- Department of ICU, The Affiliated Huaian No. 1 People’s Hospital of Nanjing Medical University, Huaian, China
Background: As a novel immune checkpoint molecular, T-cell immunoglobulin mucin 3 (TIM-3) is emerging as a therapeutic target for cancer immunotherapy. However, the predictive role of TIM-3 in cancer remains largely undetermined. This study was designed to investigate the role of TIM-3 in cancer.
Methods: Publications were searched using multiple databases. The hazard ratios (HRs) with 95% confidence intervals (CIs) were calculated. To further confirm the prognostic effect of TIM-3, The Cancer Genome Atlas (TCGA) data were applied. Functional analysis of TIM-3 was also investigated.
Results: 28 studies with 7284 patients with malignant tumors were identified. Based on multivariate Cox regression analysis, TIM-3 was an independent prognostic indicator for poor overall survival (OS) (HR= 1.54, 95% CI = 1.19-1.98, P = 0.001). However, TIM-3 was not correlated with cancer-specific survival and disease-free survival (DFS). Particularly, TIM-3 showed a worse prognosis in non-small cell lung carcinoma and gastric cancer; but it showed a favorable prognosis in breast cancer. Functional analysis showed that TIM-3 was closely correlated with immune responses such as T-cell activation and natural killer cell-mediated cytotoxicity. Moreover, TIM-3 expression was found to be related to worse OS in 9491 TCGA patients (HR = 1.2, P < 0.001), but was not associated with DFS.
Conclusions: TIM-3 was an independent prognostic factor. Meanwhile, TIM-3 played a crucial role in tumor immune responses. This supports TIM-3 as a promising target for cancer immunotherapy.
Introduction
Cancer is still a global burden issue worldwide. Cancer incidence and mortality are rapidly growing in the world. Based on the GLOBOCAN estimates, an estimated 18.1 million cancer cases were diagnosed, and approximately 9.6 million deaths were due to cancer, in 2018 (1). Until now, general treatment regimens for cancer are used, such as surgery, chemotherapy, radiotherapy, targeted molecular therapy, or immunotherapy. Despite the significant advancements made in cancer treatment, the 5‐year survival rate for cancer patients is not very high (< 70%) (2, 3). Thus, ongoing efforts to identify best practices for cancer treatment and management are needed.
Studies suggest that the development of strategies against biomarkers could be a reasonable and precise therapy approach in tumors (4–8). The immunological aspects are crucial hallmarks for tumor progression and metastasis (9–11). Cancer immunotherapy shows substantial benefits and has become a powerful treatment strategy in controlling many malignant tumors (12, 13). Programmed cell death 1 (PD-1) and programmed cell death ligand 1 (PD-L1) have the potential to become prognostic biomarkers in numerous cancers (14–17). Immune checkpoint therapy, such as PD-1 and cytotoxic T lymphocyte antigen-4 (CTLA-4), has achieved clinical success (18). T-cell immunoglobulin and mucin-dominant containing-3 (TIM-3), also known as hepatitis A virus cellular receptor 2 (HAVCR2), has been reported as an immune-checkpoint molecule (19). TIM-3 plays a vital role in suppressing cytotoxic T lymphocytes (CTL) and Th1 responses and the expression of cytokines such as tumor necrosis factor and interferon-γ (20, 21). TIM-3 regulates innate and adaptive immune responses, possibly exerting either positive or negative effects (22). Numerous studies have reported that TIM-3 is expressed in cancer (23–25). TIM-3 expression is correlated with poor prognosis in many cancers, such as oral squamous cell carcinoma (26), ovarian cancer (27), and gastric cancer (28). However, the role of TIM-3 in clinical cancer studies is still conflicting. For example, Duan 2018 et al. reported no correlation between TIM-3 expression and overall survival (OS) in esophageal squamous cell carcinoma (29). However, Hong 2019 et al. reported a significant association between TIM-3 expression and worse OS in esophageal squamous cell carcinoma (30). Thus, it is of great importance to investigate the prognostic impact of TIM-3 in malignant tumors.
The previous meta-analysis involving only seven studies with a very small population (n=869 patients) evaluated the correlation between TIM-3 expression and OS in solid tumors (31). In recent years, numerous studies (23–27, 29, 30, 32–43) were published that examined the prognosis of TIM-3 expression in various tumors. Here, the aim of the present meta-analysis was to analyze the association of TIM-3 expression with cancer survival (n=28 studies with 7284 patients). Additionally, TCGA data were further used to confirm the results of this meta-analysis, and the potential biological functions of TIM-3 were also investigated. This study will provide more evidence to suggest whether TIM-3 could be a promising target for immunotherapy.
Materials and Methods
Literature Search
The PubMed, EMBASE, and Web of Science databases were systematically searched to identify eligible publications before January 29, 2020. The following key words and search terms were applied: “T-cell immunoglobulin and mucin domain containing 3 OR TIM-3 OR TIM3 OR T-cell Ig and mucin domain 3 OR HAVCR2 OR hepatitis A virus cellular receptor 2 OR CD366”, “prognosis OR survival OR outcome OR prognostic”. The reference lists of the included publications were also scanned to find additional potential studies. This meta-analysis was carried out based on the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement (44).
Study Selection
The eligible studies were included when they met the following selection criteria: 1) the patients were diagnosed with malignant tumors; 2) studies evaluated the expression of TIM-3 from the original articles; 3) studies reported the prognosis of TIM-3 expression using multivariate Cox regression analysis; 4) the prognostic endpoints, such as OS, disease-free survival (DFS), and cancer-specific survival (CSS), were included; and 5) if the information from an eligible study is not completely reported the corresponding author is contactable via email as much as possible. Additionally, for the overlapping sample data from multiple publications, only the latest publication or the most complete study was included. We mainly excluded letters, abstracts, reviews or case reports, cell or animal studies, articles lacking sufficient information or using univariate survival analysis, and articles using RNA-resequencing or microarray data from public databases.
Data Extraction and Study Quality
The following information was extracted from the available publications, including the first author’s surname, time of publication, country, ethnicity, median/mean age, cancer type, tumor stage, antibody and its sources, detection methods, cut-off values, number of patients, expression frequency, and the survival information of multivariate Cox analysis such as CSS, OS, and DFS. The Newcastle–Ottawa Scale (NOS) was used to assess the quality of the included studies for cohort design (45, 46). Three parameters of quality consisted of the selection of participants (0–4), comparability (0–2), and assessment of outcome (0–3), including a total of nine scores. A study with ≥ six scores was defined as high quality; if s study with < 6 scores was found, it was defined as low quality. Any disagreements in the literature selection and data extraction were resolved by consensus from all authors.
Prognostic Analysis From TCGA Data in GEPIA2
Gene Expression Profiling Interactive Analysis 2 (GEPIA2) database (http://gepia2.cancer-pku.cn/) was used, including numerous tumor samples from The Cancer Genome Atlas (TCGA). In our study, the relationship between TIM-3 expression and prognosis was further validated from TCGA data.
Functional Analysis of TIM-3
Association between TIM-3 and genes was analyzed using TCGA data in the GEPIA2 database. Correlation coefficients with > 0.25 were applied for TIM-3. Finally, 733 genes were significantly correlated with TIM-3 (Table S1). The GO (Gene Ontology) analysis and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways were performed to investigate the potential biological functions of the TIM-3 gene using clusterProfiler package (R software, version 3.6.1).
Statistical Analysis
Data of this meta-analysis were obtained from the original publications. The combined hazard ratios (HRs) with 95% confidence intervals (CIs) were conducted to estimate the association between TIM-3 expression and the prognosis based on multivariate Cox analysis. The heterogeneity assumption was detected using a Cochran’s Q statistic (47). The random-effects model was applied to pool data in this meta-analysis. Substantial heterogeneity was detected when a Q test (P-value) was less than 0.1. When significant heterogeneity was found, subgroup analyses were conducted according to age, ethnicity, detection method, tumor stage, tumor type, and sample sizes. A sensitivity analysis was also performed to evaluate the change of heterogeneity and stability by removing an individual study or multiple studies. Egger’s test was applied to assess the possible publication bias (48). Meta-analysis was conducted using Stata software (version 12.0, Stata Corporation, College Station, TX, US).
