- 1Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Department of Anatomical Sciences, Faculty of Medicine, Birjand University of Medical Sciences, Birjand, Iran
- 3Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
- 4Urology and Nephrology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 5Urogenital Stem Cell Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Cell cycle is regulated by a number of proteins namely cyclin-dependent kinases (CDKs) and their associated cyclins which bind with and activate CDKs in a phase specific manner. Additionally, several transcription factors (TFs) such as E2F and p53 and numerous signaling pathways regulate cell cycle progression. Recent studies have accentuated the role of long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) in the regulation of cell cycle. Both lncRNAs and miRNAs interact with TFs participating in the regulation of cell cycle transition. Dysregulation of cell cycle regulatory miRNAs and lncRNAs results in human disorders particularly cancers. Understanding the role of lncRNAs, miRNAs, and TFs in the regulation of cell cycle would pave the way for design of anticancer therapies which intervene with the cell cycle progression. In the current review, we describe the role of lncRNAs and miRNAs in the regulation of cell cycle and their association with human malignancies.
Introduction
Cell division has a fundamental role in the development multicellular organisms. This process is accomplished through orderly sequences of happenings that together with each other make the “cell cycle”. This cycle comprises precise duplication of the genome throughout the DNA synthesis stage (S phase), and separation of whole sets of chromosomes to one of the daughter cells in the mitosis stage (M phase). Two “Gap” phases also exist in the cell cycle. The first one (G1) links the accomplishment of the M phase to the commencement of S phase in the succeeding cycle. G2 splits the S and M stages. Cells residing in the G1 phase might momentarily or enduringly exit the cell cycle and go in an inactive or blocked phase namely G0 (1). In the mammalian cells, cell cycle is regulated by a number of proteins namely cyclin-dependent kinases (CDKs) and their associated cyclins which bind with and activate CDKs in a phase specific manner (2). Cyclins A and E are activating factors for CDK2. Cyclin B binds with CDK1. Finally, CDK4/6 is activated by cyclin Ds. Binding of cyclins with CDKs leads to phosphorylation of CDKs target proteins which finally permits progression through cell cycle (2). In addition to cyclins, Wee1 kinase and CDC25 phosphatase regulate activity of CDKs by phosphorylation and dephosphorylation reactions, respectively (3). Activity of CDKs and cell cycle progression are inhibited by a number of factors such as p15ink4b, p16ink4a, p18ink4d, p21Cip1, p27Kip1, and p57Kip2. These factors have a specific binding affinity for cyclin-CDK complexes (4–6). Progression through each phase of cell cycle is regulated by CDKs and their associated proteins/pathways. For instance, MAPK pathway induced by growth factors enhances transcription of cyclin Ds in the G1 phase, leading to activation of CDK4/6 (7). The protein complex formed by cyclin Ds and CDK4/6 phosphorylates retinoblastoma protein (pRB), p107 and p130, in the final stages of G1 phase, thus releasing E2F and enhancing E2F-dependent expression of growth-stimulating genes (7). At the G1/S boundary, the complex constructed by cyclin E and CDK2 phosphorylates pRB and other proteins participating in the regulation of DNA replication to facilitate G1/S transition (8). Cyclin B-CDK1 complex has several functions such as accomplishment of the G2 phase processes (9), negative regulation of cytokinesis (10), and coordination of mitotic-related procedures in the nucleus and the cytoplasm (11). This complex has a number of targets such as the anaphase-promoting complex/cyclosome (APC/C) (10). In addition to CDKs, cyclins, and the APC/C which directly regulate cell cycle progression, other molecules are involved in this process. For instance, p53 functions in numerous stages to warrant that cells do not bring their abnormal DNA through cell division. It halts the cell cycle at the G1 checkpoint through stimulating synthesis of CDK inhibitor (CKI) proteins. These proteins attach CDK-cyclin complexes and inhibit their function extending the time for the activation of DNA repair system. It also induces DNA repair enzymes. If DNA damage cannot be fixed, p53 induces cell apoptosis to prevent transmission of the damaged DNA to the daughter cells (12).
Several lines of evidence point to the role of non-coding RNAs (ncRNAs) in the regulation of expression or activity of the above-mentioned proteins (13). Cyclins, CDKs and their inhibitors, are targets of regulation by ncRNAs at different levels including transcriptional and post-transcriptional levels (13). Being classified mainly based on their sizes, ncRNAs include micro RNAs (miRNAs) and long non-coding RNAs (lncRNAs). LncRNAs are longer than 200 nucleotides. According to their structural features, lncRNAs are categorized into different classes among them are intergenic, intronic and natural antisense lncRNAs (14). In addition to their regulatory roles on gene expression at transcriptional and post-transcriptional levels, lncRNAs act as protein scaffolds to regulate interactions between proteins (15). Expressions of a number of lncRNAs are stimulated by DNA damage. These transcripts contribute in DNA damage responses and carcinogenic processes (13). Meanwhile, there is a reciprocal interaction between miRNAs and a number of cell cycle regulators in a way that miRNAs regulate expression of cell cycle regulators, and expression of miRNA is regulated by cell-cycle-dependent transcription factors (16). Such interactive network is implicated in the pathogenesis of a number of disorders particularly cancer. Most of miRNAs are first transcribed from their encoding genes into primary miRNAs. Then, they are changed to precursor miRNAs, and eventually mature miRNAs. These steps are performed in both nucleus and cytoplasm. Mechanistically, miRNAs interact with the 3′ UTR of their target transcripts resulting in degradation their or inhibition of their translation. Yet, miRNAs binding with the 5′ UTR, coding regions, and promoters, has also been demonstrated (17). In the current review, we describe the role of lncRNAs and miRNAs in the regulation of cell cycle and their association with human malignancies.
LncRNAs and Cell Cycle Control
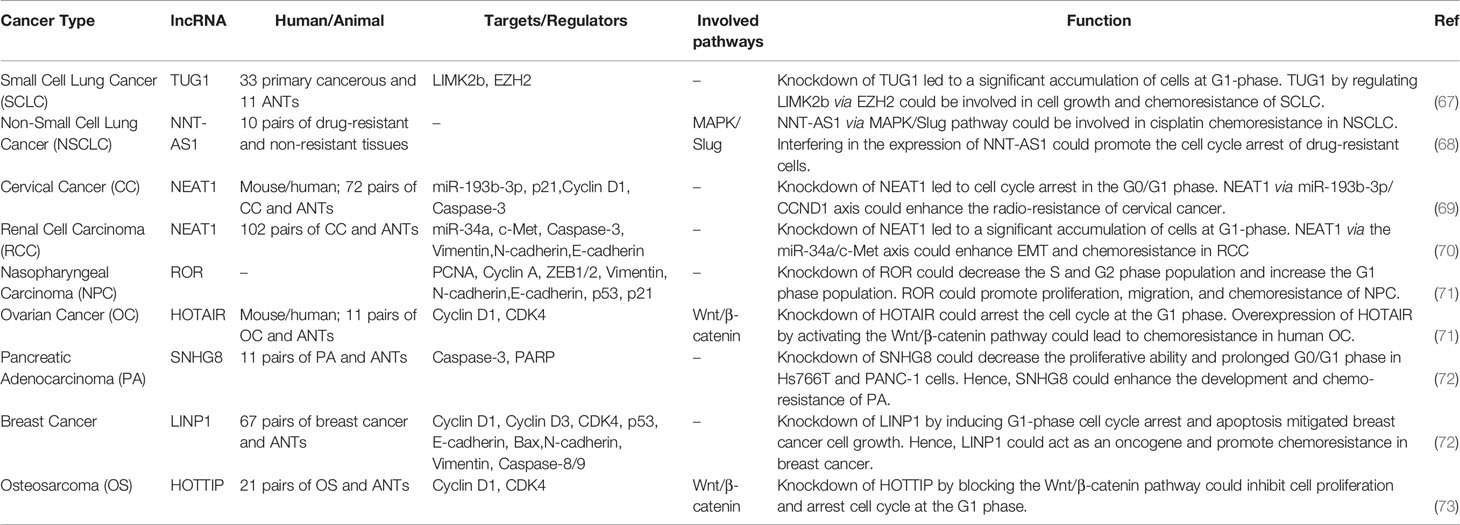
Numerous lncRNAs have been shown to regulate cell cycle progression directly through modulation of expression of CDKs/cyclins or indirectly through regulation of TFs that control expression of CDKs/cyclins. For instance, the known oncogenic lncRNA MALAT1 regulates cell cycle progression at G1 phase since its knock down has resulted in cell cycle arrest at this step and enhanced expression of cell cycle inhibitors p53, p16, p21, and p27, while suppressing expression of cyclin A2 and CDC25A (18). Moreover, this lncRNA has a crucial role in up-regulation of expression of B-Myb, an oncogenic TF participating in G2/M progression (18). Thus, MALAT1 regulates cell cycle progression in different phases through modulation of expression of cell cycle regulators.
ANRIL is an lncRNA being transcribed from the INK4 locus in an antisense direction to p15 (19). This lncRNA participates in epigenetic suppression of expression of the INK4 locus through recruitment of polycomb repression complex 2 (PRC2). Such function specifically represses expression of p15 (20). Expression of CDK inhibitors is also regulated by other lncRNAs including lncRNA-HEIH. This up-regulated lncRNA in the hepatitis B virus-associated hepatocellular carcinoma decreases expression of p15, p16, p21, and p57, through cooperation with EZH2, thus regulating cell cycle transition at G0/G1 (21).
HOTAIR has been shown to regulate expression of genes which are principally associated with cell cycle progression (22). HOTAIR silencing has led to cell cycle arrest at G0/G1 in addition to modulation of expression of cell cycle-related proteins (22). Experiment in esophageal squamous cell carcinoma cells has also verified the impact of HOTAIR silencing on suppression of cell proliferation and induction of G1 cell cycle arrest. This lncRNA serves as a molecular sponge to suppress miR-1 expression and subsequently increase expression of cyclin D1 (23). In ovarian cancer cells, HOTAIR increases expression of cyclin D1 and cyclin D2 through negatively regulating miR-206 expression (24).
A number of lncRNAs have been shown to regulate expression of p53 thus affecting cell cycle regulation by this TF. These lncRNAs include both oncogenic and tumor suppressor ones. Examples from the former group include PVT1 and ANRIL which enhance MDM2-associated degradation of p53 (25, 26). On the other hand, LOC572558 enhances expression of p53 through regulation of its phosphorylation (27). Moreover, MEG3 RNA interacts with the promoter region of p21 to increase p53 accumulation (28). Meanwhile, p53 as a TF can alter expression of several lncRNAs. For example, the lncRNA-p21 has been shown to be transcribed from a genomic region adjacent to p21Cip1. This lncRNA is a direct target of p53. LncRNA-p21 silencing has affected expression of a number of p53-target genes with the exception of p21 gene (29). Moreover, the lncRNA PANDA is transcribed from a promoter region of p21 in response to DNA damage through a p53-dependent route (30). Therefore, lncRNAs exert functional roles both at upstream and downstream of cell cycle-associated TFs such as p53.
