
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 19 May 2021
Sec. Molecular and Cellular Oncology
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.586085
Melanoma is the major lethal skin malignancy. However, the critical molecular drivers governing melanoma progression and prognosis are still not clear. By analyzing The Cancer Genome Atlas (TCGA) data, we identified FUT8-AS1 as a prognosis-related long non-coding RNA (lncRNA) in melanoma. We further confirmed that FUT8-AS1 is downregulated in melanoma. Reduced expression of FUT8-AS1 is correlated with aggressive clinical factors and inferior overall survival. Using in vitro functional assays, our findings demonstrated that ectopic expression of FUT8-AS1 represses melanoma cell proliferation, migration, and invasion. FUT8-AS1 silencing promotes melanoma cell proliferation, migration, and invasion. Furthermore, in vivo functional assays demonstrated that FUT8-AS1 represses melanoma growth and metastasis. Mechanistically, FUT8-AS1 was found to bind NF90, repress the interaction between NF90 and primary miR-145 (pri-miR-145), relieve the repressive roles of NF90 on mature miR-145-5p biogenesis, and thus promote miR-145-5p biogenesis and upregulate mature miR-145-5p level. The expression of FUT8-AS1 is positively correlated with miR-145-5p in melanoma tissues. Via upregulating miR-145-5p, FUT8-AS1 reduces the expression of NRAS, a target of miR-145-5. FUT8-AS1 further represses MAPK signaling via downregulating NRAS. Functional rescue assays demonstrated that inhibition of miR-145-5p reverses the tumor suppressive roles of FUT8-AS1 in melanoma. The oncogenic roles of FUT8-AS1 silencing are also blocked by MAPK signaling inhibitor MEK162. In conclusion, these findings demonstrate that FUT8-AS1 exerts tumor suppressive roles in melanoma via regulating NF90/miR-145-5p/NRAS/MAPK signaling axis. Targeting FUT8-AS1 and its downstream molecular signaling axis represent promising therapeutic strategies for melanoma.
Despite the total cancer incidence rate is declined since 1991, the incidence rate of skin melanoma is still increasing for both male and female since 1975 (1). Therefore, skin melanoma has been the fifth most common cancer in male, accounting for 7% of all site cancers according to 2020 cancer statistics of USA (1). In female, skin melanoma is the sixth most common cancer and accounts for 4% of all site cancers according to 2020 cancer statistics of USA (1). Although recent approval of molecule targeted therapy and immunotherapy for melanoma has greatly extended the survival of melanoma patients, most late melanomas particular for those with metastases are still incurable (2, 3). Thus, uncovering the critical molecular drivers responsible for the progression and poor survival of melanomas would enable more effective therapy for melanoma.
Recently, many high-throughput RNA sequencings and systems biology approaches have revealed the critical roles of non-coding RNA (ncRNA) in tumorigenesis and progressions (4–6). Among these ncRNAs, long non-coding RNA (lncRNA) and microRNA (miRNA) present significantly aberrant expression, clinical reverences, and important roles in various malignancies (7–10). lncRNA is a class of long ncRNAs with more than 200 nucleotides (nt) in length, and while miRNA is a class of short ncRNAs with 19–25 nt in length (11–14). Increasing knowledge of lncRNA and miRNA has revealed that aberrant expression of lncRNA and miRNA would be potential prognostic and diagnostic biomarkers for malignancies (15–17). Furthermore, many lncRNAs and miRNAs also show oncogenic or tumor suppressive roles, which are involved in almost every aspect of cancers, including the initiation, proliferation, apoptosis, cell cycle, senescence, stemness, migration, invasion, drug-resistance, and so on (18–20).
