- 1Department of Thoracic Surgery, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, China
- 2Experimental Animal Center, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, China
- 3Central Laboratory of Thoracic Surgery, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, China
Objective: The choice of adjuvant therapy for early stage lung adenocarcinoma (LUAD) remains controversial. Identifying the metabolism characteristics leading to worse prognosis may have clinical utility in offering adjuvant therapy.
Methods: The gene expression profiles of LUAD were collected from 22 public datasets. The patients were divided into a meta-training cohort (n = 790), meta-testing cohort (n = 716), and three independent validation cohorts (n = 345, 358, and 321). A metabolism-related gene pair index (MRGPI) was trained and validated in the cohorts. Subgroup analyses regarding tumor stage and adjuvant chemotherapy (ACT) were performed. To explore potential therapeutic targets, we performed in silico analysis of the MRGPI.
Results: Through machine learning, MRGPI consisting of 12 metabolism-related gene pairs was constructed. MRGPI robustly stratified patients into high- vs low-risk groups in terms of overall survival across and within subpopulations with stage I or II disease in all cohorts. Multivariable analysis confirmed that MRGPI was an independent prognostic factor. ACT could not improve prognosis in high-risk patients with stage I disease, but could improve prognosis in the high-risk patients with stage II disease. In silico analysis indicated that B3GNT3 (overexpressed in high-risk patients) and HSD17B6 (down-expressed in high-risk patients) may make synergic reaction in immune evasion by the PD-1/PD-L1 pathway. When integrated with clinical characteristics, the composite clinical and metabolism signature showed improved prognostic accuracy.
Conclusions: MRGPI could effectively predict prognosis of the patients with early stage LUAD. The patients at high risk may get survival benefit from PD-1/PD-L1 blockade (stage I) or combined with chemotherapy (stage II).
Introduction
Lung cancer is the leading cause of cancer-related death worldwide (1), and early stage lung cancer accounts for about 17% (2). Lung adenocarcinoma (LUAD) is the most common histologic subtype of lung cancer (3). Surgical resection plus lymph node dissection or sampling is the standard treatment for stage I LUAD (4). However, some patients will still suffer from disease relapse and death, and the 5-year overall survival ranges from 68 to 92% (5). According to the National Comprehensive Cancer Network (NCCN) guidelines, adjuvant systemic treatment is only considered for high-risk patients (4). The benefit of adjuvant systemic treatment for stage I LUAD remains controversial.
Biomarkers, especially gene expression, in tumor tissues are reliably related to cancer prognosis and survival (6–8). Thus, identifying the molecular features that may lead to worse prognosis may have clinical utility in offering adjuvant therapy to a subgroup of patients at high risk. The availability of large-scale public cohorts with gene expression data provides an ideal resource to identify a more individualized prognostic signature for LUAD.
Reprogramming of energy metabolism is an emerging hallmark of cancer (9) and recently has been proved to be involved in lung cancer initiation, progression, and drug resistance (10–13). Metabolic phenotypes can also be exploited to image tumors, provide prognostic information, and treat cancer (14). Therefore, understanding the metabolism characteristics by gene expression-based algorithms may be helpful for screening the patients at high risk. However, the molecular characteristics of tumor metabolism remain to be comprehensively explored regarding their prognostic potential in early stage LUAD.
In this study, we integrated multiple cohorts with gene expression profiles to develop and validate an individualized prognostic signature for early stage LUAD from metabolism-related gene pairs (MRGPs). We then explored the potential therapy regimen for the patients at high risk, which may be utilized in clinical. Further, to leverage the complementary value of molecular and clinical features, we integrated the metabolism signature with clinical factors to improve the predicted accuracy for overall survival (OS).
Methods
Patients and Datasets
This study was approved by the Ethic Committee of Shanghai Pulmonary Hospital. We retrospectively analyzed the gene expression matrixes and corresponding clinical characteristics from 22 public datasets (Supplementary Table S1), including 17 microarray and two RNAseq datasets from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), one RNAseq dataset from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov), one microarray dataset from the ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/), and one RNAseq dataset from the OncoSG database (15) (https://src.gisapps.org/OncoSG/). The patients were included according to the following criteria: (1) lung adenocarcinoma, (2) stages I–II, (3) available OS information. The patients were excluded if they met any of the exclusion criteria: (1) non-adenocarcinoma or the pathologic subtypes were unknown, (2) stage III or IV or unknown, (3) lack of OS information, (4) received neoadjuvant therapy. The gene expression matrix of normal lung tissue was downloaded from the Genotype-Tissue Expression (GTEx) database (https://www.gtexportal.org/home/). The entire tumor datasets were divided into meta-training, meta-testing, and three independent validation cohorts (TCGA, GSE68465, and GSE72094) (Supplementary Figure S1).
Data Process
All the expression level of microarray datasets was transformed by log2. For all the datasets of RNAseq, the fragments per kilobase million (FPKM) level was used as the expression value and log2(FPKM+1) transformed. If there were duplicate genes in each dataset, the mean value was calculated by the avereps function from the limma R package.
Construction of the MRGPI
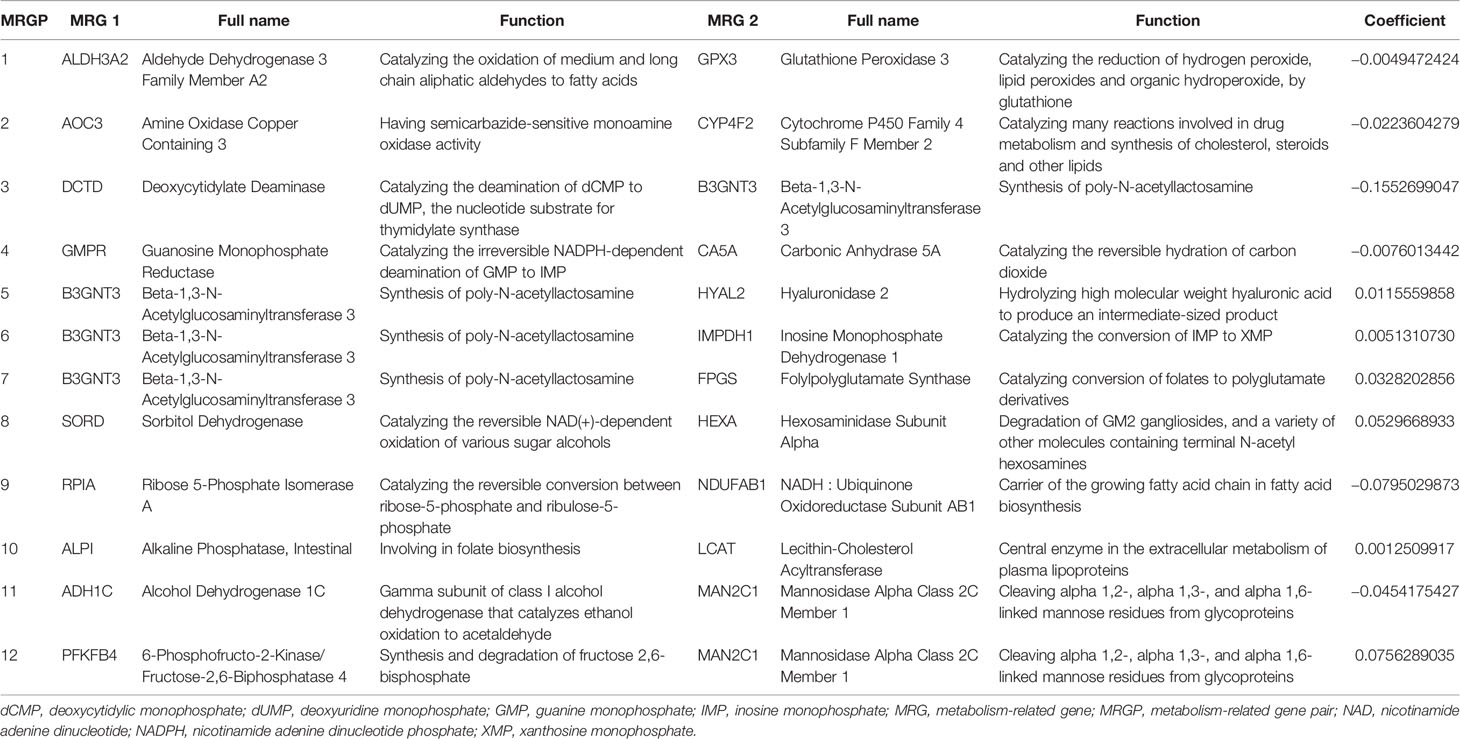
As shown in the Figure 1, we constructed a prognostic signature by focusing on metabolism-related genes (MRGs). From the c2.cp.kegg.v7.0.symbols.gmt dataset that was downloaded from the Gene Set Enrichment Analysis (GSEA) website (https://www.gsea-msigdb.org/gsea/index.jsp), 2,522 MRGs from 68 metabolism related Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were identified. Of the 2,522 MRGs, 690 MRGs were available in all datasets. The gene expression value underwent pairwise subtraction to generate a score for each metabolism-related gene pair (MRGP): MRGP score = expression value of MGP 1 − expression value of MGP 2. The score represented the log2 fold change of MGP 1 relative to MGP 2.
