
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 23 November 2020
Sec. Cancer Genetics
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.598964
This article is part of the Research Topic microRNAs and Muscle Cell Death in Cancer View all 5 articles
Skeletal muscles are the largest tissues in our body and the physiological function of muscle is essential to every aspect of life. The regulation of development, homeostasis, and metabolism is critical for the proper functioning of skeletal muscle. Consequently, understanding the processes involved in the regulation of myogenesis is of great interest. Non-coding RNAs especially microRNAs (miRNAs) are important regulators of gene expression and function. MiRNAs are small (~22 nucleotides long) noncoding RNAs known to negatively regulate target gene expression post-transcriptionally and are abundantly expressed in skeletal muscle. Gain- and loss-of function studies have revealed important roles of this class of small molecules in muscle biology and disease. In this review, we summarize the latest research that explores the role of miRNAs in skeletal muscle development, gene expression, and function as well as in muscle disorders like sarcopenia and Duchenne muscular dystrophy (DMD). Continuing with the theme of the current review series, we also briefly discuss the role of miRNAs in cancer cachexia.
Skeletal muscle consists of approximately 42 and 36% of the average body weight of male and female humans, respectively. It is a highly plastic tissue with an ability to contract and stretch. Apart from playing an important role in supporting movement and respiration, skeletal muscle also acts as a reservoir for proteins, generating body heat, and protecting other internal tissues and organs (1, 2). As a result, maintaining the proper function of skeletal muscle is indispensable and understanding the factors that regulate muscle mass is essential.
Altered expression of microRNAs has been shown to be involved in skeletal muscle homeostasis in health and disease (3–6). MicroRNAs (miRNAs) are small (~22 nucleotides long), ubiquitously expressed non-coding RNA sequences involved in regulating gene expression at the post-transcriptional level. MiRNAs are known to regulate gene expression by binding to the 3’ untranslated region (3’UTR) of targeting mRNAs resulting in either their degradation or inhibition of protein translation. A few reports have also indicated the possible binding of miRNAs to the 5’UTR or coding region of target mRNAs. MiRNAs are transcribed by RNA polymerase II or III as a long primary miRNA (pri-miRNA) from different regions of the genome, including non-coding regions (intronic or intergenic) and protein coding regions (exons) as an individual miRNA (monocistronic) or cluster (poly-cistronic) (7).
Pri-miRNAs are processed in the nucleus by the RNase III endonuclease Drosha and its cofactor Di George syndrome critical region 8 gene (DGCR8) into short ~70 nucleotide precursor miRNAs (pre-miRNAs) (8). Pre-miRNAs are exported via exportin-5 into the cytoplasm, where it is cleaved by the RNase III enzyme Dicer, yielding approximately 22 nucleotide long imperfect miRNA duplexes (ds-miRNAs) (9). Helicase unwinding of these ds-miRNAs results in a miRNA (i.e. the leading or mature strand) and a miRNA* (i.e. the passenger strand) (10). The mature strand of the miRNA/miRNA* duplex is incorporated into the RNA-induced silencing complex (RISC) containing Dicer and AGO2 (argonaute RISC catalytic component 2) along with RNA binding proteins, while the miRNA* strand is mostly degraded. The RISC is guided by the mature miRNA to the 3’UTR of target mRNAs, resulting in either degradation or translational inhibition (11, 12).
While the role of miRNAs is well documented in many biological events, including cell death, differentiation, proliferation, and cell growth (13, 14), miRNAs have also emerged as important regulators of various pathways involved in skeletal muscle development and function. The present review summarizes the latest research exploring the role of miRNAs in skeletal muscle myogenesis and describes their role in muscle disorders like sarcopenia and Duchenne muscular dystrophy (DMD). In keeping with the theme of the current review series, we have also included a section describing the role of miRNAs in cancer cachexia.
The process of generating skeletal muscle is called myogenesis. The development and growth of muscle is regulated by important muscle-specific transcription factors known as myogenic regulatory factors (MRFs). Expression of MRFs are limited to the muscle lineage leading to the formation of muscle fibers following activation of downstream signaling pathways. During embryogenesis, the paired box transcription factors Pax3 and Pax7 regulate early MRFs. MRFs include transcription factors of the MyoD family, such as myogenin, Myf5, MyoD, MRF4 (also called Myf6), as well as the MEF2 family and the serum response factor (SRF) (15–17).
Many studies have highlighted the role of muscle-specific microRNAs in myogenesis with an emphasis on their role in the signaling pathways involved in muscle cell (myocyte) proliferation and differentiation by acting either synergistically or antagonistically (18, 19). For example, a report by O’Rourke et al. demonstrated the importance of miRNAs in skeletal muscle development. In this study, the authors generated mice with a skeletal muscle-specific deletion of Dicer, which encodes an important enzyme involved in miRNA biogenesis. Dicer knock-out mice showed significantly decreased expression of many muscle-specific miRNAs resulting in muscle hypoplasia and lethality—highlighting the importance of miRNAs in muscle development (20).
There are two groups of miRNAs involved in the regulation and development of skeletal muscle. The first group is exclusively expressed in muscle, called myomiRs, while the second group is expressed in both muscle and non-muscle cells, called non-myomiRs. Much of our understanding of miRNAs in skeletal muscle initially focused on myomiRs, which are expressed or enriched in the heart and skeletal muscles. These myomiRs included eight miRNAs; miR-1, miR-133a/b, miR-208b, miR-486, and miR-499 (expressed in the myocardium and skeletal muscle), while miR-208a was expressed exclusively in the myocardium and miR-206 was restricted to skeletal muscle (21) (Table 1).
Early studies in skeletal muscle C2C12 myoblasts highlighted the importance of myomiRs in the differentiation and development of skeletal muscle (22). The myogenic regulatory factors SRF, MyoD, MyoG, MEF2, and myogenin directly regulate expression of myomiRs (34–36). The miR-1/206 family is comprised of miR-1-1, miR-1-2, and miR-206. MiR-1 and miR-6 share identical seed sequences and differ by only four nucleotides outside the seed region (23). The importance of miR-1 and miR-206 in myoblast differentiation and muscle development has been well described. For example, Kim et al. showed an increase in the expression of miR-1 and miR-206 promotes the differentiation of satellite cells (30).
MyomiRs promote muscle cell differentiation and development by targeting repressors of skeletal muscle cell differentiation. MiR-1 promotes myogenesis by targeting histone deacetylase 4 (HDAC4) and a transcriptional repressor of the muscle-specific transcription factor, MEF2; thereby, promoting expression of MEF2, which increases the expression of miR-1 and accelerates the differentiation of muscle fibers (23, 24). Similarly, miR-206 was reported to enhance the differentiation of myoblasts by repressing DNA polymerase alpha (Polα1), the gap junction protein connexin43, follistatin-like 1 protein (Fstl1), and utrophin (Utrn). Polα1 is involved in DNA synthesis, and its inhibition reduces DNA synthesis at the initial differentiation stages and promotes myocyte differentiation by restricting cell proliferation. By targeting connexin43 (Cx43), miR-206 acts to reduce intercellular communication between muscle fibers (30, 31, 37, 38). Utrn and Fstl1 are also involved in myoblast proliferation and an important target of miR-206 (31). Recently, Ma et al. reported nPTB is another key target for miR-206 and this interaction affects differentiation (25). Therefore, miR-1 and miR-206 regulate many common targets like Pax3, Pax7, HDAC4, Cx43, and Notch3 (23–25, 39), while YY1, CNN3 (calponin 3), SFRP1 are the exclusive targets of miR-1 (22, 26).
Another important member of the myomiR family is miR-133, which includes miR-133a-1, miR-133a-2, and miR-133b (40). Their expression is up-regulated during myogenesis, similar to miR-1 (23, 41). However, unlike miR-1, which promotes myogenic differentiation, miR-133 is primarily reported to be involved in muscle cell proliferation and inhibition of myocyte differentiation. MiR-133 promotes myocyte proliferation by targeting serum response factor (SRF) and decreasing its expression (23). In particular, many reports have recently examined the role of miR-133 in myocyte differentiation.
Neuronal polypyrimidine tract‐binding protein (nPTB) and uncoupling protein 2 (UCP 2) are negative regulators of myocyte differentiation. MiR-133 is reported to promote differentiation by targeting nPTB (25) and UCP-2 (27). Moreover, the inhibition of the forkhead box transcription factor 2 (Fox 2) by miR-133 can also promote differentiation (42). Fibroblast growth factor receptor 1 (FGFR1) and protein phosphatase 2A (PP2AC) expression is important for regulating the ERK1/2 signal transduction pathway. Inhibition of ERK1/2 signaling plays an important role in myocyte differentiation and miR-133 promotes muscle cell differentiation by targeting FGFR1 and PP2AC (28).
MiR-486 is another muscle enriched miRNA and it is significantly increased during muscle cell differentiation. In both satellite cells and C2C12 cells, miR-486 can promote differentiation by targeting Pax7 (33). MiR-486 also targets regulators of PI3K/AKT signaling, PTEN (phosphatase and tensin homolog), and FoxO1a (forkhead box protein O1a). PI3K/AKT signaling is involved in repression of skeletal myogenesis and the inhibition of PTEN and FoxO1a by miR-486 promotes myogenesis (43).
At the same time, miR-208 and miR-499 are involved in the differentiation of myocytes into type I slow twitch and type II fast twitch muscle myofibrils. MiR-208a is expressed exclusively in heart, whereas miR-208b and miR-499 are expressed in both the heart and skeletal muscle. They are expressed from intronic sequences embedded within the myosin heavy chain genes Myh6, Myh77, and Myh7B, respectively (32). These miRNAs broadly participate in the regulation of muscle gene expression, myofiber specification, and muscle function as double knockout mice for miR-208 and miR-499 showed increased expression of SRY-box containing gene 6 (Sox6), resulting in decreased expression of Myh7 and loss of slow type I slow twitch myofibers (32).
In addition to myomiRs many more miRs are expressed in skeletal muscle and they play a significant role in the regulation of myogenesis. These non-myomiRs (meaning their expression is not restricted to muscle cells), are known to control many processes involved in muscle, like muscle satellite cell (MSC) differentiation and proliferation, myoblast differentiation and proliferation, and myofibril formation. Consequently, the role of non-myomiRs in skeletal muscle regulation is becoming well-documented (19, 44, 45). Table 2 highlights the role of many of these miRs in skeletal muscle.
MiRNAs have been shown to play a vital role in the regulation of satellite cells and skeletal muscle myogenesis by targeting various muscle-specific transcription factors and important signaling pathways. Previous studies have highlighted the role of miRNAs in skeletal muscle cell regulation, suggesting a vital role for these miRNAs in satellite cell-mediated muscle development. The paired-box transcription factors Pax 3 and Pax 7 are crucial for satellite cell proliferation and differentiation. MiR-27b is highly expressed in activated satellite cells and it directly targets Pax3 mRNA and, consequently, promotes satellite cell differentiation while inhibiting proliferation (47). MiR-431 is another miRNA shown to suppress the proliferation of satellite cells and enhance differentiation by targeting Pax-7 mRNA. Interestingly, in miR-431 overexpressing transgenic mice, there was significant improvement in muscle regeneration compared to control mice.
Other important miRNAs include miR-195 and miR-497, which are members of the miR-15 family, which maintains the quiescent state of satellite cells. MiR-195 and miR-497 share the same seed sequences and target the cell cycle activators Ccnd2, Cdc25, cyclin D1, and cyclin D2 (49, 50). MiR-31 is also reported to maintain the quiescent state of satellite cells as it targets Myf5, an important myogenic regulator needed for the differentiation of satellite cells toward the myogenic lineage, which delays differentiation (47). Similarly, a report by Velasco et al. showed targeting of Myf5 mRNA by another miRNA (miR-106b) delays activation of the myogenic program and, subsequently, differentiation (48).
