- 1Department of Biochemistry, All India Institute of Medical Sciences, New Delhi, India
- 2Department of Laboratory Oncology, Dr. B.R. Ambedkar Institute Rotary Cancer Hospital, All India Institute of Medical Sciences, New Delhi, India
- 3Department of Medical Oncology, Dr. B.R. Ambedkar Institute Rotary Cancer Hospital, All India Institute of Medical Sciences, New Delhi, India
Around 85% of childhood Acute Lymphoblastic Leukemia (ALL) are of B-cell origin and characterized by the presence of different translocations including BCR-ABL1, ETV6-RUNX1, E2A-PBX1, and MLL fusion proteins. The current clinical investigations used to identify ETV6-RUNX1 translocation include FISH and fusion transcript specific PCR. In the current study we assessed the utility of IGF2BP1, an oncofetal RNA binding protein, that is over expressed specifically in ETV6-RUNX1 translocation positive B-ALL to be used as a diagnostic marker in the clinic. Further, public transcriptomic and Crosslinked Immunoprecipitation (CLIP) datasets were analyzed to identify the putative targets of IGF2BP1. We also studied the utility of using the mRNA expression of two such targets, MYC and EGFL7 as potential diagnostic markers separately or in conjunction with IGF2BP1. We observed that the expression of IGF2BP1 alone measured by RT-qPCR is highly sensitive and specific to be used as a potential biomarker for the presence of ETV6-RUNX1 translocation in future.
Introduction
B-cell acute lymphoblastic leukemia (B-ALL) is the commonest childhood cancer in the pediatric population which originates from the propagation of cytogenetically altered and molecularly abnormal B-lymphocyte progenitor cells (1). The defective progenitor cells, leukemic blasts, exhibit uncontrolled proliferation, get accumulated in the bone marrow and further infiltrate into various extramedullary sites, including spleen, liver, lymph node, thymus, and gonads (2). About 50% of B-ALL tumors exhibit genetic rearrangements among which translocations are most common (3). A set of sentinel genetic lesions, mostly BCR/ABL1, ETV6/RUNX1, E2A/PBX1, and MLL rearrangements, have been well recognized in B-ALL. From Western literature, we see that ETV6/RUNX1 is the commonest translocation seen in B-ALL which decreases with increasing age. The frequency of BCR/ABL1 is very low in childhood ALL (4). However, the translocation profile seen in the Asian population is quite different. ETV6-RUNX1 is present at a lower frequency while BCR-ABL1 shows a higher prevalence (5–9).
ETV6-RUNX1 fusion is formed by the fusion of the first five exons of ETV6 proximally to almost the entire RUNX1 gene depending upon the breakpoint region (10). Both ETV6 and RUNX1 are transcription factors and play an important role in haematopoiesis (11) with ETV6 known to be a strong repressor (12). The chimeric ETV6/RUNX1 transcription factor retains the essential pointed N terminal protein-protein interaction domain of ETV6 and the DNA-binding and transcriptional regulatory sequences of RUNX1 (13). ETV6-RUNX1 translocation has been seen in 1%–5% of normal new-borns (14–16) which is around hundred times the number who develop overt clinical leukemia (10, 16). This demonstrates that ETV6-RUNX1 translocation alone is insufficient for leukemogenesis and another genetic event (e.g., deletion of the other ETV6 allele) is required (3).
In situ hybridization is an important technique in the field of cytogenetics to study the presence of different chromosomal abnormalities (17). With the improvement in this technique, the identification of different translocations is now done by Fluorescence-in Situ Hybridisation (FISH) (metaphase or interphase), using fluorescent probes (18–20) which are expensive, time consuming and labor-intensive (21). Despite having several advantages, the major limiting factor from the diagnostic point of view is the availability of patient sample. Further, the process may fail occasionally due to unsuccessful culture. Alternatively, RT-PCR for fusion transcripts is also used but has variable results in comparison to FISH (22) along with a high rate of false-positive results (15) as well as false-negative results due to RNA instability (14). The success rate of these techniques depends upon the stability of the fusion transcripts, hybridization of the probe (Fluorescent probe or primer) to the DNA which further depends on the copy number of the fusion transcripts in question.
To overcome these limitations, we tried to use RT-qPCR of specific genes as an alternative to diagnose the presence of the ETV6-RUNX1 translocation. Overexpression of the RNA binding protein (RBP), Insulin like Growth Factor 2 mRNA Binding Protein 1 (IGF2BP1) has been reported previously in ETV6-RUNX1 positive B-ALL patients (23). IGF2BP1 is an oncofetal protein overexpressed in epithelial cancers including colorectal and breast cancers and is known to bind oncogenic mRNAs and increase their stability (24). In this study, we have demonstrated the utility of IGF2BP1 expression in B-ALL patient bone marrow samples to diagnose the presence of the ETV6-RUNX1 translocation. Since IGF2BP1 is an RNA binding protein (RBP), we have also used data mining of multiple high throughput public datasets to identify its binding targets and have tested the utility of the expression of two of these targets, EGFL7 and MYC, to diagnose the translocation. We conclude that all of these have the potential to be good candidates for identifying the presence of the translocation with IGF2BP1 being the best among them.
Material and Methods
Patient Samples
Inclusion/Exclusion criteria: Newly diagnosed B-ALL patients (diagnosis established by morphology, immunophenotyping, and molecular genetics), age from 0 to 18 years, and written informed consent of parents obtained prior patient enrolment. Patients treated elsewhere initially or ALL developing secondary to another malignancy were excluded from the study.
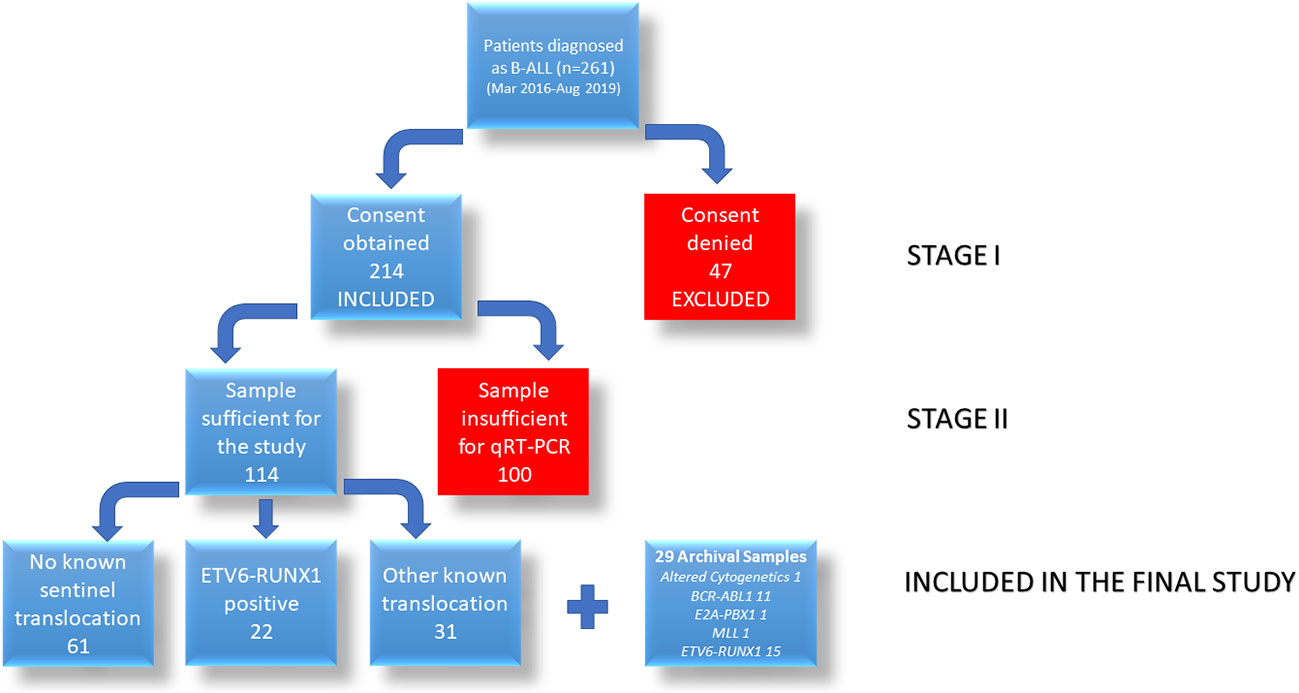
From 261 enrolled naïve B-ALL patients, we were able to collect sufficient patient bone marrow (BM) samples from 114 patients at the time of routine diagnosis from March 2016 to August 2019 at BR Ambedkar Institute Rotary Cancer Hospital at AIIMS, New Delhi after obtaining ethical clearance from the Institutional Ethics Committee and informed consent from a guardian, and assent from children above 7 years of age. To increase the power of the study, we have also included 29 [1 altered cytogenetics, 13 other known translocations (11 BCR-ABL1, 1 E2A-PBX1, 1 MLL), 15 ETV6-RUNX1 positive] archival samples (bone marrow samples collected before March 2016) for this study. Due to the limitation with the sample volume collected, we were not able to study all the genes in all the patient samples.
Cell Culture
Reh (ATCC CRL-8286) (ETV6-RUNX1 translocation positive B lymphoblastic cell line) cells were cultured in RPMI1640 (Himedia AL199S) with 10% Fetal Bovine Serum (Gibco) and 1% antibiotics (Pen-Strep, Gibco) as recommended by ATCC at 37°C and 5% CO2.
