
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 19 February 2021
Sec. Gastrointestinal Cancers
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.605801
This article is part of the Research Topic Gastrointestinal Tumor Heterogeneity and Related Anti-Cancer Strategies View all 17 articles
Extracellular and/or intracellular manipulation of pH in tumor may have noticeable potential in cancer treatment. Although the assembly factor genes of V0 domain of the V-ATPase complex are required for intracellular pH homeostasis, their significance in colorectal cancer (CRC) remains largely unknown. Here, we used bioinformatics to identify the candidates from known assembly factor genes of the V0 domain, which were further evaluated by immunohistochemistry (IHC) in CRC and adjacent normal specimens from 661 patients. Univariate and multivariate Cox analyses were used to evaluate factors contributing to prognosis. The effects of variations in the expression of VMA21 on tumor growth were assessed in vitro and in vivo. Of five known assembly factors, only VMA21 showed differential expression between CRC and adjacent normal tissues at both mRNA and protein levels. Patients with high VMA21 expression had higher differentiation grade and longer disease-specific survival (DSS) at stages I–III disease. High VMA21 expression in tumors was also an independent predictor of DSS (hazard ratio, 0.345; 95% confidence interval, 0.123–0.976), with covariates included TNM stage and differentiation grade. VMA21 overexpression decreased CRC growth, whereas VMA21 knockdown increased CRC growth in vitro and in vivo. VMA21 expression suppresses CRC growth and predicts a favorable DSS in patients with stage I-III disease.
Colorectal cancer (CRC) is one of the most common malignances worldwide, and surgical resection supplemented with chemotherapy remains the most effective treatment in patients without distant metastasis (1, 2). However, 30–50% of patients experience relapse or metachronous metastases after surgery (1, 2). Post-surgical chemotherapy and extensive surveillance are beneficial for patients who are more likely to show disease exacerbation. However, identifying patients who would benefit from these treatments is challenging because tumors are heterogenous, even among patients with the same tumor-node-metastasis (TNM) stage (1–3). The only validated marker for prognosis stratification and for selecting the appropriate chemotherapy in CRC is microsatellite instability (MSI) (3, 4), although many biomarkers have been investigated. Therefore, additional effective biomarkers are urgently required (5).
The vacuolar H+-ATPase complexes (V-ATPases) are large multisubunit protein complexes that are required to maintain the pH homeostasis of intracellular compartments (6). The complex consists of two domains, a membrane-integral V0 domain for proton translocation and a cytosolic V1 domain for ATP hydrolysis, which are assembled separately in the endoplasmic reticulum and cytosol, respectively (7, 8). The assembly of the V0 domain depends on a set of endoplasmic reticulum (ER)-resident chaperones, including TMEM199, VMA21, CCDC115, ATP6AP1, and ATP6AP2 (8). Loss-of-function mutations of the assembly factor genes are associated with a spectrum of disease symptoms (8–13), and the normal function of CCDC115, ATP6AP1, and ATP6AP2 is associated with favorable phenotypes in several cancer types (14, 15). However, the clinical significance and biological role of assembly factor genes in CRC is largely unknown.
To address these challenges, in the study we aimed to evaluate whether the five assembly factor genes of V0 domain associated with the development and progression of CRC and further to investigate the biological function of the candidate. We showed that among five V0 domain assembly factors genes, VMA21 was the only differentially expressed gene between CRC and neighboring normal tissues. Tumors with high VMA21 expression had higher differentiation grade and were associated with longer disease-specific survival (DSS) than VMA21-low tumors. Ectopic expression of VMA21 reduced the growth of CRC cells in vitro and in vivo. The present data suggest that VMA21 acts as a tumor suppressor and predicts a favorable prognosis, although VMA21 expression is elevated in CRC tissues.
The expression of five genes (TMEM199, VMA21, CCDC115, ATP6AP1, and ATP6AP2) in CRC and corresponding normal tissue specimens was firstly analyzed on the Expression Profiling Interactive Analysis (GEPIA) website based on the dataset of The Cancer Genome Atlas (TCGA) (16). Co-expression of the genes was analyzed using the Pearson Correlation Coefficient (PCC). Candidate genes showing a 1.5-fold difference between CRC and adjacent normal tissues were further investigated in other cancer types using TCGA data and in our CRC cohort by immunochemistry (IHC).
Formalin-fixed, paraffin-embedded (FFPE) specimens from 661 cancerous tissues, 26 adenoma, and 65 para-carcinoma tissues were obtained from 661 patients treated at Changhai Hospital, Second Military Medical University, Shanghai, between 2001 and 2011. None of the patients received chemotherapy or radiation therapy before surgery. All tissue samples were tested histologically by the pathologists to verify the diagnosis. Outdo Biotech Co., Ltd (Shanghai, China) constructed the tissue microarrays (TMAs) containing the FFPE specimens as previously described (17). The baseline information of each specimen donors, including age, tumor location, differentiation grade, number of examined lymph nodes, TNM stage (determined according to the American Joint Committee on Cancer Staging Manual, seventh edition), adjuvant chemotherapy, serum carcinoembryonic antigen (CEA), and carbohydrate antigen 199 (CA199) levels, were documented. The follow-up of patients was performed regularly as previously described (17). The disease-specific survival (DSS) and disease-free survival (DFS) were defined as the time from surgery to death caused by CRC and the time from surgery to relapse, respectively. Written informed consent was obtained from all patients and approved by the Ethics Committees of Changhai Hospital.
The slides of TMAs (4-μm thick) were prepared for IHC examination of the detection of VMA21 immunstaining. Each slide was deparaffinized using xylene and rehydrated in graded alcohol, and endogenous peroxidase activity was blocked using 3% H2O2. After antigen retrieval of VMA21 for 30 minutes in 10 mmol/L sodium citrate buffer (pH 6.0), slides were blocked with 5% normal goat serum for 10 min at room temperature. Then, the slides were incubated with anti-human VMA21 polyclonal antibody (PA5-42630, 1:150 dilution; Invitrogen, Carlsbad, California, USA) overnight at 4°C, followed by incubation with a secondary antibody from the ElivisionTMsuper HRP (Mouse/Rabbit) for 30 min. Then, the sections were reacted with 3-3-aminobenzidine (DAB) solution for 45 seconds and counterstained with hematoxylin for 25 seconds.