In melanoma, the knowledge of miRNAs is relatively abundant. Many oncogenic or tumor suppressive miRNAs have been identified, such as the oncogenic miR-410-3p, miR-21-5p, miR-125b-5p, and miR-378a-5p (21–24) and the tumor suppressive miR-107, miR-140-5p, miR-204-5p, and miR-128-3p (25–28). In our previous report, we also found that miR-145-5p is downregulated in melanoma and suppresses melanoma cell proliferation, migration, and invasion in vitro, and melanoma tumor growth in vivo (29). The major mechanism of action of miRNAs is to bind AGO2 and form RNA-induced silencing complex (RISC), which further binds target mRNAs and induces target mRNAs degradation and/or translation inhibition (30). In our previous report, we also identified NRAS as a direct target of miR-145-5p, which further modulates MAPK signaling (29).
In comparison, the knowledge of lncRNAs in melanoma is relative less (31). The contributions of lncRNAs to melanoma initiation and progression are only starting to be studied (32). Several cancer-related lncRNAs in melanoma were revealed, such as SLNCR1, OVAAL, EMICERI, THOR (33–36). In our previous reports, we also identified three melanoma-related lncRNAs, including PVT1, ILF3-AS1, and MHENCR (37–39). The mechanisms of action of lncRNAs are complex and various. Some lncRNAs directly bind proteins and modulates the expression and/or functions of interacted proteins, such as OVAAL, THOR, and ILF3-AS1 (34, 36, 38). Some lncRNAs directly bind miRNAs and repress the roles of interacted miRNAs, such as MHENCR (39). Some lncRNAs may also directly modulate neighboring protein-coding genes, such as EMICERI (35). Despite several lncRNAs have been elucidated in melanoma, the contributions of most other lncRNAs to melanoma are still unclear. As transcriptomic sequencings have identified more than 58,000 lncRNAs in human (40), we could not preclude the contributions of other lncRNAs to melanoma.
To further identify the lncRNAs involved in melanoma, we analyzed The Cancer Genome Atlas (TCGA) Skin Cutaneous Melanoma (SKCM) dataset, and searched the genes correlated with outcome of melanoma patients. FUT8-AS1 was identified as one of the most significantly correlated genes with melanomas’ prognosis. In this study, we further investigated the expression and clinical reverence of FUT8-AS1 in melanoma. In vitro and in vivo gain- and loss-of-function assays were performed to elucidate the biological roles of FUT8-AS1 in melanoma. Furthermore, the mechanism of action of FUT8-AS1 was explored and we identified a relative novel mechanism of action of FUT8-AS1, which is the promotion of miR-145-5p biogenesis.
A total of 68 malignant melanoma tissues and 36 age and gender-matched skin tissues with melanocytic nevi were acquired from patients who had undergone surgical resection at the 969th Hospital of PLA (Hohhot, Inner Mongolia, China). All tissue samples were diagnosed by histopathological examination. The Review Board of the 969th Hospital of PLA approved this study. Written informed consents were obtained from all patients.
Human melanoma cell lines CHL-1 and SK-MEL-2 were acquired from Cell Resource Center, Chinese Academy of Sciences. CHL-1 and SK-MEL-2 cells were cultured in DMEM and MEM medium respectively supplemented with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific). Where indicated, cells were treated with 1 µM MEK162 (Selleck) for indicated time.
Total RNA was isolated from indicated tissues and cells by Trizol reagent (Invitrogen, Thermo Fisher Scientific) in accordance with the manufacturer’s manual. The first strand cDNA was generated using the RNA and the PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Dalian, China). Real-time PCR was performed using TB Green® Premix Ex Taq™ II (Takara) on StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) with the primers 5’-GGCTCCTTGCTACTTTTAGGG-3’ (forward) and 5’-TGGGGGGGGTCTTTCTCTTC-3’ (reverse) for FUT8-AS1, 5’-GAAATACGCCAGTACCGAATG-3’ (forward) and 5’-TTCTCCTCCAGGGAAGTCAG-3’ (reverse) for NRAS, 5’-GTCGGAGTCAACGGATTTG-3’ (forward) and 5’-TGGGTGGAATCATATTGGAA-3’ (reverse) for GAPDH. GAPDH was used as endogenous control for the quantification of FUT8-AS1 and NRAS expression. For the quantification of miRNAs and pri-miRNAs expression, real-time PCR was carried out using the TaqMan™ Advanced miRNA Assay (Thermo Fisher Scientific) and TaqMan™ Pri-miRNA Assay (Thermo Fisher Scientific) respectively on StepOnePlus Real-Time PCR System in accordance with the manufacturer’s manuals.
