- 1Department of Biomedical Sciences and Human Oncology, University of Bari Medical School, Bari, Italy
- 2Department of Medical Oncology, IRCCS Istituto Tumori “Giovanni Paolo II” of Bari, Bari, Italy
- 3Bio-Proteomics Facility, Department of Translational Research, Centro di Riferimento Oncologico di Aviano (CRO) IRCCS, Aviano, Italy
Two mechanisms are involved in the immune escape of cancer cells: the immunoediting of tumor cells and the suppression of the immune system. Both processes have been revealed in multiple myeloma (MM). Complex interactions between tumor plasma cells and the bone marrow (BM) microenvironment contribute to generate an immunosuppressive milieu characterized by high concentration of immunosuppressive factors, loss of effective antigen presentation, effector cell dysfunction, and expansion of immunosuppressive cell populations, such as myeloid-derived suppressor cells, regulatory T cells and T cells expressing checkpoint molecules such as programmed cell death 1. Considering the great immunosuppressive impact of BM myeloma microenvironment, many strategies to overcome it and restore myeloma immunosurveillance have been elaborated. The most successful ones are combined approaches such as checkpoint inhibitors in combination with immunomodulatory drugs, anti-monoclonal antibodies, and proteasome inhibitors as well as chimeric antigen receptor (CAR) T cell therapy. How best to combine anti-MM therapies and what is the optimal timing to treat the patient are important questions to be addressed in future trials. Moreover, intratumor MM heterogeneity suggests the crucial importance of tailored therapies to identify patients who might benefit the most from immunotherapy, reaching deeper and more durable responses.
Introduction
Multiple myeloma (MM) is a malignant plasma cell disease mainly located in the bone marrow (BM) in multiple ‘niches’. These provide a microenvironment that promotes tumor survival and progression. Within BM niches, normal and tumor plasma cells can survive for years, even for decades. Moreover, the observation that tumor plasma cells do not grow and expand when cultured alone suggests the huge resilience of these cells within the BM microenvironment (1). The BM milieu consists of a cluster of cells such as immune cells, stromal cells, endothelial cells (ECs), and bone cells, soluble factors (cytokines, chemokines, and growth factors), and non-cellular matrix (2). It is highly vascularized by blood vessels and is a part of the lymphocyte re-circulation network. Cells re-circulating into and out of the BM have the potential to regulate tumor plasma cell growth and progression through a composite array of indirect and direct interactions involving cytokines as well as surface (3) and soluble molecules (4). In this context, the immune system plays a central and multifaceted role.
A multistep development model indicates that MM progresses from a stable premalignant plasma cell clonal expansion termed monoclonal gammopathy of undetermined significance (MGUS). This asymptomatic preneoplastic condition is characterized by a perfect equilibrium between tumor and immune system which allows disease to remain stable and does not develop to MM. Immune cells control, but not eliminate MGUS plasma cells. These findings suggest that malignant transformation depends not only on the features of the tumor cells themselves but also on the surrounding microenvironment and its effects on tumor cells. Complex cancer–immune system interactions generate both pro- and anti-tumor effects whose balance can be altered in favor of an immunosuppressive environment which promote tumor progression (5, 6). On one hand, innate and adaptive immune cells are able to detect tumor plasma cells; tumor-specific cytotoxic T cells can be found in the BM of MGUS and MM patients (7, 8). On the other hand, tumor plasma cells have the ability to promote a tolerant microenvironment and the activation of immunosuppressive mechanisms to counteract effective immune responses. These include impairment of antigen processing and presentation, and T cell response, NK and NKT cell dysfunctions, local recruitment, expansion and activation of immune suppressor cells like T regulatory cells (Tregs) and myeloid derived suppressor cells (MDSCs), and differentiation of the protumoral tumor-associated macrophages and Th17 cells (9–11) (Figure 1).
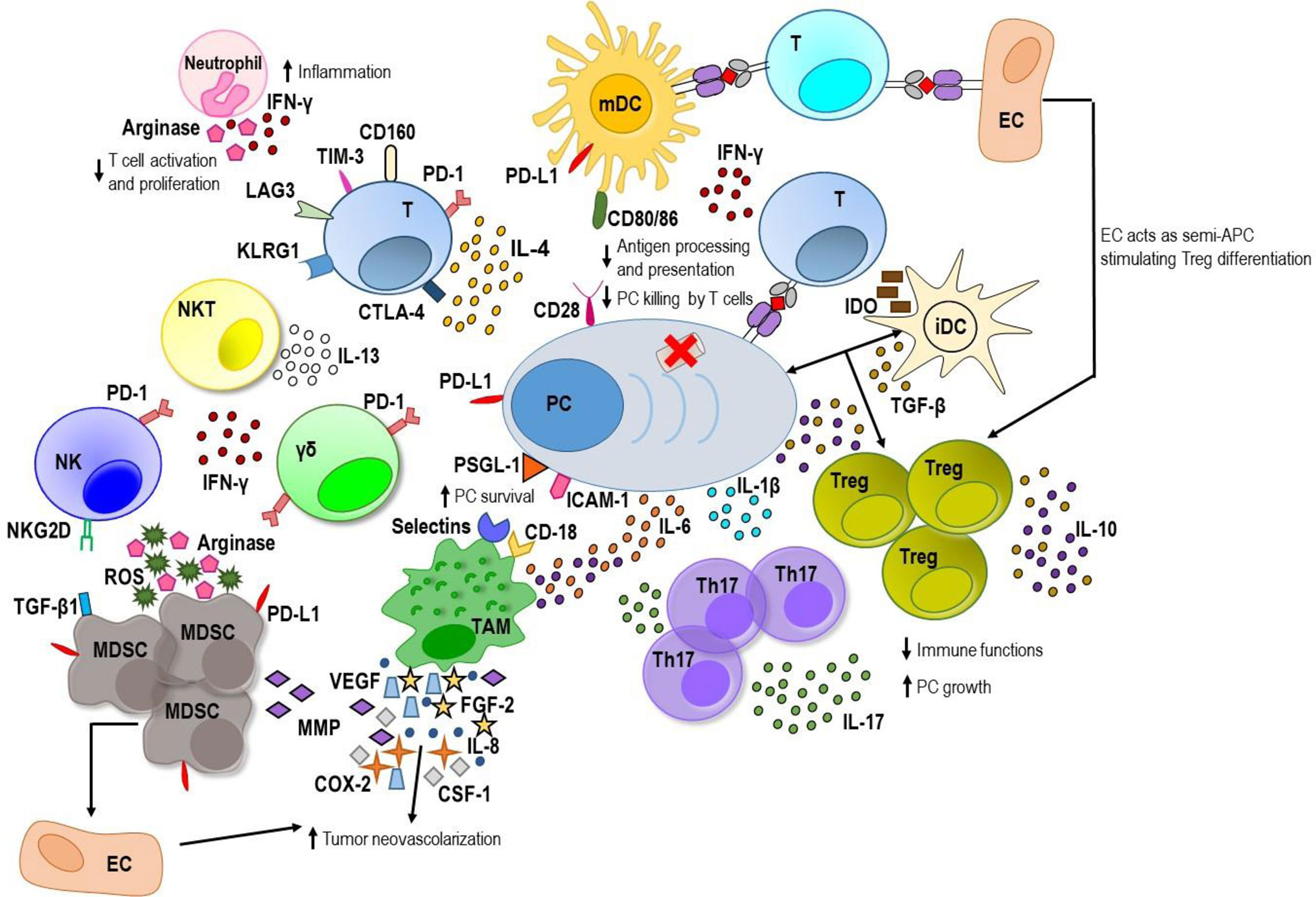
Figure 1 The MM BM microenvironment. On one hand, innate and adaptive immune cells are able to recognize myeloma plasma cells (PC) and generate a weak immune response against tumor. Mature dendritic cells (mDCs) activate tumor-specific T cells that along with natural killer (NK), NKT and gamma delta (γδ) T cells produce low amount of interferon (IFN)-γ. On the other hand, myeloma PCs are able to promote an immunosuppressive microenvironment. They produce immunosuppressive factors including transforming growth factor (TGF)-β, interleukin (IL)-10 and IL-6. PC–immature DC (iDC) interaction stimulates TGF-β production by DC inducing T regulatory (Treg) proliferation with enhancement of levels of TGF-β and IL-10. Immature DCs produce also indoleamine 2,3-dioxygenase (IDO) that causes anergy in activated T cells. The latter exhibits exhaustion markers such as programmed cell death-1 (PD-1), cytotoxic T lymphocyte antigen-4 (CTLA-4), T cell immunoglobulin-3 (TIM-3), and lymphocyte-activation gene 3 (LAG3), and high levels of the senescence markers killer-cell lectin like receptor G1 (KLRG1) and CD160. PD-1 is greatly expressed also by γδ T cells and NK cells and interacts with its ligand, programmed death ligand 1 (PD-L1), expressed by myeloma PC, DC, and myeloid derived suppressor cells (MDSCs) downregulating immune response. Myeloma PC–mature DC interaction, involving the CD28 receptor and the CD80/CD86 ligands respectively, downregulates proteasome subunit expression in tumor PC and decreases the processing and presentation of tumor antigens thus reducing myeloma PC recognition by cytotoxic CD8+ T cells. Myeloma PC-tumor-associated macrophage (TAM) interaction involving P-selectin glycoprotein ligand 1 (PSGL-1) and intercellular adhesion molecule-1 (ICAM-1) on myeloma PC and E/P selectins and CD18 on TAM confers multidrug resistance to MM PC. Within myeloma niche, TAMs release great amount of IL-6 and IL-10 and contribute to MM-associated neovascularization by vasculogenic mimicry and indirectly by secreting vascular endothelial growth factor (VEGF), IL-8, fibroblast growth factor-2 (FGF-2), metalloproteinases (MMPs), cycloxygenase-2 (COX-2), and colony-stimulating factor-1 (CSF-1). Neutrophils release high amount of IFN-γ that supports their promotion of pro-inflammatory and survival signals within the plasma cell niche and produces arginase that inhibits T cell activation and proliferation. MDSCs also produce high amounts of arginase and reactive oxygen species (ROS) that contribute to T cell suppression, induce anergy of NK cell through membrane-bound TGF-β1 and promote tumor angiogenesis by MMP secretion or direct differentiation into endothelial cells (ECs). Furthermore, ECs can act as semi-professional antigen presenting cells (APCs) stimulating a regulatory tumor-specific T cell population. Within BM, the elevated levels of IL-6, TGF-β, and IL-1β promote T helper IL-17-producing (Th17) cell polarization which release high levels of IL-17 favoring MM plasma cell growth and inhibiting immune system.
Here we describe interactions between BM tumor plasma cells and different immune cells and provide an overview of the current knowledge on immunotherapeutic strategies.
Myeloma Plasma Cell Immunogenicity
The hallmark of MGUS and MM plasma cells is the production and the surface expression of a monoclonal immunoglobulin (Ig) carrying unique antigenic (idiotypic or Id) determinants in the variable heavy (VH) (12). Thus, the Ig idiotypic structure is a tumor-specific antigen of the myeloma cell clone, distinct from normal cells or normal plasma cells that can be presented as whole molecule on the cell surface or as peptides in the groove of the of major histocompatibility complex (MHC) molecules (13, 14).
Several studies have described idiotype-specific cytotoxic T lymphocytes in MM patients with the capacity to lyse autologous primary tumor plasma cells. Many potential T cell epitopes have been identified within the tumor-derived Ig-VH region, nonetheless, the majority of them didn’t trigger high affinity T cell responses (15). Two peptide prediction algorithms, BIMAS and SYFPEITHI, have also confirmed the poor immunogenicity of human idiotypes with a low binding half-life (BIMAS) and a low/intermediate score (SYFPEITHI) on most T cell interaction human leukocyte antigen (HLA) modules (16, 17). Additionally, idiotypic vaccination in MM has been examined in clinical trials where immunologic responses occurred in <50% of patients, and clinical responses have been infrequent (18).
Within the universal tumor antigens, many myeloma-associated antigens (e.g. human telomerase reverse transcriptase (hTERT) (19), surviving (20), new york esophageal squamous cell carcinoma 1 (NY-ESO1) (21) mucin-1 (MUC-1) (22), junctional adhesion molecule-A (JAM-A) (23, 24) and the receptor for hyaluronic acid-mediated motility (RHAMM) (25) have been identified as targets recognized by T lymphocytes and used in many vaccination strategies, but in most cases failed to produce clinically meaningful responses. However, many obstacles need to be overcome. The most important one is the myeloma plasma cell escape of tumor-specific immune response. Our group demonstrated that the binding of CD28 expressed on myeloma plasma cells with its ligands CD80/CD86 expressed on BM dendritic cells (DCs) results in downregulation of the expression of proteasome subunits, alteration of the antigen repertoire displayed on myeloma plasma cell surface, and reduced recognition of tumor plasma cells by cytotoxic CD8+ T cells (10).
