- 1Urogenital Stem Cell Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Pharmacognosy Department, College of Pharmacy, Hawler Medical University, Erbil, Iraq
- 4Urology and Nephrology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Hepatocellular carcinoma (HCC) is among the utmost deadly human malignancies. This type of cancer has been associated with several environmental, viral, and lifestyle risk factors. Among the epigenetic factors which contribute in the pathogenesis of HCC is dysregulation of long non-coding RNAs (lncRNAs). These transcripts modulate expression of several tumor suppressor genes and oncogenes and alter the activity of cancer-related signaling axes. Several lncRNAs such as NEAT1, MALAT1, ANRIL, and SNHG1 have been up-regulated in HCC samples. On the other hand, a number of so-called tumor suppressor lncRNAs namely CASS2 and MEG3 are down-regulated in HCC. The interaction between lncRNAs and miRNAs regulate expression of a number of mRNA coding genes which are involved in the pathogenesis of HCC. H19/miR-15b/CDC42, H19/miR-326/TWIST1, NEAT1/miR-485/STAT3, MALAT1/miR-124-3p/Slug, MALAT1/miR-195/EGFR, MALAT1/miR-22/SNAI1, and ANRIL/miR-144/PBX3 axes are among functional axes in the pathobiology of HCC. Some genetic polymorphisms within non-coding regions of the genome have been associated with risk of HCC in certain populations. In the current paper, we describe the recent finding about the impact of lncRNAs in HCC.
Introduction
Liver cancer is among the most lethal malignancies among both sexes. More than 8% of cancer-related mortalities are due to this type of cancer (1). Hepatocellular carcinoma (HCC) includes more than 75% of the primary liver neoplasms (1). Several factors have been related with elevated risk of HCC among them are chronic infection with hepatitis B virus (HBV) B or hepatitis C virus (HCV), dietary exposure with aflatoxin, excessive alcohol use, obesity, and smoking (2). The cirrhosis-induced carcinogenic alterations have been detected in 90% of HCC patients (3). High throughput sequencing methods have shown the occurrence of several genetic changes in the HCC samples (4) among the early events are inactivating mutations in insulin-like growth factor 2 receptor (5). Catenin Beta 1 (CTNNB1) and Tumor Protein P53 (TP53) are the utmost recurrently mutated oncogene and tumor suppressor gene in HCC, respectively (4). In addition to these somatic mutations, several epigenetic factors partake in the evolution of HCC. Such involvement is further highlighted by the fact that liver is an organs that is continuously adapting to extremely various environmental factors (6). Non-coding RNAs are among epigenetic elements that contribute in the pathogenesis of HCC. Long non-coding RNAs (lncRNAs) can affect expression of genes via diverse mechanisms including recruitment of regulatory protein complexes, acting as a decoy, changing genome organization and modulating the distribution of posttranslational modifications (7). These transcripts have sizes longer than 200 nucleotides and are comparable with mRNAs in the terms of chromatin state of genome loci, their transcription by RNA polymerase II, polyadenylation, 5’ capping and being spliced, yet they do not produce large-sized polypeptides (8). However, there are several reports demonstrating the presence of stable, functional micropeptides being translated from lncRNAs (9). Several lines of evidence indicates that these transcripts contribute in the pathophysiology of HCC (10). In the present manuscript, we review the current knowledge about the partake of lncRNAs in the pathogenesis of HCC.
Up-regulated lncRNAs in HCC
The LINC01138 is located in a frequently amplified region in HCC. This lncRNA transcript is stabilized by IGF2BP1/IGF2BP3. Over-expression of LINC01138 in HCC confers malignant characteristics and is associated with poor survival of patients. Mechanistically, this lncRNA interacts with arginine methyltransferase 5 and increases the stability of this protein through inhibiting ubiquitin-mediated degradation in proteasomes (11). Over-expression of the lnc-Epidermal Growth Factor Receptor (EGFR) regulatory T cells (Tregs) has been related with tumor size and levels of EGFR/Foxp3. Its over-expression has also been negatively correlated with the levels of interferon (IFN)-γ in HCC patients and animal models. This lncRNA promotes Treg differentiation, inhibits function of cytotoxic T cells and increases HCC growth. These effects are exerted through binding of lnc-EGFR with EGFR, increasing its stability and activation of the AP-1/NF-AT1 axis (12). The oncogenic lncRNA HULC has been shown to exert its effects via modulation of phosphorylation pattern of YB-1. Notably, up-regulation of this lncRNA in HCC has been correlated with pathological grade and patients’ outcome. HULC can also increase cell proliferation, migration, and invasion and suppress cisplatin-associated cell apoptosis (13). LncRNA-MUF is another over-expressed lncRNA in HCC tissues whose up-regulation has been correlated with poor clinical outcome. This lncRNA has an indispensable impact in epithelial-mesenchymal transition (EMT). Such effects have been exerted through binding with Annexin A2 and induction of the Wnt/β-catenin signaling. Mechanistically, lncRNA-MUF serves as a competing endogenous RNA (ceRNA) for miR-34a, resulting in up-regulation of Snail1 induction of EMT process (14). GHET1 over-expression in HCC sections has been associated with vascular invasion, cirrhosis, size of tumor, histological grade, and poor clinical outcome. GHET1 silencing has suppressed cell proliferation and prompted both cell cycle arrest and cell apoptosis. GHET1 can suppress expression of KLF2 in HCC cells through recruitment of PRC2 into its promoter (15). MALAT1 is another up-regulated lncRNA in HCC, which affect neoplastic transformation through several mechanisms among them is its role as a ceRNA. Figure 1 depicts this mechanism in HCC.
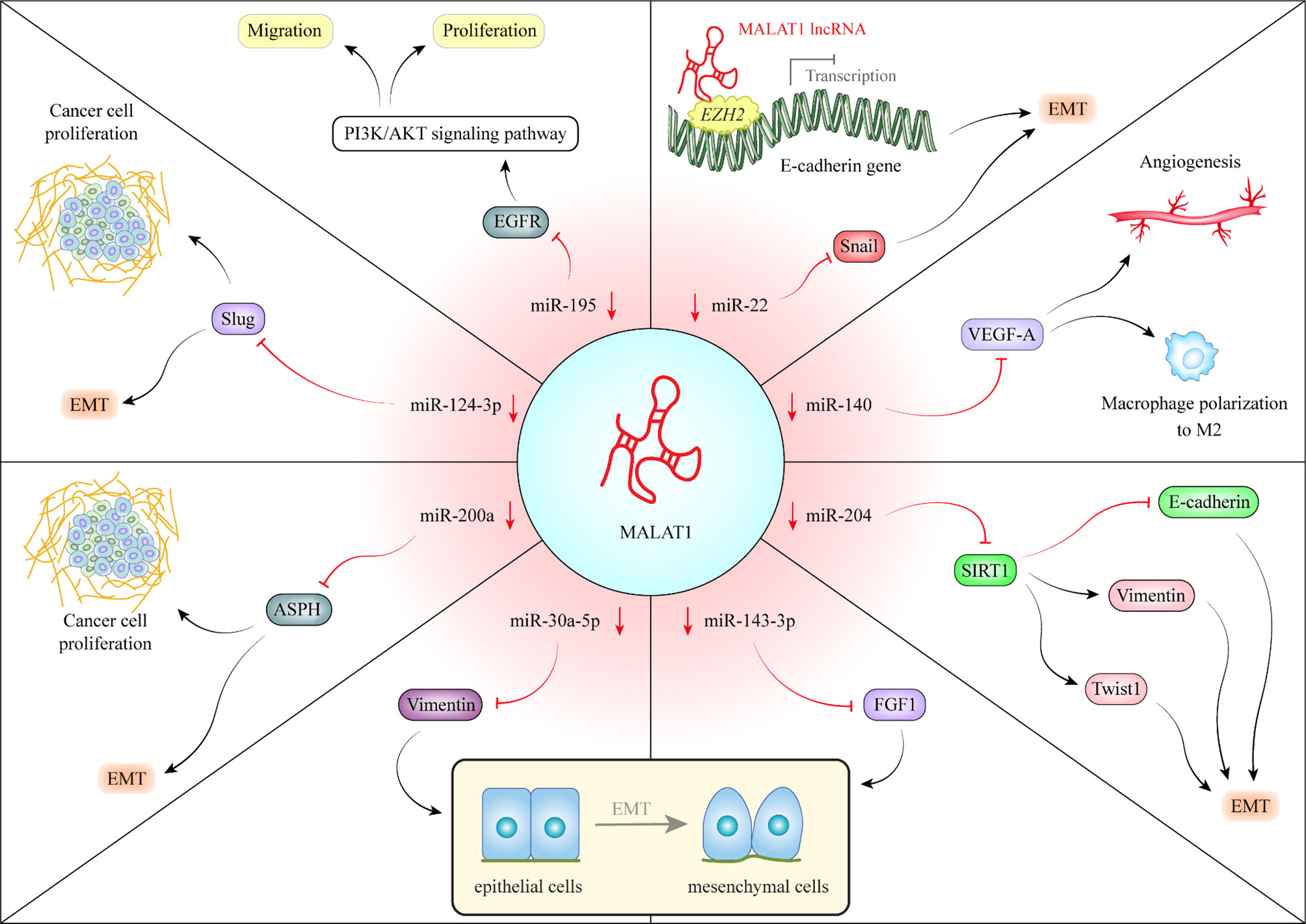
Figure 1 MALAT1 is an important oncogenic lncRNA in hepatocellular carcinoma (HCC). MALAT1 can sequester several miRNAs. For instance, MALAT1 can sequester miR-140. Through down-regulation of miR-140, MALAT1 enhances expression of VEGF-A and increases angiogenic potential. Moreover, via this route, MALAT1 enhances polarization of macrophage differentiation to M2. These macrophages facilitate tumor progression via modulation of tumor microenvironment (16). MALAT1 also reduces expression of miR-204 in HCC leading to upsurge in SIRT1 levels. SIRT1 up-regulation enhances expression of Vimentin and Twist and inhibits E-cadherin, thus facilitating epithelial-mesenchymal transition (EMT) (17). MALAT1 can also sequester miR-143-3p, thus up-regulating FGF1, N-cadherin, Vimentin, Snail, and Slug while down-regulating E-cadherin. These effects are associated with enhancement of EMT (18). Similarly, through down-regulation of miR-30a-5p, MALAT1 enhances Vimentin levels and EMT process (19). Via sequestering miR-200a, MALAT1 increases aspartate-β-hydroxylase (ASPH) levels, thus elevating expression of proteins which are involved in EMT or cell proliferation (20). MALAT1-mediated down-regulation of miR-124-3p leads to up-regulation of Slug, therefore increasing cell proliferation and EMT (21). MALAT1 can also sponge miR-195 resulting in over-expression of FGFR, activation of PI3K/AKT and enhancement of cell proliferation and invasion (22). Finally, MALAT1-mediated down-regulation of miR-22 increases Snail levels and facilitates EMT. Moreover, MALAT1 recruits EZH2 to the of promoter E-cadherin and miR-22 to decrease their expression (23). Table 1 enlists function of over-activated lncRNAs in HCC.

Table 1 Function of over-activated lncRNAs in HCC (ANT, adjacent non-cancerous tissue; HBS Ag, hepatitis B surface antigen).
Down-Regulated lncRNAs in HCC
Through a high throughput approach, Ni et al. have identified uc.134 as a novel lncRNA which is under-expressed in a highly aggressive HCC cell line. They further verified its down-regulation in clinical HCC samples compared with paired nearby tissues. Notably, down-regulation of uc.134 has been related with poor prognosis of HCC patients. Functionally, this lncRNA suppresses cell proliferation, invasion, and metastasis through binding with CUL4A suppressing its nuclear export. Besides, uc.134 suppresses the CUL4A-associted ubiquitination of LATS1 and enhances YAPS127 phosphorylation which results in down-regulation of YAP target genes of YAP (223). LncRNA-PRAL has been shown to suppress HCC growth and stimulate apoptosis via a p53-dependent route. Certain motifs at the 5’ end of this lncRNA have been identified that participate in competitive inhibition of MDM2-dependent p53 ubiquitination (224). Expression of the lncRNA-LET has been decreased in HCC. Further experiments have shown the role of hypoxia-induced histone deacetylase 3 in down-regulation of this lncRNA. Notably, repression of lncRNA-LET has been identified as an important step in the stabilization of nuclear factor 90 protein and subsequent hypoxia-associated tumor cell invasion. The association between down-regulation of lncRNA-LET and metastatic potential of HCC has also been verified in clinical samples (225). TSLNC8 is also down-regulated in HCC samples. Down-regulation of this lncRNA in HCC has been shown to confer malignant phenotype. TSLNC8 competitively interacts with transketolase and STAT3 and alters the phosphorylation patterns and transcriptional activity of STAT3 leading to suppression of the IL-6-STAT3 signaling (226). CASC2 is another down-regulated lncRNAs in HCC samples, particularly in the samples obtained patients with aggressive and recurrent forms of HCC. CASC2 suppresses migration and invasive properties of HCC cells and inhibits EMT program in these cells. Mechanistically, it serves as a competing endogenous RNA for miR-367 to increase expression of its target gene FBXW7. Notably, CASC2 down-regulation and miR-367 up-regulation have been associated with the metastasis-associated characteristics in the clinical samples (227). Table 2 displays the impact of down-regulated lncRNAs in HCC.
