- Department of Physiology and Pharmacology, Karolinska Institutet, Stockholm, Sweden
One of the most notoriously altered genes in human cancer is the tumor-suppressor TP53, which is mutated with high frequency in more cancers than any other tumor suppressor gene. Beyond the loss of wild-type p53 functions, mutations in the TP53 gene often lead to the expression of full-length proteins with new malignant properties. Among the defined oncogenic functions of mutant p53 is its effect on cell metabolism and autophagy. Due to the importance of autophagy as a stress adaptive response, it is frequently dysfunctional in human cancers. However, the role of p53 is enigmatic in autophagy regulation. While the complex action of the wild-type p53 on autophagy has extensively been described in literature, in this review, we focus on the conceivable role of distinct mutant p53 proteins in regulating different autophagic pathways and further discuss the available evidence suggesting a possible autophagy stimulatory role of mutant p53. Moreover, we describe the involvement of different autophagic pathways in targeting and degrading mutant p53 proteins, exploring the potential strategies of targeting mutant p53 in cancer by autophagy.
Introduction
Today, the tumor suppressor protein, p53, is not only known for its bona fide function as a transcription factor that controls a network of responsive genes during various cellular stress to ensure genomic stability and fidelity, but also for its key regulatory function in major signaling and metabolic adaptation, beyond preventing tumorigenesis (1–3). Correspondingly, TP53, is one of the most notoriously altered genes and tumor-associated p53 mutations are found with high frequency in more human cancers than any other tumor suppressor gene (4, 5). While mutations are found all over the TP53 gene (5), most common alterations rise from single-base protein-altering substitutions in the coding region with heavy mutational pressure of particular nucleotides (6, 7). These commonly occurring missense mutations cluster in the DNA-binding domain with often diminished ability to bind specific DNA recognition sequences (2). Consequently, the primary outcome of TP53 mutations is loss of the wild-type ability to transactivate canonical p53 target genes, which provides a fundamental advantage for cancer development. A good example of this is the mutant p53R248, which amplifies the pro-survival effects of wild-type protein by maintaining the expression of CDKN1A gene, resulting in an ability to survive glutamine and serine starvation, while this mutant no longer is able to induce cell death or senescence (8). Furthermore, unlike mutations in other tumor suppressors, the vast majority of TP53 missense mutations result in expression of stable, full-length mutant variants where cancer cells acquire selective advantages by retaining these form of the protein (9, 10). Beyond exerting dominant repression over the wild-type counterpart due to loss of heterozygosity, some mutants might exert new malignant abilities distinct from those simply caused by the loss of the wild-type function (11, 12). Such phenotypes, described as mutant p53 gain of oncogenic function(s) (GOFs) (9, 13), include increased cell proliferation, migration and invasion as well as anti-apoptotic functions, which actively contribute to various stages of tumor progression (9, 14–17). This has led to the assumption that, during development of certain tumor types, mutations leading to the expression of missense proteins appears prone to be selected for over the null mutations. In support of this, patients carrying tumors with mutant p53 proteins display higher oncogenic potential, poor prognosis, poor response to chemotherapy and accelerated tumor recurrence compared to patients with p53 null tumors (13, 14). The enhanced oncogenic GOF potential of p53 mutants, beyond the loss of p53 function, is best exemplified by studies using mice with point mutations (p53R270H/- and p53R172H/) as models for the human Li-Fraumeni syndrome, which is an autosomal dominant inherited cancer susceptibility disorder resulting from germline mutations in the TP53 gene (14, 18–21). These studies have demonstrated that knock-in mice of mutants corresponding to human R175H and R273H develop distinct tumor spectra with high frequency of metastasis, contrary to that observed in mice with p53 deletion, signifying the gain of function of the mutant p53 proteins.
Since the discovery of oncogenic feature of mutant p53 proteins, there has been a steady increase in the number of described diverse GOFs in many cancer types. This has led to reported phenotypic characteristics, culminating in several mechanisms suggested as basis for the gained mutant specific activities (4, 22, 23). A well-recognized mechanism of gained mutant p53 function is its interaction with other transcription factors (4, 24), causing profound alterations in the cancer cell transcriptome and the resulting proteome. However, there is no consensus on the molecular definition of most aspects of mutant p53 GOF(s) and their consequential effects. Distinct mutation type-dependent oncogenic activities still remain to be defined. In addition, recent methodological advances, such as integrated ‘-omics’ and p53 saturation mutagenesis screens, present new insights into the clinical outcomes in patients with myeloid malignancies, where no evidence of GOF for TP53 missense mutation could be found. Instead clonal selection, driven by the loss of canonical p53 function or the dominant-negative effect that reduced the tumor suppressor activity of wild-type p53, was suggested as the most prominent factor in the selective advantage associated with p53 mutations (25, 26). These observations argue for a fitness advantage in certain human tumors harboring missense mutations rather than the acquirement of additional functions, suggesting that GOF may be context and tumor type dependent.
It is further important to consider that although few missense substitutions (Arg175, Gly245, Arg248, Arg273, and Arg282) account for about 30% of all TP53 mutations, there are more than 1,500 types of p53 mutations reported in various cancer types (http://p53.iarc.fr/), and different mutant variants are frequently detected in different human cancers (6). Yet, not all mutant proteins accumulate at high levels in tumor cells, although such stabilization seems key for mutant p53 proteins to orchestrate its oncogenic behavior (2, 27). Further, growing evidence from in vitro studies as well as animal models signifies that the oncogenic activities of mutant p53 variants are heterogeneous and can vary with the tissue type and the genetic background of the cells (28, 29). This predicts that tissue-selective mutational activity would manifest as tissue-selective enrichment of select TP53 mutations. In fact, 25 different TP53 mutation were found to be overrepresented in specific tumor types (26). Accordingly, it has become evident that not all p53 mutants are equal or behave alike and the prognostic impact of TP53 mutations are diverse (30, 31). Therefore, generalizations about mutant p53 may not be relevant. Instead, the discrepancy of the type of mutant is important, not only as a conceptual distinction for their unique oncogenic abilities, but also for the clinical implications including diagnosis, surveillance and therapy.
For the purpose of this review, we will focus on describing and discussing the considerable distinct effects of mutant p53 proteins may exert on autophagy, although other mutant p53 activities may affect different aspects of tumor biology. Autophagy is a fundamental catabolic process by which eukaryotic cells digest macromolecules and damaged organelles in the lysosomes. A well-defined positive regulatory role of wild-type p53 on autophagy with resulting counteracting autophagy inhibitory effect caused by TP53 mutations, is widely recognized and beyond the scope of this review. Instead, the central focus will be on what we currently know about the conceivable roles of distinct mutant p53 proteins in regulating different autophagic pathways. Further, given that alterations in autophagy activity might vary in different types and stages of tumors, we will elaborate on the emerging rationale that the functional effects of distinct mutant p53 proteins on autophagy may also differ. Given that autophagy is tightly connected to dynamic changes in metabolism, we discuss the concept that in certain conditions cancer cells with mutant p53 may favor instead of counteract autophagy. Furthermore, it is well known today that autophagic pathways are reported to mediate the stability of mutant p53 proteins. In the following parts, we will describe the involvement of different autophagic pathways in controlling the cellular level and degradation of mutant p53 proteins as well as the potential therapeutic strategies for targeting mutant p53 in cancer by various autophagic pathways.
Autophagy-Lysosomal Pathways
Autophagy is a highly conserved homeostatic recycling process, where it functions to mediate the degradation of cellular macromolecules, damaged organelles or internalized pathogens in the lysosomes (32, 33). Under normal physiological conditions, autophagy is maintained at basal level, however, by responding to perturbations in the extracellular environment, e.g. when encountering nutrient deficiency, cells tune the autophagic flux to meet intracellular metabolic demands (34). Thus, beyond the fundamental significance for cellular quality control purposes and the maintenance of cellular and organismal homeostasis, activation of autophagy provides cells with cytoprotective and metabolic adaptations under stress (33–37). Its timely regulation is, therefore, finely controlled by numerous proteins. Dysregulation of autophagy with subsequent altered protein degradation and cellular metabolism, has severe consequences related to several pathophysiological conditions, such as cancer, infection, autoimmunity, inflammatory diseases, neurodegeneration and aging (38).
Multiple routes of degradation through autophagy coexist in mammalian cells that differ in the delivery mechanisms and target specificity, but converge on the same degradation site - the lysosomes (39). Beyond macroautophagy (MA), usually referred to as autophagy, which is the most extensively studied and well characterized type (39), micro- (MI) and chaperone-mediated autophagy (CMA) pathways, are key components of the cellular machinery that play important roles for lysosome-mediated protein degradation (40–42). MA is a multistep process with a nonselective seizing of cytosolic cargo or in a selective fashion that vary in target specificity and induction conditions. It involves the sequential formation of a double-membrane structure, the phagophore that ultimately fuses with lysosomes to degrade sequestered cargos via the activity of hydrolases in autolysosomes (43, 44). While Autophagy-related (Atg) proteins act on the de novo synthesis and accompanying elongation and closure of the autophagosomes that engulf the cytosolic cargo during MA (45, 46), MI involves the direct uptake of cargo material by the lysosomal or vacuolar membrane and is suggested to occur by either lysosomal protrusion, invagination or with endosomal invagination (41, 47). CMA, on the other hand, applies to select proteins with a pentapeptide motif related to KFERQ that is recognized by the heat shock cognate 71 kDa protein (Hsc70 (also known as HSPA8)) and co-chaperones (48). This interaction forms a chaperone complex that enables the translocation of the cargo protein into the lysosomal lumen via binding the lysosomal receptor, lysosome-associated membrane protein 2A (LAMP-2A) (49).
Regardless of the delivery system, the cargo of the autophagic pathways are digested by the lysosomal hydrolases and engendered building blocks are shuttled back to be reused for biosynthesis of macromolecules (50). In this way, autophagy acts as an important internal source of cellular energy through self-degradation process. Hence, the engagement of autophagic pathways confer stress resistance and sustain cell survival that benefit tumor cell growth, especially in nutrient scarce or hypoxic conditions (51). Furthermore, MA may play a critical role in tumor microenvironment and has been proposed to promote tumor dormancy (52), where cancer cells remain in a quiescent state with the potential to relapse. Consequently, autophagy is exploited by cancer cells and malignant tissues often exhibit altered MA activity (53–55), displaying autophagy addiction to sustain stress resistance. Therefore, inhibition of autophagic flux after induction of pro-survival autophagy has been suggested as a strategy to sensitize multiple human cancer types to chemotherapy. However, the role of MA in carcinogenesis is context dependent with reports indicating both pro-tumorigenic and tumor-suppressive roles (56). As a tumor-suppressing mechanism in early-stage carcinogenesis, autophagy dampens inflammation and promotes genomic stability (57). The direct evidence comes from studies using mouse models with genetic knockout of canonical autophagy-related genes, including ATG5, ATG7, and BECN1 where impaired autophagy accelerates tumorigenesis in animals (58). However, once a tumor has been established, the nature of autophagy switches and many aggressive tumors acquire reliance on autophagy for growth and survival (51, 59). Thus, in spite of the dual role of autophagy in cancer development and progression differs depending on the genetic context, type of cancer and tumor stage, it is well established today that autophagy is frequently altered in human cancers, with its activation regarded as one of the characteristic key features that contributing to malignant development. In fact, the limited penetrance of mutations in most autophagy genes across human tumors indicates that many human cancer types preserve autophagy function (60), where several well-established oncoproteins and tumor-suppressors whose depletion or mutation promote tumor formation have emerged as eminent regulators of autophagy. In addition, accumulating evidence now also supports a regulatory role for selective autophagy, including mitophagy and non-MA pathways, in human cancer (61, 62). Although CMA was initially suggested to display pro-tumorigenic functions (63), anti-tumor role for CMA is also proposed under physiological conditions in non-transformed cells (61, 64). Further, subsequent studies have demonstrated that CMA plays a more complex and context-dependent role, where cancer cells from different tissues and tumor stages may display varying CMA activity (62). Moreover, growing number of studies provide new insight as to how increased CMA activity can be beneficial for promoting the degradation of proteins displaying dominant oncogenic pro-survival activities in cancer cells (37, 65–68). However, few studies are conducted to assess the therapeutic impact of CMA activation in cancer, thus CMA-based treatment options in humans remain speculative. This is mainly due to the lack of potent chemical modulators of this process and limitation in functional CMA analysis, which mainly rely on expression levels of the known CMA component, LAMP-2A. Accordingly, while defining the major cancer-related pathways, beyond oncogenic signaling that affect autophagy and control tumorigenesis is important, the regulation and roles of selective and non-MA autophagy, such as CMA and MI, in cancer still needs further investigation, thus the subject of this review will be mainly focused on mutant p53 and MA.
