- 1Department of Gastroenterology, Affiliated Hospital of Xuzhou Medical University, Xuzhou, China
- 2Institute of Digestive Diseases, Xuzhou Medical University, Xuzhou, China
- 3Zhejiang University Kunshan Biotechnology Laboratory, Zhejiang University Kunshan Innovation Institute, Kunshan, China
- 4State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, China
- 5Department of R&D, Suzhou VersaBio Technologies Co. Ltd., Kunshan, China
- 6Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, Suzhou, China
Background: Early detection of colorectal cancer (CRC) and precancerous lesion is vitally important for mitigating CRC morbidity and mortality. Aberrant DNA methylations in certain promoter regions have been identified to be closely associated with CRC development and progression, suggesting their potential as diagnostic biomarkers for early detection. In this study, we evaluated the performance of methylated CLIP4 in stool specimens as a potential biomarker for CRC detection.
Methods: A total of 321 subjects out of 365 enrolled participants were included in the final analysis, including 154 CRC patients, 23 advanced adenoma (AA) patients, 49 small polyp (SP) patients, and 95 healthy controls. CLIP4 methylation level was examined by qPCR with bisulfite converted DNA purified from approximately 5 g stool specimen.
Results: Methylated CLIP4 test showed high sensitivities of 78.3% (95% CI: 55.8%–91.7%) and 90.3% (95% CI: 84.2%–94.3%) for detecting AA and CRC, respectively, with a specificity of 88.4% (95% CI: 79.8%–93.8%). CLIP4 methylation level discriminated AA and CRC patients from control subjects with area under the curve values of 0.892 (95% CI: 0.795–0.988) and 0.961 (95% CI: 0.938–0.983). Further analysis indicated no significant difference in sensitivities among different ages, genders, stages, locations, sides, tumor sizes and differentiation statuses.
Conclusions: Methylated CLIP4 showed a strong potential as a noninvasive biomarker for early CRC detection.
Background
Colorectal cancer (CRC) remains the third most commonly diagnosed cancer types, and the second most common cause of cancer-related deaths worldwide in 2020 (1). In China, new CRC cases and death in 2020 were 555,477 and 286,162, accounting for approximately 29% of the global disease burden. Rankings of CRC rose from the fifth most common cancer before 2015 to the second in both sexes. Incidence rate of CRC in China exhibited a substantial upward trend in the past decades, and age-standardized mortality rate also endured an upward swing (2). Meanwhile, significant and sustained declines in both incidence and mortality for adults over 50 years old have occurred in the United States, owing to increased awareness of screening, especially by colonoscopy (3, 4).
Multiple CRC screening methods have been developed over the years, and each has its own advantages and disadvantages. For adults over 50 years old in the US, routine fecal occult blood test (FOBT) or fecal immunochemical test (FIT) (5), sigmoidoscopy, colonoscopy, computed tomography colonography or stool DNA (sDNA) test are recommended for CRC screening (3, 4). Similar screening methods are recommended for high-risk group over 40 years old and low-to-average-risk group over 50 in China (6). As the gold standard, colonoscopy has higher sensitivity and specificity than stool-based tests, especially for precancerous lesion and early stage CRC. However, the population coverage in China is still insufficient. The compliance rates for colonoscopy and FOBT remained at low levels of 4.01% and 11.01%, respectively, even in Shanghai, one of the most developed Chinese cities in the past decade (7–9). A large screening campaign of 182,927 participants with high-risk for CRC from 16 Chinese provinces only increased the compliance rate for colonoscopy to 14.0%. History of FOBT or colonic polyp, family history of CRC and high level of education were found to be associated with the increased participation (10).
In the meantime, existing stool-based tests providing non-invasive and high-compliance alternatives bear their own drawbacks. FOBT or FIT for fecal hemoglobin detection is affordable, but their performance is unsatisfactory due to low sensitivity in detecting advanced colorectal neoplasia. Cologuard, the first stool-based CRC screening test approved by the US Food and Drug Administration (FDA), demonstrated relatively high sensitivity and specificity (11). However, a high list price of $649 due to its complex operations associated with multiple assays per test makes it difficult to promote among uninsured and/or low- and moderate-income population. Therefore, intensive efforts have been made to develop more accurate and cost-effective screening tests.
DNA methylation is an epigenetic mechanism of gene regulation. Aberrant DNA methylation has been observed in all cancer types including CRC (12). Therefore, it has emerged as a class of important biomarkers with more diagnostic values than mutation markers for early CRC detection (13). A number of methylated genes have been proposed as CRC biomarker candidates in previous studies. Several among them have been incorporated into one-marker or multi-marker commercial tests, such as methylated SEPT9, SDC2, SFRP2, VIM, BMP3, and NDRG4. Another new candidate, CLIP4, is a member of CAP-Gly domain containing linker protein (CLIP) family involved in plus-end binding of microtubule, and has been implicated in immune response-related biological processes, cell migration and viability in certain cancer metastases (14, 15). Hypermethylation of CLIP4 in plasma has been shown for cancer types such as CRC and gastric cancer. Further studies in multiplex blood-based methylation tests validated its potential as another promising biomarker for CRC (16–19). However, the performance of methylated CLIP4 (mCLIP4) in stool samples for CRC detection has never been reported. The aim of this study was to evaluate the feasibility of stool mCLIP4 as a biomarker for early CRC detection.
Materials and Methods
Sample Collection
The original plan was to perform stool mCLIP4 test on 400 participants at the Affiliated Hospital of Xuzhou Medical University, comprising 200 CRC patients, 100 polyp patients and 100 subjects with no evidence of disease (NED). The inclusion criteria consisted of the following: 18 years old or older, no history of CRC, no pregnant woman; and all participants must have undergone complete colonoscopies by trained physicians. Standard operation was followed for all colonoscopy examinations where endoscope reached cecum. Participants with abnormal colonoscopy results should have pathological diagnoses. Pathology analysis was first done independently by two trained pathologists. If both agreed on the same diagnosis, no further evaluation was needed. If their diagnoses did not agree, evaluation by a third pathologist was required for the final determination of diagnosis. All pathologists involved were at or above the level of associated chief pathologist. During stool sample collection, transferring urine into the collection tube was avoided, and no diarrhea sample was collected. All samples were collected before purgative bowel preparation for colonoscopy. Approximately 5 g of solid specimen was collected from whole stool and preserved in 25 mL of preservative buffer (Suzhou VersaBio Technologies Co., Ltd., Kunshan, China) in a 50 mL tube to stabilize human genomic DNA. Stool samples were stored at room temperature for at most 7 days before being transferred to −80°C for long-term preservation and storage.
