- 1Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA, United States
- 2Department of Data Science, Dana-Farber Cancer Institute, Boston, MA, United States
- 3Channing Division of Network Medicine, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, United States
- 4Division of Pulmonary and Critical Care Medicine, Brigham and Women's Hospital, Boston, MA, United States
Despite their known importance in clinical medicine, differences based on sex and gender are among the least studied factors affecting cancer susceptibility, progression, survival, and therapeutic response. In particular, the molecular mechanisms driving sex differences are poorly understood and so most approaches to precision medicine use mutational or other genomic data to assign therapy without considering how the sex of the individual might influence therapeutic efficacy. The mandate by the National Institutes of Health that research studies include sex as a biological variable has begun to expand our understanding on its importance. Sex differences in cancer may arise due to a combination of environmental, genetic, and epigenetic factors, as well as differences in gene regulation, and expression. Extensive sex differences occur genome-wide, and ultimately influence cancer biology and outcomes. In this review, we summarize the current state of knowledge about sex-specific genetic and genome-wide influences in cancer, describe how differences in response to environmental exposures and genetic and epigenetic alterations alter the trajectory of the disease, and provide insights into the importance of integrative analyses in understanding the interplay of sex and genomics in cancer. In particular, we will explore some of the emerging analytical approaches, such as the use of network methods, that are providing a deeper understanding of the drivers of differences based on sex and gender. Better understanding these complex factors and their interactions will improve cancer prevention, treatment, and outcomes for all individuals.
Introduction
Sex disparities occur in cancer incidence and mortality (Figure 1). The mortality rate of all cancer sites combined is 214 for males, and 149 for females per 100,000, age-adjusted 2000–2017 average according to the Surveillance, Epidemiology, and End Results (SEER) program explorer. These sex disparities are apparent across a range of non-reproductive cancers and vary by age and race. In general, males have a higher incidence and a higher mortality rate than females for most cancer sites, including bladder, kidney, colorectum, liver, esophagus, head and neck, brain, skin, and blood (1). For cancer sites with higher incidence in males, the age-adjusted male-to-female incidence rate ratios range from 1.036 to 9.751, with a median of 1.588. Higher cancer incidences in females are found for breast, thyroid, cranial nerves, and a few of digestive system sites including gallbladder, anus, anal canal, and anorectum. Despite females having higher incidence for these cancers, they have better survival compared to males (1).
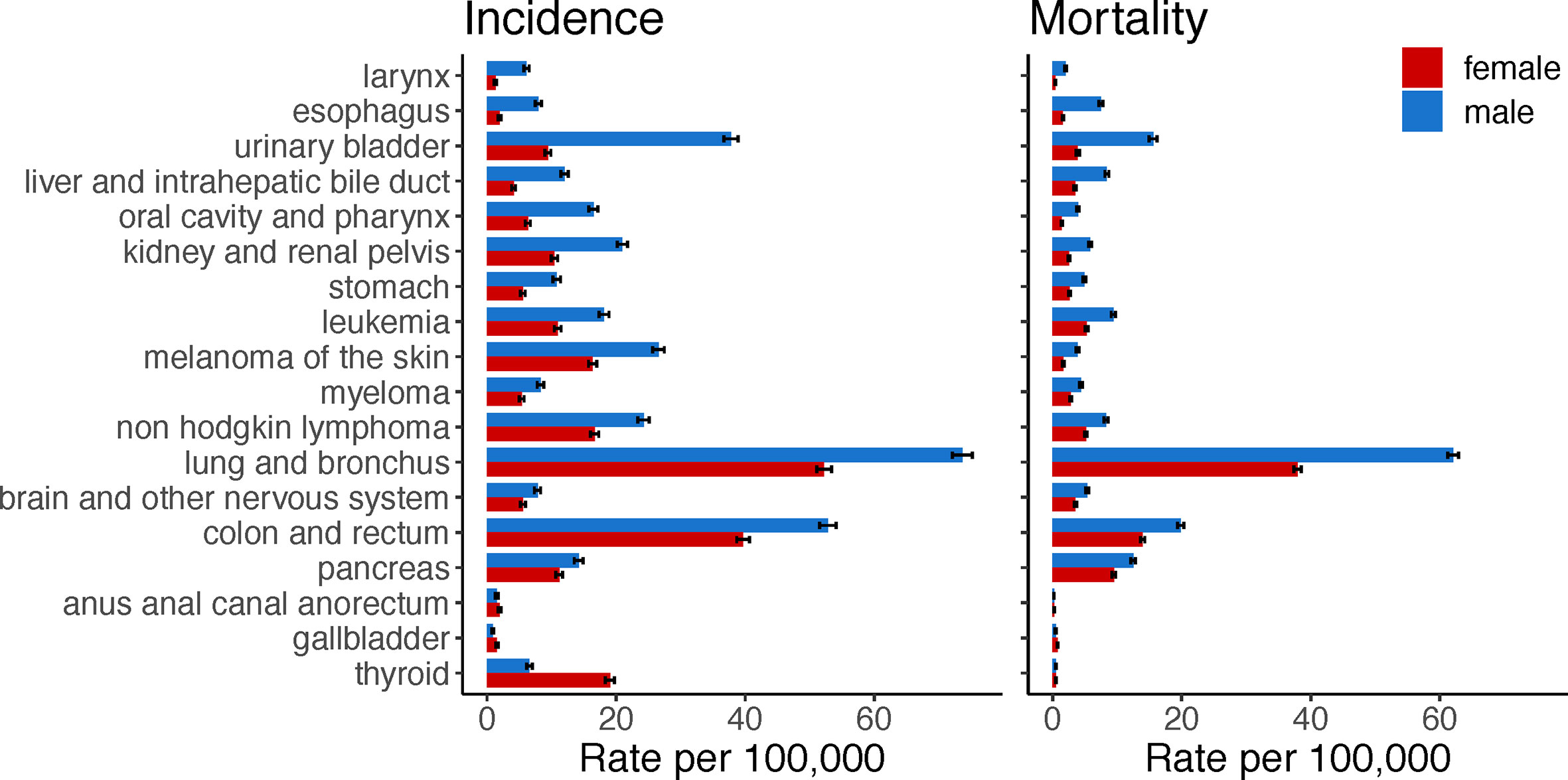
Figure 1 Cancers with sex disparity in incidence and mortality rates. Age-adjusted incidence and mortality rates per 100,000 individuals in the US were retrieved from the Surveillance, Epidemiology, and End Results explorer. Bars show the average rate from 2000 to 2017 and 95% confidence interval. Cancer sites are ordered according to the male-to-female incidence rate ratio; starting from cancer sites with higher incidence rates in males compared to females.
There are significant sex differences in therapeutic response and toxicity for many cancer types. Various chemotherapy regimens show higher toxicity, higher response rates, and longer post-treatment survival in women, including lymphoma (2), sarcoma (3), glioblastoma (4), lung (5, 6), and colorectal (7) cancers. There is also a growing literature showing cancer immunotherapy efficacy varies by sex (8–13). Poorer response rates are usually reported in females compared to males. However, controversial findings have been reported as the sex-based differences for immunotherapy response seems to depend on the cohort and cancer type analyzed as well as the immune checkpoint inhibitor agents used and whether they are used in combination with chemotherapy. Despite these known sex differences in therapeutic response, most treatment strategies, as well as drug development and selection, do not account for sex differences.
Studying both sex and gender is important to understanding differences in clinical manifestation between males and females and the molecular mechanisms involved; such considerations are now mandated as part of all NIH-funded research studies (14). Sex refers to a biological concept defined by the sex chromosome complement (15). Gender refers to a multidimensional social construct that may vary across societies and over time (16). Gender includes gender identity (the individual's self-perception and presentation), gender norms or roles (the individual's behaviors influenced by social and culture expectations) (17). The complex interplay between sex and gender is dynamic along an individual's life, influences not only cancer susceptibility and progression, but also how the individual perceives cancer, relates with health care, and adheres to treatments. Thus, it is essential to consider sex and gender in every stage of research and clinical care to improve prevention, diagnosis, and treatment for all individuals, being inclusive of gender minorities (18–20).
Sex and gender differences may influence cancer in different ways. Preventive behavior and exposure to risk factors were largely attributed to higher cancer incidence in males. However, gender-related occupational exposures (including heavy metals, and pesticides) and behavioral risk factors (including diet, tobacco, and alcohol consumption) can only partially explain higher cancer risk in males (21–23). Sex hormones play an important role in tumorigenesis and cancer susceptibility through several mechanisms likely to affect cancer stem cell self-renewal, the tumor microenvironment, the immune system, and the metabolism (24). In general, androgens have been associated with the higher cancer risk and mortality in males while estrogens observed to be protective in females (25–28). However, while sex differences in cancer may be modified by sex hormones, it is not completely explained by hormonal differences, and for most cancers over childhood and adolescent periods males also show higher incidence rates (29). Differential regulation of immune responses between males and females can also contribute to cancer susceptibility and outcomes. In general, females mount stronger immune responses than males, and sex differences in innate and adaptive immune responses occur throughout life (30). Differences in immune response and infection burden may also contribute to sex disparities of viral-related cancers. Human papillomavirus (HPV) infection rate differs by anatomic site and sex, and likely contribute to disparities in incidence of HPV-related cancers, including oral and anal (31). Other factors that might also contribute to sex differences in cancer include anatomy, physiology, body composition (lean and fat body mass), pharmacokinetics, and pharmacodynamics, which may affect drug metabolism, drug response, and drug toxicity (32, 33).
Sex differences occur genome-wide, and are not restricted to sex chromosomes, can be independent of sex hormones regulation, and ultimately impact cancer biology and outcomes. More than half of the genes targeted by FDA-approved cancer drugs show sex-biased molecular signatures, including sex differences in somatic mutation, copy number, methylation, gene expression, and protein abundance (34) (Table 1). In this review, we discuss how genetics, epigenetics, gene regulation and expression may contribute to sex differences in cancer (Figure 2). We also describe how sex can be incorporated in genomics research and investigated through model systems. We finish with an example framework in colorectal cancer showing the impact of sex- and gender-based research to help us understand sex disparities in cancer and allow us to improve cancer prevention and treatment for both sexes.
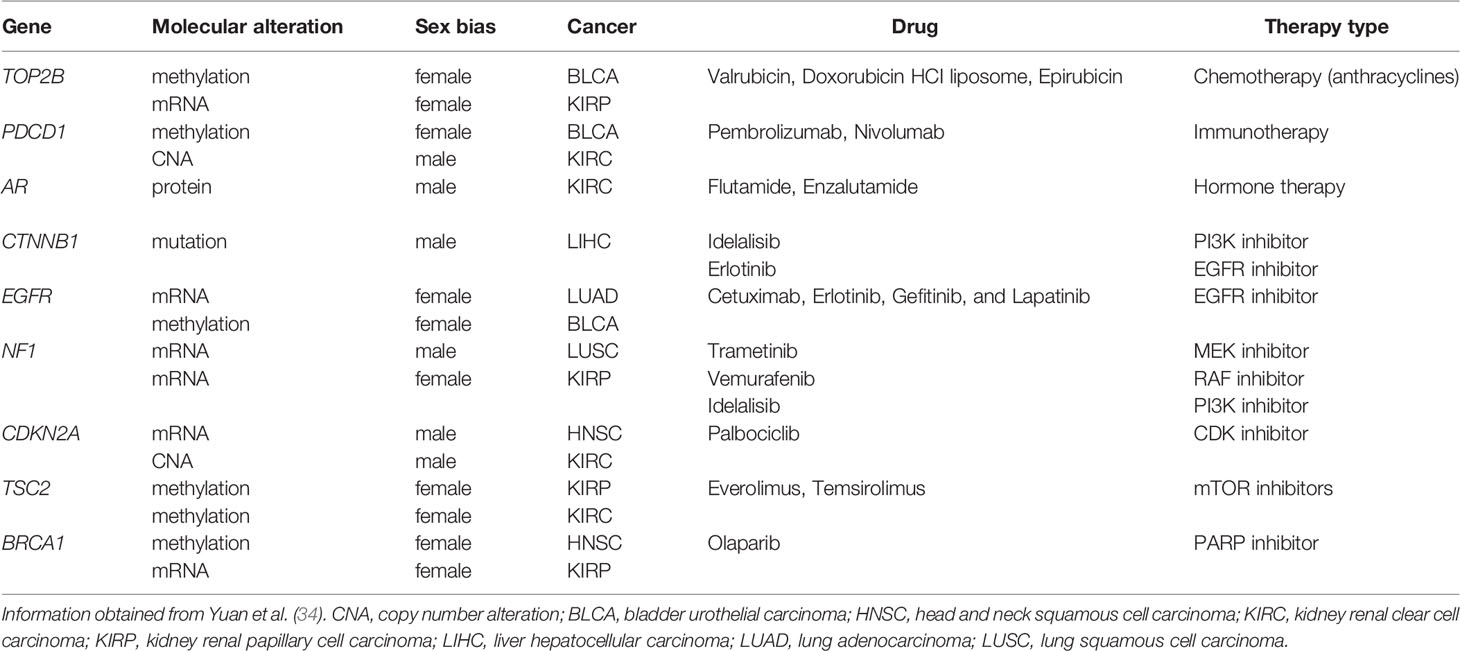
Table 1 Examples of cancer drugs and their related-actionable genes, harboring sex-biased genomic alteration.
