- Department of Bioengineering, Graduate School of Engineering, The University of Tokyo, Tokyo, Japan
Cancer stem cell (CSCs) are deemed as one of the main reasons of tumor relapse due to their resistance to standard therapies. Numerous intracellular signaling pathways along with extracellular features are crucial in regulating CSCs properties, such as heterogeneity, plasticity and differentiation. Aberrant glycosylation of these cellular signaling pathways and markers of CSCs have been directly correlated with maintaining survival, self-renewal and extravasation properties. In this review, we highlight the importance of glycosylation in promoting stemness character of CSCs, and present strategies for targeting abnormal glycosylation to eliminate the resistant CSC population.
Introduction
The emergence of drug resistance and relapse of tumors have maintained the rate of cancer related deaths despite the advances in cancer treatment. Recent studies have associated the formation of metastasis, resistance to therapy, and eventual tumor relapse with the presence of a tumorigenic subpopulation of cancer cells showing stem-like features within the cellular heterogeneity of tumors (1). These cancer stem-like cells (CSCs) are deemed as one of the main reasons of tumor relapse (2). The presence of CSCs has been reported in different type of cancers, e.g. head and neck cancer (3), liver cancer (4), breast cancer (5), brain cancer (6) and melanoma (7). While the origin of CSCs is still not clear, the source may be a result of epithelial to mesenchymal transition (EMT), which allows differentiated and non-stem cells to develop characters like CSCs (8), or the alteration of non-malignant stem cells to CSCs due to oncogenic somatic mutation (9). In recent years several upregulated markers of CSCs have been identified, such as epithelial cell-adhesion molecule (EpCAM), CD44, CD133, CD24 and aldehyde dehydrogenase (ALDH) (1). These markers are being used alone or in combination in different tumors for isolating CSCs with drug resistance and self-renewal ability (10). Moreover, several cellular signaling pathways, such as Notch, hedgehog, Wnt/β-catenin, Akt, NF-KB, JAK-STAT and PPAR, which are also present in healthy stem cells, are being studied extensively due to their effect in self-renewal, metastasis and immune evasion (11). However, it is important to note that CSCs are highly heterogeneous, showing variability between tumors and even within the same tumor tissue (12). Thus, dealing with common targets shared by CSCs could allow developing far reaching therapies capable of treating a wide range of tumors.
Altered glycosylation ensuing modifications in proteins during or after translation (13) is a trademark of almost all type of cancers regardless of the origin and stage (14), and it is a common feature of CSC population markers and signaling pathways (15). The glycan chains on cancer cells regulate a range of pathological processes, including cell-cell interaction, cell adhesion, receptor activation and signal transduction (14). The main terminal sugar of the cell surface glycan chain is neuraminic acid (sialic acid; SA), which is being studied extensively in tumor biology (16). The overexpression of terminal glycan SA has also been linked to cancer malignancy and metastasis (16, 17). Moreover, hypersialylation in cancer cells promotes cell migration (18) and apoptosis resistance (19), which lead to tumor growth and poor prognosis (16). CSCs may be further distinguished from differentiated cancer cells by the expression of carbohydrate antigens (20, 21), and the N-glycosylation of glycoproteins of CSCs has been related to drug resistance mechanisms (22). Besides, abnormal glycosylation has been shown to regulate signaling pathways and markers involved in conservation of CSCs, such as EpCAM glycosylation on cell adhesion (23) and EMT in breast cancer, or N-glycosylation on MAPK and PI3/Akt pathway (24). As the carbohydrate antigens of glycan chains are primarily located on the plasma membrane of cells, they are regarded as outstanding biomarkers for detecting altered phases of cellular differentiation (21) and could provide opportunities for generating targeted therapies.
Herein, we review the altered glycosylation of cellular pathways and markers involved in CSCs maintenance and resistance mechanisms. The possibilities to use the glycosylation of both cancer cells and CSCs as targets for tumor treatment are also presented. Finally, we discuss the future perspectives in this bourgeoning field.
CSC and Altered Glycosylation
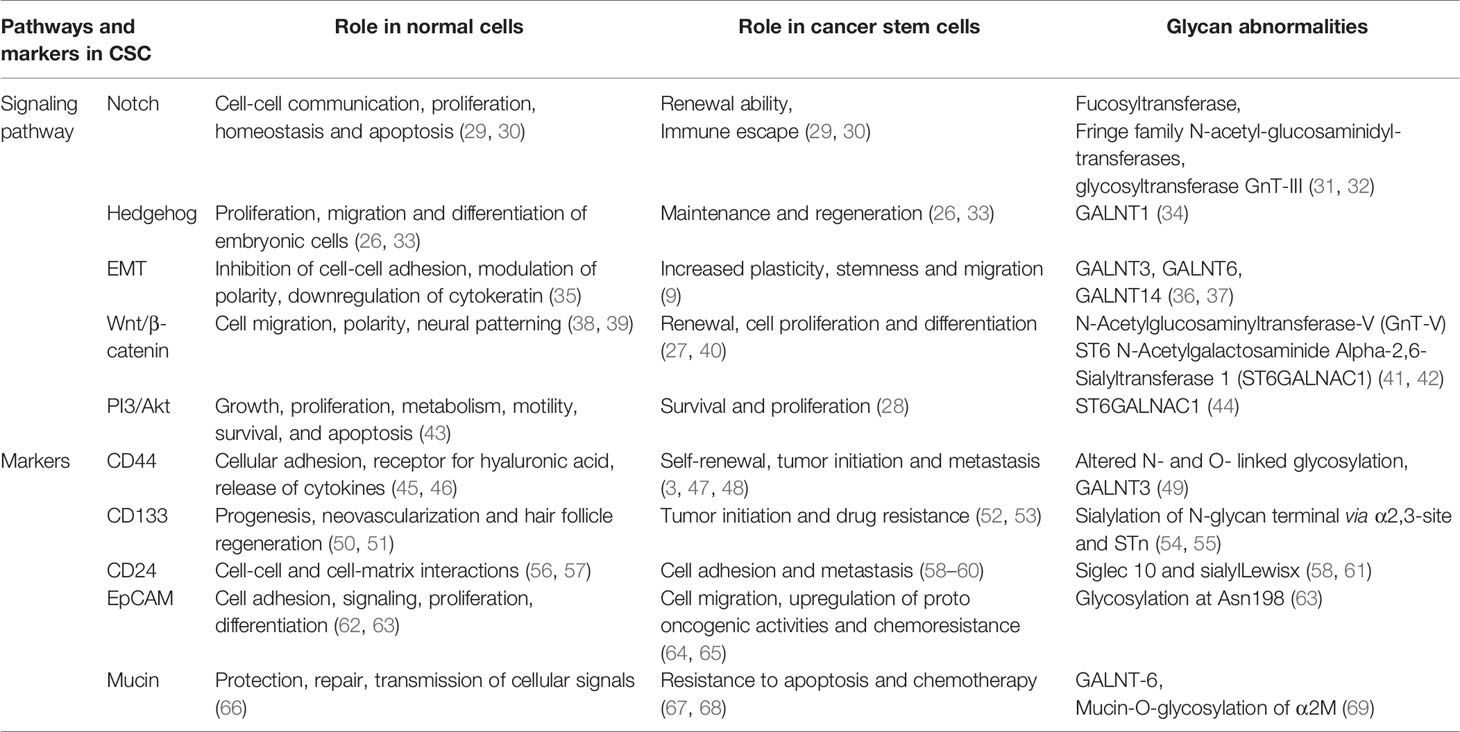
The importance of altered glycosylation of tumor in disease progression has been well documented (14). In the following sections, we focused on the aberrant glycosylation in CSC markers, such as CD44, CD133 and CD24, along with different cellular signaling and pathways, like Notch (25), hedgehog (26), Wnt/β-catenin (27) and Akt (28), in maintaining CSC properties (11, 15) (Figure 1). A summary of the role of signaling pathways and markers in normal cells and CSCs, along with their altered glycosylation reported so far, is presented in Table 1.
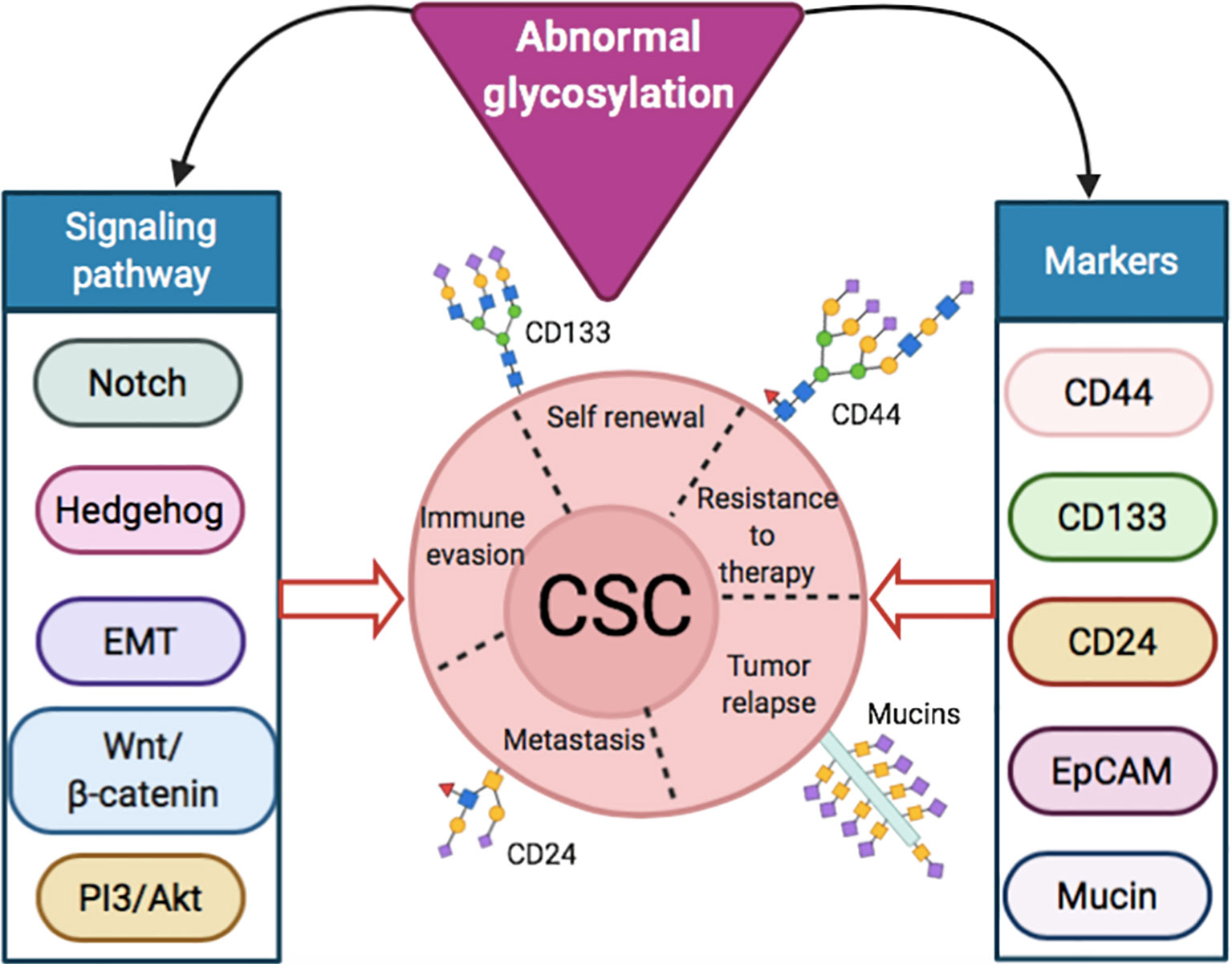
Figure 1 Abnormal glycosylation in signaling pathways and markers inducing stemness properties of CSCs.