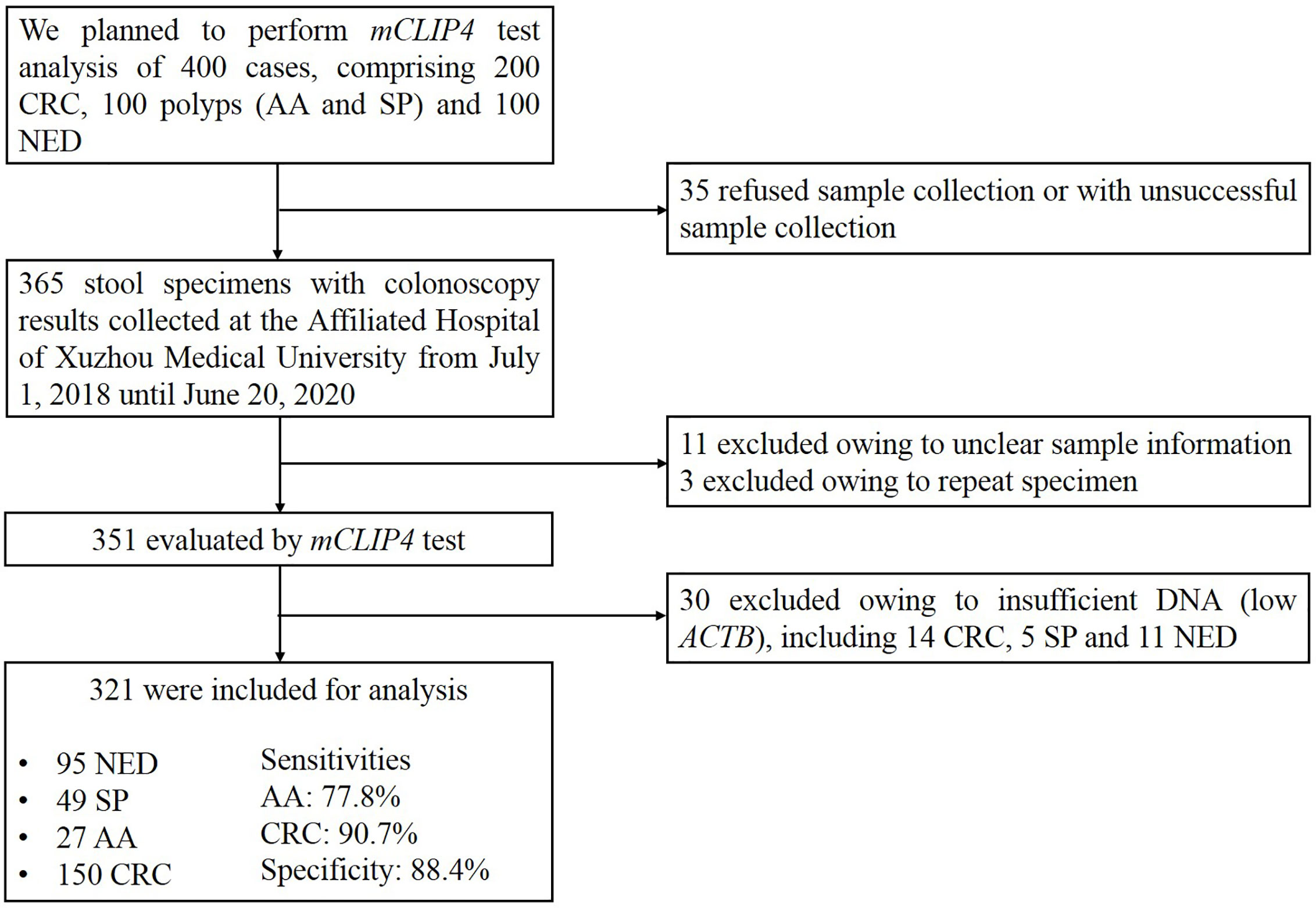
Until the submission of this manuscript, 365 stool specimens were collected, among which 11 were excluded due to insufficient sample information and another three were excluded due to repeated sampling. Of the remaining 351 specimens evaluated by mCLIP4 test, 30 samples were excluded due to insufficient DNA indicated by low ACTB levels (see data analysis). As a result, the final analysis included 321 specimens collected from 154 CRC patients, 23 patients with advanced adenomas (AA, an adenoma measuring ≥ 10 mm in size, with high-grade dysplasia, or with ≥ 25% villous features), 49 with small polyps (SP, non-advanced adenoma or hyperplastic polyp) and 95 NED control subjects (Figure 1).
Fresh-frozen CRC tissues (n=28) and paired adjacent paracancerous tissues (n=28) were collected at the time of surgery at the Affiliated Hospital of Xuzhou Medical University. The details of age and gender distribution of tissue samples were described in Supplementary Table 1. All tissue samples were stored at −80°C until use.
This study was performed according to the principles of the Helsinki Declaration and approved by the Institutional Review Board of the Affiliated Hospital of Xuzhou Medical University (Ethics Committee reference number: XYKY2020-KL156-01). All participants have acknowledged and signed the informed consent.
DNA Extraction, Bisulfite Treatment and Quantitative Real-Time PCR
Tissue genomic DNA was extracted with a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol, and purified DNA was eluted into 200 μl Buffer AE. DNA concentration was quantified with an Invitrogen NanoDrop One Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts).
Stool specimens were first thawed for approximately 30 min at 15°C to 30°C and homogenized for 1 min on a shaking device. After centrifugation at 10,000 g for 20 min, human genomic DNA was isolated with a stool DNA extraction kit (Suzhou VersaBio Technologies Co., Ltd.) from 150 μl supernatant. Bisulfite conversion of the extracted DNA and purification of the converted DNA were performed with a bisulfite conversion kit (Suzhou VersaBio Technologies Co., Ltd.). Both kits were used according to previously published protocols (20).
Converted and purified DNA was then tested by a duplex qPCR assay. Tissue genomic DNA was tested in a single PCR reaction, and stool DNA was tested in three PCR replicates for mCLIP4 and an internal control (ACTB). The primers and probes used for mCLIP4 qPCR test were showed in Supplementary Table 2. The total reaction volume was 30 μl including 15 μl DNA. qPCR was performed on an ABI 7500 instrument (Applied Biosystems, Foster City, CA, USA) under the following conditions: initial activation at 95°C for 20 min, followed by 50 cycles at 95°C for 10 sec, 60°C for 30 sec and 72°C for 15 sec, and a final cooling to 40°C for 30 sec.
The target sequences of HCT116 and Jurkat genomic DNA were determined by bisulfite Sanger sequencing. Bisulfite-treated DNA was amplified with primers flanking the target region (Supplementary Table 2), and the expected size of the PCR product was 263 bp. PCR cycling conditions were as follows: initial activation at 95°C for 15 min, followed by 50 cycles at 95°C for 1 min, 58°C for 45 sec and 72°C for 30 sec, and a final cooling to 4°C for 1 min. PCR products were excised from agarose gels and purified with AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Hangzhou, China). Purified PCR products were ligated into pUCM-T vector (Sangon Biotech) at 16°C overnight and then transformed into competent E. coli cells (Tiangen Biotech, Beijing, China) according to the manufacturers’ instructions. Randomly selected transformants were subsequently sent for Sanger sequencing with the canonical MF-13 primer by Genewiz, Inc. (Suzhou, China). The agarose gel electrophoresis and Sanger sequencing results were shown in Supplementary Figures 1 and 2.