The lncRNA EMS has been identified as a direct target of c-Myc. This lncRNA acts as an oncogenic transcript that facilitates G1/S transition. Functional studies have shown interaction between EMS and the RNA binding protein RALY to increase the stability of E2F1 transcript and enhance its expression (31). The oncogenic lncRNA MIR31HG has been shown to promote cell proliferation, facilitate cell cycle progression, and suppress cell apoptosis. This lncRNA modulates cell cycle transition through regulation of HIF1A and p21 expressions (32). In addition, SHNG3 is another dysregulated lncRNA in diverse cancers. This lncRNA has higher expression in glioma tissues and cell lines compared with normal counterparts. Forced over-expression of SNHG3 has increased proliferation, quickened cell cycle progression, and suppressed cell apoptosis via silencing KLF2 and p21 through recruitment of EZH2 to their promoter (33). FOXD2-AS1 is another oncogenic lncRNA whose knock down results in cell cycle arrest in the G0/G1 stage, inhibition of colony development, cell proliferation, and suppression of tumor growth in the xenograft model. FOXD2-AS1 and reduced expression of CDKN1B through recruitment of EZH2 to its promoter region (34). ROR1-AS1 in an up-regulated lncRNA in colon cancer tissues, particularly in stage III and IV and more massive tumors. Forced over-expression of ROR1-AS1 has increased cell proliferation, reduced the G0/G1 phase time of cell cycle, and inhibited apoptosis. This lncRNA can bind to EZH2 and suppress expression of DUSP5 (35).
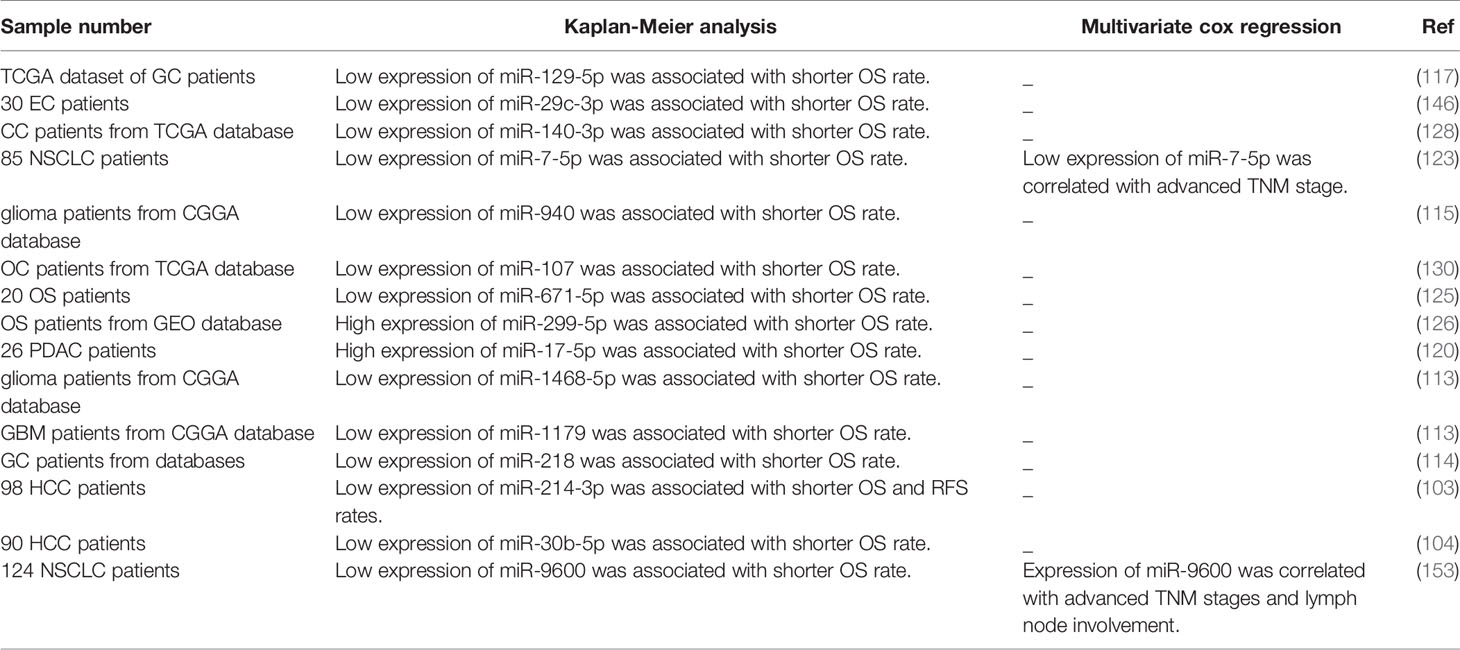
Figure 1 shows the molecular mechanism of involvement of a number of lncRNAs in cell cycle regulation. These lncRNAs recruit EZH2 to the promoter regions of their target genes.
Figure 1 The lncRNA SNHG3 recruits EZH2 to the promoter of CDKN1A to induce H3K27me3 and decrease expression of this gene. This gene encodes the p21 protein which is an inhibitor of cyclin E/CDK2 (33). The lncRNA FOXD2-AS1 enhances recruitment of EZH2 to the promoter of CDKN1B and decreases its expression via H3K27me3. Therefore, it down-regulates p27 which is an inhibitor of cyclin D (34). These two lncRNAs promote cell progression at G1/S point. Over-expression of ROR1-AS1 has increased cell proliferation, reduced the G0/G1 phase time of cell cycle, and inhibited apoptosis. This lncRNA can bind to EZH2 and suppress expression of DUSP5 (35).
Table 1 shows the results of studies which assessed the role of lncRNAs in cell cycle control.
The interaction between lncRNAs and cell cycle-related proteins can alter response of cancer cells to chemotherapeutic agents. For instance TUG1 has a role in induction of chemoresistance in small cell lung cancer cells through regulation of LIMK2b expression. Knockdown of TUG1 has resulted in the accumulation of cells at G1-phase (67). NNT-AS1 via MAPK/Slug pathway could be involved in cisplatin chemoresistance in non-small cell lung cancer (68). Table 2 summarizes the results of studies which assessed the role of lncRNAs in this regard.
The importance of cell cycle-associated lncRNAs as diagnostic/prognostic markers have been assessed in several studies. Higher expression of NR2F2-AS1, PCAT6, FOXD-AS1, SNHG3, FLVCR-AS1, and some other lncRNAs has been associated with lower OS rate. Table 3 summarizes the results of these studies.
miRNAs and Cell Cycle Control
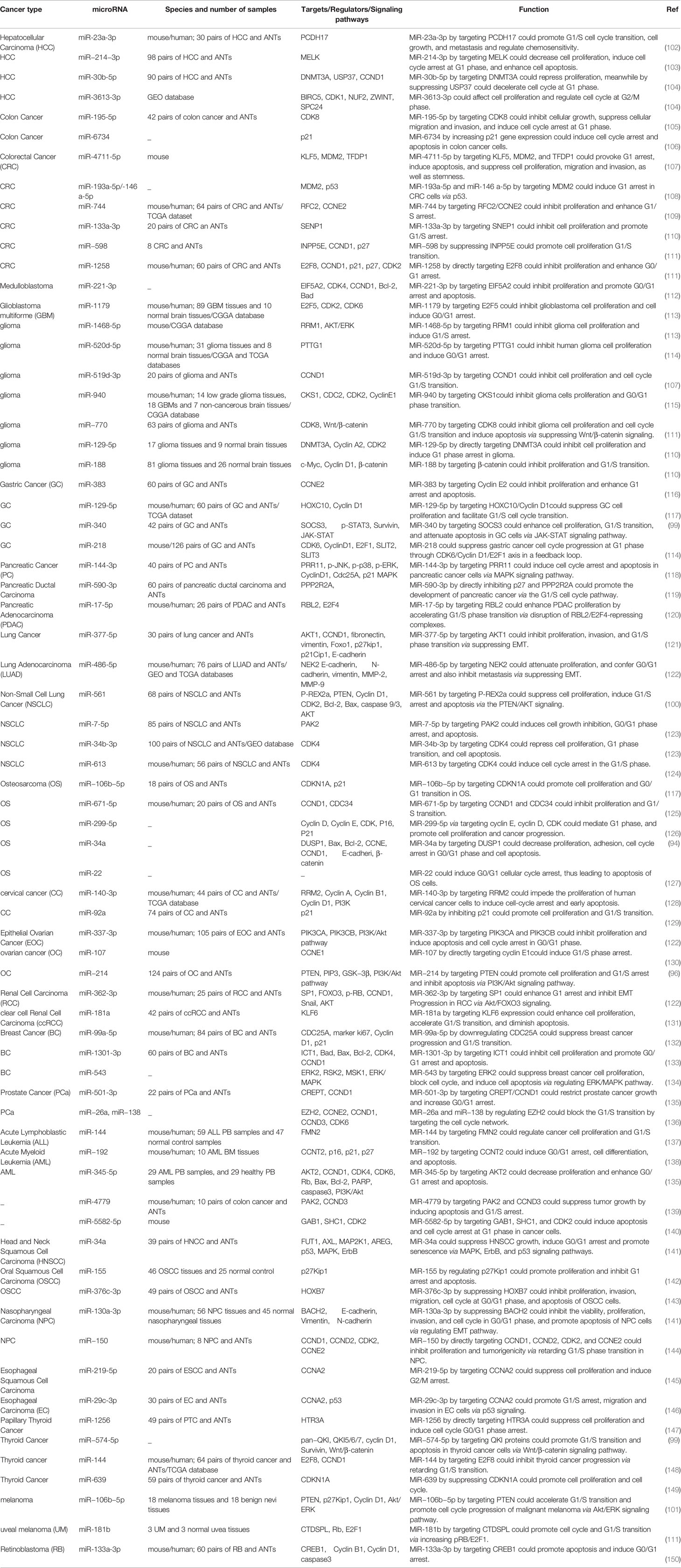
These small transcripts participate in the regulation of cell cycle control via modulation of checkpoints and DNA repair mechanisms (16, 74). Moreover, they regulate expression of cyclins, CDKs, cyclin-dependent kinase inhibitors, and TF-associated proteins such as Rb (16). For instance, the miR-15a-16-1 cluster has been shown to induce cell cycle arrest at the G1 through suppressing expression of CDK1, CDK2, and CDK6 as well as D1, D3, and E1 cyclins (75–77). miR-188 suppress cell cycle transition at G1/S through inhibition of expression of cyclins D1, D3, E1, and A2 as well as CDK4 and CDK2. This miRNA also reduces Rb phosphorylation and decreases E2F transcriptional activity (78). miR‑424 has been shown to regulate cell cycle progression in the G2/M phase through inhibition of expression of CDK1 probably via the Hippo and the extracellular signal‑regulated kinase pathways (79). Moreover, regulation of CDK5 by miRNA-26a has been shown to control cell proliferation, apoptosis and tumor growth in an animal model of diffuse large B-cell lymphoma (80). Expression of CDK5 is also regulated by the tumor suppressor miRNA-505-5p in cervical cancer cells (81). Notably, this CDK has a distinct feature from other CDKs which is that it is not activated via interaction with cyclins. Instead, it is activated through binding with p35 and p39, or their cleaved proteins namely p25 and p29 (82).
