- 1Department of Pathology and Pathophysiology, and Department of Surgical Oncology of the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Key Laboratory of Disease Proteomics of Zhejiang Province, Zhejiang University School of Medicine, Hangzhou, China
- 3Cancer Institute of Integrative Medicine, Zhejiang Academy of Traditional Chinese Medicine, Tongde Hospital of Zhejiang Province, Hangzhou, China
- 4Department of Pathophysiology, Zunyi Medical University, Zunyi, China
Background: E-cadherin, a hallmark of epithelial-mesenchymal transition (EMT), is often repressed due to Snail-mediated epigenetic modification; however, the exact mechanism remains unclear. There is an urgent need to understand the determinants of tumor aggressiveness and identify potential therapeutic targets in breast cancer.
Experimental design: We studied the association of RNF20 with Snail and G9a by co-immunoprecipitation. We employed quantitative real-time PCR, ChIP, transwell assay, colony formation assay, and mammosphere assay to dissect the molecular events associated with the repression of E-cadherin in human breast cancer. We used a proteogenomic dataset that contains 105 breast tumor samples to determine the clinical relevance of RNF20 by Kaplan-Meier analyses.
Results: In this study, we identified that Snail interacted with RNF20, an E3 ubiquitin-protein ligase responsible for monoubiquitination of H2BK120, and G9a, a methyltransferase for H3K9me2. RNF20 expression led to the inhibition of E-cadherin expression in the human breast cancer cells. Mechanically, we showed that RNF20 and H3K9m2 were enriched on the promoter of E-cadherin and knockdown of Snail reduced the enrichment of RNF20, showing a Snail-dependent manner. RNF20 expression enhanced breast cancer cell migration, invasion, tumorsphere and colony formation. Clinically, patients with high RNF20 expression had shorter overall survival.
Conclusion: RNF20 expression contributes to EMT induction and breast cancer progression through Snail-mediated epigenetic suppression of E-cadherin expression, suggesting the importance of RNF20 in breast cancer.
Introduction
Breast cancer metastasis is a major problem that causes high mortality in cancer patients (1). About 90% of cancer deaths result from the local invasion and distant metastasis of tumor cells (2–5). Therefore, a better understanding of the molecular events that contribute to tumor invasiveness is crucial to the development of therapeutic strategies.
EMT is a complex process in which epithelial cells lose contact with their neighbors, and possess the phenotype of migratory mesenchymal cells during in embryonic development, wound healing, tissue remodeling and tumor metastasis (6, 7). EMT promotes tumor progression by endowing tumor cells with the properties of CSCs. E-cadherin loss is a hallmark of EMT (5). Snail, a crucial regulators of E-cadherin, can downregulate the expression of E-cadherin by mediating histone modification (6). Histone modifications are crucial in regulating basic processes including transcription, cell differentiation, cell cycle progression, and DNA repair (8–11). RNF20 is known to be required to regulate chromosome structure by monoubiquitinating histone H2B at lysine 123 (H2BK123) in budding yeast and at lysine 120 (H2BK120) in humans (12, 13). H2BK120ub is a key histone modification that plays critical roles in gene transcriptional regulation and higher order chromatin organization in many species (8, 14–20). Ubiquitination of histone H2B (H2Bub) has been reported to be associated with highly expressed active genes in human cells (15, 21). However, recent report has shown that RNF20, presumably via H2Bub, selectively represses expression of some genes by inhibiting TFIIS binding to chromatin (22).
In this study, we have determined that RNF20 interacts with Snail and G9a and is recruited to the E-cadherin promoter to represses its expression, leading to EMT induction.
Materials and Methods
Cell Culture
All cancer cell lines were grown in DMEM/F12 supplemented with 10% FBS, except breast cancer cell lines BT549 and BT474, which were grown in RPMI-1640 plus 10% FBS. For establishing stable transfectants with overexpression of RNF20, MDA-MB231-Luc-D3H1 (with stable expression of luciferase, from Xenogene Corp.) and BT549 cells were transfected with G9a-PLVX; stable clones were selected with puromycin (300 ng/ml) for 4 weeks.
Plasmids, shRNA, and Antibodies
RNF20 shRNA expression plasmids were purchased from MISSION shRNA at Sigma-Aldrich. The catalog numbers of shRNAs were TRCN0000437632 and TRCN0000033878. The plvx-neo-RNF20 contained silent mutation which was resistant to shRNF20-1. The sequence of the mutation was 5’-CAAAGGTTAAATAGG CATCTCG-3’. Expression plasmids for RNF20, Snail were provided by Arthur Danping Wang. Human RNF20 and Snail was amplified from a MDA-MB231 cDNA library and subcloned into vector plvx(puro). Antibodies against HA, Flag, and actin were purchased from Sigma-Aldrich. Antibodies for E-cadherin and β-actin were from BD Transduction Laboratories. RNF20 antibodies and Snail antibodies were from Abcam and Cell Signaling Technology respectively.
Quantitative Real-Time PCR
Total RNA was isolated using the TRIzol Reagent (ThermoFisher scientific) according to manufacturer’s instructions. Specific quantitative real-time PCR experiments were performed using SYBR Green Power Master Mix following manufacturer’s protocol (Applied Agbio, Foster City, Changsha, CHN). The qPCR primers of E-cadherin were 5’-TGCCAACTGGCTGGAGATTA-3’ and 5’-AGTGTCCCTGTTCCAGTAGC-3’. The qPCR primers of RNF20 were 5’-AAAGCATCGCACCATGTCTC-3’ and 5’-ATCCCACTGCAGGTCATCAA-3’.
Migration and Invasion Assays
Migration and invasion assays were performed in Boyden chambers as described previously (23). The cancer cells were stained and counted with a microscope. All experiments were performed at least twice in triplicate. Statistical analysis was performed using Student’s t test; a p-value of <0.05 was considered significant.