Limit of Detection of mCLIP4 Assay
Limit of detection (19) of mCLIP4 test was evaluated with a series of mixtures between fully methylated HCT116 genomic DNA and unmethylated Jurkat genomic DNA at different ratios (0, 6.75, 12.5, 25, 50, 60, and 70 pg fully methylated DNA out of 70 pg total DNA per qPCR reaction). The test was performed in 24 replicates for each mixture. Genomic DNA concentration was measured by an Invitrogen Qubit fluorometer and a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, Massachusetts).
Data Analysis
Ct values of ACTB and mCLIP4 were obtained to validate sample processing and to determine whether mCLIP4 was detected, respectively. The results for stool specimens were considered ‘valid’ if all three replicate reactions for ACTB produced amplification signals and the mean Ct value was less than 40.0. To be scored positive by 3/3 algorithm, all three replicate mCLIP4 PCR reactions of a stool sample must have valid amplification curves and mean Ct value must be less than 39.0. Sensitivity was defined as the positive detection rate of CRC or AA and specificity was defined as 100% minus the positive detection rate of NED.
All statistical analyses were performed with IBM SPSS for Windows Version 22.0. Pearson chi-square test for sensitivity comparisons among groups was performance at a significant level of p < 0.05. And the differences in methylation levels were analyzed using the Mann–Whitney U test. ΔCt was used to determine the methylation levels of CLIP4 in tissue samples. It was defined as the difference between the Ct values of the target (mCLIP4) and the internal control gene (ACTB) to normalize for DNA amounts of tissue samples. Mean Ct values from individuals in CRC, AA and control groups were used to plot the receiver operating characteristic (21) curves and to calculate the area under the curve (22) values. Ct values of reactions returning no amplification signals were set to 50 (the maximal number of PCR cycles) for the analysis (23). Mean Ct values were also used to represent the methylation level of each plasma sample.
Results
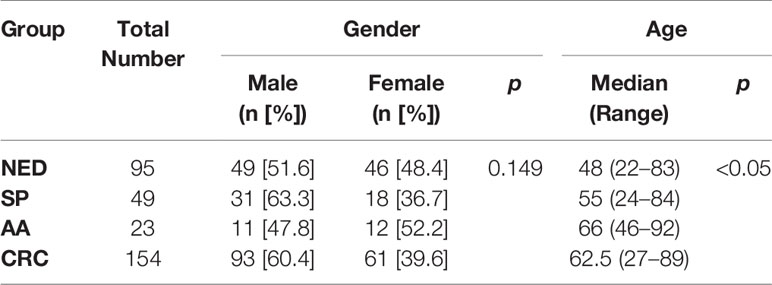
Twenty-eight colorectal cancer and paired adjacent paracancerous tissues were collected, including 16 males of 33 to 78 years old (Supplementary Table 1). Stool specimens were evaluated by mCLIP4 test for 321 subjects (Table 1), including 95 control (NED) subjects, 49 SP patients, 23 AA patients, and 154 CRC patients at median ages of 48, 55, 66 and 62.5, respectively. The CRC patients included 4 Stage 0, 26 Stage I, 48 Stage II, 48 Stage III, 10 Stage IV and 18 patients of unknown stage (Supplementary Table 4). Fifty-one point six percent of NED subjects and 60.4% of CRC patients were males. Across different groups, there was no significant difference for gender distribution, whereas age distribution showed significant difference (Table 1).
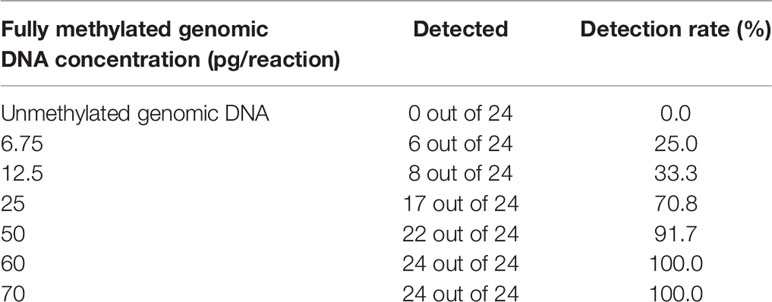
To evaluate the analytical performance of mCLIP4 test, a series of genomic DNA solutions of different methylation levels were tested in 24 replicates. As shown in Table 2, mCLIP4 test was able to detect as low as 6.75 pg fully methylated genomic DNA (~2 copies of human genome) per PCR reaction. Defined as the concentration at which more than 95% of the replicates generated amplification signals (24), LoD of mCLIP4 test was approximately 60 pg (~18 copies of human genome) per PCR reaction.
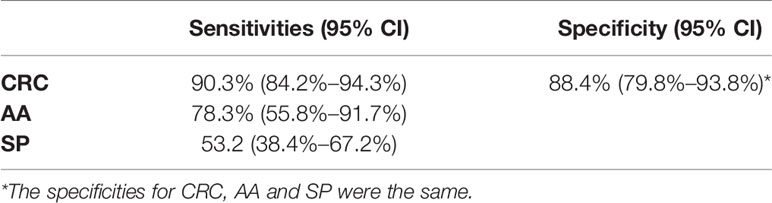
The results of tissue samples indicated that mCLIP4 levels in all cancer tissues were higher than those in their paired adjacent paracancerous tissues (p < 0.0001, Figure 2). Out of 321 subjects diagnosed by colonoscopy and further confirmed for CRC patients by pathological analysis of surgically resected specimens, mCLIP4 was detected in 11.6% of NED (11/95), 53.1% of SP (26/49), 78.3% of AA (18/23) stool specimens, as well as 75.0% of Stage 0 (3/4), 96.2% of Stage I (25/26), 95.8% of Stage II (46/48), 83.1% of Stage III (40/48), 100% of Stage IV (10/10), and 83.3% CRC samples of unknown stage (15/18) (Supplementary Table 4). The overall sensitivities for detecting AA and CRC by mCLIP4 test were 78.3% (95% CI: 55.8%–91.7%) and 90.3% (95% CI: 84.2%–94.3%), respectively, with a specificity of 88.4% (95% CI: 79.8%–93.8%) (Table 3). As shown in Figure 3, mean Ct value of each group represented the average methylation level, and a lower Ct value indicated a higher methylation level. Stool mCLIP4 levels of NED were significantly lower than those of patients with intestinal lesions, including SPs, AAs, and CRCs (p < 0.0001). There were no significant differences in mCLIP4 levels between samples of SP and stage 0 CRC patients (p > 0.05), whereas Stage I-IV CRC patient samples showed significantly higher mCLIP4 levels than those of SP patients. Differences in stool mCLIP4 levels between AA and stage III-IV CRC patients were not significant, but stage I-II CRC patients showed significantly higher mCLIP4 levels than those of AA patients. Furthermore, ROC curves of mCLIP4 test for AA and CRC detection demonstrated its ability to discriminate AA and CRC from controls with AUC values of 0.892 (95% CI: 0.795–0.988) and 0.961 (95% CI: 0.938–0.983) (Figure 4).
