- 1Tumor Immunotherapy and Microenvironment (TIME) Group, Department of Oncology, Luxembourg Institute of Health (LIH), Luxembourg City, Luxembourg
- 2Department of Hemato-oncology, Centre Hospitalier du Luxembourg, Luxembourg City, Luxembourg
- 3Université de Lorraine - UMR 7365, Ingénierie Moléculaire et Physiopathologie Articulaire (IMoPA), Vandoeuvre-lès-Nancy, France
- 4Centre François Baclesse, Esch-sur-Alzette, Luxembourg
Autophagy is a highly regulated multi-step process that occurs at the basal level in almost all cells. Although the deregulation of the autophagy process has been described in several pathologies, the role of autophagy in cancer as a cytoprotective mechanism is currently well established and supported by experimental and clinical evidence. Our understanding of the molecular mechanism of the autophagy process has largely contributed to defining how we can harness this process to improve the benefit of cancer therapies. While the role of autophagy in tumor resistance to chemotherapy is extensively documented, emerging data point toward autophagy as a mechanism of cancer resistance to radiotherapy, targeted therapy, and immunotherapy. Therefore, manipulating autophagy has emerged as a promising strategy to overcome tumor resistance to various anti-cancer therapies, and autophagy modulators are currently evaluated in combination therapies in several clinical trials. In this review, we will summarize our current knowledge of the impact of genetically and pharmacologically modulating autophagy genes and proteins, involved in the different steps of the autophagy process, on the therapeutic benefit of various cancer therapies. We will also briefly discuss the challenges and limitations to developing potent and selective autophagy inhibitors that could be used in ongoing clinical trials.
Introduction
Macroautophagy (referred to as autophagy) was first described in 1966 as a cellular process that occurs at the basal level in all cells (1). Autophagy relies on the formation of double-membraned vesicles known as autophagosomes, leading to the degradation of their cargo, such as damaged proteins or organelles (2). The autophagy process involves more than thirteen autophagy-related (ATG) proteins and requires the following major steps: (i) Initiation, (ii) Nucleation, (iii) Maturation, and (iv) Fusion with lysosome for cargo degradation (Figure 1). Originally described as a bulk degradation process, autophagy is now described as a highly selective degradation mechanism for the recycling of cellular components. Autophagy can be activated as an adaptive cellular response to external stimuli such as hypoxia, starvation, and different cancer therapies and therefore considered as a cytoprotective mechanism (1, 3–5).
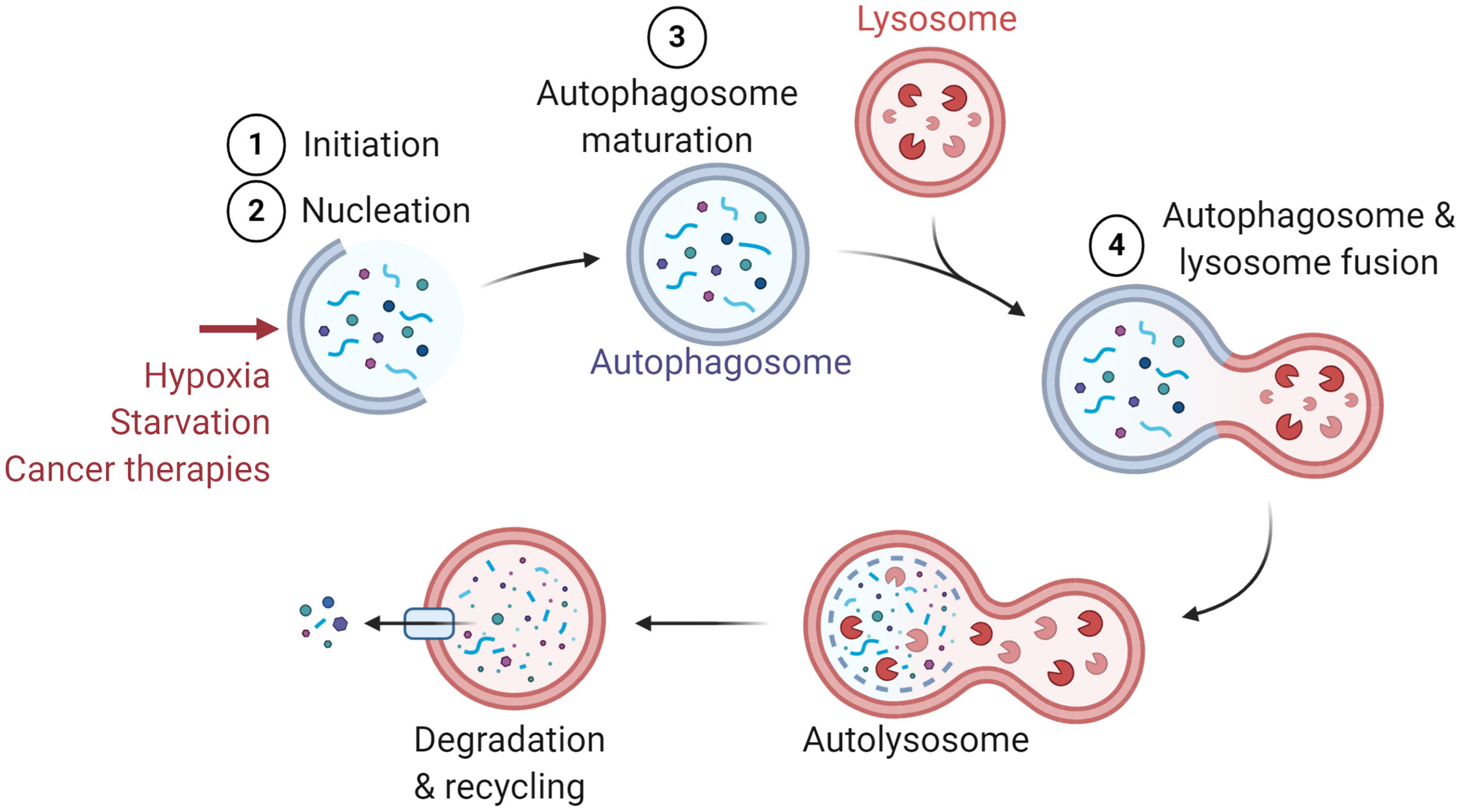
Figure 1 General presentation of the major steps of autophagy. Several stimuli have been identified to induce autophagy such as hypoxia, starvation, and cancer therapies. The major steps of autophagy are: 1) Initiation, 2) Nucleation, 3) Maturation, and 4) Fusion with lysosome for the degradation and recycling of autophagosome constituents.
Autophagy is activated under nutrient deprivation or starvation condition, which resulted in a decrease of mTOR activity and an increase of Unc-51 like autophagy activating kinase 1 (ULK1) activation. Activated ULK1 is subsequently dissociated from the 5’ adenosine monophosphate-activated protein kinase (AMPK), resulting in autophagy activation (6, 7). In addition to starvation, autophagy can also be activated in the tumor microenvironment by hypoxia through the hypoxia-inducible factor 1-α (HIF-α). The accumulation of HIF-1α in hypoxic cells activates the expression of BNIP/BNIP3L, which subsequently dissociates the complex between Bcl-2 and Beclin-1 (BECN1) to activate autophagy (8).
Autophagy was primarily considered as a tumor suppressive mechanism. Such a role was supported by studies showing that targeting BECN1, ATG5, and ATG7 promotes tumor initiation (9–11). In particular, evidences have demonstrated that Everolimus, an mTOR inhibitor and analogue of rapamycin, significantly increases mice survival in acute lymphoblastic leukemia in combination with Vincristine (12, 13). Conversely, many groups highlight the tumor supportive role of autophagy by showing its role in promoting tumor cell survival and growth in multiple tumor types (14, 15). The consensus appears to be that autophagy plays double-edged sword in suppressing tumor initiation and in promoting the survival of established tumors (16). Indeed, experimental evidence points at autophagy as a mechanism involved in cancer cell resistance to various therapies, such as chemotherapy, radiotherapy, targeted therapy, photodynamic therapy-induced apoptosis, and immunotherapy (17–21). Despite the complex interplay between the tumor suppressive and supportive role of autophagy in cancer (14), the vast majority of the clinical trials have focus on inhibiting autophagy with chloroquine (CQ) and hydroxychloroquine (HCQ) either alone or in combination with anticancer therapies (22). Therefore, autophagy inhibition has been suggested as a strategy to improve cancer therapies and has been considered in multiple clinical trials. Autophagy inhibitors have been classified according to their action on the major steps of autophagy and numerous preclinical studies have evaluated the therapeutic benefit of inhibiting autophagy.
In this review, we will summarize the impact of inhibiting the different steps of autophagy, either pharmacologically or genetically by silencing or knocking down autophagy-associated genes (Figure 2), and describe how autophagy can be leveraged to improve the therapeutic benefit of current cancer therapies and elicit a synergistic effect with antineoplastic agents.
