- 1Department of Orthopedics, The Second Xiangya Hospital, Central South University, Changsha, China
- 2Hunan Key Laboratory of Tumor Models and Individualized Medicine, The Second Xiangya Hospital, Central South University, Changsha, China
- 3Department of Orthopedics, Brain Hospital of Hunan Province (The Second People’s Hospital of Hunan Province), Changsha, China
- 4Department of Respiratory and Critical Care Medicine, Xiangya Hospital, Central South University, Changsha, China
- 5National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Changsha, China
- 6Department of General Medicine, The Second Xiangya Hospital, Central South University, Changsha, China
- 7Clinical Medicine Eight-Year Program, Central South University, Changsha, China
Background: Chromobox family genes (CBXs) are known to play roles in numerous modifications of the chromatin in order to inhibit the transcription of target genes. CBXs have been shown to be expressed at high levels in many types of cancer and can also serve as a target gene for therapeutic purposes. However, little is known about the expression and prognostic value of CBXs in human sarcomas.
Methods: The transcription level of CBXs was analyzed using the Oncomine dataset, and the differential expression of CBXs in sarcoma was reported by the Gene Expression Profiling Interactive Analysis (GEPIA) dataset. We also used the CCLE dataset to evaluate the expression of CBXs in a sarcoma cell line. The prognostic value of CBXs was analyzed using GEPIA and Kaplan–Meier analysis. In addition, the corrections between CBXs and their co-expressed genes were reported using Oncomine and GEPIA datasets. DAVID was used to perform GO function enrichment analysis for the CBXs and their co-expression genes. Finally, TIMER was used to analyze the immune cell infiltration of CBXs in patients with sarcoma.
Results: HP1-α/β/γ (CBX1/3/5) and CBX4/6/8 were found to be overexpressed in human sarcoma, and CBXs were upregulated in almost all the sarcoma cell line. The expression levels of HP1-α/β/γ (CBX1/3/5) and CBX7 were associated with overall survival (OS) in patients with sarcoma, while high expression levels of CBX7 were related to disease-free survival (DFS). In addition, the expression levels of CBX2/6/7 were related to recurrence-free survival (RFS). We also found that the CBX family was positively correlated with the infiltration of immune cells, including CD8+ T cells, CD4+ T cells, B cells, macrophages, neutrophils, and dendritic cells, in sarcoma.
Conclusions: The results from the present study indicated that CBXs were significantly associated with prognosis and immunological status in sarcoma. These data suggest that CBXs could serve as potential biomarkers for prognosis and immune infiltration in human sarcoma.
Introduction
Sarcomas are rare but aggressive bone and soft tissue malignancies that afflict patients of all ages. Sarcomas are usually incurable because chemotherapy and surgery are not effective (1); these malignant tumors also exhibit broad differentiation (2). Although some causative factors have been established for sarcomas, including environmental factors, stimulation by foreign matter, and endocrine dyscrasia, the precise underlying causes of sarcoma have yet to be elucidated. Although generally rare, sarcomas can be found in patients of any age and tend to occur more commonly in adolescents and teenagers than in the elderly (3). Osteosarcoma is the most common form of primary bone sarcoma and accounts for 1% of all tumors; the incidence of osteosarcoma in children was previously reported to be five per million (4).
Chromobox family genes (CBXs) are associated with a variety of modifications to the chromatin that inhibit the transcription of target genes as key elements of polycomb repressive complex 1 (PRC1) (5). Currently, eight members of the CBX family have been identified, based on their single N-terminal chromosomal domains, consisting of three β folds and an α helix. There are two broad categories of CBX genes: heterochromatin protein 1 and polycomb complexes. HP1-α/β/γ (CBX1/3/5) are heterochromatic proteins while CBX2/4/6/7/8 are polycomb complexes. The heterochromatin protein 1 group consists of an N-terminal chromodomain and a C-terminal chromodomain, while the polycomb group contains only a conservative N-terminal chromodomain (6). Different CBX proteins have been associated with different parts of the chromatin, leading to the specific transcription of target genes (7, 8).
According to previous studies, CBXs are known to be involved in the occurrence and development of a diverse range of tumors through various pathways (9, 10). For example, Han et al. reported that CBX2 could act as a tumor promoter in osteosarcoma by targeting miRNA let-7a (11). In addition, Ma et al. reported that CBX3 was related to an unfavorable prognosis and tumorigenesis in patients with osteosarcoma (12). In another study, Wang et al. demonstrated that targeting the CK1α/CBX4 axis may provide benefit to patients with metastasis of osteosarcoma (13). Liang et al. found that HP1-α/β/γ (CBX1/3/5) and CBX2/4/6/7 exerted an effect on breast cancer, thus indicating that CBX2 is expressed at high levels in basal-like and HER-2 subtypes (14). In addition, CBX4/7 is highly expressed in Luminal A and Luminal B subtypes of breast cancer (14, 15). The increased mRNA expression of HP1-β/γ (CBX1/3) and CBX2 has been linked to a poorer Relapse free survival (RFS) via survival analysis; better outcomes were found to be associated with higher expression levels of CBX4/5/6/7 (14). Moreover, the high expression levels of HP1-β/γ (CBX1/3) were associated with overall survival (OS) and disease-free survival (DFS) in patients with non-small-cell lung cancer. Furthermore, the elevated expression of HP1-γ (CBX3) has been demonstrated to exert impact on tumor diameter and lymph node metastasis (16). A previous study reported that the increased mRNA expression of HP1-β/γ (CBX1/3) and CBX2/6/8 was correlated with a worse OS, while the overexpression of CBX7 was related to a greater OS in patients with hepatocellular carcinoma (17). However, the role of CBXs in sarcoma remains unclear. Here, we aimed to investigate the expression of CBX in sarcomas and the relationship between CBXs and prognosis/immune cell infiltration in patients with sarcoma.
Methods
Ethics Statement
This study was approved by the Second Xiangya Hospital of Central South University Committee for Clinical Research and all methods were carried out in accordance with the Declaration of Helsinki.
ONCOMINE Analysis
The Oncomine database (https://www.oncomine.org/resource/login.html) is usually adopted to analyze DNA or RNA sequences from a comprehensive cancer microarray database and genome-wide expression data for malignant tumors. In the present study, we used the Oncomine database to identify the differential transcriptional expression of CBXs in multiple cancer tissues and corresponding normal tissues. Moreover, three databases including Detwiller sarcoma database (18), Barretina sarcoma database (19), and Quade uterus database (20) were used to analyze the differential transcriptional expression of CBXs in subtypes of sarcoma and corresponding normal tissues using p < 0.05 as the significance threshold.
GEPIA Analysis
The GEPIA dataset (http://gepia.cancer-pku.cn/) is an online service that includes a spectrum of cancer expression data. The GEPIA dataset contains 9,736 tumor samples and 8,587 normal samples from the TCGA. The GEPIA dataset is usually used to analyze data arising from the TCGA project. In this study, we used the GEPIA database to analyze the expression levels of CBX genes in sarcoma tissues and normal tissues.
CCLE Dataset Evaluation
The CCLE dataset (https://www.broadinstitute.org/ccle) is usually used to accurately depict the genetic characteristics of cancer cells. The CCLE dataset can provide information relating to DNA mutation and gene expression. We used the CCLE database to investigate the expression of CBXs in sarcoma cell lines.
Kaplan–Meier Plotter Database Analysis
The Kaplan–Meier Plotter Database (https://kmplot.com/analysis/index.php?p=service&cancer=pancancer_rnaseq) is used to evaluate the influence of genes on the survival of patients afflicted with various forms of cancers. In this study, we used the Kaplan–Meier Plotter Database to analyze the relationship between the expression of CBXs in sarcoma and associated survival rates.
TIMER Dataset Analysis
The TIMER database (https://cistrome.shinyapps.io/timer/) is an online service that can be used to investigate the infiltration of different immune cells and their clinical significance. In the present study, CBXs were input into the ‘Gene module’ tool of TIMER in order to generate scatterplots to investigate the association between CBX expression and immune infiltration in sarcoma.
GO Enrichment Analyses of DEGs
The Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.ncifcrf.gov) (version 6.7), an online biological information database that provides a comprehensive set of functional annotation information for genes and proteins, was used to perform GO enrichment analyses of CBXs and their co-expression genes (21).
Results
Transcriptional Levels of the CBX Family in Patients With Sarcoma
According to data arising from the Oncomine database, we found that HP1-α/β/γ (CBX1/3/5) and CBX4/6/8 were all highly expressed in sarcoma (Figure 1). Additionally, three databases including Detwiller sarcoma database, Barretina sarcoma database, and Quade uterus database were used to analyze the differential transcriptional expression of CBXs in subtypes of sarcoma and corresponding normal tissues. The Detwiller sarcoma database, reported by Detwiller et al. (18) identified distinctly different patterns of expression between sarcomas and normal tissues as assessed by hierarchical clustering analysis. The Barretina sarcoma database, established by Barretina et al. (19), described an integrative analysis of DNA sequence, copy number, and mRNA expression in 207 sarcoma samples encompassing seven major subtypes. The Quade uterus database, reported by Quade et al. (20), depicted RNAs profiled from four normal uterine myometria, seven uterine leiomyomas, and nine uterine leiomyosarcomas. As shown in Figure 2, the expression levels of HP1-β (CBX1) were upregulated in the Detwiller sarcoma database; the fold-changes for the expression of these genes in synovial sarcoma, pleomorphic liposarcoma, leiomyosarcoma, fibrosarcoma, and malignant fibrous histiocytoma were 4.180, 2.460, 3.155, 4.010, and 3.294, respectively. With regard to the Barretina sarcoma database, the expression levels of HP1-β (CBX1) in pleomorphic liposarcoma showed a fold-change of 2.826 when compared with normal samples and a fold-change of 3.315 when compared with normal samples in leiomyosarcoma.