Two authors (F.Z. and H.S.) who were blinded to the clinicopathological information, evaluated the VMA21 immunostaining data independently. The intensity level of each specimen dot was evaluated using the H-score method as described previously (18, 19). H-score was evaluated by multiplying the average percentage of positive cells (0–100%) by the staining intensity (0, negative; 1, weakly positive; 2, moderately positive; and 3, strongly positive staining). The IHC scores of each specimen provided by the two observers were averaged for further studies.
Four human CRC cell lines (RKO, SW620, LoVo, and CACO2) were maintained in RPMI-1640 or DEME medium with 10% heat-inactivated fetal bovine serum (FBS) (GIBCO, Grand Island, NY, USA) and 2% penicillin/streptomycin (GIBCO). All cells were obtained commercially from the Cell Bank affiliated to the Chinese Academy of Sciences (Shanghai, China). The LightCycler 480 II system (Roche, Basel, Switzerland) with SYBR Green reagent (Takara, Bio, Dalian, China) was used to detect the relative expression of VMA21 mRNA using the following primers: 5′-TGG AGC GCC CGG ATA AGG-3′ (forward) and 5′-CTG TTT GCC TTC ACG CCA C-3′ (reverse). Human GAPDH served as an internal control with the primers 5′-GGA GCG AGA TCC CTC CAA AAT-3′ (forward) and 5′-GGC TGT TGT CAT ACT TCT CAT GG-3’ (reverse). Western blot analysis of CRC cells and specimens was performed using routine protocols with antibodies against human VMA21 (1:150, PA5-42630, Invitrogen) or human GAPDH (1:1000, AP0063, Bioworld Technology, St. Louis Park, MN, USA)
Invitrogen Co., Ltd (Shanghai, China) synthesized and validated the cDNA sequence of the VMA21 transcript (NM_001017980). The products were subcloned into the pENTRTM3C vector, and the recombinant VMA21 was Gateway-recombined into the pInducer 20 vector (Addgene) (20). A Lenti-XTM HTX Packaging System (Clontech Laboratories, Inc., CA, USA) was used to produce lentiviral particles in HEK293T cells, which were titrated onto SW620 or LoVo cells cultured in media containing 1 µg/mL puromycin to reach the optimal expression of the target protein using the minimum viral load. G418 (2 mg/ml) was added to induce VMA21 gene expression, which was confirmed by qRT-PCR and western blotting.
A microRNA (miR)-30 loop and appropriate flanking sequences were added to the different miR-30-mediated shRNAs to specifically target VMA21 as described previously (21). The sequence was as followed: 5′-CAU CUA CAC UGA AGA CGC UTT AGC GUC UUC AGU GUA GAU GTT-3′. After the synthesis of single-stranded DNA templates for shRNAs, the templates were amplified with primers including Xho1 or BamH1 restriction enzymes sites. The target DNA was purified and subcloned into the Pinducer10 vector (20). Lentiviral particles were produced as described above and titrated onto RKO cells. Expression of VMA21 was induced using 1 μg/mL doxycycline (dox) and confirmed by qRT-PCR and western blotting.
CRC cells (LoVo and SW480) were seeded into 6-well plates (Corning, NY, USA) at a density of 1.5 × 103 cells/well. The cell medium was replaced every 72 h during growth. After 7–14 days of incubation, the supernatant was removed, cells were fixed with methanol, and then dyed with crystal violet. Images were captured, and colonies were counted when they contained more than 50 cells.
Male nude mice (BALB/c, 5–6 weeks old) were purchased from SLAC Laboratory Animal Co. (Shanghai, China) and were accustomed to a pathogen-free environment for 1 week. For setting up the xenograft implant tumor models, the stable inducible CRC cells (VMA21-LoVo, VMA21-SW620, and shRNA-VMA21 RKO) were subcutaneously injected into test (treated with G418 or Dox) and control groups. Tumor formation was continuously monitored using calipers every 3–4 days. The tumor volume was calculated using the formula (length × width2)/2. The mice were euthanized after 19–30 days depending on tumor size, and the tumor xenografts were removed and tested. All procedures were performed according to the National Research Council’s Guide for the Care and Use of Laboratory Animals.
Differences in VMA21 expression between CRC and adjacent non-tumorous tissues were analyzed using the Student’s t test and we used independent-sample t-tests for the differences of mRNA expression in different CRC cells. The χ2 test and Student’s t test were used to compare categorical data and continuous data separately. The R package of maxstat (22) was used to determine the optimal value for the classification of patients into subgroups according to the expression of VMA21 determined by immunostaining. The subgroups were further compared using the Kaplan-Meier method and Cox proportional hazards models. All statistical tests were performed with R 3.6.1 and SPSS (Version 23.0 for Windows), and two-tailed tests with a P <0.05 were considered significant.
To identify V-ATPase assembly factor gene candidates associated with the progression of CRC, the expression of five genes required for the V0 domain of V-ATPase (TMEM199, VMA21, CCDC115, ATP6AP1, and ATP6AP2) (8) were analyzed using TCGA-CRC cohort. Three (CCDC115, ATP6AP1 and ATP6AP2) of the five assembly factor genes are associated with several cancers (14, 15). We found that VMA21 was the only gene showing significantly higher mRNA expression in colon and rectal cancerous tissues than in adjacent normal tissues (all P < 0.05), as shown in Figure 1A. The associations between VMA21 and ATP6AP1 (r = 0.56, P < 0.001) and ATP6AP2 (r = 0.52, P < 0.001) were significant, as indicated by Pearson correlation coefficients (Supplementary Figure 1). Therefore, VMA21 was identified as a new candidate gene involved in the development of CRC.