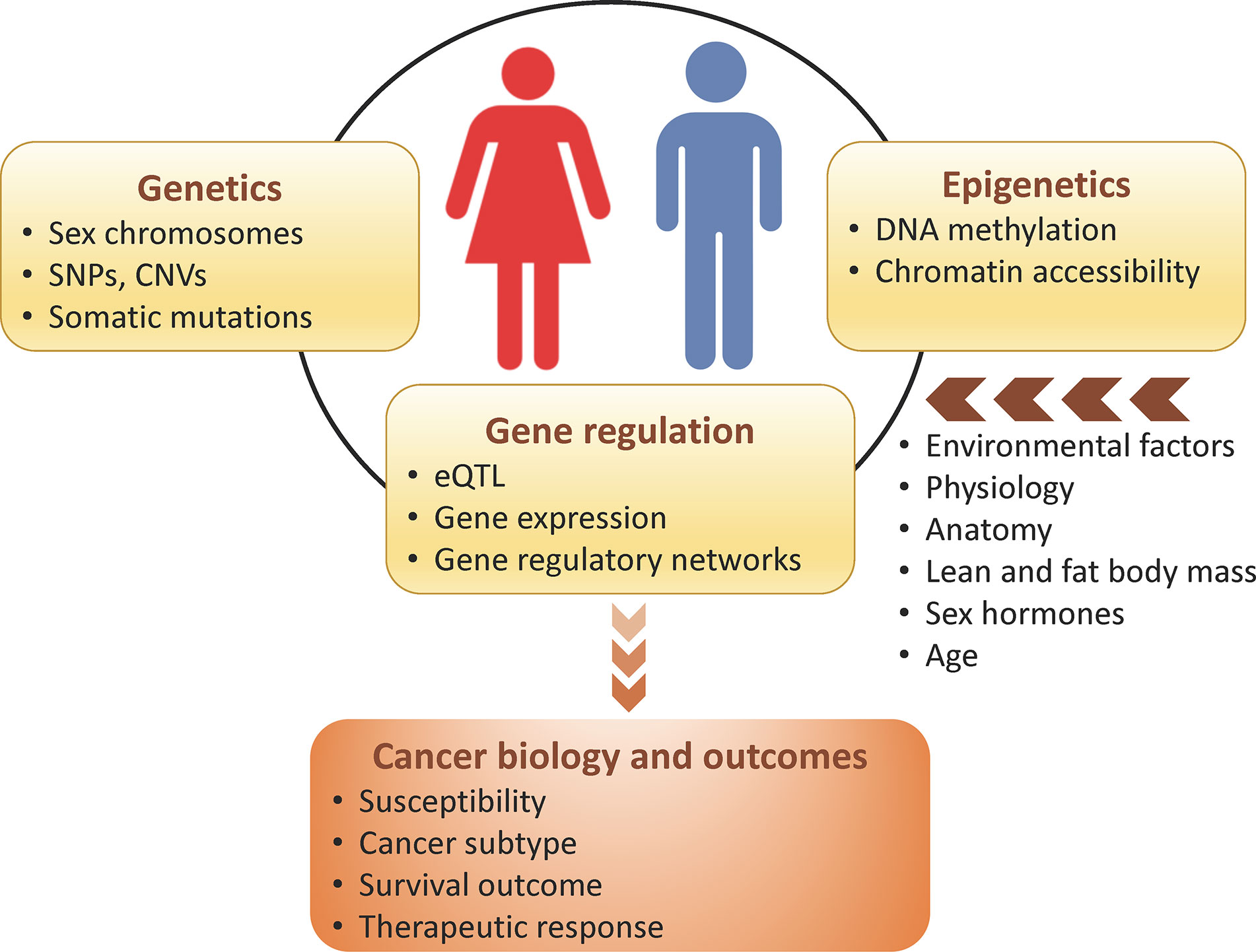
Figure 2 Sex differences in genomics, which can be modulated by host and environmental factors, influence cancer biology and outcomes.
Sex Differences in Genetics
Sex Chromosomes
Genetic sex differences exist in the inherent inequality of the sex chromosomes, resulting in differences in the numbers of X chromosome genes between males and females and the presence of Y chromosome genes exclusively in males (35). A large part of the Y chromosome was lost during evolution, and only a small segment, the pseudoautosomal region (PAR), can recombine with the X chromosome. The Y chromosome evolved to retain genes with male-specific functions, as well as genes widely expressed involved in regulation of transcription, translation, and protein stability (36). Age-related loss of the Y chromosome in peripheral blood cells is a common somatic event in males and may be associated with higher risk of cancer, including leukemia, breast, and head and neck (37–40).
To reduce transcriptional dosage imbalance of X chromosome genes between the sexes, one of the X chromosomes in females is inactivated early in embryonic development and generally remains so throughout an individual's lifetime. Abnormalities in this process and reactivation of X-linked genes can occur in some tumors (41–43). The X inactive specific transcript (XIST) is a long non-coding RNA and major player during X chromosome inactivation, and loss of Xist in mouse hematopoietic cells results in X reactivation and genome-wide changes leading to leukemia (44).
X chromosome inactivation is incomplete, and some genes are expressed from both alleles resulting in important sex bias. Genes that escape X inactivation can have significant sex differences related to mRNA expression levels (45), patterns of transcription factor targeting (46), and frequency of somatic mutations (47). Across 21 cancer types, a higher mutation rate in males was identified for six "Escape from X-Inactivation Tumor Suppressor" (EXITS) genes: ATRX, CNKSR2, DDX3X, KDM5C, KDM6A, and MAGEC3 (47). This finding showed that females might be protected for many cancers due to the biallelic expression of the EXITS genes and the need of a second hit to inactivate those. Functional experiments further support the evidence that EXITS genes can contribute to higher risk of many cancers in males, specifically for the X-linked lysine demethylase 6A (KDM6A, also known as UTX). KDM6A loss accelerates tumor initiation and progression of various in vitro and in vivo models for leukemia (48), lymphoma (49), pancreatic (50), and bladder cancer (51). Likewise, cancer risk differs between individuals with or without karyotype anomalies. Compared to the general population without karyotype anomalies, 1.34 increase in the risk of solid tumors was found in women with Turner syndrome, which is characterized by X chromosome monosomy (52). In men with Kleinfelter syndrome, characterized by the presence of two or more X chromosomes, the risk of solid tumors was reduced, but the risk of hematological tumors was increased, standardized incidence ratios of 0.66 and 2.72, respectively. This finding further supports the important role X chromosome plays in cancer etiology.
Genetic Variants
The association of genetic variants and sex effects is an understudied topic and challenging to reach power for sex-stratified or interaction analyses (53). However, evaluating genetic variants based on sex can reveal important sex disparities. Sex-biased effects of variants related to cell cycle and apoptosis have been described for many cancer types (54). Germline mutations in TP53 are associated with increased cancer risk and earlier age at first-cancer diagnosis for females compared to males; female carriers have a 2.5- to 7-fold higher odds of having cancer than male carriers (55, 56). A single nucleotide polymorphism (SNP) found in the promoter region of MDM2, a negative regulator of the p53, is associated with increased risk of various cancer types in females but not in males, which was shown in vivo to be modulated by the estrogen-signaling pathway (57–59). Many genetic variants in immune-related genes can confer cancer risk or protection exclusively in one sex, including different types of variants such as SNP (60, 61), short tandem repeat (STR) (62), and copy number variant (CNV) (63).
There are important sex differences in pharmacokinetics and pharmacodynamics affecting drug metabolism and exposure, drug sensitivity and toxicity (32, 33). In general, clearance of various anticancer drugs is reduced by approximately 20% in females compared to males (33). There are also many examples of genetic variants in genes related to drug metabolism and detoxification that are associated with cancer risk. In acute myeloblastic and lymphoblastic leukemia, the deletion of GSTT1 and a SNP in the NQO1 gene confers higher risk to males than females (64). A SNP in the detoxifying-enzyme SULT1A1 reduces the risk of bladder cancer in females but not in males (65). In lung cancer, genetic variants on MTHFR are associated with both increased or decreased risk in females, but not in males (66). Evidence also suggests interactions of MTHFR polymorphisms with both diet (vitamin B6, B12, and methionine) and tobacco smoking. In pancreatic cancer, interactions of SNPs in the drug metabolism genes CYP1A2 and NAT1 with tobacco and diet modify the risk of cancer differently in males and females (67). Taken together, these examples show that genetic variants might influence the sex divergence in cancer, and future studies need to consider the role of sex and gender in modifying both penetrance and expressivity of genetic variants.
Somatic Mutation
Sex-bias occur in mutation burden and in frequency of specific somatic mutations, which can include single nucleotide and copy number alteration (CNA). These sex differences in somatic mutation ultimately affect survival outcomes and therapeutic response.
Males have higher mutation burden (total number of mutations) for many cancer types, including lung adenocarcinoma (68), melanoma (69), urothelial cell, papillary renal cell, and hepatocellular carcinomas; difference in means by sex range from 0.16 to 2.2 (70). Among patients with high mutation count in melanoma (over 130 mutations), female patients have greater overall survival than males (69). In some cancers, mutation burden may be explained by sex differences in the efficiency of mismatch repair as suggested by the lower mRNA levels of DNA mismatch repair genes associated with high mutation burden in stomach and esophageal, kidney and liver cancers (70).
In a pan-cancer analysis using The Cancer Genome Atlas (TCGA) data, 15% of autosomal genes had sex-biased CNA, the majority of these were amplification and more prevalent in male tumors (70). Looking at each specific cancer type, eight harbored significant sex-biased frequency of CNAs: kidney clear cell, kidney papillary, head and neck, stomach and esophageal, liver, bladder, lung adenocarcinoma, and squamous cell cancers (34, 70). Sex-biased CNAs can be focal or cover long genomic segments, associated with differences in mRNA levels, and demonstrate differences in survival outcome. The authors identified 16 examples of sex-specific prognostic markers (70) (Table 2). For example, loss of LATS1 was a marker of poor prognosis in females but not in males, and UBAC1 loss was a marker of good prognosis in females but not in males.
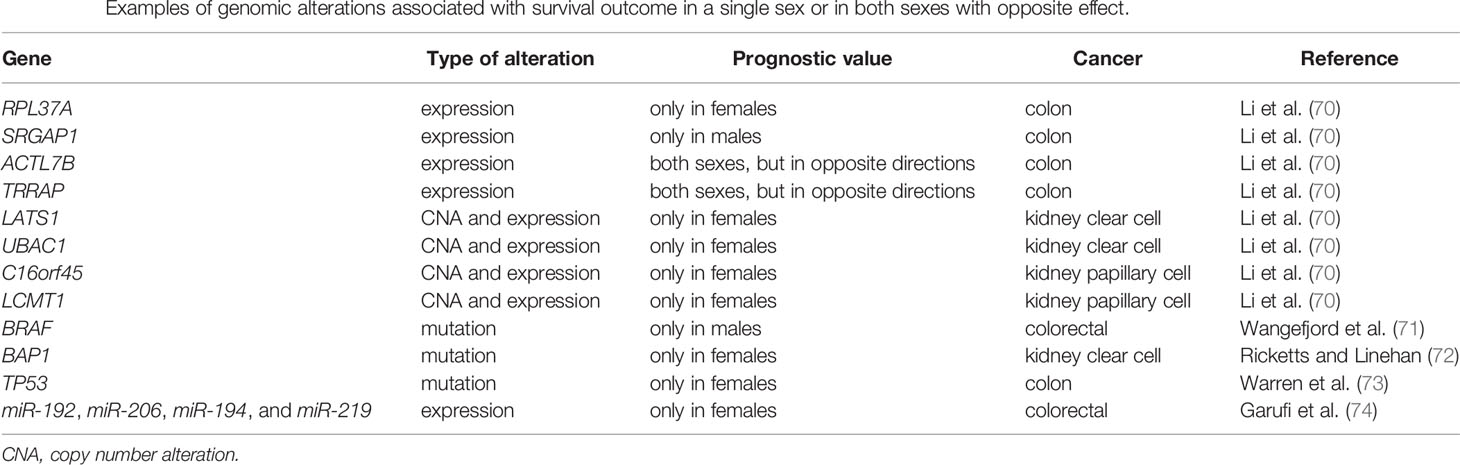
Table 2 Examples of genomic alterations associated with survival outcome in a single sex or in both sexes with opposite effect.
Specific cancer genes can also have sex-biased patterns of somatic mutation, including the lower frequency of KRAS mutation among males with colorectal cancer (75), higher mutation frequency of PBRM1 and KDM5C in males and BAP1 in females with renal carcinoma (72). Point mutations can also have sex-biased prognostic effect. BRWD3 shows higher mutation frequency in HPV-negative head and neck cancer of females and associates with poorer overall survival (76). Overall, a genome-wide analysis of somatic mutation found that 0.8% of genes were prognostic in both sexes while 1.5% were prognostic in patients of only one sex (70).
Somatic mutations in therapeutic targets can be differentially present in males and females. β-catenin (CTNNB1) is more frequently mutated in men with hepatocellular carcinoma, and the activation of this proto-oncogene can affect sensitivity to EGFR, PI3K, AKT, and WNT inhibitors (34, 70). In contrast, the incidence of EGFR mutation is higher in females with non-small cell lung cancers (34, 77). The tumor suppressor gene STK11 has higher mutation frequency in men with lung adenocarcinoma, whereas inactivating mutations in this gene may predict sensitivity to mTOR and SRC inhibitors (34). These examples highlight the need to carefully consider sex differences in cancer genomics during drug development and when defining treatment options.
Gender-related behavior and exposures can also affect the gene mutation spectrum observed in males and females. In non-small cell lung cancer, both sex and smoking influence the mutation spectrum of EGFR (78) and TP53 (79, 80). The frequency of G to T transversion mutation on TP53 is higher among females (40%) than among males (25%–28%) (79, 80), which is a mutation signature of smoking-associated cancers (81). Tobacco may be more carcinogenic in females than males. Despite the lower levels of tobacco exposure, female lung tumor tissue present higher TP53 mutation, higher level of DNA adducts, higher expression level of the carcinogen-metabolizing enzyme CYP1A1, and less efficient DNA repair (Figure 3) (79, 82–84). A combination of mechanisms can contribute to the sex differences in genetics, such as carcinogen exposures, DNA repair efficacy, and sex differential chromatin architecture (described in the Sex Differences in Epigenetics section).
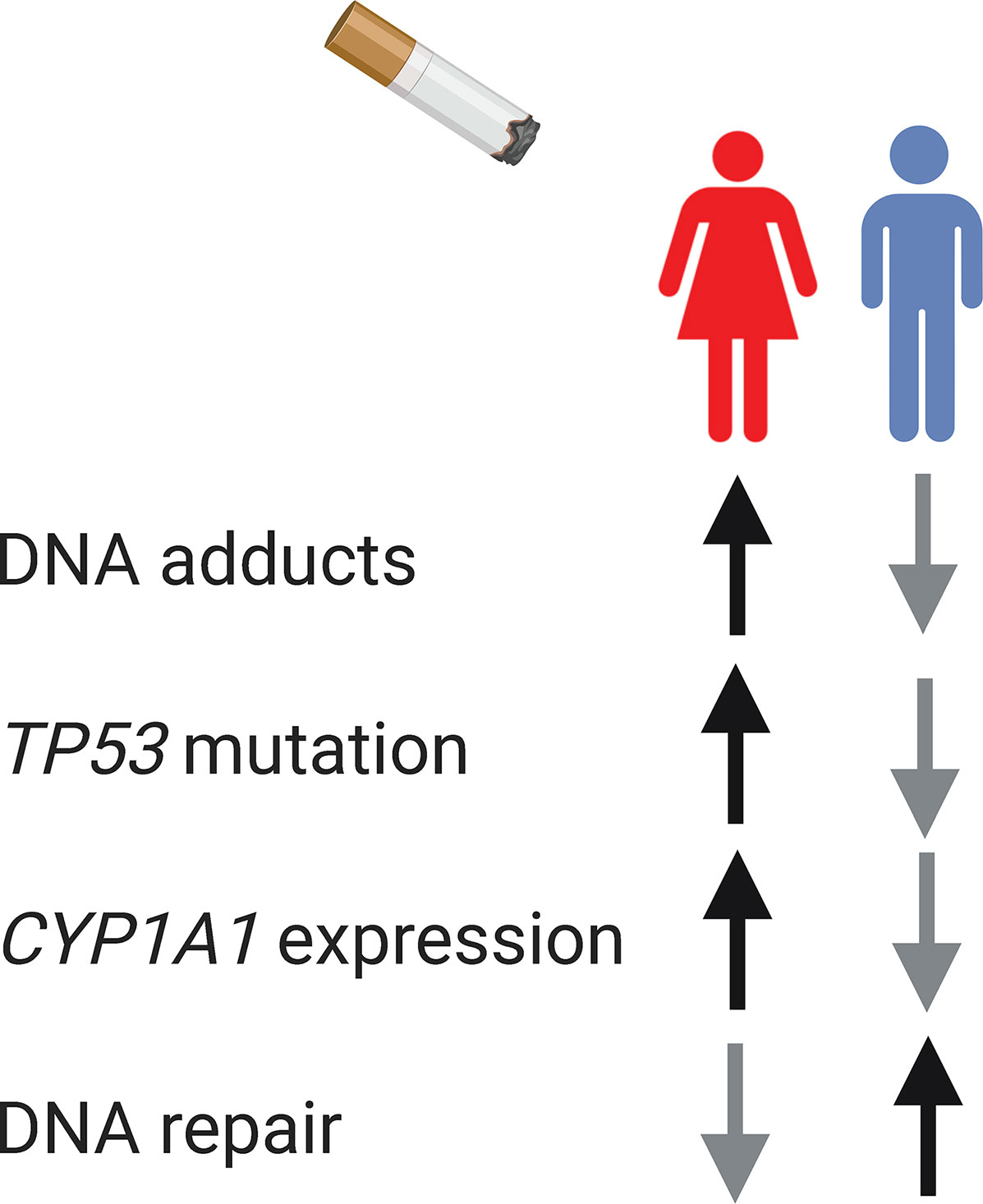
Figure 3 The interplay between sex, gender, and genomics. Biological sex and gender-influenced behavior, such as smoking, influence cancer genomics. In smoking-associated lung cancer, female lung tissue present higher level of DNA adducts, higher TP53 mutation, higher expression level of the carcinogen-metabolizing enzyme CYP1A1, and less efficient DNA repair.