Expression of cell cycle-related TFs is also regulated by miRNAs. For example, miR-17-92 cluster, miR-17-5p, miR-20a, miR-149*, miR-330, and miR-331-3p have been shown to suppress expression of E2F1, thus inducing cell cycle arrest in different tissues (83–86). On the other hand, miR-106a, miR-290, and miR-17-92 have been shown to target pRB or other proteins from this family (87–90).
A number of miRNAs such as miR-504 and miR-1285 regulate expression of p53 (91). Meanwhile, p53 has been shown to alter expression of several miRNAs such as miR-34a/b/c (92, 93). Thus, several miRNAs are implicated in the regulation of cell cycle progression through p53-mediated pathways. Figure 2 illustrates the role of a number of miRNAs in the cell cycle control.
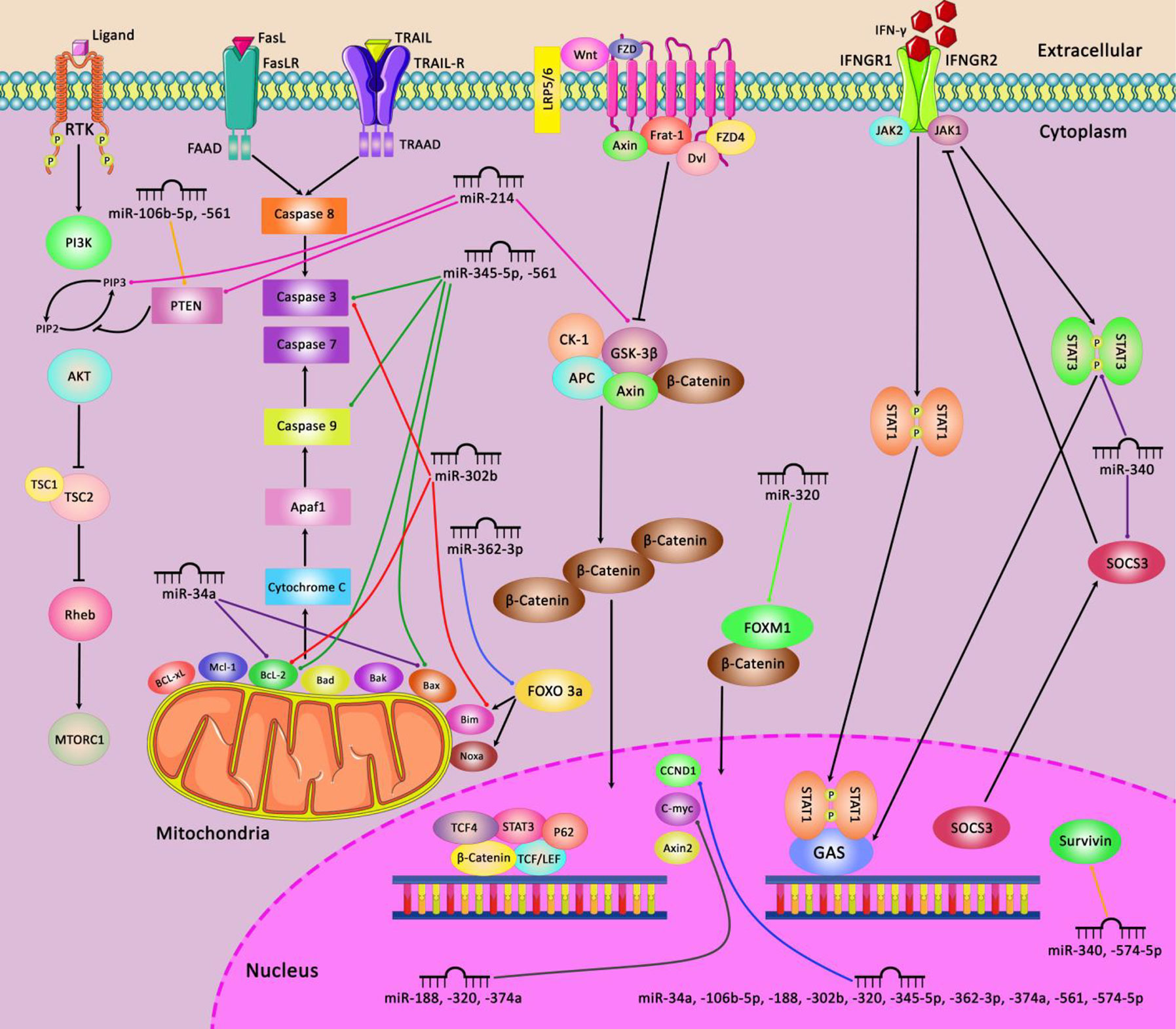
Figure 2 A schematic diagram of the regulation of mitochondrial apoptosis, Wnt/β-catenin, JAK-STAT, and PI3K/AKT signaling pathways via different miRNAs in various human cancers. Ectopic expression of some miRNAs including miR-345-5p, miR-561, miR-302b, miR-362-3p, and miR-34a could impede the mitochondrial apoptotic pathway via targeting caspase 3 and 9, Bcl-2, Bax, and Bim which can play an effective role in cell death suppression in variety of tumor cells (94, 95). Besides, miR-214, miR-320, miR-188, miR-374a, and miR-574-5p could activate the Wnt/β-catenin pathway in tumor cells through modulating GSK-3β, FOXM1, CCND1, and C-myc, and thereby promoting cell differentiation and proliferation as well as enhancing EMT and cell migration and invasion in different human cancers (96, 97). Additionally, miR-340, and miR-574-5p could regulate the JAK-STAT signaling pathway via targeting STAT3, SOCS3, and Survivin which have a significant role in regulating tumor cell growth and metastasis in various tumor cells (98, 99). In addition, aberrant expression of miR-214, miR-106b-5p, and miR-561 could negatively modulate PTEN and PIP3 in PI3K/AKT signaling pathway in different human cancers such as ovarian cancer, melanoma, and NSCLC cells (96, 100, 101).
Table 4 summarizes the function of miRNAs in cell cycle control.
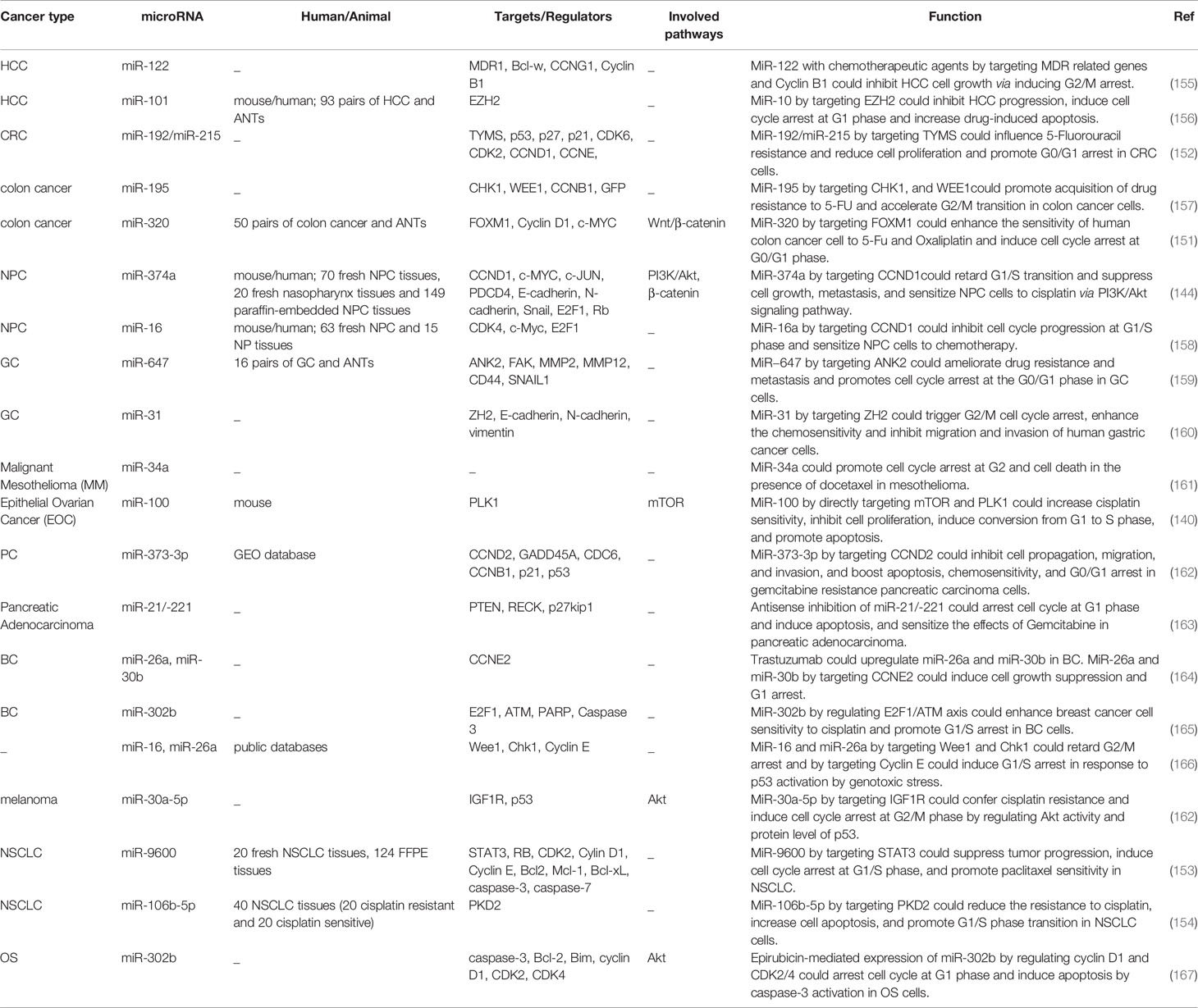
The interaction between miRNAs and cell cycle controlling proteins has implications in defining response to chemotherapeutic agents. For instance miR-192/miR-215 and miR-320 could affect response of cancer cells to 5-Fluorouracil resistance (151, 152). In addition, miR-100 could increase cisplatin sensitivity, inhibit cell proliferation, induce conversion from G1 to S phase and promote apoptosis through directly targeting mTOR and PLK1 (140). miR-374a is another miRNA which alters chemoresistance phenotype in nasopharyngeal carcinoma. In this type of cancer, miR-374a decreases proliferation, migratory aptitude, invasiveness, metastatic ability, and resistance to cisplatin. Functionally, miR-374a decreases expression of CCND1 to attenuate activity of the pPI3K/pAKT/c-JUN axis through making a negative-feedback circle. This miRNA also inhibits downstream signals associated with cell cycle transition (144). miR-9600 has been demonstrated to attenuate tumorigenesis and metastatic potential of lung cancer cells via inhibiting expression of STAT3. Besides, miR-9600 improved response of cancer cells to paclitaxel and cisplatin through this axis and enhancement of chemotherapy-associated apoptosis (153). Besides, miR-106b-5p has a crucial impact in modulation of cisplatin resistance in lung cancer through inhibiting expression of PKD2 (154). Table 5 summarizes the data regarding the role of miRNAs in conferring resistance to chemotherapeutic agents.