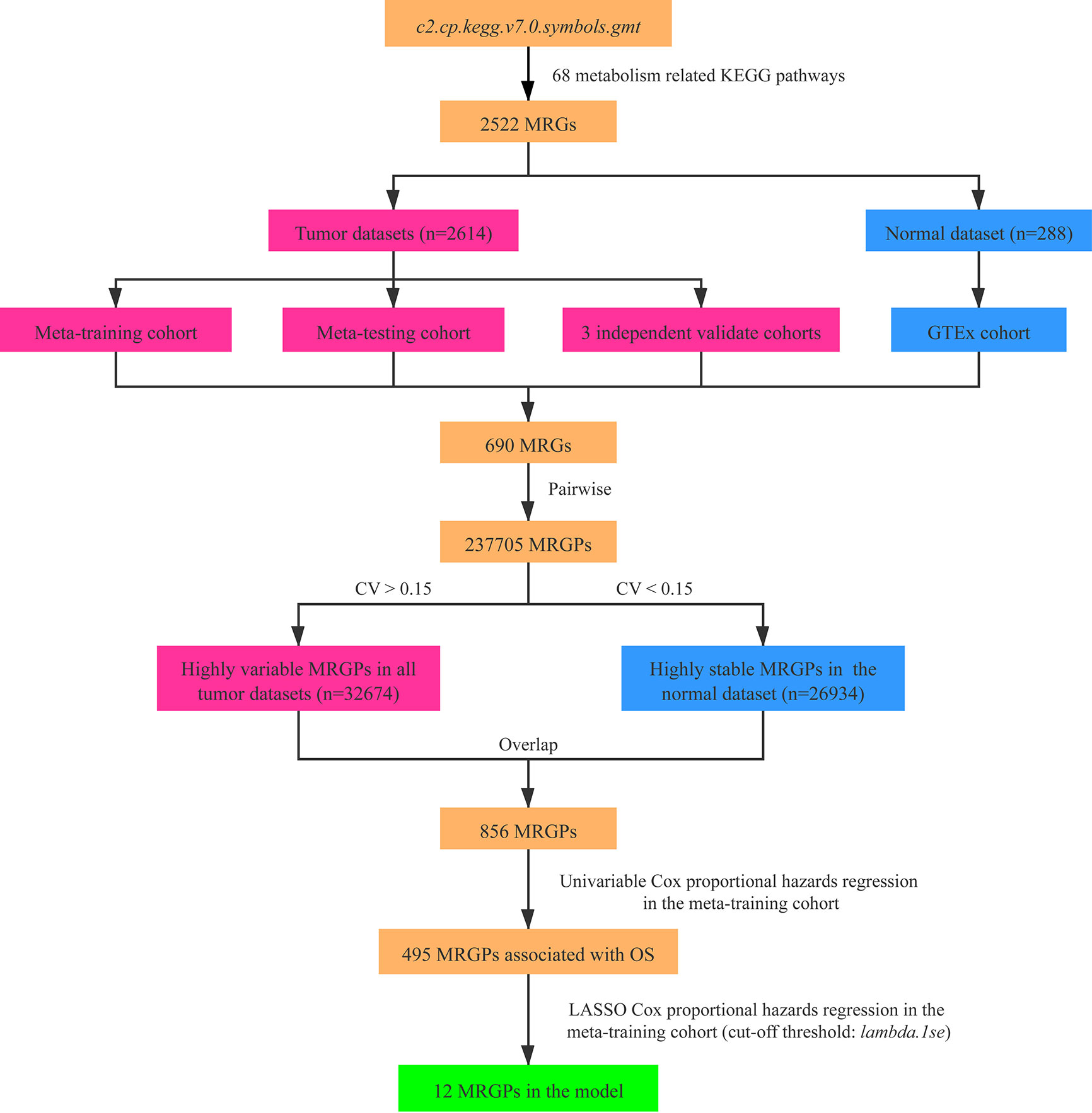
Figure 1 Flowchart of the construction process of MRGPI. CV, coefficient of variation; GTEx, the Genotype-Tissue Expression; LASSO, least absolute shrinkage and selection operator; MRGs, metabolism-related genes; MRGPs, metabolism-related gene pairs; OS, overall survival.
To screen the representative MRGPs in tumor, we identified the MRGPs that were highly variable [coefficient of variation (CV) > 0.15] in all tumor datasets and highly stable (CV < 0.15) in the normal cohort. Then the univariable Cox proportional hazards regression was used to select prognostic MRGPs in the screened MRGPs (survival R package). Finally, to minimize the risk of overfitting, a cox proportional hazards regression model combined with the least absolute shrinkage and selection operator (LASSO) was applied to identify the most important prognostic MRGPs (glmnet R package). The optimal values of the penalty parameter λ were determined by 10-fold cross-validations at 1 SE beyond the minimum partial likelihood deviance in the meta-training cohort. Based on the selected MRGPs from LASSO Cox regression model, the metabolism-related gene pair index (MRGPI) for each patient was constructed: MRGPI = . To separate patients into low- or high-risk groups, the optimal MRGPI cutoff value was determined using the surv_cutpoint function of the survminer R package.
Validation of the MRGPI
The predictive value of MRGPI for OS was evaluated in the meta-training, meta-testing and three independent validation cohorts. As described in a previous study (6), the pathologic stage was treated as continuous variable by the following converting approach: IA was coded as 1, then IB as 2, I as 1.5, I–II as 2.5, IIA as 3, IIB as 4 and II as 3.5. The univariable Cox regression model was used to evaluate the prognostic value of age, gender, smoking history, stage and MRGPI (as continuous and binary form, respectively). The multivariable Cox regression model was used to evaluate the independent prognostic value of MRGPI. Subgroup analysis was performed according to the stage.
DEGs and Gene Ontology Analysis
The gene expression differences between high and low risk were compared using the limma package, and genes with |log fold change| > 1 and false discovery rate adjusted P value <0.05 were considered to be significant differentially expressed genes (DEGs). To gain biological understanding of the MRGPI, we conducted an enrichment analysis of its component MRGs using the clusterProfiler R package. FDR-adjusted P <0.05 was used to select statistically significant gene sets.
Profiling of Infiltrating CD8 T Cells
To analyze the tumor immune microenvironment, a dataset of single cell RNAseq (scRNA-seq) with annotated cell types (16) (GSE131907) was downloaded from the GEO database. There were nine samples of stage I–II LUAD, and the cell numbers of all the samples were more than 3,200. The mean transcripts per kilobase million (TPM) value of one gene was calculated, and the log2(TPM+1) was used as the expression value of the tumor cells in each sample. Given that too less tumor cells could not reflect the characteristics of the tumor, we remove two samples whose tumor cells were less than 50, and seven samples of stage IA LUAD were included for analysis.
