
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 20 April 2021
Sec. Gastrointestinal Cancers
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.653717
This article is part of the Research TopicThe Role of non-coding RNAs in Gastrointestinal CancersView all 18 articles
 Kedeerya Aishanjiang1,2†
Kedeerya Aishanjiang1,2† Xin-dong Wei3†
Xin-dong Wei3† Yi Fu1
Yi Fu1 Xinjie Lin1
Xinjie Lin1 Yujie Ma1
Yujie Ma1 Jiamei Le1
Jiamei Le1 Qiuqin Han1
Qiuqin Han1 Xuan Wang3
Xuan Wang3 Xiaoni Kong4*
Xiaoni Kong4* Jinyang Gu2*
Jinyang Gu2* Hailong Wu1,5,6*
Hailong Wu1,5,6*Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-related death worldwide. Due to the lack of potent diagnosis and prognosis biomarkers and effective therapeutic targets, the overall prognosis of survival is poor in HCC patients. Circular RNAs (circRNAs) are a class of novel endogenous non-coding RNAs with covalently closed loop structures and implicated in diverse physiological processes and pathological diseases. Recent studies have demonstrated the involvement of circRNAs in HCC diagnosis, prognosis, development, and drug resistance, suggesting that circRNAs may be a class of novel targets for improving HCC diagnosis, prognosis, and treatments. In fact, some artificial circRNAs have been engineered and showed their therapeutic potential in treating HCV infection and gastric cancer. In this review, we introduce the potential of circRNAs as biomarkers for HCC diagnosis and prognosis, as therapeutic targets for HCC treatments and discuss the challenges in circRNA research and chances of circRNA application.
Hepatocellular carcinoma (HCC) is ranked as the sixth most common neoplasm and the third leading cause of cancer-related death worldwide (1). Curative treatments, including liver transplantation, liver resection, and ablation, are only available for early stage HCCs. But due to the absence of specific symptoms at early stages and the lack of early diagnostic biomarkers, most HCC patients are diagnosed at an advanced stage and not eligible for curative treatments (2). So far, the tyrosine kinase inhibitors (sorafenib and lenvatinib) and the combination of atezolizumab (an anti–PD-L1 antibody) with bevacizumab (a vascular endothelial growth factor inhibitor) are the only approved first-line systemic treatment for advanced HCCs (3–5). Moreover, although 5-year overall survival (OS) rate reaches up to 50%, recurrence occurs in more than 70% HCC patients after curative surgery (6), which severely impairs the prognosis of HCCs. Therefore, development of early diagnostic and/or prognostic biomarkers and identification of novel therapeutic targets are urgently required for improving HCC outcomes.
Circular RNAs (circRNAs) are a novel class of non-coding RNAs generated from back-splicing of pre-mature transcripts by forming covalently closed loop structures without 5′-caps or 3′-polyadenylated tails (7, 8) (Figure 1). Although most circRNAs were originally recognized as by-products of aberrant splicing (9), accumulating evidence has suggested their involvement in various physiological processes and pathological conditions, such as viral infection, sepsis, cardiovascular disease, diabetes, and aging and regenerative medicine (9–13). By functioning as miRNA sponges (14), acting as protein decoys, scaffolds and recruiters (15) or serving as RNA templates for short peptide synthesis (16), circRNAs play important roles in regulating gene expression and signaling transduction (8) (Figure 1).
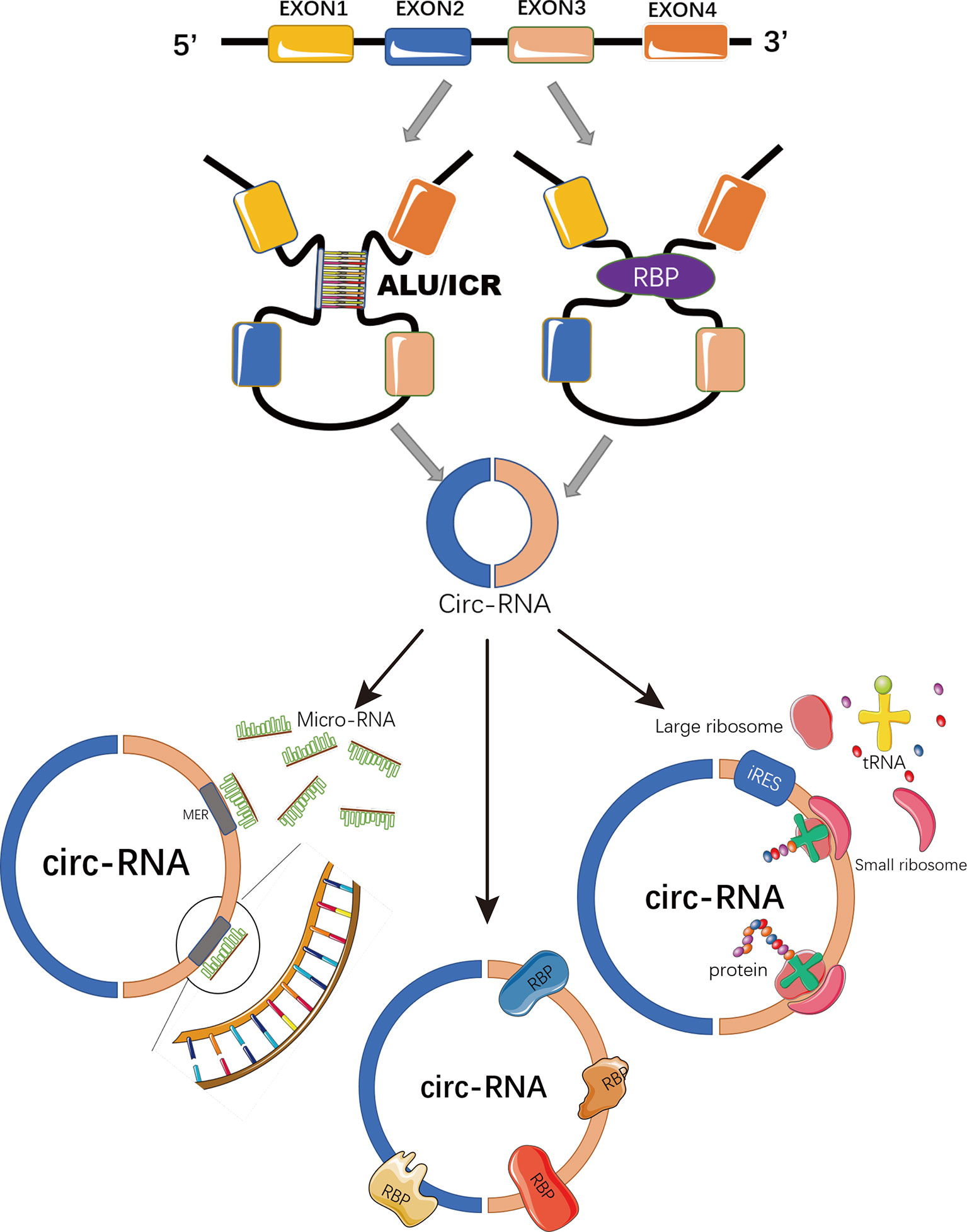
Figure 1 Schematic representation of circRNA biogenesis and functions. CircRNAs are covalently closed loop structures generated by back-splicing of pre-mature transcripts mediated by either Alu repeats, inverted complementary repeats (ICR), or RNA binding proteins (RBP). CircRNAs could function as miRNA sponges to block miRNA-mediated inhibition on target genes; or as scaffolds, recruiters or decoys of some RNA binding proteins to regulate the function of the associated proteins. Some circRNAs containing IRES and ORFs serve as RNA template for translation.