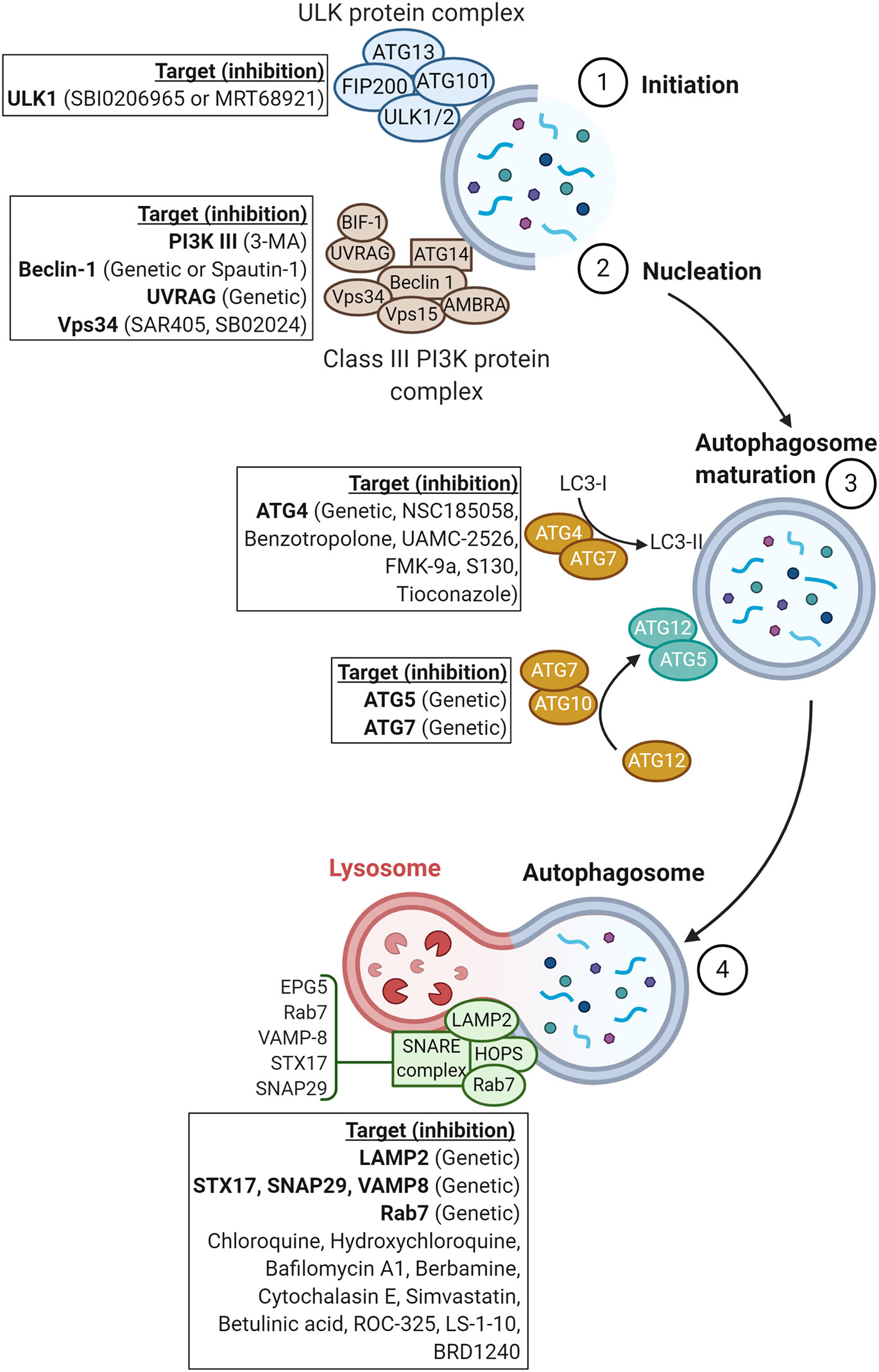
Figure 2 Schematic representation of proteins involved in the major steps of autophagy. Genetic or pharmacological approaches targeting proteins involved in each step of autophagy are reported in squares.
Impact of Inhibiting the Initiation and Nucleation Steps on Cancer Therapies
The first step of the autophagy process, so-called the initiation step, involves the ULK protein complex including ATG13, ATG101/ULK1/2, and FIP200 (2, 23). The initiation step of autophagy facilitates the recruitment of the class III PI3K or VPS34 complex containing BECN1, VPS34, regulatory subunit 4 (VPS15/P150), activating molecule in BECN1-regulated autophagy protein 1 (AMBRA), UV radiation resistance-associated gene protein (UVRAG), BIF1, and ATG14L (2, 23) to the newly formed “phagophore”. The recruitment of the class III PI3K constitutes the nucleation step. In this section, we will summarize the different drugs and/or strategies used to target the initiation and nucleation steps and briefly discuss data available on their efficacy in pre-clinical tumor mouse models.
Inhibition of ULK1
Several long noncoding RNAs (lncRNAs) have been reported to induce tumor chemoresistance to 5-fluorouracil (5-FU) such as lncRNA H19 and lncRNA small nucleolar RNA host gene 6 (SNHG6) in colorectal cancer (24, 25). SNHG6 promotes resistance of mice bearing RKO colon tumors to 5-FU. Furthermore, investigations showed that SNHG6 induced ULK1-dependent autophagy via sponging miR-26a-5p (25).
SBI0206965 is a highly selective inhibitor of ULK1 kinase (26) and it has been reported to sensitize NSCLC cells and acute myeloid leukemia (AML) cells to cisplatin- and daunorubicin-based chemotherapy, respectively, by decreasing cancer cell viability (27, 28). In pancreatic ductal adenocarcinoma (PDAC) cells, combining extracellular signal-regulated kinase (ERK) inhibitors with inhibitors of ULK1 complex (SBI0206965 or MRT68921) or with spautin-1, a specific inhibitor of two ubiquitin-specific peptidases USP10 and USP13 that control BECN1 degradation (29), decreased cell proliferation relative to ERK inhibitors alone (30). Recently, Chen et al. demonstrated that simultaneous inhibition of ULK1 (MRT68921) with NUAK1 (also known as ARK1) induces apoptosis in various cancer types (31).
Inhibition of Class III PI3K
Pre-treatment with class III PI3K inhibitors, such as 3-Methyladenine (3-MA), showed a significant improvement of the sensitivity of MDA-MB-231 and HBL-100 breast cancer cells to ionizing radiation (IR) despite an apparent low level of basal autophagy in HBL-100 cells (32). The therapeutic benefit of combining IR and 3-MA was observed in a xenograft esophageal squamous cell carcinoma mice model in vivo with a significant decrease in tumor volume relative to single treatment (33). Moreover, it has been demonstrated that 3-MA in combination with docetaxel, a semi-synthetic analog of paclitaxel, overcame docetaxel-induced autophagy and improved the sensitivity of lung adenocarcinoma (LAD) cells to docetaxel. Docetaxel-induced autophagy was mediated by High-mobility group box 1 (HMGB1) translocation, which participates in the BECN1 PI3K-III core complex formation via MEK/ERK1/2 pathway. Indeed, knockdown of HMGB1 reverted the sensitivity of LAD cells to docetaxel (34).
Sorafenib is a well-known anti-angiogenic agent and remains the standard treatment in advanced unresectable hepatocellular carcinoma (HCC) (35). Over the past decade, several studies have focused on the underlying mechanisms induced by Sorafenib and exploring new combination therapies. In various types of cancers, Sorafenib has been described as inducing autophagy (36). Yuan et al. showed that 3-MA treatment in combination with Sorafenib significantly improved growth inhibition in HepG2, Hep3B, and PLC/PRF/5 treated cells (37). In triple-negative breast cancer (TNBC) cells, dual inhibition of autophagy and the Insulin-like growth factor (IGF) signaling pathway using 3-MA and NVP-AEW541, respectively, enhanced the NVP-AEW541-induced cell growth inhibition and apoptosis (38). In addition, preclinical studies have focused on exploring the benefit of epidermal growth factor receptor (EGFR) inhibitors such as gefitinib in EGFR-positive cancers (39). In TNBC cells, an increasing concentration of gefitinib combined with 3-MA significantly decreased cell viability in vitro. Interestingly, in TNBC xenograft mice models, a gefitinib and 3-MA combination resulted in a significant decrease in tumor volume and tumor weight compared to the gefitinib treated group. Further investigations revealed that autophagy inhibition by 3-MA enhanced gefitinib-induced GO/G1 cell cycle arrest, DNA damage, and cell death via the mitochondrial apoptosis pathway (40).
Selective VPS34 kinase inhibitors have acquired great interest as potential potent drugs to inhibit early autophagy. The VPS34 kinase inhibitor SAR405 in combination with everolimus, a well-known mTOR inhibitor approved for the treatment of various tumors (41), induced efficient autophagy inhibition and reduced renal tumor cell proliferation in vitro (42). In addition, the VPS34 kinase inhibitor SB02024 in combination with sunitinib, a tyrosine kinase inhibitor, significantly decreases cell viability and multicellular spheroid (MCS) growth in both MCF-7 and MDA-MB-231 breast cancer cell lines. Notably, inhibition of MCS growth was not observed with a chloroquine (CQ) and sunitinib combination (43). We have recently shown that pharmacological targeting of VPS34 kinase activity by SB02024 (Sprint Bioscience) or SAR405 (Sanofi) significantly decreased tumor growth and improved mice survival in melanoma B16-F10 and colorectal CT26 tumor mouse models (44). We provided evidence that deep changes in the immune landscape occurred in B16-F10 and CT26 mice models treated with VPS34 inhibitors (SB02024 and SAR405), characterized by increased infiltration of immune effectors such as NK, dendritic cells (DCs), M1 macrophages, and CD8+ T cells in the tumor microenvironment. Because there was no difference in the growth of tumors engrafted in NOD scid gamma (NSG) mice upon treatment with VPS34 inhibitors, these data clearly indicated that the tumor inhibitory effect of VPS34 inhibitors involves the host immune system. Moreover, we demonstrated that pro-inflammatory chemokines such as CCL5 and CXCL10 are responsible for NK and CD8+ T cell recruitment in B16-F10 tumors and CT26 tumors treated with VPS34 inhibitors relative to control. Interestingly, SB02024 or SAR405 improved the therapeutic benefit of anti-PD-1/PD-L1 by significantly reducing tumor growth and tumor weight and improving mice survival in B16-F10 and CT26 tumors (44).
Inhibition of Beclin-1
It has been reported that genetic inhibition of BECN1 or UVRAG potentiated IR-induced DNA double-strand breaks (DSBs) and cell death in colorectal cancer cells (45). Furthermore, gene silencing of BECN-1 enhanced the efficiency of fasudil (a RhoA/ROCK inhibitor) to induce apoptosis in esophageal squamous cell carcinoma cells (46). Similarly, in vitro suppression of BECN1 reduces paclitaxel-mediated cell viability, colony formation, and induced apoptotic death in BT-474 and MDA-MB-231 breast cancer cells in dose- and time-dependent manners (47). Similar effects were observed in non-small cancer lung cancer (NSCLC) cells, endometrial carcinoma, nasopharyngeal carcinoma cells, and ovarian and renal cancers (48–53). Interestingly, the therapeutic benefit of paclitaxel was increased in a Becn1-targeted BT-474 xenograft mice model based on cleaved caspase-3 positive cells and inhibition of tumor growth (47).
In human chronic myeloid leukemia (CML) cells, co-treatment with spautin-1 and imatinib, a BCR-ABL tyrosine kinase inhibitor, potentiated imatinib-induced CML cell apoptosis in both the K562 cell line and primary cells (54). In line with this latter study, it has also been reported that imatinib in combination with microRNA-30a, identified as potent inhibitor of BECN1 and ATG5, significantly increased the imatinib-mediated cytotoxicity in CML cells (55).