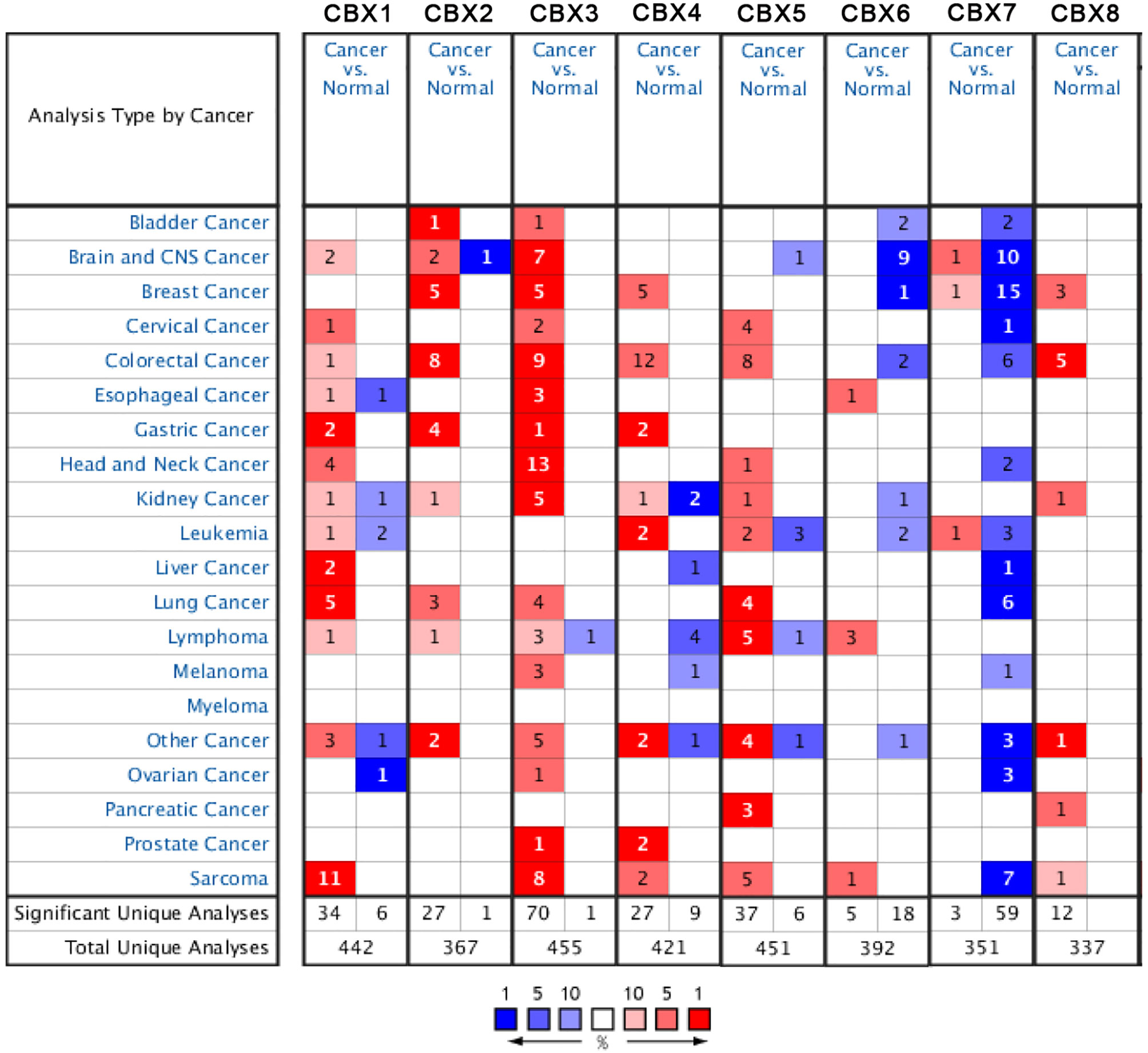
Figure 1 The expression levels of CBX genes in different types of human cancers and normal samples. The red cells represent evidence of gene overexpression. The blue cells represent evidence of reduced gene expression. The numbers in each cell represent the evidential frequencies. The deeper the color, the higher the significance.
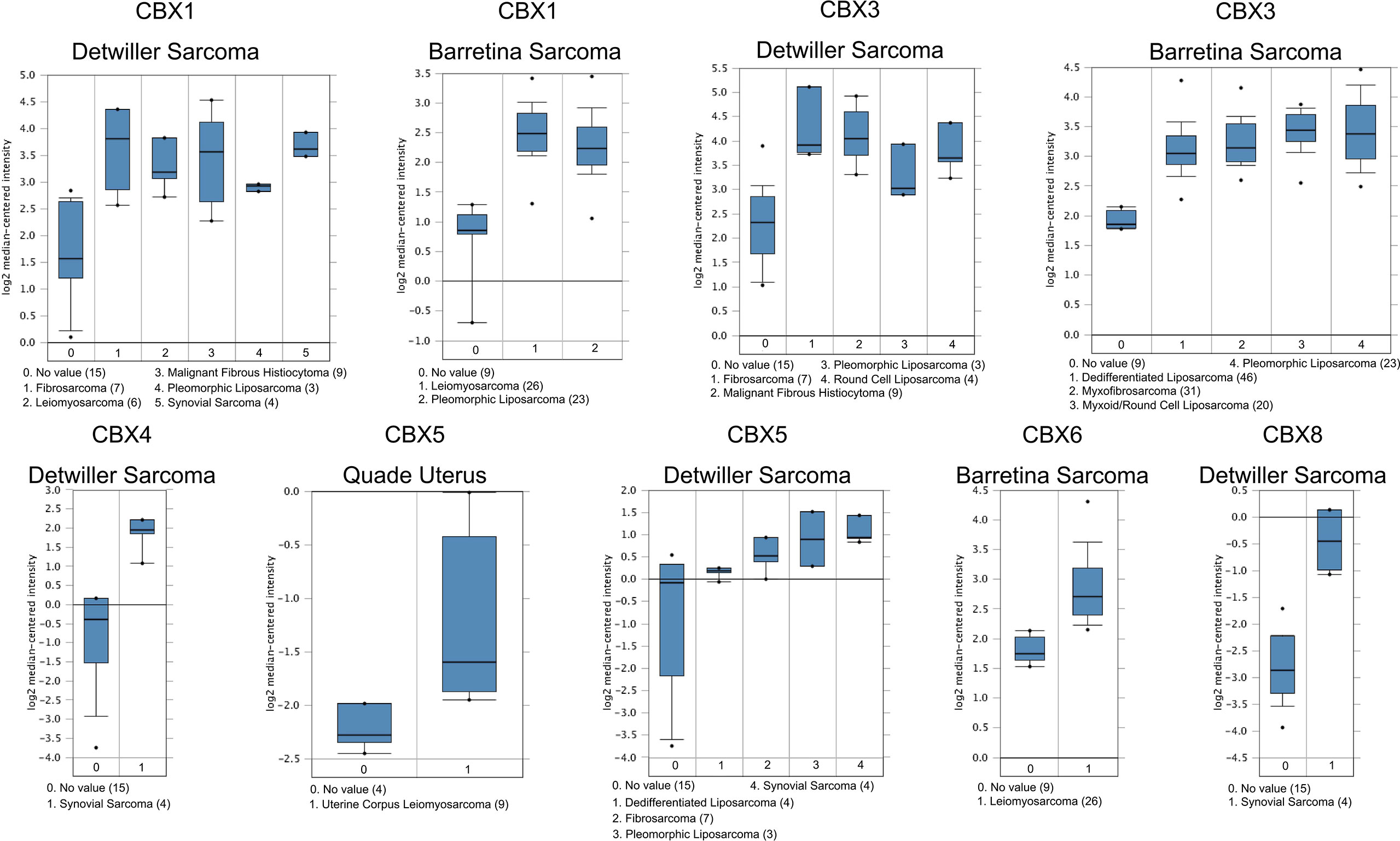
Figure 2 Significant changes of CBX gene expression at the transcription level between different types of sarcoma and normal tissues. The horizontal axis represents the type of tissue, and the vertical axis represents the expression level of CBX genes. The number next to the name of the sarcoma subtype represents the specific subtype vs ‘normal’ cases.
With regard to the Detwiller sarcoma database, the fold-changes of HP1-γ (CBX3) expression in fibrosarcoma, malignant fibrous histiocytoma, pleomorphic liposarcoma, and round cell liposarcoma were 3.515, 3.249, 2.242, and 2.645, respectively. The expression levels of HP1-γ (CBX3) in dedifferentiated liposarcoma, myxoid/round cell liposarcoma, myxofibrosarcoma, and pleomorphic liposarcoma were upregulated by 2.270, 2.803, 2.439, and 2.785, fold for the Barretina sarcoma database. The fold-change for CBX4 expression was 6.516 in synovial sarcoma when compared with normal tissues for the Detwiller sarcoma database. High expression levels of HP1-α (CBX5) were found in uterine corpus leiomyosarcoma, synovial sarcoma, pleomorphic liposarcoma, dedifferentiated liposarcoma, and fibrosarcoma, with fold-changes of 2.005, 3.713, 3.401, 2.018, and 2.673, respectively (Quade uterus database and Detwiller sarcoma database). The expression levels of CBX6/8 in leiomyosarcoma (Barretina sarcoma database) and synovial sarcoma (Detwiller sarcoma database) were raised by 2.068- and 5.159-fold, respectively.