Wild-type p53 – Dual Role in Macroautophagy Regulation
In the past decades, the mechanisms governing regulation of autophagy has been intensively investigated and the impact of p53, mainly on MA, is well described by several groups with detailed mechanisms uncovered. Collectively, the action of wild-type p53 as a pro-autophagic factor in human cancer cells is reflected by its transcriptional activity on a wide range of downstream target genes with autophagy regulatory effects that diverge on cellular functions, including; a) stimulating the nutrient energy sensor AMPK (AMP-activated protein kinase) (AMPK β1/β2 subunits, Sestrin1/2) (69, 70), b) inhibiting the signaling of mTOR (mechanistic target of rapamycin) (TSC2, IGF-BP3, REDD1) (71, 72), c) suppression of PI3K (phosphatidylinositol-3-kinase) activity (PTEN), d) promoting the expression of the MA core machinery (ULK1, ULK2, ATG7) (73, 74), e) transactivating DRAM1 (damage-regulated autophagy modulator 1) and splice variants that effects several stages of autophagy (75), f) upregulation of HIF-1 (hypoxia inducible factor 1), g) inducing regulators (Isg20L1 and HSF1 (heat shock transcription factor 1) that in turn transactivates autophagy related genes (ATG7) (76), h) induction of TGM2 (transglutaminase 2) which promotes autophagic flux by enhancing autophagic protein degradation and autolysosome clearance (77), i) interfering with the inhibitory interactions between Beclin-1 and Bcl-2 family proteins (incl. Bcl2/Bcl-XL, Bad, Bax, BNip, Mcl-1, Puma) by their direct transcriptional up or down regulation or through DAPK1 (Death-Associated Protein Kinase 1) activation, alternatively by DAPK1 mediated MAP1B interaction (78), and direct physically interacting with Bcl-XL and the p53-regulated human tumor suppressor protein p14ARF [detailed reviewed in (79, 80)]. These pro-autophagic functions of wild-type p53 are most likely credited to its tumor suppressor role under conditions of hypoxia, starvation or DNA damage, by which induction of MA assists to cope with different kind of cellular stress to prevent cell damage and maintain cellular integrity. This is further in line with the involvement of wild type p53 in several signaling pathways that promote autophagy, including MAPK (mitogen-activated protein kinase) family proteins, such as ERK (extracellular signal-related kinase) and JNK (c-Jun N-terminal kinase).
However, beyond these established pro-autophagic functions, wild-type p53 can also counteract autophagy. This inhibitory role is often attributed to the cytosolic pool of p53 under normal growth conditions, connected to G0/G1 phases of the cell cycle, and shown to be mediated through both transcription-dependent, but mainly -independent manner, involving; a) inhibition of the AMP-dependent kinase and thereby activating mTOR (81), b) induction of TIGAR (TP53-induced glycolysis and apoptosis regulator) regulating glycolysis and cellular ROS levels (82), c) transcriptional regulation of micro RNAs (miRs) (miR-34a and miRs-34a/34c-5p that targets ATG9A and ATG4B, respectively) (83, 84), d) interaction with Beclin-1 that subsequently facilitates its ubiquitination and proteasome-mediated degradation (85, 86), and e) direct molecular association with RB1CC1/FIP200, a mammalian protein homologous to Atg17 (87), and f) by reducing the accumulation of double stranded RNA and activation of PKR (protein kinase RNA-activated) (88).
Conclusively, these observations have led to the current notion that the action of wild-type p53 on MA is complex, highly context dependent, dictated by the cellular microenvironment and stress condition, along with the cell cycle progression and subcellular distribution of p53 that exert dual roles in autophagy regulation. In support of this, cumulative evidence shows that nuclear wild-type p53 can promote mitophagy by transactivation of PRKN (Parkin), a key effector of this selective autophagy, involved in degradation of impaired mitochondria (89). Cytosolic p53, on the other hand, inhibits mitophagy via direct binding to Parkin, preventing its translocation to the damaged mitochondria that cannot be removed by mitophagy (89). Accordingly, these findings are not only confirmative of the counteractive roles of p53 in autophagy regulation, but also indicative of the involvement of p53 in other autophagy pathways, beyond MA. However, despite the studies exemplified above, there are still important pending questions about the detailed molecular mechanisms that govern the role of p53 in MA. Further, the potential contribution, regulatory role, and the physiological importance of p53 in other selective macroautophagy, microautophagy or CMA is yet to be explored.
Mutant p53 as a Regulator of Autophagy
The Effect of p53 Mutant Proteins on Autophagy
Given that impairment of the wild-type function with predominant pro-autophagic role is provoked by TP53 mutations, it is expected that mutant proteins can reshape the wild-type-mediated outcomes on autophagy. Accordingly, the current accepted view is that mutant p53 displays a suppressive role in autophagy. This was initially illustrated by the assessment of the effect of ectopically overexpressing 22 different p53 mutant variants on the autophagy in p53 null colon cancer cells (90). Reintroduction of some p53 mutants, including p53A161T, S227R, E258K, R273H/L,R273L, but not the p53P151H,R282W, exhibited high correlation with efficient suppressive capacity on basal MA. However, the expression of other mutants, including p53P98S,K120D,V143A,R175C,R175D,R175H,R175P, R181H,L194F,S227K,G245C,R248L,R248W, R249S,R280K, displayed no or less suppressive effects, or in some cases even enhanced MA. This led to the awareness that certain p53 mutants may exert negative effects on autophagy. A shared feature of these mutants, including p53A161T, S227R, E258K, R273H/L,R273L, was shown to be their cytoplasmic localization, most likely with a loss-of-function to promote transactivation-dependent stimulation of autophagy (90). In support of this, it was later shown that the p53R175H or p53R273H mutants indeed suppress the formation of autophagic vesicles and their fusion with lysosomes through the transcriptional repression of key downstream p53 responsive autophagy related genes, as BECN1, DRAM1, ATG12, as well as TSC2, SESN1/2 and P-AMPK, resulting in the autophagy blockage (91, 92). Correspondingly, the knockdown of these mutants in cancer cells cause augmented autophagy by affecting signaling at various phases of the autophagic process with a concomitant stimulation of mTOR signaling. However, it should be noted that both p53 deletion and missense mutations can substantially affect the mTOR signaling, where an elevated association of Rheb with lysosomal membranes promote active mTORC1 complexes (92).
The autophagy inhibitory role of mutant p53 proteins was further ascribed to transcriptional-independent actions. Some p53 mutants, as p53R175H,L194F, R273H, were unable to form complexes with endogenous Bcl-2 or Bcl-XL, unlike the wild-type. This loss-of wild type function abolishes the capacity to interact, thus cancer cells bearing mutant p53 sustain the inhibitory interactions between Beclin-1 and Bcl-2 family proteins (93). Further, through mTOR stimulation, the aforementioned mutants also convey negative effects on Beclin-1 expression and phosphorylation, thus suppress the functionality of Beclin-1 in autophagy. Likewise, less directly through mTOR stimulation, the p53G199V mutant was demonstrated to gain regulatory function on STAT3 phosphorylation (94), with subsequent transcriptional activation of HIF-1 suggested to contribute to autophagy inhibition. In fact, several multiple mechanisms by which mutant p53 can stimulate HIF-1 have been identified. These includes increased cellular reactive oxygen species (ROS), resulting from less efficient oxidative phosphorylation, or by interference with the binding of HIF-1α to the ubiquitin-protein ligase Mdm2 in hypoxic conditions. However, the functional role of HIF-1 and hypoxia-related genes in autophagy regulation awaits further investigation.
Moreover, by engaging in protein-protein interactions with other transcription factors as a GOF, some cancer-associated p53 mutants were shown with capability of blocking autophagy indirectly by activating several growth factor receptors, such as TGFBR, EGFR, IGFR (95), contributing to sustained active PI3K/Akt/mTOR signaling that subsequently repress autophagy. In breast cancer cells a direct correlation between mutant p53R273H and Akt phosphorylation was demonstrated. Akt, in turn, propagates the effect on its direct downstream target mTOR. Taken together, regulation of the mTOR activity by either constitutive blockage of AMPK signaling or through alternative routes, appears to represent a crucial signaling that occur in cancer cells bearing mutant p53 (91, 92). Thus, regardless of the transcriptional dysregulation or GOF mediated protein-protein interaction, an important implication of these findings is that the autophagy suppressive role of mutant p53 seems mainly to merge on the canonical AMPK-mTOR signaling.
The Impact of p53 Mutants on Autophagy Through Metabolic Changes
A defining hallmark of cancer is uncontrolled cell proliferation, which is initiated once cells have accumulated adaptations in pathways that control metabolism and proliferation (96, 97). Metabolism provides the energetic and biosynthetic demands of rapid proliferation. Beyond a high glycolytic activity, the most common metabolic alteration in malignancies, rapidly proliferating cancer cells further display a sustained mitochondrial oxidative phosphorylation, as the tricarboxylic acid (TCA) cycle intermediates are important precursors for the synthesis of amino acids, lipids and nucleotides (96–99).
While, a direct interference of p53R175H, R273H mutants on MA can be denoted to LOF transcriptional repression of core autophagy genes (BECN1, ATG12) (91), most of the mutant p53-mediated autophagy inhibitory evidence stems from studies describing a gained regulatory effect of mutants on cancer metabolism (Figure 1). As stated above, autophagy is regulated by a number of effectors strictly interconnected with the metabolism as revealed by the fact that mTOR and AMPK are both master regulators of autophagy and major sensors of the cellular energy status (100). mTOR functions as a key homeostatic regulator of cell growth and orchestrates whether anabolic or catabolic reactions are favored. mTOR complex 1 (mTORC1) manages multiple biosynthetic pathways and promotes cell growth when nutrients are in plentiful supply. These include synthesis of amino acids, proteins and biogenesis of ribosomes (101). AMPK, on the other hand, is a highly conserved sensor of the cellular energy status that is activated upon low intracellular ATP levels. AMPK responds to energy stress by suppressing cell growth and biosynthetic processes, in part through its inhibition of the mTOR (mTORC1) pathway (102, 103). Thus, while p53 deletion and missense mutations can enhance mTOR, emphasizing the functional interplay between AMPK and wild-type p53, some mutants can display effects on the canonical AMPK-mTOR signaling beyond the transcriptional repression. An excellent example highlighting the difference between the wild-type function from null and missense GOF mutations, is the ability of p53R175H,G245C,R282W mutants, displaying a negatively metabolic effects on the AMPK signaling through the direct protein-protein interaction with the AMPKα subunit under conditions of energy stress (104). Now, several p53 mutants (p53P151S, E336X), but not the wild-type, have been shown to interact with AMPKα through the DNA-binding domain, where mutant p53 disrupt the interaction of AMPKα-LKB1. This causes inhibited AMPKα phosphorylation and suppressed AMPK activity. In addition, a role of p53R273H was demonstrated to control the mevalonate pathway (MVP) through the transcriptional modulation of SREBP1, a downstream target of AMPK (105). Furthermore, the p53R175H, R273H mutants were described to promote phosphorylation on the pyruvate kinase isoform M2 (PKM2) (106), a key enzyme that catalyzes the conversion of phosphoenolpyruvate (PEP) and ADP to pyruvate and ATP in glycolysis. The phosphorylation (Tyr105) on PKM2 enhances the mTOR signaling. However, these functions seem independent of the subcellular localization of p53 mutants, as mutants with acquired capability to functionally inhibit the AMPK signaling can be found both in the cytoplasm, such as p53P151S, or localize exclusively in the nucleus (p53E336X), most likely due to the lack of a C-terminal p53 nuclear export signal, whereas the cellular localization of the mutant p53G245C differs with the confluency of the cell culture. Nonetheless, based on its effect on the AMPK-mTOR axis, the cancer related expression of p53R248W,C176S,R273H,R175H, R175H mutants are shown to display a gained function of affecting metabolism, thereby inhibit autophagy irrespectively of tissue of origin or prevalence to a subcellular localization (91).
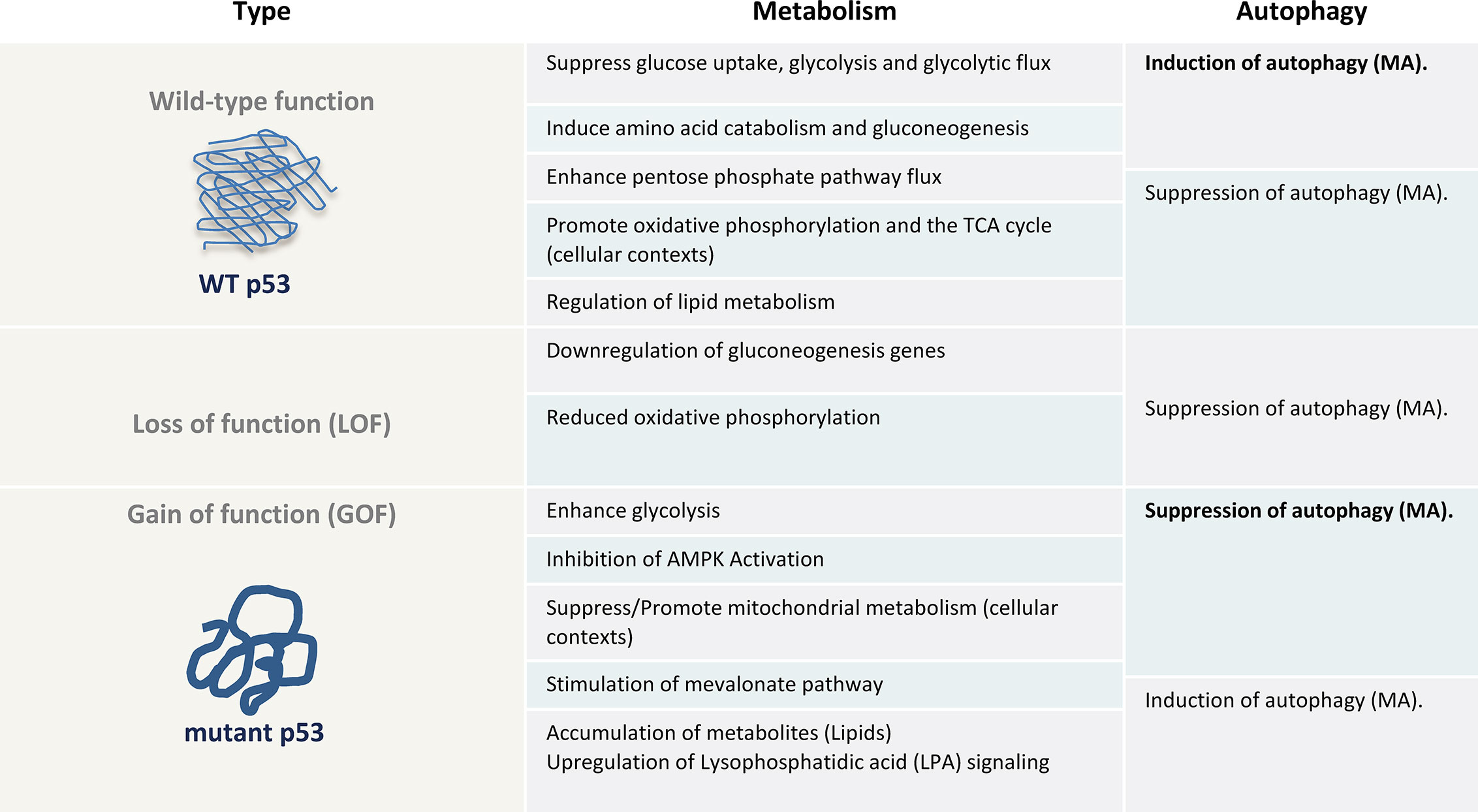
Figure 1 The metabolic and autophagy effects of wt p53, null (loss of function (LOF)) or gain of function (GOF) of mutant p53 as a table illustration. AMPK, AMP-activated protein kinase; MA, Macroutophagy.