Results
Study Characteristics
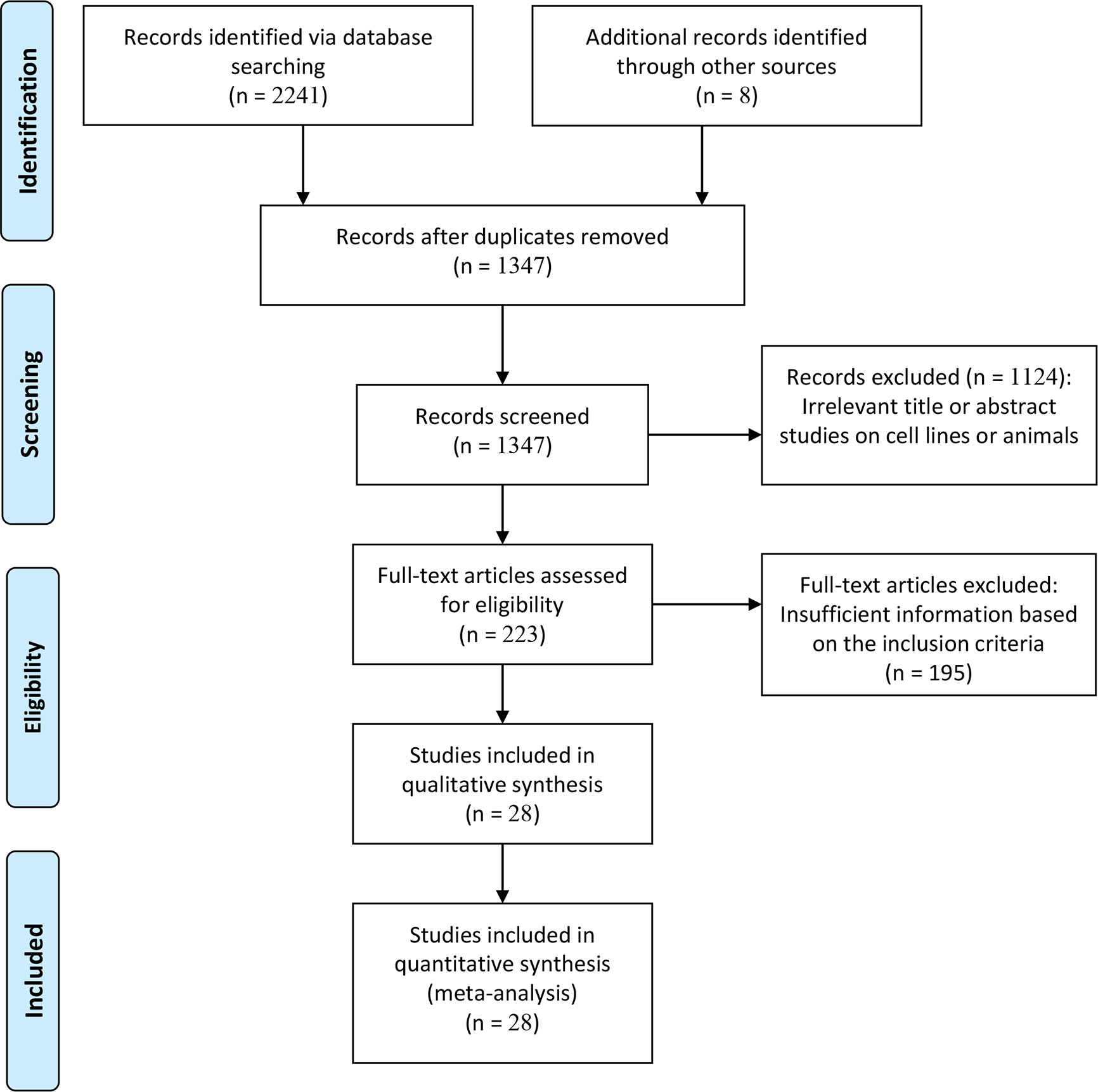
The flow diagram for the details of the study selection is presented in Figure 1. According to the inclusion criteria, inappropriate publications were excluded. Finally, 28 studies from 27 publications (23–30, 32–43, 49–55) were identified based on the multivariate Cox analysis, including 7284 patients with malignant tumors. These studies were published between 2012 and 2019. Eligible studies were conducted in China, Korea, Belgium, Canada, USA, and the Czech Republic. Various types of tumors were diagnosed, such as esophageal carcinoma, gastric cancer, sarcoma, non-small cell lung cancer, renal cell carcinoma, triple-negative breast cancer, and prostate cancer. Of these eligible studies, only three articles reported that TIM-3 expression was correlated with worse OS in cervical cancer (n= 43 cases) (54), gastric cancer (n=305 cases) (28), and colorectal cancer (n=201 cases) (50). The remaining 25 studies reported the available HR and CIs, including CSS (n = 2 studies with 3281 cases), OS (n = 22 studies with 3317 cases), and DFS (n = 7 studies with 1240 cases). All studies were considered as high quality based on NOS, with an average score of 7.9 (range, 6-9 scores). The details of the eligible publications are summarized in Table S2.
Prognostic Role of TIM-3 in Cancer
The result from 22 studies indicated that the expression of TIM-3 led to poorer OS (HR= 1.54, 95% CI = 1.19-1.98, P = 0.001) (Figure 2), including 3317 malignant tumor patients. Additionally, no significant correlation was found between TIM-3 expression and CSS (n = 2 studies with 3281 patients: HR = 1.11, 95% CI = 0.35-3.50, P = 0.863) and DFS (n = 7 studies with 1240 patients: HR = 1.61, 95% CI = 0.94-2.74, P = 0.081) (Figure 3).
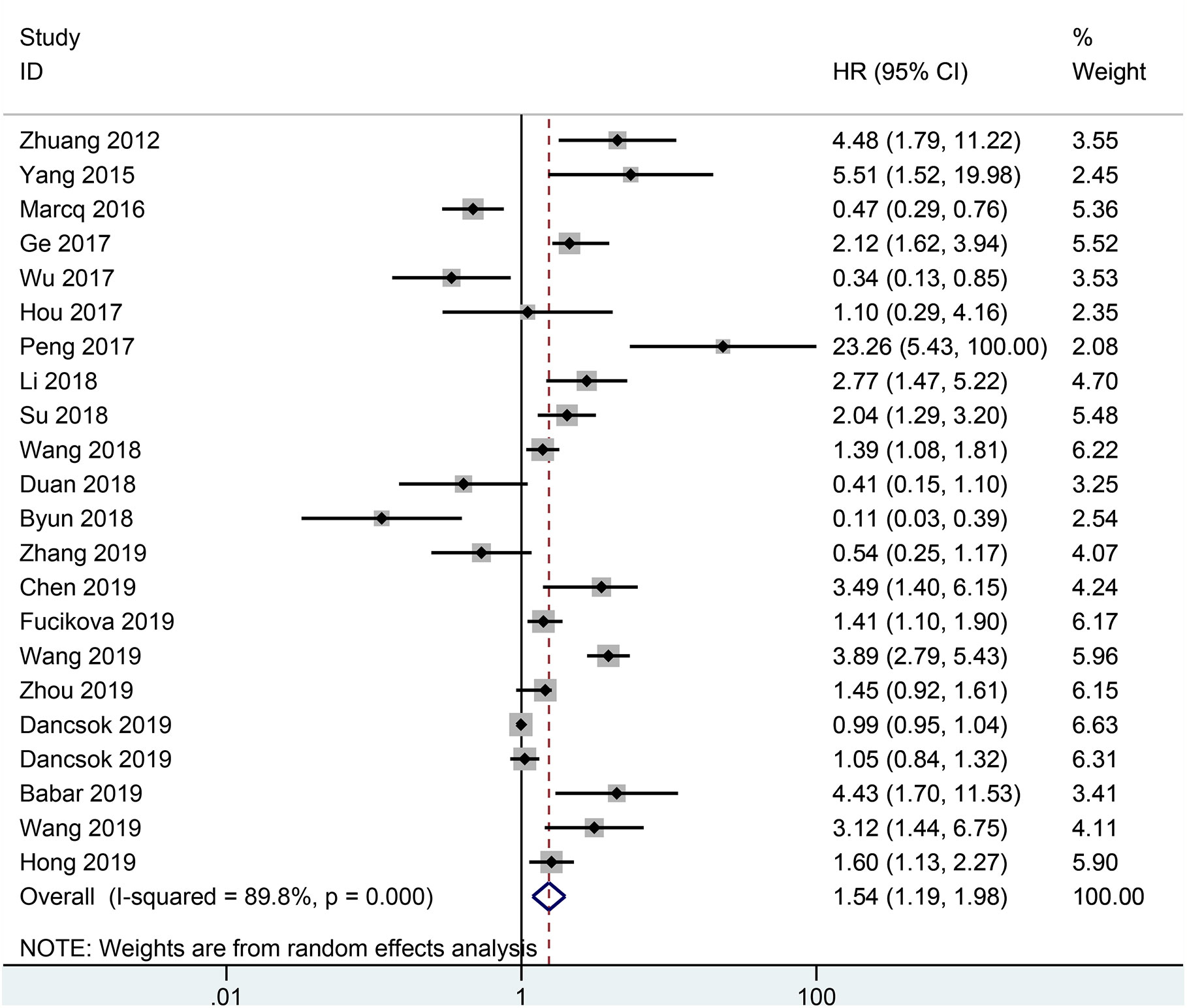
Figure 2 Forest plot of HR with 95% CI for correlation between TIM-3 expression and OS. HR, hazard ratios; CI, confidence interval; OS, overall survival.