Sex Differences in Epigenetics
DNA Methylation
Sex-biased methylation patterns have been observed across different human tissues, including blood (85–88), brain (89), liver (90, 91), rectal mucosa (92), muscle (93), and pancreas (94). Both the presence of the SRY gene and X chromosome dosage may influence sex differences in methylation level (95). Sex hormones may also influence methylation levels at specific gene regulatory regions, and in a tissue-specific manner, as observed in brain (96, 97) and liver (91). Furthermore, sex hormones can induce a stable methylation profile, establishing a sex-biased epigenetic memory maintained in the absence of the hormone.
In whole blood, 1,184 CpG sites in autosomes were found differentially methylated by sex, and further replicated in independent cohorts (86). Differences in male and female methylation patterns were enriched among imprinted genes and in CpG island shores (important for gene expression regulation), but not enriched in genes related with sex hormone biosynthesis, transport or receptors. By comparing sex-biased methylated genes in saliva and blood, an overlap of 81% was found for X chromosome genes and 8% for autosomes, confirming sex- and tissue-specific patterns of methylation (85). Genes differentially methylated in males and females were also associated with age, cigarette smoking, and alcohol consumption. As expected by the X-inactivation process, most CpG sites on the X chromosome had higher methylation levels in females compared to males. However, there are also higher methylation levels in males for some X chromosome sites. It is important to note that few studies have included the X chromosome in their analyses. A meta-analysis including 39 studies and several tissues, identified 184 autosomal CpG sites differentially methylated by sex (98). The sex-biased methylated CpG sites were located in the promotor of genes overrepresented in pathways important during cancer development, such as regulation of immune response, RNA splicing, and DNA repair. The magnitude of percent methylation differences between males and females found is usually small, with mean CpG site specific differences in methylation between males and females ranging from 3.7% to 17% (85, 98, 99).
Studies have identified sex-biased methylation patterns in cancer, including lung (100, 101) and colorectum (102). In leukemia, a genome-wide analysis identified 1,043 CpG sites differentially methylated by sex, of which 56 were located on autosomes and many known to play a role in tumor growth (103). A genome-wide analysis across 13 cancer types also reported sex-biased methylation patterns (34). On average, 236 genes with a sex-biased DNA methylation pattern were identified in the group of cancers defined as having strong sex-effect based on molecular signatures, and 10 genes in the weak sex-effect group of cancers. Most of the genes with sex-biased methylation also had sex-biased expression. The efficacy of methylation-based biomarkers for diagnosis and cancer risk prediction may be different for males and females with bladder cancer (104, 105). Moreover, radiation induces methylation changes in a sex- and tissue-specific manner (106, 107). Therefore, research focused on sex differences in DNA methylation is essential to improve treatment strategies and outcomes for all patients.
Chromatin Accessibility
Sex differences in chromatin accessibility have been found in normal (108, 109) and tumor tissues (110). In whole blood, sex-biased chromatin accessibility was enriched for genes with sex-biased gene expression, and sex-biased regulatory genetic variants (108). Sugathan et al. developed genome-wide chromatin state maps for male and female mouse liver, and observed that the patterns of chromatin accessibility and histone marks differ by sex (109). The integration of these chromatin state maps with data for five transcription factors binding sites and gene expression showed that sex-biased gene regulation is mediated by a complex interplay between sex-biased chromatin marks in regulatory regions and differential transcription factor binding by sex. Sex-biased chromatin accessibility is partly established and maintained in response to sex-specific changes in patterns of plasma growth hormones (111).
In tumor tissues, sex-bias mutation frequency is observed for chromatin remodeling genes, such as KDM6A (47), KDM5C, PBRM1, and BAP1 (72). Moreover, BAP1 mutation is associated with changes in overall survival of clear cell renal cell carcinoma only for females (72). In analyzing the TCGA ATAC-Seq dataset across 23 cancer types, 2,534 peaks differed between males and females, of which 1,035 peaks were higher in males and 1,499 peaks were higher in females (110). Sex differences in chromatin accessibility were found in the promoter of genes located in both the X chromosome and autosomes, and in promoters of sex hormone receptors. Taken together, these studies show that both genetic and epigenetic patterns influence sex-biased gene regulation, and reinforce the need of integrative data approaches to study sex differences in gene regulation.
Sex Differences in Gene Regulation
eQTL
Sex differences in genetic regulation can contribute to sex differences in prevalence, progression, and severity of diseases (112). Ober et al. suggests that sex can be used as a variable that includes information on cellular, metabolic, physiological, anatomical, and behavioral differences between males and females, and can be modelled similar to a gene by environment interaction (112). Some loci have a regulatory role in one sex but not the other. For example, SNPs shared among sexes can be in regulatory regions that are sensitive to sex hormones. The impact of sex on gene regulation can be investigated through expression quantitative trait loci (eQTL) analysis. Sex-biased eQTL refers to an association between genotype and gene expression differing between males and females. Early studies have shown that sex-biased eQTLs are widespread affecting 12%–15% of eQTLs (113). An enrichment of sex-biased eQTL on the X chromosome compared to the autosomes has been found (108). Furthermore, sex-biased chromatin accessibility was enriched for sex-biased gene expression and regulatory variants. In analyzing 11,672 complex disease-associated SNPs as a function of sex and age in whole blood, 14 sex-biased eQTLs were identified (114). Most recent studies have reported few significant sex-biased eQTLs, and many not replicating across datasets (45, 115–117). These results may reflect the underlying biology with most genes unlikely to be influenced by sex or may indicate a combination of many small effect size and low power for genotype-sex statistical interaction tests (53).
A common and interesting finding is that most of the sex-biased eQTLs do not show sex-biased mean gene expression (45, 108, 113–115, 117). Sex-biased eQTLs may also be explained by sex differences in allelic direction or sex differences in gene expression variance. Most studies have investigated whole blood or lymphoblastoid cell lines (LCLs), and have not elucidated how sex-biased eQTL are associated with cancer. In hepatocellular carcinoma, 24% of the discovered eQTLs were sex-biased (118). This set included 24 genes under genetic regulation only in males and involved genes associated with Notch and PI3K/AKT signaling. Further studies in cancer will continue to elucidate the sex differential effect of genetic variants on tumor gene regulation.
Gene Expression
Sex-biased gene expression is conserved for approximately 3,000 genes across mammals (119). Sex differential expression is found for both autosomal and sex chromosome genes across most human tissues (46, 117, 120–122), and cancer types (34). The largest expression differences are found for genes on the sex chromosomes, and the fold change of autosomal genes is generally small. The median fold change of autosomal sex-biased genes reported across 44 tissues was 1.04 (117). Despite X inactivation in females to compensate chromosome dosage, a number of X chromosome genes are differentially expressed by sex. Several X chromosome genes that escape inactivation are overexpressed in females in a tissue-specific manner (45). The opposite trend is observed for X chromosome genes that escape inactivation and are located in the pseudoautosomal region, showing higher expression in males compared to females. This pattern spans 29 normal human tissues, and it is also observed in sex-biased transcription factor targeting (46). Therefore, sex-bias mutation and expression of genes that escape from X-inactivation may contribute to higher susceptibility to certain cancers in males compared to females (47).
The number of differentially expressed genes varies by cancer type, extending up to 14% of the genes in clear cell renal cell carcinoma (34). Across cancer types, common pathways enriched for sex-biased genes include: immune response, metabolism, apoptosis and cell cycle, DNA repair and p53 (34). In hepatocellular carcinoma, sex differences are also found in several signaling pathways that are targets for anti-cancer therapies: PPAR is enriched for genes overexpressed in females, while PI3K, PI3K/AKT, EGFR, IL-2 are enriched for genes overexpressed in males (118). Other studies confirm that evaluating sex differential gene expression can help understand sex differences in cancer etiology (118, 123), distribution of molecular subtypes (124, 125), and outcomes (126–129). Sex differences in gene expression was also used to predict drug sensitivity in each sex (130).
New approaches to investigate sex effects in gene regulation and expression can provide better understanding of cancer in both sexes, which will be further exemplified by the significant sex differences in gene targeting by transcription factors discovered by gene regulatory network analysis (in the Gene Networks section). In glioblastoma, although a few number of genes are differentially expressed between males and females (34), the variance in tumor gene expression is sex biased (4). Using a join and individual variance explained (JIVE) analysis, Yang et al. identified five sex-specific molecular subtypes distinguished by gene expression and survival (4). The longest survival was associated with down-regulation of genes involved in cell cycle progression and in integrin signaling for males and females, respectively. This sex-specific pattern in expression was further associated with sex-biased chemotherapy sensitivity in a panel of male and female patient-derived glioblastoma cell lines. The authors also demonstrate the need to investigate sex-biased effect of somatic mutations to best define prognostic markers. Female patients with IDH1 mutation, known as a good prognostic marker in glioblastoma, mostly clustered in a single group with longest survival, whereas male patients with IDH1 mutation were distributed across all male clusters.
miRNA
miRNAs are small non-coding RNAs involved in post-transcriptional regulation of gene expression and thereby regulate most cellular processes (131). MiRNA dysregulation is implicated in tumor development and progression. Similar to protein-coding genes, sex hormone receptors can bind and directly regulate miRNA transcription or indirectly through intermediate hormone-responsive transcription factors. In brain development, 162 miRNAs have sex-biased expression, 92 of which are estrogen-responsive, and relevant to differences in brain organizational structure by sex (132).
The chromosome location can also indicate sex divergence in miRNAs. A higher density of miRNAs is observed in the X chromosome compared to the Y and autosomes. The X chromosome contains 10% of the miRNAs, while the Y chromosome contains only two miRNAs (133). Several X-linked miRNAs are involved in immune regulation, and can also be involved in tumorigenesis (134). For example, miR-221 and miR-222 promote cancer cell proliferation (135). The miR-506-514 cluster is overexpressed in melanoma and involved in melanocyte transformation, melanoma growth, and sensitivity to BRAF inhibitors (136, 137). X-linked miRNAs directly target PD-L1 or the transcription factors regulating PD-L1 (138). High expression levels of miR-424(322) reverses chemoresistance by blocking the PD-L1 immune checkpoint (139). These miRNAs might escape X-inactivation, and therefore present higher expression levels in females. Further investigation may show the role of X-linked miRNAs in sex differences related to immunotherapy response.
Few studies have addressed the role of miRNAs in sex differences observed in cancer. Increased expression of miR-17 and let-7a was linked with familial female breast cancer compared to males (140). In metastatic colorectal cancer, the higher expression of miRNAs that regulate clock genes (miR-192, miR-206, miR-194, and miR-219) was associated with better overall survival in females (74). miR-137, involved in cell cycle control, is frequently methylated in squamous cell carcinoma of the head and neck, and higher methylation frequency in females was found compared to males (141). In performing genome-wide miRNA differential expression, Yuan et al. reported sex-biased expression of miRNAs and their potential role as regulators of sex-biased protein expression levels (34). However, the number of miRNAs differentially expressed by sex was small, and a median of seven miRNAs was found in the group of cancer types with strong-sex effect, such as thyroid, head and neck squamous cell, lung, papillary renal cell, and clear cell renal cell carcinomas. Future studies might still find miRNAs with sex differential targeting patterns despite limited differences in the miRNA expression levels between males and females.
Gene Networks
Although we have learned a great deal about sex differences through the study of individual genes, we have come to recognize that even the effects of single-gene mutations are moderated by networks of interacting regulatory and other elements within the cell. One distinct advantage of using network-based approaches is that they allow one to go beyond a list of altered genes, and map how the various cellular components interact with altered genes and with each other (142). Analyzing the network topology and changes in network structure of males and females can provide insights into the mechanisms associated with sex differences in cancer (Figure 4). The impact of a specific genetic or epigenetic alteration is rarely restricted to the activity of one gene product, and can propagate along complex gene networks. The term "sexome" describe the combination of sex-biased effects on gene networks, creating sex differences in connectivity and activity of genes (143). Decoding the sexome will help us better understand how sex interacts with gene regulation in cancer manifestation (144). Haupt et al. described sex-bias regulation of a p53 network across 12 non-reproductive cancer types (145). TP53 mutation is not only more frequent in males compared to females, but X-linked negative regulators of p53 in wild-type TP53 cancers including UBE2A, MAGEA2, and UTP14A show higher expression, and association with reduced survival of male patients. This suggests that having two X chromosomes might protect females from many cancers, a hypothesis further supported by the high number of non-expressed mutations among p53-associated X-linked genes, including FLNA, MED12, HUWE1, and ATRX.