Construction and Validation of the MCPI
Based on the results of the multivariable Cox analysis in the all cohorts, age, stage, and MRGPI score were significantly associated with OS. Age, stage, and MRGPI score were integrated to composite a metabolism-clinical prognostic index (MCPI) by applying Cox proportional hazards regression in the meta-training cohort: MCPI score= age × coefficient + stage × coefficient + MRGPI × coefficient. The prognostic accuracy of MRGPI was estimated using the concordance index (C-index), which range from 0 to 1.0 (survcomp R package). As we mentioned above, the optimal cutoff value of MCPI score was determined by the surv_cutpoint function in the meta-training cohort. The predictive value of MCPI for OS was evaluated in the meta-testing and three independent validation cohorts.
Statistical Analysis
All statistical analyses were conducted using R software (version 3.6.2). Pearson correlation analysis was performed to determine the correlation between two variables. The Kaplan–Meier method was used to generate survival curves, and significance of differences was compared using the log-rank test. All statistical tests were two-sided, and P values less than 0.05 were considered statistically significant.
Results
Patient Characteristics of Included Cohorts
Totally, 2,614 patients with stage I–II LUAD (Table 1) and 288 heathy donors were included for analysis. The median age ranged from 62 to 70 in all cohorts, and the proportion of female were larger than male. Most patients (>48.2%) had smoking history, and the patients with stage I LUAD accounted for the major proportion, except GSE68465, in which most patients did not had specific stage (stages I–II). In the meta-training, meta-testing, and GSE68465 cohorts, the median follow-up time was more than 50 months, and the death events were observed in more than 35% patients. However, the median follow-up time of the TCGA and GSE72094 was shorter, and the events of death were less than those of other cohorts.
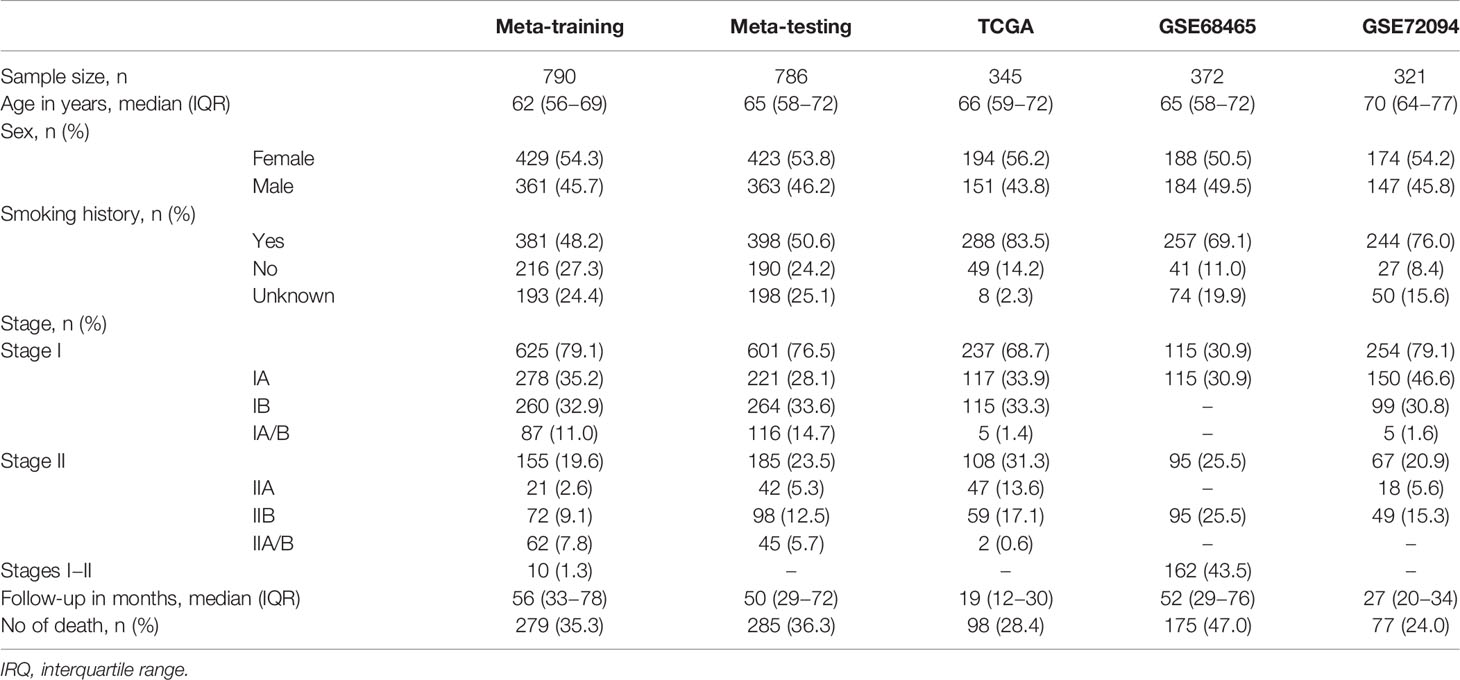
Table 1 Clinical and pathologic features of patients in meta-training, meta-testing, and independent validation cohorts.
Construction of the MRGPI
After pairwise coupling of the 690 GRPs, 237,705 MRGPs were constructed, and the corresponding scores were generated. We removed 205,031 MRGPs with CV <0.15 in all datasets and 210,771 MRGPs with CV >0.15 in the normal dataset. Between the remaining 32,674 MRGPs in the tumor cohorts and 26,934 MRGPs in the normal cohort, 856 MRGPs were overlapped. The association of the 856 MRGPs with OS was assessed in the meta-training cohort, resulting in 495 prognostic MRGPs. Finally, the LASSO Cox regression model selected 12 MRGPs in the meta-training cohort (Supplementary Figure S2A). Based on the 12 MRGPs that consisted of 20 MRGs, the MRGPI for each patient was constructed (Table 2). The optimal cutoff point (−0.261) obtained from the surv_cutpoint function served as the cutoff to assign patients into high- and low-risk groups (Supplementary Figure S2B). The Kaplan–Meier curve showed the patients in the high-risk group presented with a significantly worse OS in the meta-training cohort [hazard ratio (HR): 3.584, 95% confidence interval (CI): 2.755–4.663, P < 0.001, Supplementary Figure S2C]. Univariable Cox analysis indicated that MRGPI (both as continuous and binary form) was a prognostic factor for OS, and multivariable Cox analysis confirmed that MRGPI (as binary form) was independently associated with OS (Figure 3 and Supplementary Table S2). The C-index of the MRGPI in the meta-training cohort was 0.701 (95% CI: 0.672–0.730).
Validation of the MRGPI in Multiple Independent Cohorts
To determine whether the MRGPI was robust, the performance of the MRGPI was assessed in the meta-testing and three independent cohorts. Consistent with the outcomes of the meta-training cohort, the MRGPI significantly stratified patients into low- vs high-risk groups in terms of OS. The patients in the high-risk group had significantly worse OS in the meta-testing (HR: 2.011, 95% CI: 1.531–2.640, P < 0.001, Figure 2A), TCGA (HR: 1.657, 95% CI: 1.106–2.482, P = 0.013, Figure 2B), GSE68465 (HR: 1.626, 95% CI: 1.194–2.214, P = 0.002, Figure 2C), and GSE72094 (HR: 2.370, 95% CI: 1.514–3.714, P < 0.001, Figure 2D) cohorts. The MRGPI (both as continuous and binary form) was a prognostic factor for OS in all the validation cohorts in the univariate Cox analysis, and it remained as an independent prognostic factor in multivariate analysis, after adjusting for age, gender, smoking history, and tumor stage (Figure 3 and Supplementary Table S2). The C-index of the meta-testing, TCGA, GSE68465, GSE72094 cohort was 0.576 (95% CI: 0.541–0.612), 0.604 (95% CI: 0.535–0.673), 0.589 (95% CI: 0.543–0.634) and 0.645 (95% CI: 0.582–709), respectively.