Beyond the AMPK-mTOR signaling, several of the metabolic effects of mutant p53 oppose the metabolic functions commonly acquired by the wild-type protein, including glycolysis, lipid metabolism, the mevalonate pathway, de novo serine synthesis, urea cycle and oxidative phosphorylation (107, 108) (Figure 1). Thus, it is well known that mutant p53 rewires cancer metabolism (109). For example, wild-type p53 limits glycolysis and induces flux through the pentose phosphate pathway (82), whereas mutant proteins induce metabolic responses that include enhanced glycolysis to support tumor cell growth and proliferation. By promoting glucose uptake, mutant p53 can limit autophagy-dependent energy production. Therefore, any perturbation in cellular metabolism and redox control caused by p53 mutants can affect the autophagic outcome. However, this does not only apply to metabolic adaptation of cancer cells as a loss of function or in terms of enhanced glycolysis. For instance, mutant p53 has been shown to promote the MVP opposite to the wild-type p53, which is required for mutant p53 and Hsp40 interaction facilitating mutant p53 stabilization (110, 111). The MVP is an essential metabolic pathway that produces sterols and isoprenoids including cholesterol for the synthesis of membranes and lipids, as well as signal transduction allowing cancer cells to survive under conditions of matrix detachment (105). This in turn could promote detachment-induced autophagy (112). Further, beyond elevated glycolytic rate in cancer cells, several studies have clearly demonstrated that the majority of tumors similarly possess the capacity to sustain high fuel oxidation and ATP production in mitochondria (96–98, 113, 114). Especially, quiescent and slow proliferating tumor cells with activated MA, rather depend on oxidative phosphorylation for energy supply than glycolysis (98). Depending on the cellular context, mutant p53 have been indicated to both inhibit or promote oxidative phosphorylation (29, 115), and can thereby enhance or suppress autophagy. However, metabolic alterations are also observed in p53 null cells due to loss of wild type p53 function, such as a downregulation of genes that facilitate gluconeogenesis, which is observed in mice with an adipocyte-specific loss of p53 (116), and reduced oxidative phosphorylation in p53-null cells (117, 118). Accordingly, our understanding of the involvement of mutant p53 in direct interference of the core autophagy machinery and regulation in cancer cells as well as the detailed associated molecular mechanisms, beyond metabolic modifications, remain incomplete and need to be further assessed in human clinical specimens.
Consideration of a Potential Stimulatory Role for Mutant p53 in Autophagy
It is equally important to note that the MA inhibitory function is not shared among all mutant p53 proteins. Mutant arising from the substitution of lysine in position 382 with arginine, fails to associate with FIP200, and loose the autophagy inhibitory function (87). Moreover, the ectopic expression of p53P151H,R282W was shown not to display any efficient autophagy inhibitory behavior, apart from the fact that some mutants even show enhanced MA activities (90). Given that autophagy can sustain tumor cell metabolism, and mutant p53 can foster adaptations to nutrient deprivation, it is conceivable that certain mutant p53 proteins could therefore function in seemingly unprecedented way to respond to nutrient stress, where certain mutants may support the constitutive high levels of MA to provide selective advantage for cancer cells. Therefore, it is reasonable that some mutant p53 forms may enhance autophagy required to prevent energy crisis and maintain nucleotide pools during starvation in cancer cells caused by hypoxia and nutrition depletion in tumor microenvironment. This could be especially relevant under situations of expansion of tumor mass (Figure 1), in which some parts of the tumor starve due to insufficient nutrient availability or lack of vascularization, even when cancer cells promote metabolic pathways to support growth and proliferation. Under these conditions, numerous mechanisms, including autophagy activation and mutant p53 might converge to contribute to preserve cell viability as a supportive response. While this hypothesis remains speculative, a recent immunohistochemical study on 113 colorectal cancer specimens uncovered a significant association between high LC3B expression and mutant p53 protein expression pattern in ~35% of the patients (119). Although the type of p53 mutant remained undisclosed, the fact that a co-expression of LC3B and mutant p53 was tightly linked to aggressiveness is indicative of high rates of autophagy in malignant tumors. This feature was not observed in tumors with null expression. While further investigation is warranted, this finding provides the rationale that even when the wild-type ability to promote autophagy might be hampered by mutations, some mutant proteins enable autophagy activation in tumors. It is likely that certain point mutations may selectively retain some of the wild-type p53 pro-survival functions, including the pro-autophagic activity. An intriguing possibility is also that the pro-autophagic function of mutant p53 might be a transient phenotype under limited periods of nutrient starvation. A comparable example of this possibility is the activation of the cyclin-dependent kinase inhibitor p21 by p53. Although p21 expression generally contributes to the induction of an irreversible proliferative arrest, transient p53-mediated induction of p21 is reversible, allowing cells to re-enter the cell cycle once stress or damage has been resolved (120). Alternatively, the mutant p53-driven autophagy suppressive function might be overridden by additional signaling, mutations or epigenetic changes. For instance, in the context of proteasomal inhibition, cancer cells with mutant p53R273H display activated MA (121). Additionally, activating mutations in HRAS or KRAS elicit excessive MA, regardless of the presence of mutant p53. In contrast to normal cells, RAS-driven cancer cells display remarkably high levels of basal autophagy, and it is well acknowledged that a subset of RAS-driven human cancers shows a reliance on autophagy for their survival (54). Concomitant expression of mutant p53 and oncogenic Ras, leading to cellular transformation, and a crosstalk between Ras and various mutant p53 proteins is well documented. However, in the presence of mutant p53, some KRAS bearing tumors are still addicted to autophagy (122), indicative of that mutant p53 may not always inhibit MA (123). Perhaps a particular pathway ultimately predominates over others. While, this remains to be investigated, it was shown that different p53 mutants cooperate with H-Ras in different ways to induce a unique expression pattern of a cancer-related gene signatures (124). For instance, the p53R248Q, R273H mutants exhibited the highest level of gene expression by cooperating with NFκB, the p53R175H and p53H179R mutant induced the cancer-related gene signatures by elevating H-Ras activity. By contrast, the p53G245S displayed no effect, further emphasizing the significantly different impact and responses different mutants can exhibit.
In addition, beyond the in vitro observation that even a seemingly subtle difference of one amino acid, such as p53R248W versus p53R248Q (125), can have a large impact on the mutant p53 function. Additionally, even the same amino acid substitutions at the same position (R175H) in the p53 protein have been shown to dramatically different phenotypic effects in terms of metabolism (29). Thus, in spite of the fact most studies describe a suppressive role of mutant p53 on autophagy, there is evidence that the contribution of individual mutant p53 on autophagy might differ in a cell or tissue type, context or cancer stage-dependent manner. Considering the tumor progression promoting function of both mutant p53 and autophagy, inhibiting autophagy seems to be counterproductive for advanced tumors. Thus, it is reasonable to think that cancer cells would rather benefit from mutant p53 with enhanced autophagy activation that can serve as a cell survival mechanism during certain conditions, similar to the dual nature of autophagy which confers suppressive role in tumor initiation while aggressive cancers acquire autophagy for growth and survival. One such condition during which mutant p53 may favor instead of counteracting autophagy is Epithelial–mesenchymal transition.
Role of Autophagy in Mutant p53-Driven Epithelial-to-Mesenchymal Transition
One of the major transdifferentiation processes, through which cancer cells develop the ability to invade and disseminate is the Epithelial–mesenchymal transition (EMT) (126, 127). This process facilitates molecular and functional changes as such, cells undergoing EMT become invasive by acquiring characteristics required for cancer cells to adapt to phenotypic changes fostering capability to break out of the primary tumor. Beyond facilitating cancer dissemination, EMT can further contribute to stemness and resistance to therapy (128). However, EMT covers a complex and multifactorial spectrum, which gives rise to a variety of intermediate cell states (129). Consequently, EMT is now recognized as a dynamic and reversible process, rather than a binary state, that involves tumor microenvironment, cellular heterogeneities, as well as phenotypic plasticity, thus various metabolic reprograming occurs along with the EMT process.
It is well known that the EMT pathway is under the negative regulation of wild type p53, while mutant p53 proteins display oncogenic GOF activities with robust capacity to promote EMT by controlling the TGF-β signaling and by regulating the expression of various pro-EMT-Transcription factors (130, 131). Furthermore, considering that the metastatic potential of cancer cells increases along the EMT process, multitude of key metabolic pathways, including glycolysis, the TCA cycle, lipid and amino acid metabolism, have been attributed to contribute to EMT, tumor aggressiveness and invasiveness (132, 133). Yet, some EMT positive tumors are characterized by low proliferation rate or quiescence (134). Tumor cells with the traits of invasiveness and stemness which have undergone EMT program can manifest features of growth arrest, and cancer cell dormancy (135). The resulting stemness may drive the progression of more aggressive tumors. For instance, the TGF-β induced EMT process is related to a slow proliferation rate and cell cycle arrest in epithelial cells (136). This observation is rather counterintuitive as it is difficult to directly explain how a slow-proliferating population can lead to higher tumorigenicity and how these tumor cells can remain and exit dormancy.
One possible explanation might be that the metabolic changes elicited by mutant p53 are not mutually regulated and unidirectionally controlled in all cancer cells and may differ during the different stages of cancer progression, such as with the capacity to undergo EMT (16, 113, 137). So how could this be connected. First, several molecular mechanisms underlying the involvement of mutant p53 in malignant progression and EMT have been reported, which all converge on expansion of epithelial stem cells and induction of stem cell gene signatures, as well as mesenchymal stem cell-derived features (138–140). This suggests that mutations in TP53 not only sustain primary tumor formation, but also that mutant p53 can promote the late stage of tumorigenesis, possibly through the acquisition of an invasive ability and stem cell characteristics. Secondly, while mutant p53 have been linked to promote glycolysis through distinct mechanisms (141), emerging data supports the notion that not all mutants display enhanced glycolysis. For example, while the p53R175H, R273H mutants, are able to confer enhanced glycolysis in lung cancer cells, the stable expression of p53R175H in human breast epithelial cells displayed considerably different properties, characterized by a markedly lowered glycolytic phenotype (29) (Figure 2). These data highlight the fact that the same amino acid substitutions, in the same position of a mutant p53 protein can have dramatically different phenotypic effects in terms of glycolysis. Moreover, breast epithelial cells expressing p53R175H displayed enhanced MA, which predicts the inversely correlation between dampened glycolysis and enhanced autophagy. Thus, while mutant p53-enhanced glucose metabolism can correspondingly suppressed autophagy in proliferating cancer cells, it is reasonable that a reduced glycolysis by mutant p53 can induce autophagy in quiescent cells. Keeping in mind that a rewiring of cellular metabolism appears to precede changes in stemness, these data are supportive with metabolic changes observed in slow proliferating circulating tumor cells, which display higher mitochondrial metabolism rather than glycolysis. The observation that tumor cells with mutant p53R72 proteins show a greatly increased oxidative phosphorylation as well as increased metastatic ability further supports this (142).