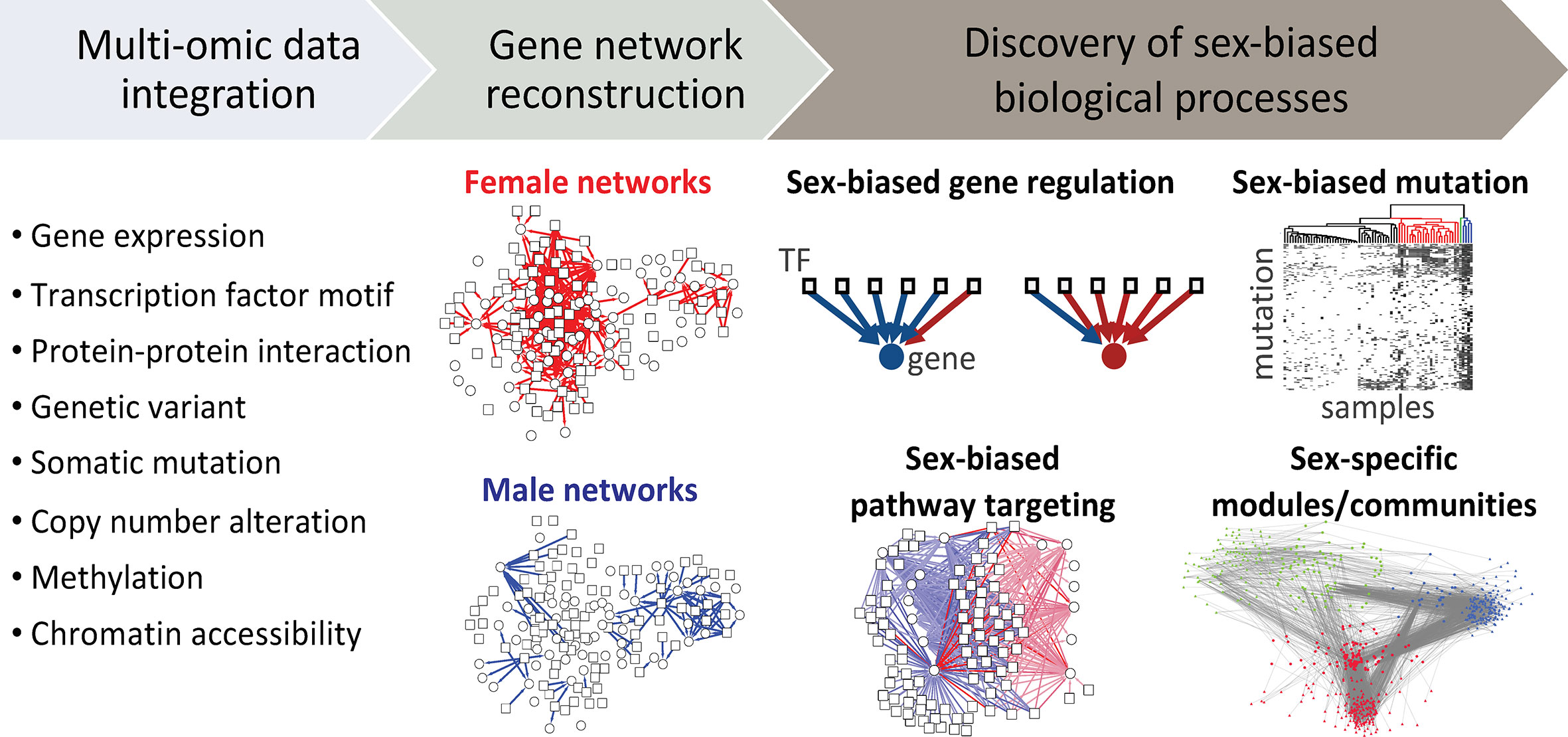
Figure 4 Using systems-based approach to study sex differences. Multi-omic data can be integrated to reconstruct gene networks that combine sex- and gender-biased effects and map how the connectivity and activity of genes differ between males and females. Analyzing the network topology and changes in network structure of males and females can provide insights into molecular mechanisms involved with sex differences in cancer. TF, transcription factor.
Co-expression networks, in which genes are linked if their expression patterns are correlated (above some threshold), have been shown to differ in biologically meaning ways between the sexes. Studies have shown sex-based patterns of gene co-expression are recurrent across many non-reproductive tissues. Several sex-specific modules are identified when using Weighted Gene Co-Expression Network Analysis (WGCNA) to compare gene co-expression networks from males and females (146). Groups of genes with correlated expression are enriched for biological processes, including spermatid differentiation and development, epidermis and ectoderm development (120). Sex-specific modules of co-expressed genes may be modulated by gonadal hormones (147) and sex-biased eQTLs (148). Moreover, co-expression networks of autosomal genes may be altered by X-chromosome dosage (149).
Using gene regulatory networks to model the interactions between transcription factors and their target genes can identify distinct regulatory processes active in males and females, even when these differences cannot be found by analysis of individual gene expression. Using methods such as Passing Attributes between Networks for Data Assimilation (PANDA) and Linear Interpolation to Obtain Network Estimates for Single Samples (LIONESS) to model gene regulatory networks, sex differences in gene regulation was observed genome-wide and across all twenty-nine normal tissues analyzed (46). Several genes are differentially targeted by transcription factors between males and females, including those that are not differentially expressed. Transcription factor differential targeting can result from differences in methylation, chromatin accessibility, post-transcriptional modification of transcription factors, association of transcription factor complexes and co-factors. The sex hormone receptors were not the sole mediators of the sex-biased regulatory processes. The tissues with most sex-biased gene targeting are breast, thyroid, and brain, all of which have strong sex biases in cancer incidence and presentation. This resource of 8,279 regulatory networks across 29 normal tissues can be further investigated on whether there are regulatory processes in normal tissues that might explain sex differences in cancer incidence and development. Another study showed that gene regulatory networks can be used to identify sex-specific modules (or communities) (150). In breast tissue, female-specific modules include not only genes enriched for estrogen receptor signaling pathway, but also processes dysregulated in cancer such as regulation of cell-substrate adhesion, ERK 1/2 cascade, and response to type I interferon.
In analyzing gene regulatory networks in colon cancer, patterns of transcriptional regulation involving genes in the drug metabolism pathway distinguished colon cancer in males and females (151). These regulatory differences were found in primary colon cancer tissues before chemotherapy, and revealed that male and female patients are primed to respond differently to therapy. Importantly, drug metabolism genes were not differentially expressed by sex in the primary tumor, but higher transcription factor targeting of the drug metabolism pathway was associated with higher overall survival in females treated with chemotherapy; this pattern was not observed in males.
Taken together, these analyses of gene regulatory networks indicate that a combination of sex-biased factors may be reflected in the regulatory networks that control gene expression in each sex. This is consistent with substantial sex differences observed in methylation and chromatin accessibility. Males and females can regulate gene expression in different ways, and will likely respond differently to perturbations, such as environmental exposures, aging, somatic mutations, and therapeutic treatment. Even in the absence of strong sex differential expression, the study of regulatory networks can uncover latent sex differences in gene regulation, which might become important drivers of sex-biased manifestations during cancer development and progression.
Incorporating Sex in Genomics Research
Many methods used in genomics research exclude sex chromosomes, and are not well-powered for sex-stratified or interaction analyses (53), largely because there are still many technical challenges to incorporate sex in genomics research. A framework to address sex differences in genomics analysis needs to (1) ensure adequate male and female ratio and sample size for powered analyses of each sex; (2) include the sex chromosomes in downstream analyses; (3) account for sequencing data mapping biases due to sequence similarity between the sex chromosomes; (4) statistically test for sex interaction effects. Sample size selection need to be carefully considered for powered analyses of each sex. For example, to study incidence in a disease with double incidence rate in males, the number of females enrolled need to be double the number of males in order to ensure similar statistical power for both sexes (152).
Sequence read mapping protocols do not account for the imbalance of sex chromosomes and the high sequence similarity between regions of the X and the Y chromosomes. This results in sequencing data that contains poor quality mapping of similar regions between the sex chromosomes, and spurious reads mapped to the Y chromosome in samples from XX genomes. A recent study showed that accounting for these artifacts in sequence mapping protocols can improve variant calling (153) and detection of sex differential gene expression (154). The XYalign is a sex-informed sequence alignment method that first identifies whether the sequencing reads derives from an XX or XY genome based on read balance, and then align the sequencing reads to a sex-appropriate reference genome (153). Sequencing reads from an XX genome are aligned against a reference genome with the Y chromosome masked, and sequencing reads from an XY genome are aligned against a reference genome, masked for regions in the Y that are identical to X (PAR1 and PAR2). It is worth noting that this is a new framework and has not yet been applied to large sequencing data efforts, such as the complete TCGA dataset. Another important quality control check is using the genomic data to annotate biological sex based on sex chromosome complement. This can be done using DNA sequencing data to define the balance of sequencing reads aligned to the X and Y chromosomes. Using RNA-Seq and methylation data, one can perform a principal component analysis using only the expression of Y chromosome genes (151, 155) or the methylation of X chromosome genes (98). A clear separation between male and female samples is expected when visualizing the first two principal components (Figure 5).
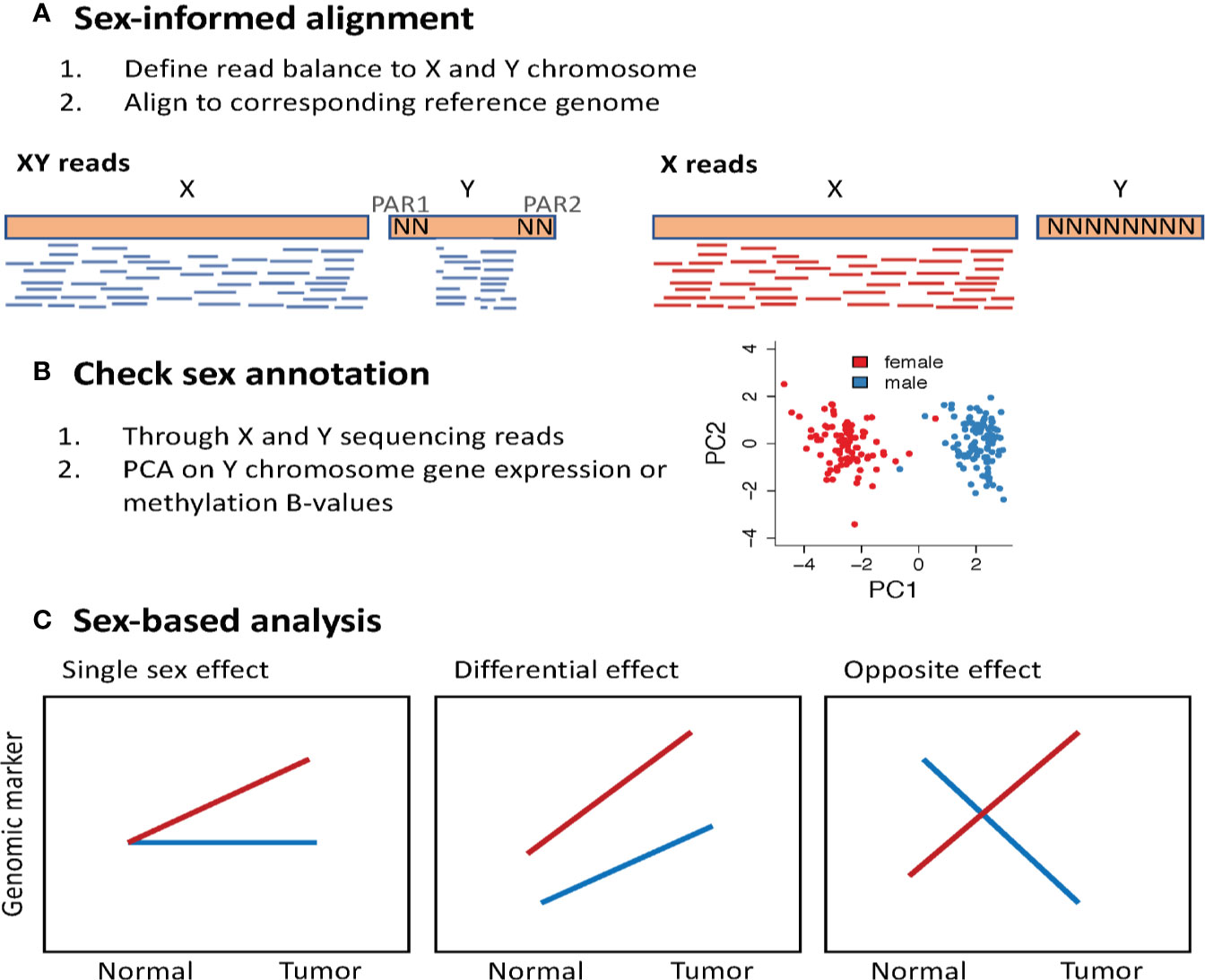
Figure 5 Methodological approach to study sex differences in genomics. (A) Perform sex-informed sequence alignment. The framework proposed in the XYalign tool (153), first identifies whether the sequencing reads are derived from an XX or XY genome, considering the balance of reads aligned to X and Y chromosomes. Next, sequencing reads from an XX genome are aligned against a reference genome with the Y chromosome masked, and sequencing reads from an XY genome are aligned against a reference genome masked for regions in the Y that are identical to X, the pseudoautosomal regions (PAR). (B) Use the genomic data to annotate biological sex based on sex chromosome complement. (C) Sex-based analysis may show how the effect of a genomic marker varies between males and females. PCA, Principal component analysis.
Statistical methods to detect differences between the sexes and interaction by sex have been reviewed elsewhere (152, 156). Sex-stratified analysis can be insufficient to appropriately estimate sex differences and detect sex interactions. Statistical interaction effects can be modeled to obtain the joint effect of two or more exposure variables on a disease outcome, such as the detection of interactions between sex and genetic variants to cause cancer.
Investigating Sex Differences in Model Systems
Pre-clinical models are important tools to investigate how sex differences in genomics influence cancer etiology and manifestation. Several mouse models have been long used in cancer research, including models of spontaneous cancer or cancers induced by carcinogenic compounds, immunocompetent and immunodeficient mice transplanted with patient-derived xenografts. Additional models include transgenic mouse in which oncogenes and tumor suppressor genes are constitutively or conditionally manipulated using conventional approaches or the clustered regularly interspaced short palindromic repeats (CRISPR)-based genome editing (157). These mouse models can be used to evaluate sex-biased effects of genomic variation (4, 158–160). For example, Bahassi et al. evaluated the risk of spontaneous cancer or cancers induced by the carcinogenic compound 7,12-dimethylbenz[a]anthracene using mouse models with a genetic variant on Chk2, a cell cycle checkpoint kinase activated in response to DNA damage and involved in cell cycle arrest and DNA damage repair (54). In wild type mice, the percentage of males that develop spontaneous tumors is higher than females. However, in mice harboring a variant on Chk2, females develop spontaneous tumors with shorter latency and higher frequency.
Mouse models that uncouple the effects from gonadal secretions and sex chromosomes have been developed and can help us better understand the mechanisms associated with sex differences (161, 162). The four core genotypes mouse model is a good example on how to achieve these goals (163). In this model, the sex-determining region Y (SRY) gene is transferred from the Y chromosome to an autosome, and four animals are produced: XX and XY mice with ovaries, as well as XX and XY mice with testes. In this model, the gonad of the animal (testes or ovaries) and the genetic sex (XX or XY) are not related and allows to disassociate phenotype influenced from the sex hormones and sex chromosomes.
Using the four core genotypes mouse model, Kaneko and Li showed that the sex chromosomes are an independent factor associated with higher risk of bladder cancer in males, and the sex hormones amplify the sex-bias effects observed (51). Loss of the EXITS gene Kdm6a increases bladder cancer risk in female mice by reducing the expression of Cdkn1a and Perp, which are targets of p53 that induce cell cycle arrest and apoptosis. This was consistent with the clinical observation that mutations or reduced expression of KDM6A is associated with worse disease-free survival in female patients with bladder cancer but not in male patients.