Patient Sample Preparation
The bone marrow sample collected was subjected to RBC Lysis Buffer for 30 min at 4°C. The samples were centrifuged at 1,500 g for 10 min at 4°C and washed with 1X Phosphate Buffer Saline (Gibco) followed by centrifugation. The pellet obtained was resuspended in 1 ml of TRIzol (Thermo Fisher) followed by RNA isolation.
RNA Isolation
For every, 1 ml TRIzol sample, 200 µl chloroform was added and mixed vigorously for 10 sec followed by 15 min incubation at room temperature. After incubation, samples were centrifuged at 12,000g for 15 min at 4°C to allow layers to separate. After centrifugation, the samples were taken out carefully and the upper aqueous layer was collected without disturbing the interface in a fresh tube. To the separated sample, 0.5 ml of isopropanol (Thermo) was added and incubated for 10 min at room temperature. After incubation, the samples were centrifuged at 12,000g for 10 min at 4°C to precipitate the RNA. The supernatant was discarded and the precipitated RNA was washed with 70% ethanol (Thermo) at 7,500g for 10 min. The supernatant was discarded and the pellet was air dried to remove the residual ethanol, dissolved in nuclease free water (Qiagen) and stored at −80°C till further use.
Real Time PCR
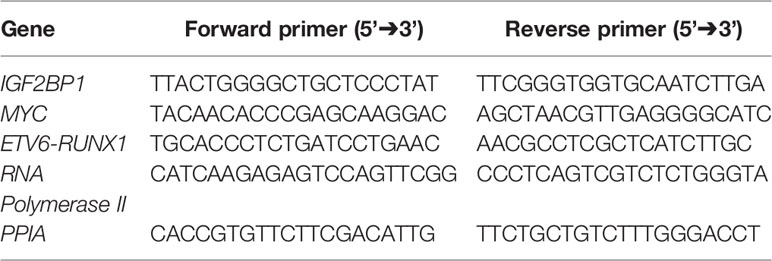
cDNA was prepared from the isolated RNA using RevertAid Reverse Transcriptase (Thermo Fisher) 10 U/µl, Random Decamers 5 µM/reaction (Sigma) and Ribolock (Thermo Fisher) 1 U/µl with 500–1,000 ng of RNA. Real time PCR (qPCR) was done (35 cycles, 95°C for 15 s, 60°C for 15 s, 72°C for 30 s) with Syto9 (Invitrogen) using specific primers for IGF2BP1, MYC, ETV6-RUNX1, and EGFL7 (Table 1). The qPCR reactions were done in technical triplicates for all genes. The expression was normalized using housekeeping genes RNA Polymerase II and PPIA. ΔΔCt method was used to compare the gene expression (25).
Statistics
All experiments were done in triplicates. Relative expression was compared between different groups using Mann Whitney (two groups)/Kruskal-Wallis (more than two groups) statistical tests using GraphPad Prism software version 5. ROC curve was made using SPSS software version 20. A p-value of <0.05 was considered to be significant.
Results
Descriptive Statistics
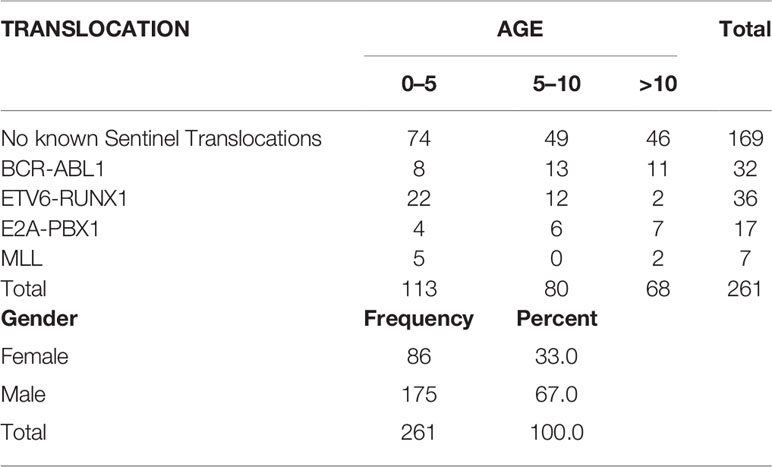
A total of 261 pediatric B-ALL patients were registered in the BRAIRCH OPD, AIIMS from March 2016 to August 2019. Out of these patients, 64.7% cases belonged to the “No known sentinel translocation” group, followed by ETV6-RUNX1 (13.8%), BCR-ABL1 (12.3%), E2A-PBX1 (6.5%), and MLL gene translocation (2.7%). A lower incidence of ETV6-RUNX1 (13%) incidence was noted as compared to the Western population (25%) in our patient cohort which is in concordance with the literature published for the Asian population (6–8). Similarly, a higher incidence of pediatric BCR-ABL1 (11.5%) translocation was also observed as reported by other studies (7–9, 26, 27) (Figure 1A, Table 2). A male preponderance (M: F = 2:1) was observed in the pediatric population with a median age of 4 years (Figures 1B, C and Table 2).
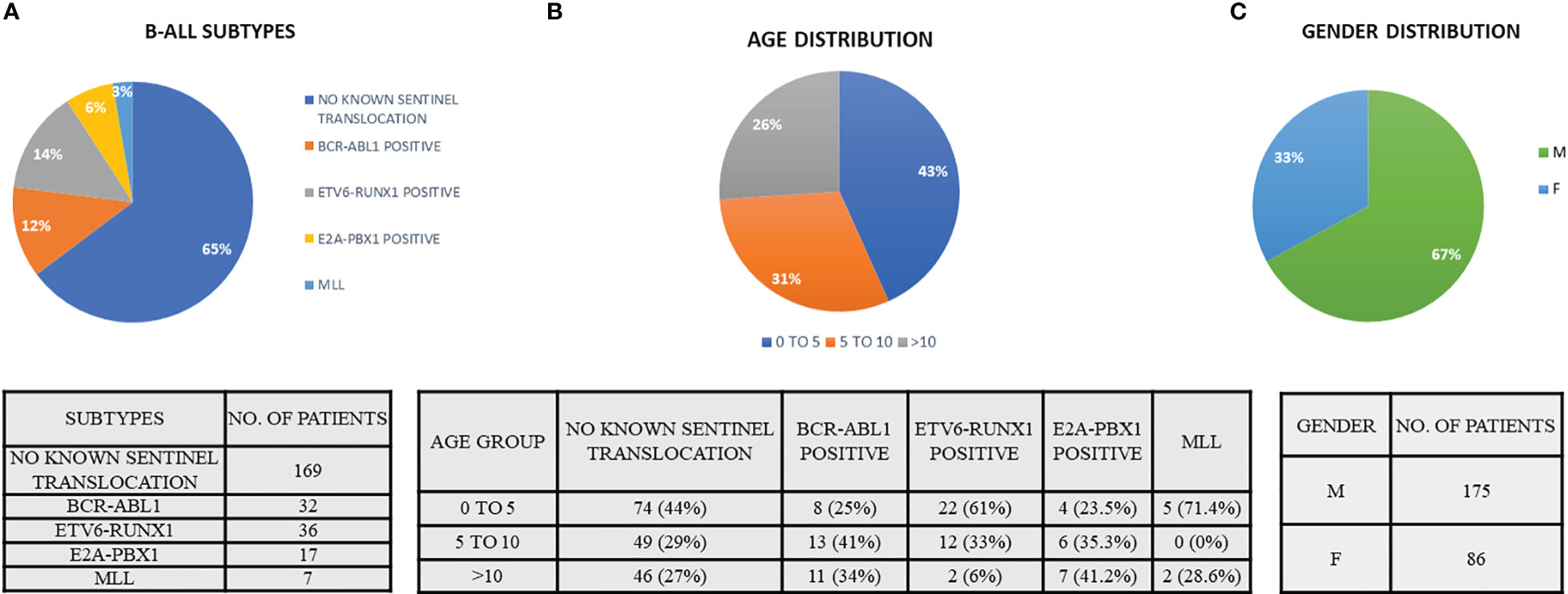
Figure 1 Demographic data indicating the (A) B-ALL subtypes, (B) age distribution of B-ALL patients, and (C) gender distribution. (Patient data reported from March 2016 to August 2019).