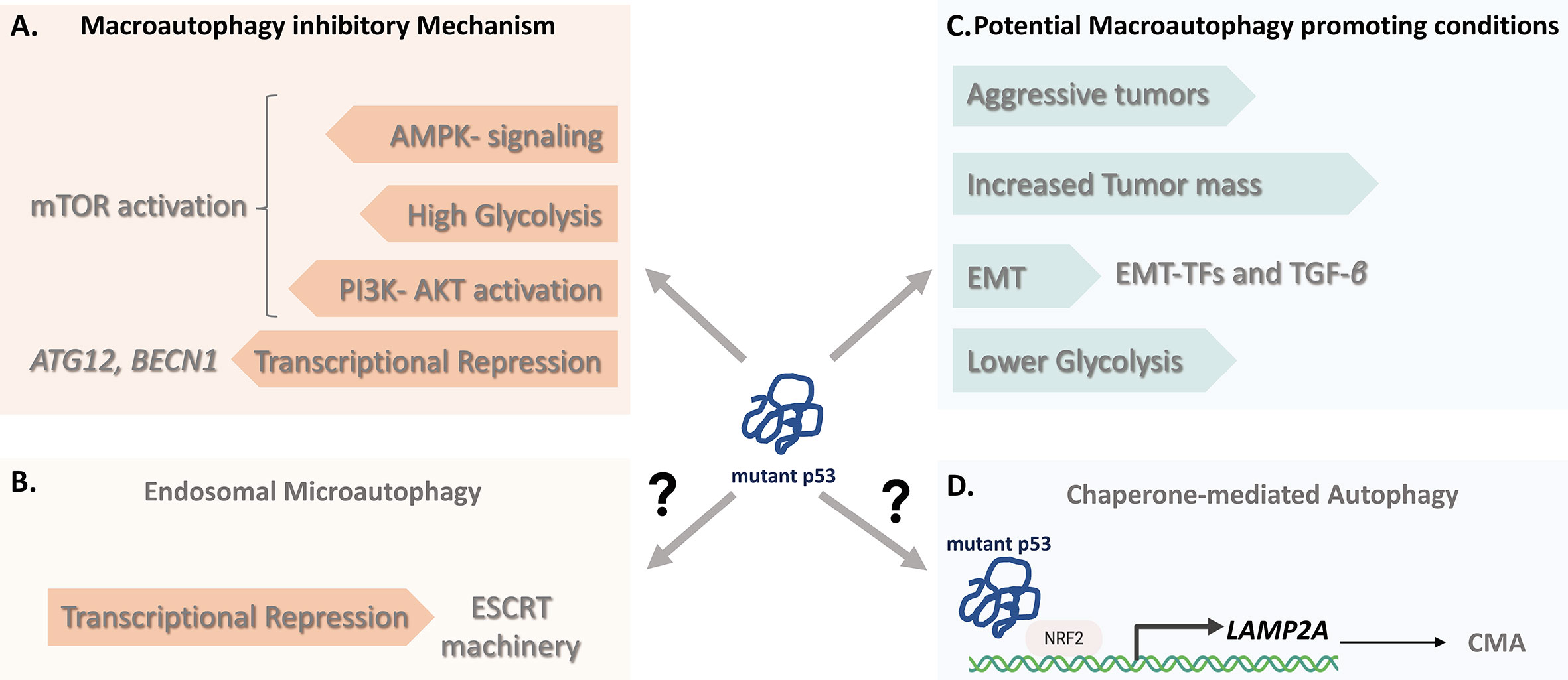
Figure 2 The impact of Mutant p53 on autophagy. (A) The macroautophagy (MA) inhibitory mechanisms of some mutant p53 proteins include the resulting transcriptional repression of core autophagy genes (BECN1, ATG12) and regulation of the mTOR activity by either constitutive blockage of AMP-activated protein kinase (AMPK) signaling or through alternative routes by affecting cancer metabolism. (B) Beyond MA, since wild-type p53 can transactivate genes promoting Endosomal Sorting Complex machinery, mutant p53, although yet to be determined, might negatively affect the signaling contributing to ESCRT-dependent mechanism involving endosomal microautophagy. (C) Cancer cells bearing some mutant p53 variants may in certain conditions favor instead of counteract autophagy. These include nutrient scarce or hypoxic conditions of aggressive tumors and with increased tumor mass, p53-mediated (EMT situations and hampering glycolysis. (D) LAMP-2A expression is an essential factor in CMA activation. Given that transcriptional control of LAMP-2A is shown to be under NRF2, it is likely that mutant p53 proteins might contribute to CMA activation through NRF2-mediated LAMP-2A transactivation, suggesting a molecular connection linking mutant p53 and CMA. CMA, Chaperone-mediated Autophagy; EMT, Epithelial–mesenchymal transition; ESCRT, Endosomal Sorting Complex Required for Transport; LAMP-2A, Lysosome-associated membrane protein 2A; NRF2, Nuclear factor erythroid 2-related factor 2.
Thirdly, we need to consider the essential role of autophagy induction in supporting cell viability during cancer progression and migration, where MA has clear positive effect on EMT (143, 144). Autophagy induction can be advantageous especially during metabolic reprogramming followed by cancer cell dormancy with a lower proliferation rate or quiescence, thus constitute an efficient adaptive strategy, which can supply of nutrients, confer stress resistance and sustain cell survival during metastatic spreading (145). Taken together, this suggest that mutant p53 may stimulate conditions of metabolic requirement for autophagy induction allowing cells to cope with a stressful or unfavorable microenvironment where cancer cells remain quiescent but may relapse (Figure 2). However, it is important to note that data on EMT plasticity and tumor dormancy are primarily derived from in vitro studies. Therefore, sophisticated animal studies are needed for tumors that have undergone mutant p53-induced EMT program to provide an in vivo correlate in preclinical models. Nevertheless, an important implication of these observations is that p53 mutants do not always acquire and possess the same metabolic consequences and may not display equal biological effects in all types of human cultured cells.
Therefore, when considering the generality of the effect of mutant p53 on autophagy, we might need to keep in mind the metabolic plasticity and different aspects of metabolism might be regulated in different cell or tissue types. The complex regulatory interaction between mutant p53 and autophagy might well be influenced by many factors, such as tissue and cell types, tumor stage, type of other oncogenic mutation, the sequential mutation appearance order, extent of damage or stress, and levels of intra-tumor oxygen or nutrients as well as on the proliferative capacity of the tumor cells. A switch between autophagy phenotypes, depending on fitness landscape or mutation-selection balance may as well applicable for mutant p53 and we need to consider this exceptional plasticity which might create significant challenges as we attempt to therapeutically intervene in these pathways.
The Role of Mutant p53 in Autophagic Pathways Beyond Macroautophagy
To date, there are no direct evidence of a select mutant p53 function in microautophagy, CMA or in selective macroautophagy, including xenophagy, ribophagy. However, as wild type p53 exerts a regulatory role in mitophagy, endosome and exosome biogenesis (146, 147), it is reasonable that mutant p53 proteins might affect undiscovered functions in multiple degradative and cellular sorting systems.
The main limitations of studying non-MA pathways in regards to mutant p53 is likely the incomplete knowledge of their regulatory mechanisms. However, while the signaling mechanisms that control CMA are currently not fully understood, a key step in the CMA process is the expression of LAMP-2A receptor at the lysosomal membrane. High lysosomal LAMP-2A levels are reported to correlate with a predisposition of CMA, whereas silencing of LAMP-2A results in inability to degrade proteins via the CMA pathway, thus increase LAMP-2A expression is an essential factor in CMA activation (148). Accordingly, a transcriptional control of LAMP-2A expression is shown to be under the control of the NFE2L2/NRF2 (Nuclear factor erythroid 2-related factor 2 (NRF2) (149), also known as nuclear factor erythroid-derived 2-like 2 (NFE2L2), which generally participates in the control of metabolic redox processes including degradation of oxidized proteins. In 2018, the missense mutant variant p53R280K was demonstrated to interact with NRF2 and to contribute to selective activation of its downstream transcriptional program (150). Thus, given that NRF2 promotes a pro-survival oxidative stress response that allow cells to cope with oxidative stress, along with the fact that CMA is induced by oxidative stress, it is likely that mutant p53 proteins might contribute to CMA activation through NRF2-mediated LAMP-2A transactivation, indicative of a molecular pathway that connects mutant p53 with CMA (Figure 2). Consistent with this indication, analysis of various human cancer cells with different mutational p53 status that either expressed wild-type, mutant p53 or null in p53 expression, revealed that Spautin-1 induced CMA in confluent growth conditions selectively induced cell death of mutant p53-expressing cancer cells. No or little effect was detected in wild-type p53 or p53-null cancer cells, suggesting that cancer cells with mutant p53 might be more susceptible to activate or undergo CMA (65).
Moreover, considering that wild-type p53 can transcribe several critical genes encoding endosomal compartment, including TSAP6, CHMP4C and CAV1 (147), provides a rationale that p53 signaling may contribute to Endosomal Sorting Complex Required for Transport (ESCRT) machinery dependent mechanism, involving endosomal microautophagy (eMI). However, it is yet to be determined whether there is an involvement of mutant p53 proteins in micro- or endosomal microautophagy (Figure 2).
Furthermore, beyond autophagic pathways, mutant p53R273H has been shown to drive alterations in endocytic membrane trafficking during which DNM1 and Myosin VI (Myo6) were upregulated in cancer cells. Apart from stimulating the expression of endosomal proteins, both the wild-type and p53R273H mutants are indicated to effect proteins involved in the secretory pathway, protein secretion via extracellular vesicles (EV) and exosomes of endosomal origin (151). Thus, mutant p53 might regulate the expression of components of the endocytic machinery and modify secretion of extracellular vesicles in multiple ways.
Mutant p53 as Target of Autophagy
Targeting Mutant p53 Proteins
Based on the high frequency of TP53 mutations in human tumors, the oncogenic effects of many missense variants with the fact that cancer-specific pathogenic stabilization of mutant proteins effectively sustains tumor progression and dissemination, mutant p53 proteins represent indisputable promising targets in cancer therapy (17, 152). Accordingly, different approaches have been explored in which targeting of mutant p53 has primarily focused on the development of therapies designed to inhibit the mutants and restore their wild-type p53 function by small molecules, targeting the gain-of-function phenotype of mutants and stimulating immunological activity directed against a mutant p53 protein (153, 154). In animal models, targeting mutant p53 functions have been shown with highly promising results that selectively kill cancer cells, with low toxicity in healthy tissues, indicating tumor-specific vulnerabilities (105, 155, 156). But in the clinics, the specific targeting of mutant p53 proteins has proven challenging, especially considering that mutations are diverse in their type, sequence context, position, and structural impact, making it difficult to identify a well-defined structure (2, 7). In fact, most desirable oncoprotein targets in cancer therapy, including mutant p53, belong to the intrinsically disordered proteins, which lack a well-defined protein structure making them challenging to pharmacologically target (157). An important factor in anticancer therapeutic failure is also associated with pharmacologic drugs that may lack response to all mutant variant or with substantial toxicity due to loss of wild-type function, or activating wildtype in normal tissue. Thus, while targeting a loss of function is difficult, growing evidence indicate that no single drug may display equal impact on all mutant proteins. Development of different drugs to target distinct mutant p53 or their activities is therefore time consuming and not cost-efficient, although such drugs could make a huge impact. Hence, exploring of alternative approaches to target mutant p53 proteins is therefore of high importance.
During recent years strategies of stimulating the cell’s own quality control mechanisms to prevent the aberrant accumulation and induce degradation of oncogenic proteins, including mutant p53, are being explored as a new therapeutic approach. Central to this idea is that oncogenic mutant p53 functions and the mutant p53 addiction of cancer cells is reliant on its sustained high levels, thus this addiction can be therapeutically exploited by targeted mutant p53 degradation strategies. Beyond pharmacological blockade of mutant p53 stabilizing mechanism to promote proteasome-dependent proteolysis, the considerable role for targeted degradation into lysosomes is suggested as a new advance to have a potentially major impact on mutant p53. The targeting of mutant p53 proteins by autophagy activation could offer promising future therapeutic option and is therefore currently investigated intensively. Below, we describe recent advances strategies that and might be potential therapeutic methods.
Targeting Mutant p53 by Macroautophagy
Although mutant p53 proteins were known to accumulate at abnormally high levels in cancer cells, the observation that lysosomal inhibitors could further stabilize mutant protein abundances strongly implied that they might be continuously degraded through the lysosomal pathway. In line with this, glucose restriction in multiple cancer types bearing the p53R175H, R280K mutants was shown to induce p53 mutant deacetylation, routing it for degradation via MA (158) (Figure 3). Accordingly, several studies now demonstrate that lysosomes indeed represent a degradation route for certain mutant p53 proteins (159–161). MA inhibition by either chemical inhibitors or downregulation of key autophagic related genes (ULK1, BCN1 or ATG5) induce stabilization of mutant p53, while, the overexpression of Ulk1 or Beclin-1 results in mutant p53 degradation (162). With MA as an emerging important pathway involved in the stability of mutant p53, several classes of small molecules enabling efficient mutant p53 degradation through the induction of autophagy has been described. These include, a) the curcumin-based zinc compound (Zn(II)-curcumin and capsaicin (8-methyl-N-vanillyl-6-noneamide)-induced macroautophagy which have been shown to deplete the expression of p53RH175 and p53R273H mutants (159, 163), b) Gambogic acid, a pro-apoptotic molecule that promotes the p53R280K and p53S241F mutant degradation by inducing autophagy (160), c) inhibition of MKK3, a dual protein MAP kinase, which reduces p53R273H mutant protein levels through ER stress-induced autophagy, d) the cruciferous-vegetable-derived phenethyl isothiocyanate (PEITC), which render the p53R175H,R273H, R248Q mutants by degradation following reactivation of the mutants, e) heat shock protein 90 (HSP90) inhibitors such as 17-allylamino-17-demethoxygeldanamycin (17-AAG) or ganetespib (155), and f) histone deacetylases inhibitors (HDACi), which have been studied as anticancer compounds based on their potential to stimulate autophagy and to degrade p53R172H,R248Q, R280K mutants (155, 164–167). Although apoptosis seems as the main route, inhibiting the HDACs, for example by the suberoylanilide hydroxamic acid (SAHA), a pan HDAC inhibitor, is shown to induce the destabilization of the HDAC6–HSP90–mutp53 complex (165), that results in mutant p53 degradation in cancer cells with pronounced autophagy induction, such as in MDA-MB-231 bearing the mutant p53R280K (164). However, while the MA stimulatory effect of SAHA on cancer cells carrying mutant p53 has been suggested, compared to null or wild-type p53 expressing cells, DLD1 cells carrying the p53S241F allele was not affected by this action. The observed degradation of p53S241F proteins upon SAHA exposure was suggested to relate on alternative degradation pathways rather than MA. Yet, ES2 cell lines bearing the same mutant (p53S241F) show difference in SAHA sensitivity. This strongly suggests that, cell type and -contexts need to be considered for SAHA-mediated cytotoxicity in cancer cells. Further, the Zn(II)-curcumin and capsaicin by acting on protein folding is able to reactivate wild-type p53 that induces its target gene DRAM to promote autophagy, while gambogic acid is shown to induce mutant p53 protein degradation through proteasome ubiquitination by carboxy terminus of Hsc70 interacting protein (CHIP). CHIP, on the other hand, known to display chaperone and E3 ligase activity, is involved in stabilizing and degrading both wild‐type and mutant p53 proteins, where the degradation of mutant p53 by CHIP was shown to be via autophagy through K63‐linked polyubiquitination. Under both normal and hypoxic conditions CHIP was shown to selectively degrade aggregation‐prone mutants p53R110L, p53R110P and p53R175H, without significant effects on the level of nonaggregating mutant p53R248W and p53R273H (168). Moreover, the p53R175 degradation by PEITC was reported to be mediated by both the proteasome and autophagy in a concentration-dependent manner, underlying the importance and need for further investigations for the selective degradation mechanism of mutant p53 in order to develop selective autophagy targeting therapeutic strategies. In addition, it is important to note that while the wild type p53 proteins are directed for proteasome-dependent degradation, autophagy-lysosome degradation is also attributed to control cellular p53 stability (169, 170). For example, Sunitinib, a small molecule multi kinase inhibitor, approved for the treatment of metastatic renal cell carcinoma, induced autophagic degradation of wild type p53 proteins in multiple cancer cell lines (169). However, the molecular mechanisms and cellular players involved in autophagic degradation of wild type p53 are still not fully known.
