- 1College of Plant Protection, Henan Agricultural University, Zhengzhou, China
- 2Key Laboratory of Forensic Toxicology of Herbal Medicines, Guizhou Education Department, School of Basic Medicine, Guizhou University of Traditional Chinese Medicine, Guiyang, China
Nuf2 participates in the regulation of cell apoptosis and proliferation by regulating the binding of centromere and spindle microtubules to achieve the correct separation of chromosomes. Previous reports have suggested that Nuf2 may play a role in various human cancers. However, the mechanism and function of Nuf2 in the development of Hepatocellular carcinoma (HCC) remains uncertain. This study investigated the prognostic potential of Nuf2 and its relation with immune cell infiltration in HCC. Nuf2 expression in tumor cells was examined using the TIMER and Oncomine databases, and its prognostic potential was assessed via the Kaplan-Meier plotter and GEPIA databases. The relationships between Nuf2 and tumor immune infiltration were analyzed using TIMER. The relationships between Nuf2 and biomarkers of tumor immune infiltration were analyzed using TIMER and GEPIA. Here we revealed that Nuf2 expression increased in tumor tissues containing HCC, and this correlated with poor relapse-free survival, disease-specific survival, progression-free survival, and overall survival in patients with HCC regardless of grades, genders, races, drinking behaviors and other clinical factors. Additionally, high expression of Nuf2 was positively correlated with differential immune cell infiltration and various immune biomarkers. Our work demonstrated that Nuf2 could be a potential prognostic biomarker and could be related to tumor immune cell infiltration in HCC.
Introduction
Hepatocellular carcinoma (HCC), caused by factors such as alcoholic hepatitis, chronic hepatitis B and hepatitis C infections, and nonalcoholic fatty liver disease, has become one of the most commonly diagnosed cancers (1–4). HCC is not only highly malignant and difficult to treat, but also has poor prognosis and high relapse rates, resulting in overall 5-year survival rates of only 5%–9% (5). Early identification of at-risk patients, exploration of new biomarkers and therapeutic targets for diagnosis, and in-depth understanding of the molecular pathogenesis of HCC are prerequisites for controlling this disease.
Nuf2, also known as CDCA1, is a key element of the Ndc80/Nuf2 complex that is required for the formation of stable kinetochore-microtubule attachments and chromosome alignment during mitosis. Nuf2 participates in the regulation of cell apoptosis and proliferation by regulating the binding of centromere and spindle microtubules to achieve the correct separation of chromosomes (6, 7). Nuf2 can be cleaved into alternative splice variants and expressed differentially between tumor tissues and the corresponding normal tissues (8). Nuf2 promotes the tumorigenesis and tumor development and is highly expressed in various human cancers, including serous adenocarcinoma, renal cell carcinoma, cholangiocarcinoma, and colorectal, lung, ovarian, gastric, and bladder cancers (8–13). Knockout of the Nuf2 gene significantly delayed cell growth and increased apoptosis in ovarian, stomach, and colorectal cancer cell lines (9, 10, 12). High Nuf2 expression has been reported to correlate with poor prognosis in non-small cell carcinoma patients (10). Additionally, Nuf2 can also function as a potential biomarker in human tumor diagnosis and immunotherapy (10). These findings strongly imply a potential function of Nuf2 in tumorigenesis. However, the mechanism and role of Nuf2 in the development of HCC remains uncertain.
This study investigated the prognostic potential of Nuf2 and its relation with immune cell infiltration in HCC. Nuf2 expression in tumor cells was examined using the TIMER and Oncomine databases, and its prognostic potential was assessed via the Kaplan-Meier plotter and GEPIA databases. The relationships between Nuf2 and tumor immune infiltration were analyzed using TIMER. The relationships between Nuf2 and biomarkers of tumor immune infiltration were analyzed using TIMER and GEPIA. Here we revealed that the expression level of Nuf2 was significantly increased in HCC, and was correlated with the prognosis of HCC patients. We suggest that Nuf2 has the potential to be a diagnostic gene in hepatocarcinogenesis and prognostic biomarkers for HCC patients.
Materials and Methods
Oncomine Analysis
Oncomine platform (https://www.oncomine.org/) is a publicly accessible online tumor related-gene microarray database, which collects the related gene expression profiles and relevant clinical information. The transcriptional levels of Nuf2 in different tumors and corresponding normal tissues were analyzed by Oncomine. The expression levels were considered different significantly when fold change is greater than 1.5, with P-value < 0.001. We set the threshold value of gene rank to “top 10%” and the data type to “all” (14).
Kaplan–Meier Plotter Analysis
The Kaplan-Meier plotter was used for analyzing the relationship between survival rate and Nuf2 expression in breast, gastric, liver, lung, and ovarian cancers based on two parameters, namely hazard ratios (HR) and log-rank P-values (15). We performed survival analysis with the parameters of Group Cutoff: Median; Hazards Ratio: Yes; 95% Confidence Interval: Yes.