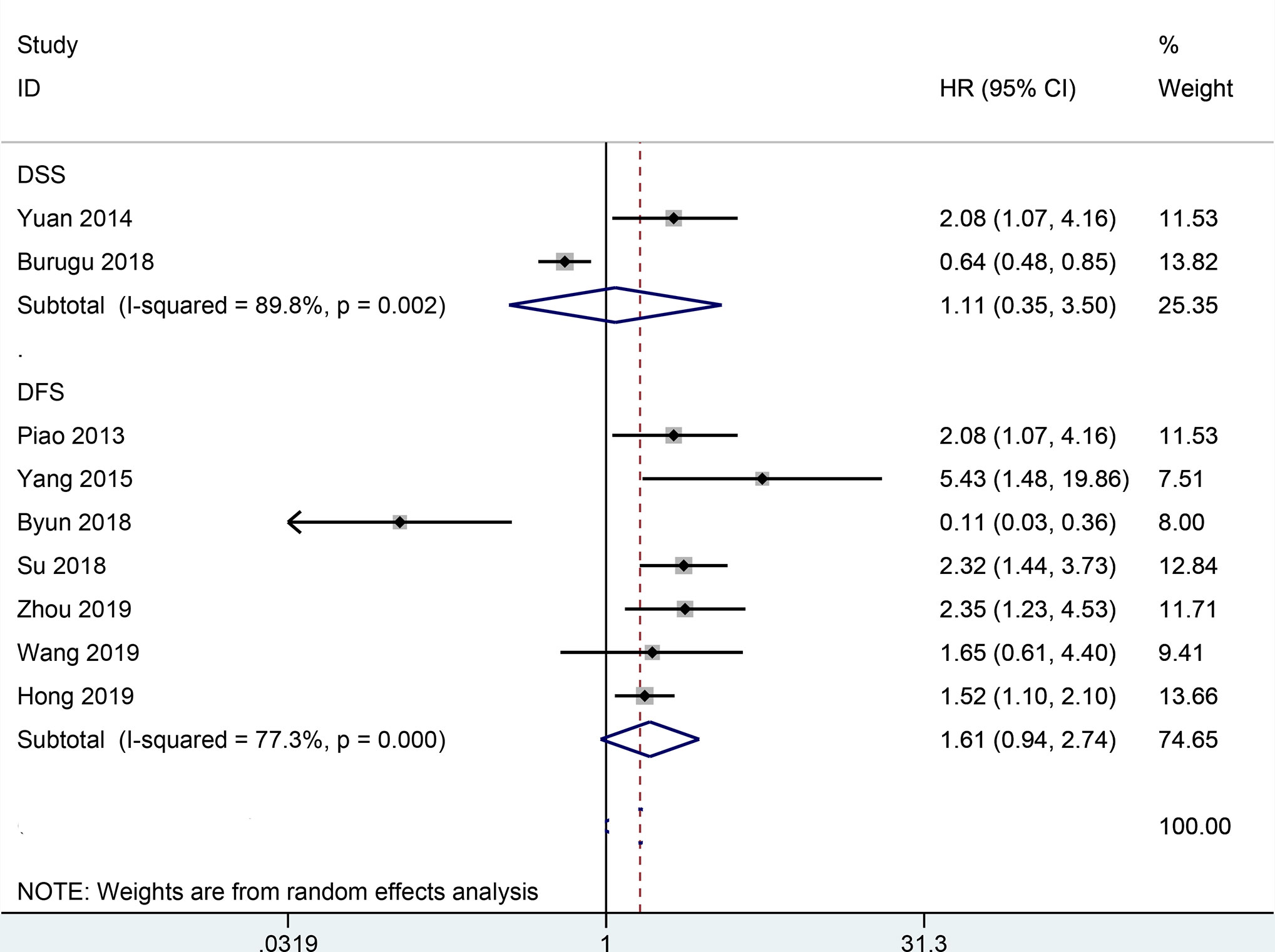
Figure 3 Forest plot of HR with 95% CI for correlation between TIM-3 expression and CSS as well as DFS. HR, hazard ratios; CI, confidence interval; CSS, cancer-specific survival; DFS, disease-free survival.
Subgroup Analyses
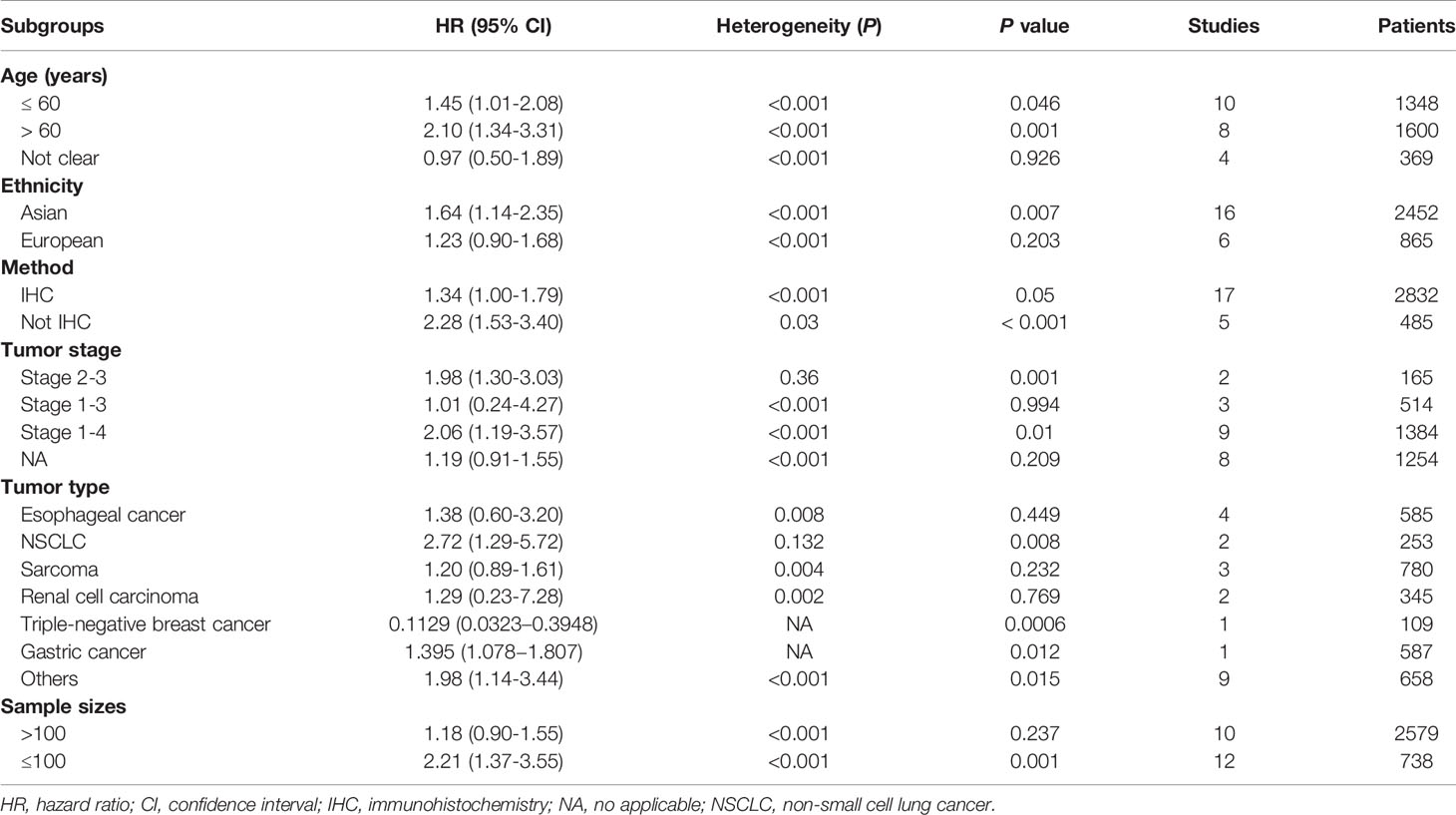
Subgroup analyses of OS were further carried out according to multiple potential factors (age, ethnicity, detection method, tumor stage tumor type, and sample sizes), which are summarized in Table 1. The results by age group showed that TIM-3 expression was correlated with worse OS in the elder age group (> 60 years: n = 8 studies with 1600 cases: HR = 2.10, 95% CI = 1.34-3.31, P = 0.001) and the younger group (≤ 60 years: n = 10 studies with 1348 cases: HR = 1.45, 95% CI = 1.01-2.08, P = 0.046). The results grouped by ethnicity showed that TIM-3 expression was associated with poor OS in Asian populations (n = 16 studies with 2452 cases: HR = 1.64, 95% CI = 1.14-2.35, P = 0.007), but not in European populations (n = 6 studies with 865 cases: P = 0.203). When stratified by detection method, immunohistochemistry (IHC) (n = 17 studies with 2832 cases: HR = 1.34, P = 0.05) and non-IHC (n = 5 studies with 485 cases: HR = 2.28, P < 0.001) were significant for survival, moreover, three articles with 549 cases reported that TIM-3 expression using IHC was correlated with worse OS (28, 50, 54), suggesting that these two methods showed a relationship with worse OS. By tumor type, TIM-3 expression was not associated with OS in esophageal carcinoma (n = 4 studies with 585 cases: P = 0.449), sarcoma (n = 3 studies with 780 cases: P = 0.232), and renal cell carcinoma (n = 2 studies with 345 cases: P = 0.769), but was related to favorable OS in triple-negative breast cancer (n = one study with 109 cases: HR = 0.1129, P = 0.0006) and prostate cancer (n = one study with 139 cases: HR = 0.336, P = 0.021); TIM-3 was related to worse OS in non-small cell lung cancer (n = 2 studies with 253 cases: HR = 2.72, P = 0.008) and gastric cancer (n = one study with 587 cases: HR = 1.395, P = 0.012).