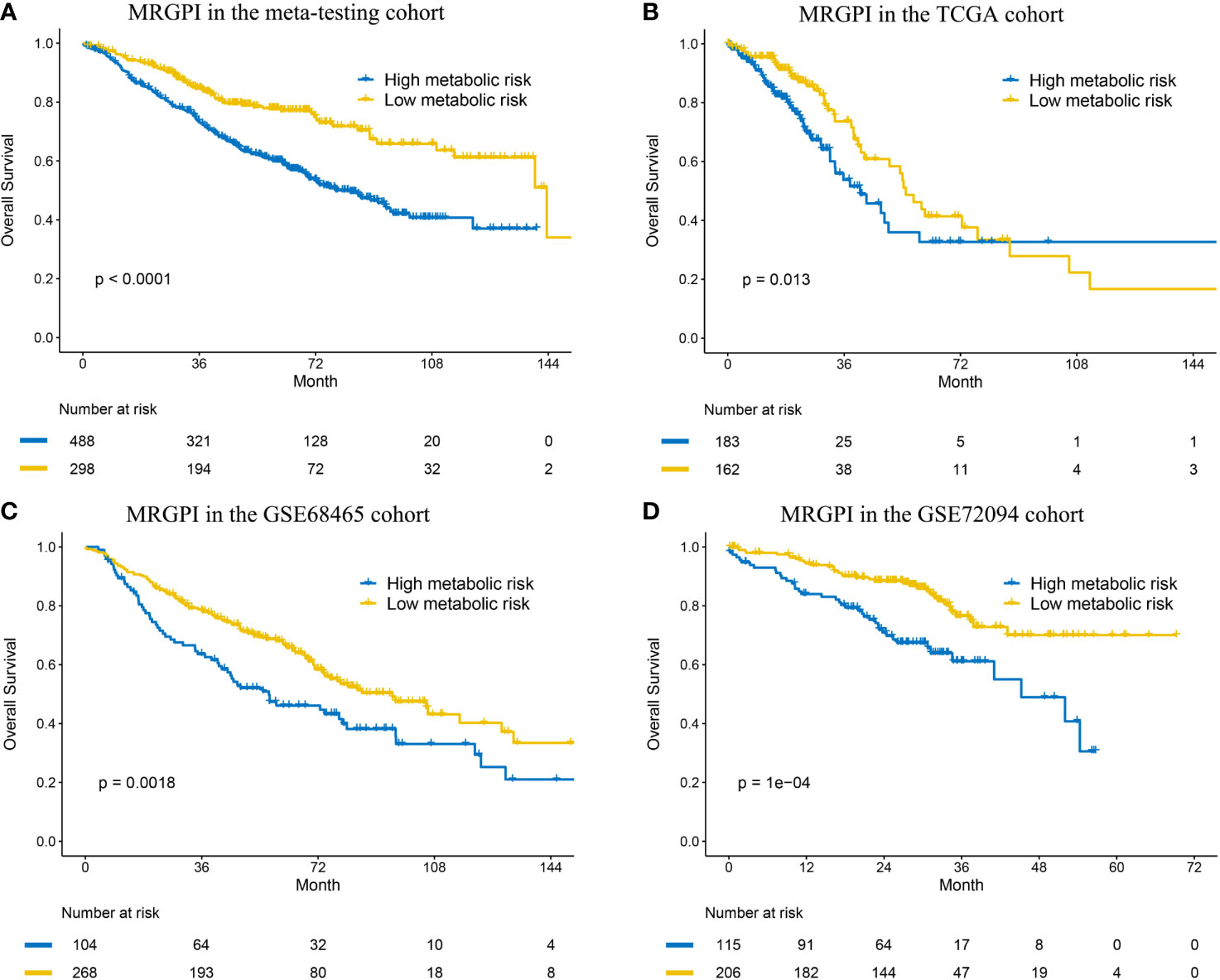
Figure 2 Kaplan–Meier curves of overall survival in the meta-testing (A), TCGA (B) and GSE68465 (C) and GSE72094 (D) cohort.
Subgroup Analysis of the MRGPI in Stage I Disease
In the patients with stage I disease, the MRGPI stratified patients in all cohorts into significantly different prognostic groups. The MRGPI remained highly prognostic for the meta-training (HR: 3.842, 95% CI: 2.801–5.270, P < 0.001), meta-testing (HR: 2.101, 95% CI: 1.499–2.945, P < 0.001), GSE68465 (HR: 2.129, 95% CI: 1.054–4.299, P = 0.031) and GSE72094 (HR: 2.260, 95% CI: 1.311–3.895, P = 0.003) cohort (Supplementary Figures S2D and S3A–D), and multivariable Cox analysis confirmed that MRGPI was independently associated with OS (Figure 3 and Supplementary Table S3). However, the result was negative in the TCGA cohort, and the short follow-up time and less death events probably accounted for it.
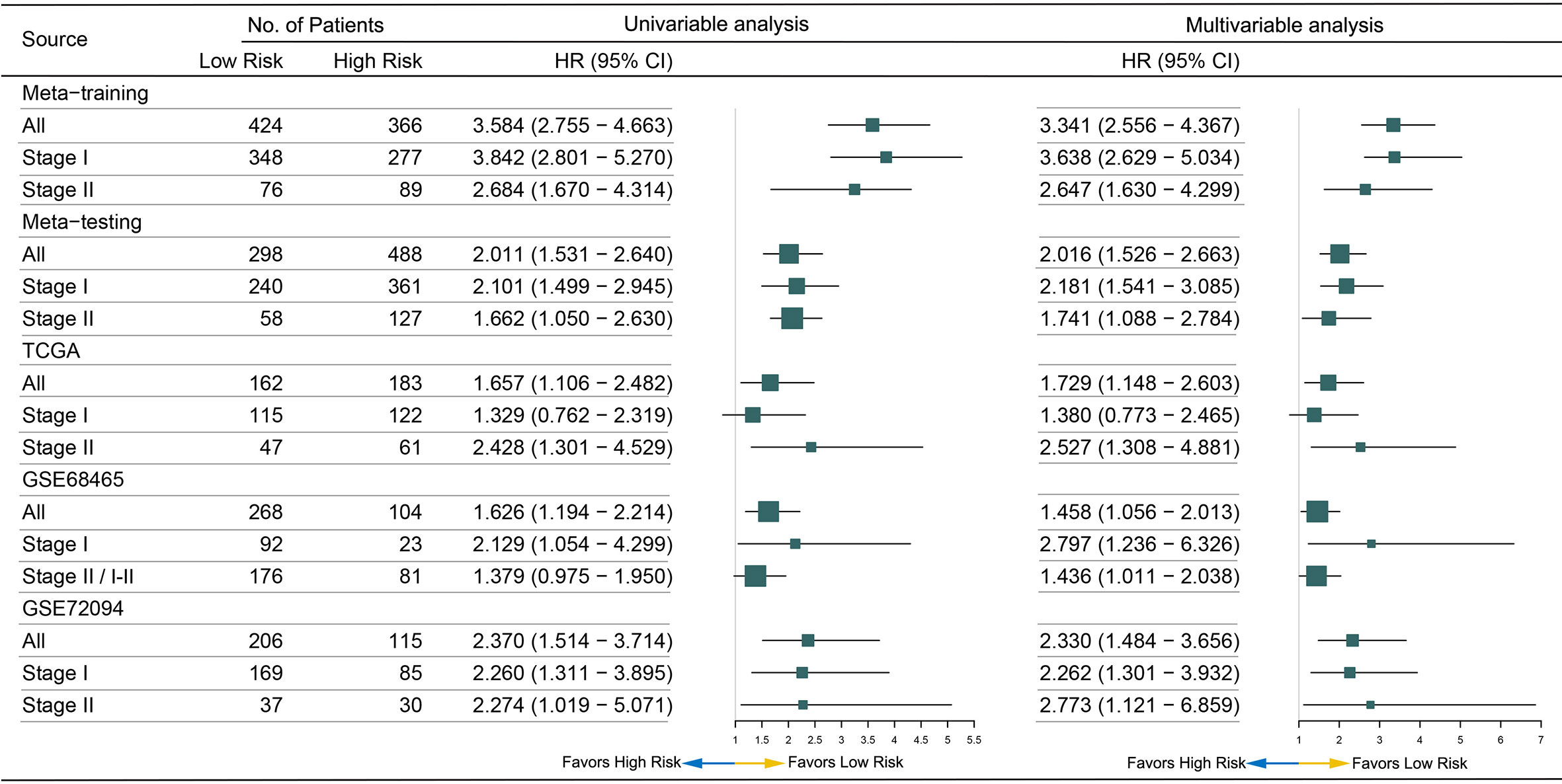
Figure 3 Forest plot for the hazard ratios (HRs) of high vs low metabolism-related gene pair index (MRGPI) risk groups.