Co-Immunoprecipitation
Co-IP assays were performed as described previously (24). Briefly, cells were lysed in lysis buffer that contained a cocktail of protease inhibitors. After being adjusted to equal protein concentration, samples were mixed with 4× SDS–PAGE sample buffer and boiled for 5 min. The pull-down complexes were examined by Western blotting.
Chromatin Immunoprecipitation
ChIP assays were performed as described previously (23). The primers for the E-cadherin promoter were 5’-ACTCCAGGCTAGAGGGTCACC-3’ and 5’-CCGCAAGCTCACAGGTGCTTTGCAGTTCC-3’ (25). The cells were prepared to perform ChIP assay with the Imprint ChIP kit (Sigma-Aldrich) according to the manufacturer’s instructions as described recently.
Colony Formation Assay
Colony formation assay was performed using double-layer soft agar in 24-well plates with a top layer of 0.35% agar and a bottom layer of 0.7% agar. Cells were seeded into 24-wellplates in the desired medium and cultured at 37°C for 20 days, and the colonies were stained and counted.
Mammosphere Assay
Mammosphere assays were performed following the protocol previously described by plating single-cell suspensions into ultralow-attachment 6-well plates (Corning) in mammosphere culturing conditions and counting after 7-14d (26).
Results
RNF20 Alters the Expression of EMT Markers and Enhances Breast Cancer Cell Migration and Invasion In Vitro
RNF20 has been found to be highly expressed in breast cancer cell lines, and is tightly associated with tumorigenic and metastatic capacity of tumor cells (27). To study the molecular function and mechanism of RNF20, we established stable transfectants with empty vector or RNF20 expression in MDA-MB231 and BT549 cells. We found that overexpression of RNF20 dramatically decreased the expression of epithelial marker E-cadherin and significantly upregulated the expression of mesenchymal markers N-cadherin and Vimentin (Figure 1A). Then we confirmed this observation by quantitative real-time PCR in the above cell lines. Consistently, RNF20 expression remarkably elevated the expression of E-cadherin mRNA (Figure 1B). We also generated stable clones with empty vector or knockdown of RNF20 expression in BT474 cells. Strikingly, knockdown of RNF20 expression restored E-cadherin expression in both mRNA and protein levels (Figures 1C, D). Collectively, these data suggest that RNF20 may induce EMT via the transcriptional suppression of E-cadherin expression in human breast cancer.
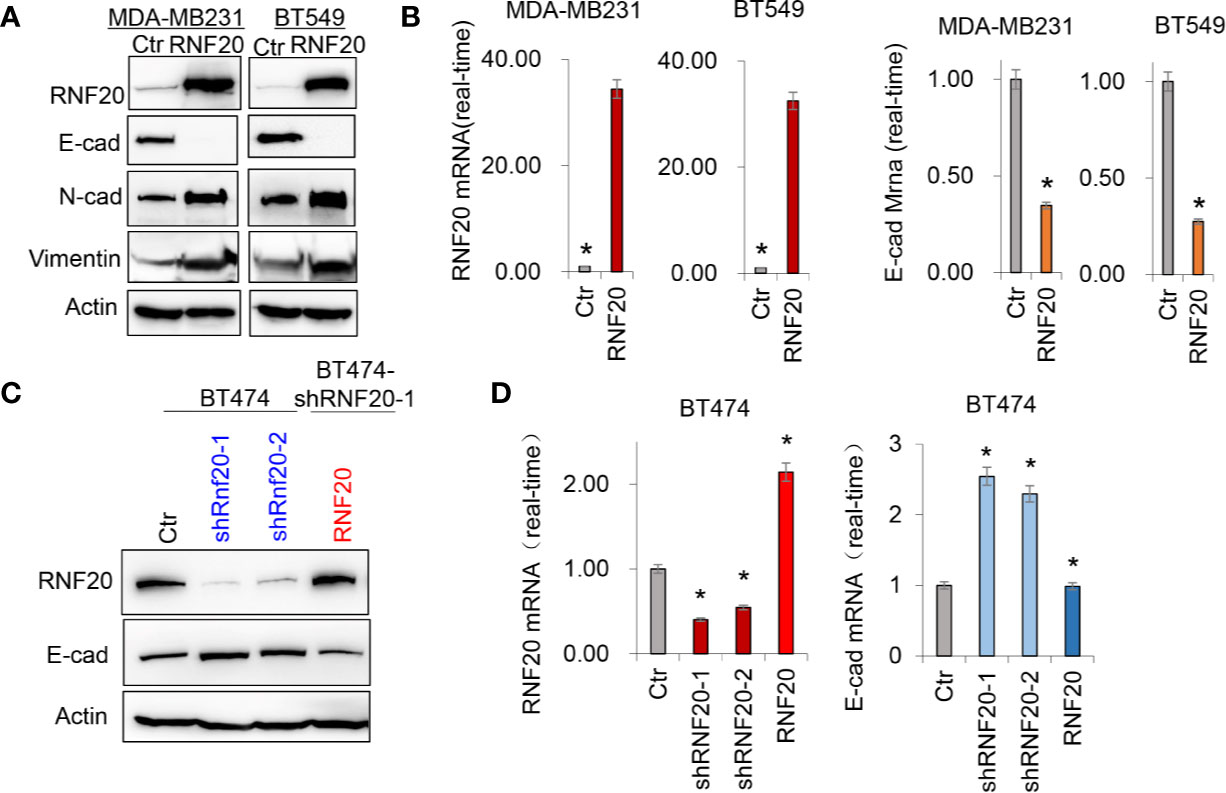
Figure 1 RNF20 alters the expression of epithelial-mesenchymal transition (EMT) markers. (A) Expression of E-cadherin, Vimentin and RNF20 in the MDA-MB231 and BT549 cells was analyzed by Western blotting. Actin was served as a loading control. (B) Expression of RNF20 and E-cadherin was analyzed by qPCR in MDA-MB231 and BT549 cells infected with empty vector (Ctr) or RNF20-expressing vector. *P < 0.01 by Student’s t test. (C) Expression of E-cadherin, and RNF20 was analyzed by Western blotting in BT474 cells with stable empty vector or knockdown of RNF20 expression as well as shRNF20-expressing BT474 cells infected with plvx-neo-RNF20. (D) Expression of RNF20 and E-cadherin was analyzed by qPCR in cells from (C). *P < 0.01 by Student’s t test.