Diagnostic and Prognostic Impact of lncRNAs in HCC
Expression patterns of several lncRNAs have been related with overall survival or disease-free survival of patients with liver neoplasm. Oncogenic lncRNAs which decrease survival of HCC patients include NEAT1, PTTG3P, UBE2CP3, LINC00461, MALAT1, MNX1-AS1, MCM3AP-AS1, ANRIL, AWPPH, PVT1, SNHG1, ENST00000429227.1, LINC00665, CRNDE, FOXD2-AS1, HULC and some other lncRNAs. Instead, low expressions of several tumor suppressor lncRNAs namely PSTAR, CASC2, lnc-FTX, LINC00472, TSLNC8, miR503HG, MEG3, LIN00607, AOC4P, uc.134, GAS8-AS1, LINC00657, MAGI2-AS3, LINC01093, GAS5, SchLAH, and NKILA predict patients’ outcome. Univariate/multivariate cox regression analyses have confirmed the role of these lncRNAs in the determination of HCC prognosis. Table 3 lists the results of studies which evaluated the prognostic roles of lncRNAs in patients with HCC.
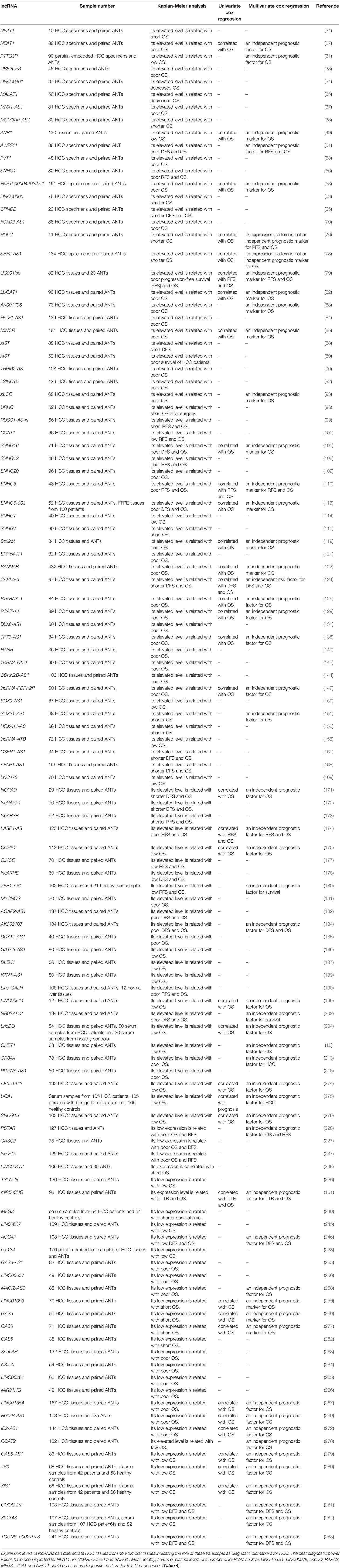
Table 3 Prognostic role of lncRNAs in HCC (ANT, adjacent non-cancerous tissue; OS, overall survival; RFS, relapse-free survival; DFS, disease-free survival; PFS, progression-free survival; TTR, time to tumor recurrence).
Genomic Variants Within lncRNAs and Risk of HCC
Genetic polymorphisms include at least four type of variations namely, single nucleotide polymorphisms, small insertion/deletion polymorphisms, polymorphic repetitive elements and microsatellites. The importance of somatic copy number variations (SCNVs) loci in non-coding regions in the development of HCC has been assessed by Zhou et al. Such investigation has led to identification of recurrent deletion of lncRNA-PRAL in HCC samples in association with poor clinical outcome (224). The lncRNA TSLNC8 on 8p12 is another tumor suppressor lncRNA which is commonly deleted in HCC tissues (226). Table 5 shows the summarized results of studies which assessed association between lncRNAs insertion/deletion or tetranucleotide repeat polymorphisms and HCC.
Discussion
LncRNAs contribute in the pathogenesis of HCC through diverse mechanisms including modulation of oncogenes and tumor suppressor genes as well as modification of tumor microenvironment. The latter route of action has been best exemplified by the lnc-EGFR which enhances differentiation of Tregs therefore increasing immune evasion (12). Moreover, certain lncRNAs such as MUF and SNHG7 facilitate EMT process through modulation of Wnt/β-catenin signaling pathway (14, 114). Other lncRNAs can modulate EMT through sponging a number of miRNAs. MAPK, PI3K/AKT and JAK/STAT signaling pathways are other cancer-related pathways that are modulated by several lncRNAs in HCC. The interactions between lncRNAs, miRNAs and mRNAs have functional importance in the pathogenesis of HCC. Examples of such trios include H19/miR-15b/CDC42, H19/miR-326/TWIST1, NEAT1/miR-485/STAT3, MALAT1/miR-124-3p/Slug, MALAT1/miR-195/EGFR, MALAT1/miR-22/SNAI1 and ANRIL/miR-144/PBX3.
Functional roles of lncRNAs in HCC have been appraised in animal models. These models have facilitated identification of lncRNAs targets and related pathways (304), which can be used as therapeutic candidates in HCC. HCC-associated lncRNAs can affect gene expression via recruiting epigenetic factors (305), regulation of transcription factors (306), modulation of protein degradation (307) and alteration of phosphorylation of proteins (308).
Genomic alterations and polymorphisms within lncRNA-coding regions have been shown to confer risk of HCC. Such variations might also predict survival of these patients. However, the observed association between these variants and HCC should be verified in independent samples from different ethnic groups. Integration of the results of genome-wide association studies with high throughput sequencing data obtained from microarray and RNA seq experiments would help in discovery of HCC-related single nucleotide polymorphisms within lncRNAs.
The biomarker role of lncRNAs in HCC has been verified by several studies indicating their importance both in the diagnosis and in the prognosis of this cancer. Expression levels of lncRNAs can differentiate HCC patients from inactive HBs Ag carriers, patients with chronic hepatitis and those with liver cirrhosis. In addition, the high diagnostic power values of peripheral levels of a number of lncRNAs such as UCA1 and NEAT1 have potentiated them as methods for non-invasive diagnosis of HCC. Moreover, lncRNAs can be regarded as therapeutic targets in HCC. The importance of lncRNAs as therapeutic targets in HCC has been noted by several experiments in animal models of HCC. Yet, such experiments wait approval in clinical settings. In vivo delivery of a number of lncRNAs such as lncRNA-PRAN, uc.134 and TSLNC8 has been shown to attenuate tumor growth and enhance lifespan of xenograft models of HCC (223, 224, 226). Moreover, a number of lncRNAs such as HULC confer resistance to chemotherapeutic agents (13), indicating the potential of targeted therapies against these transcripts in enhancement of response of HCC patients to conventional therapeutic options. Antisense oligonucleotides and small interfering RNAs are putative methods for suppression of expression of lncRNAs (309, 310) whose efficacies have been verified in animal models and cell line experiments. Yet, this knowledge has not been translated into clinical practice.
Taken together, lncRNAs as important class of regulatory transcripts can influence pathogenesis of HCC from different aspects and can be used as suitable markers for differentiation of HCC from related pathogenic conditions.
Author Contributions
SG-F and MT wrote the draft and revised it. BH and MG designed the tables and figures. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. London WT, McGlynn K, Schottenfeld D, Fraumeni J. Cancer epidemiology and prevention. Cancer Epidemiology and Prevention. 3rd edition. In: Schottenfeld D, Fraumeni JRJF, editors. New York, NY: Oxford University Press (2006) p. 763–86.
3. Zhang DY, Friedman SL. Fibrosis-dependent mechanisms of hepatocarcinogenesis. Hepatology (2012) 56(2):769–75. doi: 10.1002/hep.25670
4. Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J Carcinog (2017) 16:1–. doi: 10.4103/jcar.JCar_9_16
5. El–Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology (2007) 132(7):2557–76. doi: 10.1053/j.gastro.2007.04.061
6. Toh TB, Lim JJ, Chow EK-H. Epigenetics of hepatocellular carcinoma. Clin Transl Med (2019) 8(1):13–. doi: 10.1186/s40169-019-0230-0
7. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv (2017) 3(9):eaao2110–eaao. doi: 10.1126/sciadv.aao2110
8. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature (2009) 458(7235):223–7. doi: 10.1038/nature07672
9. Choi S-W, Kim H-W, Nam J-W. The small peptide world in long noncoding RNAs. Brief Bioinform (2019) 20(5):1853–64. doi: 10.1093/bib/bby055
10. Mai H, Zhou B, Liu L, Yang F, Conran C, Ji Y, et al. Molecular pattern of lncRNAs in hepatocellular carcinoma. J Exp Clin Cancer Res (2019) 38(1):198–. doi: 10.1186/s13046-019-1213-0
11. Li Z, Zhang J, Liu X, Li S, Wang Q, Di C, et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun (2018) 9(1):1572. doi: 10.1038/s41467-018-04006-0
12. Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun (2017) 8:15129. doi: 10.1038/ncomms15129
13. Li D, Liu X, Zhou J, Hu J, Zhang D, Liu J, et al. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology (2017) 65(5):1612–27. doi: 10.1002/hep.29010
14. Yan X, Zhang D, Wu W, Wu S, Qian J, Hao Y, et al. Mesenchymal Stem Cells Promote Hepatocarcinogenesis via lncRNA-MUF Interaction with ANXA2 and miR-34a. Cancer Res (2017) 77(23):6704–16. doi: 10.1158/0008-5472.CAN-17-1915
15. Jin L, He Y, Tang S, Huang S. LncRNA GHET1 predicts poor prognosis in hepatocellular carcinoma and promotes cell proliferation by silencing KLF2. J Cell Physiol (2018) 233(6):4726–34. doi: 10.1002/jcp.26257
16. Hou Z-H, Xu X-W, Fu X-Y, Zhou L-D, Liu S-P, Tan D-M. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am J Physiology-Cell Physiol (2020) 318(3):C649–C63. doi: 10.1152/ajpcell.00510.2018
17. Hou Z, Xu X, Zhou L, Fu X, Tao S, Zhou J, et al. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumor Biol (2017) 39(7):1010428317718135. doi: 10.1177/1010428317718135
18. Peng J, Wu H, Zhang H, Fang S, Zeng R, et al. miR-143-3p inhibits proliferation and invasion of hepatocellular carcinoma cells by regulating its target gene FGF1. Clin Trans Oncol (2021) 23:468–480. doi: 10.1007/s12094-020-02440-5
19. Pan Y, Tong S, Cui R, Fan J, Liu C, Lin Y, et al. Long non-coding MALAT1 functions as a competing endogenous RNA to regulate vimentin expression by sponging miR-30a-5p in hepatocellular carcinoma. Cell Physiol Biochem (2018) 50(1):108–20. doi: 10.1159/000493962
20. Yao W-F, Liu J-W, Huang D-S. MiR-200a inhibits cell proliferation and EMT by down-regulating the ASPH expression levels and affecting ERK and PI3K/Akt pathways in human hepatoma cells. Am J Trans Res (2018) 10(4):1117.