Tumor-Specific Cytotoxic CD8+ T Cells
The mechanisms underlying MGUS to MM progression are incompletely understood. Tumor plasma cell specific CD8+ T cells have been detected in both MGUS and MM patients (8, 26, 27), thereby establishing that there is no tolerance to plasma cell tumors. Nonetheless, in MM, myeloma plasma cell proliferation is not counteracted by CD8+ T cells. These observations have rekindled interest in the immunosurveillance mechanisms of tumor growth (28). Although MM plasma cells do not significantly differ from their premalignant MGUS precursors with respect to cytogenetic abnormalities (29, 30) and gene expression profiles (31), CD8+ T cells have been uncovered to fail to limit the clonal expansion of tumor plasma cells in MM. Our studies have shown that malignant transformation of plasma cells is associated with altered expression of HLA class I antigen processing presenting machinery (APM) components. These alterations are detectable ex vivo, occur at the transcriptional level, and, in some cases, are enhanced by IL-6, an essential MM cytokine. For some APM components, changes correlate with the extent of the plasma cells’ lysis by CD8+ T cells and with variations in the serum level of the M component in MGUS patients (8). Downregulation of proteasome subunits, in particular, is higher in plasma cells from MM patients than in those from MGUS patients and MM plasma cells are less readily lysed by autologous, in vitro-expanded cytotoxic CD8+ T cells than are MGUS plasma cells. This difference in cytotoxicity is evident at the epitope level and is not due to the intrinsic features of CD8+ T cells, given that no difference is observed when CD8+ T cells are tested against HLA-matched target cells that are not plasma cells (8). These findings support the hypothesis that proteasome subunit downregulation decreases expression of tumor antigen peptides on tumor plasma cells, enabling them to evade CD8+ T cell recognition and killing (10). Moreover, these alterations in the expression of APM components are specific of each premalignant and malignant plasma cell clone, suggesting that the myeloma-specific T cell response can differ from one patient to another. Indeed, CD8+ T cells isolated from MGUS and MM patients can be activated ex vivo by DC loaded with autologous but not allogeneic tumor lysates (7, 27, 32). The finding that the impairment of T cell response is restricted to myeloma antigens is supported also by the absence of a clinical T cell immunodeficiency. Myeloma patients show an appropriate T cell immunity against external antigens and do not show an increased incidence of mycobacterial infections or virus associated second malignancies (33).
Some shared antigens have been identified as targets of a spontaneous immune response in MGUS but not MM patients suggesting the capacity of the immune system to recognize premalignant lesions. For instance, the clonogenic CD138− compartment in MGUS patients expresses SOX2, an embryonal stem cell protein involved in the tumor-initiating potential and self-renewal of tumor cells. The expression of this antigen identifies potential MM progenitors and detection of anti-SOX2 T cells is associated with an improved clinical outcome in patients with asymptomatic plasma cell disorders. SOX2 is also expressed by CD138+ cells in patients with active MM, who do not develop anti-SOX2 immunity (34).
Recent evidence also indicates that MM-specific cytotoxic CD8+ T cells do not express CD28 and express low levels of programmed cell death-1 (PD-1), cytotoxic T lymphocyte antigen-4 (CTLA-4), lymphocyte-activation gene 3 (LAG3), and T cell immunoglobulin-3 (TIM-3) (35). These characteristics further strengthen the idea that MM-specific CD8+ T cells are not anergic or exhausted. Instead, they seem to be “senescent” given that they express high levels of CD57, CD160, and killer-cell lectin like receptor G1 (KLRG1), do not express CD27 and CD28, and display weak proliferation after antigen stimulation (35). So basically, MM-specific CD8+ T cells appear late-differentiated and suspended in a hypo-responsive, non-proliferative state. Interestingly, this state would be telomere-independent and potentially reversible (35) since MM-specific CD8+ T cells have normal-for-age telomere lengths and long-surviving MM patients retain cytotoxic T cell clones with conserved proliferative capacity (36).
CD4+ T Helper Cells
The role of CD4+ T helper cells in MM is still unclear; controversial data may be justified by differences between BM and peripheral blood, or by different quantification method (absolute count versus percentage), or by changes in Th1/Th2 polarization during the course of the disease. While some authors reported an altered Th1/Th2 balance strongly supported by IL-6, with increased production of Th2 cytokines, such as IL-10 and IL-4 and decreased production of Th1 cytokines, such as IL-2 and IFN-γ (37, 38), others described an elevated Th1/Th2 ratio in MM patients at diagnosis and in refractory phase, pointing towards a close relation to the clinical features (39–41).
Furthermore, increased levels of PD-1 on CD4+ cells have been observed in MM patients with persistent minimal residual disease (MRD) and at relapse compared with T cells of first diagnosed MGUS and MM patient (42). BM myeloma PD-1 expressing CD4+ T cells interact with plasma cells and DCs that display on their surface programmed death ligand 1 (PD-L1) promoting T cell suppression and MM progression (43).
T Helper 17 Cells
Elevated levels of interleukin (IL)-6, transforming growth factor (TGF)-β, and IL-1β in myeloma BM microenvironment promote T helper IL-17-producing (Th17) cell polarization with consequent increased of IL-17 levels in BM and peripheral blood of MM patients (36, 44–46). IL-17 induces myeloma cell growth and colony formation via IL-17 receptor and inhibits Th1 immune response (45). The amount of Th17 cells in the BM positively correlates with clinicopathological characteristics in MM, like clinical tumor stage, serum lactate dehydrogenase concentration, and serum creatinine concentration (46). In addition, IL-17 plays a role in osteoclast-mediated lytic bone disease (44). Recently, the existence of a direct immunological link between the gut and the BM in MM involving Th17 cells has been proposed. Using a Vk*MYC mouse model, it has been provided that the gut microbiota induces the differentiation of Th17 cells in the gut that are able to migrate to the BM, where they promote MM progression (47). In the BM, IL-17 activates also eosinophils involved in plasma cell homing to the BM and in their accumulation in the BM niche (47, 48).
Regulatory T Cells
There is considerable controversy regarding regulatory T (Treg) cell frequency and function in MM due to their source (peripheral blood versus BM), differences in assays, purification techniques, and markers used to identify these cells. Treg population is described as reduced and/or dysfunctional (39, 49, 50) or increased and/or functional (36, 51–57) in MM patients compared to MGUS patients or normal controls. Moreover, the increased frequency of CD4+ and CD8+ Treg cells in MM patients correlated with the active phase (54) and a reduced survival (55, 58). MM plasma cells can directly induce functional Treg in a contact dependent manner acting as immature and tolerogenic antigen presenting cells (APCs) (54) as well as in an APC independent manner by the expression of the inducible T cell co-stimulator ligand (ICOSL) (59). It is possible that the mutual and dynamic interactions among cells of the BM microenvironment along with cytokine release modulate the frequency and the suppressive activity of Treg cells. In coculture experiments, cytokines such as IL-10 and TGF-β and human myeloid immature DCs are the most efficacious for induction and expansion of Treg population (60, 61). Tumor cell–immature DC interaction stimulates TGF-β production by DC inducing Treg proliferation (62). Our study demonstrated that EC can act as semi-professional APC stimulating a regulatory tumor-specific CD8+ T cell population with suppressive function within BM of MM patients (11). Moreover, human leukocyte antigen G (HLA-G)+ T cells with an inhibitory activity comparable to natural Treg can be generated in BM of MM patients after tumor plasma cell–T cell interaction by trogocytosis of immunosuppressive molecules such as HLA-G (63).
γδ T Cells
BM MM microenvironment is extremely immunosuppressive and greatly influences gamma delta (γδ) T cells. Indeed, BM derived Vγ9Vδ2 T cells, the main subset of γδ T cells, become more dysfunctional than those isolated from the peripheral blood of MM patients. The functional exhaustion of BM Vγ9Vδ2 T cells occurs early during disease progression and does not disappear in clinical remission. Upregulation of PD-1 expression on γδ T cells is already found in MGUS patients and persists in the remission phase and further increases in the relapse (64). One possible explanation is the great concentration of phosphoantigens in the tumor microenvironment and the consequent prolonged TCR engagement. High amounts of isopentenyl pyrophosphate, the prototypic phosphoantigen recognized by Vγ9Vδ2 T cells via TCR, are produced by both BM myeloma plasma cells and stromal cells leading to chronic TCR engagement, upregulation of PD-1 expression, and functional exhaustion of γδ T cells (65). Moreover, many cell subsets including MM plasma cells, myeloid-derived suppressor cells (MDSCs), regulatory T cells (Tregs), and BM-derived stromal cells are implicated in Vγ9Vδ2 T cell hampering through the excessive expression of immune checkpoints (ICP)/ICP-ligands (65).
Dendritic Cells
The role of DC in MM progression is controversial. Some investigators reported impaired functionality and phenotypic profile, while others found that these cells are normal (9). We have demonstrated that BM DCs are functional and play a dual, but opposing role in MM. DCs are concentrated in the BM during the MGUS-to-MM progression and interact with both T cells and myeloma plasma cells. On one hand, DCs are able to uptake apoptotic myeloma plasma cells, mature and process myeloma antigens, cross-present them and successfully activate myeloma-specific BM-infiltrating CD8+ T cells. On the other hand, by using their surface CD80/86 molecules, DCs interact with non-apoptotic tumor plasma cells by the CD28 receptor that is upregulated on their surface, promoting a downregulation of proteasome subunit expression and a consequent escape of myeloma plasma cells from CD8+ T cell recognition and killing (10). Moreover, plasmacytoid DCs promote tumor plasma cell growth, survival, and drug resistance (66) and express high surface levels of programmed death-ligand 1 (PD-L1) conferring T cell and NK cell immune suppression by engaging ICP via PD1-PD-L1 signaling axis (67, 68). Myeloid CD141+ DCs also express PD-L1, and the proportion of these cells correlate with the percentage of PD-L1+ plasma cells, suggesting that both cell subsets support anti-tumor T cell response inhibition in MM (68). DCs can also indirectly favor the osteoclastogenesis process by inducing Th17 cell expansion in BM myeloma microenvironment (69) followed by IL-17 accumulation, a potent pro-osteoclastogenic factor (70).
Macrophages
Tumor-associated macrophages (TAMs) constitute an abundant component of myeloma microenvironment that induce myeloma plasma cell survival through both contact-dependent and -independent mechanisms. For instance, a direct physical interaction involving E/P selectins and CD18 on macrophages and P-selectin glycoprotein ligand 1 (PSGL-1) and intercellular adhesion molecule-1 (ICAM-1) on myeloma cells protects plasma cells from drug-induced apoptosis (71–73). Within myeloma niche, after interaction with BM-derived mesenchymal stromal cells, TAMs acquire a secretory profile characterized by a great production of IL-6 and IL-10 and poor production of IL-12 and TNF-α, providing a suitable milieu for myeloma plasma cell growth (74). TAMs also contribute to MM-associated neovascularization by vasculogenic mimicry and indirectly by secreting a wide range of proangiogenic factors, such as vascular endothelial growth factor (VEGF), IL-8, and fibroblast growth factor-2 (FGF-2) as well as metalloproteinases (MMPs), cycloxygenase-2 (COX-2), and colony-stimulating factor-1 (CSF-1). Moreover, they resemble M2-like macrophage population with a reduced cytotoxic capacity for tumor cells and a decreased antigen-presenting capability (75).
A very recent single-cell RNA sequencing study revealed that mature CD14+ monocytes/macrophages lose HLA class II surface expression as early as in the MGUS phase resulting in T cell suppression (76).
NK Cells
Natural killer (NK) cell differentiation, activation, and cytotoxic ability are strongly compromised during MM progression (77). BM myeloma plasma cells from early-stage patients display low levels of MHC class I molecules and high levels of MHC class I related chain A (MICA) and are readily recognized by NK cells (78). Nevertheless, elevated numbers of NK cells in the BM and blood of MM patients were associated with worse prognoses (79). Myeloma cell recognition and killing by NK cells involve a broad array of activating receptors including the natural killer group 2D (NKG2D), DNAX accessory molecule-1 (DNAM-1), and the natural cytotoxicity receptors (NCR) NKp46, NKp30, and NKp44 (78, 80). Changes in the expression of these NK receptors and NK cell receptor ligands have been observed in BM samples of MGUS and MM patients, suggesting a role of NK cell dysfunction during MGUS-to-MM progression (81). In addition, following an extensive interaction with cytotoxic T and NK cells, myeloma plasma cells obtained from patients with active disease exhibit the MHC class Ibright/MICAdim/−/CD95dim/− immunophenotype that compromises NK cell function (78, 82, 83). Likewise, the number of effector NK cells localized within the BM progressively decreases during MM growth and correlates with reduced BM NK cell degranulation in MM-bearing mice (84). Moreover, MICA shedding from the surface of myeloma plasma cells may promote downregulation of NKG2D expression on the surface of NK cells weakening the NK-mediated anti-tumor response (83, 85). Defective NK cell functions can be also explained by PD-1 expression on NK cells of MM patients that interact with its ligands PD-L1 on tumor plasma cells downregulating NK cell function (86). Also, the release of soluble factors in the BM microenvironment can influence NK cell activity. For instance, an inflammatory milieu rich in IFN-γ secreted by immune cells strongly increases PD-L1 expression (87). In addition, primary myeloma plasma cells express high levels of HLA-E molecules which bind to the inhibitory NK cell receptor NKG2A hampering NK cell effector functions (80, 88–90).