Heterogeneity Analysis
According to the available information, we performed subgroup analyses to explore possible sources of heterogeneity. Table 1 shows the results of subgroup analyses, and we found that all P values among each subgroup were not > 0.1 for heterogeneity. Our results suggested that these factors (age, ethnicity, detection method, tumor stage, tumor type, and sample sizes) could not explain the potential heterogeneity sources. We further performed a sensitivity analysis to assess the change of the pooled results and heterogeneity. Studies by Zhuang et al. (55), Marcq et al. (49), Wu et al. (42), Peng et al. (25), Byun et al. (36), Duan et al. (29), Chen et al. (35), Babar et al. (33), Wang et al. (26), Dancsok et al. (23), and Zhang et al. (24) were omitted. The re-calculated result from the remaining studies showed that TIM-3 expression was still significantly correlated with shorter OS (HR= 1.71, 95% CI = 1.44-2.04, P < 0.001), with no heterogeneity (P = 0.102).
Publication Bias
A slight publication bias was found between TIM-3 and OS (P = 0.025), but no publication bias was detected between TIM-3 and DFS (P = 0.75) (Figure S1).
Prognostic Value of TIM-3 From TCGA Data
To further confirm the relationship between TIM-3 expression and patients’ prognosis in cancer, we conducted the survival analysis using TCGA data from the GEPIA2 database. The results demonstrated that there was a significant association between TIM-3 expression and worse OS in 9491 cancer patients (HR = 1.2, P < 0.001), but no correlation was observed between TIM-3 expression and DFS in 9491 cancer patients (P = 0.7) (Figure 4).
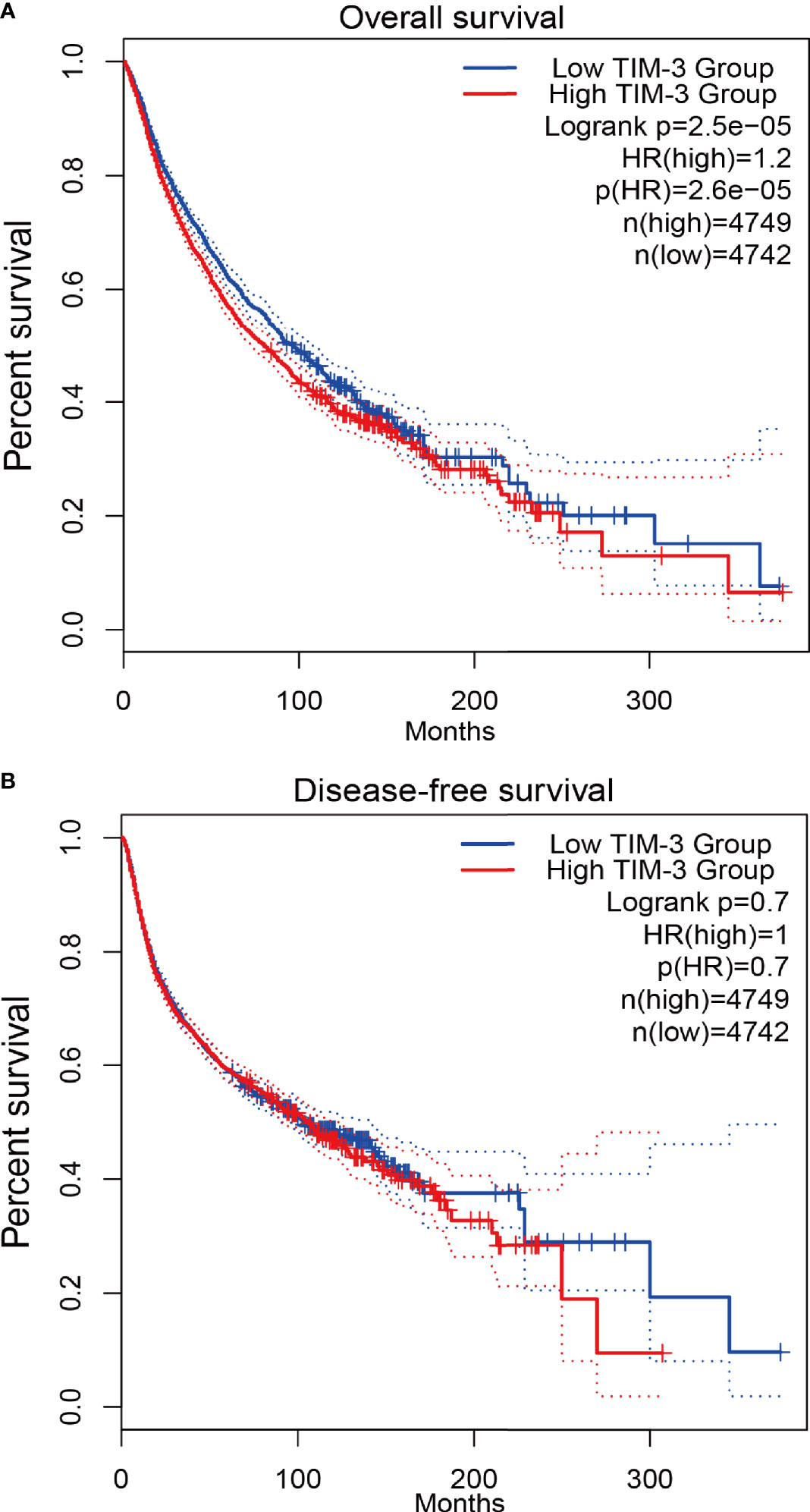
Figure 4 Survival analysis of TIM-3 expression from TCGA validation data in 9491 cancer patients. (A) Overall survival (OS). (B) Disease-free survival (DFS).