Recent emerging evidence has demonstrated that circRNAs are closely associated with tumor initiation and progression (17–20). Firstly, deregulation of circRNAs has been confirmed in many types of cancers, including breast cancer (21, 22), lung cancer (23, 24), prostate cancer (25), colorectal cancer (26), gastrointestinal cancers (27), ovarian cancer (28), thyroid cancer (29), gynecologic cancers (30), and hepatocellular carcinoma (31); Secondly, some circRNAs have been demonstrated to play either oncogenic (32, 33) or tumor suppressive (34, 35) roles in affecting multiple cancer hallmarks, including deregulating cellular energetic, self-sufficiency in growth signals, insensitivity anti-growth signals, evading cell death, limitless replicative potential, substained angiogenesis, tissue invasion and metastasis (36, 37). Moreover, given the facts that circRNAs are more resistant to exoribonuclease degradation due to their lack of free 5′- and 3′-ends (38) and are abundant in body fluids, such as saliva, blood, and urine (39–41), they have been increasingly recognized as promising tumor biomarkers (42). This review introduces recently circRNAs identified with biomarker and therapeutic potentials for HCC diagnosis, prognosis and treatments, describes circRNAs related to drug resistance and discusses the challenges in circRNA research and chances of circRNA application.
Early diagnosis is critical for the improvement of HCC prognosis and outcomes. The serum alpha-fetoprotein (AFP) (43), alpha-fetoprotein-L3 (AFP-L3) (44) and Des-gamma-carboxy prothrombin (DCP) (45) are the most commonly used non-invasive circulating biomarkers for HCC diagnosis in clinical practice, but limited evidence supports their contribution to HCC early detection or improvement in HCC outcomes (46). Therefore, identifying novel biomarkers for HCC early diagnosis is urgently required. CircRNAs are dysregulated in HCCs (47), and more resistant to nuclease degradation compared to linear RNAs due to their covalently closed loops (38), which make them potential biomarkers for HCC diagnosis. Here, we mainly introduce some recent studies indicating that circRNAs are promising non-invasive circulating diagnosis biomarkers of HCCs.
Zhang et al. demonstrated that circRNA_104075 was positively regulated by HNF4a, a HCC-promoting transcription factor, and highly expressed in HCC tissues and serum samples. CircRNA_104075 showed greater predictive performance (AUC: 0.973, sensitivity: 0.960; specificity: 0.983) for HCCs than AFP (AUC: 0.750, sensitivity: 0.693; specificity: 0.683), AFP-L3 (AUC: 0.766, sensitivity: 0.772; specificity: 0.633) and DCP (AUC: 0.771, sensitivity: 0.703; specificity: 0.750) (48).
By comparing the serum circRNA profiles between 36 healthy controls, 60 chronic hepatitis C (CHC) and 68 HCC patients, Matboli et al. found up-regulation of hsa_circ_000224 and down-regulation of hsa_circ_001565 and hsa_circ_000520 in the serum samples of HCC patients compared to those in healthy controls and CHC patients (49). The predictive performance for HCCs of hsa_circ_001565 (AUC: 0.839, sensitivity: 0.703; specificity: 0.750), hsa_circ_000520 (AUC: 0.943, sensitivity: 0.971; specificity: 0.896) and hsa_circ_000224 (AUC: 0.974, sensitivity: 0.956; specificity: 0.927) were better than that of serum AFP (AUC: 0.726, sensitivity: 0.779; specificity: 0.823) (49). Combing the three circRNAs showed remarkably high sensitivity (1.000) but did not improve the specificity (0.833). The authors failed to show the AUC value for the combined three circRNAs in diagnosing HCCs (49).
By analyzing differentially expressed circRNAs (DECs) in plasma between 40 healthy controls and 71 HCC patients, Sun et al. recently identified a three-circRNA signature, including hsa_circ_0004001, hsa_circ_0004123, and hsa_circ_0075792, which are highly expressed in HCC patients (50). This 3-circRNA signature showed a potent diagnosis value (AUC: 0.89, sensitivity: 0.905; specificity: 0.781) (50). In this study, the authors did not compared the diagnosis value of the 3-circRNA signature with that of AFP, but the AUC, sensitivity and specificity values of the 3-circRNA signature may slightly better than those of AFP from other studies (48, 49).
Comparison of circulating circRNAs, in 5 HCC patients before and after hepatectomy, 5 HBV-positive chronic hepatitis (CH) patients and 5 healthy controls, identified that the serum levels of hsa_circ_0009582, hsa_circ_0037120, and hsa_circ_0140117 were high in HCCs and decreased after hepatectomy (51). Moreover, a combination of those three circRNAs with serum AFP greatly improved the diagnosis potential for HCC in a training set (20 HCCs vs. 20 CH patients; AUC: 0.988, sensitivity: 0.960; specificity: 1.015) and a validation set (180 HCCs vs. 180 CH patients; AUC: 0.955, sensitivity: 0.915; specificity: 0.994) (51).
Down-regulation of hsa_circ_0001445 (circSMARCA5) in HCCs was identified in 3 cohorts of 208 pairs of HCC and corresponding adjacent noncancerous liver (ANL) tissues, and circSMARCA5 overexpression inhibited HCC growth and metastasis in vitro and in vivo (52). By comparing plasma circSMARCA5 levels in 103 healthy controls, 117 hepatitis (hepatitis B and C), 143 cirrhosis and 135 HCC patients, Li et al. found that plasma circSMARCA5 decreases gradually but significantly from healthy controls, hepatitis, cirrhosis to HCC patients (53). It worth noting that, in patients whose AFP levels were less than 200 ng/ml, circSMARCA5 showed satisfying accuracy in distinguishing HCCs from hepatitis (AUC = 0.847, sensitivity: 0.721; specificity: 0.882) and cirrhosis (AUC = 0.706, sensitivity: 0.721; specificity: 0.660) patients, suggesting circSMARCA5 as a putative biomarker for HCCs with low AFP levels (53).
