- 1Department of Pathology, Anhui Medical University, Hefei, China
- 2Department of Pathology, The First Affiliated Hospital of Anhui Medical University, Hefei, China
- 3Zhejiang Provincial Key Laboratory of Pancreatic Disease, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, China
- 4The Key Laboratory of Developmental Genes and Human Disease, Institute of Life Sciences, Southeast University, Nanjing, China
Introduction: Estrogen receptors (ESRs) and progesterone receptors (PGRs) are associated with the development and progression of various tumors. The feasibility of ESRs and PGRs as prognostic markers and therapeutic targets for multiple cancers was evaluated via pan-cancer analysis.
Methods: The pan-cancer mRNA expression levels, genetic variations, and prognostic values of ESR1, ESR2, and PGR were analyzed using the Gene Expression Profiling Interactive Analysis 2 (GEPIA2) and cBioPortal. The expression levels of ERa, ERb, and PGR proteins were detected by immunohistochemical staining using paraffin-embedded tissue specimens of ovarian serous cystadenocarcinoma (OV) and uterine endometrioid adenocarcinoma (UTEA). Correlation between immunomodulators and immune cells was determined based on the Tumor and Immune System Interaction Database (TISIDB).
Results: ESR1, ESR2, and PGR mRNAs were found to be differentially expressed in different cancer types, and were associated with tumor progression and clinical prognosis. ERa, ERb, and PGR proteins were further determined to be significantly differentially expressed in OV and UTEA via immunohistochemical staining. The expression of ERa protein was positively correlated with a high tumor stage, whereas the expression of PGR protein was conversely associated with a high tumor stage in patients with OV. In patients with UTEA, the expression levels of both ERa and PGR proteins were conversely associated with tumor grade and stage. In addition, the expression levels of ESR1, ESR2, and PGR mRNAs were significantly associated with the expression of immunomodulators and immune cells.
Conclusion: ESR1, ESR2, and PGR are potential prognostic markers and therapeutic targets, as well as important factors for the prediction, evaluation, and individualized treatment in several cancer types.
Introduction
Nowadays, cancer has become a leading cause of death worldwide, with continuously increasing rates of morbidity and mortality (1). In 2017, approximately 2.6 million Chinese individuals died of various types of cancer, accounting for 26.07% of the total deaths (2, 3). Multiple therapeutic strategies, including but not limited to surgery, radiotherapy, chemotherapy, and immunotherapy, have been developed for the comprehensive and individualized treatment of malignant tumors. However, overall clinical outcomes in patients with advanced cancers are still dissatisfactory, especially given the concomitant adverse effects. Therefore, there is an urgent need to identify potentially valuable molecular targets for the improvement of therapeutic efficacy and specificity.
Estrogen receptors (ESRs) belong to nuclear receptor superfamily of hormone-inducible transcription factors, which comprise ERa and ERb, encoded by ESR1 and ESR2, respectively (4, 5). PGR encodes a member of the steroid receptor superfamily, named progesterone receptors (PGRs) (6). In physiological state, the activation of ESRs and PGRs by the binding of their ligands are associated with a series of normal physical activities. However, under pathological conditions, ESR1, ESR2, and PGR have been demonstrated to be associated with tumorigenesis and tumor progression (7, 8). For instance, ESR1 is well characterized as a factor that promotes cell proliferation in breast cancer (9). In contrast, ESR2 seems to be a tumor suppressor gene (10), which is not expressed in early stages of breast cancer (11). Further, PGR is associated with the development of breast cancer (12). In addition to breast cancer, ESR1, ESR2, and PGR also mediate the progression of prostate cancer (13–15), colon cancer (16–18), ovarian cancer (19–21), and lung cancer (22–24). Accordingly, ESR1, ESR2, and PGR may be prognostic biomarkers as well as potential therapeutic targets for a variety of cancer types, necessitating further evaluation.
In this study, we conducted a comprehensive pan-cancer analysis of ESR1, ESR2, and PGR on the basis of online databases. The expression levels of ESR1, ESR2, and PGR, and the correlation of ESR1, ESR2, and PGR with overall survival (OS) and disease-free survival (RFS) in patients were assessed using Gene Expression Profiling Interactive Analysis 2 (GEPIA2). The expression levels of ERa, ERb, and PGR proteins in ovarian serous cystadenocarcinoma (OV) and uterine endometrioid adenocarcinoma (UTEA) were validated using in-house tissue specimens, and the relationship between protein levels of ERa, ERb, and PGR and clinicopathological characteristics of OV or UTEA patients was explored. Genetic alterations and immunological effects of ESR1, ESR2, and PGR were analyzed using the cBioPortal and Tumor and Immune System Interaction Database (TISIDB), respectively.
Materials and Methods
Patient Tissue Sample Collection
Forty-two paraffin-embedded OV and 51 UTEA tissue specimens were collected from patients who underwent surgery at the High-tech district of the First Affiliated Hospital of Anhui Medical University (Hefei, Anhui, China) between 2017 and 2019. We also collected 11 specimens of normal ovarian tissue from 42 patients with OV (31 specimens of tumors involving bilateral ovarian tissue were excluded) and 34 specimens of normal endometrial tissue adjacent to the cancer in 51 patients with UTEA (17 specimens of tumors involving the entire endometrial tissue were excluded). No patient had a history of other malignant tumors and no patient had undergone preoperative interventions such as radiotherapy or chemotherapy. Each patient provided written informed consent, and the study was approved by the institutional review board.
GEPIA2 Dataset Analysis
The expression levels of ESR1, ESR2, and PGR mRNAs in tumor and matched normal samples were compared using the GEPIA2 database, which is a webserver that provides cancer genomics data based on TCGA, and the GTEx database (25). In this study, differentially expressed gene analysis of tumor and matched normal samples, isoform profiling, and clinicopathological stage analysis were performed using the GEPIA2 dataset. Differentially expressed gene analysis and clinicopathological stage analysis were conducted by one-way ANOVA. Genes with |log2FC| > 1 and Q-value < 0.01 were considered to be differentially expressed. We used log2(TPM+1) for log-scaling differential expression in different clinicopathological stages, and regarded Pr(>F) < 0.05 to be statistically significant. In addition, correlative prognostic analysis of ESR1, ESR2, and PGR, including OS and RFS, was conducted to evaluate the prognostic significance using log-rank test for hypothesis evaluation at the median cutoff with 50% for either low- or high-expression cohorts.
cBioPortal Analysis
The cBioPortal for Cancer Genomics is a widely used open-access website, providing a visualization and analysis tool for multidimensional cancer genomics data (26, 27). The cBioPortal was employed to analyze the OncoPrint, mutual exclusivity, alteration frequency in multiple cancer types, and amino acid changes in proteins and for the Clinical Attribute Test. Mutual exclusivity analysis among ESR1, ESR2, and PGR was conducted using Log2 odds ratio, P-value, and Q-value, and P-value < 0.001 and Q-value < 0.001 were regarded as statistically significant.
TISIDB Analysis
The TISIDB is a user-friendly web portal containing 988 immune-related anti-tumor genes derived from 4,176 records in 2,530 publications. This database enables users to analyze the function of selected genes in the tumor–immune interplay through high-throughput data analysis or literature mining (28). In this study, we used TISIDB to construct heat maps for analyzing the spearman correlations between the expression levels of ESR1, ESR2, and PGR and immunomodulators and immune cells in multiple cancer types. A p value < 0.05 was regarded as statistically significant.
Immunohistochemical Analysis
The in situ protein expression levels of ERa, ERb, and PGR in paraffin-embedded OV and UTEA tissue sections were detected by immunohistochemistry using rabbit polyclonal antibodies against ESR1 (1:200, 21244-1-AP, Proteintech), ESR2 (1:50, 14007-1-AP, Proteintech), and PGR (1:50, 25871-1-AP, Proteintech). Five fields were randomly observed at high power under the microscope. ERa, ERb, and PGR staining intensity of the tumor cells (0, no tumor cells stained yellow; 1, light yellow stain; 2, medium depth yellow stain; and 3, dark yellow stain) and the percentage of stained cells (0, no positive tumor cells; 1, <25% positive cells, 2, 25%–50% positive cells, and 3, > 50% positive cells) were recorded, and the sum of the two group scores ranged from 0 to 6 (17). Samples with staining scores of 0–3 were designated as ERa/ERb/PGR low expression, whereas those with staining scores >3 were designated as ERa/ERb/PGR high expression.
Statistical Analysis
SPSS22.0 was used for data analysis. Chi-square test was used for variable comparison, with p < 0.05 regarded as statistically significant. Spearman’s method was used to assess the correlation between factors. p < 0.05 was regarded as statistically significant.
Results
ESR1, ESR2, and PGR mRNAs Are Differentially Expressed in Various Cancers
To explore the expression of ESR1, ESR2 and PGR in pan-cancer, we analyzed their mRNA levels via GEPIA2. We found that the ESR1 mRNA was highly expressed in breast invasive carcinoma (BRCA) and OV samples compared with their matched normal samples (Figure 1A). In contrast, low expression levels of ESR1 mRNA were found in bladder urothelial carcinoma (BLCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), liver hepatocellular carcinoma (LIHC), testicular germ cell tumors (TGCT), and uterine carcinosarcoma (UCS) samples. ESR2 mRNA was observed to be highly expressed only in the lymphoid neoplasm diffuse large B-cell lymphoma (DLBCL) samples, whereas a low expression of ESR2 mRNA was found in adrenocortical carcinoma (ACC), OV, and TGCT samples (Figure 1B). In addition, a low expression of PGR mRNA was found in CESC, colon adenocarcinoma (COAD), OV, prostate adenocarcinoma (PRAD), rectal adenocarcinoma (READ), TGCT, UCEC, and UCS samples (Figure 1C). Together, ESR1, ESR2, and PGR are differentially expressed in multiple cancer types.