Given their critical roles in the regulation of muscle gene expression as well as the myogenic program and muscle function, it is not surprising that myogenic regulatory factors (MRFs), like MyoD, Myf5, Myf6, myogenin, and members of the myocyte enhancer factor 2 (MEF2) family of transcription factors, including MEF2A, MEF2B, MEF2C, and MEF2D, are direct targets of muscle-specific miRNAs, resulting in alteration of muscle gene expression in skeletal muscle (Figure 1).
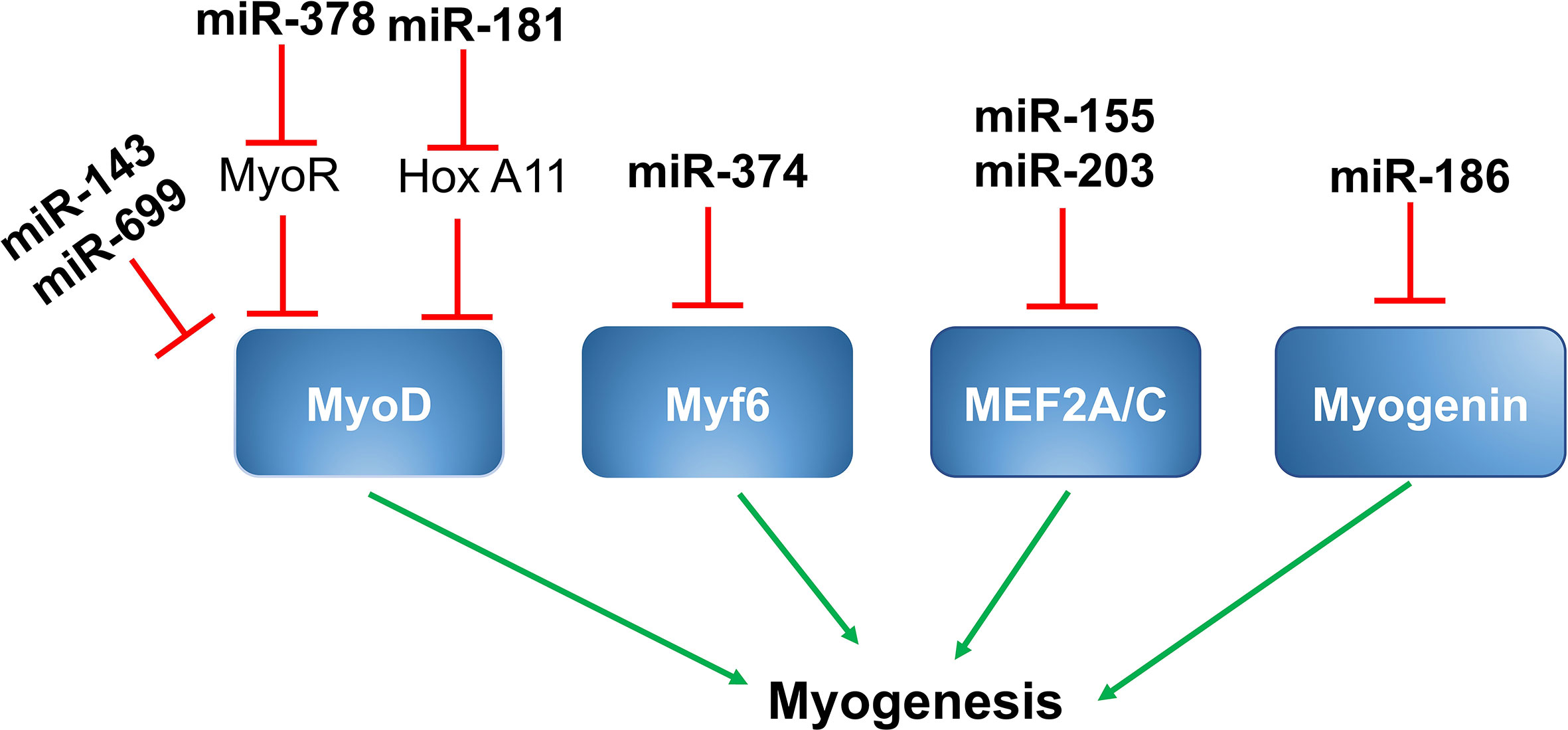
Figure 1 MiRNAs and myogenic regulatory factors. MiR-143 and miR-699 targets MyoD and suppress myogenesis. MiR-378 is up-regulated by MyoD, which targets MyoR and further upregulates MyoD through positive feedback. MiR-181 targets Hox-A11, leading to increased expression of MyoD and myogenin. MiR-376 targets Myf6 and suppresses myogenesis. MiR-155 and miR-203 suppresses myogenesis by targeting MEF2A and MEF2C respectively. MiR-186 targets myogenesis and controls myogenesis.
A study by Chen et al. showed miR‐143 was a direct target of MyoD in the skeletal muscles of Siniperca chuatsi, a type of Asian Perch. Expression of miR-143 was inversely proportional to the expression of MyoD in fast and slow twitch muscles. As suppression of miR‐143 results in a substantial increase in MyoD and fast MyhC gene expression in vivo, miR143 likely has an important role in the differentiation of different muscle fiber types in vertebrates (82).
Expression of miR-378 is induced during the differentiation of C2C12 cells in culture. MiR-378 promotes skeletal muscle cell differentiation by decreasing the expression of negative regulators. MiR-378 is encoded within an intron of the peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPARγ-c1β) that encodes for PGC-1β. Expression of PGC-1β/miR-378 is regulated by MyoD, which binds to the regulatory sequences upstream of PGC-1β/miR378. Mir-378, in turn, binds to the MyoD transcription repressor MyoR and, thereby, promotes MyoD expression in a positive feedback loop (39).
miR-181 is another microRNA that promotes skeletal muscle differentiation by decreasing the expression of negative regulators. Expression of miR-181 is elevated in differentiating myoblasts and is less expressed in terminally differentiated muscle cells. In skeletal muscle cells, increased expression of MyoD suppresses cell regeneration and promotes terminal differentiation. The target gene for miR-181 is homeobox protein called Hox-A11, which is a well-known muscle cell differentiation repressor because it suppresses the expression of MyoD. MiR-181 targets Hox-A11 and, ultimately, leads to increased expression of MyoD; thereby, promoting muscle cell terminal differentiation. In C2C12 myoblasts, inhibition of miR-181 suppresses myoblast differentiation by reducing MyoD expression (72, 83).
Two members of miR669 family, miR669a and miR669q, are involved in cardiac skeletal muscle differentiation. Both miR669a and miR669q directly target MyoD and decrease the expression of MyoD and muscle cell differentiation (21).
MiR-374 is a miRNA that regulates myoblast differentiation by targeting another myogenic regulatory factor, Myf6. Myf6 regulates the expression of genes involved in myoblast terminal differentiation. MiR-374 directly binds to the 3’ UTR region of Myf6 and decreases its expression. In mouse skeletal muscle and C2C12 myoblasts, miR-374 regulates differentiation. As shown by Ma et al., overexpression of miR-374 suppresses myoblast differentiation, while its inhibition using by 2’-O-methyl antisense oligonucleotides promotes C2C12 myoblast differentiation (25).
Myogenin is another important MRF involved in the regulation of myoblast differentiation. It plays a critical role in the regulation of terminal differentiation of the muscle cells. MiR-186 directly binds to the 3’ UTR region of myogenin and decreases its expression at both the mRNA and protein levels. MiR-186 resides between exons 8 and 9 of the ZRANB2 gene and these miRNAs are transcribed in parallel with the host gene. Overexpression and inhibition studies in C2C12 myoblasts as well as in primary muscle cells showed that inhibiting myogenin expression prevented differentiation (73).
MEF2A is a member of the MEF2 (myocyte enhancer factor 2) family of transcription factors and plays a positive role in muscle cell differentiation. MiR-155 directly binds to the 3’ UTR region of MEF2A and decreases its expression at both the mRNA and protein levels. As demonstrated by Seok et al., overexpression of miR-155 significantly down-regulates the expression of MEF2A and, thus, decreases myoblast differentiation. Also, when MEF2A is reintroduced into miR-155 overexpressing myoblasts, it partially promotes differentiation (71).
MiRNA-203 is a miRNA that plays a regulatory role in myoblast proliferation and differentiation. During differentiation of myoblasts into myotubes, miR-203 is significantly down-regulated. As shown by Lu et al., miR-203 overexpression suppresses myoblast proliferation and differentiation, and its inhibition promotes proliferation and differentiation. MEF2C and c-Jun (a transcriptional activator of the AP‐1 family) are the direct targets of miR-203. C-Jun is known to promote muscle cell proliferation and MEF2C is well-known to promote differentiation. Therefore, miR-203 can regulate both proliferation and differentiation during muscle development (76, 77).
During myogenic differentiation, expression of the myosin heavy chain (Myh) is regulated by muscle related transcription factor MEF2C (84). MiR-23a suppresses myoblast differentiation in C2C12 myoblasts. Myh gene isoforms Myh 1, 2, and 4 are the direct targets of miR-23a. Overexpression and inhibition studies in C2C12 myoblasts further confirmed that miR-23a decreases the expression of Myh isoforms and inhibits myoblast differentiation (57). A study by Mercatelli et al. showed that miR-23a and miR-23b target thioredoxin reductase 1 (Trxr1). Overexpression of Trxr1 delays differentiation by negatively modulating the expression of myogenin and Myh genes (85).
Apart from targeting MRFs, miRNAs also regulate myoblast proliferation and differentiation by targeting genes encoding key components of various signaling pathways. In the following section, we will discuss various signaling pathways like IDI (inhibitor of differentiation 1) signaling, TGF-β/SMAD/BMP signaling, insulin signaling, and NF-κB-YY1 signaling in addition to the role of associated miRNAs in the regulation of skeletal muscle development.
The miR‐17-92 cluster consists of miR-17, miR-20a, and miR-92a, and these miRNAs promote myoblast proliferation and suppress differentiation along with tube formation. Transcription of the miR-17-92 cluster is regulated by E2F1. Actin-associated protein enigma homolog 1 (ENH1) and transcription factor E2F3b (E2F transcription factor 3b) are the direct targets of microRNAs from the miR-17-92 cluster. MiR-17-92 suppresses ENH1 by directly targeting its 3’UTR and results in increased expression of ID1 in the nucleus. ID1 binds to MRF and E protein and subsequently suppresses myogenic differentiation. Another target of the miR-17-92 cluster is E2F3b. Inhibition of E2F3b further suppresses myotube formation (54, 86).
MiR-98 is involved in negatively regulating myogenic differentiation of C2C12 myoblasts. E2F5 is a transcriptional repressor and miR-98 directly targets the 3’ UTR region of E2F5. Kropp et al. showed that simultaneous inhibition of both miR-98 as well as E2F5 in C2C12 myoblasts restores differentiation. Also, E2F5 itself binds to the promoters of ID1 and HO-1 (heme oxygenase 1), both of which are inhibitors of terminal muscle cell differentiation resulting from their decreased expression (64). Therefore, miR-98 regulates myoblast differentiation via E2F5, which itself regulates ID1 and HO-1 (Figure 2).