21. Cui RJ, Fan JL, Lin YC, Pan YJ, Liu C, Wan JH, et al. miR-124-3p availability is antagonized by LncRNA-MALAT1 for Slug-induced tumor metastasis in hepatocellular carcinoma. Cancer Med (2019) 8(14):6358–69. doi: 10.1002/cam4.2482
22. Liu D, Zhu Y, Pang J, Weng X, Feng X, Guo Y. Knockdown of long non-coding RNA MALAT1 inhibits growth and motility of human hepatoma cells via modulation of miR-195. J Cell Biochem (2018) 119(2):1368–80. doi: 10.1002/jcb.26297
23. Chen S, Wang G, Tao K, Cai K, Wu K, Ye L, et al. Long noncoding RNA metastasis-associated lung adenocarcinoma transcript 1 cooperates with enhancer of zeste homolog 2 to promote hepatocellular carcinoma development by modulating the microRNA-22/Snail family transcriptional repressor 1 axis. Cancer Sci (2020) 111(5):1582. doi: 10.1111/cas.14372
24. Liu X, Liang Y, Song R, Yang G, Han J, Lan Y, et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol Cancer (2018) 17(1):1–18. doi: 10.1186/s12943-018-0838-5
25. Fang L, Sun J, Pan Z, Song Y, Zhong L, Zhang Y, et al. Long non-coding RNA NEAT1 promotes hepatocellular carcinoma cell proliferation through the regulation of miR-129-5p-VCP-IκB. Am J Physiology-Gastrointestinal Liver Physiol (2017) 313(2):G150–G6. doi: 10.1152/ajpgi.00426.2016
26. Zhang XN, Zhou J, Lu XJ. The long noncoding RNA NEAT1 contributes to hepatocellular carcinoma development by sponging miR-485 and enhancing the expression of the STAT3. J Cell Physiol (2018) 233(9):6733–41. doi: 10.1002/jcp.26371
27. Liu Z, Chang Q, Yang F, Liu B, Yao H-W, Bai Z-G, et al. Long non-coding RNA NEAT1 overexpression is associated with unfavorable prognosis in patients with hepatocellular carcinoma after hepatectomy: A Chinese population-based study. Eur J Surg Oncol (2017) 43(9):1697–703. doi: 10.1016/j.ejso.2017.06.013
28. Wang Z, Zou Q, Song M, Chen J. NEAT1 promotes cell proliferation and invasion in hepatocellular carcinoma by negative regulating miR-613 expression. Biomed Pharmacother (2017) 94:612–8. doi: 10.1016/j.biopha.2017.07.111
29. Tu J, Zhao Z, Xu M, Lu X, Chang L, Ji J. NEAT1 upregulates TGF-β1 to induce hepatocellular carcinoma progression by sponging hsa-mir-139-5p. J Cell Physiol (2018) 233(11):8578–87. doi: 10.1002/jcp.26524
30. Chen X, Zhang N. Downregulation of lncRNA NEAT1_2 radiosensitizes hepatocellular carcinoma cells through regulation of miR-101-3p/WEE1 axis. Cell Biol Int (2019) 43(1):44–55. doi: 10.1002/cbin.11077
31. Huang J-L, Cao S-w, Ou Q-s, Yang B, Zheng S-h, Tang J, et al. The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol Cancer (2018) 17(1):1–16. doi: 10.1186/s12943-018-0841-x
32. Zhou Q, Zhang W, Wang Z, Liu S. Long non-coding RNA PTTG3P functions as an oncogene by sponging miR-383 and up-regulating CCND1 and PARP2 in hepatocellular carcinoma. BMC Cancer (2019) 19(1):731. doi: 10.1186/s12885-019-5936-2
33. Lin J, Cao S, Wang Y, Hu Y, Liu H, Li J, et al. Long non-coding RNA UBE2CP3 enhances HCC cell secretion of VEGFA and promotes angiogenesis by activating ERK1/2/HIF-1α/VEGFA signalling in hepatocellular carcinoma. J Exp Clin Cancer Res (2018) 37(1):1–13. doi: 10.1186/s13046-018-0727-1
34. Ji D, Wang Y, Li H, Sun B, Luo X. Long non-coding RNA LINC00461/miR-149-5p/LRIG2 axis regulates hepatocellular carcinoma progression. Biochem Biophys Res Commun (2019) 512(2):176–81. doi: 10.1016/j.bbrc.2019.03.049
35. Chen L, Yao H, Wang K, Liu X. Long non-coding RNA MALAT1 regulates ZEB1 expression by sponging miR-143-3p and promotes hepatocellular carcinoma progression. J Cell Biochem (2017) 118(12):4836–43. doi: 10.1002/jcb.26158
36. Zhao Z-B, Chen F, Bai X-F. Long Noncoding RNA MALAT1 regulates hepatocellular carcinoma growth under hypoxia via sponging MicroRNA-200a. Yonsei Med J (2019) 60(8):727–34. doi: 10.3349/ymj.2019.60.8.727
37. Ji D, Wang Y, Sun B, Yang J, Luo X. Long non-coding RNA MNX1-AS1 promotes hepatocellular carcinoma proliferation and invasion through targeting miR-218-5p/COMMD8 axis. Biochem Biophys Res Commun (2019) 513(3):669–74. doi: 10.1016/j.bbrc.2019.04.012
38. Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu Q, et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer (2019) 18(1):28. doi: 10.1186/s12943-019-0957-7
39. Zhang H, Luo C, Zhang G. LncRNA MCM3AP-AS1 regulates epidermal growth factor receptor and autophagy to promote hepatocellular carcinoma metastasis by interacting with miR-455. DNA Cell Biol (2019) 38(8):857–64. doi: 10.1089/dna.2019.4770
40. Huang M-D, Chen W-M, Qi F-Z, Sun M, Xu T-P, Ma P, et al. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer (2015) 14(1):165. doi: 10.1186/s12943-015-0431-0
41. Lin YH, Wu MH, Huang YH, Yeh CT, Cheng ML, Chi HC, et al. Taurine up-regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology (2018) 67(1):188–203. doi: 10.1002/hep.29462
42. Wang X, Li J, Xu X, Zheng J, Li Q. miR-129 inhibits tumor growth and potentiates chemosensitivity of neuroblastoma by targeting MYO10. Biomed Pharmacother (2018) 103:1312–8. doi: 10.1016/j.biopha.2018.04.153
43. Lv J, Kong Y, Gao Z, Liu Y, Zhu P, Yu Z. LncRNA TUG1 interacting with miR-144 contributes to proliferation, migration and tumorigenesis through activating the JAK2/STAT3 pathway in hepatocellular carcinoma. Int J Biochem Cell Biol (2018) 101:19–28. doi: 10.1016/j.biocel.2018.05.010
44. Cheng Z, Lei Z, Yang P, Si A, Xiang D, Zhou J, et al. Long non-coding RNA THOR promotes cell proliferation and metastasis in hepatocellular carcinoma. Gene (2018) 678:129–36. doi: 10.1016/j.gene.2018.08.035
45. Li K, Zhao B, Wei D, Cui Y, Qian L, Wang W, et al. Long non-coding RNA ANRIL enhances mitochondrial function of hepatocellular carcinoma by regulating the MiR-199a-5p/ARL2 axis. Environ Toxicol (2020) 35(3):313–21. doi: 10.1002/tox.22867
46. Huang D, Bi C, Zhao Q, Ding X, Bian C, Wang H, et al. Knockdown long non-coding RNA ANRIL inhibits proliferation, migration and invasion of HepG2 cells by down-regulation of miR-191. BMC Cancer (2018) 18(1):1–9. doi: 10.1186/s12885-018-4831-6
47. Huang M, Chen W, Qi F, Xia R, Sun M, Xu T, et al. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell proliferation by epigenetic silencing of KLF2. J Hematol Oncol (2015) 8(1):57–. doi: 10.1186/s13045-015-0153-1
48. Ma J, Li T, Han X, Yuan H. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J Cancer Res Clin Oncol (2018) 144(2):205–14. doi: 10.1007/s00432-017-2543-y
49. Hua L, Wang C-Y, Yao K-H, Chen J-T, Zhang J-J, Ma W-L. High expression of long non-coding RNA ANRIL is associated with poor prognosis in hepatocellular carcinoma. Int J Clin Exp Pathol (2015) 8(3):3076.
50. Ma Y, Zhang H, Li G, Hu J, Liu X, Lin L. LncRNA ANRIL promotes cell growth, migration and invasion of hepatocellular carcinoma cells via sponging miR-144. Anti-Cancer Drugs (2019) 30(10):1013–21. doi: 10.1097/CAD.0000000000000807
51. Zhao X, Liu Y, Yu S. Long noncoding RNA AWPPH promotes hepatocellular carcinoma progression through YBX1 and serves as a prognostic biomarker. Biochim Biophys Acta (BBA)-Molecular Basis Dis (2017) 1863(7):1805–16. doi: 10.1016/j.bbadis.2017.04.014
52. Xu Y, Luo X, He W, Chen G, Li Y, Li W, et al. Long non-coding RNA PVT1/miR-150/HIG2 axis regulates the proliferation, invasion and the balance of iron metabolism of hepatocellular carcinoma. Cell Physiol Biochem (2018) 49(4):1403–19. doi: 10.1159/000493445
53. Lan T, Yan X, Li Z, Xu X, Mao Q, Ma W, et al. Long non-coding RNA PVT1 serves as a competing endogenous RNA for miR-186-5p to promote the tumorigenesis and metastasis of hepatocellular carcinoma. Tumor Biol (2017) 39(6):1010428317705338. doi: 10.1177/1010428317705338
54. Yang L, Peng X, Jin H, Liu J. Long non-coding RNA PVT1 promotes autophagy as ceRNA to target ATG3 by sponging microRNA-365 in hepatocellular carcinoma. Gene (2019) 697:94–102. doi: 10.1016/j.gene.2019.02.036
55. Huang D, Wei Y, Zhu J, Wang F. Long non-coding RNA SNHG1 functions as a competitive endogenous RNA to regulate PDCD4 expression by sponging miR-195-5p in hepatocellular carcinoma. Gene (2019) 714:143994. doi: 10.1016/j.gene.2019.143994
56. Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding F, et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother (2016) 80:73–9. doi: 10.1016/j.biopha.2016.02.036
57. Zhang H, Zhou D, Ying M, Chen M, Chen P, Chen Z, et al. Expression of long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) exacerbates hepatocellular carcinoma through suppressing miR-195. Med Sci Monitor: Int Med J Exp Clin Res (2016) 22:4820. doi: 10.12659/MSM.898574
58. Zhao Y, Kong C-Q, Ye J-Z, Bai T, Luo T, Wang D, et al. Upregulation of Long Non-Coding RNA ENST00000429227. 1 Is Correlated with Poor Prognosis in Human Hepatocellular Carcinoma. Med Sci Monitor: Int Med J Exp Clin Res (2019) 25:6539. doi: 10.12659/MSM.916551
59. Ding K, Liao Y, Gong D, Zhao X, Ji W. Effect of long non-coding RNA H19 on oxidative stress and chemotherapy resistance of CD133+ cancer stem cells via the MAPK/ERK signaling pathway in hepatocellular carcinoma. Biochem Biophys Res Commun (2018) 502(2):194–201. doi: 10.1016/j.bbrc.2018.05.143
60. Zhou Y, Fan R-G, Qin C-L, Jia J, Wu X-D, Zha W-Z. LncRNA-H19 activates CDC42/PAK1 pathway to promote cell proliferation, migration and invasion by targeting miR-15b in hepatocellular carcinoma. Genomics (2019) 111(6):1862–72. doi: 10.1016/j.ygeno.2018.12.009
61. Wei LQ, Li L, Lu C, Liu J, Chen Y, Wu H. Involvement of H19/miR-326 axis in hepatocellular carcinoma development through modulating TWIST1. J Cell Physiol (2019) 234(4):5153–62. doi: 10.1002/jcp.27319
62. Xu Y, Zheng Y, Liu H, Li T. Modulation of IGF2BP1 by long non-coding RNA HCG11 suppresses apoptosis of hepatocellular carcinoma cells via MAPK signaling transduction. Int J Oncol (2017) 51(3):791–800. doi: 10.3892/ijo.2017.4066
63. Shan Y, Li P. Long intergenic non-protein coding RNA 665 regulates viability, apoptosis, and autophagy via the miR-186-5p/MAP4K3 axis in hepatocellular carcinoma. Yonsei Med J (2019) 60(9):842–53. doi: 10.3349/ymj.2019.60.9.842
64. Wang H, Ke J, Guo Q, Barnabo Nampoukime KP, Yang P, Ma K. Long non-coding RNA CRNDE promotes the proliferation, migration and invasion of hepatocellular carcinoma cells through miR-217/MAPK 1 axis. J Cell Mol Med (2018) 22(12):5862–76. doi: 10.1111/jcmm.13856
65. Tang Q, Zheng X, Zhang J. Long non-coding RNA CRNDE promotes heptaocellular carcinoma cell proliferation by regulating PI3K/Akt/β-catenin signaling. Biomed Pharmacother (2018) 103:1187–93. doi: 10.1016/j.biopha.2018.04.128
66. Zhu L, Liu Y, Chen Q, Yu G, Chen J, Chen K, et al. Long-noncoding RNA colorectal neoplasia differentially expressed gene as a potential target to upregulate the expression of IRX5 by miR-136-5P to promote oncogenic properties in hepatocellular carcinoma. Cell Physiol Biochem (2018) 50(6):2229–48. doi: 10.1159/000495084
67. Ji D, Jiang C, Zhang L, Liang N, Jiang T, Yang B, et al. LncRNA CRNDE promotes hepatocellular carcinoma cell proliferation, invasion, and migration through regulating miR-203/BCAT1 axis. J Cell Physiol (2019) 234(5):6548–60. doi: 10.1002/jcp.27396
68. Tang D, Zhao L, Peng C, Ran K, Mu R, Ao Y. LncRNA CRNDE promotes hepatocellular carcinoma progression by upregulating SIX1 through modulating miR-337-3p. J Cell Biochem (2019) 120(9):16128–42. doi: 10.1002/jcb.28894
69. Chen Z, Zhang Z, Zhao D, Feng W, Meng F, Han S, et al. Long Noncoding RNA (lncRNA) FOXD2-AS1 Promotes Cell Proliferation and Metastasis in Hepatocellular Carcinoma by Regulating MiR-185/AKT Axis. Med Sci Monitor: Int Med J Exp Clin Res (2019) 25:9618. doi: 10.12659/MSM.918230