Given the tight association of RNF20 with EMT, we hypothesized that RNF20 is critical for breast cancer cells migration and invasion. As expected, RNF20 overexpression dramatically facilitated the migration and invasion of MDA-MB231 and BT549 cells in vitro (Figures 2A–C). Together, these observations indicate an important role for RNF20 in induction of EMT and acquisition of migratory and invasive ability in breast cancer cells.
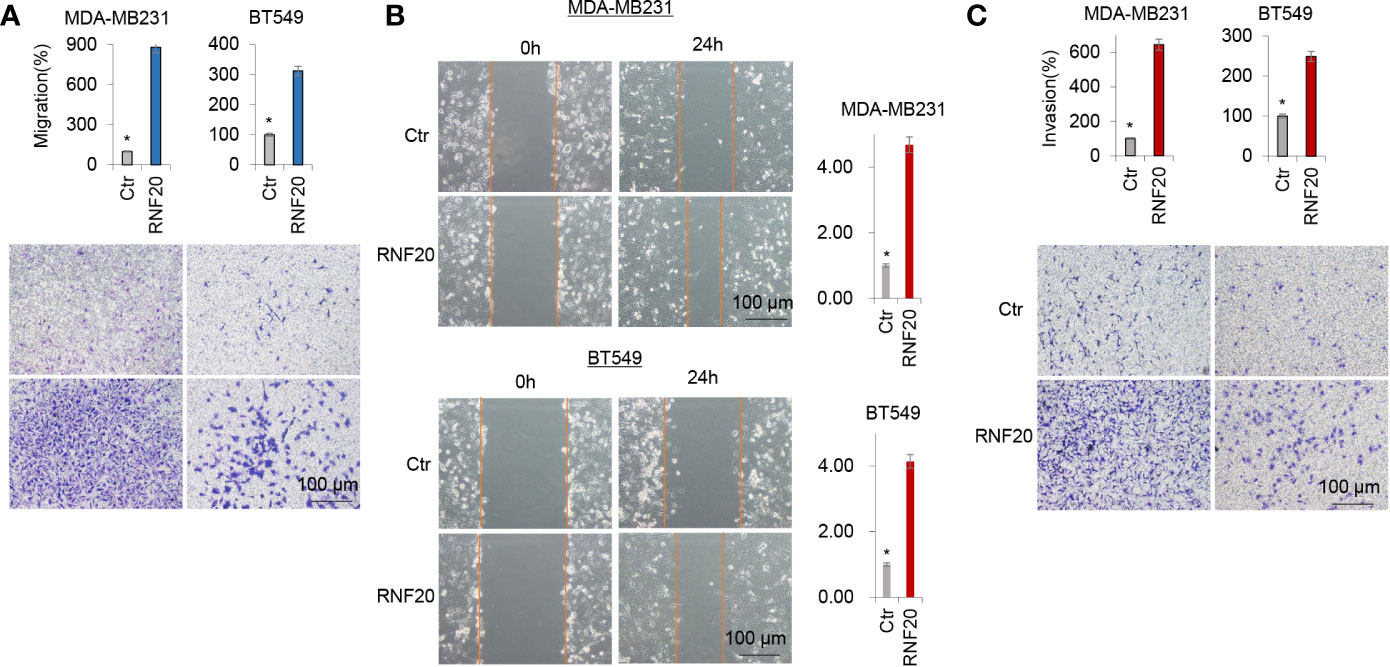
Figure 2 RNF20 enhances breast cancer cell migration and invasion in vitro. (A–C) Migratory (A, B) and invasive ability (C) of MDA-MB231 and BT549 cells with stable empty vector or RNF20 expression were analyzed. The percentage of migratory and invasive cells is shown in the bar graphs (mean ± SD in three separate experiments). *P < 0.01 by Student’s t test: 100 µm.
RNF20 Interacts With Snail
The Snail family member Snail can trigger EMT during embryonic development and tumor progression (28–31). Because RNF20 is closely associated with suppression of E-cadherin and because Snail is a key transcriptional repressor of E-cadherin, Snail may regulate E-cadherin expression by recruiting RNF20 to E-cadherin promoter. To test this notion, we co-expressed Snail-HA and Flag-RNF20 in 293T cells and performed a co-immunoprecipitation experiment. After immunoprecipitating RNF20, we detected the associated Snail, and vice versa (Figure 3A). To identify the region in Snail that associates with RNF20, we generated two deletion mutants of Snail: the N-terminal region of Snail (N-Snail; amino acids 1–153) and the C-terminal region of Snail (C-Snail; amino acids 153–264) (Figure 3B) (32). When these two deletion mutants of Snail were coexpressed with RNF20 in 293T cells, we found that C-Snail was able to interact with RNF20, indicating that the C-terminal region of Snail is responsible for its interaction with RNF20 (Figure 3C).