The mRNA Levels of CBX Genes in Sarcoma
Next, we used the GEPIA dataset to compare the expression of CBX family mRNAs between sarcoma and normal tissues. The mRNA levels of HP1-α/β/γ (CBX1/3/5) and CBX2/4/8 were significantly higher than those in normal tissues. In contrast, the mRNA expression levels of CBX6/7 in sarcoma were lower than those in normal tissues (Figure 3).
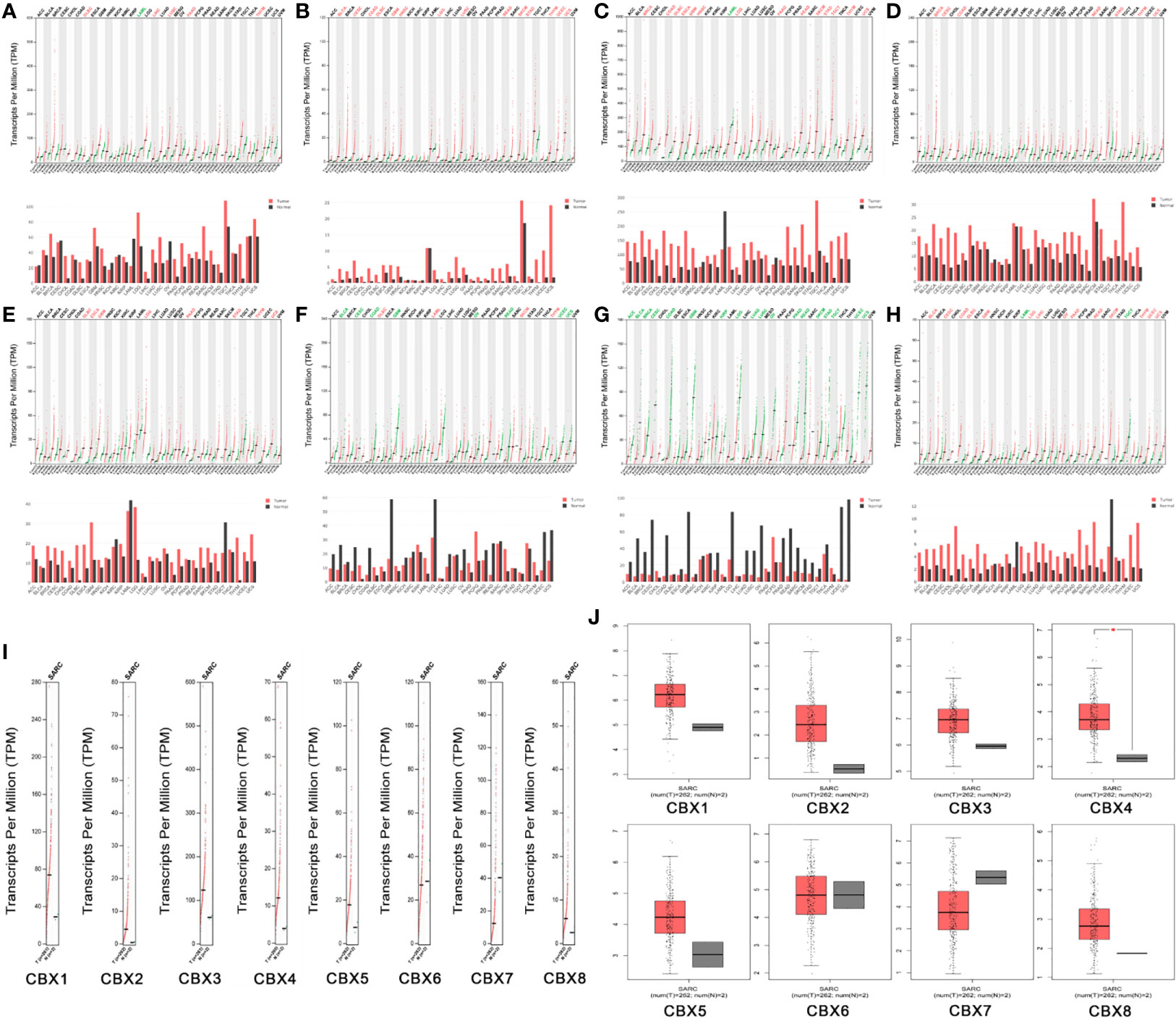
Figure 3 The expression levels of CBX genes in sarcoma. (A–H) The expression levels of CBX1-8 in pan-cancer, (I–J) The expression levels of CBX genes in sarcoma. The expression levels of CBX genes in sarcomas. Each dot represents an individual sample, *P < 0.05.
The Expression Levels of CBX Genes in Sarcoma Cell Lines
We used the CCLE dataset to investigate the expression levels of CBXs in human cancer. Data showed that all eight members of the CBX family were expressed at high levels in sarcoma cell lines (Figure 4).
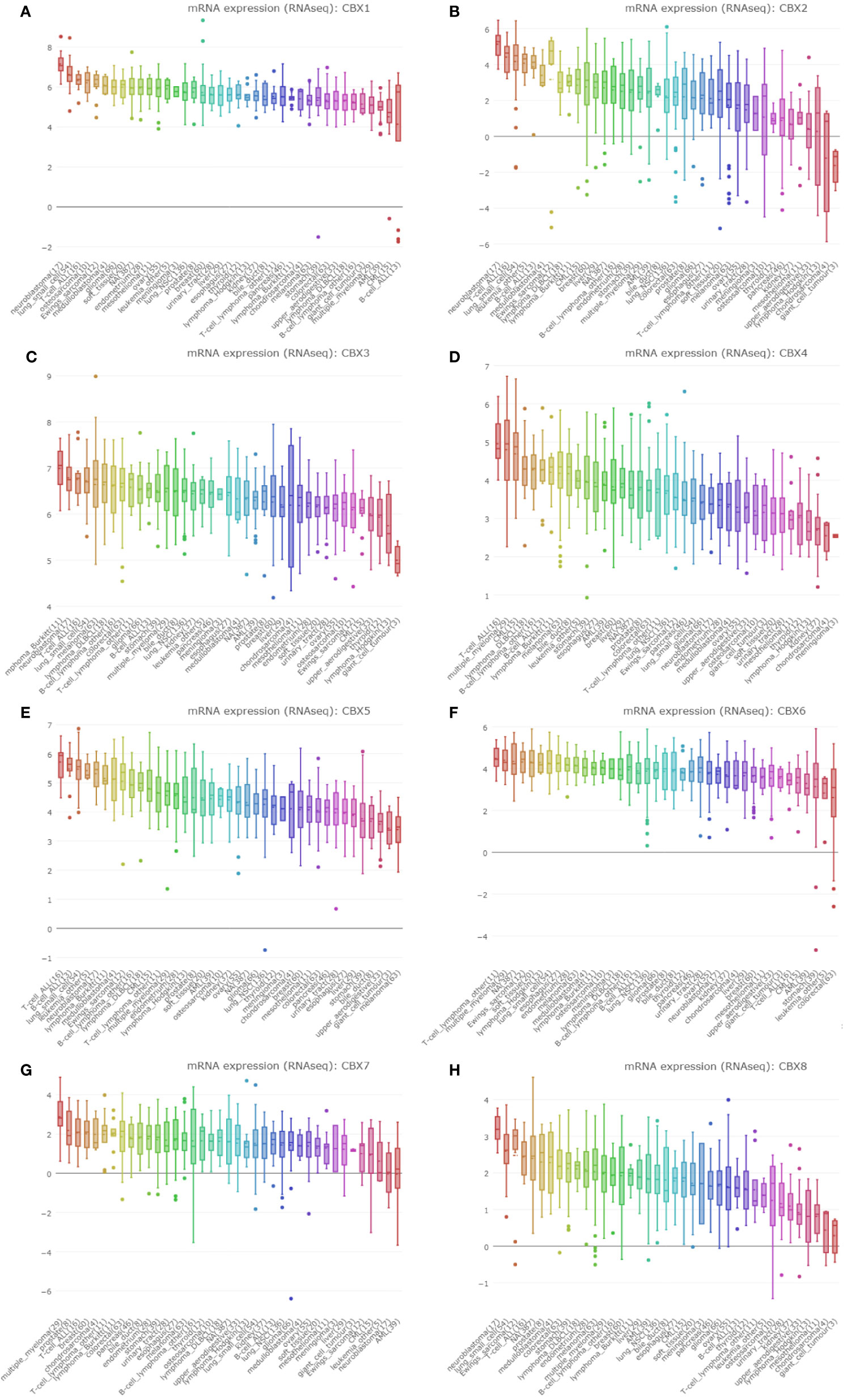
Figure 4 The expression of CBXs in sarcoma cell lines. (A–H) The expression of CBX1-8 in sarcoma cell lines. The number next to the lineage name represents number of cell lines in the lineage. The dashed line within a box is the mean.