Glycosylation of Signaling Pathways
The involvement of glycosylation in dysregulation of signaling pathways is gaining much attention due to its key roles in ligand binding, signaling of receptors, and transport control inside the cells. Besides, glycosylation affects receptor turnover on cell membrane receptors as glycans binds to galectins to form a lattice. In addition, the binding of glycosylated proteins with gangliosides can modulate the intracellular communication of receptors.
For Notch signaling, which supports the renewal ability of CSCs and plays significant roles in the immune escape of and invasion and extravasation of cancer cells, the response are mediated by 4 Notch transmembrane receptor and 5 ligands (3 Delta-like and 2 Jagged) (25). The affinity of these Notch receptors to ligands is regulated by the glycosylation of receptors (31). Specially, through insertion of fucose-N-acetylglucosamine by fucosyltransferase and Fringe family N-acetyl-glucosaminidyl-transferases (31). Various glycosyltransferases take part in the glycosylation of Notch in various tumor types (70). Regulation of Notch signaling is associated with tumor stemness and metastasis progression (25, 33).
Hedgehog (Hh) signaling is crucial for maintaining and regenerating CSCs (26, 33). The Hh signaling is a complex pathway regulated by different receptors, such as the transmembrane protein receptor PTCH and various extracellular Hh ligands and sonic Hh ligand (SHH) and the transmembrane protein SMO (71). GALNT1 a glycotransferase have been identified by researchers to play a pivotal role in SHH instigation in CSC of bladder cancer (34). GALNT1 regulated O-linked glycosylation foster activation of SHH signaling, which is vital for self-regeneration of CSCs (34).
Glycosylation also maintains tumor epithelial plasticity, including EMT driven by hexosamine biosynthetic pathway (HBP) and O-GlcNAcylation leading to chemoresistance of cancer cells (72). Increased expression of GALNT3 or GALNT6 correlates with the O-glycosylation-mediated EMT in epithelial cells of human prostate (36). Moreover, GALNT14 increase EMT genes, migration and invasion of breast cancer cells (37). EMT in breast cancer is also affected by the N-glycosylation of EpCAM via multiple cell signaling pathways (24). N-glycosylation promoted EMT also stabilizes and upregulates programmed death-ligand 1 (PD-L1), contributing to the immune escape of CSCs (73, 74).
N-acetylglucosaminyltransferase-V (GnT-V) and ST6 N-Acetylgalactosaminide alpha-2,6-Sialyltransferase 1 (ST6GALNAC1) facilitated glycosylation have been reported to control stemness of colon cancer cells through WNT signaling and Akt pathway respectively (41, 42, 44). GnT-V induces the N-glycosylation of receptors present on Wnt proteins, which alters normal Wnt/β-catenin signaling pathway (41). Such alteration causes distorted development of adenoma through regulation of CSC in colorectal cancer (41). The role of PI3K-Akt pathway in supporting proliferation and stemness characteristics of CSC in breast, brain and colorectal cancer cells have been studied thoroughly (75–77). ST6GALNAC1 may support conservation of CSCs through activation Akt pathway by facilitating recruitment of sialyl-Thomsen-nouveau (STn) antigen, which interacts with galectin-3 (44). STn is the consequence of premature sialylation of Tn antigen due to upregulation of ST6GALNAC1 and disturbance of elongation steps (78). ST6GALNAC1 facilitates recruitment of STn through addition of SA on GALNAC and backbone of protein. The STn antigen inside cells activates galectin-3 mediating phosphorylation of Akt, which decreases GSK3β and promotes protein transcription by β-catenin. Phosphorylated Akt also fosters S6 phosphorylation by mTORC1 leading to augmented protein synthesis, which triggers stemness through stimulated transcription (44). Expression of ST6 beta-galactoside alpha-2,6-sialyltransferase 1(ST6GAL1), which adds alpha2-6 lined sialic acid to glycosylated protein, is also upregulated in cancer malignancies specially in CSCs (79, 80). ST6GAL1 expression corelated with other CSC markers in colorectal carcinoma samples from patient and induced chemoresistance (81). In ovarian cancer, ST6GAL1 expression is regulated by SOX2, a well-known stem cell transcription factor (82).
Glycosylation of CSC Markers
Most CSC markers are glycoproteins, such as CD44, CD133 or CD24, expressing various glycan moieties on cell surface (15). These glycans are fundamental in many biological processes. For example, the terminal glycan SA is involved in regulating pluripotency of embryonic stem cells (20). Indeed, SA-cleaving promotes differentiation (20). Glycosylation of CSC markers regulates many functions of CSCs, including cell adhesion, extravasation, evasion of immune cells and apoptosis, self-regeneration and preservation of pluripotency (15).
CD44 is a transmembrane glycoprotein with the ability to activate EGFR and ErbB‐2 by binding to hyaluronic acid (HA) on extracellular matrix. Such activation has been linked to increased cell differentiation and relocation (45). While CD44 can be found on healthy cells, such as leukocytes, mesenchymal cells, endothelial and parenchymal liver cells (46), it is also a major player in different types of cancers, like head and neck, skin, lung, breast, prostate, pancreatic and liver cancer (47, 48). CD44 is hailed as a broad-ranging biomarker for CSCs. Moreover, isoforms of CD44 (CD44v), which are rarer than standard CD44, have also been linked with CSC subpopulations (3). Both the standard and variants of CD44 are reported to have altered N- and O- linked glycosylation (83). Such differences in glycosylation are associated with altered molecular weights (84, 85). Five probable N-linked glycosylation locations on CD44 can initiate cell binding through HA (83). Moreover, glycosylation of CD44 could control HA adhesion, as demonstrated in ovarian cancer (86), amplifying or inhibiting attachment of CD44 to HA. For example, improved binding has been reported by O-linked glycans (N-deglycosylated), N-linked N-acetylglucosamine and N-acetylgalactosamine recruitment in non-N-linked glycans on CD44. On the contrary, α2,3- linked SA on N-linked glycans have opposite effect and hinders such binding (87). The expression of SA in particularly high on CD44 positive CSCs (44, 88). In addition, the distorted glycosylation of CD44v and their expression have been linked with the aggressiveness of cancer in human patients (89).
Another extensively used CSC marker is CD133 or prominin-1 (28), which is associated with the Wnt signaling (90), and the PI3K/Akt pathway (91) in CSCs. CSCs identified with CD133+ markers in patients with ovarian cancer correspond to poor survival (92). CD133 is encoded by PROM1 gene composed of 8 N-linked glycosylation sites (93), and the glycosylation plays important roles in maintaining CSC properties (94, 95). In fact, glycosylation of CD133 has been suggested as a secondary indicator of CSCs (96–98). In particular, sialylation of CD133 N-glycan terminal via α2,3-site was found in the CSC of glioma (54), which was augmented in hypoxic conditions, correlating with the migration and survival of brain CSCs (99). Moreover, CD133 could be co-expressed with STn, as found in a subset of ovarian cancer cells (55).
CD24, which was originally identified as a B cell differentiation marker (100), has been indicated as a CSC marker in several tumors, such as ovarian (101), colorectal (102), bladder (103) and urothelial (104) cancers. CD24 has been directly correlated with disease progression and metastasis (59, 60, 105, 106). CD24 is a glycoprotein having both N- and O-linked type glycosylation and linked to glycosylphosphatidylinositol (61). CD24-expressing cells can evade the immune system by binding to Siglec-10 (a SA binding molecule) present on macrophages (61). Moreover, binding of CD24 to immune cells through a sialylation dependent way initiates a cascade facilitated by SHP-1/SHP-2 leading to inactivation of immune cells (107). Apart from Siglecs, CD24 also acts as a ligand for P-selectin, an adhesion receptor expressed on platelets and endothelial cells (108). Thus, binding of cancer cells to P-selectin via CD24 may be crucial for inducing tumor metastasis (108). Moreover, sialylLewisx (sialylLex), a SA-bearing glycan, controls CD24 facilitated rolling of cancer cells on P-selectin and extravasation during metastasis (58).
EpCAM is expressed on different types of epithelial cells, stem cells, cancer cells and CSCs (63). EpCAM is a cell surface glycoprotein with 3 N-glycosylation sites (109), with glycosylation at Asn198 being the most important for stability (63). EpCAM+, CD44+, CD24- CSCs were 10-fold more likely to induce tumors compared to EpCAM-, CD44+, CD24- CSCs in breast cancer (110). In hepatocellular carcinoma, EpCAM+ CD133+ cells demonstrated elevated colony formation ability, expression of stem cell related genes and chemoresistance compared to EpCAM- CD133+ cells (111). Moreover, inhibition of EpCAM in resistant head and neck cancer cells sensitize them to cisplatin (64). EpCAM in carcinoma tissues presented elevated levels of glycosylation compared to normal epithelia (112). Particularly, elevated glycosylation at Asn198 mediates EpCAM stability and surface retention in HEK293 cells (112). The deglycosylation of EpCAM repressed proliferation of breast cancer cells and fostered apoptosis (113). More recent research demonstrated the role of N-glycosylation of EpCAM in maintaining stemness property and EMT in hypoxic condition (114).