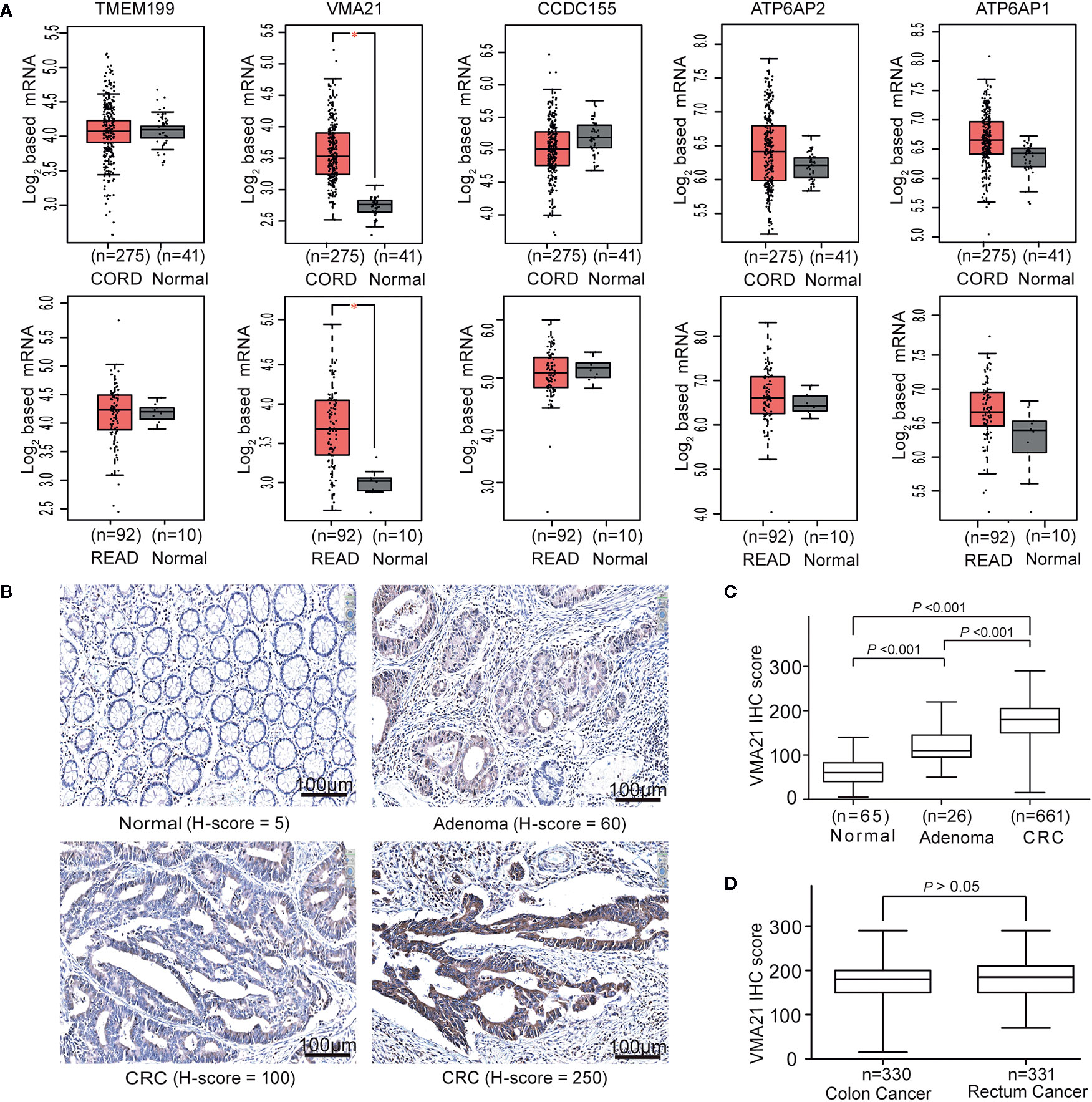
Figure 1 VMA21 is elevated in colorectal cancer. (A) Expression patterns of five assembly factor genes of the V0 domain of V-ATPase based on TCGA-CRC mRNA data. (B) Immunostaining of VMA21 in CRC and noncancerous tissues. (C) VMA21 protein expression in normal, adenoma, and CRC tissue specimens. (D) Expression pattern of the VMA21 protein in colon cancer and rectal cancer. CORD, colon cancer; READ, rectum cancer.
VMA21 expression in CRC epithelial cells may be affected by the expression profiles of whole tissues because of the potential mixture of transcripts from different cell populations (23, 24). We therefore investigated the expression pattern of VMA21 in our specimens using IHC. VMA21 protein expression was primarily detected in the cytoplasm of colorectal epithelial cells (Figure 1B). The immunostaining marks of VMA21 increased gradually in adjacent normal tissues, adenoma, and primary CRC (P trend < 0.001), as shown in Figure 1C. There was no difference in VMA21 protein expression between colon and rectal cancerous tissues (Figure 1D, P > 0.05). Taken together, these results indicate that VMA21 is upregulated in CRC at the mRNA and protein levels, suggesting that it plays a role in CRC.
Based on the 97.5% quantile (175) of VMA21 in the IHC scores from noncancerous specimens, the 661 patients were classified into two groups, the VMA21-high group (score >175) and the VMA21-low group (score ≤ 175). The associations between VMA21 expression and the clinical features of CRC patients are shown in Table 1. There were no significant associations between VMA21 protein expression and several clinical factors (all P >0.05), including age, gender, TNM stage, tumor location, and serum CEA and CA199 levels. However, VMA21-high expression was significantly associated with higher differentiation grade (P = 0.011) compared with VMA21-low expression, indicating that the expression of VMA21 may be negatively related to the progression of CRC.
The associations between the expression of VMA21 and clinical outcomes were further investigated in 639 patients with stage I-III CRC. According to the optimal IHC-score cut-off value (215) determined by maxstat software, which could most efficiently distinguish differences in clinical outcomes (Supplementary Figure 2), patients were classified into two subgroups: patients with VMA21-high (>215) and those with VMA21-low (≤215) tumors. As shown in Figure 2A, VMA21-high tumors were strongly correlated with a favorable DSS (P = 0.035) but not DFS (P = 0.390). Next, the associations between VMA21 expression and DFS and DSS in patients with stage I and II disease were estimated. The results showed a significant association between VMA21 expression and DSS (P = 0.034), but not DFS, in stage I-II CRC patients (Figure 2B). VMA21 expression was not significantly associated with DFS or DSS in patients with stage III disease (P > 0.05) (Figure 2C). Multivariate Cox analysis (Table 2) showed that VMA21 expression was associated with DSS [hazard ratio (HR), 0.345; 95% confidence interval (CI), 0.123–0.976] in patients with stage I–III disease, with the covariates including stage, lymph nodes, grade, and serum CEA levels. In patients receiving chemotherapy, the expression of VMA21 in CRC was marginally (P = 0.062) associated with DSS in stage II disease, whereas the association was not significant in patients who did not receive chemotherapy (P = 0.52) (Supplementary Figure 3).