The Prognostic Value of CBXs in Sarcoma
The prognostic value of CBX gene expression in sarcoma was evaluated by GEPIA and the Kaplan–Meier Plotter Database. As shown in Figure 5 (GEPIA), increased expression levels of HP1-α/γ (CBX3/5) were associated with a poorer OS in patients with sarcoma, while elevated expression levels of CBX7 were associated with a better OS. HP1-β (CBX1) and CBX2/4/6/8 also tended to exert impact on the OS, but without statistical significance. High expression levels of CBX6 appear to be associated with a better DFS. Similar findings were evident in our Kaplan–Meier analysis (Figure 6). The expression levels of HP1-α/β/γ (CBX1/3/5) and CBX2 were associated with a poorer OS, while the levels of CBX7 were associated with a better OS. Furthermore, high expression levels of CBX6/7 were associated with a better RFS, while high expression levels of CBX2 were associated with a worse RFS.
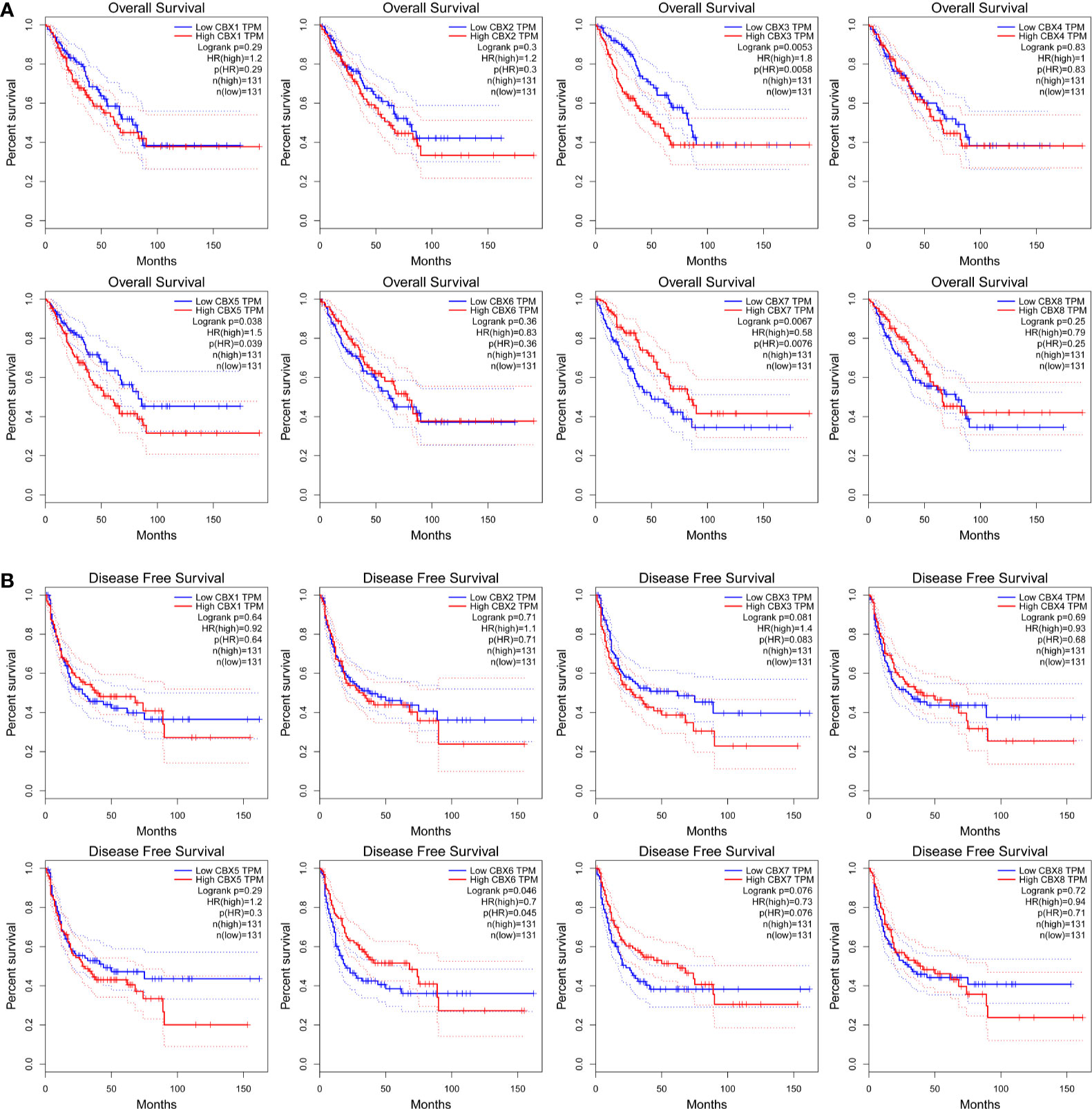
Figure 5 The prognostic significance of CBXs in sarcoma patients. (A) The association between the elevated expression of CBX genes and overall survival in patients with sarcoma. (B) The association between high expression levels of CBX genes and disease-free survival in patients with sarcoma. HR, hazard ratio; TPM, Transaction per million.
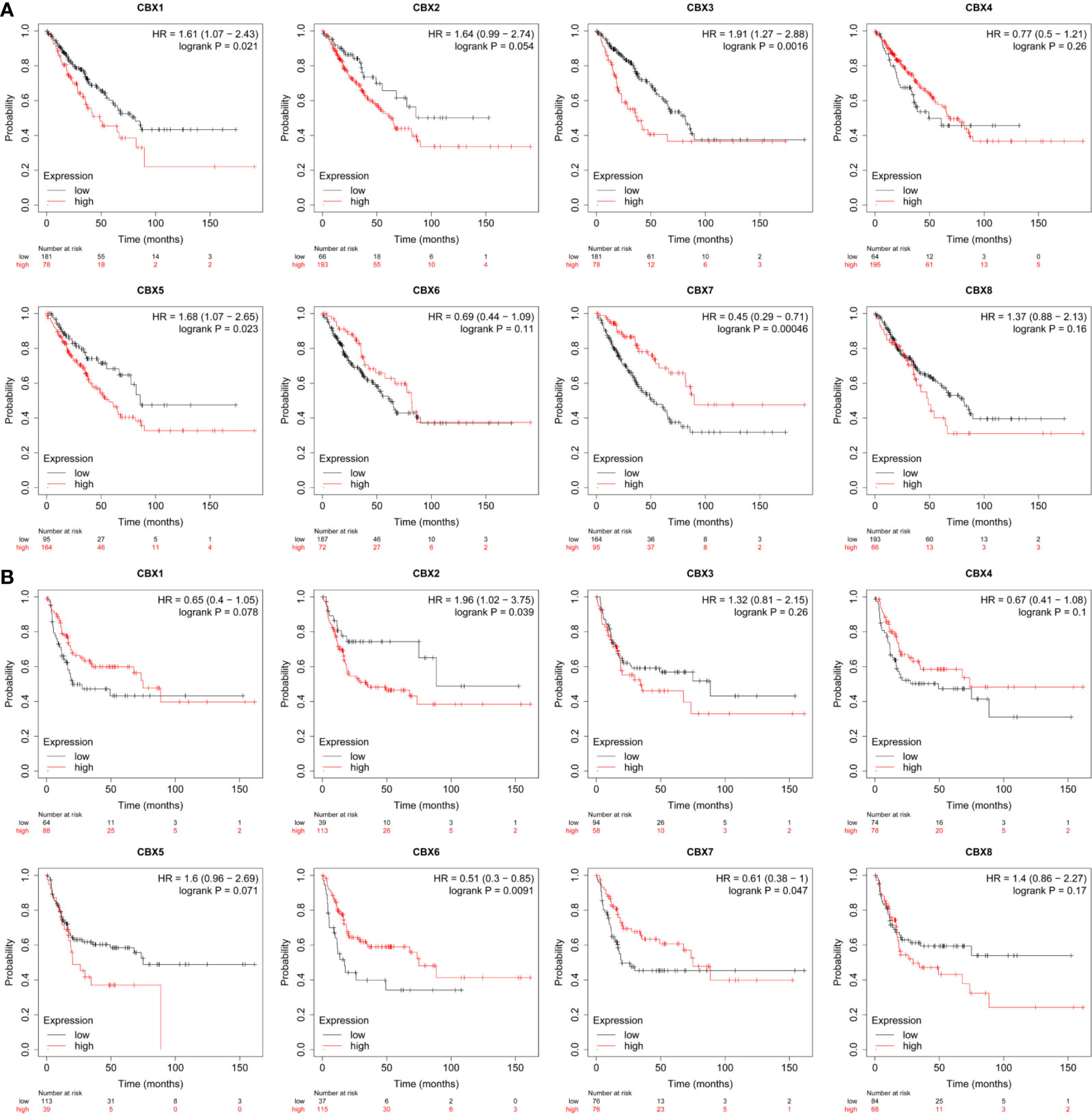
Figure 6 The prognostic value of CBX genes in sarcoma patients. (A) The association between high expression levels of CBX genes and the overall survival of patients with sarcoma. (B) The association between elevated expression levels of CBX genes and the recurrence-free survival of patients with sarcoma. HR, hazard ratio.