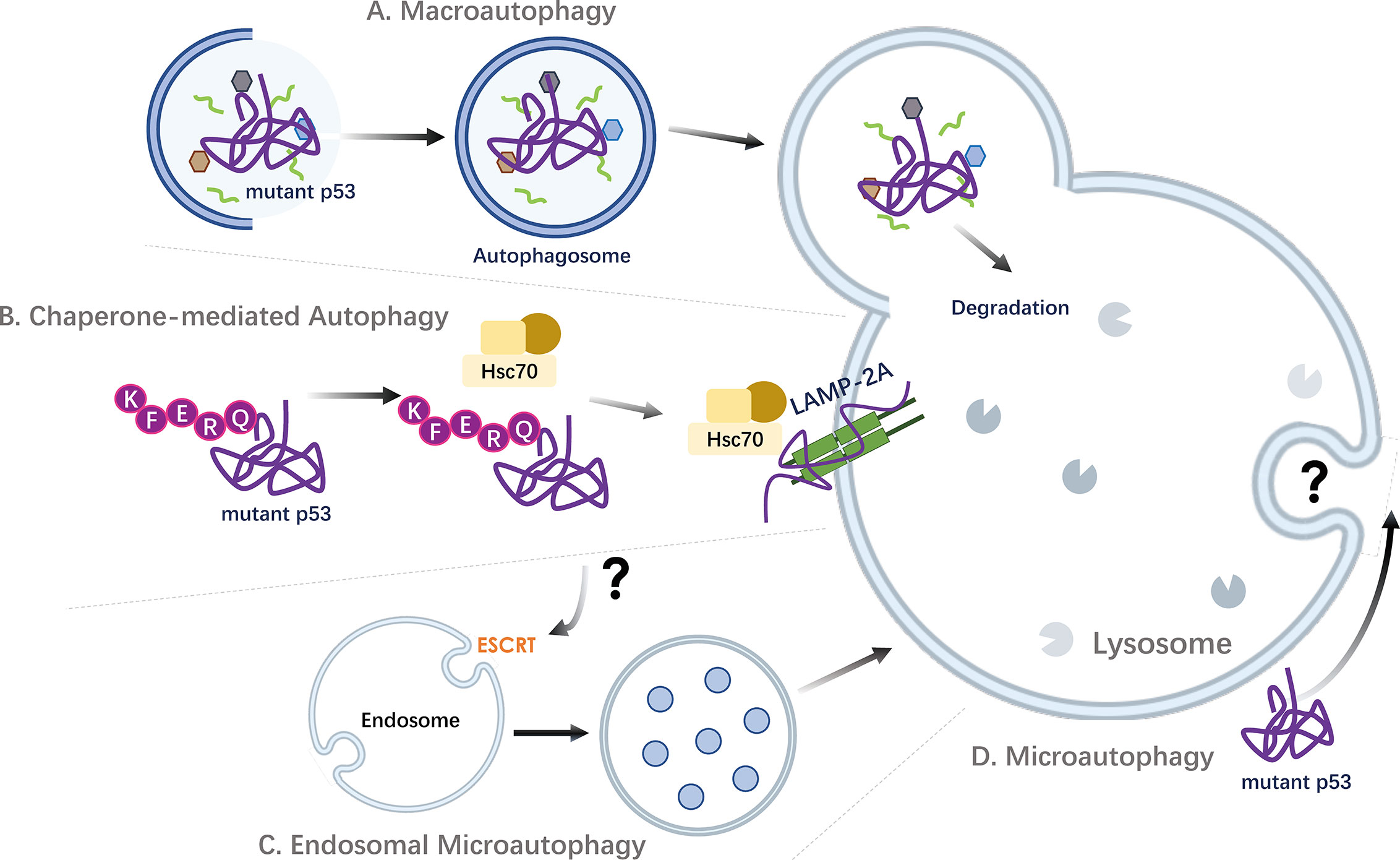
Figure 3 Targeting mutant p53 by autophagic pathways. Strategies of autophagic degradation of accumulated oncogenic mutant p53 proteins in cancer cells. (A) Mutant p53 can be engulfed and degraded via macroautophagy. P53 containing aggregates have also been implicated to undergo degradation by aggrephagy, a selective sequestration of protein aggregates by macroautophagy. (B) As p53 contains KFERQ-like motifs, mutant p53 proteins can be targeted and degraded through the stimulation of Chaperone-mediated Autophagy (CMA). (C) Via the recognition of proteins harboring KFERQ-like motifs, the molecular chaperone HSC70 and co-chaperone complex can also promote the localization of cargo proteins into endosomal compartments in an ESCRT machinery dependent mechanism, through a process called endosomal microautophagy (eMI). Thus, beyond CMA, other autophagic pathways, including endosomal eMI may mediate the degradation of mutant p53. (D) The direct uptake of mutant p53 proteins by lysosomes through microautophagy is not known. ESCRT, Endosomal Sorting Complex Required for Transport; Hsc70, Heat shock cognate 71 kDa protein (also known as HSPA8); LAMP-2A, Lysosome-associated membrane protein 2A.
Mutant p53 Proteins as Targets for Chaperone-Mediated Autophagy
Beyond contributing in lysosomal degradation of a select subset of cellular proteins, the discovery of mutant p53 proteins as CMA targets established a regulatory role for CMA in oncoprotein degradation and its potential tumor suppressive role (37, 65, 67, 171). Thus, a new degradative detour for mutant p53 via CMA was uncovered (172) (Figure 3).
As previously mentioned, CMA is a unique type of mammalian autophagy that only applies to select proteins without targeting cellular organelles (42). Its specificity relies on the recognition of a pentapeptide CMA motif (KFERQ-like) that is a prerequisite in target proteins. The cytosolic heat-shock cognate protein of 70 kDa (Hsc70/HSPA8) plays an essential role in CMA by recognizing the KFERQ-like sequence motifs in substrate proteins. Indeed, p53 harbors two pentapeptide sequences (200NLRVE204 and 341FRELN345) that are consistent with an Hsc70 recognition motif (65). The FRELN motif is on the linker region, while the 200NLRVE204 motif is exposed on the surface of the p53 protein, making it accessible for recognition.
Once activated, CMA was shown to be very effective in degrading different mutant p53 proteins, regardless of their mutational status (65), including p53P98S,P151H, A161T,R175C,R175D,R175H,L194F,S227K,S227R,G245C,R248L,R248W,E258K,R273H,R273L,R280K, R282W. This was initially illustrated by the assessment of CMA activation on the ectopically overexpressed above mentioned p53 mutants in a p53 null colon cancer cells. Subsequently, the CMA-mediated degradation of cancer associated endogenous mutant p53 proteins was shown on p53R175H,R248Q,S241F,R158InF,R280L,G266Q variants (65). This suggest that CMA-mediated mutant p53 degradation may be more efficacious than treatment with targeted mutant p53 specific reactivating small molecules and that CMA-based strategy could overcome resistance from acquired mutations. Importantly, the activation of CMA was not or less effective on wild type or p53 null expressing cancer cells. However, contrary to cancer cells, hepatitis C virus infection induced ER-stress response, which leads to CMA stimulation in untransformed primary human hepatocytes results in degradation of wild type p53 (173). The increased expression of chaperones due to unfolded protein response and ER stress associated with the CMA response, where the genetic silencing of LAMP-2A restored the observed p53 degradation. In fact, the silencing of LAMP-2A under irradiation conditions was also shown to result in increased p53 protein level (174), in some cancer cells, such as HepG2, which expresses wild type p53 (173). While these studies suggest an interplay between the CMA pathway and wild type p53, it should be kept in mind that p53 interacts with a wide range of different proteins, thus the accumulation of p53 upon LAMP-2A knockdown, may therefore depend on recognition of its molecular partners by the CMA pathway, such as HMGB1 degradation with further impact the wild type p53 protein expression (174). Nonetheless, the discovery that mutant p53 proteins are CMA substrates provided experimental evidence that CMA could be exploited as a novel approach to eliminate mutant p53 in cancer cells. Accumulating evidence now support that CMA activation plays a role in mutant p53 targeting (161). In fact, beyond mutant p53, CMA has been shown to promote the degradation of other oncoproteins, as HK2 and c-Myc (66, 67). Further, a decrease in CMA with age has been associated with higher risk of malignant transformation, and mice with hepatic blockage of CMA has been shown to develop spontaneous tumors (68). While, these findings suggest clinical implications of CMA activation, to date the role of CMA in tumorigenic conditions is not well-defined and there are no direct pharmacological CMA activators for cancer cells. Characterization of such activators would also require that it does not affect other degradation pathways. Accordingly, in order to explore the clinical implementation of CMA, development of applicable methods to measure CMA in live cells, and in vivo studies in CMA activation is needed. Thus, to date, there are no clinical studies launched to demonstrate the efficacy of CMA activation in patients. However, the knowledge of the its oncogenic targets, such as mutant p53, and understanding its selective degradation mechanism is an excellent starting point for future development of targeted therapeutic strategies involving CMA.
Mutant p53 as Possible Target for Microautophagy and CASA
Beyond CMA, recognition of proteins harboring a KFERQ-like motif by the molecular chaperone HSC70 can also lead to the endosomal localization in an ESCRT machinery dependent mechanism, through a process called endosomal microautophagy (eMI) (40, 47). Thus, selective degradation of single proteins has been described in a HSC70-driven endosomal eMI pathways (40, 42, 175). In addition, chaperone-assisted lysosomal degradation pathway CASA (chaperone-assisted selective autophagy), has been reported to require the involvement of HSC70. Keeping in mind that the amino acid sequence of p53 contains KFERQ-like motifs that is recognizable by HSC70, although yet to be proven, it is plausible that mutant p53 protein might be targeted by eMI or CASA (Figure 3). However, it is currently not known whether wild type and/or mutant p53 proteins are targeted and degraded by these pathways.
Mutant p53 Aggregates as Target for Aggreaphagy
While accumulation of protein aggregates is commonly known for their involvement in the onset of many neurodegenerative diseases, the conformation of mutant p53 with missense mutations is now known to share similarity with that of pathological mutant proteins involved in a wide range of neurodegeneration, including Alzheimer disease, Parkinson disease, and amyotrophic lateral sclerosis, the so-called protein conformation diseases that involve protein misfolding in their etiology (176). Accordingly, mutant p53 proteins display hyperstability due to acquired misfolding and partially denatured conformation with high tendency to form amyloid like micro- and macro-aggregates both in vitro and in vivo (177). The aggregation of mutant p53 (amyloid oligomers and fibrils) confers a prion-like activity on the native protein, converting it into an inactive form, thus contribute to its oncogenic function (178). The formation of aggregates largely depends on cellular chaperones and chaperone-assisted proteins. Accordingly, mutant p53 stabilization is achieved by the interaction with chaperone heat shock proteins (HSP), including HSP90, HSP40 and HSP70, that cooperate in stabilizing mutant p53.
Aggregated proteins can be degraded by the proteasome or CMA, however, only after the dissolution into soluble single peptide species, unless targeted by a process called aggrephagy, a selective sequestration of protein aggregates by macroautophagy (179). While the molecular mechanism of cargo selection during aggrephagy needs to be further elucidated, p53R175 containing aggregates have been implicated to undergo degradation by this pathway (167). This is in fact in line with the observations that CHIP, beyond targeting wild‐type p53 by K48 polyubiquitinition, preferentially degrades aggregation-prone mutant p53 proteins through K63 polyubiquitinition chains (168). Thus, although accumulation of mutant p53 occurs only in cancer cells, in which most missense mutants are shown to be more stable than wild-type p53, the aggregation of different mutants seems to correlate with individual structural characteristics, which may affect their differential recognition and degradation route.