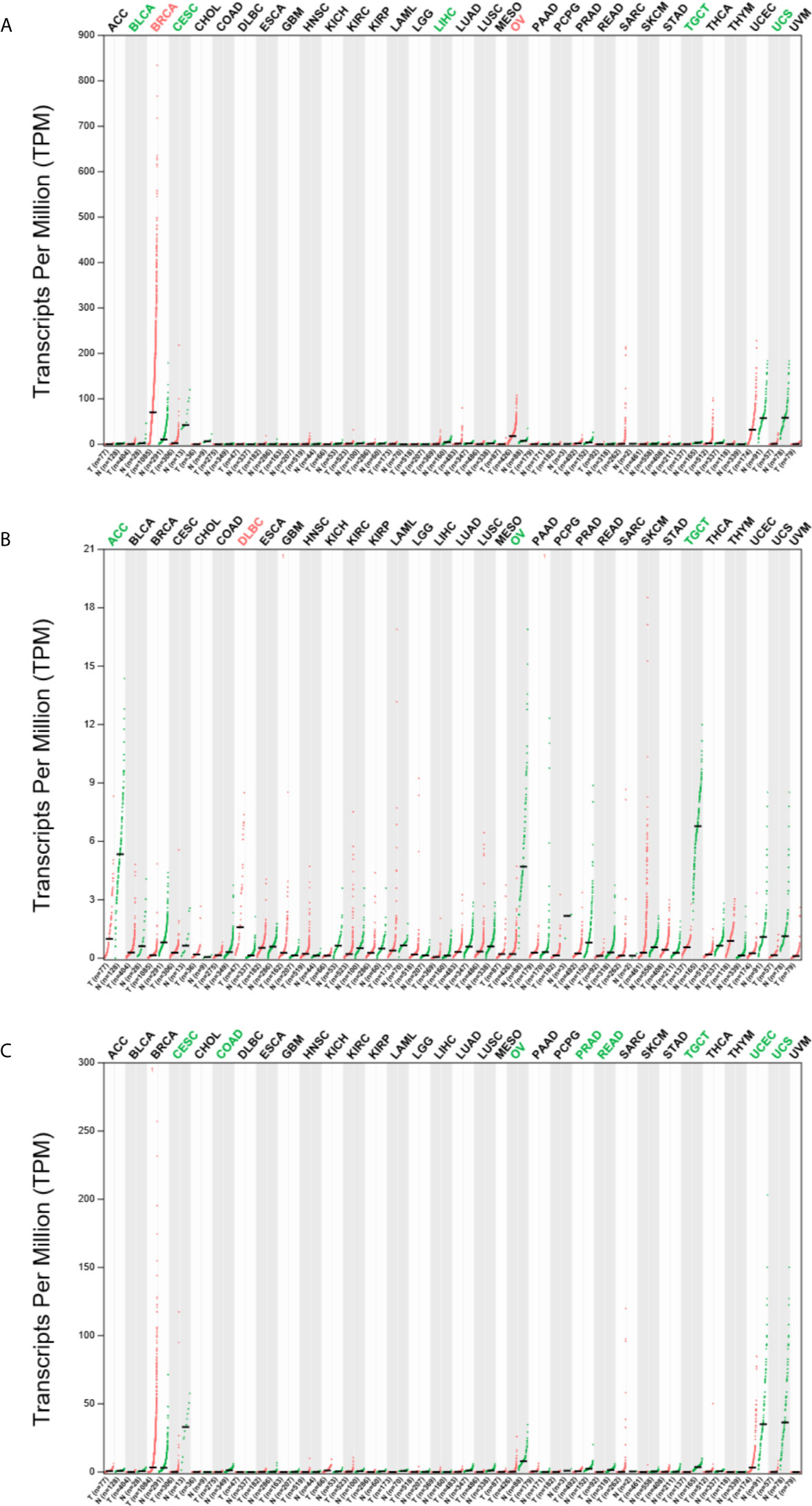
Figure 1 Expression profile of ESR1, ESR2, and PGR mRNAs across multiple cancer and matched normal samples. (A) Expression profile of ESR1 mRNA across multiple cancer and matched normal samples. (B) Expression profile of ESR2 mRNA across multiple cancer and matched normal samples. (C) Expression profile of PGR mRNA across multiple cancer and matched normal samples.
ESR1, ESR2, and PGR Isoforms Are Differentially Expressed in Different Cancer Types
To investigate the distribution of ESR1, ESR2, and PGR isoforms in pan-cancer, we compared their expression levels via GEPIA2. As shown in Figures 2A–C, the most prevalent transcripts are differentially expressed across multiple cancer types. For example, ESR-202 was the most prevalent ESR1 transcript in BRCA samples, followed by ESR-001 and ESR-201, whereas ESR-004 was the most prevalent ESR2 transcript in the same samples. In DLBCL samples, ESR-201 was the common ESR1 transcript and ESR-202 was the most prevalent ESR2 transcript. We also profiled isoform usage of these genes (Figures 3A–C). ESR1-201, ESR1-202, ESR2-004, ESR2-005 and PGR-001 were mostly commonly used transcribed isoforms in different cancer types. Thus, there exists isoform transformation during the transcription process of these genes as per the cancer type. Together, ESR1, ESR2, and PGR isoforms are differentially expressed in different cancer types.
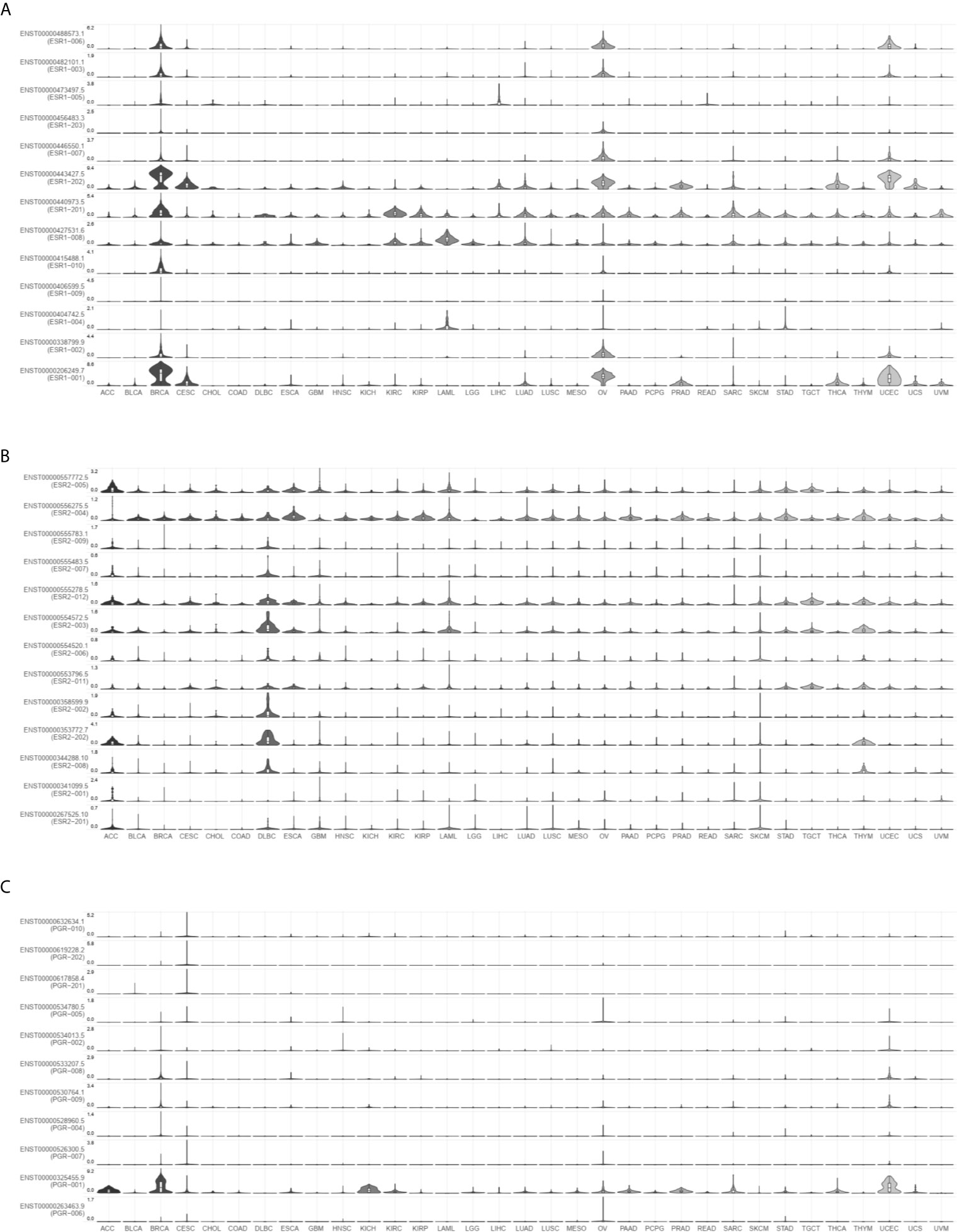
Figure 2 Expression distribution of ESR1, ESR2, and PGR mRNAs across multiple cancer types. (A) Expression distribution of ESR1 mRNA across multiple cancer types. (B) Expression distribution of ESR2 mRNA across multiple cancer types. (C) Expression distribution of PGR mRNA across multiple cancer types.

Figure 3 Isoform expression of ESR1, ESR2, and PGR genes across multiple cancer types. (A) Isoform expression of ESR1 gene across multiple cancer types. (B) Isoform expression of ESR2 gene across multiple cancer types. (C) Isoform expression of PGR gene across multiple cancer types.
Correlation Between the Expression of ESR1, ESR2, and PGR mRNAs and Tumor Stage Across Multiple Cancers
To examine the clinical relevance of ESR1, ESR2, and PGR in pan-cancer, we analyzed the correlations of their expression with tumor stage. As shown in Figures 4A–C, ESR1 and PGR transcription levels were correlated with the tumor stage (p < 0.05), and the higher expression of ESR1 and ESR2 are associated with higher tumor stage. In contrast, no significant association between ESR2 and tumor stage was observed (p > 0.05). Taken together, the expression of ESR1 and PGR was significantly associated with pan-cancer tumor stage.
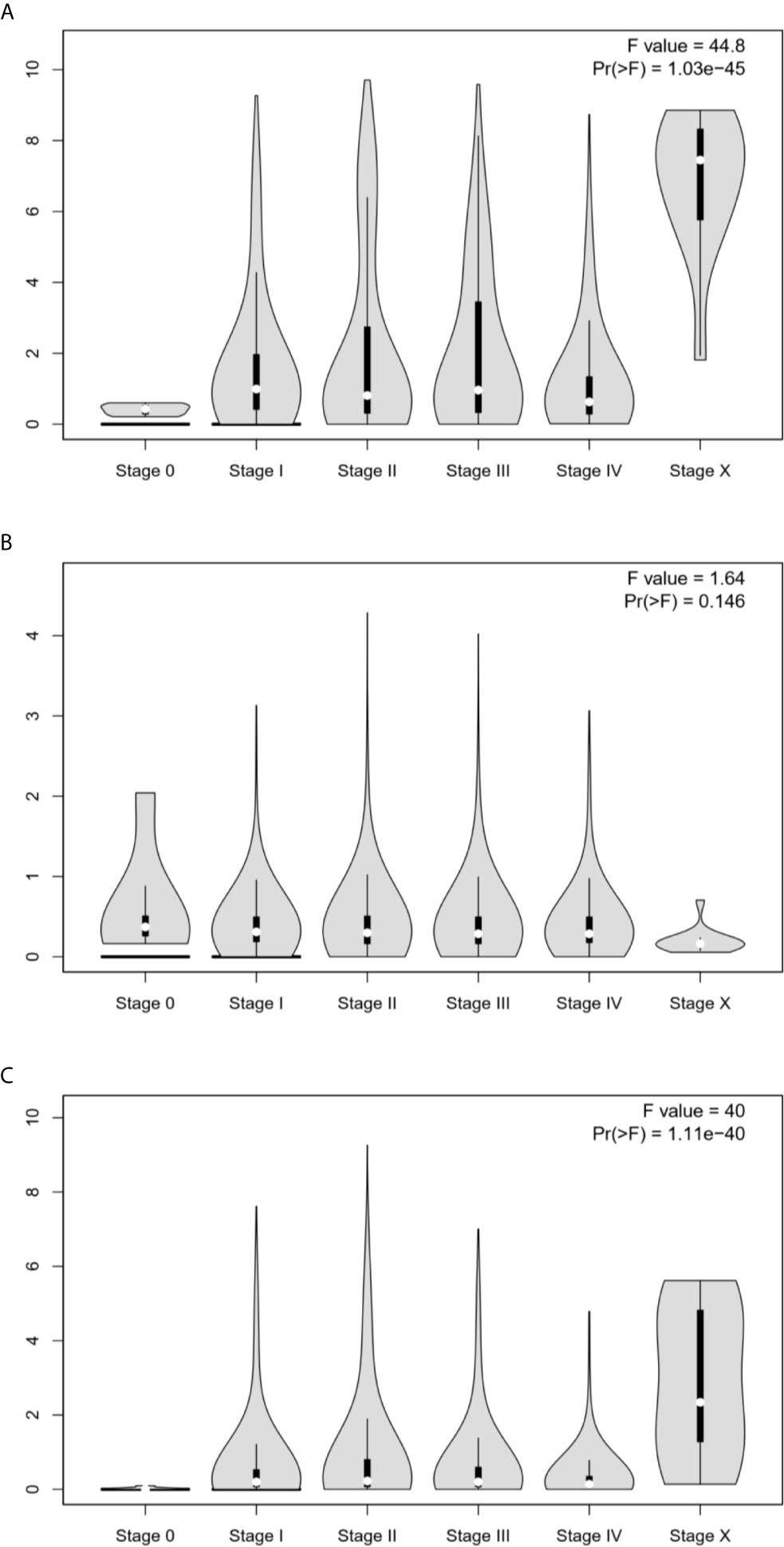
Figure 4 Correlation between ESR1, ESR2, and PGR mRNA expression levels and the tumor stage in multiple cancer samples. (A) Correlation between ESR1 mRNA expression level and tumor stage in different cancer samples. (B) Correlation between ESR2 mRNA expression level and tumor stage in different cancer samples. (C) Correlation between PGR mRNA expression level and tumor stage in different cancer samples.