The Identification of Genes That Were Co-Expressed With CBX Genes in Sarcoma
We used the Oncomine database to identify a series of genes that were co-expressed with CBX genes in sarcoma. HP1-β (CBX1) was co-expressed with S100A6, STAG1, MBD4, CHMP2B, PMM1, PCNP, C5orf54, TTC33, CEP70, and KIAA1586. CBX2 was co-expressed with USPS, TTLL4, ASRBK1, SMYDS, MDFI, BSG, MTA1, YBX1, MCM4, and GGA2. HP1-γ (CBX3) was co-expressed with HNRNPAZ2B1, CCT6A, HNRNPR, SFRS13A, DDX46, SFRS2, SFRS3, TRA2B, NONO, MSH2, and HP1-γ (CBX3). CBX4 was co-expressed with SF1, RABSB, MBOAT7, PRKACA, AKT2, WASL, CCNG1, UBL4A, PRKAR2A, and TRIB1. HP1-α (CBX5) was co-expressed with WDHD1, CDK2, ARHGAP28, PCDH19, RORB, CYP39A1, ST85IA1, PAR1GD52, SPTLC2, and ZDHHHC2. CBX6 was co-expressed with SYP11, ANKRD35, PRICKLE1, MXD4, ZBTB20, CD99L2, RNF150, JAM3, CLIP3, and MFAP4. CBX7 was co-expressed with PDCD6IP, CNOT6L, UNC80, SRI, KGFLP2, MRPS6, PCM1, S1PR1, SEPSECS, and LPP. CBX8 was co-expressed with CYP46A1, ALOX12B, C1orf109, TET2, MGC16384, MYO1A, HTR7, PNKD, C7orf28B, and ASPHD2 (Figure 7A).
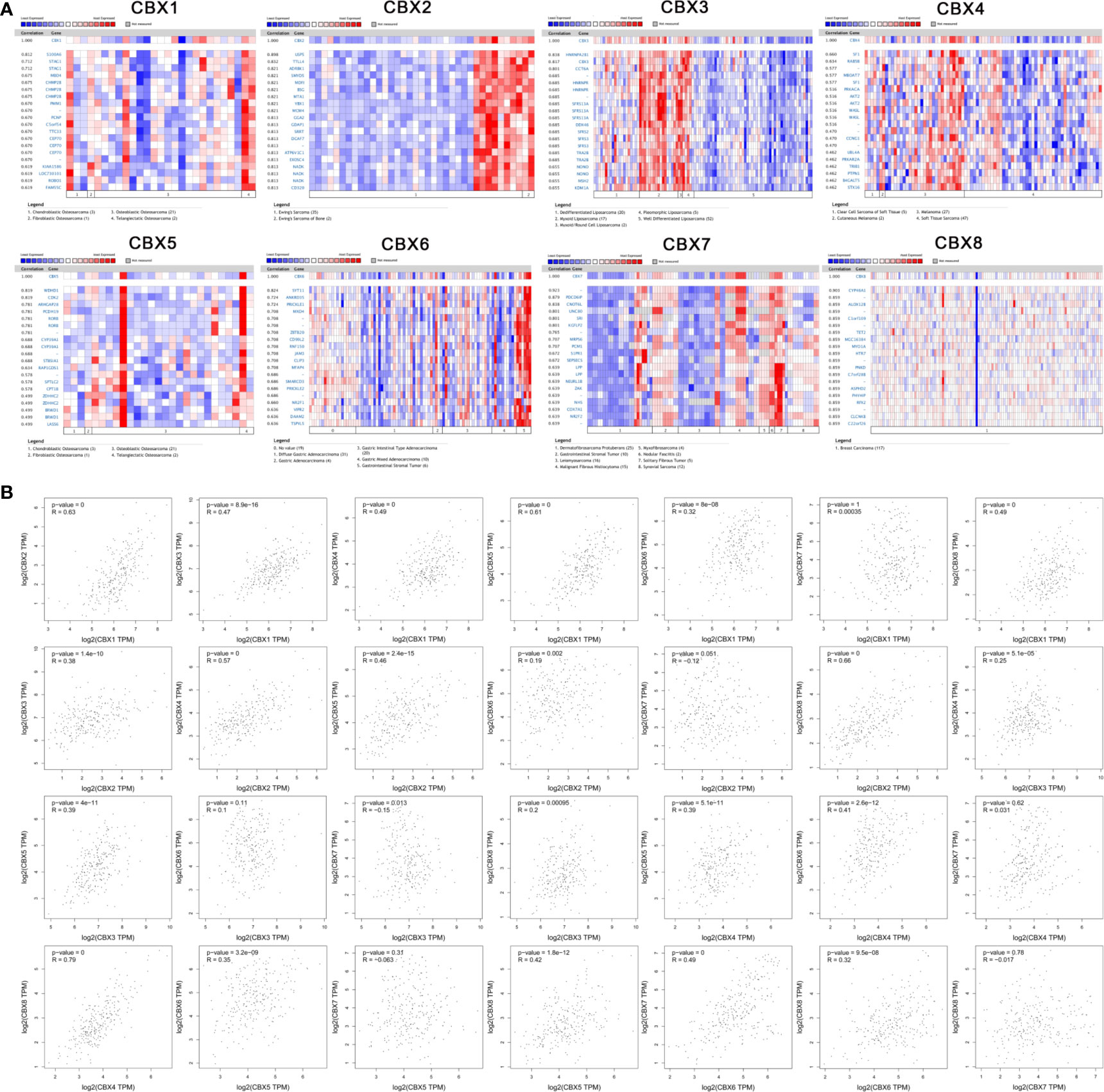
Figure 7 Co-expression analysis. (A) Genes that were co-expressed with CBX genes in sarcoma. (B) Correlation between CBX genes in sarcoma.
Similarly, we analyzed the co-expression relationships between different CBX genes. HP1-β (CBX1) was positively correlated with CBX2 (R = 0.63, P < 0.05), HP1-γ (CBX3) (R = 0.47, P < 0.05), CBX4 (R = 0.49, P < 0.05), HP1-α (CBX5) (R = 0.61, P < 0.05), CBX6 (R = 0.32, P < 0.05), and CBX8 (R = 0.49, P < 0.05), but there was no significant correlation between HP1-β (CBX1), and CBX7. CBX2 was significantly correlated with HP1-γ (CBX3) (R = 0.38, P < 0.05), CBX4 (R = 0.57, P < 0.05), HP1-α (CBX5) (R = 0.46, P < 0.05), CBX6 (R = 0.19, P < 0.05), and CBX8 (R = 0.66, P < 0.05). HP1-γ (CBX3) was significantly correlated with CBX4 (R = 0.25, P < 0.05), HP1-α (CBX5) (R = 0.39, P < 0.05), CBX7 (R = −0.15, P < 0.05) and CBX8 (R = 0.2, P < 0.05). CBX4 significantly correlated with HP1-α (CBX5) (R = 0.35, P < 0.05), CBX6 (R = 0.41, P < 0.05), and CBX8 (R = 0.79, P < 0.05). HP1-α (CBX5) was significantly correlated with CBX6 (R = 0.35, P < 0.05) and CBX8 (R = 0.42, P < 0.05). In addition, CBX6 was significantly correlated with CBX7 (R = 0.49, P < 0.05) and CBX8 (R = 0.32, P <0.05). However, there was no significant correlation between CBX7 and CBX8 (Figure 7B).
Next, we used the DAVID online tool to perform GO function enrichment analysis from three aspects: biological processes (BPs), cellular components (CCs), and molecular functions (MFs). We found that CBXs and their co-expressed genes were mainly involved in GO:0000398 (mRNA splicing, via spliceosome) for BP, GO:0005634 (nucleus) for CC, and GO:0003682 (chromatin binding) for MF. More detailed information is given in Figure 8 and Table 1.

Figure 8 GO analysis of CBX genes and co-expressed genes. GO analysis was based on three aspects: (BP) biological processes, (CC) cellular components, and (MF) molecular function.
Immune Infiltration Levels in Sarcoma
In the present study, the TIMER dataset was used to analyze whether the expression of CBX genes were correlated with the immune infiltration of sarcoma. We found that HP1-β (CBX1) expression was significantly associated with the infiltration of CD8+ T cells (Correlation coefficient (cor) = −0.131, p < 0.05), CD4+ T cells (cor = −0.362, p < 0.05), macrophages (cor = −0.27, p < 0.05), and dendritic cells (cor = −0.4, p < 0.05). CBX2 was significantly associated with the infiltration of CD4+ T cells (cor = −0.293, p < 0.05), macrophages (cor = −0.402, p < 0.05), neutrophils (cor = −0.173, p < 0.05), and dendritic cells (cor = −0.384, p < 0.05). HP1-γ (CBX3) was significantly associated with infiltration by CD4+ T cells (cor = −0.247, p < 0.05). CBX4 was significantly associated with infiltration by B cells (cor = 0.137, p < 0.05), CD4+ T cells (cor = −0.187, p < 0.05), macrophages (cor = −0.28, p < 0.05), and dendritic cells (cor = −0.197, p < 0.05). HP1-α (CBX5) was significantly associated with infiltration by CD4+ T cells (cor = −0.279, p < 0.05), macrophages (cor = −0.293, p < 0.05) and dendritic cells (cor = −0.371, p < 0.05). CBX6 was significantly associated with infiltration by CD8+ T cells (cor = 0.157 p < 0.05), CD4+ T cells (cor = −0.341 p < 0.05), macrophages (cor = −0.217 p < 0.05), and dendritic cells (cor = −0.178 p < 0.05). CBX7 was significantly associated with infiltration by CD8+ T cells (cor = 0.142 p < 0.05). Finally, CBX8 was significantly associated with infiltration by CD8+ T cells (cor = −0.146 p < 0.05), CD4+ T cells (cor = −0.182 p < 0.05), macrophages (cor = −0.357 p < 0.05), neutrophils (cor = −0.264 p < 0.05), and dendritic cells (cor = −0.284 p < 0.05) (Figure 9).