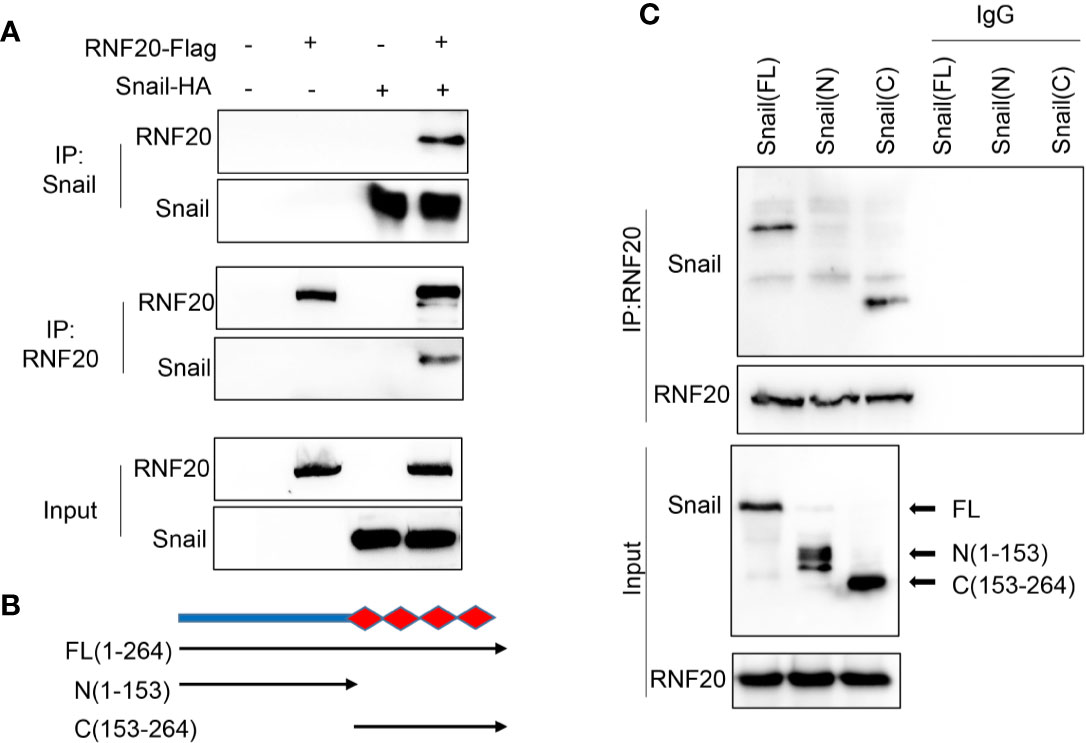
Figure 3 RNF20 interacts with Snail. (A) 293T cells were transiently coexpressed with Flag-tagged RNF20 and HA-tagged Snail. Cell extracts were immunoprecipitated separately with Flag or HA antibodies, and the associated RNF20 and Snail were examined by Western blotting, respectively. (B) Schematic diagram showing the structure of Snail and two deletion mutations. (C) Full-length and deletion mutants of Snail were coexpressed with RNF20 in 293T cells. After immunoprecipitation of RN20, associated Snail was analyzed by Western blotting.
G9a Interacts With RNF20
In the previous study, we have shown that Snail recruits G9a to the E-cadherin promoter for H3K9me2 at the E-cadherin promoter in breast cancer cells (25). RNF20 is required to regulate chromosome structure by monoubiquitinating histone H2B (15, 33). Thus, RNF20 may form complex with Snail and G9a to regulate E-cadherin expression. To examine the association of RNF20 with G9a, we co-expressed RNF20-HA and G9a-Flag in 293T cells and performed a co-immunoprecipitation experiment. After immunoprecipitating RNF20, we detected the associated G9a, and vice versa (Figure 4). These data suggest that RNF20 may inhibit E-cadherin expression by Snail-mediated epigenetic modification.
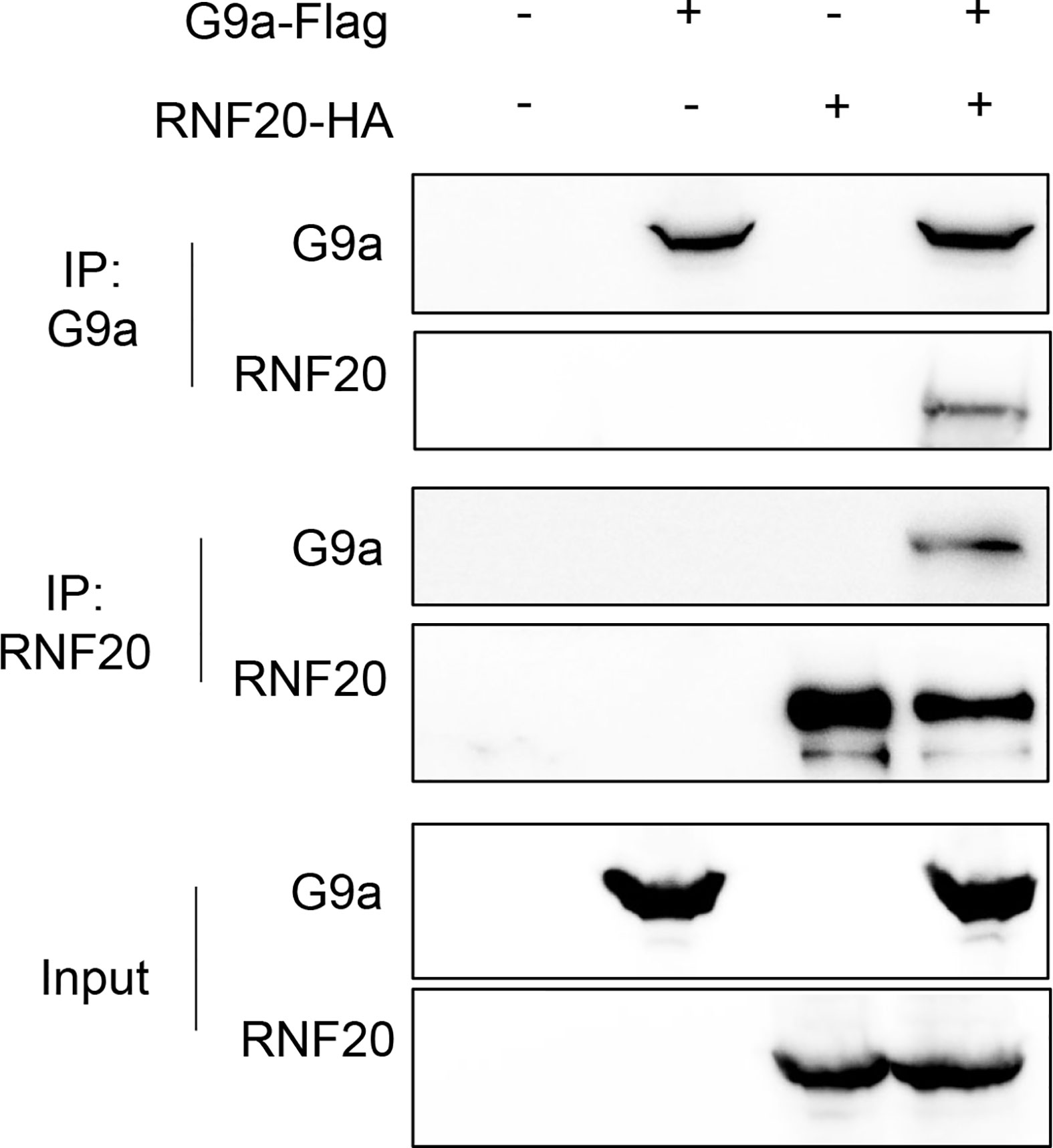
Figure 4 RNF20 interacts with G9a. 293T cells were transiently coexpressed with Flag-tagged G9a, HA-tagged RNF20. Cell extract were immunoprecipitated separately with Flag or HA antibodies, and the associated RNF20 and G9a were examined by Western blotting respectively.