Cell cycle-regulating miRNAs can be used as prognostic markers in cancer. Low expression of several miRNAs such as miR-129-5p, miR-29c-3p, miR-140-3p, miR-7-5p, miR-940, miR-107, miR-671-5p, and iR-299-5p has been associated with shorter survival rate of patients with certain types of cancers. Table 6 summarizes the results of studies which assessed this aspect of miRNAs.
Discussion
Several lncRNAs and miRNAs have been shown to regulate cell cycle at different stages thus influencing the proliferation rate. Abnormal function of these transcripts might lead to the development of human cancers. Cell cycle is regulated by several lncRNAs through epigenetic modulation of gene expression regulation of transcription factors modulation of translation mRNA stability, and enhancement of protein-protein interactions (15). Recruitment of EZH2 to the promoter region of target genes is a common mechanism of action of some of these lncRNAs. Dysregulation of cell cycle-related lncRNAs is regarded as a hallmark of cancer. A number of these lncRNAs also contribute in controlling the proliferation rate of normal differentiated cells or during organogenesis, therefore being important in the context of regenerative medicine and in cell senescence studies. For instance, Malat1 controls differentiation of myogenic cells and muscle regeneration (168). In addition, H19 regulates myoblast differentiation and muscle regeneration (169). However, most of the cell-cycle regulatory roles of lncRNAs have been assessed in the context of cancer.
miRNAs also exert functional roles in the regulation of cell cycle. A comprehensive genome-wide screen of cell cycle-associated miRNAs has led to identification of a distinctive group of miRNAs that target almost all cyclins/CDKs. These miRNAs are extremely potent in impeding cancer cell proliferation (170). Systemic administration of a number of these miRNAs using nanoparticle delivery methods has repressed tumor progression in a number of xenograft models indicating the role of these miRNAs in the treatment of cancer (170). The prominent role of cell cycle-regulatory miRNAs in the pathogenesis of cancer has been further highlighted by the observed dysregulation of these transcripts in the cancer stem cells (CSCs) (171). These miRNAs have been shown to target several genes that regulate cell cycle progression among them are PTEN (172), JAK1, SOX4, STAT3, AKT, EZH1, HMGA2 (173), CDK4/6, NOTCH1 (174), and ZEB1/2 (175). Therefore, cell-cycle regulatory miRNAs represent important targets for intervention with the invasive and metastatic properties of cancer cells which are associated with CSC phenotypes. Furthermore, identification of miRNAs with distinctive functions in CSCs and normal stem cells would facilitate design of specific targeted therapies for cancer patients with fewer side effects in normal tissues. This research avenue needs to be explored in future studies.
As lncRNAs have important roles in the regulation of activity of miRNAs through serving as molecular sponges for these small transcripts, identification of this type of interactions between cell cycle-related lncRNAs and miRNAs would pave the way for better recognition of the molecular mechanism of cell cycle progression. Several lncRNAs act as competing endogenous RNAs for miRNAs, thus regulating expression of cell cycle-associated miRNAs. Examples include (but not limited to): loc285194/miR-211 (176), HOTAIR/miR-1 (23), HOTAIR/miR-206 (24), and ANRIL/miR-384 (177). In addition to this kind of interaction between lncRNAs and miRNAs, some lncRNAs serve as precursors for miRNAs. Such situation exists between H19 and miR-675 (178). While E2F1 enhances expression of H19 lncRNA (179), miR-675 suppresses pRB expression (180). In turn, pRB inhibits E2F-associated transcription of H19 constructing a self-regulated network between H19 and pRB (13). These examples obviously show the complex network between lncRNAs, miRNAs, and TFs.
Expression of cell cycle-associated non-coding RNAs directly influences the survival of patients with diverse cancer types. This speculation is based on the obtained data from both high and low throughput studies. An example of former type of studies is a study which assessed RNA seq data of a large cohort of patients with colorectal cancer. Authors have reported several differentially expressed cell cycle genes and miRNAs which regulate expression of these genes. Subsequently, they verified correlations between expression levels of these genes/miRNAs and patients’ survival (181). Moreover, these non-coding RNAs can contribute in the construction of diagnostic panels for diverse types of cancers.
Taken together, cell cycle-associated lncRNAs/miRNAs are potential therapeutic targets for management of cancer and possible biomarkers for prediction of cancer course. However, lack of specificity in the regulatory roles of some of these transcripts limits their application in the clinical settings. Future studies should focus on identification of the network between these two kinds of transcripts and TFs using high throughput techniques. The results of these studies would fulfill the prerequisite step for design of targeted therapies.
Author Contributions
SG-F and MT wrote the manuscript and contributed in study design. HS and FTA collected the data and designed the tables. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. van den Heuvel S. Cell-cycle regulation. In: WormBook: The Online Review of C elegans Biology. Pasadena (CA, USA): WormBook (2005). doi: 10.1895/wormbook.1.28.1
2. Nurse P. Cyclin dependent kinases and cell cycle control (nobel lecture). Chembiochem. (2002) 3(7):596–603. doi: 10.1002/1439-7633(20020703)3:7<596::AID-CBIC596>3.0.CO;2-U
3. Perry JA, Kornbluth S. Cdc25 and Wee1: analogous opposites? Cell Div (2007) 2:12–. doi: 10.1186/1747-1028-2-12
4. Carnero A, Hannon G. The INK4 family of CDK inhibitors. In: Cyclin Dependent Kinase (CDK) Inhibitors. Berlin, Heidelberg: Springer (1998). p. 43–55. doi: 10.1007/978-3-642-71941-7_3
5. Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev (1999) 13(12):1501–12. doi: 10.1101/gad.13.12.1501
6. Vidal A, Koff A. Cell-cycle inhibitors: three families united by a common cause. Gene (2000) 247(1-2):1–15. doi: 10.1016/S0378-1119(00)00092-5
7. Duronio RJ, Xiong Y. Signaling pathways that control cell proliferation. Cold Spring Harb Perspect Biol (2013) 5(3):a008904–a. doi: 10.1101/cshperspect.a008904
8. Mazumder S, DuPree EL, Almasan A. A dual role of cyclin E in cell proliferation and apoptosis may provide a target for cancer therapy. Curr Cancer Drug Targets (2004) 4(1):65–75. doi: 10.2174/1568009043481669
9. van Leuken R, Clijsters L, Wolthuis R. To cell cycle, swing the APC/C. Biochim Biophys Acta (BBA)-Reviews Cancer (2008) 1786(1):49–59. doi: 10.1016/j.bbcan.2008.05.002
10. Schatten H. Mitosis. In: Maloy S, Hughes K, editors, Brenner’s Encyclopedia of Genetics (Second Edition). San Diego: Academic Press (2013) p. 448–51. doi: 10.1016/B978-0-12-374984-0.00962-1
11. Gavet O, Pines J. Activation of cyclin B1–Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J Cell Biol (2010) 189(2):247–59. doi: 10.1083/jcb.200909144
12. Chen J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb Perspect Med (2016) 6(3):a026104–a. doi: 10.1101/cshperspect.a026104
13. Kitagawa M, Kitagawa K, Kotake Y, Niida H, Ohhata T. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci (2013) 70(24):4785–94. doi: 10.1007/s00018-013-1423-0
14. Carninci P, Kasukawa T, Katayama S, Gough J, Frith M, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science (2005) 309(5740):1559–63. doi: 10.1126/science.1112014
15. Kitagawa M, Kotake Y, Ohhata T. Long non-coding RNAs involved in cancer development and cell fate determination. Curr Drug Targets (2012) 13(13):1616–21. doi: 10.2174/138945012803530026
16. Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta (BBA)-Molecular Basis Dis (2011) 1812(5):592–601. doi: 10.1016/j.bbadis.2011.02.002
17. O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (2018) 9:402. doi: 10.3389/fendo.2018.00402
18. Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PloS Genet (2013) 9(3):e1003368. doi: 10.1371/journal.pgen.1003368
19. Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res (2007) 67(8):3963–9. doi: 10.1158/0008-5472.CAN-06-2004
20. Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15 INK4B tumor suppressor gene. Oncogene (2011) 30(16):1956–62. doi: 10.1038/onc.2010.568
21. Yang F, Zhang L, Huo Xs, Yuan Jh, Xu D, Yuan Sx, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology (2011) 54(5):1679–89. doi: 10.1002/hep.24563
22. Zhang J-X, Han L, Bao Z-S, Wang Y-Y, Chen L-Y, Yan W, et al. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol (2013) 15(12):1595–603. doi: 10.1093/neuonc/not131
23. Ren K, Li Y, Lu H, Li Z, Li Z, Wu K, et al. Long noncoding RNA HOTAIR controls cell cycle by functioning as a competing endogenous RNA in esophageal squamous cell carcinoma. Trans Oncol (2016) 9(6):489–97. doi: 10.1016/j.tranon.2016.09.005
24. Chang L, Guo R, Yuan Z, Shi H, Zhang D. LncRNA HOTAIR regulates CCND1 and CCND2 expression by sponging miR-206 in ovarian cancer. Cell Physiol Biochem (2018) 49(4):1289–303. doi: 10.1159/000493408
25. Luan W, Li L, Shi Y, Bu X, Xia Y, Wang J, et al. Long non-coding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget (2016) 7(39):63901. doi: 10.18632/oncotarget.11564
26. Meseure D, Vacher S, Alsibai KD, Nicolas A, Chemlali W, Caly M, et al. Expression of ANRIL–polycomb complexes–CDKN2A/B/ARF genes in breast tumors: identification of a two-gene (EZH2/CBX7) signature with independent prognostic value. Mol Cancer Res (2016) 14(7):623–33. doi: 10.1158/1541-7786.MCR-15-0418
27. Zhu Y, Dai B, Zhang H, Shi G, Shen Y, Ye D. Long non-coding RNA LOC572558 inhibits bladder cancer cell proliferation and tumor growth by regulating the AKT–MDM2–p53 signaling axis. Cancer Lett (2016) 380(2):369–74. doi: 10.1016/j.canlet.2016.04.030
