- 1Department of Urology, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya Medicine School, Central South University, Changsha, China
- 2GloriousMed Clinical Laboratory Co., Ltd., Shanghai, China
Penile squamous cell carcinoma (PSCC) is a rare malignancy with poor survival after standard treatment. Although genomic alterations of PSCC have been characterized in several latest studies, the association between the formation of somatic landscape and regional lymph node metastasis (LNM), an important predictor for patient survival, has not been comprehensively investigated. Here, we collected formalin-fixed paraffin-embedded tumor tissue and matched normal samples of 32 PSCC patients, including 14 LNM patients and 18 clinically node-negative patients, to implement a whole-exome sequencing. Comparison of genomic features among different lymph node status subgroups was conducted after genomic profiling and its effects on patient survival were explored. Top-ranked recurrent gene mutants in our PSCC cohort were TP53 (13/32), NOTCH1 (12/32), CDKN2A (11/32), TTN (9/32) and FAT1 (8/32), mainly identified in the Notch, Hippo, cell cycle, TP53, RTK-RAS and PI3K pathways. While CDKN2A was confirmed to be the driver gene in all PSCC patients, certain gene mutants were significantly enriched in LNM involved patients, including TP53 (9/14 vs. 4/18, p = 0.029) and GBF1 (4/14 vs. 0/18, p = 0.028). Overall survival stratification of PSCC patients were found to be significantly correlated with mutations of three genes, including PIK3CA (Hazard ratio [HR] = 4.15, p = 0.029), CHD7 (HR = 4.82, p = 0.032) and LAMC3 (HR = 15.9, p < 0.001). PIK3CA and LAMC3 held a higher prevalence in patients with LNM compared to those without LNM (PIK3CA: 3/14 vs. 1/18, LAMC3: 2/14 vs. 1/18). Our finding demonstrated that genomic divergence exists across PSCC patients with different lymph node statuses, and it may be correlated with their survival outcome. It helps delineate somatic evolution during tumor progression and perfect potential therapeutic intervention in this disease.
Introduction
Penile squamous cell carcinoma (PSCC) is a rare cancer with a significantly higher incidence in developing countries compared to developed countries (1), mainly attributed to exposure to human papilloma virus (HPV) (2). For patients with advanced PSCC, standard treatment paradigm is a multimodal approach of chemotherapy combinations followed by surgical procedures (3–6). Unfortunately, more than half of the patients shortly progressed or relapsed after the treatment (6, 7).
To discover novel diagnostic and prognostic biomarkers that are capable to identify patients sensitive to specific therapy, genomic profiling of penile carcinoma has been examined (8–11) and a few PSCC cell lines were established (12). Those studies revealed that somatic alterations are associated with penile carcinogenesis, including frequent mutations in gene TP53, CDKN2A, NOTCH1 and PIK3CA (13, 14). Associated risk factors were also investigated and prediction models of patient survival were developed in the past years to achieve better management of this malignancy (15–18). In all studies, lymph node involvement was found to be the most evidential factor (19) compared to other predictors including histological subtypes (20) and high expression levels of TP53-regulated inhibitor of apoptosis 1 (21). Although it is indicated that lymph node metastasis (LNM) could be roughly inferred from lymph node staging, lymph vascular invasion or sentinel lymph node biopsy combined with sonography (22–24), accurate prediction of lymph node status is still lacking and the connections between lymphatic metastasis and potential genetic biomarkers remain unclear (25).
To investigate the evolution of somatic alterations during the process that tumors transform to a state prone to spread to the lymph node, we characterized the somatic mutation landscape and compared genetic characteristics between PSCC patients with different lymph node statuses with whole-exome sequencing. The performance of predicting patient survival with relevant variants was also evaluated.
Materials and Methods
Sample Cohort
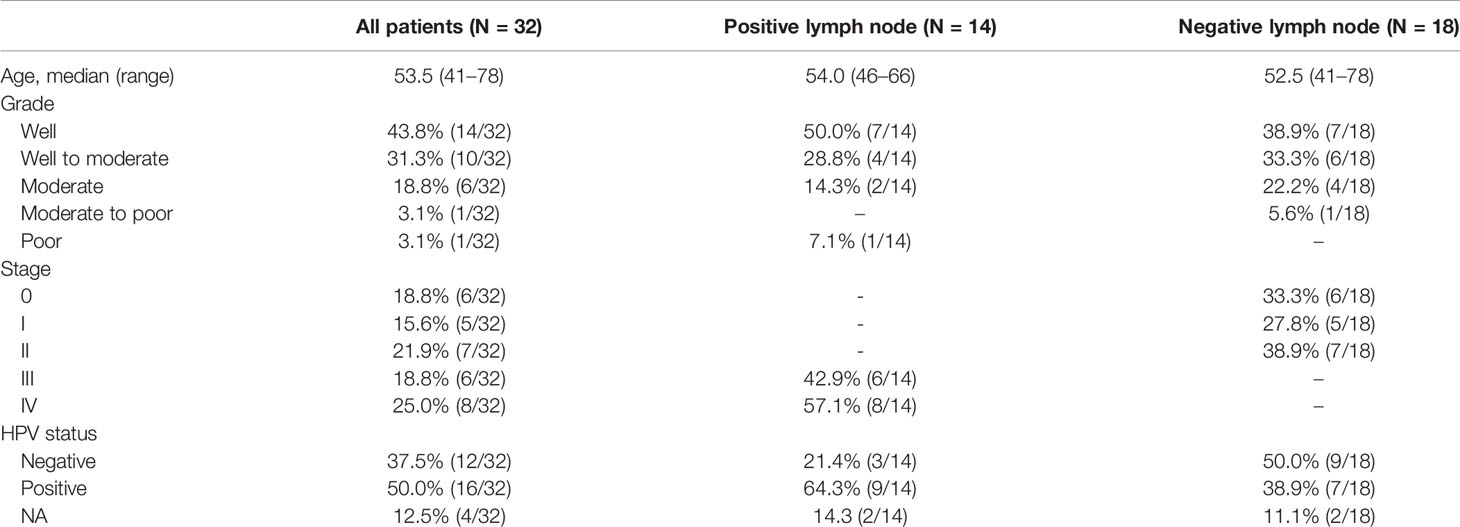
Tumor tissue and matched normal blood or tissue samples of 32 PSCC patients were collected for whole-exome sequencing. These patients were diagnosed with PSCC from June 2015 to June 2019 in Hunan Cancer Hospital and underwent surgical resection afterward. Lymph node dissection was performed and the lymph node statuses were assessed in some patients. Clinical information including age, tumor stage, pathological type, lymph node status and survival information were gathered by reviewing the electronic medical records. The study was approved by the ethics committee of Hunan Cancer Hospital and all involved human subjects have signed the informed consent.