NKT Cells
NKT dysfunction has been recognized as potentially important in disease predisposition and progression (91). A progressive decrease of NKT cells and a loss of both peripheral blood and BM NKT cell activity in MM patients have been described by many groups, with disease progression correlating with a reduction of IFN-γ production by NKT cells (26, 92, 93). Likewise, a loss of CD1d expression by myeloma plasma cells has been demonstrated during disease progression with consequent dysfunction of NKT cells (26, 92, 94, 95).
A recent study has demonstrated an enrichment of inflammation-associated lysophosphatidylcholine molecules in MM patient serum compared with healthy donors alongside with an increase of frequency of lisophosphatidylcholine-recognizing CD1d-restricted type II NKT cells. These cells release high amounts of the IL-13, an immunosuppressive cytokine involved in tumor-promoting inflammation and angiogenesis, thus supporting their role in disease progression (96). Furthermore, type II NKT cells may also promote plasma cell differentiation and play a role in the initiation of MM (97, 98).
Myeloid-Derived Suppressor Cells
In humans, two main subsets of myeloid derived suppressor cells (MDSCs) with the same level of suppressive activity can be identified based on CD14 positivity, granulocyte-MDSCs (G-MDSCs) that are CD11b+CD14−CD33+CD15+HLA-DR−/low and monocytic-MDSCs (M-MDSCs) that are CD11b+CD14+CD33+HLA-DR−/low (99). The involvement of these subsets in the pathogenesis of MM is still not clear. Several studies found a significant increase in G-MDSCs in the peripheral blood and BM of newly diagnosed, relapsed, and relapsed/refractory MM patients compared with healthy donors (100–102), while others described an increase of M-MDSCs in first diagnosed and relapsed MM patients compared with those in remission and healthy donors (103, 104). Moreover, the level of M-MDSC correlates with disease progression (104).
Because of their capacity to suppress T cell-mediated immunity, MDSCs play an important role in favoring tumor escape from immunosurveillance (101, 102). MDSCs secrete high amount of arginase which sequestrates L-arginine, an essential amino acid for T cell activity (105). Moreover, MDSCs can inhibit T cell receptor by nitrosylation and reactive oxygen species release (106) and express on their surface high levels of PD-L1 which can interact with PD-1 express on T cells (64).
In addition, MDSCs induce Treg differentiation through TGF-β-dependent and -independent mechanisms involving CD40 or IL-10 and IFN-γ, respectively (100, 106), induce NK cell anergy through membrane-bound TGF-β1 (107, 108), promote tumor angiogenesis by MMP-9 secretion or direct differentiation into EC (109), and stimulate tumor growth through the release of cytokines and growth factors (101).
Using immunocompetent mouse models, it has been demonstrated that MDSC immunosuppression occurs early in MM disease; MDSCs accumulated in the BM of mice as early as one week after tumor inoculation and when these mice were engineered to lose their ability to accumulate MDSC, growth of MM plasma cells was significantly reduced confirming the critical role of MDSC accumulation at early stages of MM progression (102).
Neutrophils
Neutrophils are essential for clearance of extracellular pathogens, as they effectively fight them by releasing cytotoxic granules, toxic enzymes, inflammatory mediators, and reactive oxygen species (110, 111). Thanks to a large number of integrins and molecules expressed on their surface, neutrophils can establish interactions with other immune cells (e.g., T, B, and NK cells, monocytes, macrophages, DCs), can act as weak antigen presenting cells, promote angiogenesis and inflammation, and regulate hematopoiesis (112). In MM, as a consequence of BM infiltration by tumor plasma cells, functional defects of neutrophils including reduced lysozyme activity and increased secretion of the amino acid degrading enzyme, arginase, have been described (113, 114). An involvement of neutrophils in immune suppression via IFN-γ signaling has been also revealed since the early asymptomatic phase of MGUS (115). Specifically, Romano et al. (115) have demonstrated that neutrophils from MGUS and MM are chronically activated because of increased signaling through IFN-γ and Toll-like receptors that trigger a chronic inflammatory response via STAT protein activation. Compared with neutrophils from healthy patients, neutrophils from MGUS and MM patients show immunosuppressive features. They display an impairment in the FC-γ-receptor I (CD64) mediated phagocytosis under control of IFN-γ and increased secretion of arginase-1, target of STAT proteins (116–118), resulting in inhibition of T cell activation and proliferation (115). Furthermore, during MGUS-to-MM progression, neutrophils progressively enhance the production of IFN-γ in response to MM soluble factors resulting in increased autophagy flux and JAK-2/STAT3 pathway activation, which support their promotion of pro-inflammatory and survival signals within MM niches (119).
In addition, the neutrophil to lymphocyte ratio (NLR) at diagnosis or after 100 days from autologous stem cell transplantation (SCT) can predict outcome in newly diagnosed MM patients treated upfront with novel agents (120–122). Interestingly, NLR could be combined with international staging system (ISS) to better evaluate the risk profile of non-elderly (<65 years) MM patients, to identify patients with poor outcome, and to personalize MM therapy (121).
Immune Checkpoints in Multiple Myeloma
The main ICP pathways CTLA-4 and PD-1/PD-L1 have emerged as major immune escape mechanisms in MM. These pathways are crucial in the physiological setting for maintaining the immune equilibrium after the initial T cell response and preventing over-activation of the immune system and tissue damage. Tumor cells upregulate these biologic mechanisms of tolerance and exploit them to elude host immunity (123). Regarding MM, contradictory results exist in this field, due mainly to the different analyzed sources (peripheral blood versus BM) suggesting a fundamental role of the local milieu in the regulation of immune ICP cell expression.
Several studies have found an increased number of CTLA4+ Treg cells in the BM of MM patients compared with MGUS patients and healthy donors (52, 56, 124), with a correlation between the proportion of cells simultaneously positive for CTLA4 and Foxp3 and the disease stage (54).
PD-1 expression is increased on NK and γδ T cells isolated from MM patients and correlates with loss of effector cell function (64, 86). CD4+ and CD8+ T cells express low level of PD-1 in MGUS and newly diagnosed MM patients, suggesting that downregulation of their effector function is partly due to senescence rather than PD-1 mediated exhaustion (35, 124, 125). Paiva et al. have reported increased PD-1 expression levels on CD4+ and CD8+ T cells only in relapsed or relapsed/refractory MM and in patients with a minimal residual disease (42).
PD-L1 is greatly expressed on plasma cells obtained from MM patients with active, relapsed, and refractory disease, whereas low expression has been found on plasma cells from MGUS patients or healthy donors, suggesting that PD-L1 expression is associated with MM progression and drug resistance (42, 68, 126–128). Moreover, soluble factors such as IFN-γ, IL-6, and indoleamine 2,3-dioxygenase (IDO), detected at high level in myeloma BM microenvironment, upregulate the expression of PD-L1 on myeloma plasma cells (126, 127, 129). PD-L1 is also expressed by other cells of myeloma BM microenvironment, including plasmacytoid DC, NK cells, and MDSCs, according to their immunoregulatory functions (67, 68, 86, 100, 127).
Immunosuppressive Factors
Along with the crosstalk between tumor plasma cells and BM niche cells, a high concentration of immunosuppressive factors including TGF-β, IL-10, IL-6, and prostaglandin E2 in the MM BM microenvironment promotes tumor propagation and survival and at the same time generates great immune dysfunction (130). In addition, the cellular contact of myeloma plasma cells with BM immature DCs, through CD47–thrombospondin-1 interaction, leads to spontaneous DC fusion and trans-differentiation into osteoclasts (131, 132), which, beside their role in bone lesions, promote suppressive immune BM microenvironment inducing T cell apoptosis by the overexpression of ICP molecules and the release of IDO and APRIL (129). Moreover, IDO causes anergy in activated T cells, induces them to become Treg, and generates a nutritionally depleted niche favoring survival of myeloma cells which have a low proliferative index and are less sensitive to tryptophan depletion (133); APRIL enhances PD-L1 expression on MM cells supplying immune suppression (129). Simultaneously, the establishment of a chronic inflammatory status contributes also to disease progression (134). Increased levels of inflammatory cytokines, such as IL-1, IL-6, IL-12, IL-15, IL-17, IL-18, IL-22, IL-23, TNF-α, and IFN-γ have been revealed in BM serum of MM patients (135), and an eight-gene signature (IL-8, IL-10, IL-17, CCL3, CCL5, VEGFA, EBI3, and NOS2) involved in B-cell inflammation has been described able to distinguish the different phases of disease progression (MGUS/smoldering/symptomatic-MM) with 84% accuracy (134). Moreover, inflammation can lead to high levels of bioactive lipids, such as several species of lysophosphatidylcholine, which can bind to CD1d molecules resulting in dysregulation of lipid-reactive immune cells, activation of CD1d-restricted type II NKT cells, and production of high amount of the immunosuppressive cytokine IL-13 (96).
Targeting Immune System as an Effective Approach to Treat Multiple Myeloma
Considering the great immunosuppressive impact of BM myeloma microenvironment, many strategies to overcome it and restore myeloma immunosurveillance have been elaborated (Figure 2). Autologous SCT following myeloablative treatment allows the introduction of a new immune system and has significantly contributed to improve survival of MM patients in the last 15 years (136). Unfortunately, the graft versus-myeloma (GvM) response is usually weak and most patients relapse. An alternative is the adoptive therapy with BM infiltrating lymphocytes enriched in myeloma-specific T cells that enhances the anti-tumor immunity, but has a poor durability of the clinical response (137), or the allogeneic SCT which provides a new T cell repertoire, triggers a potent GvM response, but it is limited by the high transplant-related mortality (138).