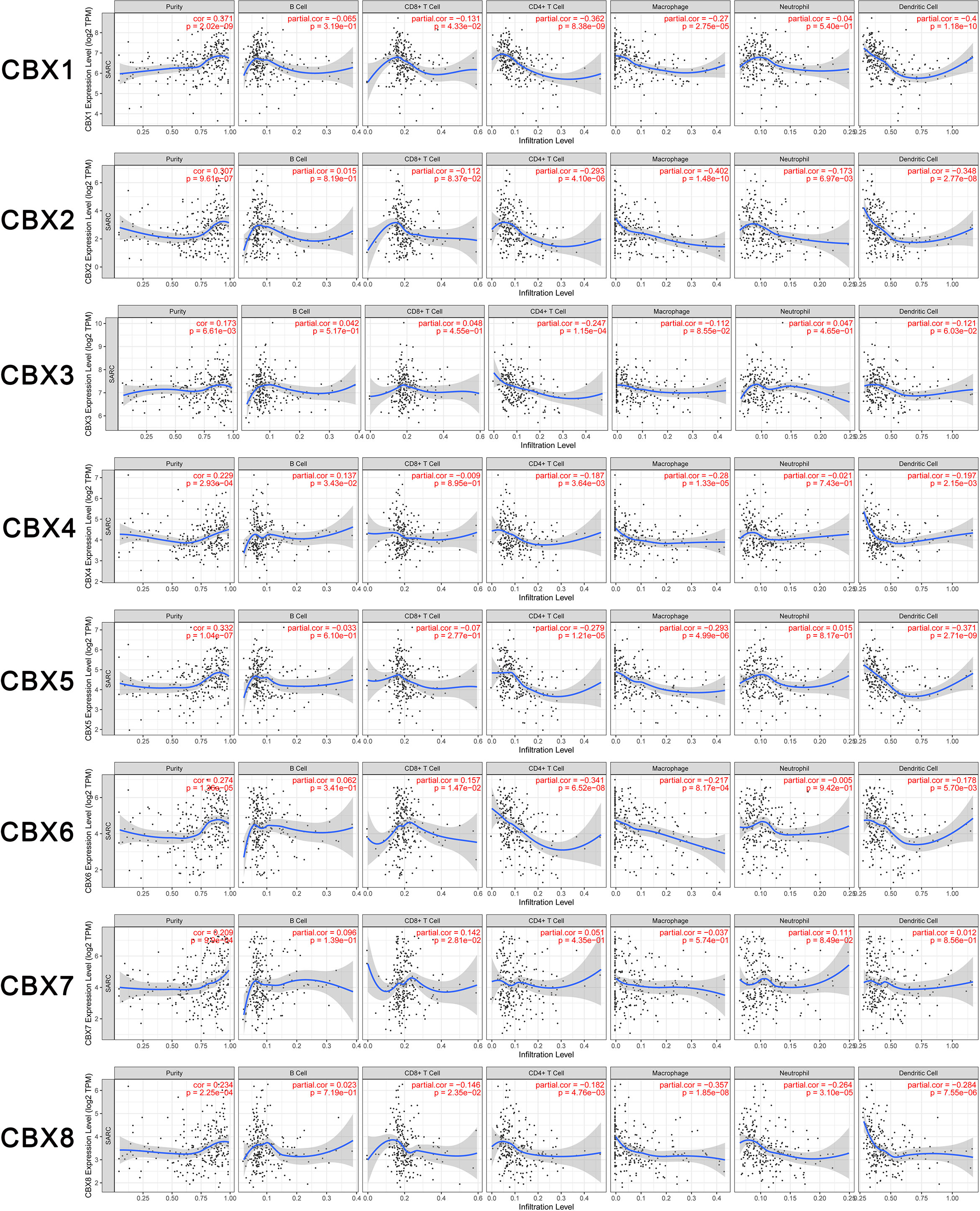
Figure 9 Relationship between differentially expressed CBX genes and immune cell infiltration. The immune cells we analyzed included B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells.
Discussion
The CBX family of genes is thought to represent vital elements in a variety of tumors (14–16). However, the association between CBXs and sarcoma has not been extensively reported. Currently, there is a renewed interest in immunotherapy for cancer. Most immunotherapy studies have focused on melanoma. These have revealed that the overexpression of GM2, GD2, and GD3 immunogenicity gangliosides can trigger an antibody response and improve the survival rate of patients with melanoma. Other studies have investigated MHC-restricted cancer-testicular antigens; these have also raised the prospect of immunotherapy (22). Subsequently, the concept of tumor vaccines was proposed for specific tumor epitopes by stimulating an immune response. Therefore, immune-related studies on sarcomas, combined with radiotherapy, chemotherapy, and several targeted therapies, have become increasingly popular (23). Therefore, we conducted this study to reveal the relationships between CBX genes and human sarcoma, particularly with respect to their prognostic value and the immune infiltration of CBXs.
The up-regulated expression of HP1-β (CBX1) has been reported for many forms of cancer. For example, Liang et al. reported high expression levels of HP1-β (CBX1) mRNA in breast cancer and found that this was associated with a poorer RFS. HP1-β (CBX1) has also been associated with chemoresistance in patients with breast cancer. Therefore, these authors defined HP1-β (CBX1) as a potential target for the treatment of breast cancer (14). In addition, Yang et al. found that the overexpression of HP1-β (CBX1) in hepatocellular carcinoma could activate the Wnt/β-Catenin signaling pathway by interacting with the transcription factor HMGA2, thus suggesting that HP1-β (CBX1) was an independent factor for hepatocellular carcinoma (24). Similar results were reported by Gang et al. (17). Higher expression levels of HP1-β/γ (CBX1/3) and CBX2/6/8 were reported to be related to a worse OS while higher expression levels of CBX7 were associated with a better OS. These results are similar to our present results which indicated that the up-regulated expression of HP1-α/γ (CBX3/5) is related to a poorer OS, while the up-regulated expression of CBX7 was associated with a better OS. Similar results were obtained from our Kaplan–Meier Plotter analyses.
According to a previous study, CBX2 is a key member of the polycomb group (PcG) family and CBX2 mRNA was predominately localized in spermatogonia and spermatocytes, as demonstrated by in situ hybridization (ISH) (25). Tatavosian et al. found that CBX2 undergoes phase separation to form condensates and that these CBX2 condensates concentrate DNA and nucleosomes (26). Plys et al. further reported that the domain of CBX2 was the same as the domain related to chromatin compaction and development, thus improving the possibility of an evolutionary or mechanistic link between these activities (27). In another study, Alexandra et al. reported that the stabilization of the testis required CBX2-mediated repression of bivalent ovary-determining genes (28). In hepatocellular carcinoma, the knockdown of CBX2 restrained the proliferation of HCC cells and increased the phosphorylation of YAP. These data suggest that CBX2 could be a potential target for hepatocellular carcinoma (29). In addition, Zheng et al. showed that higher expression levels of CBX2 were significantly and independently associated with a worse OS by affecting the PI3K/AKT signaling pathway in breast cancer (30). In our present study, we observed elevated expression levels of CBX2 and found that CBX2 was associated with a poorer OS, thus indicating that this gene might be an independent prognostic factor for human sarcoma.
High expression levels of HP1-γ (CBX3) [encoded by HP1-γ (CBX3)] are known to accelerate HCC cell proliferation, thus suggesting that HP1-γ (CBX3) is a crucial oncogene in hepatocellular carcinoma (31). Similar findings have been reported for glioma; high expression levels of HP1-γ (CBX3) are known to predict a worse prognosis (32). In gastric cancer, Lin et al. found that HP1-γ (CBX3) was overexpressed and could regulate genes associated with the cell cycle, mismatch repair, and immune-related pathways. The expression level of HP1-γ (CBX3) was also significantly and inversely related to the expression levels of TILs, PDCD1, and PDCD2, and immunotherapy responses, thus implying that HP1-γ (CBX3) could influence the efficacy of immunotherapy and chemotherapy (33). Ma et al. further confirmed the function and role of HP1-γ (CBX3) in osteosarcoma, by showing that the expression of HP1-γ (CBX3) was associated with a poorer DFS and OS, as well as a larger tumor size, a higher distant metastasis rate, and a higher clinical stage. These authors used HP1-γ (CBX3) siRNA to knockdown HP1-γ (CBX3) and thus block proliferation ability, thus resulting in increased levels of apoptosis and cell cycle arrest at the G0 and G1 phase (12); these findings were consistent with those reported in the present study. Another study reported that the elevated expression of HP1-γ (CBX3) was related to unfavorable OS in patients with human sarcoma. CBX4 is known to recruit GCN5 to the Runx2 promoter to transcriptionally upregulate Runx2; in this manner, CBX4 can promote the metastasis of osteosarcoma (13). In another paper, Hu et al. reported that the cell growth and migration of human lung cancer cell was suppressed by the knockdown of CBX4, both in vitro and in vivo. Furthermore, CBX4 has been shown to promote proliferation and metastasis by regulating the BMI-1 pathway, thus suggesting that CBX4 might be a potential therapeutic target in lung cancer (34). In the digestive system, the suppression of Runx2 by CBX4 resulted in the inhibition of cell migration, invasion, and metastasis (35). Interestingly, Ren et al. reported that CBX4 counteracts senescence in human mesenchymal stem cells (HMSC) by maintaining nucleolar homeostasis, and that CBX4 maintained nucleolar homeostasis by recruiting nucleolar protein-fibrillin and heterochromatin KRAB associated protein 1 (KAP1) within nucleolar rDNA, thereby limiting rRNA overexpression and attenuating the development of osteoarthritis in mice (36).