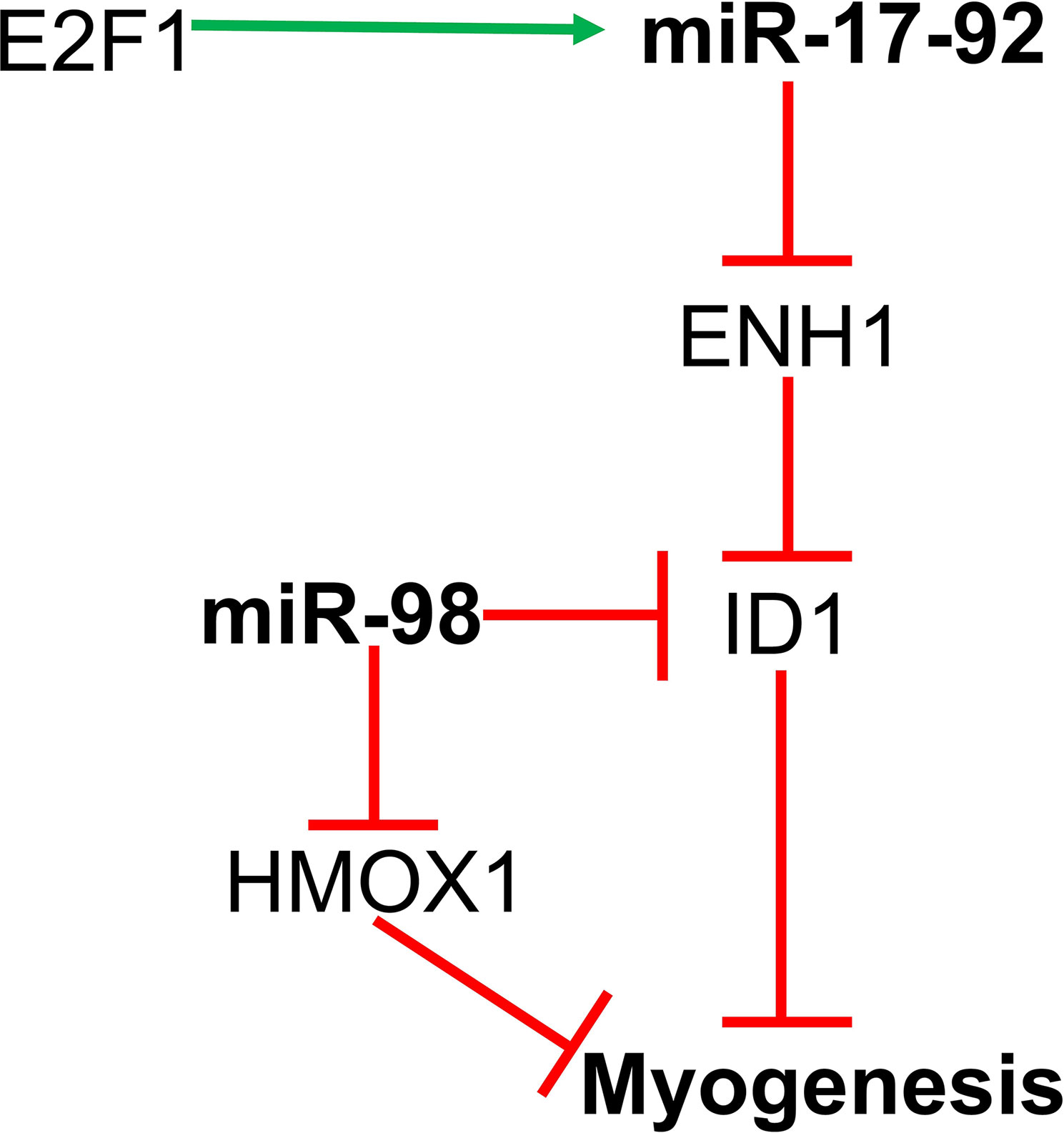
Figure 2 MiR-17-92 and miR-98 regulates myoblast proliferation and differentiation by targeting the ENH1/Id1 signaling axis.
Expression of miR-22 is up-regulated during C2C12 myoblast differentiation. MiR-22 regulates muscle differentiation by targeting transforming growth factor β receptor 1 (TGFβ-R1). TGFβ-R1 is an important receptor of the TGF-β/SMADSMAD signaling pathway. Overexpression of miR-22 in C2C12 down-regulates expression of TGFβ-R1 and promotes myoblast differentiation into myotubes; whereas, inhibition of miR-22 suppresses myoblast differentiation and promotes proliferation. Transforming Growth Factor β-1 (TGF-β1) could also regulate the expression of miR-22 in C2C12 cells as treatment of C2C12 cells with TGF-β1 decreases the level of miR-22, highlighting a significant role of TGF-β1/SMAD3 signaling in myogenic differentiation (56).
MiR-24 is another miRNA involved in myogenic differentiation. Overexpression of miR-24 in C2C12 myoblasts significantly promotes differentiation and myotube formation, and differentiation is suppressed upon treatment with TGF-β1. Recently, a report by Sun et al., suggested that miR-24 may regulate TGF-β/SMAD signaling by targeting SMAD2 (58). MiR-26 is another non-myomiR that regulates myoblast differentiation by targeting TGF-β/SMAD signaling. MiR-26 also targets the BNP (bone morphogenetic protein) of the SMAD pathway, which regulates myoblast differentiation. TGF-β/BMP signaling helps to maintain a pool of muscle stem cells by preventing myogenic differentiation.
SMAD1 and SMAD4 are direct targets of miR-26 and overexpression of miR-26 promotes differentiation of myoblasts, while miR-26 inhibition suppresses myoblast differentiation. Thus, miR-26 regulates myoblast differentiation through the TGF-β/BNP signaling pathway (60). Additionally, miR-26 targets the histone methyltransferase Ezh2 (enhancer of zeste homologue 2) and enhances differentiation of C2C12 myoblasts (87). MiR-146b is another miRNA involved in the regulation of myoblast differentiation (88). A study by Khanna et al. showed that SMAD4, Notch1, and Hmga2 are direct targets of miR-146b.
As discussed above, SMAD4 is involved in the TGFβ/BMP pathway and regulates myoblast proliferation and differentiation (89). Furthermore, Notch signaling is known to control satellite cell quiescence, and inhibition of Notch by miR-146 promotes myoblast differentiation (90). Similarly, inhibition of Hmga2 by miR-146b also promotes myoblast differentiation as Hmga2 is a key regulator of satellite cell activation and proliferation (91). Hence, miR-146b enhances myogenic differentiation through multiple targets. MiR-431 is another microRNA targeting SMAD4, which, in turn, regulates TGFβ signaling. MiR-431 is shown to be present at lower levels in the skeletal muscle of aged mice compared to young mice. Also, increased expression of miR-431 in an in vivo model of muscle regeneration significantly enhances regeneration and reduces SMAD4 levels; thereby, increasing myogenic differentiation (81).
Two miRNAs, miR-675-3p and miR675-5p, are induced during skeletal muscle differentiation. These miRNAs are encoded by exon one of the long non-coding RNA H19. H19 is known to regulate myoblast differentiation and regeneration because its knockdown in myoblasts decreases their differentiation and regeneration potential. H-19-deficient mice show decreased muscle regeneration and differentiation, which is rescued by the re-introduction of miR-675-3p and miR-675-5p. SMAD1 and SMAD5 transcription factors and the DNA replication initiation factor Cdc6 are direct targets of miR-675-3p and miR-675-5p and these miRNAs decrease expression of SMAD transcription factors involved in the anti-differentiation BMP signaling pathway (Figure 3) (92).
Expression of miR-128 is significantly up-regulated during myoblast differentiation in skeletal muscle. IR (insulin receptor), IRS-1 (insulin receptor substrate 1), and PI3K-R1 (phosphatidylinositol 3-kinases receptor 1) are the important genes involved in the insulin signaling pathway. MiR-128 targets all three of these proteins and decreases their expression at the mRNA and protein levels. As reported by Motohashi et al., tumor necrosis factor α (TNF-α) negatively regulates miR-128 and promotes myoblast proliferation and myotube hypertrophy by reducing the expression of miR-128 in myoblasts as well as myotubes (66, 93). In addition, bovine muscle miR-128 regulates myogenic differentiation through Sp1, which is an activator of MyoD and suppressor of CDKN1A (94). A study by Shi et al. showed that miR-128 also promotes myoblast differentiation and inhibits proliferation by targeting the coding domain sequence (CDS) region of myostatin, which inhibits myogenesis. Overexpression of miR-128 promotes myotube formation in C2C12 myoblasts and decreases proliferation (93) (Figure 4).
MiR-29 regulates myoblast differentiation by directly targeting YY1 (Yin Yang 1)—a ubiquitously expressed transcription factor that interacts with RYBP. YY1 recruits histone methyltransferase Ezh2 and histone deacetylase 1 (HDAC) upon activation by NF‐κB, which silences the expression of miR-29. At the beginning of myogenesis, decreased NF-κB signaling down-regulates YY1 and the repressive complex of YY1-Ezh2-HDAC is replaced by an activator complex containing MyoD and SRF. This results in increased expression of miR-29, which represses YY1 and RYBP expression and facilitates myogenesis as well as increasing differentiation of myoblasts into myotubes (61, 62). Another microRNA that targets YY1 and regulates myoblast proliferation and differentiation is miR-34c. Wang et al. showed that miR-34 is highly-expressed in differentiating cells and promotes myoblast differentiation as well as suppressing proliferation by directly targeting the YY1 transcription factor (63) (Figure 5).
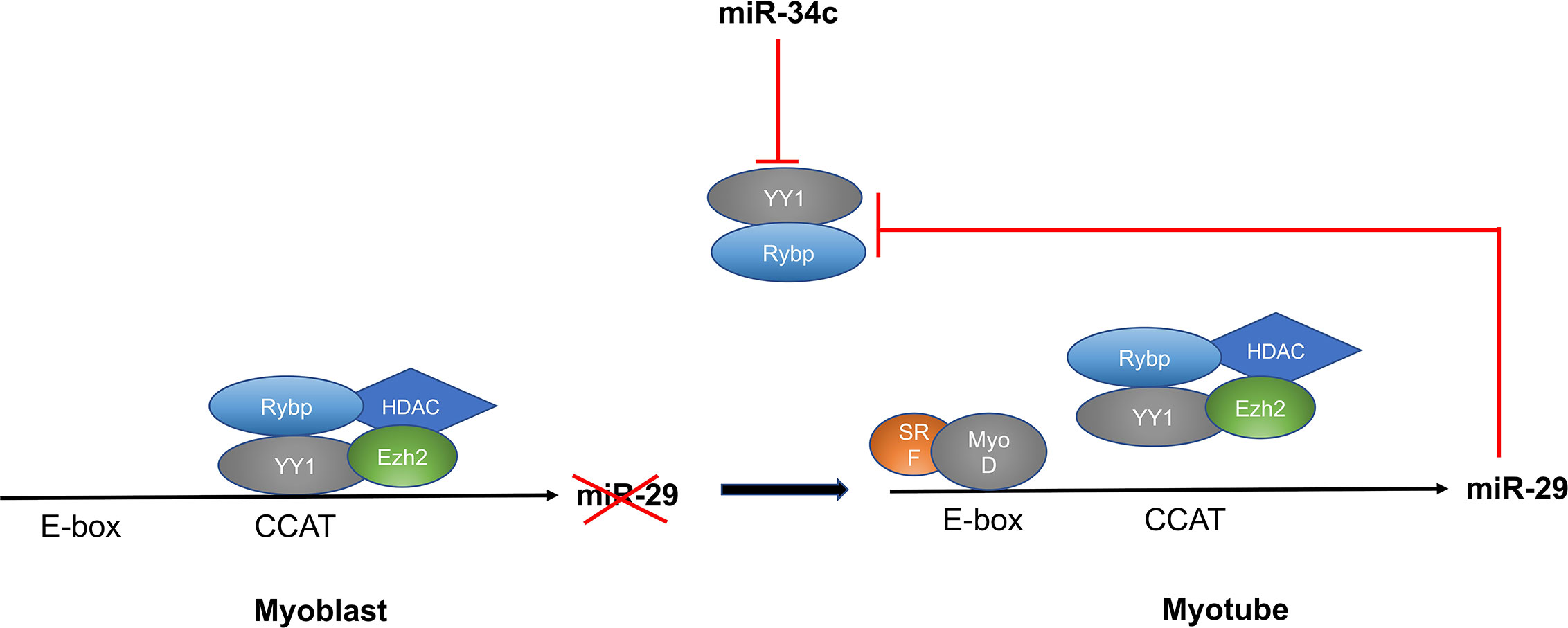
Figure 5 MiR-29 and miR-34c targets YY1 and regulates myoblast differentiation through the NF-κB/YY1 signaling axis.