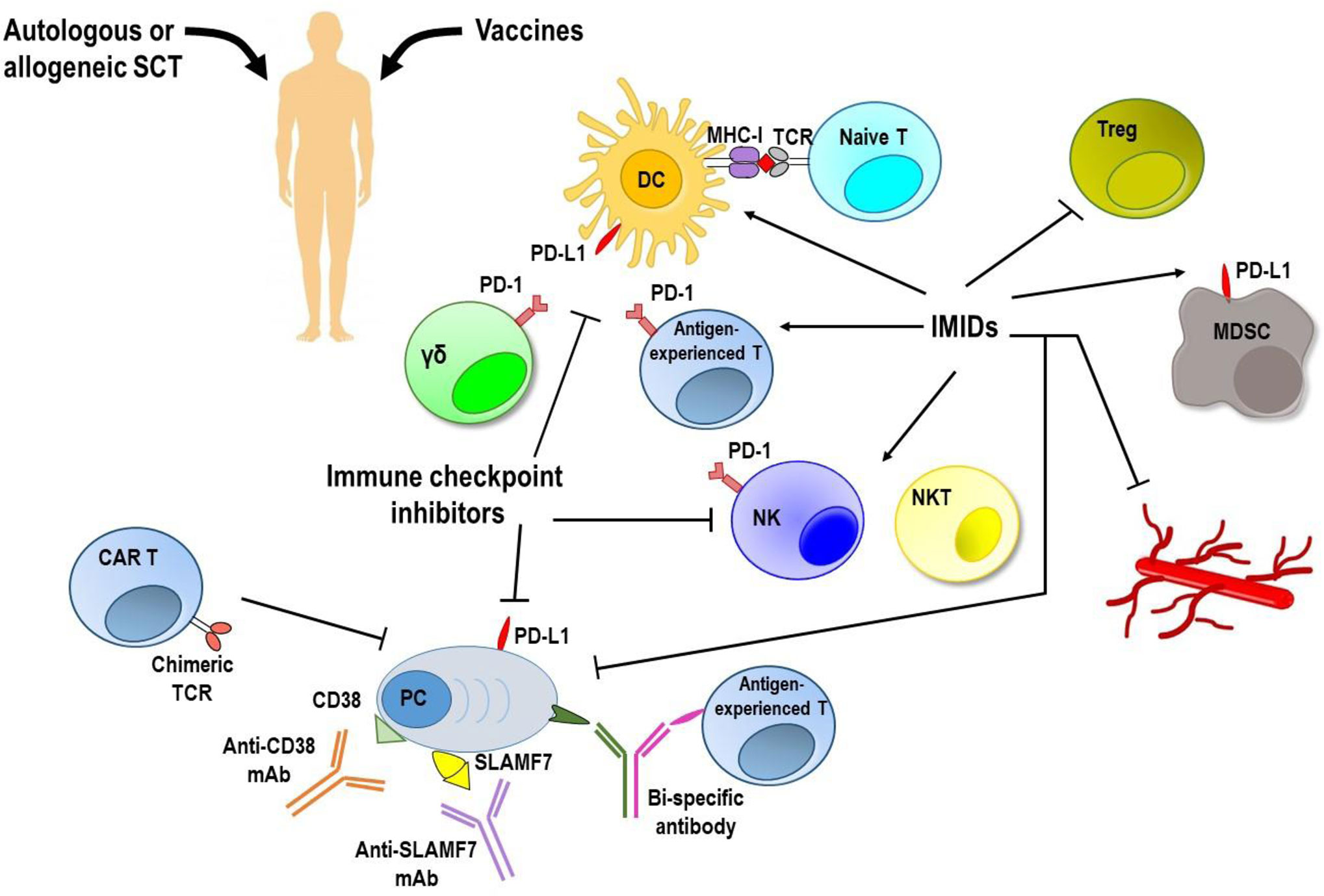
Figure 2 Targeting immune system to induce anti-MM responses. MM immunosuppressive microenvironment remains the major hurdle to achieve a long lasting response along with low toxicity. Vaccination strategies have shown no clear clinical efficacy. Autologous and allogeneic stem cell transplantation (SCT) following myeloablative treatment allows introduction of a new immune system, but generates a very weak anti-tumor immune response. Immune checkpoint inhibitors, immunomodulatory drugs (IMiDs), and monoclonal antibodies (mAbs) used as single agents provided unsatisfactory results. Immunotherapy with adoptively transferred chimeric antigen receptor (CAR) T cells and new bi-specific antibodies are currently being tested in clinical trials, and initial results have been encouraging. Moreover, newer approaches based on the combination of immunotherapeutic strategies are achieving promising results with acceptable safety and durable responses. DC, dendritic cells; γδ, gamma delta T cells; MDSC, myeloid derived suppressor cells; MHC-I, major histocompatibility complex-class I; NK, natural killer cells; NKT, natural killer T cells; PC, plasma cells; PD-1, programmed cell death-1; PD-L1, programmed death ligand 1; SLAMF7, family member 7 of the signaling lymphocytic activation molecule; TCR, T cell receptor; Treg, regulatory T cells.
The emergence of ICP blockade therapies over the last decade raised great interest also in MM. Despite at first, in vitro and in vivo studies showed that PD-1/PD-L1 blockade enhanced T and NK cell mediated anti-myeloma immune responses (42, 64, 86, 139–141) suggesting that ICP inhibition may be a promising therapeutic strategy against MM, clinical trials have provided unsatisfactory results (125). A possible explanation is that myeloma-specific T cells have an anergic or senescent phenotype rather than an exhausted phenotype, a prerequisite for the success of ICP blockade therapies.
Current lines of evidence indicate that the senescent phenotype could be reversed by immunomodulatory drugs (IMiDs), thalidomide, and its analogs lenalidomide and pomalidomide, or histone deacetylase inhibitors (35). Besides their direct anti-tumor effects (142), IMiDs promote immune activation including functional enhancement of T, NK, and NKT cells, increase of Th1 cytokine production, reduction of Treg activity, improvement of DC maturation and functions, and enhancement of anti-MM antibody dependent cell-mediated cytotoxicity (ADCC) (143–145). Therefore, IMiDs exert anti-angiogenic and anti-inflammatory effects and can disrupt plasma cell–BM microenvironment interactions (146). Interestingly, in vitro studies have demonstrated that lenalidomide treatment reduces PD-1 expression on T and NK cells and PD-L1 expression on tumor plasma cells and MDSCs (86) suggesting that IMiDs could enhance the effect of the ICP inhibitor (139). Indeed, combined therapeutic strategies with IMiDs and ICP inhibitors achieved promising results with acceptable safety and durable responses. A phase II study (NCT02289222) combining the anti-PD-1 pembrolizumab with pomalidomide and low-dose dexamethasone in 48 patients with relapsed/refractory MM resulted in an objective response of 60% including stringent complete response/complete response (8%), very good partial response (19%), and partial response (33%), with a median duration of response of 14.7 months (147). Next phase III studies of pembrolizumab in combination with pomalidomide and dexamethasone (NCT02576977) or lenalidomide and dexamethasone (NCT02579863) have been halted by the US Food and Drug Administration because of unsatisfactory results in terms of objective response and high mortality (148). A phase I trial of the anti-PD-1 nivolumab in combination with IMiDs, daratumumab, and proteasome inhibitors uncovered this combined therapy to be effective with a low toxicity profile in highly pretreated and refractory MM patients (149). Further clinical studies are ongoing to investigate the efficacy and the toxicity of nivolumab in combination with other anti-myeloma drugs in earlier stages of disease and in low-risk MM patients. In addition, clinical trials of anti-PD-L1 monoclonal antibodies (mAbs) (atezolizumab and durvalumab) alone or in combination with other anti-myeloma agents are highly expected (150).
The use of mAbs targeting CD38, daratumumab, and isatuximab is also potentially useful for treatment of relapsed/refractory MM who have received two or more prior lines of therapy (151–153). The effect of drugs alone is enhanced by the addition of IMiDs or proteasome inhibitors. Phase III trials comparing the combination of daratumumab with bortezomib and dexamethasone or lenalidomide and dexamethasone versus the drugs alone showed improved progression-free survival and overall response (154–157). The improvement of response and progression-free survival with acceptable safety has been recently achieved also in newly-diagnosed transplant-eligible patients by using daratumumab in combination with bortezomib, thalidomide, and dexamethasone (158).
The ICARIA-MM phase III study comparing the combination of isatuximab, pomalidomide, and dexamethasone versus pomalidomide and dexamethasone alone in relapsed/refractory MM patients revealed that isatuximab in the combination regimen increased the number of patients achieving a response and significantly improved the strength of response and the median progression-free survival (159).
Alternative strategies include the use of agents to disrupt BM-myeloma cell interactions. One of these agents is elotuzumab, a humanized mAb that binds to SLAMF7 (family member 7 of the signaling lymphocytic activation molecule), an immunomodulatory receptor expressed on several hematopoietic cells, including myeloma cells and NK cells (160–162). A phase I, multicenter, open-label, dose escalation study of elotuzumab showed a favorable toxicity profile but no objective responses with stable disease reported in 26% of patients (163). However, the combination of elotuzumab with pomalidomide and dexamethasone revealed a significant improvement over pomalidomide and dexamethasone alone in treatment outcomes of relapsed/refractory MM patients. Specifically, the overall response rate was higher in the elotuzumab group (53%) than in the control group (26%) with a better progressionfree survival mainly observed in patients pretreated with at least four prior lines of therapy or patients who were considered as having highrisk disease on the basis of International Myeloma Working Group Criteria (164).
Immunotherapy with adoptively transferred chimeric antigen receptor (CAR) T cells targeting myeloma-associated antigens is currently being tested in clinical trials and initial results have been encouraging. A new effective therapy for MM is the use of anti-B cell maturation antigen (BCMA) CAR T cells. Treatment of relapsed/refractory MM patients provided promising results with a high overall response. However, the durability of this response was limited and even patients with initial complete response finally relapsed. Moreover, toxicities included cytokine release syndrome, and neurotoxicity has been reported (165, 166). The main mechanism of resistance to CAR T cell therapy is the evasion of fully differentiated tumor cells expressing lower levels of BCMA. Recently, the SLAM receptor CD229/LY9 has been used as potential target for chimeric antigen receptor (CAR) T cell therapy in MM due to its strong and broader expression on the surface of BM plasma cells from MM and MGUS patients and on chemotherapy-resistant myeloma precursor cells (167–169). CD229 CAR T cells displayed a strong and persistent activity against MM in vitro and in vivo. They efficiently killed not only terminally differentiated MM plasma cells, but also memory B cells and MM propagating cells (170).
Other immunotherapies including new bi-specific antibodies, which brings tumor cells into contact with immune effector cells, e.g., T cells and NK cells, and vaccines in combination with mAbs or checkpoint inhibitors are still in early-stage clinical trials (150). To date, bi-specific antibodies have been evaluated in relapsed/refractory MM patients with promising results (171).
Future Directions
Our knowledge about mechanisms behind MM immunosuppression and sustenance of disease progression has advanced considerably. Crosstalk between immune cells and tumor endothelium regulates the entry and egress of immune cells within BM contributing to tumor immune surveillance and, at the same time, promoting angiogenesis, MM dissemination, and tumor growth (3, 11, 172, 173). Therefore, combination of canonical anti-angiogenesis treatments with immunomodulatory drugs may enhance the success of cancer immunotherapy. Moreover, it is clear that MM consists of several different genetic subtypes, and it is important to account for this when designing therapeutic regimens. A range of features including patient’s immune profile, patient’s baseline risk stratification, genetic mutations, disease biology, and imaging findings should be taken into account and integrated with each other to design tailored therapies targeting patients who might benefit the most from immunotherapy (174). New technologies for multidimensional measurement (for instance combination of single-cell RNA sequencing, genomic, immunophenotyping) of immune cells and proteins might help to build an “immunogram” to evaluate immune status and cancer-immune interactions in individual patients and thereby predict capacity to respond to immunotherapeutic strategies (76, 175).
Actually, along with immune-based approaches, the gene editing technology has emerged. Specifically, CRISPR-Cas9 technique can be used to detect necessary genes for MM plasma cells and genes involved in drug resistance, to explore the mechanism of drug action and to develop immunotherapy and screening for new drug targets (176).
Current research reveals that CRISPR/Cas9 is an efficient gene knockout platform to improve efficacy and safety of CAR T cells (177–179). Rupp et al. produced CD19-specific CAR T cells that were deficient in PD-1 using PD-1 disruption mediated by CRISPR/Cas9. The destruction of PD-1 increased CAR T cell ability to kill tumor cells in vitro (180). Based on these findings, CRISPR/Cas9 holds great promise for the treatment of MM.