Expression Levels of ERa, ERb, and PGR Proteins in OV and UTEA
To detect the expression levels of ERa, ERb, and PGR proteins in OV and UTEA, which were not reported in any previous studies, we next performed immunohistochemical staining using paraffin-embedded tissue specimens. The results showed that the expression level of ERa was significantly higher, while ERb and PGR were significantly lower in OV compared to these in their matched normal samples (p < 0.05) (Supplementary Material Figure S1A). The expression levels of ERa, ERb, and PGR proteins were significantly lower in UTEA samples compared with these in their matched normal samples (p < 0.05) (Supplementary Material Figure S1B). Moreover, ERa, ERb, and PGR proteins were highly expressed in 42.9%, 50%, and 21.4% of OV tumor samples and in 64.7%, 35.3%, and 60.8% of UTEA tumor samples, respectively (Table 1). Together, the expression trends of ERa, ERb, and PGR proteins in OV and UTEA as well as the matched normal tissue are in consistent with the findings obtained from GEPIA2 (Figure 5).
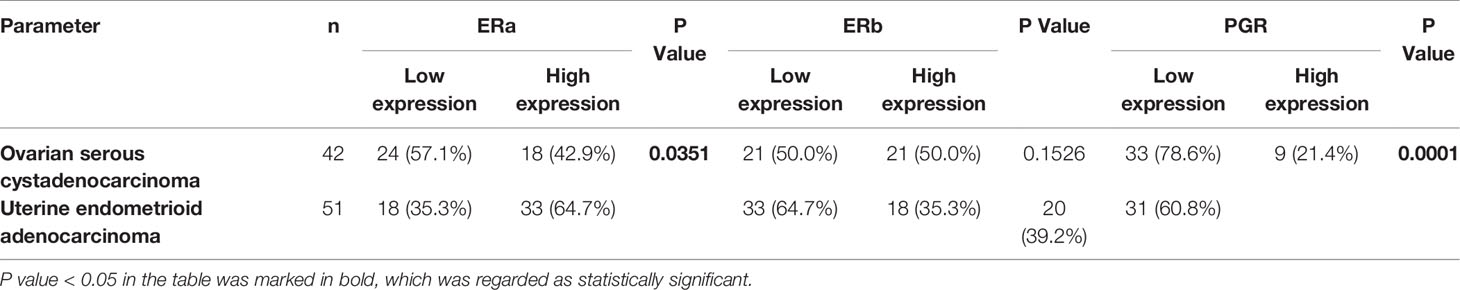
Table 1 Association among ERa, ERb, and PGR protein expression levels in tumor tissues of patients with ovarian serous cystadenocarcinoma and uterine endometrioid adenocarcinoma.
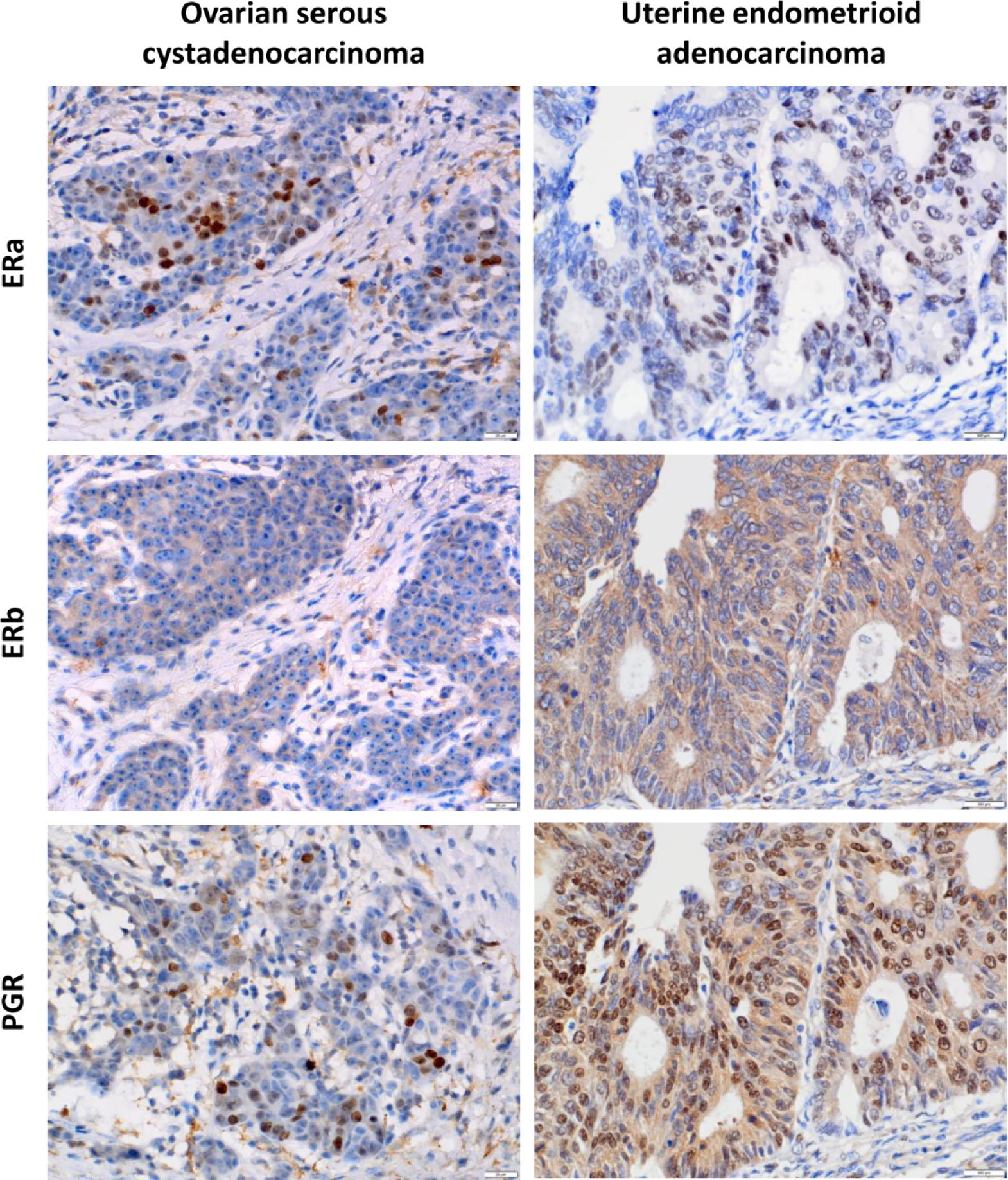
Figure 5 Immunohistochemical analysis of ERa, ERb, and PGR protein expression levels in ovarian serous cystadenocarcinoma and uterine endometrioid adenocarcinoma. Representative immunohistochemical images showed that ERa, ERb, and PGR were differentially expressed in ovarian serous carcinoma and endometrioid adenocarcinoma tissue. Left panels, low expression levels of ERa, ERb, and PGR proteins in serous ovarian carcinoma. Right panels, high expression levels of ERa, ERb, and PGR proteins in endometrioid adenocarcinoma. All micrographs were captured at ×400 magnification.
Association Between the Expressions of ERa, ERb, and PGR Proteins and the Clinicopathological Characteristics of Patients With OV or UTEA
To explore the clinical significance of ERa, ERb, and PGR expression in OV and UTEA, we further correlated their expression to clinicopathological characteristics of patients with OV or UTEA. Interestingly, the expression level of ERa protein was positively correlated to a high tumor stage, whereas the expression level of PGR protein was inversely correlated to a high tumor stage in patients with OV (Both p < 0.05) (Table 2). In UTEA, the expression levels of both ERa and PGR proteins were inversely correlated with high tumor grade and stage (all p < 0.05); this trend was not found in ERb (both p > 0.05) (Table 3). Further, the expression levels of ERb and PGR proteins were significantly correlated with patient age (both p < 0.05) (Table 3). Collectively, the expression of ERa, ERb, and PGR might be associated with the progression of OV and UTEA.
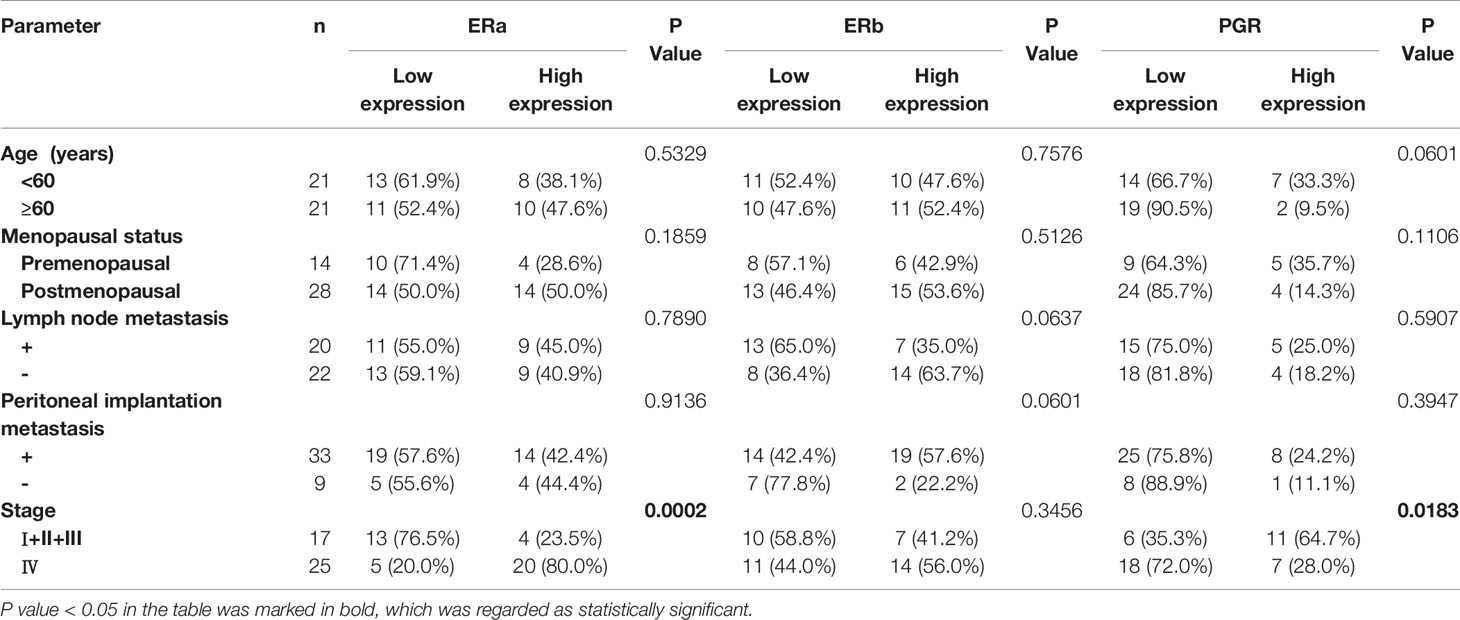
Table 2 Association of ERa, ERb, and PGR protein expression levels in tumors with the clinicopathological characteristics of patients with ovarian serous cystadenocarcinoma.