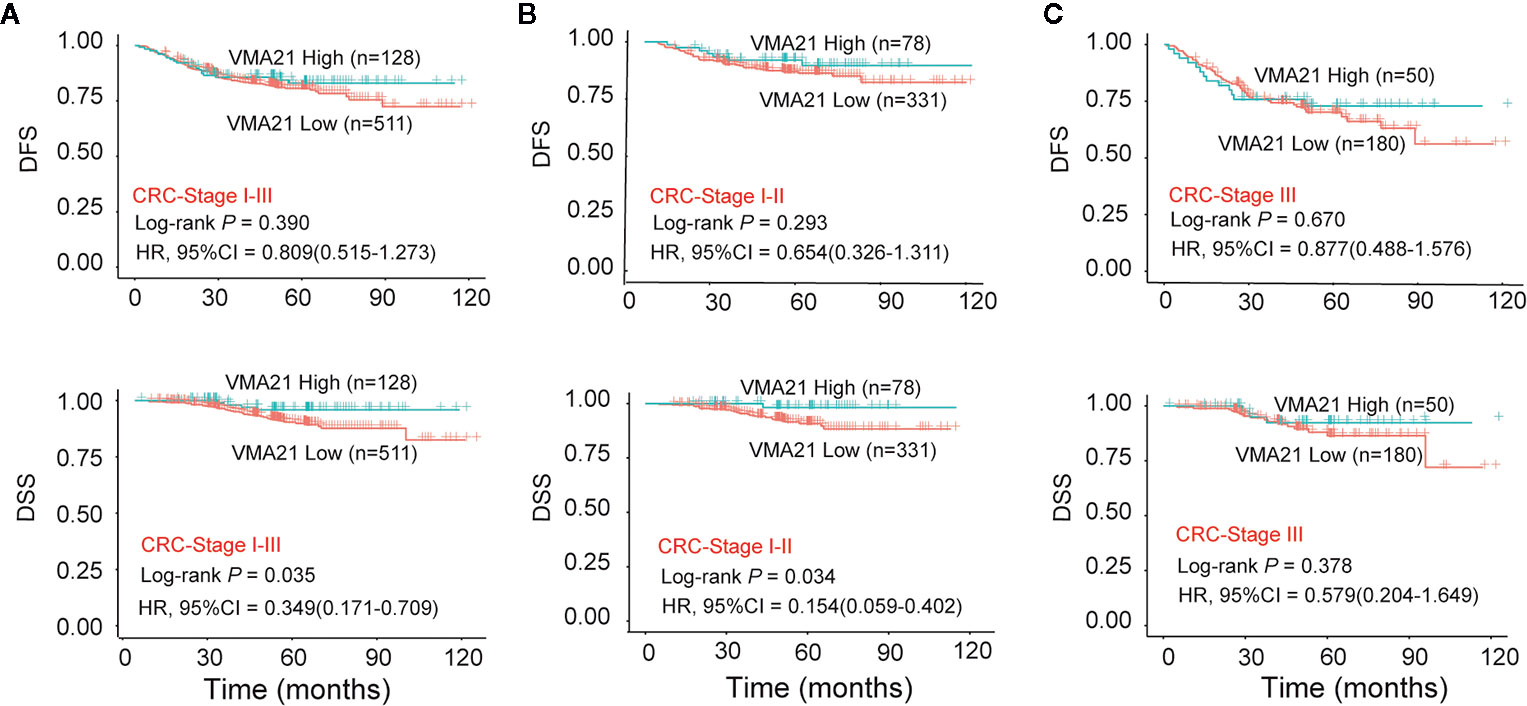
Figure 2 Relationship between VMA21 expression and patient survival. (A–C) DFS and DSS analysis in patient subgroups with high or low VMA21 protein expression in stage I-III, stage I-II, and stage III disease. DFS, disease free survival; DSS, disease specific survival.
The effect of VMA21 expression on the growth of CRC cells was tested to explore. As shown in Figure 3A, VMA21 expression is relatively low in LoVo and SW620 cells, and VMA21 was overexpressed in these cell lines for further evaluation (Figure 3B). In the colony formation assays, VMA21 overexpression decreased the number of CRC cell colonies compared with those in the control groups in both SW620 and LoVo cell models (Figure 3C). The data suggest that VMA21 is involved in the negative regulation of CRC cell proliferation.