70. Lei T, Zhu X, Zhu K, Jia F, Li S. EGR1-induced upregulation of lncRNA FOXD2-AS1 promotes the progression of hepatocellular carcinoma via epigenetically silencing DKK1 and activating Wnt/β-catenin signaling pathway. Cancer Biol Ther (2019) 20(7):1007–16. doi: 10.1080/15384047.2019.1595276
71. Gao J, Yin X, Yu X, Dai C, Zhou F. Long noncoding RNA LINC00488 functions as a ceRNA to regulate hepatocellular carcinoma cell growth and angiogenesis through miR-330-5. Digestive Liver Dis (2019) 51(7):1050–9. doi: 10.1016/j.dld.2019.03.012
72. Kang CL, Qi B, Cai QQ, Fu LS, Yang Y, Tang C, et al. LncRNA AY promotes hepatocellular carcinoma metastasis by stimulating ITGAV transcription. Theranostics (2019) 9(15):4421. doi: 10.7150/thno.32854
73. Shen Y, Liu S, Yuan H, Ying X, Fu H, Zheng X. A long non-coding RNA lncRNA-PE promotes invasion and epithelial–mesenchymal transition in hepatocellular carcinoma through the miR-200a/b-ZEB1 pathway. Tumor Biol (2017) 39(5):1010428317705756. doi: 10.1177/1010428317705756
74. Cao S-Q, Zheng H, Sun B-C, Wang Z-L, Liu T, Guo D-H, et al. Long non-coding RNA highly up-regulated in liver cancer promotes exosome secretion. World J Gastroenterol (2019) 25(35):5283. doi: 10.3748/wjg.v25.i35.5283
75. Xin X, Wu M, Meng Q, Wang C, Lu Y, Yang Y, et al. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol Cancer (2018) 17(1):1–16. doi: 10.1186/s12943-018-0843-8
76. Bai R, Yang Q, Xi R, Li L, Shi D, Chen K. miR-941 as a promising biomarker for acute coronary syndrome. BMC Cardiovasc Disord (2017) 17(1):227. doi: 10.1186/s12872-017-0653-8
77. Li Y, Liu G, Li X, Dong H, Xiao W, Lu S. Long non-coding RNA SBF2-AS1 promotes hepatocellular carcinoma progression through regulation of miR-140-5p-TGFBR1 pathway. Biochem Biophys Res Commun (2018) 503(4):2826–32. doi: 10.1016/j.bbrc.2018.08.047
78. Zhang Y, Li B, Zhang B, Ma P, Wu Q, Ming L, et al. LncRNA SBF2-AS1 promotes hepatocellular carcinoma metastasis by regulating EMT and predicts unfavorable prognosis. Eur Rev Med Pharmacol Sci (2018) 22(19):6333–41. doi: 10.26355/eurrev_201810_16044
79. Pan Y, Qin T, Yin S, Zhang X, Gao X, Mu L. Long non-coding RNA UC001kfo promotes hepatocellular carcinoma proliferation and metastasis by targeting α-SMA. Biomed Pharmacother (2017) 87:669–77. doi: 10.1016/j.biopha.2017.01.018
80. Tsang FH, Au SL, Wei L, Fan DN, Lee JM, Wong CC, et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int (2015) 35(5):1597–606. doi: 10.1111/liv.12746
81. Chang Y, Zhang J, Zhou C, Qiu G, Wang G, Wang S, et al. Long non-coding RNA FOXD2-AS1 plays an oncogenic role in hepatocellular carcinoma by targeting miR−206. Oncol Rep (2018) 40(6):3625–34. doi: 10.3892/or.2018.6752
82. Lou Y, Yu Y, Xu X, Zhou S, Shen H, Fan T, et al. Long non-coding RNA LUCAT1 promotes tumourigenesis by inhibiting ANXA2 phosphorylation in hepatocellular carcinoma. J Cell Mol Med (2019) 23(3):1873–84. doi: 10.1111/jcmm.14088
83. Han Q, Chen B, Zhang K, Xia S, Zhong W, Zhao Z. The long non-coding RNA AK001796 contributes to poor prognosis and tumor progression in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci (2019) 23(5):2013–9. doi: 10.26355/eurrev_201903_17240
84. Wang Y-D, Sun X-J, Yin J-J, Yin M, Wang W, Nie Z-Q, et al. Long non-coding RNA FEZF1-AS1 promotes cell invasion and epithelial-mesenchymal transition through JAK2/STAT3 signaling pathway in human hepatocellular carcinoma. Biomed Pharmacother (2018) 106:134–41. doi: 10.1016/j.biopha.2018.05.116
85. Jin X, Lian J, Guan Y. Overexpression of long non-coding RNA MINCR contributes to progressive clinicopathological features and poor prognosis of human hepatocellular carcinoma. Eur Rev Med Pharmacol Sci (2018) 22(23):8197–202. doi: 10.26355/eurrev_201812_16512
86. Cao J, Zhang D, Zeng L, Liu F. Long noncoding RNA MINCR regulates cellular proliferation, migration, and invasion in hepatocellular carcinoma. Biomed Pharmacother (2018) 102:102–6. doi: 10.1016/j.biopha.2018.03.041
87. Chen T, Pei J, Wang J, Luo R, Liu L, Wang L, et al. HBx-related long non-coding RNA 01152 promotes cell proliferation and survival by IL-23 in hepatocellular carcinoma. Biomed Pharmacother (2019) 115:108877. doi: 10.1016/j.biopha.2019.108877
88. Mo Y, Lu Y, Wang P, Huang S, He L, Li D, et al. Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumor Biol (2017) 39(2):1010428317690999. doi: 10.1177/1010428317690999
89. Kong Q, Zhang S, Liang C, Zhang Y, Kong Q, Chen S, et al. LncRNA XIST functions as a molecular sponge of miR-194-5p to regulate MAPK1 expression in hepatocellular carcinoma cell. J Cell Biochem (2018) 119(6):4458–68. doi: 10.1002/jcb.26540
90. Xu C, Huang Q, Zhang C, Xu W, Xu G, Zhao X, et al. Long non-coding RNA TRPM2-AS as a potential biomarker for hepatocellular carcinoma. Irish J Med Sci (1971-) (2018) 187(3):621–8. doi: 10.1007/s11845-017-1692-y
91. Huang L, Li X, Gao W. Long non-coding RNA linc-ITGB1 promotes cell proliferation, migration, and invasion in human hepatoma carcinoma by up-regulating ROCK1. Biosci Rep (2018) 38(5). doi: 10.1042/BSR20181289
92. Li O, Li Z, Tang Q, Li Y, Yuan S, Shen Y, et al. Long Stress Induced Non-Coding Transcripts 5 (LSINCT5) promotes hepatocellular carcinoma progression through interaction with high-mobility group AT-hook 2 and MiR-4516. Med Sci Monitor: Int Med J Exp Clin Res (2018) 24:8510. doi: 10.12659/MSM.911179
93. Yang F, Jiang Y, Lv L. Long non-coding RNA XLOC_010235 correlates with poor prognosis and promotes tumorigenesis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci (2017) 21(21):4867–74.
94. Wang C, Mou L, Chai H-X, Wang F, Yin Y-Z, Zhang X-Y. Long non-coding RNA HNF1A-AS1 promotes hepatocellular carcinoma cell proliferation by repressing NKD1 and P21 expression. Biomed Pharmacother (2017) 89:926–32. doi: 10.1016/j.biopha.2017.01.031
95. Liu Z, Wei X, Zhang A, Li C, Bai J, Dong J. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem Biophys Res Commun (2016) 473(4):1268–75. doi: 10.1016/j.bbrc.2016.04.054
96. Xu W-H, Zhang J-B, Dang Z, Li X, Zhou T, Liu J, et al. Long non-coding RNA URHC regulates cell proliferation and apoptosis via ZAK through the ERK/MAPK signaling pathway in hepatocellular carcinoma. Int J Biol Sci (2014) 10(7):664. doi: 10.7150/ijbs.8232
97. Xiao J-N, Yan T-H, Yu R-M, Gao Y, Zeng W-L, Lu S-W, et al. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol (2017) 143(6):981–90. doi: 10.1007/s00432-017-2370-1
98. Yang J, Li J, Liu B, Zhang R, Gu F, Zhao J, et al. Long Noncoding RNA AK021443 Promotes Cell Proliferation and Migration by Regulating Epithelial–Mesenchymal Transition in Hepatocellular Carcinoma Cells. DNA Cell Biol (2018) 37(5):481–90. doi: 10.1089/dna.2017.4030
99. Tang R, Wu J, Zheng L, Li Z, Zhou K, Zhang Z, et al. Long noncoding RNA RUSC1-AS-N indicates poor prognosis and increases cell viability in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci (2018) 22(2):388–96. doi: 10.26355/eurrev_201801_14185
100. Dou C, Sun L, Jin X, Han M, Zhang B, Li T. Long non-coding RNA colon cancer–associated transcript 1 functions as a competing endogenous RNA to regulate cyclin-dependent kinase 1 expression by sponging miR-490-3p in hepatocellular carcinoma progression. Tumor Biol (2017) 39(4):1010428317697572. doi: 10.1177/1010428317697572
101. Deng L, Yang S-B, Xu F-F, Zhang J-H. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res (2015) 34(1):1–10. doi: 10.1186/s13046-015-0136-7
102. Guo J, Ma Y, Peng X, Jin H, Liu J. LncRNA CCAT1 promotes autophagy via regulating ATG7 by sponging miR-181 in hepatocellular carcinoma. J Cell Biochem (2019) 120(10):17975–83. doi: 10.1002/jcb.29064
103. Zhang J, Cai M, Jiang D, Xu L. Upregulated LncRNA-CCAT1 promotes hepatocellular carcinoma progression by functioning as miR-30c-2-3p sponge. Cell Biochem Funct (2019) 37(2):84–92. doi: 10.1002/cbf.3375
104. Liu Y, Wang D, Li Y, Yan S, Dang H, Yue H, et al. Long noncoding RNA CCAT2 promotes hepatocellular carcinoma proliferation and metastasis through up-regulation of NDRG1. Exp Cell Res (2019) 379(1):19–29. doi: 10.1016/j.yexcr.2019.03.029
105. Guo Z, Zhang J, Fan L, Liu J, Yu H, Li X, et al. Long noncoding RNA (lncRNA) small nucleolar RNA host gene 16 (SNHG16) predicts poor prognosis and sorafenib resistance in hepatocellular carcinoma. Med Sci Monitor: Int Med J Exp Clin Res (2019) 25:2079. doi: 10.12659/MSM.915541
106. Xie X, Xu X, Sun C, Yu Z. Long intergenic noncoding RNA SNHG16 interacts with miR-195 to promote proliferation, invasion and tumorigenesis in hepatocellular carcinoma. Exp Cell Res (2019) 383(1):111501. doi: 10.1016/j.yexcr.2019.111501
107. Lan T, Yuan K, Yan X, Xu L, Liao H, Hao X, et al. LncRNA SNHG10 facilitates hepatocarcinogenesis and metastasis by modulating its homolog SCARNA13 via a positive feedback loop. Cancer Res (2019) 79(13):3220–34. doi: 10.1158/0008-5472.CAN-18-4044
108. Lan T, Ma W, Hong Z, Wu L, Chen X, Yuan Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J Exp Clin Cancer Res (2017) 36(1):11. doi: 10.1186/s13046-016-0486-9
109. Liu J, Lu C, Xiao M, Jiang F, Qu L, Ni R. Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. Biomed Pharmacother (2017) 89:857–63. doi: 10.1016/j.biopha.2017.01.011
110. Li Y, Guo D, Zhao Y, Ren M, Lu G, Wang Y, et al. Long non-coding RNA SNHG5 promotes human hepatocellular carcinoma progression by regulating miR-26a-5p/GSK3β signal pathway. Cell Death Dis (2018) 9(9):1–15. doi: 10.1038/s41419-018-0882-5
111. Chen S, Xie C, Hu X. lncRNA SNHG6 functions as a ceRNA to up-regulate c-Myc expression via sponging let-7c-5p in hepatocellular carcinoma. Biochem Biophys Res Commun (2019) 519(4):901–8. doi: 10.1016/j.bbrc.2019.09.091
112. Wu G, Ju X, Wang Y, Li Z, Gan X. Up-regulation of SNHG6 activates SERPINH1 expression by competitive binding to miR-139-5p to promote hepatocellular carcinoma progression. Cell Cycle (2019) 18(16):1849–67. doi: 10.1080/15384101.2019.1629772
113. Cao C, Zhang T, Zhang D, Xie L, Zou X, Lei L, et al. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene (2017) 36(8):1112–22. doi: 10.1038/onc.2016.278
114. Yao X, Liu C, Liu C, Xi W, Sun S, Gao Z. lncRNA SNHG7 sponges miR-425 to promote proliferation, migration, and invasion of hepatic carcinoma cells via Wnt/β-catenin/EMT signalling pathway. Cell Biochem Funct (2019) 37(7):525–33. doi: 10.1002/cbf.3429
115. Yang X, Sun L, Wang L, Yao B, Mo H, Yang W. LncRNA SNHG7 accelerates the proliferation, migration and invasion of hepatocellular carcinoma cells via regulating miR-122-5p and RPL4. Biomed Pharmacother (2019) 118:109386. doi: 10.1016/j.biopha.2019.109386
116. Dong J, Teng F, Guo W, Yang J, Ding G, Fu Z. lncRNA SNHG8 promotes the tumorigenesis and metastasis by sponging miR-149-5p and predicts tumor recurrence in hepatocellular carcinoma. Cell Physiol Biochem (2018) 51(5):2262–74. doi: 10.1159/000495871
117. Dai W, Dai J-L, Tang M-H, Ye M-S, Fang S. lncRNA-SNHG15 accelerates the development of hepatocellular carcinoma by targeting miR-490-3p/histone deacetylase 2 axis. World J Gastroenterol (2019) 25(38):5789–99. doi: 10.3748/wjg.v25.i38.5789
118. Liu Y, Yang Y, Wang T, Wang L, Wang X, Li T, et al. Long non-coding RNA CCAL promotes hepatocellular carcinoma progression by regulating AP-2α and Wnt/β-catenin pathway. Int J Biol Macromol (2018) 109:424–34. doi: 10.1016/j.ijbiomac.2017.12.110
119. Shi X-M, Teng F. Up-regulation of long non-coding RNA Sox2ot promotes hepatocellular carcinoma cell metastasis and correlates with poor prognosis. Int J Clin Exp Pathol (2015) 8(4):4008.