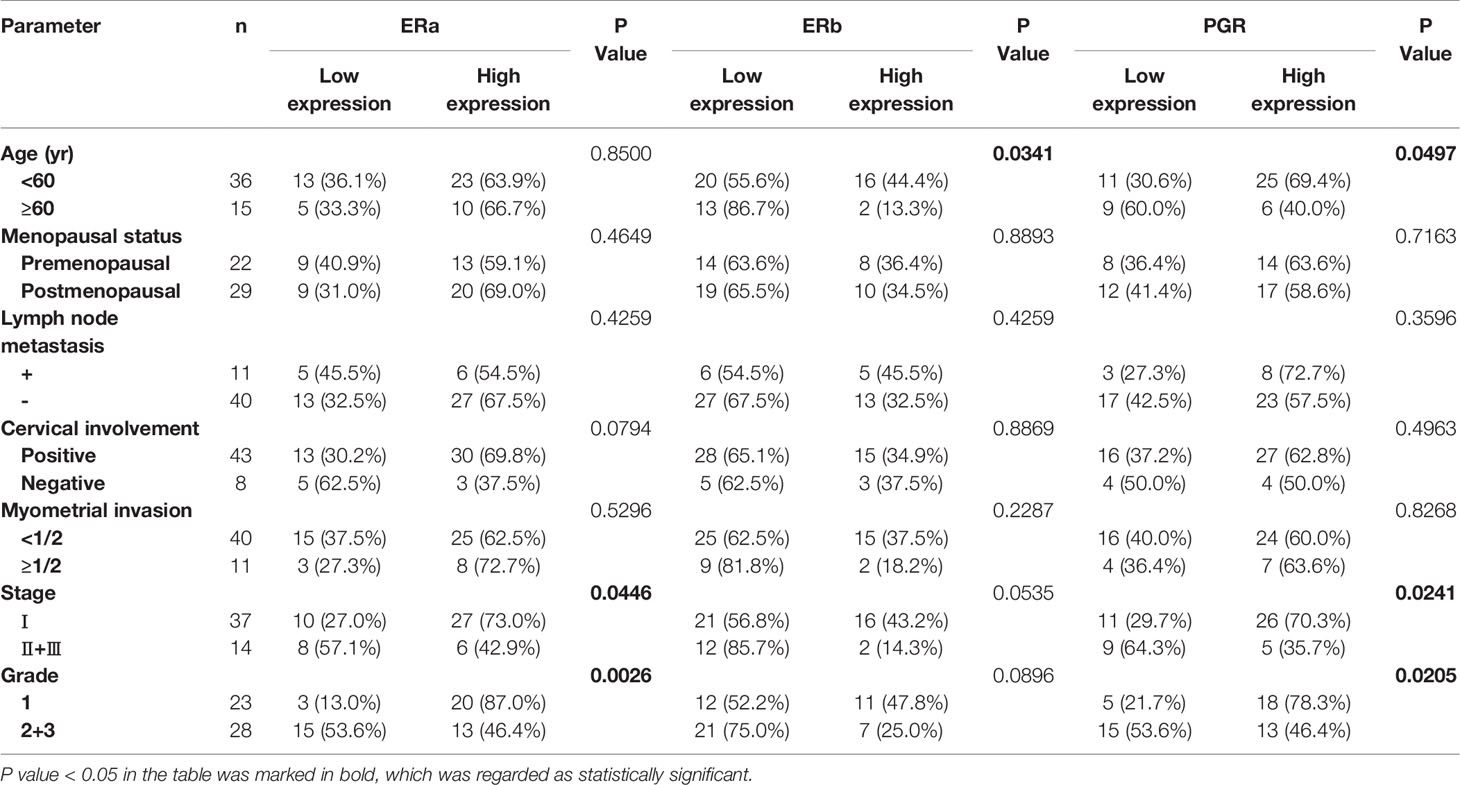
Table 3 Association of ERa, ERb, and PGR protein expression levels in tumors with the clinicopathological characteristics of patients with uterine endometrioid adenocarcinoma.
Genetic Alterations and Clinical Relevance of ESR1, ESR2, and PGR in Different Cancers
To inquiry genetic alterations of ESR1, ESR2, and PGR that may be associated with tumorigenesis, we analyzed these in pan-cancer involving a total of 10,189 patients. Genetic alterations (including amplification, fusion, deep deletion, missense mutation, and truncating mutation) were detected in 2.7%, 1.3%, and 3% of ESR1, ESR2, and PGR genes, respectively (Figure 6A). Moreover, a mutual exclusivity analysis showed the selected genes tended toward co-occurrence rather than mutual exclusivity (p < 0.05) (Figure 6B). Mutations in ESR1, ESR2, and PGR genes were the most frequent alterations in multiple cancer types, followed by amplifications and deep deletions (Figure 6C). The patients were further divided into ESR1, ESR2 and PGR altered and unaltered groups to conduct the clinical attribute test (Figure 7A). OncoTree Code was selected to indicate the ratio of cancer patients with/without genetic alterations in ESR1, ESR2, and PGR (Figure 7B), and results suggested the potentially critical roles of ESR1, ESR2 and PGR in onset of multiple cancers. Together, ESR1, ESR2, and PGR are likely closely correlated and have a role in multiple tumor genesis.
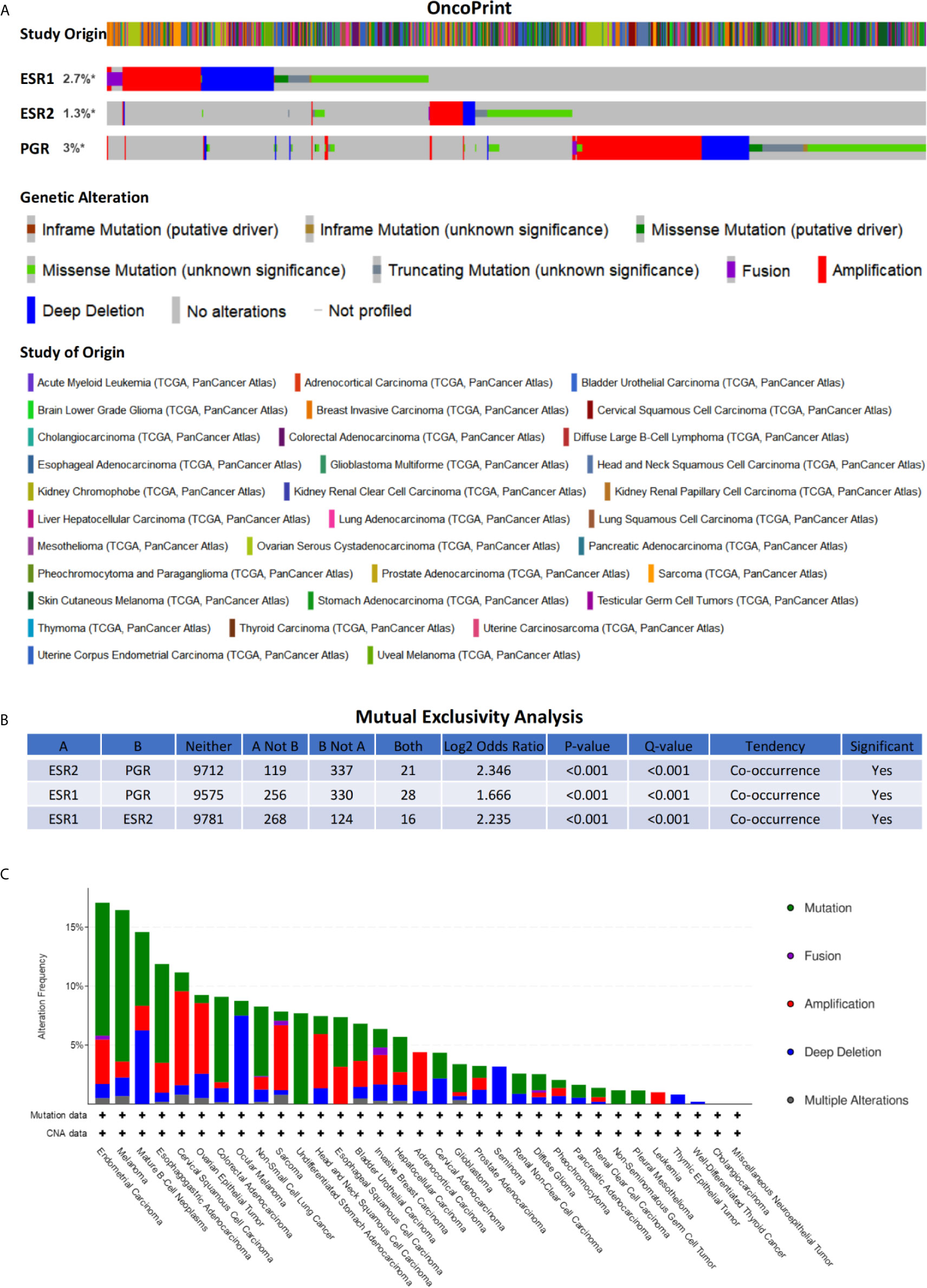
Figure 6 Genetic alterations of ESR1, ESR2, and PGR across multiple cancer types. (A) Alteration landscape for ESR1, ESR2, and PGR across multiple cancer types. (B) Mutual exclusivity analysis between alterations of ESR1, ESR2, and PGR across multiple cancer types. (C) Cancer type summary of ESR1, ESR2, and PGR alterations across multiple cancer types. * indicates not-profiled samples existing in the enquired gene.