28. Wan L, Sun M, Liu G-J, Wei C-C, Zhang E-B, Kong R, et al. Long noncoding RNA PVT1 promotes non–small cell lung cancer cell proliferation through epigenetically regulating LATS2 expression. Mol Cancer Ther (2016) 15(5):1082–94. doi: 10.1158/1535-7163.MCT-15-0707
29. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell (2010) 142(3):409–19. doi: 10.1016/j.cell.2010.06.040
30. Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet (2011) 43(7):621–9. doi: 10.1038/ng.848
31. Wang C, Yang Y, Zhang G, Li J, Wu X, Ma X, et al. Long noncoding RNA EMS connects c-Myc to cell cycle control and tumorigenesis. Proc Natl Acad Sci (2019) 116(29):14620–9. doi: 10.1073/pnas.1903432116
32. Wang R, Ma Z, Feng L, Yang Y, Tan C, Shi Q, et al. LncRNA MIR31HG targets HIF1A and P21 to facilitate head and neck cancer cell proliferation and tumorigenesis by promoting cell-cycle progression. Mol Cancer (2018) 17(1):162. doi: 10.1186/s12943-018-0916-8
33. Fei F, He Y, He S, He Z, Wang Y, Wu G, et al. Expression of Concern: LncRNA SNHG3 enhances the malignant progressof glioma through silencing KLF2 and p21. Biosci Rep (2020) 40(7):BSR–20180420_EOC. doi: 10.1042/BSR-20180420_EOC
34. Xu K, Zhang Z, Qian J, Wang S, Yin S, Xie H, et al. LncRNA FOXD2-AS1 plays an oncogenic role in hepatocellular carcinoma through epigenetically silencing CDKN1B (p27) via EZH2. Exp Cell Res (2019) 380(2):198–204. doi: 10.1016/j.yexcr.2019.04.016
35. Wang XY, Jian X, Sun BQ, Ge XS, Huang FJ, Chen YQ. LncRNA ROR1-AS1 promotes colon cancer cell proliferation bysuppressing the expression of DUSP5/CDKN1A. Eur Rev Med PharmacolSci (2020) 24(3):1116–25. doi: 10.26355/eurrev_202002_20162
36. Long B, Li N, Xu X-X, Li X-X, Xu X-J, Liu J-Y, et al. Long noncoding RNA LOXL1-AS1 regulates prostate cancer cell proliferation and cell cycle progression through miR-541-3p and CCND1. Biochem Biophys Res Commun (2018) 505(2):561–8. doi: 10.1016/j.bbrc.2018.09.160
37. Fu X, Wang D, Shu T, Cui D, Fu Q. LncRNA NR2F2-AS1 positively regulates CDK4 to promote cancer cell proliferation in prostate carcinoma. Aging male Off J Int Soc Study Aging Male (2019) 1–7. doi: 10.1080/13685538.2019.1670157
38. Li J, Zhang Z, Xiong L, Guo C, Jiang T, Zeng L, et al. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun (2017) 487(1):146–52. doi: 10.1016/j.bbrc.2017.03.169
39. Dai X, Liu L, Liang Z, Guo K, Xu S, Wang H. Silencing of lncRNA MALAT1 inhibits cell cycle progression via androgen receptor signaling in prostate cancer cells. Pathology-Research Pract (2019) 215(4):712–21. doi: 10.1016/j.prp.2019.01.011
40. Luo Y, Lin J, Zhang Y, Dai G, Li A, Liu X. LncRNA PCAT6 predicts poor prognosis in hepatocellular carcinoma and promotes proliferation through the regulation of cell cycle arrest and apoptosis. Cell Biochem Funct (2020) 38(7):895–904. doi: 10.1002/cbf.3510
41. Shao K, Shi T, Yang Y, Wang X, Xu D, Zhou P. Highly expressed lncRNA CRNDE promotes cell proliferation through Wnt/β-catenin signaling in renal cell carcinoma. Tumor Biol (2016) 37(12):15997–6004. doi: 10.1007/s13277-016-5440-0
42. Yan Z, He L, Qiu J, Deng W, Lu J, Yuan Z, et al. LncRNA HOTAIR participates in the development and progression ofadrenocortical carcinoma via regulating cell cycle. Eur Rev Med PharmacolSci (2018) 22:6640–9. doi: 10.26355/eurrev_201810_16139
43. Wei Y, Sun Q, Zhao L, Wu J, Chen X, Wang Y, et al. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol (2016) 33(8):88. doi: 10.1007/s12032-016-0804-2
44. Qi Y, Wang Z, Wu F, Yin B, Jiang T, Qiang B, et al. Long noncoding RNA HOXD-AS2 regulates cell cycle to promote glioma progression. J Cell Biochem (2019) 120(5):8343–51. doi: 10.1002/jcb.28117
45. Dai W, Tian C, Jin S. Effect of lncRNA ANRIL silencing on anoikis and cell cycle in human glioma via microRNA-203a. OncoTargets Ther (2018) 11:5103–9. doi: 10.2147/OTT.S169809
46. Sun Y, Niu X, Wang G, Qiao X, Chen L, Zhong M. A Novel lncRNA ENST00000512916 Facilitates Cell Proliferation, Migration and Cell Cycle Progression in Ameloblastoma. OncoTargets Ther (2020) 13:1519. doi: 10.2147/OTT.S236158
47. Yang G, He F, Duan H, Shen J, Dong Q. lncRNA FLVCR-AS1 promotes osteosarcoma growth by targeting miR381-3p/CCND1. OncoTargets Ther (2020) 13:163. doi: 10.2147/OTT.S214813
48. Yu X, Pang L, Yang T, Liu P. lncRNA LINC01296 regulates the proliferation, metastasis and cell cycle of osteosarcoma through cyclin D1. Oncol Rep (2018) 40(5):2507–14. doi: 10.3892/or.2018.6674
49. Qiu X, Dong J, Zhao Z, Li J, Cai X. LncRNA LINC00668 promotes the progression of breast cancer by inhibiting apoptosis and accelerating cell cycle. OncoTargets Ther (2019) 12:5615. doi: 10.2147/OTT.S188933
50. Feng T, Shao F, Wu Q, Zhang X, Xu D, Qian K, et al. miR-124 downregulation leads to breast cancer progression via LncRNA-MALAT1 regulation and CDK4/E2F1 signal activation. Oncotarget (2016) 7(13):16205–16. doi: 10.18632/oncotarget.7578
51. Hu C, Liang Y, Hu J, Liu L, Liang J, Wang R. LncRNA RUSC1-AS1 promotes the proliferation of breast cancer cellsby epigenetic silence of KLF2 and CDKN1A. Eur Rev Med Pharmacol Sci(2019) 23(15):6602–11. doi: 10.26355/eurrev_201908_18548
52. Jafari-Oliayi A, Asadi MH. SNHG6 is upregulated in primary breast cancers and promotes cell cycle progression in breast cancer-derived cell lines. Cell Oncol (Dordrecht) (2019) 42(2):211–21. doi: 10.1007/s13402-019-00422-6
53. Hui B, Xu Y, Zhao B, Ji H, Ma Z, Xu S, et al. Overexpressed long noncoding RNA TUG1 affects the cell cycle, proliferation, and apoptosis of pancreatic cancer partly through suppressing RND3 and MT2A. OncoTargets Ther (2019) 12:1043–57. doi: 10.2147/OTT.S188396
54. Huang G, He X, Wei XL. lncRNA NEAT1 promotes cell proliferation and invasion by regulating miR−365/RGS20 in oral squamous cell carcinoma. Oncol Rep (2018) 39(4):1948–56. doi: 10.3892/or.2018.6283
55. Ye LC, Chen T, Zhu DX, Lv SX, Qiu JJ, Xu J, et al. Downregulated long non-coding RNA CLMAT3 promotes the proliferation of colorectal cancer cells by targeting regulators of the cell cycle pathway. Oncotarget (2016) 7(37):58931–8. doi: 10.18632/oncotarget.10431
56. Liu J, Qian J, Mo Q, Tang L, Xu Q. LncRNA NR2F2-AS1 silencing induces cell cycle arrest in G0/G1 phase via downregulating Cyclin D1 in colorectal cancer. Cancer Manage Res (2020) 12:1835. doi: 10.2147/CMAR.S221996
57. Li F, Jiang Z, Shao X, Zou N. Downregulation of lncRNA NR2F2 Antisense RNA 1 Induces G1 Arrest of Colorectal Cancer Cells by Downregulating Cyclin-Dependent Kinase 6. Digestive Dis Sci (2020) 65(2):464–9. doi: 10.1007/s10620-019-05782-5
58. Zhao Y, Qin ZS, Feng Y, Tang XJ, Zhang T, Yang L. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1)promote cell proliferation in colorectal cancer by affecting P53. Eur RevMed Pharmacol Sci (2018)22(4):976–84. doi: 10.26355/eurrev_201802_14379
59. Na H, Li X, Zhang X, Xu Y, Sun Y, Cui J, et al. lncRNA STEAP3-AS1 Modulates Cell Cycle Progression via Affecting CDKN1C Expression through STEAP3 in Colon Cancer. Mol Ther Nucleic Acids (2020) 21:480–91. doi: 10.1016/j.omtn.2020.06.011
60. Ding S, Qu W, Jiao Y, Zhang J, Zhang C, Dang S. LncRNA SNHG12 promotes the proliferation and metastasis of papillary thyroid carcinoma cells through regulating wnt/β-catenin signaling pathway. Cancer Biomarkers section A Dis Markers (2018) 22(2):217–26. doi: 10.3233/CBM-170777
61. Li Z, Gao B, Hao S, Tian W, Chen Y, Wang L, et al. Knockdown of lncRNA-PANDAR suppresses the proliferation, cell cycleand promotes apoptosis in thyroid cancer cells. EXCLI J(2017) 16:354–62. doi: 10.17179/excli2017-113
62. Liu HT, Liu S, Liu L, Ma RR, Gao P. EGR1-Mediated Transcription of lncRNA-HNF1A-AS1 Promotes Cell-Cycle Progression in Gastric Cancer. Cancer Res (2018) 78(20):5877–90. doi: 10.1158/0008-5472.CAN-18-1011
63. Zhang L, Kang W, Lu X, Ma S, Dong L, Zou B. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle (Georgetown Tex) (2018) 17(15):1886–900. doi: 10.1080/15384101.2018.1502574
64. Liu L, Chen Y, Li Q, Duan P. lncRNA HNF1A-AS1 modulates non-small cell lung cancer progression by targeting miR-149-5p/Cdk6. J Cell Biochem (2019) 120(11):18736–50. doi: 10.1002/jcb.29186
65. Zhuang M, Li L, Ma J. LncRNA HOTTIP promotes proliferation and cell cycle progression ofacute myeloid leukemia cells. Eur Rev Med Pharmacol Sci (2019) 23(7):2908–15. doi: 10.26355/eurrev_201904_17569
66. Liu F, Xiao Y, Ma L, Wang J. Regulating of cell cycle progression by the lncrna cdkn2b-as1/mir-324-5p/rock1 axis in laryngeal squamous cell cancer. Int J Biol Markers (2020) 35(1):47–56. doi: 10.1177/1724600819898489
67. Niu Y, Ma F, Huang W, Fang S, Li M, Wei T, et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer (2017) 16(1):1–13. doi: 10.1186/s12943-016-0575-6
68. Cai Y, Dong Z, Wang J. LncRNA NNT-AS1 is a major mediator of cisplatin chemoresistance innon-small cell lung cancer through MAPK/Slug pathway. Eur Rev Med PharmacolSci (2018) 22(15):4879–87. doi: 10.26355/eurrev_201808_15624
69. Han D, Wang J, Cheng G. LncRNA NEAT1 enhances the radio-resistance of cervical cancer via miR-193b-3p/CCND1 axis. Oncotarget (2018) 9(2):2395. doi: 10.18632/oncotarget.23416
70. Liu F, Chen N, Gong Y, Xiao R, Wang W, Pan Z. The long non-coding RNA NEAT1 enhances epithelial-to-mesenchymal transition and chemoresistance via the miR-34a/c-Met axis in renal cell carcinoma. Oncotarget. (2017) 8(38):62927. doi: 10.18632/oncotarget.17757
71. Li L, Gu M, You B, Shi S, Shan Y, Bao L, et al. Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci (2016) 107(9):1215–22. doi: 10.1111/cas.12989
72. Liang Y, Li Y, Song X, Zhang N, Sang Y, Zhang H, et al. Long noncoding RNA LINP1 acts as an oncogene and promotes chemoresistance in breast cancer. Cancer Biol Ther (2018) 19(2):120–31. doi: 10.1080/15384047.2017.1394543
73. Li Z, Zhao L, Wang Q. Overexpression of long non-coding RNA HOTTIP increases chemoresistance of osteosarcoma cell by activating the Wnt/β-catenin pathway. Am J Trans Res (2016) 8(5):2385.