120. Zhou M, Zhang X-Y, Yu X. Overexpression of the long non-coding RNA SPRY4-IT1 promotes tumor cell proliferation and invasion by activating EZH2 in hepatocellular carcinoma. Biomed Pharmacother (2017) 85:348–54. doi: 10.1016/j.biopha.2016.11.035
121. Fu L, Chen Q, Yao T, Li T, Ying S, Hu Y, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget (2017) 8(27):43878. doi: 10.18632/oncotarget.16709
122. Peng W, Fan H. Long non-coding RNA PANDAR correlates with poor prognosis and promotes tumorigenesis in hepatocellular carcinoma. Biomed Pharmacother (2015) 72:113–8. doi: 10.1016/j.biopha.2015.04.014
123. Chen Y, Shen Z, Zhi Y, Zhou H, Zhang K, Wang T, et al. Long non-coding RNA ROR promotes radioresistance in hepatocelluar carcinoma cells by acting as a ceRNA for microRNA-145 to regulate RAD18 expression. Arch Biochem Biophysics (2018) 645:117–25. doi: 10.1016/j.abb.2018.03.018
124. Wang F, Xie C, Zhao W, Deng Z, Yang H, Fang Q. Long non-coding RNA CARLo-5 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Clin Exp Med (2017) 17(1):33–43. doi: 10.1007/s10238-015-0395-9
125. Wu F, Li J, Du X, Zhang W, Lei P, Zhang Q. Long non−coding RNA AB019562 promotes cell proliferation and metastasis in human hepatocellular carcinoma. Mol Med Rep (2017) 16(1):69–74. doi: 10.3892/mmr.2017.6612
126. Dong L, Ni J, Hu W, Yu C, Li H. Upregulation of long non-coding RNA PlncRNA-1 promotes metastasis and induces epithelial-mesenchymal transition in hepatocellular carcinoma. Cell Physiol Biochem (2016) 38(2):836–46. doi: 10.1159/000443038
127. Liu J, Wang Z, Yin Y, Li N, Ye N, Bao B, et al. Long noncoding RNA TPTE2P1 promotes the migration and invasion of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci (2019) 23(9):3733–41. doi: 10.26355/eurrev_201905_17799
128. Wen J, Xu J, Sun Q, Xing C, Yin W. Upregulation of long non coding RNA PCAT-1 contributes to cell proliferation, migration and apoptosis in hepatocellular carcinoma. Mol Med Rep (2016) 13(5):4481–6. doi: 10.3892/mmr.2016.5075
129. Wang Y, Hu Y, Wu G, Yang Y, Tang Y, Zhang W, et al. Long noncoding RNA PCAT-14 induces proliferation and invasion by hepatocellular carcinoma cells by inducing methylation of miR-372. Oncotarget (2017) 8(21):34429. doi: 10.18632/oncotarget.16260
130. Peng Y, Leng W, Duan S, Hong M. Long noncoding RNA BLACAT1 is overexpressed in hepatocellular carcinoma and its downregulation suppressed cancer cell development through endogenously competing against hsa-miR-485-5p. Biomed Pharmacother (2019) 116:109027. doi: 10.1016/j.biopha.2019.109027
131. Zhang L, He X, Jin T, Gang L, Jin Z. Long non-coding RNA DLX6-AS1 aggravates hepatocellular carcinoma carcinogenesis by modulating miR-203a/MMP-2 pathway. Biomed Pharmacother (2017) 96:884–91. doi: 10.1016/j.biopha.2017.10.056
132. Koo JI, Lee H-J, Jung JH, Im E, Kim J-H, Shin N, et al. The Pivotal Role of Long Noncoding RNA RAB5IF in the Proliferation of Hepatocellular Carcinoma via LGR5 Mediated β-Catenin and c-Myc Signaling. Biomolecules (2019) 9(11):718. doi: 10.3390/biom9110718
133. Xu J-H, Chang W-H, Fu H-W, Shu W-Q, Yuan T, Chen P. Upregulated long non-coding RNA LOC90784 promotes cell proliferation and invasion and is associated with poor clinical features in HCC. Biochem Biophys Res Commun (2017) 490(3):920–6. doi: 10.1016/j.bbrc.2017.06.141
134. Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du C, et al. Long non-coding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. Int J Mol Sci (2014) 15(3):4060–76. doi: 10.3390/ijms15034060
135. Cheng D, Deng J, Zhang B, He X, Meng Z, Li G, et al. LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine (2018) 36:159–70. doi: 10.1016/j.ebiom.2018.08.055
136. Wang W, Chen G, Wang B, Yuan Z, Liu G, Niu B, et al. Long non-coding RNA BZRAP1-AS1 silencing suppresses tumor angiogenesis in hepatocellular carcinoma by mediating THBS1 methylation. J Trans Med (2019) 17(1):1–15. doi: 10.1186/s12967-019-02145-6
137. Li Y, Guo D, Ren M, Zhao Y, Wang X, Chen Y, et al. Long non-coding RNA SNAI3-AS1 promotes the proliferation and metastasis of hepatocellular carcinoma by regulating the UPF1/Smad7 signalling pathway. J Cell Mol Med (2019) 23(9):6271–82. doi: 10.1111/jcmm.14513
138. Li S, Huang Y, Huang Y, Fu Y, Tang D, Kang R, et al. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res (2017) 36(1):1–12. doi: 10.1186/s13046-017-0519-z
139. Song W, Zhang J, Xia Q, Sun M. Down-regulated lncRNA TP73-AS1 reduces radioresistance in hepatocellular carcinoma via the PTEN/Akt signaling pathway. Cell Cycle (2019) 18(22):3177–88. doi: 10.1080/15384101.2019.1671089
140. Xiao J, Lv Y, Jin F, Liu Y, Ma Y, Xiong Y, et al. LncRNA HANR promotes tumorigenesis and increase of chemoresistance in hepatocellular carcinoma. Cell Physiol Biochem (2017) 43(5):1926–38. doi: 10.1159/000484116
141. Huang X, Gao Y, Qin J, Lu S. lncRNA MIAT promotes proliferation and invasion of HCC cells via sponging miR-214. Am J Physiology-Gastrointestinal Liver Physiol (2018) 314(5):G559–G65. doi: 10.1152/ajpgi.00242.2017
142. Zhao L, Hu K, Cao J, Wang P, Li J, Zeng K, et al. lncRNA miat functions as a ceRNA to upregulate sirt1 by sponging miR-22-3p in HCC cellular senescence. Aging (Albany NY) (2019) 11(17):7098. doi: 10.18632/aging.102240
143. Li B, Mao R, Liu C, Zhang W, Tang Y, Guo Z. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci (2018) 197:122–9. doi: 10.1016/j.lfs.2018.02.006
144. Huang Y, Xiang B, Liu Y, Wang Y, Kan H. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett (2018) 437:56–66. doi: 10.1016/j.canlet.2018.08.024
145. Zhuang H, Cao G, Kou C, Li D. Overexpressed lncRNA CDKN2B-AS1 is an independent prognostic factor for liver cancer and promotes its proliferation. J BU ON (2019) 24(4):1441–8.
146. Chen J, Huang X, Wang W, Xie H, Li J, Hu Z, et al. LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR-153-5p/ARHGAP18 signaling axis. Aging (Albany NY) (2018) 10(11):3371. doi: 10.18632/aging.101645
147. Pan W, Li W, Zhao J, Huang Z, Zhao J, Chen S, et al. lnc RNA-PDPK 2P promotes hepatocellular carcinoma progression through the PDK 1/AKT/Caspase 3 pathway. Mol Oncol (2019) 13(10):2246–58. doi: 10.1002/1878-0261.12553
148. Li X, Zhao Q, Qi J, Wang W, Zhang D, Li Z, et al. lncRNA Ftx promotes aerobic glycolysis and tumor progression through the PPARγ pathway in hepatocellular carcinoma. Int J Oncol (2018) 53(2):551–66. doi: 10.3892/ijo.2018.4418
149. Kong Q, Liang C, Jin Y, Pan Y, Tong D, Kong Q, et al. The lncRNA MIR4435-2HG is upregulated in hepatocellular carcinoma and promotes cancer cell proliferation by upregulating miRNA-487a. Cell Mol Biol Lett (2019) 24(1):26. doi: 10.1186/s11658-019-0148-y
150. Zhang W, Wu Y, Hou B, Wang Y, Deng D, Fu Z, et al. A SOX9-AS1/miR-5590-3p/SOX9 positive feedback loop drives tumor growth and metastasis in hepatocellular carcinoma through the Wnt/β-catenin pathway. Mol Oncol (2019) 13(10):2194–210. doi: 10.1002/1878-0261.12560
151. Wang H, Liang L, Dong Q, Huan L, He J, Li B, et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics (2018) 8(10):2814. doi: 10.7150/thno.23012
152. Zhang W-L, Zhao Y-N, Shi Z-Z, Gu G-Y, Cong D, Wei C, et al. HOXA11-AS promotes the migration and invasion of hepatocellular carcinoma cells by inhibiting miR-124 expression by binding to EZH2. Hum Cell (2019) 32(4):504–14. doi: 10.1007/s13577-019-00269-x
153. Zhang Y, Xu J, Zhang S, An J, Zhang J, Huang J, et al. HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition via the miR-520c-3p/GPC3 axis in hepatocellular carcinoma. Cell Physiol Biochem (2018) 50(6):2124–38. doi: 10.1159/000495056
154. Zhang X, Chen H, Zhou B, Zhang Q, Liao Y, Wang J, et al. lncRNA HOXB-AS3 promotes hepatoma by inhibiting p53 expression. Eur Rev Med Pharmacol Sci (2018) 22(20):6784–92. doi: 10.26355/eurrev_201810_16145
155. Xu X, Gu J, Ding X, Ge G, Zang X, Ji R, et al. LINC00978 promotes the progression of hepatocellular carcinoma by regulating EZH2-mediated silencing of p21 and E-cadherin expression. Cell Death Dis (2019) 10(10):1–15. doi: 10.1038/s41419-019-1990-6
156. Wang C-Z, Yan G-X, Dong D-S, Xin H, Liu Z-Y. LncRNA-ATB promotes autophagy by activating Yes-associated protein and inducing autophagy-related protein 5 expression in hepatocellular carcinoma. World J Gastroenterol (2019) 25(35):5310. doi: 10.3748/wjg.v25.i35.5310
157. Huang H, Chen J, Ding CM, Jin X, Jia ZM, Peng J. Lnc RNA NR 2F1-AS 1 regulates hepatocellular carcinoma oxaliplatin resistance by targeting ABCC 1 via miR-363. J Cell Mol Med (2018) 22(6):3238–45. doi: 10.1111/jcmm.13605
158. Guo D, Li Y, Chen Y, Zhang D, Wang X, Lu G, et al. DANCR promotes HCC progression and regulates EMT by sponging miR-27a-3p via ROCK1/LIMK1/COFILIN1 pathway. Cell Proliferation (2019) 52(4):e12628. doi: 10.1111/cpr.12628
159. Wang J, Pu J, Zhang Y, Yao T, Luo Z, Li W, et al. DANCR contributed to hepatocellular carcinoma malignancy via sponging miR-216a-5p and modulating KLF12. J Cell Physiol (2019) 234(6):9408–16. doi: 10.1002/jcp.27625
160. Zhang L, Wang Y, Sun J, Ma H, Guo C. LINC00205 promotes proliferation, migration and invasion of HCC cells by targeting miR-122-5p. Pathology-Research Pract (2019) 215(9):152515. doi: 10.1016/j.prp.2019.152515
161. Fan J, Zhang J, Huang S, Li P. lncRNA OSER1-AS1 acts as a ceRNA to promote tumorigenesis in hepatocellular carcinoma by regulating miR-372-3p/Rab23 axis. Biochem Biophys Res Commun (2020) 521(1):196–203. doi: 10.1016/j.bbrc.2019.10.105
162. Guo Y, Bai M, Lin L, Huang J, An Y, Liang L, et al. LncRNA DLEU2 aggravates the progression of hepatocellular carcinoma through binding to EZH2. Biomed Pharmacother (2019) 118:109272. doi: 10.1016/j.biopha.2019.109272
163. Huang J-L, Ren T-Y, Cao S-W, Zheng S-H, Hu X-M, Hu Y-W, et al. HBx-related long non-coding RNA DBH-AS1 promotes cell proliferation and survival by activating MAPK signaling in hepatocellular carcinoma. Oncotarget (2015) 6(32):33791. doi: 10.18632/oncotarget.5667
164. Bao J, Chen X, Hou Y, Kang G, Li Q, Xu Y. LncRNA DBH-AS1 facilitates the tumorigenesis of hepatocellular carcinoma by targeting miR-138 via FAK/Src/ERK pathway. Biomed Pharmacother (2018) 107:824–33. doi: 10.1016/j.biopha.2018.08.079
165. Wang J, Zhang Y, Lu L, Lu Y, Tang Q, Pu J. Insight into the molecular mechanism of LINC00152/miR-215/CDK13 axis in hepatocellular carcinoma progression. J Cell Biochem (2019) 120(11):18816–25. doi: 10.1002/jcb.29197
