- 1Laboratory of Comparative and Translational Oncology (LOCT), Department of Veterinary Medicine, Faculty of Animal Science and Food Engineering, University of Sao Paulo, Pirassununga, Brazil
- 2Structural Genomics Consortium and Institute of Pharmaceutical Chemistry, Buchmann Institute for Molecular Life Sciences, Johann Wolfgang Goethe University, Frankfurt am Main, Germany
A plethora of data has highlighted the role of epigenetics in the development of cancer. Initiation and progression of different cancer types are associated with a variety of changes of epigenetic mechanisms, including aberrant DNA methylation, histone modifications, and miRNA expression. At the same time, advances in the available epigenetic tools allow to investigate and reverse these epigenetic changes and form the basis for the development of anticancer drugs in human oncology. Although human and canine cancer shares several common features, only recently that studies emerged investigating the epigenetic landscape in canine cancer and applying epigenetic modulators to canine cancer. This review focuses on the existing studies involving epigenetic changes in different types of canine cancer and the use of small-molecule inhibitors in canine cancer cells.
Introduction
The epigenome consists of a set of complex, dynamic, and reversible information comprising chemical modifications of the DNA and histone proteins, which are directly associated with the regulation of gene expression within the genome. These modifications, described as “Epigenetic Marks”, are heritable and occur without changes in the DNA sequence, playing a key role in biological processes such as embryonic development, differentiation, gene imprinting and silencing of the X chromosome. Furthermore, epigenetic modifications can affect DNA accessibility having a major influence on DNA-based processes including transcription, DNA repair, and replication (1).
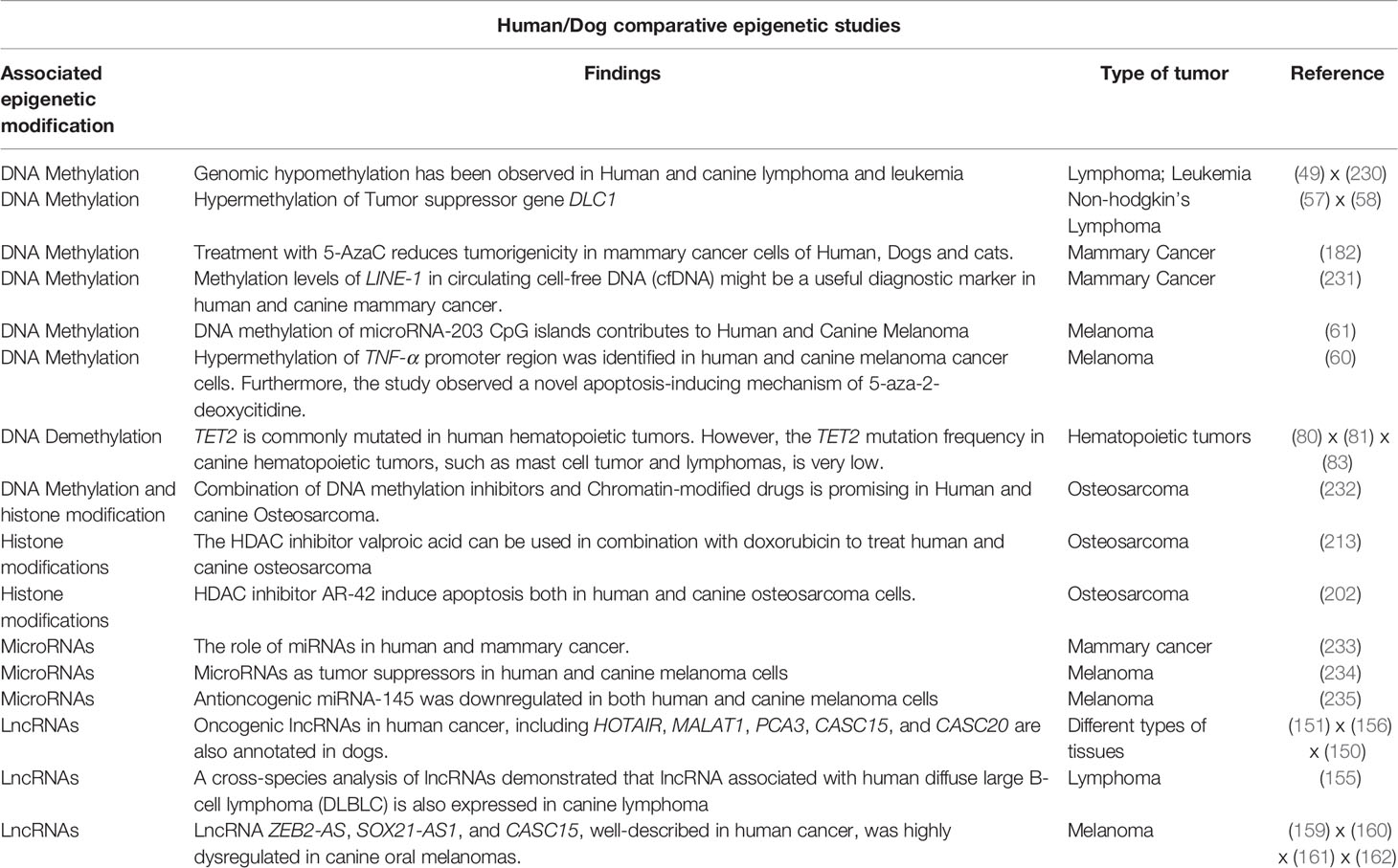
The epigenetic process is driven by a machinery of proteins, responsible for adding, removing or recognizing modifications of the DNA and histones. Epigenetic ‘writers’ are responsible for adding epigenetic marks and include DNA and histone methyltransferases as well as histone acetyltransferases (HATs); the ‘erasers’ such as histone deacetylases (HDACs) and histone demethylases (HKDMs) remove the corresponding epigenetic marks. Finally, a class of proteins exists, which recognizes and ‘interprets’ epigenetic modifications, referred to as ‘readers,’ a large class of proteins with reader domains for residues such as acetyllysine residues, including bromodomains, and methyllysine residues, such as chromodomains, Tudor domains, PhD domains, and others (2) (Figure 1). All these proteins play important functions in regulating gene expression, acting directly on DNA accessibility or indirectly recruiting non-coding RNAs and chromatin remodelers (3). Therefore, abnormal expression or mutations in these chromatin regulators can alter the pattern of gene expression and, consequently, lead to the induction and maintenance of diverse types of diseases, including cancer. Besides, these epigenetic alterations represent disease biomarkers with diagnosis and/or prognosis potential (4). Several of these epigenetic protein classes have been shown to contain druggable targets opening up possibilities to treat epigenetic-associated diseases (5–7).
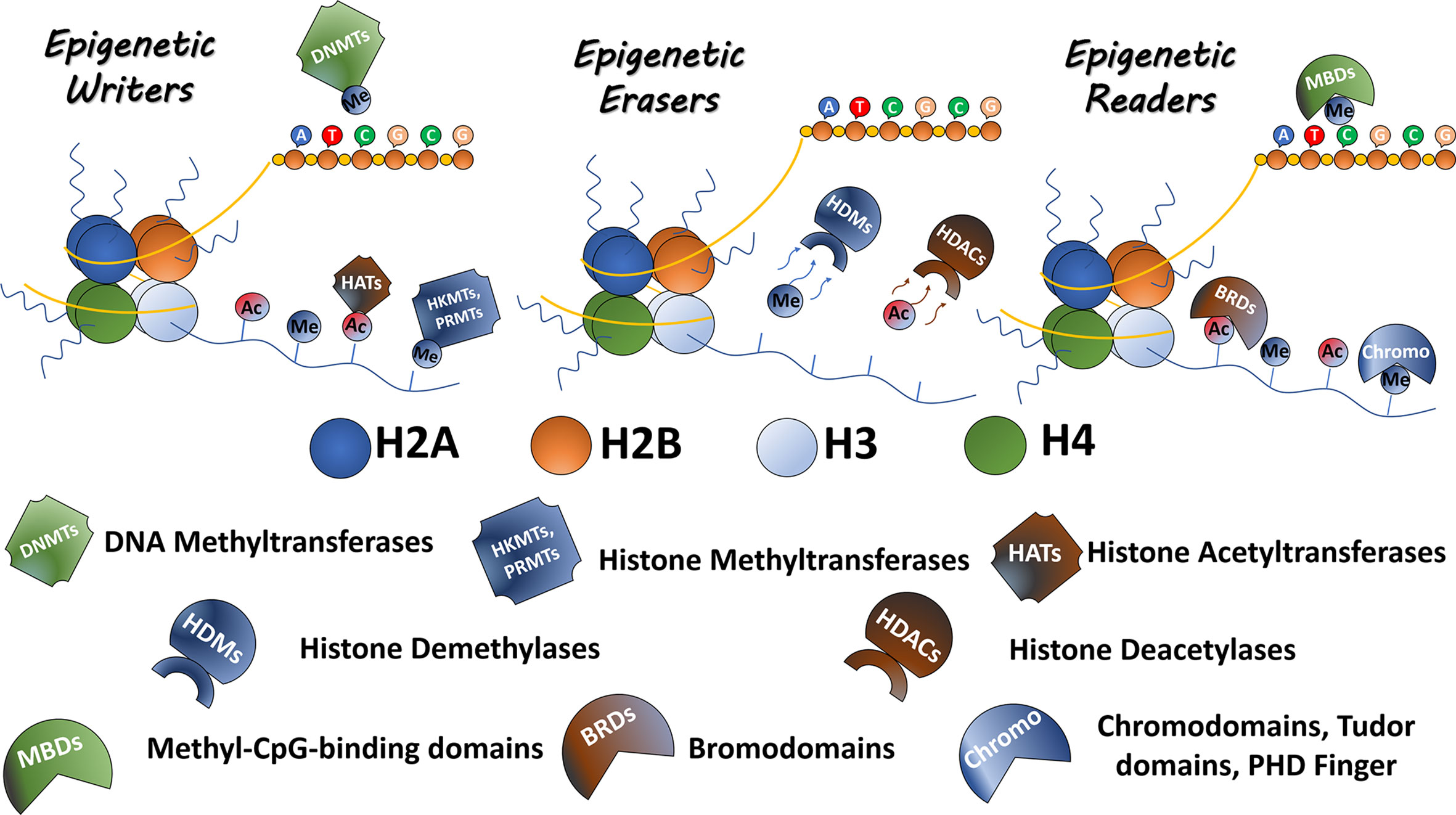
Figure 1 Epigenetic modulators. The epigenetic proteins are classified as Readers Writers and Erasers. Writers including DNMTs, HKMTs, PRMTs, and HATs are responsible to mark residues in DNA or histone tails. Erasers, including HKDMs and HDACs, remove epigenetic marks. Readers such as proteins containing bromodomains, chromodomains, Tudor domains, and PHD fingers recognize and bind to the epigenetic marks.
The dog is probably the best model for human disease and has several advantages in comparison to other animal models, such as natural development of several different tumors similar to humans; generally shares the same environment and exposure to the same carcinogens as humans thereby influencing the epigenetic make-up (8). However, in comparison to human cancer, the number of studies investigating the epigenetic landscape in canine cancer is still restricted and the potential of epigenetic drugs in the treatment of canine cancer remains widely unexplored. In this review, we highlight epigenetic studies of canine cancer and provide information about how alterations of epigenetic regulators can influence diverse types of canine cancer. We also highlight potential drugs aimed at targeting these epigenetic regulators.
An Overview of Canine Cancer
Over the past years, the animal-owner relationship has been changing and pets have genuinely become part of the human family. Therefore, advances in veterinary care have emerged, and, just like in humans, an increased life expectancy of dogs is observed (9). Consequently, age-related diseases, mainly cancer, are becoming the main causes of deaths in dogs worldwide (10–15). The epidemiologic studies of canine cancer are largely retrospective and usually present varying results depending on the region where they were performed. The incidence of cancer in dogs is 99.3 per 100,000 in male dogs and 272.1 in female dogs according to a study performed in Genoa, Italy (16). A large set of studies has shown that most common types of canine tumors are located in the skin followed by mammary tumors. Some studies have registered skin tumors frequencies as high as 40 or 50% (17, 18). However, depending on the proportion between females and males, mammary tumors have been observed around in about 36% of total cases (19), since mammary tumors account for more than 50% of the diagnosed tumors in females (20–23). Other common types of cancer in dogs are located in the soft tissues (14), hematopoietic, and lymphoid tissues (18), digestive organs (19), and bones (24).
Most canine cancer share a common pattern with the corresponding human disease including incidence, spontaneous development, associated risk factors, response to treatment, and expression of molecular targets. Non-Hodgkin lymphoma (NHL), for example, presents an incidence between 15.5 and 29.9 per 100,000 in humans and 15-30 per 100,000 in dogs (25), while mammary tumors incidence in female dogs is around 25% compared to 12% in women. Interestingly, some studies even showed that the average age at onset of mammary tumors is approximately the same for women and female dogs and the peak incidence to mammary cancer is comparable if their age is calculated proportionally (26, 27). Some strategies for cancer treatment can be applied to humans and dogs. CHOP therapy (vincristine, cyclophosphamide, prednisone, and doxorubicin), for instance, exhibits favorable outcomes in patients with lymphoma in both species, with median survival times of 8–13 months for dogs (28, 29).
Regarding genetic and signaling-pathway alterations, many types of canine cancer show similarities with their respective types of cancer in humans. Both human and canine osteosarcoma tumors carry mutations in tumor suppressor genes such as p53 (24, 30, 31) and RB1 (32) besides alterations in oncogenes expression including MYC and MET and constitutive expression of STAT3 (24, 33). Overexpression of MYC, a consequence of copy number aberrations, can also be observed in both human and canine lymphomas (34, 35). Diffuse large B-cell lymphoma in both species exhibit alterations in NF-κB and B-cell receptors pathway signaling (36). In leukemia, a classical chromosomal rearrangement on the Philadelphia chromosome is present in 95% of cases, producing a constitutively active cytoplasmic tyrosine kinase fusion protein, BCR-ABL (37). Although less constant, the same translocation was observed in dogs with leukemia (38, 39). Canine mammary tumors and breast cancer in women share many clinical and molecular similarities, such as hormonal dependence, age of onset, and identical course of the disease (22). At the molecular level, the disease in both species also exhibits equivalent features. Despite not having a consistent molecular classification based on specific molecular markers (estrogen receptor, progesterone receptor, and HER2) like in breast cancer, canine mammary tumors can present germline mutations in BRCA1 and BRCA2 (40), important tumor suppressor genes inherited mutated in women breast tumors (41). Likewise, overexpression of HER2 is observed between 20 and 29.7% of canine malignant mammary tumors (42), overlapping the increase of HER2 expression exhibited by breast cancer in women (43).
