- 1National Clinical Research Center for Hematologic Diseases, Jiangsu Institute of Hematology, The First Affiliated Hospital of Soochow University, Soochow University, Suzhou, China
- 2Institute of Blood and Marrow Transplantation, Collaborative Innovation Center of Hematology, Soochow University, Suzhou, China
- 3Department of Hematology, The Affiliated People’s Hospital of Jiangsu University, Zhenjiang, China
Adult acute myeloid leukemia (AML) patients with biallelic mutations of CEBPA (biCEBPA) displays a favorable clinical outcome, and is defined as a unique entity in the 2016 World Health Organization classification. However, due to the intrinsic characteristics of the mutation, existence of co-occurring mutations and diversified gene expression signature, the prognosis of these patients needs to be analyzed in a more systematic way. In this study we evaluated the genetic characteristics and clinical outcome in a cohort of 137 biCEBPA AML cases, and proposed a prognostic nomogram to predict the overall survival (OS) of based on the clinical variables selected by multivariate Cox regression model in training cohort, including age, white blood cell count, co-existence of DNMT3A and CSF3R mutation and whether patients could achieve complete remission after induction therapy. The area under the receiver operating characteristic (ROC) curves for 3 and 5-year OS were 0.833 and 0.863, respectively. RNA sequencing of 4 relapsed patients showed that over-expression of VMP1 was an indicator of poor prognosis of biCEBPA AML patients. In conclusion, this prognostic nomogram might provide a more accurate prediction of the clinical outcomes of biCEBPA AML patients.
Introduction
CCAAT/enhancer binding protein α (CEBPA) plays a pivotal role as a transcription factor in both self-renewal of hematopoietic stem cells (HSCs) and proliferation and differentiation of myeloid progenitor cells. Major CEBPA mutated AML cases carry two mutations, one in the N-terminal of the protein and the other one in the basic leucine zipper (bZIP) domain. N-terminal nonsense and frameshift mutations truncate the CEBPA protein and lead to a dominant negative effect, while mutations in the bZIP domain at the C terminus are generally in-frame insertions or deletions which brings disrupted DNA binding and dimerization (1, 2). Biallelic CEBPA (biCEBPA) mutations are detected in 2-15% of de novo acute myeloid leukemia (AML) patients, and are associated with a favorable clinical outcome compared to wildtype or monoallelic CEBPA mutation (3). Due to its biological and clinical significance, AML with biCEBPA mutations has been classified as a distinct entity with an excellent overall prognosis in the World Health Organization (WHO) 2016 edition of classification of tumors of hematopoietic and lymphoid tissues (4–6). However, around 40% patients could relapse after conventional chemotherapy, indicating a blatant heterogeneity within this disease entity (7, 8).
To date, several studies have reported genetic heterogeneity in biCEBPA AML cases, the number of the genes and patients being analyzed was limited. In the present study, we aimed to evaluate the role of concurrent mutations and their prognostic value in biCEBPA AML patients. We will also investigate on underlying reasons of treatment failures in these patients.
Materials And Methods
Clinical Patients
From June 2016 to November 2018, a total of 137 de novo AML patients who were detected with CEBPA mutations and received treatment were enrolled in the study. The diagnosis of these patients fulfilled the criteria of the WHO 2016 edition of myeloid neoplasms and acute leukemia. The study was approved by the Ethics Committee of the First Affiliated Hospital of Soochow University [No. 221 of 2019 LSP (application)] and was conducted following the Declaration of Helsinki. All patients carried biallelic CEBPA mutations, involving both the N-terminal TAD1 region and the C-terminal bZIP domain. Major (99/137, 72.3%) patients were treated with standard “3+7” regimen for initial induction therapy (darubicin/idarubicin + cytarabine). In some elderly and severe underlying diseases patients, pre excitation scheme [cytarabine + aclarubicin + granulocyte-colony stimulating factor (G-CSF)] were administered. The first consolidation therapy was generally the same as that used to achieve CR or high/medium-dose cytarabine at 2-3 g/m (2) were administered for consolidation therapy. High-risk patients, or those with a matched sibling, were treated with hematopoietic stem cell transplantation (HSCT).
DNA Sequencing and Mutation Analysis
Genomic DNA was extracted from bone marrow or peripheral blood samples at the onset of disease diagnosis by using Invitrogen DNA Extraction Kit. The mutational hotspots or whole coding regions of 51 genes (Supplementary Table 1) that were recurrently mutated in hematological malignancies were sequenced. The procedures were in accordance with an amplicon-based Next Generation Sequencing (NGS) protocol with Ion Torrent PGM sequencer (Thermo Fisher Scientific, Waltham, MA, USA). An allele frequency threshold of 2% was defined for mutation detection. All CEBPA mutations were confirmed by Sanger sequencing. Bone marrow or peripheral blood samples in complete remission (CR) or fingernail samples were also detected for exclusion of CEBPA germline mutations.
RNA Sequencing
RNA was extracted from 16 bone marrow samples of biCEBPA AML cases using TRIzol reagent. These libraries were set up through TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA). Then these libraries were detected on the Illumina sequencing platform (HiSeq TM 2500 or Illumina HiSeq X Ten) and 125bp/150bp paired-end reads were amplificated.