166. Li S-Q, Chen Q, Qin H-X, Yu Y-Q, Weng J, Mo Q-R, et al. Long intergenic nonprotein coding RNA 0152 promotes hepatocellular carcinoma progression by regulating phosphatidylinositol 3-Kinase/Akt/Mammalian target of rapamycin signaling pathway through miR-139/PIK3CA. Am J Pathol (2020) 190(5):1095–107. doi: 10.1016/j.ajpath.2019.11.010
167. Ma P, Wang H, Sun J, Liu H, Zheng C, Zhou X, et al. LINC00152 promotes cell cycle progression in hepatocellular carcinoma via miR-193a/b-3p/CCND1 axis. Cell Cycle (2018) 17(8):974–84. doi: 10.1080/15384101.2018.1464834
168. Lu X, Zhou C, Li R, Liang Z, Zhai W, Zhao L, et al. Critical role for the long non-coding RNA AFAP1-AS1 in the proliferation and metastasis of hepatocellular carcinoma. Tumor Biol (2016) 37(7):9699–707. doi: 10.1007/s13277-016-4858-8
169. Chen H, Yang F, Li X, Gong Z-J, Wang L-W. Long noncoding RNA LNC473 inhibits the ubiquitination of survivin via association with USP9X and enhances cell proliferation and invasion in hepatocellular carcinoma cells. Biochem Biophys Res Commun (2018) 499(3):702–10. doi: 10.1016/j.bbrc.2018.03.215
170. Li Y, Li Y, Xu X. The long noncoding RNA cardiac hypertrophy-related factor plays oncogenic roles in hepatocellular carcinoma by downregulating microRNA-211. J Cell Biochem (2019) 120(8):13361–71. doi: 10.1002/jcb.28611
171. Yang X, Cai JB, Peng R, Wei CY, Lu JC, Gao C, et al. The long noncoding RNA NORAD enhances the TGF-β pathway to promote hepatocellular carcinoma progression by targeting miR-202-5p. J Cell Physiol (2019) 234(7):12051–60. doi: 10.1002/jcp.27869
172. Qi H, Lu Y, Lv J, Wu H, Lu J, Zhang C, et al. The long noncoding RNA lncPARP1 contributes to progression of hepatocellular carcinoma through up-regulation of PARP1. Biosci Rep (2018) 38(3):BSR20180703. doi: 10.1042/BSR20180703
173. Li Y, Ye Y, Feng B, Qi Y. Long noncoding RNA lncARSR promotes doxorubicin resistance in hepatocellular carcinoma via modulating PTEN-PI3K/Akt pathway. J Cell Biochem (2017) 118(12):4498–507. doi: 10.1002/jcb.26107
174. Shin VY, Chen J, Cheuk IW-Y, Siu M-T, Ho C-W, Wang X, et al. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Disease (2019) 10(4):1–10. doi: 10.1038/s41419-019-1513-5
175. Peng W, Fan H. Long noncoding RNA CCHE1 indicates a poor prognosis of hepatocellular carcinoma and promotes carcinogenesis via activation of the ERK/MAPK pathway. Biomed Pharmacother (2016) 83:450–5. doi: 10.1016/j.biopha.2016.06.056
176. Jin W, Chen L, Cai X, Zhang Y, Zhang J, Ma D, et al. Long non-coding RNA TUC338 is functionally involved in sorafenib-sensitized hepatocarcinoma cells by targeting RASAL1. Oncol Rep (2017) 37(1):273–80. doi: 10.3892/or.2016.5248
177. Sui C-J, Zhou Y-M, Shen W-F, Dai B-H, Lu J-J, Zhang M-F, et al. Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. J Mol Med (2016) 94(11):1281–96. doi: 10.1007/s00109-016-1442-z
178. Huang G, Jiang H, Lin Y, Wu Y, Cai W, Shi B, et al. lncAKHE enhances cell growth and migration in hepatocellular carcinoma via activation of NOTCH2 signaling. Cell Death Dis (2018) 9(5):1–11. doi: 10.1038/s41419-018-0554-5
179. Ren W, Guan W, Zhang J, Wang F, Xu G. Pyridoxine 5′-phosphate oxidase is correlated with human breast invasive ductal carcinoma development. Aging (Albany NY) (2019) 11(7):2151. doi: 10.18632/aging.101908
180. Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene (2016) 35(12):1575–84. doi: 10.1038/onc.2015.223
181. Yu J, Ou Z, Lei Y, Chen L, Su Q, Zhang K. LncRNA MYCNOS facilitates proliferation and invasion in hepatocellular carcinoma by regulating miR-340. Hum Cell (2020) 33(1):148–58. doi: 10.1007/s13577-019-00303-y
182. Liu Z, Wang Y, Wang L, Yao B, Sun L, Liu R, et al. Long non-coding RNA AGAP2-AS1, functioning as a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16-5p and promotes proliferation and metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res (2019) 38(1):194. doi: 10.1186/s13046-019-1188-x
183. Tran D, Kessler C, Niehus S, Mahnkopf M, Koch A, Tamura T. Myc target gene, long intergenic noncoding RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and cell survival by titrating tumor suppressor microRNAs. Oncogene (2018) 37(1):75–85. doi: 10.1038/onc.2017.312
184. Tang YH, He GL, Huang SZ, Zhong KB, Liao H, Cai L, et al. The long noncoding RNA AK002107 negatively modulates miR-140-5p and targets TGFBR1 to induce epithelial–mesenchymal transition in hepatocellular carcinoma. Mol Oncol (2019) 13(5):1296–310. doi: 10.1002/1878-0261.12487
185. Li Y, Zhuang W, Huang M, Li X. Long noncoding RNA DDX11-AS1 epigenetically represses LATS2 by interacting with EZH2 and DNMT1 in hepatocellular carcinoma. Biochem Biophys Res Commun (2019) 514(4):1051–7. doi: 10.1016/j.bbrc.2019.05.042
186. Luo X, Zhou N, Wang L, Zeng Q, Tang H. Long Noncoding RNA GATA3-AS1 Promotes Cell Proliferation and Metastasis in Hepatocellular Carcinoma by Suppression of PTEN, CDKN1A, and TP53. Can J Gastroenterol Hepatol (2019) 2019:1389653. doi: 10.1155/2019/1389653
187. Zhang W, Liu S, Liu K, Liu Y. Long non-coding RNA deleted in lymphocytic leukaemia 1 promotes hepatocellular carcinoma progression by sponging miR-133a to regulate IGF-1R expression. J Cell Mol Med (2019) 23(8):5154–64. doi: 10.1111/jcmm.14384
188. Xu X, Yin Y, Tang J, Xie Y, Han Z, Zhang X, et al. Long non-coding RNA Myd88 promotes growth and metastasis in hepatocellular carcinoma via regulating Myd88 expression through H3K27 modification. Cell Death Dis (2017) 8(10):e3124–e. doi: 10.1038/cddis.2017.519
189. Tang T, Guo C, Xia T, Zhang R, Zen K, Pan Y, et al. LncCCAT1 promotes breast Cancer stem cell function through activating WNT/β-catenin signaling. Theranostics (2019) 9(24):7384. doi: 10.7150/thno.37892
190. Xu X, Lou Y, Tang J, Teng Y, Zhang Z, Yin Y, et al. The long non-coding RNA Linc-GALH promotes hepatocellular carcinoma metastasis via epigenetically regulating Gankyrin. Cell Death Dis (2019) 10(2):1–13. doi: 10.1038/s41419-019-1348-0
191. Ma M, Xu H, Liu G, Wu J, Li C, Wang X, et al. Metabolism-induced tumor activator 1 (MITA1), an Energy Stress–Inducible Long Noncoding RNA, Promotes Hepatocellular Carcinoma Metastasis. Hepatology (2019) 70(1):215–30. doi: 10.1002/hep.30602
192. Wang YL, Liu JY, Yang JE, Yu XM, Chen ZL, Chen YJ, et al. Lnc-UCID promotes G1/S transition and hepatoma growth by preventing DHX9-mediated CDK6 down-regulation. Hepatology (2019) 70(1):259–75. doi: 10.1002/hep.30613
193. Yang X, Yao B, Niu Y, Chen T, Mo H, Wang L, et al. Hypoxia-induced lncRNA EIF3J-AS1 accelerates hepatocellular carcinoma progression via targeting miR-122–5p/CTNND2 axis. Biochem Biophys Res Commun (2019) 518(2):239–45. doi: 10.1016/j.bbrc.2019.08.039
194. Fan H, Lv P, Mu T, Zhao X, Liu Y, Feng Y, et al. LncRNA n335586/miR-924/CKMT1A axis contributes to cell migration and invasion in hepatocellular carcinoma cells. Cancer Lett (2018) 429:89–99. doi: 10.1016/j.canlet.2018.05.010
195. Zhuang J, He S, Wang G, Wang G, Ni J, Zhang S, et al. Long Noncoding RNA FGFR3-AS1 Promotes Hepatocellular Carcinoma Carcinogenesis via Modulating the PI3K/AKT Pathway. Oncol Res Featuring Preclinical Clin Cancer Ther (2018) 26(8):1257–65. doi: 10.3727/096504018X15172756878992
196. Mo J, Li B, Zhou Y, Xu Y, Jiang H, Cheng X, et al. LINC00473 promotes hepatocellular carcinoma progression via acting as a ceRNA for microRNA-195 and increasing HMGA2 expression. Biomed Pharmacother (2019) 120:109403. doi: 10.1016/j.biopha.2019.109403
197. Gao J, Yin X, Yu X, Dai C, Zhou F. Long noncoding LINC01551 promotes hepatocellular carcinoma cell proliferation, migration, and invasion by acting as a competing endogenous RNA of microRNA-122-5p to regulate ADAM10 expression. J Cell Biochem (2019) 120(10):16393–407. doi: 10.1002/jcb.28549
198. Yu S, Li N, Huang Z, Chen R, Yi P, Kang R, et al. A novel lncRNA, TCONS_00006195, represses hepatocellular carcinoma progression by inhibiting enzymatic activity of ENO1. Cell Death Dis (2018) 9(12):1–13. doi: 10.1038/s41419-018-1231-4
199. Wang R, Jiang J, Jiang T, Wang Y, Chen L. Increased long noncoding RNA LINC00511 is correlated with poor prognosis and contributes to cell proliferation and metastasis by modulating miR-424 in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci (2019) 23(8):3291–301. doi: 10.26355/eurrev_201904_17691
200. Hu W-Y, Wei H-Y, Li K-M, Wang R-B, Xu X-Q, Feng R. LINC00511 as a ceRNA promotes cell malignant behaviors and correlates with prognosis of hepatocellular carcinoma patients by modulating miR-195/EYA1 axis. Biomed Pharmacother (2020) 121:109642. doi: 10.1016/j.biopha.2019.109642
201. Gong J, Qi X, Zhang Y, Yu Y, Lin X, Li H, et al. Long noncoding RNA linc00462 promotes hepatocellular carcinoma progression. Biomed Pharmacother (2017) 93:40–7. doi: 10.1016/j.biopha.2017.06.004
202. Chen Z, Zhou Z, He C, Zhang J, Wang J, Xiao Z. Down-regulation of LncRNA NR027113 inhibits cell proliferation and metastasis via PTEN/PI3K/AKT signaling pathway in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci (2018) 22(21):7222–32. doi: 10.26355/eurrev_201811_16256
203. Chen Z, Xu D, Zhang T. Inhibition of proliferation and invasion of hepatocellular carcinoma cells by lncRNA-ASLNC02525 silencing and the mechanism. Int J Oncol (2017) 51(3):851–8. doi: 10.3892/ijo.2017.4069
204. Zeng B, Lin Z, Ye H, Cheng D, Zhang G, Zhou J, et al. Upregulation of LncDQ is associated with poor prognosis and promotes tumor progression via epigenetic regulation of the EMT pathway in HCC. Cell Physiol Biochem (2018) 46(3):1122–33. doi: 10.1159/000488841
205. Wu J, Tian X, An Q, Guan X, Hao C. LINC00963 promotes hepatocellular carcinoma progression by activating PI3K/AKT pathway. Eur Rev Med Pharmacol Sci (2018) 22(6):1645–52. doi: 10.26355/eurrev_201803_14574
206. Chen J, Wu D, Zhang Y, Yang Y, Duan Y, An Y. LncRNA DCST1-AS1 functions as a competing endogenous RNA to regulate FAIM2 expression by sponging miR-1254 in hepatocellular carcinoma. Clin Sci (2019) 133(2):367–79. doi: 10.1042/CS20180814
207. Chen H, Liu J, Hu G, Shi L, Lan T. Promotion of proliferation and metastasis of hepatocellular carcinoma by LncRNA00673 based on the targeted-regulation of notch signaling pathway. Eur Rev Med Pharmacol Sci (2017) 21(15):3412–20.