TIMER Analysis
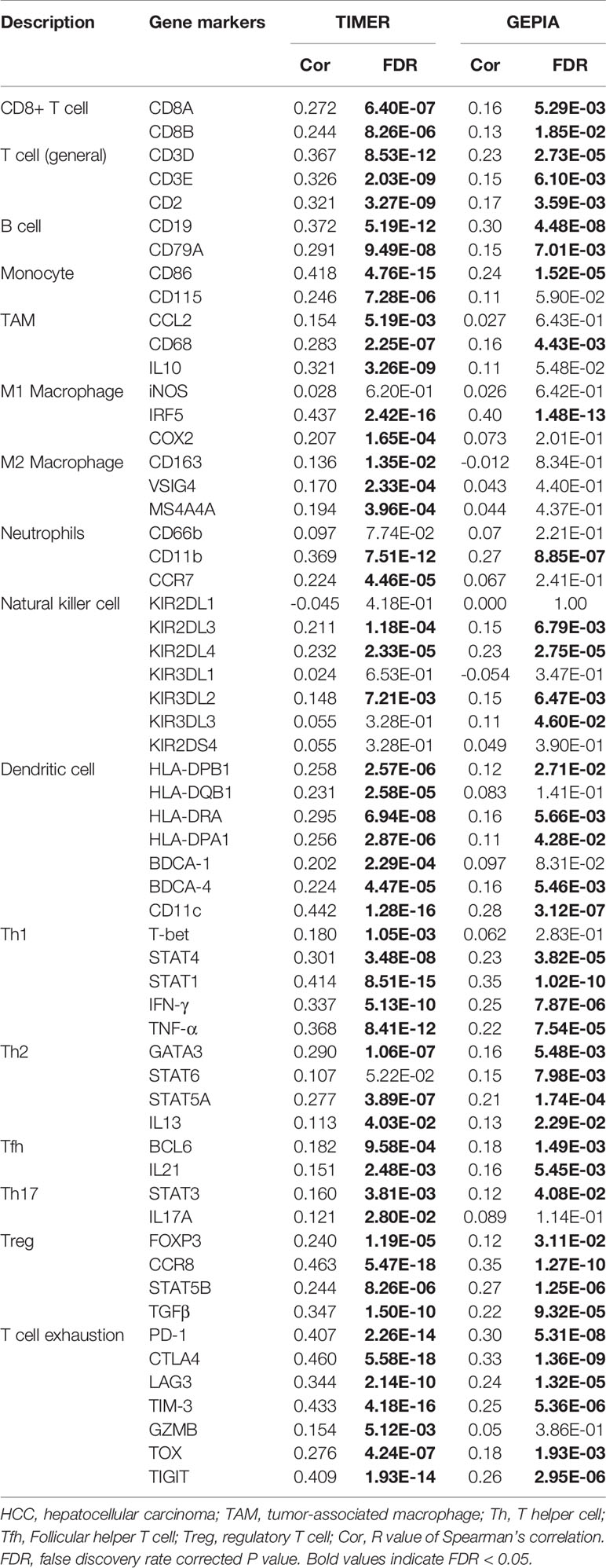
The TIMER database (http://timer.comp-genomics.org/) is a consolidated database to analyze the immune infiltration in different tumor types. Information from 32 types of tumors with more than 10,000 samples from TCGA database was used for immune infiltration analysis via the TIMER database. TIMER ascertains the abundance of tumor infiltrates based on gene expression (16). The correlations between Nuf2 expression and immune cell infiltration levels in different tumors were analyzed according to biomarker gene expression in tumors. The biomarker genes of tumor-infiltrating immune cells including B cells, CD8+T cells, dendritic cells, T cells (general), TAMs, M1 macrophages, M2 macrophages, monocytes, neutrophils, natural killer cells, T-helper cells (Th), Tregs, follicular helper T cells (Tfh), and exhausted T cells were investigated in this study (17, 18).
GEPIA Analysis
GEPIA (http://gepia.cancer-pku.cn/index.html) is a web server for analyzing the RNA sequencing expression data from the TCGA and the GTEx projects, using a standard processing pipeline (19). We analyzed the correlation between Nuf2 and different immune cell biomarkers via GEPIA. The correlation coefficient was determined by the Spearman method with default parameters.
Results
The Expression of Nuf2 in HCC and Other Cancers
Analysis using the Oncomine platform revealed a significant increase in the transcription level of Nuf2 in a variety of cancerous tissues compared to normal tissues, including liver and other 14 types of cancers (bladder, brain and central nervous system, breast, cervical, colorectal, esophageal, gastric, head and neck, lung, lymphoma, melanoma, ovarian, pancreatic, and prostate cancers) (Figure 1A). Additionally, analysis of TCGA RNA-seq data in TIMER database, we found consistent results, i.e. the expression of Nuf2 in HCC was significantly higher than that in normal tissue, as well as other 16 tumor tissues (Figure 1B), suggesting that Nuf2 may play a role in tumorigenesis, especially in HCC, and has the potential to be a diagnostic gene for liver cancer and other cancers.