Tamoxifen is one of the most efficient endocrine treatments in estrogen receptor (ER) positive breast cancers, which account for 70% of the breast cancer subtypes. However, the therapeutic benefit of Tamoxifen is negatively impacted by the development of drug resistance (56). Gu et al. discovered that tamoxifen resistance was associated with an increased BECN1 and human epidermal growth factor receptor 2 (HER2) expression in breast cancer cells. BECN1 silencing enhanced the sensitivity of breast cancer cells to tamoxifen by reducing tumor cell proliferation, migration, and invasion capabilities. These data highlight a novel role of BECN1 in HER2 regulation that contributes to tamoxifen resistance (57).
Using several ovarian cancer cells, Zhang et al. showed that cancer cell resistance to cisplatin relied on autophagy-dependent induction of nuclear accumbens-1 (NAC1). Indeed, targeting NAC1 or autophagy, via 3-MA or BECN1 silencing, enhanced ovarian cancer cell sensitivity to cisplatin (58). Similar results were reported in adenoid cystic carcinoma of the salivary glands, glioma, and urothelial carcinoma (59–61).
Bevacizumab, a monoclonal anti-vascular endothelial growth factor (VEGF) antibody, is widely used to treat metastatic colorectal cancer, lung cancer and renal cell carcinoma (62–64). Knowing that bevacizumab induces autophagy, it has been reported that genetic inhibition of BECN1 improves the anticancer effects of this drug in colorectal cancer cells (65). Likewise, improved clinical response to trastuzumab was observed in HER2+ breast cancer displaying loss of BECN1 gene (38, 66). The role of autophagy, including BECN1, in tumor resistance to targeted therapy is comprehensively reviewed by Mele et al. (67).
We have previously reported that genetic targeting of Becn1 in melanoma cells prevents the degradation of Natural killer (NK)-derived Granzyme B and enhances melanoma susceptibility to NK-mediated killing (68). Furthermore, we showed that the infiltration of NK cells into Becn1 defective melanoma is increased, which results in significant inhibition of tumor growth (69). Importantly, the impact of inhibiting autophagy on the infiltration of cytotoxic immune cells into the tumors and the decrease in tumor growth is also reported by other studies (70–73).
Targeting Autophagosome Maturation Genes ATG4B, ATG5, and ATG7 Potentiates Anticancer Therapies
The third major step of autophagy involves two key complexes that promote the expansion of the phagophore membrane and result in the formation of a double-membraned vesicle named autophagosome. The first complex involves cooperation between ATG4B and ATG7, allowing for the conjugation of LC3I with phosphatidylethanolamine (PE) to form LC3II. LC3II is subsequently incorporated into the newly formed autophagosome (74). The second complex includes ATG7 and the E2-like enzyme ATG10, which are involved in ATG5-ATG12 conjugation (2). In this section, we will describe the therapeutic benefit of inhibiting autophagy genes involved in the maturation of autophagosomes.
Inhibition of ATG4B
The serine/threonine protein kinase MST4, also known as mammalian STE20-like protein kinase 4 (MST4) (75), was reported to facilitate p-ERK pathway and promote epithelial to mesenchymal transition (EMT) and cancer metastasis in gastric cancer (76). MST4 is associated with prostate cancer, hepatocellular carcinoma, and breast cancer progression (77, 78). It has been reported that MST4 directly phosphorylates ATG4B at S383 position (79). Furthermore, ATG4B inhibition, by NSC185058 (80), improves the anti-tumor effect of radiotherapy in intracranial glioblastoma (GBM) patient derived xenograft (PDX) mice models (79). These data suggest a potential interconnection between MST4, autophagy and malignancy in GBM; however, the value of direct targeting MST4 as a strategy to modulate autophagy remains to be defined.
Recently, benzotropolone derivatives were synthetized and tested for ATG4B inhibition. UAMC-2526 was selected as the best candidate according to its efficiency to reduce basal autophagy and its high stability in the plasma. A combination of UAMC-2526 with oxaliplatin-based chemotherapy reduced colorectal cell proliferation and promoted tumor growth inhibition in HT29 colorectal tumor-bearing mice (81).
FMK-9a is another ATG4B inhibitor, reported to attenuate the pro-LC3 cleavage process and the LC3-PE delipidation. FMK-9a could also induce autophagy independent of its inhibition on ATG4B activity (82). Recently, S130 has been identified to bind and inhibit ATG4B, hence attenuating the delipidation of LC3-II and suppressing the recycling of LC3-I in colorectal cancer cells. Therefore, S130 has been described as a novel small-molecule to improve cancer therapy (83).
Moreover, an FDA-approved drug screening identified tioconazole as a new ATG4 inhibitor. Tioconazole treatment enhanced doxorubicin efficiency by decreasing cell viability in H4, HCT116, and MDA-MB-231 cells. Interestingly, the combination of tioconazole and doxorubicin showed an enhanced antitumor effect in HCT116 xenografted mice relative to each drug alone (84). In this context, it has been reported that the HER2 status was positively correlated with the expression of ATG4B protein. Interestingly, ATG4B silencing was associated with reduced viability of trastuzumab treated HER2+ cells compared to trastuzumab treatment alone (85).
Inhibition of ATG5 and ATG7
In A549 human lung cancer, overcoming the cytoprotective effect of autophagy induced by cisplatin, via ATG5 silencing, improves cancer cell apoptosis, as compared to cisplatin treatment alone (86). Dual combination of Epirubicin, a structural analog of doxorubicin, with ATG5- or ATG7-silencing, significantly reduced cell viability in anthracycline-sensitive and resistant TNBC cells (87). O’Donovan et al. showed that combining both siRNA BECN1 and ATG7 decreased cell survival in 5-FU-treated esophageal cancer cells while targeting BECN1 or ATG7 alone had no impact (88). Therefore, it appears that targeting different steps of autophagy may be a more appropriate strategy to improve chemotherapy efficacy. In TBNC cells, Wu et al. demonstrated that dual inhibition of ATG7, genetically, and IGF-1R pharmacologically, promotes apoptosis and cell growth inhibition (38). In PDAC cells, genetic inhibition of ATG5 or ATG7 significantly improved the effect of ERK inhibitors on inhibiting cell proliferation relative to ERK inhibitors alone (30). Dual combination of ATG5 siRNA and docetaxel, a well-known second-line approved treatment in NSCLC, decreased cell proliferation together with increasing cytotoxicity and apoptosis in LAD cells (34). In renal cell carcinoma cells, ATG5 silencing or 3-MA treatment in combination with Sorafenib enhanced the sensitivity of RCC cells to Sorafenib (89).
Impact of Inhibiting Autophagosome-Lysosome Fusion on the Response to Various Anticancer Therapies
The final step of autophagy consists of fusion between autophagosomes and lysosomes for the degradation, and recycling of damaged proteins and organelles. Thus, the outer membrane of autophagosomes merges with the lysosomal membrane, and then the inner membrane is degraded (90). The principal factors involved in autophagosome-lysosome fusion are the homotypic fusion and protein sorting (HOPS) complex, RAB7, and the N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) (91). Other proteins, such as the lysosomal-associated membrane protein 2 (LAMP2), are also required for proper fusion (92). This part will focus on the impact of targeting these factors and the fusion step on the therapeutic benefit of radiotherapy, chemotherapy, targeted therapy, and immunotherapy.
Targeting the Soluble N-Ethylmaleimide-Sensitive Factor Attachment Protein Receptors (SNAREs) to Enhance Response to Conventional Treatments
Syntaxin 17 (STX17), SNAP29, and vesicle-associated membrane protein 8 (VAMP8) are N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) involved in the autophagosome-lysosome fusion. During this process, STX17 is recruited to the outer membrane of autophagosomes through HOPS complex and interacts with VAMP8, located on lysosomes. This interaction is enhanced by SNAP29, which forms a SNARE complex with STX17 and VAMP8 (93). Knockdown of STX17 causes a blockade of the fusion between autophagosomes and lysosomes and results in the accumulation of autophagosomes (93). Therefore, targeting SNARE proteins is considered a strategy for preventing the late step of autophagy. Moreover, SNAP29-STX17-VAMP8 complex formation can be enhanced by the down-regulation of O-GlcNAc transferase (OGT). This was correlated with the resistance of ovarian cancer to cisplatin treatment (94), and highlights that targeting SNAP29-STX17-VAMP8 complex by overexpression of OGT could improve the sensitivity to cisplatin treatment. Overexpression of VAMP8 has also been associated with resistance to temozolomide in human glioma cells, and knockdown of STX17 in glioma cells overexpressing VAMP8 led to increased chemosensitivity (95).
In addition, Berbamine, a natural product isolated from traditional Chinese medicine, inhibits autophagosome-lysosome fusion by preventing the interaction between VAMP8 and SNAP29. Berbamine was proposed as a new potential inhibitor of autophagy that could enhance the effects of chemotherapy treatment (96). Moreover, Berbamine has been investigated as a potential anticancer drug in several studies and seems to act on multiple pathways such as MEK/ERK (97) and WNT/β-catenin pathways (98).
Targeting Lysosomal-Associated Membrane Protein 2 (LAMP2) as a Potential Target to Inhibit Autophagosome-Lysosome Fusion and Improve the Response to Anti-Cancer Therapies
LAMP2 is a glycosylated protein ubiquitously expressed, and mostly located on lysosome membranes. LAMP2 is required for the proper fusion between autophagosomes and lysosomes (92). In neuroendocrine prostate cancer, knockdown of LAMP2 by siRNA induced an autophagy blockade and decreased both cancer cell proliferation and neuroendocrine markers. These results indicate that LAMP2 plays a dual role in cell survival, by inducing autophagy and in the differentiation of androgen-sensitive human prostate adenocarcinoma cells into neuroendocrine prostate cancer cells (99). In addition, a recent in silico approach showed that the expression of LAMP2 was decreased in prostate cancer tissues as compared to normal prostate tissues (100), indicating that the expression level of LAMP2 could act as a regulatory element in cancer progression. Another study compared the expression level of LAMP2 in salivary adenoid cystic carcinoma and pleomorphic adenoma and/or a normal salivary gland (101). The results showed an increased expression of LAMP2 in salivary adenoid cystic carcinoma, which was associated with cancer progression. Although the expression level of LAMP2 seems to be different in various cancer types, several data suggest that LAMP2 is a potential target for cancer therapy in combination with conventional treatments. This statement was supported by data showing that the silencing of LAMP2 by siRNAs led to a radiosensitization of prostate cancer cell lines (102). In addition, a reduced expression of LAMP2 has been associated with a decreased resistance to both cisplatin in human ovarian carcinoma cells (103) and azacitidine in acute myeloid leukemia (104). Nevertheless, the sensitization to radiotherapy and chemotherapy by LAMP2 targeting should be investigated in other types of cancer cells.