Author Contributions
Conceptualization: PL, AGS, and VR. Writing: PL. Data curation: PL, AGS, EM, RF, AB, FP, VDR, and AA. Funding: VR. Supervision: NS, AV, and VR. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Italian Association for Cancer Research (AIRC) through an Investigator Grant no. 20441 to VR. The sponsors of this study are non-profit organizations that support science in general; they had no role in gathering, analyzing, or interpreting the data.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Hughes V. Microenvironment: Neighbourhood watch. Nature (2011) 480(7377):S48–9. doi: 10.1038/480S48a
2. Kawano Y, Moschetta M, Manier S, Glavey S, Görgün GT, Roccaro AM, et al. Targeting the bone marrow microenvironment in multiple myeloma. Immunol Rev (2015) 263(1):160–72. doi: 10.1111/imr.12233
3. Solimando AG, Da Via MC, Leone P, Borrelli P, Croci GA, Tabares P, et al. Halting the vicious cycle within the multiple myeloma ecosystem: blocking JAM-A on bone marrow endothelial cells restores the angiogenic homeostasis and suppresses tumor progression. Haematologica (2020) haematol.2019.239913. doi: 10.3324/haematol.2019.239913
4. Balakumaran A, Robey PG, Fedarko N, Landgren O. Bone marrow microenvironment in myelomagenesis: its potential role in early diagnosis. Expert Rev Mol Diagn (2010) 10(4):465–80. doi: 10.1586/erm.10.31
5. Quezada SA, Peggs KS, Simpson TR, Allison JP. Shifting the equilibrium in cancer immunoediting: from tumor tolerance to eradication. Immunol Rev (2011) 241(1):104–18. doi: 10.1111/j.1600-065X.2011.01007.x
6. Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science (N Y NY) (2011) 331(6024):1565–70. doi: 10.1126/science.1203486
7. Dhodapkar MV, Krasovsky J, Osman K, Geller MD. Vigorous premalignancy-specific effector T cell response in the bone marrow of patients with monoclonal gammopathy. J Exp Med (2003) 198(11):1753–7. doi: 10.1084/jem.20031030
8. Racanelli V, Leone P, Frassanito MA, Brunetti C, Perosa F, Ferrone S, et al. Alterations in the antigen processing-presenting machinery of transformed plasma cells are associated with reduced recognition by CD8+ T cells and characterize the progression of MGUS to multiple myeloma. Blood (2010) 115(6):1185–93. doi: 10.1182/blood-2009-06-228676
9. Guillerey C, Nakamura K, Vuckovic S, Hill GR, Smyth MJ. Immune responses in multiple myeloma: role of the natural immune surveillance and potential of immunotherapies. Cell Mol Life Sci: CMLS (2016) 73(8):1569–89. doi: 10.1007/s00018-016-2135-z
10. Leone P, Berardi S, Frassanito MA, Ria R, De Re V, Cicco S, et al. Dendritic cells accumulate in the bone marrow of myeloma patients where they protect tumor plasma cells from CD8+ T-cell killing. Blood (2015) 126(12):1443–51. doi: 10.1182/blood-2015-01-623975
11. Leone P, Di Lernia G, Solimando AG, Cicco S, Saltarella I, Lamanuzzi A, et al. Bone marrow endothelial cells sustain a tumor-specific CD8+ T cell subset with suppressive function in myeloma patients. Oncoimmunology (2019) 8(1):e1486949. doi: 10.1080/2162402X.2018.1486949
12. Mellstedt H, Holm G, Björkholm M. Multiple myeloma, Waldenström’s macroglobulinemia, and benign monoclonal gammopathy: characteristics of the B cell clone, immunoregulatory cell populations and clinical implications. Adv Cancer Res (1984) 41:257–89. doi: 10.1016/s0065-230x(08)60018-4
13. Bogen B, Weiss S. Processing and presentation of idiotypes to MHC-restricted T cells. Int Rev Immunol (1993) 10(4):337–55. doi: 10.3109/08830189309061709
14. Weiss S. Bogen B. B-lymphoma cells process and present their endogenous immunoglobulin to major histocompatibility complex-restricted T cells. Proc Natl Acad Sci U S A (1989) 86(1):282–6. doi: 10.1073/pnas.86.1.282
15. Brown RD, Ho PJ. Detection of malignant plasma cells in the bone marrow and peripheral blood of patients with multiple myeloma. Methods Mol Biol (Clifton NJ) (2002) 179:85–91. doi: 10.1385/1-59259-238-4:085
16. Hansson L, Rabbani H, Fagerberg J, Osterborg A. Mellstedt H. T-cell epitopes within the complementarity-determining and framework regions of the tumor-derived immunoglobulin heavy chain in multiple myeloma. Blood (2003) 101(12):4930–6. doi: 10.1182/blood-2002-04-1250
17. Sze DMY, Brown RD, Yuen E, Gibson J, Ho J, Raitakari M, et al. Clonal cytotoxic T cells in myeloma. Leuk Lymphoma (2003) 44(10):1667–74. doi: 10.1080/1042819031000097438
18. Bogen B, Ruffini PA, Corthay A, Fredriksen AB, Frøyland M, Lundin K, et al. Idiotype-specific immunotherapy in multiple myeloma: suggestions for future directions of research. Haematologica (2006) 91(7):941–8.
19. Maecker B, von Bergwelt-Baildon MS, Anderson KS, Vonderheide RH, Anderson KC, Nadler LM, et al. Rare naturally occurring immune responses to three epitopes from the widely expressed tumour antigens hTERT and CYP1B1 in multiple myeloma patients. Clin Exp Immunol (2005) 141(3):558–62. doi: 10.1111/j.1365-2249.2005.02879.x
20. Grube M, Moritz S, Obermann EC, Rezvani K, Mackensen A, Andreesen R, et al. CD8+ T cells reactive to survivin antigen in patients with multiple myeloma. Clin Cancer Res (2007) 13(3):1053–60. doi: 10.1158/1078-0432.CCR-06-1722
21. Szmania S, Tricot G, van Rhee F. NY-ESO-1 immunotherapy for multiple myeloma. Leuk Lymphoma (2006) 47(10):2037–48. doi: 10.1080/10428190600742292
22. Treon SP, Maimonis P, Bua D, Young G, Raje N, Mollick J, et al. Elevated soluble MUC1 levels and decreased anti-MUC1 antibody levels in patients with multiple myeloma. Blood (2000) 96(9):3147–53.
23. Kelly KR, Espitia CM, Zhao W, Wendlandt E, Tricot G, Zhan F, et al. Junctional adhesion molecule-A is overexpressed in advanced multiple myeloma and determines response to oncolytic reovirus. Oncotarget (2015) 6(38):41275–89. doi: 10.18632/oncotarget.5753
24. Solimando AG, Brandl A, Mattenheimer K, Graf C, Ritz M, Ruckdeschel A, et al. JAM-A as a prognostic factor and new therapeutic target in multiple myeloma. Leukemia (2018) 32(3):736–43. doi: 10.1038/leu.2017.287
25. Schmitt M, Schmitt A, Rojewski MT, Chen J, Giannopoulos K, Fei F, et al. RHAMM-R3 peptide vaccination in patients with acute myeloid leukemia, myelodysplastic syndrome, and multiple myeloma elicits immunologic and clinical responses. Blood (2008) 111(3):1357–65. doi: 10.1182/blood-2007-07-099366
26. Dhodapkar MV, Geller MD, Chang DH, Shimizu K, Fujii S-I, Dhodapkar KM, et al. A reversible defect in natural killer T cell function characterizes the progression of premalignant to malignant multiple myeloma. J Exp Med (2003) 197(12):1667–76. doi: 10.1084/jem.20021650
27. Dhodapkar MV, Krasovsky J, Olson K. T cells from the tumor microenvironment of patients with progressive myeloma can generate strong, tumor-specific cytolytic responses to autologous, tumor-loaded dendritic cells. Proc Natl Acad Sci United States America (2002) 99(20):13009–13. doi: 10.1073/pnas.202491499
28. Finn OJ. Premalignant lesions as targets for cancer vaccines. J Exp Med (2003) 198(11):1623–6. doi: 10.1084/jem.20031787
29. Fonseca R, Bailey RJ, Ahmann GJ, Rajkumar SV, Hoyer JD, Lust JA, et al. Genomic abnormalities in monoclonal gammopathy of undetermined significance. Blood (2002) 100(4):1417–24.
30. Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer (2002) 2(3):175–87. doi: 10.1038/nrc746
31. Zhan F, Hardin J, Kordsmeier B, Bumm K, Zheng M, Tian E, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood (2002) 99(5):1745–57. doi: 10.1182/blood.v99.5.1745
32. Wen Y-J, Min R, Tricot G, Barlogie B, Yi Q. Tumor lysate-specific cytotoxic T lymphocytes in multiple myeloma: promising effector cells for immunotherapy. Blood (2002) 99(9):3280–5. doi: 10.1182/blood.v99.9.3280
33. Joshua D, Suen H, Brown R, Bryant C, Ho PJ, Hart D, et al. The T Cell in Myeloma. Clin Lymphoma Myeloma Leuk (2016) 16(10):537–42. doi: 10.1016/j.clml.2016.08.003
34. Spisek R, Kukreja A, Chen L-C, Matthews P, Mazumder A, Vesole D, et al. Frequent and specific immunity to the embryonal stem cell-associated antigen SOX2 in patients with monoclonal gammopathy. J Exp Med (2007) 204(4):831–40. doi: 10.1084/jem.20062387
35. Suen H, Brown R, Yang S, Weatherburn C, Ho PJ, Woodland N, et al. Multiple myeloma causes clonal T-cell immunosenescence: identification of potential novel targets for promoting tumour immunity and implications for checkpoint blockade. Leukemia (2016) 30(8):1716–24. doi: 10.1038/leu.2016.84
36. Bryant C, Suen H, Brown R, Yang S, Favaloro J, Aklilu E, et al. Long-term survival in multiple myeloma is associated with a distinct immunological profile, which includes proliferative cytotoxic T-cell clones and a favourable Treg/Th17 balance. Blood Cancer J (2013) 3:e148. doi: 10.1038/bcj.2013.34
37. Frassanito MA, Cusmai A, Dammacco F. Deregulated cytokine network and defective Th1 immune response in multiple myeloma. Clin Exp Immunol (2001) 125(2):190–7. doi: 10.1046/j.1365-2249.2001.01582.x
38. Sharma A, Khan R, Joshi S, Kumar L, Sharma M. Dysregulation in T helper 1/T helper 2 cytokine ratios in patients with multiple myeloma. Leuk Lymphoma (2010) 51(5):920–7. doi: 10.3109/10428191003699563
39. Feng P, Yan R, Dai X, Xie X, Wen H, Yang S. The alteration and clinical significance of Th1/Th2/Th17/Treg cells in patients with multiple myeloma. Inflammation (2015) 38(2):705–9. doi: 10.1007/s10753-014-9980-4
40. Murakami H, Ogawara H, Hiroshi H. Th1/Th2 cells in patients with multiple myeloma. Hematol (Amsterdam Netherlands) (2004) 9(1):41–5. doi: 10.1080/10245330310001652437
41. Ogawara H, Handa H, Yamazaki T, Toda T, Yoshida K, Nishimoto N, et al. High Th1/Th2 ratio in patients with multiple myeloma. Leuk Res (2005) 29(2):135–40. doi: 10.1016/j.leukres.2004.06.003
42. Paiva B, Azpilikueta A, Puig N, Ocio EM, Sharma R, Oyajobi BO, et al. PD-L1/PD-1 presence in the tumor microenvironment and activity of PD-1 blockade in multiple myeloma. Leukemia (2015) 29(10):2110–3. doi: 10.1038/leu.2015.79
43. Dhodapkar MV, Sexton R, Das R, Dhodapkar KM, Zhang L, Sundaram R, et al. Prospective analysis of antigen-specific immunity, stem-cell antigens, and immune checkpoints in monoclonal gammopathy. Blood (2015) 126(22):2475–8. doi: 10.1182/blood-2015-03-632919
44. Noonan K, Marchionni L, Anderson J, Pardoll D, Roodman GD, Borrello I. A novel role of IL-17-producing lymphocytes in mediating lytic bone disease in multiple myeloma. Blood (2010) 116(18):3554–63. doi: 10.1182/blood-2010-05-283895
45. Prabhala RH, Pelluru D, Fulciniti M, Prabhala HK, Nanjappa P, Song W, et al. Elevated IL-17 produced by TH17 cells promotes myeloma cell growth and inhibits immune function in multiple myeloma. Blood (2010) 115(26):5385–92. doi: 10.1182/blood-2009-10-246660
46. Shen C-J, Yuan Z-H, Liu Y-X, Hu G-Y. Increased Numbers of T Helper 17 Cells and the Correlation with Clinicopathological Characteristics in Multiple Myeloma. J Int Med Res (2012) 40(2):556–64. doi: 10.1177/147323001204000217
47. Calcinotto A, Brevi A, Chesi M, Ferrarese R, Garcia Perez L, Grioni M, et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat Commun (2018) 9(1):4832. doi: 10.1038/s41467-018-07305-8
48. Chu VT, Fröhlich A, Steinhauser G, Scheel T, Roch T, Fillatreau S, et al. Eosinophils are required for the maintenance of plasma cells in the bone marrow. Nat Immunol (2011) 12(2):151–9. doi: 10.1038/ni.1981
49. Gupta R, Ganeshan P, Hakim M, Verma R, Sharma A, Kumar L. Significantly reduced regulatory T cell population in patients with untreated multiple myeloma. Leuk Res (2011) 35(7):874–8. doi: 10.1016/j.leukres.2010.11.010