Figure 1 Nuf2 expression in different human tumor cell types. (A) Nuf2 expression in cancer tissue types compared to that in normal tissues (data from Oncomine). (B) Level of Nuf2 expression in various cancers (data from TCGA via TIMER) (***P<0.001).
Prognostic Potential of Nuf2 Expression in Hepatocellular Carcinoma
Overall survival (OS), disease specific survival (DSS), relapse free survival (RFS) and progression free survival (PFS) are four common prognostic monitoring indexes. As the name suggests, these four indicators can basically summarize the prognosis and survival of cancer patients. Using the Kaplan-Meier plotter, we found that the expression level of Nuf2 gene was significantly correlated with the prognostic survival rate. For example, poor first progression survival and overall survival in lung cancer; poor post progression survival, overall survival and progression free survival in ovarian cancer; poor post progression survival, first progression survival and overall survival in gastric cancer; and poor relapse-free survival in breast cancer (Supplementary Figure S1).
In particular, for HCC, we found that the DSS (Figure 2A, HR=2.99, 95% CI=1.89 to 4.75, P=1e-06), PFS (Figure 2B, HR=1.94, 95% CI=1.41 to 2.66, P=3.7e-05), OS (Figure 2C, HR=2.32, 95% CI=1.61 to 3.34, P=3.9e-06), and RFS (Figure 2D, HR=1.95, 95% CI=1.36 to 2.78, P=2e-04) were significantly reduced when the expression level of Nuf2 was high, indicating that active transcription of Nuf2 might cause health risks, and these genes could be potential prognostic biomarkers for HCC patients.
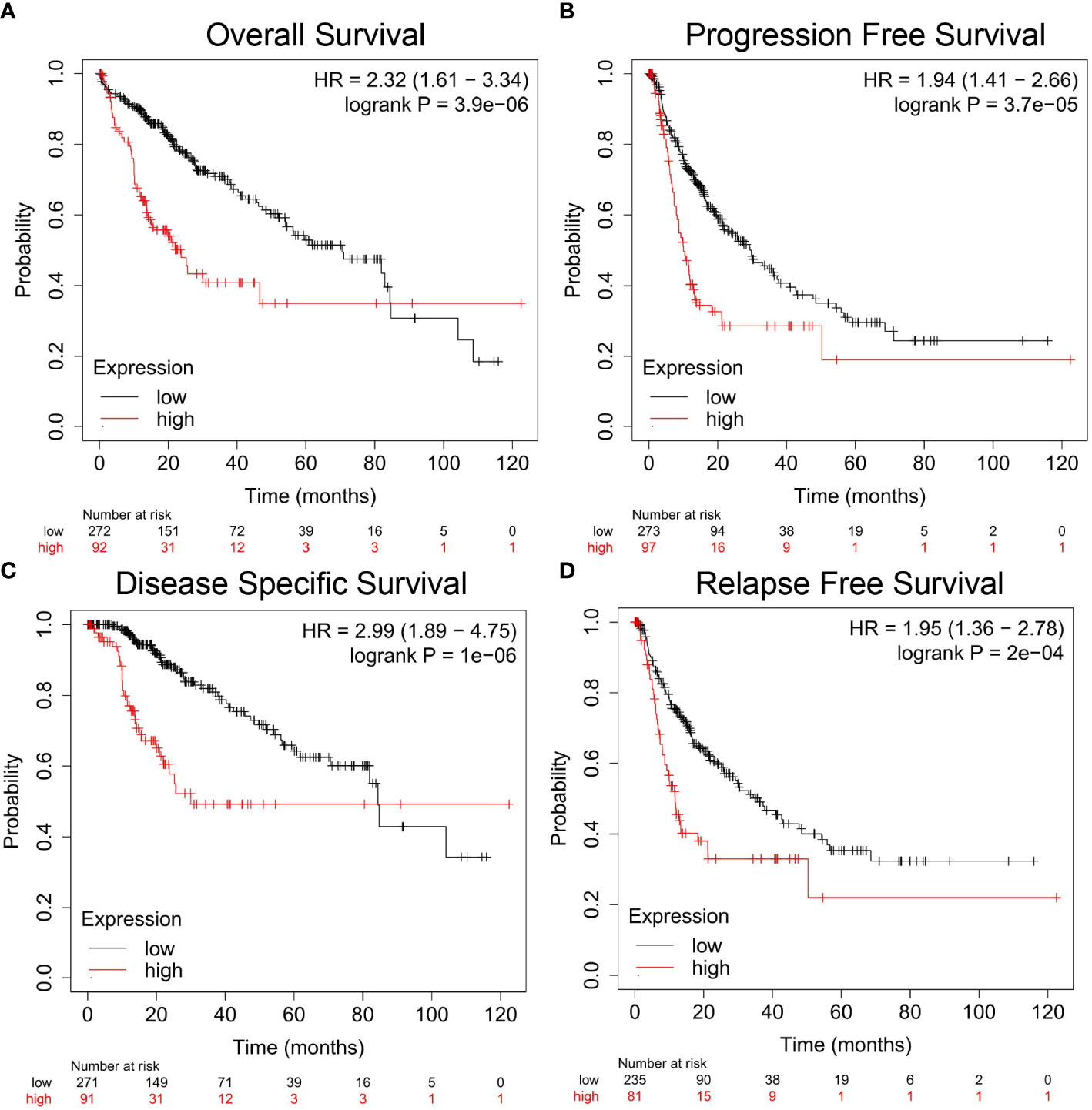
Figure 2 Correlation analysis between Nuf2 expression and prognostic survival in HCC patients via Kaplan-Meier plotter analysis. (A) Overall survival, n = 364; (B) Progression-free survival, n = 370; (C) Disease-specific survival, n = 362; (D) Relapse-free survival, n = 316.