Other important non-myomiRs include members of the miR-15 family miR-195 and miR-497, which are involved in maintaining the quiescent state of satellite cells by targeting the cell cycle activators Ccnd2, Cdc25, cyclin D1, and cyclin D2 (49, 50). MiR-195/497 also plays a role in regulating myoblast development. MiR-195/497 promotes myoblast proliferation by directly targeting IGF‐1R and IR genes and, during muscle growth and differentiation, the expression of miR-195 and miR-497 was significantly up-regulated. Also, expression of miR-195/497 was negatively regulated by NF-κB in C2C12 myoblasts and mouse skeletal muscle. Previous studies showed that NF-κB is constitutively increased in myoblasts and, after their differentiation, its expression is decreased (95). Taken together, these results show the role of NF-κB/IGF-1R/IR signaling in regulation of proliferation in myoblasts (50).
MiR‐139 is significantly increased during proliferation of C2C12 myoblasts and its ectopic introduction decreases their differentiation. The Wnt/β-catenin signaling pathway is an important pathway in myogenesis. MiR-139-5p binds to the 3’UTR region of Wnt-1, a critical regulator of Wnt/β-catenin signaling pathway. Inhibition of Wnt-1 leads to up-regulation of glycogen synthase kinase 3β (GSK-3β) and down-regulation of phospho-GSK-3β, β-catenin, and nuclear β-catenin, leading to decreased activation of downstream genes involved in myogenesis (67, 68). Another non-myomiR involved in regulation of myogenesis through the Wnt signaling pathway is miR-145-5p, which is up-regulated during C2C12 myoblast differentiation. The target of miR-145 is not yet clear but overexpression of miR-145 enhances the expression of genes involved in the endogenous Wnt signaling pathway like Wnt5a, LRP5, Axin2, and β-catenin and promotes myoblast differentiation (69).
MiR‐199a‐3p negatively regulates differentiation of C2C12 myoblasts by directly targeting members of the IGF‐1/AKT/mTOR signaling pathway, which includes IGF-1 and mTOR. This pathway plays a critical role in muscle development and IGF-1 activates both ERK1/2 and PI3K/Akt/mTOR pathways upon binding to its receptor IGF-R1. ERK1/2 regulates myoblast proliferation (96) and IGF‐1/AKT/mTOR signaling aids in protein synthesis, myotube formation, and muscle hypertrophy (97). Expression of miR-199-3p is also regulated by TGF-β, which is a well-known inhibitor of skeletal muscle differentiation (98). Overexpression and inhibition studies confirmed the role of miR-199-3p in regulating IGF‐1/AKT/mTOR signaling as overexpression of miR-199-3p decreases expression of IGF and mTOR in C2C12 myoblasts, whereas inhibition of miR-199-3p rescues IGF-1 and mTOR. Also, overexpression of miR-199-3p decreases the phosphorylation levels of AKT and mTOR, which confirms the role of IGF‐1/AKT/mTOR signaling in miR-199-3p-mediated down-regulation of differentiation in myoblasts (74).
Apart from the role of miRNAs in humans many miRNAs have been reported in other species including chickens, goats, and cattle—highlighting the importance of microRNAs in muscle development across species. Recently, Yin et al., explored the role of miR-9-5p in muscle development. MiR-9-5p is abundantly expressed in chicken skeletal muscles and regulates the proliferation and differentiation of myoblasts via the PI3K/Akt signaling pathway. This study also reports that insulin-like growth factor 2 mRNA-binding protein 3 (IGF2-BP3), is a direct target of miR-9-5p and IGF2-BP3 is important activator of IGF-2, which activates AKT and promotes the proliferation and differentiation of satellite cells. Overexpression of miR-9-5p inhibits satellite cell proliferation and differentiation (46).
Another microRNA, miR-16-5p, was found to be associated with skeletal muscle growth in chickens. MiR-16-5p directly targets the 3’UTR of SESN1, which is associated with increased proliferation and differentiation of myoblasts, along with decreased apoptosis. Also, SESN1 regulates the p53 pathway through a feedback mechanism. MiR-16-5p targets SESN1 and, thereby, regulates the p53 pathway and promotes apoptosis, along with decreasing proliferation and differentiation of myoblasts (53).
MiR-99a-5p is also reported to regulate proliferation and differentiation of satellite cells in chickens. MiR-99a-5p targets the 3’ UTR region of myotubularin-related protein 3 (MTMR3) and knockdown of MTMR3 by miR-99a-5p promotes proliferation and inhibits differentiation of chicken satellite cells (99). Recently, miR-146b-3p was shown to regulate myogenesis in the chicken. AKT1 and MDFIC are the direct targets of miR-146b-3p and overexpression of miR-146b-3p decreases the expression of AKT1 and MDFIC; thereby, inhibiting PI3K/AKT activation, resulting in decreased proliferation and differentiation as well as increased apoptosis of myoblasts in the chicken (29).
MiRNAs have also been investigated for their role in goat skeletal muscle development. Recently, miR-125b was reported to target and inhibit the expression of insulin-like growth factor 2 (IGF-2) and inhibit differentiation of SMSC in this species. In the same study, the authors found that the long non-coding RNA lncR-125b directly targets miR-125b and acts as a molecular sponge for miR-125b, which decreases its expression and promotes myoblast differentiation and myotube formation (100). MiR-101a was also shown to promote goat satellite cell differentiation by targeting Ezh2 (101). In other studies, miR-487b-3p was shown to play a significant role in myogenesis in goats and overexpression of miR-487b-3p decreases C2C12 myoblast proliferation and differentiation. IRS1 is a direct target of miR-487b-3p and it is a critical regulator of PI3K/AKT and MAPK/ERK signaling pathways. Consequently, inhibition of IRS1 expression by miR-487b-3p results in decreased PI3K/AKT and MAPK/ERK signaling and inhibition of myoblast proliferation and differentiation (102).
The role of microRNAs in skeletal muscle development in cattle is also well explored. Recently, miR-885 was shown to promote proliferation and inhibit differentiation in these animals. MiR-885 directly binds to the 3’UTR region of MyoD1 and overexpression of miR-885 in primary bovine myoblasts upregulates proliferation and suppresses differentiation (103). Another microRNA, miR-216a, was shown to suppress myoblast differentiation and proliferation in primary bovine myoblasts by targeting SNIP1 and SMAD7 (104). MiR-148a-3p was also shown to regulate myoblast proliferation and apoptosis in bovine muscle cells.
KLF6 is the direct target of miR-148a-3p and its overexpression inhibits myoblast proliferation and promotes apoptosis, while knock-down of miR-148a-3p has the opposite effect (105). MiR-107 is another microRNA reported to be highly expressed in bovine myoblasts that inhibits differentiation and apoptosis by targeting Wnt3a. In the same study, the circular RNA circFGFR4 was shown to inhibit expression of miR-107, which promotes myoblast differentiation (106). MiR-378 also plays a role in bovine skeletal muscle development as it is highly expressed in differentiating satellite cells by directly targeting the 3’UTR regions of POLA2 (52). Zhang et al., reported that miR-2400 promotes bovine skeletal muscle satellite cell proliferation by targeting myogenin (107) and miR-2425-5p also targeted myogenin and RAD9A (RAD9 homolog A) to promote proliferation and suppress differentiation (52).
In summary, miRNAs are highly conserved and exploring the role of these molecules in a variety of species may advance our understanding of miRNA-facilitated regulation in various physiological and pathophysiological processes.
Although the role of miRNAs in skeletal muscle cell regulation and development is well documented; a better understanding their function in muscle will provide important insights about the regulation of gene expression in health and disease. This section of the review will discuss the current understanding of miRNA regulation in skeletal muscle and their role in myopathies including sarcopenia and Duchenne muscular dystrophy (DMD). We will also discuss how miRNAs are involved in cancer cachexia, a muscle wasting syndrome associated with cancer progression.
As we age, there is gradual loss of muscle mass, function, and strength along with reduced regenerative capacity and protein synthesis. This age-associated muscle wasting is termed sarcopenia (108, 109) and miRNAs are emerging as important regulators of skeletal muscle development and homeostasis during aging.
MiRNAs can be used as biomarkers or therapeutic targets for muscle wasting as a consequence of age or disease. Several microarray-based and sequencing studies using muscle from young and old animal models and humans highlights the importance of dysregulated microRNAs in age-related muscle wasting. A study by Drummond et al. performed microarray analyses using muscles from young and old humans and found 18 differentially expressed miRNAs. In muscle samples from older individuals, eight microRNAs were significantly up-regulated (let-7a, -b, -e, and -f, as well as miR-25, miR-98, miR-195, and miR-1268) and ten microRNAs were significantly down-regulated (miR-22, miR-24, miR-27a, miR-27b, miR-30d, miR-133a, miR-133b, miR-223, miR-378, and miR-378*) when compared to young individuals. Expression of let-7b and let-7e were validated using qRT-PCR along with the expression of cell cycle regulators (CDK6, CDC25A, and CDC34) and PAX7 expression. Expression of CDK6, CDC25A, CDC34, and PAX7 was significantly lower in muscle biopsies from older individuals, suggesting the involvement let7 in muscle cell renewal and regeneration through the regulation of the cell cycle (110).
Another large-scale miRNA screening study investigated the role of resistance exercise on miRNA expression in skeletal muscle biopsies from young and old individuals. It was found that 26 miRNAs were differentially expressed in older muscle when compared to young. Nine miRNAs from the miR99/100 family were predicted to target the Akt-mTOR signaling pathway—highlighting the role of this family in regulating skeletal muscle mass during aging. Also, apart from miR-499a, there was no change in the expression levels of myomiRs (miR-1, miR-133a, miR-133b, miR-206, and miR-486-5p) in old individuals when compared to young individuals (111).
Rivas et al. conducted expression profiling of miRNAs in young and old men following an acute bout of resistance exercise and found that 21 miRNAs were altered in young men with no change in miRNA expression in older men. MiR-126 expression was significantly decreased in young men compared to older men after resistance exercise. Overexpression and inhibition studies in C2C12 myoblasts showed miR-126 regulates the expression of Foxo1, MyoD, Myf5, and IGF-1. Therefore, regulation of miR-126 in older patients might improve the function of skeletal muscle in aged individuals (112).
Kim et al. reported that 34 miRNAs (15 up-regulated and 19 down-regulated) were dysregulated in genome-wide miRNA profiles of gastrocnemius muscle biopsies from young and old mice. Using qRT-PCR, up-regulated miRNAs (miR-34a-5p, miR-146a-5p, miR-92b-3p, miR-155-5p, and miR-203-3p) and down-regulated miRNAs (miR-337-3p*, miR-434-3p, miR-434-5p*, miR-136-5p, and miR-148a-3p) were validated (113). In addition, miRNA profiling using a TaqMan miRNA array of quadricep muscles from young and old mice showed 57 miRNAs were differentially altered (36 down-regulated and 21 miRNAs were up-regulated.). Validation of up-regulated miRNAs (miR-7, miR-542, miR-486, and miR-686) and down-regulated miRNAs (miR-181, miR-434, miR-455, and miR-221) were also performed using qRT-PCR (114).
MiRNA profiling using next generation sequencing (NGS) of skeletal muscle tissue from young and old Rhesus monkeys was also performed. It was found that 35 miRNAs were differentially expressed in skeletal muscles of old versus young monkeys. Most of miRNAs were up-regulated, with only five miRNAs down-regulated, including miR-181a and miR-181b in old monkeys compared to young monkeys (115). In another study, miRNA profiling using NGS in myoblasts isolated from hind limb muscles of young and old mice found that miR-431 expression is significantly reduced in aged myoblasts. Overexpression of miR-431 in aged myoblasts improved regeneration, and inhibition of miR-431 also prevented myogenesis. MiR-431 directly targets the 3’UTR of SMAD4 and effects TGF-β signaling. In aged myoblasts, SMAD4 is up-regulated, resulting in increased TGF-β signaling (an inhibitor of myogenesis), signifying miR-431 might play an important role in regulating the myogenic potential of aged myoblasts (81).
