
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 20 November 2020
Sec. Molecular and Cellular Oncology
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.589849
This article is part of the Research Topic Tumor Microenvironment: Molecular Mechanisms and Signaling Pathways Involved in Metastatic Progression View all 43 articles
 Giuseppe Broggi1*†
Giuseppe Broggi1*† Antonio Ieni2†
Antonio Ieni2† Daniela Russo3
Daniela Russo3 Silvia Varricchio3
Silvia Varricchio3 Lidia Puzzo1
Lidia Puzzo1 Andrea Russo4
Andrea Russo4 Michele Reibaldi4,5
Michele Reibaldi4,5 Antonio Longo4
Antonio Longo4 Giovanni Tuccari2
Giovanni Tuccari2 Stefania Staibano3‡
Stefania Staibano3‡ Rosario Caltabiano1‡
Rosario Caltabiano1‡Uveal melanoma, in spite of its rarity, represents the most common primitive intraocular malignant neoplasm of the adults; it affects choroid, ciliary bodied and iris and remains clinically silent for a long time, being accidentally discovered by routine ophthalmic exams. Prognosis of uveal melanoma is poor and frequently characterized by liver metastases, within 10–15 years from diagnosis. Autophagy is a multi-step catabolic process by which cells remove damaged organelles and proteins and recycle nutrients. It has been hypothesized that in early stages of tumorigenesis autophagy has a tumor suppressor role while, in more advanced stages, it may represent a survival mechanism of neoplastic cells in response to stress. Several proteins related to autophagy cascade have been investigated in numerous subtypes of human cancer, with overall controversal results. In this paper we studied the immunohistochemical expression of 3 autophagy related proteins (Beclin-1, p62 and ATG7) in a cohort of 85 primary uveal melanoma treated by primary enucleation (39 with metastasis and 46 non metastatic) and correlated their expression with clinico-pathological parameters and blood vascular microvessel density, in order to investigate the potential prognostic role of autophagy in this rare neoplasm. We found that high immunohistochemical levels of Beclin-1 correlated with a lower risk of metastasis and higher disease-free survival times, indicating a positive prognostic role for Beclin-1 in uveal melanoma. No statistically significative differences regarding the expression of ATG7 and p62 between metastatic and non metastatic patients was detected.
Although uveal melanoma (UM) is traditionally considered a rare tumor, it is the most frequent primitive intraocular malignancy of the adults and the second most frequent melanoma not associated with epithelial structures (1–3). UM is extremely rare in children in whom it recognizes a congenital origin and is often diagnosed in advanced stages of disease with extraocular spreading (4). Choroid is the most common intraocular site involved, followed by ciliary bodies and, less frequently, iris (3). The occurrence of uveal melanoma is 200 times higher in Caucasians, Fitzpatrick phototypes I-II (scores 1 to 13), than in the black population (5, 6). Nevertheless, the ultimate role of UV-B and UV-A exposure as a risk factor for uveal melanoma is still to be fully clarified. Recent reports evidenced that the use of sun-tanning devices is a well-recognized risk factor for uveal melanoma, as opposed to sunlight exposure, hypothesizing that this phenomenon could be correlated to the inability of artificial tanning to increase systemic level of vitamin D3 (7). Presence of choroidal nevus, syndromic/congenital diseases such as cutaneous dysplastic nevus syndrome, oculo-dermal melanocytosis (Ota nevus) and type 1 neurofibromatosis, have traditionally been included among risk factors associated with the onset of UM (8). Sudden retinal detachment is the most frequent presentation of disease; however, this tumor is often clinically silent for a long time, and it is frequently accidentally discovered by routine ophthalmic exams (9, 10). Although several steps have been made in the knowledge and treatment of this neoplasm, the prognosis of UM remains poor with about half of patient developing distant metastases, especially in the liver, within 10-15 years from primary enucleation (10).
To date, the peculiar interplay between the molecular routes that underlie the clinical behavior of UM has still to be fully clarified (11). Deregulation of cell death programs is a hallmark of cancer development, progression, and resistance to therapies (12, 13). A negative regulation of pro-apoptotic molecules by oncogenic signaling (14) and the involvement of several anti-apoptotic proteins involved in the regulation of extrinsic and intrinsic apoptotic pathways have been largely reported in melanoma (15, 16). Recently, also non-apoptotic, autophagy-dependent cell death pathways necroptosis, ferroptosis, pyroptosis, and parthanatos have been found involved in skin melanoma cell response to therapy. Melanoma cells use autophagy to counteract drug activity and tumor microenvironment drug-induced changes, conditioning clinical outcome and resistance to target therapy and immunotherapy (17, 18).
Autophagy is a catabolic process through which damaged organelles and proteins are removed and nutrients recycled by cells (19, 20). During the initiation and development of tumors, autophagy is likely to mainly play a tumor suppressor role while, in well-established cancers, it may constitute a survival mechanism in response to stress (21). Up to now, its role in cancer remains controversial, particularly for its relationship with prognosis. Three main dynamic autophagy pathways exist: chaperone-mediated autophagy (CMA), microautophagy, and macroautophagy (22). CMA is a selective multistep autophagy pathway that modulates the turnover of soluble cytosolic proteins, delivering them directly to lysosomes for degradation, without formation of vesicles (23). Microautophagy leads to the degradation of intracellular proteins and organelles directly engulfed by lysosomes or endosomes. The extent and role of microautophagy in mammalian cells have been yet poorly understood (24). Macroautophagy is a multistep process characterized by the formation of autophagosomes (double-membraned coated vesicles), that fuse with the membrane of lysosome, that degrade and recycle their contents (25). The macroautophagic process is regulated by six autophagy-related (ATG) protein classes and/or complexes (26). Autophagy cannot be detected in routine hematoxylin and eosin-stained and formalin-fixed and paraffin-embedded tissues. Biomarkers detection by IHC constitutes at present the best way to examine this process on routine tissue sections (27). Many of more than 30 autophagy-related genes (ATG) encoded proteins in human cells are detectable by immunohistochemistry (28, 29).
In this study, we choose to immunohistochemically investigate the expression of three proteins involved in the macroautophagic step of the autophagy cascade, Beclin-1, the autophagy-related gene 7 (ATG7), and p62, in a cohort of 62 patients affected by UM at different clinical stage.
BECN1 (Beclin-1) is the mammalian ortholog of yeast autophagy-related gene 6 (ATG6), which induces autophagosome formation. The human Beclin-1 gene is located on chromosome 17q21, and in the early stages of autophagy interacts with various cofactors forming a complex necessary for the recruitment of other ATG proteins and for the progression of the autophagic process (29–32). The evidence that Beclin-1 expression is monoallelicaly suppressed in some human breast, ovarian and prostatic cancer cell lines, suggests that it acts as a tumor suppressor gene (30–32). A decreased expression of Beclin-1 at mRNA and protein levels has been found in human brain tissues (33) and in estrogen receptor-negative, HER2-positive breast cancer, in which the reduced BECN1mRNA expression has been found associated with poor prognosis (34). In addition, upstream positive regulators of Beclin-1, such as UV radiation and Bax interacting factor-1 (Bif-1), have been found downregulated in several types of human cancers (35, 36) and an increased tumor incidence has been reported in Beclin1 heterozygous mice.
All these evidences support the tumor-suppressive function of macroautophagy (37). By converse, macroautophagy can be induced by cancer therapy, and in this eveniences, it has been found to support tumor cell survival, suggesting the gain of oncogenic function (38, 39). The role of macroautophagy in tumorigenesis, then, looks very complex and controversial, with sensible variation among different tumor types and/or different tumor stage (40). ATG7, located on chromosome 3p25.3, encodes an E1-like activating enzyme essential for cytoplasmic to vacuole transport (41). The encoded protein also has modulating function on p53-dependent cell cycle pathways during sustained metabolic stress phases (41). The expression of ATG7 has been recently found to correlate with resistance to anticancer drugs (42). p62 acts as an adaptor protein, through the binding of ubiquitylated protein aggregates and the deliver to the autophagosomes (32, 43, 44). A p62-dependent response triggered to counteract oxidative stress has been reported in melanoma cells exposed to ultraviolet A (UVA) (45).
The aim of this paper is to investigate any relationship among the expression of these three macroautophagy-related proteins and clinical-pathological parameters and behavior of UM.
Clinical data and histological samples of 85 patients affected by primary UM, who underwent primary enucleation at the Eye Clinic of both University of Catania and University of Naples “Federico II” during the ten years up to October 2019, were retrospectively analyzed. All cases were not eligible for non-surgical procedures, such as plaque brachytherapy or proton beam radiotherapy. The research protocol for this retrospective study was approved by our Institutional Medical Ethical Committee (according to the ethical guidelines of the Declaration of Helsinki). Paraffin-embedded tissue samples from each case were selected from the surgical pathology archive of the Sections of Anatomic Pathology, Department G.F. Ingrassia, University of Catania, and Department of Advanced Biomedical Sciences, University of Naples “Federico II”. The following exclusion criteria were used for the selection of cases: i) paraffin blocks in which there was not enough tissue to get additional slides for immunohistochemistry; ii) representative neoplastic tissue was absent; iii) necrosis was the major component of the tumor blocks; iv) the tumor underwent preoperative radiotherapy. Five or more sections were obtained from each paraffin block. Five pathologists (GB, LP, DR, SS and RC), not aware of the clinical and prognostic data of the corresponding patients, evaluated all histological specimens separately. Discrepancies in the evaluation were resolved by consensus.
The cohort of cases included 39 UMs with metastasis and 46 non metastatic UMs. We collected the following clinical data: tumor size and location, determined with standard methods, such as ophthalmoscopy and A and B scan ultrasonography, and onset of metastases, evaluated with physical examination, liver ultrasound and total body computed tomography (CT). The A-scan ultrasound is a mono-dimensional amplitude modulation scan, mainly used in ophthalmology to study routine visual disorders and/or to investigate the size and ultrasound features of intraocular masses; conversely, B-scan ultrasound refers to a two-dimensional, cross-section brightness scan, that, if combined to A-scan imaging, allows to get more details about the tumor, including exact anatomic site, borders, shape, and size, and to obtain a more detailed preoperative diagnosis.
MVD was evaluated by five pathologists (GB, LP, DR, SS and RC), who identified vascular hotspots on immunohistochemical sections of the UM cohort stained for anti-CD31 antibody by a light microscope at 4× and 10× magnifications, as previously described (46). MVD consisted of the amount of vessels per mm2 (conversion factor: 1 mm2 = 4 high power fields (HPFs) at 40× magnification inside vascular hotspots. Areas with ≥50 of viable tumor cells were considered usable for counting; Presence of diffuse necrotic, hemorrhagic and totally pigmented areas were considered as exclusion factors. Each CD31+ endothelial cell and/or lumen for long branched vessels were counted. Positive staining of small clusters of ≥2 endothelial cells was assessed as one single vascular structure. MVD levels were considered high if > the median value, low if < the median value.