Statistical Analysis
Overall survival (OS) is defined as the time from diagnosis to death or to the time of the last follow-up. Disease-free survival (DFS) is defined as the time from CR to relapse、death or the time of last follow-up. All alive patients were followed on December 31, 2019.
The SPSS software (version 23.0; SPSS Inc., Chicago, IL) was applied in statistical analysis. The significance between categorical data was calculated by Chi-square test. Kaplan–Meier method was employed for overall survival analysis, and log-rank test was used to compare differential survival rates between groups. A two-sided P < 0.05 was considered as statistical significance.
Results
Clinical Characteristics
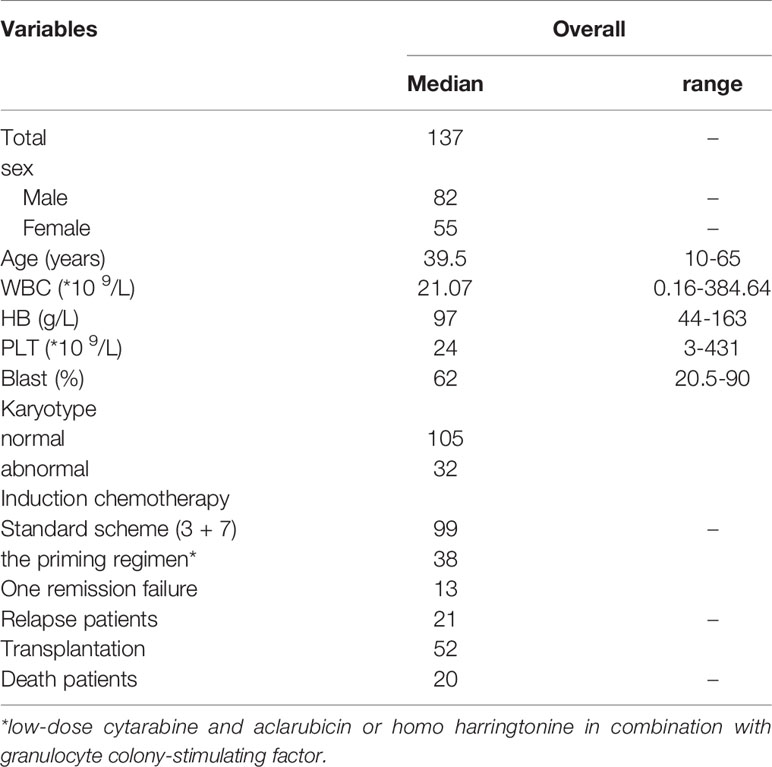
The clinical characteristics of patients in the study were summarized in Table 1 (N=137). The median age of biCEBPA mutated patients in the study was 39.5 years old (range, 10-65 years old), including 82 male and 55 female patients. The median WBC count and PLT count was 21.07×109/L (range, 0.16-384.64×109/L) and 24×109/L (range, 3-431×109/L), while the median bone marrow blast cell percentage was 62% (range, 20.5-90%). Only 32 (23.3%) patients demonstrated abnormal karyotype, among which +21 and del(9q) were the most commonly seen aberrations. In those patients, GATA2 mutations were the most frequently observed additional mutation, occurring in 21.9% (30/137) patients. Other commonly mutated genes were WT1 (25/137), FLT3-ITD (23/137), NRAS (19/137), CSF3R (16/137), C-kit (14/137) and DNMT3A (10/137).
Cox Regression Analysis of Training Cohort
Univariate Cox proportional hazard regression analysis for OS showed that there were significant difference in OS of age, WBC, bone marrow blast cell percentage, one course complete remission and transplantation, which were further included in multivariate Cox regression analysis (Supplementary Table 2). GATA2 mutation, although seen in over 20% of biCEBPA AML cases, did not impact the clinical outcome of these patients (p = 0.914, Supplementary Figure 1) Multivariate Cox proportional hazard regression models demonstrated that age, whether CR is achieved, transplantation, CSF3R mutation were independent prognostic factors for biCEBPA AML (Figure 1). We found that CSF3R mutation improved the survival of biCEBPA AML patients (p = 0.005). In addition, biCEBPA AML patients harboring DNMT3A mutation also showed better outcome, although failing to reach statistical significance (p = 0.057).