RT-qPCR Analysis for IGF2BP1 Expression
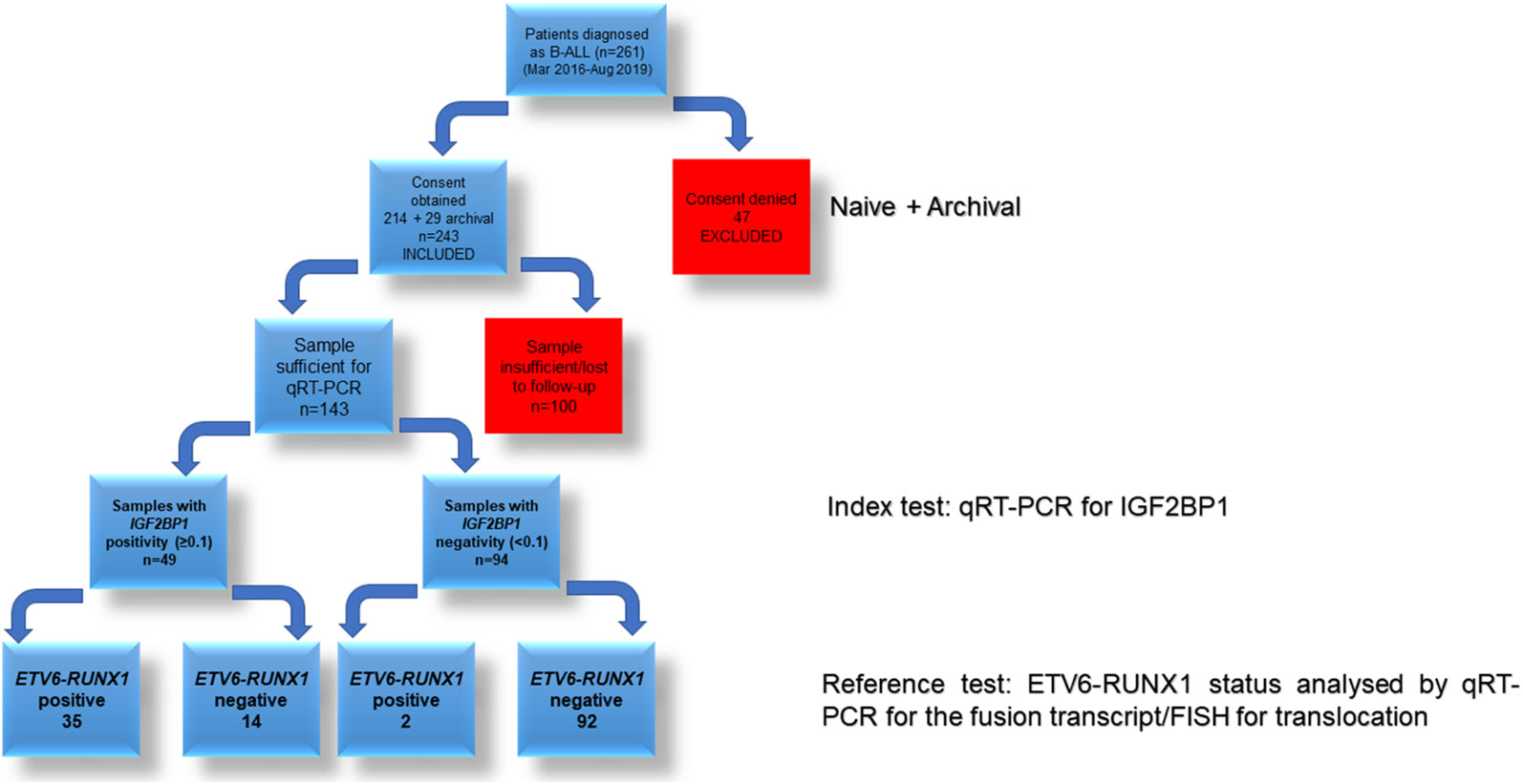
From the 261 patients, sufficient bone marrow sample for further analysis could be obtained for 114 patients (Figure 2). These patients were diagnosed on the basis of morphology, cytochemistry and immunophenotyping. IGF2BP1 mRNA expression was studied by real time PCR using cDNA prepared from BM mononuclear cells using TRIzol. We also included 29 archival patient samples to increase the power of the study. The final analysis was done on 143 patient samples including 37 ETV6-RUNX1 positive samples, 44 samples with other translocations (E2A-PBX1 (n = 15), BCR-ABL1 (n = 24), MLL (n = 5) fusion proteins), and 62 samples with no known translocations. The last group also included 13 patients with altered cytogenetics [Hyper diploidy (n = 8), hypodiploidy (n = 1), chromosomal deletions (n = 1) (chr 6q21)]. The relative expression of IGF2BP1 was normalized using RNA Pol II and PPIA. Similar results were obtained using either of the housekeeping genes.
IGF2BP1 mRNA expression was significantly higher in ETV6-RUNX1 positive B-ALL (n = 37), (median value = 2.74) compared to the negative samples (n = 106), (median value = 0.0022) (p < 0.0001, > 1,000 fold overexpression) (Figure 3A). This group also showed a significant downregulation in wild type ETV6 expression levels, which is concordant with the loss of the other wild type ETV6 allele reported in this subtype of B-ALL (28) (Figure 3B). The expression of IGF2BP1 appeared to be specific to the ETV6-RUNX1 translocation positive sub-group. Interestingly, some patients (12/62) in the “No known sentinel translocations” group had a high expression of IGF2BP1. In addition, 4/12 of them had altered cytogenetics. One of them had hyperploidy (53–54 chromosomes) including chromosome 17 which harbors the IGF2BP1 gene. One patient showed extra signals for 21q (RUNX1 locus, iamp21) on FISH and one patient’s sample was positive for extra signals for both 12p as well as 21q. The other 8 were cytogenetically normal.
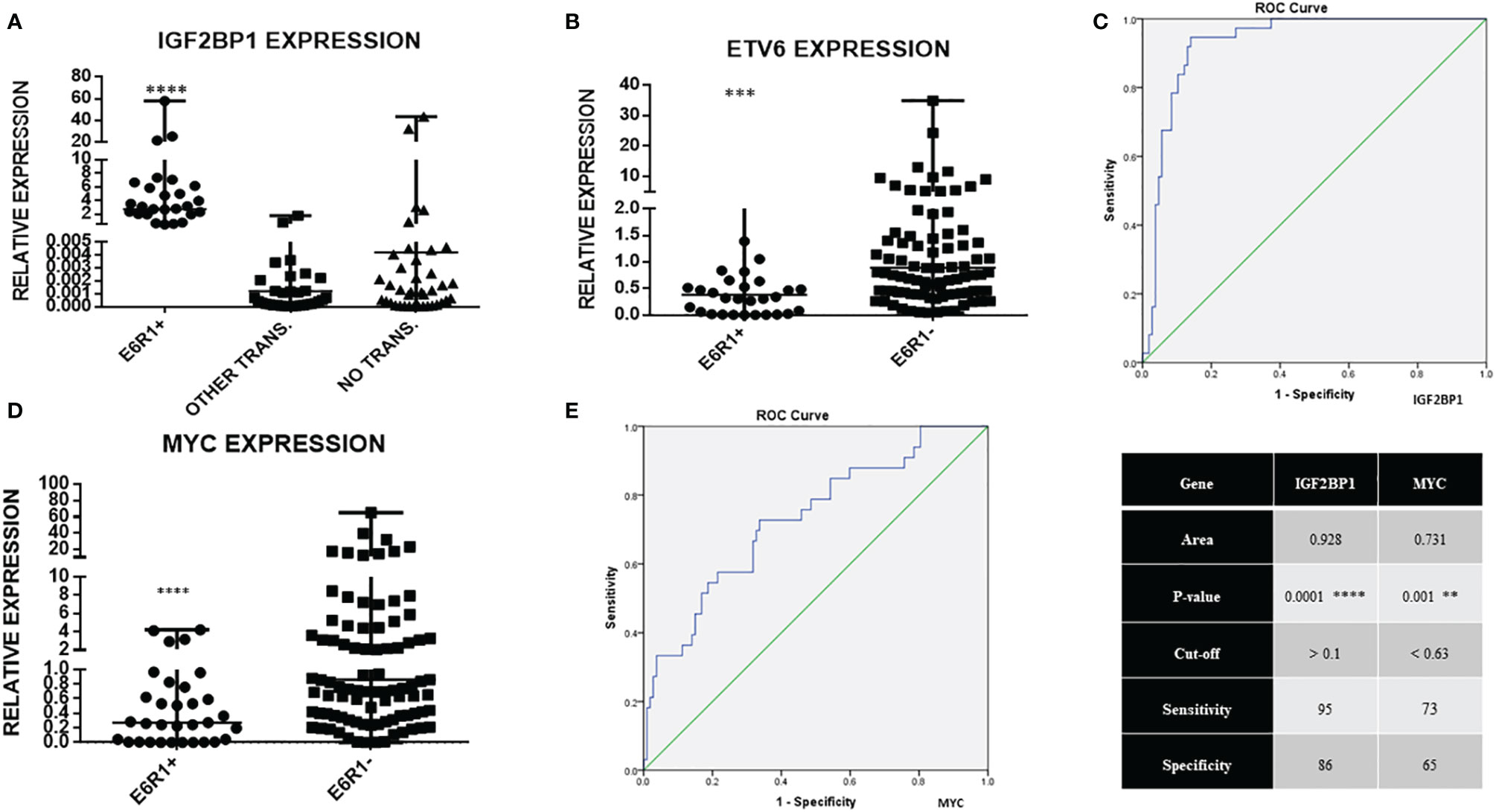
Figure 3 Real time expression data showing (A) IGF2BP1 overexpression in ETV6-RUNX1 translocation positive patients (N = 37), patients with other translocations (N = 44, BCR-ABL1, E2A-PBX1, MLL), and patients with no known translocation (N = 62, includes patients with altered cytogenetics, N = 13); (B) ETV6 expression in ETV6-RUNX1 translocation positive (N = 32) and translocation negative (N = 101) patients; (C) ROC curve for IGF2BP1 comparing it to FISH/RT-PCR for ETV6-RUNX1; (D) MYC expression in ETV6-RUNX1 translocation positive (N = 34) and ETV6-RUNX1 translocation negative (N = 106) patients, and (E) ROC curve for MYC expression. ****p < 0.0001; ***p < 0.001, **p < 0.01. ETV6-RUNX1 positive is denoted as E6R1+ and ETV6-RUNX1 negative as E6R1-.