Whole-Exome Sequencing
DNA was extracted from formalin-fixed paraffin-embedded tissues and white blood cells using QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) and Blood Genomic DNA Mini Kit (Cwbiotech, Beijing, China). The whole exome was captured according to the standard procedures of xGen Exome Research Panel v1.0 (Integrated DNA Technologies, Coralville, IA). The captured DNA fragments were then used for library preparation and quantification guided by KAPA Hyper Prep protocols (Kapa Biosystems, Wilmington, MA), followed by purification with AMPure XP (Beckman Coulter, Brea, CA) and quantification using Qubit™ dsDNA HS Assay Kit (ThermoFisher, Waltham, MA). Pooled library was finally sequenced using Novoseq6000 (Illumina, San Diego, CA).
Variant Calling and Annotation
After adapter trimming with Trimmomatic, the sequencing reads were then aligned to the human reference genome hg19 using Burrows-Wheeler Aligner (BWA). Reads were then realigned using Genome Analysis Tool Kit (GATK) after duplicated reads were flagged with Picard. Mutect2 was used to identify somatic mutations, which were then annotated with ANNOVAR. Human identity consistency of paired samples was verified using an in-house script. Somatic mutations were filtered out under the following conditions (1): base quality value under 20; (2) mutation reads depth less than 10; (3) variant allele frequency less than 5%; (4) variant supporting reads more than 4 or variant allele frequency above 2% in the paired normal sample. Then synonymous and benign mutations were removed from the remaining variants. OncodriveCLUSTL was used to detect significant clusters of variation across genomic regions to identify candidate driver genes (26). Visualization of gene alterations in oncogenic signaling pathways was conducted using the PathwayMapper tool (http://pathwaymapper.org).
HPV Genotyping
HPV status of PSCC patients was assessed by HPV genotyping (17 high risk HPV: 16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73 and 82; 6 low risk HPV: 6, 11, 42, 43, 81 and 83), which was performed with a polymerase chain reaction reverse dot blot (PCR-RDB) approach (Yaneng Bio, Shenzhen, China) using DNA extracted from tumor tissue samples.
Tumor Mutation Burden, Heterogeneity and Genomic Stability
Tumor mutational burden (TMB), heterogeneity and genomic stability were assessed to evaluate the genomic status of their tumor samples for each patient (27). TMB was defined as the number of nonsynonymous somatic mutations per million bases, and heterogeneity was estimated with mutant-allele tumor heterogeneity (MATH) calculated by R package maftools (28, 29). Genomic instability was represented by the weighted genome integrity index (wGII), which denotes the chromosome-weighted proportion of genomic fragments with abnormal copy number (30).
Statistical Analysis
All statistical analyses were performed with R v3.6.0. Prevalence comparison of gene mutant was conducted using Fisher’s exact test. TMB, MATH and wGII among lymph node subgroups were compared with Wilcoxon rank-sum test. Kaplan-Meier estimate was implemented for survival analysis and the log-rank test was used to determine the mutated gene that correlated with patient survival. Hazard ratio (HR) was reported by the univariate and multivariable cox proportional hazard regression models. Two-sided P<0.05 was considered statistically significant.
Results
Cohort Characteristics
32 PSCC patients including 14 (43.75%) lymph node-positive patients and 18 (56.25%) negative node patients were enrolled in our investigation, summarized in Table 1. The median age of this cohort was 53.5 years (41–78 years). Among all patients, 8 (25%) of them were diagnosed with low, low-to-moderate or moderate grade cancer while 24 (75%) of them were evaluated as moderate-to-high or high grade cancer. 14 (43.75%) patients were assessed at stage III or higher. 28 (87.5%) patients were tested for HPV genotyping, and 16 (50%) of them were found to be HPV-positive. 7 (21.88%) patients experienced metastases or relapse and 9 (28.13%) patients deceased during follow-up.
Somatic Mutation Landscape of PSCC in Chinese Patients
A total of 3,026 somatic mutations were identified in 2,418 genes, including single nucleotide variants and small insertions/deletions. The most common variant type was missense mutation (80.3%), followed by frameshift insertion/deletion (8.5%), nonsense mutation (7.3%), in-frame insertion/deletion (2.2%), splicing mutation (1.5%) and nonstop mutation (0.2%) (Figure 1A). TP53 (13, 40.63%), NOTCH1 (12, 37.50%), CDKN2A (11, 34.38%), TTN (9, 28.13%) and FAT1 (8, 25.00%) were found to be the most common repeatedly mutated genes in this cohort (Figure 1B), which have been reported to be frequently mutated in penile carcinoma. CASP8, which was previously reported to be frequently altered in the Chinese PSCC population (11), was mutated in 6 (18.75%) patients in this study. Although CSN1 mutant was reported in a Caucasian cohort (9), it was not found in any patient in our cohort. Other reported frequent gene mutants, like PIK3CA and HRAS, were mutated in a few patients but not among the top mutated genes.