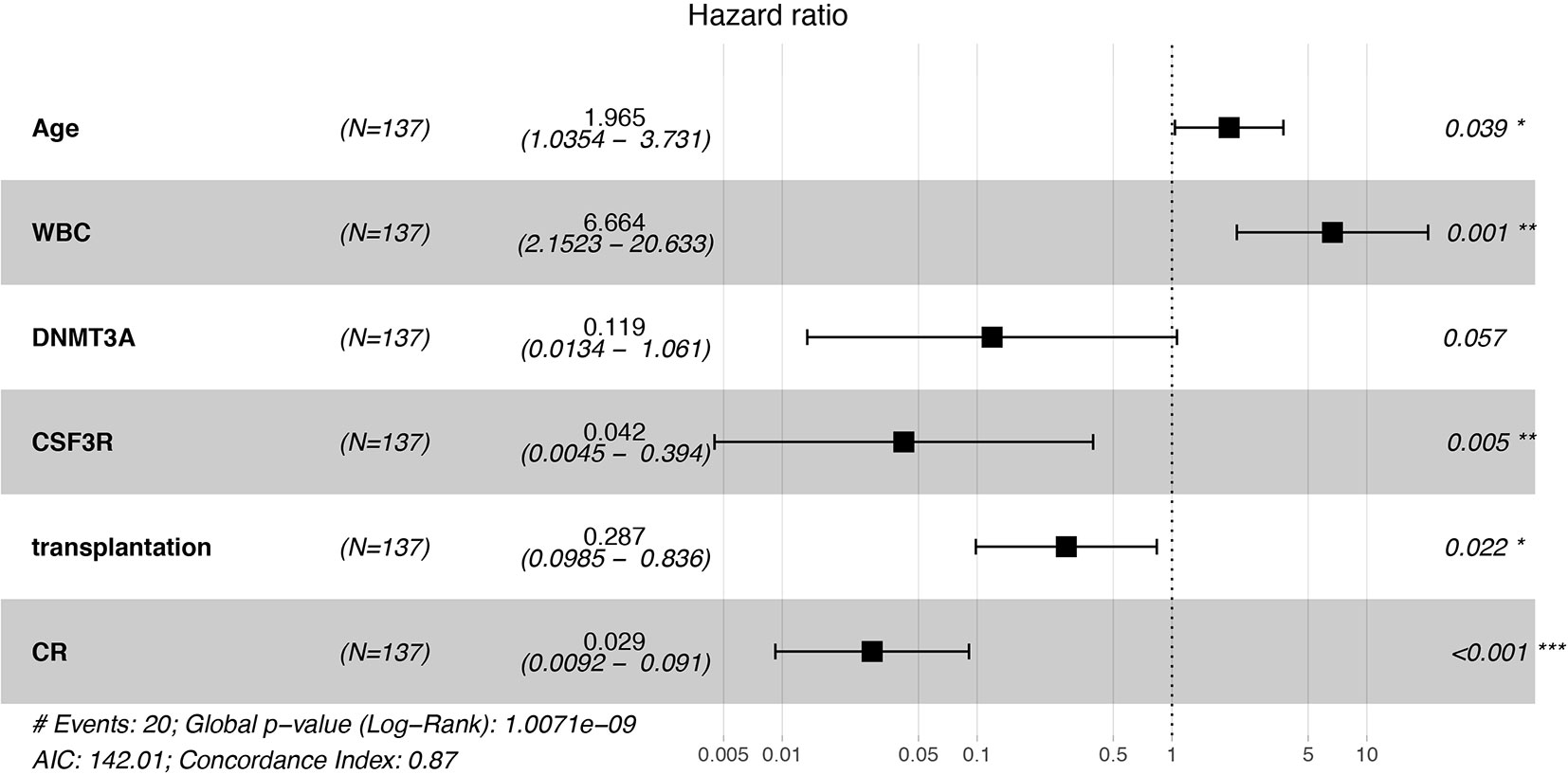
Figure 1 Forest figure for AML patients with biCEBPA mutations. Old age and high WBC count predicted lower overall survival, while CSF3R mutation, transplantation and the achievement of complete remission (CR) was associated with higher overall survival. *p < 0.05, **p < 0.01, ***p < 0.001.
Nomograms of biCEBPA AML Predicting Survival
Clinical categorical data after multivariate Cox regression were taken into the construction of training cohort nomogram (Figure 2). However, due to P < 0.05 of multivariate Cox regression in the OS, WBC and bone marrow blast could not be applied in nomogram.