A new subtype of B-ALL known as “ETV6-RUNX1-like leukemia” has been identified recently based on a gene expression profile similar to that of ETV6-RUNX1 translocation positive leukemia in the absence of the translocation. This subtype also reported a functional loss of ETV6 in the form of mutation or deletions (29, 30). A lower relative expression of ETV6 was noticeable in the patients with high IGF2BP1 expression in the “No known sentinel translocation” group. It is interesting to speculate that these few patients might belong to the newly identified ETV6-RUNX1 like leukemia subgroup. Loss or perturbation of the ETV6 or its regulatory loci might be triggering an ETV6-RUNX1-like leukemia in these patients.
Testing the Efficacy of IGF2BP1 as a Diagnostic Marker
The potential of IGF2BP1 expression to identify the ETV6-RUNX1 translocation was studied using an ROC curve. ETV6-RUNX1 positivity confirmed either by FISH for the t(12;21) translocation or PCR for ETV6-RUNX1 fusion transcript or by both (Figure 3C) was utilized as the reference or gold standard (STARD flowchart in Figure 4). The area under the curve was 0.91 having a sensitivity of 95% and specificity of 86%, with a cut-off of >0.1 with respect to RNA Pol II. Various papers have previously identified MYC as a known target of IGF2BP1 (24, 31, 32). Interestingly, MYC expression showed significant downregulation in the ETV6-RUNX1 positive group (Figure 3D), contradictory to the existing literature which suggests a MYC stabilizing role for IGF2BP1 (24). This might imply the tissue-specific function of IGF2BP1. ROC curves also suggested that reduced MYC expression, with a cut-off value of <0.63 with respect to RNA Pol II, has the potential to be used as a diagnostic marker to detect ETV6-RUNX1 translocation, albeit with a lesser sensitivity (73%) and specificity (65%) than IGF2BP1 expression (Figure 3E).
The sensitivity of IGF2BP1 expression to identify this translocation was compared against studying the expression of the ETV6-RUNX1 fusion mRNA itself. Varying mixtures of Reh (ETV6-RUNX1 translocation positive cell line: representing the blasts) and control PBMCs (no ETV6-RUNX1) were subjected to Real Time RT-PCR for IGF2BP1 as well as ETV6-RUNX1. The correlation coefficient demonstrated the linearity and accuracy of IGF2BP1 expression with an inferior correlation seen for ETV6-RUNX1 expression. While the expression of both ETV6-RUNX1 and IGF2BP1 correlated significantly with the blast percentage, the relative expression of ETV6-RUNX1 was much lower than IGF2BP1 levels contributing to the higher sensitivity of IGF2BP1 expression (Figures 5A, B).
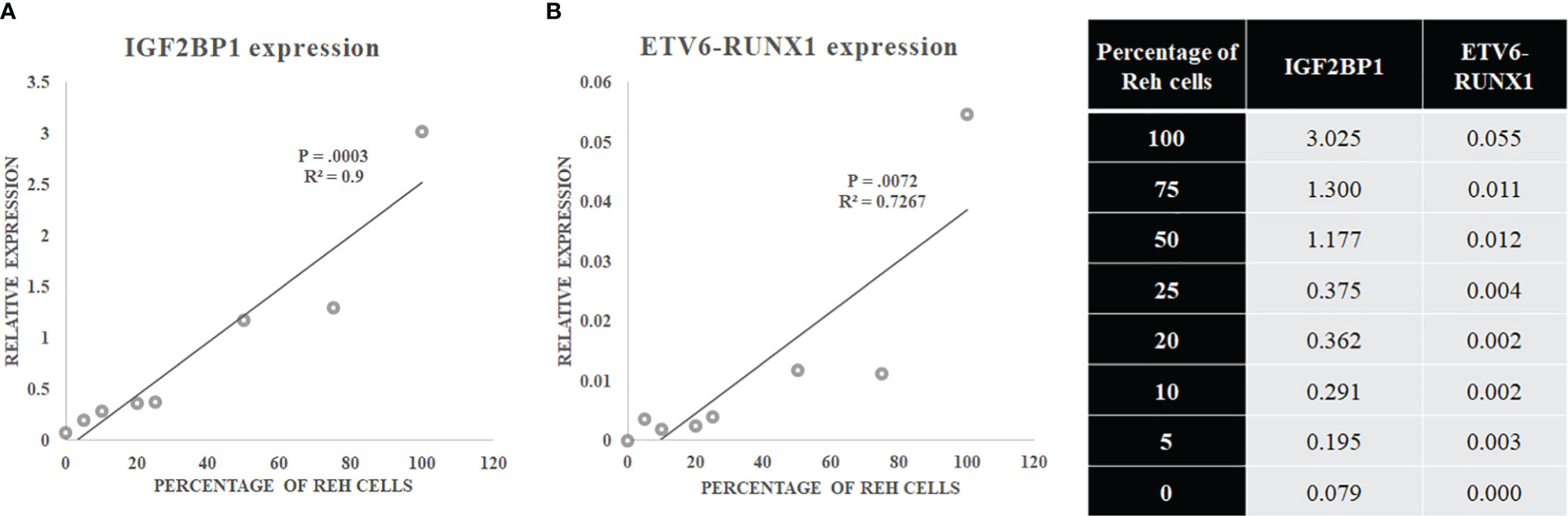
Figure 5 (A) IGF2BP1 and (B) ETV6-RUNX1 expression in a mixture of Reh (corresponding to blasts) and PBMCs.
Validating the Efficacy of Putative IGF2BP1 Targets in Diagnosing ETV6-RUNX1 Positive Leukemia
Since IGF2BP1 is an RNA binding protein, we next proceeded on elucidating its cognate targets. To identify ETV6-RUNX1 positive B-ALL specific putative targets of IGF2BP1, we compared three high-throughput gene expression datasets from published articles:
1. Microarray analysis of ETV6-RUNX1 translocation positive patients (n = 4) compared to normal CD-19 positive B-cells from healthy donors (n = 2) (33);
2. Differential gene expression analysis among subtypes of B-ALL comprising of ETV6-RUNX1, E2A-PBX1, and MLL positive patients (n = 44) GSE65647 (34).
3. MILE (Microarray Innovations in LEukemia) study of B-ALL patient samples (n= 3242) (35, 36).
The first study (33) compared the gene expression levels of ETV6-RUNX1 positive patients’ bone marrow with respect to the MACS sorted CD19 + B cells’ RNA using a microarray. We identified the top 100 overexpressed genes from this differentially expressed list. These are the genes which are specifically overexpressed due to the presence of the ETV6-RUNX1 fusion protein.
The second dataset (34) compared the gene expression pattern between patient samples with three different translocations in B-ALL: ETV6-RUNX1, E2A-PBX1, and MLL translocations by microarray. From this data, we compared the three groups among each other. The genes specifically overexpressed in the comparison between these two different groups [ETV6-RUNX1 vs. E2A-PBX1 (203 genes), ETV6-RUNX1 vs. MLL (329 genes)] were identified. These are the ETV6-RUNX1 positive B-ALL specific genes from this study. Dataset 1 and 2 were overlapped to identify common genes.
The third dataset we used is from the MILE (Microarray Innovations in Leukemia) study (35, 36) which was used for corroboration. We used this study to check the expression of all the common genes from Dataset 1 and 2. We then selected only those genes whose expression levels were high in the ETV6-RUNX1 cohort in the MILE study.
Using the three datasets, we had constructed a list of genes commonly overexpressed in ETV6-RUNX1 positive B-ALL. We then intersected this list with the dataset from an eCLIP experiment done for IGF2BP1 in K562 cell line (37). CLIP (Cross-linked Immunoprecipitation) is a technique where cells are UV-crosslinked, pulled down with an IGF2BP1 specific antibody followed by RNA isolation and sequencing. The peaks from this dataset were visualized on the UCSC browser. The binding of IGF2BP1 to its targets was quantified as weak (+), medium (++) or strong (+++/++++).