To validate that the Snail-RNF20-G9a complex was associated with the E-cadherin promoter and mediated the transcriptional expression of the E-cadherin in breast cancer, we subsequently established stable clones with empty vector or RNF20 expression in MDA-MB231 and BT549. To test whether RNF20 is involved in the dynamic process of EMT induction, we investigated the association of RNF20 and the G9a-mediated H3K9me2 on the E-cadherin promoter using ChIP assays. We observed that RNF20 and H3K9me2 were enriched on the promoter of E-cadherin promoter, whereas knockdown of Snail expression significantly reduced the enrichment of RNF20 andH3K9me2, and increased level of H3K9 acetylation on the E-cadherin promoter (Figure 5A). Similar results were identified by the quantitative real-time PCR (Figure 5B). Together, these results suggest that RNF20 is recruited to the E-cadherin promoter for epigenetic silencing of E-cadherin expression in a Snail-dependent manner.
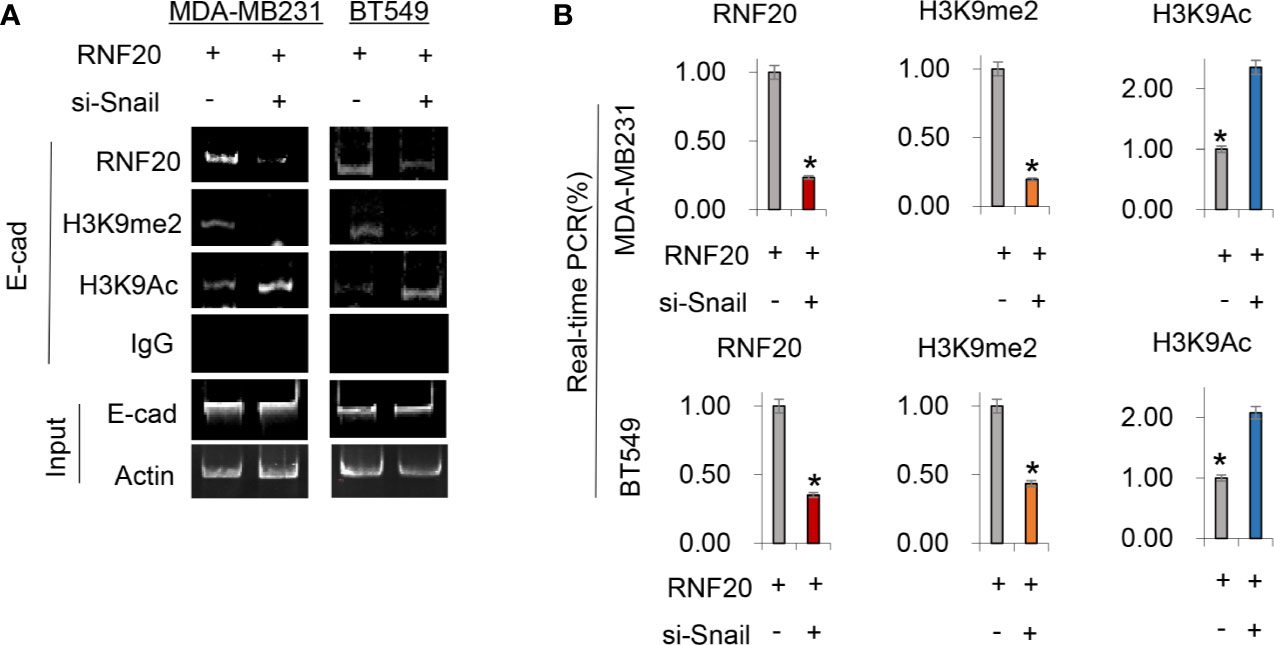
Figure 5 RNF20 is recruited to the E-cadherin promoter for epigenetic silencing of E-cadherin expression by Snail. (A, B) RNF20 was expressed in MDA-MB231 and BT549 cells, and RNF20, H3K9me2, and H3K9 acetylation at the E-cadherin promoter were analyzed by the ChIP assay. Quantitative real-time PCR was also performed to analyze ChIP samples in (A) (mean ± SD from three separate experiments). *P < 0.01 by Student’s t test.