Sections were treated for immunohistochemical analyses with standard methods (47), using the streptavidin/biotin-based system for immunoperoxidase; briefly, after appropriate deparaffinization and pre-treatments, sections were incubated for 30 min at 37°C with primary polyclonal rabbit anti-human antisera against Beclin-1 (working dilution 1:250; Abcam, Cambridge, MA, USA), p62 (working dilution 1:200; Abcam, Cambridge, MA, USA), ATG7 (working dilution 1:100; Abcam, San Francisco, CA, USA) and with primary monoclonal mouse anti-human antiserum against CD31 (JC70A; working dilution 1:40; DAKO, Glostrup, Denmark). The secondary biotinylated antibody was applied for 30 min at room temperature, followed by the streptavidin–biotin–peroxidase complex for a further 30 min at room temperature. The immunoreaction was visualized by incubating the sections for 4 min in a 0.1% 3,3’-diaminobenzidine (DAB) and 0.02% hydrogen peroxide solution (DAB substrate kit, Vector Laboratories, CA, USA).
p62 and Beclin-1 were immunoexpressed both in the cytoplasm and in the nucleus of neoplastic cells, while ATG-7 only showed cytoplasmic staining. Negative controls were additionally used by omitting the primary antibody. Intensity of staining (IS) was scored on a scale of 0–3 (47), as follows: absence of staining = 0, weak staining = 1, moderate staining = 2, strong staining = 3. The percentage of immunopositive cells (Extent Score, ES) was graded by five groups (47): <5% (0); 5–30% (+); 31–50% (++); 51–75% (+++), and >75% (++++). Counting was performed at 200× magnification. The intensity reactivity score (IRS) was obtained by multiplying IS and ES (47): IRS < 6 was interpreted as low expression (L-IRS), IRS > 6 as high expression (H-IRS).
The rates of high and low levels of beclin-1, p62, ATG7 expression in melanoma of patients with and without metastasis were non parametrically compared by chi-square test. Agreement among observers was tested by Cohen K.
Univariate and multivariate analyses, based on a Cox proportional hazards regression model (time free from metastasis as outcome), were performed; gender, age, melanoma location (choroid or ciliary body), temporal or nasal location, cells type (epithelioid, spindle cells or mixed), echographic parameters (height, greatest diameter), and expression level (low and high) of beclin-1, p62, ATG7 and MVD were all included in this model. If a predictor had a P value <0.15 (cut off) in the univariate analysis, it was included in the multivariate one. Survival analysis according to beclin-1, p62, ATG7 and MVD expression levels (high and low) was performed by Kaplan-Meyer test; survival rates were compared by log-rank (Mantel-Cox) test. P values < 0.05 were considered as statistically significant.
Eighty-five patients were part of the study (44 males; 41 females); median age was 67 years (range 29–85). Choroid was the only affected site in 64 cases, while choroid and ciliary body were simultaneously involved in 21 cases. Extrascleral extension was identified in 3 cases. Histologically, 20 cases were diagnosed as epithelioid cell, 25 as spindle cell, while 40 cases as mixed epithelioid and spindle cell UMs. According to the “TNM classification of malignant tumors”, pathological T stage was: pT1a in 15 patients, pT1b in 4 patients, pT2a in 45 patients, pT2b in 16 patients, pT2d in 1 patient, pT3a in 21 patients, pT3b in 10 patients, pT3d in 1 patient, pT4a in 8 patients, pT4b in 8 patients and pT4d in 1 patient. Liver metastases were present in 39 patients. Median follow-up period was 58 months (range 8–138).
Among the 46 non metastatic patients, 25 were males and 21 females; median age was 64 years (range 19–84). Out of 39 metastatic patients, 19 were males and 20 females; median age was 71 (range 50–85). 25/39 patients with metastases died during the follow-up time for disease progression (Figures 1, 2). No significant difference was seen in median age, location of the melanoma (choroid or choroid/ciliary body), tumor thickness, histological type, extrascleral invasion and pathological T stage between metastatic and non metastatic patients; UMs with greater median largest diameter (15.4 mm vs 12.4 mm, p = 0.009), lower median Beclin-1 expression (6 vs 8, p = 0.018), lower median p62 expression (6 vs 8, p = 0.025), lower median ATG7 expression (6 vs 9, p = 0.026) and higher median MVD levels (54 vs 26, p < 0.001) were observed in metastatic patients, who also presented lower median disease free survival (25 months vs 73 months, p < 0.001) (Figure 3).
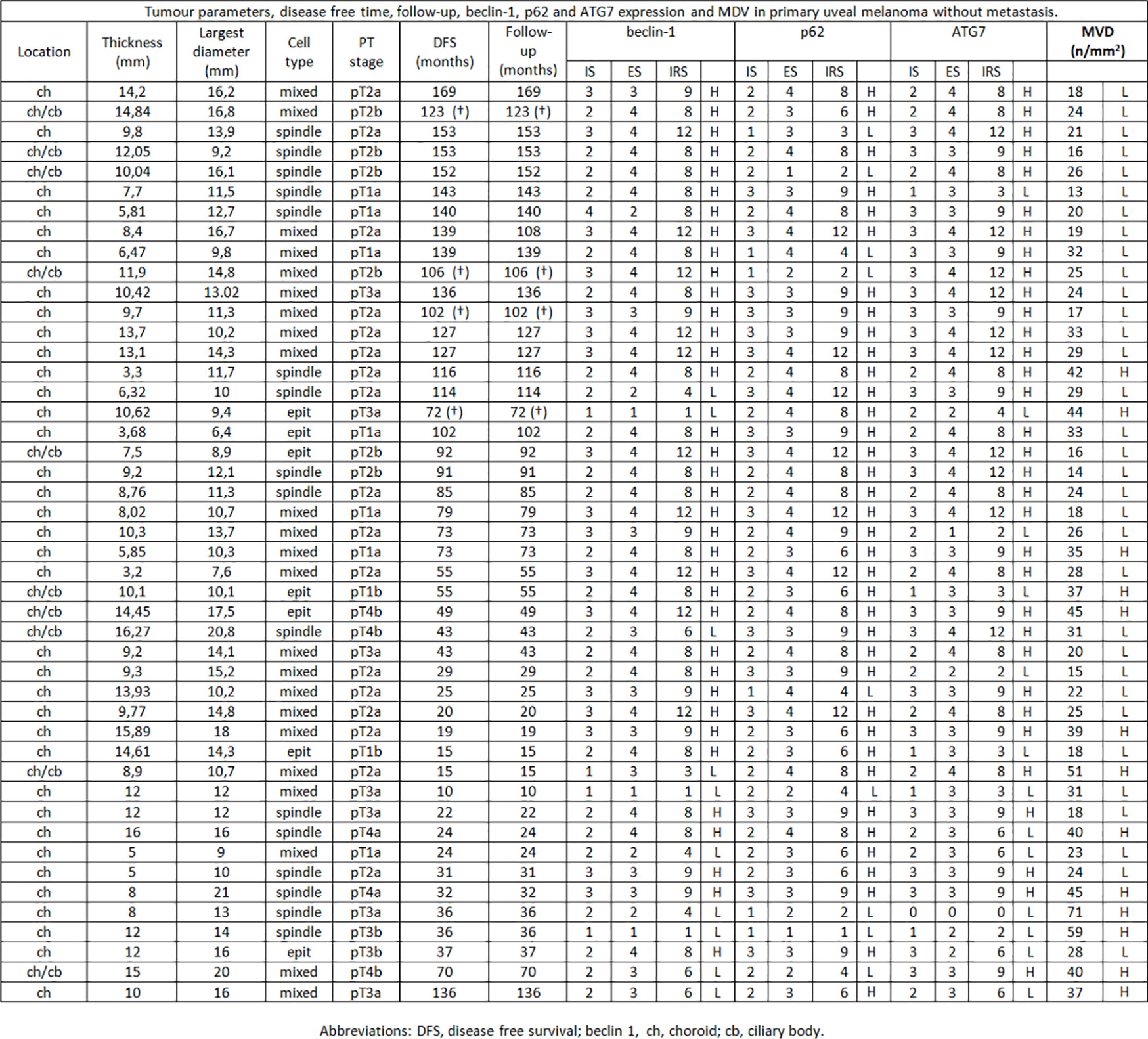
Figure 1 Tumor parameters, disease free time, follow-up, beclin-1, p62 and ATG7 expression and MVD in primary uveal melanoma without metastasis.
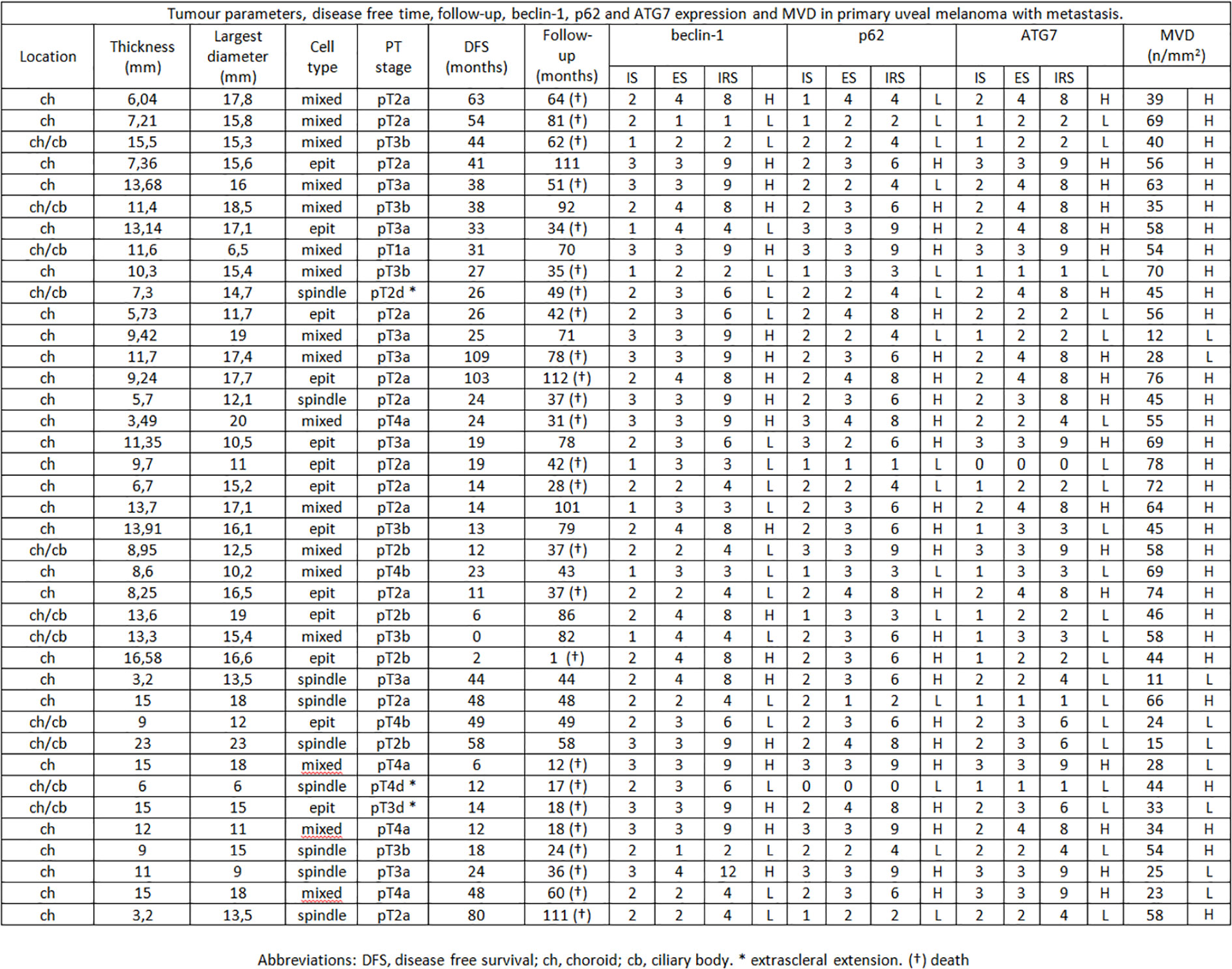
Figure 2 Tumor parameters, disease free time, follow-up, beclin-1, p62 and ATG7 expression and MVD in primary uveal melanoma with metastasis.