Overexpression of RAB7 as a Potential Strategy to Improve Sensitivity to Anti-Cancer Treatments
RAB7 is a small GTPase localized to late endosomes and lysosomes and has multiple functions in autophagy. In mammalian, RAB7 is not directly involved in the autophagosome-lysosome fusion but rather in autolysosome maturation under nutrient-rich conditions (105). The significance of RAB7 as a target for autophagy modulation is not well defined so far.
The role of RAB7 in cancer progression has recently been described as a protein involved in promoting the proliferation, invasion, and migration of gastric cancer cells (106). RAB7 has also been associated with chemoresistance to cisplatin. Indeed, RAB7 was downregulated in cisplatin-resistant cervical cancer cell lines as compared to chemosensitive ones (107). Additionally, RAB7 overexpression induced chemosensitization of cisplatin-resistant cells, while depletion of RAB7 by siRNA induced resistance to cisplatin in chemosensitive cells (108). Furthermore, RUBICON (Run domain Beclin-1 interacting and cysteine-rich containing), a negative regulator of autophagy, inhibits autophagosome-lysosome fusion and interacts with RAB7-GTP via a RUBICON homology (RH) domain (109).
Impact of Using Drugs Inhibiting the Last Step of Autophagy Process on Various Cancer Therapies
CQ and its derivate, hydroxychloroquine (HCQ), are the only drugs having shown their ability to block the last step of autophagy and being approved by the Food and Drug Administration (FDA) for clinical use. Indeed, CQ blocks the autophagic flux by altering autophagosome fusion with lysosomes, presumably by interfering with SNAP29 recruitment (110). In addition, CQ cytotoxicity induced autophagy-associated cell death associated with nuclei abnormalities, lipofuscinogenesis, and senescence (111). As reported in clinicaltrials.gov, CQ is currently being investigated as a potent anticancer drug in small cell lung cancer, breast cancer, pancreatic cancer, glioblastoma, melanoma, and other types of cancers.
Because CQ and HCQ are the only autophagy inhibitors available and approved for clinical use, multiple studies evaluated the potential effects of CQ in vitro, in combination with anticancer treatments. In fact, CQ was responsible for radio-sensitizing bladder cancer cells and bladder cancer xenografts in mice (112). CQ also had a synergetic effect with radiotherapy on glioma initiating cells by inducing apoptosis and inhibiting autophagy induced by ionizing radiation (113). The same synergistic effects were observed on glioblastoma cell lines (114). CQ administered after radiation is also capable of increasing the death of breast cancer cells and tumor regression in vivo (115). The use of CQ as a potent enhancer of radiotherapy is currently being evaluated in clinical trials involving small cell lung cancer (NCT01575782) and glioblastoma (NCT04397679), and in patients with brain metastases from solid tumors (NCT01894633). It should be highlighted that CQ sensitized various breast cancer cell lines to cisplatin and LY294002, reported to induce autophagy in these cells. However, CQ sensitization in these cells occurred independent of autophagy inhibition. Therefore, the autophagy independent sensitizing effects of CQ should be considered in clinical trials where CQ or its derivatives are used in the treatment of cancer (116, 117). In HCT-116 and HT-29 colorectal cancer cell lines, it has been reported that CQ sensitized these cell lines to radiation and 5-FU treatment and resulted in a significant decrease in clonogenic survival of HT-29 cell line without any impact on cell cycle progression or cell death (118). However, in GBM, CQ was found to induce P53-independent cell deaths that do not require caspase-mediated apoptosis. The CQ derivatives, Quinacrine and Mefloquine, are more potent and displayed superior blood-brain barrier penetration compared to CQ (119).
Maycotte et al. evaluated the effects of combining CQ with chemotherapeutic drugs such as the DNA damaging agent cisplatin, the mTOR inhibitor Rapamycin, and the PtdIns3K inhibitor LY294002 in two mouse breast cancer cell lines (117). While the combination of CQ and cisplatin had no significant effect on the viability of both cell lines, CQ combined with PtdIns3K and mTOR inhibition sensitized both cell lines. However, the CQ-mediated sensitization seems to be independent of autophagy, since this sensitization was not observed following Atg12 and Becn1 knockdown (117). A similar result was observed in KRAS-driven cancer cell lines where the antiproliferative effects of CQ were similar between ATG7-deficient tumor cell lines with undetectable autophagic flux and ATG7-efficient tumor cell lines (120). In addition, CQ sensitizes bladder cancer cells to cisplatin treatment by inhibiting cisplatin-induced autophagy (121). Similar results were observed in nasopharyngeal carcinoma cells (122) and hypopharyngeal squamous cell carcinoma xenografted mice (123). This suggests that CQ effects, in combination with chemotherapy, depend on the type of cancer and therefore require further investigation.
Several studies have been conducted on CQ in combination with targeted therapies. Erlotinib and Rapamycin are two tyrosine kinase inhibitors targeting the EGFR and the mammalian target of Rapamycin, respectively. These anticancer drugs are particularly used for NSCLC treatment where EGFR and PI3K/AKT/mTOR pathways are often dysregulated. It has been shown that the combination of Erlotinib and Rapamycin with Monensin, a polyether antibiotic inhibiting autophagosome-lysosome fusion, improved Erlotinib and Rapamycin induced tumor growth inhibition and apoptosis in NSCLC (124). Similar results were observed in prostate cancer cells by Monensin, although the involvement of autophagy inhibition was not clearly suggested in this study (125). Furthermore, the combination of the tyrosine kinase inhibitor, sunitinib, with CQ or LAMP2 knockdown also showed promising results in a metastatic pancreatic neuroendocrine tumor mice model (126). The combination of the monoclonal antibody trastuzumab with CQ in HER2+ breast cancer (127) also led to promising results. Indeed, CQ sensitized both trastuzumab-resistant breast cancer cells and trastuzumab-resistant xenografts, resulting in increased cell death in vitro and decreased tumor growth in vivo.
In addition to its effects in combination with radiotherapy, chemotherapy, and targeted therapy, CQ seems to be responsible for various effects on the immune system. CQ resets tumor-associated M2 macrophages to the tumor-inhibiting M1 phenotype in B16 melanoma and H22 hepatocarcinoma mouse tumor models, and ameliorates the immunosuppressive tumor immune microenvironment through a lysosomal calcium-TFEB pathway (91). Another recent article showed that CQ in combination with 5-FU increased the driving of DC maturation by HCT-116 colon cancer cells, and in this way stimulates T cell responses induced by tumor cell lysates (128). Considering the impact of CQ on the immune system, CQ was tested in combination with dual CTLA4 and PD-1 immune checkpoint blockade in orthotopic tumors established from pancreatic ductal adenocarcinoma cells. This study revealed an increased CD8+ T cell infiltration within the tumors and a tumor sensitization to anti-PD-1 and anti-CTLA4 therapy when combined with CQ (129). These data support further investigations on the potential effect of combining CQ with other immunotherapy such as new immune checkpoint inhibitors, T-cell transfer therapy, or monoclonal antibodies, on the immunosuppressive tumor microenvironment.
Besides CQ, Bafilomycin A1 (BafA1) is another drug acting at the last step of autophagy. BafA1 is an antibiotic targeting the vacuolar H+-ATPase enzyme, thus inducing acidification of lysosomal pH, therefore inhibiting lysosomal degradation capacity. BafA1 has also shown promising effects in combination with chemotherapy, since BafA1 increased cisplatin cytotoxicity in tongue squamous cell carcinoma cells and bladder cancer cells (121, 130). BafA1 also increased chemosensitivity to 5-FU in gastric cancer cells (131). Other drugs have been described to target autophagosome-lysosome fusion, such as LS-1-10, Cytochalasin E and Simvastatin. LS-1-10 is a novel acridine derivative sharing structure with CQ and HCQ. LS-1-10 has a dual function, it is able to both induce DNA damage and block autophagosome-lysosome fusion. In addition, LS-1-10 is significantly more potent in reducing cell viability than CQ (>50%) in colon cancer cells (132).
Cytochalasin E is a fungal toxin found in Aspergillus clavatus, which binds to actin filaments. Cytochalasin E can inhibit autophagosome-lysosome fusion in the same way as CQ. In addition, cytochalasin enhances the effect of bortezomib in human lung cancer cells (133). Simvastatin is a powerful inhibitor of hydroxymethylglutaryl CoA reductase, an enzyme involved in cholesterol biosynthesis. Interestingly, Simvastatin can induce cell death in astrocytoma, neuroblastoma, glioblastoma, breast cancer, and lung adenocarcinoma (134). Moreover, this drug appears to inhibit the fusion between autophagosomes and lysosomes and increase the effects of Temozolomide in glioblastoma cell lines (135). Finally, the combination of Simvastatin with Vorinostat, a histone deacetylase inhibitor, inhibits autophagy by preventing RAB7 prenylation and decreases tumor growth in mice bearing triple-negative breast tumors (136).
Conclusion
Despite the dual role of autophagy in suppressing tumor initiation and in promoting the survival of established tumors (16), the studies reported in this review highlight the pivotal role of autophagy as a cytoprotective and therapy resistance mechanism in cancer. Therefore, strategies used to modulate autophagy for enhancing the therapeutic benefit of current anticancer therapies are an area of intense investigation. Based on data reported in this review, we strongly believe that inhibiting autophagy represents a new paradigm for overcoming therapy resistance and enhancing drug sensitivity in multiple tumor cell types. Because autophagy inhibition is currently used in many clinical trials along with different therapeutic strategies, it is reasonable to consider that several cancer treatments themselves induce autophagy in tumor cells.