Several of these similarities underscore dogs as an excellent model to study novel biological patterns and therapeutic targets in diverse types of cancer. Furthermore, the advances in veterinary oncology research has promoted and verified a substantial interest and discoveries of epigenetic alterations that assist in the development of many canine tumors, several of which are also seen in human oncology. In the following section, we highlight these epigenetic alterations and potential drugs aimed at targeting these epigenetic regulators.
DNA Methylation and Canine Cancer
DNA methylation is probably the most studied epigenetic modification in animals and plants, playing a fundamental role in development, differentiation, and reproduction. DNA methylation occurs when a DNA n-methyltransferase (DNMT) adds a methyl group to cytosine residues in CpG dinucleotides. These CpG dinucleotides are occasionally enriched in some regions of the DNA called to CpG islands (CGIs), which in turn are preferentially located at gene promoters. DNA methylation results in the silencing of gene expression by essentially two different mechanisms: 1) DNA methylation can provide binding sites for methyl-binding domain proteins, which in turn can interact with histone deacetylases (HDACs), reducing chromatin accessibility and repressing gene activation; 2) methylation can prevent gene expression by blocking transcription factors to bind to the promoter regions of genes impeding transcription activation (44). Beyond gene promoters, CpG methylation is also found in repetitive sequences, gene bodies, and intergenic regions, which can influence gene expression of different approaches (45). CpG hypermethylation in gene bodies, for example, is associated with increased gene expression (46).
Under normal conditions, most CpG sites in the genome are methylated while the CGIs are usually unmethylated. In contrast, cancer cells exhibit a genome-wide hypomethylation and CGIs promoter hypermethylation (47). Genome-wide hypomethylation usually occurs in genomic regions including repetitive sequences, retrotransposons, and CpG poor promoters, resulting in chromosomal rearrangement, activation, and translocation of retrotransposons and, consequently, induce genomic instability. Besides, loss of methylation may lead to activation of proto-oncogenes, such as RAS, S-100, and MAGE (48). Genomic hypomethylation has likewise been observed in canine leukemia and lymphoma. In canine leukemia and lymphoma cases, 30 and 69% respectively were found to be genome-wide hypomethylated. Furthermore, these unusual methylation patterns are associated with the early phases of tumor transformation and progression in canine leukemia and lymphoma (49), just as has been observed in different types of human cancer (50–52). These findings were the first to report global hypomethylation in canine cancer and, consequently, to show similarities between the epigenetic landscape in canine and human cancer.
Also, other canine cancer types display global hypomethylation. A recent study has shown that genome-wide hypomethylation was frequently found in grade III canine mast cell tumor, which is the most common skin tumor in dogs, thus correlating DNA hypomethylation with the aggressiveness of this type of cancer (53). In addition, dogs bearing non-Hodgkin lymphoma (NHL) exhibit higher DNA global hypomethylation of circulating leukocytes in comparison with healthy dogs (54). DNA hypomethylation was also observed in canine lung cancer samples and in metastatic osteosarcoma from the primary lung cancer (55). Thus, albeit a still low number of reports, genome-wide hypomethylation seems to be a common feature of at least some types of canine cancer.
Hypermethylation of CGIs also contributes to the development and promotion of cancer through the silencing of tumor suppressor genes. In human cancer, many tumor suppressor genes such as Rb, p16, RASSF1, CDH1, TIMP3, and BRCA1 have been shown to possess hypermethylated promoter regions (56). These genes are associated with processes including cell cycle, apoptosis, metastasis and DNA repair and, consequently, silencing might induce cancer. Until 2008, no article has shown the presence of promoter hypermethylation in dogs. The first report showing hypermethylation of a tumor suppressor gene in canine cancer was in canine NHL, describing the profile of DLC1 gene methylation in this cancer. DLC1 is a tumor suppressor gene found to be highly methylated in human NHL (57). Bryan and others performed methylation-specific PCR (MSP) and combined bisulfite restriction analysis (COBRA) to demonstrate the presence of DNA methylated in DLC1 in six of 13 canine NHL samples and two of three canine chronic lymphocytic leukemia, providing, for the first time, information regarding hypermethylation in canine cancer (58). In addition, there was an association between DLC1 hypermethylation and the malignant phenotype of NHL. However, hypermethylation of the DLC1 promoter was not associated with silencing of DLC1 expression and did not correlate with survival (59). Hypermethylation of TNF-α has been shown in human and canine melanoma cells by performing MSP. In addition, it has been observed that the methylation status and the level of TNF-α expression were inversely correlated in canine melanoma cell lines and melanoma tissues (60). Both human and canine melanoma cells have hypermethylated DNA in the CpG islands of the microRNA-203 (61), a suppressor of growth in melanoma cells, as shown by bisulfite sequencing and MSP (62). Hypermethylation and epigenetic silencing has also be observed for several other important tumor suppressor genes such as tissue factor pathway inhibitor 2 (TFPI-2) (63), death-associated protein kinase (DAPK) (64), cyclin-dependent kinase inhibitor 2A (CDKN2A/p16), HOXD10, FGFR2, ITIH5, and RASAL3 in B-cell lymphoma (65–67). In addition, DAPK hypermethylation is associated with overall survival and considered a negative prognostic factor in canine high-grade B-cell lymphoma (68). In canine acute myeloid leukemia, a heterogeneous pattern of DNA methylation was observed with subsets of cases hypermethylated or hypomethylated when compared with healthy tissues (69). Genome-wide analysis of DNA methylation in canine lymphomas revealed that lymphoma cells have gained methylation at CpG sites located in CGIs that were unmethylated in normal peripheral blood mononuclear cells (PBMCs), used as controls. In contrast, CpG sites outside CGIs lose methylation in lymphoma cells compared to the healthy PBMCs (70).
Some evidence points to downregulation of BRCA1, an important tumor suppressor in mammary cancer, in canine mammary samples (71, 72). However, the mechanism responsible for the decrease of BRCA1 expression is not well understood. Recently, a study showed BRCA1 hypermethylation in canine mammary tumors. However, the rate of BRCA1-hypermethylated samples was very low (1/15, 6.7%), making it difficult to conclude that BRCA1 downregulation is a consequence of BRCA1 promoter hypermethylation (73). Attempts to studying both DNA methylation and histone modifications in canine cancer have been performed. Canine lymphoid tumor cell lines with different drug-sensibility were analyzed using bisulfite sequencing and chromatin immunoprecipitation, and it was found that DNA methylation and histone H3 acetylation are involved in ABCB1 gene expression (74). ABCB1 is a P-glycoprotein highly expressed in several different types of human cancer and has been recognized to be a key player in the multidrug resistance phenotype (75). Another study observed that CGIs of the ABCB1 gene were hypomethylated in dogs with lymphoma. However, the authors did not find a correlation between the methylation status and levels of ABCB1 mRNA expression in these samples (76). Both human and canine mammary carcinomas can present deregulation of estrogen receptor α (ERα), and its expression levels guide the prognosis and kind of therapy for the respective patient. In human breast cancer, the most aggressive type of cancer is triple-negative breast cancer, featured by the lack of ERα expression, which is mainly attributed to ERα promoter methylation (77, 78). However, no significant variation in methylation patterns were found between ERα-positive canine mammary carcinomas and ERα-negative canine mammary carcinomas pointing to a difference of ERα regulation mechanisms between human and dogs (79).
TET proteins are responsible to catalyze the successive oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC), promoting DNA demethylation. TET2 is considered an important tumor suppressor gene being commonly mutated in hematopoietic tumors but rarely in solid tumors (80). In hematopoietic canine tumors, TET2 mutations have also been observed in canine mast cell tumors but in very low frequency (2.7%) (81), contrary the TET2 mutations rate in human systemic mastocytosis, which are observed in 40% of the cases (82). TET2 was also found mutated in canine T-cell lymphoma samples but in low frequency as well (83). All these information regarding DNA methylation and canine cancer are summarizing in Supplementary Table 1.
Histone Modifications and Canine Cancer
In the nucleus, DNA is compacted and complexed by proteins called histones resulting in a DNA-protein complex named chromatin. Histone proteins can be divided in two groups: core histones (H2A, H2B, H3, and H4) and linker histones (H1 and H5). Together, these proteins make up the nucleosome (the unit of chromatin), which is ‘coated’ with 146 base pairs of DNA. Histones contain a C-terminal domain and an unstructured N-terminal domain, commonly referred to as histone tail (84). These histone tails are susceptible to different post-translational covalent modifications such as methylation, acetylation, phosphorylation, ubiquitylation, sumoylation, deamination, propionylation, and butyrylation (85, 86), which directly affect the chromatin structure by recruiting enzymes able to remodel chromatin. Consequently, histone modifications are tightly linked to cellular processes including replication, repair, and recombination (87). Furthermore, because of their influence on regulating the accessibility of chromatin to the transcriptional machinery, some modifications are responsible to regulate gene expression (88). For instance, histone acetylation neutralizes the positive charge of histone lysines and consequently results in loosening the DNA packing around histones, making it more accessible to transcriptional activation. On the other hand, histone methylation is usually associated with both transcriptional activation and repressions, depending on which residue is modified or the degree of methylation (mono-di or thimethylation).
Just as aberrations in DNA methylation patterns can lead to cancer development, altered histone modifications and chromatin changes can be observed in cancer cells. For example, some studies revealed a global loss of acetylated H4-lysine 16 (H4K16ac) and H4-lysine 20 trimethylation (H4K20me3) in different cancer cell lines and primary tumors, such as leukemia, breast, lung, and colon cancer (89). In addition, alterations of H3K9me and H3K27me patterns are also observed in different types of human cancer, including bladder, colorectal, glioma, breast, and lung cancer (90–94). Accordingly, the expression of enzymes responsible for these modifications, histone acetylases/deacetylases (HATs and HDACs) and histone methyltransferases (HMTs), is observed to be dysregulated in cancer. HDACs are often overexpressed in human cancer (95), but HATs can be also altered (96). HMTs such as EZH2 and G9a have be found overexpressed in different types of cancer (97–99).
Some studies have shown dysregulation of histone modifications exhibiting key roles in the development and progression of canine cancer (Supplementary Table 2). Recently, in canine urothelial carcinomas samples, significant deacetylation of histones compared to normal samples was observed, and these lower acetylation levels were associated with a poor prognosis of the animals (100). SETD2 gene, a histone methyltransferase and an important tumor-suppressor, was found to be mutated in 21% of canine osteosarcoma samples and showed a variety of mutation types including frameshift, nonsense, splice, and missense mutations (101). Another study detects SETD2 somatic point mutations, deletions, and chromosomal translocations in 42% of canine osteosarcoma samples (102). Just like in human cancers, overexpression of EZH2 was found in canine lymphoma, melanoma, basal cell tumors, squamous cells carcinoma, and prostate and mammary cancer (103, 104).
Along with the simple carcinomas, complex carcinomas present the most common type of canine mammary cancer and are characterized by the presence of epithelial and myoepithelial cells (105). These types of mammary cancers seem to be profoundly influenced by the epigenetic landscape, and recent findings suggest that they originate from epigenomic rather than genomic alterations. Analysis of whole-genome sequencing, whole-exome sequencing, RNA-seq, and/or high-density arrays on twelve canine mammary cancer cases, including seven simple carcinomas and four complex carcinomas showed that, contrary to simple carcinomas, complex carcinomas did not have any copy number abnormalities and also low mutation rates. Conversely, complex canine mammary carcinomas displayed a number of epigenetic dysregulations, such as downregulation of 35 chromatin-modification genes or abnormally enriched activating histone modification H4-acetylation while showing a reduction in the repressive histone modification H3K9me3 (106).
Non-Coding RNAs andCanine Cancer
Non-coding RNAs (ncRNAs) are defined as RNA molecules that are not translated into a protein and, for a long time, their functions in the genome were not well understood. However, with the recent advances in cell biology, transcriptomic and bioinformatic tools, it has become possible to elucidate the role of these molecules regulating biological pathways and processes. These ncRNAs are classified as microRNAs (miRNAs), transfer RNAs (tRNAs), PIWI-interacting RNAs (piRNAs), long non-coding RNAs (lncRNAs), pseudogenes, and circular RNAs (circRNAs) (107). In this review, miRNAs and lncRNAs are addressed (Supplementary Table 3).
MicroRNAs (miRNAs) are a class of non-coding RNAs encoded in the genome. The first miRNA description was in the nematode C. elegans (Lee et al, 1993). Since then more than 15,000 miRNAs have been identified (www.mirbase.org). Several miRNAs are expressed in many different species and are highly conserved amongst them. These molecules were found to develop a key role in many biological processes including cell proliferation, metabolism, development, differentiation, apoptosis and stress response (108–110). They control these processes by regulating gene expression through post-transcriptional mechanisms. MiRNAs bind to their target mRNA and downregulate it via one of two different mechanisms: 1) when miRNA and mRNA have a full complementary, the miRNA triggers the degradation of the target mRNA; 2) miRNAs can also bind to the mRNA 3’UTR regions with incomplete complementarity, leading to translational repression (111) (Figure 2).