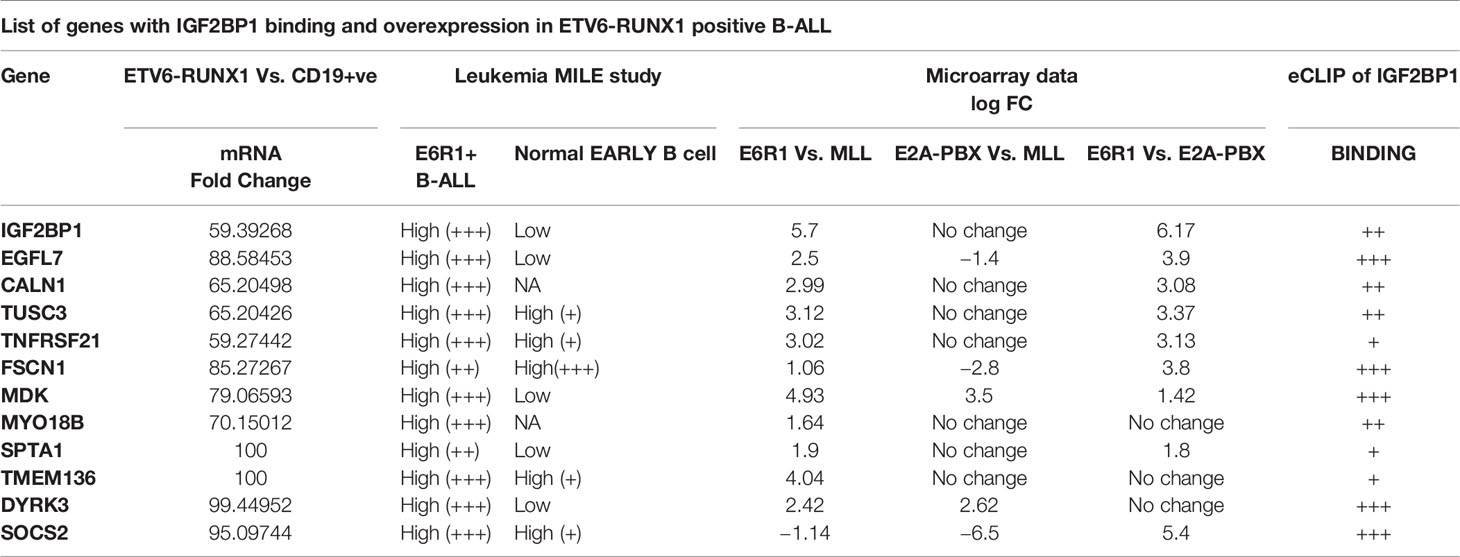
The final list consisted of only 12 such genes which we further segregated based on their known function and putative role in any hematological malignancy (Figure 6 and Table 3). Out of these 12 genes, EGFL7, CALN1, TUSC3 and TNFRSF21 were the top genes that were found to be highly expressed (>2 log fold change) in ETV6-RUNX1 positive group in comparison to MLL and E2A-PBX1 positive groups as well as were bound strongly by IGF2BP1 from the eCLIP dataset. CALN1 has previously known to be overexpressed in ETV6-RUNX1 leukemia but its exact function is still unknown (38), while TUSC3 is reported to be a tumor suppressor gene (39). Among these gene, EGFL7 has been shown to play a role previously in the pathogenesis of Acute Myeloid Leukemia (AML) and hematopoiesis and is also known to be overexpressed in ETV6-RUNX1 leukemia (40, 41). Hence, we tested the potential of using EGFL7 expression to diagnose the ETV6-RUNX1 translocation.
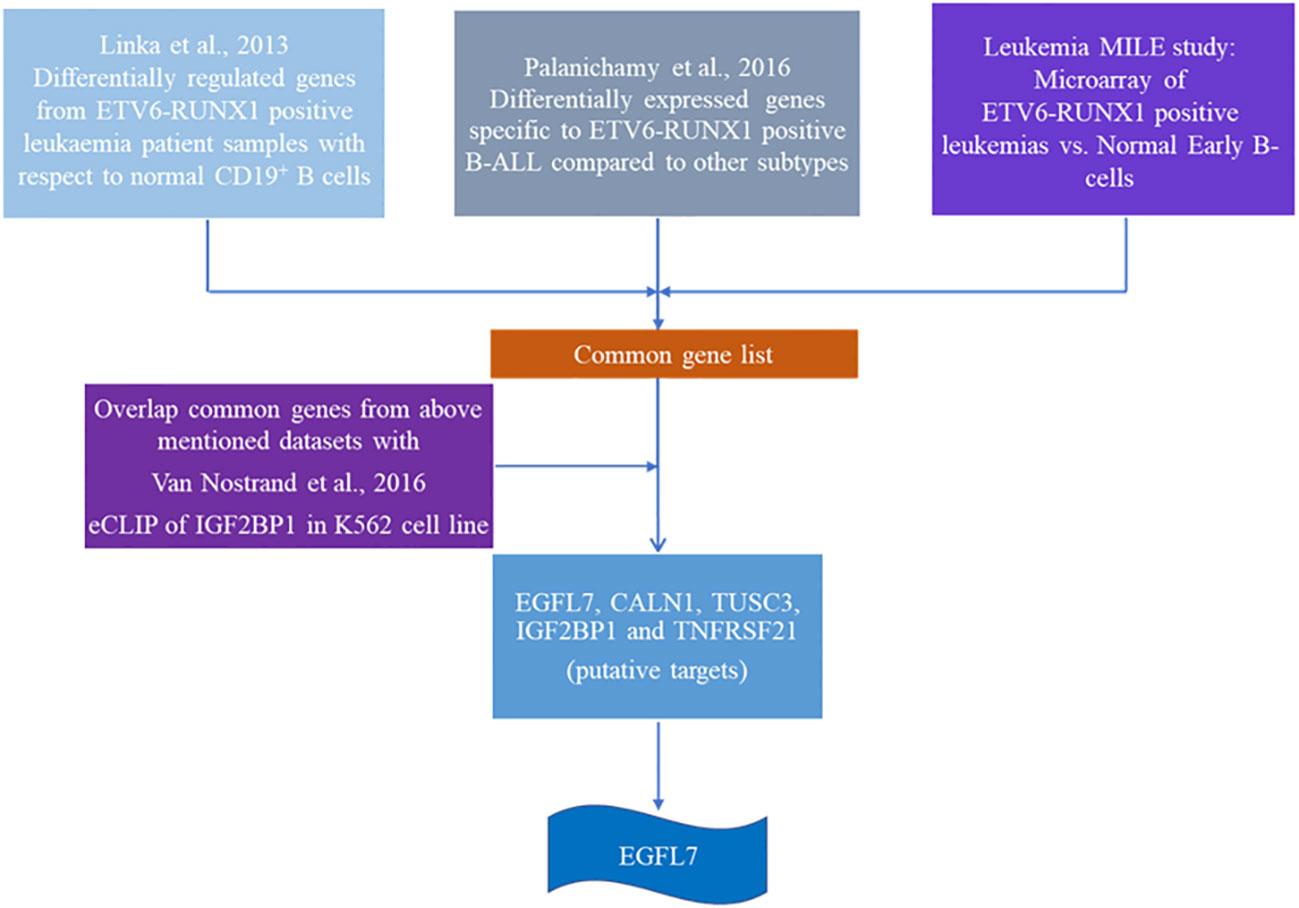
Figure 6 Schematic showing the algorithm used to identify putative targets of IGF2BP1 in ETV6-RUNX1 positive B-ALL.
EGFL7 mRNA was also found to be significantly over expressed in the ETV6-RUNX1 translocation positive group in our cohort (Figure 7A). ROC curves for EGFL7 also showed potential of being a biomarker to diagnose ETV6-RUNX1 translocation but still slightly inferior to IGF2BP1 expression (AUC = 0.865 with a sensitivity of 88.9% and specificity of 81.6%, with cut-off >0.309 with respect to RNA Pol II) (Figure 7B). We tried using the expression levels of IGF2BP1, MYC, and EGFL7 in various combinations to study their diagnostic efficiency. All the combinations had a good AUC but IGF2BP1 expression alone was the best with the highest sensitivity and specificity at a relative expression cut-off >0.1 (Figures 8A–C).
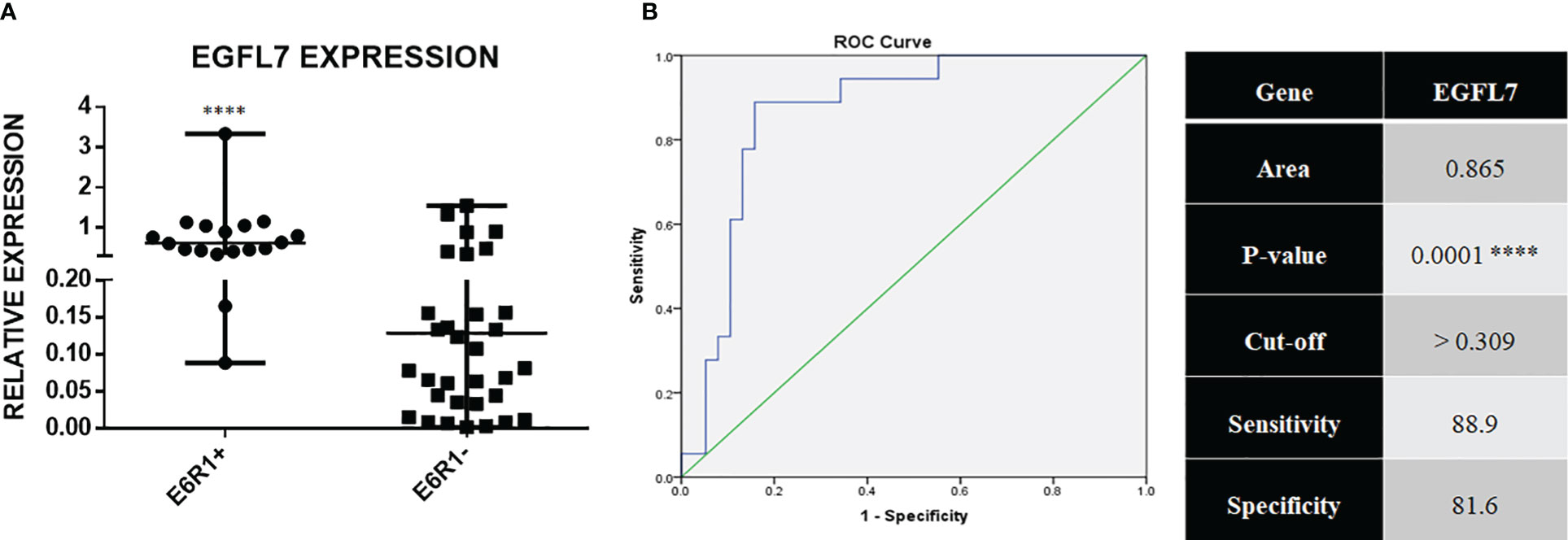
Figure 7 (A) Real time expression data showing EGFL7 overexpression in ETV6-RUNX1 translocation positive patients (N = 18) and ETV6-RUNX1 translocation negative patients (N = 38); and (B) ROC curve for EGFL7 ****p < 0.001.
