasia. Faraday Discuss (2016) 187(0):377–92. doi: 10.1039/C5FD00151J
131. Bergholt MS, Lin K, Wang J, Zheng W, Xu H, Huang Q, et al. Simultaneous Fingerprint and High-Wavenumber Fiber-Optic Raman Spectroscopy Enhances Real-Time In Vivo Diagnosis of Adenomatous Polyps During Colonoscopy. J Biophotonics (2016) 9(4):333–42. doi: 10.1002/jbio.201400141
132. Almond LM, Hutchings J, Lloyd G, Barr H, Shepherd N, Day J, et al. Endoscopic Raman Spectroscopy Enables Objective Diagnosis of Dysplasia in Barrett’s Esophagus. Gastrointest Endosc (2014) 79(1):37–45. doi: 10.1016/j.gie.2013.05.028
133. Zavaleta CL, Smith BR, Walton I, Doering W, Davis G, Shojaei B, et al. Multiplexed Imaging of Surface Enhanced Raman Scattering Nanotags in Living Mice Using Noninvasive Raman Spectroscopy. Proc Natl Acad Sci (2009) 106(32):13511–6. doi: 10.1073/pnas.0813327106
134. Bohndiek SE, Wagadarikar A, Zavaleta CL, Sompel de DV, Garai E, Jokerst JV, et al. A Small Animal Raman Instrument for Rapid, Wide-Area, Spectroscopic Imaging. Proc Natl Acad Sci (2013) 110(30):12408–13. doi: 10.1073/pnas.1301379110
135. McVeigh PZ, Mallia RJ, Veilleux I, Wilson BC. Widefield Quantitative Multiplex Surface Enhanced Raman Scattering Imaging In Vivo. J BioMed Opt (2013) 18(4):046011. doi: 10.1117/1.JBO.18.4.046011
136. Garai E, Sensarn S, Zavaleta CL, Van de Sompel D, Loewke NO, Mandella MJ, et al. High-Sensitivity, Real-Time, Ratiometric Imaging of Surface-Enhanced Raman Scattering Nanoparticles With a Clinically Translatable Raman Endoscope Device. J BioMed Opt (2013) 18(9):096008. doi: 10.1117/1.JBO.18.9.096008
137. Zavaleta CL, Garai E, Liu JTC, Sensarn S, Mandella MJ, Sompel de DV, et al. A Raman-based Endoscopic Strategy for Multiplexed Molecular Imaging. Proc Natl Acad Sci (2013) 110(25):2288–97. doi: 10.1073/pnas.1211309110
138. Ji H, Rolnick JA, Haker S, Barish MA. Multislice CT Colonography: Current Status and Limitations. Eur J Radiol (2003) 47(2):123–34. doi: 10.1016/S0720-048X(03)00169-4
139. Pineau BC, Paskett ED, Chen GJ, Espeland MA, Phillips K, Han JP, et al. Virtual Colonoscopy Using Oral Contrast Compared With Colonoscopy for the Detection of Patients With Colorectal Polyps. Gastroenterology (2003) 125(2):304–10. doi: 10.1016/S0016-5085(03)00885-0
140. van der Paardt MP, Zijta FM, Stoker J. MRI of the Colon. Imaging Med (2010) 2(2):195–209. doi: 10.2217/iim.10.12
141. Hu J, Whiting N, Constantinou PE, Farach-Carson MC, Carson DD, Bhattacharya PK. Real-Time Molecular MRI With Hyperpolarized Silicon Particles. In: Kumar CSSR, editor. Nanotechnology Characterization Tools for Biosensing and Medical Diagnosis. Berlin, Heidelberg: Springer Berlin Heidelberg (2018). p. 127–61. doi: 10.1007/978-3-662-56333-5_3
142. Cassidy MC, Chan HR, Ross BD, Bhattacharya PK, Marcus CM. In Vivo Magnetic Resonance Imaging of Hyperpolarized Silicon Particles. Nat Nanotechnol (2013) 8(5):363–8. doi: 10.1038/nnano.2013.65
143. Whiting N, Hu J, Zacharias NM, Lokesh GLR, Volk DE, Menter DG, et al. Developing Hyperpolarized Silicon Particles for In Vivo MRI Targeting of Ovarian Cancer. J Med Imaging (2016) 3(3):36001. doi: 10.1117/1.JMI.3.3.036001
144. Whiting N, Hu J, Pudakalakatti S, McCowan C, Carson D, Davis J, et al. In Vivo Molecular Imaging of MUC1-Expressing Colorectal Tumors Using Targeted Hyperpolarized Silicon Particles [Internet]. Poster Presented At: Joint Annual Meeting Ismrm-Esmrmb. Paris, France: International Society for Magnetic Resonance in Medicine. (2018). Available at: https://www.ismrm.org/18/program_files/PP11.htm.