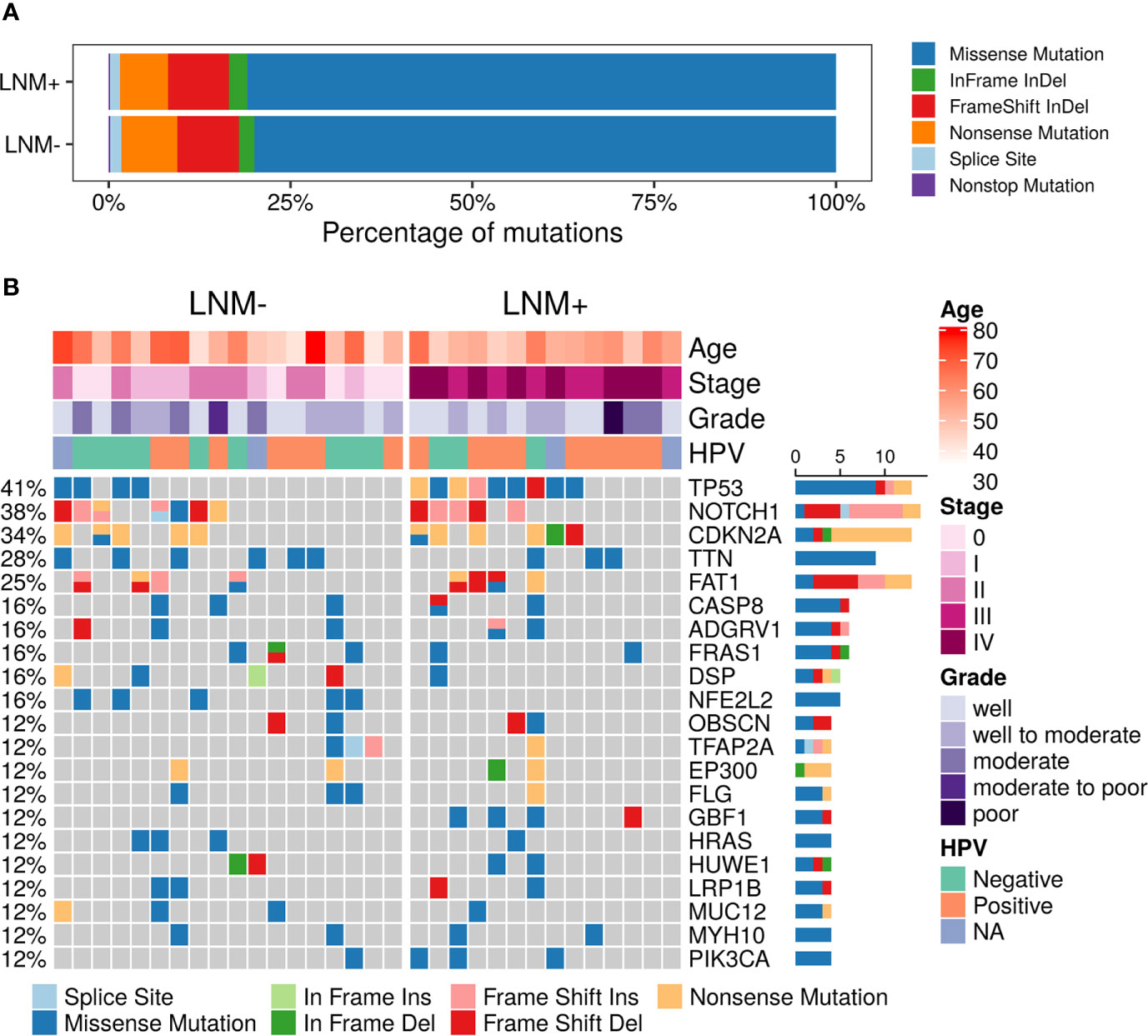
Figure 1 Summary of somatic mutations in 32 PSCC patients. (A) The frequencies of different variant types in lymph node metastasis (LNM) involved patients and negative-node patients. (B) Mutational landscape of all 32 PSCC patients. Each row represents one gene while each column represents one patient. The frequencies of gene mutants and clinical characteristics are labeled by the side of the heatmap. (NA, not assessed).
Pathway Alterations and Driver Mutant
Somatic mutations of 10 commonly altered pathways in cancer were characterized and variants were found in all these pathways with varying frequencies (Figure 2A). Notch (20, 62.50%), Hippo (18, 56.25%), TP53 (15, 46.88%), cell cycle (13, 40.63%), RTK-RAS (12, 37.50%) and PI3K (7, 21.88%) were the most frequently mutated pathways. The prevalence of each pathway was contributed by different dominant gene mutants, as exemplified for FAT1 in the Hippo pathway, CDKN2A in the cell cycle pathway and HRAS in the RTK-RAS pathway.
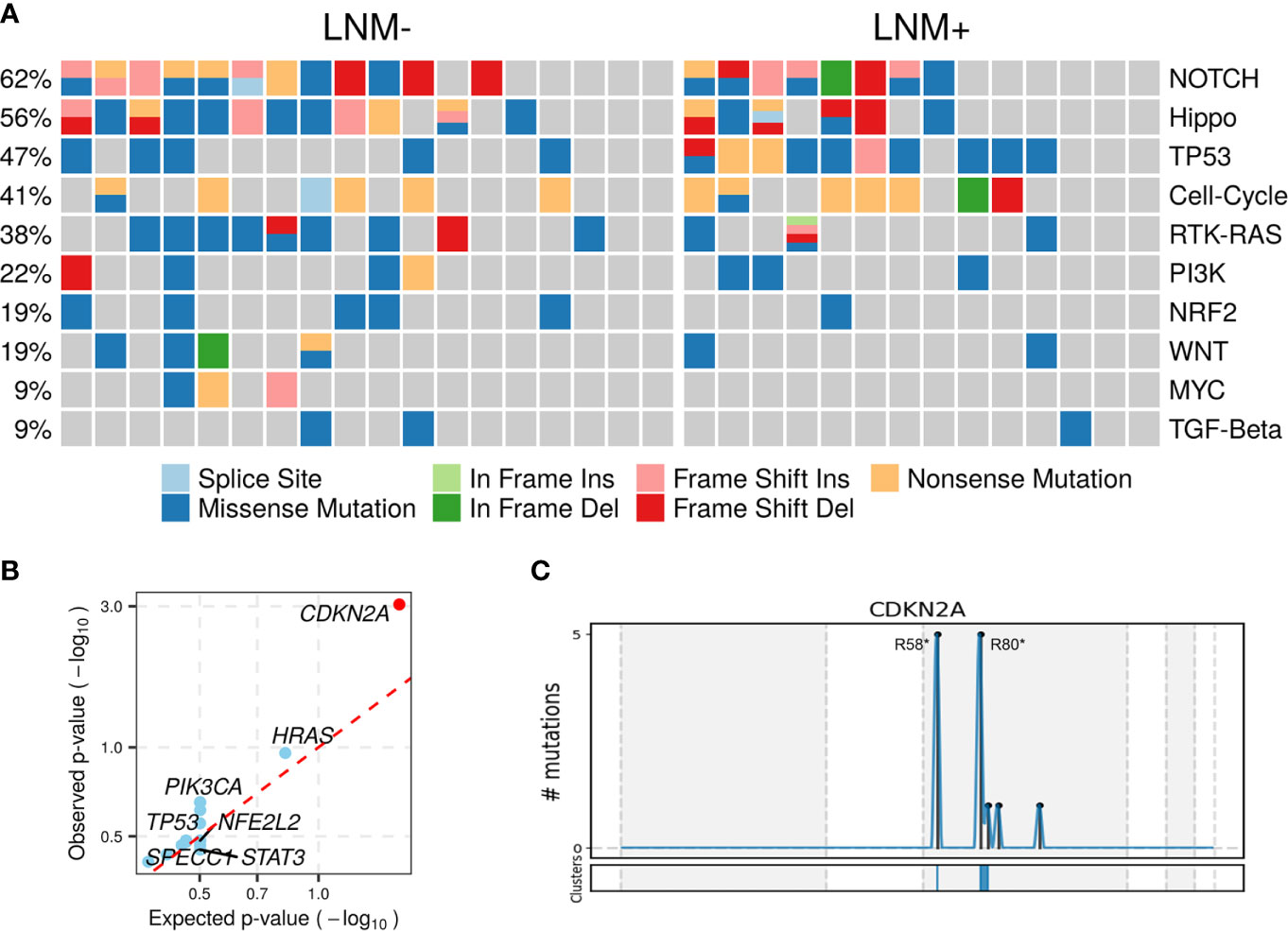
Figure 2 Altered pathways and diver mutant in penile carcinoma. (A) The frequencies of ten common cancer-related pathways altered in 32 PSCC patients, (B) PSCC candidate driver genes identified by OncodriveCLUSTL. Significant gene (observed p-value < 0.01) is highlighted with red circle. (C) Distribution of mutations across CDKN2A region in 32 PSCC patients. The mutations, mainly two hotspots labeled in the figure, are enriched in two clusters (shown at the bottom) that span 1 base and 16 bases respectively.
To further investigate tumorigenesis-associated pathways in penile carcinoma, somatic mutations were used to identify candidate driver genes with OncodriveCLUSTL, which has been proven to be a state-of-the-art method in the field of driver gene prediction. The only gene that showed significance was CDKN2A (adjusted p < 0.001, Figure 2B), whose two prevailing hotspot mutations enriched in its ankyrin repeat-containing domain (Figure 2C), indicating alterations in the cell cycle pathway may be involved in triggering this malignancy.
LNM-Associated Somatic Alterations Correlate With Patient Survival in Penile Carcinoma
Regional lymph node involvement was considered to be a key predictor of patient survival in PSCC. We assessed the overall survival (OS) by lymph node statuses in our cohort and confirmed that the positive lymph node was associated with shorter survival (HR = 4.92, log-rank p = 0.028; Figure 3A). Only 7 genes appeared in the intersection of the first 20 mutated genes in LNM positive and negative patients, including NOTCH1, TTN, CDKN2A, FAT1, TP53, CASP8 and AFDGRV1 (Supplementary Figure 1). Furthermore, the