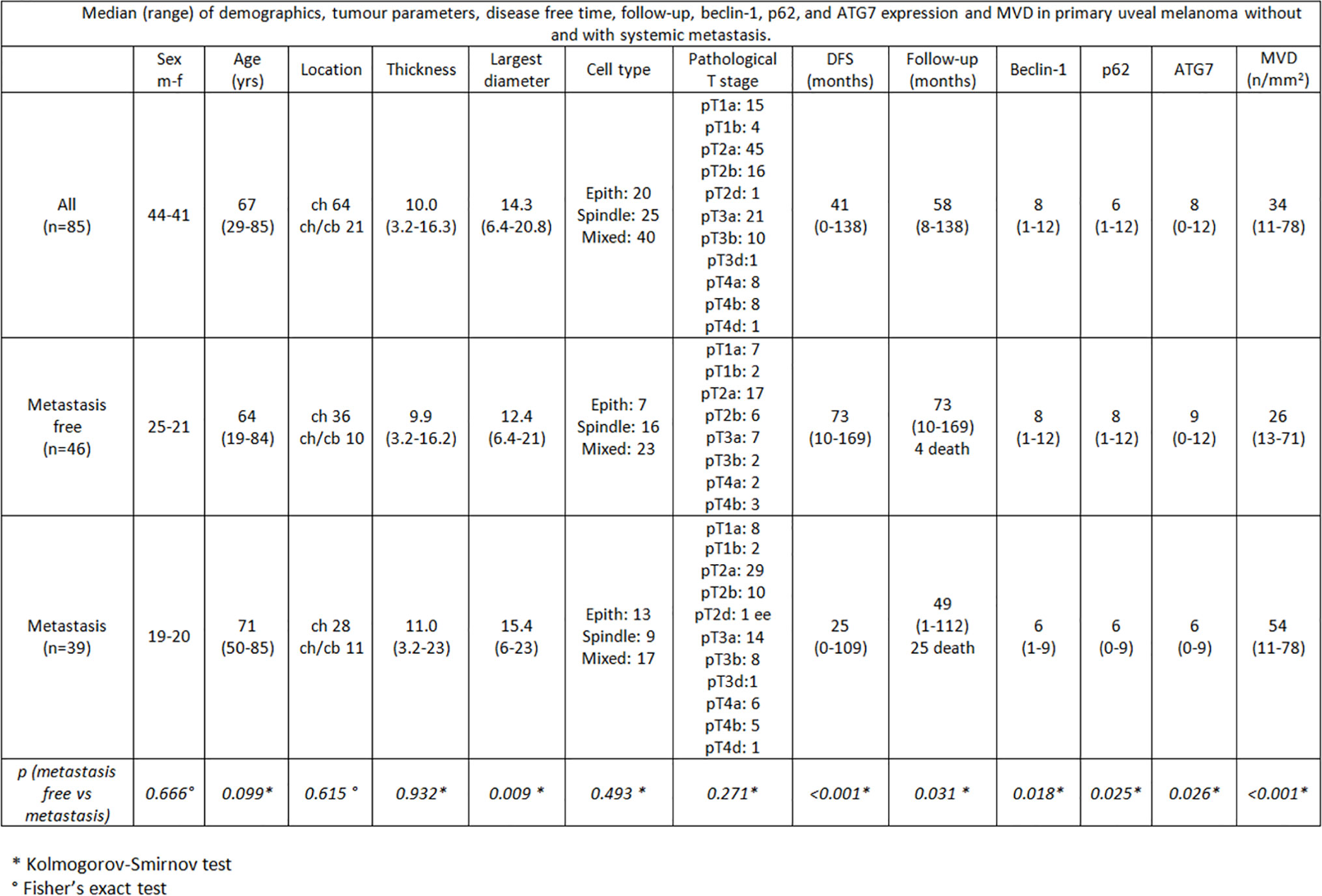
Figure 3 Median (range) of demographics, tumor parameters, disease free time, follow-up, beclin-1, p62, and ATG7 expression and MVD in primary uveal melanoma without and with systemic metastasis.
In the overall cohort of patients included in the study (n = 85) the median Beclin-1 value was 8. Beclin-1 expression was high in 55 Iand low in 20 UMs (Figures 4A, B). Among 46 primary non metastatic UMs, only 10/46 cases (21.7%) showed L-IRS, while the other 36 UMs showed H-IRS (78.3%) (Fisher’s exact test, p = 0.007, Figure 5). In 39 primary metastatic UMs 19/39 cases (48.7%) had H-IRS, while L-IRS was found in 20/39 UMs (51.3%) (Fisher’s exact test, p = 0.007, Figure 5).
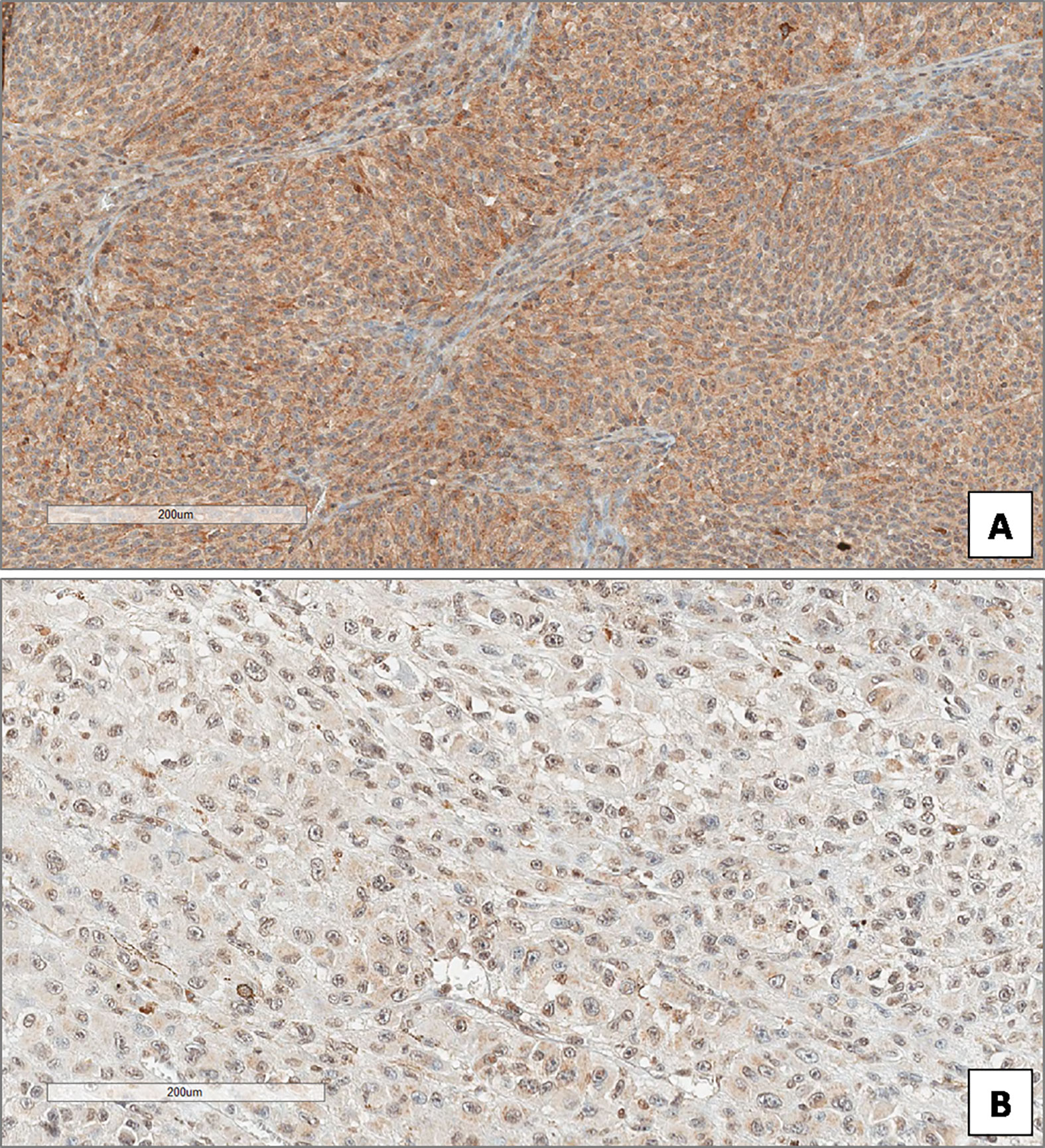
Figure 4 High (A) and low (B) expression of Beclin-1 in uveal melanoma. (Immunoperoxidase stain; original magnifications 100×).
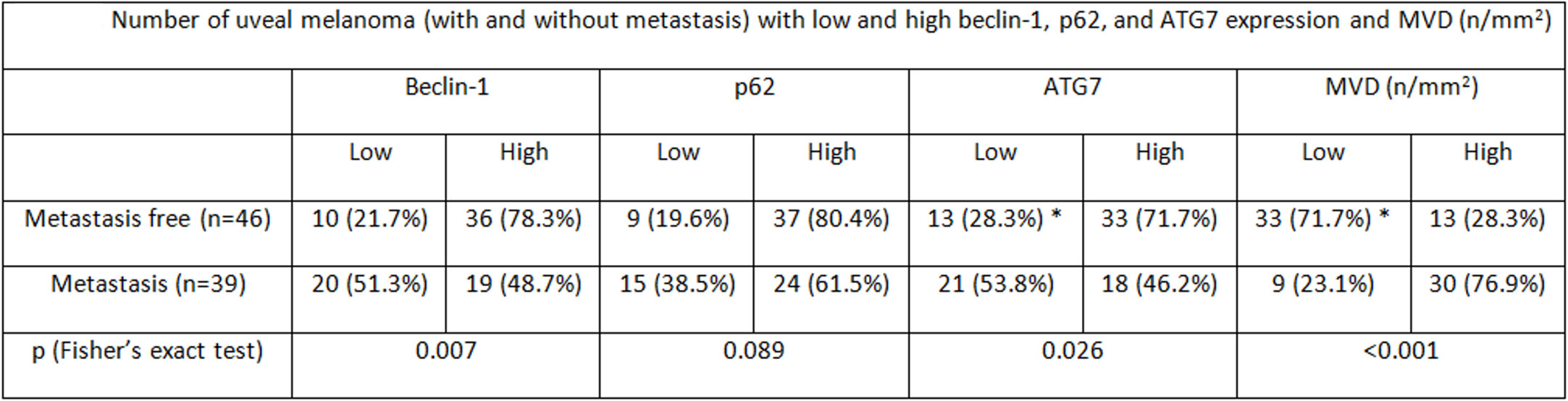
Figure 5 Number of uveal melanoma (with and without metastasis) with low and high beclin-1, p62, and ATG7 expression and MVD (n/mm2).