Relationship Between Nuf2 Expression and Clinical Features in Hepatocellular Carcinoma Patients
The correlation between Nuf2 expression and various clinical features in HCC patients was evaluated via the Kaplan-Meier plotter. High expression of Nuf2 was associated with poor OS and PFS for HCC patients regardless of genders (female and male), races (white and Asian), HCC grades or alcohol consumption. Particularly, high Nuf2 expression was correlation with poor OS and PFS in grades 1 to 3 in HCC patients, indicating that high Nuf2 expression might be harmful to the prognosis of HCC patients (Table 1). Notably, Nuf2 expression was only associated with survival in the absence of hepatitis virus, but not in the presence of hepatitis virus. Interestingly, when there was vascular invasion in HCC, Nuf2 expression and PFS show a significant negative correlation; while when there is no vascular invasion, Nuf2 and OS show a significant negative correlation (Table 1). The differences in clinical features suggest that the application of Nuf2 as an indicator gene should be combined with the patient’s condition.
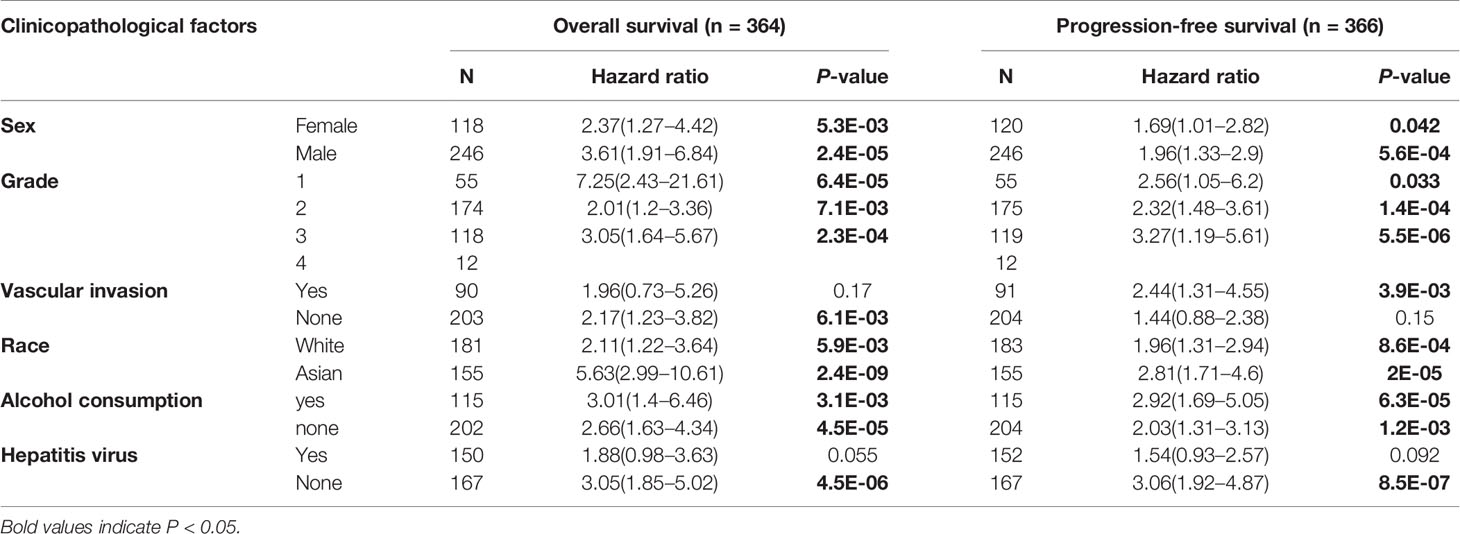
Table 1 Correlation of Nuf2 expression and prognosis in HCC with diverse clinicopathological factors by Kaplan-Meier plotter.