Hu et al. reported that expression of miR-29 is significantly up-regulated in the muscles of aged rodents compared to young rodents. MiR-29 targets mediators of myoblast proliferation (e.g. p85a, IGF-1, and B-Myb). Overexpression of miR-29 in muscle progenitor cells leads to reduced proliferation and increased senescence. Also, injection of miR-29 into the muscles of young mice resulted in an aged muscle phenotype (116). Another microRNA miR-195 was up-regulated in myoblasts isolated from aged mice compared to young mice. MiR-195 targets the anti-senescence proteins Sirtuin1 (SIRT1), which promotes senescence. Blockade of miR-195 significantly up-regulated the reprograming potential of old myoblasts. Blocking miR-195 in induced pluripotent stem cells derived from old myoblasts does not affect pluripotency in transformed myoblasts, providing evidence that iPSCs with expression of miR-195 blocked, have the potential to be transplanted in elderly patients (117).
Recently, a study identified the role of miR-658-5p in regulating expression of the striated muscle activator of Rho signaling (STARS) in young and old individuals following acute resistance exercise. MiR-658-5p expression was significantly up-regulated in older subjects as compared to young subjects. MiR-658-5p targets the 3’UTR of STARS mRNA and, therefore, decreases the expression of STARS in old subjects compared to young (118). MiR-143 is significantly decreased in satellite cells during aging and it is proposed to target the 3’UTR of the insulin growth factor-binding protein 5 (Igfbp5). The authors demonstrated that miR-143-3p:IGF-BP5 interactions play an vital role in age-associated changes in satellite cells (119).
Duchenne muscular dystrophy (DMD) is progressive muscle wasting X-linked recessive genetic disorder caused by out of frame mutations in the dystrophin gene resulting in severe cellular and molecular changes. MiRNAs involved in the pathological pathways activated in skeletal muscle damage and regeneration are exhibited to hasten and intensify the dystrophic phenotype (19). MiRNAs can be used as a biomarker for the early detection as well as a therapeutic target in the treatment of DMD.
MyomiRs (miR-1, miR-31, miR-133, miR-206) are also known as dystromirs as these microRNAs, which are involved in myogenesis, are also linked to muscular dystrophies. These dystromirs show potential as biomarkers as they are highly-expressed in the serum of DMD patients and are correlated with disease severity (116, 120–122). MiR-206 is a skeletal muscle-specific miRNA shown to play an important role in skeletal muscle regeneration in response to injury. Mice lacking miR-206 show hindered skeletal muscle regenerative capability following cardiotoxin injury. MiR-206 expression is significantly higher in the mdx mouse model of DMD and plays a protective role by slowing progression of DMD in these mice because miR-206 promotes satellite cell differentiation and the formation of new myofibers (123).
MiR-486 is another muscle-specific microRNA involved in myogenic differentiation. Expression of miR-486 is significantly down-regulated in DMD patients as well as in mdx mice. MiR-486 targets dedicator of cytokinesis 3 (DOCK3) whose expression is elevated in dystrophic muscle and regulates the PTEN/AKT signaling pathway by decreasing expression of PTEN, resulting in increased phosphorylated Akt. Overexpression of miR-486 in mdx mice improves sarcolemmal integrity and performance, enhances fiber size, and decreases nuclear centralization (124).
MiR-199a-5p is also significantly down-regulated in dystrophin-deficient zebrafish, mdx mice, and DMD human muscle biopsies. MiR-199a-5p regulates myoblast differentiation by controlling various factors involved in WNT signaling such as FZD4, JAG1, and WNT2 (75). MiR-499 is reported as a serum biomarker in both human patients and in the Golden Retriever model of DMD (121, 125, 126). Another report by Liu et al. showed that reinstatement of miR-499 in mdx mice reduces the severity of DMD. MiR-499 targets Fnip1, a well-known negative regulator of AMPK, which is an activator of PGC‐1α. Inhibition of Fnip1 in myocytes activates AMPK/PGC‐1α signaling and mitochondrial function (127).
MiR-127 is a miRNA predominantly expressed in skeletal muscle and its overexpression promotes myoblast differentiation in C2C12 cells. Moreover, in transgenic mice overexpressing miR-127 there was an increase in satellite cell differentiation and enhanced muscle regeneration compared with wild-type mice. MiR-127 promotes differentiation by targeting the gene encoding sphingosine-1-phosphate receptor 3 (S1PR3), a G-protein-coupled receptor for sphingosine-1-phosphate. The same study showed that overexpression of miR-127 in mdx mice significantly ameliorates disease severity by promoting satellite cell differentiation (128).
Another miRNA, miR-200c, shows significantly increased expression in mdx mice. MiR-200c was reported to regulate myoblast differentiation as overexpression of miR-200c inhibits myoblast differentiation. MiR-200c was also reported to enhance ROS production and phosphorylation of p66Shc in Ser-36 by targeting FOXO1 and eNOS; thereby, down-regulating skeletal muscle differentiation. Increased expression of miR-200c in mdx mice might be responsible decreased differentiation and enhanced muscle wasting and myotube loss in mdx mice (129).
Another important feature of DMD is fibrosis and collagen deposition. It is shown that decreased expression of miR-29 in mdx mice promotes fibrosis as inhibition of miR-29 in myoblasts promotes differentiation of myoblasts into myofibroblasts. MiR-29, apart from promoting myoblast differentiation via Akt3/NFkB/YY1 signaling, also decreases fibrosis by targeting fibrotic genes (COL3A1, FBN1, and COL1A1). In mdx mice, miR-29 is significantly down-regulated, along with increased muscle fibrosis and restoration of miR-29 in mdx mice by intramuscular and intravenous injection, improved the dystrophic pathology by promoting muscle regeneration and decreasing fibrosis (130). In mdx mice, muscle strength was completely restored when both mir-29 and micro-dystrophin were administered together (131).
MiR-21 is another miRNA that is significantly increased in mdx mice, and its expression is significantly correlated with increased expression of fibrotic genes such as COL1A1 and COL6A. Mir-21 expression is also increased in DMD fibroblasts and further enhanced by TGF-β1 treatment. MiR-21 directly targets the 3’UTR of PTEN (phosphatase and tensin homolog deleted on chromosome 10) and SPRY-1 (Sprouty homolog 1). Expression of PTEN and SPRY-1 was reduced significantly in DMD fibroblasts along with increased expression of COL1A1 and COL6A. Inhibition of miR-21 in mdx mice as well as in DMD fibroblasts reinstates expression of PTEN and SPRY-1 along with decreases in the expression of COL1A1 and COL6A; thereby, improving the disease phenotype (132). Recently, Daria et al. explored the role of synthetic preimplantation factor (sPIF) in regulating miR-21 in DMD. In DMD patient-derived myoblasts, sPIF overexpression promotes myoblast differentiation and decreases expression of COL1A1, COL1A2, and TGF-β, along with increases in the expression of the dystrophin homolog, utrophin protein. It was reported that sPIF mediated the protective effects in DMD myoblasts by upregulating lncRNA H19 and miR-675, and downregulating let-7 and miR-21 (133).
Another microRNA studied extensively in DMD is miR-31. MiR-31 maintains satellite cells quiescence by directly targeting Myf5. An additional target of miR-31 is dystrophin, and its expression is significantly up-regulated in mdx mice along with increased expression in human DMD biopsies compared to healthy controls. Human DMD myoblasts treated with antisense oligonucleotides to induce exon skipping showed greater dystrophin rescue upon inhibition of miR-31 (122). Also, inhibition of miR-31 using CRISPR/Cas9 editing in myotubes derived from induced pluripotent stem cells (iPSCs) of DMD patients showed restoration of dystrophin (134).
Another important syndrome associated with muscle wasting is cachexia. It is a multifactorial syndrome that leads to significant loss of skeletal muscle mass with or without the loss of fat mass (135–137). Cachexia is associated with the pathology of many diseases including cancer. Cancer-associated cachexia (CAX) affects 50–80% of cancer patients and is linked to 20–40% of patient mortality (138–140). Recently studies have highlighted the role of miRNAs in the pathophysiology of cancer cachexia.
A report by Mubaid et al. highlights the role of the RNA binding protein HuR in cancer cachexia. HuR plays a dual role as a promoter of muscle fiber formation and an inducer of muscle loss by interacting with the STAT3 mRNA. Activated STAT3 promotes transcription of STAT3-dependent pro-cachectic genes, resulting in increased muscle wasting. HuR binds to the 3’UTR of the STAT3mRNA and prevents binding of miR-300 to the 3’UTR of STAT3, resulting in increased expression of STAT3, which leads to augmented muscle wasting (141).
MiR-203 was significantly up-regulated in colorectal cancer (CRC) patients when compared to control subjects, and miR-203 levels were correlated with the incidence of myopenia in CRC. Overexpression and inhibition studies of miR-203 in human skeletal muscle cells (SkMCs) identified BIRC5 (Survivin) as a target of miR‐203 in skeletal muscle cells. BIRC5 is well known to inhibit apoptosis through inhibition of caspases 3, 7, and 9. Therefore, mir-203 promotes myopenia in CRC by inhibiting miR-203 and promoting apoptosis and muscle wasting (142).
In another study, Wei et al. showed severe cachexia in muscle cells from Lewis lung carcinoma (LLC) tumor-bearing mice compared with control mice. LLC tumor-bearing mice secrete significantly increased amounts of miR-21 through micro vesicles (MVs) and promotes apoptosis of skeletal muscle cells. MiR-21 encourages apoptosis via c-Jun N-terminal kinase (JNK) signaling following activation of TLR-7 receptors present on myoblasts. Moreover, there is significant increase of apoptotic muscle cell along with decreased muscle nuclei resulting in muscle wasting.
In colorectal cancer (CRC) patients, miR-21 expression was significantly increased in both tissue and serum of CRC patients compared to control subjects. Increased expression of miR-21 was negatively correlated with decreased psoas muscle mass index (PMI) of CRC patients (143). Finally, a recent study by Wouter et al. aimed to identify differentially expressed microRNAs in cachectic non-small cell lung cancer (NSCLC) patients. Using miRNA arrays, the authors reported that five miRNAs were significantly increased and 23 were significantly decreased in NSCLC patients compared to controls (144).
Over the last decade, numerous studies have highlighted the importance of miRNAs as critical regulators of myogenesis along with playing a significant role in muscle-associated diseases like DMD and sarcopenia. It is quite interesting how the same transcription factors that regulate muscle-associated protein-coding genes also control the expression of muscle-specific miRNAs, suggesting interplay between miRNAs, protein-coding genes, and transcription factors. Tremendous progress has been made in recognizing muscle-specific miRNAs and their associated functional mechanisms in myogenesis. Still, there is a need to further explore the role these miRNAs in muscle-associated myopathies including DMD, sarcopenia, and cancer cachexia.
In this review, we have also discussed various molecular mechanisms associated with muscle-associated miRNAs. Attempts to unravel the function of these miRNAs not only improves our knowledge about miRNAs in muscle development and homeostasis, but may facilitate the development of miRNA-based therapies in muscle-related diseases, including DMD, sarcopenia and cancer cachexia. In the near future, microRNAs have significant potential as diagnostic, prognostic, or therapeutic tools for skeletal muscle-associated pathologies.
Despite this potential, there are substantial hurdles to address before miRNAs can be used as a therapeutic or diagnostic tool. As a therapeutics targeting miRNAs may ameliorate the symptoms of muscle-associated myopathies, the heterogeneity of miRNA expression, target specificity, and off target effects could hamper the prospects for miRNA-based therapeutics. Moreover, microRNAs may behave in a tissue-specific manner and miRNAs with opposite effects in different tissues would need to be carefully scrutinized before they can be considered for therapeutic use. Also, miRNAs often exist as a families and may contain many miRNAs with similar targets and functions. So, silencing of an individual miRNA might be rescued by other miRNAs with similar or overlapping functions. Apart from identifying targets for miRNA-based therapeutics, the development of an appropriate and stable delivery system needs to be addressed.