In the present study, we found that CBX4 was highly expressed in human sarcoma tissues. Guo et al. were the first to confirm the high expression levels of HP1-α (CBX5) in gastric cancer tissues, and then revealed that HP1-α (CBX5) could promote the proliferation, migration, and invasion, of gastric cancer cells in vitro (37). In lung cancer, Yu et al. used a panel of tumor stem-like cells (hESCs) to verify the materiality of HP1-α (CBX5) (38). In our study, increased levels of HP1-α (CBX5) expression were related to a poorer OS, both in the GEPIA dataset and the Kaplan–Meier Plotter dataset. Wang et al. reported that the level of anti-HP1-α (CBX5) antibody was associated with age, cigarette-smoking habits, and blood pressure, in patients who had suffered from transient ischemic attack, thus indicating that serum levels of antibodies against HP1-α (CBX5) could potentially serve as tools for diagnosing transient ischemic attack (39). The elevated expression of CBX6 has been reported in HCC tissues and cell lines; CBX6 was also correlated with a larger tumor size (≥5 cm; p = 0.011). Moreover, HCC patients with higher expression levels of CBX6 showed a worse OS and RFS than patients with lower expression levels of CBX6 (40). Deng et al. identified CBX6 as a downregulated gene in breast cancer via a comprehensive analysis of The Cancer Genome Atlas (TCGA); their final findings supported CBX6 as a cancer suppressor in breast cancer (41). Similarly, our present study suggested that higher expression levels of CBX6 were related to DFS in patients with sarcoma.
Reduced expression levels of CBX7 have been found to be associated with a poorer OS and the aggressiveness of thyroid cancers, colorectal cancers and breast cancers (42–44), In HCC, the downregulation of CBX7 may be related to a short OS (17). However, high expression levels of CBX7 were associated with a reduced OS and DFS in patient with prostate cancers and ovarian cancers (45, 46). In sarcoma, we found that the expression level of CBX7 was downregulated in sarcoma, and that the elevated expression of CBX7 was associated with a better OS and DFS. Therefore, further studies are still needed to fully evaluate the role of CBX7 in cancers. CBX8 was found to have the ability to promote invasion and migration in breast cancer, lung cancer, and glioblastoma (47), and in HCC. A previous study showed that CBX8 could act as an oncogene and play an important role in upregulating the Akt/β-catenin pathway stimulated by EGR1 and miR-365-3p. The upregulated expression of CBX8 was also correlated with a poorer OS (48). In our study, high expression levels of CBX8 tended to exert impact on the survival rate but without statistical significance.
The tumor microenvironment can affect the progression and recurrence of multiple cancers. Immune cells within the tumor microenvironment have been shown to promote or suppress cancer activities and are considered as an important determining factor in clinical outcome and immune therapy. In the present study, we found that the expression levels of CBX genes were significantly related to immune cell infiltration, thus indicating that CBXs could reflect the immune status of sarcoma. Our study can provide more detailed immune information to sustain immune therapy for patients with sarcoma. Our study has several limitations that need to be considered. First, the data used for analysis were obtained from online services. We need to carry out more cell-based studies and clinical experiments to confirm our findings and to further explore interactions between relevant molecules, the precise mechanisms involved, and the potential clinical applications of CBX genes in sarcoma. In addition, the performance of immune infiltration should be validated by co-localization with different members of the CBX family and immune infiltration markers.
Conclusion
In conclusion, our study showed that HP1-α/β/γ (CBX1/3/5) and CBX4/6/8 were highly overexpressed in human sarcoma tissues. The high expression levels of HP1-α/γ (CBX3/5) were closely associated with a poorer OS while the high expression levels of CBX7 were associated with a greater OS. CBX genes were positively correlated with the infiltration of immune cells, including CD8+ T cells, CD4+ T cells, B cells, macrophages, neutrophils, and dendritic cells, in sarcoma. These results indicated the crucial value of CBX genes in the prognosis and immune therapy of human sarcoma. Our findings may provide new insight and comprehensive analysis to select novel prognostic and immune biomarkers for sarcoma.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author Contributions
JZ and WW conceived, designed, and conducted the experiments. JZ and ZC wrote the paper. JZ and YL collected the data. JZ and WW edited the paper. JZ and GW provided the research guide. RW, JZ, MZ, and TW revised the manuscript. YL, WW, and TL supervised this project. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities of Central South University (Grant No in application system. 1053320210251) and the Scientific Research Project of Hunan Health Committee (Grant No. 20200357).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Wilky BA, Goldberg JM. From Vision to Reality: Deploying the Immune System for Treatment of Sarcoma. Discovery Med (2017) 23:61–74.
2. Reid R. Update on Sarcomas of Bone and Soft Tissue. Scott Med J (2007) 52:31–5. doi: 10.1258/rsmsmj.52.2.31
3. Reed DR, Naghavi A, Binitie O. Sarcoma as a Model for Adolescent and Young Adult Care. J Oncol Pract (2019) 15:239–47. doi: 10.1200/JOP.18.00684
4. Anderson ME. Update on Survival in Osteosarcoma. Orthop Clin North Am (2016) 47:283–92. doi: 10.1016/j.ocl.2015.08.022
5. Gil J, O’Loghlen A. Prc1 Complex Diversity: Where is It Taking Us? Trends Cell Biol (2014) 24:632–41. doi: 10.1016/j.tcb.2014.06.005
6. Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, et al. Nonoverlapping Functions of the Polycomb Group Cbx Family of Proteins in Embryonic Stem Cells. Cell Stem Cell (2012) 10:47–62. doi: 10.1016/j.stem.2011.12.006
7. Ma RG, Zhang Y, Sun TT, Cheng B. Epigenetic Regulation by Polycomb Group Complexes: Focus on Roles of CBX Proteins. J Zhejiang Univ Sci B (2014) 15:412–28. doi: 10.1631/jzus.B1400077
8. Zhen CY, Tatavosian R, Huynh TN, Duc HN, Das R, Kokotovic M, et al. Live-Cell Single-Molecule Tracking Reveals Co-Recognition of H3k27me3 and DNA Targets Polycomb Cbx7-PRC1 to Chromatin. ELIFE (2016) 5:e17667. doi: 10.7554/eLife.17667
9. Jangal M, Lebeau B, Witcher M. Beyond EZH2: Is the Polycomb Protein CBX2 an Emerging Target for Anti-Cancer Therapy? Expert Opin Ther Targets (2019) 23:565–78. doi: 10.1080/14728222.2019.1627329
10. Vad-Nielsen J, Nielsen AL. Beyond the Histone Tale: HP1alpha Deregulation in Breast Cancer Epigenetics. Cancer Biol Ther (2015) 16:189–200. doi: 10.1080/15384047.2014.1001277
11. Han Q, Li C, Cao Y, Bao J, Li K, Song R, et al. CBX2 is a Functional Target of Mirna Let-7a and Acts as a Tumor Promoter in Osteosarcoma. Cancer Med (2019) 8:3981–91. doi: 10.1002/cam4.2320
12. Ma C, Nie XG, Wang YL, Liu XH, Liang X, Zhou QL, et al. Cbx3 Predicts an Unfavorable Prognosis and Promotes Tumorigenesis in Osteosarcoma. Mol Med Rep (2019) 19:4205–12. doi: 10.3892/mmr.2019.10104
13. Wang X, Qin G, Liang X, Wang W, Wang Z, Liao D, et al. Targeting the CK1alpha/CBX4 Axis for Metastasis in Osteosarcoma. Nat Commun (2020) 11:1141. doi: 10.1038/s41467-020-14870-4
14. Liang YK, Lin HY, Chen CF, Zeng. Prognostic Values of Distinct Cbx Family Members in Breast Cancer. Oncotarget (2017) 8:92375–87. doi: 10.18632/oncotarget.21325
15. Li X, Gou J, Li H, Yang X. Bioinformatic Analysis of the Expression and Prognostic Value of Chromobox Family Proteins in Human Breast Cancer. Sci Rep (2020) 10:17739. doi: 10.1038/s41598-020-74792-5