Recently, a large-scale, multicenter study identified a plasma circRNA panel (CircPanel) containing three circRNAs (hsa_circ_0000976, hsa_circ_0007750, and hsa_circ_0139897), which had greater accuracy than AFP in distinguishing individuals with HCC from Non-HCC (54). The performance of CircPanel vs. AFP was AUC 0.863 vs. 0.790 (p=0.036) in the training set (53 healthy controls, 52 with CHB, 50 with liver cirrhosis and 158 with HCC), AUC 0.843 vs. 0.747 (p=0.011) in Validation Set 1 (152 HCC patients, 50 healthy controls, 54 CHB patients and 50 HBV-induced liver cirrhosis patients), and AUC 0.864 vs. 0.769 (p<0.001) in Validation Set 2 (290 HCC patients, 76 healthy controls, 80 CHB patients and 80 HBV-induced liver cirrhosis patients) (54). Moreover, CircPanel showed a higher accuracy than AFP in diagnosis of small-HCC (solitary, diameter ≤ 3 cm) (AUC of CircPanel vs. AFP: 0.862 vs. 0.680, p=0.001 in the training set; 0.838 vs. 0.699, p=0.011 in Validation Set 1 and 0.851 vs. 0.738, p=0.009 in Validation Set 2) (54). More significantly, CircPanel has been identified to be able to diagnose AFP-negative HCC and AFP-negative Small-HCC (54).
Recurrence occurs in more than 70% HCC patients post curative surgery (6). Even for the most effective liver transplantation (LT), the recurrence rate is near 20% (55). Although the TNM staging system of the American Joint Committee on Cancer (AJCC), the Barcelona Clinic Liver Cancer (BCLC) classification, and the Cancer of the Liver Italian Program (CLIP) staging system, have been employed to evaluate the prognosis of HCC patients (56), their prognostic predictive performance was unsatisfactory partly because those assessments do not take the critical and complicated molecular pathogenesis of HCCs into account (57). Recent studies demonstrated that some circRNAs could serve as prognostic biomarkers for HCCs.
Hsa_circ_0000267 was up-regulated in HCC tumor tissues and positively associated aggressive clinicopathological features such as tumor size and TNM stage (58). Multivariable analysis identified enhanced hsa_circ_0000267 as an independent prognostic factor for the OS in HCC patients (58).
Chromosomal amplification at 7q21–7q31 was closely related to tumor recurrence in various types of cancers including HCCs (59–62). By assessing the expression of 43 putative circRNAs in this chromosomal region in 4 pairs of matched HCC and normal tissues, Huang et al. identified that hsa_circ_0082002 (circMET) was the most significantly and consistently up-regulated circRNA in HCCs (63). By evaluating circMET expression in 90 paired HCC and normal tissues and in a tissue microarray (TMA) consisting 209 HCC tissues, levels of circMET were significantly and positively correlated with microvascular invasion, absent tumor encapsulation, multiple tumors and late HCC stage. Multivariate analysis identified circMET as an independent predictor for both OS and postoperative recurrence in HCC patients (63).
Sun et al. have demonstrated that hsa_circ_0027345 (circLRIG3), a nucleus-enriched circRNA, was highly expressed in HCCs and positively associated with aggressive clinicopathological features, such as tumor size, vascular invasion, Edmondson’s grade and later TNM stage, and was identified as an independent risk prognostic factor for OS in HCC patients (64).
The down-regulation of hsa_circ_0001727 (circZKSCAN1) in HCCs were determined by two independent groups (65, 66). Expression of circZKSCAN1 was associated with multiple clinicopathologic factors, such as tumor numbers, cirrhosis, vascular invasion, microscopic vascular invasion, or tumor grade (65), and was determined as an independent and significant factor affecting both OS and RFS in HCCs (66).
In addition to serving as a non-invasive biomarker, decreased expression of circSMARCA5 in HCC tumor tissues was significantly correlated with poorer tumor differentiation, more advanced tumor stage, larger tumor size and presence of microvascular invasion. Multivariate analysis demonstrated that circSMARCA5 expression level was an independent risk factor for OS and RFS in HCCs (52).
A recent study has demonstrated that an up-regulated plasma circRNA, hsa_cic_0005397, was positively correlated with tumor size (p = 0.020) and TNM stage (p = 0.006) (67). Meanwhile, a dynamic monitoring of plasma hsa_cic_0005397 in HCC patients who had undergone surgical resection revealed that plasma hsa_cic_0005397 level was drastically dropped in HCC patients after operation, but prominently elevated in recurrent or metastatic HCC patients, suggesting that plasma hsa_cic_0005397 could serve as a prognostic biomarker for post-operational recurrence and metastasis in HCC patients (67). Moreover, the plasma hsa_cic_0005397 level was negatively associated with OS in HCC patients (67).
However, due to the limited sample size and the lack of multicenter validation, the diagnosis and prognosis potential of those circRNAs should be further determined in the future.
Like many other non-coding RNAs, such as lncRNAs and miRNAs, circRNAs have been demonstrated to function as either oncogenes or tumor suppressors in HCCs.
By comparing circRNA expression profiles in primary HCCs with or without post-surgery pulmonary metastasis, Hu et al. identified that hsa_circ_0085616 (circASAP1) is highly expressed in HCCs with greater metastatic potential (31). Mechanistically, circASAP1 functions as a sponge for miR-326 and miR-532-5p and enhances the expression of CSF-1 and MAPK1, resulting in an increase in intra-tumor infiltration of tumor associated macrophages and activation of the MAPK-ERK signaling pathway (31). Consistent with this study, Li et al. also demonstrated that circASAP1 plays an oncogenic role in HCCs possibly by activating the β-catenin, ERK and AKT signaling pathways (68).
Through bioinformatics analysis and experimental validation, Li et al. demonstrated that up-regulation of hsa_circ_0074854 (circMAT2B) in HCCs was correlated with poor prognosis and could serve as an independent prognostic factor (69). By functioning as a sponge of miR-338-3p, circMAT2B released miR-338-3p-mediated inhibition on PKM2, resulting in enhanced glycolysis, proliferation and metastasis in HCCs (69).
In addition to acting as a miRNA sponge, some circRNAs have been reported to function as protein decoys, scaffolds and recruiters to promote HCC progression. Wang et al. demonstrated that hsa_circ_102034 (circRHOT1) was highly expressed in HCCs and promoted proliferation, migration and invasion in HCC cells by acting as a protein recruiter to recruit TIP60 onto the promoter region of NR2F6 to initiate NR2F6 expression (70).
By performing a circRNA microarray assay specifically targeting human circRNA splicing sites in seven paired HCC tumor and normal samples, Han et al. demonstrated that hsa_circ_0007874 (circMTO1) was significantly decreased in HCCs and correlated with poor prognosis of HCC patients (71). CircMTO1 could serve as a miR-9 sponge to release miR-9–mediated inhibition on p21, resulting in inhibition on HCC growth both in vitro and in vivo (71).