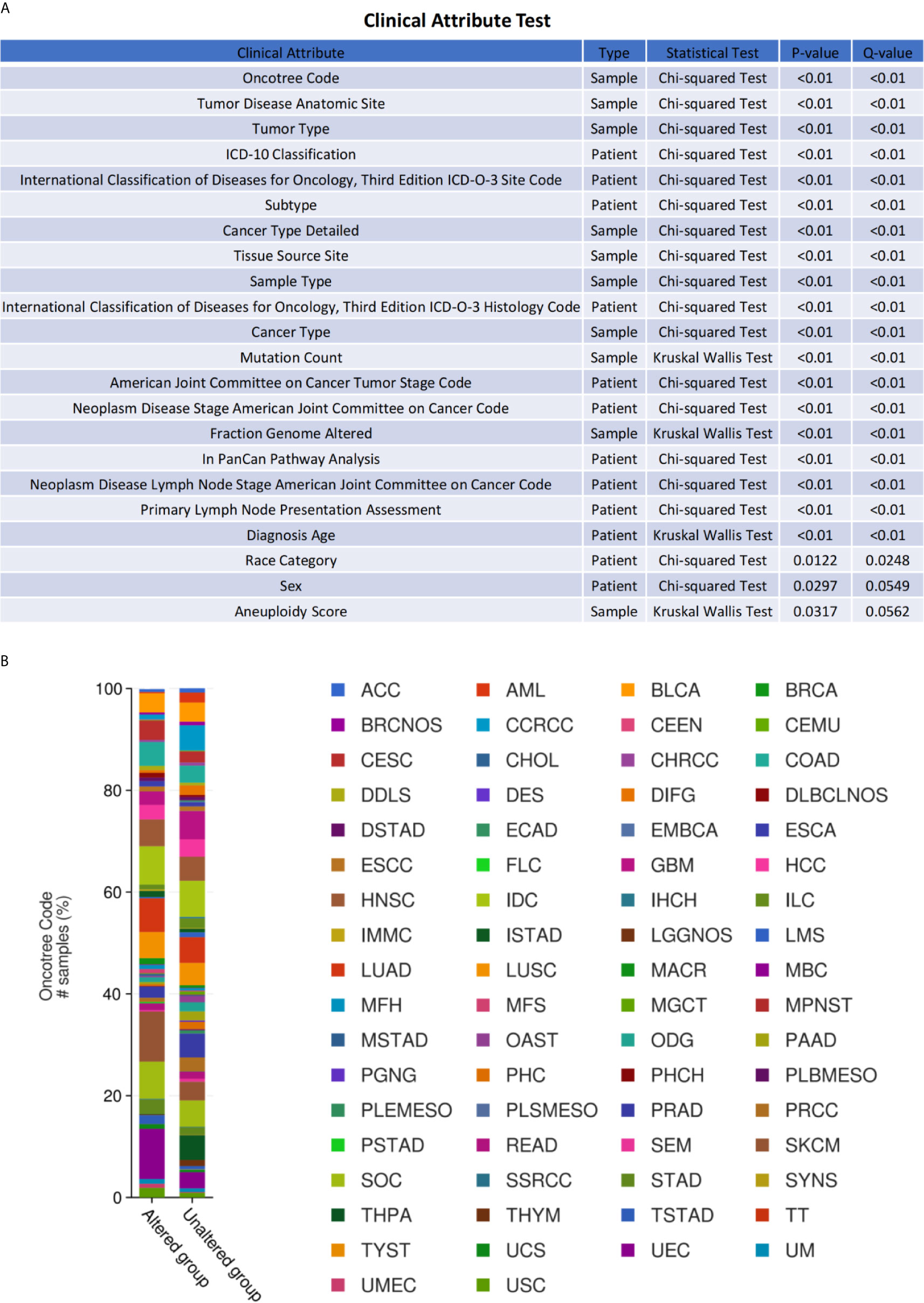
Figure 7 Clinical relevance of ESR1, ESR2, and PGR alterations across multiple cancer types. (A) OncoTree code of ESR1, ESR2, and PGR in different cancer types. (B) Clinical attribute test of ESR1, ESR2, and PGR in different cancer types.
Mutation Site Analysis of ESR1, ESR2, and PGR in Multiple Cancer Types
To identify mutation sites in the ESR1, ESR2, and PGR genes, we assessed 10,189 samples from multiple cancer types. The mutation sites were most commonly located within the Oest_recep, zf-C4, Hormone_recep, and ESR1_C domains (Figure 8A). Specifically, 169 mutations of ESR1 were detected, consisting of 131 missense mutations, 19 truncating mutations, 3 inframe mutations, and 16 other types of mutations. Seven of these mutations were E247K/D, a hotspot for protein activation. In addition, 112 ESR2 and 214 PGR nonsynonymous mutation sites were detected in different cancers, with the highest frequency mutations in R227H/C/L and R740Q/* (Figures 8B, C). Together, there are an abundance of mutation sites of ESR1, ESR2 and PGR, suggesting the complexity of their mutations.
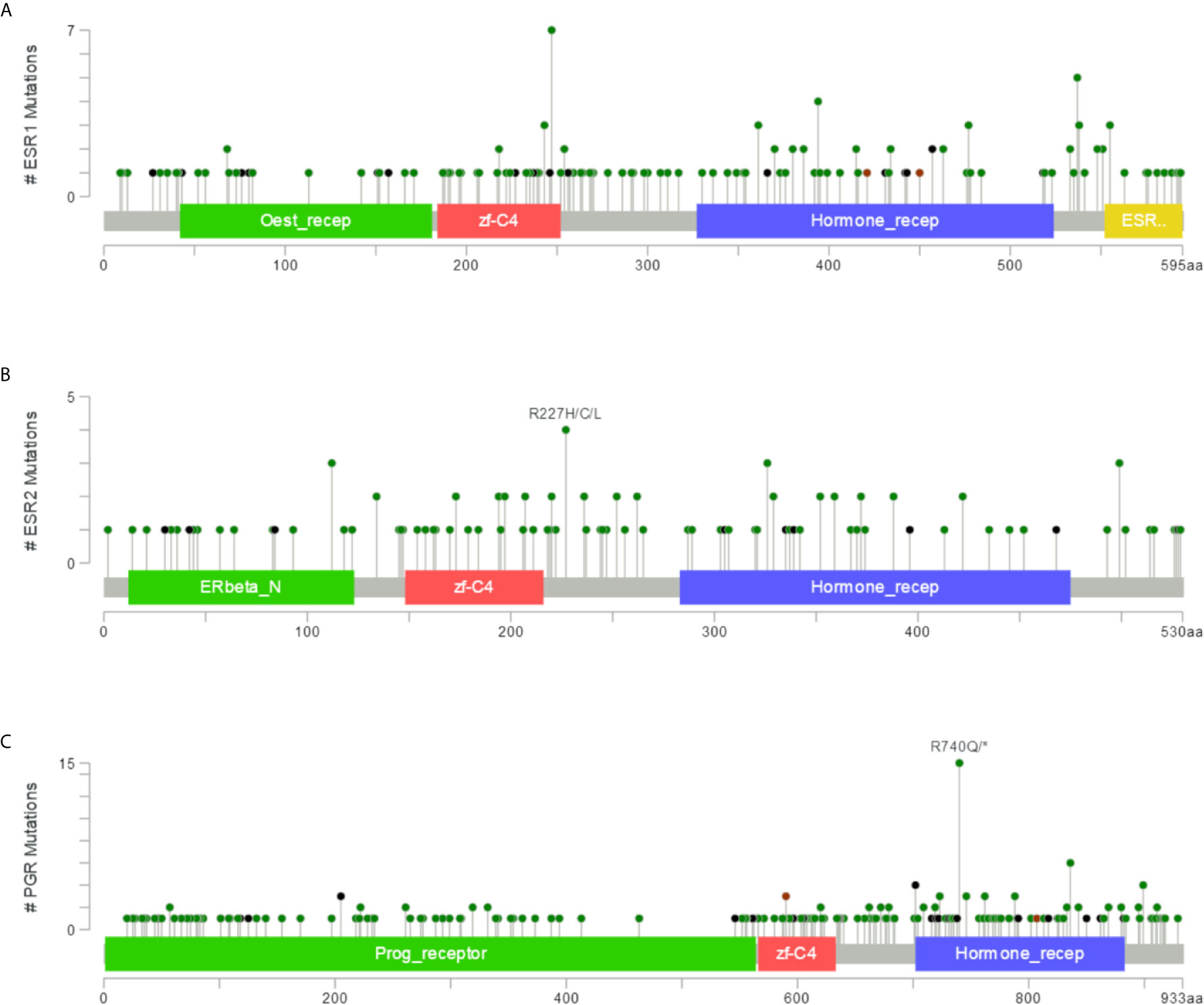
Figure 8 Mutations in ESR1, ESR2, and PGR across multiple cancer types. (A) Mutation frequency in ESR1 across multiple cancer types. (B) Mutation frequency in ESR2 across multiple cancer types. (C) Mutation frequency in PGR across multiple cancer types.
Immunological Correlation Between ESR1, ESR2, and PGR and Immune Modulatory Factors Across Multiple Cancer Types
To assess the relevance of ESR1, ESR2, and PGR with immune system that plays critical roles in cancer progression (28), we first compared their co-expression with the abundance of immunomodulators. As shown in Figures 9A–C, there is a positive correlation of the expression levels of ESR1, ESR2, and PGR genes with multiple immune-inhibitors (such as CD274, TIGIT, and CTLA4). Moreover, the expression levels of ESR1, ESR2, and PGR were observed to have a positive correlation with several immune-stimulators (such as CD27, CD28, and CXCL12) (Figures 9D–F). These findings suggest ESR1, ESR2 and PGR might be associate with both immune stimulation and inhibition. Together, the potential roles of ESR1, ESR2 and PGR in cancer are likely in an immune system-dependent manner.
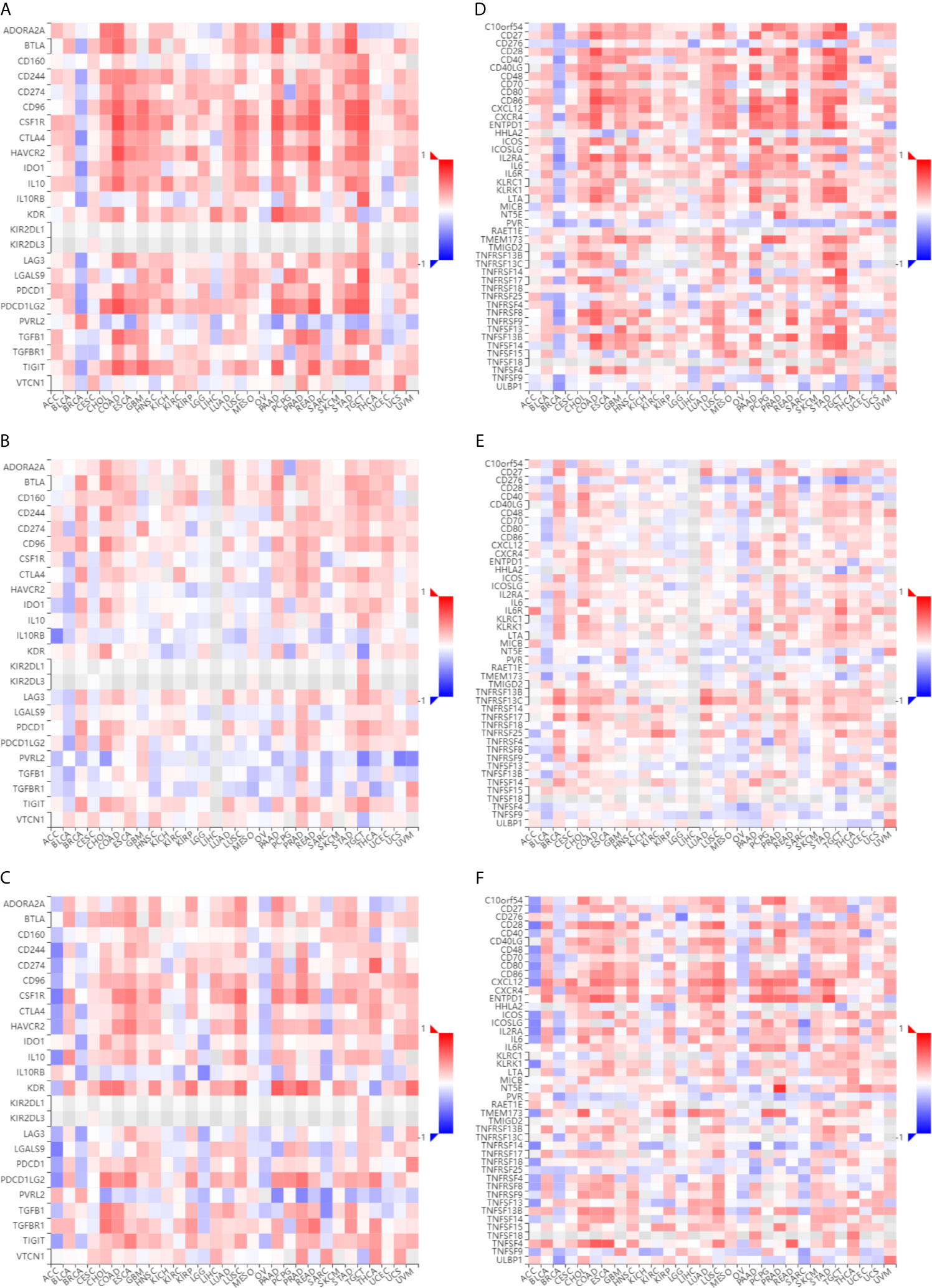
Figure 9 Immunological correlation of ESR1, ESR2, and PGR with various cancer immunomodulatory factors. (A) Correlations between ESR1 and immunoinhibitors across multiple cancer types. (B) Correlations between ESR2 and immunoinhibitors across multiple cancer types. (C) Correlations between PGR and immunoinhibitors across multiple cancer types. (D) Correlations between ESR1 and immunostimulators across multiple cancer types. (E) Correlations between ESR2 and immunostimulators across multiple cancer types. (F) Correlations between PGR and immunostimulators across multiple cancer types.