Given the prognosis differences between high- and low- risk patients, we analyzed the benefit of adjuvant chemotherapy (ACT) in the two groups. Of all the validation datasets, five datasets (OncoSG, GSE42127, GSE14814, TCGA, and GSE68465) recorded the information of ACT. Compared to surgery alone, ACT did not improve OS in the low-risk group (HR: 1.817, 95% CI: 0.871–3.791, P = 0.111; Figure 4A). We also did not observe that patients in the high-risk group could get OS benefit from ACT (HR: 0.959, 95% CI: 0.521–1.765, P = 0.893; Figure 4B), which indicated that ACT may be not suitable for the patients. To improve the prognosis, other adjuvant therapy regimens should be explored.
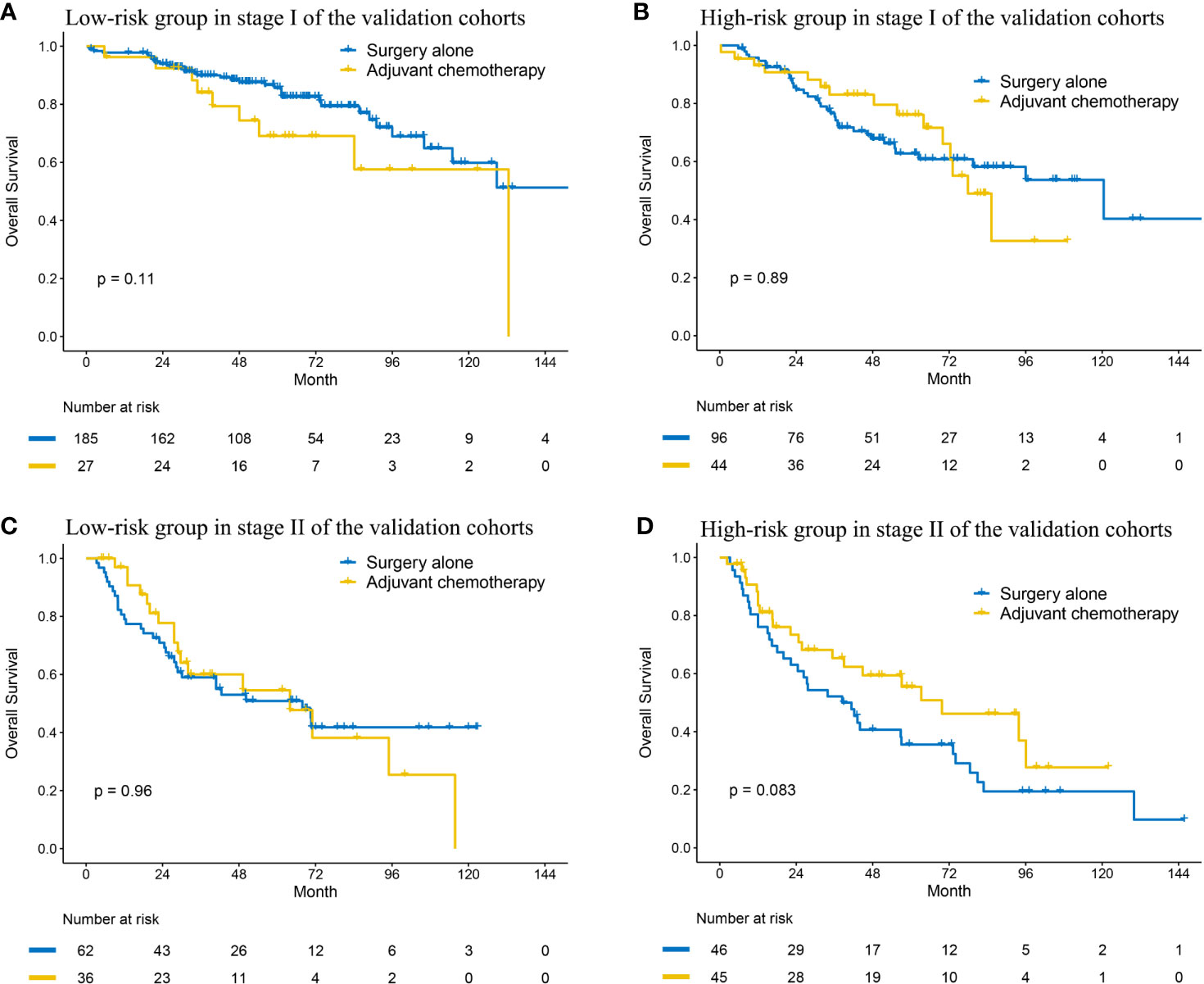
Figure 4 Kaplan–Meier curves of overall survival regarding adjuvant chemotherapy in patients with stage I (A, B) and stage II (C, D) disease at low and high risk in the validation cohort.
Subgroup Analysis of the MRGPI in Stage II Disease
The MRGPI could also stratified patients in all cohorts into significantly different prognostic groups in the patients with stage II disease. The patients in the high-risk group had significantly worse OS in the meta-training (HR: 2.684, 95% CI: 1.670–4.314, P < 0.001), meta-testing (HR: 1.662, 95% CI: 1.050–2.630, P = 0.030), TCGA (HR: 2.428, 95% CI: 1.301–4.529, P < 0.001), and GSE72094 (HR: 2.274, 95% CI: 2.274, P = 0.045) cohort (Supplementary Figures S2E and S4A–D). The MRGPI remained an independent risk factor in multivariable analysis (Figure 3 and Supplementary Table S4). A margin positive result (HR: 1.379, 95% CI: 0.975–1.950, P = 0.069) was observed in the GSE68465 cohort (including stages I–II, Supplementary Figure S4C); however, the result of multivariable analysis showed that the MRGPI was an independent risk factor (Supplementary Table S4).
Then, we also explored the effect of ACT in the two groups. The Kaplan–Meier curve indicated that ACT could not improve OS in the low-risk group (HR: 1.013, 95% CI: 0.561–1.829, P = 0.965; Figure 4C). In the high-risk group, although the result was negative (HR: 0.621, 95% CI: 0.360–1.070, P = 0.086; Figure 4D), the curves had an obvious tendency to separate and the small sample size probably accounted for it.
Biological Phenotypes Associated With the MRGPI
Enrichment analysis of the 20 unique MRGs in the MRGPI identified two overrepresented biological processes (organic acid catabolic process and carboxylic acid catabolic process) in the gene ontology (Supplementary Figure S5A). To explore the potential survival mechanism related to the MRGPI, we analyzed the DEGs between the high and low-risk groups in the three independent validation cohorts, and we focused on the differentially expressed MRGs. Among the DEGs from the three cohorts, three MRGs (B3GNT3, ADH1B, and HSD17B6) were overlapped (Figures 5A–C), and their expression levels were significantly correlated with MRGPI (Supplementary Figure S6). The three MRGs had been reported to be associated with other cancers (17–19), but few studies reported their role in LUAD.