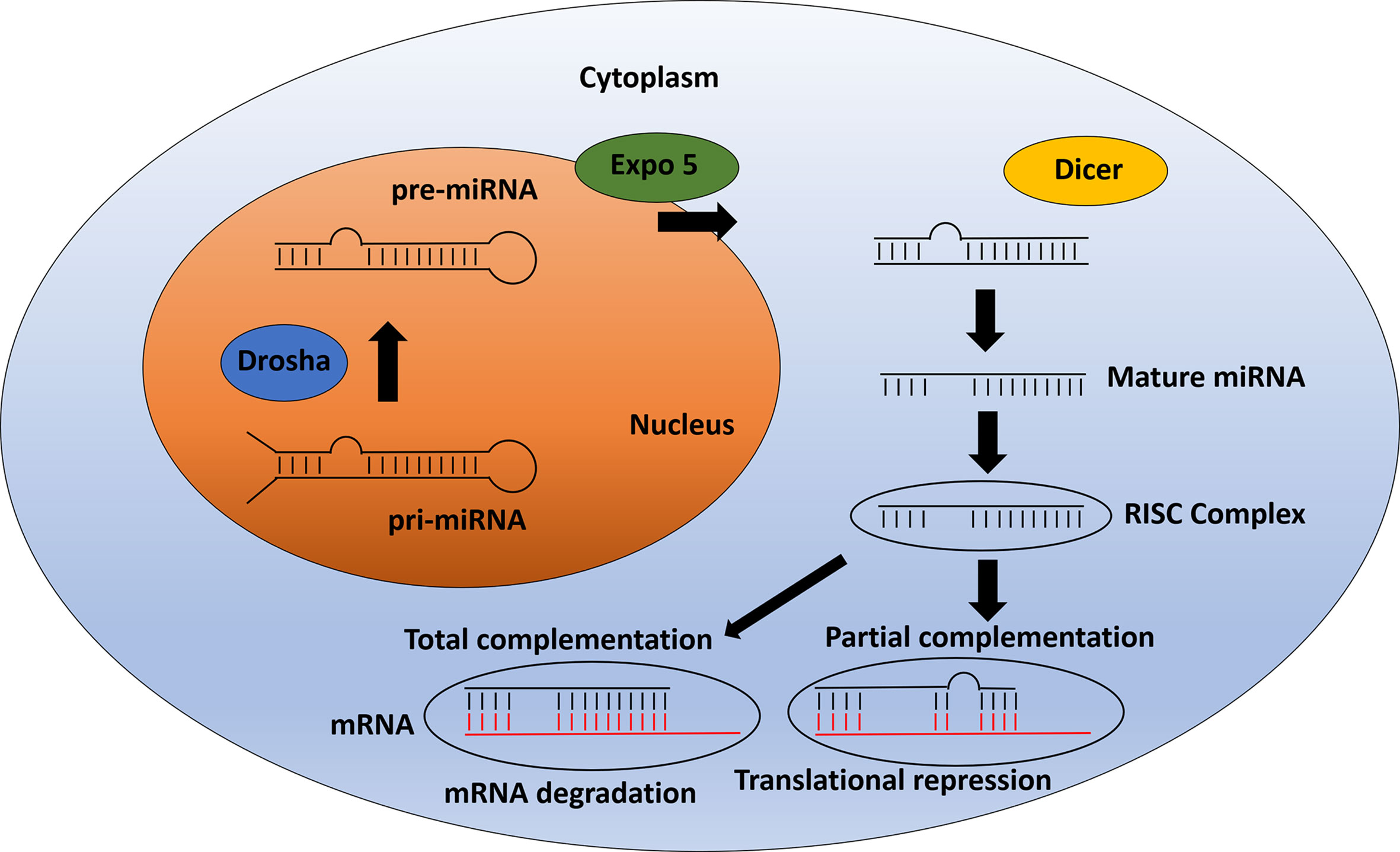
Figure 2 miRNA pathway: From biogenesis to mRNA inhibition. After pri-miRNA generation by transcription, the microprocessor complex Drosha processes and cleaves the pri-miRNA to produce the precursor-miRNA (pre-miRNA). Then, the pre-miRNA is transported from the nucleus to cytoplasm by Exportin 5. In the cytoplasm, pre-miRNA is processed by Dicer to produce the mature miRNA. The mature miRNA is incorporated into a protein complex termed RISC. Finally, this complex induces gene inhibition in two different ways. 1) The mRNA can be degraded if there is total complementation between the miRNA and the mRNA. 2) In the case of partial complementation, there is a translational repression.
Several lines of evidence have shown that miRNAs play an important role in the development of diseases in humans including cardiovascular diseases (112), neurodegenerative diseases (113) and several types of cancer. The mechanisms responsible for miRNA dysregulation in human cancer has been well elucidated and include amplification or deletion of miRNA genes, aberrant transcriptional control of miRNAs due to the dysregulation of some transcription factors such as C-Myc and P53, dysregulated epigenetic changes with some studies showing aberrant patterns of DNA methylation and histone acetylation in miRNA genes, and defects in miRNA biogenesis machinery (114–119).
The first evidence of miRNA dysregulation in canine cancer was described for canine mammary cancer. Both miR-29 and miR-29b were found to be upregulated in canine mammary cancer samples. Furthermore, the same study showed that miR-15a and miR-16 are significantly downregulated in canine ductal carcinomas while miR-181b, -21, -29b, and let-7f showed a significant upregulation in canine tubular papillary carcinomas (120). Thenceforth, some studies have described miRNA profiling in different types of canine cancer such as mast cell tumors (121), osteosarcoma (122), hemangiosarcoma (123), prostate cancer (124), canine multicentric lymphoma (125), and melanoma (126, 127). Interestingly, important and well-described miRNAs in human cancer including miR-9, miR-18a, miR-126, miR-383, and miR-204 were found to be dysregulated in canine cancer. The presence of MiR-181 and miR-17-5p were observed in B- and T-cell lymphomas compared to non-neoplastic cells (128). Furthermore, miRNA dysregulation appears to have a key role in regulating metastasis in some canine cancer. The expression of 14 miRNAs were significantly different between metastasizing and non-metastasizing uveal melanomas, highlighting cfa-miR-362, cfa-miR-155, cfa-miR-182, and cfa-miR-124 as strongly associated with the metastasizing class in this type of cancer (129). Ten miRNA (cfa-let-7c, cfa-miR-10b, cfa-miR-26a, cfa-miR-26b, cfa-miR-29c, cfa-miR-30a, cfamiR-30b, cfa-miR-30c, cfa-miR-148a, and cfa-miR-299) were validated and showed significant different expression in metastatic and non-metastatic mammary tumors (130). MiR-9 was found to be overexpressed and associated with metastasis in mast cells tumors and osteosarcoma (121, 122), while miR-34a also appeared to be associated with invasion ability in canine osteosarcoma cell lines (131). Some miRNAs also correlated with tumor grading in canine splenic lymphoma (132).
Circulating miRNAs detected on liquid biopsies such as blood and urine may provide diagnostic and prognostic information regarding cancer (133). MiRNA-214 and -126 have been considered potential diagnostic and prognostic biomarkers for canine neoplastic diseases. In a recent study, using 181 cases of canine neoplastic diseases and healthy controls, circulating miRNA-214 was considered a good diagnostic marker in sarcomas, whereas levels of the circulating miRNA-126 was high in most of the types of canine tumors (134). These same miRNAs were demonstrated to have a strong potential to predict the outcome of canine appendicular osteosarcoma patients receiving amputation and chemotherapy (135). Dogs with disseminated histiocytic sarcoma and carcinomas showed downregulation of circulating Let-7g (136). Another study described the profile of circulating serum miRNAs in dogs with lymphoma. Four miRNAs (let-7b, miR-223, miR-25, and miR-92a) were significantly reduced in dogs with lymphoma, whereas miR-423a levels were significantly increased compared to the controls (137). MiR-99a was also differentially expressed in the plasma of dogs with lymphoma (125). Circulating miRNAs detected in the urine are also been detected in canine cancers. In canine bladder transitional cell carcinomas, miR-103b and miR-16 were considered as potential diagnostic urine biomarkers (138). Analysis of the miRNA profiles within the exosomes released from canine tumors has also been made. A recent study observed that canine mammary epithelial cancer cells shed exosomes that contained differentially expressed miRNAs in comparison with normal cells (139). In a study performed in canine lymphoma, three miRNAs (miR-151, miR-8908a-3p, and miR-486) derived from exosomes demonstrated to be differently expressed between vincristine-sensitive and resistant lymphoma cell lines supporting a role of these miRNAs in the resistance of this cancer (140).
LncRNAs are non-coding transcripts greater than 200 bp in length and some studies demonstrated the influence of these molecules in gene expression at the epigenetic, transcriptional, and post-transcriptional levels. One of the most classical mechanisms through which lncRNAs regulate gene expression is by association with chromatin modeling complexes and transcription factors, influencing transcriptional repression and activation of gene promoters. For example, the well-characterized lncRNA HOTAIR can bind to epigenetic complexes such as PRC2 and LSD1/CoREST/REST, modulating histone methylation (141). In addition, lncRNAs binds directly to DNA, mRNAs, and/or miRNAs affecting and regulating their respective functions and levels (142–144).
The lncRNAs play a fundamental role in the development and physiology of the human organism but can be also associated with disease evolution, especially cancer. HOTAIR overexpression, for example, has been associated with an increase of metastasis, invasiveness, and, consequently, to poor outcomes in breast and other types of cancer (145, 146). Many oncogenic lncRNAs including THOR (147), ARLNC1 (143), SAMSOON (148), and EPIC1 have also been associated with different types of cancer such as lung, prostate, melanomas, ovarian, and pancreatic cancer (149).
Due to the importance of lncRNAs in the genome and their association with different human diseases, the canine lncRNA profile has also been described (150). An alignment-free program that accurately annotates lncRNAs, FEELnc, was used on a real data set of 20 RNA-Seq data from 16 different canine tissues produced by the European LUPA consortium to expand the canine genome annotation including 10.374 novel lncRNAs and 58.640 mRNAs transcripts (151). This study particularly highlighted duplications of lncRNAs in dog. Interestingly, among the novel lncRNAs genes, around 15% were also described as non-protein coding genes in the human GENCODE. Finally, with this set of data, it was possible to annotate three new cancer susceptibility candidate lncRNAs in dogs, which are well described in human cancer, including CASC9, associated with esophageal squamous cell carcinoma (152), MALAT1, associated with metastasis in lung cancer (153), and IFNG-AS that plays an important role in T-cell differentiation (154). Another study, observed more than 900 dog-human conserved lncRNAs using comparative genomics. The authors confirmed the annotation of well-studied lncRNAs in dogs, such as HOTAIR, MALAT, NEAT_1, PCA3, CASC15, CASC17, CASC18, CASC20, and INHBA-AS1. In addition, 44% of the canine lncRNAs are expressed in a tissue-specific manner, which is also widely seen in humans (155). Finally, co-expression analysis suggested that these lncRNAs function as regulatory elements in the dog genome (156). Despite the increase of the lncRNA number and description, few lncRNA are functionally and experimentally characterized in dogs, and only few of them have been found to be associated with diseases. For instance, the lncRNA GDNF-AS was observed to be involved in a Hereditary Sensory Autonomic Neuropathy (HSAN) in hunting dogs (157).
LncRNAs is also associated with some types of canine cancer. Cross-species analysis of lncRNAs demonstrated that a non-negligible fraction of lncRNA associated with human diffuse large B-cell lymphoma (DLBLC) is also expressed in canine lymphoma (155). A recent study has also developed a methodology to identify lncRNAs in canine DLBCL. The authors concluded that this methodology was able to quantify the expression of novel and annotated lncRNAs and, interestingly, subclassified the DLBCL in two main groups. Furthermore, these two DLBCL groups showed statistically different survival rates, pointing to the potential of using lncRNAs as prognostic markers using this methodology (158). In canine oral melanomas, 417 differentially expressed lncRNAs were identified in comparison with control samples, using deep transcriptome sequencing. Most of these lncRNAs have not yet been functionally characterized; however, lncRNA ZEB2-AS, a lncRNA involved in the regulation of the transcription factor ZEB2 during epithelial-mesenchymal transition (EMT) in human colon, pancreatic, and breast cancer cell lines (159), was highly expressed in canine oral melanomas compared to control samples. Other examples of lncRNAs dysregulated in these tumors that are well described in human cancer, were SOX21-AS1 (160), and CASC15 (161). Finally, using co-expression network analysis (WGCNA), the differentially expressed lncRNAs were associated with Gene Ontology (GO) biological process including cancer-related genes, cell cycle, cellular response to stress, DNA metabolic process, and carbohydrate metabolism (162).
Epigenetic Drugs to Treat Canine Cancer
Contrary to genetic mutations, epigenetic changes occurring in cancer are potentially reversible. There is thus the possibility of treating cancer with epigenetic drugs and, consequently, reverse some malignant phenotypes including metastasis potential (163, 164), tumorigenicity (165) and multidrug resistance (166). Several efforts have been undertaken for the development of epigenetic drugs targeting defective DNMTs and histone modifying enzymes as well as reader domains in cancer, but, unlike in human oncology, epigenetic drugs are still little in use in veterinary oncology, as we outlined below. However, it is important to emphasize that dogs are used as models in most preclinical tests for these drugs providing an overview of the possible side effects of these anticancer agents in this species.
DNA Methyltransferases Inhibitors
The first two epigenetic cancer drugs, the 5-azacytidine (5-azaC or azacitidine) (167) and the 5-aza-2′-deoxycytidine (5-aza-dC or decitabine) (168) were synthesized in 1964 but only approved by the FDA in 2004 and 2006, respectively (169, 170). Both are DNMT inhibitors and are currently first-line therapy for myelodysplastic syndrome (MDS), a bone marrow disorder that can progress to acute myeloid leukemia (AML). Furthermore, 5-azaC and 5-aza-dC are administrated to treat hematological malignancies including chronic myelomonocytic leukemia (CMML) and AML in elderly patients ineligible for intensive chemotherapy (171–173). Although efficient, this first generation of DNMT inhibitors presents some issues including lack of specificity, which could trigger some side effects, poor bioavailability and limited half-life (174). Thus, second-generation DNMT inhibitors have been developed including zebularine (175), CP-4200 (176) and guadecitabine (177). For solid tumors, azanucleoside-based therapies are also being tested in phase I/II clinical trials in several types of cancer (178, 179).
Due to their promising results in human cancer, DNMT inhibitors have been tested, although at a low scale, in canine cancer (Supplementary Table 4). The first 5-aza-dC test report in dogs was published in 1983. Dogs were used to investigate the plasma and cerebrospinal fluid pharmacokinetics of 5-aza-dC and the results showed that the compound could be rapidly cleared from plasma and cross the blood-CSF barrier resulting in potentially and cytotoxic concentrations by infusion (180). However, in this study, dogs were only used as experimental models, not aiming treatment of canine cancer. Dogs with naturally occurring invasive urothelial carcinoma were treated with subcutaneous 5-aza-C. Of the 18 dogs in the study, partial remission was achieved in 22%; 50% showed stable disease, whereas in 22%, the cancer progressed. The subcutaneous 5-azaC strategy in dogs was considered promising and important for the translation and design of human urothelial carcinoma clinical trials (181).