prevalence of mutated genes between different lymph node statuses was compared to explore the association between somatic variants and the LNM process in PSCC. The candidate genes for this comparison were limited to 58 genes that altered in at least 3 patients. Alterations of two genes, TP53 (9/14 vs. 4/18, p = 0.029) and GBF1 (4/14 vs. 0/18, p = 0.028), were found to be significantly enriched in lymph node-positive samples (Figure 3B), suggesting the occurrence of such genomic events during tumor progression may potentially promote regional lymph node metastasis. Although NFE2L2 mutations tended to serve a protective role of LNM, no significant difference was observed (0/14 vs. 5/18, p = 0.052).
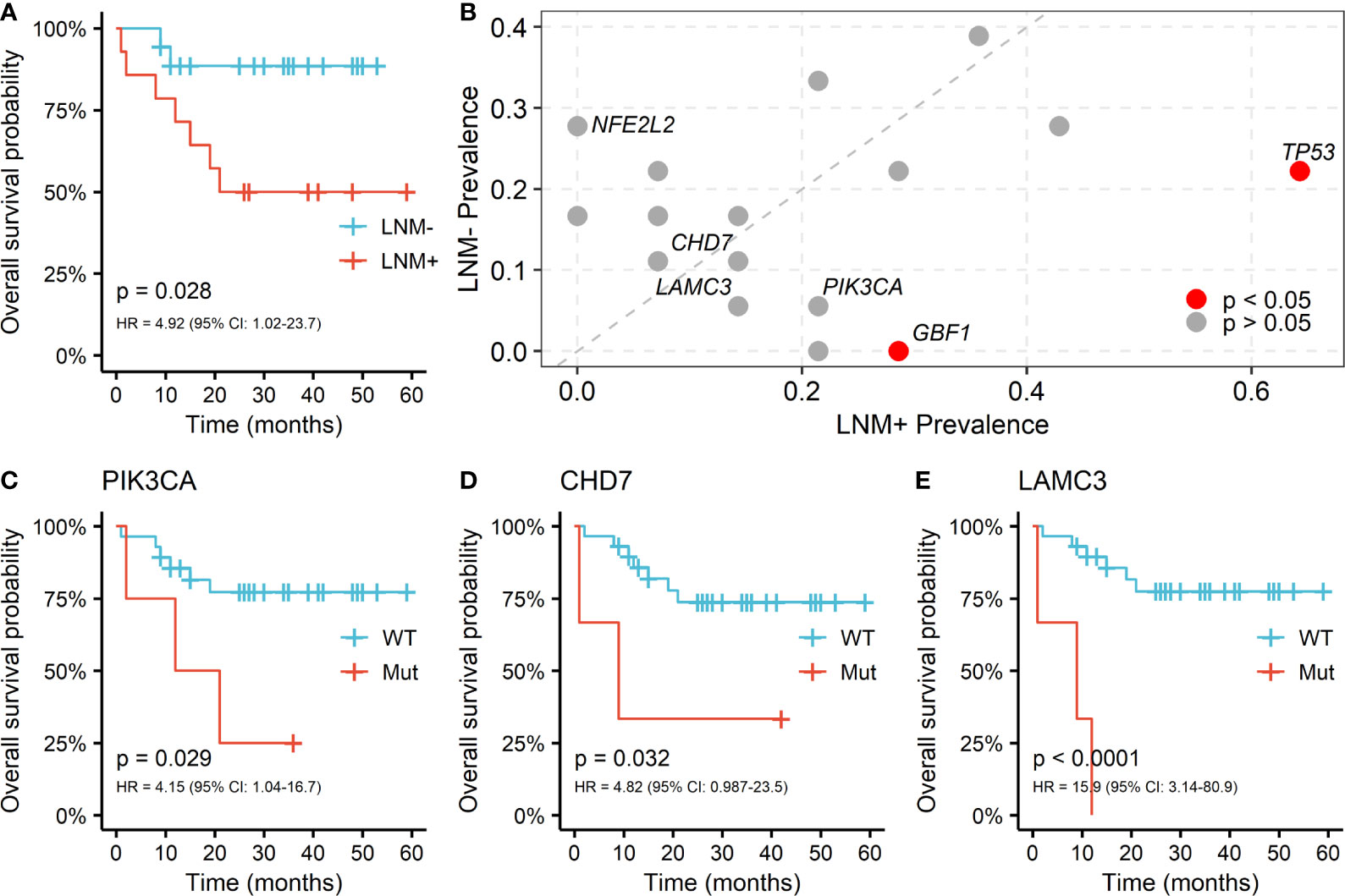
Figure 3 Somatic alterations between different lymph node metastasis (LNM) subgroups and its correlation with OS in PSCC. (A) Kaplan-Meier curve of overall survival by lymph node metastasis (LNM). (B) Enrichment of somatic alterations by lymph node status. Kaplan-Meier curves of overall survival by the mutation status of gene (C) PIK3CA, (D) CHD7 and (E) LAMC3 in PSCC patients. P values of the log-rank test and hazard ratios are shown at the bottom left for each curve.
The associations between somatic mutations and survival outcome of PSCC patients were further investigated. Mutants of TP53 and GBF1, which were significantly enriched in positive lymph node patients, indicated shorter OS but the difference was not statistically significant (TP53: HR = 1.27; GBF1: HR = 1.94; Supplementary Figure 2). However, fine performance of stratifying overall survival of patients was observed in three other genes, including PIK3CA (HR = 4.15, p = 0.029; Figure 3C), CHD7 (HR = 4.82, p = 0.032; Figure 3D) and LAMC3 (HR = 15.9, p < 0.001; Figure 3E). Furthermore, these genes together with age and LNM status, were included as covariates in cox multivariable regression to verify their significance. Independent associations with OS were confirmed in CHD7 (HR = 29.4, p = 0.009) and LAMC3 (HR = 11.9, p = 0.003), except for PIK3CA (HR = 3.65, p = 0.1) (Supplementary Table 1). Moreover, PIK3CA and LAMC3, held a higher frequency of mutation in patients with LNM but did not reach the significant level (PIK3CA: 3/14 vs. 1/18, LAMC3: 2/14 vs. 1/18; Figure 3B).
LNM-Related Somatic Alterations in Pathway Level