Degradation of Mutant p53 Proteins by Multiple Autophagic Pathways
Autophagy pathways are mechanistically and functionally linked such that blockage to either one can lead to upregulation of the other in a way. The degradation of distinct mutant variant can therefore vary between the different types of autophagy, when one pathway is blocked or inhibited, or in response to different stresses. However, it is important to note that although MA and CMA are both operational under normal nutritional conditions, their basal activities are not sufficient for efficient removal of mutant p53. Rather, as described above, mutant proteins can undergo degradation through MA induced by glucose restriction or by proteasomal inhibition, however when MA is inhibited, which significantly accelerates the activation of CMA that in turn promotes the degradation of mutant p53. This differential degradation route was demonstrated for the p53R248Q mutant in a context dependent manner. In tumors growing in normoxia, with no stress, the treatment with Hsp90 inhibitor (17-AAG) was able to induce the degradation of p53R248Q through MA (161). However, during metabolic stress caused by the pyruvate dehydrogenase kinase-1 (PDK1) inhibitor dichloroacetate (DCA), p53R248Q proteins were stabilized by increased interaction with the Hsp90 chaperone machinery. Thus, in this condition, the co-treatment of 17-AAG instead promotes the association of p53R248Q with Hsc70 and CMA activation, resulting in p53R248Q degradation via the CMA pathway (161). Thus, different metabolic contexts and stressors induce diverse autophagy mechanisms that can degrade mutant proteins. In fact, beyond enabling efficient p53R248Q degradation by either MA or CMA, the HSP90 inhibitor, geldanamycin, has been suggested with an unspecific ability to activate CMA.
Furthermore, it is unclear whether different autophagic pathways may display any preference to degrade certain mutants. Since mutant p53 proteins encoded by different mutant alleles exhibit a distinctive tendency to misfold and aggregate, it may affect their susceptibility for recognition and targetability, thus it is reasonable that the mutational status may play a determinant role in its ability to be degraded through the distinct autophagic system. For instance, this may be due to the diverse ability of certain mutant to aggregates into prion-like amyloid oligomers, including p53R175H, R249S, which can form larger multimeric assemblies, while p53R248Q mutant displays significantly increased amyloidogenic potential, whereas p53M237I mutant is shown to co-localize with amyloid oligomers (180). Thus, beyond defects in degradation and recognition mechanisms, the accumulation of mutant p53 proteins to different levels in cancer cells may depend on their targetability by multiple vs certain degradation pathways.
Concluding Remarks
To conclude, the role and impact of mutant p53 in autophagy regulation is complex, context-dependent and far from fully elucidated. Growing evidence along with rapidly developing genome editing and omics techniques are likely to revolutionize new roles and autophagic activities of different mutant p53 proteins that may vary according to changes within tumors or in the tumor microenvironment. These new technologies may shed new insights for a knowledge-based discovery to identify knowledge gaps and analyze scenarios that require a reconsideration for the function of mutant p53 on autophagy.
Further, since it is clearly demonstrated that mutant p53 stabilization is a tumor-specific vulnerability, strategies to promote the degradation of mutant p53 by autophagy represents an attractive anti-cancer approach. Yet the effective therapeutic use of autophagy induction requires detailed knowledge of how the autophagy-lysosome pathway might be affected in cancer diseases. This is especially important given that disease-related genetic defects may affect autophagic pathway e.g., when lysosomal fusion or degradation is impaired. Thus, the stimulation of autophagy may rather worsen the disease progression. While autophagy modulation is an exciting area of clinical development, the effects of autophagy upregulation may vary substantially depending on the precise nature of the tumor state. Further comprehensive understanding of the roles of autophagic pathways throughout different stages of carcinogenesis has potential to guide development of novel therapeutic strategies to eradicate cancer cells with mutant p53. Furthermore, most if not all autophagy modulating drugs in clinical trials are inhibitors of the process, with the effectiveness of inhibiting autophagy to enhance chemotherapy cytotoxicity. Accordingly, pharmacological methods are not currently available to selectively and solely activate and target oncoproteins, including mutant p53, by autophagic pathways. While CMA can be directed to target oncogenic proteins, such as mutant p53, molecular mechanisms of its selective cargo recognition remain largely uncharacterized.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the review and writing, and approved it for publication.
Funding
This work was supported by grants from Karolinska Institutet, the Swedish Research Council (VR), the Ragnar Söderberg Foundation and the Swedish Cancer Society (Cancerfonden). Special thanks to Dr. Zhang for the graphic design of figures.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
AMPK, AMP-activated protein kinase; CDKN1A, Cyclin Dependent Kinase Inhibitor 1A; Chmp4C, Charged multivesicular body protein 4c; CMA, chaperon-mediated autophagy; DAPK1, Death-Associated Protein Kinase 1; DNM1, Dynamin 1; DRAM1, damage-regulated autophagy modulator 1; EGFR, epidermal growth factor receptor; EMT, Epithelial–mesenchymal transition; ESCRT, endosomal sorting complexes required for transport; GOF, gain of function; HIF-1, hypoxia inducible factor 1; IGF-BP3, Insulin-like growth factor-binding protein 3; IGFR, Insulin-like growth factor receptor; LAMP-2A, lysosome-associated membrane protein 2A; LKB1, liver kinase B1; MDM2, murine double minute 2; MKK3, Mitogen-activated protein kinase kinase 3; mTOR, mechanistic target of rapamycin; PI3K, phosphatidylinositol 3-kinase; PKM2, pyruvate kinase isoform M2; RB1CC1, Retinoblastoma coiled coil protein 1; REDD1, protein regulated in development and DNA damage responses 1; ROS, reactive oxygen species; SREBP1, sterol regulatory element-binding protein 1; TGFBR, transforming growth factor-beta receptor; TIGAR, TP53-induced glycolysis and apoptosis regulator; TSAP6, tumor suppressor-activated pathway 6; TSC, Tuberous Sclerosis Complex.
References
1. Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol (2007) 8(4):275–83. doi: 10.1038/nrm2147
2. Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev (2012) 26(12):1268–86. doi: 10.1101/gad.190678.112
3. Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene (2005) 24(17):2899–908. doi: 10.1038/sj.onc.1208615
4. Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol (2013) 15(1):2–8. doi: 10.1038/ncb2641
5. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature (2013) 502(7471):333–9. doi: 10.1038/nature12634
6. Leroy B, Anderson M, Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat (2014) 35(6):672–88. doi: 10.1002/humu.22552
7. Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell (2017) 170(6):1062–78. doi: 10.1016/j.cell.2017.08.028
8. Tran TQ, Lowman XH, Reid MA, Mendez-Dorantes C, Pan M, Yang Y, et al. Tumor-associated mutant p53 promotes cancer cell survival upon glutamine deprivation through p21 induction. Oncogene (2017) 36(14):1991–2001. doi: 10.1038/onc.2016.360
9. Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol (2010) 2(2):a001107. doi: 10.1101/cshperspect.a001107
10. Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer (2009) 9(10):701–13. doi: 10.1038/nrc2693
11. Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature (2015) 518(7540):495–501. doi: 10.1038/nature14169
12. Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer (2002) 2(8):594–604. doi: 10.1038/nrc864
13. Dittmer D, Pati S, Zambetti G, Chu S, Teresky AK, Moore M, et al. Gain of function mutations in p53. Nat Genet (1993) 4(1):42–6. doi: 10.1038/ng0593-42
14. Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell (2004) 119(6):847–60. doi: 10.1016/j.cell.2004.11.004
15. Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol (1994) 4(1):1–7. doi: 10.1016/S0960-9822(00)00002-6
16. Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell (2009) 139(7):1327–41. doi: 10.1016/j.cell.2009.11.026
17. Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell (2014) 25(3):304–17. doi: 10.1016/j.ccr.2014.01.021
18. Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell (2004) 119(6):861–72. doi: 10.1016/j.cell.2004.11.006
19. Liu G, McDonnell TJ, Montes de Oca Luna R, Kapoor M, Mims B, El-Naggar AK, et al. High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc Natl Acad Sci USA (2000) 97(8):4174–9. doi: 10.1073/pnas.97.8.4174
20. Lozano G. Mouse models of p53 functions. Cold Spring Harb Perspect Biol (2010) 2(4):a001115. doi: 10.1101/cshperspect.a001115
21. Kenzelmann Broz D, Attardi LD. In vivo analysis of p53 tumor suppressor function using genetically engineered mouse models. Carcinogenesis (2010) 31(8):1311–8. doi: 10.1093/carcin/bgp331
22. Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell (2006) 10(3):191–202. doi: 10.1016/j.ccr.2006.08.013
23. Weisz L, Damalas A, Liontos M, Karakaidos P, Fontemaggi G, Maor-Aloni R, et al. Mutant p53 enhances nuclear factor kappaB activation by tumor necrosis factor alpha in cancer cells. Cancer Res (2007) 67(6):2396–401. doi: 10.1158/0008-5472.CAN-06-2425
24. Kim MP, Lozano G. Mutant p53 partners in crime. Cell Death Differ (2018) 25(1):161–8. doi: 10.1038/cdd.2017.185
25. Boettcher S, Miller PG, Sharma R, McConkey M, Leventhal M, Krivtsov AV, et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science (2019) 365(6453):599–604. doi: 10.1126/science.aax3649
26. Giacomelli AO, Yang X, Lintner RE, McFarland JM, Duby M, Kim J, et al. Mutational processes shape the landscape of TP53 mutations in human cancer. Nat Genet (2018) 50(10):1381–7. doi: 10.1038/s41588-018-0204-y
27. Walerych D, Lisek K, Del Sal G. Mutant p53: One, No One, and One Hundred Thousand. Front Oncol (2015) 5:289. doi: 10.3389/fonc.2015.00289
28. Jackson JG, Lozano G. The mutant p53 mouse as a pre-clinical model. Oncogene (2013) 32(37):4325–30. doi: 10.1038/onc.2012.610
29. Eriksson M, Ambroise G, Ouchida AT, Lima Queiroz A, Smith D, Gimenez-Cassina A, et al. Effect of Mutant p53 Proteins on Glycolysis and Mitochondrial Metabolism. Mol Cell Biol (2017) 37(24):e00328-17. doi: 10.1128/MCB.00328-17
30. Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res (2006) 12(4):1157–67. doi: 10.1158/1078-0432.CCR-05-1029
31. Silwal-Pandit L, Vollan HK, Chin SF, Rueda OM, McKinney S, Osako T, et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res (2014) 20(13):3569–80. doi: 10.1158/1078-0432.CCR-13-2943
32. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell (2011) 147(4):728–41. doi: 10.1016/j.cell.2011.10.026
33. Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell (2010) 40(2):280–93. doi: 10.1016/j.molcel.2010.09.023
34. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet (2009) 43:67–93. doi: 10.1146/annurev-genet-102808-114910
35. Kristensen AR, Schandorff S, Hoyer-Hansen M, Nielsen MO, Jaattela M, Dengjel J, et al. Ordered organelle degradation during starvation-induced autophagy. Mol Cell Proteomics (2008) 7(12):2419–28. doi: 10.1074/mcp.M800184-MCP200
36. Muller M, Schmidt O, Angelova M, Faserl K, Weys S, Kremser L, et al. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. Elife (2015) 4:e07736. doi: 10.7554/eLife.07736
37. Hao Y, Kacal M, Ouchida AT, Zhang B, Norberg E, Vakifahmetoglu-Norberg H. Targetome analysis of chaperone-mediated autophagy in cancer cells. Autophagy (2019) 15(9):1558–71. doi: 10.1080/15548627.2019.1586255
38. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell (2008) 132(1):27–42. doi: 10.1016/j.cell.2007.12.018
39. Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal (2014) 20(3):460–73. doi: 10.1089/ars.2013.5371
40. Tekirdag K, Cuervo AM. Chaperone-mediated autophagy and endosomal microautophagy: Joint by a chaperone. J Biol Chem (2018) 293(15):5414–24. doi: 10.1074/jbc.R117.818237