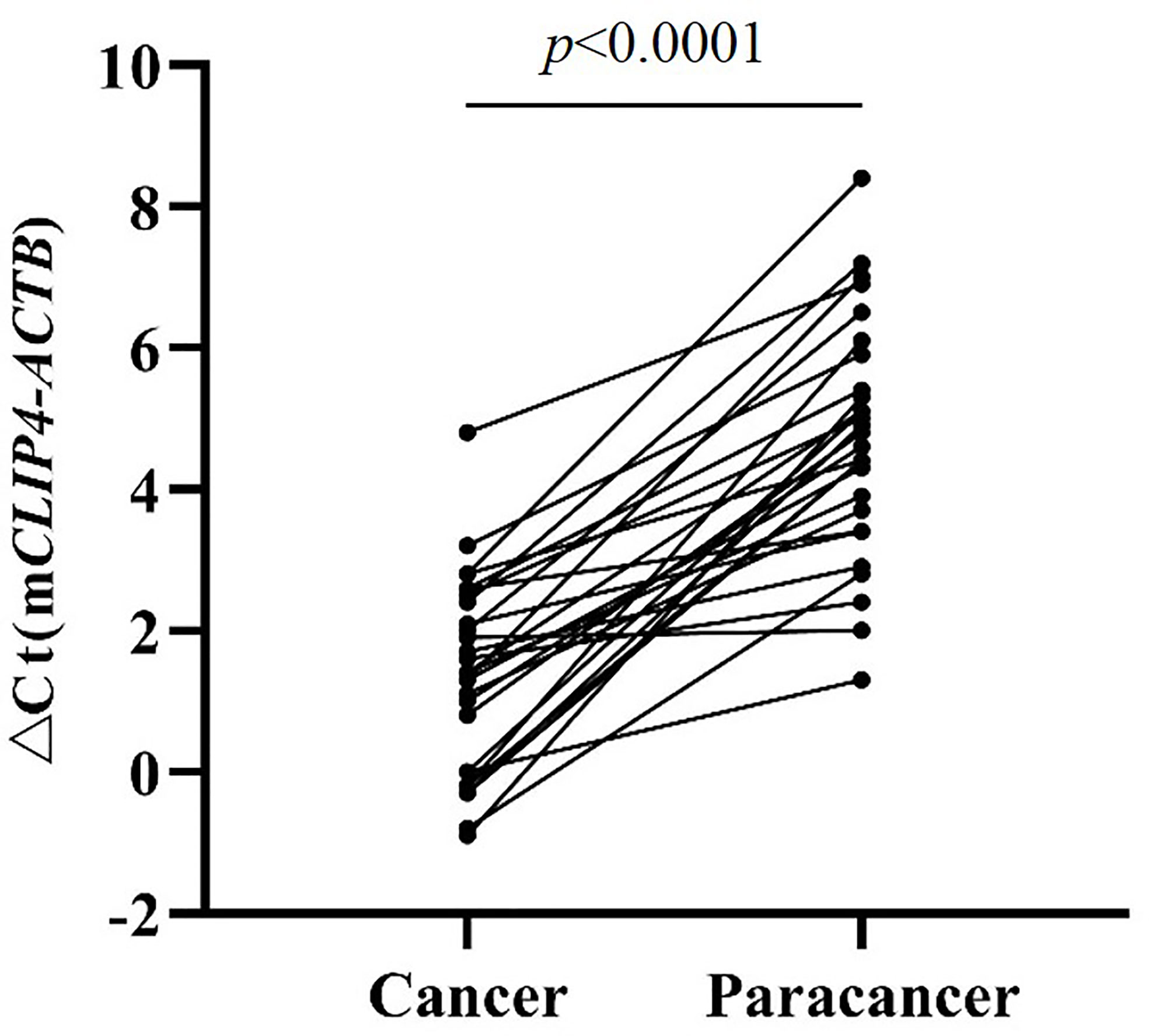
Figure 2 The Methylation levels of CLIP4 gene in CRC tissues and paired adjacent paracancerous tissues.
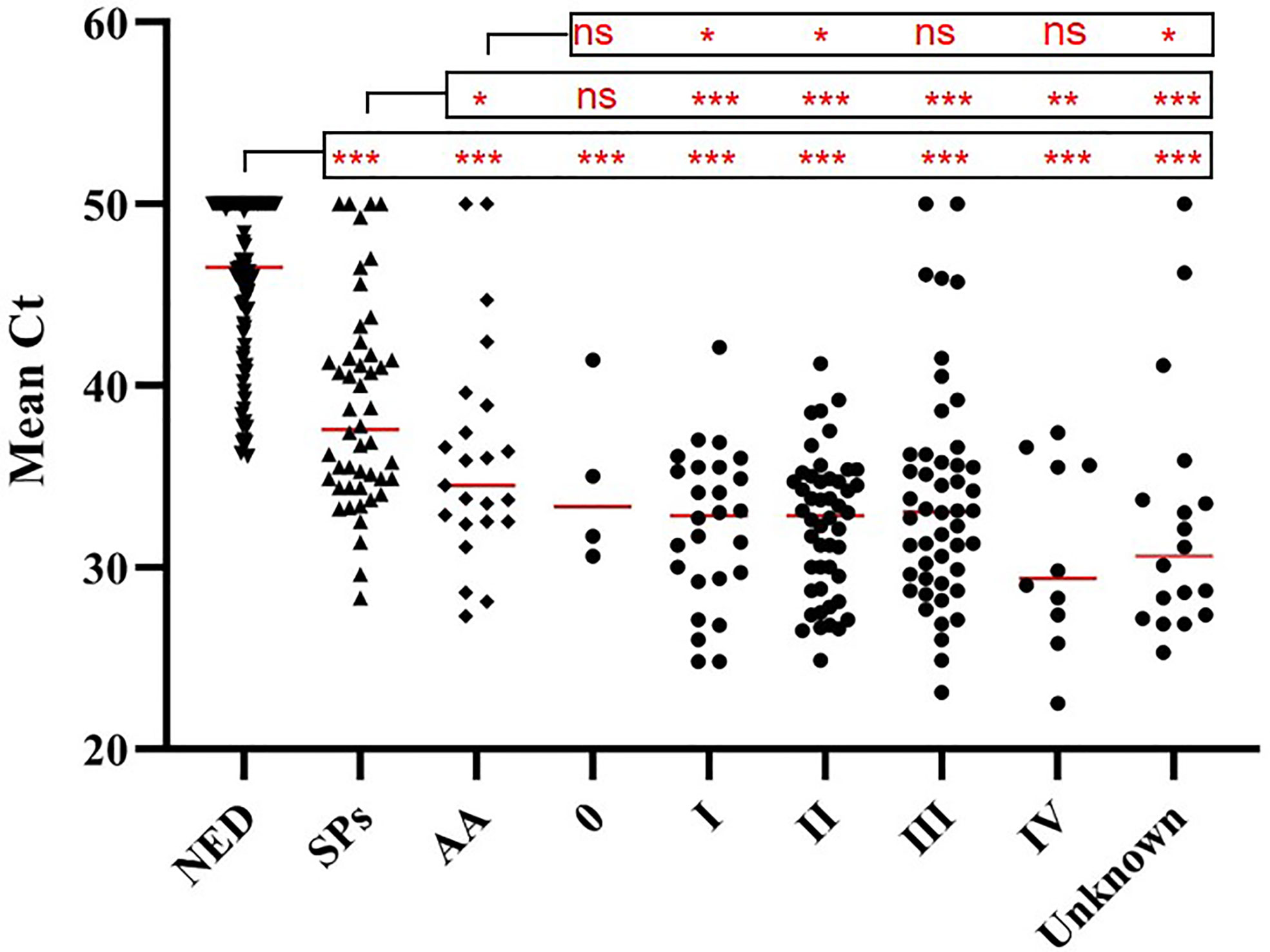
Figure 3 Methylation levels (mean Ct values) of CLIP4 gene in stool samples from NED subjects, as well as SP, AA and CRC patients of different stages. Horizontal red bars denote the median value of mean Ct values of all samples within the group. ns, not significant. *p < 0.05, **p < 0.01, ***p < 0.0001.
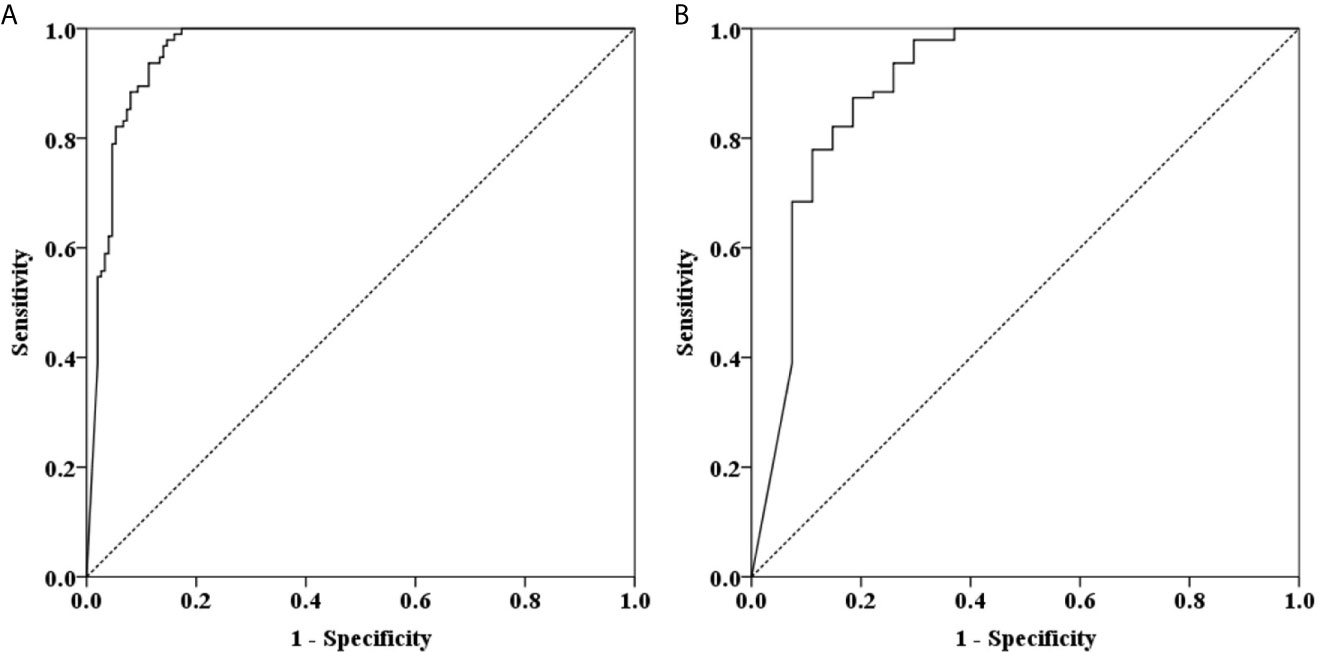