Among the 85 patients studied, the median p62 value was 6. p62 levels were high in 61 and low in 24 cases (Figures 6A, B). Among metastasis-free patients, only 9/46 cases (19.6%) showed L-IRS, while H-IRS was observed in the remaining 37/46 cases (80.4%) (Fisher’s exact test, p = 0.089, Figure 5). 15/39 metastatic patients (38.5%) showed L-IRS, while 24/39 (61.5%) had H-IRS (Fisher’s exact test, p = 0.089, Figure 5).
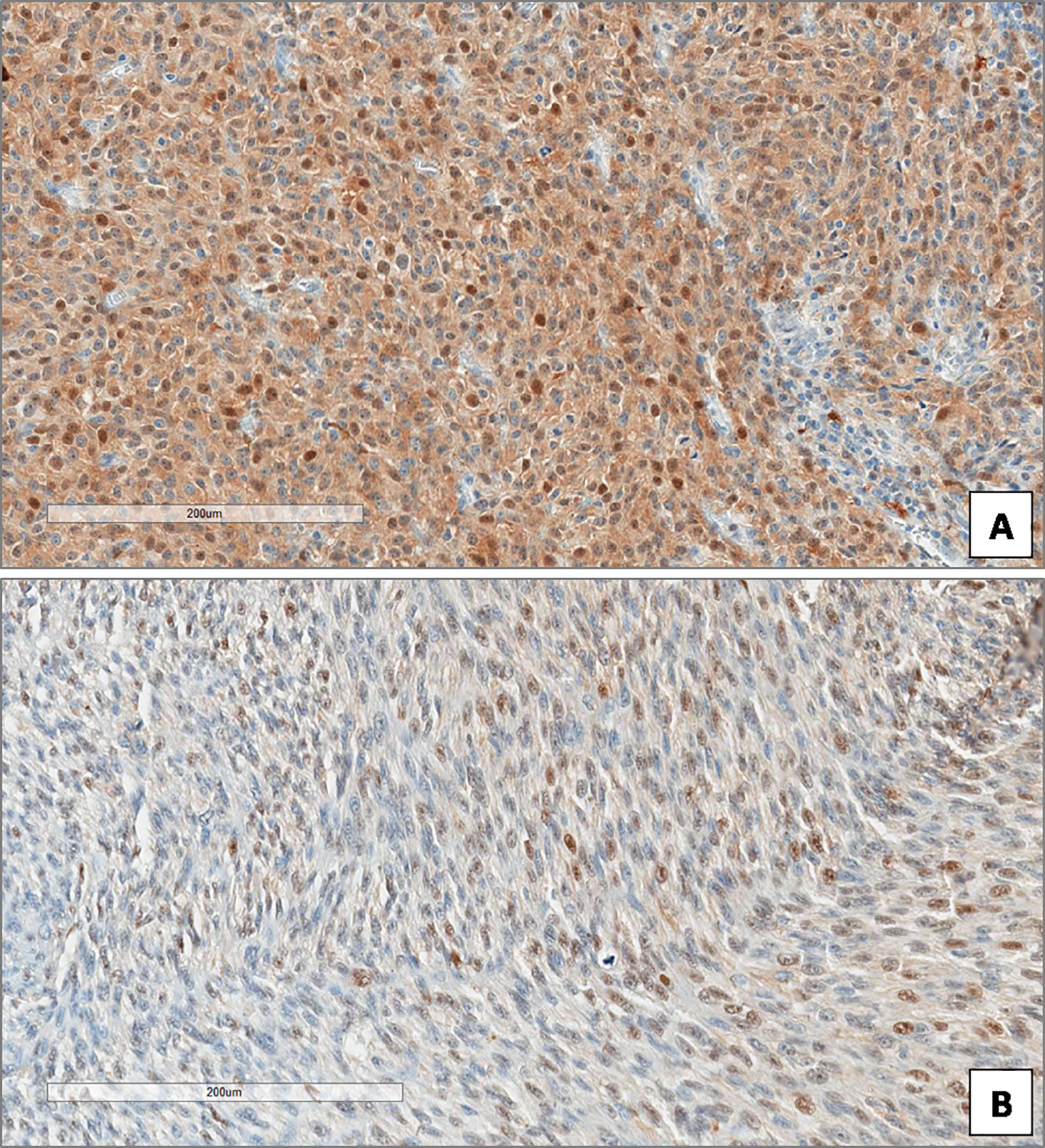
Figure 6 High (A) and low (B) expression of p62 in uveal melanoma. (Immunoperoxidase stain; original magnifications 100×).
In the whole group of UMs, the median ATG7 value was 8. ATG7 immunoexpression was high in 51 patients and low in 34 (Figures 7A, B). Out of the 46 patients without metastatic disease, ATG7 L-IRS was found only in 13/46 non metastatic UMs (28.3%), while 33/46 (71.7%) had ATG7 H-IRS (Fisher’s exact test, p = 0.026, Figure 5). 21/39 metastatic patients (53.8%) showed ATG7 L-IRS, while the remaining 18/39 (46.2%) ATG7 H-IRS (Fisher’s exact test, p = 0.026, Figure 5).
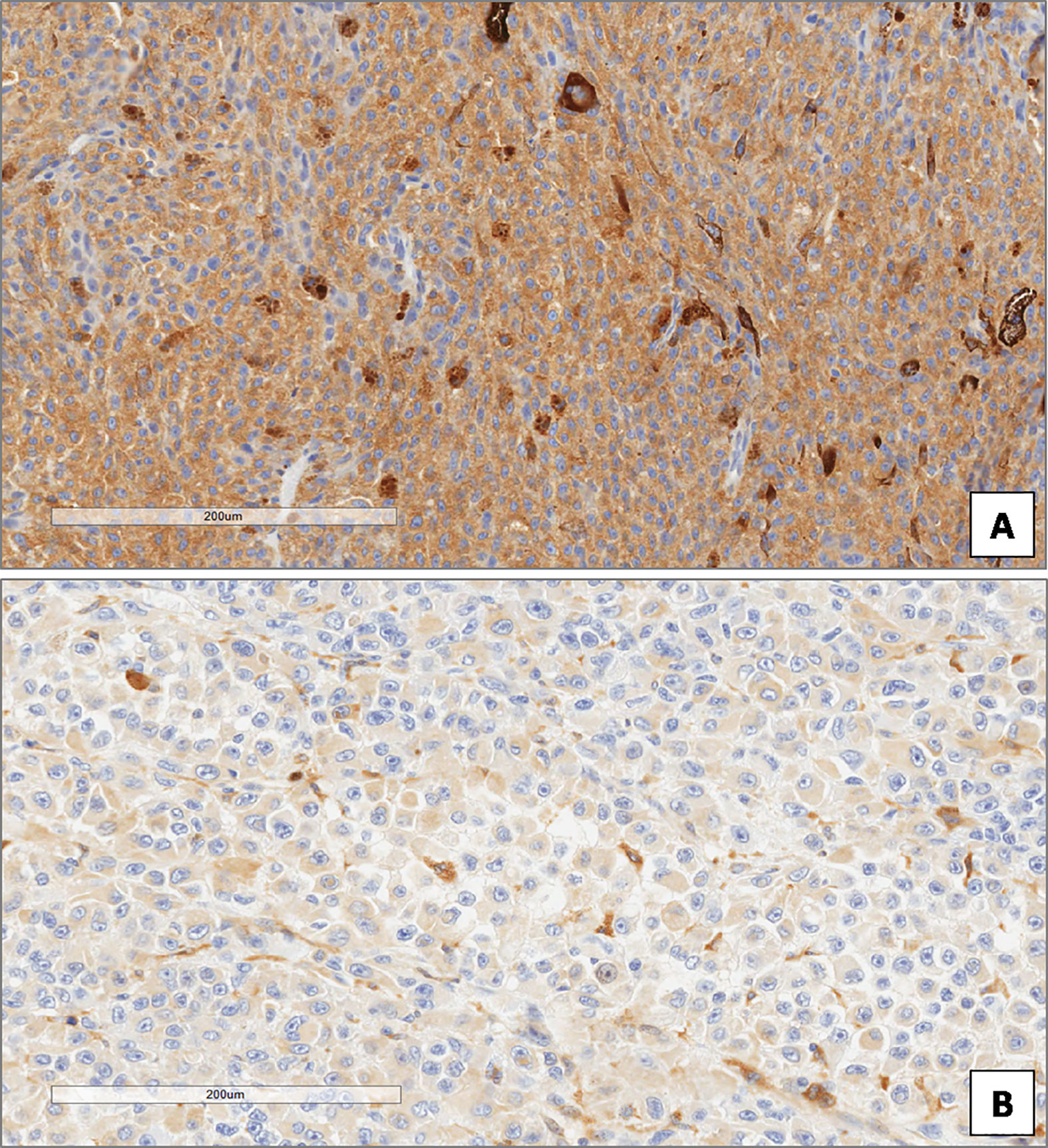
Figure 7 High (A) and low (B) expression of ATG7 in uveal melanoma. (Immunoperoxidase stain; original magnifications 100×).
Out of 85 patients studied, the median MVD value was 34 (range 11–78). MVD levels were high in 43 and low in 42 cases. Among metastasis-free patients, MVD was low in 33/46 (71.7%), while high in the remaining 13/46 cases (28.3%) (Fisher’s exact test, p < 0.001, Figure 5). 30/39 metastatic patients (76.9%) showed high MVD, while only 9/39 (23.1%) showed low MVD (Fisher’s exact test, p < 0.001, Figure 5).
Factors related to the presence of metastasis at univariate analysis on a Cox proportional hazards regression model were: age (p=0.011), diameter (p=0.044), epithelioid cell type (p=0.017), pT stage (p=0.023), and beclin-1 level (p=0.001), p62 level (p=0.079), ATG7 (p=0.002) and MVD (p<0.001).
At multivariate analysis MVD (p=0.009), epithelioid cell type (p=0.014), diameter (0.026) and beclin-1 level (p=0.035) were significant.
No correlation was found between histological type and beclin-1 expression (Spearman’s rho p=0.289), p62 expression (Spearman’s rho p=0.568), ATG7 (Spearman’s rho p=0.127), and MVD (Spearman’s rho p=0.087).
Figure 8 shows the results of Kaplan–Meier survival analyses in patients with uveal melanomas with low and high beclin-1 expression. The mean survival time free from metastasis (SE, with 95% CI) estimated were respectively: 50.7 (9.1) (CI: 32.9 to 68.5) and 113.9 (9.9) (CI: 94.5 to 133.4). The log-rank test showed a significant difference (p=0.001) between the two groups.