Compared to oncogenic circRNAs, the tumor suppressive circRNAs are less studied probably due to their low expression in HCCs. Only countable circRNAs have been identified as tumor suppressors in HCCs. For example, circSMARCA5, a down-regulated circRNA in HCC, has been identified as a tumor suppressor for HCC progression. Over-expression of circSMARCA5 retained miR-17-3p and miR-181b-5p to release their common target, TIMP3, a well-known tumor suppressor in HCCs, resulting in enhanced HCC growth and metastasis both in vitro and in vivo (52, 72). By functioning as a decoy of fragile X mental retardation protein (FMBP), an RNA binding protein, circZKSCAN1 prevented the binding of FMBP with cell cycle and apoptosis regulator 1 (CCAR1) mRNA and in turn inhibited CCAR1 expression and CCAR1-mediated activation of the Wnt/β-catenin signaling pathway, resulting in compromised stemness in HCC cells (66).
Advanced HCCs are not legible for curative treatments. Although chemotherapy, targeted therapy and immunotherapy are commonly employed for those advanced HCCs, the therapeutic efficacy is unsatisfied due to low objective response rate and acquired resistance (73–75). In addition to playing oncogenic or tumor suppressive roles in HCC progression, some circRNAs have been recently reported to be related to drug resistance in HCC treatments.
For HCC chemotherapy, hsa_circ_0001001 (circFBXO11), an up-regulated circRNA in HCCs, could serve as a sponge for miR-605 to induce FOXO3-mediated ABCB1 expression, resulting in HCC oxaliplatin resistance (76). CircRNA_101505 was down-regulated in cisplatin-resistant HCC tissues and cell lines, and over-expression of circRNA_101505 trapped miR-103 and relieved its target, oxidored-nitro domain-containing protein 1 (NOR1), a putative tumor suppressor in HCCs (77), consequently resulting in cisplatin sensitization (78).
For sorafenib-mediated targeted therapy, Wu et al. have identified thousands of deregulated circRNAs in sorafenib-resistant HCC cells compared to sorafenib-sensitive cells (79). Yang et al. have demonstrated that hsa_circ_0058124 (circFN1) was up-regulated in both sorafenib-resistant HCC tumor tissues and cell lines (80). Overexpression of circFN1 conferred HCC cell sorafenib resistance by restricting miR-1205–mediated E2F1 inhibition (80).
For PD1 antibody mediated immunotherapy, circMET has been demonstrated to confer HCC cells resistance to anti-PD1 treatment by enhancing the immunosuppressive tumor microenvironment (63). Mechanistically, circMET functions as a sponge of miR-30-5p to induce SNAIL-mediated Dipeptidyl 4 (DPP4) expression, resulting in degradation of CXCL10, an important chemokine promoting intra-tumor infiltration of effector T cells, and leading to subsequent resistance to anti-PD1 treatment (63).
Exosome is a type of nano-sized secreted vesicles, which are present in all body fluids under both physiological and pathological conditions (81). By carrying various biomolecules, including proteins, nucleic acids, lipids, and transferring from one cell to another, exomes play an important role in mediating intercellular communication (81). In 2015, Li, et al. for the first time demonstrated the presence of abundant circRNAs in exosomes (82). By RNA-seq analysis, they demonstrated enrichment of circRNAs in exosome compared to producer cells (82). A number of exosomal circRNAs have been identified to function as either diagnosis/prognosis biomarkers or oncogenic/tumor suppressive factors in HCCs (83, 84). For example, an exosomal circRNA, hsa_circ_0070396, was identified as a better diagnostic biomarker than AFP in distinguishing HCCs from healthy controls (AUC: 0.8574 vs. 0.781), and differentiating early HCC patients (with BCLC are 0 or A) from advanced ones (with BCLC are B+C) (AUC: 0.7132 vs. 0.6535) (85). Higher circulating exosomal circAKT3 was identified in HCC patients compared to healthy controls, and positively correlated with tumor recurrence rates and mortality, suggesting that circAKT3 could serve as a prognostic biomarker of HCCs (86). A recent study has demonstrated that three exosomal circPTGR1 isoforms secreted from HCC cells with higher metastasis potential to promote the metastasis of HCC cells with lower metastasis potential via regulating the miR449a-MET pathway (87). Su et al. have demonstrated that circRNA Cdr1as competitively bound to miR-1270 to upregulate AFP level, thereafter accelerating proliferative and migratory abilities of HCC cells (88). Meanwhile, exosome-transmitted circRNA Cdr1as stimulated malignant behaviors of surrounding normal cells and finally contributed to the progression of HCC (88). An interesting study showed that a pro-invasive exosomal circRNA-100338 secreted from HCC cells could transfer to HUVECs to affect cell proliferation, angiogenesis, permeability, and vasculogenic mimicry (VM) formation ability of HUVECs, and in turn promote HCC metastasis (89).
Instead of transporting exosomal circRNAs from HCC cells to surrounding normal cells, the exosomal circRNA can also be generated and transported from surrounding normal tissues to HCC cells, resulting in altered HCC progression. For example, an exosomal circRNA, circ-0051443, which was generated in normal cells and transferred to HCC cells, could inhibit HCC progression by competitive binding to miR-331-3p, resulting in enhanced apoptosis and cell cycle arrest in HCC cells (90).
It is clear that exosomal circular RNAs play a significant role in HCC progression. Therefore, identifying novel exosomal circRNAs and understanding their biological functions can not only help us better understand the new mechanisms of potential HCC development, but also improve the clinical diagnosis, prognosis and treatment of HCCs.
We have summarized all the circRNAs mentioned in this review in Table 1.
With the advent of high-throughput sequencing and high-efficiency big data analysis, circular RNAs have been emerged as a novel class of non-coding RNAs in eukaryotes. Accumulating evidence indicates that circRNAs are novel non-coding players the involved in various biological processes and diseases. However, some fundamental questions regarding the function and regulation of circRNAs remain unknown.
First, the biological role of circRNAs is less defined. Only less than 3% (> 7000 circRNAs) of recorded circRNAs (295,526 circRNAs integrated from circBase, circNet, and circRNAdb) (91) were curated functional (92), which at least in part suggests that the most circRNAs may be the non-functional by-products of RNA splicing events. In a previous study, Guo et al. claimed that compared to mRNAs, most circRNAs are less abundant, less cell-type specific and less conserved (93). Moreover, ribosome profiling provides no evidence for their translation (93).
Second, serving as miRNA sponges may not be the general role of circRNAs. Although abundant studies support the role of circRNAs as miRNA sponges in regulating gene expression and executing their biological functions, the existence of numerous circRNAs in P. falciparum and S. cerevisiae (94, 95), who lack known siRNA and miRNA pathways, suggests that as miRNA sponges may not be a general role for most circRNAs. This is consistent with a previous study showing that there is no particular enrichment of AGO2 binding on exons involved in circRNAs compared to their neighboring linear exons (93).
Third, the cis and trans regulators for circRNAs remain illuminated. Some studies demonstrated that the significantly longer introns and the inverted repeat sequence bracketing the regions that produce circRNAs are essential cis regulators for circRNA formation in humans and flies (38, 96–98). Meanwhile, canonical splicing signals are required for the circularization of most circRNAs (93, 99), suggesting that canonical spliceosomal machinery may serve as trans regulators for circRNA formation. However, accumulating strong evidence indicates that the expression of circRNAs does not simply correlate with their linear host genes (100, 101), suggesting that circRNA formation is a complicated process which may be regulated by additional circRNA-specific cis and trans regulators.