Further investigations were carried out by exploring correlations between somatic alterations and lymphatic metastasis in the pathway level. TP53 pathway is the only significantly enriched pathway in node-positive patients (p = 0.031, Supplementary Figure 3), which is mainly caused by the mutations in tumor suppressor gene TP53 and ATM (Figure 4A). There also is a tendency that alterations in RAS pathway preferentially occurred in LNM negative patients (HRAS and BRAF, Figure 4B). In the cell cycle pathway (CDKN2A and E2F3, Figure 4C) and NRF2 pathway (NFE2L2 and KEAP1, Figure 4D), oncogene mutations in lymph node-negative patients and tumor suppressor gene mutations in lymph node-positive patients can be observed in similar patterns. The opposite phenomena were found in the PI3K pathway (PIK3CA in node-positive patients and PTEN in node-negative patients, Figure 4E).
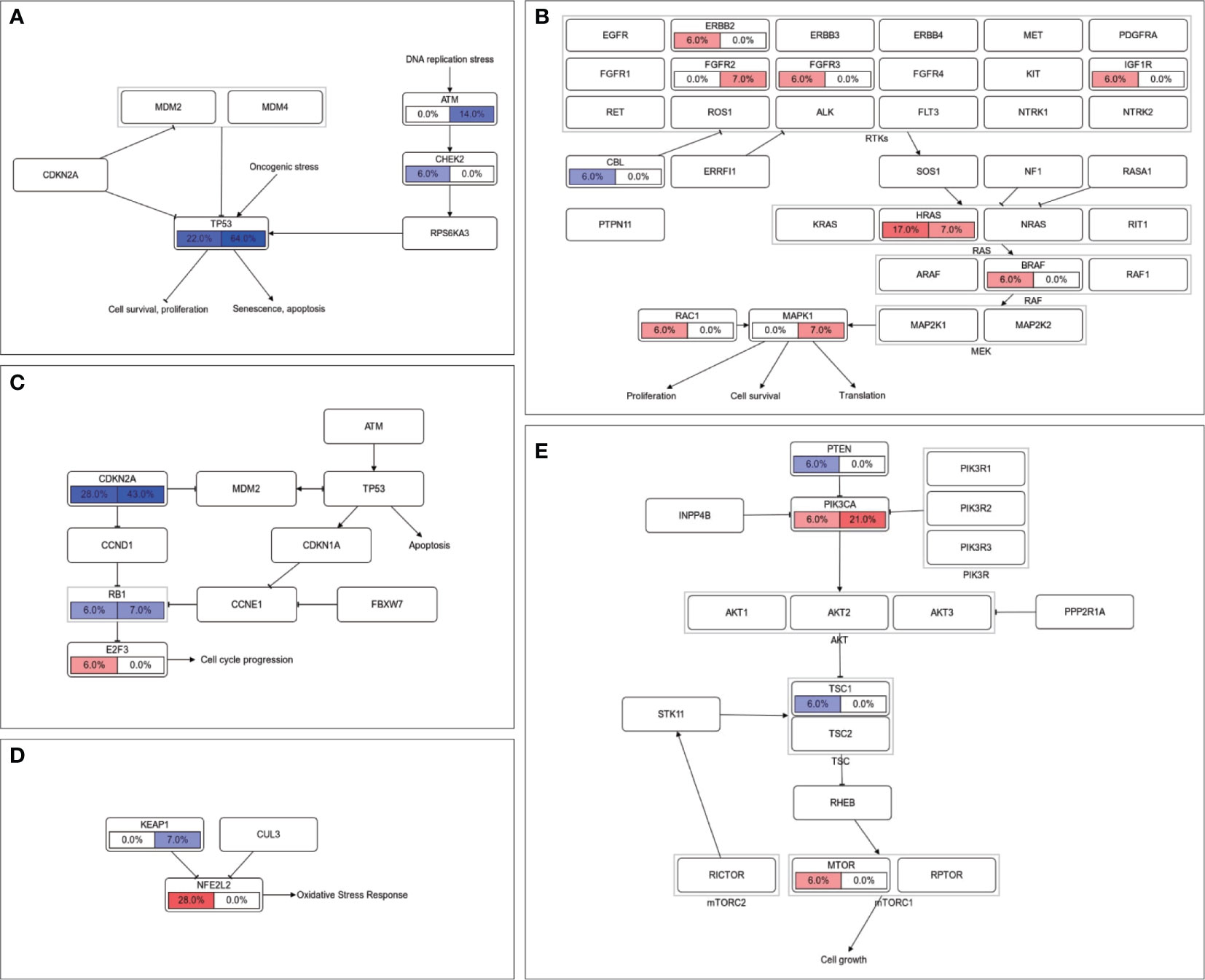
Figure 4 Comparison of somatic alterations between LNM subgroups in signaling pathway level. Mutation frequencies of genes in (A) TP53 pathway, (B) RTK-RAS pathway, (C) cell cycle pathway, (D) NRF2 pathway and (E) PI3K pathway were labeled with LNM negative on the left and LNM positive on the right. Oncogenes were filled with red color and tumor suppressor genes were filled with blue.
Except for certain mutants, the relationship between LNM and other tumor genomic features including tumor mutational burden, tumor heterogeneity and genomic stability were also investigated (Supplementary Figure 4). No significant differences were found between node-negative and positive lymph node patients in TMB (3.4 vs. 2.2, p = 0.44), MATH (31.9 vs. 21.6, p = 0.67) and wGII (0.15 vs. 0.12, p = 0.28). The similarity of different lymph node statuses further indicated that lymph node metastasis may be driven by key alterations rather than advanced tumor status.
Discussion
Risk stratification of lymph node metastasis is essential for both clinical intervention and prognosis prediction of PSCC. Given the high-risk lymph node micrometastases in node-negative tumors (31) and high false-negative rates of modified inguinal lymph nodes dissection and dynamic sentinel lymph node biopsy (32, 33), the molecular drivers of metastasis and novel biomarkers for risk assessment of LNM need to be urgently uncovered.
The advancements within genomic characterization of PSCC were mostly constrained in a form of targeted panel strategy, except for two (9, 11). In this study, we implemented a whole-exome sequencing to perform comprehensive somatic alteration profiling of 32 PSCC patients. The observation that TP53, CDKN2A, NOTCH1, TTN and FAT1 being the most frequently mutated genes was in concordance with previous studies and similar result was found in the pathway level. We also confirmed that CDKN2A plays a critical role in tumorigenesis of PSCC, which has been reported to be preferentially occurred in lichen sclerosus-external genital carcinoma (34).