RNF20 Promotes Tumorsphere Formation and Tumorigenicity In Vitro
Growing evidence has shown that the EMT program endows cells with stem cell-like properties, promoting tumor initiation and metastasis. As RNF20 expression contributes to the induction of EMT, we speculated that RNF20 expression might confer stem cell–like properties to breast cancer cells. To test this notion, we examined tumorsphere formation of these cells. Strikingly, RNF20 expression enhanced tumorsphere formation in MDA-MB231 and BT549 cells (Figure 6A). We then examined the effect of RNF20 expression on the in vitro tumorigenicity using soft agar assay. Indeed, RNF20 expression led to a marked increase of colony formation in MDA-MB231 and BT549 cells, whereas knockdown of RNF20 expression caused a dramatic decrease of colonies in BT474 cells (Figure 6B). Given the importance of RNF20 expression in breast cancer, we performed Kaplan-Meier analyses to determine the clinical relevance of RNF20 by analyzing a proteogenomic dataset that contains 65 breast tumor samples (34). Patients were divided into two groups according to RNF20 expression, with high RNF20 expression having shorter overall survival (OS) (Figure 6C). These data support the critical role of RNF20 in breast cancer.
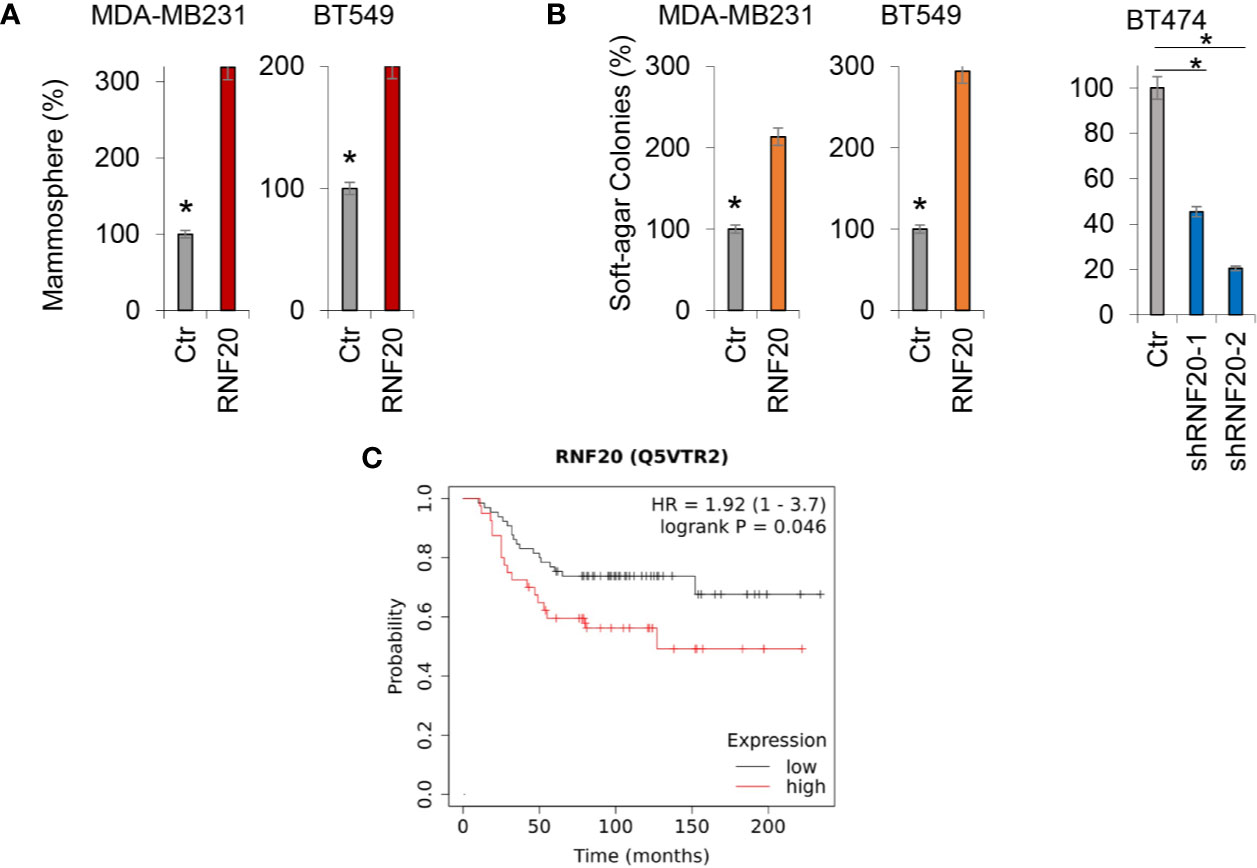
Figure 6 RNF20 promotes tumorsphere and colony formation. (A) Tumorsphere formation of MDA-MB231 and BT549 cells with stable empty vector (Ctr) or overexpression of RNF20 was measured. (B) The formation of colonies was measured from MDA-MB231 and BT549 cells with stable empty vector (Ctr) or overexpression of RNF20, as well as BT474 cells with stable empty vector (Ctr) or knockdown of RNF20 expression. Data are presented as a percentage of empty vector cells lines (mean ± SD in three separate experiments). *P < 0.01 by Student’s t test (C) Kaplan-Meier survival analysis for overall survival (OS) of patients in a breast cancer dataset according to RNF20 protein expression status. The p value was determined using the log-rank test.
Discussion
The histone code hypothesis predicts that the post-translational modification of histones can alter the chromatin state, and therefore contributes to a key regulatory role in chromatin biology (35–37). Ubiquitination of histone H2B(H2Bub), catalyzed by the heterodimeric ubiquitin ligase complex RNF20, regulates multiple molecular and biological processes (15, 27, 38–41). H2Bub alters nucleosome stability, nucleosome reassembly and higher order compaction of the chromatin. There is growing evidence that H2Bub is a cotranscriptional event regulating histone H3 methylation at lysines 4 and 79 (10, 14, 42, 43). Recent report has shown that RNF20 can cause transcriptional suppression of some genes (21, 41, 42). Our study identified that RNF20 formed a complex with Snail and G9a. Snail-RNF20-G9a complex might regulate the expression of E-cadherin and then mediate the EMT progress. Indeed, we found that RNF20 was recruited to the promoter of E-cadherin by the interaction with Snail, which may drive histone H2B monoubiquitylation. Ubiquitination of histone H2B by RNF20 is a prerequisite for DNA methylation and gene silencing (10, 14, 42, 43). G9a also can be recruited to the E-cadherin promoter by Snail for H3K9me2 that is a marker for gene silencing (24). Together, the epigenetic regulation mediated by Snail-RNF20-G9a complex results in the suppression of E-cadherin expression.