145. Huang S-W, Hsu C-M, Jeng W-J, Yen T-C, Su M-Y, Chiu C-T. A Comparison of Positron Emission Tomography and Colonoscopy for the Detection of Advanced Colorectal Neoplasms in Subjects Undergoing a Health Check-Up. PloS One (2013) 8(7):e69111. doi: 10.1371/journal.pone.0069111
146. Vikram R, Iyer RB. PET/CT Imaging in the Diagnosis, Staging, and Follow-Up of Colorectal Cancer. Cancer Imaging Off Publ Int Cancer Imaging Soc (2008) 8 Spec No A(Spec Iss A):S46–51. doi: 10.1102/1470-7330.2008.9009
147. Griffiths GL, Chang C-H, McBride WJ, Rossi EA, Sheerin A, Tejada GR, et al. Reagents and Methods for PET Using Bispecific Antibody Pretargeting and 68Ga-Radiolabeled Bivalent Hapten-Peptide-Chelate Conjugates. J Nucl Med Off Publ Soc Nucl Med (2004) 45(1):30–9.
148. Carrasquillo JA, Pandit-Taskar N, O’Donoghue JA, Humm JL, Zanzonico P, Smith-Jones PM, et al. 124I-Hua33 Antibody PET of Colorectal Cancer. J Nucl Med (2011) 52(8):1173–80. doi: 10.2967/jnumed.110.086165
149. Shi X, Gao K, Huang H, Gao R. Pretargeted Immuno-PET Based on Bioorthogonal Chemistry for Imaging EGFR Positive Colorectal Cancer. Bioconjug Chem (2018) 29(2):250–4. doi: 10.1021/acs.bioconjchem.8b00023
Keywords: colorectal cancer, biomarkers, colonoscopy, cell surface protein, adenocarcinoma, early detection
Citation: Ramezani S, Parkhideh A, Bhattacharya PK, Farach-Carson MC and Harrington DA (2021) Beyond Colonoscopy: Exploring New Cell Surface Biomarkers for Detection of Early, Heterogenous Colorectal Lesions. Front. Oncol. 11:657701. doi: 10.3389/fonc.2021.657701
Received: 23 January 2021; Accepted: 09 April 2021;
Published: 05 July 2021.
Edited by:
Yun Dai, Peking University First Hospital, ChinaReviewed by:
Xueliang Wu, First Affiliated Hospital of Hebei North University, ChinaTD Wang, University of Michigan, United States
Copyright © 2021 Ramezani, Parkhideh, Bhattacharya, Farach-Carson and Harrington. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel A. Harrington, RGFuaWVsLkhhcnJpbmd0b25AdXRoLnRtYy5lZHU=
†These authors have contributed equally to this work
 Saleh Ramezani
Saleh Ramezani Arianna Parkhideh
Arianna Parkhideh Pratip K. Bhattacharya
Pratip K. Bhattacharya Mary C. Farach-Carson
Mary C. Farach-Carson Daniel A. Harrington
Daniel A. Harrington