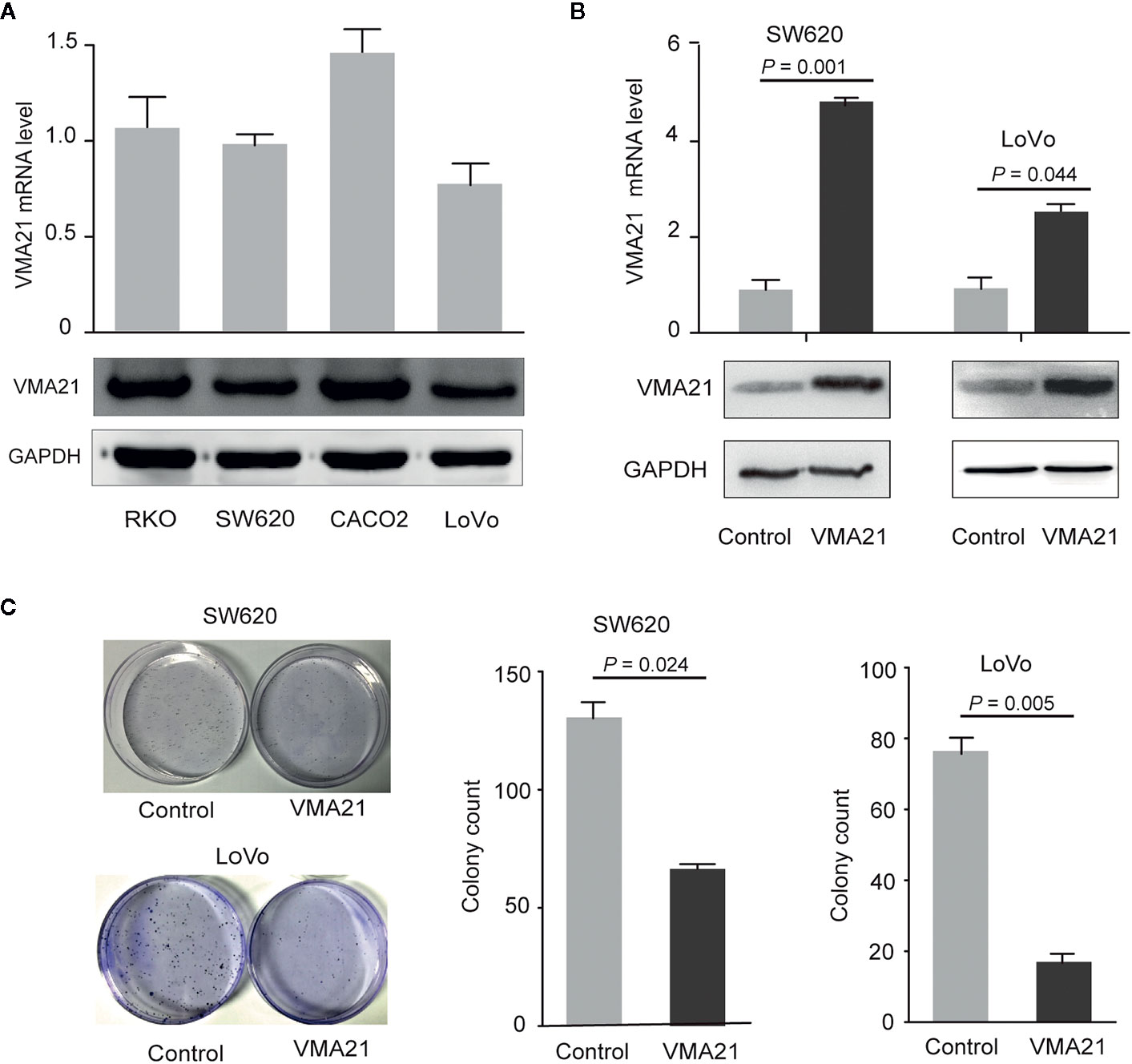
Figure 3 VMA21 overexpression suppresses colony formation of CRC cells. (A) Baseline mRNA and protein expression of VMA21 (lower panel) in four CRC cell lines. (B) The efficiency of VMA21 overexpression in the indicated CRC cells at the mRNA and protein levels (lower panel). (C) The colony formation abilities are affected by overexpressed VMA21 in CRC cells.
VMA21-SW620 or VMA21-LoVo cells were subcutaneously injected into BALB/c nude mice (supplemented with or without G418 in drinking water) to determine whether VMA21 overexpression reduces CRC growth in animal models. In the first 10 days for VMA21-SW620 and the first 25 days for VMA21-LoVo models, tumor size did not differ significantly between the test (G418+) and control (G418-) groups in both cell models (Figure 4A). However, xenograft tumors were significantly smaller in the test groups than in the control groups after the indicated days (Figure 4A). By the end of the observation period, both the size (Figure 4B) and weight (Figure 4C) of isolated tumors were significantly lower in the test animals than in the control animals for both cell models. In xenograft tumors from RKO cells, knockdown of VMA21 promoted the development of CRC, as shown in Figure 5. Therefore, high VMA21 expression suppresses tumor growth in vivo, indicating that VMA21 is a negative regulator of CRC tumorigenicity.
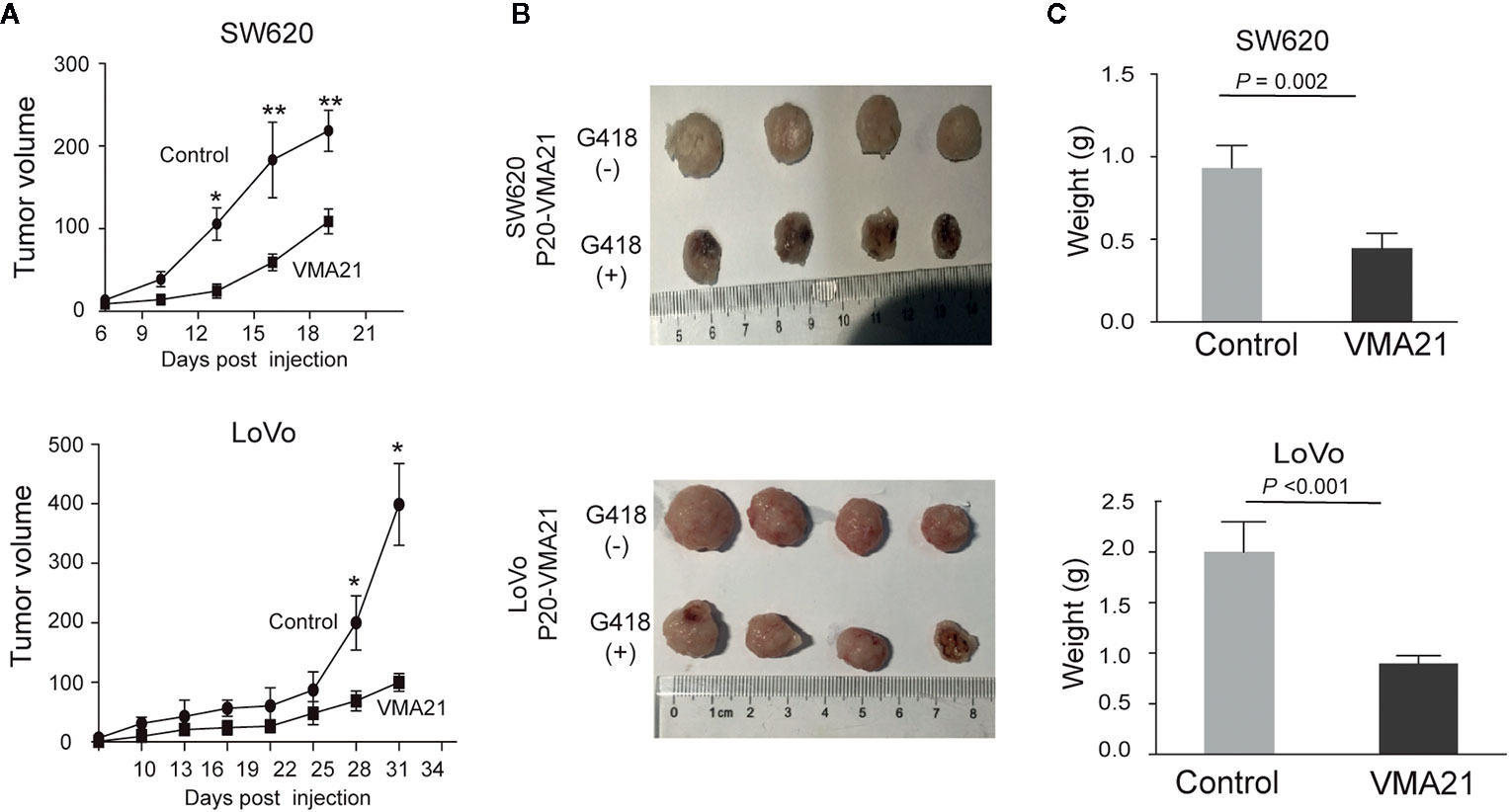
Figure 4 Effects of VMA21 overexpression on xenograft tumor growth. (A) Dynamic effect of VMA21 overexpression on the volume of xenograft CRC models. (B) Isolated tumors after surgical excision. (C) Comparison of the weights of isolated tumors between the subgroups of VMA21 overexpression and the control. (*P < 0.05; **P < 0.01; ***P < 0.001).