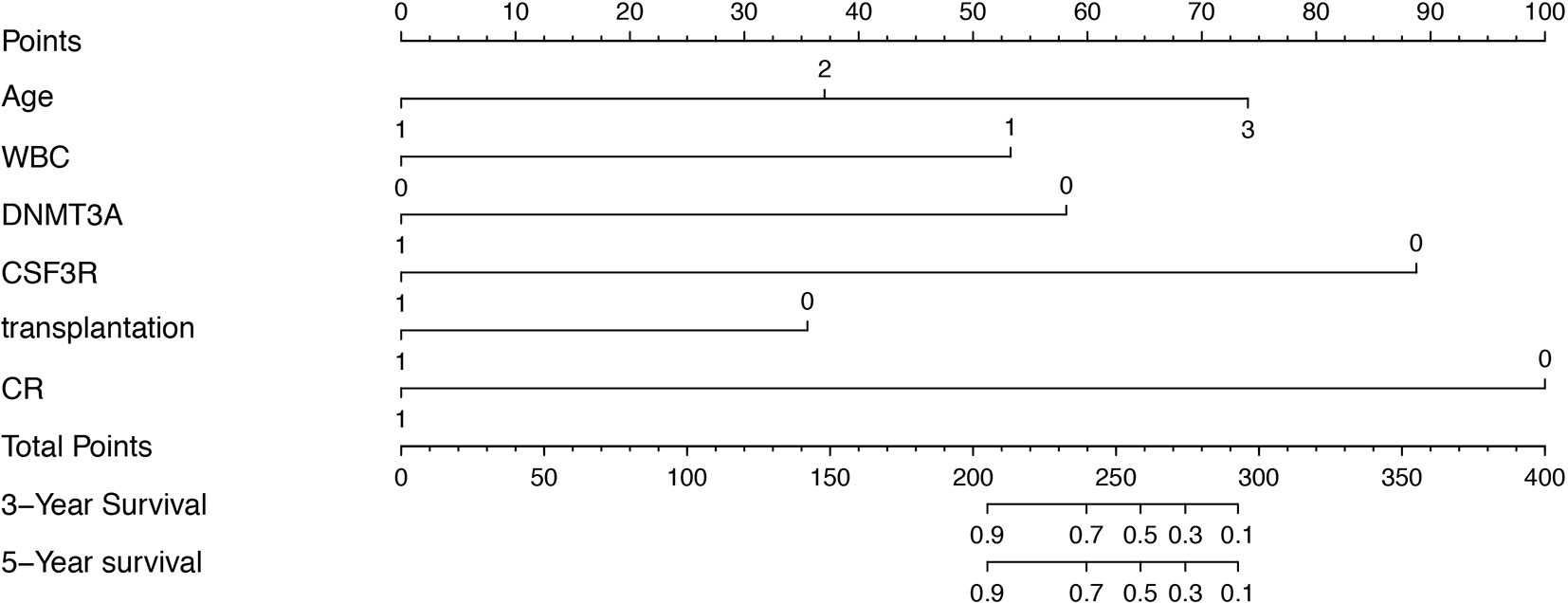
Figure 2 Prognostic nomogram for AML patients with biCEBPA mutations. Age: 1, age < 35 years; 2, age between 35-50 years; 3, age > 51 years. WBC: 1, WBC>25*109/L; 0 WBC<25*109/L. DNMT3A: 1, DNMT3A mutation positive; 0, DNMT3A mutation negative. CSF3R: 1, CSF3R mutation positive; 0, CSF3R mutation negative. Transplantation: 1, HSCT; 0, non-HSCT. CR: 1, complete remission after one course of induction therapy; 0, failure to achieve complete remission after one course of induction therapy.
Internal Validation
The calibration plot for the probabilities of 3 and 5-year survival rate displayed a great correlation between the actual observed and prediction outcome by this study nomogram (Supplementary Figure 2). The predictive ability for OS in training cohort is using ROC curves. The area under the curve (AUC) of ROC curves for 3 and 5-year survival rates were 0.833 and 0.863, respectively (Figure 3).
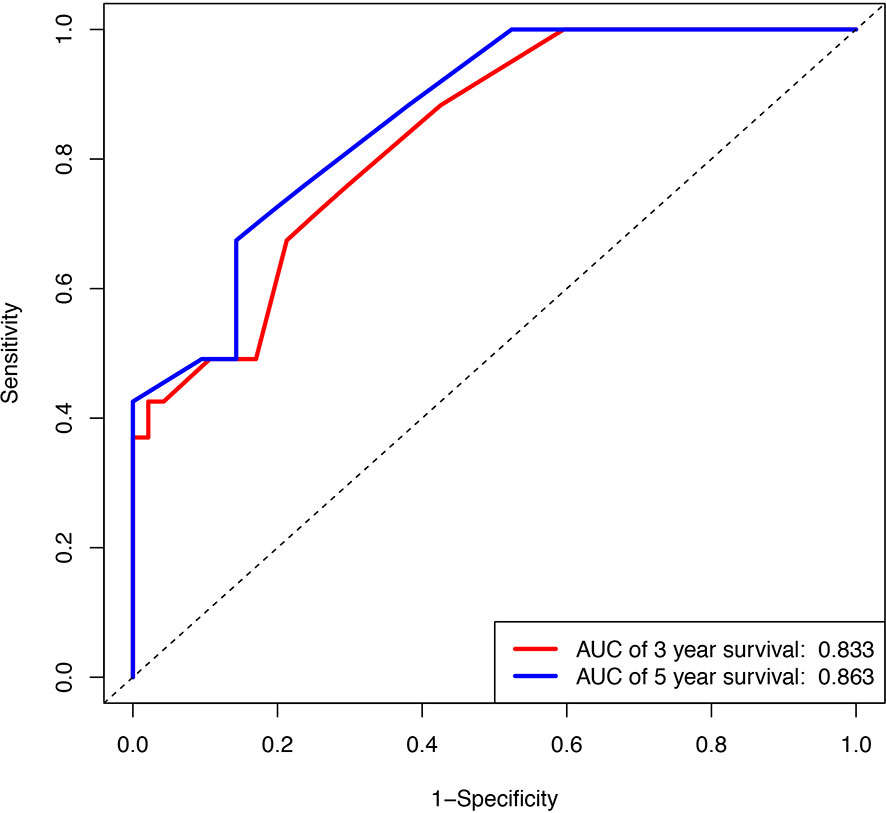
Figure 3 Internal validation of the nomogram to predict OS likelihoods in patients using ROC curve. Blue line, ROC curve of three years survival; Red line, ROC curve of five years survival.