Although it remains controversial on the general roles of circRNAs, most functional circRNAs identified so far have been demonstrated to serve as miRNA sponges in regulating gene expression. By employing this feature, some artificial circRNAs have been engineered to sequester miRNAs relevant in human disease and shown promising potential for application in molecular medicine and biology. For example, an artificial circRNA carrying an array of miRNA-122 binding sites could sequester liver specific miR-122 from HCV RNA, resulting in inhibition on HCV viral protein production and HCV replication (102). Liu et al. generated an artificial circRNA sponge for miR-21, an oncomiR in many types of cancers, to block miR-21–mediated proliferation in gastric carcinoma cells (103). Therefore, in companion of growing understanding of circRNAs, expended application of circRNAs could be established for both research and clinic purposes.
In summary, circRNAs are emerging as a class of novel non-coding RNAs participating diverse physiological processes and pathological conditions. Although it remains controversial on the general roles of circRNAs, accumulating evidence indicates that circRNAs are functional by serving as either miRNA sponges, protein decoys, or translational templates. Like other types of non-coding RNAs, deregulation of circRNAs has been demonstrated in HCCs. A number of deregulated circRNAs have been identified as either oncogenic or tumor suppressive regulators in HCC progression. Moreover, some deregulated circRNAs could serve as non-invasive circulating biomarkers for HCC early diagnosis with great specificity and sensitivity superior to clinically used serum AFP. Meanwhile, some circRNAs have been involved in drug resistance in HCC treatments. In this review, we summarized the recent findings of circRNAs in HCC diagnosis, prognosis, progression and drug resistance and proposed that circRNAs may have great potential to serve as novel biomarkers for HCC early diagnosis, prognosis and therapeutic targets for HCC treatments.
Despite the advancements achieved in the circRNA field, several fundamental questions remain elusive. Given the fact that only a few percentage functional circRNAs (< 3%) were identified from the great circRNA popularity, whether circRNAs are a group of functional non-coding RNAs is still an open question. The existence of numerous circRNAs in species deficient with RNAi and miRNA pathways raises the question whether as miRNA sponges is the general role of circRNAs or just is an exception. Although some studies have demonstrated that the formation of circRNAs depends on the cis elements, such as the inverted repeats, and the trans regulators, such as spliceosomes, of linear host genes, the lack of correlation at expression levels and cell-type specificity between circRNAs and their linear host genes strongly suggests the existence of circRNA specific regulators. Therefore, novel techniques and concepts regarding those fundamental questions are warranted in the future to expand our understanding of circRNAs and improve our investigation on circRNAs.
KA, X-DW, YF, XL, YM, JL, XW, and QH performed extensive literature search and discussion. KA and X-DW drafted the manuscript. XK, JG, and HW edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (31870905 to HW, 82072645 and 81772507 to JG, 82070633 and 81873582 to XK), the Scientific Program of Shanghai Municipal Health Commission (SHWJ2019211 to HW), the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (No. 20191910), the Foundation for Shanghai Jiao Tong University for SMC-morning Star Youth Scholars Program, the Clinical Research Plan of SHDC (SHDC2020CR3005A), the Program of Medical Engineering Cross Research Fund of Shanghai Jiao Tong University (YG2017MS50), Clinical innovation project of Xinhua Hospital Affiliated to Medical College of Shanghai Jiaotong University (19XHCR02A), “Teaching incentive plan” project of Xinhua Hospital Affiliated to Medical College of Shanghai Jiaotong University (XH001.006.010.020), National Health and Family Planning Commission medical and health science and technology development research center key projects (NHC2018RWS01007), the Construction project of Shanghai Key Laboratory of Molecular Imaging(18DZ2260400), Shanghai Municipal Education Commission (Class II Plateau Disciplinary Construction Program of Medical Technology of SUMHS, 2018-2020), and the Key Program of National Natural Science Foundation of China (grant 81830052).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AFP, alpha-fetoprotein; AFP-L3, alpha-fetoprotein-L3; AJCC, the American Joint Committee on Cancer; ANL, adjacent noncancerous liver; AUC, Area Under Curve; BCLC, the Barcelona Clinic Liver Cancer; CCAR1, cell cycle and apoptosis regulator 1; CH, chronic hepatitis; CHC, chronic hepatitis C; circRNAs, Circular RNAs; CLIP, the Cancer of the Liver Italian Program; CSF-1, colony-stimulating factor 1; CXCL10, C-X-C motif chemokine ligand 10; DCP, Des-gamma-carboxy prothrombin; DECs, differentially expressed Circular RNAs; DPP4, SNAIL-mediated Dipeptidyl 4; FMBP, fragile X mental retardation protein; HCC, Hepatocellular carcinoma; HCV, Hepatitis C Virus; HNF4a, hepatocyte nuclear factor 4 alpha; LT, liver transplantation; MAPK1, mitogen-activated protein kinase 1; NOR1, Oxidored-nitro domain-containing protein 1; OS, Overall Survival; RFS, relapse-free survival; TMA, tissue microarray; Cdr1as, Cerebellar degeneration-related protein 1 antisense RNA; HUVECs, human umbilical vein endothelial cells; AKT3, AKT serine/threonine kinase 3.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
2. Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut (2019) 68:1014–23. doi: 10.1136/gutjnl-2017-315084
3. Casak SJ, Donoghue M, Fashoyin-Aje L, Jiang X, Rodriguez L, Shen YL, et al. FDA Approval Summary: Atezolizumab plus bevacizumab for the treatment of patients with advanced unresectable or metastatic hepatocellular carcinoma. Clin Cancer Res Off J Am Assoc Cancer Res (2021) 27(7):1836–41. doi: 10.1158/1078-0432.CCR-20-3407
4. Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol (2009) 10:25–34. doi: 10.1016/S1470-2045(08)70285-7
5. Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. New Engl J Med (2008) 359:378–90. doi: 10.1056/NEJMoa0708857
6. Sherman M. Recurrence of hepatocellular carcinoma. New Engl J Med (2008) 359:2045–7. doi: 10.1056/NEJMe0807581
7. Chen L, Huang C, Wang X, Shan G. Circular RNAs in Eukaryotic Cells. Curr Genomics (2015) 16:312–8. doi: 10.2174/1389202916666150707161554
8. Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet TIG (2016) 32:309–16. doi: 10.1016/j.tig.2016.03.002
9. Han B, Chao J, Yao H, Circular RNA. and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther (2018) 187:31–44. doi: 10.1016/j.pharmthera.2018.01.010
10. Braicu C, Zimta AA, Gulei D, Olariu A, Berindan-Neagoe I. Comprehensive analysis of circular RNAs in pathological states: biogenesis, cellular regulation, and therapeutic relevance. Cell Mol Life Sci CMLS (2019) 76:1559–77. doi: 10.1007/s00018-019-03016-5
11. Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, et al. The emerging landscape of circular RNA in life processes. RNA Biol (2017) 14:992–9. doi: 10.1080/15476286.2016.1220473
12. Hashemian SM, Pourhanifeh MH, Fadaei S, Velayati AA, Mirzaei H, Hamblin MR. Non-coding RNAs and Exosomes: Their Role in the Pathogenesis of Sepsis. Mol Ther Nucleic Acids (2020) 21:51–74. doi: 10.1016/j.omtn.2020.05.012
13. Nahand JS, Jamshidi S, Hamblin MR, Mahjoubin-Tehran M, Vosough M, Jamali M, et al. Circular RNAs: New Epigenetic Signatures in Viral Infections. Front Microbiol (2020) 11:1853:1853. doi: 10.3389/fmicb.2020.01853
14. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol (2016) 238:42–51. doi: 10.1016/j.jbiotec.2016.09.011
15. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics (2020) 10:3503–17. doi: 10.7150/thno.42174
16. Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun (2018) 9:4475. doi: 10.1038/s41467-018-06862-2
17. Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res (2015) 5:472–80.