While cancer cell lines are an indispensable research model, caution is needed when using cell lines to study sex effects. Several male-derived cancer cell lines have lost their Y chromosome, including many commercial cell lines, and therefore have eliminated the effect of sex chromosome complement on the investigational model (164). Using primary cancer cells can be a better alternative. For example, sex-biased chemotherapy sensitivity were observed in a panel of male and female patient-derived glioblastoma cell lines (4). Patient-derived cancer organoids can also be a potential model for studying sex differences. Three-dimensional organoids derived from individual patient's tumor have a high success rate and better recapitulate tumor complexity than two-dimensional cultured cancer cells, showing genomic, transcriptomic, and epigenomic concordance between organoids and their corresponding patient tumors (165–167). Moreover, organoid models have been used effectively for drug testing (168–170).
Colorectal Cancer and The Impact of Conducting Sex-Based Research
Here, we will focus on colorectal cancer as an example to emphasize the need to increase awareness of how sex influences cancer, and the importance of conducting sex-based research to better achieve the goals of precision medicine (Figure 6). Sex differences exist in the incidence, clinical and pathological presentations of colorectal cancer. However, these sex differences are not considered during prevention and treatment.
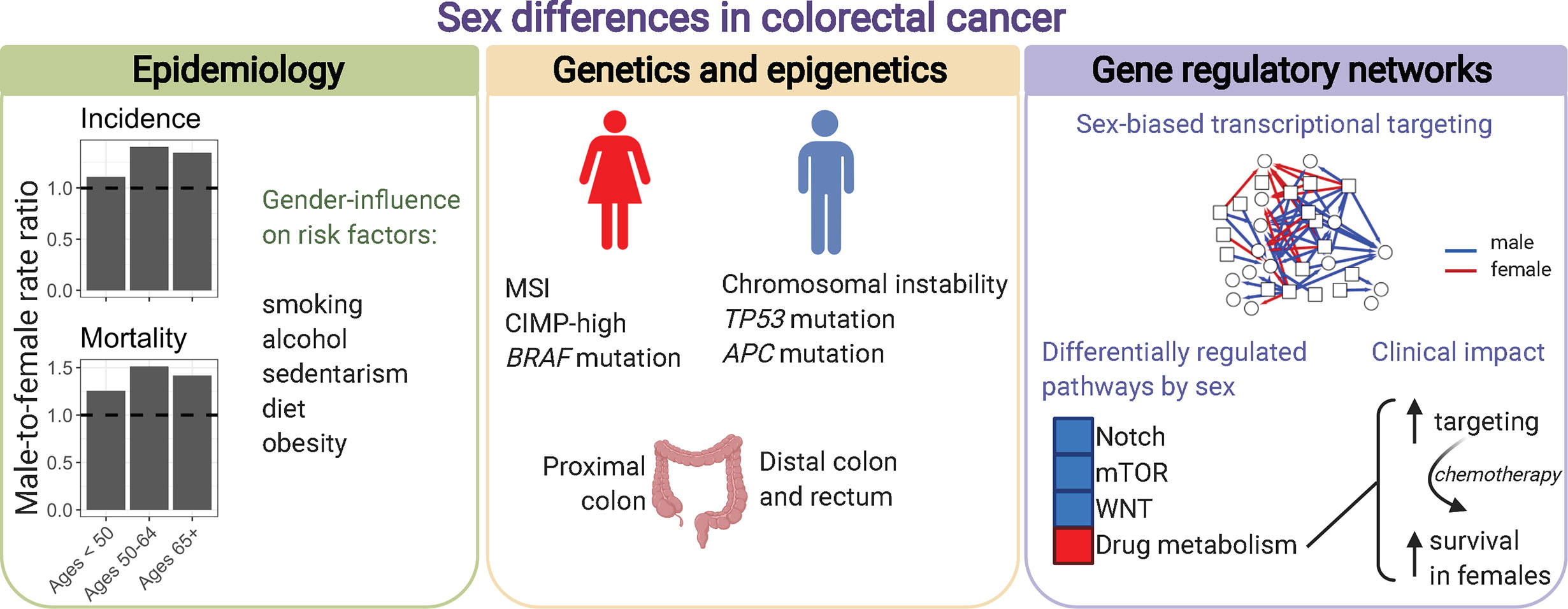
Figure 6 Sex differences in colorectal cancer. Males and females present differences in colorectal cancer incidence, mortality, anatomic site location, genetics, epigenetics, and transcriptional regulatory processes characterized by gene regulatory networks. Incidence and mortality rates per 100,000 individuals in the US were retrieved from the Surveillance, Epidemiology, and End Results explorer (2000–2017).
Colorectal cancer is the third most common cancer, and the third leading cause of cancer death among males and females (171). Overall, males with colorectal cancer have worse survival than females (age-adjusted male-to-female hazard ratio of 1.124) (1). Across races and geographical regions, males have higher incidence of colorectal cancer than females (172). In the United States, the age-adjusted male-to-female incidence rate ratio (IRR) is 1.287 (1). Incidence rates vary according to anatomic subsite and age, with significant changes observed for distal colon cancers (1, 173). Specifically, for sigmoid colon male-to-female IRR increases from 0.869 at 15-39 years, to 1.335 at 40-64 years, and to 1.526 after 65 years (1). Compared to distal colon, patients with proximal colon cancer are predominantly females (174). Biological sex differences, and gender-related behavior can contribute to observed differences in incidence by sex. Known risk factors for colorectal cancer include smoking, alcohol consumption, sedentarism, diet high in red and processed meat, and obesity (175). For example, overall obesity, as measured by high body mass index, is a known risk factor for colorectal cancer with larger associations observed in males compared to females. Studies have shown that not only obesity but the pattern of weight gain throughout life influences the cancer risk. Early life obesity is the primary risk factor for colorectal cancer in females, and adult weight gain in males (176). Sex hormones can also influence colorectal cancer incidence. In general, estrogens are shown to be a protective factor for colorectal cancer in females, whereas testosterone is associated with increased risk in males (25, 28, 177, 178). A high estrogen/testosterone ratio is associated with lower relative risk of colorectal cancer in postmenopausal women, but higher risk in men (179).
Sex differences are observed for many molecular features that are associated with the presentation, progression, and treatment response of colorectal cancer. In general, female sex is associated with microsatellite instability (MSI), CpG island methylator phenotype (CIMP)-high, and BRAF mutation (180). Male sex is associated with chromosomal instability, and mutation on TP53, and APC. MSI, observed in approximately 15% of colorectal cancers, is characterized by a hypermutable phenotype due to a deficient DNA mismatch repair system (181). This molecular phenotype, more frequently observed in females, is a marker of better prognosis and resistance to chemotherapy (182, 183). MSI in sporadic cancer is generally caused by hypermethylation of MLH1. MSI is also the main molecular feature to identify the most frequent form of hereditary colorectal cancer, Lynch syndrome, caused by germline mutation in one or more DNA mismatch repair genes. Germline mutation in these genes is associated with greater lifetime colorectal cancer risk in males compared to females (74% versus 30%) (184). Another example of sex-biased genetic feature is observed for KRAS, a major driver of colorectal tumorigenesis, and marker of resistance to EGFR-targeted therapy (185). While 30%–40% of colorectal cancers carry a KRAS mutation, a higher mutation frequency is found in females compared to males, with pronounced differences for tumors in the proximal colon (75, 186, 187). Moreover, carcinogens may act in a sex-biased manner, supported by an enrichment of a specific type of mutation (G to C transversion) in rectal tumors from females (187).
When comparing large-scale genomic features between males and females with colorectal cancer, including mutation, CNA, methylation, protein, miRNA and gene expression, only a small number of features show significantly sex differences (34, 70). However, similar genomic backgrounds can have different effects in males and females. There are many examples of prognostic and predictive biomarkers that are sex-specific or of greater value to one sex (71, 73, 160, 188). Therefore, the combination of environmental exposures and small size-effect genomic factors can work together on modifying the risk and disease presentation in each sex, and consequently be reflected in the organization of gene networks. In a study modeling gene regulatory networks for each of 1,308 patients with colon cancer, although gene expression only differed significantly in sex chromosome genes, gene regulatory networks exhibited marked sex differences in transcriptional regulatory processes (151). Genes more strongly targeted by transcription factors in males were enriched for pathways with key roles in colon cancer development, including the Notch, mTOR, and WNT signaling pathways. Moreover, genes in the drug metabolism pathway presented differential transcriptional regulation by sex. These sex-linked regulatory patterns were found in primary colon cancer tissues before chemotherapy, and were associated with higher overall survival in females treated with chemotherapy, but not in males. The genes with the largest regulatory sex differences belong to the glutathione S-transferase (GST) family involved in removing xenobiotics. Indeed, females with colorectal cancer have better survival benefit from adjuvant chemotherapy (7), but also higher toxicity (189, 190). Sex differences in drug metabolism and elimination, not only in renal and metabolic clearance (33) but also in gene transcriptional targeting in the tumor (151), suggest adjustment of drug doses by sex. Clinical trials are not designed to identify different optimal doses for both males and females, but dose modification by sex should be further evaluated for targeted therapies and some checkpoint inhibitors that are administered at fixed doses, and also for chemotherapy and antibodies that are dosed according to body surface area and weight.
Discussion
There is an extensive body of clinical and epidemiological evidence for variability in cancer associated with sex and gender and a growing body of multi-omic data that demonstrate the presence of genome-wide sex differences in cancer beyond those affecting reproductive tissues. Males and females respond differently to environmental exposures and genetic and epigenetic alterations. Thus, genomic features associated with cancer etiology, prognosis, and therapeutic response may have differential effects in males and females. For example, changes in gene expression or mutation can be prognostic and predictive markers in only one sex or in opposite directions between the sexes (Table 2). Differences between male and female cancers are observed when analyzing several omic data types, including mutation, copy number, DNA methylation, chromatin accessibility, and expression of mRNA, miRNA, and protein. Integrating multi-omic information can better inform molecular mechanisms involved with sex differences in cancer. For example, analysis of sex-specific gene regulatory networks can identify biological processes differentially regulated by sex, and how sex-biased patterns associate with sex differences observed during tumorigenesis and clinical outcomes.
Despite the overwhelming evidence that sex influences cancer incidence, progression, and therapeutic response, the widely used paradigm in precision medicine generally ignores the sex of the individual. Further, there remain conceptual and methodological gaps to incorporating sex in research and in clinical practice (191, 192). Although the growing literature on sex differences in cancer manifestation and in cancer genomics provide strong arguments for implementation of sex-aware precision therapy, additional data are needed to fill the gaps in our understanding. This will require that sex be explicitly considered as a key variable in the design and conduct of both preclinical and translational research. Continuing to investigate sex and gender differences in cancer will inform sex-specific strategies for cancer prevention and early diagnosis and will lead to more refined precision medicine therapeutic strategies that will improve treatment and outcomes including survival.
Author Contributions
CL-R wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
CL-R, and JQ are supported by a grant from the National Cancer Institute, National Institutes of Health, R35 CA220523. JQ is also supported by U24 CA231846. DLD is supported by grants from the National Heart, Lung, and Blood Institute (5P01HL105339, 5R01HL111759, and 5P01HL114501) and by a BWH First-In-Women Precision Medicine IGNITE award.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Figures 3 and 6 were created with BioRender.com.