Comparison between different lymph node status subgroups showed that LNM is associated with alterations of certain genes, like TP53 and GBF1 in our study. GBF1 is required for the trans-Golgi network localization of HPV16 infection (35), which inactivates tumor suppressor protein p53 in penile carcinoma. It has been found that specific mutations or changes in expression of TP53 are correlated with LNM in various cutaneous squamous cell carcinomas (36–38), and alterations in TP53 were significantly associated with shorter event-free survival (10). In addition, higher prevalence in positive-node patients along with the tendency towards shorter survival were observed in PIK3CA and LAMC3 mutants. Notably, it has been demonstrated that lymphatic metastasis in PSCC was correlated with the elevated expression of LAMC2 (39), another heterotrimer of the laminin gamma family. The mutations within all these genes during tumor progression of PSCC could promote its spread to lymph nodes, leading to a poor prognosis.
There were some limitations in this study. Firstly, it is unrealistic to harbor both the pre-LNM sample and samples in a state with positive-node from the same patient. This leads to the lack of direct evidence for our findings, which may need to be resolved after the establishment of animal models. Due to the low incidence of penile carcinoma, partial results did not reach the significance level with a small number of enrolled samples, like in most PSCC studies. It will be further validated by a larger cohort in the upcoming future.
In summary, we reproduced an accordant genomic landscape in penile carcinoma and depicted the formation of somatic alterations while the tumor evolved to the status liable to spread to lymph nodes. The findings also proposed candidate genetic biomarkers for both the management of low-risk primary penile tumors and prognosis prediction of patients with this malignancy.
Data Availability Statement
The datasets presented in this article are not readily available because of restrictions by national legislation/guidelines, specifically the Administrative Regulations of the People's Republic of China on Human Genetic Resources. Requests to access the datasets should be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethics committee of Hunan Cancer Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JC, C-HY, W-QH, YX, and S-SJ conceived and designed the study. JC, C-HY, W-QH, YX, and Z-ZL conducted data collection, analysis, and interpretation. JC and C-HY wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research is supported by Natural Science Foundation of Changsha Science and Technology Bureau (Basic Research Program: kq2004134) and Wu Jie-Ping Medical Foundation (Clinical Research Special Funding: 320.6750.2020-14-14).