Mucins are heavily glycosylated extracellular proteins, which are mostly O-linked type, that have major impact on cell differentiation, adhesion and metastasis (67). For example, GALNT-6 elevated mucin-O-glycosylation of α2M, which activates downstream PI3/Akt signaling pathway, fosters metastasis of breast cancer (69). CD44+, CD24+ and CD133+ CSC populations of pancreatic cancer also express Mucin-1 (115) and Mucin 5AC also plays significant role in maintaining stemness properties of pancreatic CSC (116). Increased fraction of CD44+, CD24- CSC and Mucin-1 expression was also reported in breast cancer MCF-7 cells when exposed to tumor associated macrophages (117). Mucin-1 have also been identified as an important factor in effectiveness of colorectal CSC vaccine (118). In case of gastric cancer metastasis, expression of Mucin-1 was detected in peripheral blood and bone marrow signifying its importance (119). Abnormal expression of Mucin-4 has also been reported in many cancers (120), and correlated with metastasis (121) and chemoresistance (122). Downregulation of Mucin-4 sensitizes CSCs of pancreatic cells to gemcitabine (123). Mucin-4 also helps to maintain CSC population of ovarian cancer cells by stabilizing expression of Her2 (124). Mucin-16 (CA125) enhancement is present in CSCs of both pancreatic (125) and ovarian (126, 127) cancer. In ALDH+ CSCs of pancreatic cancer, MUC16 (carboxyl terminal fragment) facilitates augmentation of JAK2, which upregulates LMO2 and NANOG genes to provide stemness properties (128).
Glycosylation in the Resistance of CSCs to Conventional Treatments
Eliminating CSCs represents a major challenge, as CSCs present less reactive oxygen species (129, 130) and are resistant to radio- and chemotherapy (129, 130) by increased efflux pumps, DNA repair and scavenger agents (129, 130). CSCs show phenotypical heterogeneities both at inter- and intratumoral levels (131, 132), which are associated with genetic mutations and epigenetic changes, or differences in tumor microenvironment, e.g. cytokines and hypoxia (131, 132). The heterogeneity of CSCs represents a major challenge for targeted therapies. Moreover, CSCs could exist in specialized environments within tumors by forming niches that promote their survival (133, 134). CSCs can also adjust their niche and keep homeostatic processes, such as EMT and angiogenesis (133, 134). The CSC niche is dynamic, changing with tumor progression, and adapting to the treatments. Moreover, the niche has the ability to revert non-tumorigenic cancer cells into CSCs through EMT-related processes (8, 9, 135). This fluctuation between cancer cells and CSCs populations implies that targeting only CSCs may not be sufficient, as residual cancer cells may repopulate CSCs. Thus, it can be envisioned that for developing effective therapies, the elimination of both cancer cells and CSCs should be considered for achieving safe and robust long-term responses.
The functions of glycans present on CSC markers in inducing chemoresistance are not fully understood. Nonetheless, studies have indicated that glycosylation may play key roles in the resistance of CSCs. For example, O-GlcNAcylation of GNB2L1 facilitates chemoresistance of gastric cancer by regulating EMT (136). Furthermore, resistance to chemotherapeutic drug doxorubicin in the CD44+/CD24- CSC population of breast tumors was reversed by silencing expression of glucosylceramide synthase, which is involved in glycolipid biosynthesis (137). Also, in breast cancer, treatment with doxorubicin induces resistance through glycosylation of O-GlcNAc (138). Doxorubicin activates Akt pathway leading to upregulation of HBP and eventually O-GlcNAcylation, which promotes survival of cells through deterring apoptotic function of caspases and stimulating pro-survival factors like NF-KB and Akt (138). Elevated expression of Mucin-4 has also been reported in CD133+ CSCs in pancreatic cancer, which facilitated resistance to gemcitabine (123). Moreover, CD133+ CSC population isolated from patients malignant primary neoplasm was also more resistant to gemcitabine than CD133- cancer cells (52). Overexpression of Mucin-4 in two pancreatic cell lines Panc-1 and Mia-PaCa-2 was correlated with aggressiveness and resistance to gemcitabine compared to non Mucin-4 expressing Panc-1- and Mia-PaCa-pSectag C (123). In addition, the overexpression of Mucin-4/sialomucin complex (SMC) may protect cancer cells from the antibody Herceptin, as well as immune cells, by covering surface antigens, thus, hindering their availability (139–141). Therefore, therapies capable of dealing with the altered glycosylation of CSCs could allow developing strategies overcoming the resistance of CSC to conventional treatments.
Targeting Altered Glycosylation in Cancer Cells and CSCs
Treatment approaches against abnormal glycosylation are emerging as appealing options for effective tumor therapy (142). Inhibition of glycans can decrease CSCs ability for maintaining stemness, thereby, decreasing tumor proliferation. For example, by siRNA-mediated silencing of GALNT1, which control O-linked glycosylation activation of SHH signaling in CD44+ CSCs in bladder cancer to mediate stemness, the tumor growth was suppressed in a similar extent to cyclopamine in an orthotopic mouse model of bladder cancer (34). Moreover, inhibiting glycosylation cannot only make changes in CSCs, but also in the tumor microenvironment. For example, targeting the SA-siglec interaction can convert the immunosuppressive tumor microenvironment into an immunoactive environment, as overexpression of SA has been deemed as a major mechanism for cancer cells to evade detection by immune cells (143). The sialoglycan-siglec glycol-immune checkpoint can be targeted in CSCs. T cells inside tumors, specially CD8+ T cells, present elevated amount of Siglecs, which engage with heavily sialylated CSCs (143, 144). Besides, cancer cells expressing CD24 can escape detection by the immune system by interacting with the inhibitory receptor SA-binding Ig-like lectin 10 (Siglec-10). Blocking the CD24-Siglec-10 binding by monoclonal antibodies improves mice survival outcome by increasing phagocytosis of CD24+ cells (61).
Targeting glycosylated CSC markers with specific antibodies is another approach to aim CSC precisely (145). Among them, CD44 targeting appear as a promising approach for enhancing antitumor efficacy. For example, KMP-1 antibody targeting CD44 can inhibit cell division, relocation and adhesion, allowing tumor growth suppression in a mouse model of bladder cancer (146). The anti-CD44 antibody RG7356 have shown also the possibility to activate the immune system by initiating phagocytosis through macrophage recruitment in breast cancer model (147). Antibodies, such as L2A5, can also be used for targeting tumor specific STn and short glycans expressing terminal α2–6 SA (78), and promote tumor inhibition (148, 149). As STn is co-expressed with CD133, anti-STn antibody drug conjugates could suppress tumor growth, effectively diminishing CSCs (55). Other antibodies, such as the anti-sialyl-di-Lewisa antibody FG129, are being tested for targeting tumor-associated glycans toward the development of tumor-selective treatments and diagnosis modalities (150).
In most cases, the abnormal glycosylation in tumor leads to overexpression of SA at the terminal carbohydrate of the glycan chains, providing a useful target (14). Thus, efforts are being made to develop therapeutic and diagnostic approaches directed towards aberrant sialylation (151). These tactics includes blocking SA by using glycomimetics (152, 153) or targeting overexpressed SA in tumor to deliver therapeutic agents (154–157). Glycomimetic drugs, such as Uproleselan (GMI 1271) and Rivipansel (GMI 1070), which are both selectin (SA binding molecule) inhibitors, are being tested in clinical trials for treating acute myeloid leukemia (NCT03616470) (158, 159). Another molecule GMI 1359 which inhibits both E-selectin and CXCR4 have recently started Phase 1b clinical trial for metastatic breast cancer (NCT04197999) (160–162). Intratumoral injection of glycomimetic agents has shown enhanced antitumor responses, which are mediated by CD8+ T cell attack to cancer cells (152). However, the systemic application of such glycomimetic strategies blocking SA is difficult due to increased side effects.
SA is also a promising target for developing targeted therapies. Lectins can target receptors containing sialic acid. For example, a Phase I clinical trial with Maackia amurensis seed lectin (MASL), which can target podoplanin, is set to start for the treatment of squamous cell carcinoma of head and neck (NCT04188665) (163, 164). Podoplanin is a mucin like sialoglycoprotein, co-expressed with CSC marker CD44 and CD44v (165). However, lectins may also bind to healthy cells presenting SA. Moreover, treatment with the SA-cleaving enzyme, neuraminidase, for removing the SA on cancer cells has been explored as a therapeutic approach (166). By conjugating neuraminidase with the HER2-binding antibody trastuzumab (167, 168), it was possible to selectively remove SA from the surface of cancer cells and avoid damaging normal cells. However, systemic injection of such systems could pose major toxicity issues due to the vital role of SA on normal cells and tissues. Thus, it is essential to develop smart ligand-based systems that can only be activated at the tumor microenvironment for specifically targeting SA overexpressed on cancer cells. We have recently reported a novel phenylboronic acid derivative, i.e. 5-boronopicolinic acid (5-BPA), with a unique pH-dependent binding profile to SA (169). The binding of 5-BPA to SA and glucose follows opposite trend with reduced pH (169). In other words, 5-BPA molecule favors binding with glucose at pH 7.4, but strongly binds to SA at the acidic tumor microenvironment (pH 7.2-6.5) (170). Thus, 5-BPA can be used to develop pH triggered smart ligand systems, which will only activate in the low pH microenvironment of tumors, avoiding unspecific binding to normal cells. We have recently demonstrated the potential of 5-BPA as a ligand for targeting highly sialylated CD44+ CSCs (Figure 2A). Installing 5-BPA molecules on the surface of polymeric micelles loading (1,2-diaminocyclohexane)platinum(II), which is the parent complex of the anticancer drug oxaliplatin, allow reducing CSCs in vitro (Figure 2B) and also in an orthotopic model of head and neck cancer (Figure 2D), improving overall survival (Figure 2C) (88). Importantly, as SA is broadly available at the end of glycan chains of different tumors, and tumor acidosis is a hallmark of cancer, 5-BPA could be applied for developing targeted treatments against CSCs in a wide range of tumors.