Among the various autophagy inhibitors, CQ or its derivative HCQ are the major drugs used in clinical trials, with mitigated success. While CQ provides promising results in combination with conventional anticancer therapies, in some studies CQ sensitization appears to be independent of autophagy inhibition. Therefore, more selective and potent autophagy inhibitors must be designed to definitively endorse the therapeutic benefit of targeting autophagy in cancer patients. While CQ and HCQ inhibits the last step of autophagy (110), other druggable autophagy proteins have recently been proposed, which include the early autophagy protein BECN1 (45) and its interacting protein VPS34 (or PI3K class III) (41, 42, 44). Thus, more potent and selective autophagy specific inhibitors are currently in pre-clinical development; these include drugs targeting earlier steps in the autophagy process, such as ULK1, VPS34, and ATG4B (27, 28) (79). Other factors interacting with the autophagy process could also be considered as potential targets to inhibit autophagy and overcome therapy resistance, such as AMPK and HIF-1α, which are reported as key inducers of autophagy through negative regulation of the mTOR pathways and inducing hypoxic conditions, respectively (6, 7, 137). Other drugs are able to target the late step of autophagy by interacting with lysosomes. ROC-325 has been described as a potent autophagy inhibitor exhibiting superior in vitro and in vivo anticancer effects compared to CQ. In Renal cell carcinoma RCC, ROC-325 induced an accumulation of autophagosomes in vitro and inhibited RCC growth and survival in an ATG5/7-dependent manner in vivo by disrupting autophagic degradation (138). In acute myeloid leukemia, ROC-325 improved the anti-leukemic activity of azacitidine through inhibiting autophagy (139). BRD1240 is a small-molecule suppressing the V-ATPase function and therefore inhibiting the lysosomal acidification property. It has been reported that, similar to BafA1, BRD1240 inhibited autolysosome formation and subsequently triggering a significant accumulation of autophagosomes (140). Similar to ROC-325 and LS-1-10, Betulinic acid (BA) disrupt the degradative lysosomal function (74, 141–143), leading to the accumulation of mitochondria inside dysfunctional autolysosomes. Such a lysosome-mitochondrial stress axis is responsible for the induction of lipofuscinogenesis and ageing (143).
It should be highlighted that even if a selective and potent autophagy inhibitor is identified, the challenging task is to demonstrate that the therapeutic benefit that could be observed is indeed related to inhibition of the autophagy process, as almost all autophagy-related genes have non-autophagic functions. Another challenge is to maintain a balance between the benefits gained by autophagy inhibition and the deleterious effects of this inhibition in cancer patients. Indeed, the process of autophagy seems to both activates and inhibits cellular senescence (144), and chronic inhibition of autophagy appears to increase permanently the risk of cancer (145). Finally, considering the controversial role of autophagy regarding its cytoprotective or cytotoxic function, it is more likely obvious that the clinical outcome of combination treatment between an inhibitor of autophagy with chemotherapy, radiotherapy, targeted therapy, or immunotherapy would lead to contradictory or equivocal results (146). Therefore, the last challenge to overcome would be to determine whether patients would benefit from autophagy inhibition prior to conventional therapies, with for example the use of novel biomarkers of cytoprotective autophagy.
Author Contributions
MX, AB, MH, CD, GV, GB, MN, and BJ have contributed in writing and editing the manuscript. BJ has generated figures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Luxembourg Institute of Health and grants from the Luxembourg National Research Fund (PRIDE15/10675146/CANBIO and C18/BM/12670304/COMBATIC); FNRS Televie (grants 7.4606.18 and 7.4579.20); Fondation Cancer, Luxembourg (FC/2018/06); Kriibskrank Kanner Foundation, Luxembourg (2019); RCMS foundation, and Action LIONS Vaincre le Cancer Luxembourg. The authors also declare that this study received funding from Janssen Cilag Pharma and Roche Pharma. These funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol (1966) 28:435–92. doi: 10.1146/annurev.ph.28.030166.002251
2. Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer (2017) 17(9):528–42. doi: 10.1038/nrc.2017.53
3. Noman MZ, Janji B, Kaminska B, Van Moer K, Pierson S, Przanowski P, et al. Blocking hypoxia-induced autophagy in tumors restores cytotoxic T-cell activity and promotes regression. Cancer Res (2011) 71(18):5976–86. doi: 10.1158/0008-5472.CAN-11-1094
4. Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis (2013) 4:e838. doi: 10.1038/cddis.2013.350
5. Palorini F, Cozzarini C, Gianolini S, Botti A, Carillo V, Iotti C, et al. First application of a pixel-wise analysis on bladder dose-surface maps in prostate cancer radiotherapy. Radiother Oncol (2016) 119(1):123–8. doi: 10.1016/j.radonc.2016.02.025
6. Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett (2010) 584(7):1287–95. doi: 10.1016/j.febslet.2010.01.017
7. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol (2011) 13(2):132–41. doi: 10.1038/ncb2152
8. Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol (2009) 29(10):2570–81. doi: 10.1128/MCB.00166-09
9. Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest (2003) 112(12):1809–20. doi: 10.1172/JCI20039
10. Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA (2003) 100(25):15077–82. doi: 10.1073/pnas.2436255100
11. Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev (2011) 25(8):795–800. doi: 10.1101/gad.2016211
12. Crazzolara R, Cisterne A, Thien M, Hewson J, Baraz R, Bradstock KF, et al. Potentiating effects of RAD001 (Everolimus) on vincristine therapy in childhood acute lymphoblastic leukemia. Blood (2009) 113(14):3297–306. doi: 10.1182/blood-2008-02-137752
13. Singh SS, Vats S, Chia AY, Tan TZ, Deng S, Ong MS, et al. Dual role of autophagy in hallmarks of cancer. Oncogene (2018) 37(9):1142–58. doi: 10.1038/s41388-017-0046-6
14. Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev (2011) 25(19):1999–2010. doi: 10.1101/gad.17558811
15. Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell (2013) 155(6):1216–9. doi: 10.1016/j.cell.2013.11.019
16. Janji B, Viry E, Moussay E, Paggetti J, Arakelian T, Mgrditchian T, et al. The multifaceted role of autophagy in tumor evasion from immune surveillance. Oncotarget (2016) 7(14):17591–607. doi: 10.18632/oncotarget.7540
17. Ito H, Daido S, Kanzawa T, Kondo S, Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol (2005) 26(5):1401–10. doi: 10.3892/ijo.26.5.1401
18. Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest (2007) 117(2):326–36. doi: 10.1172/JCI28833
19. Ertmer A, Huber V, Gilch S, Yoshimori T, Erfle V, Duyster J, et al. The anticancer drug imatinib induces cellular autophagy. Leukemia (2007) 21(5):936–42. doi: 10.1038/sj.leu.2404606
20. Wei MF, Chen MW, Chen KC, Lou PJ, Lin SY, Hung SC, et al. Autophagy promotes resistance to photodynamic therapy-induced apoptosis selectively in colorectal cancer stem-like cells. Autophagy (2014) 10(7):1179–92. doi: 10.4161/auto.28679
21. Janji B, Berchem G, Chouaib S. Targeting Autophagy in the Tumor Microenvironment: New Challenges and Opportunities for Regulating Tumor Immunity. Front Immunol (2018) 9:887. doi: 10.3389/fimmu.2018.00887
22. Towers CG, Thorburn A. Therapeutic Targeting of Autophagy. EBioMedicine (2016) 14:15–23. doi: 10.1016/j.ebiom.2016.10.034
23. Xiao M, Noman MZ, Menard L, Chevigne A, Szpakowska M, Bosseler M, et al. Driving Cytotoxic Natural Killer Cells into Melanoma: If CCL5 Plays the Music, Autophagy Calls the Shots. Crit Rev Oncog (2018) 23(5-6):321–32. doi: 10.1615/CritRevOncog.2018027526
24. Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang R, et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis (2018) 9(12):1149. doi: 10.1038/s41419-018-1187-4
25. Wang X, Lan Z, He J, Lai Q, Yao X, Li Q, et al. LncRNA SNHG6 promotes chemoresistance through ULK1-induced autophagy by sponging miR-26a-5p in colorectal cancer cells. Cancer Cell Int (2019) 19:234. doi: 10.1186/s12935-019-0951-6
26. Egan DF, Chun MG, Vamos M, Zou H, Rong J, Miller CJ, et al. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol Cell (2015) 59(2):285–97. doi: 10.1016/j.molcel.2015.05.031
27. Tang F, Hu P, Yang Z, Xue C, Gong J, Sun S, et al. SBI0206965, a novel inhibitor of Ulk1, suppresses non-small cell lung cancer cell growth by modulating both autophagy and apoptosis pathways. Oncol Rep (2017) 37(6):3449–58. doi: 10.3892/or.2017.5635