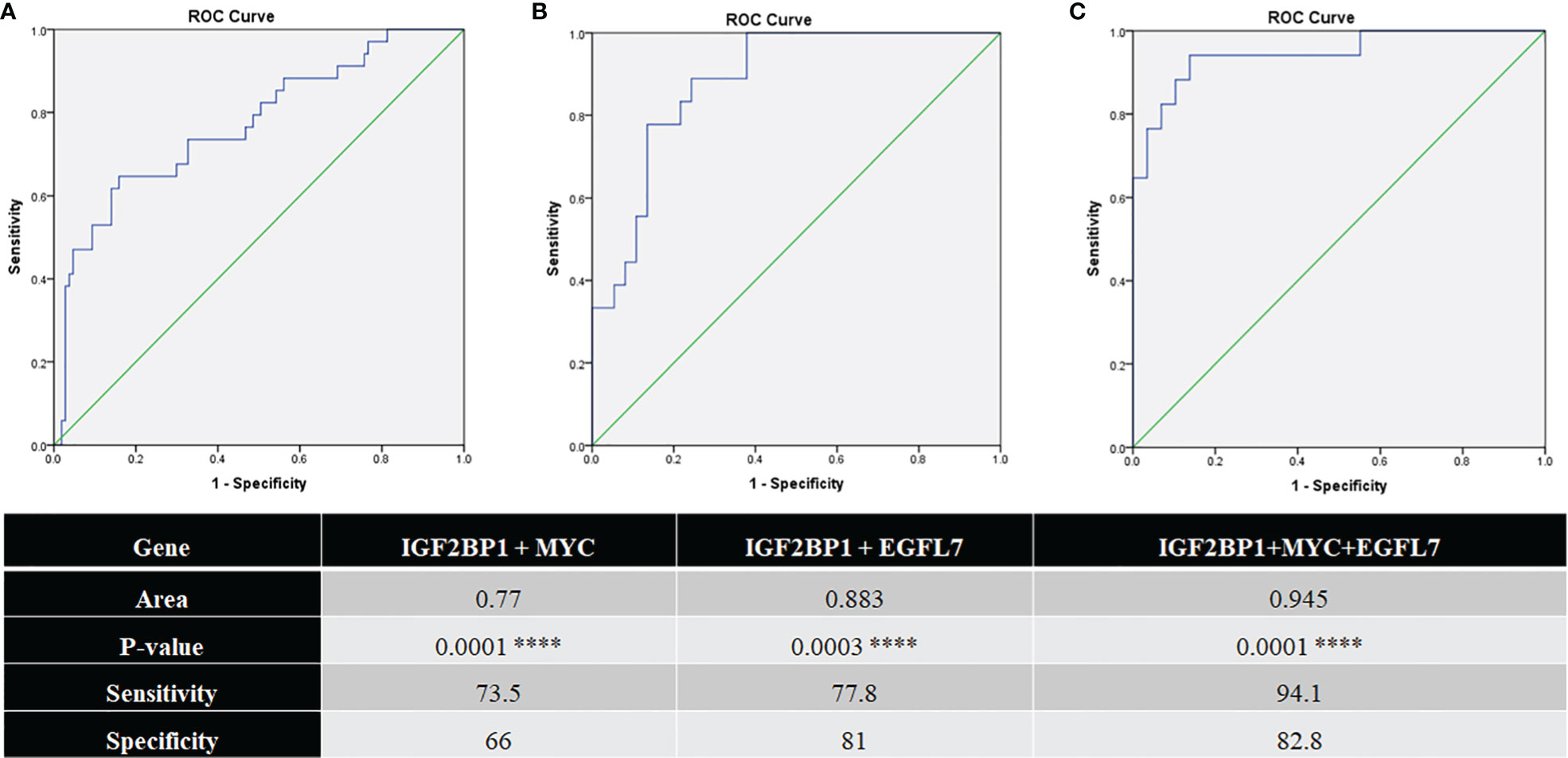
Figure 8 ROC curves for (A) IGF2BP1 + MYC (N = 141; pos = 34, neg = 107); (B) IGF2BP1 + EGFL7 (N = 55; pos = 18, neg = 37); and (C) IGF2BP1 + MYC + EGFL7 (N = 46; pos = 17, neg = 29) comparing them to FISH/RT-PCR for ETV6-RUNX1 ****p < 0.001.
The heatmap showing the expression of IGF2BP1, MYC, and EGFL7 in ETV6-RUNX1 translocation positive and negative patients highlights the discriminatory power of IGF2BP1 to identify this translocation (Figure 9). Previous studies have identified various genes including EPOR, TCFL5, and TERF2 to be specifically overexpressed in ETV6-RUNX1 leukemia (38). We compared the expression of IGF2BP1 as well as these genes in ALL patient samples from the publicly available Leukemia MILE study (36) on the BloodSpot database (35). Although all these genes are overexpressed in ETV6-RUNX1 positive B-ALL, it is evident that IGF2BP1 expression stands out in being extremely specific to this subtype (Figure 10).
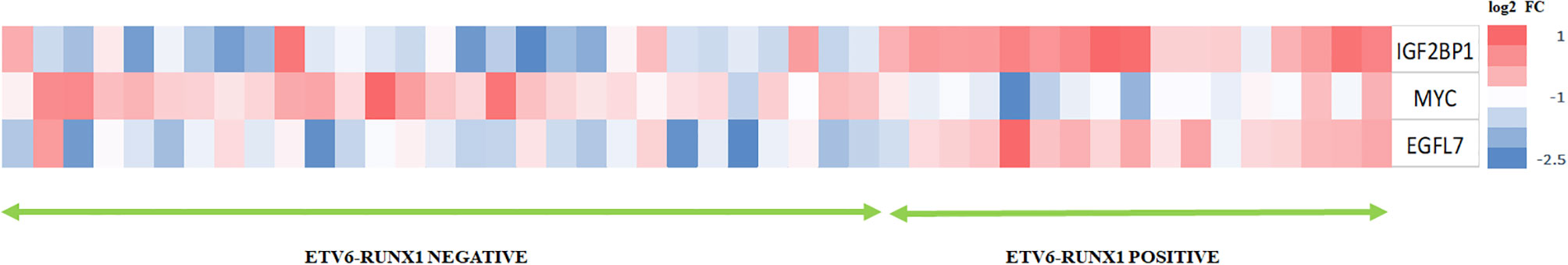
Figure 9 Heat map showing the expression of IGF2BP1, MYC, and EGFL7 in ETV6-RUNX1 translocation positive patients with respect to ETV6-RUNX1 translocation negative patients (N = 46).