VMP1 High Expression Predicts Poorer OS
RNA sequencing of samples onset of diagnosis was performed in four patients who finally relapsed as well as 12 patients who survived without detectable genetic aberration. Results showed that autophagy related genes clustered in relapsed samples (Figure 4), and the main differences lay in the NKX2-3 and VMP1 genes. Furthermore, overexpression of VMP1 may negatively impact the survival of biCEBPA AML patients (Figure 5A, p = 0.00014).
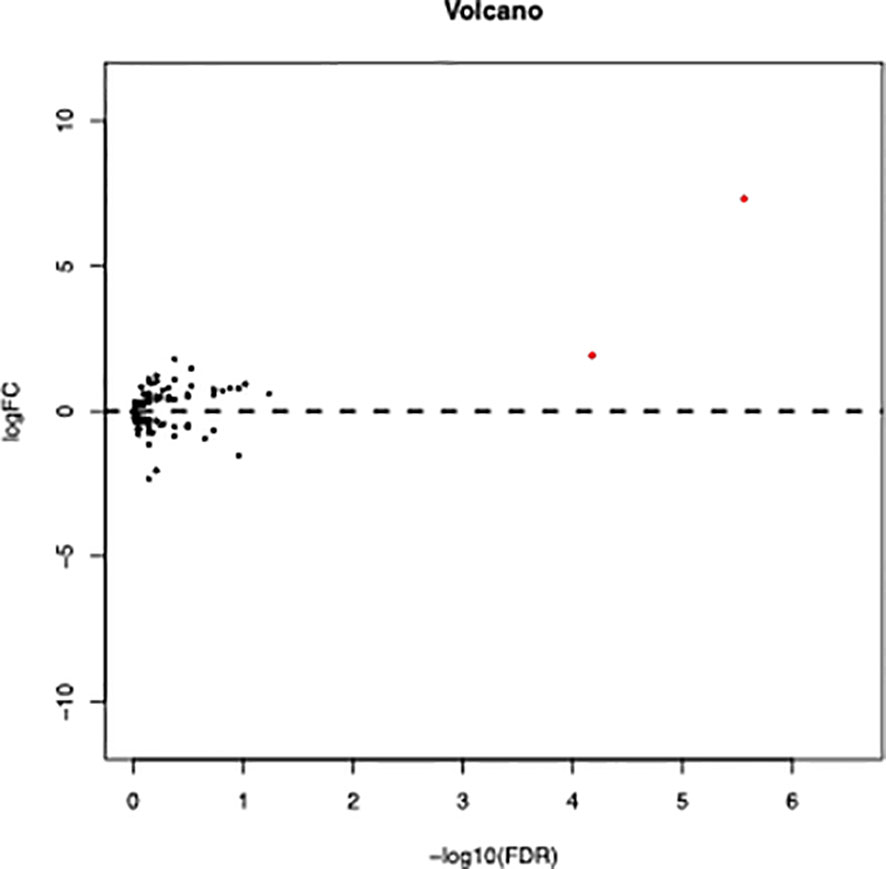
Figure 4 Volcano plot of differentially expressed autophagy related genes between relapse and remission patients with biCEBPA mutation. (FDR < 0.05, P < 0.05 and |log2 FC|>1.5).
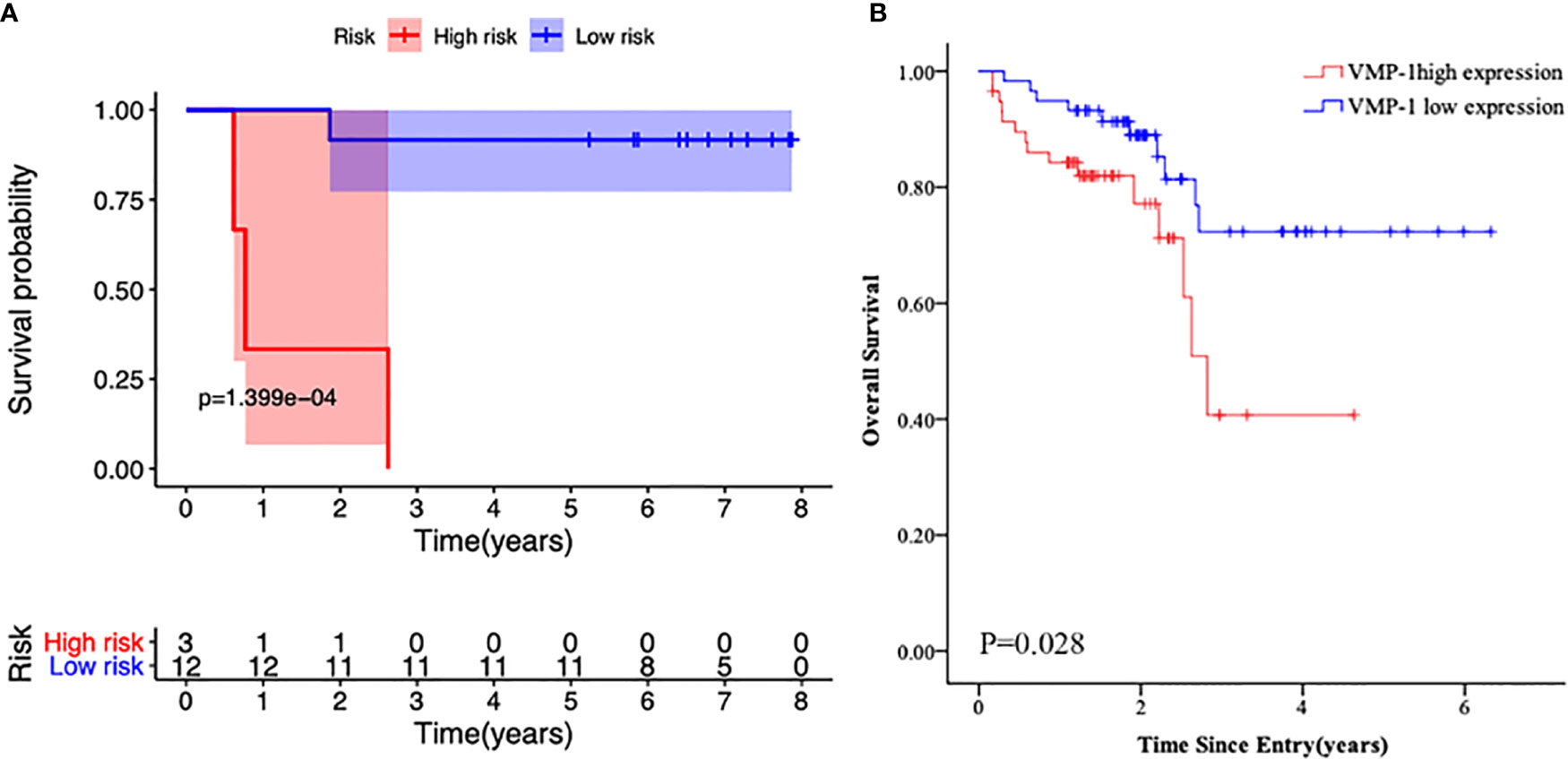
Figure 5 Kaplan-Meier survival curves of the primary cohort according to VMP1 gene expression. (A) Survival probability in biCEBPA AML patients with VMP1 high vs. low VMP1 expression. (B) Survival probability in normal karyotype AML patients with VMP1 high vs. low VMP1 expression.
To further explore the role of VMP1 expression in AML patients, we evaluated its expression in another 116 normal karyotype AML patients by real time PCR. We found that high VMP1 gene expression was an apparent correlation with poor overall survival rate (Figure 5B, p = 0.028).
Discussion
Although AML patients with biCEBPA mutation were associated with longer survival, the heterogeneity of these patients was reported in recent years (9). In the present study, we aimed to estimate the probability of 5-year OS based on a multivariate Cox proportional hazards model that included five clinical variables at the onset of diagnosis, including age, white blood cell count, co-existence of DNMT3A mutation and/or CSF3R mutation and complete remission is achieved after induction therapy.
The best characterized concurrent mutation was GATA2 mutation. In line with previous studies, we found that GATA2 mutation, although comprising the highest additional lesion, did not significantly influence the outcome of biCEBPA AML patients showed poor relativity (10–12). Several other mutations such as FLT3-ITD (13), WT1 (14), CSF3R mutation (15) are indicators of unfavorable clinical outcome in AML patients