Conflict of Interest
C-HY was employed by the company GloriousMed Clinical Laboratory (Shanghai) Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.641869/full#supplementary-material
References
1. Montes Cardona CE, Garcia-Perdomo HA. Incidence of Penile Cancer Worldwide: Systematic Review and Meta-Analysis. Rev Panam Salud Publica (2017) 41:e117. doi: 10.26633/RPSP.2017.117
2. Olesen TB, Sand FL, Rasmussen CL, Albieri V, Toft BG, Norrild B, et al. Prevalence of Human Papillomavirus DNA and p16(INK4a) in Penile Cancer and Penile Intraepithelial Neoplasia: A Systematic Review and Meta-Analysis. Lancet Oncol (2019) 20(1):145–58. doi: 10.1016/S1470-2045(18)30682-X
3. Haas GP, Blumenstein BA, Gagliano RG, Russell CA, Rivkin SE, Culkin DJ, et al. Cisplatin, Methotrexate and Bleomycin for the Treatment of Carcinoma of the Penis: A Southwest Oncology Group Study. J Urol (1999) 161(6):1823–5. doi: 10.1016/S0022-5347(05)68815-5
4. Hakenberg OW, Drager DL, Erbersdobler A, Naumann CM, Junemann KP, Protzel C. The Diagnosis and Treatment of Penile Cancer. Dtsch Arztebl Int (2018) 115(39):646–52. doi: 10.3238/arztebl.2018.0646
5. Pagliaro LC, Williams DL, Daliani D, Williams MB, Osai W, Kincaid M, et al. Neoadjuvant Paclitaxel, Ifosfamide, and Cisplatin Chemotherapy for Metastatic Penile Cancer: A Phase II Study. J Clin Oncol (2010) 28(24):3851–7. doi: 10.1200/JCO.2010.29.5477
6. Nicholson S, Hall E, Harland SJ, Chester JD, Pickering L, Barber J, et al. Phase II Trial of Docetaxel, Cisplatin and 5FU Chemotherapy in Locally Advanced and Metastatic Penis Cancer (CRUK/09/001). Br J Cancer (2013) 109(10):2554–9. doi: 10.1038/bjc.2013.620
7. Wang J, Pettaway CA, Pagliaro LC. Treatment for Metastatic Penile Cancer After First-line Chemotherapy Failure: Analysis of Response and Survival Outcomes. Urology (2015) 85(5):1104–10. doi: 10.1016/j.urology.2014.12.049
8. Ali SM, Pal SK, Wang K, Palma NA, Sanford E, Bailey M, et al. Comprehensive Genomic Profiling of Advanced Penile Carcinoma Suggests a High Frequency of Clinically Relevant Genomic Alterations. Oncologist (2016) 21(1):33–9. doi: 10.1634/theoncologist.2015-0241
9. Feber A, Worth DC, Chakravarthy A, de Winter P, Shah K, Arya M, et al. Csn1 Somatic Mutations in Penile Squamous Cell Carcinoma. Cancer Res (2016) 76(16):4720–7. doi: 10.1158/0008-5472.CAN-15-3134
10. McDaniel AS, Hovelson DH, Cani AK, Liu CJ, Zhai Y, Zhang Y, et al. Genomic Profiling of Penile Squamous Cell Carcinoma Reveals New Opportunities for Targeted Therapy. Cancer Res (2015) 75(24):5219–27. doi: 10.1158/0008-5472.CAN-15-1004
11. Wang Y, Wang K, Chen Y, Zhou J, Liang Y, Yang X, et al. Mutational Landscape of Penile Squamous Cell Carcinoma in a Chinese Population. Int J Cancer (2019) 145(5):1280–9. doi: 10.1002/ijc.32373
12. Zhou QH, Deng CZ, Li ZS, Chen JP, Yao K, Huang KB, et al. Molecular Characterization and Integrative Genomic Analysis of a Panel of Newly Established Penile Cancer Cell Lines. Cell Death Dis (2018) 9(6):684. doi: 10.1038/s41419-018-0736-1
13. Chahoud J, Pickering CR, Pettaway CA. Genetics and Penile Cancer: Recent Developments and Implications. Curr Opin Urol (2019) 29(4):364–70. doi: 10.1097/MOU.0000000000000640
14. Aydin AM, Chahoud J, Adashek JJ, Azizi M, Magliocco A, Ross JS, et al. Understanding Genomics and the Immune Environment of Penile Cancer to Improve Therapy. Nat Rev Urol (2020) 17(10):555–70. doi: 10.1038/s41585-020-0359-z
15. Reddy JP, Pettaway CA, Levy LB, Pagliaro LC, Tamboli P, Rao P, et al. Factors Associated With Regional Recurrence After Lymph Node Dissection for Penile Squamous Cell Carcinoma. BJU Int (2017) 119(4):591–7. doi: 10.1111/bju.13686
16. Zheng W, Li K, Zhu W, Ding Y, Wu Q, Tang Q, et al. Nomogram Prediction of Overall Survival Based on Log Odds of Positive Lymph Nodes for Patients With Penile Squamous Cell Carcinoma. Cancer Med (2020) 9(15):5425–35. doi: 10.1002/cam4.3232
17. Necchi A, Lo Vullo S, Mariani L, Zhu Y, Ye DW, Ornellas AA, et al. Nomogram-Based Prediction of Overall Survival After Regional Lymph Node Dissection and the Role of Perioperative Chemotherapy in Penile Squamous Cell Carcinoma: A Retrospective Multicenter Study. Urol Oncol (2019) 37(8):531.e7–15. doi: 10.1016/j.urolonc.2019.04.003
18. Bandini M, Spiess PE, Pederzoli F, Marandino L, Brouwer OR, Albersen M, et al. A Risk Calculator Predicting Recurrence in Lymph Node Metastatic Penile Cancer. BJU Int (2020) 126(5):577–85. doi: 10.1111/bju.15177
19. Rieken M, Djajadiningrat RS, Kluth LA, Favaretto RL, Xylinas E, Guimaraes GC, et al. Predictors of Cancer-Specific Mortality After Disease Recurrence in Patients With Squamous Cell Carcinoma of the Penis. Eur Urol (2014) 66(5):811–4. doi: 10.1016/j.eururo.2014.05.032
20. Reyes ME, Borges H, Adjao MS, Vijayakumar N, Spiess PE, Schabath MB. Novel Prognostic Models for Patients With Penile Carcinoma. Cancer Control (2020) 27(1):1–12. doi: 10.1177/1073274820924728. 1073274820924728.