50. Prabhala RH, Neri P, Bae JE, Tassone P, Shammas MA, Allam CK, et al. Dysfunctional T regulatory cells in multiple myeloma. Blood (2006) 107(1):301–4. doi: 10.1182/blood-2005-08-3101
51. Beyer M, Kochanek M, Giese T, Endl E, Weihrauch MR, Knolle PA, et al. In vivo peripheral expansion of naive CD4+CD25high FoxP3+ regulatory T cells in patients with multiple myeloma. Blood (2006) 107(10):3940–9. doi: 10.1182/blood-2005-09-3671
52. Braga WMT, da Silva BR, de Carvalho AC, Maekawa YH, Bortoluzzo AB, Rizzatti EG, et al. FOXP3 and CTLA4 overexpression in multiple myeloma bone marrow as a sign of accumulation of CD4(+) T regulatory cells. Cancer Immunol Immunother: CII (2014) 63(11):1189–97. doi: 10.1007/s00262-014-1589-9
53. Feyler S, von Lilienfeld-Toal M, Jarmin S, Marles L, Rawstron A, Ashcroft AJ, et al. CD4(+)CD25(+)FoxP3(+) regulatory T cells are increased whilst CD3(+)CD4(-)CD8(-)alphabetaTCR(+) Double Negative T cells are decreased in the peripheral blood of patients with multiple myeloma which correlates with disease burden. Br J Haematol (2009) 144(5):686–95. doi: 10.1111/j.1365-2141.2008.07530.x
54. Frassanito MA, Ruggieri S, Desantis V, Di Marzo L, Leone P, Racanelli V, et al. Myeloma cells act as tolerogenic antigen-presenting cells and induce regulatory T cells in vitro. Eur J Haematol (2015) 95(1):65–74. doi: 10.1111/ejh.12481
55. Giannopoulos K, Kaminska W, Hus I, Dmoszynska A. The frequency of T regulatory cells modulates the survival of multiple myeloma patients: detailed characterisation of immune status in multiple myeloma. Br J Cancer (2012) 106(3):546–52. doi: 10.1038/bjc.2011.575
56. Muthu Raja KR, Kubiczkova L, Rihova L, Piskacek M, Vsianska P, Hezova R, et al. Functionally suppressive CD8 T regulatory cells are increased in patients with multiple myeloma: a cause for immune impairment. PLoS One (2012) 7(11):e49446. doi: 10.1371/journal.pone.0049446
57. Wang J-N, Cao X-X, Zhao A-L, Cai H, Wang X, Li J. Increased activated regulatory T cell subsets and aging Treg-like cells in multiple myeloma and monoclonal gammopathy of undetermined significance: a case control study. Cancer Cell Int (2018) 18:187. doi: 10.1186/s12935-018-0687-8
58. Alrasheed N, Lee L, Ghorani E, Henry JY, Conde L, Chin M, et al. Marrow-Infiltrating Regulatory T Cells Correlate with the Presence of Dysfunctional CD4+PD-1+ Cells and Inferior Survival in Patients with Newly Diagnosed Multiple Myeloma. Clin Cancer Res (2020) 26(13):3443–54. doi: 10.1158/1078-0432.CCR-19-1714
59. Feyler S, Scott GB, Parrish C, Jarmin S, Evans P, Short M, et al. Tumour cell generation of inducible regulatory T-cells in multiple myeloma is contact-dependent and antigen-presenting cell-independent. PLoS One (2012) 7(5):e35981. doi: 10.1371/journal.pone.0035981
60. Banerjee DK, Dhodapkar MV, Matayeva E, Steinman RM, Dhodapkar KM. Expansion of FOXP3high regulatory T cells by human dendritic cells (DCs) in vitro and after injection of cytokine-matured DCs in myeloma patients. Blood (2006) 108(8):2655–61. doi: 10.1182/blood-2006-03-011353
61. Levings MK, Gregori S, Tresoldi E, Cazzaniga S, Bonini C, Roncarolo MG. Differentiation of Tr1 cells by immature dendritic cells requires IL-10 but not CD25+CD4+ Tr cells. Blood (2005) 105(3):1162–9. doi: 10.1182/blood-2004-03-1211
62. Ghiringhelli F, Puig PE, Roux S, Parcellier A, Schmitt E, Solary E, et al. Tumor cells convert immature myeloid dendritic cells into TGF-beta-secreting cells inducing CD4+CD25+ regulatory T cell proliferation. J Exp Med (2005) 202(7):919–29. doi: 10.1084/jem.20050463
63. Brown R, Kabani K, Favaloro J, Yang S, Ho PJ, Gibson J, et al. CD86+ or HLA-G+ can be transferred via trogocytosis from myeloma cells to T cells and are associated with poor prognosis. Blood (2012) 120(10):2055–63. doi: 10.1182/blood-2012-03-416792
64. Castella B, Foglietta M, Sciancalepore P, Rigoni M, Coscia M, Griggio V, et al. Anergic bone marrow Vγ9Vδ2 T cells as early and long-lasting markers of PD-1-targetable microenvironment-induced immune suppression in human myeloma. Oncoimmunology (2015) 4(11):e1047580. doi: 10.1080/2162402X.2015.1047580
65. Castella B, Foglietta M, Riganti C, Massaia M. Vγ9Vδ2 T Cells in the Bone Marrow of Myeloma Patients: A Paradigm of Microenvironment-Induced Immune Suppression. Front Immunol (2018) 9:1492. doi: 10.3389/fimmu.2018.01492
66. Chauhan D, Singh AV, Brahmandam M, Carrasco R, Bandi M, Hideshima T, et al. Functional interaction of plasmacytoid dendritic cells with multiple myeloma cells: a therapeutic target. Cancer Cell (2009) 16(4):309–23. doi: 10.1016/j.ccr.2009.08.019
67. Ray A, Das DS, Song Y, Richardson P, Munshi NC, Chauhan D, et al. Targeting PD1-PDL1 immune checkpoint in plasmacytoid dendritic cell interactions with T cells, natural killer cells and multiple myeloma cells. Leukemia (2015) 29(6):1441–4. doi: 10.1038/leu.2015.11
68. Sponaas A-M, Moharrami NN, Feyzi E, Standal T, Holth Rustad E, Waage A, et al. PDL1 Expression on Plasma and Dendritic Cells in Myeloma Bone Marrow Suggests Benefit of Targeted anti PD1-PDL1 Therapy. PLoS One (2015) 10(10):e0139867. doi: 10.1371/journal.pone.0139867
69. Dhodapkar KM, Barbuto S, Matthews P, Kukreja A, Mazumder A, Vesole D, et al. Dendritic cells mediate the induction of polyfunctional human IL17-producing cells (Th17-1 cells) enriched in the bone marrow of patients with myeloma. Blood (2008) 112(7):2878–85. doi: 10.1182/blood-2008-03-143222
70. Kotake S, Udagawa N, Takahashi N, Matsuzaki K, Itoh K, Ishiyama S, et al. IL-17 in synovial fluids from patients with rheumatoid arthritis is a potent stimulator of osteoclastogenesis. J Clin Invest (1999) 103(9):1345–52. doi: 10.1172/JCI5703
71. Kim J, Denu RA, Dollar BA, Escalante LE, Kuether JP, Callander NS, et al. Macrophages and mesenchymal stromal cells support survival and proliferation of multiple myeloma cells. Br J Haematol (2012) 158(3):336–46. doi: 10.1111/j.1365-2141.2012.09154.x
72. Zheng Y, Cai Z, Wang S, Zhang X, Qian J, Hong S, et al. Macrophages are an abundant component of myeloma microenvironment and protect myeloma cells from chemotherapy drug-induced apoptosis. Blood (2009) 114(17):3625–8. doi: 10.1182/blood-2009-05-220285
73. Zheng Y, Yang J, Qian J, Qiu P, Hanabuchi S, Lu Y, et al. PSGL-1/selectin and ICAM-1/CD18 interactions are involved in macrophage-induced drug resistance in myeloma. Leukemia (2013) 27(3):702–10. doi: 10.1038/leu.2012.272
74. Kim J, Hematti P. Mesenchymal stem cell-educated macrophages: a novel type of alternatively activated macrophages. Exp Hematol (2009) 37(12):1445–53. doi: 10.1016/j.exphem.2009.09.004
75. Berardi S, Ria R, Reale A, De Luisi A, Catacchio I, Moschetta M, et al. Multiple myeloma macrophages: pivotal players in the tumor microenvironment. J Oncol (2013) 2013:183602. doi: 10.1155/2013/183602
76. Zavidij O, Haradhvala NJ, Mouhieddine TH, Sklavenitis-Pistofidis R, Cai S, Reidy M, et al. Single-cell RNA sequencing reveals compromised immune microenvironment in precursor stages of multiple myeloma. Nat Cancer (2020) 1:493–506. doi: 10.1038/s43018-020-0053-3
77. Dosani T, Carlsten M, Maric I, Landgren O. The cellular immune system in myelomagenesis: NK cells and T cells in the development of myeloma [corrected] and their uses in immunotherapies. Blood Cancer J (2015) 5:e306. doi: 10.1038/bcj.2015.32
78. Carbone E, Neri P, Mesuraca M, Fulciniti MT, Otsuki T, Pende D, et al. NKG2D, and natural cytotoxicity receptors regulate multiple myeloma cell recognition by natural killer cells. Blood (2005) 105(1):251–8. doi: 10.1182/blood-2004-04-1422
79. García-Sanz R, González M, Orfão A, Moro MJ, Hernández JM, Borrego D, et al. Analysis of natural killer-associated antigens in peripheral blood and bone marrow of multiple myeloma patients and prognostic implications. Br J Haematol (1996) 93(1):81–8. doi: 10.1046/j.1365-2141.1996.4651006.x
80. El-Sherbiny YM, Meade JL, Holmes TD, McGonagle D, Mackie SL, Morgan AW, et al. The requirement for DNAM-1, NKG2D, and NKp46 in the natural killer cell-mediated killing of myeloma cells. Cancer Res (2007) 67(18):8444–9. doi: 10.1158/0008-5472.CAN-06-4230
81. Costello RT, Boehrer A, Sanchez C, Mercier D, Baier C, Le Treut T, et al. Differential expression of natural killer cell activating receptors in blood versus bone marrow in patients with monoclonal gammopathy. Immunology (2013) 139(3):338–41. doi: 10.1111/imm.12082
82. Bernal M, Garrido P, Jiménez P, Carretero R, Almagro M, López P, et al. Changes in activatory and inhibitory natural killer (NK) receptors may induce progression to multiple myeloma: implications for tumor evasion of T and NK cells. Hum Immunol (2009) 70(10):854–7. doi: 10.1016/j.humimm.2009.07.004
83. Jinushi M, Vanneman M, Munshi NC, Tai Y-T, Prabhala RH, Ritz J, et al. MHC class I chain-related protein A antibodies and shedding are associated with the progression of multiple myeloma. Proc Natl Acad Sci U S A (2008) 105(4):1285–90. doi: 10.1073/pnas.0711293105
84. Ponzetta A, Benigni G, Antonangeli F, Sciumè G, Sanseviero E, Zingoni A, et al. Multiple Myeloma Impairs Bone Marrow Localization of Effector Natural Killer Cells by Altering the Chemokine Microenvironment. Cancer Res (2015) 75(22):4766–77. doi: 10.1158/0008-5472.CAN-15-1320
85. von Lilienfeld-Toal M, Frank S, Leyendecker C, Feyler S, Jarmin S, Morgan R, et al. Reduced immune effector cell NKG2D expression and increased levels of soluble NKG2D ligands in multiple myeloma may not be causally linked. Cancer Immunol Immunother: CII (2010) 59(6):829–39. doi: 10.1007/s00262-009-0807-3
86. Benson DM, Bakan CE, Mishra A, Hofmeister CC, Efebera Y, Becknell B, et al. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti-PD-1 antibody. Blood (2010) 116(13):2286–94. doi: 10.1182/blood-2010-02-271874
87. Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep (2017) 19(6):1189–201. doi: 10.1016/j.celrep.2017.04.031
88. Fauriat C, Just-Landi S, Mallet F, Arnoulet C, Sainty D, Olive D, et al. Deficient expression of NCR in NK cells from acute myeloid leukemia: Evolution during leukemia treatment and impact of leukemia cells in NCRdull phenotype induction. Blood (2007) 109(1):323–30. doi: 10.1182/blood-2005-08-027979
89. Sarkar S, van Gelder M, Noort W, Xu Y, Rouschop KMA, Groen R, et al. Optimal selection of natural killer cells to kill myeloma: the role of HLA-E and NKG2A. Cancer Immunol Immunother: CII (2015) 64(8):951–63. doi: 10.1007/s00262-015-1694-4
90. Tognarelli S, Wirsching S, von Metzler I, Rais B, Jacobs B, Serve H, et al. Enhancing the Activation and Releasing the Brakes: A Double Hit Strategy to Improve NK Cell Cytotoxicity Against Multiple Myeloma. Front Immunol (2018) 9:2743. doi: 10.3389/fimmu.2018.02743
91. Dhodapkar MV. MGUS to myeloma: a mysterious gammopathy of underexplored significance. Blood (2016) 128(23):2599–606. doi: 10.1182/blood-2016-09-692954
92. Nur H, Fostier K, Aspeslagh S, Renmans W, Bertrand E, Leleu X, et al. Preclinical evaluation of invariant natural killer T cells in the 5T33 multiple myeloma model. PLoS One (2013) 8(5):e65075. doi: 10.1371/journal.pone.0065075
93. Pasiarski M, Grywalska E, Kosmaczewska A, Góźdź S, Steckiewicz P, Garus B, et al. Assessment of peripheral blood and bone marrow T, NK, NKT and dendritic cells in patients with multiple myeloma. Postepy Higieny I Medycyny Doswiadczalnej (Online) (2015) 69:1435–42.