Blood Cell Type-Specific Expression Profiles of ESR1, ESR2, and PGR Across Multiple Cancer Types
To further explore the correlation of ESR1, ESR2 and PGR with immunity, we analyzed their expression in peripheral blood cell types. ESR1 was found to be expressed in classical monocyte, MAIT T-cell, naive CD4 T cells, memory CD4 T cells, memory CD8 T-cell, naïve CD8 T cell, memory B cell and myeloid dendritic cells (DC) (Figure 10A). Similarly, the expression of ESR2 was observed in multiple peripheral blood cells, with the highest expression level in plasma cell-like DC (Figure 10B). In contrast, PGR expression was not observed in peripheral blood cells (Figure 10C). Together, ESR1 and ESR2 might potentially influence multiple immune cells.
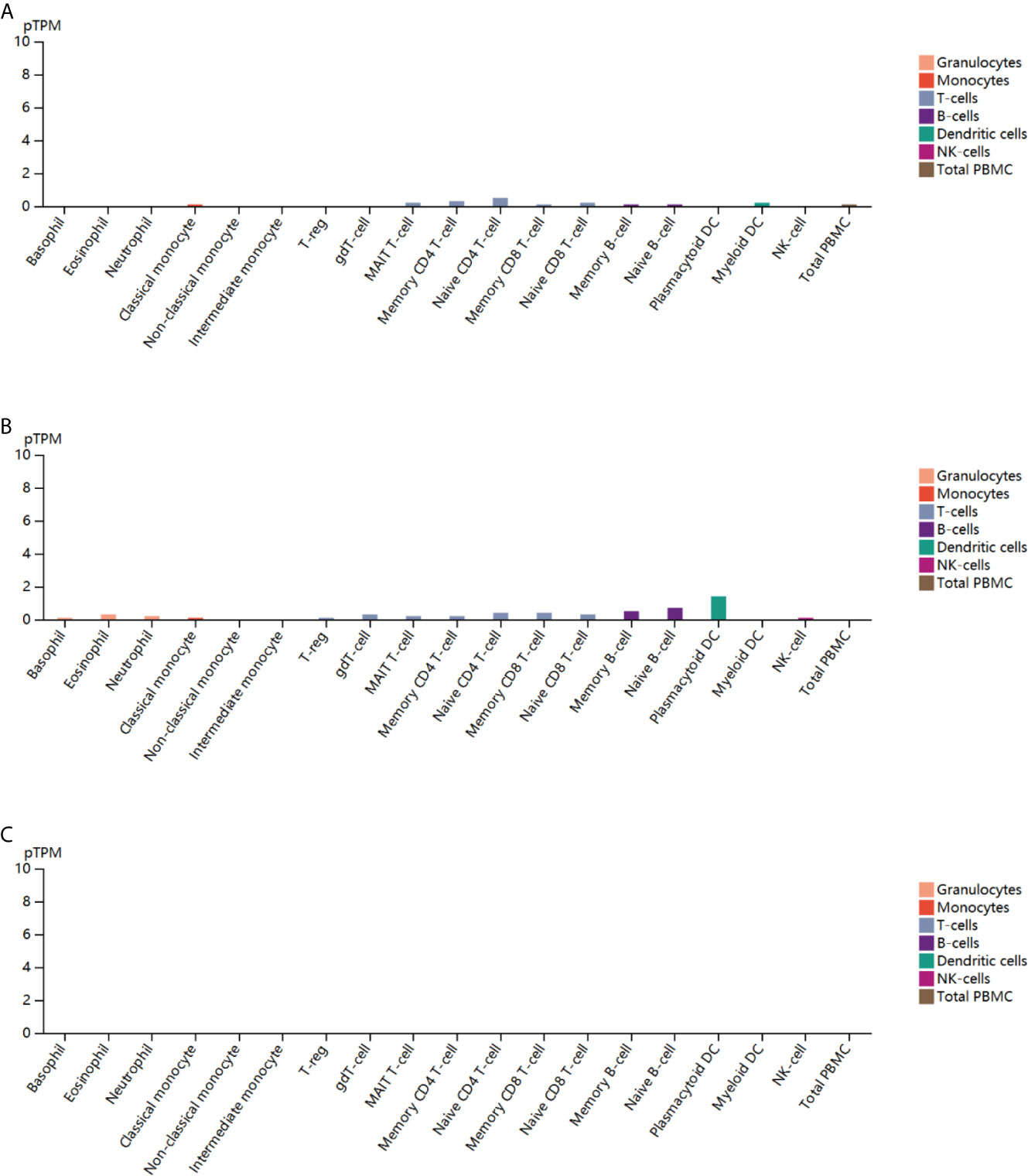
Figure 10 Blood cell type-specific expression profile of ESR1, ESR2, and PGR across multiple cancer types. (A) Blood cell type-specific distribution of ESR1 across multiple cancer types. (B) Blood cell type-specific distribution of ESR2 across multiple cancer types. (C) Blood cell type-specific distribution of PGR across multiple cancer types.
Contribution of ESR1, ESR2, and PGR to Survival Across Multiple Cancer Types
To investigate the potential roles of ESR1, ESR2, and PGR in prognosis, we analyzed the correlation of their expression with pan-cancer survival. As excepted, the expression of these genes was significantly associated with the OS and RFS in several cancers (Figures 11A, B). For instance, the higher expression of ESR1 is significantly related with superior OS in head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), liver hepatocellular carcinoma (LIHC) and skin cutaneous melanoma (SKCM). In contrast, the down-regulation of ESR1 is suggestive of better OS in acute myeloid leukemia (LAML), brain lower grade glioma (LGG), lung squamous cell carcinoma (LUSC) and stomach adenocarcinoma (STAD). Together, these findings suggest that ESR1, ESR2 and PGR are potential prognostic factor in multiple cancers.
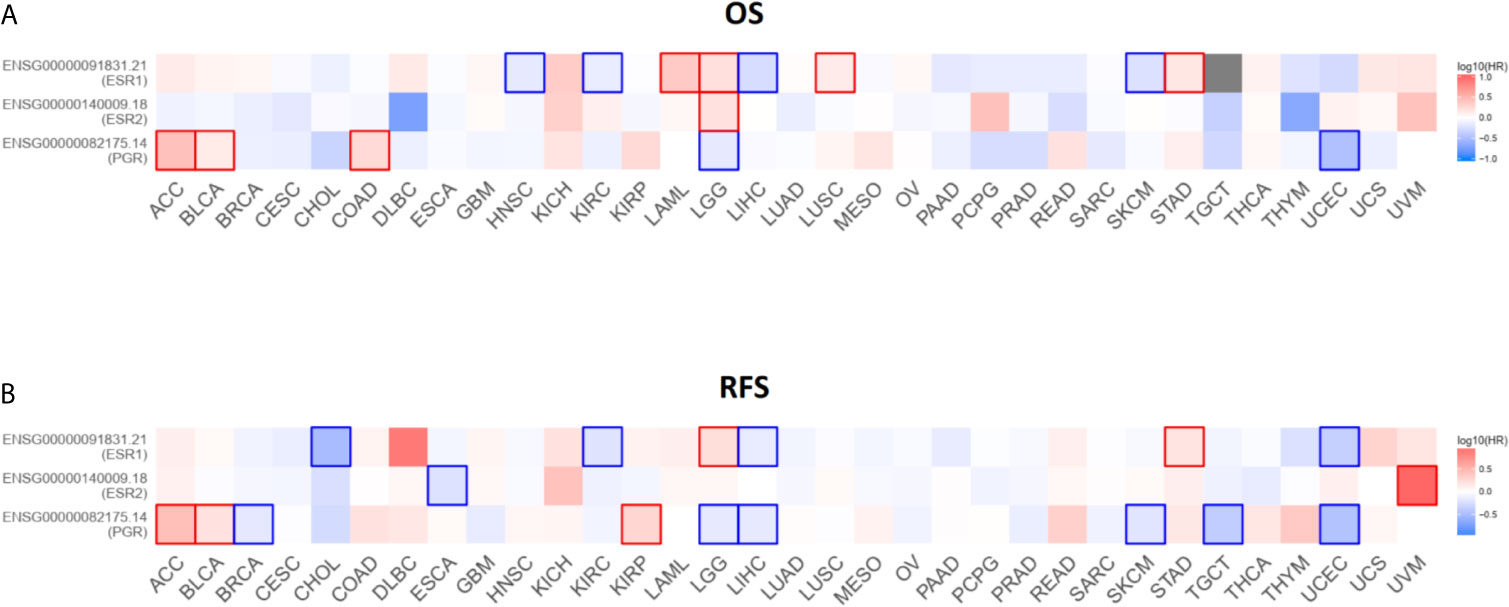
Figure 11 Survival contribution of ESR1, ESR2, and PGR across multiple cancer types. (A) Contribution analysis of ESR1, ESR2, and PGR to OS in multiple cancer types. (B) Contribution analysis of ESR1, ESR2, and PGR to RFS in multiple cancer types.