74. Chen D, Farwell MA, Zhang B. MicroRNA as a new player in the cell cycle. J Cell Physiol (2010) 225(2):296–301. doi: 10.1002/jcp.22234
75. Takeshita F, Patrawala L, Osaki M, Takahashi R-u, Yamamoto Y, Kosaka N, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther (2010) 18(1):181–7. doi: 10.1038/mt.2009.207
76. Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res (2008) 36(16):5391. doi: 10.1093/nar/gkn522
77. Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol (2007) 27(6):2240–52. doi: 10.1128/MCB.02005-06
78. Wu J, Lv Q, He J, Zhang H, Mei X, Cui K, et al. MicroRNA-188 suppresses G1/S transition by targeting multiple cyclin/CDK complexes. Cell Commun Signal (2014) 12:66–. doi: 10.1186/PREACCEPT-1482274339133333
79. Xie D, Song H, Wu T, Li D, Hua K, Xu H, et al. MicroRNA−424 serves an anti−oncogenic role by targeting cyclin−dependent kinase 1 in breast cancer cells. Oncol Rep (2018) 40(6):3416–26. doi: 10.3892/or.2018.6741
80. Farina FM, Inguscio A, Kunderfranco P, Cortesi A, Elia L, Quintavalle M. MicroRNA-26a/cyclin-dependent kinase 5 axis controls proliferation, apoptosis and in vivo tumor growth of diffuse large B-cell lymphoma cell lines. Cell Death Dis (2017) 8(6):e2890–e. doi: 10.1038/cddis.2017.291
81. Kapora E, Feng S, Liu W, Sakhautdinova I, Gao B, Tan W. MicroRNA-505-5p functions as a tumor suppressor by targetingcyclin-dependent kinase 5 in cervical cancer. Biosci Rep (2019) 39(7):BSR20191221. doi: 10.1042/BSR20191221
82. Contreras-Vallejos E, Utreras E, Gonzalez-Billault C. Going out of the brain: non-nervous system physiological and pathological functions of Cdk5. Cell Signalling (2012) 24(1):44–52. doi: 10.1016/j.cellsig.2011.08.022
83. Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene. (2009) 28(1):140–5. doi: 10.1038/onc.2008.372
84. Lin RJ, Lin YC, Yu AL. miR-149* induces apoptosis by inhibiting Akt1 and E2F1 in human cancer cells. Mol carcinogenesis (2010) 49(8):719–27. doi: 10.1002/mc.20647
85. Lee K-H, Chen Y-L, Yeh S, Hsiao M, Lin J, Goan Y, et al. MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene (2009) 28(38):3360–70. doi: 10.1038/onc.2009.192
86. Guo X, Guo L, Ji J, Zhang J, Zhang J, Chen X, et al. miRNA-331-3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochem Biophys Res Commun (2010) 398(1):1–6. doi: 10.1016/j.bbrc.2010.05.082
87. Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci (2006) 103(7):2257–61. doi: 10.1073/pnas.0510565103
88. Benetti R, Gonzalo S, Jaco I, Muñoz P, Gonzalez S, Schoeftner S, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol (2008) 15(3):268. doi: 10.1038/nsmb.1399
89. Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci (2008) 105(8):2889–94. doi: 10.1073/pnas.0800178105
90. Cloonan N, Brown MK, Steptoe AL, Wani S, Chan WL, Forrest AR, et al. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol (2008) 9(8):1–14. doi: 10.1186/gb-2008-9-8-r127
91. Tian S, Huang S, Wu S, Guo W, Li J, He X. MicroRNA-1285 inhibits the expression of p53 by directly targeting its 3′ untranslated region. Biochem Biophys Res Commun (2010) 396(2):435–9. doi: 10.1016/j.bbrc.2010.04.112
92. Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, Appella E, et al. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res (2008) 68(9):3193–203. doi: 10.1158/0008-5472.CAN-07-2780
93. He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer (2007) 7(11):819–22. doi: 10.1038/nrc2232
94. Gang L, Qun L, Liu W-D, Li Y-S, Xu Y-Z, Yuan D-T. MicroRNA-34a promotes cell cycle arrest and apoptosis and suppresses cell adhesion by targeting DUSP1 in osteosarcoma. Am J Trans Res (2017) 9(12):5388.
95. Zhu H, Wang S, Shen H, Zheng X, Xu X. SP1/AKT/FOXO3 Signaling Is Involved in miR-362-3p-Mediated Inhibition of Cell-Cycle Pathway and EMT Progression in Renal Cell Carcinoma. Front Cell Dev Biol (2020) 8:297. doi: 10.3389/fcell.2020.00297
96. Liu J, Chen W, Zhang H, Liu T, Zhao L. miR−214 targets the PTEN−mediated PI3K/Akt signaling pathway and regulates cell proliferation and apoptosis in ovarian cancer. Oncol Lett (2017) 14(5):5711–8. doi: 10.3892/ol.2017.6953
97. Chen L, Ma D, Li Y, Li X, Zhao L, Zhang J, et al. Effect of long non-coding RNA PVT1 on cell proliferation and migration in melanoma. Int J Mol Med (2018) 41(3):1275–82. doi: 10.3892/ijmm.2017.3335
98. Xiao C, Hong H, Yu H, Yuan J, Guo C, Cao H, et al. MiR-340 affects gastric cancer cell proliferation, cycle, and apoptosis through regulating SOCS3/JAK-STAT signaling pathway. Immunopharmacol Immunotoxicol (2018) 40(4):278–83. doi: 10.1080/08923973.2018.1455208
99. Zhang Z, Li X, Xiao Q, Wang Z. MiR-574-5p mediates the cell cycle and apoptosis in thyroid cancer cells via Wnt/β-catenin signaling by repressing the expression of Quaking proteins. Oncol Lett (2018) 15(4):5841–8. doi: 10.3892/ol.2018.8067
100. Liao Z, Zheng Q, Wei T, Zhang Y, Ma J, Zhao Z, et al. MicroRNA-561 affects proliferation and cell cycle transition through PTEN/AKT signaling pathway by targeting P-REX2a in NSCLC. Oncol Res Featuring Preclinical Clin Cancer Ther (2020) 28(2):147–59. doi: 10.3727/096504019X15732109856009
101. Chen X-E, Chen P, Chen S-S, Ma T, Shi G, Zhou Y, et al. miR−106b−5p promotes cell cycle progression of malignant melanoma by targeting PTEN. Oncol Rep (2018) 39(1):331–7. doi: 10.3892/or.2017.6099
102. Xiang Y, Yang Y, Lin C, Wu J, Zhang X. MiR-23a-3p promoted G1/S cell cycle transition by targetingprotocadherin17 in hepatocellular carcinoma. J Physiol Biochem (2020) 76:123–34. doi: 10.1007/s13105-020-00726-4
103. Qin X, Chen J, Wu L, Liu Z. MiR-30b-5p acts as a tumor suppressor, repressing cell proliferation and cell cycle in human hepatocellular carcinoma. Biomedicine Pharmacother (2017) 89:742–50. doi: 10.1016/j.biopha.2017.02.062
104. Zhang D, Liu E, Kang J, Yang X, Liu H. MiR-3613-3p affects cell proliferation and cell cycle in hepatocellular carcinoma. Oncotarget (2017) 8(54):93014. doi: 10.18632/oncotarget.21745
105. Luo Q, Zhang Z, Dai Z, Basnet S, Li S, Xu B, et al. Tumor-suppressive microRNA-195-5p regulates cell growth and inhibits cell cycle by targeting cyclin dependent kinase 8 in colon cancer. Am J Trans Res (2016) 8(5):2088.