Relationship Between Nuf2 Expression and Immune Cell Infiltration in Hepatocellular Carcinoma
TIMER was used to investigate the correlation between the expression of Nuf2 and infiltration levels of immune cells. In HCC, on the whole, high Nuf2 transcripts were associated with high immune cell infiltration (Figure 3). Specifically, Nuf2 expression was positively correlated with infiltration of B cells (r=0.451, P=1.28e-18); DCs (r=0.417, P=9.35e-16); macrophages (r=0.408, P=4.30e-15); neutrophils (r=0.329, P=3.72e-10); CD4+ T cells (r=0.307, P=6.38e-09); and CD8+ T cells (r=0.298, P=1.83e-08) in HCC (Figure 3).
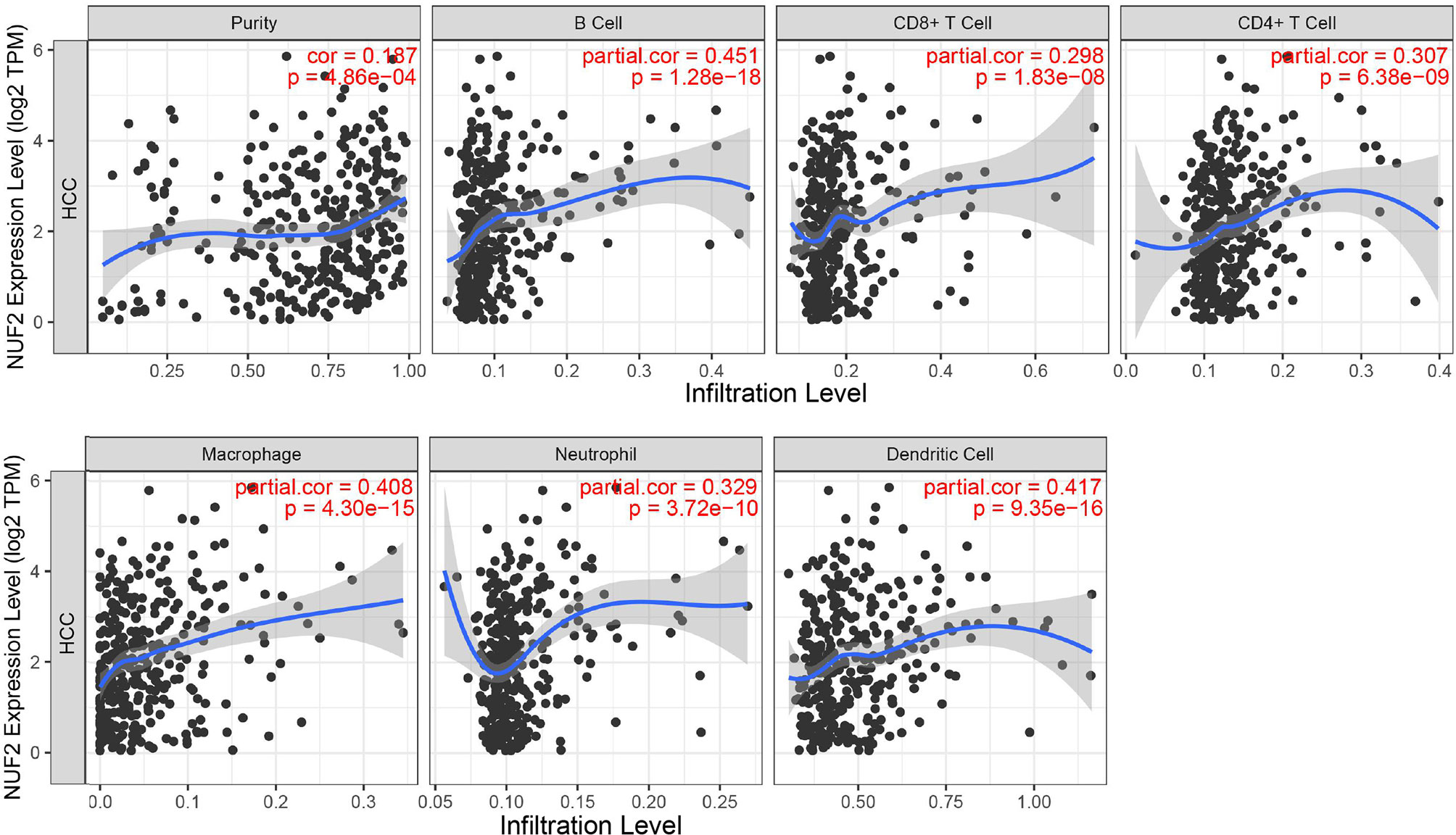
Figure 3 Correlation between Nuf2 expression and immune cell infiltration levels in HCC tissues analyzed via TIMER (n = 371).