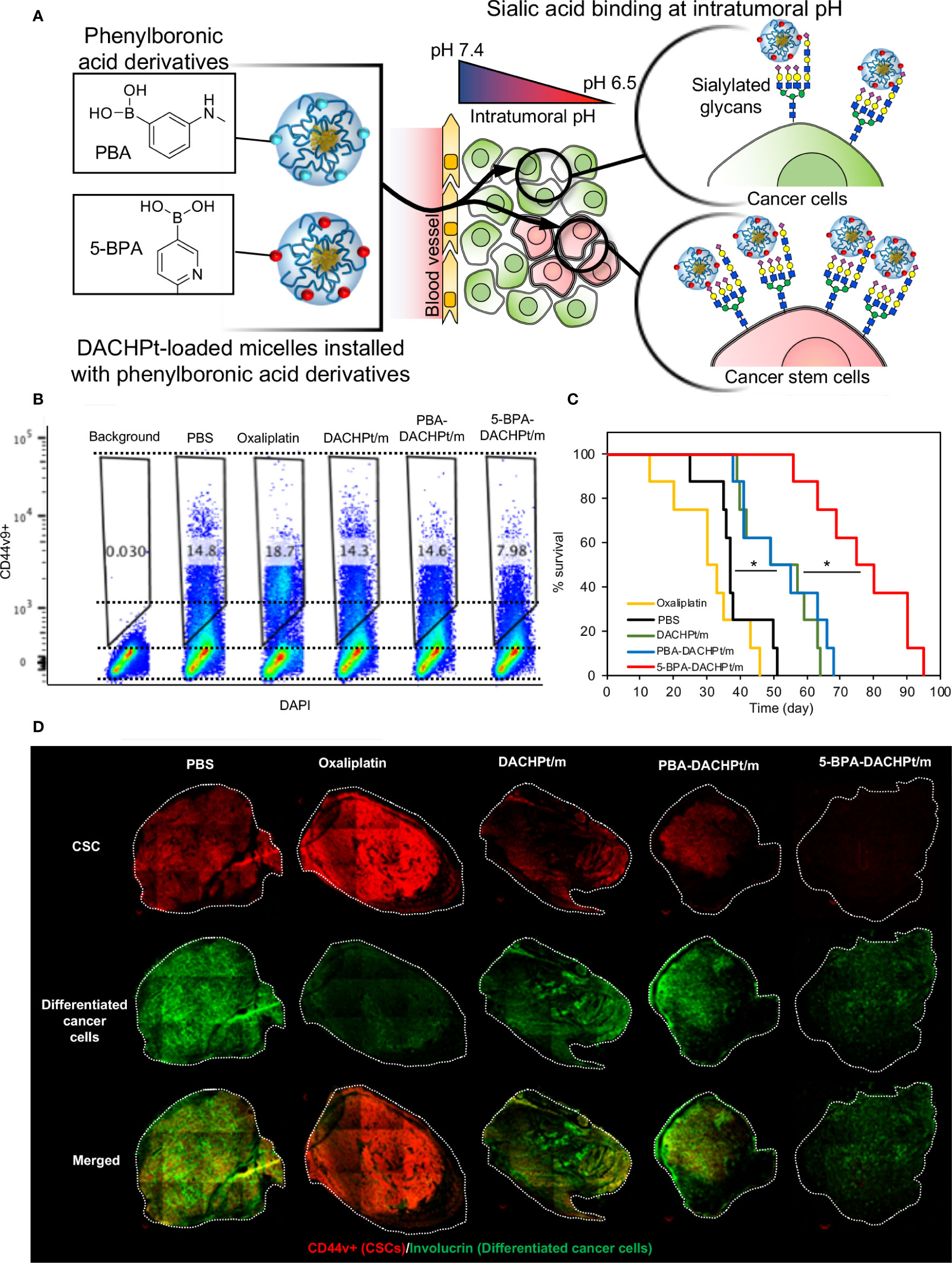
Figure 2 Targeting sialylated epitopes on cancer cells by using pH-activated 5-boronopicolinic acid (5-BPA) as a ligand. (A) Design strategy and targeting of overexpressed SA in CSC using pH-sensitive 5-BPA installed polymeric micelles. (B) Reduction of CD44+ CSCs in head and neck cancer cells (HSC-2) in vitro. (C) Improved survival of animal treated with 5-BPA installed polymeric micelles (D) Significant reduction in CD44+ CSC population (red) after treatment with 5-BPA installed polymeric micelles. Adapted with permission from reference (88). Copyright 2020 American Chemical Society.
Conclusion and Future Perspectives
Here, we briefly presented the effects of glycosylation on CSCs and the possibility to develop therapeutic approaches against such aberrant glycosylation. While glycosylation has been identified to be essential in the signaling pathways and markers of CSC regulating self-renewal, stemness and extravasation, more studies are necessary to unveil differences in glycome and glycosylation of normal cells, stem cells, CSCs and non-CSCs. Such information will allow researchers to develop biomarkers for detecting cancer progression, and precisely target cancer cells and resistant CSCs. Abnormal glycosylation of CSCs plays key role and contributes to their resistance to chemotherapy and ability to metastasize via several pathways. While inhibition or manipulation of the glycosylation in CSCs has been shown to be therapeutic, further exploration into the glycosylation associated processes is required to develop effective strategies targeting specific altered markers or signaling pathways without affecting healthy cells. Some approaches, like selectively cleaving surface glycan of tumors, or drugs with affinity for tumor associated glycans, have already demonstrated differential toxicity to tumor cells compared to normal cells, suggesting therapeutic potential through therapeutic window optimization. Moreover, even though CSC markers can present intratumoral and intertumoral heterogeneities, glycosylation could provide relevant targets that are preserved throughout tumors, such as SA, thereby, facilitating the development effective and wide-ranging treatment strategies.
Author Contributions
TK and HC wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Grants-in-Aid for Challenging Research (Exploratory) (18K19901; HC) and Grants-in-Aid for Scientific Research B (20H04524 and JP16H03179; HC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Zhou H-M, Zhang J-G, Zhang X, Li Q. Targeting cancer stem cells for reversing therapy resistance: mechanism, signaling, and prospective agents. Signal Transduct Target Ther (2021) 6(1). doi: 10.1038/s41392-020-00430-1
2. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med (2017) 23(10):1124–34. doi: 10.1038/nm.4409
3. Yoshikawa M, Tsuchihashi K, Ishimoto T, Yae T, Motohara T, Sugihara E, et al. XCT inhibition depletes CD44v-expressing tumor cells that are resistant to EGFR-targeted therapy in head and neck squamous cell carcinoma. Cancer Res (2013) 73(6):1855–66. doi: 10.1158/0008-5472.CAN-12-3609-T
4. Tomuleasa C, Soritau O, Rus-Ciuca D, Pop T, Todea D, Mosteanu O, et al. Isolation and characterization of hepatic cancer cells with stem-like properties from hepatocellular carcinoma. J Gastrointest Liver Dis (2010) 19(1):61–7.
5. Quinn HM, Vogel R, Popp O, Mertins P, Lan L, Messerschmidt C, et al. YAP and β-catenin cooperate to drive oncogenesis in basal breast cancer. Cancer Res (2021). doi: 10.1101/2020.06.05.115881
6. Ishii H, Mimura Y, Zahra MH, Katayama S, Hassan G, Afify SM, et al. Isolation and characterization of cancer stem cells derived from human glioblastoma. Am J Cancer Res (2021) 11(2):441–57.
7. Fattore L, Mancini R, Ciliberto G. Cancer Stem Cells and the Slow Cycling Phenotype: How to Cut the Gordian Knot Driving Resistance to Therapy in Melanoma. Cancers (Basel) (2020) 12(11):3368. doi: 10.3390/cancers12113368
8. Li N, Babaei-Jadidi R, Lorenzi F, Spencer-Dene B, Clarke P, Domingo E, et al. An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis (2019) 8(3):13. doi: 10.1038/s41389-019-0125-3
9. Tanabe S, Quader S, Cabral H, Ono R. Interplay of EMT and CSC in Cancer and the Potential Therapeutic Strategies. Front Pharmacol (2020) 11:904. doi: 10.3389/fphar.2020.00904
10. Miranda A, Hamilton PT, Zhang AW, Pattnaik S, Becht E, Mezheyeuski A, et al. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc Natl Acad Sci (2019) 116(18):9020–9. doi: 10.1073/pnas.1818210116
11. Clara JA, Monge C, Yang Y, Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells — a clinical update. Nature Reviews Clinical Oncology. Nat Res (2019) (4):204–32. doi: 10.1038/s41571-019-0293-2
12. Jung E, Osswald M, Ratliff M, Dogan H, Xie R, Weil S, et al. Tumor cell plasticity, heterogeneity, and resistance in crucial microenvironmental niches in glioma. Nat Commun (2021) 12(1):1014. doi: 10.1038/s41467-021-21117-3
13. Hedlund M, Ng E, Varki A, Varki NM. α2-6–Linked Sialic Acids on N-Glycans Modulate Carcinoma Differentiation In vivo. Cancer Res (2008) 68(2):388–94. doi: 10.1158/0008-5472.CAN-07-1340
14. Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer (2015) 15(9):540–55. doi: 10.1038/nrc3982
15. Barkeer S, Chugh S, Batra SK, Ponnusamy MP. Glycosylation of Cancer Stem Cells: Function in Stemness, Tumorigenesis, and Metastasis. Neoplasia (2018) 20(8):813–25. doi: 10.1016/j.neo.2018.06.001
16. Dobie C, Skropeta D. Insights into the role of sialylation in cancer progression and metastasis. Br J Cancer (2021) 124(1):76–90. doi: 10.1038/s41416-020-01126-7
17. Du J, Hong S, Dong L, Cheng B, Lin L, Zhao B, et al. Dynamic Sialylation in Transforming Growth Factor-β (TGF-β)-induced Epithelial to Mesenchymal Transition. J Biol Chem (2015) 290(19):12000–13. doi: 10.1074/jbc.M115.636969
18. Nagasundaram M, Horstkorte R, Gnanapragassam VS. Sialic Acid Metabolic Engineering of Breast Cancer Cells Interferes with Adhesion and Migration. Molecules (2020) 25(11):2632. doi: 10.3390/molecules25112632
19. Suzuki O, Abe M, Hashimoto Y. Caspase-dependent drug-induced apoptosis is regulated by cell surface sialylation in human B-cell lymphoma. Oncol Lett (2015) 10(2):687–90. doi: 10.3892/ol.2015.3320
20. Alisson-Silva F, de Carvalho Rodrigues D, Vairo L, Asensi KD, Vasconcelos-dos-Santos A, Mantuano NR, et al. Evidences for the involvement of cell surface glycans in stem cell pluripotency and differentiation. Glycobiology (2014) 24(5):458–68. doi: 10.1093/glycob/cwu012
21. Lanctot PM, Gage FH, Varki AP. The glycans of stem cells. Curr Opin Chem Biol (2007) 11(4):373–80. doi: 10.1016/j.cbpa.2007.05.032
22. Xu F, Wang Y, Xiao K, Hu Y, Tian Z, Chen Y. Quantitative site- and structure-specific N-glycoproteomics characterization of differential N-glycosylation in MCF-7/ADR cancer stem cells. Clin Proteomics (2020) 17(1):3. doi: 10.1186/s12014-020-9268-7
23. Liu X, Gao J, Sun Y, Zhang D, Liu T, Yan Q, et al. Mutation of N-linked glycosylation in EpCAM affected cell adhesion in breast cancer cells. Biol Chem (2017) 398(10):1119–26. doi: 10.1515/hsz-2016-0232
24. Liu X, Yang L, Zhang D, Liu T, Yan Q, Yang X. Deglycosylation of epithelial cell adhesion molecule affects epithelial to mesenchymal transition in breast cancer cells. J Cell Physiol (2019) 234(4):4504–14. doi: 10.1002/jcp.27256