106. Kang MR, Park KH, Yang J-O, Lee CW, Oh SJ, Yun J, et al. miR-6734 up-regulates p21 gene expression and induces cell cycle arrest and apoptosis in colon cancer cells. PloS One (2016) 11(8):e0160961. doi: 10.1371/journal.pone.0160961
107. Morimoto Y, Mizushima T, Wu X, Okuzaki D, Yokoyama Y, Inoue A, et al. miR-4711-5p regulates cancer stemness and cell cycle progression via KLF5, MDM2 and TFDP1 in colon cancer cells. Br J Cancer (2020) 122(7):1037–49. doi: 10.1038/s41416-020-0758-1
108. Noorolyai S, Baghbani E, Maleki LA, Kojabad AB, Shanehbansdi D, Shahgoli VK, et al. Restoration of miR-193a-5p and miR-146 a-5p expression induces G1 arrest in colorectal cancer through targeting of MDM2/p53. Adv Pharm Bull (2020) 10(1):130. doi: 10.15171/apb.2020.017
109. Lähteenvuo J, Hätinen OP, Kuivanen A, Huusko J, Paananen J, Lähteenvuo M, et al. Susceptibility to Cardiac Arrhythmias and Sympathetic Nerve Growthin VEGF-B Overexpressing Myocardium. Mol Ther (2020) 28(7):1731–40. doi: 10.1016/j.ymthe.2020.03.011
110. Zhou G-Q, Han F, Shi Z-L, Yu L, Li X-F, Yu C, et al. miR-133a-3p targets SUMO-specific protease 1 to inhibit cell proliferation and cell cycle progress in colorectal cancer. Oncol Res Featuring Preclinical Clin Cancer Ther (2018) 26(5):795–800. doi: 10.3727/096504017X15004613574679
111. Zhang L, He X, Li F, Pan H, Huang X, Wen X, et al. The miR-181 family promotes cell cycle by targeting CTDSPL, a phosphatase-like tumor suppressor in uveal melanoma. J Exp Clin Cancer Res (2018) 37(1):15. doi: 10.1186/s13046-018-0679-5
112. Yang Y, Cui H, Wang X. Downregulation of EIF5A2 by miR-221-3p inhibits cell proliferation, promotes cell cycle arrest and apoptosis in medulloblastoma cells. Biosci Biotechnol Biochem (2019) 83(3):400–8. doi: 10.1080/09168451.2018.1553604
113. Jiang K, Zhi T, Xu W, Xu X, Wu W, Yu T, et al. MicroRNA-1468-5p inhibits glioma cell proliferation and induces cellcycle arrest by targeting RRM1. Am J Cancer Res (2017) 7(4):784–800.
114. Deng M, Zeng C, Lu X, He X, Zhang R, Qiu Q, et al. miR-218 suppresses gastric cancer cell cycle progression through the CDK6/Cyclin D1/E2F1 axis in a feedback loop. Cancer Lett (2017) 403:175–85. doi: 10.1016/j.canlet.2017.06.006
115. Luo H, Xu R, Chen B, Dong S, Zhou F, Yu T, et al. MicroRNA-940 inhibits glioma cells proliferation and cell cycleprogression by targeting CKS1. Am J Trans Res (2019) 11(8):4851–65.
116. Zhu Y, Tang H, Zhang L, Gong L, Wu G, Ni J, et al. Suppression of miR-21-3p enhances TRAIL-mediated apoptosis in liver cancer stem cells by suppressing the PI3K/Akt/Bad cascade via regulating PTEN. Cancer Manage Res (2019) 11:955. doi: 10.2147/CMAR.S183328
117. He J, Ge Q, Lin Z, Shen W, Lin R, Wu J, et al. MiR-129-5p induces cell cycle arrest through modulatingHOXC10/Cyclin D1 to inhibit gastric cancer progression. FASEB J (2020) 34(6):8544–57. doi: 10.1096/fj.201903217R
118. Li J, Sun P, Yue Z, Zhang D, You K, Wang J. miR-144-3p induces cell cycle arrest and apoptosis in pancreatic cancer cells by targeting proline-rich protein 11 expression via the mitogen-activated protein kinase signaling pathway. DNA Cell Biol (2017) 36(8):619–26. doi: 10.1089/dna.2017.3656
119. Shi X, Sheng W, Jia C, Tang J, Dong M. Hsa-MiR-590-3p promotes the malignancy progression of pancreaticductal carcinoma cell by inhibiting the expression of p27 and PPP2R2A via G1/S cell cyclepathway. Onco Targets Ther (2020) 13:11045–58. doi: 10.2147/OTT.S260499
120. Zhu Y, Gu J, Li Y, Peng C, Shi M, Wang X, et al. MiR-17-5p enhances pancreatic cancer proliferation by altering cell cycle profiles via disruption of RBL2/E2F4-repressing complexes. Cancer Lett (2018) 412:59–68. doi: 10.1016/j.canlet.2017.09.044
121. Wu H, Liu HY, Liu WJ, Shi YL, Bao D. miR-377-5p inhibits lung cancer cell proliferation, invasion, and cell cycle progression by targeting AKT1 signaling. J Cell Biochem (2019) 120(5):8120–8. doi: 10.1002/jcb.28091
122. Hu H, Xu H, Lu F, Zhang J, Xu L, Xu S, et al. Exosome-derived miR-486-5p regulates cell cycle, proliferation and metastasis in lung adenocarcinoma via targeting NEK2. Front Bioeng Biotechnol (2020) 8:259. doi: 10.3389/fbioe.2020.00259
123. Feng H, Ge F, Du L, Zhang Z, Liu D. MiR-34b-3p represses cell proliferation, cell cycle progression and cell apoptosis in non-small-cell lung cancer (NSCLC) by targeting CDK4. J Cell Mol Med (2019) 23(8):5282–91. doi: 10.1111/jcmm.14404
124. Li D, Li D-Q, Liu D, Tang X-J. MiR-613 induces cell cycle arrest by targeting CDK4 in non-small cell lung cancer. Cell Oncol (2016) 39(2):139–47. doi: 10.1007/s13402-015-0262-4
125. Xin C, Lu S, Li Y, Zhang Y, Tian J, Zhang S, et al. miR-671-5p Inhibits Tumor Proliferation by Blocking Cell Cycle in Osteosarcoma. DNA Cell Biol (2019) 38(9):996–1004. doi: 10.1089/dna.2019.4870
126. Zhang C, Li L, She C, Xie Y, Ge D, Dong Q. MiR-299-5p targets cell cycle to promote cell proliferation andprogression of osteosarcoma. Eur Rev Med Pharmacol Sci (2018) 22(9):2606–13. doi: 10.26355/eurrev_201805_14954
127. Gai P, Sun H, Wang G, Xu Q, Qi X, Zhang Z, et al. miR-22 promotes apoptosis of osteosarcoma cells via inducing cell cycle arrest. Oncol Lett (2017) 13(4):2354–8. doi: 10.3892/ol.2017.5674
128. Ma J, Zhang F, Sun P. miR-140-3p impedes the proliferation of human cervical cancer cells by targeting RRM2 to induce cell-cycle arrest and early apoptosis. Bioorg Med Chem (2020) 28(3):115283. doi: 10.1016/j.bmc.2019.115283
129. Su Z, Yang H, Zhao M, Wang Y, Deng G, Chen R. MicroRNA-92a promotes cell proliferation in cervical cancer via inhibiting p21 expression and promoting cell cycle progression. Oncol Res Featuring Preclinical Clin Cancer Ther (2017) 25(1):137–45. doi: 10.3727/096504016X14732772150262
130. Tang Z, Fang Y, Du R. MicroRNA-107 induces cell cycle arrests by directly targeting cyclin E1 in ovarian cancer. Biochem Biophys Res Commun (2019) 512(2):331–7. doi: 10.1016/j.bbrc.2019.03.009
131. Lei Z, Ma X, Li H, Zhang Y, Gao Y, Fan Y, et al. Up-regulation of miR-181a in clear cell renal cell carcinoma isassociated with lower KLF6 expression, enhanced cell proliferation, accelerated cell cycle transition, and diminished apoptosis. In: Urologic Oncology: Seminars and Original Investigations. Elsevier Inc (USA): Elsevier (2018). doi: 10.1016/j.urolonc.2017.09.019
132. Qin H, Liu W. MicroRNA-99a-5p suppresses breast cancer progression and cell-cycle pathway through downregulating CDC25A. J Cell Physiol (2019) 234(4):3526–37. doi: 10.1002/jcp.26906
133. Peng X, Yan B, Shen Y. MiR-1301-3p inhibits human breast cancer cell proliferation by regulating cell cycle progression and apoptosis through directly targeting ICT1. Breast Cancer (2018) 25(6):742–52. doi: 10.1007/s12282-018-0881-5
134. Chen P, Xu W, Luo Y, Zhang Y, He Y, Yang S, et al. MicroRNA 543 suppresses breast cancer cell proliferation, blocks cell cycle and induces cell apoptosis via direct targeting of ERK/MAPK. OncoTargets Ther (2017) 10:1423. doi: 10.2147/OTT.S118366
135. Ying X, Zhang W, Fang M, Zhang W, Wang C, Han L. miR-345-5p regulates proliferation, cell cycle, and apoptosis of acute myeloid leukemia cells by targeting AKT2. J Cell Biochem (2019) 120(2):1620–9. doi: 10.1002/jcb.27461
136. Erdmann K, Kaulke K, Rieger C, Salomo K, Wirth MP, Fuessel S. MiR-26a and miR-138 block the G1/S transition by targeting the cell cycle regulating network in prostate cancer cells. J Cancer Res Clin Oncol (2016) 142(11):2249–61. doi: 10.1007/s00432-016-2222-4
137. Jin J, Wang Y, Xu Y, Zhou X, Liu Y, Li X, et al. MicroRNA-144 regulates cancer cell proliferation and cell-cycle transition in acute lymphoblastic leukemia through the interaction of FMN2. J Gene Med (2017) 19(6-7):e2898. doi: 10.1002/jgm.2898
138. Ke S, R-c Li, Lu J, Meng F-k, Feng Y-k, Fang M-h. MicroRNA-192 regulates cell proliferation and cell cycle transition in acute myeloid leukemia via interaction with CCNT2. Int J Hematol (2017) 106(2):258–65. doi: 10.1007/s12185-017-2232-2
139. Koo KH, Kwon H. MicroRNA miR-4779 suppresses tumor growth by inducing apoptosis and cell cycle arrest through direct targeting of PAK2 and CCND3. Cell Death Dis (2018) 9(2):1–14. doi: 10.1038/s41419-017-0100-x
140. Guo P, Xiong X, Zhang S, Peng D. miR-100 resensitizes resistant epithelial ovarian cancer to cisplatin. Oncol Rep (2016) 36(6):3552–8. doi: 10.3892/or.2016.5140
141. Chen X, Yue B, Zhang C, Qi M, Qiu J, Wang Y, et al. MiR-130a-3p inhibits the viability, proliferation, invasion, and cell cycle, and promotes apoptosis of nasopharyngeal carcinoma cells by suppressing BACH2 expression. Biosci Rep (2017) 37(3):BSR20160576. doi: 10.1042/BSR20160576
142. Fu S, Chen HH, Cheng P, Zhang CB, Wu Y. MiR-155 regulates oral squamous cell carcinoma Tca8113 cellproliferation, cycle, and apoptosis via regulating p27Kip1. Eur Rev Med Pharmacol Sci (2017) 21(5):937–44.