Correlation Between Nuf2 Expression and Biomarkers of Different Immune Cell Subsets
The association between Nuf2 expression and tumor-infiltrating immune cell status was investigated based on immune biomarker gene expression levels in HCC. Immune cells in HCC tissues contained dendritic cells, B cells, monocytes, CD4+ T cells, natural killer cells (NKs), CD8+ T cells, neutrophils, M1 macrophages, M2 macrophages, and tumor-associated macrophages (TAMs). Furthermore, different T cell subsets, including T-helper 17 (Th17), T-helper 2 (Th2), T-helper 1 (Th1), exhausted T cells, follicular helper T cells (Tfh), and regulatory T cells (Tregs) were analyzed. Analysis by GEPIA and TIMER indicated a significant positive correlation between Nuf2 expression and most of biomarkers expression in immune cells in HCC (Supplementary Figure S2 and Table 2).
Through the analysis of TIMER and GEPIA databases, a notable positive correlation was found between Nuf2 expression and specific immune cell biomarkers, namely CD8+ T cell biomarkers (CD8A, CD8B), T cell (general) biomarkers (CD2, CD3E, CD3D), B cell biomarkers (CD19, CD79A), Monocyte biomarkers (CD86), TAM biomarkers (CD68), M1 Macrophage biomarkers (IRF5), Neutrophil biomarkers (CD11b), NK biomarkers (KIR2DL3, KIR2DL4, KIR3DL2), DC biomarkers (CD11c, BDCA-4, HLA-DRA, HLA-DQB1, HLA-DPB1, HLA-DPA1), Th1 biomarkers (TNF-α, IFN-γ, STAT1, STAT4), Th2 biomarkers (IL13, GATA3, STAT5A), Tfh biomarkers (BCL6, IL21), Th17 biomarkers (STAT3), Treg biomarkers (TGF-β, STAT5B, CCR8, FOXP3), T-cell exhaustion biomarkers (TIGIT, TOX, TIM-3, LAG3, CTLA4, PD-1).
Discussion
According to the 2018 global cancer statistics report, the incidence and mortality rates of HCC ranked sixth and fourth, respectively (20). Although surgical resection, tumor vascular embolization, and radiofrequency ablation can improve survival rates, the probability that most patients will eventually encounter the invasion or progression of liver cancer is high, and the prognosis is usually poor (21–23). Immune escape, invasion, and metastasis further reduce the long-term survival rate of HCC patients (24). Through a series of bioinformatics analysis with the publicly accessible online databases, we investigated the expression levels of Nuf2 in HCC and corresponding normal tissues, and the effect of Nuf2 expression on survival of prognosis and immune cell infiltration. We showed that Nuf2 expression increased in tumor tissues containing HCC, and this correlated with poor relapse free survival, disease specific survival, progression free survival, and overall survival in patients with HCC regardless of grades, genders, races, drinking behaviors and other clinical factors. Additionally, high expression of Nuf2 was positively correlated with differential immune cell infiltration and various immune biomarkers. Our works demonstrated that Nuf2 could be a potential diagnostic gene in hepatocarcinogenesis and prognostic biomarkers for HCC patients.
In the course of tumorigenesis and development, due to the post-transcriptional regulation mediated by small non-coding RNAs such as miRNA and lncRNA, the transcription level of mRNA sometimes inconsistent with the final protein expression. However, mRNA was still selected as our main research object in this study, because many physiological and biochemical interactions take place at the mRNA level. In addition, compared with protein, the transcriptional alterations of mRNA can be detected on a large scale by simple operation and low cost. To clarify the role of Nuf2 in HCC, by multiple database analysis we revealed that Nuf2 was highly expressed in HCC tissue, indicating that Nuf2 has potential as a diagnostic gene for the occurrence and development of HCC. Similarly, Nuf2 has been proved to be highly expressed in many other cancer types, indicating its wide applicability and functional conservation. However, there was no significant up-regulation of Nuf2 was found in leukemia, suggesting that it is necessary to distinguish the types of cancer when they were used as diagnostic genes.
According to previous reports, Nuf2, also known as CDCA1, is mainly responsible for regulating cell mitosis (25). Down-regulation of Nuf2 expression can inhibit the proliferation of tumor cells, while over expression of Nuf2 is associated with poor prognosis (6, 13). This is consistent with the results of our study. In particular, there is a significant negative correlation between Nuf2 and prognostic survival of OS, DSS, RFS, and PFS, and this correlation is generally applicable to HCC patients with different clinical conditions, indicating that high Nuf2 expression may be one of the causes of poor prognosis. Cancer patients need careful observation after treatment, and Nuf2 may be used, to some extent, as a prognostic marker to reduce the risk of recurrence.
There has been considerable progress in immune therapy for cancer in recent years, and people have begun to pay attention to the effects of the immune system in tumorigenesis and development (26). The study of tumor microenvironment is an active research field of tumor diagnosis, treatment targets, and prognostic biomarkers (27). Many subtypes of immune cells, for example of Th1, Th2, and tumor-associated macrophages (TAMs) have been reported in tumor microenvironment (28). Nuf2 showed a significant positive correlation with various immune cells, indicating that Nuf2-mediated hepatocarcinogenesis might mobilize the activity of these immune cells and make them play an anti-tumor role. Macrophage was divided into M1 macrophage and M2 macrophage. M1 macrophages are mainly related to the recognition and attack of tumor cells, while M2 macrophages are related to tumor progression and immunosuppression (29–31). In this study we revealed that Nuf2 expression was related to the biomarker genes of M1 macrophages, not M2 macrophages. We therefore hypothesized that Nuf2 might not be involved in the mechanism of tumorigenesis mediated by M2 macrophage, but mainly play the anti-tumor role via M1 macrophages pathway. Our findings, to a certain extent, indicated the future research direction.