In conclusion, miRNAs have been established as key regulators of muscle development and function. There is ample evidence for the contribution of miRNAs in muscle development including differentiation, proliferation, and regeneration. In many skeletal muscle-associated diseases, including DMD, sarcopenia, and cancer cachexia, dysregulation of miRNAs have been reported. Altered levels of miRNAs might serve as useful diagnostic tools for early detection of myopathies. Unlike most RNAs, miRNAs are quite stable in most body fluids and abnormal expression of miRNAs in the circulation may serve as an important way to detect muscle diseases. Understanding the target genes associated with muscle miRNAs could offer useful insights into the regulation and function of muscle-related signaling pathways. MiRNAs play a significant role in skeletal muscle development and pathophysiology and may provide a means for the development of new therapeutics to treat muscle disorders.
GS: Review conception, design, manuscript writing and preparation. D-ZW: review conception, design and editing. DC: manuscript editing. All authors contributed to the article and approved the submitted version.
Research in the D-ZW laboratory is supported by the NIH (HL125925 and HL093242).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank former and current D-ZW lab members for their contribution and helpful discussions. The authors claim there is no conflict to be disclosed.
1. Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev (2006) 16(5):525–32. doi: 10.1016/j.gde.2006.08.008
2. Güller I, Russell AP. MicroRNAs in skeletal muscle: their role and regulation in development, disease and function. J Physiol (2010) 588(Pt 21):4075–87. doi: 10.1113/jphysiol.2010.194175
3. Paul PK, Bhatnagar S, Mishra V, Srivastava S, Darnay BG, Choi Y, et al. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol Cell Biol (2012) 32(7):1248–59. doi: 10.1128/MCB.06351-11
4. McGregor RA, Poppitt SD, Cameron-Smith D. Role of microRNAs in the age-related changes in skeletal muscle and diet or exercise interventions to promote healthy aging in humans. Ageing Res Rev (2014) 17:25–33. doi: 10.1016/j.arr.2014.05.001
5. Nystrom G, Pruznak A, Huber D, Frost RA, Lang CH. Local insulin-like growth factor I prevents sepsis-induced muscle atrophy. Metabolism. (2009) 58(6):787–97. doi: 10.1016/j.metabol.2009.01.015
6. Wang H, Lai YJ, Chan YL, Li TL, Wu CJ. Epigallocatechin-3-gallate effectively attenuates skeletal muscle atrophy caused by cancer cachexia. Cancer Lett (2011) 305(1):40–9. doi: 10.1016/j.canlet.2011.02.023
7. Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature (2004) 432(7014):231–5. doi: 10.1038/nature03049
8. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol (2014) 15(8):509–24. doi: 10.1038/nrm3838
9. Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev (2003) 17(24):3011–6. doi: 10.1101/gad.1158803
10. Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science (2001) 294(5543):858–62. doi: 10.1126/science.1065062
11. Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature (2005) 436(7051):740–4. doi: 10.1038/nature03868
12. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet (2008) 9(2):102–14. doi: 10.1038/nrg2290
13. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet (2004) 5(7):522–31. doi: 10.1038/nrg1379
14. O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) (2018) 9:402. doi: 10.3389/fendo.2018.00402
15. Buckingham M. Skeletal muscle formation in vertebrates. Curr Opin Genet Dev (2001) 11(4):440–8. doi: 10.1016/S0959-437X(00)00215-X
16. Zhang S, Chen N. Regulatory Role of MicroRNAs in Muscle Atrophy during Exercise Intervention. Int J Mol Sci (2018) 19(2):405. doi: 10.3390/ijms19020405
17. Gasparini P, Ferrari A, Casanova M, Limido F, Massimino M, Sozzi G, et al. MiRNAs as Players in Rhabdomyosarcoma Development. Int J Mol Sci (2019) 20(22):5818. doi: 10.3390/ijms20225818
18. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer (2006) 6(11):857–66. doi: 10.1038/nrc1997
19. Diniz GP, Wang DZ. Regulation of Skeletal Muscle by microRNAs. Compr Physiol (2016) 6(3):1279–94. doi: 10.1002/cphy.c150041
20. O’Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, et al. Essential role for Dicer during skeletal muscle development. Dev Biol (2007) 311(2):359–68. doi: 10.1016/j.ydbio.2007.08.032
21. Zilahi E, Adamecz Z, Bodoki L, Griger Z, Póliska S, Nagy-Vincze M, et al. Dysregulated expression profile of myomiRs in the skeletal muscle of patients with polymyositis. Ejifcc (2019) 30(2):237–45.
22. Lu L, Zhou L, Chen EZ, Sun K, Jiang P, Wang L, et al. A Novel YY1-miR-1 regulatory circuit in skeletal myogenesis revealed by genome-wide prediction of YY1-miRNA network. PloS One (2012) 7(2):e27596. doi: 10.1371/journal.pone.0027596
23. Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet (2006) 38(2):228–33. doi: 10.1038/ng1725
24. Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol (2010) 190(5):867–79. doi: 10.1083/jcb.200911036
25. Ma G, Wang Y, Li Y, Cui L, Zhao Y, Zhao B, et al. MiR-206, a key modulator of skeletal muscle development and disease. Int J Biol Sci (2015) 11(3):345–52. doi: 10.7150/ijbs.10921
26. Yang Y, Sun W, Wang R, Lei C, Zhou R, Tang Z, et al. Wnt antagonist, secreted frizzled-related protein 1, is involved in prenatal skeletal muscle development and is a target of miRNA-1/206 in pigs. BMC Mol Biol (2015) 16:4. doi: 10.1186/s12867-015-0035-7
27. Chen X, Wang K, Chen J, Guo J, Yin Y, Cai X, et al. In vitro evidence suggests that miR-133a-mediated regulation of uncoupling protein 2 (UCP2) is an indispensable step in myogenic differentiation. J Biol Chem (2009) 284(8):5362–9. doi: 10.1074/jbc.M807523200
28. Feng Y, Niu LL, Wei W, Zhang WY, Li XY, Cao JH, et al. A feedback circuit between miR-133 and the ERK1/2 pathway involving an exquisite mechanism for regulating myoblast proliferation and differentiation. Cell Death Dis (2013) 4(11):e934. doi: 10.1038/cddis.2013.462
29. Huang W, Guo L, Zhao M, Zhang D, Xu H, Nie Q. The Inhibition on MDFIC and PI3K/AKT Pathway Caused by miR-146b-3p Triggers Suppression of Myoblast Proliferation and Differentiation and Promotion of Apoptosis. Cells. (2019) 8(7):656. doi: 10.3390/cells8070656
30. Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol (2006) 174(5):677–87. doi: 10.1083/jcb.200603008
31. Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol (2006) 175(1):77–85. doi: 10.1083/jcb.200603039
32. van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell (2009) 17(5):662–73. doi: 10.1016/j.devcel.2009.10.013
33. Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol (2011) 31(1):203–14. doi: 10.1128/MCB.01009-10
34. Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A (2007) 104(52):20844–9. doi: 10.1073/pnas.0710558105
35. Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A (2006) 103(23):8721–6. doi: 10.1073/pnas.0602831103
36. Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, et al. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol (2008) 321(2):491–9. doi: 10.1016/j.ydbio.2008.06.019
37. Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res (2006) 34(20):5863–71. doi: 10.1093/nar/gkl743
38. Hirai H, Verma M, Watanabe S, Tastad C, Asakura Y, Asakura A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J Cell Biol (2010) 191(2):347–65. doi: 10.1083/jcb.201006025
39. Gagan J, Dey BK, Layer R, Yan Z, Dutta A. Notch3 and Mef2c proteins are mutually antagonistic via Mkp1 protein and miR-1/206 microRNAs in differentiating myoblasts. J Biol Chem (2012) 287(48):40360–70. doi: 10.1074/jbc.M112.378414
40. Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell (2010) 18(4):510–25. doi: 10.1016/j.devcel.2010.03.010
41. Luo W, Chen J, Li L, Ren X, Cheng T, Lu S, et al. c-Myc inhibits myoblast differentiation and promotes myoblast proliferation and muscle fibre hypertrophy by regulating the expression of its target genes, miRNAs and lincRNAs. Cell Death Differ (2019) 26(3):426–42. doi: 10.1038/s41418-018-0129-0
42. Luo Y, Wu X, Ling Z, Yuan L, Cheng Y, Chen J, et al. microRNA133a targets Foxl2 and promotes differentiation of C2C12 into myogenic progenitor cells. DNA Cell Biol (2015) 34(1):29–36. doi: 10.1089/dna.2014.2522
43. Xu M, Chen X, Chen D, Yu B, Huang Z. FoxO1: a novel insight into its molecular mechanisms in the regulation of skeletal muscle differentiation and fiber type specification. Oncotarget (2017) 8(6):10662–74. doi: 10.18632/oncotarget.12891
44. Xu M, Chen X, Chen D, Yu B, Li M, He J, et al. Regulation of skeletal myogenesis by microRNAs. J Cell Physiol (2020) 235(1):87–104. doi: 10.1002/jcp.28986
45. Gonçalves TJM, Armand AS. Non-coding RNAs in skeletal muscle regeneration. Noncoding RNA Res (2017) 2(1):56–67. doi: 10.1016/j.ncrna.2017.03.003
46. Yin H, He H, Shen X, Zhao J, Cao X, Han S, et al. miR-9-5p Inhibits Skeletal Muscle Satellite Cell Proliferation and Differentiation by Targeting IGF2BP3 through the IGF2-PI3K/Akt Signaling Pathway. Int J Mol Sci (2020) 21(5):1655. doi: 10.3390/ijms21051655
47. Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell (2012) 11(1):118–26. doi: 10.1016/j.stem.2012.03.011
48. Lozano-Velasco E, Vallejo D, Esteban FJ, Doherty C, Hernández-Torres F, Franco D, et al. A Pitx2-MicroRNA Pathway Modulates Cell Proliferation in Myoblasts and Skeletal-Muscle Satellite Cells and Promotes Their Commitment to a Myogenic Cell Fate. Mol Cell Biol (2015) 35(17):2892–909. doi: 10.1128/MCB.00536-15
49. Sato T, Yamamoto T, Sehara-Fujisawa A. miR-195/497 induce postnatal quiescence of skeletal muscle stem cells. Nat Commun (2014) 5:4597. doi: 10.1038/ncomms5597
50. Wei W, Zhang WY, Bai JB, Zhang HX, Zhao YY, Li XY, et al. The NF-κB-modulated microRNAs miR-195 and miR-497 inhibit myoblast proliferation by targeting Igf1r, Insr and cyclin genes. J Cell Sci (2016) 129(1):39–50. doi: 10.1242/jcs.174235
51. Wu R, Li H, Li T, Zhang Y, Zhu D. Myostatin regulates miR-431 expression via the Ras-Mek-Erk signaling pathway. Biochem Biophys Res Commun (2015) 461(2):224–9. doi: 10.1016/j.bbrc.2015.03.150