Many solid tumors including breast cancer contain a small population of CSCs, which contribute to tumor progression. Increasing evidence has shown that EMT program confers tumor cells with CSC properties (44–46). Consistent with this notion, RNF20 expression caused the increased CSC properties by activating the EMT program in breast cancer, indicating the importance of RNF20 in controlling the viability of CSCs. CSCs are tightly associated with tumor progression (47–49). Indeed, our studies showed that RNF20 expression significantly increased tumorigenicity in vitro, migration and invasion of breast cancer cells. Clinically, high RNF20 expression predicted shorter survival in breast cancer patients. Clearly, our study supports that RNF20 is crucial for the aggressiveness of breast cancer cells.
Identification of the molecular targets may contribute to the development of tailored therapies to improve breast cancer patient outcomes. Given the key roles of RNF20 in EMT, CSCs, and breast cancer, RNF20 may provide potential targets for controlling breast cancer progression.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/gene/56254, Gene ID: 56254.
Author Contributions
DW designed and performed most of experiments. YW generated the G9a plasmids. XW generated the Snail plasmids. JL and XK performed the bioinformatic analysis. CD and JL supervised the work. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from Key Program of Zhejiang Province Natural Science Foundation (No. LZ17H160002 to CD), Natural Science Foundation of China (No. 81972456, 81772801 and 81472455 to CD; No. 32060163 to XW), and National Key R&D Program of China (No. 2016YFC1303200 to CD), and the Initial Research Funds for the PhD of Zunyi Medical University (No. F-932 to XW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Michelle D Althuis JMD, Anderson WF, Devesa SS, Brinton LA. Global trends in breast cancer incidence and mortality 1973-1997. Int J Epidemiol (2005) 34:405–12. doi: 10.1093/ije/dyh414
2. Duffy MJ, McGowan PM, Gallagher WM. Cancer invasion and metastasis: changing views. J Pathol (2010) 214:283–93. doi: 10.1002/path.2282
3. Chambers AF, Groom AC, MacDonald IC. Metastasis: Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer (2002) 2:563–72. doi: 10.1038/nrc865
4. Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer (2006) 6:449–58. doi: 10.1038/nrc1886
5. Douglas H, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
6. Thiery JP, Acloque H, Huang RYJ, Angela Nieto M. Epithelial-Mesenchymal Transitions in Development and Disease. Cell (2009) 139:871–90. doi: 10.1016/j.cell.2009.11.007
7. Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev (2009) 28:15–33. doi: 10.1007/s10555-008-9169-0
8. Osley MA, Fleming AB, Kao C-F. Histone Ubiquitylation and the Regulation of Transcription. Results Probl Cell Differ (2006) 41:47–75. doi: 10.1007/400_006
9. Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem (2006) 75:243–69. doi: 10.1146/annurev.biochem.75.103004.142422
10. Shilatifard A. Molecular Implementation and Physiological Roles for Histone H3 Lysine 4 (H3K4) Methylation. Curr Opin Cell Biol (2008) 20:341–8. doi: 10.1016/j.ceb.2008.03.019
11. Escargueil AE, Soares DG, Salvador M, Larsen AK, Pegas Henriques JA. What histone code for DNA repair? Mutat Res/Reviews Mutat Res (2008) 658:259–70. doi: 10.1016/j.mrrev.2008.01.004
12. William WMB. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res (1980) 8:4671–80. doi: 10.1093/nar/8.20.4671
13. Robzyk K, Recht J, Osley MA. Rad6-Dependent Ubiquitination of Histone H2B in Yeast. Science (2000) 287:501–4. doi: 10.1126/science.287.5452.501
14. Sun Z-W, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature (2002) 418:104–8. doi: 10.1038/nature00883
15. Kim J, Hake SB, Roeder RG. The Human Homolog of Yeast BRE1 Functions as a Transcriptional Coactivator through Direct Activator Interactions. Mol Cell (2006) 20:759–70. doi: 10.1016/j.molcel.2005.11.012
16. Shema E, Torish I, Aylon Y, Huang J, Ye C, Moskovits N, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev (2008) 22:2664–76. doi: 10.1101/gad.1703008
17. McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature (2008) 453:812–6. doi: 10.1038/nature06906
18. H.P. WM, Bonner WM. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res (1980) 20. doi: 10.1093/nar/8.20.4671
19. Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev (2007) 21:835–47. doi: 10.1101/gad.1516207
20. Sihao Zheng DL, Lu Z, Chen X. Bre1-dependent H2B ubiquitination promotes homologous recombination by stimulating histone eviction at DNA breaks. Nucleic Acids Res (2018) 46:11326–39. doi: 10.1093/nar/gky918
21. Rushad Pavri BZ, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B Monoubiquitination Functions Cooperatively with FACT to Regulate Elongation by RNA Polymerase II. Cell (2006) 125:703–17. doi: 10.1016/j.cell.2006.04.029
22. Efrat Shema JK, Roeder RG, Oren M. RNF20 Inhibits TFIIS-Facilitated Transcriptional Elongation to Suppress Pro-oncogenic Gene Expression. Mol Cell (2011) 42:477–88. doi: 10.1016/j.molcel.2011.03.011
23. Chenfang Dong YW, Wang Y, Wang C, Kang T, Rychahou PG, Chi Y-I, et al. Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene (2013) 32:1351–62. doi: 10.1038/onc.2012.169
24. Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, et al. Dual regulation of Snail by GSK-3β-mediated phosphorylation in control of epithelial–mesenchymal transition. Nat Cell Biol (2004) 6:931–40. doi: 10.1038/ncb1173
25. Chenfang Dong YW, Yao J, Wang Y, Yu Y, Rychahou PG, Evers BM, et al. G9a interacts with SNAIL and is critical for SNAIL-mediated E-cadherin repression in human breast cancer. J Clin Invest (2012) 122:1469–86. doi: 10.1172/JCI57349
26. Grimshaw MJ, Cooper L, Papazisis K, Coleman JA, Bohnenkamp HR, Burchell JM, et al. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res (2008) 10. doi: 10.1186/bcr2106
27. Gautam Sethi MK, Arfuso SF, Kumar AP. Role of RNF20 in cancer development and progression – a comprehensive review. Bioence Rep (2018) 38:BSR20171287. doi: 10.1042/BSR20171287
28. Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development (2005) 132:3151–61. doi: 10.1242/dev.01907
29. Hongyan Jin YY, Zhang T, Zhou X, Zhou J, Jia L, Wu Y, et al. Snail is critical for tumor growth and metastasis of ovarian carcinoma. Int J Cancer (2009) 126:2102–11. doi: 10.1002/ijc.24901
30. Héctor Peinado DO, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer (2007) 7:415–28. doi: 10.1038/nrc2131
31. Alberga A, Kempe E, Dennefeld C, Haenlin M. J. L. BThe snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development (1991) 111:983–92.