Biological Functions of the TIM-3 Gene
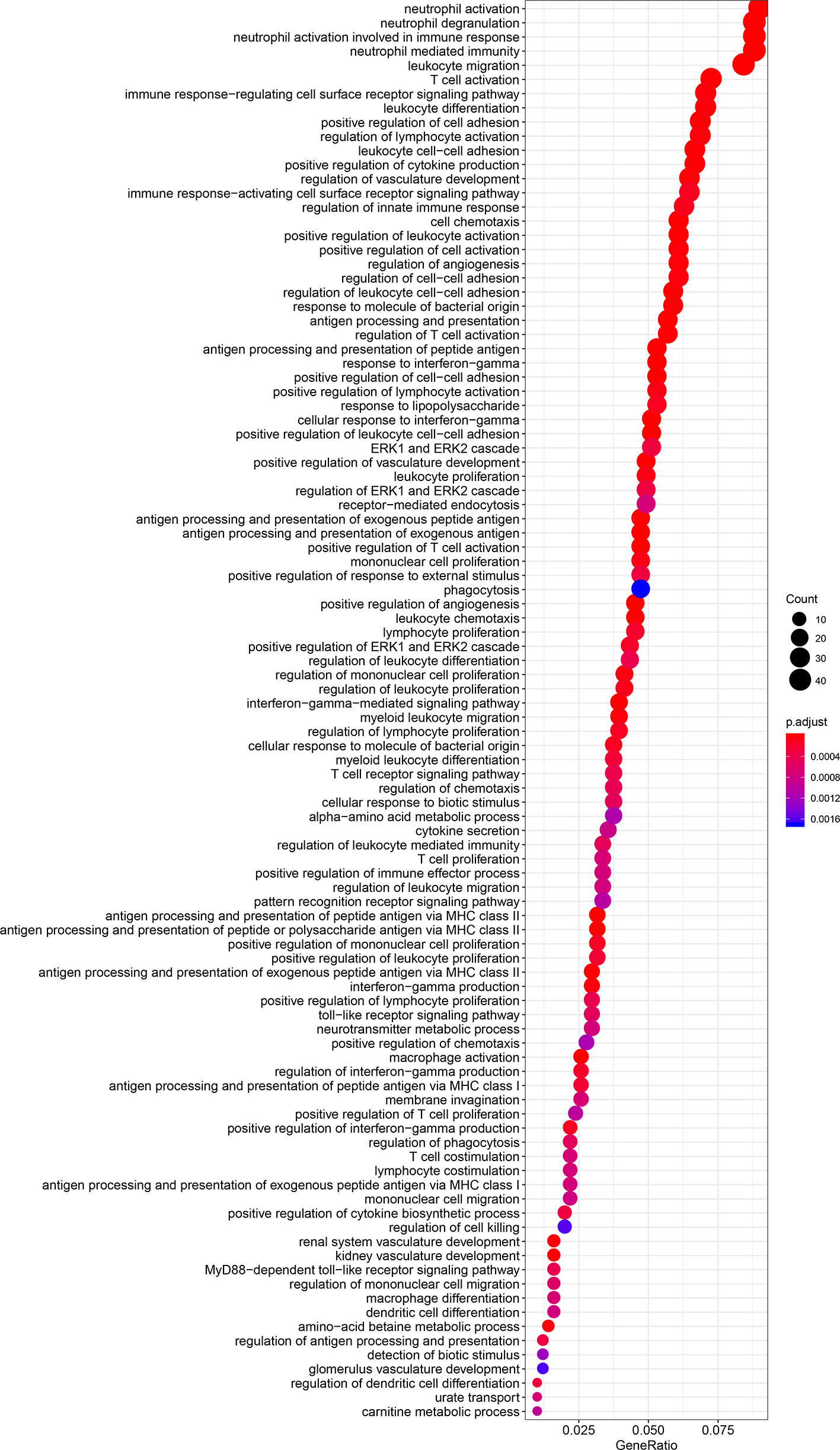
The results of GO and KEGG analyses demonstrated that TIM-3 was related to a wide variety of immune responses, such as T-cell activation, response to interferon-gamma, neutrophil-mediated immunity, macrophage differentiation, regulation of the cell surface receptor signaling pathway, innate immune response and dendritic cell differentiation, antigen processing and presentation of peptide antigen via MHC class I/II, toll−like receptor signaling, cytokine-cytokine receptor interaction, Th1 and Th17 cell differentiation, Th2 cell differentiation, and natural killer cell-mediated cytotoxicity (Figures 5 and 6). These results suggested that TIM-3 had a vital role in immune regulation in cancer.
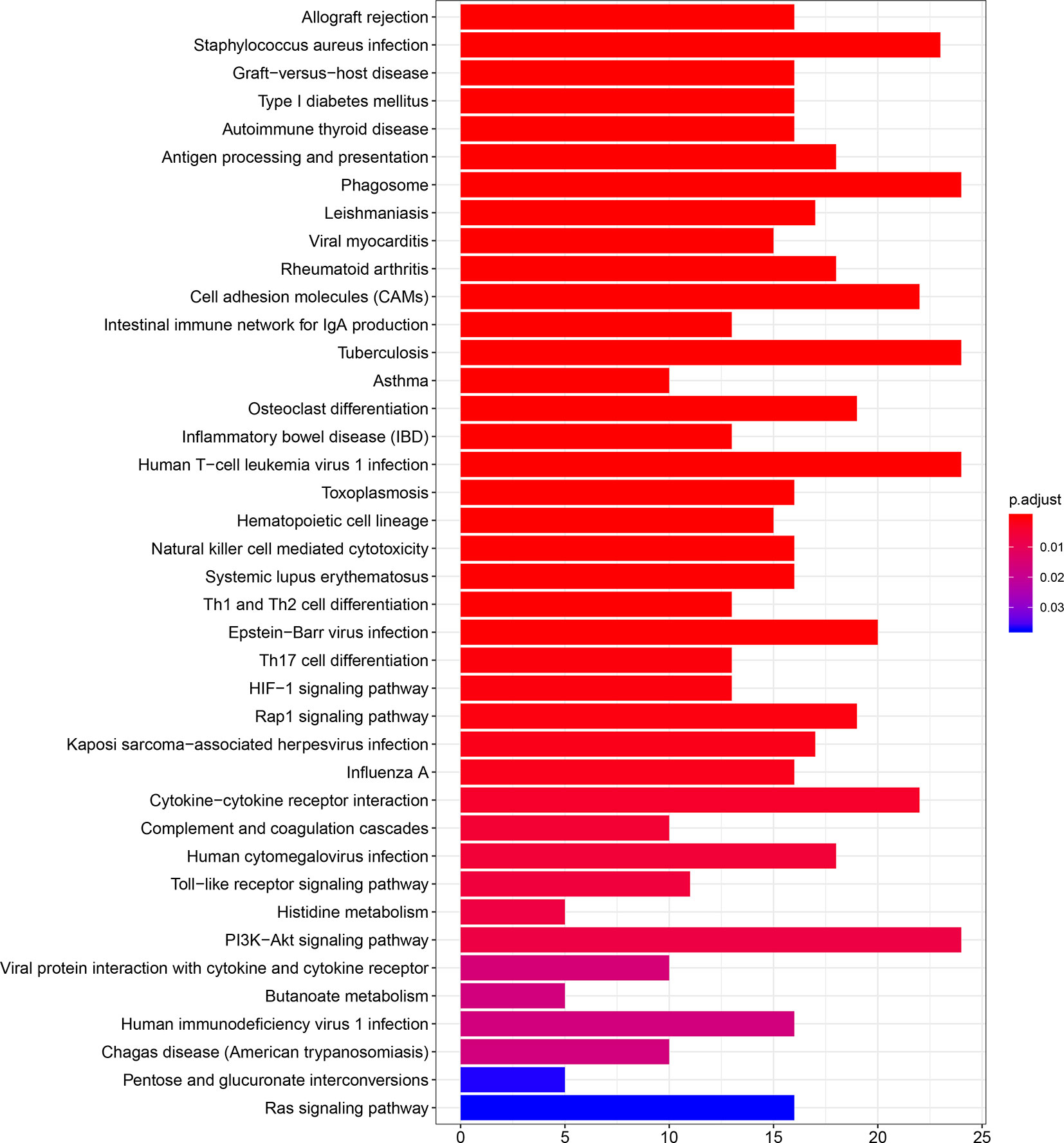
Figure 6 TIM-3 involves in immune regulation from the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways.
Discussion
Cancer is a major public health problem worldwide, with high incidence and mortality rates (1). The mechanism of tumor development is complicated. This is generally thought to be closely related to the immune system. The immune system has a crucial role in the development, progression, and metastasis of cancer (9–11, 56). Immune checkpoints are inhibitory signals used by the immune system that have a functional role in the regulation of the immune response and maintaining self-tolerance (57, 58). Immune checkpoint blockade (ICB) PD-1 and CTLA-4 have been shown as promising approaches for tumor immunotherapy and are used to treat some cancers (59). TIM-3 is identified as a crucial immune checkpoint molecule and has been demonstrated as a marker of T-cell exhaustion (20, 57). TIM-3 could also shift the immune responses by the regulation of Th1 CD4 T cells and cytotoxic CD8 T cells, which may dampen the anti-tumor immune response through modulating the T-cell activity (57, 60). TIM-3 may play an essential role in the development and progression of cancer (40, 61–63). Studies have reported that the expression of TIM-3 can be frequently detected in malignant tumors (23–25). TIM-3 expression is related to worse prognosis in some cancers, such as gastric cancer (38) and ovarian cancer (27), but is not correlated with prognosis in some cancers, such as renal cell carcinoma (24) and sarcoma (23). Therefore, the role of TIM-3 in malignant tumors is still largely uncertain. In the current work, we determined the expression of TIM-3 on the prognostic impact of patients with malignant tumors.