In human and canine melanoma cells, a recent study has shown a new apoptosis-inducing mechanism of 5-aza-dC through demethylation and induction of cytotoxic cytokines such as TNF-α in in vitro and in vivo experiments, suggesting a potential therapeutic agent for human and canine melanomas (60). 5-aza-C reduced in vitro growth, invasion, tumorigenicity, and mitochondrial activity and increased the susceptibility to apoptosis of breast cancer cells from human, canine, and feline species. In addition, 5-aza-C was toxic to mammary cancer cells but not to healthy mammary cells lines from these species, indicating a therapeutic window and sustaining animals as useful models for pre-clinical evaluation of new drugs targeting breast cancer (182). Likewise, second-generation of DNMT inhibitors have been tested in canine models. Zebularine was able to inhibit DNMT1 and promote global demethylation of canine malignant lymphoid cells resulting in dose-dependent apoptosis (183). Toxicological and pharmacokinetic studies with Zebularine were performed using laboratory animals and dogs with natural occurring tumors. Plasma zebularine clearance was constant. Laboratory dogs treated with a daily oral zebularine dose of 4 mg kg-1 developed some side effects including neutropenia, found in all dogs, thrombocytopenia in one dog, anorexia in four dogs, and dermatological changes. In the dogs with tumors, thrombocytopenia was observed in one dog. No other hematologic abnormalities, serum biochemical abnormalities or dermatologic changes were detected. Despite important information regarding plasma pharmacokinetics and toxicity of zebularine in dogs, more studies should still be performed to observe the anticancer activity of zebularine in this specie (184).
Histone Deacetylase Inhibitors (HDACi)
Since HDACs are often dysregulated in different types of cancer, many efforts have been made to develop efficient HDAC inhibitors. In human multiple myeloma, ovarian, gastric, breast, pancreatic, and prostate cancer, for example, HDACs are overexpressed and associated with poor outcome (185–192). Different patterns of HDAC1 have been established as prognostic marker in osteosarcoma. Whereas in primary osteosarcoma, cells showed a high expression of HDAC1 and 2, low levels of HDAC1 were associated with the presence of initial metastasis (193). Considering the relevance of this target class, four HDACs inhibitors have been approved for cancer treatment by the U.S Food and Drug Administration (FDA): vorinostat, romidepsin, belinostat, and panobinostat. The first HDACi, SAHA (Vorinostat, Zolinza™, Merck & Co, Inc., USA), was approved by FDA in 2006, and since then, HDACi are being developed for the treatment of T-cell lymphoma (194–196) and multiple myeloma (197). Furthermore, other studies and clinical trials showed the effects of Vorinostat in hematological and solid cancers including pancreatic (198), ovarian (199), prostate (200), and breast cancer (201).
HDACis have been also shown good effects in canine cancer (Supplementary Table 5). Vorinostat reduced the viability and increased apoptosis in a dose-dependent manner besides decreasing phosphorylation in oncogenic pathways including Akt-Ser473 and mTOR in canine osteosarcoma cell lines (202). In canine urothelial carcinoma cells, Vorinostat inhibited the growth and induced G0/G1 cell cycle arrest through the upregulation of p21 and dephosphorylation of Rb in these cancer cells (100). Both studies showed that Vorinostat was able to induce histone H3 acetylation in these canine cancer cells. The effects of another HDACi, sulforaphane, has been shown on canine osteosarcoma cells, significantly decreasing cell invasion and downregulating focal adhesion kinase (FAK) signaling (203).
A panel of seven HDACis were tested, in a well-established canine B-cell lymphoma cell line, CLBL-1 using in vitro and in vivo (xenograft) models. All HDACis tested exhibited dose-dependent inhibitory effects on the proliferation of CLBL-1 cells. Furthermore, Panobinostat, the most potent HDACi tested in vitro, inhibited CLBL-1 xenograft tumor growth, triggering acetylation of H3 and apoptosis in vivo (204). Panobinostat also efficiently inhibited the growth of tumors in xenograft models inoculated with a modified and bioluminescent canine B-cell lymphoma cell line (205). Trichostatin A (TSA), an antifugical agent with properties to selectively inhibit histone deacetylase activity in mammalian cells has shown inhibitory effects of proliferation and apoptosis in cancer cells (206). In vitro inhibitory effects of TSA were also shown on canine grade 3 mast cell tumor, decreasing cell viability, by increasing apoptosis and the number of cells in sub-G1 phase of cell cycle, indicating cell death (207). TSA also inhibited the proliferation of one canine mammary cancer cell line (208). A novel HDAC inhibitor AR-42, recently in phase I/Ib trials for multiple myeloma and T- and B-cell lymphomas (209), has shown effects in canine osteosarcoma, prostate, and malignant mast cancer cells. Cell viability inhibition and induction of apoptosis via activation of the intrinsic mitochondrial pathway were observed in canine osteosarcoma cells treated with AR-42. In addition, AR-42 showed synergistic effects when combined with doxorubicin (202). In canine prostate cancer, AR-42 inhibited in vitro proliferation in a time- and dose-dependent manner and decreased migration and the incidence of bone metastasis in xenograft models (210). AR-42 treatment of canine malignant mast cells induced proliferation inhibition, cell cycle arrest, apoptosis, and activation of caspases-3/7. Downregulation of KIT, a commonly mutated gene in malignant mast cells, via inhibition of KIT transcription was also observed. Finally, AR-42 treatment downregulated several important cancer molecules including p-AKT, total AKT, phosphorylated STAT3/5, and total STAT3/5 (211).
The effects of HDACs inhibition in combination with other therapies has also been studied in canine cancer. A phase I pharmacokinetic and pharmacodynamic study of combined valproic acid (VPA) and doxorubicin was performed in spontaneous canine cancers. Of the 21 dogs treated in this study, two presented complete responses (10%) (both lymphomas), three presented partial responses (14%) (lymphoma, melanoma, and lung carcinoma), five showed stable disease after treatment (24%) (osteosarcoma, renal cell carcinoma, apocrine gland adenocarcinoma, melanoma, and soft-tissue sarcoma), and 11 exhibited progressive disease (58%) (212). In another study from the same group using both human and canine osteosarcoma (OS) cells, pre-incubation with VPA followed by doxorubicin increased the growth inhibition and apoptosis rates in both human and canine OS cells, associated with a dose-dependent increase in nuclear doxorubicin accumulation, supporting a potential addition of HDACis for treatments of human and canine OS (213).
Alternative Epigenetic Targets
Due to successful development of DNMT and HDAC inhibitors and the use of these molecules in the treatment of diseases, new classes of epigenetic drugs has been developed to target epigenetic writers, erasers and even epigenetic readers. Several examples are reported including histone methyltransferases inhibitors such as DOT1L (214), EZH2 (215), and G9A inhibitors (216). Many studies are also observing the effects of lysine demethylases inhibition including LSD1 and LSD2 (217) and epigenetic readers such as bromodomains with the BET (Bromo and Extra terminal) family comprising BRD2, BRD3, BRD4, and BRDT as the most prominent targets for drug discovery (218). During the past years, the Structural Genomic Consortium (SGC), a public-private partnership that supports the discovery of new medicines through open access research, has designed and developed a set of tool compounds for epigenetic targets with clearly defined properties (219, 220).
Our group recently screened a small-molecule library containing 27 of these developed epigenetic inhibitors in canine mammary cancer cell lines (CMCs). We observed three inhibitors inducing significant reduction of cell viability in CMCs including (+)-JQ1 (BET family inhibitor), NVS-CECR2-1 (CECR2 inhibitor), and UNC1999 (EZH2/1 inhibitor). Furthermore, BET inhibition by (+)-JQ1 was very efficient to inhibit CMCs colony and tumorsphere formation, demonstrating an effect on tumorigenicity and self-renewal (Figure 3) (222). Inhibition of SET methyltransferase by shRNA and FTY720, reported to directly interact with SET proteins, suppressed cell proliferation, colony formation, and in vivo tumor growth of canine mammary and osteosarcoma cell lines. Furthermore, SET knockdown repressed mTOR and NF-kB signaling in both types of canine cancer (223, 224). Using BB-Cl-Amidine (BB-CLA) to inhibit protein-arginine deiminases (PADs) resulted in the decrease of viability and tumorigenicity of canine mammary cancer cells, activating endoplasmic reticulum stress pathway in these cells (225).
Figure 3 Effects of some alternative epigenetic inhibitors in canine mammary cancer cells. A small library of 27 epigenetic inhibitors was screened in order to determine effects regarding cell viability, tumorigenicity, and self-renewal assessed by 3D cell culture models such as colony formation in soft-agar and tumorspheres formation in low-adherent plates (221). The (+)-JQ1 (BET family inhibitor), NVS-CECR2-1 (CECR2 inhibitor), and UNC1999 (EZH2/1 inhibitor) decreased cell viability of CF41.Mg canine mammary cancer cell line. Furthermore, (+)-JQ1 exhibits a strong impact on colony and tumorspheres formation, demonstrating effects on tumorigenicity and self-renewal phenotypes (222).
Dogs have also been used as models for pre-clinical trials of LSD1 inhibitors. A recent study, showed that the LSD1 inhibitor GSK2879552 caused severe but reversible toxicities in dogs including thrombocytopenia, neutropenia, myelofibrosis, and congestion with and without lymphoid necrosis in lymphoid organs (217). However, studies demonstrating the effects of these new alternative epigenetic drugs in dogs are very scarce (Supplementary Table 6).
BarkBase: A Canine Epigenomic Resource
The abundance of information acquired in recent years on human genomics both in healthy and diseased tissues enabled the construction of powerful platforms of data that can be used for the deep investigation of different phenotypes. Resources such as ENCODE (226), GTEx (227), Cancer Genome Atlas (TCGA) (https://www.cancer.gov/tcga) and NIH reference human epigenome (228), are constantly fueled with large numbers of information generated with next-generation bioinformatic tools and are extremely important for the elucidation of the most diverse diseases that affect humans and their therapeutic advances. With the same purpose, a ~7 gigabytes genomic data platform, the BarkBase resource, has been recently developed (229). BarkBase contains data for 27 adult tissue types, with biological replicates, from five adult dogs, paired with 30x whole genome sequence data. RNA sequencing data are complemented by whole genomic sequencing and assay data for transposable-accessible chromatin using sequencing (ATAC-seq). All these genomic and epigenomic data from healthy canine tissues will be highly important and useful for future studies in canine cancer providing the basis of a high-quality tool to compare the findings found in canine cancer tissues with healthy tissues. Furthermore, BarkBase introduces a reliable and solid resource to support comparative studies between canine and human species (http://www.barkbase.org/).
Conclusion and Perspectives
Epigenetic alterations are present and possibly regulating several types of canine cancer. Furthermore, many of these epigenetic alterations in canine cancer are also observed in human cancer including genome-wide hypomethylation, hypermethylation of tumor suppressor genes, aberrant histone modifications, and dysregulation of non-coding RNA (Table 1). These data suggest a potential approach using the canine model to determine new epigenetic mechanisms regulating cancer, diagnostic/prognostic markers, and targets for the development of new anticancer drugs. Interestingly, these findings increase the possibility to investigate which environmental factors play a role in epigenetic alterations both in human and dogs, since both species are exposed to the same carcinogens in the environments during their life, and, surprisingly, few studies aimed to observe the environmental risk factors in canine cancer (236–240). Despite these similarities, some differences regarding the epigenetic landscape can be observed in human and canine cancer such as the epigenetic regulation of estrogen receptor α between breast cancer in women and mammary cancer in dogs and the epigenetic regulation of ABCB1 gene in lymphomas. In addition, most studies aiming to elucidate the epigenetic profile of canine cancer and to determine possible targets and therapies for this disease in these animals are performed using in vitro models. Thus, further investigations are needed to confirm the potential of use dogs as a comparative and translational model to study epigenetics.
Currently, therapeutic options to treat canine cancer are basically surgery, radiotherapy, hyperthermia, photodynamic therapy, immunotherapy, and chemotherapy (241). Thus far, there is no epigenetic drug specific to treat canine cancer or being used in veterinary oncology clinics. This fact is probably a consequence of the lack of solid studies determining the main epigenetic targets in canine cancer. Studies of targeted therapy in dogs using appropriate protocols and models inhibiting epigenetic targets are missing to investigate the potential of epigenetic modulation for the treatment of canine cancer in clinics. Furthermore, all current epigenetic drugs were designed for human treatment. Thus, despite promising in vitro results of epigenetic drugs in canine cancer cells, the effect of the compounds was not optimized for canine cancers, and the side effects may be present due to differences in physiology between man and dog.
Following the exciting development of studying the role of epigenetic reprogramming in human cancer, this area is also emerging in veterinary oncology. Several studies have unveiled epigenetic alterations in canine cancer types, and importantly, some common features corroborate findings observed in human cancer. There are several important similarities such as spontaneous tumor development and the influence of environmental factors that entail for more thoroughly designed comparative studies of human and dog cancer. Databases such as CCOGC (Canine and Comparative Oncology and Genomics Consortium) and BarkBase provide promising first steps and tools to elucidate the mechanisms behind canine cancer and support comparative studies between dogs and humans. However, these are only first steps and more research is necessary in order to better understand dogs as models to study epigenetics in cancer and drug development. We hope that the advancement of knowledge and technologies of epigenetic tools will aid the development of new targets and the advancement of drugs in the area of veterinary oncology.