52. Tong H, Jiang R, Liu T, Wei Y, Li S, Yan Y. bta-miR-378 promote the differentiation of bovine skeletal muscle-derived satellite cells. Gene. (2018) 668:246–51. doi: 10.1016/j.gene.2018.03.102
53. Cai B, Ma M, Chen B, Li Z, Abdalla BA, Nie Q, et al. MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation. Cell Death Dis (2018) 9(3):367. doi: 10.1038/s41419-018-0403-6
54. Qiu H, Liu N, Luo L, Zhong J, Tang Z, Kang K, et al. MicroRNA-17-92 regulates myoblast proliferation and differentiation by targeting the ENH1/Id1 signaling axis. Cell Death Differ (2016) 23(10):1658–69. doi: 10.1038/cdd.2016.56
55. Luo W, Li G, Yi Z, Nie Q, Zhang X. E2F1-miR-20a-5p/20b-5p auto-regulatory feedback loop involved in myoblast proliferation and differentiation. Sci Rep (2016) 6:27904. doi: 10.1038/srep27904
56. Wang H, Zhang Q, Wang B, Wu W, Wei J, Li P, et al. miR-22 regulates C2C12 myoblast proliferation and differentiation by targeting TGFBR1. Eur J Cell Biol (2018) 97(4):257–68. doi: 10.1016/j.ejcb.2018.03.006
57. Wang L, Chen X, Zheng Y, Li F, Lu Z, Chen C, et al. MiR-23a inhibits myogenic differentiation through down regulation of fast myosin heavy chain isoforms. Exp Cell Res (2012) 318(18):2324–34. doi: 10.1016/j.yexcr.2012.06.018
58. Sun Y, Wang H, Li Y, Liu S, Chen J, Ying H. miR-24 and miR-122 Negatively Regulate the Transforming Growth Factor-β/Smad Signaling Pathway in Skeletal Muscle Fibrosis. Mol Ther Nucleic Acids (2018) 11:528–37. doi: 10.1016/j.omtn.2018.04.005
59. Sun Q, Zhang Y, Yang G, Chen X, Zhang Y, Cao G, et al. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res (2008) 36(8):2690–9. doi: 10.1093/nar/gkn032
60. Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem (2008) 283(15):9836–43. doi: 10.1074/jbc.M709614200
61. Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell (2008) 14(5):369–81. doi: 10.1016/j.ccr.2008.10.006
62. Zhou L, Wang L, Lu L, Jiang P, Sun H, Wang H. A novel target of microRNA-29, Ring1 and YY1-binding protein (Rybp), negatively regulates skeletal myogenesis. J Biol Chem (2012) 287(30):25255–65. doi: 10.1074/jbc.M112.357053
63. Wang M, Liu C, Su Y, Zhang K, Zhang Y, Chen M, et al. miRNA-34c inhibits myoblasts proliferation by targeting YY1. Cell Cycle (2017) 16(18):1661–72. doi: 10.1080/15384101.2017.1281479
64. Kropp J, Degerny C, Morozova N, Pontis J, Harel-Bellan A, Polesskaya A. miR-98 delays skeletal muscle differentiation by down-regulating E2F5. Biochem J (2015) 466(1):85–93. doi: 10.1042/BJ20141175
65. Cardinali B, Castellani L, Fasanaro P, Basso A, Alemà S, Martelli F, et al. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PloS One (2009) 4(10):e7607. doi: 10.1371/journal.pone.0007607
66. Motohashi N, Alexander MS, Shimizu-Motohashi Y, Myers JA, Kawahara G, Kunkel LM. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J Cell Sci (2013) 126(Pt 12):2678–91. doi: 10.1242/jcs.119966
67. Mi L, Li Y, Zhang Q, Zhao C, Peng Y, Yang G, et al. MicroRNA-139-5p regulates C2C12 cell myogenesis through blocking Wnt/β-catenin signaling pathway. Biochem Cell Biol (2015) 93(1):8–15. doi: 10.1139/bcb-2014-0079
68. Xu M, Chen X, Huang Z, Chen D, Yu B, Chen H, et al. MicroRNA-139-5p suppresses myosin heavy chain I and IIa expression via inhibition of the calcineurin/NFAT signaling pathway. Biochem Biophys Res Commun (2018) 500(4):930–6. doi: 10.1016/j.bbrc.2018.04.202
69. Du J, Li Q, Shen L, Lei H, Luo J, Liu Y, et al. miR-145a-5p Promotes Myoblast Differentiation. BioMed Res Int (2016) 2016:5276271. doi: 10.1155/2016/5276271
70. Zhang J, Ying ZZ, Tang ZL, Long LQ, Li K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J Biol Chem (2012) 287(25):21093–101. doi: 10.1074/jbc.M111.330381
71. Seok HY, Tatsuguchi M, Callis TE, He A, Pu WT, Wang DZ. miR-155 inhibits expression of the MEF2A protein to repress skeletal muscle differentiation. J Biol Chem (2011) 286(41):35339–46. doi: 10.1074/jbc.M111.273276
72. Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol (2006) 8(3):278–84. doi: 10.1038/ncb1373
73. Antoniou A, Mastroyiannopoulos NP, Uney JB, Phylactou LA. miR-186 inhibits muscle cell differentiation through myogenin regulation. J Biol Chem (2014) 289(7):3923–35. doi: 10.1074/jbc.M113.507343
74. Jia L, Li YF, Wu GF, Song ZY, Lu HZ, Song CC, et al. MiRNA-199a-3p regulates C2C12 myoblast differentiation through IGF-1/AKT/mTOR signal pathway. Int J Mol Sci (2013) 15(1):296–308. doi: 10.3390/ijms15010296
75. Alexander MS, Kawahara G, Motohashi N, Casar JC, Eisenberg I, Myers JA, et al. MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell Death Differ (2013) 20(9):1194–208. doi: 10.1038/cdd.2013.62
76. Luo W, Wu H, Ye Y, Li Z, Hao S, Kong L, et al. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death Dis (2014) 5(7):e1347. doi: 10.1038/cddis.2014.289
77. Ouyang H, Chen X, Li W, Li Z, Nie Q, Zhang X. Circular RNA circSVIL Promotes Myoblast Proliferation and Differentiation by Sponging miR-203 in Chicken. Front Genet (2018) 9:172. doi: 10.3389/fgene.2018.00172
78. Li G, Luo W, Abdalla BA, Ouyang H, Yu J, Hu F, et al. miRNA-223 upregulated by MYOD inhibits myoblast proliferation by repressing IGF2 and facilitates myoblast differentiation by inhibiting ZEB1. Cell Death Dis (2017) 8(10):e3094. doi: 10.1038/cddis.2017.479
79. Sarkar S, Dey BK, Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell (2010) 21(13):2138–49. doi: 10.1091/mbc.e10-01-0062
80. Wei X, Li H, Zhang B, Li C, Dong D, Lan X, et al. miR-378a-3p promotes differentiation and inhibits proliferation of myoblasts by targeting HDAC4 in skeletal muscle development. RNA Biol (2016) 13(12):1300–9. doi: 10.1080/15476286.2016.1239008
81. Lee KP, Shin YJ, Panda AC, Abdelmohsen K, Kim JY, Lee SM, et al. miR-431 promotes differentiation and regeneration of old skeletal muscle by targeting Smad4. Genes Dev (2015) 29(15):1605–17. doi: 10.1101/gad.263574.115
82. Chen L, Wu P, Guo XH, Hu Y, Li YL, Shi J, et al. miR-143: a novel regulator of MyoD expression in fast and slow muscles of Siniperca chuatsi. Curr Mol Med (2014) 14(3):370–5. doi: 10.2174/1566524014666140228100250
83. Bem J, Grabowska I, Daniszewski M, Zawada D, Czerwinska AM, Bugajski L, et al. Transient MicroRNA Expression Enhances Myogenic Potential of Mouse Embryonic Stem Cells. Stem Cells (2018) 36(5):655–70. doi: 10.1002/stem.2772
84. Potthoff MJ, Arnold MA, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol Cell Biol (2007) 27(23):8143–51. doi: 10.1128/MCB.01187-07
85. Mercatelli N, Fittipaldi S, De Paola E, Dimauro I, Paronetto MP, Jackson MJ, et al. MiR-23-TrxR1 as a novel molecular axis in skeletal muscle differentiation. Sci Rep (2017) 7(1):7219. doi: 10.1038/s41598-017-07575-0
86. Tang Z, Liu N, Luo L, Kang K, Li L, Ni R, et al. MicroRNA-17-92 Regulates the Transcription Factor E2F3b during Myogenesis In Vitro and In Vivo. Int J Mol Sci (2017) 18(4):727. doi: 10.3390/ijms18040727
87. Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell (2009) 36(1):61–74. doi: 10.1016/j.molcel.2009.08.008
88. Khanna N, Ge Y, Chen J. MicroRNA-146b promotes myogenic differentiation and modulates multiple gene targets in muscle cells. PloS One (2014) 9(6):e100657. doi: 10.1371/journal.pone.0100657
89. Wang H, Noulet F, Edom-Vovard F, Tozer S, Le Grand F, Duprez D. Bmp signaling at the tips of skeletal muscles regulates the number of fetal muscle progenitors and satellite cells during development. Dev Cell (2010) 18(4):643–54. doi: 10.1016/j.devcel.2010.02.008
90. Mourikis P, Sambasivan R, Castel D, Rocheteau P, Bizzarro V, Tajbakhsh S. A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells (2012) 30(2):243–52. doi: 10.1002/stem.775
91. Li Z, Gilbert JA, Zhang Y, Zhang M, Qiu Q, Ramanujan K, et al. An HMGA2-IGF2BP2 axis regulates myoblast proliferation and myogenesis. Dev Cell (2012) 23(6):1176–88. doi: 10.1016/j.devcel.2012.10.019
92. Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev (2014) 28(5):491–501. doi: 10.1101/gad.234419.113
93. Shi L, Zhou B, Li P, Schinckel AP, Liang T, Wang H, et al. MicroRNA-128 targets myostatin at coding domain sequence to regulate myoblasts in skeletal muscle development. Cell Signal (2015) 27(9):1895–904. doi: 10.1016/j.cellsig.2015.05.001
94. Guo CS, Degnin C, Fiddler TA, Stauffer D, Thayer MJ. Regulation of MyoD activity and muscle cell differentiation by MDM2, pRb, and Sp1. J Biol Chem (2003) 278(25):22615–22. doi: 10.1074/jbc.M301943200
95. Bakkar N, Wang J, Ladner KJ, Wang H, Dahlman JM, Carathers M, et al. IKK/NF-kappaB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J Cell Biol (2008) 180(4):787–802. doi: 10.1083/jcb.200707179
96. Jones NC, Fedorov YV, Rosenthal RS, Olwin BB. ERK1/2 is required for myoblast proliferation but is dispensable for muscle gene expression and cell fusion. J Cell Physiol (2001) 186(1):104–15. doi: 10.1002/1097-4652(200101)186:1<104::AID-JCP1015>3.0.CO;2-0
97. Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol (2001) 3(11):1014–9. doi: 10.1038/ncb1101-1014
98. Mungunsukh O, Day RM. Transforming growth factor-β1 selectively inhibits hepatocyte growth factor expression via a micro-RNA-199-dependent posttranscriptional mechanism. Mol Biol Cell (2013) 24(13):2088–97. doi: 10.1091/mbc.e13-01-0017
99. Cao X, Tang S, Du F, Li H, Shen X, Li D, et al. miR-99a-5p Regulates the Proliferation and Differentiation of Skeletal Muscle Satellite Cells by Targeting MTMR3 in Chicken. Genes (Basel) (2020) 11(4):369. doi: 10.3390/genes11040369
100. Zhan S, Qin C, Li D, Zhao W, Nie L, Cao J, et al. A Novel Long Noncoding RNA, lncR-125b, Promotes the Differentiation of Goat Skeletal Muscle Satellite Cells by Sponging miR-125b. Front Genet (2019) 10:1171. doi: 10.3389/fgene.2019.01171
101. Li D, Zhan S, Wang Y, Wang L, Zhong T, Li L, et al. Role of microRNA-101a in the regulation of goat skeletal muscle satellite cell proliferation and differentiation. Gene (2015) 572(2):198–204. doi: 10.1016/j.gene.2015.07.010
102. Wang J, Tan J, Qi Q, Yang L, Wang Y, Zhang C, et al. miR-487b-3p Suppresses the Proliferation and Differentiation of Myoblasts by Targeting IRS1 in Skeletal Muscle Myogenesis. Int J Biol Sci (2018) 14(7):760–74. doi: 10.7150/ijbs.25052