25. Xiao W, Gao Z, Duan Y, Yuan W, Ke Y. Notch signaling plays a crucial role in cancer stem-like cells maintaining stemness and mediating chemotaxis in renal cell carcinoma. J Exp Clin Cancer Res (2017) 36(1):41. doi: 10.1186/s13046-017-0507-3
26. Zhu R, Gires O, Zhu L, Liu J, Li J, Yang H, et al. TSPAN8 promotes cancer cell stemness via activation of sonic Hedgehog signaling. Nat Commun (2019) 10(1):2863. doi: 10.1038/s41467-019-10739-3
27. Jang G-B, Kim J-Y, Cho S-D, Park K-S, Jung J-Y, Lee H-Y, et al. Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci Rep (2015) 5(1):12465. doi: 10.1038/srep12465
28. Yoon C, Lu J, Yi BC, Chang KK, Simon MC, Ryeom S, et al. PI3K/Akt pathway and Nanog maintain cancer stem cells in sarcomas. Oncogenesis (2021) 10(1):12. doi: 10.1038/s41389-020-00300-z
29. Lee SH, Do SI, Lee HJ, Kang HJ, Koo BS, Lim YC. Notch1 signaling contributes to stemness in head and neck squamous cell carcinoma. Lab Investig (2016) 96(5):508–16. doi: 10.1038/labinvest.2015.163
30. Kumar S, Srivastav RK, Wilkes DW, Ross T, Kim S, Kowalski J, et al. Estrogen-dependent DLL1-mediated Notch signaling promotes luminal breast cancer. Oncogene (2019) 38(12):2092–107. doi: 10.1038/s41388-018-0562-z
31. Miele L, Espinoza I, Pochampally, Watabe, Xing F. Notch signaling: targeting cancer stem cells and epithelial-to-mesenchymal transition. Onco Targets Ther (2013) 6:1249. doi: 10.2147/OTT.S36162
32. Allam H, Johnson BP, Zhang M, Lu Z, Cannon MJ, Abbott KL. The glycosyltransferase GnT-III activates Notch signaling and drives stem cell expansion to promote the growth and invasion of ovarian cancer. J Biol Chem (2017) 292(39):16351–9. doi: 10.1074/jbc.M117.783936
33. Chang WH, Lai AG. Aberrations in Notch-Hedgehog signalling reveal cancer stem cells harbouring conserved oncogenic properties associated with hypoxia and immunoevasion. Br J Cancer (2019) 121(8):666–78. doi: 10.1038/s41416-019-0572-9
34. Li C, Du Y, Yang Z, He L, Wang Y, Hao L, et al. GALNT1-Mediated Glycosylation and Activation of Sonic Hedgehog Signaling Maintains the Self-Renewal and Tumor-Initiating Capacity of Bladder Cancer Stem Cells. Cancer Res (2016) 76(5):1273–83. doi: 10.1158/0008-5472.CAN-15-2309
35. Lambert AW, Weinberg RA. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat Rev Cancer (2021). doi: 10.1038/s41568-021-00332-6
36. Freire-de-Lima L, Gelfenbeyn K, Ding Y, Mandel U, Clausen H, Handa K, et al. Involvement of O-glycosylation defining oncofetal fibronectin in epithelial-mesenchymal transition process. Proc Natl Acad Sci (2011) 108(43):17690–5. doi: 10.1073/pnas.1115191108
37. Huanna T, Tao Z, Xiangfei W, Longfei A, Yuanyuan X, Jianhua W, et al. GALNT14 mediates tumor invasion and migration in breast cancer cell MCF-7. Mol Carcinog (2015) 54(10):1159–71. doi: 10.1002/mc.22186
38. Yamashita S, Miyagi C, Carmany-Rampey A, Shimizu T, Fujii R, Schier AF, et al. Stat3 Controls Cell Movements during Zebrafish Gastrulation. Dev Cell (2002) 2(3):363–75. doi: 10.1016/S1534-5807(02)00126-0
39. Boitard M, Bocchi R, Egervari K, Petrenko V, Viale B, Gremaud S, et al. Wnt Signaling Regulates Multipolar-to-Bipolar Transition of Migrating Neurons in the Cerebral Cortex. Cell Rep (2015) 10(8):1349–61. doi: 10.1016/j.celrep.2015.01.061
40. Chen K, Chen L, Li L, Qu S, Yu B, Sun Y, et al. A positive feedback loop between Wnt/β-catenin signaling and hTERT regulates the cancer stem cell-like traits in radioresistant nasopharyngeal carcinoma cells. J Cell Biochem (2020) 121(11):jcb.29681. doi: 10.1002/jcb.29681
41. Guo H, Nagy T, Pierce M. Post-translational Glycoprotein Modifications Regulate Colon Cancer Stem Cells and Colon Adenoma Progression in Apc min/+ Mice through Altered Wnt Receptor Signaling. J Biol Chem (2014) 289(45):31534–49. doi: 10.1074/jbc.M114.602680
42. Regan JL, Schumacher D, Staudte S, Steffen A, Haybaeck J, Keilholz U, et al. Non-Canonical Hedgehog Signaling Is a Positive Regulator of the WNT Pathway and Is Required for the Survival of Colon Cancer Stem Cells. Cell Rep (2017) 21(10):363–75. doi: 10.1016/j.celrep.2017.11.025
43. Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: Its functions and alterations in human cancer. Apoptosis (2004) 9(6):667–76. doi: 10.1023/B:APPT.0000045801.15585.dd
44. Ogawa T, Hirohashi Y, Murai A, Nishidate T, Okita K, Wang L, et al. ST6GALNAC1 plays important roles in enhancing cancer stem phenotypes of colorectal cancer via the Akt pathway. Oncotarget (2017) 8(68):112550–64. doi: 10.18632/oncotarget.22545
45. Misra S, Toole BP, Ghatak S. Hyaluronan Constitutively Regulates Activation of Multiple Receptor Tyrosine Kinases in Epithelial and Carcinoma Cells. J Biol Chem (2006) 281(46):34936–41. doi: 10.1074/jbc.C600138200
46. Vermeulen L, De Sousa E Melo F, van der Heijden M, Cameron K, de Jong JH, Borovski T, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol (2010) 12(5):468–76. doi: 10.1038/ncb2048
47. Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of Pancreatic Cancer Stem Cells. Cancer Res (2007) 67(3):1030–7. doi: 10.1158/0008-5472.CAN-06-2030
48. Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci (2007) 104(3):973–8. doi: 10.1073/pnas.0610117104
49. Barkeer S, Chugh S, Karmakar S, Kaushik G, Rauth S, Rachagani S, et al. Novel role of O-glycosyltransferases GALNT3 and B3GNT3 in the self-renewal of pancreatic cancer stem cells. BMC Cancer (2018) 18(1):1157. doi: 10.1186/s12885-018-5074-2
50. Herrmann M, Binder A, Menzel U, Zeiter S, Alini M, Verrier S. CD34/CD133 enriched bone marrow progenitor cells promote neovascularization of tissue engineered constructs in vivo. Stem Cell Res (2014) 13(3):465–77. doi: 10.1016/j.scr.2014.10.005
51. Zhou L, Xu M, Yang Y, Yang K, Wickett RR, Andl T, et al. Activation of β-Catenin Signaling in CD133-Positive Dermal Papilla Cells Drives Postnatal Hair Growth. PloS One (2016) 11(7):e0160425. doi: 10.1371/journal.pone.0160425
52. Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell Stem Cell (2007) 1(3):313–23. doi: 10.1016/j.stem.2007.06.002
53. Wang H, Gong P, Li J, Fu Y, Zhou Z, Liu L. Role of CD133 in human embryonic stem cell proliferation and teratoma formation. Stem Cell Res Ther (2020) 11(1):208. doi: 10.1186/s13287-020-01729-0
54. Zhou F, Cui C, Ge Y, Chen H, Li Q, Yang Z, et al. 2,3-Sialylation regulates the stability of stem cell marker CD133. J Biochem (2010) 148(3):273–80. doi: 10.1093/jb/mvq062
55. Starbuck K, Al-Alem L, Eavarone DA, Hernandez SF, Bellio C, Prendergast JM, et al. Treatment of ovarian cancer by targeting the tumor stem cell-associated carbohydrate antigen, Sialyl-Thomsen-nouveau. Oncotarget (2018) 9(33):23289–305. doi: 10.18632/oncotarget.25289
56. Sammar M, Gulbins E, Hilbert K, Lang F, Altevogt P. Mouse CD24 as a Signaling Molecule for Integrin-Mediated Cell Binding: Functional and Physical Association with src-Kinases. Biochem Biophys Res Commun (1997) 234(2). doi: 10.1006/bbrc.1997.6639
57. Salamone MC, Rosselot C, Salamone GV, Barboza M, Kado M, Fainboim L. Antibodies recognizing CD24 LAP epitope on human T cells enhance CD28 and IL-2 T cell proliferation. J Leukoc Biol (2001) 69(2):215–23 doi: 10.1189/jlb.69.2.215.
58. Friederichs J, Zeller Y, Hafezi-Moghadam A, Gröne H-J, Ley K, Altevogt P. The CD24/P-selectin Binding Pathway Initiates Lung Arrest of Human A125 Adenocarcinoma Cells. Cancer Res (2000) 60(23):6714 LP – 6722.