Author Contributions
PX performed the literature review, wrote the manuscript, and produced figures and tables. SM wrote and reviewed the manuscript, figures, and tables. HF reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This review has been supported by grants from the Sao Paulo Research Foundation (FAPESP) (grant and PX scholarship numbers: 2014/02493-7, 2017/11966-4, and 2019/05778-6). SM is grateful for funding received from the SGC, a registered charity (no: 1097737) that receives funds from; AbbVie, Bayer AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genentech, Genome Canada through Ontario Genomics Institute [OGI-196], EU/EFPIA/OICR/McGill/KTH/Diamond, Innovative Medicines Initiative 2 Joint Undertaking [EUbOPEN grant 875510], Janssen, Merck KGaA (aka EMD in Canada and US), Merck & Co (aka MSD outside Canada and US), Pfizer, São Paulo Research Foundation-FAPESP, Takeda and Wellcome [106169/ZZ14/Z].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.591843/full#supplementary-material
References
1. Baylin SB, Jones PA. A decade of exploring the cancer epigenome-biological and translational implications. Nat Rev Cancer (2011) 11:726–34. doi: 10.1038/nrc3130
2. Biswas S, Rao CM. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur J Pharmacol (2018) 837:8–24. doi: 10.1016/j.ejphar.2018.08.021
3. Dawson MA. The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Sci (80 ) (2017) 355:1147–52. doi: 10.1126/science.aam7304
4. Dawson MA, Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell (2012) 150:12–27. doi: 10.1016/j.cell.2012.06.013
5. Wu Q, Heidenreich D, Zhou S, Ackloo S, Krämer A, Nakka K, et al. A chemical toolbox for the study of bromodomains and epigenetic signaling. Nat Commun (2019) 10:1–14. doi: 10.1038/s41467-019-09672-2
6. Scheer S, Ackloo S, Medina TS, Schapira M, Li F, Ward JA, et al. A chemical biology toolbox to study protein methyltransferases and epigenetic signaling. Nat Commun (2019) 10:1–14. doi: 10.1038/s41467-018-07905-4
7. Huston A, Arrowsmith CH, Knapp S, Schapira M. Probing the epigenome. Nat Chem Biol (2015) 11:542–5. doi: 10.1038/nchembio.1871
8. Khanna C, Lindblad-Toh K, Vail D, London C, Bergman P, Barber L, et al. The dog as a cancer model. Nat Biotechnol (2006) 24:1065–6. doi: 10.1038/nbt0906-1065b
9. Cozzi B, Ballarin C, Mantovani R, Rota A. Aging and veterinary care of cats, dogs, and horses through the records of three university veterinary hospitals. Front Vet Sci (2017) 4:14. doi: 10.3389/fvets.2017.00014
10. Inoue M, Hasegawa A, Hosoi Y, Sugiura K. A current life table and causes of death for insured dogs in Japan. Prev Vet Med (2015) 120:210–8. doi: 10.1016/j.prevetmed.2015.03.018
11. Adams VJ, Evans KM, Sampson J, Wood JLN. Methods and mortality results of a health survey of purebred dogs in the UK. J Small Anim Pract (2010) 51:512–24. doi: 10.1111/j.1748-5827.2010.00974.x
12. Egenvall A, Bonnett BN, Hedhammar Å, Olson P. Mortality in over 350,000 insured Swedish dogs from 1995-2000: II. Breed-specific age and survival patterns and relative risk for causes of death. Acta Vet Scand (2005) 46:121–36. doi: 10.1186/1751-0147-46-121
13. Fleming JM, Creevy KE, Promislow DEL. Mortality in North American Dogs from 1984 to 2004: An Investigation into Age-, Size-, and Breed-Related Causes of Death. J Vet Intern Med (2011) 25:187–98. doi: 10.1111/j.1939-1676.2011.0695.x
14. Grüntzig K, Graf R, Hässig M, Welle M, Meier D, Lott G, et al. The Swiss canine cancer registry: A retrospective study on the occurrence of tumours in dogs in Switzerland from 1955 to 2008. J Comp Pathol (2015) 152:161–71. doi: 10.1016/j.jcpa.2015.02.005
15. Fighera RA, Souza TM, Silva MC, Brum JS, Graça DL, Kommers GD, et al. Causas de morte e razões para eutanásia de cães da Mesorregião do Centro Ocidental Rio-Grandense (1965-2004). Pesqui Veterineria Bras (2008) 28:223–30. doi: 10.1590/s0100-736x2008000400005
16. Merlo DF, Rossi L, Pellegrino C, Ceppi M, Cardellino U, Capurro C, et al. Cancer Incidence in Pet Dogs: Findings of the Animal Tumor Registry of Genoa, Italy. J Vet Intern Med (2008) 22:976–84. doi: 10.1111/j.1939-1676.2008.0133.x
17. García E, Alpízar A, Fajardo R, Córdova D, Pérez L, Martínez S. Epidemiology of tumors in dogs in the capital of the state of Mexico from 2002-2016. Arq Bras Med Vet E Zootec (2019) 71:1085–92. doi: 10.1590/1678-4162-10534
18. Brønden LB, Nielsen SS, Toft N, Kristensen AT. Data from the Danish veterinary cancer registry on the occurrence and distribution of neoplasms in dogs in Denmark. Vet Rec (2010) 166:586–90. doi: 10.1136/vr.b4808
19. Choi JW, Yoon HY, Jeong SW. Clinical outcomes of surgically managed spontaneous tumors in 114 client-owned dogs. Immune Netw (2016) 16:116–25. doi: 10.4110/in.2016.16.2.116
20. Sorenmo K. Canine mammary gland tumors. Vet Clin North Am Small Anim Pract (2003) 33:573–96. doi: 10.1016/S0195-5616(03)00020-2
21. Pastor N, Caballé NC, Santella M, Ezquerra LJ, Tarazona R, Duran E. Epidemiological study of canine mammary tumors: Age, breed, size and malignancy. Austral J Vet Sci (2018) 50:143–7. doi: 10.4067/s0719-81322018000300143
22. Gupta K, Sood N, Uppal S. Epidemiological studies on canine mammary tumour and its relevance for breast cancer studies. IOSR J Pharm (2012) 2:322–33. doi: 10.9790/3013-0220322333
23. Baioni E, Scanziani E, Vincenti MC, Leschiera M, Bozzetta E, Pezzolato M, et al. Estimating canine cancer incidence: Findings from a population-based tumour registry in northwestern Italy. BMC Vet Res (2017) 13:1–9. doi: 10.1186/s12917-017-1126-0
24. Fenger JM, London CA, Kisseberth WC. Canine osteosarcoma: A naturally occurring disease to inform pediatric oncology. ILAR J (2014) 55:69–85. doi: 10.1093/ilar/ilu009
25. Gardner HL, Fenger JM, London CA. Dogs as a Model for Cancer. Annu Rev Anim Biosci (2016) 4:199–222. doi: 10.1146/annurev-animal-022114-110911
26. Queiroga FL, Raposo T, Carvalho MI, Prada J, Pires I. Canine mammary tumours as a model to study human breast cancer: most recent findings. Vivo (2011) 25:455–65.
28. Chun R. Lymphoma: Which Chemotherapy Protocol and Why? Top Companion Anim Med (2009) 24:157–62. doi: 10.1053/j.tcam.2009.03.003
29. Rebhun RB, Kent MS, Borrofka SAEB, Frazier S, Skorupski K, Rodriguez CO. CHOP chemotherapy for the treatment of canine multicentric T-cell lymphoma. Vet Comp Oncol (2011) 9:38–44. doi: 10.1111/j.1476-5829.2010.00230.x
30. Kirpensteijn J, Kik M, Teske E, Rutteman GR. TP53 gene mutations in canine osteosarcoma. Vet Surg (2008) 37:454–60. doi: 10.1111/j.1532-950X.2008.00407.x
31. Van Leeuwen IS, Cornelisse CJ, Misdorp W, Goedegebuure SA, Kirpensteijn J, Rutteman GR. P53 gene mutations in osteosarcomas in the dog. Cancer Lett (1997) 111:173–8. doi: 10.1016/S0304-3835(96)04529-6
32. Thomas R, Wang HJ, Tsai PC, Langford CF, Fosmire SP, Jubala CM, et al. Influence of genetic background on tumor karyotypes: Evidence for breed-associated cytogenetic aberrations in canine appendicular osteosarcoma. Chromosom Res (2009) 17:365–77. doi: 10.1007/s10577-009-9028-z
33. Fossey SL, Liao AT, McCleese JK, Bear MD, Lin J, Li PK, et al. Characterization of STAT3 activation and expression in canine and human osteosarcoma. BMC Cancer (2009) 9:1–15. doi: 10.1186/1471-2407-9-81
34. Thomas R, Seiser EL, Motsinger-Reif A, Borst L, Valli VE, Kelley K, et al. Refining tumor-associated aneuploidy through ‘genomic recoding’ of recurrent DNA copy number aberrations in 150 canine non-Hodgkin lymphomas. Leuk Lymphoma (2011) 52:1321–35. doi: 10.3109/10428194.2011.559802
35. Pophali PA, Marinelli LM, Ketterling RP, Meyer RG, McPhail ED, Kurtin PJ, et al. High level MYC amplification in B-cell lymphomas: is it a marker of aggressive disease? Blood Cancer J (2020) 10:1–9. doi: 10.1038/s41408-019-0271-z
36. Mudaliar MAV, Haggart RD, Miele G, Sellar G, Tan KAL, Goodlad JR, et al. Comparative Gene Expression Profiling Identifies Common Molecular Signatures of NF-κB Activation in Canine and Human Diffuse Large B Cell Lymphoma (DLBCL). PloS One (2013) 8:1–17. doi: 10.1371/journal.pone.0072591
37. Quintás-Cardama A, Cortes J. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood (2009) 113:1619–30. doi: 10.1182/blood-2008-03-144790
38. Cruz Cardona JA, Milner R, Alleman AR, Williams C, Vernau W, Breen M, et al. BCR-ABL translocation in a dog with chronic monocytic leukemia. Vet Clin Pathol (2011) 40:40–7. doi: 10.1111/j.1939-165X.2010.00277.x
39. Figueiredo JF, Culver S, Behling-Kelly E, Breen M, Friedrichs KR. Acute myeloblastic leukemia with associated BCR-ABL translocation in a dog. Vet Clin Pathol (2012) 41:362–8. doi: 10.1111/j.1939-165X.2012.00450.x
40. Rivera PJ, Melin M, Biagi T, Fall T, Häggström J, Lindblad-Toh K, et al. Mammary tumor development in dogs is associated with BRCA1 and BRCA2. Cancer Res (2009) 69:8770–4. doi: 10.1158/0008-5472.CAN-09-1725
41. Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet (1998) 62:676–89. doi: 10.1086/301749
42. Hsu WL, Huang HM, Liao JW, Wong ML, Chang SC. Increased survival in dogs with malignant mammary tumours overexpressing HER-2 protein and detection of a silent single nucleotide polymorphism in the canine HER-2 gene. Vet J (2009) 180:116–23. doi: 10.1016/j.tvjl.2007.10.013
43. Iqbal N, Iqbal N. Human Epidermal Growth Factor Receptor 2 (HER2) in Cancers: Overexpression and Therapeutic Implications. Mol Biol Int (2014) 2014:1–9. doi: 10.1155/2014/852748
44. Greenberg MVC, Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol (2019) 20:590–607. doi: 10.1038/s41580-019-0159-6
45. Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet (2012) 13:484–92. doi: 10.1038/nrg3230
46. Yang X, Han H, DeCarvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell (2014) 26:577–90. doi: 10.1016/j.ccr.2014.07.028
47. Ehrlich M. DNA methylation in cancer: Too much, but also too little. Oncogene (2002) 21:5400–13. doi: 10.1038/sj.onc.1205651
48. Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics (2009) 1:239–59. doi: 10.2217/epi.09.33
49. Pelham JT, Irwin PJ, Kay PH. Genomic hypomethylation in neoplastic cells from dogs with malignant lymphoproliferative disorders. Res Vet Sci (2003) 74:101–4. doi: 10.1016/S0034-5288(02)00179-0
50. Alvarez H, Opalinska J, Zhou L, Sohal D, Fazzari MJ, Yu Y, et al. Widespread hypomethylation occurs early and synergizes with gene amplification during esophageal carcinogenesis. PloS Genet (2011) 7:1–14. doi: 10.1371/journal.pgen.1001356
51. Narayan A, Ji W, Zhang XY, Marrogi A, Graff JR, Baylin SB, et al. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int J Cancer (1998) 77:833–8. doi: 10.1002/(SICI)1097-0215(19980911)77:6<833::AID-IJC6>3.0.CO;2-V
52. Jackson K, Yu MC, Arakawa K, Fiala E, Youn B, Fiegl H, et al. DNA hypomethylation is prevalent even in low-grade breast cancers. Cancer Biol Ther (2004) 3:1225–31. doi: 10.4161/cbt.3.12.1222
53. Morimoto CY, Tedardi MV, da Fonseca IIM, Kimura KC, Sanches DS, Epiphanio TF, et al. Evaluation of the global DNA methylation in canine mast cell tumour samples by immunostaining of 5-methyl cytosine. Vet Comp Oncol (2017) 15:1014–8. doi: 10.1111/vco.12241
54. Epiphanio TMF, de Azevedo Fernandes NCC, de Oliveira TF, Lopes PA, Réssio RA, Gonçalves S, et al. Global DNA methylation of peripheral blood leukocytes from dogs bearing multicentric non-Hodgkin lymphomas and healthy dogs: A comparative study. PloS One (2019) 14:1–22. doi: 10.1371/journal.pone.0211898
55. Herrera CL, Kim DY, Kumar SR, Bryan JN. Peroxisome proliferator activated receptor γ protein expression is asymmetrically distributed in primary lung tumor and metastatic to lung osteosarcoma samples and does not correlate with gene methylation. BMC Vet Res (2015) 11:1–11. doi: 10.1186/s12917-015-0547-x
56. Cheung H-H, Lee T-L, Rennert OM, Chan W-Y. DNA methylation of cancer genome. Birth Defects Res Part C Embryo Today Rev (2009) 87:335–50. doi: 10.1002/bdrc.20163
57. Shi H, Guo J, Duff DJ, Rahmatpanah F, Chitima-Matsiga R, Al-Kuhlani M, et al. Discovery of novel epigenetic markers in non-Hodgkin’s lymphoma. Carcinogenesis (2007) 28:60–70. doi: 10.1093/carcin/bgl092
58. Bryan JN, Taylor KH, Henry CJ, Selting KA, Rahmatpanah F, Lewis MR, et al. DNA methylation in cancer: techniques and preliminary evidence of hypermethylation in canine lymphoma. Cancer Ther (2008) 6:137–48.