32. Yadi Wu BME, Zhou BP. Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J Biol Chem (2009) 284:640–8. doi: 10.1074/jbc.M806916200
33. Bing Zhu YZ, Pham A-D, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of Human Histone H2B: The Factors Involved and Their Roles in HOX Gene Regulation. Mol Cell (2005) 20:601–11. doi: 10.1016/j.molcel.2005.09.025
34. Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat (2010) 123:725–31. doi: 10.1007/s10549-009-0674-9
35. Kouzarides T. Chromatin Modifications and Their Function. Cell (2007) 128:693–705. doi: 10.1016/j.cell.2007.02.005
36. Berger SL. Histone modification in transcriptional regulation. Curr Opin Genet Dev (2002) 12:142–8. doi: 10.1016/S0959-437X(02)00279-4
37. Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol (2001) 2:195–201. doi: 10.1038/35056583
38. Martina Foglizzo AJM, Day CL. Structure and Function of the RING Domains of RNF20 and RNF40, Dimeric E3 Ligases that Monoubiquitylate Histone H2B. J Mol Biol (2016) 428:4073–86. doi: 10.1016/j.jmb.2016.07.025
39. Chernikova SB, Razorenova OV, Higgins JP, Sishc BJ, Nicolau M, Dorth JA, et al. Deficiency in Mammalian Histone H2B Ubiquitin Ligase Bre1(Rnf20/Rnf40) Leads to Replication Stress and Chromosomal Instability. Cancer Res (2012) 72:2111–9. doi: 10.1158/0008-5472.CAN-11-2209
40. Rushad Pavri BZ, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell (2006) 125:703–17. doi: 10.1016/j.cell.2006.04.029
41. Ohad Tarcic ISP, Cooks T, Shema E, Kanterman J, Ashkenazi H, Boocholez H, et al. RNF20 Links Histone H2B Ubiquitylation with Inflammation and Inflammation-Associated Cancer. Cell Rep (2016) 14:1462–76. doi: 10.1016/j.celrep.2016.01.020
42. Jim Dover JS, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of Histone H3 by COMPASS Requires Ubiquitination of Histone H2B by Rad6. J Biol Chem (2002) 277:28368–71. doi: 10.1074/jbc.C200348200
43. Nevan J Krogan JD, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a Histone H3 (Lysine 4) Methyltransferase Required for Telomeric Silencing of Gene Expression. J Biol Chem (2002) 277:10753–5. doi: 10.1074/jbc.C200023200
44. Nauseef JT, Henry MD. Epithelial-to-mesenchymal transition in prostate cancer: paradigm or puzzle? Nat Rev Urol (2011) 8:428–39. doi: 10.1038/nrurol.2011.85
45. Medici D, Muñoz-Cánoves P, Yang P-C, Brunelli S. Mesenchymal Transitions in Development and Disease. Stem Cells Int (2016) 2016. doi: 10.1155/2016/5107517
46. Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer (2009) 9:265–73. doi: 10.1038/nrc2620
47. Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science (2009) 324:1670–3. doi: 10.1126/science.1171837
48. Liu S, Wicha MS. Targeting breast cancer stem cells. J Clin Oncol (2010) 28:4006–12. doi: 10.1200/JCO.2009.27.5388
Keywords: epithelial-mesenchymal transition, snail, RNF20, ubiquitination, G9a
Citation: Wang D, Wang Y, Wu X, Kong X, Li J and Dong C (2020) RNF20 Is Critical for Snail-Mediated E-Cadherin Repression in Human Breast Cancer. Front. Oncol. 10:613470. doi: 10.3389/fonc.2020.613470
Received: 02 October 2020; Accepted: 09 November 2020;
Published: 08 December 2020.
Edited by:
Binhua (Peter) Zhou, University of Kentucky, United StatesReviewed by:
Weijia Cai, Thomas Jefferson University, United StatesBolin Liu, Louisiana State University, United States
Copyright © 2020 Wang, Wang, Wu, Kong, Li and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Li, MjMwNzAxNkB6anUuZWR1LmNu; Chenfang Dong, Y2hlbmZhbmdkb25nQHpqdS5lZHUuY24=
†These authors have contributed equally to this work
 Danping Wang
Danping Wang Yifan Wang
Yifan Wang Xuebiao Wu
Xuebiao Wu Xiangxing Kong1
Xiangxing Kong1 Jun Li
Jun Li Chenfang Dong
Chenfang Dong