28. Qiu L, Zhou G, Cao S. Targeted inhibition of ULK1 enhances daunorubicin sensitivity in acute myeloid leukemia. Life Sci (2020) 243:117234. doi: 10.1016/j.lfs.2019.117234
29. Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell (2011) 147(1):223–34. doi: 10.1016/j.cell.2011.08.037
30. Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med (2019) 25(4):628–40. doi: 10.1038/s41591-019-0368-8
31. Chen Y, Xie X, Wang C, Hu Y, Zhang H, Zhang L, et al. Dual targeting of NUAK1 and ULK1 using the multitargeted inhibitor MRT68921 exerts potent antitumor activities. Cell Death Dis (2020) 11(8):712. doi: 10.1038/s41419-020-02885-0
32. Chaachouay H, Ohneseit P, Toulany M, Kehlbach R, Multhoff G, Rodemann HP. Autophagy contributes to resistance of tumor cells to ionizing radiation. Radiother Oncol (2011) 99(3):287–92. doi: 10.1016/j.radonc.2011.06.002
33. Chen Y, Li X, Guo L, Wu X, He C, Zhang S, et al. Combining radiation with autophagy inhibition enhances suppression of tumor growth and angiogenesis in esophageal cancer. Mol Med Rep (2015) 12(2):1645–52. doi: 10.3892/mmr.2015.3623
34. Pan B, Chen D, Huang J, Wang R, Feng B, Song H, et al. HMGB1-mediated autophagy promotes docetaxel resistance in human lung adenocarcinoma. Mol Cancer (2014) 13:165. doi: 10.1186/1476-4598-13-165
35. Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med (2008) 359(4):378–90. doi: 10.1056/NEJMoa0708857
36. Prieto-Dominguez N, Ordonez R, Fernandez A, Garcia-Palomo A, Muntane J, Gonzalez-Gallego J, et al. Modulation of Autophagy by Sorafenib: Effects on Treatment Response. Front Pharmacol (2016) 7:151. doi: 10.3389/fphar.2016.00151
37. Yuan H, Li AJ, Ma SL, Cui LJ, Wu B, Yin L, et al. Inhibition of autophagy signi fi cantly enhances combination therapy with sorafenib and HDAC inhibitors for human hepatoma cells. World J Gastroenterol (2014) 20(17):4953–62. doi: 10.3748/wjg.v20.i17.4953
38. Wu W, Ma J, Shao N, Shi Y, Liu R, Li W, et al. Co-Targeting IGF-1R and Autophagy Enhances the Effects of Cell Growth Suppression and Apoptosis Induced by the IGF-1R Inhibitor NVP-AEW541 in Triple-Negative Breast Cancer Cells. PloS One (2017) 12(1):e0169229. doi: 10.1371/journal.pone.0169229
39. Xu H, Yu Y, Marciniak D, Rishi AK, Sarkar FH, Kucuk O, et al. Epidermal growth factor receptor (EGFR)-related protein inhibits multiple members of the EGFR family in colon and breast cancer cells. Mol Cancer Ther (2005) 4(3):435–42. doi: 10.1158/1535-7163.MCT-04-0280
40. Liu Z, He K, Ma Q, Yu Q, Liu C, Ndege I, et al. Autophagy inhibitor facilitates gefitinib sensitivity in vitro and in vivo by activating mitochondrial apoptosis in triple negative breast cancer. PloS One (2017) 12(5):e0177694. doi: 10.1371/journal.pone.0177694
41. Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discovery (2011) 10(11):868–80. doi: 10.1038/nrd3531
42. Kim TS. Hope as a mode of coping in amyotrophic lateral sclerosis. J Neurosci Nurs (1989) 21(6):342–7. doi: 10.1097/01376517-198912000-00003
43. Dyczynski M, Yu Y, Otrocka M, Parpal S, Braga T, Henley AB, et al. Targeting autophagy by small molecule inhibitors of vacuolar protein sorting 34 (Vps34) improves the sensitivity of breast cancer cells to Sunitinib. Cancer Lett (2018) 435:32–43. doi: 10.1016/j.canlet.2018.07.028
44. Noman MZ, Parpal S, Van Moer K, Xiao M, Yu Y, Viklund J, et al. Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti-PD-1/PD-L1 immunotherapy. Sci Adv (2020) 6(18):eaax7881. doi: 10.1126/sciadv.aax7881
45. Park JM, Tougeron D, Huang S, Okamoto K, Sinicrope FA. Beclin 1 and UVRAG confer protection from radiation-induced DNA damage and maintain centrosome stability in colorectal cancer cells. PloS One (2014) 9(6):e100819. doi: 10.1371/journal.pone.0100819
46. Xie FJ, Zheng QQ, Qin J, Zhang LL, Han N, Mao WM. Autophagy Inhibition Stimulates Apoptosis in Oesophageal Squamous Cell Carcinoma Treated with Fasudil. J Cancer (2018) 9(6):1050–6. doi: 10.7150/jca.23388
47. Wu CL, Liu JF, Liu Y, Wang YX, Fu KF, Yu XJ, et al. Beclin1 inhibition enhances paclitaxelmediated cytotoxicity in breast cancer in vitro and in vivo. Int J Mol Med (2019) 43(4):1866–78. doi: 10.3892/ijmm.2019.4089
48. Xi G, Hu X, Wu B, Jiang H, Young CY, Pang Y, et al. Autophagy inhibition promotes paclitaxel-induced apoptosis in cancer cells. Cancer Lett (2011) 307(2):141–8. doi: 10.1016/j.canlet.2011.03.026
49. Zhang Q, Si S, Schoen S, Chen J, Jin XB, Wu G. Suppression of autophagy enhances preferential toxicity of paclitaxel to folliculin-deficient renal cancer cells. J Exp Clin Cancer Res (2013) 32:99. doi: 10.1186/1756-9966-32-99
50. Liu S, Li X. Autophagy inhibition enhances sensitivity of endometrial carcinoma cells to paclitaxel. Int J Oncol (2015) 46(6):2399–408. doi: 10.3892/ijo.2015.2937
51. Zhang SF, Wang XY, Fu ZQ, Peng QH, Zhang JY, Ye F, et al. TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy (2015) 11(2):225–38. doi: 10.1080/15548627.2014.998931
52. Song Y, Li W, Peng X, Xie J, Li H, Tan G. Inhibition of autophagy results in a reversal of taxol resistance in nasopharyngeal carcinoma by enhancing taxol-induced caspase-dependent apoptosis. Am J Transl Res (2017) 9(4):1934–42. doi: 10.1097/cad.0b013e3283363980
53. Zhen Z, Yang K, Ye L, You Z, Chen R, Liu Y, et al. Suberoylanilide hydroxamic acid sensitizes neuroblastoma to paclitaxel by inhibiting thioredoxin-related protein 14-mediated autophagy. Cancer Sci (2017) 108(7):1485–92. doi: 10.1111/cas.13279
54. Shao S, Li S, Qin Y, Wang X, Yang Y, Bai H, et al. Spautin-1, a novel autophagy inhibitor, enhances imatinib-induced apoptosis in chronic myeloid leukemia. Int J Oncol (2014) 44(5):1661–8. doi: 10.3892/ijo.2014.2313
55. Yu Y, Yang L, Zhao M, Zhu S, Kang R, Vernon P, et al. Targeting microRNA-30a-mediated autophagy enhances imatinib activity against human chronic myeloid leukemia cells. Leukemia (2012) 26(8):1752–60. doi: 10.1038/leu.2012.65
56. Clark GM, Osborne CK, McGuire WL. Correlations between estrogen receptor, progesterone receptor, and patient characteristics in human breast cancer. J Clin Oncol (1984) 2(10):1102–9. doi: 10.1200/JCO.1984.2.10.1102
57. Gu Y, Chen T, Li G, Xu C, Xu Z, Zhang J, et al. Lower Beclin 1 downregulates HER2 expression to enhance tamoxifen sensitivity and predicts a favorable outcome for ER positive breast cancer. Oncotarget (2017) 8(32):52156–77. doi: 10.18632/oncotarget.11044
58. Zhang Y, Cheng Y, Ren X, Zhang L, Yap KL, Wu H, et al. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene (2012) 31(8):1055–64. doi: 10.1038/onc.2011.290
59. Ma B, Liang LZ, Liao GQ, Liang YJ, Liu HC, Zheng GS, et al. Inhibition of autophagy enhances cisplatin cytotoxicity in human adenoid cystic carcinoma cells of salivary glands. J Oral Pathol Med (2013) 42(10):774–80. doi: 10.1111/jop.12066
60. Zhang R, Wang R, Chen Q, Chang H. Inhibition of autophagy using 3-methyladenine increases cisplatin-induced apoptosis by increasing endoplasmic reticulum stress in U251 human glioma cells. Mol Med Rep (2015) 12(2):1727–32. doi: 10.3892/mmr.2015.3588
61. Schlutermann D, Skowron MA, Berleth N, Bohler P, Deitersen J, Stuhldreier F, et al. Targeting urothelial carcinoma cells by combining cisplatin with a specific inhibitor of the autophagy-inducing class III PtdIns3K complex. Urol Oncol (2018) 36(4):160 e161–160 e113. doi: 10.1016/j.urolonc.2017.11.021
62. Giampieri R, Caporale M, Pietrantonio F, De Braud F, Negri FV, Giuliani F, et al. Second-line angiogenesis inhibition in metastatic colorectal cancer patients: Straightforward or overcrowded? Crit Rev Oncol Hematol (2016) 100:99–106. doi: 10.1016/j.critrevonc.2016.02.005
63. Stratigos M, Matikas A, Voutsina A, Mavroudis D, Georgoulias V. Targeting angiogenesis in small cell lung cancer. Transl Lung Cancer Res (2016) 5(4):389–400. doi: 10.21037/tlcr.2016.08.04
64. Jain RK, Gandhi S, George S. Second-line systemic therapy in metastatic renal-cell carcinoma: A review. Urol Oncol (2017) 35(11):640–6. doi: 10.1016/j.urolonc.2017.08.010
65. Zhao Z, Xia G, Li N, Su R, Chen X, Zhong L. Autophagy Inhibition Promotes Bevacizumab-induced Apoptosis and Proliferation Inhibition in Colorectal Cancer Cells. J Cancer (2018) 9(18):3407–16. doi: 10.7150/jca.24201
66. Peeters M, Price T. Biologic therapies in the metastatic colorectal cancer treatment continuum–applying current evidence to clinical practice. Cancer Treat Rev (2012) 38(5):397–406. doi: 10.1016/j.ctrv.2011.08.002
67. Mele L, Del Vecchio V, Liccardo D, Prisco C, Schwerdtfeger M, Robinson N, et al. The role of autophagy in resistance to targeted therapies. Cancer Treat Rev (2020) 88:102043. doi: 10.1016/j.ctrv.2020.102043
68. Baginska J, Viry E, Berchem G, Poli A, Noman MZ, van Moer K, et al. Granzyme B degradation by autophagy decreases tumor cell susceptibility to natural killer-mediated lysis under hypoxia. Proc Natl Acad Sci USA (2013) 110(43):17450–5. doi: 10.1073/pnas.1304790110