59. Bryan JN, Jabbes M, Berent LM, Arthur GL, Taylor KH, Rissetto KC, et al. Hypermethylation of the DLC1 CpG island does not alter gene expression in canine lymphoma. BMC Genet (2009) 10:1–12. doi: 10.1186/1471-2156-10-73
60. Noguchi S, Mori T, Igase M, Mizuno T. A novel apoptosis-inducing mechanism of 5-aza-2’-deoxycitidine in melanoma cells: Demethylation of TNF-α and activation of FOXO1. Cancer Lett (2015) 369:344–53. doi: 10.1016/j.canlet.2015.08.023
61. Noguchi S, Mori T, Nakagawa T, Itamoto K, Haraguchi T, Mizuno T. DNA methylation contributes toward silencing of antioncogenic microRNA-203 in human and canine melanoma cells. Melanoma Res (2015) 25:390–8. doi: 10.1097/CMR.0000000000000183
62. Noguchi S, Mori T, Otsuka Y, Yamada N, Yasui Y, Iwasaki J, et al. Anti-oncogenic microRNA-203 induces senescence by targeting E2F3 protein in human melanoma cells. J Biol Chem (2012) 287:11769–77. doi: 10.1074/jbc.M111.325027
63. Ferraresso S, Bresolin S, Aricò A, Comazzi S, Gelain ME, Riondato F, et al. Epigenetic silencing of TFPI-2 in canine diffuse large B-cell lymphoma. PloS One (2014) 9:1–10. doi: 10.1371/journal.pone.0092707
64. Sato M, Mochizuki H, Goto-Koshino Y, Fujiwara-Igarashi A, Takahashi M, Fujino Y, et al. Hypermethylation of the death-associated protein kinase CpG island in canine B-cell lymphoid tumors. Vet Immunol Immunopathol (2014) 161:222–31. doi: 10.1016/j.vetimm.2014.08.011
65. Fujiwara-Igarashi A, Goto-Koshino Y, Mochizuki H, Sato M, Fujino Y, Ohno K, et al. Inhibition of p16 tumor suppressor gene expression via promoter hypermethylation in canine lymphoid tumor cells. Res Vet Sci (2014) 97:60–3. doi: 10.1016/j.rvsc.2014.04.008
66. Ferraresso S, Aricò A, Sanavia T, Da Ros S, Milan M, Cascione L, et al. DNA methylation profiling reveals common signatures of tumorigenesis and defines epigenetic prognostic subtypes of canine Diffuse Large B-cell Lymphoma. Sci Rep (2017) 7:1–11. doi: 10.1038/s41598-017-11724-w
67. Da Ros S, Aresu L, Ferraresso S, Zorzan E, Gaudio E, Bertoni F, et al. Validation of epigenetic mechanisms regulating gene expression in canine B-cell lymphoma: An in vitro and in vivo approach. PloS One (2018) 13:1–19. doi: 10.1371/journal.pone.0208709
68. Sato M, Mochizuki H, Goto-Koshino Y, Fujiwara-Igarashi A, Takahashi M, Ohno K, et al. Prognostic significance of hypermethylation of death-associated protein kinase (DAPK) gene CpG island in dogs with high-grade B-cell lymphoma. Vet Comp Oncol (2018) 16:409–15. doi: 10.1111/vco.12395
69. Bronzini I, Aresu L, Paganin M, Marchioretto L, Comazzi S, Cian F, et al. DNA methylation and targeted sequencing of methyltransferases family genes in canine acute myeloid leukaemia, modelling human myeloid leukaemia. Vet Comp Oncol (2017) 15:910–8. doi: 10.1111/vco.12231
70. Yamazaki J, Jelinek J, Hisamoto S, Tsukamoto A, Inaba M. Dynamic changes in DNA methylation patterns in canine lymphoma cell lines demonstrated by genome-wide quantitative DNA methylation analysis. Vet J (2018) 231:48–54. doi: 10.1016/j.tvjl.2017.11.007
71. Im KS, Kim IH, Kim NH, Lim HY, Kim JH, Sur JH. Breed-related differences in altered BRCA1 expression, phenotype and subtype in malignant canine mammary tumors. Vet J (2013) 195:366–72. doi: 10.1016/j.tvjl.2012.07.014
72. Qiu HB, Sun WD, Yang X, Jiang QY, Chen S, Lin DG. Promoter mutation and reduced expression of BRCA1 in canine mammary tumors. Res Vet Sci (2015) 103:143–8. doi: 10.1016/j.rvsc.2015.10.003
73. Qiu H, Lin D. Roles of DNA mutation in the coding region and DNA methylation in the 5′ flanking region of BRCA1 in canine mammary tumors. J Vet Med Sci (2016) 78:943–9. doi: 10.1292/jvms.15-0557
74. Tomiyasu H, Goto-Koshino Y, Fujino Y, Ohno K, Tsujimoto H. Epigenetic regulation of the ABCB1 gene in drug-sensitive and drug-resistant lymphoid tumour cell lines obtained from canine patients. Vet J (2014) 199:103–9. doi: 10.1016/j.tvjl.2013.10.022
75. Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE, Gottesman MM. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer (2018) 18:452–64. doi: 10.1038/s41568-018-0005-8
76. Tomiyasu H, Fujiwara-Igarashi A, Goto-Koshino Y, Fujino Y, Ohno K, Tsujimoto H. Evaluation of DNA methylation profiles of the CpG island of the ABCB1 gene in dogs with lymphoma. Am J Vet Res (2014) 75:835–41. doi: 10.2460/ajvr.75.9.835
77. Ottaviano YL, Issa JP, Pari FF, Smith HS, Baylin SB, Davidson NE. Methylation of the Estrogen Receptor Gene CpG Island Marks Loss of Estrogen Receptor Expression in Human Breast Cancer Cells. Cancer Res (1994) 54:2552–5.
78. Prabhu JS, Wahi K, Korlimarla A, Correa M, Manjunath S, Raman N, et al. The epigenetic silencing of the estrogen receptor (ER) by hypermethylation of the ESR1 promoter is seen predominantly in triple-negative breast cancers in Indian women. Tumor Biol (2012) 33:315–23. doi: 10.1007/s13277-012-0343-1
79. Brandão Y de O, Toledo MB, Chequin A, Cristo TG, Sousa RS, Ramos EAS, et al. DNA Methylation Status of the Estrogen Receptor α Gene in Canine Mammary Tumors. Vet Pathol (2018) 55:510–6. doi: 10.1177/0300985818763711
80. Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev (2016) 30:733–50. doi: 10.1101/gad.276568.115
81. Zorzan E, Hanssens K, Giantin M, Dacasto M, Dubreuil P. Mutational hotspot of TET2, IDH1, IDH2, SRSF2, SF3B1, KRAS, and NRAS from human systemic mastocytosis are not conserved in canine mast cell tumors. PloS One (2015) 10:1–11. doi: 10.1371/journal.pone.0142450
82. Tefferi A, Levine RL, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, et al. Frequent TET2 mutations in systemic mastocytosis: Clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia (2009) 23:900–4. doi: 10.1038/leu.2009.37
83. McDonald JT, Kritharis A, Beheshti A, Pilichowska M, Burgess K, Ricks-Santi L, et al. Correction: Comparative oncology DNA sequencing of canine T cell lymphoma via human hotspot panel. Oncotarget (2018) 9:33441. doi: 10.18632/oncotarget.26114
84. Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature (1997) 389:251–60. doi: 10.1038/38444
85. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res (2011) 21:381–95. doi: 10.1038/cr.2011.22
86. Kebede AF, Schneider R, Daujat S. Novel types and sites of histone modifications emerge as players in the transcriptional regulation contest. FEBS J (2015) 282:1658–74. doi: 10.1111/febs.13047
87. Kouzarides T. Chromatin Modifications and Their Function. Cell (2007) 128:693–705. doi: 10.1016/j.cell.2007.02.005
88. Lawrence M, Daujat S, Schneider R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet (2016) 32:42–56. doi: 10.1016/j.tig.2015.10.007
89. Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet (2005) 37:391–400. doi: 10.1038/ng1531
90. Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JCY, Liang G, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res (2002) 62:6456–61.
91. Wan YCE, Liu J, Chan KM. Histone H3 Mutations in Cancer. Curr Pharmacol Rep (2018) 4:292–300. doi: 10.1007/s40495-018-0141-6
92. Nichol JN, Dupéré-Richer D, Ezponda T, Licht JD, Miller WH Jr. H3K27 Methylation: A Focal Point of Epigenetic Deregulation in Cancer. Adv Cancer Res (2016) 131:59–95. doi: 10.1016/bs.acr.2016.05.001
93. Michalak EM, Visvader JE. Dysregulation of histone methyltransferases in breast cancer – Opportunities for new targeted therapies? Mol Oncol (2016) 10:1497. doi: 10.1016/j.molonc.2016.09.003
94. Rowbotham SP, Li F, Dost AFM, Louie SM, Marsh BP, Pessina P, et al. H3K9 methyltransferases and demethylases control lung tumor-propagating cells and lung cancer progression. Nat Commun (2018) 9:1–13. doi: 10.1038/s41467-018-07077-1
95. Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene (2007) 26:5420–32. doi: 10.1038/sj.onc.1210610
96. Iyer NG, Özdag H, Caldas C. p300/CBP and cancer. Oncogene (2004) 23:4225–31. doi: 10.1038/sj.onc.1207118
98. Casciello F, Windloch K, Gannon F, Lee JS. Functional role of G9a histone methyltransferase in cancer. Front Immunol (2015) 6:487. doi: 10.3389/fimmu.2015.00487
99. Tu WB, Shiah YJ, Lourenco C, Mullen PJ, Dingar D, Redel C, et al. MYC Interacts with the G9a Histone Methyltransferase to Drive Transcriptional Repression and Tumorigenesis. Cancer Cell (2018) 34:579–595.e8. doi: 10.1016/j.ccell.2018.09.001
100. Eto S, Saeki K, Yoshitake R, Yoshimoto S, Shinada M, Ikeda N, et al. Anti-tumor effects of the histone deacetylase inhibitor vorinostat on canine urothelial carcinoma cells. PloS One (2019) 14:1–15. doi: 10.1371/journal.pone.0218382
101. Sakthikumar S, Elvers I, Kim J, Arendt ML, Thomas R, Turner-Maier J, et al. SETD2 is recurrently mutated in whole-exome sequenced canine osteosarcoma. Cancer Res (2018) 78:3421–31. doi: 10.1158/0008-5472.CAN-17-3558
102. Gardner HL, Sivaprakasam K, Briones N, Zismann V, Perdigones N, Drenner K, et al. Canine osteosarcoma genome sequencing identifies recurrent mutations in DMD and the histone methyltransferase gene SETD2. Commun Biol (2019) 2:1–13. doi: 10.1038/s42003-019-0487-2
103. Choi HJ, Jang S, Ryu JE, Lee HJ, Lee HB, Ahn WS, et al. Significance of EZH2 expression in canine mammary tumors. BMC Vet Res (2016) 12:1–7. doi: 10.1186/S12917-016-0789-2
104. Choi HJ, Lee HB, Park HK, Cho SM, Han HJ, Lee SJ, et al. EZH2 expression in naturally occurring canine tumors. Comp Med (2018) 68:148–55.
105. Goldschmidt MH, Peña L, Rasotto R, Zappulli V. Classification and grading of canine mammary tumors. Vet Pathol (2011) 48:117–31. doi: 10.1177/0300985810393258
106. Liu D, Xiong H, Ellis AE, Northrup NC, Rodriguez CO, O’Regan RM, et al. Molecular homology and difference between spontaneous canine mammary cancer and human breast cancer. Cancer Res (2014) 74:5045–56. doi: 10.1158/0008-5472.CAN-14-0392
107. Slack FJ, Chinnaiyan AM. The Role of Non-coding RNAs in Oncology. Cell (2019) 179:1033–55. doi: 10.1016/j.cell.2019.10.017
108. Alberti C, Cochella L. A framework for understanding the roles of miRNAs in animal development. Dev (2017) 144:2548–59. doi: 10.1242/dev.146613
109. Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev (2005) 15:563–8. doi: 10.1016/j.gde.2005.08.005
110. Kloosterman WP, Plasterk RHA. The Diverse Functions of MicroRNAs in Animal Development and Disease. Dev Cell (2006) 11:441–50. doi: 10.1016/j.devcel.2006.09.009
111. Wilczynska A, Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ (2015) 22:22–33. doi: 10.1038/cdd.2014.112
112. Zhou SS, Jin JP, Wang JQ, Zhang ZG, Freedman JH, Zheng Y, et al. MiRNAS in cardiovascular diseases: Potential biomarkers, therapeutic targets and challenges review-article. Acta Pharmacol Sin (2018) 39:1073–84. doi: 10.1038/aps.2018.30
113. Maciotta S, Meregalli M, Torrente Y. The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci (2013) 7:265. doi: 10.3389/fncel.2013.00265
114. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA (2002) 99:15524–9. doi: 10.1073/pnas.242606799
115. O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature (2005) 435:839–43. doi: 10.1038/nature03677
116. Han L, Witmer PD, Casey E, Valle D, Sukumar S. DNA methylation regulates microRNA expression. Cancer Biol Ther (2007) 6:1284–8. doi: 10.4161/cbt.6.8.4486
117. Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev (2006) 20:2202–7. doi: 10.1101/gad.1444406
118. Iliou MS, Da Silva-Diz V, Carmona FJ, Ramalho-Carvalho J, Heyn H, Villanueva A, et al. Impaired DICER1 function promotes stemness and metastasis in colon cancer. Oncogene (2014) 33:4003–15. doi: 10.1038/onc.2013.398
119. Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther (2016) 1:1–9. doi: 10.1038/sigtrans.2015.4
120. Boggs RM, Wright ZM, Stickney MJ, Porter WW, Murphy KE. MicroRNA expression in canine mammary cancer. Mamm Genome (2008) 19:561–9. doi: 10.1007/s00335-008-9128-7
121. Fenger JM, Bear MD, Volinia S, Lin TY, Harrington BK, London CA, et al. Overexpression of miR-9 in mast cells is associated with invasive behavior and spontaneous metastasis. BMC Cancer (2014) 14:1–16. doi: 10.1186/1471-2407-14-84