with biCEBPA mutations. However, Julia E reported that pediatric AML patients with CSF3R mutation had a trend towards low risk disease (p = 0.055) (16, 17). Furthermore, Shigeo Masuda also reported that in biCEBPA AML patients, DNMT3A mutations did not impact neither OS nor DFS (18). In our study, CSF3R mutation was significantly correlated a better clinical outcome and DNMT3A showed a trend. Further studies with a greater number of patients are warranted to test the results.
Relapse is the major cause of treatment failure and final death. In order to probe into the reason why some patients still relapse after remission, RNA sequencing was performed on samples onset of diagnosis in 4 relapsed, with12 patients who survived without detectable genetic aberration as the control. Results showed that the relapsed patients had high expression of blood-related gold and silver VMP1 gene.
VMP1 is a transmembrane protein that is related to endoplasmic reticulum, Golgi and intracellular vesicles (19), and is functionally important for cell adhesion, cellular membrane biology and early autophagosome formation (20). It has been shown to be highly expressed in ovarian tumors and is linked to malignant cell proliferation and metastasis (21). Nevertheless, overexpression of VMP1 gene may decrease the proliferation, invasion and metastasis of tumor cells in colorectal (22) and hepatocellular cancer (23),. This contradiction may be partially explained by different types of tumor, but more importantly, by the role of VMP1 in autophagy. Hypoxia inducible factors (HIFs) are activated in regions of rapidly growing tumors that are often poorly oxygenated (24). HIF1α expression increases VMP1-induced autophagy that results in less cell death in response to photodynamic therapy (25). To date, the impact of VMP1 in AML is poorly clarified. In this study, we found that biCEBPA AML patients with high VPM1 gene expression had a poor survival, and this may be partially explained by more autophagosome formation, which provides building blocks for cell replication and survival. Taken together, our findings implicated that high VMP1 expression may be a predictable marker for prognosis of biCEBPA-mutated AML patients.