Discussion
ESRs and PGR promote cell proliferation in breast cancer (29). Further, ESRs and PGR, which are associated with tumorigenesis and progression under pathological conditions, have become ideal molecular treatment targets (30–34). Accordingly, previous studies have demonstrated that drugs targeting ESR1, ESR2, and PGR are effective in the treatment of breast cancer and improve promote clinical outcomes (Supplementary Material Tables S1–S3). In addition, it has already been described the role of ESR1, ESR2, and PGR in promoting ovarian, lung, and prostate tumorigenesis (35–37). However, the roles of ESR1, ESR2, and PGR in other cancer types have rarely been studied and further investigations are needed to reach a consensus. In our study, we selected ESR1, ESR2, and PGR for an in-depth analysis of mRNA expression, genetic alternations, and clinical outcomes as well as the co-expression of these genes with immunomodulatory factors in a variety of cancer types. We also validated the expression levels of ERa, ERb, and PGR proteins in OV and UTEA using paraffin-embedded tissue specimens, and explored the relationship between ERa, ERb, and PGR proteins and clinicopathological characteristics of patients. To the best of our knowledge, this is the first such study based on integrated bioinformatics analysis. Through this comprehensive pan-cancer analysis, the feasibility of using ESR1, ESR2, and PGR as prognostic markers and therapeutic targets for multiple cancers was evaluated.
The results based on GEPIA2 dataset analysis were partly consistent with those reported previously (38–40). Hishida et al. showed that ESR1 gene transcripts were absent or decreased in more than 90% of liver cancer (n = 24) samples compared with their matched normal liver tissue counterparts. These results highlighted ESR1 as a tumor suppressor gene in liver cancer, and indicated that lower cellular estrogen levels stimulated liver cancer cell growth (41). The results of the present study revealed that the expression of ESR1 and PGR correlated with the tumor stage, whereas the expression of ESR2 did not. Additionally, prognostic analysis suggested that ESR1, ESR2, and PGR were significantly correlated with OS and RFS in patients with specific cancer types. However, due to the heterogeneity, subtypes, and sample size of cancers, or limited length of follow-up in TCGA datasets, there were discrepancies between the results of this study and those of previously published studies. For example, analysis using TCGA database showed that patients had shorter OS and RFS with higher ERb expression levels in renal cell carcinoma (42). This is in contrast to the results of the present study, which showed that ESR2 expression was not a risk factor for kidney cancer. Generally, the results of this study indicated that ESR1, ESR2, and PGR can be regarded as predictive and prognostic biomarkers across different cancer types.
The progression of cancer is influenced by multiple factors including, but not limited to, somatically acquired genetic, epigenetic, transcriptomic and proteomic alterations (43). Some alterations in particular genomic regions exhibit potential pro- and anti-tumor effects (44). Therefore, we used the cBioPortal web tool for further analysis of the genetic mutations in ESR1, ESR2, and PGR in multiple cancer types. Our results revealed genetic alterations in ESR1, ESR2, and PGR in multiple cancer types, including amplification, fusion, deep deletion, missense mutation, and truncating mutation. In addition, we identified a trend for co-occurrence of genetic alterations in ESR1, ESR2, and PGR. Based on these results, combining the expressions of ESR1, ESR2, and PGR may provide a better prognostic value in cancer patients. Yi et al. proposed that higher ESR1 expression and a higher ESR ratio (ESR1/ESR2) were associated with worse overall survival in female papillary thyroid carcinoma patients (45). There were also differences in the types and frequencies of genetic alterations in ESR1, ESR2, and PGR in multiple cancer types. Furthermore, mutations in ESR1, ESR2, and PGR could result in the amino acid changes in several sites. Considering these results, we hypothesized that genetic alterations in ESR1, ESR2, and PGR play an essential role in cancer progression and combining the expression levels of ESR1, ESR2, and PGR provide prognostic value.
It has been reported that genomic diversity increases with the rate of genetic alterations result in cancers, resulting in an increased frequency of neoantigens and greater immune cell infiltration (46). Understanding the effects of immune cells on cancers will lead to a new era in oncotherapy. Therefore, the effectiveness and efficiency of immune checkpoint-target agents, which direct the host immune system to target cancer cells, has become a focus of research. However, results showed relatively low response rates of immune checkpoint-target agents in some tumors (47–49). To overcome this challenge, further understanding of immunotherapy is needed to select the patients who will benefit most from this type of therapy. Studies have found that immune checkpoint proteins (PD-L1, VISTA) are more frequently expressed in certain ESR-negative breast cancers (50, 51). Liu et al. demonstrated an inverse correlation between ESRs and PD-L1 in breast cancer cells, indicating that PD-L1 gene transcription is negatively regulated by ESRs (52), which is consistent with the results of the current study. Hence, in the present study, we investigated the potential of ESR1, ESR2, and PGR as predictive and prognostic biomarkers in multiple cancer types from an immuno-oncological perspective based on bioinformatics analysis to provide a reference for future studies and the application of immunotherapies. In this study, we explored the relationship between ESR1, ESR2, and PGR and immunomodulators or immune cells using the TISIDB database. The results demonstrated that ESR1 had the greatest correlation with immunoinhibitors (such as CD274, CD96, CFS1R, and CTLA-4) and immunostimulators (such as CD27, CD28, and CXCL12). It is noteworthy that the role of ESR1 in the function of immunomodulators is context-dependent. ESR2 and PGR showed similar results. In addition, we found that there was a certain relationship between the expression levels of ESR1, ESR2, and PGR and peripheral blood cells. Therefore, this preliminary analysis of the association between ESR1, ESR2, and PGR and immune function highlights the importance of future research to elucidate the potential roles of ESR1, ESR2, and PGR as predictive and prognostic biomarkers as well as therapeutic targets for immunotherapy across multiple cancer types.
Our study also has some limitations. The results derived from different online databases are inevitably accompanied by background heterogeneity. Moreover, our immunohistochemical verification experiment was conducted only in OV and UTEA, with inadequate prognostic studies of patient cohorts. More cancer types need to be included in subsequent verification experiments, which can be further verified by adding cytological function studies and patient cohort studies.
Conclusions
In summary, we identified significant differences in the expression levels of ESR1, ESR2, and PGR mRNAs in different cancer types, which associated with tumor progression and clinical prognosis. Our study provides comprehensive evidence that ESR1, ESR2, and PGR are feasible prognostic markers and therapeutic targets for multiple cancers and that they could be a factor for disease prediction, disease evaluation, and individualized treatment in various types of cancer.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Anhui Medical University, Hefei 230032, Anhui, China. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
XH and Z-sW conceived the study. Y-tS, XH, GZ, BJ, and C-jL collected the data. XH and Z-sW analyzed and interpreted the data. Y-tS, XH, and GZ wrote and revised the manuscript. All authors discussed and revised the manuscript. All authors contributed to the article and approved the submitted version. XH, Y-tS, and GZ contributed equally to the study. XH and Z-sW supervised the study and share the senior authorship.
Funding
This study was funded by grants from the National Natural Science Foundation of China (31970696 and 81502975 to XH; 81972472 to Z-sW), China Postdoctoral Science Foundation (2016T90413 and 2015M581693 to XH), and Natural Science Foundation of Anhui (2008085MH276 to Z-sW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to sincerely thank the open-access databases and web servers for data collection and for the provision of data processing and customization functions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.636365/full#supplementary-material
Abbreviations
ESR1, estrogen receptor 1; ESR2, estrogen receptor 2; PGR, progesterone receptor; GEPIA2, the Gene Expression Profiling Interactive Analysis2; TISIDB, the Tumor and Immune System Interaction Database; OS, overall survival; RFS, disease-free survival; BRCA, breast invasive carcinoma; OV, ovarian serous cystadenocarcinoma; UTEA, uterine endometrioid adenocarcinoma; BLCA, bladder urothelial carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; LIHC, liver hepatocellular carcinoma; TGCT, testicular germ cell tumors; UCS, uterine carcinosarcoma; DLBCL, lymphoid neoplasm diffuse large B-cell lymphoma; ACC, adrenocortical carcinoma; COAD, colon adenocarcinoma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; UCEC, uterine corpus endometrial carcinoma; DC, dendritic cells; HNSC, neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; LIHC, liver hepatocellular carcinoma; SKCM, skin cutaneous melanoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LUSC, lung squamous cell carcinoma; STAD, stomach adenocarcinoma.
References
1. Feng RM, Zong YN, Cao SM, Xu RH. Current Cancer Situation in China: Good or Bad News From the 2018 Global Cancer Statistics? Cancer Commun (Lond) (2019) 39:22. doi: 10.1186/s40880-019-0368-6
2. Chen WQ, Xia CF, Zheng RS, Zhou MG, Lin CQ, Zeng HM, et al. Disparities by Province, Age, and Sex in Site-Specific Cancer Burden Attributable to 23 Potentially Modifiable Risk Factors in China: A Comparative Risk Assessment. Lancet Global Health (2019) 7:e257–69. doi: 10.1016/s2214-109x(18)30488-1
3. GBD 2017 Causes of Death Collaborators. Global, Regional, and National Age-Sex-Specific Mortality for 282 Causes of Death in 195 Countries and Territories, 1980–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet (2018) 392:1736–88. doi: 10.1016/s0140-6736(18)32203-7
4. Gibson DA, Saunders PT. Estrogen Dependent Signaling in Reproductive Tissues - a Role for Estrogen Receptors and Estrogen Related Receptors. Mol Cell Endocrinol (2012) 348:361–72. doi: 10.1016/j.mce.2011.09.026