18. Zhang X, Lu N, Wang L, Wang Y, Li M, Zhou Y, et al. Circular RNAs and esophageal cancer. Cancer Cell Int (2020) 20:362. doi: 10.1186/s12935-020-01451-0
19. Song H, Liu Q, Liao Q, Circular RNA. and tumor microenvironment. Cancer Cell Int (2020) 20:211. doi: 10.1186/s12935-020-01301-z
20. Yang X, Mei J, Wang H, Gu D, Ding J, Liu C. The emerging roles of circular RNAs in ovarian cancer. Cancer Cell Int (2020) 20:265. doi: 10.1186/s12935-020-01367-9
21. Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R, et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer (2020) 19:73. doi: 10.1186/s12943-020-01183-9
22. Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W, et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer (2020) 19:65. doi: 10.1186/s12943-020-01152-2
23. Wang C, Tan S, Liu WR, Lei Q, Qiao W, Wu Y, et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer (2019) 18:134. doi: 10.1186/s12943-019-1061-8
24. Van Der Steen N, Lyu Y, Hitzler AK, Becker AC, Seiler J, Diederichs S. The Circular RNA Landscape of Non-Small Cell Lung Cancer Cells. Cancers (2020) 12(5):1091. doi: 10.3390/cancers12051091
25. Xia Q, Ding T, Zhang G, Li Z, Zeng L, Zhu Y, et al. Circular RNA Expression Profiling Identifies Prostate Cancer- Specific circRNAs in Prostate Cancer. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol (2018) 50:1903–15. doi: 10.1159/000494870
26. Artemaki PI, Scorilas A, Kontos CK. Circular RNAs: A New Piece in the Colorectal Cancer Puzzle. Cancers (2020) 12(9):2464. doi: 10.3390/cancers12092464
27. Naeli P, Pourhanifeh MH, Karimzadeh MR, Shabaninejad Z, Movahedpour A, Tarrahimofrad H, et al. Circular RNAs and gastrointestinal cancers: Epigenetic regulators with a prognostic and therapeutic role. Crit Rev Oncol Hematol (2020) 145:102854. doi: 10.1016/j.critrevonc.2019.102854
28. Shabaninejad Z, Vafadar A, Movahedpour A, Ghasemi Y, Namdar A, Fathizadeh H, et al. Circular RNAs in cancer: new insights into functions and implications in ovarian cancer. J Ovarian Res (2019) 12:84. doi: 10.1186/s13048-019-0558-5
29. Borran S, Ahmadi G, Rezaei S, Anari MM, Modabberi M, Azarash Z, et al. Circular RNAs: New players in thyroid cancer. Pathol Res Pract (2020) 216:153217. doi: 10.1016/j.prp.2020.153217
30. Razavi ZS, Tajiknia V, Majidi S, Ghandali M, Mirzaei HR, Rahimian N, et al. Gynecologic cancers and non-coding RNAs: Epigenetic regulators with emerging roles. Crit Rev Oncol Hematol (2021) 157:103192. doi: 10.1016/j.critrevonc.2020.103192
31. Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology (2020) 72(3):906–22. doi: 10.1002/hep.31068
32. Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell (2016) 165:289–302. doi: 10.1016/j.cell.2016.03.020
33. Li L, Zhang Q, Lian K. Circular RNA circ_0000284 plays an oncogenic role in the progression of non-small cell lung cancer through the miR-377-3p-mediated PD-L1 promotion. Cancer Cell Int (2020) 20:247. doi: 10.1186/s12935-020-01310-y
34. Han D, Wang Y, Wang Y, Dai X, Zhou T, Chen J, et al. The Tumor-Suppressive Human Circular RNA CircITCH Sponges miR-330-5p to Ameliorate Doxorubicin-Induced Cardiotoxicity Through Upregulating SIRT6, Survivin, and SERCA2a. Circ Res (2020) 127:e108–25. doi: 10.1161/CIRCRESAHA.119.316061
35. Liu M, Luo C, Dong J, Guo J, Luo Q, Ye C, et al. CircRNA_103809 Suppresses the Proliferation and Metastasis of Breast Cancer Cells by Sponging MicroRNA-532-3p (miR-532-3p). Front Genet (2020) 11:485:485. doi: 10.3389/fgene.2020.00485
36. Mirzaei H, Hamblin MR. Regulation of Glycolysis by Non-coding RNAs in Cancer: Switching on the Warburg Effect. Mol Ther Oncol (2020) 19:218–39. doi: 10.1016/j.omto.2020.10.003
37. Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y, et al. Circular RNAs in Cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer (2019) 18:90. doi: 10.1186/s12943-019-1002-6
38. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna (2013) 19:141–57. doi: 10.1261/rna.035667.112
39. Jafari Ghods F. Circular RNA in Saliva. Adv Exp Med Biol (2018) 1087:131–9. doi: 10.1007/978-981-13-1426-1_11
40. Vea A, Llorente-Cortes V, de Gonzalo-Calvo D. Circular RNAs in Blood. Adv Exp Med Biol (2018) 1087:119–30. doi: 10.1007/978-981-13-1426-1_10
41. Kölling M, Haddad G, Wegmann U, Kistler A, Bosakova A, Seeger H, et al. Circular RNAs in Urine of Kidney Transplant Patients with Acute T Cell-Mediated Allograft Rejection. Clin Chem (2019) 65:1287–94. doi: 10.1373/clinchem.2019.305854
42. Wang M, Yang Y, Xu J, Bai W, Ren X, Wu H. CircRNAs as biomarkers of cancer: a meta-analysis. BMC Cancer (2018) 18:303. doi: 10.1186/s12885-018-4213-0
43. Di Bisceglie AM, Sterling RK, Chung RT, Everhart JE, Dienstag JL, Bonkovsky HL, et al. Serum alpha-fetoprotein levels in patients with advanced hepatitis C: results from the HALT-C Trial. J Hepatol (2005) 43:434–41. doi: 10.1016/j.jhep.2005.03.019
44. Sterling RK, Jeffers L, Gordon F, Sherman M, Venook AP, Reddy KR, et al. Clinical utility of AFP-L3% measurement in North American patients with HCV-related cirrhosis. Am J Gastroenterol (2007) 102:2196–205. doi: 10.1111/j.1572-0241.2007.01405.x
45. Marrero JA, Su GL, Wei W, Emick D, Conjeevaram HS, Fontana RJ, et al. Des-gamma carboxyprothrombin can differentiate hepatocellular carcinoma from nonmalignant chronic liver disease in american patients. Hepatology (2003) 37:1114–21. doi: 10.1053/jhep.2003.50195
46. Bruix J, Reig M, Sherman M. Evidence-Based Diagnosis, Staging, and Treatment of Patients With Hepatocellular Carcinoma. Gastroenterology (2016) 150:835–53. doi: 10.1053/j.gastro.2015.12.041