In conclusion, we found that although AML patients with biCEBPA mutation were generally correlated with an excellent survival, a significant portion of those patients such as those with VMP1 mutation are still at high risk for relapse. In future studies we will investigate on how VMP1 mutation will impact the prognosis of biCEBPA AML patients.
Data Availability Statement
The data presented in the study are deposited in the GSA repository, accession number HDAC000675, https://ngdc.cncb.ac.cn/gsa-human/browse/HRA001135.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of The First Affiliated Hospital of Soochow University [No. 221 of 2019 LSP (application)] and was conducted following the Declaration of Helsinki. All study participants or their statutory guardian signed informed consent.
Author Contributions
XX and WC wrote the manuscript. SC developed the treatment concept. XX and PC edited the manuscript and assisted with methods and figures. LZ, TZ, HY, HS, and SC edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grant from the National Key R&D Program of China (2019YFA0111000), the National Natural Science Foundation of China (82000158), the Natural Science Foundation of the Jiangsu Higher Education Institution of China (18KJA320005), the Natural Science Foundation of Jiangsu Province (BK20190180), China Postdoctoral Science Foundation (2018M632372), the priority academic program development of Jiangsu Higher Education Institution, Translational Research Grant of NCRCH (2020WSB11, 2020WSB13).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.628248/full#supplementary-material
References
1. Frohling S, Schlenk RF, Stolze I, Bihlmayr J, Benner A, Kreitmeier S, et al. CEBPA Mutations in Younger Adults With Acute Myeloid Leukemia and Normal Cytogenetics: Prognostic Relevance and Analysis of Cooperating Mutations. J Clin Oncol (2004) 22(4):624–33. doi: 10.1200/JCO.2004.06.060
2. Asou H, Gombart AF, Takeuchi S, Tanaka H, Tanioka M, Matsui H, et al. Establishment of the Acute Myeloid Leukemia Cell Line Kasumi-6 From a Patient With a Dominant-Negative Mutation in the DNA-Binding Region of the C/EBPalpha Gene. Genes Chromosomes Cancer (2003) 36(2):167–74. doi: 10.1002/gcc.10161
3. Wilhelmson AS, Porse BT. CCAAT Enhancer Binding Protein Alpha (CEBPA) Biallelic Acute Myeloid Leukaemia: Cooperating Lesions, Molecular Mechanisms and Clinical Relevance. Br J Haematol (2020) 190(4):495–507. doi: 10.1111/bjh.16534
4. Dufour A, Schneider F, Metzeler KH, Hoster E, Schneider S, Zellmeier E, et al. Acute Myeloid Leukemia With Biallelic CEBPA Gene Mutations and Normal Karyotype Represents a Distinct Genetic Entity Associated With a Favorable Clinical Outcome. J Clin Oncol (2010) 28(4):570–7. doi: 10.1200/JCO.2008.21.6010
5. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood (2016) 127(20):2391–405. doi: 10.1182/blood-2016-03-643544
6. Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and Management of AML in Adults: 2017 ELN Recommendations From an International Expert Panel. Blood (2017) 129(4):424–47. doi: 10.1182/blood-2016-08-733196
7. Tallman MS, Wang ES, Altman JK, Appelbaum FR, Bhatt VR, Bixby D, et al. Acute Myeloid Leukemia, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw (2019) 17(6):721–49. doi: 10.6004/jnccn.2019.0028
8. Pastore F, Kling D, Hoster E, Dufour A, Konstandin NP, Schneider S, et al. Long-Term Follow-Up of Cytogenetically Normal CEBPA-Mutated Aml. J Hematol Oncol (2014) 7:55. doi: 10.1186/s13045-014-0055-7
9. Mannelli F, Ponziani V, Bencini S, Bonetti MI, Benelli M, Cutini I, et al. CEBPA-Double-Mutated Acute Myeloid Leukemia Displays a Unique Phenotypic Profile: A Reliable Screening Method and Insight Into Biological Features. Haematologica (2017) 102(3):529–40. doi: 10.3324/haematol.2016.151910
10. Theis F, Corbacioglu A, Gaidzik VI, Paschka P, Weber D, Bullinger L, et al. Clinical Impact of GATA2 Mutations in Acute Myeloid Leukemia Patients Harboring CEBPA Mutations: A Study of the AML Study Group. Leukemia (2016) 30(11):2248–50. doi: 10.1038/leu.2016.185