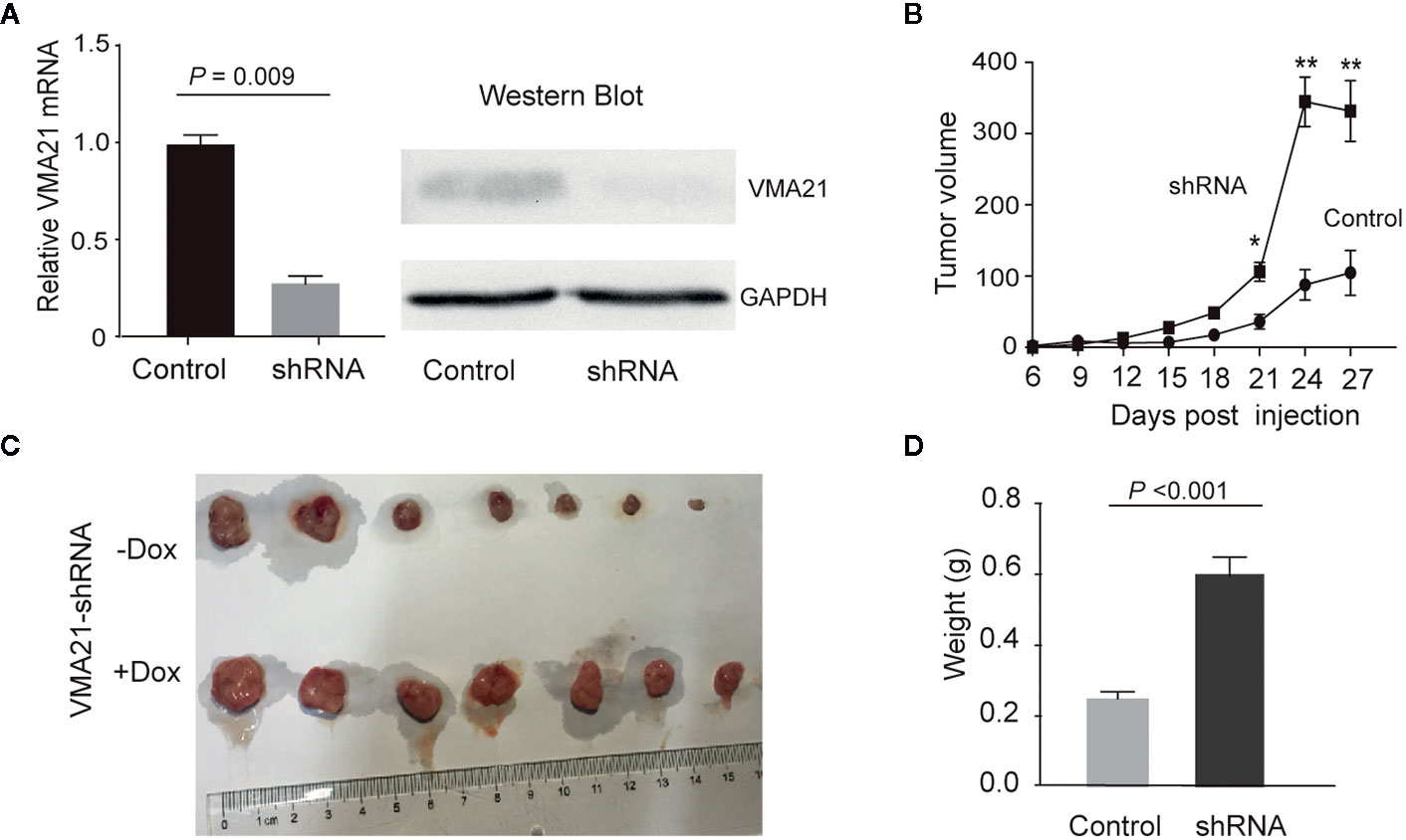
Figure 5 Effects of VMA21 knockdown on xenograft tumor growth. (A) The efficiency of VMA21 knockdown in RKO cells at the mRNA and protein levels. (B) Dynamic effect of VMA21 knockdown on the volume of CRC xenograft models. (C) Isolated tumors after surgical excision. (C) Comparison of the isolated tumor weights from the subgroups with VMA21 knockdown or not. (*P < 0.05; **P < 0.01; ***P < 0.001).