59. Zheng J, Li Y, Yang J, Liu Q, Shi M, Zhang R, et al. NDRG2 inhibits hepatocellular carcinoma adhesion, migration and invasion by regulating CD24 expression. BMC Cancer (2011) 11(1):251. doi: 10.1186/1471-2407-11-251
60. Gilad N, Zukerman H, Pick M, Gatt ME. The role of CD24 in multiple myeloma tumorigenicity and effects of the microenvironment on its expression. Oncotarget (2019) 10(52):5480–91. doi: 10.18632/oncotarget.27190
61. Barkal AA, Brewer RE, Markovic M, Kowarsky M, Barkal SA, Zaro BW, et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature (2019) 572(7769):392–6. doi: 10.1038/s41586-019-1456-0
62. Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol (2009) 11(2):162–171. doi: 10.1038/ncb1824
63. Pavšič M, Gunčar G, Djinović-Carugo K, Lenarčič B. Crystal structure and its bearing towards an understanding of key biological functions of EpCAM. Nat Commun (2014) 5(1):4764. doi: 10.1038/ncomms5764
64. Noman ASM, Parag RR, Rashid MI, Islam S, Rahman MZ, Chowdhury AA, et al. Chemotherapeutic resistance of head and neck squamous cell carcinoma is mediated by EpCAM induction driven by IL-6/p62 associated Nrf2-antioxidant pathway activation. Cell Death Dis (2020) 11(8):663. doi: 10.1038/s41419-020-02907-x
65. Sun X, Martin RCG, Zheng Q, Farmer R, Pandit H, Li X, et al. Drug-induced expression of EpCAM contributes to therapy resistance in esophageal adenocarcinoma. Cell Oncol (2018) 41(6):651–62. doi: 10.1007/s13402-018-0399-z
66. Radicioni G, Cao R, Carpenter J, Ford AA, Wang TT, Li Y, et al. The innate immune properties of airway mucosal surfaces are regulated by dynamic interactions between mucins and interacting proteins: the mucin interactome. Mucosal Immunol (2016) 9(6):1442–54. doi: 10.1038/mi.2016.27
67. Remmers N, Anderson JM, Linde EM, DiMaio DJ, Lazenby AJ, Wandall HH, et al. Aberrant Expression of Mucin Core Proteins and O-Linked Glycans Associated with Progression of Pancreatic Cancer. Clin Cancer Res (2013) 19(8):1981–93. doi: 10.1158/1078-0432.CCR-12-2662
68. Ham SY, Kwon T, Bak Y, Yu J-H, Hong J, Lee SK, et al. Mucin 1-mediated chemo-resistance in lung cancer cells. Oncogenesis (2016) 5(1):e185. doi: 10.1038/oncsis.2015.47
69. Liu C, Li Z, Xu L, Shi Y, Zhang X, Shi S, et al. GALNT6 promotes breast cancer metastasis by increasing mucin-type O-glycosylation of α2M. Aging (Albany NY) (2020) 12(12):11794–811. doi: 10.18632/aging.103349
70. Takeuchi H, Haltiwanger RS. Significance of glycosylation in Notch signaling. Biochem Biophys Res Commun (2014) 453(2):235–42. doi: 10.1016/j.bbrc.2014.05.115
71. Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther (2020) 5(1):8. doi: 10.1038/s41392-020-0110-5
72. Lucena MC, Carvalho-Cruz P, Donadio JL, Oliveira IA, de Queiroz RM, Marinho-Carvalho MM, et al. Epithelial Mesenchymal Transition Induces Aberrant Glycosylation through Hexosamine Biosynthetic Pathway Activation. J Biol Chem (2016) 291(25):e185. doi: 10.1074/jbc.M116.729236
73. Li C-W, Lim S-O, Xia W, Lee H-H, Chan L-C, Kuo C-W, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun (2016) 7(1):12632. doi: 10.1038/ncomms12632
74. Hsu J-M, Xia W, Hsu Y-H, Chan L-C, Yu W-H, Cha J-H, et al. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat Commun (2018) 9(1):1908. doi: 10.1038/s41467-018-04313-6
75. Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, et al. CD44v6 Is a Marker of Constitutive and Reprogrammed Cancer Stem Cells Driving Colon Cancer Metastasis. Cell Stem Cell (2014) 14(3):342–56. doi: 10.1016/j.stem.2014.01.009
76. Gargini R, Cerliani JP, Escoll M, Antón IM, Wandosell F. Cancer Stem Cell-Like Phenotype and Survival Are Coordinately Regulated by Akt/FoxO/Bim Pathway. Stem Cells (2015) 33(3):646–60. doi: 10.1002/stem.1904
77. Gargini R, Escoll M, García E, García-Escudero R, Wandosell F, Antón IM. WIP Drives Tumor Progression through YAP/TAZ-Dependent Autonomous Cell Growth. Cell Rep (2016) 17(8):1962–77. doi: 10.1016/j.celrep.2016.10.064
78. Loureiro LR, Sousa DP, Ferreira D, Chai W, Lima L, Pereira C, et al. Novel monoclonal antibody L2A5 specifically targeting sialyl-Tn and short glycans terminated by alpha-2–6 sialic acids. Sci Rep (2018) 8(1):12196. doi: 10.1038/s41598-018-30421-w
79. Schultz MJ, Holdbrooks AT, Chakraborty A, Grizzle WE, Landen CN, Buchsbaum DJ, et al. The Tumor-Associated Glycosyltransferase ST6Gal-I Regulates Stem Cell Transcription Factors and Confers a Cancer Stem Cell Phenotype. Cancer Res (2016) 76(13):3978–88. doi: 10.1158/0008-5472.CAN-15-2834
80. Dorsett KA, Marciel MP, Hwang J, Ankenbauer KE, Bhalerao N, Bellis SL. Regulation of ST6GAL1 sialyltransferase expression in cancer cells. Glycobiology (2020). doi: 10.1093/glycob/cwaa110
81. Cui H, Yang S, Jiang Y, Li C, Zhao Y, Shi Y, et al. The glycosyltransferase ST6Gal-I is enriched in cancer stem-like cells in colorectal carcinoma and contributes to their chemo-resistance. Clin Transl Oncol (2018) 20(9):1175–84. doi: 10.1007/s12094-018-1840-5
82. Dorsett KA, Jones RB, Ankenbauer KE, Hjelmeland AB, Bellis SL. Sox2 promotes expression of the ST6Gal-I glycosyltransferase in ovarian cancer cells. J Ovarian Res (2019) 12(1). doi: 10.1186/s13048-019-0574-5
83. Bartolazzi A, Nocks A, Aruffo A, Spring F, Stamenkovic I. Glycosylation of CD44 is implicated in CD44-mediated cell adhesion to hyaluronan. J Cell Biol (1996) 132(6):1199–208. doi: 10.1083/jcb.132.6.1199
84. Camp RL, Kraus TA, Puré E. Variations in the cytoskeletal interaction and posttranslational modification of the CD44 homing receptor in macrophages. J Cell Biol (1991) 115(5):1283–92. doi: 10.1083/jcb.115.5.1283
86. Catterall JB, Jones LMH, Turner GA. Membrane protein glycosylation and CD44 content in the adhesion of human ovarian cancer cells to hyaluronan. Clin Exp Metastasis (1999) 17(7):583–91. doi: 10.1023/A:1006756518500
87. Skelton TP, Zeng C, Nocks A, Stamenkovic I. Glycosylation Provides Both Stimulatory and Inhibitory Effects on Cell Surface and Soluble CD44 Binding to Hyaluronan. J Cell Biol (1998) 140(2):431–46. doi: 10.1083/jcb.140.2.431
88. Khan T, Igarashi K, Tanabe A, Miyazawa T, Fukushima S, Miura Y, et al. Structural Control of Boronic Acid Ligands Enhances Intratumoral Targeting of Sialic Acid To Eradicate Cancer Stem-like Cells. ACS Appl Bio Mater (2020) 3(8):5030–9. doi: 10.1021/acsabm.0c00530
89. Hakomori S. Tumor Malignancy Defined by Aberrant Glycosylation and Sphingo(glyco)lipid Metabolism. Cancer Res (1996) 56(23):5309 LP – 5318.
90. Katoh Y, Katoh M. Comparative genomics on PROM1 gene encoding stem cell marker CD133. Int J Mol Med (2007) 19:967–70. doi: 10.3892/ijmm.19.6.967
91. Jamal SM El, Alamodi A, RU W, Grada Z, MA S, Hassan S-Y, et al. Melanoma stem cell maintenance and chemo-resistance are mediated by CD133 signal to PI3K-dependent pathways. Oncogene (2020) 39(32):5468–78. doi: 10.1038/s41388-020-1373-6
92. Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, et al. Aldehyde Dehydrogenase in Combination with CD133 Defines Angiogenic Ovarian Cancer Stem Cells That Portend Poor Patient Survival. Cancer Res (2011) 71(11):3991–4001. doi: 10.1158/0008-5472.CAN-10-3175
93. Jang J-W, Song Y, Kim S-H, Kim J, Seo HR. Potential mechanisms of CD133 in cancer stem cells. Life Sci (2017) 184:25–9. doi: 10.1016/j.lfs.2017.07.008
94. Mak AB, Blakely KM, Williams RA, Penttilä P-A, Shukalyuk AI, Osman KT, et al. CD133 Protein N -Glycosylation Processing Contributes to Cell Surface Recognition of the Primitive Cell Marker AC133 Epitope. J Biol Chem (2011) 286(47):41046–56. doi: 10.1074/jbc.M111.261545
95. Liu Y, Ren S, Xie L, Cui C, Xing Y, Liu C, et al. Mutation of N-linked glycosylation at Asn548 in CD133 decreases its ability to promote hepatoma cell growth. Oncotarget (2015) 6(24):20650–60. doi: 10.18632/oncotarget.4115
96. Li Y, Shi D, Yang F, Chen X, Xing Y, Liang Z, et al. Complex N-glycan promotes CD 133 mono-ubiquitination and secretion. FEBS Lett (2019) 593(7):719–31. doi: 10.1002/1873-3468.13358
97. Sakaue T, Koga H, Iwamoto H, Nakamura T, Ikezono Y, Abe M, et al. Glycosylation of ascites-derived exosomal CD133: a potential prognostic biomarker in patients with advanced pancreatic cancer. Med Mol Morphol (2019) 52(4):198–208. doi: 10.1007/s00795-019-00218-5
98. Gardelli C, Russo L, Cipolla L, Moro M, Andriani F, Rondinone O, et al. Differential glycosylation of collagen modulates lung cancer stem cell subsets through β1 integrin-mediated interactions. Cancer Sci (2021) 112(1):217–30. doi: 10.1111/cas.14700
99. Lehnus KS, Donovan LK, Huang X, Zhao N, Warr TJ, Pilkington GJ, et al. CD133 glycosylation is enhanced by hypoxia in cultured glioma stem cells. Int J Oncol (2013) 42(3):1011–7. doi: 10.3892/ijo.2013.1787
100. Pirruccello SJ, LeBien TW. The human B cell-associated antigen CD24 is a single chain sialoglycoprotein. J Immunol (1986) 136(10):3779 LP – 3784.