103. Elsaeid Elnour I, Dong D, Wang X, Zhansaya T, Khan R, Jian W, et al. Bta-miR-885 promotes proliferation and inhibits differentiation of myoblasts by targeting MyoD1. J Cell Physiol (2020) 235(10):6625–36. doi: 10.1002/jcp.29559
104. Yang Z, Song C, Jiang R, Huang Y, Lan X, Lei C, et al. Micro-Ribonucleic Acid-216a Regulates Bovine Primary Muscle Cells Proliferation and Differentiation via Targeting SMAD Nuclear Interacting Protein-1 and Smad7. Front Genet (2019) 10:1112. doi: 10.3389/fgene.2019.01112
105. Song C, Yang J, Jiang R, Yang Z, Li H, Huang Y, et al. miR-148a-3p regulates proliferation and apoptosis of bovine muscle cells by targeting KLF6. J Cell Physiol (2019). doi: 10.1002/jcp.28232
106. Li H, Wei X, Yang J, Dong D, Hao D, Huang Y, et al. circFGFR4 Promotes Differentiation of Myoblasts via Binding miR-107 to Relieve Its Inhibition of Wnt3a. Mol Ther Nucleic Acids (2018) 11:272–83. doi: 10.1016/j.omtn.2018.02.012
107. Zhang WW, Tong HL, Sun XF, Hu Q, Yang Y, Li SF, et al. Identification of miR-2400 gene as a novel regulator in skeletal muscle satellite cells proliferation by targeting MYOG gene. Biochem Biophys Res Commun (2015) 463(4):624–31. doi: 10.1016/j.bbrc.2015.05.112
108. Ferrucci L, Baroni M, Ranchelli A, Lauretani F, Maggio M, Mecocci P, et al. Interaction between bone and muscle in older persons with mobility limitations. Curr Pharm Des (2014) 20(19):3178–97. doi: 10.2174/13816128113196660690
109. Phillips BE, Hill DS, Atherton PJ. Regulation of muscle protein synthesis in humans. Curr Opin Clin Nutr Metab Care (2012) 15(1):58–63. doi: 10.1097/MCO.0b013e32834d19bc
110. Drummond MJ, McCarthy JJ, Sinha M, Spratt HM, Volpi E, Esser KA, et al. Aging and microRNA expression in human skeletal muscle: a microarray and bioinformatics analysis. Physiol Genomics (2011) 43(10):595–603. doi: 10.1152/physiolgenomics.00148.2010
111. Zacharewicz E, Della Gatta P, Reynolds J, Garnham A, Crowley T, Russell AP, et al. Identification of microRNAs linked to regulators of muscle protein synthesis and regeneration in young and old skeletal muscle. PloS One (2014) 9(12):e114009. doi: 10.1371/journal.pone.0114009
112. Rivas DA, Lessard SJ, Rice NP, Lustgarten MS, So K, Goodyear LJ, et al. Diminished skeletal muscle microRNA expression with aging is associated with attenuated muscle plasticity and inhibition of IGF-1 signaling. FASEB J (2014) 28(9):4133–47. doi: 10.1096/fj.14-254490
113. Kim JY, Park YK, Lee KP, Lee SM, Kang TW, Kim HJ, et al. Genome-wide profiling of the microRNA-mRNA regulatory network in skeletal muscle with aging. Aging (Albany NY) (2014) 6(7):524–44. doi: 10.18632/aging.100677
114. Hamrick MW, Herberg S, Arounleut P, He HZ, Shiver A, Qi RQ, et al. The adipokine leptin increases skeletal muscle mass and significantly alters skeletal muscle miRNA expression profile in aged mice. Biochem Biophys Res Commun (2010) 400(3):379–83. doi: 10.1016/j.bbrc.2010.08.079
115. Mercken EM, Majounie E, Ding J, Guo R, Kim J, Bernier M, et al. Age-associated miRNA alterations in skeletal muscle from rhesus monkeys reversed by caloric restriction. Aging (Albany NY) (2013) 5(9):692–703. doi: 10.18632/aging.100598
116. Hu Z, Klein JD, Mitch WE, Zhang L, Martinez I, Wang XH. MicroRNA-29 induces cellular senescence in aging muscle through multiple signaling pathways. Aging (Albany NY) (2014) 6(3):160–75. doi: 10.18632/aging.100643
117. Kondo H, Kim HW, Wang L, Okada M, Paul C, Millard RW, et al. Blockade of senescence-associated microRNA-195 in aged skeletal muscle cells facilitates reprogramming to produce induced pluripotent stem cells. Aging Cell (2016) 15(1):56–66. doi: 10.1111/acel.12411
118. Russell AP, Wallace MA, Kalanon M, Zacharewicz E, Della Gatta PA, Garnham A, et al. Striated muscle activator of Rho signalling (STARS) is reduced in ageing human skeletal muscle and targeted by miR-628-5p. Acta Physiol (Oxf) (2017) 220(2):263–74. doi: 10.1111/apha.12819
119. Soriano-Arroquia A, McCormick R, Molloy AP, McArdle A, Goljanek-Whysall K. Age-related changes in miR-143-3p:Igfbp5 interactions affect muscle regeneration. Aging Cell (2016) 15(2):361–9. doi: 10.1111/acel.12442
120. Matsuzaka Y, Kishi S, Aoki Y, Komaki H, Oya Y, Takeda S, et al. Three novel serum biomarkers, miR-1, miR-133a, and miR-206 for Limb-girdle muscular dystrophy, Facioscapulohumeral muscular dystrophy, and Becker muscular dystrophy. Environ Health Prev Med (2014) 19(6):452–8. doi: 10.1007/s12199-014-0405-7
121. Li X, Li Y, Zhao L, Zhang D, Yao X, Zhang H, et al. Circulating Muscle-specific miRNAs in Duchenne Muscular Dystrophy Patients. Mol Ther Nucleic Acids (2014) 3(7):e177. doi: 10.1038/mtna.2014.29
122. Cacchiarelli D, Legnini I, Martone J, Cazzella V, D’Amico A, Bertini E, et al. miRNAs as serum biomarkers for Duchenne muscular dystrophy. EMBO Mol Med (2011) 3(5):258–65. doi: 10.1002/emmm.201100133
123. Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, Richardson JA, et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest (2012) 122(6):2054–65. doi: 10.1172/JCI62656
124. Alexander MS, Casar JC, Motohashi N, Vieira NM, Eisenberg I, Marshall JL, et al. MicroRNA-486-dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy-associated symptoms. J Clin Invest (2014) 124(6):2651–67. doi: 10.1172/JCI73579
125. Zaharieva IT, Calissano M, Scoto M, Preston M, Cirak S, Feng L, et al. Dystromirs as serum biomarkers for monitoring the disease severity in Duchenne muscular Dystrophy. PloS One (2013) 8(11):e80263. doi: 10.1371/journal.pone.0080263
126. Jeanson-Leh L, Lameth J, Krimi S, Buisset J, Amor F, Le Guiner C, et al. Serum profiling identifies novel muscle miRNA and cardiomyopathy-related miRNA biomarkers in Golden Retriever muscular dystrophy dogs and Duchenne muscular dystrophy patients. Am J Pathol (2014) 184(11):2885–98. doi: 10.1016/j.ajpath.2014.07.021
127. Liu J, Liang X, Zhou D, Lai L, Xiao L, Liu L, et al. Coupling of mitochondrial function and skeletal muscle fiber type by a miR-499/Fnip1/AMPK circuit. EMBO Mol Med (2016) 8(10):1212–28. doi: 10.15252/emmm.201606372
128. Zhai L, Wu R, Han W, Zhang Y, Zhu D. miR-127 enhances myogenic cell differentiation by targeting S1PR3. Cell Death Dis (2017) 8(3):e2707. doi: 10.1038/cddis.2017.128
129. D’Agostino M, Torcinaro A, Madaro L, Marchetti L, Sileno S, Beji S, et al. Role of miR-200c in Myogenic Differentiation Impairment via p66Shc: Implication in Skeletal Muscle Regeneration of Dystrophic mdx Mice. Oxid Med Cell Longev (2018) 2018:4814696. doi: 10.1155/2018/4814696
130. Wang L, Zhou L, Jiang P, Lu L, Chen X, Lan H, et al. Loss of miR-29 in myoblasts contributes to dystrophic muscle pathogenesis. Mol Ther (2012) 20(6):1222–33. doi: 10.1038/mt.2012.35
131. Perkins KJ, Davies KE. Alternative utrophin mRNAs contribute to phenotypic differences between dystrophin-deficient mice and Duchenne muscular dystrophy. FEBS Lett (2018) 592(11):1856–69. doi: 10.1002/1873-3468.13099
132. Zanotti S, Gibertini S, Curcio M, Savadori P, Pasanisi B, Morandi L, et al. Opposing roles of miR-21 and miR-29 in the progression of fibrosis in Duchenne muscular dystrophy. Biochim Biophys Acta (2015) 1852(7):1451–64. doi: 10.1016/j.bbadis.2015.04.013
133. Morgoulis D, Berenstein P, Cazacu S, Kazimirsky G, Dori A, Barnea ER, et al. sPIF promotes myoblast differentiation and utrophin expression while inhibiting fibrosis in Duchenne muscular dystrophy via the H19/miR-675/let-7 and miR-21 pathways. Cell Death Dis (2019) 10(2):82. doi: 10.1038/s41419-019-1307-9
134. Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, et al. A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell Stem Cell (2016) 18(4):533–40. doi: 10.1016/j.stem.2016.01.021
135. Fearon K, Strasser F, Anker SD, Bosaeus I, Bruera E, Fainsinger RL, et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol (2011) 12(5):489–95. doi: 10.1016/S1470-2045(10)70218-7
136. Fearon K, Arends J, Baracos V. Understanding the mechanisms and treatment options in cancer cachexia. Nat Rev Clin Oncol (2013) 10(2):90–9. doi: 10.1038/nrclinonc.2012.209
137. Arthur ST, Noone JM, Van Doren BA, Roy D, Blanchette CM. One-year prevalence, comorbidities and cost of cachexia-related inpatient admissions in the USA. Drugs Context (2014) 3:212265. doi: 10.7573/dic.212265
138. Hauser CA, Stockler MR, Tattersall MH. Prognostic factors in patients with recently diagnosed incurable cancer: a systematic review. Support Care Cancer (2006) 14(10):999–1011. doi: 10.1007/s00520-006-0079-9
139. Laviano A, Meguid MM, Inui A, Muscaritoli M, Rossi-Fanelli F. Therapy insight: Cancer anorexia-cachexia syndrome–when all you can eat is yourself. Nat Clin Pract Oncol (2005) 2(3):158–65. doi: 10.1038/ncponc0112
140. Tisdale MJ. Cachexia in cancer patients. Nat Rev Cancer (2002) 2(11):862–71. doi: 10.1038/nrc927
141. Mubaid S, Ma JF, Omer A, Ashour K, Lian XJ, Sanchez BJ, et al. HuR counteracts miR-330 to promote STAT3 translation during inflammation-induced muscle wasting. Proc Natl Acad Sci U S A (2019) 116(35):17261–70. doi: 10.1073/pnas.1905172116
142. Okugawa Y, Toiyama Y, Hur K, Yamamoto A, Yin C, Ide S, et al. Circulating miR-203 derived from metastatic tissues promotes myopenia in colorectal cancer patients. J Cachexia Sarcopenia Muscle (2019) 10(3):536–48. doi: 10.1002/jcsm.12403
143. Okugawa Y, Yao L, Toiyama Y, Yamamoto A, Shigemori T, Yin C, et al. Prognostic impact of sarcopenia and its correlation with circulating miR-21 in colorectal cancer patients. Oncol Rep (2018) 39(4):1555–64. doi: 10.3892/or.2018.6270
Keywords: microRNA, skeletal muscle, cachexia, Duchenne muscular dystrophy, sarcopenia
Citation: Singh GB, Cowan DB and Wang DZ (2020) Tiny Regulators of Massive Tissue: MicroRNAs in Skeletal Muscle Development, Myopathies, and Cancer Cachexia. Front. Oncol. 10:598964. doi: 10.3389/fonc.2020.598964
Received: 26 August 2020; Accepted: 29 October 2020;
Published: 23 November 2020.
Edited by:
Federica Calore, The Ohio State University, United StatesReviewed by:
Kotb Abdelmohsen, National Institutes of Health (NIH), United StatesCopyright © 2020 Singh, Cowan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Da-Zhi Wang, RGEtWmhpLldhbmdAY2hpbGRyZW5zLmhhcnZhcmQuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.