69. Mgrditchian T, Arakelian T, Paggetti J, Noman MZ, Viry E, Moussay E, et al. Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc Natl Acad Sci USA (2017) 114(44):E9271–9. doi: 10.1073/pnas.1703921114
70. Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature (2013) 504(7479):296–300. doi: 10.1038/nature12865
71. Maes H, Kuchnio A, Peric A, Moens S, Nys K, De Bock K, et al. Tumor vessel normalization by chloroquine independent of autophagy. Cancer Cell (2014) 26(2):190–206. doi: 10.1016/j.ccr.2014.06.025
72. Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun (2014) 5:3056. doi: 10.1038/ncomms4056
73. Poillet-Perez L, Xie X, Zhan L, Yang Y, Sharp DW, Hu ZS, et al. Autophagy maintains tumour growth through circulating arginine. Nature (2018) 563(7732):569–73. doi: 10.1038/s41586-018-0697-7
74. Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy (2016) 12(1):1–222. doi: 10.1080/15548627.2015.1100356
75. Qian Z, Lin C, Espinosa R, LeBeau M, Rosner MR. Cloning and characterization of MST4, a novel Ste20-like kinase. J Biol Chem (2001) 276(25):22439–45. doi: 10.1074/jbc.M009323200
76. Li T, Deng L, He X, Jiang G, Hu F, Ye S, et al. MST4 Predicts Poor Prognosis And Promotes Metastasis By Facilitating Epithelial-Mesenchymal Transition In Gastric Cancer. Cancer Manag Res (2019) 11:9353–69. doi: 10.2147/CMAR.S219689
77. Madsen CD, Hooper S, Tozluoglu M, Bruckbauer A, Fletcher G, Erler JT, et al. STRIPAK components determine mode of cancer cell migration and metastasis. Nat Cell Biol (2015) 17(1):68–80. doi: 10.1038/ncb3083
78. Thompson BJ, Sahai E. MST kinases in development and disease. J Cell Biol (2015) 210(6):871–82. doi: 10.1083/jcb.201507005
79. Huang T, Kim CK, Alvarez AA, Pangeni RP, Wan X, Song X, et al. MST4 Phosphorylation of ATG4B Regulates Autophagic Activity, Tumorigenicity, and Radioresistance in Glioblastoma. Cancer Cell (2017) 32840-855(6):e848. doi: 10.1016/j.ccell.2017.11.005
80. Akin D, Wang SK, Habibzadegah-Tari P, Law B, Ostrov D, Li M, et al. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy (2014) 10(11):2021–35. doi: 10.4161/auto.32229
81. Kurdi A, Cleenewerck M, Vangestel C, Lyssens S, Declercq W, Timmermans JP, et al. ATG4B inhibitors with a benzotropolone core structure block autophagy and augment efficiency of chemotherapy in mice. Biochem Pharmacol (2017) 138:150–62. doi: 10.1016/j.bcp.2017.06.119
82. Chu J, Fu Y, Xu J, Zheng X, Gu Q, Luo X, et al. ATG4B inhibitor FMK-9a induces autophagy independent on its enzyme inhibition. Arch Biochem Biophys (2018) 644:29–36. doi: 10.1016/j.abb.2018.03.001
83. Fu Y, Hong L, Xu J, Zhong G, Gu Q, Gu Q, et al. Discovery of a small molecule targeting autophagy via ATG4B inhibition and cell death of colorectal cancer cells in vitro and in vivo. Autophagy (2019) 15(2):295–311. doi: 10.1080/15548627.2018.1517073
84. Liu PF, Tsai KL, Hsu CJ, Tsai WL, Cheng JS, Chang HW, et al. Drug Repurposing Screening Identifies Tioconazole as an ATG4 Inhibitor that Suppresses Autophagy and Sensitizes Cancer Cells to Chemotherapy. Theranostics (2018) 8(3):830–45. doi: 10.7150/thno.22012
85. Bortnik S, Choutka C, Horlings HM, Leung S, Baker JH, Lebovitz C, et al. Identification of breast cancer cell subtypes sensitive to ATG4B inhibition. Oncotarget (2016) 7(41):66970–88. doi: 10.18632/oncotarget.11408
86. Chen J, Zhang L, Zhou H, Wang W, Luo Y, Yang H, et al. Inhibition of autophagy promotes cisplatin-induced apoptotic cell death through Atg5 and Beclin 1 in A549 human lung cancer cells. Mol Med Rep (2018) 17(5):6859–65. doi: 10.3892/mmr.2018.8686
87. Chittaranjan S, Bortnik S, Dragowska WH, Xu J, Abeysundara N, Leung A, et al. Autophagy inhibition augments the anticancer effects of epirubicin treatment in anthracycline-sensitive and -resistant triple-negative breast cancer. Clin Cancer Res (2014) 20(12):3159–73. doi: 10.1158/1078-0432.CCR-13-2060
88. O’Donovan TR, O’Sullivan GC, McKenna SL. Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy (2011) 7(5):509–24. doi: 10.4161/auto.7.6.15066
89. Zheng B, Zhu H, Gu D, Pan X, Qian L, Xue B, et al. MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem Biophys Res Commun (2015) 459(2):234–9. doi: 10.1016/j.bbrc.2015.02.084
90. Yu L, Chen Y, Tooze SA. Autophagy pathway: Cellular and molecular mechanisms. Autophagy (2018) 14(2):207–15. doi: 10.1080/15548627.2017.1378838
91. Chen D, Xie J, Fiskesund R, Dong W, Liang X, Lv J, et al. Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nat Commun (2018) 9(1):873. doi: 10.1038/s41467-018-03225-9
92. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell (2008) 132(1):27–42. doi: 10.1016/j.cell.2007.12.018
93. Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell (2012) 151(6):1256–69. doi: 10.1016/j.cell.2012.11.001
94. Zhou F, Yang X, Zhao H, Liu Y, Feng Y, An R, et al. Down-regulation of OGT promotes cisplatin resistance by inducing autophagy in ovarian cancer. Theranostics (2018) 8(19):5200–12. doi: 10.7150/thno.27806
95. Chen Y, Meng D, Wang H, Sun R, Wang D, Wang S, et al. VAMP8 facilitates cellular proliferation and temozolomide resistance in human glioma cells. Neuro Oncol (2015) 17(3):407–18. doi: 10.1093/neuonc/nou219
96. Fu R, Deng Q, Zhang H, Hu X, Li Y, Liu Y, et al. A novel autophagy inhibitor berbamine blocks SNARE-mediated autophagosome-lysosome fusion through upregulation of BNIP3. Cell Death Dis (2018) 9(2):243. doi: 10.1038/s41419-018-0276-8
97. Mou L, Liang B, Liu G, Jiang J, Liu J, Zhou B, et al. Berbamine exerts anticancer effects on human colon cancer cells via induction of autophagy and apoptosis, inhibition of cell migration and MEK/ERK signalling pathway. J BUON (2019) 24(5):1870–5.
98. Zhang H, Jiao Y, Shi C, Song X, Chang Y, Ren Y, et al. Berbamine suppresses cell proliferation and promotes apoptosis in ovarian cancer partially via the inhibition of Wnt/beta-catenin signaling. Acta Biochim Biophys Sin (Shanghai) (2018) 50(6):532–9. doi: 10.1093/abbs/gmy036
99. Morell C, Bort A, Vara-Ciruelos D, Ramos-Torres A, Altamirano-Dimas M, Diaz-Laviada I, et al. Up-Regulated Expression of LAMP2 and Autophagy Activity during Neuroendocrine Differentiation of Prostate Cancer LNCaP Cells. PloS One (2016) 11(9):e0162977. doi: 10.1371/journal.pone.0162977
100. Jamali L, Moradi A, Ganji M, Ayati M, Kazeminezhad B, Fazeli Attar Z, et al. Potential Prognostic Role for SPOP, DAXX, RARRES1, and LAMP2 as an Autophagy Related Genes in Prostate Cancer. Urol J (2020) 17(2):156–63. doi: 10.22037/uj.v0i0.4935
101. Huang CF, Deng WW, Zhang L, Zhang WF, Sun ZJ. Expression of LC3, LAMP2, KEAP1 and NRF2 in Salivary Adenoid Cystic Carcinoma. Pathol Oncol Res (2016) 22(1):109–14. doi: 10.1007/s12253-015-9981-0
102. Koukourakis MI, Kalamida D, Mitrakas A, Pouliliou S, Kalamida S, Sivridis E, et al. Intensified autophagy compromises the efficacy of radiotherapy against prostate cancer. Biochem Biophys Res Commun (2015) 461(2):268–74. doi: 10.1016/j.bbrc.2015.04.014
103. Safaei R, Larson BJ, Cheng TC, Gibson MA, Otani S, Naerdemann W, et al. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther (2005) 4(10):1595–604. doi: 10.1158/1535-7163.MCT-05-0102
104. Robert G, Auberger P. Azacitidine resistance caused by LAMP2 deficiency: a therapeutic window for the use of autophagy inhibitors in MDS/AML patients? Autophagy (2019) 15(5):927–9. doi: 10.1080/15548627.2019.1586259
105. Kuchitsu Y, Fukuda M. Revisiting Rab7 Functions in Mammalian Autophagy: Rab7 Knockout Studies. Cells (2018) 7(11):215. doi: 10.3390/cells7110215
106. Liu H, Xu J, Yao Q, Zhang Z, Guo Q, Lin J. Rab7 Is Associated with Poor Prognosis of Gastric Cancer and Promotes Proliferation, Invasion, and Migration of Gastric Cancer Cells. Med Sci Monit (2020) 26:e922217. doi: 10.12659/MSM.922217
107. Guerra F, Bucci C. Role of the RAB7 Protein in Tumor Progression and Cisplatin Chemoresistance. Cancers (Basel) (2019) 11(8):1096. doi: 10.3390/cancers11081096
108. Guerra F, Paiano A, Migoni D, Girolimetti G, Perrone AM, De Iaco P, et al. Modulation of RAB7A Protein Expression Determines Resistance to Cisplatin through Late Endocytic Pathway Impairment and Extracellular Vesicular Secretion. Cancers (Basel) (2019) 11(1):52. doi: 10.3390/cancers11010052
109. Bhargava HK, Tabata K, Byck JM, Hamasaki M, Farrell DP, Anishchenko I, et al. Structural basis for autophagy inhibition by the human Rubicon-Rab7 complex. Proc Natl Acad Sci USA (2020) 117(29):17003–10. doi: 10.1073/pnas.2008030117
110. Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy (2018) 14(8):1435–55. doi: 10.1080/15548627.2018.1474314
111. Tsubone T, Rocha C, Tonolli P, II-Sei W, Stolf B. In vitro Autophagy Modulation with Chloroquine: Some Lessons to Learn. Adv Biochem Biotechnol (2020) 5:1098. doi: 10.29011/2574-7258.001098