47. Qiu L, Xu H, Ji M, Shang D, Lu Z, Wu Y, et al. Circular RNAs in hepatocellular carcinoma: Biomarkers, functions and mechanisms. Life Sci (2019) 231:116660. doi: 10.1016/j.lfs.2019.116660
48. Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis (2018) 9:1091. doi: 10.1038/s41419-018-1132-6
49. Matboli M, Shafei AE, Ali MA, Ashry AM, Kamal KM, Agag MA, et al. circRNAs (hsa_circ_00156, hsa_circ _000224, and hsa_circ _000520) are novel potential biomarkers in hepatocellular carcinoma. J Cell Biochem (2019) 120:7711–24. doi: 10.1002/jcb.28045
50. Sun XH, Wang YT, Li GF, Zhang N, Fan L. Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int (2020) 20:226. doi: 10.1186/s12935-020-01302-y
51. Wu C, Deng L, Zhuo H, Chen X, Tan Z, Han S, et al. Circulating circRNA predicting the occurrence of hepatocellular carcinoma in patients with HBV infection. J Cell Mol Med (2020) 24(17):10216–22. doi: 10.1111/jcmm.15635
52. Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol (2018) 68:1214–27. doi: 10.1016/j.jhep.2018.01.012
53. Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang L, et al. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin Chim Acta Int J Clin Chem (2019) 492:37–44. doi: 10.1016/j.cca.2019.02.001
54. Yu J, Ding WB, Wang MC, Guo XG, Xu J, Xu QG, et al. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: A large-scale, multicenter study. Int J Cancer (2020) 146:1754–63. doi: 10.1002/ijc.32647
55. Maccali C, Chagas AL, Boin I, Quiñonez E, Marciano S, Vilatobá M, et al. Recurrence of hepatocellular carcinoma after liver transplantation: Prognostic and predictive factors of survival in a Latin American cohort. Liver Int Off J Int Assoc Study Liver (2021) 41(4):851–62. doi: 10.1111/liv.14736
56. Liu PH, Hsu CY, Hsia CY, Lee YH, Su CW, Huang YH, et al. Prognosis of hepatocellular carcinoma: Assessment of eleven staging systems. J Hepatol (2016) 64:601–8. doi: 10.1016/j.jhep.2015.10.029
57. Zhu J, Tang B, Li J, Shi Y, Chen M, Lv X, et al. Identification and validation of the angiogenic genes for constructing diagnostic, prognostic, and recurrence models for hepatocellular carcinoma. Aging (2020) 12:7848–73. doi: 10.18632/aging.103107
58. Pan H, Tang L, Jiang H, Li X, Wang R, Gao J, et al. Enhanced expression of circ_0000267 in hepatocellular carcinoma indicates poor prognosis and facilitates cell progression by sponging miR-646. J Cell Biochem (2019) 120:11350–7. doi: 10.1002/jcb.28411
59. Rao UN, Gollin SM, Beaves S, Cieply K, Nalesnik M, Michalopoulos GK. Comparative genomic hybridization of hepatocellular carcinoma: correlation with fluorescence in situ hybridization in paraffin-embedded tissue. Mol Diagn J Devoted Underst Hum Dis Clin Appl Mol Biol (2001) 6:27–37. doi: 10.1054/modi.2001.22021
60. Glukhova L, Lavialle C, Fauvet D, Chudoba I, Danglot G, Angevin E, et al. Mapping of the 7q31 subregion common to the small chromosome 7 derivatives from two sporadic papillary renal cell carcinomas: increased copy number and overexpression of the MET proto-oncogene. Oncogene (2000) 19:754–61. doi: 10.1038/sj.onc.1203397
61. Jenkins R, Takahashi S, DeLacey K, Bergstralh E, Lieber M. Prognostic significance of allelic imbalance of chromosome arms 7q, 8p, 16q, and 18q in stage T3N0M0 prostate cancer. Genes Chromosomes Cancer (1998) 21:131–43. doi: 10.1002/(SICI)1098-2264(199802)21:2<131::AID-GCC9>3.0.CO;2-1
62. Li Y, Zhai Y, Song Q, Zhang H, Cao P, Ping J, et al. Genome-Wide Association Study Identifies a New Locus at 7q21.13 Associated with Hepatitis B Virus-Related Hepatocellular Carcinoma. Clin Cancer Res Off J Am Assoc Cancer Res (2018) 24:906–15. doi: 10.1158/1078-0432.CCR-17-2537
63. Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer (2020) 19:92. doi: 10.1186/s12943-020-01213-6
64. Sun S, Gao J, Zhou S, Li Y, Wang Y, Jin L, et al. A novel circular RNA circ-LRIG3 facilitates the malignant progression of hepatocellular carcinoma by modulating the EZH2/STAT3 signaling. J Exp Clin Cancer Res CR (2020) 39:252. doi: 10.1186/s13046-020-01779-5
65. Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol (2017) 11:422–37. doi: 10.1002/1878-0261.12045
66. Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics (2019) 9:3526–40. doi: 10.7150/thno.32796
67. Liu R, Li Y, Wu A, Kong M, Ding W, Hu Z, et al. Identification of Plasma hsa_circ_0005397 and Combined With Serum AFP, AFP-L3 as Potential Biomarkers for Hepatocellular Carcinoma. Front Pharmacol (2021) 12:639963:639963. doi: 10.3389/fphar.2021.639963
68. Li W, Zhou X, Wu X, Wei J, Huang Z. The role of circular RNA hsa_circ_0085616 in proliferation and migration of hepatocellular carcinoma cells. Cancer Manage Res (2019) 11:7369–76. doi: 10.2147/CMAR.S211020
69. Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B Promotes Glycolysis and Malignancy of Hepatocellular Carcinoma Through the miR-338-3p/PKM2 Axis Under Hypoxic Stress. Hepatology (2019) 70:1298–316. doi: 10.1002/hep.30671
70. Wang L, Long H, Zheng Q, Bo X, Xiao X, Li . Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer (2019) 18:119. doi: 10.1186/s12943-019-1046-7
71. Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology (2017) 66:1151–64. doi: 10.1002/hep.29270
72. Gu X, Fu M, Ding Y, Ni H, Zhang W, Zhu Y, et al. TIMP-3 expression associates with malignant behaviors and predicts favorable survival in HCC. PloS One (2014) 9:e106161. doi: 10.1371/journal.pone.0106161
73. Ikeda M, Mitsunaga S, Ohno I, Hashimoto Y, Takahashi H, Watanabe K, et al. Systemic Chemotherapy for Advanced Hepatocellular Carcinoma: Past, Present, and Future. Diseases (2015) 3:360–81. doi: 10.3390/diseases3040360