The lysosome mediates the degradation and recycling of macromolecules and signals to the cytosol and nucleus by releasing metabolites and ions; it is implicated in several malignancies (25). These lysosomal behaviors depend on a low intraluminal pH. Although the V0 domain of V-ATPases is required for proton translocation and lysosome acidification (6–8), the role of the assembly factor genes of the domain in cancer remains largely unknown. In this study, we identified VMA21 as the only candidate gene showing differential expression between cancer and noncancerous colorectal tissues among five known assembly factor genes (TMEM199, VMA21, CCDC115, ATP6AP1, and ATP6AP2) of the V0 domain. IHC data confirmed that VMA21 expression is higher in CRC than in adjacent normal tissues. The data consistently suggested that VMA21 plays a critical role in the development of CRC.
The genes encoding CCDC115, ATP6AP1, and ATP6AP2 may play a tumor suppressor role in different cancer types (14, 15). In this study, the expression of VMA21 was correlated with the expression of ATP6AP1 and ATP6AP2, although which were not differentially expressed between cancer and noncancer tissues. These data suggest that VMA21 plays a negative role in CRC development. Similar findings were reported for the gene encoding GUCY2C, which is elevated in CRC but serves as a tumor suppressor (26). Further, we observed that high expression of VMA21 was associated with the well differentiation of CRC. Survival analysis showed that high expression of VMA21 was associated with longer DSS in stage I–III disease and served as an independent risk factor, and this observation was similar in patients with early CRC and in those receiving chemotherapy for stage II disease. Although we observed no associations between DFS and the expression of VMA21 in the subgroups with different disease stages or chemotherapy regimens, the evidence indicated that elevated VMA21 may be a important factor involved in controlling the progression of CRC.
To further elucidate the biological role of VMA21 in CRC, we modulated the expression of VMA21 in different CRC cells and evaluated its function. Overexpression of VMA21 in human colon cancer LoVo and SW620 cells significantly suppressed colony formation ability. Consistently, ectopic expression or knockdown of VMA21 significantly inhibited or increased CRC development in animal models, confirming the clinical data and in vitro results, as well as the suppressive effect of VMA21 on CRC growth. Previous investigation of the underlying molecular mechanism showed that VMA21 deficiency decreases lysosomal-mediated degradation and blocks autophagy (27), and increased autophagy inhibits tumor progression (28, 29). However, the recent evidence also shows that VMA21 may have a positive role to promote the growth of ovarian cancer and lung cancer cells (30, 31). Therefore, tumorigenesis is an extremely complex process involving several factors and many biological phenomena, and it reflects a competition between carcinogens and tumor suppressors (32). Defining the exact role of VMA21 in CRC requires further investigation.
The present study had several limitations. First, the inconsistencies in the prognostic role of VMA21 in DFS and DSS cannot be explained. Second, other critical prognostic factors, such as MSI (4) were not included in our studies because the information was difficult to obtain. Finally, the potential mechanism about the role VMA21 as a tumor suppressor was not explored in the present study. In summary, we showed that VMA21 is upregulated in CRC and associated with a high differentiation grade and favorable DFS. Ectopic expression of VMA21 in CRC significantly inhibited the growth of CRC in cell culture models and animal models. Although the mechanisms involved in VMA21-mediated tumor suppression remain unclear, the current study provides the first evidence of the clinical and biological significance of VMA21 in CRC, as well as their application in the targeted drugs and chemotherapy will be further explored based on mechanism research. The expression of VMA21 may represent a potential diagnostic and prognostic marker for CRC, especially for patients with early-stage CRC.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by the Ethics Committees of the Changhai Hospital. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Ethics Committees of the Changhai Hospital.
As for the authorship, FZ, HS, and YF analyzed whole data independently and presented the same results. GY and FC were responsible for the follow-up of CRC patients. FZ, HS, and YF were responsible for pathological analysis. GY, FC, and ZX were involved in the pathological diagnosis and recruitment of the patients in the hospital. FZ, WC, and ZX were responsible for the statistical analysis. WC, FZ, and ZX designed and organized the study and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (81972302 and 81572451 to WC) and the Wenzhou Science & technological Project (Y2020927 to ZX).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.605801/full#supplementary-material
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin (2015) 65:87–108. doi: 10.3322/caac.21262
2. Schmoll HJ, Van Cutsem E, Stein A, Valentini V, Glimelius B, Haustermans K, et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer :a personalized approach to clinical decision making. Ann Oncol (2012) 23:2479–516. doi: 10.1093/annonc/mds236
3. Van Schaeybroeck S, Allen WL, Turkington RC, Johnston PG. Implementing prognostic and predictive biomarkers in CRC clinical trials. Nat Rev Clin Oncol (2011) 8:222–32. doi: 10.1038/nrclinonc.2011.15
4. Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol (2005) 23:609–18. doi: 10.1200/JCO.2005.01.086
5. Brown K, Solomon MJ, Mahon K, O’Shannassy S. Management of colorectal cancer. BMJ (2019) 366:l4561. doi: 10.1136/bmj.l4561
6. Breton S, Brown D. Regulation of Luminal Acidification by the V-ATPase. Physiology (2013) 28:318–29. doi: 10.1152/physiol.00007.2013