References
1. Dong M, Cioffi G, Wang J, Waite KA, Ostrom QT, Kruchko C, et al. Sex Differences in Cancer Incidence and Survival: A Pan-Cancer Analysis. Cancer Epidemiol Biomarkers Prev (2020) 29:1389–97. doi: 10.1158/1055-9965.EPI-20-0036
2. Klimm B, Reineke T, Haverkamp H, Behringer K, Eich HT, Josting A, et al. German Hodgkin Study Group. Role of hematotoxicity and sex in patients with Hodgkin’s lymphoma: an analysis from the German Hodgkin Study Group. J Clin Oncol (2005) 23:8003–11. doi: 10.1200/JCO.2005.205.60
3. van den Berg H, Paulussen M, Le Teuff G, Judson I, Gelderblom H, Dirksen U, et al. Impact of gender on efficacy and acute toxicity of alkylating agent -based chemotherapy in Ewing sarcoma: secondary analysis of the Euro-Ewing99-R1 trial. Eur J Cancer (2015) 51:2453–64. doi: 10.1016/j.ejca.2015.06.123
4. Yang W, Warrington NM, Taylor SJ, Whitmire P, Carrasco E, Singleton KW, et al. Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Sci Transl Med (2019) 11:eaao5253. doi: 10.1126/scitranslmed.aao5253
5. Singh S, Parulekar W, Murray N, Feld R, Evans WK, Tu D, et al. Influence of sex on toxicity and treatment outcome in small-cell lung cancer. J Clin Oncol (2005) 23:850–6. doi: 10.1200/JCO.2005.03.171
6. Wakelee HA, Wang W, Schiller JH, Langer CJ, Sandler AB, Belani CP, et al. Survival Differences by Sex for Patients with Advanced Non-small Cell Lung Cancer on Eastern Cooperative Oncology Group Trial 1594. J Thorac Oncol (2006) 1:441–6. doi: 10.1016/S1556-0864(15)31609-9
7. Elsaleh H, Joseph D, Grieu F, Zeps N, Spry N, Iacopetta B. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet (2000) 355:1745–50. doi: 10.1016/S0140-6736(00)02261-3
8. Conforti F, Pala L, Bagnardi V, De Pas T, Martinetti M, Viale G, et al. Cancer immunotherapy efficacy and patients’ sex: a systematic review and meta-analysis. Lancet Oncol (2018) 19:737–46. doi: 10.1016/S1470-2045(18)30261-4
9. Conforti F, Pala L, Bagnardi V, Viale G, De Pas T, Pagan E, et al. Sex-Based Heterogeneity in Response to Lung Cancer Immunotherapy: A Systematic Review and Meta-Analysis. J Natl Cancer Inst (2019) 111:772–81. doi: 10.1093/jnci/djz094
10. Wallis CJD, Butaney M, Satkunasivam R, Freedland SJ, Patel SP, Hamid O, et al. Association of Patient Sex With Efficacy of Immune Checkpoint Inhibitors and Overall Survival in Advanced Cancers: A Systematic Review and Meta-analysis. JAMA Oncol (2019) 5:529–36. doi: 10.1001/jamaoncol.2018.5904
11. Wang S, Cowley LA, Liu X-S. Sex Differences in Cancer Immunotherapy Efficacy, Biomarkers, and Therapeutic Strategy. Molecules (2019) 24:3214. doi: 10.3390/molecules24183214
12. Klein SL, Morgan R. The impact of sex and gender on immunotherapy outcomes. Biol Sex Differ (2020) 11:24. doi: 10.1186/s13293-020-00301-y
13. Castro A, Pyke RM, Zhang X, Thompson WK, Day C-P, Alexandrov LB, et al. Strength of immune selection in tumors varies with sex and age. Nat Commun (2020) 11:4128. doi: 10.1038/s41467-020-17981-0
14. Clayton JA, Collins FS. Policy: NIH to balance sex in cell and animal studies. Nature (2014) 509:282–3. doi: 10.1038/509282a
15. Clayton JA, Tannenbaum C. Reporting Sex, Gender, or Both in Clinical Research? JAMA (2016) 316:1863. doi: 10.1001/jama.2016.16405
16. Polderman TJC, Kreukels BPC. The Biological Contributions to Gender Identity and Gender Diversity: Bringing Data to the Table on behalf of the International Gender Diversity Genomics Consortium. Behav Genet (2018) 48:23. doi: 10.1007/s10519-018-9889-z
17. Vaught JB, Henderson MK, Compton CC. Biospecimens and Biorepositories: From Afterthought to Science. Cancer Epidemiol Biomarkers Prev (2012) 21:253–5. doi: 10.1158/1055-9965.EPI-11-1179
18. Bartz D, Chitnis T, Kaiser UB, Rich-Edwards JW, Rexrode KM, Pennell PB, et al. Clinical Advances in Sex- and Gender-Informed Medicine to Improve the Health of All. JAMA Intern Med (2020) 180:574. doi: 10.1001/jamainternmed.2019.7194
19. Safer JD, Tangpricha V. Care of Transgender Persons. N Engl J Med (2019) 381:2451–60. doi: 10.1056/NEJMcp1903650
20. Quinn GP, Sanchez JA, Sutton SK, Vadaparampil ST, Nguyen GT, Green BL, et al. Cancer and lesbian, gay, bisexual, transgender/transsexual, and queer/questioning (LGBTQ) populations. CA Cancer J Clin (2015) 65:384–400. doi: 10.3322/caac.21288
21. Edgren G, Liang L, Adami H-O, Chang ET. Enigmatic sex disparities in cancer incidence. Eur J Epidemiol (2012) 27:187–96. doi: 10.1007/s10654-011-9647-5
22. Bertin M, Thébaud-Mony A, Counil E. Giscop93 study group. Do Women and Men Have the Same Patterns of Multiple Occupational Carcinogenic Exposures? Results from a Cohort of Cancer Patients. Ann Work Expo Heal (2018) 62:450–64. doi: 10.1093/annweh/wxx116
23. Pukkala E, Martinsen JI, Lynge E, Gunnarsdottir HK, Sparén P, Tryggvadottir L, et al. Occupation and cancer - follow-up of 15 million people in five Nordic countries. Acta Oncol (2009) 48:646–790. doi: 10.1080/02841860902913546
24. Clocchiatti A, Cora E, Zhang Y, Dotto GP. Sexual dimorphism in cancer. Nat Rev Cancer (2016) 16:330–9. doi: 10.1038/nrc.2016.30
25. Chlebowski RT, Wactawski-Wende J, Ritenbaugh C, Hubbell FA, Ascensao J, Rodabough RJ, et al. Estrogen plus progestin and colorectal cancer in postmenopausal women. N Engl J Med (2004) 350:991–1004. doi: 10.1056/NEJMoa032071
26. Hsu J-W, Hsu I, Xu D, Miyamoto H, Liang L, Wu X-R, et al. Decreased tumorigenesis and mortality from bladder cancer in mice lacking urothelial androgen receptor. Am J Pathol (2013) 182:1811–20. doi: 10.1016/j.ajpath.2013.01.018
27. Miyamoto H, Yang Z, Chen Y, Ishiguro H, Uemura H, Kubota Y, et al. Promotion of Bladder Cancer Development and Progression by Androgen Receptor Signals. J Natl Cancer Inst (2007) 99:558–68. doi: 10.1093/JNCI/DJK113
28. Amos-Landgraf JM, Heijmans J, Wielenga MCB, Dunkin E, Krentz KJ, Clipson L, et al. Sex disparity in colonic adenomagenesis involves promotion by male hormones, not protection by female hormones. Proc Natl Acad Sci USA (2014) 111:16514–9. doi: 10.1073/pnas.1323064111
29. Williams LA, Richardson M, Marcotte EL, Poynter JN, Spector LG. Sex ratio among childhood cancers by single year of age. Pediatr Blood Cancer (2019) 66:e27620. doi: 10.1002/pbc.27620
30. Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol (2016) 16:626–38. doi: 10.1038/nri.2016.90
31. Giuliano AR, Nyitray AG, Kreimer AR, Pierce Campbell CM, Goodman MT, Sudenga SL, et al. EUROGIN 2014 roadmap: differences in human papillomavirus infection natural history, transmission and human papillomavirus-related cancer incidence by gender and anatomic site of infection. Int J Cancer (2015) 136:2752–60. doi: 10.1002/ijc.29082
32. Özdemir BC, Csajka C, Dotto G-P, Wagner AD. Sex Differences in Efficacy and Toxicity of Systemic Treatments: An Undervalued Issue in the Era of Precision Oncology. J Clin Oncol (2018) 36:2680–3. doi: 10.1200/JCO.2018.78.3290
33. Wagner A, Oertelt-Prigione S, Adjei A, Buclin T, Cristina V, Csajka C, et al. Gender Medicine and Oncology: Report and Consensus of an ESMO Workshop. Ann Oncol (2019) 30:1914–24. doi: 10.1093/ANNONC/MDZ414
34. Yuan Y, Liu L, Chen H, Wang Y, Xu Y, Mao H, et al. Comprehensive Characterization of Molecular Differences in Cancer between Male and Female Patients. Cancer Cell (2016) 29:711–22. doi: 10.1016/j.ccell.2016.04.001
35. Arnold A. A general theory of sexual differentiation. J Neurosci Res (2017) 95:291–300. doi: 10.1002/jnr.23884
36. Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho T-J, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature (2014) 508:494–9. doi: 10.1038/nature13206
37. Forsberg LA, Rasi C, Malmqvist N, Davies H, Pasupulati S, Pakalapati G, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet (2014) 46:624–8. doi: 10.1038/ng.2966
38. Noveski P, Madjunkova S, Sukarova Stefanovska E, Matevska Geshkovska N, Kuzmanovska M, Dimovski A, et al. Loss of Y Chromosome in Peripheral Blood of Colorectal and Prostate Cancer Patients. PLoS One (2016) 11:e0146264. doi: 10.1371/journal.pone.0146264
39. Hollows R, Wei W, Cazier J-B, Mehanna H, Parry G, Halford G, et al. Association between loss of Y chromosome and poor prognosis in male head and neck squamous cell carcinoma. Head Neck (2019) 41:993–1006. doi: 10.1002/hed.25537
40. Agahozo MC, Timmermans MAM, Sleddens HFBM, Foekens R, Trapman-Jansen AMAC, Schröder CP, et al. Loss of Y-Chromosome during Male Breast Carcinogenesis. Cancers (Basel) (2020) 12:631. doi: 10.3390/cancers12030631
41. Weakley SM, Wang H, Yao Q, Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg (2011) 35:1751–6. doi: 10.1007/s00268-010-0951-0
42. Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell (2006) 9:121–32. doi: 10.1016/j.ccr.2006.01.013
43. Chaligné R, Heard E. X-chromosome inactivation in development and cancer. FEBS Lett (2014) 588:2514–22. doi: 10.1016/j.febslet.2014.06.023
44. Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell (2013) 152:727–42. doi: 10.1016/j.cell.2013.01.034
45. Tukiainen T, Villani A-C, Yen A, Rivas MA, Marshall JL, Satija R, et al. Landscape of X chromosome inactivation across human tissues. Nature (2017) 550:244–8. doi: 10.1038/nature24265
46. Lopes-Ramos CM, Chen C-Y, Kuijjer ML, Paulson JN, Sonawane AR, Fagny M, et al. Sex Differences in Gene Expression and Regulatory Networks across 29 Human Tissues. Cell Rep (2020) 31:107795. doi: 10.1016/j.celrep.2020.107795
47. Dunford A, Weinstock DM, Savova V, Schumacher SE, Cleary JP, Yoda A, et al. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat Genet (2017) 49:10–6. doi: 10.1038/ng.3726
48. Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood (2015) 125:13–21. doi: 10.1182/blood-2014-05-577270
49. Li X, Zhang Y, Zheng L, Liu M, Chen CD, Jiang H. UTX is an escape from X-inactivation tumor-suppressor in B cell lymphoma. Nat Commun (2018) 9:2720. doi: 10.1038/s41467-018-05084-w
50. Andricovich J, Perkail S, Kai Y, Casasanta N, Peng W, Tzatsos A. Loss of KDM6A Activates Super-Enhancers to Induce Gender-Specific Squamous-like Pancreatic Cancer and Confers Sensitivity to BET Inhibitors. Cancer Cell (2018) 33:512–526.e8. doi: 10.1016/j.ccell.2018.02.003
51. Kaneko S, Li X. X chromosome protects against bladder cancer in females via a KDM6A -dependent epigenetic mechanism. Sci Adv (2018) 4:eaar5598. doi: 10.1126/sciadv.aar5598
52. Ji J, Zöller B, Sundquist J, Sundquist K. Risk of solid tumors and hematological malignancy in persons with Turner and Klinefelter syndromes: A national cohort study. Int J Cancer (2016) 139:754–8. doi: 10.1002/ijc.30126
53. Khramtsova EA, Davis LK, Stranger BE. The role of sex in the genomics of human complex traits. Nat Rev Genet (2019) 20:173–90. doi: 10.1038/s41576-018-0083-1
54. Bahassi EM, Robbins SB, Yin M, Boivin GP, Kuiper R, van Steeg H, et al. Mice with the CHEK2*1100delC SNP are predisposed to cancer with a strong gender bias. Proc Natl Acad Sci USA (2009) 106:17111–6. doi: 10.1073/pnas.0909237106
55. Hwang S-J, Lozano G, Amos CI, Strong LC. Germline p53 mutations in a cohort with childhood sarcoma: sex differences in cancer risk. Am J Hum Genet (2003) 72:975–83. doi: 10.1086/374567
56. Wu C-C, Shete S, Amos CI, Strong LC. Joint effects of germ-line p53 mutation and sex on cancer risk in Li-Fraumeni syndrome. Cancer Res (2006) 66:8287–92. doi: 10.1158/0008-5472.CAN-05-4247
57. Bond GL, Levine AJ. A single nucleotide polymorphism in the p53 pathway interacts with gender, environmental stresses and tumor genetics to influence cancer in humans. Oncogene (2007) 26:1317–23. doi: 10.1038/sj.onc.1210199
58. Bond GL, Menin C, Bertorelle R, Alhopuro P, Aaltonen LA, Levine AJ. MDM2 SNP309 accelerates colorectal tumour formation in women. J Med Genet (2006) 43:950–2. doi: 10.1136/jmg.2006.043539
59. Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H, et al. MDM2 SNP309 Accelerates Tumor Formation in a Gender-Specific and Hormone-Dependent Manner. Cancer Res (2006) 66:5104–10. doi: 10.1158/0008-5472.CAN-06-0180
60. Singh SK, Lupo PJ, Scheurer ME, Saxena A, Kennedy AE, Ibrahimou B, et al. A childhood acute lymphoblastic leukemia genome-wide association study identifies novel sex-specific risk variants. Med (Baltimore) (2016) 95:e5300. doi: 10.1097/MD.0000000000005300
61. Do TN, Ucisik-Akkaya E, Davis CF, Morrison BA, Dorak MT. An intronic polymorphism of IRF4 gene influences gene transcription in vitro and shows a risk association with childhood acute lymphoblastic leukemia in males. Biochim Biophys Acta (2010) 1802:292–300. doi: 10.1016/j.bbadis.2009.10.015
62. Tian W, Zeng X-M, Li L-X, Jin H-K, Luo Q-Z, Wang F, et al. Gender-specific associations between MICA-STR and nasopharyngeal carcinoma in a southern Chinese Han population. Immunogenetics (2006) 58:113–21. doi: 10.1007/s00251-006-0093-6
63. Tse K-P, Su W-H, Yang M, Cheng H-Y, Tsang N-M, Chang K-P, et al. A gender-specific association of CNV at 6p21.3 with NPC susceptibility. Hum Mol Genet (2011) 20:2889–96. doi: 10.1093/hmg/ddr191
64. Bolufer P, Collado M, Barragán E, Cervera J, Calasanz M-J, Colomer D, et al. The potential effect of gender in combination with common genetic polymorphisms of drug-metabolizing enzymes on the risk of developing acute leukemia. Haematologica (2007) 92:308–14. doi: 10.3324/haematol.10752
65. Zheng L, Wang Y, Schabath M, Grossman H, Wu X. Sulfotransferase 1A1 (SULT1A1) Polymorphism and Bladder Cancer Risk: A Case-Control Study. Cancer Lett (2003) 202:61–9. doi: 10.1016/J.CANLET.2003.08.007
66. Shi Q, Zhang Z, Li G, Pillow PC, Hernandez LM, Spitz MR, et al. Sex differences in risk of lung cancer associated with methylene-tetrahydrofolate reductase polymorphisms. Cancer Epidemiol Biomarkers Prev (2005) 14:1477–84. doi: 10.1158/1055-9965.EPI-04-0905
67. Suzuki H, Morris JS, Li Y, Doll MA, Hein DW, Liu J, et al. Interaction of the cytochrome P4501A2, SULT1A1 and NAT gene polymorphisms with smoking and dietary mutagen intake in modification of the risk of pancreatic cancer. Carcinogenesis (2008) 29:1184–91. doi: 10.1093/carcin/bgn085
68. Xiao D, Pan H, Li F, Wu K, Zhang X, He J. Analysis of ultra-deep targeted sequencing reveals mutation burden is associated with gender and clinical outcome in lung adenocarcinoma. Oncotarget (2016) 7:22857–64. doi: 10.18632/oncotarget.8213
69. Gupta S, Artomov M, Goggins W, Daly M, Tsao H. Gender Disparity and Mutation Burden in Metastatic Melanoma. J Natl Cancer Inst (2015) 107:djv221. doi: 10.1093/JNCI/DJV221
70. Li CH, Haider S, Shiah Y-J, Thai K, Boutros PC. Sex Differences in Cancer Driver Genes and Biomarkers. Cancer Res (2018) 78:5527–37. doi: 10.1158/0008-5472.CAN-18-0362
71. Wangefjord S, Sundström M, Zendehrokh N, Lindquist KE, Nodin B, Jirström K, et al. Sex differences in the prognostic significance of KRAS codons 12 and 13, and BRAF mutations in colorectal cancer: a cohort study. Biol Sex Differ (2013) 4:17. doi: 10.1186/2042-6410-4-17
72. Ricketts CJ, Linehan WM. Gender Specific Mutation Incidence and Survival Associations in Clear Cell Renal Cell Carcinoma (CCRCC). PLoS One (2015) 10:e0140257. doi: 10.1371/journal.pone.0140257
73. Warren RS, Atreya CE, Niedzwiecki D, Weinberg VK, Donner DB, Mayer RJ, et al. Association of TP53 mutational status and gender with survival after adjuvant treatment for stage III colon cancer: results of CALGB 89803. Clin Cancer Res (2013) 19:5777–87. doi: 10.1158/1078-0432.CCR-13-0351
74. Garufi C, Giacomini E, Torsello A, Sperduti I, Melucci E, Mottolese M, et al. Gender effects of single nucleotide polymorphisms and miRNAs targeting clock-genes in metastatic colorectal cancer patients (mCRC). Sci Rep (2016) 6:34006. doi: 10.1038/srep34006
75. Breivik J, Lothe RA, Meling GI, Rognum TO, Børresen-Dale A-L, Gaudernack G. Different genetic pathways to proximal and distal colorectal cancer influenced by sex-related factors. Int J Cancer (1997) 74:664–9. doi: 10.1002/(SICI)1097-0215(19971219)74:6<664::AID-IJC18>3.0.CO;2-5
76. Mundi N, Ghasemi F, Zeng PYF, Prokopec SD, Patel K, Kim HAJ, et al. Sex disparities in head and neck cancer driver genes: An analysis of the TCGA dataset. Oral Oncol (2020) 104:104614. doi: 10.1016/J.ORALONCOLOGY.2020.104614
77. Shigematsu H, Gazdar A. Somatic Mutations of Epidermal Growth Factor Receptor Signaling Pathway in Lung Cancers. Int J Cancer (2006) 118:257–62. doi: 10.1002/IJC.21496
78. Toyooka S, Matsuo K, Shigematsu H, Kosaka T, Tokumo M, Yatabe Y, et al. The impact of sex and smoking status on the mutational spectrum of epidermal growth factor receptor gene in non small cell lung cancer. Clin Cancer Res (2007) 13:5763–8. doi: 10.1158/1078-0432.CCR-07-0216
79. Kure EH, Ryberg D, Hewer A, Phillips DH, Skaug V, Bæera R, et al. p53 mutations in lung tumours: relationship to gender and lung DNA adduct levels. Carcinogenesis (1996) 17:2201–5. doi: 10.1093/carcin/17.10.2201
80. Guinee DG, Travis WD, Trivers GE, De Benedetti VM, Cawley H, Welsh JA, et al. Gender comparisons in human lung cancer: analysis of p53 mutations, anti-p53 serum antibodies and C-erbB-2 expression. Carcinogenesis (1995) 16:993–1002. doi: 10.1093/carcin/16.5.993
81. Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene (2002) 21:7435–51. doi: 10.1038/sj.onc.1205803
82. Mollerup S, Ryberg D, Hewer A, Phillips DH, Haugen A. Sex Differences in Lung CYP1A1 Expression and DNA Adduct Levels among Lung Cancer Patients. Cancer Res (1999) 59:3317–20.