Nuf2 was positively associated with the biomarkers of T-cell exhaustion (PD-1, CTLA4, TIM-3, LAG3, TOX, TIGIT). TIM-3 is a T-cell suppressor molecule that can cause CD8+T cell (exhausted CD8+ T cells, TEX) to fail in chronic conditions and tumors (32). TOX is one of the most common immunotherapeutic targets. Wherry et al. have reported that TOX+ cells can express inhibitory receptors such as CD160, LAG3, TIGIT, and PD-1, and suggest that TOX is a major molecule which regulates the differentiation of TEX at the transcriptional and epigenetic levels (33). In our study, high level of Nuf2 expression was significantly correlated with TOX and TIM-3, thus clarifying the potential function of Nuf2 in the induction of TEX via the TOX and TIM-3 pathways. This may explain the reason underlying the relationship between high Nuf2 expression or high levels of immune cell infiltration and low survival rate in patients with HCC. This association may lead to the development of new immunotherapy for patients with HCC who do not respond to existing immunosuppressive checkpoint inhibitors. But for most immune cells and their subsets, Nuf2 was only related to some (not all) of the biomarker genes, indicating that there is a certain specificity and selectivity in this interaction, which also provides some basis for immunotherapy in the future.
It should be emphasized that although big data analysis can comprehensively and rapidly mine potential data and functional biomolecules, various false-positive results are inevitable. Our work was mainly to provide a fast and simple method for functional genes screening, and point out a direction for future research. However, accurate conclusions need further experimental analysis and clinical verification.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by all the research data based on the bioinformatics analysis of the open resources from the TIMER, Oncomine, TCGA, Kaplan–Meier plotter, and GEPIA databases. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
XX and XL contributed to the concept and wrote the manuscript. XX and SJ designed the experiments, performed the experiments, and analyzed the data. XX and SJ contributed equally to this study. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the 2019 Doctor Initiation Fund of Guizhou University of Chinese Medicine (3043-043190019) and Research Initiation Foundation for Doctor of Henan Agricultural University (30602107).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.621373/full#supplementary-material
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
2. Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol (2018) 15:599–616. doi: 10.1038/s41571-018-0073-4
3. Makarova-Rusher OV, Altekruse SF, McNeel TS, Ulahannan S, Duffy AG, Graubard BI, et al. Population attributable fractions of risk factors for hepatocellular carcinoma in the United States. Cancer (2016) 122:1757–65. doi: 10.1002/cncr.29971
4. Zhang BH, Yang BH, Tang ZY. Randomized controlled trial of screening for hepatocellular carcinoma. J Cancer Res Clin Oncol (2004) 130:417–22. doi: 10.1007/s00432-004-0552-0
5. Li C, Chen J, Zhang K, Feng B, Wang R, Chen L. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem (2015) 36:423–34. doi: 10.1159/000430109
6. Hu P, Chen X, Sun J, Bie P, Zhang LD. siRNA-mediated knockdown against NUF2 suppresses pancreatic cancer proliferation in vitro and in vivo. Biosci Rep (2015) 87:1183–9. doi: 10.1042/BSR20140124
7. Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y. A conserved protein, NUF2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma (2001) 110:322–34. doi: 10.1007/s004120100153
8. Ohnuma S, Miura K, Horii A, Fujibuchi W, Kaneko N, Gotoh O, et al. Cancer-associated splicing variants of the CDCA1 and MSMB genes expressed in cancer cell lines and surgically resected gastric cancer tissues. Surgery (2009) 145:57–68. doi: 10.1016/j.surg.2008.08.010
9. Kaneko N, Miura K, Gu Z, Karasawa H, Ohnuma S, Sasaki H, et al. siRNA-mediated knockdown against CDCA1 and KNTC2, both frequently overexpressed in colorectal and gastric cancers, suppresses cell proliferation and induces apoptosis. Biochem Biophys Res Commun (2009) 390:1235–40. doi: 10.1016/j.bbrc.2009.10.127
10. Hayama S, Daigo Y, Kato T, Ishikawa N, Yamabuki T, Miyamoto M, et al. Activation of CDCA1-KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis. Cancer Res (2006) 66:10339–48. doi: 10.1158/0008-5472.CAN-06-2137