208. Liu W, Huai R, Zhang Y, Rao S, Ding R, Mao C, et al. Down-regulation expression of TGFB2-AS1 inhibits the proliferation, migration, invasion and induces apoptosis in HepG2 cells. Genes Genomics (2019) 41(8):951–9. doi: 10.1007/s13258-019-00826-6
209. Zhang K, Zhao Z, Yu J, Chen W, Xu Q, Chen L. LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate cancer cell proliferation, migration, and invasion in hepatocellular carcinoma. J Cell Biochem (2018) 119(7):6045–56. doi: 10.1002/jcb.26802
210. Tu J, Zhao Z, Xu M, Chen M, Weng Q, Wang J, et al. LINC00707 contributes to hepatocellular carcinoma progression via sponging miR-206 to increase CDK14. J Cell Physiol (2019) 234(7):10615–24. doi: 10.1002/jcp.27737
211. Chen Z, Liu Y, Yao L, Guo S, Gao Y, Zhu P. The long noncoding RNA lncZic2 drives the self-renewal of liver tumor–initiating cells via the protein kinase C substrates MARCKS and MARCKSL1. J Biol Chem (2018) 293(21):7982–92. doi: 10.1074/jbc.RA117.001321
212. Li S, Sui M, Sun Z, Zhang W. LncRNA 00152 promotes the development of hepatocellular carcinoma by activating JAK2/STAT3 pathway. Eur Rev Med Pharmacol Sci (2019) 23(3):1038–46. doi: 10.26355/eurrev_201902_16991
213. Liu L, Zhu Y, Liu A, Feng Y, Chen Y. Long noncoding RNA LINC00511 involves in breast cancer recurrence and radioresistance by regulating STXBP4 expression via miR-185. Eur Rev Med Pharmacol Sci (2019) 23(17):7457–68. doi: 10.26355/eurrev_201909_18855
214. Ma J, Qin C, Yuan Z, Liu S. LncRNA PAPAS promotes hepatocellular carcinoma by interacting with miR-188-5p. J Cell Biochem (2019) 120(8):13494–500. doi: 10.1002/jcb.28623
215. Huang H, Bu YZ, Zhang XY, Liu J, Zhu LY, Fang Y. LINC01433 promotes hepatocellular carcinoma progression via modulating the miR-1301/STAT3 axis. J Cell Physiol (2019) 234(5):6116–24. doi: 10.1002/jcp.27366
216. Sun J, Zhang Y, Li B, Dong Y, Sun C, Zhang F, et al. PITPNA-AS1 abrogates the inhibition of miR-876-5p on WNT5A to facilitate hepatocellular carcinoma progression. Cell Death Dis (2019) 10(11):1–15. doi: 10.1038/s41419-019-2067-2
217. Tan N, Zhu B, Shu H, Tao YF, Wu JR, Fang M, et al. Effect of lncRNA−BC200 on proliferation and migration of liver cancer cells in vitro and in vivo. Oncol Rep (2020) 43(2):461–70. doi: 10.3892/or.2019.7447
218. Huang W, Liu J, Yan J, Huang Z, Zhang X, Mao Y, et al. LncRNA LINC00470 promotes proliferation through association with NF45/NF90 complex in hepatocellular carcinoma. Hum Cell (2020) 33(1):131–9. doi: 10.1007/s13577-019-00288-8
219. García-Venzor A, Mandujano-Tinoco EA, Lizarraga F, Zampedri C, Krötzsch E, Salgado RM, et al. Microenvironment-regulated lncRNA-HAL is able to promote stemness in breast cancer cells. Biochim Biophys Acta (BBA)-Molecular Cell Res (2019) 1866(12):118523. doi: 10.1016/j.bbamcr.2019.118523
220. Tu J, Zhao Z, Xu M, Chen M, Weng Q, Ji J. LINC00460 promotes hepatocellular carcinoma development through sponging miR-485-5p to up-regulate PAK1. Biomed Pharmacother (2019) 118:109213. doi: 10.1016/j.biopha.2019.109213
221. Hu M, Han Y, Zhang Y, Zhou Y, Ye L. lncRNA TINCR sponges miR-214-5p to upregulate ROCK1 in hepatocellular carcinoma. BMC Med Genet (2020) 21(1):1–6. doi: 10.1186/s12881-019-0940-6
222. Fen H, Hongmin Z, Wei W, Chao Y, Yang Y, Bei L, et al. RHPN1-AS1 drives the progression of hepatocellular carcinoma via regulating miR-596/IGF2BP2 axis. Curr Pharm Des (2019) 25(43):4630–40. doi: 10.2174/1381612825666191105104549
223. Ni W, Zhang Y, Zhan Z, Ye F, Liang Y, Huang J, et al. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J Hematol Oncol (2017) 10(1):91. doi: 10.1186/s13045-017-0449-4
224. Zhou CC, Yang F, Yuan SX, Ma JZ, Liu F, Yuan JH, et al. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology (2016) 63(3):850–63. doi: 10.1002/hep.28393
225. Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell (2013) 49(6):1083–96. doi: 10.1016/j.molcel.2013.01.010
226. Zhang J, Li Z, Liu L, Wang Q, Li S, Chen D, et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology (2018) 67(1):171–87. doi: 10.1002/hep.29405
227. Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang C, et al. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer (2017) 16(1):123. doi: 10.1186/s12943-017-0702-z
228. Qin G, Tu X, Li H, Cao P, Chen X, Song J, et al. Long Noncoding RNA p53-Stabilizing and Activating RNA Promotes p53 Signaling by Inhibiting Heterogeneous Nuclear Ribonucleoprotein K deSUMOylation and Suppresses Hepatocellular Carcinoma. Hepatology (2020) 71(1):112–29. doi: 10.1002/hep.30793
229. Ding H, Liu J, Zou R, Cheng P, Su Y. Long non-coding RNA TPTEP1 inhibits hepatocellular carcinoma progression by suppressing STAT3 phosphorylation. J Exp Clin Cancer Res (2019) 38(1):189. doi: 10.1186/s13046-019-1193-0
230. Sun J, Liu L, Zou H, Yu W. The Long Non-Coding RNA CASC2 Suppresses Cell Viability, Migration, and Invasion in Hepatocellular Carcinoma Cells by Directly Downregulating miR-183. Yonsei Med J (2019) 60(10):905–13. doi: 10.3349/ymj.2019.60.10.905
231. Gan Y, Han N, He X, Yu J, Zhang M, Zhou Y, et al. Long non-coding RNA CASC2 regulates cell biological behaviour through the MAPK signalling pathway in hepatocellular carcinoma. Tumor Biol (2017) 39(6):1010428317706229. doi: 10.1177/1010428317706229
232. Zhao L, Zhang Y, Zhang Y. Long noncoding RNA CASC2 regulates hepatocellular carcinoma cell oncogenesis through miR-362-5p/Nf-κB axis. J Cell Physiol (2018) 233(10):6661–70. doi: 10.1002/jcp.26446
233. Fan JC, Zeng F, Le YG, Xin L. LncRNA CASC2 inhibited the viability and induced the apoptosis of hepatocellular carcinoma cells through regulating miR-24-3p. J Cell Biochem (2018) 119(8):6391–7. doi: 10.1002/jcb.26479
234. Wang Y-G, Wang T, Shi M, Zhai B. Long noncoding RNA EPB41L4A-AS2 inhibits hepatocellular carcinoma development by sponging miR-301a-5p and targeting FOXL1. J Exp Clin Cancer Res (2019) 38(1):1–13. doi: 10.1186/s13046-019-1128-9
235. Cai K, Li T, Guo L, Guo H, Zhu W, Yan L, et al. Long non-coding RNA LINC00467 regulates hepatocellular carcinoma progression by modulating miR-9-5p/PPARA expression. Open Biol (2019) 9(9):190074. doi: 10.1098/rsob.190074
236. Wang X, Sun W, Shen W, Xia M, Chen C, Xiang D, et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J Hepatol (2016) 64(6):1283–94. doi: 10.1016/j.jhep.2016.01.019
237. Liu F, Yuan J, Huang J, Yang F, Wang T, Ma J, et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene (2016) 35(41):5422–34. doi: 10.1038/onc.2016.80
238. Chen C, Zheng Q, Kang W, Yu C. Long non-coding RNA LINC00472 suppresses hepatocellular carcinoma cell proliferation, migration and invasion through miR-93-5p/PDCD4 pathway. Clinics Res Hepatol Gastroenterol (2019) 43(4):436–45. doi: 10.1016/j.clinre.2018.11.008
239. Zhu P, Li Y, Li P, Zhang Y, Wang X. c-Myc induced the regulation of long non-coding RNA RHPN1-AS1 on breast cancer cell proliferation via inhibiting P53. Mol Genet Genomics (2019) 294(5):1219–29. doi: 10.1007/s00438-019-01572-w
240. Dong H, Zhang Y, Xu Y, Ma R, Liu L, Luo C, et al. Downregulation of long non-coding RNA MEG3 promotes proliferation, migration, and invasion of human hepatocellular carcinoma cells by upregulating TGF-β1. Acta Biochim Biophys Sinica (2019) 51(6):645–52. doi: 10.1093/abbs/gmz046
241. Xu F, Wang B, Liu M, Liu T, Zhang R. A long non-coding RNA TSLD8 inhibits hepatocellular carcinoma by stabilizing WWOX. Biochem Biophys Res Commun (2019) 516(2):526–32. doi: 10.1016/j.bbrc.2019.06.043
242. Wu J, Zhou X, Fan Y, Cheng X, Lu B, Chen Z. Long non-coding RNA 00312 downregulates cyclin B1 and inhibits hepatocellular carcinoma cell proliferation in vitro and in vivo. Biochem Biophys Res Commun (2018) 497(1):173–80. doi: 10.1016/j.bbrc.2018.02.049
243. Yao Z, Xiong Z, Li R, Liang H, Jia C, Deng M. Long non-coding RNA NRON is downregulated in HCC and suppresses tumour cell proliferation and metastasis. Biomed Pharmacother (2018) 104:102–9. doi: 10.1016/j.biopha.2018.05.006
244. Chen C-L, Tseng Y-W, Wu J-C, Chen G-Y, Lin K-C, Hwang S-M, et al. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials (2015) 44:71–81. doi: 10.1016/j.biomaterials.2014.12.023
245. Sun Q-M, Hu B, Fu P-Y, Tang W-G, Zhang X, Zhan H, et al. Long non-coding RNA 00607 as a tumor suppressor by modulating NF-κB p65/p53 signaling axis in hepatocellular carcinoma. Carcinogenesis (2018) 39(12):1438–46. doi: 10.1093/carcin/bgy113
246. Wang T-H, Lin Y-S, Chen Y, Yeh C-T, Huang Y-L, Hsieh T-H, et al. Long non-coding RNA AOC4P suppresses hepatocellular carcinoma metastasis by enhancing vimentin degradation and inhibiting epithelial-mesenchymal transition. Oncotarget (2015) 6(27):23342. doi: 10.18632/oncotarget.4344
247. Sui J, Yang X, Qi W, Guo K, Gao Z, Wang L, et al. Long non-coding RNA Linc-USP16 functions as a tumour suppressor in hepatocellular carcinoma by regulating PTEN expression. Cell Physiol Biochem (2017) 44(3):1188–98. doi: 10.1159/000485449
248. Sun X, Zheng G, Li C, Liu C. Long non−coding RNA Fer−1−like family member 4 suppresses hepatocellular carcinoma cell proliferation by regulating PTEN in vitro and in vivo. Mol Med Rep (2019) 19(1):685–92. doi: 10.3892/mmr.2018.9629
249. Wu J, Huang J, Wang W, Xu J, Yin M, Cheng N, et al. Long non-coding RNA Fer-1-like protein 4 acts as a tumor suppressor via miR-106a-5p and predicts good prognosis in hepatocellular carcinoma. Cancer Biomarkers (2017) 20(1):55–65. doi: 10.3233/CBM-170090
250. Wang X, Dong K, Jin Q, Ma Y, Yin S, Wang S. Upregulation of lncRNA FER1L4 suppresses the proliferation and migration of the hepatocellular carcinoma via regulating PI3K/AKT signal pathway. J Cell Biochem (2019) 120(4):6781–8. doi: 10.1002/jcb.27980
251. Peng C, Hu W, Weng X, Tong R, Cheng S, Ding C, et al. Over expression of long non-coding RNA PANDA promotes hepatocellular carcinoma by inhibiting senescence associated inflammatory factor IL8. Sci Rep (2017) 7(1):1–11. doi: 10.1038/s41598-017-04045-5
252. Bo C, Li X, He L, Zhang S, Li N, An Y. A novel long noncoding RNA HHIP-AS1 suppresses hepatocellular carcinoma progression through stabilizing HHIP mRNA. Biochem Biophys Res Commun (2019) 520(2):333–40. doi: 10.1016/j.bbrc.2019.09.137
253. Lin X-Q, Huang Z-M, Chen X, Wu F, Wu W. XIST induced by JPX suppresses hepatocellular carcinoma by sponging miR-155-5p. Yonsei Med J (2018) 59(7):816–26. doi: 10.3349/ymj.2018.59.7.816
254. Zhang G, Li H, Sun R, Li P, Yang Z, Liu Y, et al. Long non-coding RNA ZEB2-AS1 promotes the proliferation, metastasis and epithelial mesenchymal transition in triple-negative breast cancer by epigenetically activating ZEB2. J Cell Mol Med (2019) 23(5):3271–9. doi: 10.1111/jcmm.14213
255. Pan W, Zhang N, Liu W, Liu J, Zhou L, Liu Y, et al. The long noncoding RNA GAS8-AS1 suppresses hepatocarcinogenesis by epigenetically activating the tumor suppressor GAS8. J Biol Chem (2018) 293(44):17154–65. doi: 10.1074/jbc.RA118.003055
256. Hu B, Cai H, Zheng R, Yang S, Zhou Z, Tu J. Long non-coding RNA 657 suppresses hepatocellular carcinoma cell growth by acting as a molecular sponge of miR-106a-5p to regulate PTEN expression. Int J Biochem Cell Biol (2017) 92:34–42. doi: 10.1016/j.biocel.2017.09.008
257. Gao Y, Wang G, Zhang C, Lin M, Liu X, Zeng Y, et al. Long non-coding RNA linc-cdh4-2 inhibits the migration and invasion of HCC cells by targeting R-cadherin pathway. Biochem Biophys Res Commun (2016) 480(3):348–54. doi: 10.1016/j.bbrc.2016.10.048
258. Li Y, Han X, Li Q, Wang C, Lou Z, Wang X. Long noncoding RNA HOXD-AS1 induces epithelial-mesenchymal transition in breast cancer by acting as a competing endogenous RNA of miR-421. J Cell Biochem (2019) 120(6):10633–42. doi: 10.1002/jcb.28353
259. Sun W, Xu X, Jiang Y, Jin X, Zhou P, Liu Y, et al. Transcriptome analysis of luminal breast cancer reveals a role for LOL in tumor progression and tamoxifen resistance. Int J Cancer (2019) 145(3):842–56. doi: 10.1002/ijc.32185
260. Chang L, Li C, Lan T, Wu L, Yuan Y, Liu Q, et al. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol Med Rep (2016) 13(2):1541–50. doi: 10.3892/mmr.2015.4716
261. Hu L, Ye H, Huang G, Luo F, Liu Y, Liu Y, et al. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumor Biol (2016) 37(2):2691–702. doi: 10.1007/s13277-015-4111-x
262. Zhao P, Cui X, Zhao L, Liu L, Wang D. Overexpression of Growth-Arrest-Specific Transcript 5 Improved Cisplatin Sensitivity in Hepatocellular Carcinoma Through Sponging miR-222. DNA Cell Biol (2020) 39(4):724–32. doi: 10.1089/dna.2019.5282
263. Ge Z, Cheng Z, Yang X, Huo X, Wang N, Wang H, et al. Long noncoding RNA SchLAH suppresses metastasis of hepatocellular carcinoma through interacting with fused in sarcoma. Cancer Sci (2017) 108(4):653–62. doi: 10.1111/cas.13200
264. Yu X, Tang W, Yang Y, Tang L, Dai R, Pu B, et al. Long noncoding RNA NKILA enhances the anti-cancer effects of baicalein in hepatocellular carcinoma via the regulation of NF-κB signaling. Chem Biol Interactions (2018) 285:48–58. doi: 10.1016/j.cbi.2018.02.027
265. Zhang H-F, Li W, Han Y-D. LINC00261 suppresses cell proliferation, invasion and Notch signaling pathway in hepatocellular carcinoma. Cancer Biomarkers (2018) 21(3):575–82. doi: 10.3233/CBM-170471
266. Yan S, Tang Z, Chen K, Liu Y, Yu G, Chen Q, et al. Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J Exp Clin Cancer Res (2018) 37(1):214. doi: 10.1186/s13046-018-0853-9
267. Zheng Y-L, Li L, Jia Y-X, Zhang B-Z, Li J-C, Zhu Y-H, et al. LINC01554-mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR signaling pathway. Theranostics (2019) 9(3):796. doi: 10.7150/thno.28992
268. Mo M, Liu S, Ma X, Tan C, Wei L, Sheng Y, et al. A liver-specific lncRNA, FAM99B, suppresses hepatocellular carcinoma progression through inhibition of cell proliferation, migration, and invasion. J Cancer Res Clin Oncol (2019) 145(8):2027–38. doi: 10.1007/s00432-019-02954-8
269. Sheng N, Li Y, Qian R, Li Y. The clinical significance and biological function of lncRNA RGMB-AS1 in hepatocellular carcinoma. Biomed Pharmacother (2018) 98:577–84. doi: 10.1016/j.biopha.2017.12.067
270. Yan S, Shan X, Chen K, Liu Y, Yu G, Chen Q, et al. LINC00052/miR-101-3p axis inhibits cell proliferation and metastasis by targeting SOX9 in hepatocellular carcinoma. Gene (2018) 679:138–49. doi: 10.1016/j.gene.2018.08.038
271. Wang YG, Liu J, Shi M, Chen FX. LncRNA DGCR5 represses the development of hepatocellular carcinoma by targeting the miR-346/KLF14 axis. J Cell Physiol (2019) 234(1):572–80. doi: 10.1002/jcp.26779
272. Zhou Y, Huan L, Wu Y, Bao C, Chen B, Wang L, et al. LncRNA ID2-AS1 suppresses tumor metastasis by activating the HDAC8/ID2 pathway in hepatocellular carcinoma. Cancer Lett (2020) 469:399–409. doi: 10.1016/j.canlet.2019.11.007
273. Du J, Chen M, Liu J, Hu P, Guan H, Jiao X. Lncrna f11-as1 suppresses liver hepatocellular carcinoma progression by competitively binding with mir-3146 to regulate pten expression. J Cell Biochem (2019) 120(10):18457–64. doi: 10.1002/jcb.29163
274. Wu J, Shuang Z, Zhao J, Tang H, Liu P, Zhang L, et al. Linc00152 promotes tumorigenesis by regulating DNMTs in triple-negative breast cancer. Biomed Pharmacother (2018) 97:1275–81. doi: 10.1016/j.biopha.2017.11.055
275. Zheng Z-K, Pang C, Yang Y, Duan Q, Zhang J, Liu W-C. Serum long noncoding RNA urothelial carcinoma-associated 1: A novel biomarker for diagnosis and prognosis of hepatocellular carcinoma. J Int Med Res (2018) 46(1):348–56. doi: 10.1177/0300060517726441
276. Zhang J, Wei H, Yang H. Long noncoding RNA SNHG15, a potential prognostic biomarker for hepatocellular carcinoma. Eur Rev Med Pharmacol Sci (2016) 20(9):1720–4.