Figure 4 ROC curves for stool mCLIP4 test for detecting AA (A) and CRC (B). AUC values for AA (A) and CRC (B) were 0.892 (95% CI: 0.795–0.988) and 0.961 (95% CI: 0.938–0.983), respectively.
Further analysis showed no significant sensitivity difference among different age groups, genders, stages, locations, sides, tumor sizes and differentiation statuses (p > 0.05, Table 4).
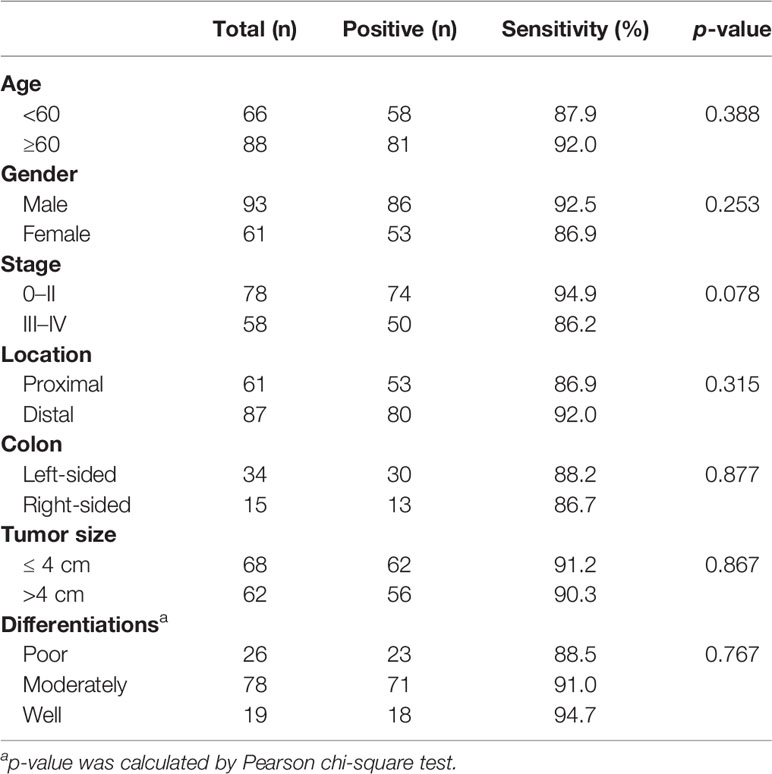
Table 4 Sensitivities of stool mCLIP4 test for detecting CRC for different age groups, genders, stages, tumor locations, sides, tumor sizes and differentiation statuses.