143. Wang K, Jin J, Ma T, Zhai H. MiR-376c-3p regulates the proliferation, invasion, migration, cell cycle and apoptosis of human oral squamous cancer cells by suppressing HOXB7. Biomedicine Pharmacother (2017) 91:517–25. doi: 10.1016/j.biopha.2017.04.050
144. Zhen Y, Fang W, Zhao M, Luo R, Liu Y, Fu Q, et al. miR-374a-CCND1-pPI3K/AKT-c-JUN feedback loop modulated by PDCD4 suppresses cell growth, metastasis, and sensitizes nasopharyngeal carcinoma to cisplatin. Oncogene (2017) 36(2):275–85. doi: 10.1038/onc.2016.201
145. Ma Q. MiR-219-5p suppresses cell proliferation and cell cycle progression in esophageal squamous cell carcinoma by targeting CCNA2. Cell Mol Biol Lett (2019) 24(1):4. doi: 10.1186/s11658-018-0129-6
146. Wang H, Fu L, Wei D, Wang B, Zhang C, Zhu T, et al. MiR-29c-3p Suppresses the Migration, Invasion and Cell Cycle in Esophageal Carcinoma via CCNA2/p53 Axis. Front Bioeng Biotechnol (2020) 8:75. doi: 10.3389/fbioe.2020.00075
147. Wu C, Ma L, Wei H, Nie F, Ning J, Jiang T. MiR-1256 inhibits cell proliferation and cell cycle progression inpapillary thyroid cancer by targeting 5-hydroxy tryptamine receptor 3A. Hum Cell (2020) 33:630–40. doi: 10.1007/s13577-020-00325-x
148. Sun J, Shi R, Zhao S, Li X, Lu S, Bu H, et al. E2F8, a direct target of miR-144, promotes papillary thyroid cancer progression via regulating cell cycle. J Exp Clin Cancer Res (2017) 36(1):1–14. doi: 10.1186/s13046-017-0504-6
149. Lei S-t, Shen F, Chen J-w, Feng J-h, Cai W-s, Shen L, et al. MiR-639 promoted cell proliferation and cell cycle in human thyroid cancer by suppressing CDKN1A expression. Biomedicine Pharmacother (2016) 84:1834–40. doi: 10.1016/j.biopha.2016.10.087
150. Li J, Liu X, Wang W, Li C. miR-133a-3p promotes apoptosis and induces cell cycle arrest by targeting CREB1 in retinoblastoma. Arch Med Sci: AMS (2020) 16(4):941. doi: 10.5114/aoms.2019.86901
151. Wan L-Y, Deng J, Xiang X-J, Zhang L, Yu F, Chen J, et al. miR-320 enhances the sensitivity of human colon cancer cells to chemoradiotherapy in vitro by targeting FOXM1. Biochem Biophys Res Commun (2015) 457(2):125–32. doi: 10.1016/j.bbrc.2014.11.039
152. Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, Maiello E, et al. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol Cancer Ther (2010) 9(8):2265–75. doi: 10.1158/1535-7163.MCT-10-0061
153. Sun C-C, Li S-J, Zhang F, Zhang Y-D, Zuo Z-Y, Xi Y-Y, et al. The novel miR-9600 suppresses tumor progression and promotes paclitaxel sensitivity in non–small-cell lung cancer through altering STAT3 expression. Mol Therapy-Nucleic Acids (2016) 5:e387. doi: 10.1038/mtna.2016.96
154. Yu S, Qin X, Chen T, Zhou L, Xu X, Feng J. MicroRNA-106b-5p regulates cisplatin chemosensitivity by targeting polycystic kidney disease-2 in non-small-cell lung cancer. Anti-cancer Drugs (2017) 28(8):852–60. doi: 10.1097/CAD.0000000000000524
155. Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z, et al. MicroRNA-122 sensitizes HCC cancer cells to adriamycin andvincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett (2011) 310(2):160–9. doi: 10.1016/j.canlet.2011.06.027
156. Xu L, Beckebaum S, Iacob S, Wu G, Kaiser GM, Radtke A, et al. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol (2014) 60(3):590–8. doi: 10.1016/j.jhep.2013.10.028
157. Kim C, Hong Y, Lee H, Kang H, Lee EK. MicroRNA-195 desensitizes HCT116 human colon cancer cells to 5-fluorouracil. Cancer Lett (2018) 412:264–71. doi: 10.1016/j.canlet.2017.10.022
158. Jiang Q, Zhang Y, Zhao M, Li Q, Chen R, Long X, et al. miR-16 induction after CDK4 knockdown is mediated by c-Myc suppression and inhibits cell growth as well as sensitizes nasopharyngeal carcinoma cells to chemotherapy. Tumor Biol (2016) 37(2):2425–33. doi: 10.1007/s13277-015-3966-1
159. Cao W, Wei W, Zhan Z, Xie D, Xie Y, Xiao Q. Regulation of drug resistance and metastasis of gastric cancer cells via the microRNA647-ANK2 axis. Int J Mol Med (2018) 41(4):1958–66. doi: 10.3892/ijmm.2018.3381
160. Sun K-k, Shen X-j, Yang D, Gan M-q, Liu G, Zhang Y-f, et al. MicroRNA-31 triggers G2/M cell cycle arrest, enhances the chemosensitivity and inhibits migration and invasion of human gastric cancer cells by downregulating the expression of zeste homolog 2 (ZH2). Arch Biochem Biophys (2019) 663:269–75. doi: 10.1016/j.abb.2019.01.023
161. Ghawanmeh T, Thunberg U, Castro J, Murray F, Laytragoon-Lewin N. miR-34a expression, cell cycle arrest and cell death of malignant mesothelioma cells upon treatment with radiation, docetaxel or combination treatment. Oncology (2011) 81(5-6):330–5. doi: 10.1159/000334237
162. Li Y, Zhang J, Liu Y, Zhang B, Zhong F, Wang S, et al. MiR-30a-5p confers cisplatin resistance by regulating IGF1R expression in melanoma cells. BMC Cancer (2018) 18(1):404. doi: 10.1186/s12885-018-4233-9
163. Park J-K, Lee EJ, Esau C, Schmittgen TD. Antisense inhibition of microRNA-21 or-221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. (2009) 38(7):e190–e9. doi: 10.1097/MPA.0b013e3181ba82e1
164. Ichikawa T, Sato F, Terasawa K, Tsuchiya S, Toi M, Tsujimoto G, et al. Trastuzumab produces therapeutic actions by upregulating miR-26a and miR-30b in breast cancer cells. PloS One (2012) 7(2):e31422. doi: 10.1371/journal.pone.0031422
165. Cataldo A, Cheung DG, Balsari A, Tagliabue E, Coppola V, Iorio MV, et al. miR-302b enhances breast cancer cell sensitivity to cisplatin by regulating E2F1 and the cellular DNA damage response. Oncotarget (2016) 7(1):786. doi: 10.18632/oncotarget.6381
166. Lezina L, Purmessur N, Antonov A, Ivanova T, Karpova E, Krishan K, et al. miR-16 and miR-26a target checkpoint kinases Wee1 and Chk1 in response to p53 activation by genotoxic stress. Cell Death Dis (2013) 4(12):e953–e. doi: 10.1038/cddis.2013.483
167. Zhang Y, Hu H, Song L, Cai L, Wei R, Jin W. Epirubicin-mediated expression of miR-302b is involved in osteosarcoma apoptosis and cell cycle regulation. Toxicol Lett (2013) 222(1):1–9. doi: 10.1016/j.toxlet.2013.06.242
168. Chen X, He L, Zhao Y, Li Y, Zhang S, Sun K, et al. Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discovery (2017) 3:17002–. doi: 10.1038/celldisc.2017.2
169. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell (2013) 52(1):101–12. doi: 10.1016/j.molcel.2013.08.027
170. Hydbring P, Wang Y, Fassl A, Li X, Matia V, Otto T, et al. Cell-cycle-targeting MicroRNAs as therapeutic tools against refractory cancers. Cancer Cell (2017) 31(4):576–90. doi: 10.1016/j.ccell.2017.03.004
171. Mens MMJ, Ghanbari M. Cell Cycle Regulation of Stem Cells by MicroRNAs. Stem Cell Rev Rep (2018) 14(3):309–22. doi: 10.1007/s12015-018-9808-y
172. Liu X, Feng J, Tang L, Liao L, Xu Q, Zhu S. The regulation and function of miR-21-FOXO3a-miR-34b/c signaling in breast cancer. Int J Mol Sci (2015) 16(2):3148–62. doi: 10.3390/ijms16023148
173. Liu S, Patel SH, Ginestier C, Ibarra I, Martin-Trevino R, Bai S, et al. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PloS Genet (2012) 8(6):e1002751. doi: 10.1371/journal.pgen.1002751
174. Engkvist M, Stratford E, Lorenz S, Meza-Zepeda L, Myklebost O, Munthe E. Analysis of the miR-34 family functions in breast cancer reveals annotation error of miR-34b. Sci Rep (2017) 7(1):1–10. doi: 10.1038/s41598-017-10189-1
175. Mekala JR, Naushad SM, Ponnusamy L, Arivazhagan G, Sakthiprasad V, Pal-Bhadra M. Epigenetic regulation of miR-200 as the potential strategy for the therapy against triple-negative breast cancer. Gene (2018) 641:248–58. doi: 10.1016/j.gene.2017.10.018
176. Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res (2013) 41(9):4976–87. doi: 10.1093/nar/gkt182
177. Ji Y, Sun H, Liang H, Wang Y, Lu M, Guo Z, et al. Evaluation of LncRNA ANRIL Potential in Hepatic CancerProgression. J Environ Pathol Toxicol Oncol (2019) 38(2):119–31. doi: 10.1615/JEnvironPatholToxicolOncol.2019028282
178. Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol (2012) 14(7):659–65. doi: 10.1038/ncb2521
179. Berteaux N, Lottin S, Monté D, Pinte S, Quatannens B, Coll J, et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem (2005) 280(33):29625–36. doi: 10.1074/jbc.M504033200
180. Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis (2010) 31(3):350–8. doi: 10.1093/carcin/bgp181
Keywords: cell cycle, microRNA, long non coding RNA, expression, polymorphism
Citation: Ghafouri-Fard S, Shoorei H, Anamag FT and Taheri M (2020) The Role of Non-Coding RNAs in Controlling Cell Cycle Related Proteins in Cancer Cells. Front. Oncol. 10:608975. doi: 10.3389/fonc.2020.608975
Received: 22 September 2020; Accepted: 27 October 2020;
Published: 30 November 2020.
Edited by:
Yueming Sun, The First Affiliated Hospital of Nanjing Medical University, ChinaReviewed by:
Fabio Luis Forti, University of São Paulo, BrazilAndrea Perra, University of Cagliari, Italy
Alessandra Cataldo, Istituto Nazionale dei Tumori (IRCCS), Italy
Eulàlia De Nadal, Pompeu Fabra University, Spain
Copyright © 2020 Ghafouri-Fard, Shoorei, Anamag and Taheri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==
 Soudeh Ghafouri-Fard1
Soudeh Ghafouri-Fard1 Hamed Shoorei
Hamed Shoorei Mohammad Taheri
Mohammad Taheri