101. Gao M-Q, Choi Y-P, Kang S, Youn JH, Cho N-H. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene (2010) 29(18):2672–80. doi: 10.1038/onc.2010.35
102. Yeung TM, Gandhi SC, Wilding JL, Muschel R, Bodmer WF. Cancer stem cells from colorectal cancer-derived cell lines. Proc Natl Acad Sci U S A (2010) 107(8):3722–7. doi: 10.1073/pnas.0915135107
103. Ooki A, VandenBussche CJ, Kates M, Hahn NM, Matoso A, McConkey DJ, et al. CD24 regulates cancer stem cell (CSC)-like traits and a panel of CSC-related molecules serves as a non-invasive urinary biomarker for the detection of bladder cancer. Br J Cancer (2018) 119(8):961–70. doi: 10.1038/s41416-018-0291-7
104. Ooki A, Del Carmen Rodriguez Pena M, Marchionni L, Dinalankara W, Begum A, Hahn NM, et al. YAP1 and COX2 Coordinately Regulate Urothelial Cancer Stem-like Cells. Cancer Res (2018) 78(1):168–81. doi: 10.1158/0008-5472.CAN-17-0836
105. Kristiansen G, Winzer K-J, Mayordomo E, Bellach J, Schlüns K, Denkert C, et al. CD24 Expression Is a New Prognostic Marker in Breast Cancer. Clin Cancer Res (2003) 9(13):4906–4913.
106. Overdevest JB, Thomas S, Kristiansen G, Hansel DE, Smith SC, Theodorescu D. CD24 Offers a Therapeutic Target for Control of Bladder Cancer Metastasis Based on a Requirement for Lung Colonization. Cancer Res (2011) 71(11):3802–11. doi: 10.1158/0008-5472.CAN-11-0519
107. Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol (2007) 7(4):255–66. doi: 10.1038/nri2056
108. Aigner S, Ramos CL, Hafezi-Moghadam A, Lawrence MB, Friederichs J, Altevogt P, et al. CD24 mediates rolling of breast carcinoma cells on P-selectin. FASEB J (1998) 12(12):1241–51. doi: 10.1096/fasebj.12.12.1241
109. Pauli C, Münz M, Kieu C, Mack B, Breinl P, Wollenberg B, et al. Tumor-specific glycosylation of the carcinoma-associated epithelial cell adhesion molecule EpCAM in head and neck carcinomas. Cancer Lett (2003) 193(1):25–32. doi: 10.1016/S0304-3835(03)00003-X
110. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci (2003) 100(7):3983–8. doi: 10.1073/pnas.0530291100
111. Chen Y, Yu D, Zhang H, He H, Zhang C, Zhao W, et al. CD133 + EpCAM + Phenotype Possesses More Characteristics of Tumor Initiating Cells in Hepatocellular Carcinoma Huh7 Cells. Int J Biol Sci (2012) 8(7):992–1004. doi: 10.7150/ijbs.4454
112. Munz M. Glycosylation is crucial for stability of tumour and cancer stem cell antigen EpCAM. Front Biosci (2008) 13:5195. doi: 10.2741/3075
113. Zhang D, Liu X, Gao J, Sun Y, Liu T, Yan Q, et al. The role of epithelial cell adhesion molecule N-glycosylation on apoptosis in breast cancer cells. Tumor Biol (2017) 39(3):101042831769597. doi: 10.1177/1010428317695973
114. Zhang D, Yang L, Liu X, Gao J, Liu T, Yan Q, et al. Hypoxia modulates stem cell properties and induces EMT through N -glycosylation of EpCAM in breast cancer cells. J Cell Physiol (2020) 235(4):3626–33. doi: 10.1002/jcp.29252
115. Curry JM, Thompson KJ, Rao SG, Besmer DM, Murphy AM, Grdzelishvili VZ, et al. The use of a novel MUC1 antibody to identify cancer stem cells and circulating MUC1 in mice and patients with pancreatic cancer. J Surg Oncol (2013) 107(7):713–22. doi: 10.1002/jso.23316
116. Ganguly K, Krishn SR, Rachagani S, Jahan R, Shah A, Nallasamy P, et al. Secretory mucin 5AC promotes neoplastic progression by augmenting KLF4-mediated pancreatic cancer cell stemness. Cancer Res (2020) 81(1):91–102. doi: 10.1158/0008-5472.CAN-20-1293
117. Zhou N, Zhang Y, Zhang X, Lei Z, Hu R, Li H, et al. Exposure of Tumor-Associated Macrophages to Apoptotic MCF-7 Cells Promotes Breast Cancer Growth and Metastasis. Int J Mol Sci (2015) 16(12):11966–82. doi: 10.3390/ijms160611966
118. Guo M, Luo B, Pan M, Li M, Zhao F, Dou J. MUC1 plays an essential role in tumor immunity of colorectal cancer stem cell vaccine. Int Immunopharmacol (2020) 85:106631. doi: 10.1016/j.intimp.2020.106631
119. Bali P, Lozano−Pope I, Pachow C, Obonyo M. Early detection of tumor cells in bone marrow and peripheral blood in a fast−progressing gastric cancer model. Int J Oncol (2021) 58(3):388–96. doi: 10.3892/ijo.2021.5171
120. Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer (2004) 4(1):45–60. doi: 10.1038/nrc1251
121. Momi N, Ponnusamy MP, Kaur S, Rachagani S, Kunigal SS, Chellappan S, et al. Nicotine/cigarette smoke promotes metastasis of pancreatic cancer through α7nAChR-mediated MUC4 upregulation. Oncogene (2013) 32(11):1384–95. doi: 10.1038/onc.2012.163
122. Skrypek N, Duchêne B, Hebbar M, Leteurtre E, van Seuningen I, Jonckheere N. The MUC4 mucin mediates gemcitabine resistance of human pancreatic cancer cells via the Concentrative Nucleoside Transporter family. Oncogene (2013) Mar 1432(13):1714–23. doi: 10.1038/onc.2012.179
123. Mimeault M, Johansson SL, Senapati S, Momi N, Chakraborty S, Batra SK. MUC4 down-regulation reverses chemoresistance of pancreatic cancer stem/progenitor cells and their progenies. Cancer Lett (2010) 295(1):69–84. doi: 10.1016/j.canlet.2010.02.015
124. Ponnusamy MP, Seshacharyulu P, Vaz A, Dey P, Batra SK. MUC4 stabilizes HER2 expression and maintains the cancer stem cell population in ovarian cancer cells. J Ovarian Res (2011) 4(1):7. doi: 10.1186/1757-2215-4-7
125. Das S, Rachagani S, Torres-Gonzalez MP, Lakshmanan I, Majhi PD, Smith LM, et al. Carboxyl-terminal domain of MUC16 imparts tumorigenic and metastatic functions through nuclear translocation of JAK2 to pancreatic cancer cells. Oncotarget (2015) 6(8):5772–87. doi: 10.18632/oncotarget.3308
126. Aithal A, Junker WM, Kshirsagar P, Das S, Kaur S, Orzechowski C, et al. Development and characterization of carboxy-terminus specific monoclonal antibodies for understanding MUC16 cleavage in human ovarian cancer. PloS One (2018) 13(4):e0193907. doi: 10.1371/journal.pone.0193907
127. Zhang H, Yang Y, Wang Y, Gao X, Wang W, Liu H, et al. Relationship of tumor marker CA125 and ovarian tumor stem cells: preliminary identification. J Ovarian Res (2015) 8(1):19. doi: 10.1186/s13048-015-0132-8
128. Das S, Batra SK. Understanding the Unique Attributes of MUC16 (CA125): Potential Implications in Targeted Therapy. Cancer Res (2015) 75(22):4669–74. doi: 10.1158/0008-5472.CAN-15-1050
129. Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer (2008) 8(10):755–68. doi: 10.1038/nrc2499
130. Magee JA, Piskounova E, Morrison SJ. Cancer Stem Cells: Impact, Heterogeneity, and Uncertainty. Cancer Cell (2012) 21(3):283–96. doi: 10.1016/j.ccr.2012.03.003
131. Medema JP. Cancer stem cells: The challenges ahead. Nat Cell Biol (2013) 15(4):338–44. doi: 10.1038/ncb2717
132. Diehn M, Majeti R. Metastatic Cancer Stem Cells: An Opportunity for Improving Cancer Treatment? Cell Stem Cell (2010) 6(6):502–3. doi: 10.1016/j.stem.2010.05.001
133. Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer (2005) 5(4):275–84. doi: 10.1038/nrc1590
134. Dreesen O, Brivanlou AH. Signaling Pathways in Cancer and Embryonic Stem Cells. Stem Cell Rev (2007) 3(1):7–17. doi: 10.1007/s12015-007-0004-8
135. Dubrovska A, Kim S, Salamone RJ, Walker JR, Maira S-M, Garcia-Echeverria C, et al. The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations. Proc Natl Acad Sci (2009) 106(1):268–73. doi: 10.1073/pnas.0810956106
136. Cheng S, Mao Q, Dong Y, Ren J, Su L, Liu J, et al. GNB2L1 and its O-GlcNAcylation regulates metastasis via modulating epithelial-mesenchymal transition in the chemoresistance of gastric cancer. PloS One (2017) 12(8):e0182696. doi: 10.1371/journal.pone.0182696
137. Gupta V, Bhinge KN, Hosain SB, Xiong K, Gu X, Shi R, et al. Ceramide Glycosylation by Glucosylceramide Synthase Selectively Maintains the Properties of Breast Cancer Stem Cells. J Biol Chem (2012) 287(44):37195–205. doi: 10.1074/jbc.M112.396390
138. Liu Y, Cao Y, Pan X, Shi M, Wu Q, Huang T, et al. O-GlcNAc elevation through activation of the hexosamine biosynthetic pathway enhances cancer cell chemoresistance. Cell Death Dis (2018) 9(5):485. doi: 10.1038/s41419-018-0522-0
139. Komatsu M, Yee L, Carraway KL. Overexpression of Sialomucin Complex, a Rat Homologue of MUC4, Inhibits Tumor Killing by Lymphokine-activated Killer Cells. Cancer Res (1999) 59(9):2229 LP – 2236.