112. Wang F, Tang J, Li P, Si S, Yu H, Yang X, et al. Chloroquine Enhances the Radiosensitivity of Bladder Cancer Cells by Inhibiting Autophagy and Activating Apoptosis. Cell Physiol Biochem (2018) 45(1):54–66. doi: 10.1159/000486222
113. Ye H, Chen M, Cao F, Huang H, Zhan R, Zheng X. Chloroquine, an autophagy inhibitor, potentiates the radiosensitivity of glioma initiating cells by inhibiting autophagy and activating apoptosis. BMC Neurol (2016) 16(1):178. doi: 10.1186/s12883-016-0700-6
114. Roy LO, Poirier MB, Fortin D. Chloroquine inhibits the malignant phenotype of glioblastoma partially by suppressing TGF-beta. Invest New Drugs (2015) 33(5):1020–31. doi: 10.1007/s10637-015-0275-x
115. Ratikan JA, Sayre JW, Schaue D. Chloroquine engages the immune system to eradicate irradiated breast tumors in mice. Int J Radiat Oncol Biol Phys (2013) 87(4):761–8. doi: 10.1016/j.ijrobp.2013.07.024
116. Yoon YH, Cho KS, Hwang JJ, Lee SJ, Choi JA, Koh JY. Induction of lysosomal dilatation, arrested autophagy, and cell death by chloroquine in cultured ARPE-19 cells. Invest Ophthalmol Vis Sci (2010) 51(11):6030–7. doi: 10.1167/iovs.10-5278
117. Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy (2012) 8(2):200–12. doi: 10.4161/auto.8.2.18554
118. Schonewolf CA, Mehta M, Schiff D, Wu H, Haffty BG, Karantza V, et al. Autophagy inhibition by chloroquine sensitizes HT-29 colorectal cancer cells to concurrent chemoradiation. World J Gastrointest Oncol (2014) 6(3):74–82. doi: 10.4251/wjgo.v6.i3.74
119. Geng Y, Kohli L, Klocke BJ, Roth KA. Chloroquine-induced autophagic vacuole accumulation and cell death in glioma cells is p53 independent. Neuro Oncol (2010) 12(5):473–81. doi: 10.1093/neuonc/nop048
120. Eng CH, Wang Z, Tkach D, Toral-Barza L, Ugwonali S, Liu S, et al. Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc Natl Acad Sci USA (2016) 113(1):182–7. doi: 10.1073/pnas.1515617113
121. Lin JF, Lin YC, Tsai TF, Chen HE, Chou KY, Hwang TI. Cisplatin induces protective autophagy through activation of BECN1 in human bladder cancer cells. Drug Des Devel Ther (2017) 11:1517–33. doi: 10.2147/DDDT.S126464
122. Aga T, Endo K, Tsuji A, Aga M, Moriyama-Kita M, Ueno T, et al. Inhibition of autophagy by chloroquine makes chemotherapy in nasopharyngeal carcinoma more efficient. Auris Nasus Larynx (2019) 46(3):443–50. doi: 10.1016/j.anl.2018.10.013
123. Zhao XG, Sun RJ, Yang XY, Liu DY, Lei DP, Jin T, et al. Chloroquine-enhanced efficacy of cisplatin in the treatment of hypopharyngeal carcinoma in xenograft mice. PloS One (2015) 10(4):e0126147. doi: 10.1371/journal.pone.0126147
124. Choi HS, Jeong EH, Lee TG, Kim SY, Kim HR, Kim CH. Autophagy Inhibition with Monensin Enhances Cell Cycle Arrest and Apoptosis Induced by mTOR or Epidermal Growth Factor Receptor Inhibitors in Lung Cancer Cells. Tuberc Respir Dis (Seoul) (2013) 75(1):9–17. doi: 10.4046/trd.2013.75.1.9
125. Ketola K, Vainio P, Fey V, Kallioniemi O, Iljin K. Monensin is a potent inducer of oxidative stress and inhibitor of androgen signaling leading to apoptosis in prostate cancer cells. Mol Cancer Ther (2010) 9(12):3175–85. doi: 10.1158/1535-7163.MCT-10-0368
126. Wiedmer T, Blank A, Pantasis S, Normand L, Bill R, Krebs P, et al. Autophagy Inhibition Improves Sunitinib Efficacy in Pancreatic Neuroendocrine Tumors via a Lysosome-dependent Mechanism. Mol Cancer Ther (2017) 16(11):2502–15. doi: 10.1158/1535-7163.MCT-17-0136
127. Cufi S, Vazquez-Martin A, Oliveras-Ferraros C, Corominas-Faja B, Cuyas E, Lopez-Bonet E, et al. The anti-malarial chloroquine overcomes primary resistance and restores sensitivity to trastuzumab in HER2-positive breast cancer. Sci Rep (2013) 3:1–13. 2469. doi: 10.1038/srep02469
128. Zamame Ramirez JA, Romagnoli GG, Falasco BF, Gorgulho CM, Sanzochi Fogolin C, Dos Santos DC, et al. Blocking drug-induced autophagy with chloroquine in HCT-116 colon cancer cells enhances DC maturation and T cell responses induced by tumor cell lysate. Int Immunopharmacol (2020) 84:106495. doi: 10.1016/j.intimp.2020.106495
129. Yamamoto K, Venida A, Yano J, Biancur DE, Kakiuchi M, Gupta S, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature (2020) 581(7806):100–5. doi: 10.1038/s41586-020-2229-5
130. Chu HY, Wang W, Chen X, Jiang YE, Cheng R, Qi X, et al. Bafilomycin A1 increases the sensitivity of tongue squamous cell carcinoma cells to cisplatin by inhibiting the lysosomal uptake of platinum ions but not autophagy. Cancer Lett (2018) 423:105–12. doi: 10.1016/j.canlet.2018.03.003
131. Li LQ, Xie WJ, Pan D, Chen H, Zhang L. Inhibition of autophagy by bafilomycin A1 promotes chemosensitivity of gastric cancer cells. Tumour Biol (2016) 37(1):653–9. doi: 10.1007/s13277-015-3842-z
132. Fu W, Li X, Lu X, Zhang L, Li R, Zhang N, et al. A novel acridine derivative, LS-1-10 inhibits autophagic degradation and triggers apoptosis in colon cancer cells. Cell Death Dis (2017) 8(10):e3086. doi: 10.1038/cddis.2017.498
133. Takanezawa Y, Nakamura R, Kojima Y, Sone Y, Uraguchi S, Kiyono M. Cytochalasin E increased the sensitivity of human lung cancer A549cells to bortezomib via inhibition of autophagy. Biochem Biophys Res Commun (2018) 498(3):603–8. doi: 10.1016/j.bbrc.2018.03.029
134. Alizadeh J, Zeki AA, Mirzaei N, Tewary S, Rezaei Moghadam A, Glogowska A, et al. Mevalonate Cascade Inhibition by Simvastatin Induces the Intrinsic Apoptosis Pathway via Depletion of Isoprenoids in Tumor Cells. Sci Rep (2017) 7:44841. doi: 10.1038/srep44841
135. Shojaei S, Koleini N, Samiei E, Aghaei M, Cole LK, Alizadeh J, et al. Simvastatin increases temozolomide-induced cell death by targeting the fusion of autophagosomes and lysosomes. FEBS J (2020) 287(5):1005–34. doi: 10.1111/febs.15069
136. Kou X, Yang Y, Jiang X, Liu H, Sun F, Wang X, et al. Vorinostat and Simvastatin have synergistic effects on triple-negative breast cancer cells via abrogating Rab7 prenylation. Eur J Pharmacol (2017) 813:161–71. doi: 10.1016/j.ejphar.2017.08.022
137. Daskalaki I, Gkikas I, Tavernarakis N. Hypoxia and Selective Autophagy in Cancer Development and Therapy. Front Cell Dev Biol (2018) 6:104. doi: 10.3389/fcell.2018.00104
138. Carew JS, Espitia CM, Zhao W, Han Y, Visconte V, Phillips J, et al. Disruption of Autophagic Degradation with ROC-325 Antagonizes Renal Cell Carcinoma Pathogenesis. Clin Cancer Res (2017) 23(11):2869–79. doi: 10.1158/1078-0432.CCR-16-1742
139. Nawrocki ST, Han Y, Visconte V, Przychodzen B, Espitia CM, Phillips J, et al. The novel autophagy inhibitor ROC-325 augments the antileukemic activity of azacitidine. Leukemia (2019) 33(12):2971–4. doi: 10.1038/s41375-019-0529-2
140. Aldrich LN, Kuo SY, Castoreno AB, Goel G, Kuballa P, Rees MG, et al. Discovery of a Small-Molecule Probe for V-ATPase Function. J Am Chem Soc (2015) 137(16):5563–8. doi: 10.1021/jacs.5b02150
141. Carew JS, Espitia CM, Esquivel JA,2, Mahalingam D, Kelly KR, Reddy G, et al. Lucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis. J Biol Chem (2011) 286(8):6602–13. doi: 10.1074/jbc.M110.151324
142. Martins WK, Costa ET, Cruz MC, Stolf BS, Miotto R, Cordeiro RM, et al. Parallel damage in mitochondrial and lysosomal compartments promotes efficient cell death with autophagy: The case of the pentacyclic triterpenoids. Sci Rep (2015) 5:12425. doi: 10.1038/srep12425
143. Martins WK, Gomide AB, Costa ET, Junqueira HC, Stolf BS, Itri R, et al. Membrane damage by betulinic acid provides insights into cellular aging. Biochim Biophys Acta Gen Subj (2017) 1861(1 Pt A):3129–43. doi: 10.1016/j.bbagen.2016.10.018
144. Kang C, Elledge SJ. How autophagy both activates and inhibits cellular senescence. Autophagy (2016) 12(5):898–9. doi: 10.1080/15548627.2015.1121361
145. Cassidy LD, Young ARJ, Young CNJ, Soilleux EJ, Fielder E, Weigand BM, et al. Temporal inhibition of autophagy reveals segmental reversal of ageing with increased cancer risk. Nat Commun (2020) 11(1):307. doi: 10.1038/s41467-019-14187-x
Keywords: autophagy, cancer resistance, chemotherapy, radiotherapy, targeted therapy, immunotherapy
Citation: Xiao M, Benoit A, Hasmim M, Duhem C, Vogin G, Berchem G, Noman MZ and Janji B (2021) Targeting Cytoprotective Autophagy to Enhance Anticancer Therapies. Front. Oncol. 11:626309. doi: 10.3389/fonc.2021.626309
Received: 05 November 2020; Accepted: 15 January 2021;
Published: 25 February 2021.
Edited by:
Waleska Kerllen Martins, Universidade Anhanguera de São Paulo, BrazilReviewed by:
Keith R. Laderoute, SRI International, United StatesYi Ding, Geisinger Health System, United States
Copyright © 2021 Xiao, Benoit, Hasmim, Duhem, Vogin, Berchem, Noman and Janji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bassam Janji, YmFzc2FtLmphbmppQGxpaC5sdQ==
†These authors have contributed equally to this work
 Malina Xiao
Malina Xiao Alice Benoit
Alice Benoit Meriem Hasmim
Meriem Hasmim Caroline Duhem2
Caroline Duhem2 Muhammad Zaeem Noman
Muhammad Zaeem Noman Bassam Janji
Bassam Janji