Fang et al. reported that TIM-3 single nucleotide polymorphisms (SNPs) were correlated with an increased cancer risk (case-control studies with 4852 participants) (64). Zhang et al. reported that TIM-3 expression was associated with shorter OS in solid tumors (n=869 patients) (31). Qin et al. reported that TIM-3 expression was relevant to poor OS in solid tumors (n=3072 patients) (65). Data of the prognostic evaluation from the previous meta-analyses included univariate and multivariate Cox analysis (31, 65). To our knowledge, the present work was a comprehensive meta-analysis of 28 studies with a very large population for the assessment of TIM-3 expression with survival in malignant tumors (n=7284 patients). Moreover, data using multivariate Cox analysis could reduce the effect of confounding factors (4). Thus, our meta-analysis only included the prognostic data based on multivariate analysis. We found that TIM-3 expression was significantly correlated with reduced OS in cancer. However, TIM-3 expression was not correlated with DFS and CSS, which were consistent with the previous publications for OS (25–28, 30, 32, 33, 35, 38–40, 43, 50, 51, 54, 55) and for DFS (32). Moreover, further TCGA data (n=9491 cases) also confirmed a significant relationship between TIM-3 expression and poor OS, and no relationship between TIM-3 expression and DFS in cancer. TIM-3 may have a function in promoting tumor cell proliferation, migration, and invasion (66, 67). TIM-3 facilitates the initiation of tumor and tumor-promoting activities (66, 68). The interactions between TIM-3 and its ligands inhibit Th1 and Th17 responses and NK cell-mediated cytotoxicity, resulting in immune tolerance (68–70). The TIM-3-galectin-9 pathway could be involved in the prevention of anti-tumor immunity (71). TIM-3 expression is correlated with resistance to PD-1 blockade in preclinical models (72). TIM-3 regulates immune responses in cancer (20). We also observed a similar finding; TIM-3 was closely related to immune regulation, such as T-cell activation, response to interferon-gamma, neutrophil-mediated immunity, macrophage differentiation, regulation of the cell surface receptor signaling pathway, innate immune response and dendritic cell differentiation, antigen processing and presentation of peptide antigen via MHC class I/II, toll−like receptor signaling, cytokine-cytokine receptor interaction, Th1, Th17 and Th2 cell differentiation, and natural killer cell-mediated cytotoxicity. These analyses suggest that TIM-3 could become an independent prognostic marker for predicting worse OS, and targeting TIM-3 is a potentially effective approach for cancer immunotherapy.
Although no association was found between TIM-3 expression and CSS in our work, the result should be considered with caution. Among these two studies, Yuan 2014 et al. reported that TIM-3 expression was correlated with worse CSS in clear cell renal cell carcinoma (n=137 cases) (52), but Burugu 2018 et al. reported that TIM-3 expression was related to favorable CSS in a large cohort of breast cancer (> 3000 cases) (37). Additionally, we also observed similar findings in other prognostic endpoints. TIM-3 expression was associated with worse OS in clear cell renal cell carcinoma (n= 182 cases) by Wang et al. (32). TIM-3 expression was correlated with better OS and DFS in triple-negative breast cancer (n= 109 cases) by Byun et al. (36). These results showed that TIM-3 might be an independent positive prognostic factor in breast cancer and be an independent negative prognostic factor in clear cell renal cell carcinoma. Further relevant studies are necessary to confirm these findings in the future.
Stratification by age showed that TIM-3 was significantly correlated with poor OS in the elder group (P = 0.001). A slight association between TIM-3 and OS was observed in the younger group (P = 0.046). Some studies demonstrate an age effect going in different directions (73–75). For example, Zhang et al. reported that age (the elder group) was associated with a poorer prognosis (73). In the future, additional studies with larger sample sizes are essential to further confirm whether TIM-3 is closely correlated with worse prognosis in younger patients. Subgroup analysis by ethnicity demonstrated that TIM-3 expression was related to worse OS in Asian populations, but not in European populations. When stratified by tumor type, TIM-3 expression was not associated with OS in esophageal carcinoma, sarcoma, and renal cell carcinoma, but was correlated with favorable OS in triple-negative breast cancer; TIM-3 was related to worse OS in non-small cell lung cancer and gastric cancer. Evidence from some previous publications is consistent with the present results, such as esophageal carcinoma (no association) (29, 41), sarcoma (no association) (23), renal cell carcinoma (no association) (24), non-small cell lung cancer (worse OS) (39, 55), gastric cancer (worse OS) (28, 38), and breast cancer (better CSS) (37). We found the correlation between TIM-3 expression and worse prognosis among most cancers, such as lung, gastric, cervical, ovarian, colorectal, bladder, pancreatic, hepatocellular, and oral squamous cell carcinomas, as well as diffuse large B-cell lymphoma. Moreover, there was an association between TIM-3 expression and favorable prognosis among several cancers, such as malignant pleural mesothelioma and breast cancer, suggesting that TIM-3 may be either a negative prognostic factor or a positive prognostic factor based on different tumor types. Studies show TIM-3 regulates immune responses, possibly exerting either positive or negative effects (22). Thus, more studies are necessary among different tumor types in the future.
The current study had some limitations. First, most studies (n=22) used Asian participants; the remaining six studies used Europeans. Other ethnic groups, such as Africans, were inadequately represented. Second, slight publication bias was measured between TIM-3 and OS (P = 0.025), possibly because three publications only reported positive results, but did not provide available HR values with 95% CI. Thus, the pooled HR was not included in these three publications. Third, although subgroup analyses and sensitivity analyses were performed to explore the potential sources of heterogeneity, factors such as age, ethnicity, detection method, tumor stage, tumor type, and sample sizes failed to explain the possible heterogeneity sources. The detailed reasons for heterogeneity were not very certain. The unavoidable reasons, such as different or unclear cut-off values of TIM-3 expression and different follow-up time points, may cause the potential sources of heterogeneity. Fourthly, only one study used a prospective design (32); more prospective studies are needed.
Although our study had some limitations, this work was still the largest meta-analysis that incorporated 28 studies with over 7000 patients with malignant tumors. Moreover, over 9000 cancer patients from TCGA data were also applied to confirm our results.
The present study provided more evidence that TIM-3 expression was significantly associated with worse OS, and it might be a useful prognosticator in malignant tumors. Biological functions also showed that TIM-3 played a key role in immune regulation. Immune checkpoint TIM-3 could be a valuable immunotherapy target. Future prospective studies are required to validate the results.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
LH and KZ provided the study conception and design. KZ and LH contributed to the drafting of the article and final approval of the submitted version. All authors provided the analyses and interpretation of the data and completion of figures and tables. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge The Cancer Genome Atlas.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.579351/full#supplementary-material
Supplementary Figure 1 | Publication bias by using Egger’s test.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Han S, Huang T, Hou F, Yao L, Wang X, Wu X. The prognostic value of hypoxia-inducible factor-1alpha in advanced cancer survivors: a meta-analysis with trial sequential analysis. Ther Adv Med Oncol (2019) 11:1758835919875851. doi: 10.1177/1758835919875851
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin (2019) 69(1):7–34. doi: 10.3322/caac.21551
4. Han S, Huang T, Wu X, Wang X, Li W, Liu S, et al. Prognostic value of ALDH1 and Nestin in advanced cancer: a systematic meta-analysis with trial sequential analysis. Ther Adv Med Oncol (2019) 11:1758835919830831. doi: 10.1177/1758835919830831
5. Han S, Huang T, Li W, Wang X, Wu X, Liu S, et al. Prognostic Value of CD44 and Its Isoforms in Advanced Cancer: A Systematic Meta-Analysis With Trial Sequential Analysis. Front Oncol (2019) 9:39:39. doi: 10.3389/fonc.2019.00039
6. Luo D, Liu H, Lin D, Lian K, Ren H. The Clinicopathologic and Prognostic Value of Hypoxia-Inducible Factor-2alpha in Cancer Patients: A Systematic Review and Meta-Analysis. Cancer Epidemiol Biomarkers Prev (2019) 28(5):857–66. doi: 10.1158/1055-9965.EPI-18-0881
7. Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med (2015) 21(8):938–45. doi: 10.1038/nm.3909