122. Fenger JM, Roberts RD, Iwenofu OH, Bear MD, Zhang X, Couto JI, et al. MiR-9 is overexpressed in spontaneous canine osteosarcoma and promotes a metastatic phenotype including invasion and migration in osteoblasts and osteosarcoma cell lines. BMC Cancer (2016) 16:1–19. doi: 10.1186/s12885-016-2837-5
123. Grimes JA, Prasad N, Levy S, Cattley R, Lindley S, Boothe HW, et al. Smith BF. A comparison of microRNA expression profiles from splenic hemangiosarcoma, splenic nodular hyperplasia, and normal spleens of dogs. BMC Vet Res (2016) 12:1–12. doi: 10.1186/s12917-016-0903-5
124. Kobayashi M, Saito A, Tanaka Y, Michishita M, Kobayashi M, Irimajiri M, et al. Microrna expression profiling in canine prostate cancer. J Vet Med Sci (2017) 79:719–25. doi: 10.1292/jvms.16-0279
125. Craig KKL, Wood GA, Keller SM, Mutsaers AJ, Wood RD. MicroRNA profiling in canine multicentric lymphoma. PloS One (2019) 14:1–24. doi: 10.1371/journal.pone.0226357
126. Rahman MM, Lai YC, Husna AA, Chen HW, Tanaka Y, Kawaguchi H, et al. Micro RNA transcriptome profile in canine oral melanoma. Int J Mol Sci (2019) 20:1–19. doi: 10.3390/ijms20194832
127. Ushio N, Rahman MM, Maemura T, Lai YC, Iwanaga T, Kawaguchi H, et al. Identification of dysregulated microRNAs in canine malignant melanoma. Oncol Lett (2019) 17:1080–8. doi: 10.3892/ol.2018.9692
128. Mortarino M, Gioia G, Gelain ME, Albonico F, Roccabianca P, Ferri E, et al. Identification of suitable endogenous controls and differentially expressed microRNAs in canine fresh-frozen and FFPE lymphoma samples. Leuk Res (2010) 34:1070–7. doi: 10.1016/j.leukres.2009.10.023
129. Starkey MP, Compston-Garnett L, Malho P, Dunn K, Dubielzig R. Metastasis-associated microRNA expression in canine uveal melanoma. Vet Comp Oncol (2018) 16:81–9. doi: 10.1111/vco.12315
130. Bulkowska M, Rybicka A, Senses KM, Ulewicz K, Witt K, Szymanska J, et al. MicroRNA expression patterns in canine mammary cancer show significant differences between metastatic and non-metastatic tumours. BMC Cancer (2017) 17:1–17. doi: 10.1186/s12885-017-3751-1
131. Lopez CM, Yu PY, Zhang X, Yilmaz AS, London CA, Fenger JM. MiR-34a regulates the invasive capacity of canine osteosarcoma cell lines. PloS One (2018) 13:1–23. doi: 10.1371/journal.pone.0190086
132. Albonico F, Mortarino M, Avallone G, Gioia G, Comazzi S, Roccabianca P. The expression ratio of miR-17-5p and miR-155 correlates with grading in canine splenic lymphoma. Vet Immunol Immunopathol (2013) 155:117–23. doi: 10.1016/j.vetimm.2013.06.018
133. Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui R, et al. Circulating MicroRNAs in Cancer: Potential and Challenge. Front Genet (2019) 10:1–11. doi: 10.3389/fgene.2019.00626
134. Heishima K, Ichikawa Y, Yoshida K, Iwasaki R, Sakai H, Nakagawa T, et al. Circulating microRNA-214 and -126 as potential biomarkers for canine neoplastic disease. Sci Rep (2017) 7:1–14. doi: 10.1038/s41598-017-02607-1
135. Heishima K, Meuten T, Yoshida K, Mori T, Thamm DH. Prognostic significance of circulating microRNA-214 and -126 in dogs with appendicular osteosarcoma receiving amputation and chemotherapy. BMC Vet Res (2019) 15:1–13. doi: 10.1186/s12917-019-1776-1
136. Børresen B, Nielsen LN, Jessen LR, Kristensen AT, Fredholm M, Cirera S. Circulating let-7g is down-regulated in Bernese Mountain dogs with disseminated histiocytic sarcoma and carcinomas – a prospective study. Vet Comp Oncol (2017) 15:525–33. doi: 10.1111/vco.12196
137. Fujiwara-Igarashi A, Igarashi H, Mizutani N, Goto-Koshino Y, Takahashi M, Ohno K, et al. Expression profile of circulating serum microRNAs in dogs with lymphoma. Vet J (2015) 205:317–21. doi: 10.1016/j.tvjl.2015.04.029
138. Kent MS, Zwingenberger A, Westropp JL, Barrett LE, Durbin-Johnson BP, Ghosh P, et al. MicroRNA profiling of dogs with transitional cell carcinoma of the bladder using blood and urine samples. BMC Vet Res (2017) 13:1–13. doi: 10.1186/s12917-017-1259-1
139. Fish EJ, Irizarry KJ, DeInnocentes P, Ellis CJ, Prasad N, Moss AG, et al. Malignant canine mammary epithelial cells shed exosomes containing differentially expressed microRNA that regulate oncogenic networks. BMC Cancer (2018) 18:1–20. doi: 10.1186/s12885-018-4750-6
140. Asada H, Tomiyasu H, Uchikai T, Ishihara G, Goto-Koshino Y, Ohno K, et al. Comprehensive analysis of miRNA and protein profiles within exosomes derived from canine lymphoid tumour cell lines. PloS One (2019) 14:1–15. doi: 10.1371/journal.pone.0208567
141. Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Sci (80 ) (2010) 329:689–93. doi: 10.1126/science.1192002
142. Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat Commun (2015) 6:1–17. doi: 10.1038/ncomms8743
143. Zhang Y, Pitchiaya S, Cieślik M, Niknafs YS, Tien JC-Y, Hosono Y, et al. Analysis of the androgen receptor–regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet (2018) 50:814–24. doi: 10.1038/s41588-018-0120-1
144. Du Z, Sun T, Hacisuleyman E, Fei T, Wang X, Brown M, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun (2016) 7:1–10. doi: 10.1038/ncomms10982
145. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature (2010) 464:1071–6. doi: 10.1038/nature08975
146. Li Y, Wang Z, Shi H, Li H, Li L, Fang R, et al. HBXIP and LSD1 scaffolded by lncRNA hotair mediate transcriptional activation by c-Myc. Cancer Res (2016) 76:293–304. doi: 10.1158/0008-5472.CAN-14-3607
147. Hosono Y, Niknafs YS, Prensner JR, Iyer MK, Dhanasekaran SM, Mehra R, et al. Oncogenic Role of THOR, a Conserved Cancer/Testis Long Non-coding RNA. Cell (2017) 171:1559–1572.e20. doi: 10.1016/j.cell.2017.11.040
148. Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature (2016) 531:518–22. doi: 10.1038/nature17161
149. Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y, et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell (2018) 33:706–20. doi: 10.1016/j.ccell.2018.03.006
150. Hoeppner MP, Lundquist A, Pirun M, Meadows JRS, Zamani N, Johnson J, et al. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PloS One (2014) 9:1–11. doi: 10.1371/journal.pone.0091172
151. Wucher V, Legeai F, Hédan B, Rizk G, Lagoutte L, Leeb T, et al. FEELnc: A tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res (2017) 45:1–12. doi: 10.1093/nar/gkw1306
152. Hao Y, Wu W, Shi F, Dalmolin RJS, Yan M, Tian F, et al. Prediction of long noncoding RNA functions with co-expression network in esophageal squamous cell carcinoma. BMC Cancer (2015) 15:1–10. doi: 10.1186/s12885-015-1179-z
153. Gutschner T, Hämmerle M, Eißmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res (2013) 73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850
154. Muljo SA, Zhu J, Zhao K. Expression and regulation of lincRNAs during T cell development and differentiation. Nat Immunol (2013) 14:1190–8. doi: 10.1038/ni.2712.Expression
155. Verma A, Jiang Y, Du W, Fairchild L, Melnick A, Elemento O. Transcriptome sequencing reveals thousands of novel long non-coding RNAs in B cell lymphoma. Genome Med (2015) 7:1–17. doi: 10.1186/s13073-015-0230-7
156. Le Béguec C, Wucher V, Lagoutte L, Cadieu E, Botherel N, Hédan B, et al. Characterisation and functional predictions of canine long non-coding RNAs. Sci Rep (2018) 8:1–12. doi: 10.1038/s41598-018-31770-2
157. Plassais J, Lagoutte L, Correard S, Paradis M, Guaguère E, Hédan B, et al. A Point Mutation in a lincRNA Upstream of GDNF Is Associated to a Canine Insensitivity to Pain: A Spontaneous Model for Human Sensory Neuropathies. PloS Genet (2016) 12:1–21. doi: 10.1371/journal.pgen.1006482
158. Cascione L, Giudice L, Ferraresso S, Marconato L, Giannuzzi D, Napoli S, et al. Long non-coding RNAs as molecular signatures for canine B-cell lymphoma characterization. Non Coding RNA (2019) 5:1–12. doi: 10.3390/ncrna5030047
159. Beltran M, Puig I, Peña C, García JM, Álvarez AB, Peña R, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev (2008) 22:756–69. doi: 10.1101/gad.455708
160. Yang CM, Wang TH, Chen HC, Li SC, Lee MC, Liou HH, et al. Aberrant DNA hypermethylation-silenced SOX21-AS1 gene expression and its clinical importance in oral cancer. Clin Epigenet (2016) 8:129. doi: 10.1186/s13148-016-0291-5
161. Lessard L, Liu M, Marzese DM, Wang H, Chong K, Kawas N, et al. The CASC15 Long Intergenic Noncoding RNA Locus Is Involved in Melanoma Progression and Phenotype Switching. J Invest Dermatol (2015) 135:2464–74. doi: 10.1038/jid.2015.200
162. Hitte C, Le Béguec C, Cadieu E, Wucher V, Primot A, Prouteau A, et al. Genome-wide analysis of long non-coding RNA profiles in canine oral melanomas. Genes (Basel) (2019) 10:1–14. doi: 10.3390/genes10060477
163. Hendrix MJC, Seftor EA, Seftor REB, Kasemeier-Kulesa J, Kulesa PM, Postovit LM. Reprogramming metastatic tumour cells with embryonic microenvironments. Nat Rev Cancer (2007) 7:246–55. doi: 10.1038/nrc2108
164. Bockhorn J, Prat A, Chang YF, Liu X, Huang S, Shang M, et al. Differentiation and loss of malignant character of spontaneous pulmonary metastases in patient-derived breast cancer models. Cancer Res (2014) 74:7406–17. doi: 10.1158/0008-5472.CAN-14-1188
165. Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Sci (80 ) (2017) 357:1–10. doi: 10.1126/science.aal2380
166. Brown R, Curry E, Magnani L, Wilhelm-Benartzi CS, Borley J. Poised epigenetic states and acquired drug resistance in cancer. Nat Rev Cancer (2014) 14:747–53. doi: 10.1038/nrc3819
167. Tkaczynski T, Šmejkal J, Šorm F. Nucleic acids components and their analogues. L. Synthesis of 1-β-D-xylofuranosyl-6-azauracil. Collect Czechoslov Chem Commun (1964) 29:1736–8. doi: 10.1135/cccc19641736
168. Pliml J, Šorm F. Synthesis of a 2-deoxy-d-ribofuranosyl-s-azacytosine. Collect Czechoslov Chem Commun (1964) 29:2576–8. doi: 10.1135/cccc19642576
169. Kaminskas E, Farrell AT, Wang Y, Sridhara R, Pazdur R. FDA Drug Approval Summary: Azacitidine (5-azacytidine, Vidaza TM ) for Injectable Suspension. Oncologist (2005) 10:176–82. doi: 10.1634/theoncologist.10-3-176
170. Saba HI. Decitabine in the treatment of myelodysplastic syndromes. Ther Clin Risk Manag (2007) 3:807–17.
171. Ossenkoppele G, Lowenberg B. How I treat the older patient with AML. Blood (2015) 125:767–74. doi: 10.1182/blood-2014-08-551499.c
172. Fenaux P, Mufti GJ, Hellström-Lindberg E, Santini V, Gattermann N, Germing U, et al. Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. J Clin Oncol (2010) 28:562–9. doi: 10.1200/JCO.2009.23.8329
173. Diesch J, Zwick A, Garz AK, Palau A, Buschbeck M. Götze KS. A clinical-molecular update on azanucleoside-based therapy for the treatment of hematologic cancers. Clin Epigenet (2016) 8:1–11. doi: 10.1186/s13148-016-0237-y
174. Ganesan A, Arimondo PB, Rots MG, Jeronimo C, Berdasco M. The timeline of epigenetic drug discovery: from reality to dreams. Clin Epigenet (2019) 11:174. doi: 10.1186/s13148-019-0776-0
175. Yoo C, Cheng J, Jones PA. Zebularine: A new drug for epigenetic therapy. Biochem Soc Trans (2005) 32:910–2. doi: 10.1042/BST0320910
176. Singh V, Capalash PS. and N. DNA Methyltransferase-1 Inhibitors as Epigenetic Therapy for Cancer. Curr Cancer Drug Targets (2013) 13:379–99. doi: 10.2174/15680096113139990077
177. Kantarjian HM, Roboz GJ, Kropf PL, Yee KWL, O’Connell CL, Tibes R, et al. Guadecitabine (SGI-110) in treatment-naive patients with acute myeloid leukaemia: phase 2 results from a multicentre, randomised, phase 1/2 trial. Lancet Oncol (2017) 18:1317–26. doi: 10.1016/S1470-2045(17)30576-4
178. Nervi C, De Marinis E, Codacci-Pisanelli G. Epigenetic treatment of solid tumours: A review of clinical trials. Clin Epigenet (2015) 7:1–20. doi: 10.1186/s13148-015-0157-2
179. Fan H, Lu X, Wang X, Liu Y, Guo B, Zhang Y, et al. Low-dose decitabine-based chemoimmunotherapy for patients with refractory advanced solid tumors: A phase I/II report. J Immunol Res (2014) 2014:1–14. doi: 10.1155/2014/371087
180. Chabot GG, Rivard GE, Momparler RL. Plasma and Cerebrospinal Fluid Pharmacokinetics of 5-Aza-2 ‘ -deoxycytidine in Rabbits and Dogs. Cancer Res (1983) 43:592–7.