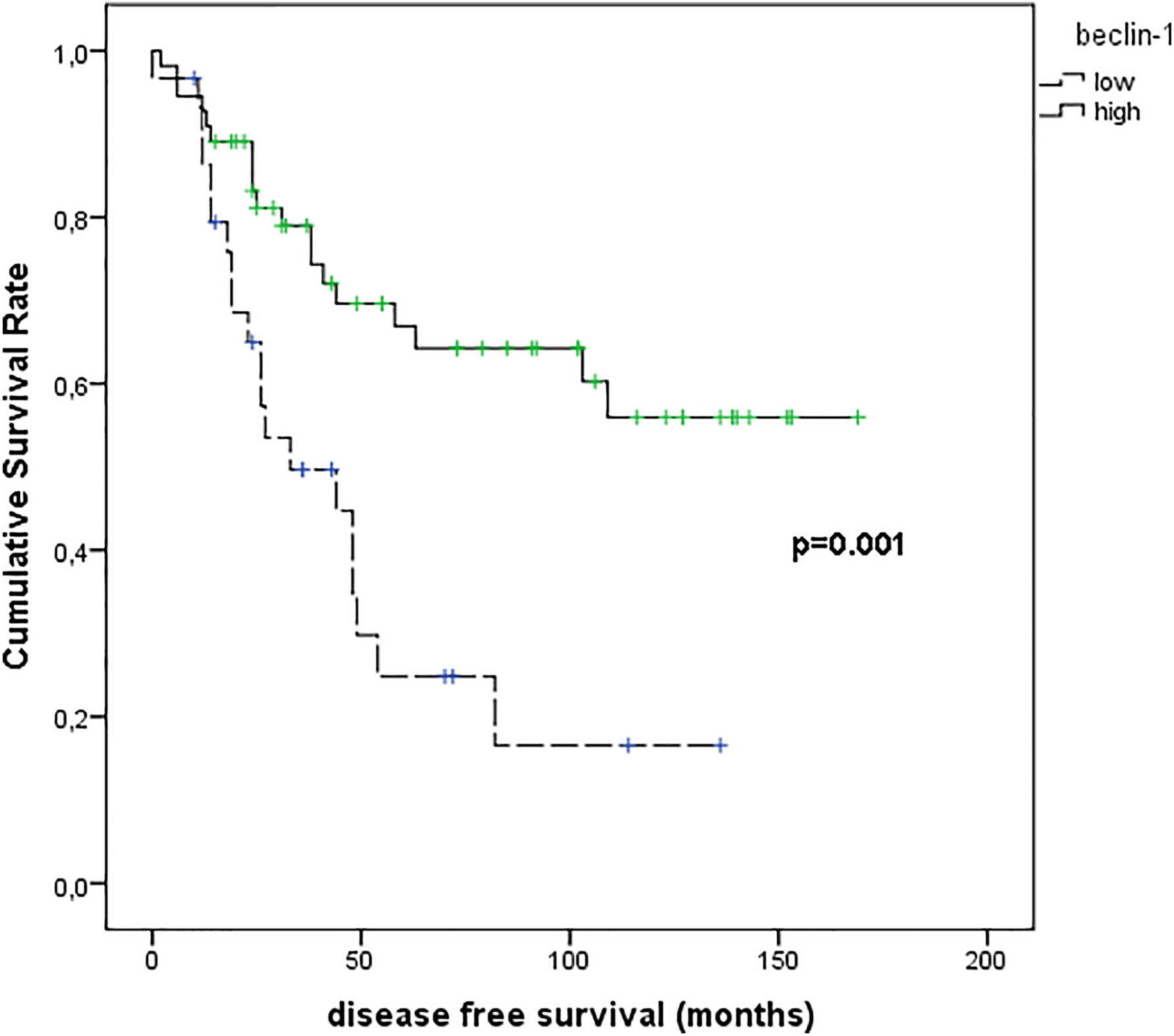
Figure 8 Kaplan–Meier survival analyses in patients with uveal melanomas with low and high Beclin-1 expression.
Figure 9 shows the results of Kaplan–Meier survival analyses in patients with uveal melanomas with low and high p62 expression. The mean survival time free from metastasis (SE, with 95% CI) estimated were respectively: 66.8 (12.3) (CI: 42.7 to 90.9) and 104.9 (9.8) (CI: 85.6 to 124.1). The log-rank test showed no significant difference (p=0.073) between the two groups.
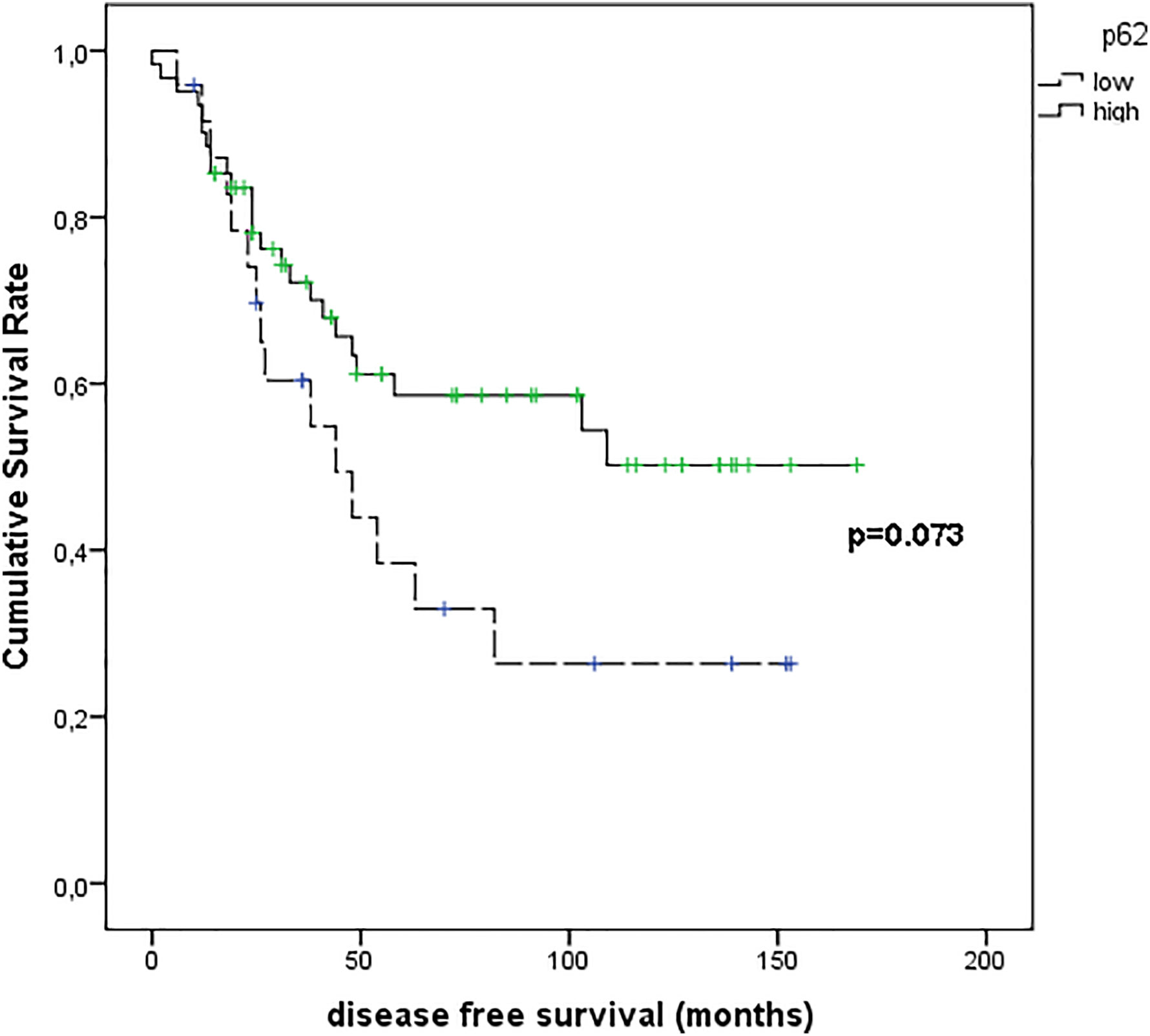
Figure 9 Kaplan–Meier survival analyses in patients with uveal melanomas with low and high p62 expression.
Figure 10 shows the results of Kaplan–Meier survival analyses in patients with uveal melanomas with low and high ATG7 expression. The mean survival time free from metastasis (SE, with 95% CI) estimated were respectively: 54.8 (9.7) (CI: 35.8 to 73.9) and 114.6 (10.1) (CI: 94.9 to 134.3). The log-rank test showed a significant difference (p=0.002) between the two groups.
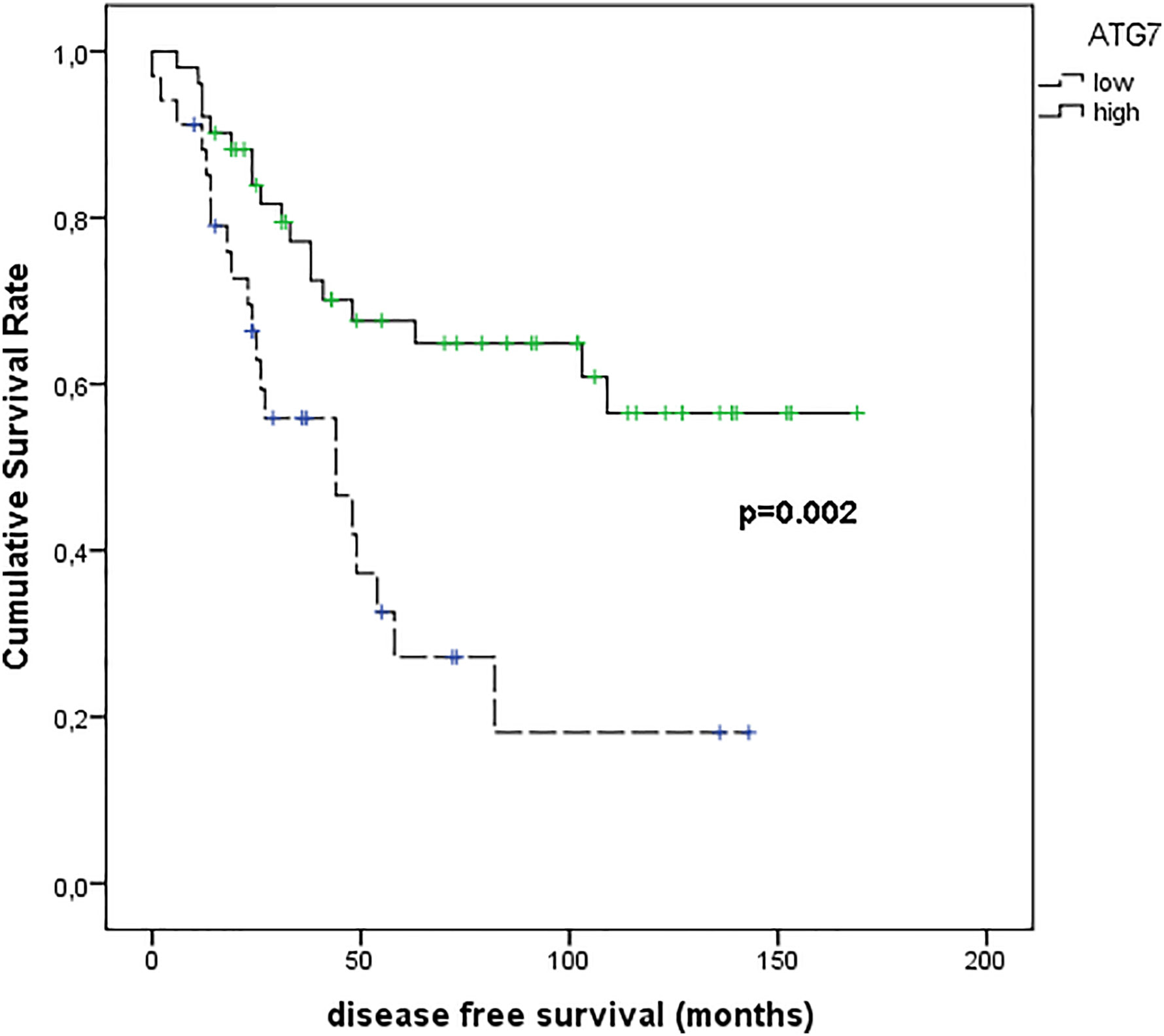
Figure 10 Kaplan–Meier survival analyses in patients with uveal melanomas with low and high ATG7 expression.