94. Chan AC, Neeson P, Leeansyah E, Tainton K, Quach H, Prince HM, et al. Natural killer T cell defects in multiple myeloma and the impact of lenalidomide therapy. Clin Exp Immunol (2014) 175(1):49–58. doi: 10.1111/cei.12196
95. Spanoudakis E, Hu M, Naresh K, Terpos E, Melo V, Reid A, et al. Regulation of multiple myeloma survival and progression by CD1d. Blood (2009) 113(11):2498–507. doi: 10.1182/blood-2008-06-161281
96. Chang DH, Deng H, Matthews P, Krasovsky J, Ragupathi G, Spisek R, et al. Inflammation-associated lysophospholipids as ligands for CD1d-restricted T cells in human cancer. Blood (2008) 112(4):1308–16. doi: 10.1182/blood-2008-04-149831
97. Nair S, Boddupalli CS, Verma R, Liu J, Yang R, Pastores GM, et al. Type II NKT-TFH cells against Gaucher lipids regulate B-cell immunity and inflammation. Blood (2015) 125(8):1256–71. doi: 10.1182/blood-2014-09-600270
98. Nair S, Sng J, Boddupalli CS, Seckinger A, Chesi M, Fulciniti M, et al. Antigen-mediated regulation in monoclonal gammopathies and myeloma. JCI Insight (2018) 3(8):e98259. doi: 10.1172/jci.insight.98259
99. Peranzoni E, Zilio S, Marigo I, Dolcetti L, Zanovello P, Mandruzzato S, et al. Myeloid-derived suppressor cell heterogeneity and subset definition. Curr Opin Immunol (2010) 22(2):238–44. doi: 10.1016/j.coi.2010.01.021
100. Favaloro J, Liyadipitiya T, Brown R, Yang S, Suen H, Woodland N, et al. Myeloid derived suppressor cells are numerically, functionally and phenotypically different in patients with multiple myeloma. Leuk Lymphoma (2014) 55(12):2893–900. doi: 10.3109/10428194.2014.904511
101. Görgün GT, Whitehill G, Anderson JL, Hideshima T, Maguire C, Laubach J, et al. Tumor-promoting immune-suppressive myeloid-derived suppressor cells in the multiple myeloma microenvironment in humans. Blood (2013) 121(15):2975–87. doi: 10.1182/blood-2012-08-448548
102. Ramachandran IR, Martner A, Pisklakova A, Condamine T, Chase T, Vogl T, et al. Myeloid-derived suppressor cells regulate growth of multiple myeloma by inhibiting T cells in bone marrow. J Immunol (Baltimore Md: 1950) (2013) 190(7):3815–23. doi: 10.4049/jimmunol.1203373
103. Brimnes MK, Vangsted AJ, Knudsen LM, Gimsing P, Gang AO, Johnsen HE, et al. Increased level of both CD4+FOXP3+ regulatory T cells and CD14+HLA-DR−/low myeloid-derived suppressor cells and decreased level of dendritic cells in patients with multiple myeloma. Scand J Immunol (2010) 72(6):540–7. doi: 10.1111/j.1365-3083.2010.02463.x
104. Wang Z, Zhang L, Wang H, Xiong S, Li Y, Tao Q, et al. Tumor-induced CD14+HLA-DR (-/low) myeloid-derived suppressor cells correlate with tumor progression and outcome of therapy in multiple myeloma patients. Cancer Immunol Immunother: CII (2015) 64(3):389–99. doi: 10.1007/s00262-014-1646-4
105. Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res (2004) 64(16):5839–49. doi: 10.1158/0008-5472.CAN-04-0465
106. Serafini P, Borrello I, Bronte V. Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Semin Cancer Biol (2006) 16(1):53–65. doi: 10.1016/j.semcancer.2005.07.005
107. Lazarova M, Steinle A. Impairment of NKG2D-Mediated Tumor Immunity by TGF-beta. Front Immunol (2019) 10:2689. doi: 10.3389/fimmu.2019.02689
108. Li H, Han Y, Guo Q, Zhang M, Cao X. Cancer-Expanded Myeloid-Derived Suppressor Cells Induce Anergy of NK Cells through Membrane-Bound TGF-β1. J Immunol (2009) 182(1):240–9. doi: 10.4049/jimmunol.182.1.240
109. Yang L, DeBusk LM, Fukuda K, Fingleton B, Green-Jarvis B, Shyr Y, et al. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell (2004) 6(4):409–21. doi: 10.1016/j.ccr.2004.08.031
110. Cossio I, Lucas D, Hidalgo A. Neutrophils as regulators of the hematopoietic niche. Blood (2019) 133(20):2140–8. doi: 10.1182/blood-2018-10-844571
111. Tamassia N, Bianchetto-Aguilera F, Arruda-Silva F, Gardiman E, Gasperini S, Calzetti F, et al. Cytokine production by human neutrophils: Revisiting the “dark side of the moon”. Eur J Clin Invest (2018) 48(Suppl 2):e12952. doi: 10.1111/eci.12952
112. Mayadas TN, Cullere X, Lowell CA. The multifaceted functions of neutrophils. Annu Rev Pathol (2014) 9:181–218. doi: 10.1146/annurev-pathol-020712-164023
113. Karle H, Hansen NE, Plesner T. Neutrophil defect in multiple myeloma. Studies on intraneutrophilic lysozyme in multiple myeloma and malignant lymphoma. Scand J Haematol (1976) 17(1):62–70.
114. Romano A, Parrinello NL, La Cava P, Tibullo D, Giallongo C, Camiolo G, et al. PMN-MDSC and arginase are increased in myeloma and may contribute to resistance to therapy. Expert Rev Mol Diagn (2018) 18(7):675–83. doi: 10.1080/14737159.2018.1470929
115. Romano A, Parrinello NL, Simeon V, Puglisi F, La Cava P, Bellofiore C, et al. High-density neutrophils in MGUS and multiple myeloma are dysfunctional and immune-suppressive due to increased STAT3 downstream signaling. Sci Rep (2020) 10(1):1983. doi: 10.1038/s41598-020-58859-x
116. Munera V, Popovic PJ, Bryk J, Pribis J, Caba D, Matta BM, et al. Stat 6-dependent induction of myeloid derived suppressor cells after physical injury regulates nitric oxide response to endotoxin. Ann Surg (2010) 251(1):120–6. doi: 10.1097/SLA.0b013e3181bfda1c
117. Vasquez-Dunddel D, Pan F, Zeng Q, Gorbounov M, Albesiano E, Fu J, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest (2013) 123(4):1580–9. doi: 10.1172/JCI60083
118. Waight JD, Netherby C, Hensen ML, Miller A, Hu Q, Liu S, et al. Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J Clin Invest (2013) 123(10):4464–78. doi: 10.1172/JCI68189
119. Puglisi F, Parrinello NL, Giallongo C, Cambria D, Camiolo G, Bellofiore C, et al. Plasticity of High-Density Neutrophils in Multiple Myeloma is Associated with Increased Autophagy Via STAT3. Int J Mol Sci (2019) 20(14):3548. doi: 10.3390/ijms20143548
120. Romano A, Laura Parrinello N, Cerchione C, Letizia Consoli M, Parisi M, Calafiore V, et al. and LMR ratio in newly diagnosed MM patients treated upfront with novel agents. Blood Cancer J (2017) 7(12):649. doi: 10.1038/s41408-017-0019-6
121. Romano A, Parrinello NL, Consoli ML, Marchionni L, Forte S, Conticello C, et al. Neutrophil to lymphocyte ratio (NLR) improves the risk assessment of ISS staging in newly diagnosed MM patients treated upfront with novel agents. Ann Hematol (2015) 94(11):1875–83. doi: 10.1007/s00277-015-2462-4
122. Solmaz Medeni S, Acar C, Olgun A, Acar A, Seyhanli A, Taskiran E, et al. Can Neutrophil-to-Lymphocyte Ratio, Monocyte-to-Lymphocyte Ratio, and Platelet-to-Lymphocyte Ratio at Day +100 be used as a prognostic marker in Multiple Myeloma patients with autologous transplantation? Clin Transplant (2018) 32(9):e13359. doi: 10.1111/ctr.13359
123. Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol (2013) 14(10):1014–22. doi: 10.1038/ni.2703
124. Zelle-Rieser C, Thangavadivel S, Biedermann R, Brunner A, Stoitzner P, Willenbacher E, et al. T cells in multiple myeloma display features of exhaustion and senescence at the tumor site. J Hematol Oncol (2016) 9(1):116. doi: 10.1186/s13045-016-0345-3
125. Suen H, Brown R, Yang S, Ho PJ, Gibson J, Joshua D. The failure of immune checkpoint blockade in multiple myeloma with PD-1 inhibitors in a phase 1 study. Leukemia (2015) 29(7):1621–2. doi: 10.1038/leu.2015.104
126. Liu J, Hamrouni A, Wolowiec D, Coiteux V, Kuliczkowski K, Hetuin D, et al. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-{gamma} and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood (2007) 110(1):296–304. doi: 10.1182/blood-2006-10-051482
127. Tamura H, Ishibashi M, Yamashita T, Tanosaki S, Okuyama N, Kondo A, et al. Marrow stromal cells induce B7-H1 expression on myeloma cells, generating aggressive characteristics in multiple myeloma. Leukemia (2013) 27(2):464–72. doi: 10.1038/leu.2012.213
128. Yousef S, Marvin J, Steinbach M, Langemo A, Kovacsovics T, Binder M, et al. Immunomodulatory molecule PD-L1 is expressed on malignant plasma cells and myeloma-propagating pre-plasma cells in the bone marrow of multiple myeloma patients. Blood Cancer J (2015) 5:e285. doi: 10.1038/bcj.2015.7
129. An G, Acharya C, Feng X, Wen K, Zhong M, Zhang L, et al. Osteoclasts promote immune suppressive microenvironment in multiple myeloma: therapeutic implication. Blood (2016) 128(12):1590–603. doi: 10.1182/blood-2016-03-707547
130. Pratt G, Goodyear O, Moss P. Immunodeficiency and immunotherapy in multiple myeloma. Br J Haematol (2007) 138(5):563–79. doi: 10.1111/j.1365-2141.2007.06705.x
131. Kukreja A, Radfar S, Sun B-H, Insogna K, Dhodapkar MV. Dominant role of CD47-thrombospondin-1 interactions in myeloma-induced fusion of human dendritic cells: implications for bone disease. Blood (2009) 114(16):3413–21. doi: 10.1182/blood-2009-03-211920
132. Tucci M, Ciavarella S, Strippoli S, Brunetti O, Dammacco F, Silvestris F. Immature dendritic cells from patients with multiple myeloma are prone to osteoclast differentiation in vitro. Exp Hematol (2011) 39(7):773–83.e1. doi: 10.1016/j.exphem.2011.04.006
133. Nair JR, Rozanski CH, Lee KP. Under one roof: The bone marrow survival niche for multiple myeloma and normal plasma cells. Oncoimmunology (2012) 1(3):388–9. doi: 10.4161/onci.18746
134. Botta C, Di Martino MT, Ciliberto D, Cucè M, Correale P, Rossi M, et al. A gene expression inflammatory signature specifically predicts multiple myeloma evolution and patients survival. Blood Cancer J (2016) 6(12):e511. doi: 10.1038/bcj.2016.118
135. Musolino C, Allegra A, Innao V, Allegra AG, Pioggia G, Gangemi S. Inflammatory and Anti-Inflammatory Equilibrium, Proliferative and Antiproliferative Balance: The Role of Cytokines in Multiple Myeloma. Mediators Inflamm (2017) 2017:1852517. doi: 10.1155/2017/1852517
136. Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am J Hematol (2020) 95(5):548–67. doi: 10.1002/ajh.25791
137. Noonan KA, Huff CA, Davis J, Lemas MV, Fiorino S, Bitzan J, et al. Adoptive transfer of activated marrow-infiltrating lymphocytes induces measurable antitumor immunity in the bone marrow in multiple myeloma. Sci Trans Med (2015) 7(288):288ra78. doi: 10.1126/scitranslmed.aaa7014
138. Bensinger W. Allogeneic stem cell transplantation for multiple myeloma. Hematology/Oncology Clinics North Am (2014) 28(5):891–902. doi: 10.1016/j.hoc.2014.06.001
139. Görgün G, Samur MK, Cowens KB, Paula S, Bianchi G, Anderson JE, et al. Lenalidomide Enhances Immune Checkpoint Blockade-Induced Immune Response in Multiple Myeloma. Clin Cancer Res (2015) 21(20):4607–18. doi: 10.1158/1078-0432.CCR-15-0200
140. Hallett WHD, Jing W, Drobyski WR, Johnson BD. Immunosuppressive effects of multiple myeloma are overcome by PD-L1 blockade. Biol Blood Marrow Transplant (2011) 17(8):1133–45. doi: 10.1016/j.bbmt.2011.03.011
141. Kearl TJ, Jing W, Gershan JA, Johnson BD. Programmed death receptor-1/programmed death receptor ligand-1 blockade after transient lymphodepletion to treat myeloma. J Immunol (Baltimore Md: 1950) (2013) 190(11):5620–8. doi: 10.4049/jimmunol.1202005
142. Hideshima T, Chauhan D, Shima Y, Raje N, Davies FE, Tai YT, et al. Thalidomide and its analogs overcome drug resistance of human multiple myeloma cells to conventional therapy. Blood (2000) 96(9):2943–50.