8. Ocana A, Vera-Badillo F, Seruga B, Templeton A, Pandiella A, Amir E. HER3 overexpression and survival in solid tumors: a meta-analysis. J Natl Cancer Inst (2013) 105(4):266–73. doi: 10.1093/jnci/djs501
9. Aragon-Sanabria V, Kim GB, Dong C. From Cancer Immunoediting to New Strategies in Cancer Immunotherapy: The Roles of Immune Cells and Mechanics in Oncology. Adv Exp Med Biol (2018) 1092:113–38. doi: 10.1007/978-3-319-95294-9_7
10. Janssen LME, Ramsay EE, Logsdon CD, Overwijk WW. The immune system in cancer metastasis: friend or foe? J Immunother Cancer (2017) 5(1):79. doi: 10.1186/s40425-017-0283-9
11. Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol (2015) 15(2):73–86. doi: 10.1038/nri3789
12. Sanmamed MF, Chen L. A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell (2019) 176(3):677. doi: 10.1016/j.cell.2019.01.008
13. Riley RS, June CH, Langer R, Mitchell MJ. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discovery (2019) 18(3):175–96. doi: 10.1038/s41573-018-0006-z
14. Lecerf C, Kamal M, Vacher S, Chemlali W, Schnitzler A, Morel C, et al. Immune gene expression in head and neck squamous cell carcinoma patients. Eur J Cancer (2019) 121:210–23. doi: 10.1016/j.ejca.2019.08.028
15. Yeong J, Lim JCT, Lee B, Li H, Ong CCH, Thike AA, et al. Prognostic value of CD8 + PD-1+ immune infiltrates and PDCD1 gene expression in triple negative breast cancer. J Immunother Cancer (2019) 7(1):34. doi: 10.1186/s40425-019-0499-y
16. Ma LJ, Feng FL, Dong LQ, Zhang Z, Duan M, Liu LZ, et al. Clinical significance of PD-1/PD-Ls gene amplification and overexpression in patients with hepatocellular carcinoma. Theranostics (2018) 8(20):5690–702. doi: 10.7150/thno.28742
17. Aghajani M, Graham S, McCafferty C, Shaheed CA, Roberts T, DeSouza P, et al. Clinicopathologic and Prognostic Significance of Programmed Cell Death Ligand 1 Expression in Patients with Non-Medullary Thyroid Cancer: A Systematic Review and Meta-Analysis. Thyroid (2018) 28(3):349–61. doi: 10.1089/thy.2017.0441
18. Goswami S, Walle T, Cornish AE, Basu S, Anandhan S, Fernandez I, et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat Med (2020) 26(1):39–46. doi: 10.1038/s41591-019-0694-x
19. Phong BL, Avery L, Sumpter TL, Gorman JV, Watkins SC, Colgan JD, et al. Tim-3 enhances FcepsilonRI-proximal signaling to modulate mast cell activation. J Exp Med (2015) 212(13):2289–304. doi: 10.1084/jem.20150388
20. Das M, Zhu C, Kuchroo VK. Tim-3 and its role in regulating anti-tumor immunity. Immunol Rev (2017) 276(1):97–111. doi: 10.1111/imr.12520
21. Freeman GJ, Casasnovas JM, Umetsu DT, DeKruyff RH. TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunol Rev (2010) 235(1):172–89. doi: 10.1111/j.0105-2896.2010.00903.x
22. Solinas C, De Silva P, Bron D, Willard-Gallo K, Sangiolo D. Significance of TIM3 expression in cancer: From biology to the clinic. Semin Oncol (2019) 46(4-5):372–9. doi: 10.1053/j.seminoncol.2019.08.005
23. Dancsok AR, Setsu N, Gao D, Blay JY, Thomas D, Maki RG, et al. Expression of lymphocyte immunoregulatory biomarkers in bone and soft-tissue sarcomas. Mod Pathol (2019) 32(12):1772–85. doi: 10.1038/s41379-019-0312-y
24. Zhang X, Yin X, Zhang H, Sun G, Yang Y, Chen J, et al. Differential expression of TIM-3 between primary and metastatic sites in renal cell carcinoma. BMC Cancer (2019) 19(1):49. doi: 10.1186/s12885-019-5273-5
25. Peng PJ, Li Y, Sun S. On the significance of Tim-3 expression in pancreatic cancer. Saudi J Biol Sci (2017) 24(8):1754–7. doi: 10.1016/j.sjbs.2017.11.006
26. Wang H, Mao L, Zhang T, Zhang L, Wu Y, Guo W, et al. Altered expression of TIM-3, LAG-3, IDO, PD-L1, and CTLA-4 during nimotuzumab therapy correlates with responses and prognosis of oral squamous cell carcinoma patients. J Oral Pathol Med (2019) 48(8):669–76. doi: 10.1111/jop.12883
27. Fucikova J, Rakova J, Hensler M, Kasikova L, Belicova L, Hladikova K, et al. TIM-3 Dictates Functional Orientation of the Immune Infiltrate in Ovarian Cancer. Clin Cancer Res (2019) 25(15):4820–31. doi: 10.1158/1078-0432.CCR-18-4175
28. Jiang J, Jin MS, Kong F, Cao D, Ma HX, Jia Z, et al. Decreased galectin-9 and increased Tim-3 expression are related to poor prognosis in gastric cancer. PloS One (2013) 8(12):e81799. doi: 10.1371/journal.pone.0081799
29. Duan J, Xie Y, Qu L, Wang L, Zhou S, Wang Y, et al. A nomogram-based immunoprofile predicts overall survival for previously untreated patients with esophageal squamous cell carcinoma after esophagectomy. J Immunother Cancer (2018) 6(1):100. doi: 10.1186/s40425-018-0418-7
30. Hong MH, Shin SJ, Shin SK, Kim DJ, Zo JI, Shim YM, et al. High CD3 and ICOS and low TIM-3 expression predict favourable survival in resected oesophageal squamous cell carcinoma. Sci Rep (2019) 9(1):20197. doi: 10.1038/s41598-019-56828-7
31. Zhang Y, Cai P, Liang T, Wang L, Hu L. TIM-3 is a potential prognostic marker for patients with solid tumors: A systematic review and meta-analysis. Oncotarget (2017) 8(19):31705–13. doi: 10.18632/oncotarget.15954
32. Wang Q, Zhang J, Tu H, Liang D, Chang DW, Ye Y, et al. Soluble immune checkpoint-related proteins as predictors of tumor recurrence, survival, and T cell phenotypes in clear cell renal cell carcinoma patients. J Immunother Cancer (2019) 7(1):334. doi: 10.1186/s40425-019-0810-y
33. Babar L, Kosovec JE, Jahangiri V, Chowdhury N, Zheng P, Omstead AN, et al. Prognostic immune markers for recurrence and survival in locally advanced esophageal adenocarcinoma. Oncotarget (2019) 10(44):4546–55. doi: 10.18632/oncotarget.27052
34. Zhou J, Jiang Y, Zhang H, Chen L, Luo P, Li L, et al. Clinicopathological implications of TIM3(+) tumor-infiltrating lymphocytes and the miR-455-5p/Galectin-9 axis in skull base chordoma patients. Cancer Immunol Immunother (2019) 68(7):1157–69. doi: 10.1007/s00262-019-02349-1
35. Chen BJ, Dashnamoorthy R, Galera P, Makarenko V, Chang H, Ghosh S, et al. The immune checkpoint molecules PD-1, PD-L1, TIM-3 and LAG-3 in diffuse large B-cell lymphoma. Oncotarget (2019) 10(21):2030–40. doi: 10.18632/oncotarget.26771
36. Byun KD, Hwang HJ, Park KJ, Kim MC, Cho SH, Ju MH, et al. T-Cell Immunoglobulin Mucin 3 Expression on Tumor Infiltrating Lymphocytes as a Positive Prognosticator in Triple-Negative Breast Cancer. J Breast Cancer (2018) 21(4):406–14. doi: 10.4048/jbc.2018.21.e61
37. Burugu S, Gao D, Leung S, Chia SK, Nielsen TO. TIM-3 expression in breast cancer. Oncoimmunology (2018) 7(11):e1502128. doi: 10.1080/2162402X.2018.1502128
38. Wang Y, Zhao E, Zhang Z, Zhao G, Cao H. Association between Tim3 and Gal9 expression and gastric cancer prognosis. Oncol Rep (2018) 40(4):2115–26. doi: 10.3892/or.2018.6627
39. Su H, Xie H, Dai C, Ren Y, She Y, Xu L, et al. Characterization of TIM-3 expression and its prognostic value in patients with surgically resected lung adenocarcinoma. Lung Cancer (2018) 121:18–24. doi: 10.1016/j.lungcan.2018.04.009
40. Li F, Li N, Sang J, Fan X, Deng H, Zhang X, et al. Highly elevated soluble Tim-3 levels correlate with increased hepatocellular carcinoma risk and poor survival of hepatocellular carcinoma patients in chronic hepatitis B virus infection. Cancer Manag Res (2018) 10:941–51. doi: 10.2147/CMAR.S162478
41. Hou N, Ma J, Li W, Zhao L, Gao Q, Mai L. T-cell immunoglobulin and mucin domain-containing protein-3 and galectin-9 protein expression: Potential prognostic significance in esophageal squamous cell carcinoma for Chinese patients. Oncol Lett (2017) 14(6):8007–13. doi: 10.3892/ol.2017.7188
42. Wu J, Lin G, Zhu Y, Zhang H, Shi G, Shen Y, et al. Low TIM3 expression indicates poor prognosis of metastatic prostate cancer and acts as an independent predictor of castration resistant status. Sci Rep (2017) 7(1):8869. doi: 10.1038/s41598-017-09484-8
43. Ge W, Li J, Fan W, Xu D, Sun S. Tim-3 as a diagnostic and prognostic biomarker of osteosarcoma. Tumour Biol (2017) 39(7):1010428317715643. doi: 10.1177/1010428317715643
44. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PloS Med (2009) 6(7):e1000097. doi: 10.1371/journal.pmed.1000097
45. Piovezan JM, Premaor MO, Comim FV. Negative impact of polycystic ovary syndrome on bone health: a systematic review and meta-analysis. Hum Reprod Update (2019) 25(5):633–45. doi: 10.1093/humupd/dmz020
46. Kantor R, Kim A, Thyssen JP, Silverberg JI. Association of atopic dermatitis with smoking: A systematic review and meta-analysis. J Am Acad Dermatol (2016) 75(6):1119–25 e1. doi: 10.1016/j.jaad.2016.07.017
47. Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics (2005) 21(18):3672–3. doi: 10.1093/bioinformatics/bti536
48. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (1997) 315(7109):629–34. doi: 10.1136/bmj.315.7109.629
49. Marcq E, Siozopoulou V, De Waele J, van Audenaerde J, Zwaenepoel K, Santermans E, et al. Prognostic and predictive aspects of the tumor immune microenvironment and immune checkpoints in malignant pleural mesothelioma. Oncoimmunology (2017) 6(1):e1261241. doi: 10.1080/2162402X.2016.1261241
50. Zhou E, Huang Q, Wang J, Fang C, Yang L, Zhu M, et al. Up-regulation of Tim-3 is associated with poor prognosis of patients with colon cancer. Int J Clin Exp Pathol (2015) 8(7):8018–27.