Discussion
Appropriate screening and surveillance for precancerous lesion and early stage CRC can significantly mitigate CRC mortality, and AA is the preferred target stage. Coverage of guideline-recommended screening in the US has increased to 67%: approximately 61% and 11% of US adults over 50 underwent a colonoscopy or a stool test, respectively, contributing to substantial reduction in morbidity and mortality (4). However, the invasiveness of colonoscopy and limited medical resource per capita have resulted in low compliance rate especially in average-risk and young adults in most countries. In this study, we provided a convenient stool DNA (mCLIP4) test as an alternative screening and potential surveillance method.
The cost-effectiveness of FIT and gFOBT, two guideline-compliant screening methods, has been intensively investigated. Previous studies showed the sensitivities of selected commercial FIT and high-sensitivity gFOBT kits for detecting advanced colorectal neoplasia and CRC varied from 7.4% to 57.1%, with relatively high specificities between 96.8% and 98.6% (25, 26). Studies suggested that FIT in consecutive 3 years could play a role in significant cost savings by replacing colonoscopy, with a risk of missing 40% to 70% AAs and 30% to 40% CRCs (27). In comparison, our case-control study showed a much higher sensitivity of 78.3% and 90.3%, respectively, for detecting AA and CRC. As a result, the risk of missed diagnosis would dramatically decrease to approximately 21.7%, 4.7% or 1.0% for AA, and 9.7%, 0.9% or 0.1% for CRC, respectively, if mCLIP4 tests were performed 1, 2 or 3 times in consecutive years, indicating its potential for early CRC screening.
Numerous studies indicated better performance of DNA methylation markers in stool than those in plasma due to a limited amount of circulating tumor DNA (ctDNA) in plasma and a substantial background of circulating free DNA (cfDNA) from other sources. This was particularly true for the ability to detect precancerous lesions and CRC at early stage. In addition to better performance, stool DNA test offered a feasible solution of at-home cancer screening in populous countries with limited medical resources per capita (28). Furthermore, unlike stool DNA test, blood-based DNA test has not been included in the guideline for routine CRC screening, since its effectiveness has yet to be demonstrated in asymptomatic screening population (29). Comparisons of the performance of the same methylated DNA markers in stool to those in plasma were conducted in several studies. Epi proColon 2.0 assay, the first blood-based mSEPT9 assay approved by FDA, showed a limited sensitivity of 22% for AA and 68.2% for CRC with a specificity of 78.2% (30). A direct comparison study found significantly higher mSEPT9 level in stool samples than in plasma. Whereas the performance of both tests in detecting all stage CRC was similar, stool mSEPT9 test achieved improvement of 35.9% and 7.9% in sensitivity for detecting AA and stage I-II CRC when compared to plasma test (31). Jensen et al. identified three methylation markers, C9orf50, KCNQ5, and CLIP4, and evaluated their performance for CRC detection with plasma samples. Hypermethylation of CLIP4 by itself showed a 77% sensitivity to discriminate CRC patients from healthy individuals. Multiplex methylation assay of all three markers showed an improved sensitivity of 85% at 99% specificity, and sensitivities of 80%, 85%, 89% and 88% for stage I, stage II, stage III and stage IV CRC, respectively, while lacking data for AA patients (16). Compared to the above plasma multiplex test, our stool mCLIP4 test demonstrated sensitivities of 78.3% for AA and 90.3% for CRC (96.2% for stage I, 95.8% for stage II, 83.1% for stage III and 100% for stage IV) with a slight compromise in specificity, suggesting it as a promising tool for early CRC screening.
Single-and multi-target stool DNA assays have been developed and evaluated over the past decades. For example, studies on stool-based mSDC2 tests showed sensitivities ranging from 42.1% to 66.7% for AA and 81.1% to 90.2% for CRC with a 90.2% to 98.0% specificity (24, 32–34). In comparison, mCLIP4 assay in this study showed better performance for detecting AAs by an increase of 12% to 36% in sensitivity with similar specificity for CRC detection. Furthermore, Wang et al. showed significantly lower sensitivity of 75.6% for detecting stage IV CRC with stool mSDC2 test, implying the possible preference of mSDC2 by stages. For mCLIP4 test, no such preference was observed for stage IV CRC in our limited study. In general, multi-target methylation or methylation-mutation assays were considered capable of reducing false negative rate and improving sensitivity. Cologuard, another FDA-approved molecular diagnostic test for early CRC screening, included assays for 7 K-RAS point mutations, aberrant NDRG4 and BMP3 methylation with β-actin as a reference gene and a hemoglobin immunoassay. It demonstrated sensitivities of 42.4% for AA and 92.3% for CRC with 86.6% specificity in an extensive study (11). Our previous study also evaluated a combined assay of mSEPT9 and mSDC2, ColoDefence test, resulting in sensitivities of 66.7% for AA and 92.3% for CRC with 93.2% specificity (20). Compared to these two multiplex tests, mCLIP4 test achieved an even higher sensitivity for AA by an increase of 12% to 36% with a similarly high specificity. In addition, similar to ColoDefence test, only 5 g of stool sample was required for mCLIP4 test, a single-tube multiplex qPCR assay, leading to reduction of cost and complexity of the procedure.
Stool mCLIP4 test demonstrated the feasibility for CRC detection, especially in detecting precancerous lesions and early stage CRC in our study. However, this case-control study had several limitations. First, the main purpose of this study was to evaluate the feasibility of stool mCLIP4 test for CRC detection in a training cohort of a limited number of participants. Further validation and comparison with other existing molecular diagnostic tests in future studies could provide additional support to its potential for CRC screening and prevention. Second, due to limited enrollment, characteristics of the subjects, such as age distribution in different groups, did not reflect the true distribution in a larger population. Although mCLIP4 level did not correlate with patient age in our study, future studies with larger cohorts may better define this relationship. Third, hypermethylation of CLIP4 in plasma and/or tissues was also found in other gastrointestinal (GI) cancers, possibly leading to false positives in CRC detection in the presence of other GI cancers. Although degradation of DNA from upper gastrointestinal tract via intestine would be expected to significantly reduce the false positive results due to other GI cancers, including stool samples from such patients as control in future studies would help address this concern. Nonetheless, our findings demonstrated that mCLIP4 stool test may be a promising tool for early CRC detection.