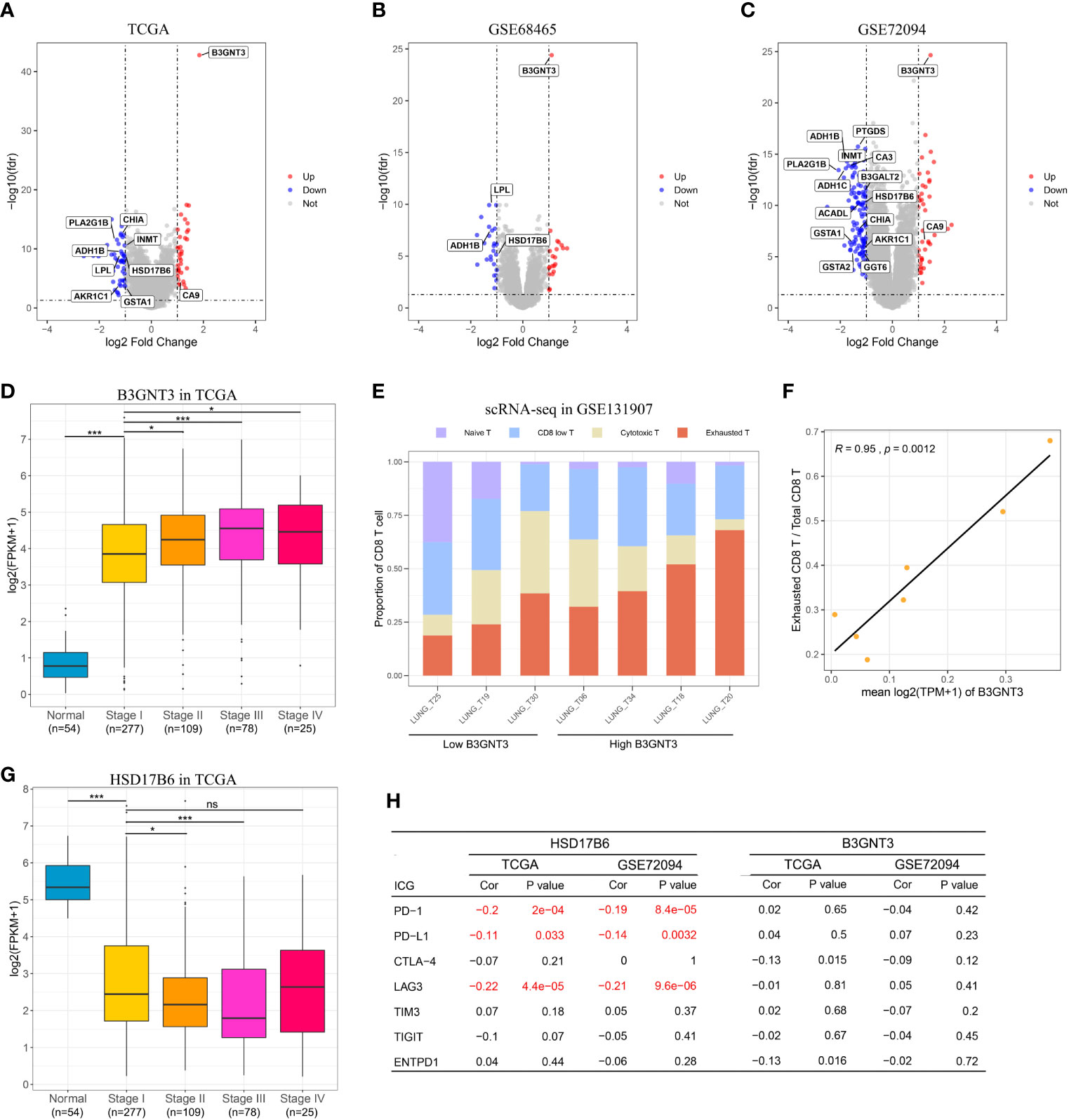
Figure 5 In silico analysis of the MRGPI. (A–C): Volcano plot showing fold changes for genes differentially expressed between high- and low-risk patients in the TCGA, GSE68465, and GSE72094 cohort. (D) Boxplots of the expression level of B3GNT3 in the normal tissue and different tumor stages showing that upregulation of B3GNT correlated with tumor progression. (E) Proportion of different CD8+ T cell in each patient, and the patients were divided into low and high B3GNT3 group based on the median value. (F) The expression level of B3GNT3 was significantly associated with the proportion of the exhausted CD8+ T cell. (G) Boxplots of the expression level of HSD17B6 in the normal tissue and different tumor stages showing that down-regulation of HSD17B6 correlated with tumor progression. (H) Pearson’s correlation test between B3GNT3, HSD17B6, and immune checkpoint genes. ns, not significant (P > 0.05), **P < 0.01, ***P < 0.001.
B3GNT3 was overexpressed in LUAD, and its expression level was positively associated with tumor stage (Figure 5D), which suggested that B3GNT3 played an important role in tumor carcinogenesis and prognosis. Previous study reported that N-linked glycosylation of PD-L1 that was catalyzed by B3GNT3 was required for physical contact between PD-L1 and PD-1 in triple-negative breast cancer, and then caused CD8+ T cell exhausted (18). We then explored whether there was a similar mechanism in LUAD. From the scRNA-seq result, we noticed that the expression level of B3GNT3 in tumor cell was positively correlated with the proportion of the exhausted CD8+ T cell (r = 0.95, P = 0.0012, Figures 5E, F). However, the expression level of B3GNT3 was not correlated with immune checkpoint genes (ICGs) in the TCGA and GSE72094 cohorts (most ICGs were not available in the GSE68465 dataset), especially PD-1 and PD-L1 (Figure 5H). The results demonstrated that there may be the same mechanism of B3GNT3 in LUAD.
HSD17B6 was down-expressed in LUAD, and the expression level of HSD17B6 was negatively associated with tumor stage (Figure 5G). HSD17B6 could convert 3 alpha-adiol to dihydrotestosterone that was closely related to the development of many tumors (20). Lv et al. (17) reported that low expression of HSD17B6 correlated with multiple ICGs expression in hepatocellular carcinoma. In this study, we observed that the expression level of HSD17B6 was negatively correlated with PD-1 (r = −0.20 and P < 0.001 in TCGA, r = −0.19 and P < 0.001 in GSE72094), PD-L1 (r = −0.11 and P = 0.033 in TCGA, r = −0.14 and P = 0.003 in GSE72094), and LAG3 (r = −0.22 and P < 0.001 in TCGA, r = -0.21 and P < 0.001 in GSE72094) (Figure 5H), suggesting that low HSD17B6 expression potentially played an important role in mediating immune evasion. ADH1B was also down-expressed in LUAD (Supplementary Figure S5B); however, its expression level was not negatively correlated with ICGs as HSD17B6 (Supplementary Figure S5C), which suggested that there may be other mechanisms behind it.
Together, these results indicated that B3GNT3 and HSD17B6 may make synergic reaction in immune evasion, with HSD17B6 up-regulating PD-L1 and B3GNT3 stabilizing the PD-L/PD-L1 ligation. Immune checkpoint inhibitors, especially PD-1/PD-L1 anti-body may be a therapeutic choice. Combined with the results of ACT in LUAD at different stages and risks, we thought that patients at high risk may get survival benefit from PD-1/PD-L1 blockade (stage I) or combined with chemotherapy (stage II). Although PD-1/PD-L1 anti-body as neoadjuvant therapy has been used in early stage NSCLC in clinical trials recently (21–24), there are no transcriptomic data of the tumor before treatment at present, so the regimen we proposed could not be validated in this study.