Figure 11 shows the results of Kaplan–Meier survival analyses in patients with uveal melanomas with low and high MVD expression. The mean survival time free from metastasis (SE, with 95% CI) estimated were respectively: 50.0 (7.4) (CI: 35.5 to 64.6) and 134.3 (10.0) (CI: 114.7 to 153.9). The log-rank test showed a significant difference (p<0.001) between the two groups.
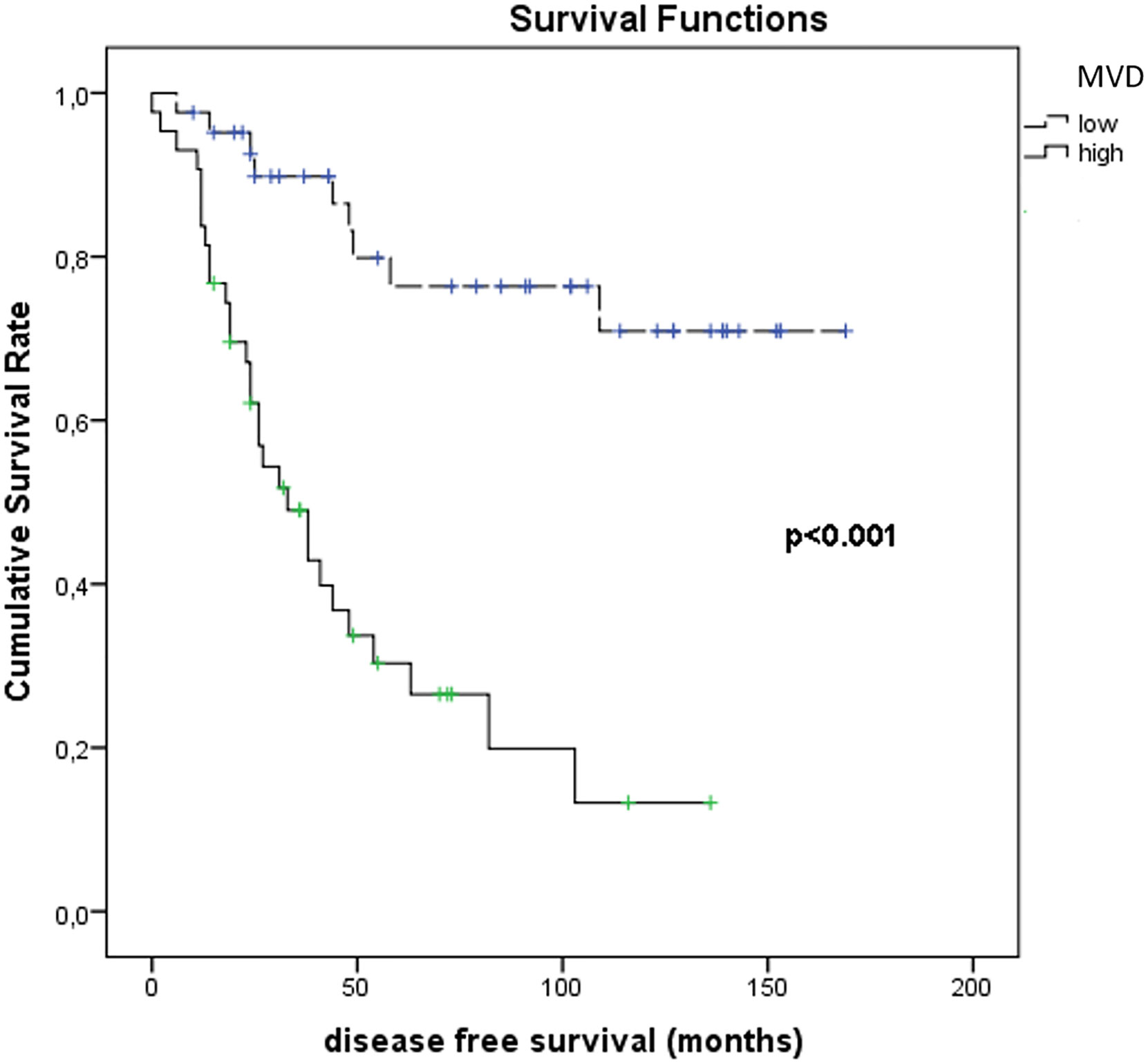
Figure 11 Kaplan–Meier survival analyses in patients with uveal melanomas with low and high MVD count.
The results of the correlation analysis between Beclin-1 IRS, MVD values and DFS are displayed in Figure 12.
Due to its apparent “indolence” and slow progression, UM may be considered as a tumor with ambiguous clinical course, marked by the occurrence of liver metastases in almost all cases even after 10–15 years after the first diagnosis. Several clinical and histopathological factors have been traditionally included among the ones negatively affecting the prognosis of UM: advanced age and stage at diagnosis, male gender, tumor thickness and largest diameter, ocular/cutaneous melanocytosis, localization to ciliary bodies, extrascleral invasion. By a morphological microscopic point of view, the prevalence of an epithelioid cytotype (48), high mitotic index and tumor associated angiogenesis (expressed as MVD), necrosis, are considered factors linked to a more aggressive tumor behaviour. As well, tumor infiltrating lymphocytes and/or histiocytes (49) and cytogenetic factors including monosomy 3, chromosome 8q-gain or 8p-loss, chromosome 1p-loss, chromosome 6q-loss, and high expression of insulin-like growth factor-1 receptor (IGF-1R) (50), also contribute to the definition of the class risk of each case of UM. In particular, the reduction/loss of the nuclear immunostaining for the BRCA1 associated protein-1 (BAP-1), reflecting the presence of inactivating mutations of the corresponding gene, represents a poor prognostic factor for of UM (51, 52).
We previously identified the immunohistochemical expression of proteins, such as RKIP, pRKIP, ADAM10, ABCB5, SPANX-C, and MacroH2A, as possible useful prognostic markers in UM (47, 53–56). However, despite the efforts of the scientific community and the increase of possible new biomarkers, the response to therapy of patients with advanced disease are yet dismal, so the identification of new prognostic and predictive tissue markers for UM is still an urgent need.
Autophagy looks to act as “double-edged sword” in cancers (57), either contributing to the induction of apoptosis in aberrantly proliferating or damaged cells in early tumor stages, or contributing to the survival of advanced cancer cells upon hostile events, as hypoxia and reduced availability of nutrients, or antineoplastic drugs.
Beclin-1, has a central role in the autophagic process as a major member of the macro-autophagic phase, and ranks above the most studied proteins. A great variability in the expression of this macroautophagy-related protein in human tumor tissues has been documented, sometimes with frankly divergent results. The loss/reduction of Beclin-1 expression has been shown to be closely related either to a better survival, as in endometrial, renal, gastric and colorectal cancers (29, 58–60), or to a reduced disease-free survival (DFS) and poorer prognosis in ovarian cancer patients receiving combined therapy with platinum and taxanes (61), and in a cohort of non-small cell lung cancer (NSCLC), in which the expression of the protein showed an independent positive prognostic factor (62). In squamous cell carcinomas of the uterine cervix the overexpression of Beclin-1 has been reported to inhibit metastatic progression in vitro (63), and in non-Hodgkin lymphomas (NHL), the increased expression of Beclin-1 has been found related to an increase in LC3-positive autophagic vacuoles and a better outcome of patients after chemotherapy (64). In melanocytic skin lesions, a gradual decrease of cytoplasmic expression of Beclin-1 has been found correlated with the progressive gain and increase of malignancy (65). The immunohistochemical expression of Beclin-1 has been detected, in fact, in about 100% of benign nevi and 86.4% of dysplastic nevi, decreasing to 54.3% in primary melanomas and up to 26.7% in melanoma metastases. Further studies showed instead that vemurafenib reduced miR-216b level resulting in upregulation of Beclin-1 (66), has been reported as a mechanism of drug-induced autophagy, allowing vemurafenib-resistant melanoma cells to employ autophagosomes to secret ATP to enhance cell migration and invasion (67).
The prognostic role of autophagy and Beclin-1 in UM has been investigated by Giatromanolaki et al. (68), who found either the overexpression and the reduced expression of BECN1 associated with poor prognosis.
Our results indicated a prognostic role of Beclin-1 in UM, with a lower risk of metastasis and higher disease-free survival times observed in UM cases with higher immunohistochemical expression of the protein. By converse, our population of UM with low expression of Beclin-1 was characterized by a higher metastatic risk. In our study, no statistically significative difference in the immunohistochemical expression of ATG7 and p62 proteins between metastasizing and non-metastasizing primary UM was detected. In our series of cases, the expression of these two macro-autophagy related proteins didn’t show any predictive value for metastatic risk of UM. Among the autophagy-related proteins analyzed in our study, then, only Beclin-1 resulted promising as a new possible immunohistochemical marker able to predict metastatic risk in patients with primary UM.
Some considerations have to be made, before any conclusion at this regard may be got. To date, there is still the lack of robust markers, improved staining protocols, and standardized interpretation to measure autophagy activity in archived tissue, and we must interpret our data with caution, strictly integrating them with clinical and follow-up data (22).
New specific markers identifying the core autophagy components and its upstream and downstream regulators, and the identification of specific gene “signatures” are needed for a more accurate assessment of the autophagic activity in FFPE tumor samples, at diagnostic and/or prognostics level, and to predict response to new autophagy-targeted treatment. Moreover, controversies remain regarding whether to inhibit or enhance autophagy in cancer, considering that, besides activation of endocytosis, the deregulation of autophagy-related genes has been associated also with cell death pathways and DNA repair responses (non-autophagy functions) (69, 70). Nevertheless, our finding of a strict correlation between Beclin-1 expression and the clinical outcome of UM support the idea that alteration in autophagy may be a particularly attracting targetable way to treat UM progression, as it has been proposed for skin melanoma. In cutaneous melanoma, even considering the large patient-to-patient variability of drug resistance mechanisms, non-apoptotic cell death has been considered as alternative therapeutic target when induction of apoptosis is impaired (71).
In addition, the finding of an altered expression of Beclin-1 in our study population sounds particularly attractive, if we consider that recent evidence suggests that alterations in autophagy may be a major mechanism of tumor escape from immune surveillance also by interfering with signaling pathways in tumor and immune cells (72) and autophagy-associated cell death has emerged as a key immunogenic mechanism able to potentiate tumor response to therapy in several human malignancies and in skin melanoma (29, 58–68). In particular, recent studies indicate that targeting autophagy in melanoma cells in combination with immunotherapy could gain results in promoting tumor regression, and autophagy has been shown to act a pivotal role in dendritic cell and T-lymphocyte infiltration (73, 74) in immune-competent animal models. These observations sound particularly interesting, that targeting autophagy could offer a new chance for UM patients non-responder to targeted and immunotherapeutic protocols active on most of human solid malignancies.
Further considerations emerge, in addition, from the analysis of several existing data indicating additional non-autophagic roles of beclin-1 expression in cancer. Besides inducing autophagy, this protein has been documented to be involved in parallel in growth signaling pathways AKT (protein kinase B) and ERK (kinase regulated by extracellular signals). It is therefore plausible that the increased expression of Beclin-1 inhibits these signaling pathways, limiting neoplastic growth (75).