7. Cotter K, Stransky L, McGuire C, Forgac M. Recent Insights into the Structure, Regulation, and Function of the V-ATPases. Trends Biochem Sci (2015) 40:611–22. doi: 10.1016/j.tibs.2015.08.005
8. Whitton B, Okamoto H, Packham G, Crabb SJ. Vacuolar ATPase as a potential therapeutic target and mediator of treatment resistance in cancer. Cancer Med (2018) 7:3800–11. doi: 10.1002/cam4.1594
9. Cannata Serio M, Graham LA, Ashikov A, Larsen LE, Raymond K, Timal S, et al. sMutations in the V-ATPase assembly factor VMA21 cause a congenital disorder of glycosylation with autophagic liver disease. Hepatology (2020) 72:1968–86. doi: 10.1002/hep.31218
10. Jansen EJR, Timal S, Ryan M, Ashikov A, van Scherpenzeel M, Graham LA, et al. ATP6AP1 deficiency causes an immunodeficiency with hepatopathy, cognitive impairment and abnormal protein glycosylation. Nat Commun (2016) 7:11600. doi: 10.1038/ncomms11600
11. Rujano MA, Cannata Serio M, Panasyuk G, Péanne R, Reunert J, Rymen D, et al. Mutations in the X-linked ATP6AP2 cause a glycosylation disorder with autophagic defects. J Exp Med (2017) 214:3707–29. doi: 10.1084/jem.20170453
12. Guida MC, Hermle T, Graham LA, Hauser V, Ryan M, Stevens TH, et al. ATP6AP2 functions as a V-ATPase assembly factor in the endoplasmic reticulum. Mol Biol Cell (2018) 29:2156–64. doi: 10.1091/mbc.E18-04-0234
13. Stransky L, Cotter K, Forgac M. The Function of V-ATPases in Cancer. Physiol Rev (2016) 96:1071–91. doi: 10.1152/physrev.00035.2015
14. Winter JM, Curry NL, Gildea DM, Williams KA, Lee M, Hu Y, et al. Modifier locus mapping of a transgenic F2 mouse population identifies CCDC115 as a novel aggressive prostate cancer modifier gene in humans. BMC Genomics (2018) 19:450. doi: 10.1186/s12864-018-4827-2
15. Pareja F, Brandes AH, Basili T, Selenica P, Geyer FC, Fan D, et al. Loss-of-function mutations in ATP6AP1 and ATP6AP2 in granular cell tumors. Nat Commun (2018) 9:3533. doi: 10.1038/s41467-018-05886-y
16. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) 45:W98–W102. doi: 10.1093/nar/gkx247
17. Chang W, Gao X, Han Y, Du Y, Liu Q, Wang L, et al. Gene expression profiling-derived immunohistochemistry signature with high prognostic value in colorectal carcinoma. Gut (2014) 63:1457–67. doi: 10.1136/gutjnl-2013-305475
18. Finn RS, Press MF, Dering J, Arbushites M, Koehler M, Oliva C, et al. Estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 (HER2), and epidermal growth factor receptor expression and benefit from lapatinib in a randomized trial of paclitaxel with lapatinib or placebo as first-line treatment in HER2-negative or unknown metastatic breast cancer. J Clin Oncol (2009) 27:3908–15. doi: 10.1200/JCO.2008.18.1925
19. Dziadziuszko R, Merrick DT, Witta SE, Mendoza AD, Szostakiewicz B, Szymanowska A, et al. Insulin-like growth factor receptor 1 (IGF1R) gene copy number is associated with survival in operable non-small-cell lung cancer: a comparison between IGF1R fluorescent in situ hybridization, protein expression, and mRNA expression. J Clin Oncol (2010) 28:2174–80. doi: 10.1200/JCO.2009.24.6611
20. Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci USA (2011) 108:3665–70. doi: 10.1073/pnas.1019736108
21. Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, et al. A pipeline for the generation of shRNA transgenic mice. Nat Protoc (2012) 7:374–93. doi: 10.1038/nprot.2011.446
22. Choi YH, Lee JW, Lee SH, Choi JH, Kang J, Lee BS, et al. A High Monocyte-to-Lymphocyte Ratio Predicts Poor Prognosis in Patients with Advanced Gallbladder Cancer Receiving Chemotherapy. Cancer Epidemiol Biomarkers Prev (2019) 28:1045–51. doi: 10.1158/1055-9965.EPI-18-1066
23. Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods (2010) 7:287–9. doi: 10.1038/nmeth.1439
24. Zhong Y, Wan YW, Pang K, Chow LM, Liu Z. Digital sorting of complex tissues for cell type-specific gene expression profiles. BMC Bioinf (2013) 14:89. doi: 10.1186/1471-2105-14-89
25. McCarty MF, Whitaker J. Manipulating tumor acidification as a cancer treatment strategy. Altern Med Rev (2010) 15:264–72.
26. Bashir B, Merlino DJ, Rappaport JA, Gnass E, Palazzo JP, Feng Y, et al. Silencing the GUCA2A-GUCY2C tumor suppressor axis in CIN, serrated, and MSI colorectal neoplasia. Hum Pathol (2019) 87:103–14. doi: 10.1016/j.humpath.2018.11.032
27. Ramachandran N, Munteanu I, Wang P, Ruggieri A, Rilstone JJ, Israelian N, et al. VMA21 deficiency prevents vacuolar ATPase assembly and causes autophagic vacuolar myopathy. Acta Neuropathol (2013) 125:439–57. doi: 10.1007/s00401-012-1073-6
28. Nassour J, Radford R, Correia A, Fusté JM, Schoell B, Jauch A, et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature (2019) 565:659–63. doi: 10.1038/s41586-019-0885-0
29. Delaney JR, Patel CB, Bapat J, Jones CM, Ramos-Zapatero M, Ortell KK, et al. Autophagy gene haploinsufficiency drives chromosome instability, increases migration, and promotes early ovarian tumors. PloS Genet (2020) 16:e1008558. doi: 10.1371/journal.pgen.1008558
30. Xue M, Tao W, Yu S, Yan Z, Peng Q, Jiang F, et al. LncRNA ZFPM2-AS1 promotes proliferation via miR-18b-5p/VMA21 axia in lung adenocarcinoma. J Cell Biochem (2020) 121:313–21. doi: 10.1002/jcb.29176
31. Xue F, Xu YH, Shen CC, Qin ZL, Zhou HB. Non-coding RNA LOXL1-AS1 exhibits oncogenic activity in ovarian cancer via regulation of miR-18b-5p/VMA21 axia. BioMed Pharmacother (2020) 125:109568. doi: 10.1016/j.biopha.2019.109568
Keywords: vacuolar membrane ATPase activity 21 (VMA21), vacuolar ATPases, prognosis, tumor suppressor, colorectal carcinoma
Citation: Zhang F, Shen H, Fu Y, Yu G, Cao F, Chang W and Xie Z (2021) Vacuolar Membrane ATPase Activity 21 Predicts a Favorable Outcome and Acts as a Suppressor in Colorectal Cancer. Front. Oncol. 10:605801. doi: 10.3389/fonc.2020.605801
Received: 13 September 2020; Accepted: 31 December 2020;
Published: 19 February 2021.
Edited by:
Yujun Shi Sichuan University, ChinaReviewed by:
Hui Guo, First Affiliated Hospital of Xi’an Jiaotong University, ChinaCopyright © 2021 Zhang, Shen, Fu, Yu, Cao, Chang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjun Chang, Y3dqY3dqMTk3NkBzbW11LmVkdS5jbg==; Zhongdong Xie, bTE1ODY4NTAzOTMxQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.