5. Hewitt SC, Winuthayanon W, Korach KS. What's New in Estrogen Receptor Action in the Female Reproductive Tract. J Mol Endocrinol (2016) 56:R55–71. doi: 10.1530/JME-15-0254
6. Graham JD, Clarke CL. Physiological Action of Progesterone in Target Tissues. Endocr Rev (1997) 18:502–19. doi: 10.1210/edrv.18.4.0308
7. Edwards DP. Regulation of Signal Transduction Pathways by Estrogen and Progesterone. Annu Rev Physiol (2005) 67:335–76. doi: 10.1146/annurev.physiol.67.040403.120151
8. Allison KH, Hammond MEH, Dowsett M, McKernin SE, Carey LA, Fitzgibbons PL, et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J Clin Oncol (2020) 38:1346–66. doi: 10.1200/JCO.19.02309
9. Yager JD, Davidson NE. Estrogen Carcinogenesis in Breast Cancer. N Engl J Med (2006) 354:270–82. doi: 10.1056/NEJMra050776
10. Rizza P, Barone I, Zito D, Giordano F, Lanzino M, De Amicis F, et al. Estrogen Receptor Beta as a Novel Target of Androgen Receptor Action in Breast Cancer Cell Lines. Breast Cancer Res (2014) 16:R21. doi: 10.1186/bcr3619
11. Huang B, Omoto Y, Iwase H, Yamashita H, Toyama T, Coombes RC, et al. Differential Expression of Estrogen Receptor α, β1, and β2 in Lobular and Ductal Breast Cancer. Proc Natl Acad Sci (2014) 111:1933–8. doi: 10.1073/pnas.1323719111
12. Cui X, Schiff R, Arpino G, Osborne CK, Lee AV. Biology of Progesterone Receptor Loss in Breast Cancer and its Implications for Endocrine Therapy. J Clin Oncol (2005) 23:7721–35. doi: 10.1200/JCO.2005.09.004
13. Cheng J, Lee EJ, Madison LD, Lazennec G. Expression of Estrogen Receptor Beta in Prostate Carcinoma Cells Inhibits Invasion and Proliferation and Triggers Apoptosis. FEBS Lett (2004) 566:169–72. doi: 10.1016/j.febslet.2004.04.025
14. Prins GS, Korach KS. The Role of Estrogens and Estrogen Receptors in Normal Prostate Growth and Disease. Steroids (2008) 73:233–44. doi: 10.1016/j.steroids.2007.10.013
15. Grindstad T, Richardsen E, Andersen S, Skjefstad K, Rakaee Khanehkenari M, Donnem T, et al. Progesterone Receptors in Prostate Cancer: Progesterone Receptor B Is the Isoform Associated With Disease Progression. Sci Rep (2018) 8:11358. doi: 10.1038/s41598-018-29520-5
16. Liu S, Fan W, Gao X, Huang K, Ding C, Ma G, et al. Estrogen Receptor Alpha Regulates the Wnt/Beta-Catenin Signaling Pathway in Colon Cancer by Targeting the NOD-Like Receptors. Cell Signal (2019) 61:86–92. doi: 10.1016/j.cellsig.2019.05.009
17. Peng J, Ou Q, Wu X, Zhang R, Zhao Q, Jiang W, et al. Expression of Voltage-Gated Sodium Channel Nav1.5 in Non-Metastatic Colon Cancer and Its Associations With Estrogen Receptor (ER)-Beta Expression and Clinical Outcomes. Chin J Cancer (2017) 36:89. doi: 10.1186/s40880-017-0253-0
18. Slattery ML, Samowitz WS, Holden JA. Estrogen and Progesterone Receptors in Colon Tumors. Am J Clin Pathol (2000) 113:364–8. doi: 10.1309/5MHB-K6XX-QV50-PCJQ
19. Deroo BJ, Korach KS. Estrogen Receptors and Human Disease. J Clin Invest (2006) 116:561–70. doi: 10.1172/JCI27987
20. Treeck O, Pfeiler G, Mitter D, Lattrich C, Piendl G, Ortmann O. Estrogen Receptor {Beta}1 Exerts Antitumoral Effects on SK-OV-3 Ovarian Cancer Cells. J Endocrinol (2007) 193:421–33. doi: 10.1677/JOE-07-0087
21. Pedernera E, Gomora MJ, Morales-Vasquez F, Perez-Montiel D, Mendez C. Progesterone Reduces Cell Survival in Primary Cultures of Endometrioid Ovarian Cancer. J Ovarian Res (2019) 12:15. doi: 10.1186/s13048-019-0486-4
22. Hiramitsu S, Ishikawa T, Lee WR, Khan T, Crumbley C, Khwaja N, et al. Estrogen Receptor Beta-Mediated Modulation of Lung Cancer Cell Proliferation by 27-Hydroxycholesterol. Front Endocrinol (Lausanne) (2018) 9:470. doi: 10.3389/fendo.2018.00470
23. Rades D, Setter C, Dahl O, Schild SE, Noack F. The Prognostic Impact of Tumor Cell Expression of Estrogen Receptor-Alpha, Progesterone Receptor, and Androgen Receptor in Patients Irradiated for Nonsmall Cell Lung Cancer. Cancer (2012) 118:157–63. doi: 10.1002/cncr.26282
24. Kawprasertsri S, Pietras RJ, Marquez-Garban DC, Boonyaratanakornkit V. Progesterone Receptor (PR) Polyproline Domain (PPD) Mediates Inhibition of Epidermal Growth Factor Receptor (EGFR) Signaling in non-Small Cell Lung Cancer Cells. Cancer Lett (2016) 374:279–91. doi: 10.1016/j.canlet.2016.02.014
25. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: An Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res (2019) 47:W556–60. doi: 10.1093/nar/gkz430
26. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The Cbio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery (2012) 2:401–4. doi: 10.1158/2159-8290.CD-12-0095
27. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the Cbioportal. Sci Signaling (2013) 6:pl1. doi: 10.1126/scisignal.2004088
28. Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: An Integrated Repository Portal for Tumor-Immune System Interactions. Bioinformatics (2019) 35:4200–2. doi: 10.1093/bioinformatics/btz210
29. White MM, Zamudio S, Stevens T, Tyler R, Lindenfeld J, Leslie K, et al. Estrogen, Progesterone, and Vascular Reactivity: Potential Cellular Mechanisms. Endocr Rev (1995) 16:739–51. doi: 10.1210/edrv-16-6-739
30. DeCensi A, Pruneri G, Guerrieri-Gonzaga A. Estrogen Receptor in Breast Ductal Carcinoma in Situ: Good Cop, Bad Cop? J Clin Oncol (2012) 30:1384–6. doi: 10.1200/JCO.2011.40.7494
31. Brisken C. Progesterone Signalling in Breast Cancer: A Neglected Hormone Coming Into the Limelight. Nat Rev Cancer (2013) 13:385–96. doi: 10.1038/nrc3518
32. Tanos T, Sflomos G, Echeverria PC, Ayyanan A, Gutierrez M, Delaloye JF, et al. Progesterone/RANKL Is a Major Regulatory Axis in the Human Breast. Sci Transl Med (2013) 5:182ra155. doi: 10.1126/scitranslmed.3005654
33. Jordan VC, O'Malley BW. Selective Estrogen-Receptor Modulators and Antihormonal Resistance in Breast Cancer. J Clin Oncol (2007) 25:5815–24. doi: 10.1200/JCO.2007.11.3886
34. Fabian CJ, Kimler BF. Selective Estrogen-Receptor Modulators for Primary Prevention of Breast Cancer. J Clin Oncol (2005) 23:1644–55. doi: 10.1200/JCO.2005.11.005
35. Smida T, Bruno TC, Stabile LP. Influence of Estrogen on the NSCLC Microenvironment: A Comprehensive Picture and Clinical Implications. Front Oncol (2020) 10:137. doi: 10.3389/fonc.2020.00137
36. He M, Yu W, Chang C, Miyamoto H, Liu X, Jiang K, et al. Estrogen Receptor Alpha Increases Lung Cancer Cell Invasion via Increase of and Cross-Talk With Infiltrated Macrophages Through the CCL2/CCR2/MMP9 and CXCL12/CXCR4 Signaling Pathways. Mol Oncol (2020) 14(8):1779–99. doi: 10.1002/1878-0261.12701
37. Warner M, Huang B, Gustafsson JA. Estrogen Receptor Beta as a Pharmaceutical Target. Trends Pharmacol Sci (2017) 38:92–9. doi: 10.1016/j.tips.2016.10.006
38. Salvati A, Gigantino V, Nassa G, Giurato G, Alexandrova E, Rizzo F, et al. The Histone Methyltransferase DOT1L Is a Functional Component of Estrogen Receptor Alpha Signaling in Ovarian Cancer Cells. Cancers (Basel) (2019) 11(11):1720. doi: 10.3390/cancers11111720
39. Hirvonen-Santti SJ, Rannikko A, Santti H, Savolainen S, Nyberg M, Jänne OA, et al. Down-Regulation of Estrogen Receptor Beta and Transcriptional Coregulator SNURF/RNF4 in Testicular Germ Cell Cancer. Eur Urol (2003) 44:742–747; discussion 747. doi: 10.1016/s0302-2838(03)00382-8
40. Zhao Y, Li Z. Interplay of Estrogen Receptors and FOXA Factors in the Liver Cancer. Mol Cell Endocrinol (2015) 418 Pt 3:334–9. doi: 10.1016/j.mce.2015.01.043
41. Hishida M, Nomoto S, Inokawa Y, Hayashi M, Kanda M, Okamura Y, et al. Estrogen Receptor 1 Gene as a Tumor Suppressor Gene in Hepatocellular Carcinoma Detected by Triple-Combination Array Analysis. Int J Oncol (2013) 43:88–94. doi: 10.3892/ijo.2013.1951
42. Ding J, Yeh CR, Sun Y, Lin C, Chou J, Ou Z, et al. Estrogen Receptor Beta Promotes Renal Cell Carcinoma Progression via Regulating LncRNA HOTAIR-miR-138/200c/204/217 Associated CeRNA Network. Oncogene (2018) 37:5037–53. doi: 10.1038/s41388-018-0175-6
43. Karim MA, Samad A, Adhikari UK, Kader MA, Kabir MM, et al. A Multi-Omics Analysis of Bone Morphogenetic Protein 5 (BMP5) mRNA Expression and Clinical Prognostic Outcomes in Different Cancers Using Bioinformatics Approaches. Biomedicines (2020) 8(2):19. doi: 10.3390/biomedicines8020019
44. Klonowska K, Czubak K, Wojciechowska M, Handschuh L, Zmienko A, Figlerowicz M, et al. Oncogenomic Portals for the Visualization and Analysis of Genome-Wide Cancer Data. Oncotarget (2016) 7:176–92. doi: 10.18632/oncotarget.6128
45. Yi JW, Kim SJ, Kim JK, Seong CY, Yu HW, Chai YJ, et al. Upregulation of the ESR1 Gene and ESR Ratio (ESR1/ESR2) Is Associated With a Worse Prognosis in Papillary Thyroid Carcinoma: The Impact of the Estrogen Receptor Alpha/Beta Expression on Clinical Outcomes in Papillary Thyroid Carcinoma Patients. Ann Surg Oncol (2017) 24:3754–62. doi: 10.1245/s10434-017-5780-z
46. Karn T, Jiang T, Hatzis C, Sänger N, El-Balat A, Rody A, et al. Association Between Genomic Metrics and Immune Infiltration in Triple-Negative Breast Cancer. JAMA Oncol (2017) 3:1707–11. doi: 10.1001/jamaoncol.2017.2140
47. Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med (2016) 375:1856–67. doi: 10.1056/NEJMoa1602252
48. Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee JL, Fong L, et al. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. N Engl J Med (2017) 376:1015–26. doi: 10.1056/NEJMoa1613683
49. Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Pembrolizumab Versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med (2016) 375:1823–33. doi: 10.1056/NEJMoa1606774
50. Zong L, Mo S, Yu S, Zhou Y, Zhang M, Chen J, et al. Expression of the Immune Checkpoint VISTA in Breast Cancer. Cancer Immunol Immunother (2020) 69:1437–46. doi: 10.1007/s00262-020-02554-3
51. Soliman H, Khalil F, Antonia S. PD-L1 Expression Is Increased in a Subset of Basal Type Breast Cancer Cells. PloS One (2014) 9:e88557. doi: 10.1371/journal.pone.0088557
Keywords: estrogen receptor, progesterone receptor, expression profile, pathological correlation, genetic alteration, clinical relevance, immunological correlation, survival contribution
Citation: Shen Y-t, Huang X, Zhang G, Jiang B, Li C-j and Wu Z-s (2021) Pan-Cancer Prognostic Role and Targeting Potential of the Estrogen-Progesterone Axis. Front. Oncol. 11:636365. doi: 10.3389/fonc.2021.636365
Received: 01 December 2020; Accepted: 23 June 2021;
Published: 12 July 2021.
Edited by:
Vincenzo Quagliariello, Istituto Nazionale Tumori Fondazione G. Pascale (IRCCS), ItalyReviewed by:
Ravi Manoharan, University of Madras, IndiaMuriel Le Romancer, INSERM U1052 Centre de Recherche en Cancerologie de Lyon, France
Copyright © 2021 Shen, Huang, Zhang, Jiang, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng-sheng Wu, d29venNvbkB5YWhvby5jb20=; Xing Huang, aHVhbmd4aW5nNjZAemp1LmVkdS5jbg==
†ORCID: Xing Huang, orcid.org/0000-0002-8886-2777
‡These authors have contributed equally to this work
 Yu-ting Shen
Yu-ting Shen Xing Huang
Xing Huang Gang Zhang
Gang Zhang Bo Jiang
Bo Jiang Cheng-jun Li
Cheng-jun Li Zheng-sheng Wu
Zheng-sheng Wu