21. Zhang J, Cong R, Zhao K, Wang Y, Song N, Gu M. High TRIAP1 Expression in Penile Carcinoma Is Associated With High Risk of Recurrence and Poor Survival. Ann Transl Med (2019) 7(14):330. doi: 10.21037/atm.2019.06.47
22. Wang JY, Gao MZ, Yu DX, Xie DD, Wang Y, Bi LK, et al. Histological Subtype Is a Significant Predictor for Inguinal Lymph Node Metastasis in Patients With Penile Squamous Cell Carcinoma. Asian J Androl (2018) 20(3):265–9. doi: 10.4103/aja.aja_60_17
23. Peak TC, Russell GB, Dutta R, Rothberg MB, Chapple AG, Hemal AK. A National Cancer Database-Based Nomogram to Predict Lymph Node Metastasis in Penile Cancer. BJU Int (2019) 123(6):1005–10. doi: 10.1111/bju.14652
24. Lutzen U, Zuhayra M, Marx M, Zhao Y, Colberg C, Knupfer S, et al. Value and Efficiency of Sentinel Lymph Node Diagnostics in Patients With Penile Carcinoma With Palpable Inguinal Lymph Nodes as a New Multimodal, Minimally Invasive Approach. Eur J Nucl Med Mol Imaging (2016) 43(13):2313–23. doi: 10.1007/s00259-016-3482-6
25. Khalil MI, Kamel MH, Dhillon J, Master V, Davis R, Hajiran AJ, et al. What You Need to Know: Updates in Penile Cancer Staging. World J Urol (2020). doi: 10.1007/s00345-020-03302-z
26. Arnedo-Pac C, Mularoni L, Muinos F, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUSTL: A Sequence-Based Clustering Method to Identify Cancer Drivers. Bioinformatics (2019) 35(24):5396. doi: 10.1093/bioinformatics/btz588
27. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N Engl J Med (2014) 371(23):2189–99. doi: 10.1056/NEJMoa1406498
28. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: Efficient and Comprehensive Analysis of Somatic Variants in Cancer. Genome Res (2018) 28(11):1747–56. doi: 10.1101/gr.239244.118
29. Mroz EA, Rocco JW. MATH, a Novel Measure of Intratumor Genetic Heterogeneity, Is High in Poor-Outcome Classes of Head and Neck Squamous Cell Carcinoma. Oral Oncol (2013) 49(3):211–5. doi: 10.1016/j.oraloncology.2012.09.007
30. Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, et al. Replication Stress Links Structural and Numerical Cancer Chromosomal Instability. Nature (2013) 494(7438):492–6. doi: 10.1038/nature11935
31. Graafland NM, Lam W, Leijte JA, Yap T, Gallee MP, Corbishley C, et al. Prognostic Factors for Occult Inguinal Lymph Node Involvement in Penile Carcinoma and Assessment of the High-Risk EAU Subgroup: A Two-Institution Analysis of 342 Clinically Node-Negative Patients. Eur Urol (2010) 58(5):742–7. doi: 10.1016/j.eururo.2010.08.015
32. Kirrander P, Andren O, Windahl T. Dynamic Sentinel Node Biopsy in Penile Cancer: Initial Experiences At a Swedish Referral Centre. BJU Int (2013) 111(3 Pt B):E48–53. doi: 10.1111/j.1464-410X.2012.11437.x
33. Peyraud F, Allenet C, Gross-Goupil M, Domblides C, Lefort F, Daste A, et al. Current Management and Future Perspectives of Penile Cancer: An Updated Review. Cancer Treat Rev (2020) 90:102087. doi: 10.1016/j.ctrv.2020.102087
34. Soufir N, Queille S, Liboutet M, Thibaudeau O, Bachelier F, Delestaing G, et al. Inactivation of the CDKN2A and the p53 Tumour Suppressor Genes in External Genital Carcinomas and Their Precursors. Br J Dermatol (2007) 156(3):448–53. doi: 10.1111/j.1365-2133.2006.07604.x
35. Day PM, Thompson CD, Schowalter RM, Lowy DR, Schiller JT. Identification of a Role for the trans-Golgi Network in Human Papillomavirus 16 Pseudovirus Infection. J Virol (2013) 87(7):3862–70. doi: 10.1128/JVI.03222-12
36. Biswas NK, Das C, Das S, Maitra A, Nair S, Gupta T, et al. Lymph Node Metastasis in Oral Cancer Is Strongly Associated With Chromosomal Instability and DNA Repair Defects. Int J Cancer (2019) 145(9):2568–79. doi: 10.1002/ijc.32305
37. Lerias S, Esteves S, Silva F, Cunha M, Cochicho D, Martins L, et al. Cd274 (Pd-L1), CDKN2A (P16), TP53, and EGFR Immunohistochemical Profile in Primary, Recurrent and Metastatic Vulvar Cancer. Mod Pathol (2020) 33(5):893–904. doi: 10.1038/s41379-019-0429-z
38. Li YY, Hanna GJ, Laga AC, Haddad RI, Lorch JH, Hammerman PS. Genomic Analysis of Metastatic Cutaneous Squamous Cell Carcinoma. Clin Cancer Res (2015) 21(6):1447–56. doi: 10.1158/1078-0432.CCR-14-1773
Keywords: penile squamous cell carcinoma, lymph node metastasis, somatic alteration, patient survival, whole-exome sequencing
Citation: Cao J, Yang C-H, Han W-Q, Xie Y, Liu Z-Z and Jiang S-S (2021) Correlation Between the Evolution of Somatic Alterations During Lymphatic Metastasis and Clinical Outcome in Penile Squamous Cell Carcinoma. Front. Oncol. 11:641869. doi: 10.3389/fonc.2021.641869
Received: 15 December 2020; Accepted: 29 April 2021;
Published: 02 June 2021.
Edited by:
Qiyuan Li, Xiamen University, ChinaCopyright © 2021 Cao, Yang, Han, Xie, Liu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Cao, Y2FvamlhbkBobmNhLm9yZy5jbg==; Shu-Suan Jiang, amlhbmdzaHVzdWFuQGhuY2Eub3JnLmNu
 Jian Cao
Jian Cao Chun-He Yang
Chun-He Yang Wei-Qing Han1
Wei-Qing Han1 Shu-Suan Jiang
Shu-Suan Jiang