Conclusions
Stool methylated CLIP4 test demonstrated high sensitivities in detecting SP, AA and CRC with a high specificity. Its performance on precancerous lesions and early stage CRCs made it a promising biomarker for the early detection of colorectal neoplasms. Small amount of sample needed and single-biomarker assay may also reduce screening cost. Therefore, stool mCLIP4 test has the potential to become a convenient alternative method for early CRC screening.
Data Availability Statement
The data sets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
This study was performed according to the principles of the Helsinki Declaration and approved by the Institutional Review Board of the Affiliated Hospital of Xuzhou Medical University (Ethics Committee reference number: XYKY2020-KL156-01). All participants have acknowledged and signed the informed consent. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YC, GZ, YPC, SX, and YM performed the statistical analyses and drafted the manuscript. YC, YPC, ZC, XL, MY, JY, XW, ZL, MZ, and SF participated in sample collection and data analysis. GZ, SX, MZ, and SF conceived of the study and participated in the design and coordination of the study. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by the grants from the Suzhou Technology Entrepreneur Angel Project (grant CYTS2018051), Key Technologies R & D Program for Social Development of Jiangsu Province (grant BE2019688), Kunshan Leading Talent Project (grant 00311), Suzhou Innovation and Entrepreneurship Leading Talent Program (grant ZXL2020046), and Key Technologies R & D Program for Social Development of Xuzhou (grant KC17184).
Conflict of Interest
GZ and SX are employees of Suzhou VersaBio Technologies Co. Ltd. SX is a shareholder of Suzhou VersaBio Technologies Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Qiwei Li at School of Biological Science and Medical Engineering, Southeast University for his technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.647066/full#supplementary-material
Abbreviations
CRC, colorectal cancer; AA, advanced adenomas; SP, small polyps; AUC, area under curve; FOBT, fecal occult blood test; FIT, fecal immunochemical test; sDNA, stool DNA; FDA, Food and Drug Administration; CLIP, CAP-Gly domain containing linker protein; mCLIP4, methylated CLIP4; NED, no evidence of disease; LoD, Limit of detection; ROC, receiver operating characteristic; ctDNA, circulating tumor DNA; cfDNA, circulating free DNA.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021). doi: 10.3322/caac.21660
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer Statistics in China, 2015. CA Cancer J Clin (2016) 66(2):115–32. doi: 10.3322/caac.21338
3. Siegel RL, Ward EM, Jemal A. Trends in Colorectal Cancer Incidence Rates in the United States by Tumor Location and Stage, 1992-2008. Cancer Epidemiol Biomarkers Prev (2012) 21(3):411–6. doi: 10.1158/1055-9965.EPI-11-1020
4. Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal Cancer Statistics, 2020. CA Cancer J Clin (2020) 70(3):145–64. doi: 10.3322/caac.21601
5. Mansukhani S, Barber LJ, Kleftogiannis D, Moorcraft SY, Davidson M, Woolston A, et al. Ultra-Sensitive Mutation Detection and Genome-Wide Dna Copy Number Reconstruction by Error-Corrected Circulating Tumor Dna Sequencing. Clin Chem (2018) 64(11):1626–35. doi: 10.1373/clinchem.2018.289629
6. National Cancer Center C. Expert Group of the Development of China Guideline for the Screening, Early Detection and Early Treatment of Colorectal Cancer. China Guideline for the Screening, Early Detection and Early Treatment of Colorectal Cancer (2020, Beijing). China Cancer (2021) 30(1):1–28. doi: 10.11735/j.issn.1004-0242.2021.01.A001
7. Goss PE, Strasser-Weippl K, Lee-Bychkovsky BL, Fan L, Li J, Chavarri-Guerra Y, et al. Challenges to Effective Cancer Control in China, India, and Russia. Lancet Oncol (2014) 15(5):489–538. doi: 10.1016/s1470-2045(14)70029-4
8. Feng RM, Zong YN, Cao SM, Xu RH. Current Cancer Situation in China: Good or Bad News From the 2018 Global Cancer Statistics? Cancer Commun (Lond) (2019) 39(1):22. doi: 10.1186/s40880-019-0368-6
9. Zhang L, Cao F, Zhang G, Shi L, Chen S, Zhang Z, et al. Trends in and Predictions of Colorectal Cancer Incidence and Mortality in China From 1990 to 2025. Front Oncol (2019) 9:98. doi: 10.3389/fonc.2019.00098
10. Chen H, Li N, Ren J, Feng X, Lyu Z, Wei L, et al. Participation and Yield of a Population-Based Colorectal Cancer Screening Programme in China. Gut (2019) 68(8):1450–7. doi: 10.1136/gutjnl-2018-317124
11. Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget Stool DNA Testing for Colorectal-Cancer Screening. N Engl J Med (2014) 370(14):1287–97. doi: 10.1056/NEJMoa1311194
12. Klutstein M, Nejman D, Greenfield R, Cedar H. Dna Methylation in Cancer and Aging. Cancer Res (2016) 76(12):3446–50. doi: 10.1158/0008-5472.CAN-15-3278
13. Hua Yang B-QX, Jiang BO, Wang G, Yang Y-P, Chen H, Li B-S, et al. Diagnostic Value of Stool DNA Testing for Multiple Markers of Colorectal Cancer and Advanced Adenoma: A Meta-Analysis. Can J Gastroenterol (2013) 27(8):467–75. doi: 10.1155/2013/258030
14. Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-Based Propagation of Functional Annotations Within the Gene Ontology Consortium. Brief Bioinform (2011) 12(5):449–62. doi: 10.1093/bib/bbr042
15. Ahn J, Han KS, Heo JH, Bang D, Kang YH, Jin HA, et al. FOXC2 and CLIP4 : A Potential Biomarker for Synchronous Metastasis of ≤7-Cm Clear Cell Renal Cell Carcinomas. Oncotarget (2016) 7(32):51423–34. doi: 10.18632/oncotarget.9842