Integrated Prognostic Index by Combining the MRGPI With Clinical Factors
To further improve accuracy, we combined age, stage, and MRGPI score to fit a Cox proportional hazards regression model in the meta-training cohort and derived a MCPI: MCPI = age × 0.028 + stage × 0.312 + MRGPI × 1.726. The optimal cutoff value of the MCPI for stratifying patients was determined to be 2.007 (Supplementary Figure S7A). Improved estimation of OS was achieved by the binary form of MCPI compared with MRGPI (Supplementary Figures S7B−F), and the C-index for the meta-training, meta-testing, TCGA, GSE68465, GSE72094 cohort was 0.729 (95% CI 0.700–0.757), 0.648 (95% CI 0.613–0.682), 0.641 (95% CI 0.567–0.709), 0.665 (95% CI 0.634–0.709), and 0.666 (95% CI 0.602–0.731), respectively (Supplementary Figure S7G).
Discussion
When diagnosed at early stages, LUAD could be effectively treated with surgical resection. However, the use of ACT for stage I LUAD in the setting of standard therapy remains controversial because several clinical trials fail to show a survival benefit among unselected patients, and the toxic effects of chemotherapy are inevitable (25). The strategy is to identify of the subset of patients at high risk for recurrence and death. A prognostic signature beyond the current staging system is desired to accurately identify the patients at high risk and to better guide adjuvant treatment (7). In this study, we developed a prognostic signature based on 12 MRGPs to predict prognosis of early stage LUAD and validated it in multiple independent cohorts across different platforms. The MRGPI was extremely robust in stratifying the patients into the low- and high-risk groups with different survival outcomes. Several models based on the expression value have already been reported to present with the ability for predicting prognosis in lung cancer (26–29). However, the models based on the absolute value of the expression level could not avoid the technical biases inherent across different platforms. The gene pairs signature proposed by Li et al. (6) is based on the relative value of gene expression level, which only refers to the pairwise comparison of the gene expression profile within a sample. Li et al. constructed a gene pair signature based on 25 immune-related gene pairs consisting of 40 immune-related genes in non-squamous lung cancer (6). Our prognostic signature was derived from MRGs in LUAD and MRGPI consisted of 12 gene pairs involving 20 MRGs. With less gene pairs, MRGPI performed comparable accuracy to Li and colleagues’ model in the TCGA (C-index: 0.60 vs 0.62) cohort.
After identifying the patients at different risks, we explored the benefit of ACT. Not surprisingly, ACT could not bring survival benefit in stage I LUAD at low risk. However, ACT also could not improve OS in stage I LUAD at high risk, suggesting that chemotherapy may be not suitable for the patients. For stage II LUAD, ACT may improve OS in the patients at high risk, which was in accordance with the clinical practice. However, we also noticed that the patients at low risk could not get survival benefit from ACT, suggesting that ACT should also be used selectively in a subset of patients with stage II LUAD. According to the NCCN guidelines, ACT should be performed in stage IIB LUAD with R0 resection, but it is alternative in stage IIA LUAD and just required for high-risk patients (4). Besides identifying high-risk patients with stage I LUAD, MRGPI could also identify a subset of patients in stage II who may be free from ACT. However, the sample size of ACT was small in this study, and more studies were needed to validate the results.
To explore potential therapeutic targets for the patients with poor prognosis based on the MRGPI, we performed DEG analysis using the three independent datasets. Three MRGs were identified, and B3GNT3 and HSD17B6 may make synergic reaction in immune evasion by the PD-1/PD-L1 pathway. Thus, PD-1/PD-L1 blockade was an optimal therapy regimen for the patients at high risk. Compared with conventional ACT, adjuvant immunotherapy could improve prognosis in resectable solid tumor (30, 31), and neoadjuvant therapy may get more survival benefit than adjuvant therapy (32). Recently, PD-1/PD-L1 anti-body as neoadjuvant therapy has been proved to be feasible in resectable lung cancer (21−24). Thus, the patients with stage I LUAD at high risk may be get survival benefit from PD-1/PD-L1 blockade. For the patients with stage II LUAD at high risk, both chemotherapy and PD-1/PD-L1 blockade may improve prognosis, so PD-1/PD-L1 anti-body plus chemotherapy as neoadjuvant therapy may be optimal. However, there are no transcriptomic data of the tumor before immunotherapy available at present to validate it. For the patients at low risk, surgery alone may be optimal, but the benefit of immunotherapy should also be explored in future studies.
There were some limitations in our study. First, some biases were inevitable because of the retrospective nature of this study. Second, the mutation status was not considered due to lack of information of most datasets. Since driver genes like EGFR and ALK mutation were common in LUAD, the benefit of targeted therapy in the patients at risk could not be evaluated, and adjuvant targeted therapy was proved to be better than ACT in clinical trials (33, 34). Third, as we mentioned above, the sample size of ACT was small, and more studies were needed to validate the results. Last, the therapy regimens we proposed were warranted to validate in clinical studies.
In conclusion, this study identified metabolism-related gene pair-based signature that can effectively predict survival outcomes of the patients with early stage LUAD. The patients at high risk may get survival benefit from PD-1/PD-L1 blockade (stage I) or combined with chemotherapy (stage II). Prospective studies are needed to further validate its analytical accuracy for estimating prognosis and test its clinical utility in individualized management of early stage LUAD.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Tongji University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
Conception and design: PZ. Administrative support: GJ. Provision of study materials or patients: JH, HY, and LZ. Collection and assembly of data: JH, LS, and YY. Data analysis and interpretation: JH, LS, and HY. Manuscript writing: all authors. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2019YFC1315803), National Natural Science Foundation of China (Grant No. 81972172), the Shanghai Municipal Health Commission (Grant No. 20174Y0111), the Shanghai Science and Technology Committee (Grant No. 19XD1423200, 18140903900, 201409001000), and Programs of Shanghai Pulmonary Hospital (No. fkcx1904).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.650853/full#supplementary-material
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Ca-Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Cancer Stat Facts: Lung and Bronchus Cancer. Available at: https://seer.cancer.gov/statfacts/html/lungb.html (Accessed Sep 13, 2020).
3. Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thoracic Oncol (2011) 6(2):244–85. doi: 10.1097/JTO.0b013e318206a221
4. National Comprehensive Cancer Network. Nccn Clinical Practice Guidelines in Oncology: non-Small Cell Lung Cancer. Version 7. 2020 - September 11, 2020. Available at: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf (Accessed Sep 13, 2020).