74. Chen S, Cao Q, Wen W, Wang H. Targeted therapy for hepatocellular carcinoma: Challenges and opportunities. Cancer Lett (2019) 460:1–9. doi: 10.1016/j.canlet.2019.114428
75. Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. New Engl J Med (2017) 377:2500–1. doi: 10.1056/NEJMc1713444
76. Li J, Qin X, Wu R, Wan L, Zhang L, Liu R. Circular RNA circFBXO11 modulates hepatocellular carcinoma progress and oxaliplatin resistance through miR-605/FOXO3/ABCB1 axis. J Cell Mol Med (2020) 24:5152–61. doi: 10.1111/jcmm.15162
77. Li DQ, Qiu M, Nie XM, Gui R, Huang MZ. Oxidored-nitro domain-containing protein 1 expression is associated with the progression of hepatocellular carcinoma. Oncol Lett (2016) 11:3003–8. doi: 10.3892/ol.2016.4362
78. Luo Y, Fu Y, Huang R, Gao M, Liu F, Gui R, et al. CircRNA_101505 sensitizes hepatocellular carcinoma cells to cisplatin by sponging miR-103 and promotes oxidored-nitro domain-containing protein 1 expression. Cell Death Discovery (2019) 5:121. doi: 10.1038/s41420-019-0202-6
79. Wu MY, Tang YP, Liu JJ, Liang R, Luo XL. Global transcriptomic study of circRNAs expression profile in sorafenib resistant hepatocellular carcinoma cells. J Cancer (2020) 11:2993–3001. doi: 10.7150/jca.39854
80. Yang C, Dong Z, Hong H, Dai B, Song F, Geng L, et al. circFN1 Mediates Sorafenib Resistance of Hepatocellular Carcinoma Cells by Sponging miR-1205 and Regulating E2F1 Expression. Mol Ther Nucleic Acids (2020) 22:421–33. doi: 10.1016/j.omtn.2020.08.039
81. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem (2019) 88:487–514. doi: 10.1146/annurev-biochem-013118-111902
82. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res (2015) 25:981–4. doi: 10.1038/cr.2015.82
83. Sukowati CHC, Cabral LKD, Tiribelli C, Pascut D. Circulating Long and Circular Noncoding RNA as Non-Invasive Diagnostic Tools of Hepatocellular Carcinoma. Biomedicines (2021) 9(1):90. doi: 10.3390/biomedicines9010090
84. Li LM, Liu ZX, Cheng QY. Exosome plays an important role in the development of hepatocellular carcinoma. Pathol Res Pract (2019) 215:152468. doi: 10.1016/j.prp.2019.152468
85. Lyu L, Yang W, Yao J, Wang H, Zhu J, Jin A, et al. The diagnostic value of plasma exosomal hsa_circ_0070396 for hepatocellular carcinoma. Biomarkers Med (2021) 15(5):359–71. doi: 10.2217/bmm-2020-0476
86. Luo Y, Liu F, Gui R. High expression of circulating exosomal circAKT3 is associated with higher recurrence in HCC patients undergoing surgical treatment. Surg Oncol (2020) 33:276–81. doi: 10.1016/j.suronc.2020.04.021
87. Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine (2019) 40:432–45. doi: 10.1016/j.ebiom.2018.12.062
88. Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (2019) 11:8183–203. doi: 10.18632/aging.102312
89. Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res CR (2020) 39:20. doi: 10.1186/s13046-020-1529-9
90. Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett (2020) 475:119–28. doi: 10.1016/j.canlet.2020.01.022
91. Zhao M, Qu H. circVAR database: genome-wide archive of genetic variants for human circular RNAs. BMC Genomics (2020) 21:750. doi: 10.1186/s12864-020-07172-y
92. Meng X, Hu D, Zhang P, Chen Q, Chen M. CircFunBase: a database for functional circular RNAs. Database J Biol Database Curation (2019) 2019:baz003. doi: 10.1093/database/baz003
93. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol (2014) 15:409. doi: 10.1186/s13059-014-0409-z
94. Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, et al. RNAi in budding yeast. Science (2009) 326:544–50. doi: 10.1126/science.1176945
95. Baum J, Papenfuss AT, Mair GR, Janse CJ, Vlachou D, Waters AP, et al. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res (2009) 37:3788–98. doi: 10.1093/nar/gkp239
96. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
97. Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep (2014) 9:1966–80. doi: 10.1016/j.celrep.2014.10.062
98. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep (2015) 10:170–7. doi: 10.1016/j.celrep.2014.12.019
99. Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, et al. Exon circularization requires canonical splice signals. Cell Rep (2015) 10:103–11. doi: 10.1016/j.celrep.2014.12.002
100. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PloS Genet (2013) 9:e1003777. doi: 10.1371/journal.pgen.1003777
101. Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife (2015) 4:e07540. doi: 10.7554/eLife.07540
102. Jost I, Shalamova LA, Gerresheim GK, Niepmann M, Bindereif A, Rossbach O. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol (2018) 15:1032–9. doi: 10.1080/15476286.2018.1435248
Keywords: circular RNAs, hepatocellular carcinoma, biomarkers for diagnosis and prognosis, oncogenic circRNAs, tumor suppressive circRNAs, drug resistance
Citation: Aishanjiang K, Wei X-d, Fu Y, Lin X, Ma Y, Le J, Han Q, Wang X, Kong X, Gu J and Wu H (2021) Circular RNAs and Hepatocellular Carcinoma: New Epigenetic Players With Diagnostic and Prognostic Roles. Front. Oncol. 11:653717. doi: 10.3389/fonc.2021.653717
Received: 15 January 2021; Accepted: 22 March 2021;
Published: 20 April 2021.
Edited by:
Rongxin Zhang, Guangdong Pharmaceutical University, ChinaReviewed by:
Hamed Mirzaei, Kashan University of Medical Sciences, IranCopyright © 2021 Aishanjiang, Wei, Fu, Lin, Ma, Le, Han, Wang, Kong, Gu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hailong Wu, d3VobEBzdW1ocy5lZHUuY24=; Jinyang Gu, Z2p5bnlkQDEyNi5jb20=; Xiaoni Kong, eGlhb25pLWtvbmdAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.