83. Ryberg D, Hewer A, Phillips DH, Haugen A. Different Susceptibility to Smoking-induced DNA Damage among Male and Female Lung Cancer Patients. Cancer Res (1994) 54:5801–3.
84. Qingyi Wei Q, Cheng L, Amos CI, Wang L-E, Guo Z, Hong WK, et al. Repair of Tobacco Carcinogen-Induced DNA Adducts and Lung Cancer Risk: a Molecular Epidemiologic Study. J Natl Cancer Inst (2000) 92:1764–72. doi: 10.1093/jnci/92.21.1764
85. Liu J, Morgan M, Hutchison K, Calhoun V. A study of the influence of sex on genome wide methylation. PLoS One (2010) 5:e10028. doi: 10.1371/JOURNAL.PONE.0010028
86. Singmann P, Shem-Tov D, Wahl S, Grallert H, Fiorito G, Shin S-Y, et al. Characterization of whole-genome autosomal differences of DNA methylation between men and women. Epigenet Chromatin (2015) 8:43. doi: 10.1186/s13072-015-0035-3
87. Maschietto M, Bastos LC, Tahira AC, Bastos EP, Euclydes VLV, Brentani A, et al. Sex differences in DNA methylation of the cord blood are related to sex-bias psychiatric diseases. Sci Rep (2017) 7:44547. doi: 10.1038/srep44547
88. Yousefi P, Huen K, Davé V, Barcellos L, Eskenazi B, Holland N. Sex differences in DNA methylation assessed by 450 K BeadChip in newborns. BMC Genomics (2015) 16:911. doi: 10.1186/s12864-015-2034-y
89. Xu H, Wang F, Liu Y, Yu Y, Gelernter J, Zhang H. Sex-biased methylome and transcriptome in human prefrontal cortex. Hum Mol Genet (2014) 23:1260. doi: 10.1093/HMG/DDT516
90. García-Calzón S, Perfilyev A, de Mello VD, Pihlajamäki J, Ling C. Sex Differences in the Methylome and Transcriptome of the Human Liver and Circulating HDL-Cholesterol Levels. J Clin Endocrinol Metab (2018) 103:4395–408. doi: 10.1210/jc.2018-00423
91. Reizel Y, Spiro A, Sabag O, Skversky Y, Hecht M, Keshet I, et al. Gender-specific postnatal demethylation and establishment of epigenetic memory. Genes Dev (2015) 29:923. doi: 10.1101/GAD.259309.115
92. Tapp HS, Commane DM, Bradburn DM, Arasaradnam R, Mathers JC, Johnson IT, et al. Nutritional factors and gender influence age-related DNA methylation in the human rectal mucosa. Aging Cell (2013) 12:148. doi: 10.1111/ACEL.12030
93. Davegårdh C, Hall Wedin E, Broholm C, Henriksen TI, Pedersen M, Pedersen BK, et al. Sex influences DNA methylation and gene expression in human skeletal muscle myoblasts and myotubes. Stem Cell Res Ther (2019) 10:26. doi: 10.1186/s13287-018-1118-4
94. Hall E, Volkov P, Dayeh T, Esguerra JLS, Salö S, Eliasson L, et al. Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol (2014) 15:522. doi: 10.1186/s13059-014-0522-z
95. Ho B, Greenlaw K, Al Tuwaijri A, Moussette S, Martínez F, Giorgio E, et al. X chromosome dosage and presence of SRY shape sex-specific differences in DNA methylation at an autosomal region in human cells. Biol Sex Differ (2018) 9:10. doi: 10.1186/s13293-018-0169-7
96. Ghahramani N, Ngun T, Chen P, Tian Y, Krishnan S, Muir S, et al. The effects of perinatal testosterone exposure on the DNA methylome of the mouse brain are late-emerging. Biol Sex Differ (2014) 5:8. doi: 10.1186/2042-6410-5-8
97. Nugent BM, Wright CL, Shetty AC, Hodes GE, Lenz KM, Mahurkar A, et al. Brain feminization requires active repression of masculinization via DNA methylation. Nat Neurosci (2015) 18:690–7. doi: 10.1038/nn.3988
98. McCarthy NS, Melton PE, Cadby G, Yazar S, Franchina M, Moses EK, et al. Meta-analysis of human methylation data for evidence of sex-specific autosomal patterns. BMC Genomics (2014) 15:981. doi: 10.1186/1471-2164-15-981
99. Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, et al. The Relationship of DNA Methylation with Age, Gender and Genotype in Twins and Healthy Controls. PLoS One (2009) 4:e6767. doi: 10.1371/journal.pone.0006767
100. Vaissiè T, Hung RJ, Zaridze D, Moukeria A, Cuenin C, Fasolo V, et al. Quantitative Analysis of DNA Methylation Profiles in Lung Cancer Identifies Aberrant DNA Methylation of Specific Genes and Its Association with Gender and Cancer Risk Factors. Cancer Res (2009) 69:243–52. doi: 10.1158/0008-5472.CAN-08-2489
101. Wu J-Y, Wang J, Lai J-C, Cheng Y-W, Yeh K-T, Wu T-C, et al. Association of O6-Methylguanine-DNA Methyltransferase (MGMT) Promoter Methylation with p53 Mutation Occurrence in Non-Small Cell Lung Cancer with Different Histology, Gender, and Smoking Status. Ann Surg Oncol (2008) 15:3272–7. doi: 10.1245/s10434-008-0078-9
102. Bi H, Liu Y, Pu R, Xia T, Sun H, Huang H, et al. CHST7 Gene Methylation and Sex-Specific Effects on Colorectal Cancer Risk. Dig Dis Sci (2019) 64:2158–66. doi: 10.1007/s10620-019-05530-9
103. Lin S, Liu Y, Goldin LR, Lyu C, Kong X, Zhang Y, et al. Sex-related DNA methylation differences in B cell chronic lymphocytic leukemia. Biol Sex Differ (2019) 10:2. doi: 10.1186/s13293-018-0213-7
104. Köhler CU, Bonberg N, Ahrens M, Behrens T, Hovanec J, Eisenacher M, et al. Noninvasive diagnosis of urothelial cancer in urine using DNA hypermethylation signatures-Gender matters. Int J Cancer (2019) 145(10):2861–72. doi: 10.1002/ijc.32356
105. Wilhelm CS, Kelsey KT, Butler R, Plaza S, Gagne L, Zens MS, et al. Implications of LINE1 Methylation for Bladder Cancer Risk in Women. Clin Cancer Res (2010) 16:1682–9. doi: 10.1158/1078-0432.CCR-09-2983
106. Kovalchuk O, Burke P, Besplug J, Slovack M, Filkowski J, Pogribny I. Methylation changes in muscle and liver tissues of male and female mice exposed to acute and chronic low-dose X-ray-irradiation. Mutat Res Mol Mech Mutagen (2004) 548:75–84. doi: 10.1016/J.MRFMMM.2003.12.016
107. Pogribny I, Raiche J, Slovack M, Kovalchuk O. Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochem Biophys Res Commun (2004) 320:1253–61. doi: 10.1016/J.BBRC.2004.06.081
108. Kukurba KR, Parsana P, Balliu B, Smith KS, Zappala Z, Knowles DA, et al. Impact of the X Chromosome and sex on regulatory variation. Genome Res (2016) 26:768–77. doi: 10.1101/gr.197897.115
109. Sugathan A, Waxman D. Genome-wide Analysis of Chromatin States Reveals Distinct Mechanisms of Sex-Dependent Gene Regulation in Male and Female Mouse Liver. Mol Cell Biol (2013) 33:3594–610. doi: 10.1128/MCB.00280-13
110. Liu Y. Clinical implications of chromatin accessibility in human cancers. Oncotarget (2020) 11:1666–78. doi: 10.18632/oncotarget.27584
111. Ling G, Sugathan A, Mazor T, Fraenkel E, Waxman DJ. Unbiased, genome-wide in vivo mapping of transcriptional regulatory elements reveals sex differences in chromatin structure associated with sex-specific liver gene expression. Mol Cell Biol (2010) 30:5531–44. doi: 10.1128/MCB.00601-10
112. Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet (2008) 9:911–22. doi: 10.1038/nrg2415
113. Dimas AS, Nica AC, Montgomery SB, Stranger BE, Raj T, Buil A, et al. Sex-biased genetic effects on gene regulation in humans. Genome Res (2012) 22:2368–75. doi: 10.1101/gr.134981.111
114. Yao C, Joehanes R, Johnson AD, Huan T, Esko T, Ying S, et al. Sex- and age-interacting eQTLs in human complex diseases. Hum Mol Genet (2014) 23:1947–56. doi: 10.1093/hmg/ddt582
115. Shen JJ, Wang Y-F, Yang W. Sex-Interacting mRNA- and miRNA-eQTLs and Their Implications in Gene Expression Regulation and Disease. Front Genet (2019) 10:313. doi: 10.3389/fgene.2019.00313
116. Kassam I, Lloyd-Jones L, Holloway A, Small KS, Zeng B, Bakshi A, et al. Autosomal genetic control of human gene expression does not differ across the sexes. Genome Biol (2016) 17:248. doi: 10.1186/s13059-016-1111-0
117. Oliva M, Muñoz-Aguirre M, Kim-Hellmuth S, Wucher V, Gewirtz ADH, Cotter DJ, et al. The impact of sex on gene expression across human tissues. Science (2020) 369:eaba3066. doi: 10.1126/science.aba3066
118. Natri HM, Wilson MA, Buetow KH. Distinct molecular etiologies of male and female hepatocellular carcinoma. BMC Cancer (2019) 19:951. doi: 10.1186/s12885-019-6167-2
119. Naqvi S, Godfrey AK, Hughes JF, Goodheart ML, Mitchell RN, Page DC. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science (2019) 365:eaaw7317. doi: 10.1126/science.aaw7317
120. Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, et al. The human transcriptome across tissues and individuals. Science (2015) 348:660–5. doi: 10.1126/science.aaa0355
121. Gershoni M, Pietrokovski S. The landscape of sex-differential transcriptome and its consequent selection in human adults. BMC Biol (2017) 15:7. doi: 10.1186/s12915-017-0352-z
122. Kassam I, Wu Y, Yang J, Visscher PM, McRae AF. Tissue-specific sex differences in human gene expression. Hum Mol Genet (2019) 28:2976–86. doi: 10.1093/hmg/ddz090
123. Li Y, He C, Li W, Zhang R, Duan Y. Transcriptome Analysis Reveals Gender-Specific Differences in Overall Metabolic Response of Male and Female Patients in Lung Adenocarcinoma. PLoS One (2020) 15:e0230796. doi: 10.1371/JOURNAL.PONE.0230796
124. de Jong JJ, Boormans JL, van Rhijn BWG, Seiler R, Boorjian SA, Konety B, et al. Distribution of Molecular Subtypes in Muscle-invasive Bladder Cancer Is Driven by Sex-specific Differences. Eur Urol Oncol (2020) 3(4):420–3. doi: 10.1016/j.euo.2020.02.010
125. Callari M, Cappelletti V, De Cecco L, Musella V, Miodini P, Veneroni S, et al. Gene expression analysis reveals a different transcriptomic landscape in female and male breast cancer. Breast Cancer Res Treat (2011) 127:601–10. doi: 10.1007/s10549-010-1015-8
126. Wu Y, Yao N, Feng Y, Tian Z, Yang Y, Zhao Y. Identification and characterization of sexual dimorphism-linked gene expression profile in hepatocellular carcinoma. Oncol Rep (2019) 42:937–52. doi: 10.3892/or.2019.7217
127. Mostertz W, Stevenson M, Acharya C, Chan I, Walters K, Lamlertthon W, et al. Age- and Sex-Specific Genomic Profiles in Non–Small Cell Lung Cancer. JAMA (2010) 303:535. doi: 10.1001/jama.2010.80
128. Wierinckx A, Delgrange E, Bertolino P, François P, Chanson P, Jouanneau E, et al. Sex-Related Differences in Lactotroph Tumor Aggressiveness Are Associated With a Specific Gene-Expression Signature and Genome Instability. Front Endocrinol (Lausanne) (2018) 9:706. doi: 10.3389/fendo.2018.00706
129. Ippolito JE, Yim AK-Y, Luo J, Chinnaiyan P, Rubin JB. Sexual dimorphism in glioma glycolysis underlies sex differences in survival. JCI insight (2017) 2:e92142. doi: 10.1172/jci.insight.92142
130. Ma J, Malladi S, Beck AH. Systematic Analysis of Sex-Linked Molecular Alterations and Therapies in Cancer. Sci Rep (2016) 6:19119. doi: 10.1038/srep19119
131. Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov (2016) 6:235–46. doi: 10.1158/2159-8290.CD-15-0893
132. Morgan C, Bale T. Sex Differences in microRNA-mRNA Networks: Examination of Novel Epigenetic Programming Mechanisms in the Sexually Dimorphic Neonatal Hypothalamus. Biol Sex Differ (2017) 8:27. doi: 10.1186/S13293-017-0149-3
133. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature (2010) 466:835–40. doi: 10.1038/nature09267
134. Pinheiro I, Dejager L, Libert C. X-chromosome-located microRNAs in Immunity: Might They Explain male/female Differences? The X Chromosome-Genomic Context May Affect X-located miRNAs and Downstream Signaling, Thereby Contributing to the Enhanced Immune Response of Females. Bioessays (2011) 33:791–802. doi: 10.1002/BIES.201100047
135. le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J (2007) 26:3699–708. doi: 10.1038/sj.emboj.7601790
136. Streicher K, Zhu W, Lehmann K, Georgantas R, Morehouse C, Brohawn P, et al. A Novel Oncogenic Role for the miRNA-506-514 Cluster in Initiating Melanocyte Transformation and Promoting Melanoma Growth. Oncogene (2012) 31:1558–70. doi: 10.1038/ONC.2011.345
137. Stark MS, Bonazzi VF, Boyle GM, Palmer JM, Symmons J, Lanagan CM, et al. miR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget (2015) 6:17753. doi: 10.18632/ONCOTARGET.3924