181. Hahn NM, Bonney PL, Dhawan D, Jones DR, Balch C, Guo Z, et al. Subcutaneous 5-Azacitidine Treatment of Naturally Occurring Canine Urothelial Carcinoma: A Novel Epigenetic Approach to Human Urothelial Carcinoma Drug Development. J Urol (2012) 187:302–9. doi: 10.1016/j.juro.2011.09.010
182. Harman RM, Curtis TM, Argyle DJ, Coonrod SA, Van de Walle GR. A Comparative Study on the In Vitro Effects of the DNA Methyltransferase Inhibitor 5-Azacytidine (5-AzaC) in Breast/Mammary Cancer of Different Mammalian Species. J Mammary Gland Biol Neoplasia (2016) 21:51–66. doi: 10.1007/s10911-016-9350-y
183. Flesner BK, Kumar SR, Bryan JN. 6-Thioguanine and zebularine down-regulate DNMT1 and globally demethylate canine malignant lymphoid cells. BMC Vet Res (2014) 10:1–7. doi: 10.1186/s12917-014-0290-8
184. Fulkerson CM, Dhawan D, Jones DR, Marquez VE, Jones PA, Wang Z, et al. Pharmacokinetics and toxicity of the novel oral demethylating agent zebularine in laboratory and tumor bearing dogs. Vet Comp Oncol (2017) 15:226–36. doi: 10.1111/vco.12159
185. Damaskos C, Valsami S, Kontos M, Spartalis E, Kalampokas T, Kalampokas E, et al. Histone deacetylase inhibitors: An attractive therapeutic strategy against breast cancer. Anticancer Res (2017) 37:35–46. doi: 10.21873/anticanres.11286
186. Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, et al. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast. Breast Cancer Res Treat (2005) 94:11–6. doi: 10.1007/s10549-005-6001-1
187. Li A, Liu Z, Li M, Zhou S, Xu Y, Xiao Y, et al. Correction: HDAC5, a potential therapeutic target and prognostic biomarker, promotes proliferation, invasion and migration in human breast cancer. Oncotarget (2017) 8:30619–20. doi: 10.18632/oncotarget.17542
188. Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and Nuclear Recruitment of HDACl in Hormone Refractory Prostate Cancer. Prostate (2004) 59:177–89. doi: 10.1002/pros.20022
189. Kim JH, Choi YK, Kwon HJ, Yang HK, Choi JH, Kim DY. Downregulation of gelsolin and retinoic acid receptor β expression in gastric cancer tissues through histone deacetylase 1. J Gastroenterol Hepatol (2004) 19:218–24. doi: 10.1111/j.1440-1746.2004.03336.x
190. Weichert W, Röske A, Gekeler V, Beckers T, Ebert MP, Pross M, et al. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: a retrospective analysis. Lancet Oncol (2008) 9:139–48. doi: 10.1016/S1470-2045(08)70004-4
191. Weichert W, Denkert C, Noske A, Darb-Esfahani S, Dietel M, Kalloger SE, et al. Expression of class I histone deacetylases indicates poor prognosis in endometrioid subtypes of ovarian and endometrial carcinomas. Neoplasia (2008) 10:1021–7. doi: 10.1593/neo.08474
192. Mithraprabhu S, Kalff A, Chow A, Khong T, Spencer A. Dysregulated Class I histone deacetylases are indicators of poor prognosis in multiple myeloma. Epigenetics (2014) 9:1511–20. doi: 10.4161/15592294.2014.983367
193. Chaiyawat P, Pruksakorn D, Phanphaisarn A, Teeyakasem P, Klangjorhor J, Settakorn J. Expression patterns of class i histone deacetylases in osteosarcoma: A novel prognostic marker with potential therapeutic implications. Mod Pathol (2018) 31:264–74. doi: 10.1038/modpathol.2017.125
194. Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA Approval Summary: Vorinostat for Treatment of Advanced Primary Cutaneous T-Cell Lymphoma. Oncologist (2007) 12:1247–52. doi: 10.1634/theoncologist.12-10-1247
195. Lee HZ, Kwitkowski VE, Del Valle PL, Ricci MS, Saber H, Habtemariam BA, et al. FDA approval: Belinostat for the treatment of patients with relapsed or refractory peripheral T-cell lymphoma. Clin Cancer Res (2015) 21:2666–70. doi: 10.1158/1078-0432.CCR-14-3119
196. Grant C, Rahman F, Piekarz R, Peer C, Frye R, Robey RW, et al. Romidepsin: a new therapy for cutaneous T-cell lymphoma and a potential therapy for solid tumors. Expert Rev Anticancer Ther (2010) 10:997–1008. doi: 10.1586/era.10.88
197. Raedler B, Writer M. Monoclonal Antibody Approved for Patients with Relapsed Multiple Myeloma. Am Heal Drug Benefits (2016) 9:84–9.
198. Kumagai T, Wakimoto N, Yin D, Gery S, Kawamata N, Takai N, et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (Vorinostat, SAHA) profoundly inhibits the growth of human pancreatic cancer cells. Int J Cancer (2007) 121:656–65. doi: 10.1002/ijc.22558
199. Ma X, Wang J, Liu J, Mo Q, Yan X, Ma D, et al. Targeting CD146 in combination with vorinostat for the treatment of ovarian cancer cells. Oncol Lett (2017) 13:1681–7. doi: 10.3892/ol.2017.5630
200. Kaushik D, Vashistha V, Isharwal S, Sediqe SA, Lin MF. Histone deacetylase inhibitors in castration-resistant prostate cancer: Molecular mechanism of action and recent clinical trials. Ther Adv Urol (2015) 7:388–95. doi: 10.1177/1756287215597637
201. Luu T, Kim Kp, Blanchard S, Anyang B, Hurria A, Yang L, et al. Phase IB trial of ixabepilone and vorinostat in metastatic breast cancer. Breast Cancer Res Treat (2018) 167:469–78. doi: 10.1007/s10549-017-4516-x
202. Murahari S, Jalkanen AL, Kulp SK, Chen CS, Modiano JF, London CA, et al. Sensitivity of osteosarcoma cells to HDAC inhibitor AR-42 mediated apoptosis. BMC Cancer (2017) 17:1–11. doi: 10.1186/s12885-017-3046-6
203. Rizzo VL, Levine CB, Wakshlag JJ. The effects of sulforaphane on canine osteosarcoma proliferation and invasion. Vet Comp Oncol (2017) 15:718–30. doi: 10.1111/vco.12212
204. Dias JNR, Aguiar SI, Pereira DM, André AS, Gano L, Correia JDG, et al. The histone deacetylase inhibitor panobinostat is a potent antitumor agent in canine diffuse large B-cell lymphoma. Oncotarget (2018) 9:28586–98. doi: 10.18632/oncotarget.25580
205. Dias JNR, André AS, Aguiar SI, Ministro J, Oliveira J, Peleteiro MC, et al. Establishment of a bioluminescent canine B-cell lymphoma xenograft model for monitoring tumor progression and treatment response in preclinical studies. PloS One (2018) 13:1–14. doi: 10.1371/journal.pone.0208147
206. Vigushin DM, Ali S, Pace PE, Mirsaidi N, Ito K, Adcock I, et al. Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clin Cancer Res (2001) 7:971–6.
207. Nagamine MK, Sanches DS, Pinello KC, Torres LN, Mennecier G, Latorre AO, et al. In vitro inhibitory effect of trichostatin A on canine grade 3 mast cell tumor. Vet Res Commun (2011) 35:391–9. doi: 10.1007/s11259-011-9474-x
208. Watanabe M, Chen IYJ, Ishikura Y, Nakagawa T, Mochizuki M, Nishimura R, et al. Tumor cell growth inhibition and cell differentiation analysis in a canine mammary tumor cell line (MCM-B2) treated with four chemical reagents. J Vet Med Sci (2009) 71:1413–7. doi: 10.1292/jvms.001413
209. Sborov DW, Canella A, Hade EM, Mo X, Khountham S, Wang J, et al. A phase 1 trial of the HDAC inhibitor AR-42 in patients with multiple myeloma and T- and B-cell lymphomas. Leuk Lymphoma (2017) 58:2310–8. doi: 10.1080/10428194.2017.1298751
210. Elshafae SM, Kohart NA, Altstadt LA, Dirksen WP, Rosol TJ. The Effect of a Histone Deacetylase Inhibitor (AR-42) on Canine Prostate Cancer Growth and Metastasis. Prostate (2017) 77:776–93. doi: 10.1002/pros.23318
211. Lin TY, Fenger J, Murahari S, Bear MD, Kulp SK, Wang D, et al. AR-42, a novel HDAC inhibitor, exhibits biologic activity against malignant mast cell lines via down-regulation of constitutively activated Kit. Blood (2010) 115:4217–25. doi: 10.1182/blood-2009-07-231985
212. Wittenburg LA, Gustafson DL, Thamm DH. Phase I pharmacokinetic and pharmacodynamic evaluation of combined valproic acid/doxorubicin treatment in dogs with spontaneous cancer. Clin Cancer Res (2010) 16:4832–42. doi: 10.1158/1078-0432.CCR-10-1238
213. Wittenburg LA, Bisson L, Rose BJ, Korch C, Thamm DH. The histone deacetylase inhibitor valproic acid sensitizes human and canine osteosarcoma to doxorubicin. Cancer Chemother Pharmacol (2011) 67:83–92. doi: 10.1007/s00280-010-1287-z
214. Stein EM, Garcia-Manero G, Rizzieri DA, Tibes R, Berdeja JG, Savona MR, et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood (2018) 131:2662–9. doi: 10.1182/blood-2017-12-818948
215. Gulati N, Béguelin W, Giulino-Roth L. Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk Lymphoma (2018) 59:1574–85. doi: 10.1080/10428194.2018.1430795
216. Charles MRC, Dhayalan A, Hsieh HP, Coumar MS. Insights for the design of protein lysine methyltransferase G9a inhibitors. Future Med Chem (2019) 11:993–1014. doi: 10.4155/fmc-2018-0396
217. Fang Y, Liao G, Yu B. LSD1/KDM1A inhibitors in clinical trials: Advances and prospects. J Hematol Oncol (2019) 12:1–14. doi: 10.1186/s13045-019-0811-9
218. Doroshow DB, Eder JP, LoRusso PM. BET inhibitors: A novel epigenetic approach. Ann Oncol (2017) 28:1776–87. doi: 10.1093/annonc/mdx157
219. Brown PJ, Müller S. Open access chemical probes for epigenetic targets. Future Med Chem (2015) 7:1901–17. doi: 10.4155/fmc.15.127
220. Müller S, Brown PJ. Epigenetic chemical probes. Clin Pharmacol Ther (2012) 92:689–93. doi: 10.1038/clpt.2012.154
221. Weiswald LB, Bellet D, Dangles-Marie V. Spherical Cancer Models in Tumor Biology. Neoplasia (U S) (2015) 17:1–15. doi: 10.1016/j.neo.2014.12.004
222. Xavier PLP, Cordeiro YG, Alexandre PA, Pires PRL, Saranholi BH, Silva ER, et al. An epigenetic screening determines BET proteins as targets to suppress self-renewal and tumorigenicity in canine mammary cancer cells. Sci Rep (2019) 9:1–15. doi: 10.1038/s41598-019-53915-7
223. Kake S, Tsuji S, Enjoji S, Hanasaki S, Hayase H, Yabe R, et al. The role of SET/I2PP2A in canine mammary tumors. Sci Rep (2017) 7:1–11. doi: 10.1038/s41598-017-04291-7
224. Tsuji S, Ohama T, Nakagawa T, Sato K. Efficacy of an anti-cancer strategy targeting set in canine osteosarcoma. J Vet Med Sci (2019) 81:1424–30. doi: 10.1292/jvms.19-0311
225. Ledet MM, Anderson R, Harman R, Muth A, Thompson PR, Coonrod SA, et al. BB-Cl-Amidine as a novel therapeutic for canine and feline mammary cancer via activation of the endoplasmic reticulum stress pathway. BMC Cancer (2018) 18:1–13. doi: 10.1186/s12885-018-4323-8
226. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature (2012) 489:57–74. doi: 10.1038/nature11247
227. Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet (2013) 45:580–5. doi: 10.1038/ng.2653
228. Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature (2015) 518:317–29. doi: 10.1038/nature14248
229. Megquier K, Genereux DP, Hekman J, Swofford R, Turner-Maier J, Johnson J, et al. Barkbase: Epigenomic annotation of canine genomes. Genes (Basel) (2019) 10:1–28. doi: 10.3390/genes10060433
230. Yang X, Wong MPM, Ng RK. Aberrant DNA methylation in acute myeloid leukemia and its clinical implications. Int J Mol Sci (2019) 20:1–20. doi: 10.3390/ijms20184576
231. Lee KH, Shin TJ, Kim WH, Lee SY, Cho JY. Methylation of LINE-1 in cell-free DNA serves as a liquid biopsy biomarker for human breast cancers and dog mammary tumors. Sci Rep (2019) 9:1–10. doi: 10.1038/s41598-018-36470-5
232. Thayanithy V, Park CW, Sarver AL, Kartha RV, Korpela DM, Graef AJ, et al. Combinatorial Treatment of DNA and Chromatin-Modifying Drugs Cause Cell Death in Human and Canine Osteosarcoma Cell Lines. PloS One (2012) 7:1–10. doi: 10.1371/journal.pone.0043720
233. Yu RMC, Cheah YK. The roles of miRNAs in human breast cancer and canine mammary tumor. Appl Cancer Res (2017) 37:1–11. doi: 10.1186/s41241-017-0043-7
234. Noguchi S, Mori T, Hoshino Y, Yamada N, Maruo K, Akao Y. MicroRNAs as tumour suppressors in canine and human melanoma cells and as a prognostic factor in canine melanomas. Vet Comp Oncol (2013) 11:113–23. doi: 10.1111/j.1476-5829.2011.00306.x
235. Noguchi S, Mori T, Hoshino Y, Yamada N, Nakagawa T, Sasaki N, et al. Comparative study of anti-oncogenic MicroRNA-145 in canine and human malignant melanoma. J Vet Med Sci (2012) 74:1–8. doi: 10.1292/jvms.11-0264
236. Bukowski JA, Wartenberg D, Goldschmidt M. Environmental causes for sinonasal cancers in pet dogs, and their usefulness as sentinels of indoor cancer risk. J Toxicol Environ Heal Part A (1998) 54:579–91. doi: 10.1080/009841098158719
237. Gavazza A, Presciuttini S, Barale R, Lubas G, Gugliucci B. Association between canine malignant lymphoma, living in industrial areas, and use of chemicals by dog owners. J Vet Intern Med (2001) 15:190–5. doi: 10.1111/j.1939-1676.2001.tb02310.x
238. Hayes HM, Hoover R, Tarone RE. Bladder cancer in pet dogs: A sentinel for environmental cancer? Am J Epidemiol (1981) 114:229–33. doi: 10.1093/oxfordjournals.aje.a113186
239. Kelsey JL, Moore AS, Glickman LT. Epidemiologic studies of risk factors for cancer in pet dogs. Epidemiol Rev (1998) 20:204–17. doi: 10.1093/oxfordjournals.epirev.a017981
240. Zanini DA, Kimura KC, Nishiya AT, Ubukata R, Leandro RM, de Brito CP, et al. Environmental risk factors related to the development of canine non-Hodgkin’s lymphoma. Ciec Rural (2013) 43:1302–8. doi: 10.1590/s0103-84782013005000089
Keywords: comparative oncology, epigenetics, DNA methylation, histone modifications, canine cancer, non-coding RNAs
Citation: Xavier PLP, Müller S and Fukumasu H (2020) Epigenetic Mechanisms in Canine Cancer. Front. Oncol. 10:591843. doi: 10.3389/fonc.2020.591843
Received: 05 August 2020; Accepted: 05 October 2020;
Published: 23 October 2020.
Edited by:
Attila A. Seyhan, Brown University, United StatesReviewed by:
Reo Maruyama, Japanese Foundation For Cancer Research, JapanHong Zan, The University of Texas Health Science Center at San Antonio, United States
Copyright © 2020 Xavier, Müller and Fukumasu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heidge Fukumasu, ZnVrdW1hc3VAdXNwLmJy
 Pedro Luiz Porfirio Xavier
Pedro Luiz Porfirio Xavier Susanne Müller
Susanne Müller Heidge Fukumasu
Heidge Fukumasu