Likewise, interesting consideration could derive from the analysis of the possible correlation between the expression of Beclin-1and uveal melanoma angiogenesis. As before outlined, increased angiogenesis, expressed as micro-vascular density (MVD) evaluated immunohistochemically, has been associated with poor prognosis (high metastasis and mortality rate) in uveal melanoma (76, 77). This finding it is not surprisingly, if we consider that UM metastasizes solely via the haematogenous route. In a previous study, we found that the chemokine receptor CXCR4, a prognostic factor in cutaneous melanoma being involved in angiogenesis and metastasis formation, is commonly expressed in uveal melanoma and correlates to the epithelioid-mixed cell type (78). In addition, Brouwer NJ and colleagues analyzed the correlation between several angiogenesis-related cytokines and the development of tumor vessels of primary UM, founding that a high MVD is associated with an increased expression of angiopoietin 2, Von Willebrand Factor, and a decreased expression of vascular endothelial growth factor B (VEGF-B) (79). It has been found that the expression of endothelial growth factors (VEGF, MMP-9) is negatively modulated by high levels of Beclin-1 (80).
Basing on this finding, the results concerning Beclin-1 expression in our study population lead us to hypothesize that a similar scenario could have important effects on tumor angiogenesis in UM. This could at least in part address the many still unanswered questions concerning the complex interplay between the regulation angiogenesis pathways and its correlation with UM biological behavior.
The mechanisms by which Beclin-1 negatively modulates tumor growth are not fully understood. A recent study outlined that Beclin-1 may interact also with members of the bcl-2 protein, acting as a tumor suppressor protein (81), and this finding could explain, at least partially, its positive prognostic role in different types of human cancers. The conflicting data in literature indicate in addition different patterns and roles of Beclin-1 expression depending on neoplastic cell types.
To date, many concerns need to be still overcome, to fully clarify the ultimate role of Beclin-1 expression in cancer progression and cancer response to therapy in UM, and a further study on larger, multi-institutional series of cases is currently in progress to definitively validate our results. Nevertheless, the results of our study indicate that a relationship between the immunohistochemical expression of Beclin-1 and the biological behavior of UM exists, and this could open up new prognostic and therapeutic strategies for this peculiar, deadly malignant tumor.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by Comitato Etico Catania 1, Azienda Ospedaliero Universitaria Policlinico “G.Rodolico - San Marco” via S.Sofia, 78 - 95123 Catania. The patients/participants provided their written informed consent to participate in this study.
GB, AI, and AR contributed to implementation of the study and to the writing of the manuscript. SS and RC, as co-senior authors, contributed to the design of the study. GT, DR, AR, AL, MR, and SV were involved in planning and supervised the study. GB, RC, AI, SS, DR, LP, and GT performed the histopathologic diagnoses and the complete analysis of the surgery material. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Spagnolo F, Caltabiano G, Queirolo P. Uveal melanoma. Cancer Treat Rev (2012) 38:549–53. doi: 10.1016/j.ctrv.2012.01.002
2. Jager MJ, Shields CL, Cebulla CM, Abdel-Rahman MH, Grossniklaus HE, Stern MH, et al. Uveal melanoma. Nat Rev Dis Primers (2020) 6:24. doi: 10.1038/s41572-020-0158-0
3. Mahendraraj K, Lau CS, Lee I, Chamberlain RS. Trends in incidence, survival, and management of uveal melanoma: A population-based study of 7516 patients from the surveillance, epidemiology, and end results database (1973–2012). Clin Ophthalmol (2016) 10:2113–9. doi: 10.2147/OPTH.S113623
4. Pukrushpan P, Tulvatana W, Pittayapongpat R. Congenital uveal malignant melanoma. J Am Assoc Pediatr Ophthalmol Strabismus (2014) 18:199–201. doi: 10.1016/j.jaapos.2013.11.020
5. Singh AD, Turell ME, Topham AK. Uveal melanoma: Trends in incidence, treatment, and survival. Ophthalmology (2011) 118:1881–5. doi: 10.1016/j.ophtha.2011.01.040
6. Margo CE, Mulla Z, Billiris K. Incidence of surgically treated uveal melanoma by race and ethnicity. Ophthalmology (1998) 105:1087–90. doi: 10.1016/S0161-6420(98)96012-2
7. Mallet JD, Gendron SP, Drigeard Desgarnier MC, Rochette PJ. Implication of ultraviolet light in the etiology of uveal melanoma: A review. Photochem Photobiol (2014) 90:15–21. doi: 10.1111/php.12161
8. Krantz BA, Dave N, Komatsubara KM, Marr BP, Carvajal RD. Uveal melanoma: Epidemiology, etiology, and treatment of primary disease. Clin Ophthalmol (2017) 11:279–89. doi: 10.2147/OPTH.S89591
9. Eskelin S, Kivelä T. Mode of presentation and time to treatment of uveal melanoma in Finland. Br J Ophthalmol (2002) 86:333–8. doi: 10.1136/bjo.86.3.333
10. Griewank KG, van de Nes J, Schilling B, Moll I, Sucker A, Kakavand H, et al. Genetic and clinico-pathologic analysis of metastatic uveal melanoma. Mod Pathol (2014) 27:175–83. doi: 10.1038/modpathol.2013.138
11. Hartman ML. Non-Apoptotic Cell Death Signaling Pathways in Melanoma. Int J Mol Sci (2020) 21:2980. doi: 10.3390/ijms21082980
13. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
14. Cartlidge RA, Thomas GR, Cagnol S, Jong KA, Molton SA, Finch AJ, et al. Oncogenic BRAF(V600E) inhibits BIM expression to promote melanoma cell survival. Pigment Cell Melanoma Res (2008) 21:534–44. doi: 10.1111/j.1755-148X.2008.00491.x
15. Hartman ML, Czyz M. Anti-apoptotic proteins on guard of melanoma cell survival. Cancer Lett (2013) 331:24–34. doi: 10.1016/j.canlet.2013.01.010
16. Mielczarek-Lewandowska A, Hartman ML, Czyz M. Inhibitors of HSP90 in melanoma. Apoptosis (2020) 25:12–28. doi: 10.1007/s10495-019-01577-1
17. Antunes F, Pereira GJS, Saito RF, Buri MV, Gagliardi M, Bincoletto C, et al. Effective synergy of sorafenib and nutrient shortage in inducing melanoma cell death through energy stress. Cells (2020) 9:640. doi: 10.3390/cells9030640
18. Liu X, Wu J, Qin H, Xu J. The role of autophagy in the resistance to BRAF inhibition in BRAF-mutated melanoma. Target Oncol (2018) 13:437–46. doi: 10.1007/s11523-018-0565-2
19. Ravanan P, Srikumar IF, Talwar P. Autophagy: The spotlight for cellular stress responses. Life Sci (2017) 188:53–67. doi: 10.1016/j.lfs.2017.08.029
20. Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol (2010) 221:3–12. doi: 10.1002/path.2697
21. Chen HY, White E. Role of Autophagy in Cancer Prevention. Cancer Prev Res (2011) 4:973–83. doi: 10.1158/1940-6207.CAPR-10-0387
22. Humbert M, Morán M, de la Cruz-Ojeda P, Muntané J, Wiedmer T, Apostolova N, et al. Assessing Autophagy in Archived Tissue or How to Capture Autophagic Flux from a Tissue Snapshot. Biology (2020) 9:59. doi: 10.3390/biology9030059
23. Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol (2018) 19:365–81. doi: 10.1038/s41580-018-0001-6
24. Olsvik HL, Svenning S, Abudu YP, Brech A, Stenmark H, Johansen T, et al. Endosomal microautophagy is an integrated part of the autophagic response to amino acid starvation. Autophagy (2019) 15:182–3. doi: 10.1080/15548627.2018.1532265
25. Tanida I. Autophagy basics. Microbiol Immunol (2011) 55:1–11. doi: 10.1111/j.1348-0421.2010.00271.x
26. Yu L, Chen Y, Tooze SA. Autophagy pathway: Cellular and molecular mechanisms. Autophagy (2018) 14:207–15. doi: 10.1080/15548627.2017.1378838
27. Langer R, Streutker CJ, Swanson PE. Autophagy and its current relevance to the diagnosis and clinical management of esophageal diseases. Ann N Y Acad Sci (2016) 1381:113–21. doi: 10.1111/nyas.13190
28. Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, et al. ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol (2019) 218:1787–98. doi: 10.1083/jcb.201811139
29. Ieni A, Cardia R, Giuffrè G, Rigoli L, Caruso RA, Tuccari G. Immunohistochemical Expression of Autophagy-Related Proteins in Advanced Tubular Gastric Adenocarcinomas and Its Implications. Cancers (2019) 11:389. doi: 10.3390/cancers11030389
30. Chen W, Li Z, Liu H, Jiang S, Wang G, Sun L, et al. MicroRNA-30a targets BECLIN-1 to inactivate autophagy and sensitizes gastrointestinal stromal tumor cells to imatinib. Cell Death Dis (2020) 11:198. doi: 10.1038/s41419-020-2390-7
31. Karantza-Wadsworth V, White E. Role of autophagy in breast cancer. Autophagy (2007) 3:610–3. doi: 10.4161/auto.4867
32. Masuda GO, Yashiro M, Kitayama K, Miki Y, Kasashima H, Kinoshita H, et al. Clinicopathological Correlations of Autophagy-related Proteins LC3, Beclin 1 and p62 in Gastric Cancer. Anticancer Res (2016) 36:129–36.