277. Tu Z-Q, Li R-J, Mei J-Z, Li X-H. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol (2014) 7(7):4303.
278. Fu C, Xu X, Lu W, Nie L, Yin T, Wu D. Increased expression of long non-coding RNA CCAT2 predicts poorer prognosis in patients with hepatocellular carcinoma. Medicine (2019) 98(42):e17412. doi: 10.1097/MD.0000000000017412
279. Wang Y, Jing W, Ma W, Liang C, Chai H, Tu J. Down-regulation of long non-coding RNA GAS5-AS1 and its prognostic and diagnostic significance in hepatocellular carcinoma. Cancer Biomarkers (2018) 22(2):227–36. doi: 10.3233/CBM-170781
280. Ma W, Wang H, Jing W, Zhou F, Chang L, Hong Z, et al. Downregulation of long non-coding RNAs JPX and XIST is associated with the prognosis of hepatocellular carcinoma. Clinics Res Hepatol Gastroenterol (2017) 41(2):163–70. doi: 10.1016/j.clinre.2016.09.002
281. Li S-Y, Wang H, Mai H-F, Li G-F, Chen S-J, Li G-S, et al. Down-regulated long non-coding RNA RNAZFHX4-AS1 suppresses invasion and migration of breast cancer cells via FAT4-dependent Hippo signaling pathway. Cancer Gene Ther (2019) 26(11):374–87. doi: 10.1038/s41417-018-0066-6
282. Zeng Z, Dong J, Li Y, Dong Z, Liu Z, Huang J, et al. The expression level and clinical significance of lncRNA X91348 in hepatocellular carcinoma. Artif Cells Nanomed Biotechnol (2019) 47(1):3067–71. doi: 10.1080/21691401.2019.1640228
283. Chen Q, Tian G, Wang C. Expression of lncRNA TCONS_00027978 in hepatocellular carcinoma and its influence on prognosis and survival. Eur Rev Med Pharmacol Sci (2017) 21(24):5655–60. doi: 10.26355/eurrev_201712_14009
284. Gu J-X, Zhang X, Miao R-C, Xiang X-H, Fu Y-N, Zhang J-Y, et al. Six-long non-coding RNA signature predicts recurrence-free survival in hepatocellular carcinoma. World J Gastroenterol (2019) 25(2):220. doi: 10.3748/wjg.v25.i2.220
285. Refai NS, Louka ML, Halim HY, Montasser I. Long non-coding RNAs (CASC2 and TUG1) in hepatocellular carcinoma: Clinical significance. J Gene Med (2019) 21(9):e3112. doi: 10.1002/jgm.3112
286. Zhao Q-J, Zhang J, Xu L, Liu F-F. Identification of a five-long non-coding RNA signature to improve the prognosis prediction for patients with hepatocellular carcinoma. World J Gastroenterol (2018) 24(30):3426. doi: 10.3748/wjg.v24.i30.3426
287. Shaker OG, Abdelwahed MY, Ahmed NA, Hassan EA, Ahmed TI, Abousarie MA, et al. Evaluation of serum long noncoding RNA NEAT and MiR-129-5p in hepatocellular carcinoma. IUBMB Life (2019) 71(10):1571–8. doi: 10.1002/iub.2096
288. Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong M, et al. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol (2015) 8(5):5395.
289. Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget (2015) 6(6):4505. doi: 10.18632/oncotarget.2934
290. Ma Y, Luo T, Dong D, Wu X, Wang Y. Characterization of long non-coding RNAs to reveal potential prognostic biomarkers in hepatocellular carcinoma. Gene (2018) 663:148–56. doi: 10.1016/j.gene.2018.04.053
291. Xu H, Chen Y, Dong X, Wang X. Serum exosomal long noncoding RNAs ENSG00000258332. 1 and LINC00635 for the diagnosis and prognosis of hepatocellular carcinoma. Cancer Epidemiol Prev Biomarkers (2018) 27(6):710–6. doi: 10.1158/1055-9965.EPI-17-0770
292. Chao Y, Zhou D. lncRNA-D16366 Is a potential biomarker for diagnosis and prognosis of hepatocellular carcinoma. Med Sci Monitor: Int Med J Exp Clin Res (2019) 25:6581. doi: 10.12659/MSM.915100
293. Habieb A, Matboli M, El-Tayeb H, El-Asmar F. Potential role of lncRNA-TSIX, miR-548-a-3p, and SOGA1 mRNA in the diagnosis of hepatocellular carcinoma. Mol Biol Rep (2019) 46(4):4581–90. doi: 10.1007/s11033-019-04810-x
294. Zeng Y-L, Guo Z-Y, Su H-Z, Zhong F-D, Jiang K-Q, Yuan G-D. Diagnostic and prognostic value of lncRNA cancer susceptibility candidate 9 in hepatocellular carcinoma. World J Gastroenterol (2019) 25(48):6902. doi: 10.3748/wjg.v25.i48.6902
295. Luo P, Liang C, Zhang X, Liu X, Wang Y, Wu M, et al. Identification of long non-coding RNA ZFAS1 as a novel biomarker for diagnosis of HCC. Biosci Rep (2018) 38(4):BSR20171359. doi: 10.1042/BSR20171359
296. Wang C, Ren T, Wang K, Zhang S, Liu S, Chen H, et al. Identification of long non-coding RNA p34822 as a potential plasma biomarker for the diagnosis of hepatocellular carcinoma. Sci China Life Sci (2017) 60(9):1047. doi: 10.1007/s11427-017-9054-y
297. Xie Z, Zhou F, Yang Y, Li L, Lei Y, Lin X, et al. Lnc-PCDH9-13: 1 is a hypersensitive and specific biomarker for early hepatocellular carcinoma. EBioMedicine (2018) 33:57–67. doi: 10.1016/j.ebiom.2018.06.026
298. Liu X-F, Thin KZ, Ming X-L, -Li S, -Luo P, -Zhu M, et al. Small nucleolar RNA host gene 18 acts as a tumor suppressor and a diagnostic indicator in hepatocellular carcinoma. Technol Cancer Res Treat (2018) 17:1533033818794494. doi: 10.1177/1533033818794494
299. Gao S, Xu X, Wang Y, Zhang W, Wang X. Diagnostic utility of plasma lncRNA small nucleolar RNA host gene 1 in patients with hepatocellular carcinoma. Mol Med Rep (2018) 18(3):3305–13. doi: 10.3892/mmr.2018.9336
300. Zhu S, Huang X, Zhang K, Tan W, Lin Z, He Q, et al. Low expression of long noncoding RNA CTC-297N7. 9 predicts poor prognosis in patients with hepatocellular carcinoma. Cancer Med (2019) 8(18):7679–92. doi: 10.1002/cam4.2618
301. Motawi TM, El-Maraghy SA, Sabry D, Mehana NA. The expression of long non coding RNA genes is associated with expression with polymorphisms of HULC rs7763881 and MALAT1 rs619586 in hepatocellular carcinoma and HBV Egyptian patients. J Cell Biochem (2019) 120(9):14645–56. doi: 10.1002/jcb.28726
302. Tao R, Hu S, Wang S, Zhou X, Zhang Q, Wang C, et al. Association between indel polymorphism in the promoter region of lncRNA GAS5 and the risk of hepatocellular carcinoma. Carcinogenesis (2015) 36(10):1136–43. doi: 10.1093/carcin/bgv099
303. Wan J, Huang M, Zhao H, Wang C, Zhao X, Jiang X, et al. A novel tetranucleotide repeat polymorphism within KCNQ1OT1 confers risk for hepatocellular carcinoma. DNA Cell Biol (2013) 32(11):628–34. doi: 10.1089/dna.2013.2118
304. O’Brien A, Zhou T, Tan C, Alpini G, Glaser S. Role of Non-Coding RNAs in the Progression of Liver Cancer: Evidence from Experimental Models. Cancers (Basel) (2019) 11(11):1652. doi: 10.3390/cancers11111652
305. Zhang H, Xing Z, Mani SKK, Bancel B, Durantel D, Zoulim F, et al. RNA helicase DEAD box protein 5 regulates Polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology (2016) 64(4):1033–48. doi: 10.1002/hep.28698
306. Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP, Gao YF. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat Commun (2016) 7:12598. doi: 10.1038/ncomms12598
307. Zhang H-T, Zeng L-F, He Q-Y, Tao WA, Zha Z-G, Hu C-D. The E3 ubiquitin ligase CHIP mediates ubiquitination and proteasomal degradation of PRMT5. Biochim Biophys Acta (BBA)-Molecular Cell Res (2016) 1863(2):335–46. doi: 10.1016/j.bbamcr.2015.12.001
308. Ding C-H, Yin C, Chen S-J, Wen L-Z, Ding K, Lei S-J, et al. The HNF1α-regulated lncRNA HNF1A-AS1 reverses the malignancy of hepatocellular carcinoma by enhancing the phosphatase activity of SHP-1. Mol Cancer (2018) 17(1):1–14. doi: 10.1186/s12943-018-0813-1
309. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell (2018) 172(3):393–407. doi: 10.1016/j.cell.2018.01.011
Keywords: lncRNA, biomarker, hepatocellular carcinoma, expression, polymorphism
Citation: Ghafouri-Fard S, Gholipour M, Hussen BM and Taheri M (2021) The Impact of Long Non-Coding RNAs in the Pathogenesis of Hepatocellular Carcinoma. Front. Oncol. 11:649107. doi: 10.3389/fonc.2021.649107
Received: 03 January 2021; Accepted: 22 March 2021;
Published: 21 April 2021.
Edited by:
Xing-Xing He, Huazhong University of Science and Technology, ChinaReviewed by:
Ying Chang, Wuhan University, ChinaMin Wang, Huazhong University of Science and Technology, China
Copyright © 2021 Ghafouri-Fard, Gholipour, Hussen and Taheri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==
 Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard Mahdi Gholipour
Mahdi Gholipour Bashdar Mahmud Hussen3
Bashdar Mahmud Hussen3 Mohammad Taheri
Mohammad Taheri