11. Green CL, Tawana K, Hills RK, Bodor C, Fitzgibbon J, Inglott S, et al. GATA2 Mutations in Sporadic and Familial Acute Myeloid Leukaemia Patients With CEBPA Mutations. Br J Haematol (2013) 161(5):701–5. doi: 10.1111/bjh.12317
12. Greif PA, Dufour A, Konstandin NPK, sienzyk B, Zellmeier E, Tizazu B, et al. GATA2 Zinc Finger 1 Mutations Associated With Biallelic CEBPA Mutations Define a Unique Genetic Entity of Acute Myeloid Leukemia. Blood (2012) 120(2):395–403. doi: 10.1182/blood-2012-01-403220
13. Green CL, Koo KK, Hills RK, Burnett AK, Linch DC, Gale RE. Prognostic Significance of CEBPA Mutations in a Large Cohort of Younger Adult Patients With Acute Myeloid Leukemia: Impact of Double CEBPA Mutations and the Interaction With FLT3 and NPM1 Mutations. J Clin Oncol (2010) 28(16):2739–47. doi: 10.1200/JCO.2009.26.2501
14. Tien FM, Hou HA, Tang JL, Kuo YY, Chen CY, Tsai CH, et al. Concomitant WT1 Mutations Predict Poor Prognosis in Acute Myeloid Leukemia Patients With Double Mutant CEBPA. Haematologica (2018) 103(11):e510–3. doi: 10.3324/haematol.2018.189043
15. Su L, Gao S, Tan Y, Lin H, Liu X, Liu S, et al. CSF3R Mutations Were Associated With an Unfavorable Prognosis in Patients With Acute Myeloid Leukemia With CEBPA Double Mutations. Ann Hematol (2019) 98(7):1641–6. doi: 10.1007/s00277-019-03699-7
16. Tarlock K, Alonzo T, Wang YC, Gerbing RB, Ries RE, Hylkema T, et al. Prognostic Impact of CSF3R Mutations in Favorable Risk Childhood Acute Myeloid Leukemia. Blood (2020) 135(18):1603–6. doi: 10.1182/blood.2019004179
17. Maxson JE, Ries RE, Wang YC, Gerbing RB, Kolb EA, Thompson SL, et al. CSF3R Mutations Have a High Degree of Overlap With CEBPA Mutations in Pediatric AML. Blood (2016) 127(24):3094–8. doi: 10.1182/blood-2016-04-709899
18. Masuda S. DNMT3A Mutations in Acute Myeloid Leukemia: Impact on Low-Risk Patients With CEBPA Mutations. J Clin Oncol (2011) 29(34):4592–93; author reply 4593-4594. doi: 10.1200/JCO.2011.38.2127
19. Tabara LC, Vicente JJ, Biazik J, Eskelinen EL, Vincent O, Escalante R. Vacuole Membrane Protein 1 Marks Endoplasmic Reticulum Subdomains Enriched in Phospholipid Synthesizing Enzymes and Is Required for Phosphoinositide Distribution. Traffic (2018) 19(8):624–38. doi: 10.1111/tra.12581
20. Tabara LC, Escalante R. Vmp1 Establishes Er-Microdomains That Regulate Membrane Contact Sites and Autophagy. PloS One (2016) 11(11):e0166499. doi: 10.1371/journal.pone.0166499
21. Zheng L, Chen L, Zhang X, Zhan J, Chen J. TMEM49-Related Apoptosis and Metastasis in Ovarian Cancer and Regulated Cell Death. Mol Cell Biochem (2016) 416(1-2):1–9. doi: 10.1007/s11010-016-2684-3
22. Guo XZ, Ye XL, Xiao WZ, Wei XN, You QH, Che XH, et al. Downregulation of VMP1 Confers Aggressive Properties to Colorectal Cancer. Oncol Rep (2015) 34(5):2557–66. doi: 10.3892/or.2015.4240
23. Guo L, Yang LY, Fan C, Chen GD, Wu F. Novel Roles of Vmp1: Inhibition Metastasis and Proliferation of Hepatocellular Carcinoma. Cancer Sci (2012) 103(12):2110–9. doi: 10.1111/cas.12025
24. Amelio I, Melino G. The p53 Family and the Hypoxia-Inducible Factors (Hifs): Determinants of Cancer Progression. Trends Biochem Sci (2015) 40(8):425–34. doi: 10.1016/j.tibs.2015.04.007
Keywords: acute myeloid leukemia, biallelic CEBPA mutation, prognostic nomogram, VMP1 expression, CSF3R mutation
Citation: Xu X, Cai W, Cai P, Zhang L, Yao H, Zhang T, Shen H and Chen S (2021) Prognostic Nomogram for Acute Myeloid Leukemia Patients With Biallelic CEBPA Mutations. Front. Oncol. 11:628248. doi: 10.3389/fonc.2021.628248
Received: 11 November 2020; Accepted: 15 July 2021;
Published: 26 August 2021.
Edited by:
Marcos De Lima, The Ohio State University, United StatesReviewed by:
Mohamed Mahmoud Moussa, Ain Shams University, EgyptSarah Wall, The Ohio State University, United States
Copyright © 2021 Xu, Cai, Cai, Zhang, Yao, Zhang, Shen and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suning Chen, Y2hlbnN1bmluZ0BzaW5hLmNvbQ==
†These authors have contributed equally to this work
 Xiaoyu Xu
Xiaoyu Xu Wenzhi Cai1†
Wenzhi Cai1† Hongjie Shen
Hongjie Shen Suning Chen
Suning Chen