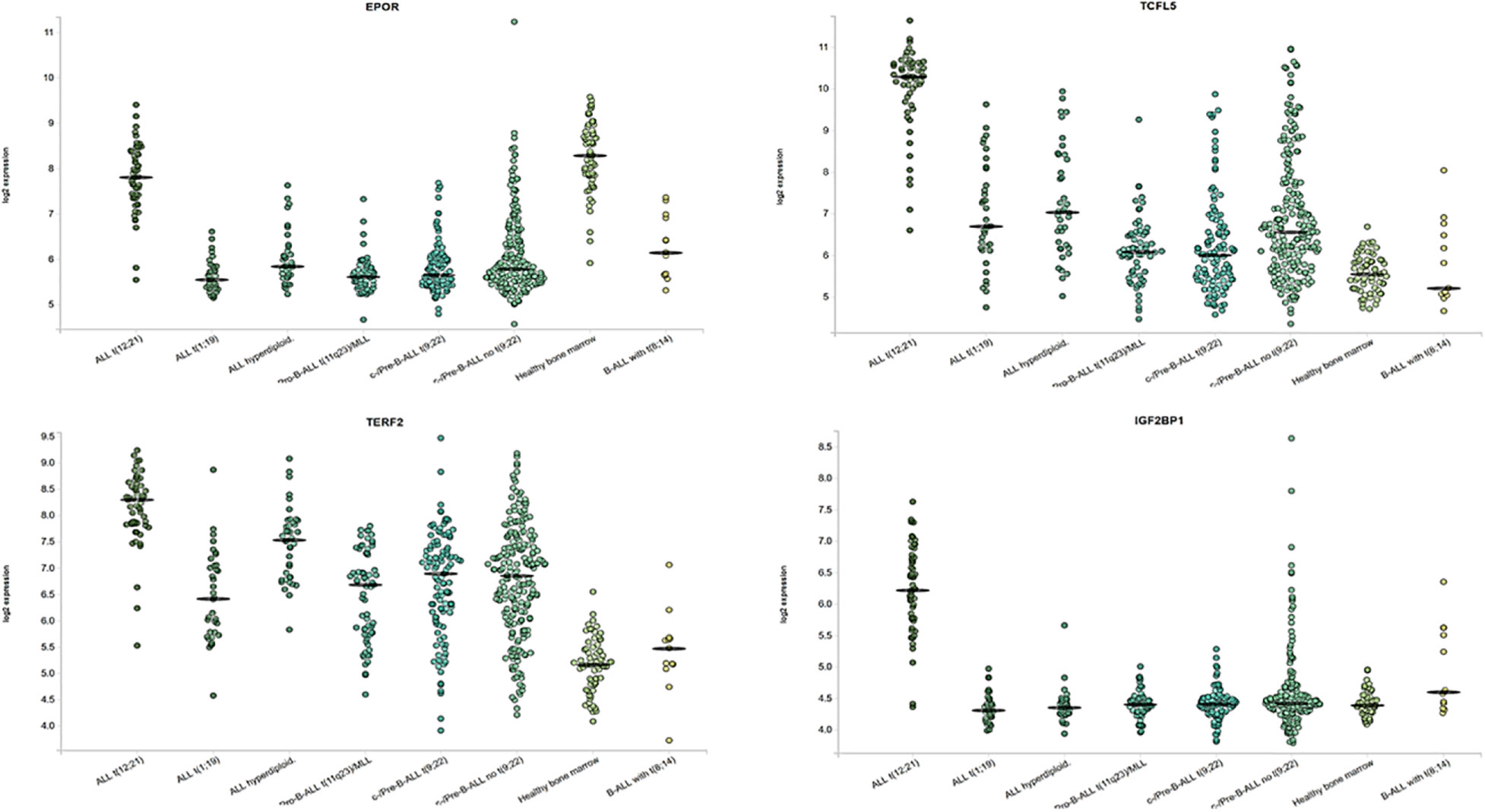
Figure 10 Expression of ETV6-RUNX1 positive B-ALL specific genes and IGF2BP1 from the MILE study. ALL with hyperdiploid karyotype (n=40), ALL with t(1;19) (n=36), ALL with t(12;21) (n=58), Mature B-ALL with t(8;14) (n=13), c- ALL/Pre-B-ALL with no t(9;22) (n=237), c- ALL/Pre-B-ALL with t(9;22) (n=122), Non-leukemic Healthy bone marrow (n=73), Pro-B-ALL with t(11q23)/MLL (n=70).
Discussion
Routine diagnostic techniques used in ALL diagnosis and management include flowcytometry (for immunophenotyping), RT-PCR, Karyotyping, and FISH. FISH is the gold standard technique which is commonly used to diagnose the presence of leukemic translocations and it has distinct advantages (21, 42). The average turnaround time (TAT) for FISH is 3 days (43) and cytogenetics is 7 days (44) in standard laboratories. We have investigated the utility of studying the expression of IGF2BP1, an RNA binding protein known to be overexpressed in ETV6-RUNX1 leukemia, by qPCR, to be used as a cheaper and quicker alternative to diagnose the presence of the translocation. This would facilitate risk stratification using low amounts of patient bone marrow sample. This would be especially useful in pediatric patients where the sample obtained during the bone marrow aspiration is not sufficient enough to even complete the panel of routine diagnostic tests (45). If utilized, the TAT for gene expression analysis is ~6 h and can be done easily in house without incurring extra cost to the patient. Based on the sensitivity and specificity of the gene as demonstrated in the results, we could potentially add the qPCR for IGF2BP1 as a diagnostic marker for the presence of ETV6-RUNX1 translocation. This study has utilized the most commonly observed sentinel translocations (BCR-ABL1, E2A-PBX1, MLL-AF4). This study has not addressed the newer gene expression based minor subtypes including the “ETV6-RUNX1-like” leukemia which would be part of the “no known sentinel translocations” group.
This is the first study to analyze the utility of gene expression analysis of IGF2BP1 for the diagnosis of ETV6-RUNX1 positive B-ALL. It is also the first time that such an exercise has been carried out in an Indian patient cohort of over 100 patients. Since IGF2BP1 is an RNA binding protein, we have used data mining from public high throughput datasets to identify its targets in B-ALL. We have utilized three datasets to identify an ETV6-RUNX1 specific gene signature which was then overlapped with an eCLIP dataset of IGF2BP1. The overlapped dataset gave us a novel list of ETV6-RUNX1 specific genes bound and stabilized by IGF2BP1.
From this list we have further characterized two genes (EGFL7 and MYC) for their sensitivity and specificity in identifying the translocation. Both these genes were also found to have the potential to be used as biomarkers to identify the translocation. However, IGF2BP1 mRNA expression by RT qPCR was found to be an excellent candidate to be used as a biomarker to diagnose ETV6-RUNX1 translocation showing 95% sensitivity and 86% specificity at a relative expression cut off of 0.1. IGF2BP1 expression in the bone marrow as well as peripheral blood can be utilized as a rapid test to diagnose the presence of the ETV6-RUNX1 translocation in the future.
This increase in IGF2BP1 expression might be due to the loss of ETV6 which might be repressing IGF2BP1 or the presence of the ETV6-RUNX1 fusion protein or it might be a combination of both factors. Interestingly, the de-repression of IGF2BP1 in ETV6-RUNX1 positive ALL is so strong that its gene expression is ~100 fold higher than the expression of the ETV6-RUNX1 fusion RNA. This contributes to the extremely high sensitivity and specificity of this assay. Future work could also involve identifying the mechanistic role of IGF2BP1 in leukemogenesis and developing it into a therapeutic target in this particular subset of B-ALL.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Ethics Committee, All India Institute of Medical Sciences, New Delhi, India. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
GS, EB, and TN carried out the experiments. AJ, GS, JS, and EB contributed to sample collection and analysis. SB and AC provided patient samples and clinical data. GS and JP wrote the manuscript with inputs from AS, SB, PC, and AC. JP conceived the study. All authors contributed to the article and approved the submitted version.
Funding
This work is being supported by the Wellcome Trust/DBT India Alliance Early Career Fellowship to JP (IA/CPHE/15/1/502050). GS is supported by DBT Senior Research Fellowship as well as SERB Overseas Visiting Doctoral Research Fellowship. JS is being supported by a DBT Senior Research Fellowship and AJ by a CSIR Senior Research Fellowship. We thank Dr Kalaivani for her help with the statistical analysis. We thank all our collaborators for vibrant discussions regarding the experiments and data.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Carroll WL, Raetz EA. Clinical and laboratory biology of childhood acute lymphoblastic leukemia. J Pediatr (2012) 160(1):10–8. doi: 10.1016/j.jpeds.2011.08.006
2. Malouf C, Ottersbach K. Molecular processes involved in B cell acute lymphoblastic leukaemia. Cell Mol Life Sci (2018) 75(3):417–46. doi: 10.1007/s00018-017-2620-z
3. Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest (2012) 122(10):3407–15. doi: 10.1172/JCI61203
4. Harrison CJ. Cytogenetics of paediatric and adolescent acute lymphoblastic leukaemia. Br J Haematol (2009) 144(2):147–56. doi: 10.1111/j.1365-2141.2008.07417.x
5. Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer (2019) 144(8):1941–53. doi: 10.1002/ijc.31937
6. Aljamaan K, Aljumah TK, Aloraibi S, Absar M, Iqbal Z. Low Frequency of ETV6-RUNX1 (t 12; 21) in Saudi Arabian Pediatric Acute Lymphoblastic Leukemia Patients: Association with Clinical Parameters and Early Remission. Asian Pac J Cancer Prev (2015) 16(17):7523–7. doi: 10.7314/APJCP.2015.16.17.7523
7. Chopra A, Soni S, Verma D, Kumar D, Dwivedi R, Vishwanathan A, et al. Prevalence of common fusion transcripts in acute lymphoblastic leukemia: A report of 304 cases. Asia Pac J Clin Oncol (2015) 11(4):293–8. doi: 10.1111/ajco.12400
8. Pandita A, Harish R, Digra SK, Raina A, Sharma AA, Koul A. Molecular cytogenetics in childhood acute lymphoblastic leukemia: a hospital-based observational study. Clin Med Insights Oncol (2015) 9:39–42. doi: 10.4137/CMO.S24463
9. Sugapriya D, Preethi S, Shanthi P, Chandra N, Jeyaraman G, Sachdanandam P, et al. BCR-ABL Translocation in Pediatric Acute Lymphoblastic Leukemia in Southern India. Indian J Hematol Blood Transfus (2012) 28(1):37–41. doi: 10.1007/s12288-011-0096-9
10. Zelent A, Greaves M, Enver T. Role of the TEL-AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia. Oncogene (2004) 23(24):4275–83. doi: 10.1038/sj.onc.1207672
11. Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer (2002) 2(7):502–13. doi: 10.1038/nrc840
12. Green SM, Coyne HJ, McIntosh LP, Graves BJ. DNA Binding by the ETS Protein TEL (ETV6) Is Regulated by Autoinhibition and Self-association. J Biol Chem (2010) 285(24):18496–504. doi: 10.1074/jbc.M109.096958
13. Loh ML, Rubnitz JE. TEL/AML1-positive pediatric leukemia: prognostic significance and therapeutic approaches. Curr Opin Hematol (2002) 9(4):345–52. doi: 10.1097/00062752-200207000-00013