51. Yang M, Yu Q, Liu J, Fu W, Cao Y, Yu L, et al. T-cell immunoglobulin mucin-3 expression in bladder urothelial carcinoma: Clinicopathologic correlations and association with survival. J Surg Oncol (2015) 112(4):430–5. doi: 10.1002/jso.24012
52. Yuan J, Jiang B, Zhao H, Huang Q. Prognostic implication of TIM-3 in clear cell renal cell carcinoma. Neoplasma (2014) 61(1):35–40. doi: 10.4149/neo_2014_006
53. Piao YR, Piao LZ, Zhu LH, Jin ZH, Dong XZ. Prognostic value of T cell immunoglobulin mucin-3 in prostate cancer. Asian Pac J Cancer Prev (2013) 14(6):3897–901. doi: 10.7314/apjcp.2013.14.6.3897
54. Cao Y, Zhou X, Huang X, Li Q, Gao L, Jiang L, et al. Tim-3 expression in cervical cancer promotes tumor metastasis. PloS One (2013) 8(1):e53834. doi: 10.1371/journal.pone.0053834
55. Zhuang X, Zhang X, Xia X, Zhang C, Liang X, Gao L, et al. Ectopic expression of TIM-3 in lung cancers: a potential independent prognostic factor for patients with NSCLC. Am J Clin Pathol (2012) 137(6):978–85. doi: 10.1309/AJCP9Q6OVLVSHTMY
56. Muenst S, Laubli H, Soysal SD, Zippelius A, Tzankov A, Hoeller S. The immune system and cancer evasion strategies: therapeutic concepts. J Intern Med (2016) 279(6):541–62. doi: 10.1111/joim.12470
57. Borcherding N, Kolb R, Gullicksrud J, Vikas P, Zhu Y, Zhang W. Keeping Tumors in Check: A Mechanistic Review of Clinical Response and Resistance to Immune Checkpoint Blockade in Cancer. J Mol Biol (2018) 430(14):2014–29. doi: 10.1016/j.jmb.2018.05.030
58. Lee L, Gupta M, Sahasranaman S. Immune Checkpoint inhibitors: An introduction to the next-generation cancer immunotherapy. J Clin Pharmacol (2016) 56(2):157–69. doi: 10.1002/jcph.591
59. Soularue E, Lepage P, Colombel JF, Coutzac C, Faleck D, Marthey L, et al. Enterocolitis due to immune checkpoint inhibitors: a systematic review. Gut (2018) 67(11):2056–67. doi: 10.1136/gutjnl-2018-316948
60. Sabins NC, Chornoguz O, Leander K, Kaplan F, Carter R, Kinder M, et al. TIM-3 Engagement Promotes Effector Memory T Cell Differentiation of Human Antigen-Specific CD8 T Cells by Activating mTORC1. J Immunol (2017) 199(12):4091–102. doi: 10.4049/jimmunol.1701030
61. Zhang H, Xiang R, Wu B, Li J, Luo G. T-cell immunoglobulin mucin-3 expression in invasive ductal breast carcinoma: Clinicopathological correlations and association with tumor infiltration by cytotoxic lymphocytes. Mol Clin Oncol (2017) 7(4):557–63. doi: 10.3892/mco.2017.1360
62. Kikushige Y, Miyamoto T, Yuda J, Jabbarzadeh-Tabrizi S, Shima T, Takayanagi S, et al. A TIM-3/Gal-9 Autocrine Stimulatory Loop Drives Self-Renewal of Human Myeloid Leukemia Stem Cells and Leukemic Progression. Cell Stem Cell (2015) 17(3):341–52. doi: 10.1016/j.stem.2015.07.011
63. Komohara Y, Morita T, Annan DA, Horlad H, Ohnishi K, Yamada S, et al. The Coordinated Actions of TIM-3 on Cancer and Myeloid Cells in the Regulation of Tumorigenicity and Clinical Prognosis in Clear Cell Renal Cell Carcinomas. Cancer Immunol Res (2015) 3(9):999–1007. doi: 10.1158/2326-6066.CIR-14-0156
64. Fang H, Yuan C, Gu X, Chen Q, Huang D, Li H, et al. Association between TIM-3 polymorphisms and cancer risk: a meta-analysis. Ann Transl Med (2019) 7(20):550. doi: 10.21037/atm.2019.09.101
65. Qin S, Dong B, Yi M, Chu Q, Wu K. Prognostic Values of TIM-3 Expression in Patients With Solid Tumors: A Meta-Analysis and Database Evaluation. Front Oncol (2020) 10:1288. doi: 10.3389/fonc.2020.01288
66. Cong Y, Cui Y, Zhu S, Cao J, Zou H, Martin TA, et al. Tim-3 promotes cell aggressiveness and paclitaxel resistance through NF-kappaB/STAT3 signalling pathway in breast cancer cells. Chin J Cancer Res (2020) 32(5):564–79. doi: 10.21147/j.issn.1000-9604.2020.05.02
67. Xiao Y, Qing J, Li B, Chen L, Nong S, Yang W, et al. TIM-3 Participates in the Invasion and Metastasis of Nasopharyngeal Carcinoma via SMAD7/SMAD2/SNAIL1 Axis-Mediated Epithelial-Mesenchymal Transition. Onco Targets Ther (2020) 13:1993–2006. doi: 10.2147/OTT.S237222
68. Saleh R, Toor SM, Elkord E. Targeting TIM-3 in solid tumors: innovations in the preclinical and translational realm and therapeutic potential. Expert Opin Ther Targets (2020) 24(12):1251–62. doi: 10.1080/14728222.2020.1841750
69. Zhao J, Lin B, Deng H, Zhi X, Li Y, Liu Y, et al. Decreased Expression of TIM-3 on Th17 Cells Associated with Ophthalmopathy in Patients with Graves’ Disease. Curr Mol Med (2018) 18(2):83–90. doi: 10.2174/1566524018666180705105753
70. Ndhlovu LC, Lopez-Verges S, Barbour JD, Jones RB, Jha AR, Long BR, et al. Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity. Blood (2012) 119(16):3734–43. doi: 10.1182/blood-2011-11-392951
71. Yasinska IM, Sakhnevych SS, Pavlova L, Teo Hansen Selno A, Teuscher Abeleira AM, Benlaouer O, et al. The Tim-3-Galectin-9 Pathway and Its Regulatory Mechanisms in Human Breast Cancer. Front Immunol (2019) 10:1594. doi: 10.3389/fimmu.2019.01594
72. Barrueto L, Caminero F, Cash L, Makris C, Lamichhane P, Deshmukh RR. Resistance to Checkpoint Inhibition in Cancer Immunotherapy. Transl Oncol (2020) 13(3):100738. doi: 10.1016/j.tranon.2019.12.010
73. Zhang H, Han H, He T, Labbe KE, Hernandez AV, Chen H, et al. Clinical Characteristics and Outcomes of COVID-19-Infected Cancer Patients: A Systematic Review and Meta-Analysis. J Natl Cancer Inst (2020) djaa168:1–10. doi: 10.1093/jnci/djaa168
74. Li Q, Huang LY, Xue HP. Comparison of prognostic factors in different age groups and prognostic significance of neutrophil-lymphocyte ratio in patients with gastric cancer. World J Gastrointest Oncol (2020) 12(10):1146–66. doi: 10.4251/wjgo.v12.i10.1146
Keywords: immune checkpoint, TIM-3, tumor, prognosis, immune response
Citation: Zang K, Hui L, Wang M, Huang Y, Zhu X and Yao B (2021) TIM-3 as a Prognostic Marker and a Potential Immunotherapy Target in Human Malignant Tumors: A Meta-Analysis and Bioinformatics Validation. Front. Oncol. 11:579351. doi: 10.3389/fonc.2021.579351
Received: 18 August 2020; Accepted: 22 January 2021;
Published: 22 February 2021.
Edited by:
Pedro Berraondo, Cima Universidad de Navarra, SpainReviewed by:
Mario Ostrowski, University of Toronto, CanadaFernando Aranda, Instituto de Investigación Sanitaria de Navarra (IdiSNA), Spain
Copyright © 2021 Zang, Hui, Wang, Huang, Zhu and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangliang Hui, aHVpbGlhbmdsaWFuZ0AxNjMuY29t
 Kui Zang
Kui Zang Liangliang Hui
Liangliang Hui