41. Oku M, Sakai Y. Three Distinct Types of Microautophagy Based on Membrane Dynamics and Molecular Machineries. Bioessays (2018) 40(6):e1800008. doi: 10.1002/bies.201800008
42. Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol (2018) 19(6):365–81. doi: 10.1038/s41580-018-0001-6
43. Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, et al. Molecular definitions of autophagy and related processes. EMBO J (2017) 36(13):1811–36. doi: 10.15252/embj.201796697
44. Klionsky DJ, Cregg JM, Dunn WA Jr., Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell (2003) 5(4):539–45. doi: 10.1016/S1534-5807(03)00296-X
45. Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol (2007) 9(10):1102–9. doi: 10.1038/ncb1007-1102
46. Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, et al. A comprehensive glossary of autophagy-related molecules and processes (2nd edition). Autophagy (2011) 7(11):1273–94. doi: 10.4161/auto.7.11.17661
47. Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell (2011) 20(1):131–9. doi: 10.1016/j.devcel.2010.12.003
48. Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci (1990) 15(8):305–9. doi: 10.1016/0968-0004(90)90019-8
49. Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science (1996) 273(5274):501–3. doi: 10.1126/science.273.5274.501
50. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature (2008) 451(7182):1069–75. doi: 10.1038/nature06639
51. Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell (2006) 10(1):51–64. doi: 10.1016/j.ccr.2006.06.001
52. Yang X, Yu DD, Yan F, Jing YY, Han ZP, Sun K, et al. The role of autophagy induced by tumor microenvironment in different cells and stages of cancer. Cell Biosci (2015) 5:14. doi: 10.1186/s13578-015-0005-2
53. Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer (2007) 7(12):961–7. doi: 10.1038/nrc2254
54. Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev (2011) 25(5):460–70. doi: 10.1101/gad.2016311
55. Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discovery (2013) 3(11):1272–85. doi: 10.1158/2159-8290.CD-13-0397
56. White E. The role for autophagy in cancer. J Clin Invest (2015) 125(1):42–6. doi: 10.1172/JCI73941
57. White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol (2010) 22(2):212–7. doi: 10.1016/j.ceb.2009.12.008
58. Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev (2011) 25(8):795–800. doi: 10.1101/gad.2016211
59. Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell (2013) 155(6):1216–9. doi: 10.1016/j.cell.2013.11.019
60. Lebovitz CB, Robertson AG, Goya R, Jones SJ, Morin RD, Marra MA, et al. Cross-cancer profiling of molecular alterations within the human autophagy interaction network. Autophagy (2015) 11(9):1668–87. doi: 10.1080/15548627.2015.1067362
61. Tang Y, Wang XW, Liu ZH, Sun YM, Tang YX, Zhou DH. Chaperone-mediated autophagy substrate proteins in cancer. Oncotarget (2017) 8(31):51970–85. doi: 10.18632/oncotarget.17583
62. Arias E, Cuervo AM. Pros and Cons of Chaperone-Mediated Autophagy in Cancer Biology. Trends Endocrinol Metab (2020) 31(1):53–66. doi: 10.1016/j.tem.2019.09.007
63. Kon M, Kiffin R, Koga H, Chapochnick J, Macian F, Varticovski L, et al. Chaperone-mediated autophagy is required for tumor growth. Sci Transl Med (2011) 3(109):109ra17. doi: 10.1126/scitranslmed.3003182
64. Galan-Acosta L, Xia H, Yuan J, Vakifahmetoglu-Norberg H. Activation of chaperone-mediated autophagy as a potential anticancer therapy. Autophagy (2015) 11(12):2370–1. doi: 10.1080/15548627.2015.1106666
65. Vakifahmetoglu-Norberg H, Kim M, Xia HG, Iwanicki MP, Ofengeim D, Coloff JL, et al. Chaperone-mediated autophagy degrades mutant p53. Genes Dev (2013) 27(15):1718–30. doi: 10.1101/gad.220897.113
66. Gomes LR, Menck CFM, Cuervo AM. Chaperone-mediated autophagy prevents cellular transformation by regulating MYC proteasomal degradation. Autophagy (2017) 13(5):928–40. doi: 10.1080/15548627.2017.1293767
67. Xia HG, Najafov A, Geng J, Galan-Acosta L, Han X, Guo Y, et al. Degradation of HK2 by chaperone-mediated autophagy promotes metabolic catastrophe and cell death. J Cell Biol (2015) 210(5):705–16. doi: 10.1083/jcb.201503044
68. Schneider JL, Villarroya J, Diaz-Carretero A, Patel B, Urbanska AM, Thi MM, et al. Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell (2015) 14(2):249–64. doi: 10.1111/acel.12310
69. Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res (2007) 67(7):3043–53. doi: 10.1158/0008-5472.CAN-06-4149
70. Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell (2008) 134(3):451–60. doi: 10.1016/j.cell.2008.06.028
71. Drakos E, Atsaves V, Li J, Leventaki V, Andreeff M, Medeiros LJ, et al. Stabilization and activation of p53 downregulates mTOR signaling through AMPK in mantle cell lymphoma. Leukemia (2009) 23(4):784–90. doi: 10.1038/leu.2008.348
72. Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger BR, et al. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature (1995) 377(6550):646–9. doi: 10.1038/377646a0
73. Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev (2013) 27(9):1016–31. doi: 10.1101/gad.212282.112
74. Gao W, Shen Z, Shang L, Wang X. Upregulation of human autophagy-initiation kinase ULK1 by tumor suppressor p53 contributes to DNA-damage-induced cell death. Cell Death Differ (2011) 18(10):1598–607. doi: 10.1038/cdd.2011.33
75. Crighton D, Wilkinson S, Ryan KM. DRAM links autophagy to p53 and programmed cell death. Autophagy (2007) 3(1):72–4. doi: 10.4161/auto.3438
76. Desai S, Liu Z, Yao J, Patel N, Chen J, Wu Y, et al. Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of autophagy-related protein 7 (ATG7). J Biol Chem (2013) 288(13):9165–76. doi: 10.1074/jbc.M112.422071
77. Yeo SY, Itahana Y, Guo AK, Han R, Iwamoto K, Nguyen HT, et al. Transglutaminase 2 contributes to a TP53-induced autophagy program to prevent oncogenic transformation. Elife (2016) 5:e07101. doi: 10.7554/eLife.07101
78. Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell (2005) 122(6):927–39. doi: 10.1016/j.cell.2005.07.002
79. Hu W, Chen S, Thorne RF, Wu M. TP53, TP53 Target Genes (DRAM, TIGAR), and Autophagy. Adv Exp Med Biol (2019) 1206:127–49. doi: 10.1007/978-981-15-0602-4_6
80. Mrakovcic M, Frohlich LF. p53-Mediated Molecular Control of Autophagy in Tumor Cells. Biomolecules (2018) 8(2):14. doi: 10.3390/biom8020014
81. Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol (2008) 10(6):676–87. doi: 10.1038/ncb1730
82. Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell (2006) 126(1):107–20. doi: 10.1016/j.cell.2006.05.036
83. Xu J, Wang Y, Tan X, Jing H. MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy (2012) 8(6):873–82. doi: 10.4161/auto.19629
84. Liao JM, Cao B, Zhou X, Lu H. New insights into p53 functions through its target microRNAs. J Mol Cell Biol (2014) 6(3):206–13. doi: 10.1093/jmcb/mju018
85. Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell (2011) 147(1):223–34. doi: 10.1016/j.cell.2011.08.037
86. Tripathi R, Ash D, Shaha C. Beclin-1-p53 interaction is crucial for cell fate determination in embryonal carcinoma cells. J Cell Mol Med (2014) 18(11):2275–86. doi: 10.1111/jcmm.12386
87. Morselli E, Shen S, Ruckenstuhl C, Bauer MA, Marino G, Galluzzi L, et al. p53 inhibits autophagy by interacting with the human ortholog of yeast Atg17, RB1CC1/FIP200. Cell Cycle (2011) 10(16):2763–9. doi: 10.4161/cc.10.16.16868
88. Galluzzi L, Kepp O, Kroemer G. A new role for cytoplasmic p53: binding and destroying double-stranded RNA. Cell Cycle (2010) 9(13):2491–2. doi: 10.4161/cc.9.13.12191
89. Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T, et al. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun (2013) 4:2308. doi: 10.1038/ncomms3308
90. Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A, et al. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle (2008) 7(19):3056–61. doi: 10.4161/cc.7.19.6751
91. Cordani M, Oppici E, Dando I, Butturini E, Dalla Pozza E, Nadal-Serrano M, et al. Mutant p53 proteins counteract autophagic mechanism sensitizing cancer cells to mTOR inhibition. Mol Oncol (2016) 10(7):1008–29. doi: 10.1016/j.molonc.2016.04.001
92. Agarwal S, Bell CM, Taylor SM, Moran RG. p53 Deletion or Hotspot Mutations Enhance mTORC1 Activity by Altering Lysosomal Dynamics of TSC2 and Rheb. Mol Cancer Res (2016) 14(1):66–77. doi: 10.1158/1541-7786.MCR-15-0159
93. Tomita Y, Marchenko N, Erster S, Nemajerova A, Dehner A, Klein C, et al. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J Biol Chem (2006) 281(13):8600–6. doi: 10.1074/jbc.M507611200
94. Kim TH, Lee SY, Rho JH, Jeong NY, Soung YH, Jo WS, et al. Mutant p53 (G199V) gains antiapoptotic function through signal transducer and activator of transcription 3 in anaplastic thyroid cancer cells. Mol Cancer Res (2009) 7(10):1645–54. doi: 10.1158/1541-7786.MCR-09-0117
95. Aschauer L, Muller PA. Novel targets and interaction partners of mutant p53 Gain-Of-Function. Biochem Soc Trans (2016) 44(2):460–6. doi: 10.1042/BST20150261
96. DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv (2016) 2(5):e1600200. doi: 10.1126/sciadv.1600200
97. Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell (2017) 168(4):657–69. doi: 10.1016/j.cell.2016.12.039
98. Stanley IA, Ribeiro SM, Gimenez-Cassina A, Norberg E, Danial NN. Changing appetites: the adaptive advantages of fuel choice. Trends Cell Biol (2014) 24(2):118–27. doi: 10.1016/j.tcb.2013.07.010
99. Vakifahmetoglu-Norberg H, Ouchida AT, Norberg E. The role of mitochondria in metabolism and cell death. Biochem Biophys Res Commun (2017) 482(3):426–31. doi: 10.1016/j.bbrc.2016.11.088
100. Rabinowitz JD, White E. Autophagy and metabolism. Science (2010) 330(6009):1344–8. doi: 10.1126/science.1193497
101. Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol (2020) 21(4):183–203. doi: 10.1038/s41580-019-0199-y
102. DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell (2012) 148(6):1132–44. doi: 10.1016/j.cell.2012.02.032
103. Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol (2018) 19(2):121–35. doi: 10.1038/nrm.2017.95
104. Zhou G, Wang J, Zhao M, Xie TX, Tanaka N, Sano D, et al. Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol Cell (2014) 54(6):960–74. doi: 10.1016/j.molcel.2014.04.024
105. Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell (2012) 148(1-2):244–58. doi: 10.1016/j.cell.2011.12.017
106. Dando I, Cordani M, Donadelli M. Mutant p53 and mTOR/PKM2 regulation in cancer cells. IUBMB Life (2016) 68(9):722–6. doi: 10.1002/iub.1534
107. Humpton T, Vousden KH. Taking up the reins of power: metabolic functions of p53. J Mol Cell Biol (2019) 11(7):610–4. doi: 10.1093/jmcb/mjz065
108. Itahana Y, Itahana K. Emerging Roles of p53 Family Members in Glucose Metabolism. Int J Mol Sci (2018) 19(3):776. doi: 10.3390/ijms19030776
109. Labuschagne CF, Zani F, Vousden KH. Control of metabolism by p53 - Cancer and beyond. Biochim Biophys Acta Rev Cancer (2018) 1870(1):32–42. doi: 10.1016/j.bbcan.2018.06.001
110. Ingallina E, Sorrentino G, Bertolio R, Lisek K, Zannini A, Azzolin L, et al. Mechanical cues control mutant p53 stability through a mevalonate-RhoA axis. Nat Cell Biol (2018) 20(1):28–35. doi: 10.1038/s41556-017-0009-8
111. Parrales A, Ranjan A, Iyer SV, Padhye S, Weir SJ, Roy A, et al. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat Cell Biol (2016) 18(11):1233–43. doi: 10.1038/ncb3427
112. Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell (2008) 19(3):797–806. doi: 10.1091/mbc.e07-10-1092
113. Ahn CS, Metallo CM. Mitochondria as biosynthetic factories for cancer proliferation. Cancer Metab (2015) 3(1):1. doi: 10.1186/s40170-015-0128-2
114. Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB, et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell (2012) 22(4):547–60. doi: 10.1016/j.ccr.2012.08.014
115. Wang PY, Ma W, Park JY, Celi FS, Arena R, Choi JW, et al. Increased oxidative metabolism in the Li-Fraumeni syndrome. N Engl J Med (2013) 368(11):1027–32. doi: 10.1056/NEJMoa1214091
116. Minamino T, Orimo M, Shimizu I, Kunieda T, Yokoyama M, Ito T, et al. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat Med (2009) 15(9):1082–7. doi: 10.1038/nm.2014
117. Ibrahim MM, Razmara M, Nguyen D, Donahue RJ, Wubah JA, Knudsen TB. Altered expression of mitochondrial 16S ribosomal RNA in p53-deficient mouse embryos revealed by differential display. Biochim Biophys Acta (1998) 1403(3):254–64. doi: 10.1016/S0167-4889(98)00066-4
118. Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science (2006) 312(5780):1650–3. doi: 10.1126/science.1126863
119. Sakanashi F, Shintani M, Tsuneyoshi M, Ohsaki H, Kamoshida S. Apoptosis, necroptosis and autophagy in colorectal cancer: Associations with tumor aggressiveness and p53 status. Pathol Res Pract (2019) 215(7):152425. doi: 10.1016/j.prp.2019.04.017
120. Warfel NA, El-Deiry WS. p21WAF1 and tumourigenesis: 20 years after. Curr Opin Oncol (2013) 25(1):52–8. doi: 10.1097/CCO.0b013e32835b639e
121. Saini H, Hakeem I, Mukherjee S, Chowdhury S, Chowdhury R. Autophagy Regulated by Gain of Function Mutant p53 Enhances Proteasomal Inhibitor-Mediated Cell Death through Induction of ROS and ERK in Lung Cancer Cells. J Oncol (2019) 2019:6164807. doi: 10.1155/2019/6164807
122. Zhang X, Cheng Q, Yin H, Yang G. Regulation of autophagy and EMT by the interplay between p53 and RAS during cancer progression (Review). Int J Oncol (2017) 51(1):18–24. doi: 10.3892/ijo.2017.4025
123. Schofield HK, Zeller J, Espinoza C, Halbrook CJ, Del Vecchio A, Magnuson B, et al. Mutant p53R270H drives altered metabolism and increased invasion in pancreatic ductal adenocarcinoma. JCI Insight (2018) 3(2):e97422. doi: 10.1172/jci.insight.97422
124. Solomon H, Buganim Y, Kogan-Sakin I, Pomeraniec L, Assia Y, Madar S, et al. Various p53 mutant proteins differently regulate the Ras circuit to induce a cancer-related gene signature. J Cell Sci (2012) 125(Pt 13):3144–52. doi: 10.1242/jcs.099663
125. Hanel W, Marchenko N, Xu S, Yu SX, Weng W, Moll U. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ (2013) 20(7):898–909. doi: 10.1038/cdd.2013.17
126. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol (2019) 20(2):69–84. doi: 10.1038/s41580-018-0080-4
127. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest (2009) 119(6):1420–8. doi: 10.1172/JCI39104
128. Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol (2017) 14(10):611–29. doi: 10.1038/nrclinonc.2017.44
129. Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell (2017) 171(7):1611–24 e24. doi: 10.1016/j.cell.2017.10.044
130. Ambroise G, Yu TT, Zhang B, Kacal M, Hao Y, Queiroz AL, et al. Systematic analysis reveals a functional role for STAMBPL1 in the epithelial-mesenchymal transition process across multiple carcinomas. Br J Cancer (2020) 123(7):1164–77. doi: 10.1038/s41416-020-0972-x
131. Tang Q, Su Z, Gu W, Rustgi AK. Mutant p53 on the Path to Metastasis. Trends Cancer (2020) 6(1):62–73. doi: 10.1016/j.trecan.2019.11.004
132. Kang H, Kim H, Lee S, Youn H, Youn B. Role of Metabolic Reprogramming in Epithelial(-)Mesenchymal Transition (EMT). Int J Mol Sci (2019) 20(8):2042. doi: 10.3390/ijms20082042
133. Sciacovelli M, Frezza C. Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer. FEBS J (2017) 284(19):3132–44. doi: 10.1111/febs.14090
134. Brabletz T. To differentiate or not–routes towards metastasis. Nat Rev Cancer (2012) 12(6):425–36. doi: 10.1038/nrc3265
135. Brabletz T, Jung A, Reu S, Porzner M, Hlubek F, Kunz-Schughart LA, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci USA (2001) 98(18):10356–61. doi: 10.1073/pnas.171610498
136. Barr S, Thomson S, Buck E, Russo S, Petti F, Sujka-Kwok I, et al. Bypassing cellular EGF receptor dependence through epithelial-to-mesenchymal-like transitions. Clin Exp Metastasis (2008) 25(6):685–93. doi: 10.1007/s10585-007-9121-7
137. Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell (2009) 137(1):87–98. doi: 10.1016/j.cell.2009.01.039
138. Lu X, Liu DP, Xu Y. The gain of function of p53 cancer mutant in promoting mammary tumorigenesis. Oncogene (2013) 32(23):2900–6. doi: 10.1038/onc.2012.299
139. Solomon H, Dinowitz N, Pateras IS, Cooks T, Shetzer Y, Molchadsky A, et al. Mutant p53 gain of function underlies high expression levels of colorectal cancer stem cells markers. Oncogene (2018) 37(12):1669–84. doi: 10.1038/s41388-017-0060-8
140. Koifman G, Shetzer Y, Eizenberger S, Solomon H, Rotkopf R, Molchadsky A, et al. A Mutant p53-Dependent Embryonic Stem Cell Gene Signature Is Associated with Augmented Tumorigenesis of Stem Cells. Cancer Res (2018) 78(20):5833–47. doi: 10.1158/0008-5472.CAN-18-0805
141. Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun (2013) 4:2935. doi: 10.1038/ncomms3935
142. Basu S, Gnanapradeepan K, Barnoud T, Kung CP, Tavecchio M, Scott J, et al. Mutant p53 controls tumor metabolism and metastasis by regulating PGC-1alpha. Genes Dev (2018) 32(3–4):230–43. doi: 10.1101/gad.309062.117
143. Li J, Yang B, Zhou Q, Wu Y, Shang D, Guo Y, et al. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis (2013) 34(6):1343–51. doi: 10.1093/carcin/bgt063
144. Colella B, Faienza F, Di Bartolomeo S. EMT Regulation by Autophagy: A New Perspective in Glioblastoma Biology. Cancers (Basel) (2019) 11(3):312. doi: 10.3390/cancers11030312
145. Chen HT, Liu H, Mao MJ, Tan Y, Mo XQ, Meng XJ, et al. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol Cancer (2019) 18(1):101. doi: 10.1186/s12943-019-1030-2
146. Schuldner M, Dorsam B, Shatnyeva O, Reiners KS, Kubarenko A, Hansen HP, et al. Exosome-dependent immune surveillance at the metastatic niche requires BAG6 and CBP/p300-dependent acetylation of p53. Theranostics (2019) 9(21):6047–62. doi: 10.7150/thno.36378
147. Yu X, Riley T, Levine AJ. The regulation of the endosomal compartment by p53 the tumor suppressor gene. FEBS J (2009) 276(8):2201–12. doi: 10.1111/j.1742-4658.2009.06949.x
148. Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic (2000) 1(7):570–83. doi: 10.1034/j.1600-0854.2000.010707.x
149. Pajares M, Rojo AI, Arias E, Diaz-Carretero A, Cuervo AM, Cuadrado A. Transcription factor NFE2L2/NRF2 modulates chaperone-mediated autophagy through the regulation of LAMP2A. Autophagy (2018) 14(8):1310–22. doi: 10.1080/15548627.2018.1474992
150. Lisek K, Campaner E, Ciani Y, Walerych D, Del Sal G. Mutant p53 tunes the NRF2-dependent antioxidant response to support survival of cancer cells. Oncotarget (2018) 9(29):20508–23. doi: 10.18632/oncotarget.24974
151. Pavlakis E, Stiewe T. p53’s Extended Reach: The Mutant p53 Secretome. Biomolecules (2020) 10(2):307. doi: 10.3390/biom10020307
152. Schulz-Heddergott R, Moll UM. Gain-of-Function (GOF) Mutant p53 as Actionable Therapeutic Target. Cancers (Basel) (2018) 10(6):188. doi: 10.3390/cancers10060188
153. Blandino G, Di Agostino S. New therapeutic strategies to treat human cancers expressing mutant p53 proteins. J Exp Clin Cancer Res (2018) 37(1):30. doi: 10.1186/s13046-018-0705-7
154. Zhao D, Tahaney WM, Mazumdar A, Savage MI, Brown PH. Molecularly targeted therapies for p53-mutant cancers. Cell Mol Life Sci (2017) 74(22):4171–87. doi: 10.1007/s00018-017-2575-0
155. Alexandrova EM, Yallowitz AR, Li D, Xu S, Schulz R, Proia DA, et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature (2015) 523(7560):352–6. doi: 10.1038/nature14430
156. Weissmueller S, Manchado E, Saborowski M, Morris JP, Wagenblast E, Davis CA, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell (2014) 157(2):382–94. doi: 10.1016/j.cell.2014.01.066
157. Uversky VN. p53 Proteoforms and Intrinsic Disorder: An Illustration of the Protein Structure-Function Continuum Concept. Int J Mol Sci (2016) 17(11):1874. doi: 10.3390/ijms17111874
158. Rodriguez OC, Choudhury S, Kolukula V, Vietsch EE, Catania J, Preet A, et al. Dietary downregulation of mutant p53 levels via glucose restriction: mechanisms and implications for tumor therapy. Cell Cycle (2012) 11(23):4436–46. doi: 10.4161/cc.22778
159. Garufi A, Pucci D, D’Orazi V, Cirone M, Bossi G, Avantaggiati ML, et al. Degradation of mutant p53H175 protein by Zn(II) through autophagy. Cell Death Dis (2014) 5:e1271. doi: 10.1038/cddis.2014.217
160. Foggetti G, Ottaggio L, Russo D, Monti P, Degan P, Fronza G, et al. Gambogic acid counteracts mutant p53 stability by inducing autophagy. Biochim Biophys Acta Mol Cell Res (2017) 1864(2):382–92. doi: 10.1016/j.bbamcr.2016.11.023
161. Allende-Vega N, Villalba M. Metabolic stress controls mutant p53 R248Q stability in acute myeloid leukemia cells. Sci Rep (2019) 9(1):5637. doi: 10.1038/s41598-019-42220-y
162. Choudhury S, Kolukula VK, Preet A, Albanese C, Avantaggiati ML. Dissecting the pathways that destabilize mutant p53: the proteasome or autophagy? Cell Cycle (2013) 12(7):1022–9. doi: 10.4161/cc.24128
163. Garufi A, Pistritto G, Cirone M, D’Orazi G. Reactivation of mutant p53 by capsaicin, the major constituent of peppers. J Exp Clin Cancer Res (2016) 35(1):136. doi: 10.1186/s13046-016-0417-9
164. Foggetti G, Ottaggio L, Russo D, Mazzitelli C, Monti P, Degan P, et al. Autophagy induced by SAHA affects mutant P53 degradation and cancer cell survival. Biosci Rep (2019) 39(2):BSR20181345. doi: 10.1042/BSR20181345
165. Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ (2011) 18(12):1904–13. doi: 10.1038/cdd.2011.71
166. Baldari S, Ubertini V, Garufi A, D’Orazi G, Bossi G. Targeting MKK3 as a novel anticancer strategy: molecular mechanisms and therapeutical implications. Cell Death Dis (2015) 6:e1621. doi: 10.1038/cddis.2014.591
167. Aggarwal M, Saxena R, Sinclair E, Fu Y, Jacobs A, Dyba M, et al. Reactivation of mutant p53 by a dietary-related compound phenethyl isothiocyanate inhibits tumor growth. Cell Death Differ (2016) 23(10):1615–27. doi: 10.1038/cdd.2016.48
168. Maan M, Pati U. CHIP promotes autophagy-mediated degradation of aggregating mutant p53 in hypoxic conditions. FEBS J (2018) 285(17):3197–214. doi: 10.1111/febs.14602
169. Luo P, Xu Z, Li G, Yan H, Zhu Y, Zhu H, et al. HMGB1 represses the anti-cancer activity of sunitinib by governing TP53 autophagic degradation via its nucleus-to-cytoplasm transport. Autophagy (2018) 14(12):2155–70. doi: 10.1080/15548627.2018.1501134
170. Patergnani S, Guzzo S, Mangolini A, dell’Atti L, Pinton P, Aguiari G. The induction of AMPK-dependent autophagy leads to P53 degradation and affects cell growth and migration in kidney cancer cells. Exp Cell Res (2020) 395(1):112190. doi: 10.1016/j.yexcr.2020.112190
171. Park C, Suh Y, Cuervo AM. Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat Commun (2015) 6:6823. doi: 10.1038/ncomms7823
172. Vakifahmetoglu-Norberg H, Yuan J. A degradative detour for mutant TP53. Autophagy (2013) 9(12):2158–60. doi: 10.4161/auto.26338
173. Aydin Y, Chatterjee A, Chandra PK, Chava S, Chen W, Tandon A, et al. Interferon-alpha-induced hepatitis C virus clearance restores p53 tumor suppressor more than direct-acting antivirals. Hepatol Commun (2017) 1(3):256–69. doi: 10.1002/hep4.1025
174. Wu JH, Guo JP, Shi J, Wang H, Li LL, Guo B, et al. CMA down-regulates p53 expression through degradation of HMGB1 protein to inhibit irradiation-triggered apoptosis in hepatocellular carcinoma. World J Gastroenterol (2017) 23(13):2308–17. doi: 10.3748/wjg.v23.i13.2308
175. Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol (2012) 22(8):407–17. doi: 10.1016/j.tcb.2012.05.006
176. Silva JL, De Moura Gallo CV, Costa DC, Rangel LP. Prion-like aggregation of mutant p53 in cancer. Trends Biochem Sci (2014) 39(6):260–7. doi: 10.1016/j.tibs.2014.04.001
177. Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53 and cancer-associated mutants. Adv Cancer Res (2007) 97:1–23. doi: 10.1016/S0065-230X(06)97001-8
178. Rangel LP, Costa DC, Vieira TC, Silva JL. The aggregation of mutant p53 produces prion-like properties in cancer. Prion (2014) 8(1):75–84. doi: 10.4161/pri.27776
179. Yamamoto A, Simonsen A. The elimination of accumulated and aggregated proteins: a role for aggrephagy in neurodegeneration. Neurobiol Dis (2011) 43(1):17–28. doi: 10.1016/j.nbd.2010.08.015
Keywords: autophagy, chaperone-mediated autophagy, cancer, mutant p53, TP53
Citation: Shi Y, Norberg E and Vakifahmetoglu-Norberg H (2021) Mutant p53 as a Regulator and Target of Autophagy. Front. Oncol. 10:607149. doi: 10.3389/fonc.2020.607149
Received: 16 September 2020; Accepted: 15 December 2020;
Published: 03 February 2021.
Edited by:
Marco Cordani, IMDEA Nanociencia, SpainReviewed by:
Silvia Di Agostino, Magna Graecia University of Catanzaro, ItalyTomoo Iwakuma, University of Kansas Medical Center Research Institute, United States
Alfredo Criollo, University of Chile, Chile
Copyright © 2021 Shi, Norberg and Vakifahmetoglu-Norberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Helin Vakifahmetoglu-Norberg, SGVsaW4ubm9yYmVyZ0BraS5zZQ==
 Yong Shi
Yong Shi Erik Norberg
Erik Norberg Helin Vakifahmetoglu-Norberg
Helin Vakifahmetoglu-Norberg