138. Carè A, Bellenghi M, Matarrese P, Gabriele L, Salvioli S, Malorni W. Sex Disparity in Cancer: Roles of microRNAs and Related Functional Players. Cell Death Differ (2018) 25:477–85. doi: 10.1038/S41418-017-0051-X
139. Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, et al. miR-424(322) Reverses Chemoresistance via T-cell Immune Response Activation by Blocking the PD-L1 Immune Checkpoint. Nat Commun (2016) 7:11406. doi: 10.1038/NCOMMS11406
140. Pinto R, Pilato B, Ottini L, Lambo R, Simone G, Paradiso A, et al. Different Methylation and microRNA Expression Pattern in Male and Female Familial Breast Cancer. J Cell Physiol (2013) 228:1264–9. doi: 10.1002/JCP.24281
141. Langevin SM, Stone RA, Bunker CH, Grandis JR, Sobol RW, Taioli E. MicroRNA-137 promoter methylation in oral rinses from patients with squamous cell carcinoma of the head and neck is associated with gender and body mass index. Carcinogenesis (2010) 31:864–70. doi: 10.1093/carcin/bgq051
142. Barabási A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet (2004) 5:101–13. doi: 10.1038/nrg1272
143. Arnold AP, Lusis AJ. Understanding the sexome: measuring and reporting sex differences in gene systems. Endocrinology (2012) 153:2551–5. doi: 10.1210/en.2011-2134
144. Arnold AP, Disteche CM. Sexual Inequality in the Cancer Cell. Cancer Res (2018) 78:5504–5. doi: 10.1158/0008-5472.CAN-18-2219
145. Haupt S, Caramia F, Herschtal A, Soussi T, Lozano G, Chen H, et al. Identification of cancer sex-disparity in the functional integrity of p53 and its X chromosome network. Nat Commun (2019) 10:5385. doi: 10.1038/s41467-019-13266-3
146. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf (2008) 9:559. doi: 10.1186/1471-2105-9-559
147. van Nas A, Guhathakurta D, Wang SS, Yehya N, Horvath S, Zhang B, et al. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology (2009) 150:1235–49. doi: 10.1210/en.2008-0563
148. Mozhui K, Lu L, Armstrong WE, Williams RW. Sex-Specific Modulation of Gene Expression Networks in Murine Hypothalamus. Front Neurosci (2012) 6:63. doi: 10.3389/FNINS.2012.00063
149. Raznahan A, Parikshak N, Chandran V, Blumenthal J, Clasen L, Alexander-Bloch A, et al. Sex-chromosome Dosage Effects on Gene Expression in Humans. Proc Natl Acad Sci USA (2018) 115:145(10):7398–403. doi: 10.1073/PNAS.1802889115
150. Padi M, Quackenbush J. Detecting phenotype-driven transitions in regulatory network structure. NPJ Syst Biol Appl (2018) 4:16. doi: 10.1038/s41540-018-0052-5
151. Lopes-Ramos CM, Kuijjer ML, Ogino S, Fuchs CS, DeMeo DL, Glass K, et al. Gene Regulatory Network Analysis Identifies Sex-Linked Differences in Colon Cancer Drug Metabolism. Cancer Res (2018) 78:5538–47. doi: 10.1158/0008-5472.CAN-18-0454
152. Rich-Edwards JW, Kaiser UB, Chen GL, Manson JE, Goldstein JM. Sex and Gender Differences Research Design for Basic, Clinical, and Population Studies: Essentials for Investigators. Endocr Rev (2018) 39:424–39. doi: 10.1210/er.2017-00246
153. Webster TH, Couse M, Grande BM, Karlins E, Phung TN, Richmond PA, et al. Identifying, understanding, and correcting technical artifacts on the sex chromosomes in next-generation sequencing data. Gigascience (2019) 8:giz074. doi: 10.1093/gigascience/giz074
154. Olney KC, Brotman SM, Andrews JP, Valverde-Vesling VA, Wilson MA. Reference genome and transcriptome informed by the sex chromosome complement of the sample increase ability to detect sex differences in gene expression from RNA-Seq data. Biol Sex Differ (2020) 11:42. doi: 10.1186/s13293-020-00312-9
155. Paulson JNJN, Chen C-YC-Y, Lopes-Ramos CM, Kuijjer MLML, Platig J, Sonawane ARAR, et al. Tissue-aware RNA-Seq processing and normalization for heterogeneous and sparse data. BMC Bioinf (2017) 18:437. doi: 10.1186/s12859-017-1847-x
156. Rubin JB, Lagas JS, Broestl L, Sponagel J, Rockwell N, Rhee G, et al. Sex differences in cancer mechanisms. Biol Sex Differ (2020) 11:17. doi: 10.1186/s13293-020-00291-x
157. Lampreht Tratar U, Horvat S, Cemazar M. Transgenic Mouse Models in Cancer Research. Front Oncol (2018) 8:268. doi: 10.3389/fonc.2018.00268
158. Lim H, Kim SY, Lee E, Lee S, Oh S, Jung J, et al. Sex-Dependent Adverse Drug Reactions to 5-Fluorouracil in Colorectal Cancer. Biol Pharm Bull (2019) 42:594–600. doi: 10.1248/bpb.b18-00707
159. Ciucci A, Meco D, De Stefano I, Travaglia D, Zannoni GF, Scambia G, et al. Gender effect in experimental models of human medulloblastoma: does the estrogen receptor β signaling play a role? PLoS One (2014) 9:e101623. doi: 10.1371/journal.pone.0101623
160. Crnčec I, Modak M, Gordziel C, Svinka J, Scharf I, Moritsch S, et al. STAT1 is a sex-specific tumor suppressor in colitis-associated colorectal cancer. Mol Oncol (2018) 12:514–28. doi: 10.1002/1878-0261.12178
161. Arnold AP. Conceptual frameworks and mouse models for studying sex differences in physiology and disease: why compensation changes the game. Exp Neurol (2014) 259:2–9. doi: 10.1016/j.expneurol.2014.01.021
162. Cox KH, Bonthuis PJ, Rissman EF. Mouse model systems to study sex chromosome genes and behavior: relevance to humans. Front Neuroendocrinol (2014) 35:405–19. doi: 10.1016/j.yfrne.2013.12.004
163. Arnold AP, Chen X. What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Front Neuroendocrinol (2009) 30:1–9. doi: 10.1016/j.yfrne.2008.11.001
164. Shah K, McCormack CE, Bradbury NA. Do you know the sex of your cells? Am J Physiol - Cell Physiol (2014) 306:C3. doi: 10.1152/AJPCELL.00281.2013
165. Lancaster M, Knoblich J. Organogenesis in a Dish: Modeling Development and Disease Using Organoid Technologies. Science (2014) 345:1247125. doi: 10.1126/SCIENCE.1247125
166. Aberle M, Burkhart R, Tiriac, Olde Damink S, Dejong C, Tuveson D, et al. Patient-derived Organoid Models Help Define Personalized Management of Gastrointestinal Cancer. Br J Surg (2018) 105:e48–60. doi: 10.1002/BJS.10726
167. Linkous A, Balamatsias D, Snuderl M, Edwards L, Miyaguchi K, Milner T, et al. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep (2019) 26:3203–3211.e5. doi: 10.1016/J.CELREP.2019.02.063
168. Weeber F, Ooft S, Dijkstra K, Voest E. Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell Chem Biol (2017) 24:1092–100. doi: 10.1016/J.CHEMBIOL.2017.06.012
169. Granat L, Kambhampati O, Klosek S, Niedzwecki B, Parsa K, Zhang D. The Promises and Challenges of Patient-Derived Tumor Organoids in Drug Development and Precision Oncology. Anim Model Exp Med (2019) 2:150–61. doi: 10.1002/AME2.12077
170. Boretto M, Maenhoudt N, Luo X, Hennes A, Boeckx B, Bui B, et al. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat Cell Biol (2019) 21:1041–51. doi: 10.1038/s41556-019-0360-z
171. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin (2020) 70:7–30. doi: 10.3322/caac.21590
172. Koo JH, Jalaludin B, Wong SKC, Kneebone A, Connor SJ, Leong RWL. Improved Survival in Young Women With Colorectal Cancer. Am J Gastroenterol (2008) 103:1488–95. doi: 10.1111/j.1572-0241.2007.01779.x
173. Murphy G, Devesa SS, Cross AJ, Inskip PD, McGlynn KA, Cook MB. Sex disparities in colorectal cancer incidence by anatomic subsite, race and age. Int J Cancer (2011) 128:1668–75. doi: 10.1002/ijc.25481
174. Baran B, Mert Ozupek N, Yerli Tetik N, Acar E, Bekcioglu O, Baskin Y. Difference Between Left-Sided and Right-Sided Colorectal Cancer: A Focused Review of Literature. Gastroenterol Res (2018) 11:264–73. doi: 10.14740/gr1062w
175. Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol (2019) 16:713–32. doi: 10.1038/s41575-019-0189-8
176. Kim H, Giovannucci EL. Sex differences in the association of obesity and colorectal cancer risk. Cancer Causes Control (2017) 28:1–4. doi: 10.1007/s10552-016-0831-5
177. Murphy N, Strickler HD, Stanczyk FZ, Xue X, Wassertheil-Smoller S, Rohan TE, et al. A Prospective Evaluation of Endogenous Sex Hormone Levels and Colorectal Cancer Risk in Postmenopausal Women. J Natl Cancer Inst (2015) 107:djv210. doi: 10.1093/jnci/djv210
178. Barzi A, Lenz AM, Labonte MJ, Lenz H-J. Molecular pathways: Estrogen pathway in colorectal cancer. Clin Cancer Res (2013) 19:5842–8. doi: 10.1158/1078-0432.CCR-13-0325
179. Lin JH, Zhang SM, Rexrode KM, Manson JE, Chan AT, Wu K, et al. Association Between Sex Hormones and Colorectal Cancer Risk in Men and Women. Clin Gastroenterol Hepatol (2013) 11:419–24.e1. doi: 10.1016/j.cgh.2012.11.012
180. Kim S-E, Paik HY, Yoon H, Lee JE, Kim N, Sung M-K. Sex- and gender-specific disparities in colorectal cancer risk. World J Gastroenterol (2015) 21:5167–75. doi: 10.3748/wjg.v21.i17.5167
181. Setaffy L, Langner C. Microsatellite instability in colorectal cancer: clinicopathological significance. Pol J Pathol (2015) 66:203–18. doi: 10.5114/pjp.2015.54953
182. Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol (2010) 7:153–62. doi: 10.1038/nrclinonc.2009.237
183. Gelsomino F, Barbolini M, Spallanzani A, Pugliese G, Cascinu S. The evolving role of microsatellite instability in colorectal cancer: A review. Cancer Treat Rev (2016) 51:19–26. doi: 10.1016/j.ctrv.2016.10.005
184. Dunlop MG, Farrington SM, Carothers AD, Wyllie AH, Sharp L, Burn J, et al. Cancer risk associated with germline DNA mismatch repair gene mutations. Hum Mol Genet (1997) 6:105–10. doi: 10.1093/hmg/6.1.105
185. Zhao B, Wang L, Qiu H, Zhang M, Sun L, Peng P, et al. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget (2017) 8:3980. doi: 10.18632/ONCOTARGET.14012
186. Lorentzen JA, Grzyb K, De Angelis PM, Hoff G, Eide TJ, Andresen PA. Oncogene Mutations in Colorectal Polyps Identified in the Norwegian Colorectal Cancer Prevention (NORCCAP) Screening Study. Clin Med Insights Pathol (2016) 9:19–28. doi: 10.4137/CPath.s40143
187. Breivik J, Meling GI, Spurkland A, Rognum TO, Gaudernack G. K-ras mutation in colorectal cancer: relations to patient age, sex and tumour location. Br J Cancer (1994) 69:367–71. doi: 10.1038/bjc.1994.67
188. Ning Y, Gerger A, Zhang W, Hanna DL, Yang D, Winder T, et al. Plastin polymorphisms predict gender- and stage-specific colon cancer recurrence after adjuvant chemotherapy. Mol Cancer Ther (2014) 13:528–39. doi: 10.1158/1535-7163.MCT-13-0646
189. Ilich AI, Danilak M, Kim CA, Mulder KE, Spratlin JL, Ghosh S, et al. Effects of gender on capecitabine toxicity in colorectal cancer. J Oncol Pharm Pract (2016) 22:454–60. doi: 10.1177/1078155215587345
190. Sloan JA, Goldberg RM, Sargent DJ, Vargas-Chanes D, Nair S, Cha SS, et al. Women experience greater toxicity with fluorouracil-based chemotherapy for colorectal cancer. J Clin Oncol (2002) 20:1491–8. doi: 10.1200/JCO.2002.20.6.1491
191. Woitowich NC, Beery A, Woodruff T. A 10-year follow-up study of sex inclusion in the biological sciences. Elife (2020) 9:e56344. doi: 10.7554/eLife.56344
Keywords: sex, gender, cancer, genomics, genetics, epigenetics, gene networks
Citation: Lopes-Ramos CM, Quackenbush J and DeMeo DL (2020) Genome-Wide Sex and Gender Differences in Cancer. Front. Oncol. 10:597788. doi: 10.3389/fonc.2020.597788
Received: 22 August 2020; Accepted: 19 October 2020;
Published: 23 November 2020.
Edited by:
Sen Peng, Translational Genomics Research Institute, United StatesReviewed by:
Dake Zhang, Beihang University, ChinaEdenir Inez Palmero, Barretos Cancer Hospital, Brazil
Copyright © 2020 Lopes-Ramos, Quackenbush and DeMeo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John Quackenbush, am9obnFAaHNwaC5oYXJ2YXJkLmVkdQ==; Dawn L. DeMeo, ZGF3bi5kZW1lb0BjaGFubmluZy5oYXJ2YXJkLmVkdQ==
 Camila M. Lopes-Ramos
Camila M. Lopes-Ramos