33. Miracco C, Cosci E, Oliveri G, Luzi P, Pacenti L, Monciatti I, et al. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int J Oncol (2007) 30:429–36. doi: 10.3892/ijo.30.2.429
34. Tang H, Sebti S, Titone R, Zhou Y, Isidoro C, Ross TS, et al. Decreased BECN1mRNAExpression in Human Breast Cancer is Associated with Estrogen Receptor-Negative Subtypes and Poor Prognosis. EBioMedicine (2015) 2:255–63. doi: 10.1016/j.ebiom.2015.01.008
35. Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol (2007) 9:1142–51. doi: 10.1038/ncb1634
36. Coppola D, Khalil F, Eschrich SA, Boulware D, Yeatman T, Wang HG. Down-regulation of Bax-interacting factor-1 in colorectal adenocarcinoma. Cancer (2008) 113:2665–70. doi: 10.1002/cncr.23892
37. Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest (2003) 112:1809–20. doi: 10.1172/JCI20039
38. Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, et al. Autophagy in malignant transformation and cancer progression. EMBO J (2015) 34:856–80. doi: 10.15252/embj.201490784
39. Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G. Pharmacological modulation of autophagy: Therapeutic potential and persisting obstacles. Nat Rev Drug Discovery (2017) 16:487–511. doi: 10.1038/nrd.2017.22
40. Hoare M, Young ARJ, Narita M. Autophagy in cancer: Having your cake and eating it. Semin Cancer Biol (2011) 21:397–404. doi: 10.1016/j.semcancer.2011.09.004
41. Yamaguchi M, Satoo K, Suzuki H, Fujioka Y, Ohsumi Y, Inagaki F, et al. Atg7 Activates an Autophagy-Essential Ubiquitin-like Protein Atg8 through Multi-Step Recognition. J Mol Biol (2018) 430:249–57. doi: 10.1016/j.jmb.2017.12.002
42. Kim MJ, Woo SJ, Yoon CH, Lee JS, An S, Choi YH, et al. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J Biol Chem (2011) 286:12924–32. doi: 10.1074/jbc.M110.138958
43. Jiang T, Harder B, Rojo de la Vega M, Wong PK, Chapman E, Zhang DD. p62 links autophagy and Nrf2 signaling. Free Radic Biol Med (2015) 88:199–204. doi: 10.1016/j.freeradbiomed.2015.06.014
44. Amaya C, Fader CM, Colombo MI. Autophagy and proteins involved in vesicular trafficking. FEBS Lett (2015) 589:3343–53. doi: 10.1016/j.febslet.2015.09.021
45. Sample A, Zhao B, Wu C, Qian S, Shi X, Aplin A, et al. The Autophagy Receptor Adaptor p62 is Up-regulated by UVA Radiation in Melanocytes and in Melanoma Cells. Photochem Photobiol (2018) 94:432–7. doi: 10.1111/php.12809
46. Barbagallo D, Caponnetto A, Brex D, Mirabella F, Barbagallo C, Lauretta G, et al. CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme Through the Binding of SRSF1. Cancers (2019) 11:194. doi: 10.3390/cancers11020194
47. Broggi G, Musumeci G, Puzzo L, Russo A, Reibaldi M, Ragusa M, et al. Immunohistochemical Expression of ABCB5 as a Potential Prognostic Factor in Uveal Melanoma. Appl Sci (2019) 9:1316. doi: 10.3390/app9071316
48. McLean IW, Foster WD, Zimmerman LE, Gamel JW. Modifications of Callender’s classification of uveal melanoma at the Armed Forces Institute of Pathology. Am J Ophthalmol (2018) 195:lvi–lx. doi: 10.1016/j.ajo.2018.08.025
49. Staibano S, Mascolo M, Tranfa F, Salvatore G, Mignogna C, Bufo P, et al. Tumor infiltrating lymphocytes in uveal melanoma: a link with clinical behavior? Int J Immunopathol Pharmacol (2006) 19:171–9. doi: 10.1177/205873920601900117
50. Jovanovic P, Mihajlovic M, Djordjevic-Jocic J, Vlajkovic S, Cekic S, Stefanovic V. Ocular melanoma: An overview of the current status. Int J Clin Exp Pathol (2013) 6:1230–44.
51. Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science (2010) 330:1410–3. doi: 10.1126/science.1194472
52. Szalai E, Wells JR, Ward L, Grossniklaus HE. Uveal melanoma nuclear BRCA1-associated protein-1 immunoreactivity is an indicator of metastasis. Ophthalmology (2018) 125:203–9. doi: 10.1016/j.ophtha.2017.07.018
53. Caltabiano R, Puzzo L, Barresi V, Cardile V, Loreto C, Ragusa M, et al. Expression of Raf Kinase Inhibitor Protein (RKIP) is a predictor of uveal melanoma metastasis. Histol Histopathol (2014) 29:1325–34. doi: 10.14670/HH-29.1325
54. Caltabiano R, Puzzo L, Barresi V, Ieni A, Loreto C, Musumeci G, et al. ADAM 10 expression in primary uveal melanoma as prognostic factor for risk of metastasis. Pathol Res Pract (2016) 212:980–7. doi: 10.1016/j.prp.2016.08.003
55. Salvatorelli L, Puzzo L, Russo A, Reibaldi M, Longo A, Ragusa M, et al. Immunoexpression of SPANX-C in metastatic uveal melanoma. Pathol Res Pract (2019) 29:152431. doi: 10.1016/j.prp.2019.04.023
56. Salvatorelli L, Puzzo L, Bartoloni G, Palmucci S, Longo A, Russo A, et al. Immunoexpression of Macroh2a in Uveal Melanoma. Appl Sci (2019) 9:3244. doi: 10.3390/app9163244
58. Giatromanolaki A, Koukourakis MI, Koutsopoulos A, Chloropoulou P, Liberis V, Sivridis E. High Beclin 1 expression defines a poor prognosis in endometrial adenocarcinomas. Gynecol Oncol (2011) 123:147–51. doi: 10.1016/j.ygyno.2011.06.023
59. Nishikawa M, Miyake H, Liu B, Fujisawa M. Expression pattern of autophagy-related markers in non-metastatic clear cell renal cell carcinoma: association with disease recurrence following radical nephrectomy. J Cancer Res Clin Oncol (2015) 141:1585–91. doi: 10.1007/s00432-015-1923-4
60. Han Y, Xue XF, Shen HG, Guo XB, Wang X, Yuan B, et al. Prognostic significance of Beclin-1 expression in colorectal cancer: a meta-analysis. Asian Pac J Cancer Prev (2014) 15:4583–7. doi: 10.7314/apjcp.2014.15.11.4583
61. Minamoto T, Nakayama K, Nakamura K, Katagiri H, Sultana R, Ishibashi T, et al. Loss of beclin 1 expression in ovarian cancer: A potential biomarker for predicting unfavorable outcomes. Oncol Lett (2018) 15:1170–6. doi: 10.3892/ol.2017.7379
62. Zhou W, Yue C, Deng J, Hu R, Xu J, Feng L, et al. Autophagic Protein Beclin 1 Serves as an Independent Positive Prognostic Biomarker for Non-Small Cell Lung Cancer. PloS One (2013) 8:e80338. doi: 10.1371/journal.pone.0080338
63. Sun Y, Liu JH, Sui YX, Jin L, Yang Y, Lin SM, et al. Beclin1 overexpression inhibits proliferation, invasion and migration of CaSki cervical cancer cells. Asian Pac J Cancer Prev (2011) 12:1269–73.
64. Nicotra G, Mercalli F, Peracchio C, Castino R, Follo C, Valente G, et al. Autophagy-active beclin-1 correlates with favourable clinical outcome in non-Hodgkin lymphomas. Mod Pathol (2010) 23:937–50. doi: 10.1038/modpathol.2010.80
65. Miracco C, Cevenini G, Franchi A, Luzi P, Cosci E, Mourmouras V, et al. Beclin 1 and LC3 autophagic gene expression in cutaneous melanocytic lesions. Hum Pathol (2010) 41:503–12. doi: 10.1016/j.humpath.2009.09.004
66. Luo M, Wu L, Zhang K, Wang H, Wu S, O’Connell D, et al. miR-216b enhances the e_cacy of vemurafenib by targeting Beclin-1, UVRAG and ATG5 in melanoma. Cell Signal (2018) 42:30–43. doi: 10.1016/j.cellsig.2017.09.024
67. Martin S, Dudek-Peric AM, Garg AD, Roose H, Demirsoy S, Van Eygen S, et al. An autophagy-driven pathway of ATP secretion supports the aggressive phenotype of BRAFV600E inhibitor-resistant metastatic melanoma cells. Autophagy (2017) 13:1512–27. doi: 10.1080/15548627.2017.1332550
68. Giatromanolaki AN, Charitoudis GS, Bechrakis NE, Kozobolis VP, Koukourakis MI, Foerster MH, et al. Autophagy patterns and prognosis in uveal melanomas. Mod Pathol (2011) 24:1036–45. doi: 10.1038/modpathol.2011.63
69. Galluzzi L, Green DR. Autophagy-Independent Functions of the Autophagy Machinery. Cell (2019) 177:1682–99. doi: 10.1016/j.cell.2019.05.026
70. Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell (2019) 176:11–42. doi: 10.1016/j.cell.2018.09.048
71. Hartman ML, Sztiller-Sikorska M, Gajos-Michniewicz A, Czyz M. Dissecting mechanisms of melanoma resistance to BRAF and MEK inhibitors revealed genetic and non-genetic patient- and drug-specific alterations and remarkable phenotypic plasticity. Cells (2020) 9:142. doi: 10.3390/cells9010142
72. Mulcahy Levy JM, Thorburn A. Autophagy in cancer: Moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ (2020) 27:843–57. doi: 10.1038/s41418-019-0474-7
73. Bustos SO, da Silva Pereira GJ, de Freitas Saito R, Gil CD, Zanatta DB, Smaili SS, et al. Galectin-3 sensitized melanoma cell lines to vemurafenib (PLX4032) induced cell death through prevention of autophagy. Oncotarget (2018) 9:14567–79. doi: 10.18632/oncotarget.24516
74. Sun T, Jiao L, Wang Y, Yu Y, Ming L. SIRT1 induces epithelial-mesenchymal transition by promoting autophagic degradation of E-cadherin in melanoma cells. Cell Death Dis (2018) 9:136. doi: 10.1038/s41419-017-0167-4
75. Sosna J, Philipp S, Chico JF, Saggau C, Fritsch J, Föll A, et al. Differences and Similarities in TRAIL and Tumor Necrosis Factor-Mediated Necroptotic Signaling in Cancer Cells. Mol Cell Biol (2016) 36:2626–44. doi: 10.1128/MCB.00941-15
76. Makitie T, Summanen P, Tarkkanen A, Kivela T. Microvascular density in predicting survival of patients with choroidal and ciliary body melanoma. Invest Ophthalmol Vis Sci (1999) 40:2471–80.
77. Foss AJ, Alexander RA, Jefferies LW, Hungerford JL, Harris AL, Lightman S. Microvessel count predicts survival in uveal melanoma. Cancer Res (1996) 56:2900–3.
78. Scala S, Ieranò C, Ottaiano A, Franco R, La Mura A, Liguori G, et al. CXC chemokine receptor 4 is expressed in uveal malignant melanoma and correlates with the epithelioid-mixed cell type. Cancer Immunol Immunother (2007) 56:1589–95. doi: 10.1007/s00262-007-0303-6
79. Brouwer NJ, Gezgin G, Wierenga APA, Bronkhorst IHG, Marinkovic M, Luyten GPM, et al. Tumour Angiogenesis in Uveal Melanoma Is Related to Genetic Evolution.1. Cancers (Basel) (2019) 11:979. doi: 10.3390/cancers11070979
80. Bennett PE, Bemis L, Norris DA, Shellman YG. miR in melanoma development: miRNAs and acquired hallmarks of cancer in melanoma. Physiol Genom (2013) 45:1049–59. doi: 10.1152/physiolgenomics.00116.2013
Keywords: uveal melanoma, autophagy, immunohistochemistry, prognosis, Beclin-1
Citation: Broggi G, Ieni A, Russo D, Varricchio S, Puzzo L, Russo A, Reibaldi M, Longo A, Tuccari G, Staibano S and Caltabiano R (2020) The Macro-Autophagy-Related Protein Beclin-1 Immunohistochemical Expression Correlates With Tumor Cell Type and Clinical Behavior of Uveal Melanoma. Front. Oncol. 10:589849. doi: 10.3389/fonc.2020.589849
Received: 31 July 2020; Accepted: 20 October 2020;
Published: 20 November 2020.
Edited by:
Antonella Zannetti, Italian National Research Council, ItalyReviewed by:
Renato Franco, University of Campania Luigi Vanvitelli, ItalyCopyright © 2020 Broggi, Ieni, Russo, Varricchio, Puzzo, Russo, Reibaldi, Longo, Tuccari, Staibano and Caltabiano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giuseppe Broggi, Z2l1c2VwcGUuYnJvZ2dpQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
‡These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.