11. Harao M, Hirata S, Irie A, Senju S, Nakatsura T, Komori H, et al. HLA-A2-restricted CTL epitopes of a novel lung cancer-associated cancer testis antigen, cell division cycle associated 1, can induce tumorreactive CTL. Int J Cancer (2008) 123:2616–25. doi: 10.1002/ijc.23823
12. Sethi G, Pathak HB, Zhang H, Zhou Y, EMargret B, Vathipadiekal V, et al. An RNA interference lethality screen of the human druggable genome to identify molecular vulnerabilities in epithelial ovarian cancer. PloS One (2012) 7:470–86. doi: 10.1371/journal.pone.0047086
13. Hu P, Shangguan J, Zhang L. Downregulation of NUF2 inhibits tumor growth and induces apoptosis by regulating lncRNA AF339813. Int J Clin Exp Pathol (2015) 8:2638–48. doi: 10.1287/orsc.12.5.599.10094
14. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia (2007) 9:166–80. doi: 10.1593/neo.07112
15. Lánczky A, Nagy Á, Bottai G, Munkácsy G, Szabó A, Santarpia L, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat (2016) 160:439–46. doi: 10.1007/s10549-016-4013-7
16. Danaher P, Warren S, Dennis L, D’Amico L, White A, Disis ML, et al. Gene expression biomarkers of Tumor Infiltrating Leukocytes. J Immunother Cancer (2017) 5:18–32. doi: 10.1101/068940
17. Sousa S, Määttä J. The role of tumour-associated macrophages in bone metastasis. J Bone Oncol (2016) 5:135–8. doi: 10.1016/j.jbo.2016.03.004
18. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) 45:98–102. doi: 10.1093/nar/gkx247
19. Siegel RL, Miller KD, Jemal A. CA: A Cancer Journal for Clinicians. Cancer Statistics (2019) 69:7–34. doi: 10.3322/caac.21551
20. Gaetano B, Shirin D, Annalisa A, Maria P, Giulia M, Nicoletta B, et al. The immune system in hepatocellular carcinoma and potential new immunotherapeutic strategies. BioMed Res Int (2015) 2015:1–12. doi: 10.1155/2015/731469
21. Ikeda K, Inoue S. TRIM proteins as RING finger E3 ubiquitin ligases. Adv Exp Med Biol (2012) 770:27–37. doi: 10.1007/978-1-4614-5398-7_3
22. Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci (2017) 42:297–311. doi: 10.1016/j.tibs.2017.01.002
23. You K, Sun P, Yue Z, Li J, Xiong W, Wang J, et al. NOR1 promotes hepatocellular carcinoma cell proliferation and migration through modulating the Notch signaling pathway. Exp Cell Res (2017) 352:375–81. doi: 10.1016/j.yexcr.2017.02.032
24. Zhang T, Zhou Y, Qi ST, Wang ZB, Qian WP, Ouyang YC, et al. Nuf2 is required for chromosome segregation during mouse oocyte meiotic maturation. Cell Cycle (2015) 14:2701–10. doi: 10.1080/15384101.2015.1058677
25. Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol (2019) 16:341–55. doi: 10.1038/s41571-019-0173-9
26. Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer (2019) 19:9–31. doi: 10.1038/s41568-018-0081-9
27. Ino Y, Yamazaki-Itoh R, Shimada K, Iwasaki M, Kosuge T, Kanai Y, et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer (2013) 108:914–23. doi: 10.1038/bjc.2013.32
28. Rodell CB, Arlauckas SP, Cuccarese MF, Garris CS, Li R, Ahmed MS, et al. TLR7/8-agonist-loadednanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat BioMed Eng (2018) 2:578–88. doi: 10.1038/s41551-018-0236-8
29. Jayasingam SD, Citartan M, Thang TH, Mat Zin AA, Ang KC, Ch’ng ES. Evaluating the polarization of tumor-associated macrophages into M1 and M2 phenotypes in human cancer tissue: technicalities and challenges in routine clinical practice. Front Oncol (2019) 9:1512–20. doi: 10.3389/fonc.2019.01512
30. Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature (2013) 496:445–55. doi: 10.1038/nature12034
31. Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature (2006) 38:682–7. doi: 10.1038/nature04444
32. Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol (2015) 5:486–99. doi: 10.1038/nri3862
Keywords: Nuf2, hepatocellular carcinoma, biomarkers, tumor immunity, prognosis
Citation: Xie X, Jiang S and Li X (2021) Nuf2 Is a Prognostic-Related Biomarker and Correlated With Immune Infiltrates in Hepatocellular Carcinoma. Front. Oncol. 11:621373. doi: 10.3389/fonc.2021.621373
Received: 26 October 2020; Accepted: 01 February 2021;
Published: 09 March 2021.
Edited by:
Zhenyu Jia, University of California, Riverside, United StatesReviewed by:
Tolga Turan, AbbVie, United StatesXueliang Xu, Jiangxi Academy of Agricultural Sciences (CAAS), China
Copyright © 2021 Xie, Jiang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Li, bGl4aWFuZ0BoZW5hdS5lZHUuY24=
†These authors have contributed equally to this work
 Xingwei Xie
Xingwei Xie Shanshan Jiang
Shanshan Jiang Xiang Li
Xiang Li