5. Goldstraw P, Chansky K, Crowley J, Rami-Porta R, Asamura H, Eberhardt WEE, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thoracic Oncol (2016) 11(1):39–51. doi: 10.1016/j.jtho.2015.09.009
6. Li B, Cui Y, Diehn M, Li R. Development and Validation of an Individualized Immune Prognostic Signature in Early-Stage Nonsquamous Non-Small Cell Lung Cancer. JAMA Oncol (2017) 3(11):1529–37. doi: 10.1001/jamaoncol.2017.1609
7. Cai WY, Dong ZN, Fu XT, Lin LY, Wang L, Ye GD, et al. Identification of a Tumor Microenvironment-Relevant Gene Set-Based Prognostic Signature and Related Therapy Targets in Gastric Cancer. Theranostics (2020) 10(19):8633–47. doi: 10.7150/thno.47938
8. Kandimalla R, Tomihara H, Banwait JK, Yamamura K, Singh G, Baba H, et al. A 15-Gene Immune, Stromal, and Proliferation Gene Signature That Significantly Associates With Poor Survival in Patients With Pancreatic Ductal Adenocarcinoma. Clin Cancer Res (2020) 26(14):3641–8. doi: 10.1158/1078-0432.Ccr-19-4044
9. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
10. Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell (2016) 164(4):681–94. doi: 10.1016/j.cell.2015.12.034
11. Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, et al. Lactate Metabolism in Human Lung Tumors. Cell (2017) 171(2):358. doi: 10.1016/j.cell.2017.09.019
12. Chen PH, Cai L, Huffman K, Yang CD, Kim J, Faubert B, et al. Metabolic Diversity in Human non-Small Cell Lung Cancer Cells. Mol Cell (2019) 76(5):838. doi: 10.1016/j.molcel.2019.08.028
13. Kim SM, Yun MR, Hong YK, Solca F, Kim JH, Kim HJ, et al. Glycolysis Inhibition Sensitizes Non-Small Cell Lung Cancer With T790M Mutation to Irreversible Egfr Inhibitors Via Translational Suppression of Mcl-1 by AMPK Activation. Mol Cancer Ther (2013) 12(10):2145–56. doi: 10.1158/1535-7163.Mct-12-1188
14. Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections Between Metabolism and Cancer Biology. Cell (2017) 168(4):657–69. doi: 10.1016/j.cell.2016.12.039
15. Chen JB, Yang HC, Teo ASM, Amer LB, Sherbaf FG, Tan CQ, et al. Genomic Landscape of Lung Adenocarcinoma in East Asians. Nat Genet (2020) 52(2):177. doi: 10.1038/s41588-019-0569-6
16. Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW, et al. Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma. Nat Commun (2020) 11(1):2285. doi: 10.1038/s41467-020-16164-1
17. Lv L, Zhao YJ, Wei QQ, Zhao Y, Yi QY. Downexpression of HSD17B6 Correlates With Clinical Prognosis and Tumor Immune Infiltrates in Hepatocellular Carcinoma. Cancer Cell Int (2020) 20:210. doi: 10.1186/s12935-020-01298-5
18. Li CW, Lim SO, Chung EM, Kim YS, Park AH, Yao J, et al. Eradication of Triple-Negative Breast Cancer Cells by Targeting Glycosylated PD-L1. Cancer Cell (2018) 33(2):187. doi: 10.1016/j.ccell.2018.01.009
19. Suo C, Yang YJ, Yuan ZY, Zhang TJ, Yang XR, Qing T, et al. Alcohol Intake Interacts With Functional Genetic Polymorphisms of Aldehyde Dehydrogenase (ALDH2) and Alcohol Dehydrogenase (ADH) to Increase Esophageal Squamous Cell Cancer Risk. J Thoracic Oncol (2019) 14(4):712–25. doi: 10.1016/j.jtho.2018.12.023
20. Chan YX, Yeap BB. Dihydrotestosterone and Cancer Risk. Curr Opin Endocrinol (2018) 25(3):209–17. doi: 10.1097/Med.0000000000000411
21. Forde PM, Chaft JE, Smith KN, Anagnostou V, Cottrell TR, Hellmann MD, et al. Neoadjuvant PD-1 Blockade in Resectable Lung Cancer. N Engl J Med (2018) 378(21):1976–86. doi: 10.1056/NEJMoa1716078
22. Gao S, Li N, Gao S, Xue Q, Ying J, Wang S, et al. Neoadjuvant PD-1 Inhibitor (Sintilimab) in NSCLC. J Thorac Oncol (2020) 15(5):816–26. doi: 10.1016/j.jtho.2020.01.017
23. Shu CA, Gainor JF, Awad MM, Chiuzan C, Grigg CM, Pabani A, et al. Neoadjuvant Atezolizumab and Chemotherapy in Patients With Resectable non-Small-Cell Lung Cancer: An Open-Label, Multicentre, Single-Arm, Phase 2 Trial. Lancet Oncol (2020) 21(6):786–95. doi: 10.1016/S1470-2045(20)30140-6
24. Provencio M, Nadal E, Insa A, Garcia-Campelo MR, Casal-Rubio J, Domine M, et al. Neoadjuvant Chemotherapy and Nivolumab in Resectable non-Small-Cell Lung Cancer (NADIM): An Open-Label, Multicentre, Single-Arm, Phase 2 Trial. Lancet Oncol (2020) 21(11):1413–22. doi: 10.1016/S1470-2045(20)30453-8
25. Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Stephens RJ, et al. Lung Adjuvant Cisplatin Evaluation: A Pooled Analysis by the LACE Collaborative Group. J Clin Oncol (2008) 26(21):3552–9. doi: 10.1200/Jco.2007.13.9030
26. Zuo SG, Wei M, Zhang HL, Chen AX, Wu JH, Wei JW, et al. A Robust Six-Gene Prognostic Signature for Prediction of Both Disease-Free and Overall Survival in non-Small Cell Lung Cancer. J Transl Med (2019) 17(1):152. doi: 10.1186/s12967-019-1899-y
27. Liu Y, Wu LG, Ao HJ, Zhao M, Leng X, Liu MD, et al. Prognostic Implications of Autophagy-Associated Gene Signatures in non-Small Cell Lung Cancer. Aging-Us (2019) 11(23):11440–62. doi: 10.18632/aging.102544
28. Li Y, Sun N, Lu ZL, Sun SG, Huang JB, Chen ZL, et al. Prognostic Alternative mRNA Splicing Signature in non-Small Cell Lung Cancer. Cancer Lett (2017) 393:40–51. doi: 10.1016/j.canlet.2017.02.016
29. Shukla S, Evans JR, Malik R, Feng FY, Dhanasekaran SM, Cao XH, et al. Development of a RNA-Seq Based Prognostic Signature in Lung Adenocarcinoma. JNCI-J Natl Cancer I (2017) 109(1):djw200. doi: 10.1093/jnci/djw200
30. Eggermont AM, Chiarion-Sileni V, Grob JJ, Dummer R, Wolchok JD, Schmidt H, et al. Adjuvant Ipilimumab Versus Placebo After Complete Resection of High-Risk Stage III Melanoma (EORTC 18071): A Randomised, Double-Blind, Phase 3 Trial. Lancet Oncol (2015) 16(5):522–30. doi: 10.1016/S1470-2045(15)70122-1
31. Weber J, Mandala M, Del Vecchio M, Gogas HJ, Arance AM, Cowey CL, et al. Adjuvant Nivolumab Versus Ipilimumab in Resected Stage III or IV Melanoma. N Engl J Med (2017) 377(19):1824–35. doi: 10.1056/NEJMoa1709030
32. Liu J, Blake SJ, Yong MCR, Harjunpaa H, Ngiow SF, Takeda K, et al. Improved Efficacy of Neoadjuvant Compared to Adjuvant Immunotherapy to Eradicate Metastatic Disease. Cancer Discov (2016) 6(12):1382–99. doi: 10.1158/2159-8290.Cd-16-0577
33. Zhong WZ, Wang Q, Mao WM, Xu ST, Wu L, Shen Y, et al. Gefitinib Versus Vinorelbine Plus Cisplatin as Adjuvant Treatment for Stage II-IIIA (N1-N2) EGFR-mutant NSCLC (ADJUVANT/CTONG1104): A Randomised, Open-Label, Phase 3 Study. Lancet Oncol (2018) 19(1):139–48. doi: 10.1016/S1470-2045(17)30729-5
Keywords: lung adenocarcinoma, early stage, metabolism genes, prognostic signature, adjuvant therapy
Citation: Hu J, Yu H, Sun L, Yan Y, Zhang L, Jiang G and Zhang P (2021) Identification of an Individualized Metabolism Prognostic Signature and Related Therapy Regimens in Early Stage Lung Adenocarcinoma. Front. Oncol. 11:650853. doi: 10.3389/fonc.2021.650853
Received: 08 January 2021; Accepted: 30 March 2021;
Published: 28 April 2021.
Edited by:
Junji Uchino, Kyoto Prefectural University of Medicine, JapanReviewed by:
Jiajun Wang, Fudan University, ChinaAzhar Ali, National University of Singapore, Singapore
Copyright © 2021 Hu, Yu, Sun, Yan, Zhang, Jiang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Zhang, zhangpeng1121@tongji.edu.cn
†These authors have contributed equally to this work
 Junjie Hu
Junjie Hu Huansha Yu2†
Huansha Yu2† Lele Zhang
Lele Zhang Peng Zhang
Peng Zhang