140. Price-Schiavi SA, Jepson S, Li P, Arango M, Rudland PS, Yee L, et al. Rat Muc4 (sialomucin complex) reduces binding of anti-ErbB2 antibodies to tumor cell surfaces, a potential mechanism for herceptin resistance. Int J Cancer (2002) 99(6):783–91. doi: 10.1002/ijc.10410
141. Komatsu M, Jepson S, Arango ME, Carothers Carraway CA, Carraway KL. Muc4/sialomucin complex, an intramembrane modulator of ErbB2/HER2/Neu, potentiates primary tumor growth and suppresses apoptosis in a xenotransplanted tumor. Oncogene (2001) 20(4):461–70. doi: 10.1038/sj.onc.1204106
142. Vajaria BN, Patel KR, Begum R, Patel PS. Sialylation: an Avenue to Target Cancer Cells. Pathol Oncol Res (2016) 22(3):443–7. doi: 10.1007/s12253-015-0033-6
143. Stanczak MA, Siddiqui SS, Trefny MP, Thommen DS, Boligan KF, Von Gunten S, et al. Self-associated molecular patterns mediate cancer immune evasion by engaging Siglecs on T cells. J Clin Invest (2018) 128(11):4912–23. doi: 10.1172/JCI120612
144. Haas Q, Boligan KF, Jandus C, Schneider C, Simillion C, Stanczak MA, et al. Siglec-9 Regulates an Effector Memory CD8 + T-cell Subset That Congregates in the Melanoma Tumor Microenvironment. Cancer Immunol Res (2019) 7(5):707–18. doi: 10.1158/2326-6066.CIR-18-0505
145. Deonarain MP, Kousparou CA, Epenetos AA. Antibodies targeting cancer stem cells: A new paradigm in immunotherapy? MAbs (2009) 1(1):12–25. doi: 10.4161/mabs.1.1.7347
146. Chen Y, Wang H, Zuo Y, Li N, Ding M, Li C. A novel monoclonal antibody KMP1 has potential antitumor activity of bladder cancer by blocking CD44 in vivo and in vitro. Cancer Med (2018) 7(5):2064–77. doi: 10.1002/cam4.1446
147. Maisel D, Birzele F, Voss E, Nopora A, Bader S, Friess T, et al. Targeting Tumor Cells with Anti-CD44 Antibody Triggers Macrophage-Mediated Immune Modulatory Effects in a Cancer Xenograft Model. PloS One (2016) 11(7):e0159716. doi: 10.1371/journal.pone.0159716
148. Prendergast JM, Galvao da Silva AP, Eavarone DA, Ghaderi D, Zhang M, Brady D, et al. Novel anti-Sialyl-Tn monoclonal antibodies and antibody-drug conjugates demonstrate tumor specificity and anti-tumor activity. MAbs (2017) 9(4):615–27. doi: 10.1080/19420862.2017.1290752
149. Eavarone DA, Al-Alem L, Lugovskoy A, Prendergast JM, Nazer RI, Stein JN, et al. Humanized anti-Sialyl-Tn antibodies for the treatment of ovarian carcinoma. PloS One (2018) 13(7):e0201314. doi: 10.1371/journal.pone.0201314
150. Tivadar ST, McIntosh RS, Chua JX, Moss R, Parsons T, Zaitoun AM, et al. Monoclonal Antibody Targeting Sialyl-di-Lewis a –Containing Internalizing and Noninternalizing Glycoproteins with Cancer Immunotherapy Development Potential. Mol Cancer Ther (2020) 19(3):790–801. doi: 10.1158/1535-7163.MCT-19-0221
151. Munkley J, Scott E. Targeting Aberrant Sialylation to Treat Cancer. Medicines (2019) 6(4):102. doi: 10.3390/medicines6040102
152. Büll C, Boltje TJ, Balneger N, Weischer SM, Wassink M, Van Gemst JJ, et al. Sialic acid blockade suppresses tumor growth by enhancing t-cell-mediated tumor immunity. Cancer Res (2018) 78(13):3574–88. doi: 10.1158/0008-5472.CAN-17-3376.
153. Rillahan CD, Antonopoulos A, Lefort CT, Sonon R, Azadi P, Ley K, et al. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat Chem Biol (2012) 8(7):661–8. doi: 10.1038/nchembio.999
154. Deshayes S, Cabral H, Ishii T, Miura Y, Kobayashi S, Yamashita T, et al. Phenylboronic Acid-Installed Polymeric Micelles for Targeting Sialylated Epitopes in Solid Tumors. J Am Chem Soc (2013) 135(41):15501–7. doi: 10.1021/ja406406h
155. Zhao D, Xu J-Q, Yi X-Q, Zhang Q, Cheng S-X, Zhuo R-X, et al. pH-Activated Targeting Drug Delivery System Based on the Selective Binding of Phenylboronic Acid. ACS Appl Mater Interfaces (2016) 8(23):14845–54. doi: 10.1021/acsami.6b04737
156. Elgohary MM, Helmy MW, Abdelfattah EZA, Ragab DM, Mortada SM, Fang JY, et al. Targeting sialic acid residues on lung cancer cells by inhalable boronic acid-decorated albumin nanocomposites for combined chemo/herbal therapy. J Control Release (2018) 285:230–43. doi: 10.1016/j.jconrel.2018.07.014
157. Ji M, Li P, Sheng N, Liu L, Pan H, Wang C, et al. Sialic Acid-Targeted Nanovectors with Phenylboronic Acid-Grafted Polyethylenimine Robustly Enhance siRNA-Based Cancer Therapy. ACS Appl Mater Interfaces (2016) 8(15):9565–76. doi: 10.1021/acsami.5b11866
158. Ernst B, Magnani JL. From carbohydrate leads to glycomimetic drugs. Nat Rev Drug Discovery (2009) 8(8):661–77. doi: 10.1038/nrd2852
159. Mereiter S, Balmaña M, Campos D, Gomes J, Reis CA. Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading? Cancer Cell (2019) 36(1):6–16. doi: 10.1016/j.ccell.2019.06.006
160. Festuccia C, Mancini A, Gravina GL, Colapietro A, Vetuschi A, Pompili S, et al. Dual CXCR4 and E-Selectin Inhibitor, GMI-1359, Shows Anti-Bone Metastatic Effects and Synergizes with Docetaxel in Prostate Cancer Cell Intraosseous Growth. Cells (2019) 9(1):32. doi: 10.3390/cells9010032
161. Muz B, Azab F, Fiala M, King J, Kohnen D, Fogler WE, et al. Inhibition of E-Selectin (GMI-1271) or E-selectin together with CXCR4 (GMI-1359) re-sensitizes multiple myeloma to therapy. Blood Cancer J (2019) 9(9):68. doi: 10.1038/s41408-019-0227-3
162. Zhang W, Chang KH, Basyal M, Jia Y, Ostermann LB, Fogler WE, et al. Combined Blockage of E-Selectin and CXCR4 (GMI-1359) Enhances Anti-Leukemia Effect of FLT3 Inhibition (Sorafenib) and Protects Hematopoiesis in Pre-Clinical AML Models. Blood (2020) 136(Supplement 1):17–18. doi: 10.1182/blood-2020-140984
163. Ochoa-Alvarez JA, Krishnan H, Shen Y, Acharya NK, Han M, McNulty DE, et al. Plant Lectin Can Target Receptors Containing Sialic Acid, Exemplified by Podoplanin, to Inhibit Transformed Cell Growth and Migration. PloS One (2012) 7(7):e41845. doi: 10.1371/journal.pone.0041845
164. Hamilton KL, Sheehan SA, Retzbach EP, Timmerman CA, Gianneschi GB, Tempera PJ, et al. Effects of Maackia amurensis seed lectin (MASL) on oral squamous cell carcinoma (OSCC) gene expression and transcriptional signaling pathways. J Cancer Res Clin Oncol (2021) 147(2):445–57. doi: 10.1007/s00432-020-03456-8
165. Montero-Montero L, Renart J, Ramírez A, Ramos C, Shamhood M, Jarcovsky R, et al. Interplay between Podoplanin, CD44s and CD44v in Squamous Carcinoma Cells. Cells (2020) 9(10):2200. doi: 10.3390/cells9102200
166. Bagshawe KD, Currie GA. Immunogenicity of L 1210 Murine Leukaemia Cells after Treatment with Neuraminidase. Nature (1968) 218(5148):1254–5. doi: 10.1038/2181254a0
167. Xiao H, Woods EC, Vukojicic P, Bertozzi CR. Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc Natl Acad Sci (2016) 113(37):10304–9. doi: 10.1073/pnas.1608069113
168. Gray MA, Stanczak MA, Mantuano NR, Xiao H, Pijnenborg JFA, Malaker SA, et al. Targeted glycan degradation potentiates the anticancer immune response in vivo. Nat Chem Biol (2020) 16:1376–84. doi: 10.1038/s41589-020-0622-x
169. Matsumoto A, Stephenson-Brown AJ, Khan T, Miyazawa T, Cabral H, Kataoka K, et al. Heterocyclic boronic acids display sialic acid selective binding in a hypoxic tumor relevant acidic environment. Chem Sci (2017) 8(9):6165–70. doi: 10.1039/C7SC01905J
Keywords: sialic acid (N-acetyl neuraminic acid), phenylboronic acid chemistry, sialyltransferase (ST), CSC markers, stemness signaling pathway
Citation: Khan T and Cabral H (2021) Abnormal Glycosylation of Cancer Stem Cells and Targeting Strategies. Front. Oncol. 11:649338. doi: 10.3389/fonc.2021.649338
Received: 04 January 2021; Accepted: 15 March 2021;
Published: 06 April 2021.
Edited by:
Cecilia Ana Suarez, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Qingbin Cui, University of Toledo Medical Center, United StatesDaniela Di Marcantonio, Fox Chase Cancer Center, United States
Copyright © 2021 Khan and Cabral. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Horacio Cabral, aG9yYWNpb0BibXcudC51LXRva3lv
 Thahomina Khan
Thahomina Khan Horacio Cabral
Horacio Cabral