16. Xie X, Ning Y, Long J, Wang H, Chen X. Diverse CBX Family Members as Potential Prognostic Biomarkers in Non-Small-Cell Lung Cancer. FEBS Open BIO (2020) 10:2206–15. doi: 10.1002/2211-5463.12971
17. Ning G, Huang YL, Zhen LM, Xu WX, Jiao Q, Yang FJ, et al. Transcriptional Expressions of Chromobox 1/2/3/6/8 as Independent Indicators for Survivals in Hepatocellular Carcinoma Patients. Aging (Albany NY) (2018) 10:3450–73. doi: 10.18632/aging.101658
18. Detwiller KY, Fernando NT, Segal NH, Ryeom SW, D’Amore PA, Yoon SS. Analysis of Hypoxia-Related Gene Expression in Sarcomas and Effect of Hypoxia on RNA Interference of Vascular Endothelial Cell Growth Factor a. Cancer Res (2005) 65:5881–9. doi: 10.1158/0008-5472.CAN-04-4078
19. Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, et al. Subtype-Specific Genomic Alterations Define New Targets for Soft-Tissue Sarcoma Therapy. Nat Genet (2010) 42:715–21. doi: 10.1038/ng.619
20. Quade BJ, Wang TY, Sornberger K, Dal Cin P, Mutter GL, Morton CC. Molecular Pathogenesis of Uterine Smooth Muscle Tumors From Transcriptional Profiling. Genes Chromosomes Cancer (2004) 40:97–108. doi: 10.1002/gcc.20018
21. Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene Functional Classification Tool: a Novel Biological Module-Centric Algorithm to Functionally Analyze Large Gene Lists. Genome Biol (2007) 8:R183. doi:10.1186/gb-2007-8-9-r183
22. Linehan DC, Bowne WB, Lewis JJ. Immunotherapeutic Approaches to Sarcoma. Semin Surg Oncol (1999) 17:72–7. doi: 10.1002/(sici)1098-2388(199907/08)17:1<72::aid-ssu9>3.0.co;2-a
23. Burgess M, Gorantla V, Weiss K, Tawbi H. Immunotherapy in Sarcoma: Future Horizons. Curr Oncol Rep (2015) 17:52. doi: 10.1007/s11912-015-0476-7
24. Yang YF, Pan YH, Tian QH, Wu DC, Su SG. Cbx1 Indicates Poor Outcomes and Exerts Oncogenic Activity in Hepatocellular Carcinoma. Transl Oncol (2018) 11:1110–8. doi: 10.1016/j.tranon.2018.07.002
25. Chao Q, Shen F, Xue Y, Wu J, Zhang J. Cbx2, a Pcg Family Gene, Plays a Regulatory Role in Medaka Gonadal Development. Int J Mol Sci (2020) 21:1288. doi: 10.3390/ijms21041288
26. Tatavosian R, Kent S, Brown K, Yao T, Duc HN, Huynh TN, et al. Nuclear Condensates of the Polycomb Protein Chromobox 2 (Cbx2) Assemble Through Phase Separation. J Biol Chem (2019) 294:1451–63. doi: 10.1074/jbc.RA118.006620
27. Plys AJ, Davis CP, Kim J, Rizki G, Keenen MM, Marr SK, et al. Phase Separation of Polycomb-Repressive Complex 1 is Governed by a Charged Disordered Region of CBX2. Genes Dev (2019) 33:799–813. doi: 10.1101/gad.326488.119
28. Garcia-Moreno SA, Lin YT, Futtner CR, Salamone IM, Capel B, Maatouk DM. CBX2 is Required to Stabilize the Testis Pathway by Repressing Wnt Signaling. PloS Genet (2019) 15:e1007895. doi: 10.1371/journal.pgen.1007895
29. Mao J, Tian Y, Wang C, Jiang K, Li R, Yao Y, et al. Cbx2 Regulates Proliferation and Apoptosis Via the Phosphorylation of YAP in Hepatocellular Carcinoma. J Cancer (2019) 10:2706–19. doi: 10.7150/jca.31845
30. Zheng S, Lv P, Su J, Miao K, Xu H, Li M. Overexpression of CBX2 in Breast Cancer Promotes Tumor Progression Through the PI3K/AKT Signaling Pathway. Am J Transl Res (2019) 11:1668–82.
31. Zhong X, Kan A, Zhang W, Zhou J, Zhang H, Chen J, et al. Cbx3/Hp1gamma Promotes Tumor Proliferation and Predicts Poor Survival in Hepatocellular Carcinoma. Aging (Albany NY) (2019) 11:5483–97. doi: 10.18632/aging.102132
32. Zhao SP, Wang F, Yang M, Wang XY, Jin CL, Ji QK, et al. Cbx3 Promotes Glioma U87 Cell Proliferation and Predicts an Unfavorable Prognosis. J Neurooncol (2019) 145:35–48. doi: 10.1007/s11060-019-03286-w
33. Lin H, Lian J, Xia L, Guan G, You J. Cbx3 Promotes Gastric Cancer Progression and Affects Factors Related to Immunotherapeutic Responses. Cancer MANAG Res (2020) 12:10113–25. doi: 10.2147/CMAR.S271807
34. Hu C, Zhang Q, Tang Q, Zhou H, Liu W, Huang J, et al. Cbx4 Promotes the Proliferation and Metastasis Via Regulating BMI-1 in Lung Cancer. J Cell Mol Med (2020) 24:618–31. doi: 10.1111/jcmm.14771
35. Wang X, Li L, Wu Y, Zhang R, Zhang M, Liao D, et al. Cbx4 Suppresses Metastasis Via Recruitment of HDAC3 to the Runx2 Promoter in Colorectal Carcinoma. Cancer Res (2016) 76:7277–89. doi: 10.1158/0008-5472.CAN-16-2100
36. Ren X, Hu B, Song M, Ding Z, Dang Y, Liu Z, et al. Maintenance of Nucleolar Homeostasis by CBX4 Alleviates Senescence and Osteoarthritis. Cell Rep (2019) 26:3643–56.e7. doi: 10.1016/j.celrep.2019.02.088
37. Guo J, Zhang Z, Pan L, Zhou Y. Identification of Mir-758-3p as Potential Modulator of CBX5 Expression in Gastric Cancer. Technol Cancer Res Treat (2018) 17:1533033818816061. doi: 10.1177/1533033818816061
38. Yu YH, Chiou GY, Huang PI, Lo WL, Wang CY, Lu KH, et al. Network Biology of Tumor Stem-Like Cells Identified a Regulatory Role of CBX5 in Lung Cancer. Sci Rep (2012) 2:584. doi: 10.1038/srep00584
39. Wang H, Zhang XM, Tomiyoshi G, Nakamura R, Shinmen N, Kuroda H, et al. Association of Serum Levels of Antibodies Against MMP1, CBX1, and CBX5 With Transient Ischemic Attack and Cerebral Infarction. Oncotarget (2018) 9:5600–13. doi: 10.18632/oncotarget.23789
40. Zheng H, Jiang WH, Tian T, Tan HS, Chen Y, Qiao GL, et al. Cbx6 Overexpression Contributes to Tumor Progression and is Predictive of a Poor Prognosis in Hepatocellular Carcinoma. Oncotarget (2017) 8:18872–84. doi: 10.18632/oncotarget.14770
41. Deng H, Guan X, Gong L, Zeng J, Zhang H, Chen MY, et al. CBX6 is Negatively Regulated by EZH2 and Plays a Potential Tumor Suppressor Role in Breast Cancer. Sci Rep (2019) 9:197. doi: 10.1038/s41598-018-36560-4
42. Pallante P, Federico A, Berlingieri MT, Bianco M, Ferraro A, Forzati F, et al. Loss of the CBX7 Gene Expression Correlates With a Highly Malignant Phenotype in Thyroid Cancer. Cancer Res (2008) 68:6770–8. doi: 10.1158/0008-5472.CAN-08-0695
43. Mansueto G, Forzati F, Ferraro A, Pallante P, Bianco M, Esposito F, et al. Identification of a New Pathway for Tumor Progression: Microrna-181b Up-Regulation and CBX7 Down-Regulation by HMGA1 Protein. Genes Cancer (2010) 1:210–24. doi: 10.1177/1947601910366860
44. Pallante P, Terracciano L, Carafa V, Schneider S, Zlobec I, Lugli A, et al. The Loss of the CBX7 Gene Expression Represents an Adverse Prognostic Marker for Survival of Colon Carcinoma Patients. Eur J Cancer (2010) 46:2304–13. doi: 10.1016/j.ejca.2010.05.011
45. Shinjo K, Yamashita Y, Yamamoto E, Akatsuka S, Uno N, Kamiya A, et al. Expression of Chromobox Homolog 7 (CBX7) is Associated With Poor Prognosis in Ovarian Clear Cell Adenocarcinoma Via Trail-Induced Apoptotic Pathway Regulation. Int J Cancer (2014) 135:308–18. doi: 10.1002/ijc.28692
46. Bernard D, Martinez-Leal JF, Rizzo S, Martinez D, Hudson D, Visakorpi T, et al. Cbx7 Controls the Growth of Normal and Tumor-Derived Prostate Cells by Repressing the Ink4a/Arf Locus. Oncogene (2005) 24:5543–51. doi: 10.1038/sj.onc.1208735
47. Jia Y, Wang Y, Zhang C, Chen MY. Upregulated CBX8 Promotes Cancer Metastasis Via the WNK2/MMP2 Pathway. Mol Ther Oncolytics (2020) 19:188–96. doi: 10.1016/j.omto.2020.09.012
Keywords: CBX family genes, prognosis, immune infiltration, sarcoma, survival
Citation: Zhou J, Chen Z, Zou M, Wan R, Wu T, Luo Y, Wu G, Wang W and Liu T (2021) Prognosis and Immune Infiltration of Chromobox Family Genes in Sarcoma. Front. Oncol. 11:657595. doi: 10.3389/fonc.2021.657595
Received: 25 January 2021; Accepted: 13 April 2021;
Published: 11 May 2021.
Edited by:
Lori Rink, Fox Chase Cancer Center, United StatesReviewed by:
Xiaojun Ren, University of Colorado Denver, United StatesShuai Ye, Fox Chase Cancer Center, United States
Copyright © 2021 Zhou, Chen, Zou, Wan, Wu, Luo, Wu, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gen Wu, d3UuZ2VuQGNzdS5lZHUuY24=; Wanchun Wang, d2FuY2h1bi53YW5nQGNzdS5lZHUuY24=; Tang Liu, bGl1dGFuZzEyMDRAY3N1LmVkdS5jbg==
†These authors have contributed equally to this work
 Jian Zhou
Jian Zhou Ziyuan Chen
Ziyuan Chen Ming Zou3
Ming Zou3 Wanchun Wang
Wanchun Wang