16. Jensen S, Øgaard N, Ørntoft MW, Rasmussen MH, Bramsen JB, Kristensen H, et al. Novel DNA Methylation Biomarkers Show High Sensitivity and Specificity for Blood-Based Detection of Colorectal Cancer-a Clinical Biomarker Discovery and Validation Study. Clin Epigenet (2019) 11(1):158. doi: 10.1186/s13148-019-0757-3
17. Pirini F, Noazin S, Jahuira-Arias MH, Rodriguez-Torres S, Friess L, Michailidi C, et al. Early Detection of Gastric Cancer Using Global, Genome-Wide and IRF4, Elmo1, CLIP4 and MSC DNA Methylation in Endoscopic Biopsies. Oncotarget (2017) 8(24):38501–16. doi: 10.18632/oncotarget.16258
18. Hu C, Zhou Y, Liu C, Kang Y. A Novel Scoring System for Gastric Cancer Risk Assessment Based on the Expression of Three CLIP4 DNA Methylation-Associated Genes. Int J Oncol (2018) 53(2):633–43. doi: 10.3892/ijo.2018.4433
19. Bacolod MD, Mirza AH, Huang J, Giardina SF, Feinberg PB, Soper SA, et al. Application of Multiplex Bisulfite PCR-Ligase Detection Reaction-Real-Time Quantitative PCR Assay in Interrogating Bioinformatically Identified, Blood-Based Methylation Markers for Colorectal Cancer. The Journal of Molecular Diagnostics. JMD (2020) 22(7):885–900. doi: 10.1016/j.jmoldx.2020.03.009
20. Zhao G, Liu X, Liu Y, Li H, Ma Y, Li S, et al. Aberrant DNA Methylation of SEPT9 and SDC2 in Stool Specimens as an Integrated Biomarker for Colorectal Cancer Early Detection. Front Genet (2020) 11:643. doi: 10.3389/fgene.2020.00643
21. Yizhak K, Aguet F, Kim J, Hess JM, Kübler K, Grimsby J, et al. RNA Sequence Analysis Reveals Macroscopic Somatic Clonal Expansion Across Normal Tissues. Science (2019) 364(6444):1–9. doi: 10.1126/science.aaw0726
22. Aucamp J, Bronkhorst AJ, Badenhorst CPS, Pretorius PJ. The Diverse Origins of Circulating Cell-Free DNA in the Human Body: A Critical Re-Evaluation of the Literature. Biol Rev Camb Philos Soc (2018) 93(3):1649–83. doi: 10.1111/brv.12413
23. Wu D, Zhou G, Jin P, Zhu J, Li S, Wu Q, et al. Detection of Colorectal Cancer Using a Simplified Sept9 Gene Methylation Assay is a Reliable Method for Opportunistic Screening. The Journal of Molecular Diagnostics. JMD (2016) 18(4):535–45. doi: 10.1016/j.jmoldx.2016.02.005
24. Oh TJ, Oh HI, Seo YY, Jeong D, Kim C, Kang HW, et al. Feasibility of Quantifying SDC2 Methylation in Stool DNA for Early Detection of Colorectal Cancer. Clin Epigenet (2017) 9:126. doi: 10.1186/s13148-017-0426-3
25. Tinmouth J, Lansdorp-Vogelaar I, Allison JE. Faecal Immunochemical Tests Versus Guaiac Faecal Occult Blood Tests: What Clinicians and Colorectal Cancer Screening Programme Organisers Need to Know. Gut (2015) 64(8):1327–37. doi: 10.1136/gutjnl-2014-308074
26. Shapiro JA, Bobo JK, Church TR, Rex DK, Chovnick G, Thompson TD, et al. A Comparison of Fecal Immunochemical and High-Sensitivity Guaiac Tests for Colorectal Cancer Screening. Am J Gastroenterol (2017) 112(11):1728–35. doi: 10.1038/ajg.2017.285
27. Cross AJ, Wooldrage K, Robbins EC, Kralj-Hans I, MacRae E, Piggott C, et al. Faecal Immunochemical Tests (FIT) Versus Colonoscopy for Surveillance After Screening and Polypectomy: A Diagnostic Accuracy and Cost-Effectiveness Study. Gut (2019) 68(9):1642–52. doi: 10.1136/gutjnl-2018-317297
28. Qian C. At-Home Cancer Screening: A Solution for China and Other Developing Countries With a Large Population and Limited Number of Healthcare Practitioners. Chin J Cancer (2017) 36(68):1–3. doi: 10.1186/s40880-017-0235-2
29. Wolf AMD, Fontham ETH, Church TR, Flowers CR, Guerra CE, LaMonte SJ, et al. Colorectal Cancer Screening for Average-Risk Adults: 2018 Guideline Update From the American Cancer Society. CA Cancer J Clin (2018) 68(4):250–81. doi: 10.3322/caac.21457
30. Lamb YN, Dhillon S. Epi proColon(®) 2.0 Ce: A Blood-Based Screening Test for Colorectal Cancer. Mol Diagnosis Ther (2017) 21(2):225–32. doi: 10.1007/s40291-017-0259-y
31. Liu Y, Zhao G, Miao J, Li H, Ma Y, Liu X, et al. Performance Comparison Between Plasma and Stool Methylated SEPT9 Tests for Detecting Colorectal Cancer. Front Genet (2020) 11:324. doi: 10.3389/fgene.2020.00324
32. Niu F, Wen J, Fu X, Li C, Zhao R, Wu S, et al. Stool DNA Test of Methylated Syndecan-2 for the Early Detection of Colorectal Neoplasia. Cancer Epidemiol Biomarkers Prev (2017) 26(9):1411–9. doi: 10.1158/1055-9965.EPI-17-0153
33. Han YD, Oh TJ, Chung TH, Jang HW, Kim YN, An S, et al. Early Detection of Colorectal Cancer Based on Presence of Methylated Syndecan-2 (SDC2) in Stool DNA. Clin Epigenet (2019) 11(1):51. doi: 10.1186/s13148-019-0642-0
Keywords: colorectal cancer, early detection, stool, CLIP4, DNA methylation
Citation: Cao Y, Zhao G, Cao Y, Chen Z, Liu X, Yuan M, Yang J, Wang X, Ma Y, Liu Z, Xiong S, Zheng M and Fei S (2021) Feasibility of Methylated CLIP4 in Stool for Early Detection of Colorectal Cancer: A Training Study in Chinese Population. Front. Oncol. 11:647066. doi: 10.3389/fonc.2021.647066
Received: 29 December 2020; Accepted: 01 April 2021;
Published: 22 April 2021.
Edited by:
Zhaohui Huang, Affiliated Hospital of Jiangnan University, ChinaReviewed by:
Jessica Yang, Memorial Sloan Kettering Cancer Center, United StatesMohammad Amin Kerachian, Mashhad University of Medical Sciences, Iran
Copyright © 2021 Cao, Zhao, Cao, Chen, Liu, Yuan, Yang, Wang, Ma, Liu, Xiong, Zheng and Fei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shangmin Xiong, c2hhbmdtaW5feGlvbmdAaG90bWFpbC5jb20=; Minxue Zheng, bWlueHVlLnpoZW5nQHNpYmV0LmFjLmNu; Sujuan Fei, eHlmeWZlaXNqOTlAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Yang Cao1,2†
Yang Cao1,2† Guodong Zhao
Guodong Zhao Yaping Cao
Yaping Cao Mufa Yuan
Mufa Yuan Shangmin Xiong
Shangmin Xiong