143. Costa F, Vescovini R, Bolzoni M, Marchica V, Storti P, Toscani D, et al. Lenalidomide increases human dendritic cell maturation in multiple myeloma patients targeting monocyte differentiation and modulating mesenchymal stromal cell inhibitory properties. Oncotarget (2017) 8(32):53053–67. doi: 10.18632/oncotarget.18085
144. Galustian C, Meyer B, Labarthe M-C, Dredge K, Klaschka D, Henry J, et al. The anti-cancer agents lenalidomide and pomalidomide inhibit the proliferation and function of T regulatory cells. Cancer Immunol Immunother: CII (2009) 58(7):1033–45. doi: 10.1007/s00262-008-0620-4
145. Luptakova K, Rosenblatt J, Glotzbecker B, Mills H, Stroopinsky D, Kufe T, et al. Lenalidomide enhances anti-myeloma cellular immunity. Cancer Immunol Immunother: CII (2013) 62(1):39–49. doi: 10.1007/s00262-012-1308-3
146. Quach H, Ritchie D, Stewart AK, Neeson P, Harrison S, Smyth MJ, et al. Mechanism of action of immunomodulatory drugs (IMiDS) in multiple myeloma. Leukemia (2010) 24(1):22–32. doi: 10.1038/leu.2009.236
147. Badros A, Hyjek E, Ma N, Lesokhin A, Dogan A, Rapoport AP, et al. Pembrolizumab, pomalidomide, and low-dose dexamethasone for relapsed/refractory multiple myeloma. Blood (2017) 130(10):1189–97. doi: 10.1182/blood-2017-03-775122
148. Costa F, Das R, Kini Bailur J, Dhodapkar K, Dhodapkar MV. Checkpoint Inhibition in Myeloma: Opportunities and Challenges. Front Immunol (2018) 9:2204. doi: 10.3389/fimmu.2018.02204
149. Thanendrarajan S, Puryear J, Schinke CD, van Rhee F, Zangari M, Mathur P, et al. Nivolumab for Treatment of Advanced, Refractory, High-Risk Multiple Myeloma. Blood (2017) 130.
150. Soekojo CY, Ooi M, de Mel S, Chng WJ. Immunotherapy in Multiple Myeloma. Cells (2020) 9(3):601. doi: 10.3390/cells9030601
151. Lokhorst HM, Plesner T, Laubach JP, Nahi H, Gimsing P, Hansson M, et al. Targeting CD38 with Daratumumab Monotherapy in Multiple Myeloma. N Engl J Med (2015) 373(13):1207–19. doi: 10.1056/NEJMoa1506348
152. Lonial S, Weiss BM, Usmani SZ, Singhal S, Chari A, Bahlis NJ, et al. Daratumumab monotherapy in patients with treatment-refractory multiple myeloma (SIRIUS): an open-label, randomised, phase 2 trial. Lancet (Lond Engl) (2016) 387(10027):1551–60. doi: 10.1016/S0140-6736(15)01120-4
153. Mikhael J, Richter J, Vij R, Cole C, Zonder J, Kaufman JL, et al. A dose-finding Phase 2 study of single agent isatuximab (anti-CD38 mAb) in relapsed/refractory multiple myeloma. Leukemia (2020). doi: 10.1038/s41375-020-0857-2
154. Dimopoulos MA, Oriol A, Nahi H, San-Miguel J, Bahlis NJ, Usmani SZ, et al. Daratumumab, Lenalidomide, and Dexamethasone for Multiple Myeloma. N Engl J Med (2016) 375(14):1319–31. doi: 10.1056/NEJMoa1607751
155. Mateos M-V, González-Calle V. Timing of treatment of smoldering myeloma: early treatment. Blood Adv (2018) 2(21):3045–9. doi: 10.1182/bloodadvances.2018021220
156. Palumbo A, Chanan-Khan A, Weisel K, Nooka AK, Masszi T, Beksac M, et al. Daratumumab, Bortezomib, and Dexamethasone for Multiple Myeloma. N Engl J Med (2016) 375(8):754–66. doi: 10.1056/NEJMoa1606038
157. Spencer A, Lentzsch S, Weisel K, Avet-Loiseau H, Mark TM, Spicka I, et al. Daratumumab plus bortezomib and dexamethasone versus bortezomib and dexamethasone in relapsed or refractory multiple myeloma: updated analysis of CASTOR. Haematologica (2018) 103(12):2079–87. doi: 10.3324/haematol.2018.194118
158. Moreau P, Attal M, Hulin C, Arnulf B, Belhadj K, Benboubker L, et al. Bortezomib, thalidomide, and dexamethasone with or without daratumumab before and after autologous stem-cell transplantation for newly diagnosed multiple myeloma (CASSIOPEIA): a randomised, open-label, phase 3 study. Lancet (Lond Engl) (2019) 394(10192):29–38. doi: 10.1016/S0140-6736(19)31240-1
159. Attal M, Richardson PG, Rajkumar SV, San-Miguel J, Beksac M, Spicka I, et al. Isatuximab plus pomalidomide and low-dose dexamethasone versus pomalidomide and low-dose dexamethasone in patients with relapsed and refractory multiple myeloma (ICARIA-MM): a randomised, multicentre, open-label, phase 3 study. Lancet (Lond Engl) (2019) 394(10214):2096–107. doi: 10.1016/S0140-6736(19)32556-5
160. Balasa B, Yun R, Belmar NA, Fox M, Chao DT, Robbins MD, et al. Elotuzumab enhances natural killer cell activation and myeloma cell killing through interleukin-2 and TNF-α pathways. Cancer Immunol Immunother: CII (2015) 64(1):61–73. doi: 10.1007/s00262-014-1610-3
161. Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol (2011) 29:665–705. doi: 10.1146/annurev-immunol-030409-101302
162. Collins SM, Bakan CE, Swartzel GD, Hofmeister CC, Efebera YA, Kwon H, et al. Elotuzumab directly enhances NK cell cytotoxicity against myeloma via CS1 ligation: evidence for augmented NK cell function complementing ADCC. Cancer Immunol Immunother: CII (2013) 62(12):1841–9. doi: 10.1007/s00262-013-1493-8
163. Zonder JA, Mohrbacher AF, Singhal S, van Rhee F, Bensinger WI, Ding H, et al. A phase 1, multicenter, open-label, dose escalation study of elotuzumab in patients with advanced multiple myeloma. Blood (2012) 120(3):552–9. doi: 10.1182/blood-2011-06-360552
164. Dimopoulos MA, Dytfeld D, Grosicki S, Moreau P, Takezako N, Hori M, et al. Elotuzumab plus Pomalidomide and Dexamethasone for Multiple Myeloma. N Engl J Med (2018) 379(19):1811–22. doi: 10.1056/NEJMoa1805762
165. Cohen AD, Garfall AL, Stadtmauer EA, Melenhorst JJ, Lacey SF, Lancaster E, et al. B cell maturation antigen-specific CAR T cells are clinically active in multiple myeloma. J Clin Invest (2019) 129(6):2210–21. doi: 10.1172/JCI126397
166. Raje N, Berdeja J, Lin Y, Siegel D, Jagannath S, Madduri D, et al. Anti-BCMA CAR T-Cell Therapy bb2121 in Relapsed or Refractory Multiple Myeloma. New Engl J Med (2019) 380(18):1726–37. doi: 10.1056/NEJMoa1817226
167. Carulli G, Buda G, Azzarà A, Ciancia EM, Sammuri P, Domenichini C, et al. CD229 Expression on Bone Marrow Plasma Cells from Patients with Multiple Myeloma and Monoclonal Gammopathies of Uncertain Significance. Acta Haematol (2016) 135(1):11–4. doi: 10.1159/000380939
168. Muccio VE, Saraci E, Gilestro M, Gattei V, Zucchetto A, Astolfi M, et al. Multiple myeloma: New surface antigens for the characterization of plasma cells in the era of novel agents. Cytometry Part B Clin Cytometry (2016) 90(1):81–90. doi: 10.1002/cyto.b.21279
169. Yousef S, Kovacsovics-Bankowski M, Salama ME, Bhardwaj N, Steinbach M, Langemo A, et al. CD229 is expressed on the surface of plasma cells carrying an aberrant phenotype and chemotherapy-resistant precursor cells in multiple myeloma. Hum Vaccines Immunother (2015) 11(7):1606–11. doi: 10.1080/21645515.2015.1046658
170. Radhakrishnan SV, Luetkens T, Scherer SD, Davis P, Vander Mause ER, Olson ML, et al. CD229 CAR T cells eliminate multiple myeloma and tumor propagating cells without fratricide. Nat Commun (2020) 11(1):798. doi: 10.1038/s41467-020-14619-z
171. Zhou X, Einsele H, Danhof S. Bispecific Antibodies: A New Era of Treatment for Multiple Myeloma. J Clin Med (2020) 9(7). doi: 10.3390/jcm9072166
172. Da Via MC, Solimando AG, Garitano-Trojaola A, Barrio S, Munawar U, Strifler S, et al. CIC Mutation as a Molecular Mechanism of Acquired Resistance to Combined BRAF-MEK Inhibition in Extramedullary Multiple Myeloma with Central Nervous System Involvement. Oncologist (2020) 25(2):112–8. doi: 10.1634/theoncologist.2019-0356
173. Lamanuzzi A, Saltarella I, Desantis V, Frassanito MA, Leone P, Racanelli V, et al. Inhibition of mTOR complex 2 restrains tumor angiogenesis in multiple myeloma. Oncotarget (2018) 9(29):20563–77. doi: 10.18632/oncotarget.25003
174. Solimando AG, Da Vià MC, Cicco S, Leone P, Di Lernia G, Giannico D, et al. High-Risk Multiple Myeloma: Integrated Clinical and Omics Approach Dissects the Neoplastic Clone and the Tumor Microenvironment. J Clin Med (2019) 8(7). doi: 10.3390/jcm8070997
175. Blank CU, Haanen JB, Ribas A, Schumacher TN. CANCER IMMUNOLOGY. The “cancer immunogram”. Science (2016) 352(6286):658–60. doi: 10.1126/science.aaf2834
176. Hong L, Zhang C, Jiang Y, Liu H, Huang H, Guo D. Therapeutic status and the prospect of CRISPR/Cas9 gene editing in multiple myeloma. Future Oncol (Lond Engl) (2020) 16(16):1125–36. doi: 10.2217/fon-2019-0822
177. Beauvais D, Danhof S, Hayden PJ, Einsele H, Yakoub-Agha I. Clinical data, limitations and perspectives on chimeric antigen receptor T-cell therapy in multiple myeloma. Curr Opin Oncol (2020) 32(5):418–26. doi: 10.1097/CCO.0000000000000667
178. Liu X, Zhang Y, Cheng C, Cheng AW, Zhang X, Li N, et al. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res (2017) 27(1):154–7. doi: 10.1038/cr.2016.142
179. Nerreter T, Letschert S, Götz R, Doose S, Danhof S, Einsele H, et al. Super-resolution microscopy reveals ultra-low CD19 expression on myeloma cells that triggers elimination by CD19 CAR-T. Nat Commun (2019) 10(1):3137. doi: 10.1038/s41467-019-10948-w
Keywords: multiple myeloma, microenvironment, immune cells, immune checkpoints, immunotherapy
Citation: Leone P, Solimando AG, Malerba E, Fasano R, Buonavoglia A, Pappagallo F, De Re V, Argentiero A, Silvestris N, Vacca A and Racanelli V (2020) Actors on the Scene: Immune Cells in the Myeloma Niche. Front. Oncol. 10:599098. doi: 10.3389/fonc.2020.599098
Received: 26 August 2020; Accepted: 05 October 2020;
Published: 29 October 2020.
Edited by:
Cirino Botta, Unit of Clinical Hematology, Cosenza Hospital, ItalyReviewed by:
Alessandra Romano, University of Catania, ItalyRocco Savino, University of Catanzaro, Italy
Copyright © 2020 Leone, Solimando, Malerba, Fasano, Buonavoglia, Pappagallo, De Re, Argentiero, Silvestris, Vacca and Racanelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vito Racanelli, dml0by5yYWNhbmVsbGkxQHVuaWJhLml0
 Patrizia Leone
Patrizia Leone Antonio Giovanni Solimando
Antonio Giovanni Solimando Eleonora Malerba
Eleonora Malerba Rossella Fasano
Rossella Fasano Alessio Buonavoglia
Alessio Buonavoglia Fabrizio Pappagallo
Fabrizio Pappagallo Valli De Re
Valli De Re Antonella Argentiero
Antonella Argentiero Nicola Silvestris
Nicola Silvestris Angelo Vacca
Angelo Vacca Vito Racanelli
Vito Racanelli