14. Schäfer D, Olsen M, Lähnemann D, Stanulla M, Schmiegelow K, Borkhardt A, et al. 5% of healthy newborns have an ETV6-RUNX1 fusion as revealed by DNA-based GIPFEL screening. Blood (2018) 131(7):821–6. doi: 10.1182/blood-2017-09-808402
15. Lausten-Thomsen U, Madsen HO, Vestergaard TR, Hjalgrim H, Nersting J, Schmiegelow K. Prevalence of t(12,21)[ETV6-RUNX1]-positive cells in healthy neonates. Blood (2011) 117(1):186–9. doi: 10.1182/blood-2010-05-282764
16. Mori H, Colman SM, Xiao Z, Ford AM, Healy LE, Donaldson C, et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Natl Acad Sci U S A (2002) 99(12):8242–7. doi: 10.1073/pnas.112218799
17. Pardue ML, Gall JG. Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc Natl Acad Sci U S A (1969) 64(2):600–4. doi: 10.1073/pnas.64.2.600
18. Rudkin GT, Stollar BD. High resolution detection of DNA–RNA hybrids in situ by indirect immunofluorescence. Nature (1977) 265(5593):472–3. doi: 10.1038/265472a0
19. Trask BJ. Human cytogenetics: 46 chromosomes, 46 years and counting. Nat Rev Genet (2002) 3(10):769–78. doi: 10.1038/nrg905
20. Speicher MR, Carter NP. The new cytogenetics: blurring the boundaries with molecular biology. Nat Rev Genet (2005) 6(10):782–92. doi: 10.1038/nrg1692
21. Huber D, Voith von Voithenberg L, Kaigala GV. Fluorescence in situ hybridization (FISH): History, limitations and what to expect from micro-scale FISH? Micro Nano Eng (2018) 1:15–24. doi: 10.1016/j.mne.2018.10.006
22. Akhter A, Mughal MK, Elyamany G, Sinclair G, Azma RZ, Masir N, et al. Multiplexed automated digital quantification of fusion transcripts: comparative study with fluorescent in-situ hybridization (FISH) technique in acute leukemia patients. Diagn Pathol (2016) 11(1):89. doi: 10.1186/s13000-016-0541-z
23. Stoskus M, Eidukaite A, Griskevicius L. Defining the significance of IGF2BP1 overexpression in t (12,21)(p13;q22)-positive leukemia REH cells. Leukemia Res (2016) 47:16–21. doi: 10.1016/j.leukres.2016.05.009
24. Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci (2013) 70(15):2657–75. doi: 10.1007/s00018-012-1186-z
25. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
26. Golub TR, Barker GF, Bohlander SK, Hiebert SW, Ward DC, Bray-Ward P, et al. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci (1995) 92(11):4917–21. doi: 10.1073/pnas.92.11.4917
27. Bernt KM, Hunger SP. Current concepts in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia. Front Oncol (2014) 4:54. doi: 10.3389/fonc.2014.00054
28. Attarbaschi A, Mann G, König M, Dworzak MN, Trebo MM, Mühlegger N, et al. Incidence and relevance of secondary chromosome abnormalities in childhood TEL/AML1+ acute lymphoblastic leukemia: an interphase FISH analysis. Leukemia (2004) 18(10):1611–6. doi: 10.1038/sj.leu.2403471
29. Zaliova M, Kotrova M, Bresolin S, Stuchly J, Stary J, Hrusak O, et al. ETV6/RUNX1-like acute lymphoblastic leukemia: A novel B-cell precursor leukemia subtype associated with the CD27/CD44 immunophenotype. Genes Chromosomes Cancer (2017) 56(8):608–16. doi: 10.1002/gcc.22464
30. Lilljebjörn H, Fioretos T. New oncogenic subtypes in pediatric B-cell precursor acute lymphoblastic leukemia. Blood (2017) 130(12):1395–401. doi: 10.1182/blood-2017-05-742643
31. Noubissi FK, Nikiforov MA, Colburn N, Spiegelman VS. Transcriptional Regulation of CRD-BP by c-myc: Implications for c-myc Functions. Genes Cancer (2010) 1(10):1074–82. doi: 10.1177/1947601910395581
32. Weidensdorfer D, Stöhr N, Baude A, Lederer M, Köhn M, Schierhorn A, et al. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA (2009) 15(1):104–15. doi: 10.1261/rna.1175909
33. Linka Y, Ginzel S, Krüger M, Novosel A, Gombert M, Kremmer E, et al. The impact of TEL-AML1 (ETV6-RUNX1) expression in precursor B cells and implications for leukaemia using three different genome-wide screening methods. Blood Cancer J (2013) 3(10):e151–1. doi: 10.1038/bcj.2013.48
34. Palanichamy JK, Tran TM, Howard JM, Contreras JR, Fernando TR, Sterne-Weiler T, et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J Clin Invest (2016) 126(4):1495–511. doi: 10.1172/JCI80046
35. Bagger FO, Kinalis S, Rapin N. BloodSpot: a database of healthy and malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res (2018) 47(D1):D881–5. doi: 10.1093/nar/gky1076
36. Haferlach T, Kohlmann A, Basso G, Béné M-C, Chiaretti S, Downing JR, et al. The Clinical Utility of Microarray-Based Gene Expression Profiling in the Diagnosis and Sub-Classification of Leukemia: Final Report on 3252 Cases from the International MILE Study Group. Blood (2008) 112(11):753–3. doi: 10.1182/blood.V112.11.753.753
37. Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods (2016) 13(6):508–14. doi: 10.1038/nmeth.3810
38. Fuka G, Kauer M, Kofler R, Haas OA, Panzer-Grümayer R. The Leukemia-Specific Fusion Gene ETV6/RUNX1 Perturbs Distinct Key Biological Functions Primarily by Gene Repression. PLoS One (2011) 6(10):e26348. doi: 10.1371/journal.pone.0026348
39. Coccé MC, Alonso CN, Rossi JG, Bernasconi AR, Rampazzi MA, Felice MS, et al. Cytogenetic and Molecular Findings in Children with Acute Lymphoblastic Leukemia: Experience of a Single Institution in Argentina. Mol Syndromol (2015) 6(4):193–203. doi: 10.1159/000441046
40. Papaioannou D, Shen C, Nicolet D, McNeil B, Bill M, Karunasiri M, et al. Prognostic and biological significance of the proangiogenic factor EGFL7 in acute myeloid leukemia. Proc Natl Acad Sci U S A (2017) 114(23):E4641–7. doi: 10.1073/pnas.1703142114
41. Gandemer V, Rio A-G, de Tayrac M, Sibut V, Mottier S, Ly Sunnaram B, et al. Five distinct biological processes and 14 differentially expressed genes characterize TEL/AML1-positive leukemia. BMC Genomics (2007) 8(1):385. doi: 10.1186/1471-2164-8-385
42. Bishop R. Applications of fluorescence in situ hybridization (FISH) in detecting genetic aberrations of medical significance. Biosci Horizons: Int J Student Res (2010) 3(1):85–95. doi: 10.1093/biohorizons/hzq009
43. Spector DL, Goldman RD, Leinwand LA. “Fluorescence in situ hybridization to DNA”. In: Cells: A Laboratory Manual. Cold Spring Harbor, New York, USA: Harbor Laboratory Press (1998) 111.26–.36. doi: 10.1038/nmeth0305-237
44. Hammer RD, Doll D, Layfield L. Is It Time for a New Gold Standard? FISH vs Cytogenetics in AML Diagnosis. Am J Clin Pathol (2016) 145(3):430–2. doi: 10.1093/ajcp/aqw008
Keywords: IGF2BP1, EGFL7, ETV6-RUNX1 translocation, B-ALL, receiver operating characteristic curve (ROC)
Citation: Sharma G, Boby E, Nidhi T, Jain A, Singh J, Singh A, Chattopadhyay P, Bakhshi S, Chopra A and Palanichamy JK (2021) Diagnostic Utility of IGF2BP1 and Its Targets as Potential Biomarkers in ETV6-RUNX1 Positive B-Cell Acute Lymphoblastic Leukemia. Front. Oncol. 11:588101. doi: 10.3389/fonc.2021.588101
Received: 28 July 2020; Accepted: 08 January 2021;
Published: 23 February 2021.
Edited by:
Jose-Maria Ribera, Germans Trias i Pujol Health Science Research Institute (IGTP), SpainReviewed by:
Vito Carlo Alberto Caponio, University of Foggia, ItalyRanganatha R. Somasagara, North Carolina Central University, United States
Copyright © 2021 Sharma, Boby, Nidhi, Jain, Singh, Singh, Chattopadhyay, Bakhshi, Chopra and Palanichamy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jayanth Kumar Palanichamy, ZHJqYXlhbnRoQGFpaW1zLmVkdQ==
 Gunjan Sharma
Gunjan Sharma Elza Boby1
Elza Boby1 Thakur Nidhi
Thakur Nidhi Ayushi Jain
Ayushi Jain Jay Singh
Jay Singh Archna Singh
Archna Singh Anita Chopra
Anita Chopra Jayanth Kumar Palanichamy
Jayanth Kumar Palanichamy