To construct melanoma cells stably overexpressing FUT8-AS1, FUT8-AS1 overexpressing lentivirus (LV11/CMV/Neo) were acquired from GenePharma (Shanghai, China) and infected into CHL-1 and SK-MEL-2 cells. Next, the cells were treated with neomycin for 4 weeks to select CHL-1 and SK-MEL-2 cells overexpressing FUT8-AS1. Two pairs of cDNA oligonucleotides inhibiting FUT8-AS1 expression were designed and generated by GenePharma. After annealing, double-strand oligonucleotides were inserted into the shRNA lentiviral vector pLV6/EF-1a/Puro to produce shRNA lentivirus inhibiting FUT8-AS1 expression. A scrambled non-targeting shRNA was used as negative control (NC). Next, CHL-1 and SK-MEL-2 cells were infected with the shRNA lentivirus. The cells were treated with puromycin for 4 weeks to select FUT8-AS1 silenced cells. The shRNA sequences were as follows: 5’-GATCCGCCCTACTTTATCTTGTAAGATTCAAGAGATCTTACAAGATAAAGTAGGGCTTTTTTG-3’ (forward) and 5’-AATTCAAAAAAGCCCTACTTTATCTTGTAAGATCTCTTGAATCTTACAAGATAAAGTAGGGCG-3’ (reverse) for LV-shRNA-1, 5’-GATCCGCGGAAGTTTATTTAGTACGGTTCAAGAGACCGTACTAAATAAACTTCCGCTTTTTTG-3’ (forward) and 5’-AATTCAAAAAAGCGGAAGTTTATTTAGTACGGTCTCTTGAACCGTACTAAATAAACTTCCGCG-3’ (reverse) for LV-shRNA-2, 5’-GATCCGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAACTTTTTTG-3’ (forward) and 5’-AATTCAAAAAAGTTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAACG-3’ (reverse) for LV-shNC. To construct melanoma cells overexpressing FUT8-AS1 and concurrently inhibiting miR-145-5p, FUT8-AS1 overexpressed CHL-1 cells were infected with miR-145-5p inhibiting lentivirus (Genechem Co. Ltd., Shanghai, China) and treated with neomycin and puromycin for 4 weeks to select FUT8-AS1 overexpressed and concurrently miR-145-5p inhibited cells.
Glo cell viability assay was performed to measure cell viability as previously described (38). Briefly, 3,000 indicated melanoma cells per well were plated into 96-well plates. At the indicated time, the luminescence values were detected by the Cell Titer-Glo Luminescent Cell Viability Assay (Promega) to record cell viability. Ethynyl deoxyuridine (EdU) incorporation assay was performed using the EdU Kit (Roche) to measure cell proliferation as previously described (38). Transwell migration assay was performed to measure cell migration as previously described (38). Transwell invasion assay was performed to measure cell invasion as previously described (38).
Athymic BALB/c nude mice were purchased from Chinese Academy of Sciences and maintained in pathogen-free condition. The use of animals was approved by the Review Board of the 969th Hospital of PLA (Hohhot, Inner Mongolia, China). A total of 3.0 × 106 indicated melanoma cells were subcutaneously injected into the flanks of nude mice. Subcutaneous xenograft volumes were detected using caliper every 7 days and calculated following the formula V = 0.5 × LW2 (L, tumor length; W, tumor width). At the 28th day after injection, subcutaneous xenografts were resected and weighed. Subcutaneous xenografts were further used to carry out immunohistochemistry (IHC) staining with the primary antibody against Ki67 (ab15580, 1 µg/ml, Abcam). Subcutaneous xenografts were also used to perform TdT-mediated dUTP Nick-End Labeling (TUNEL) staining using the One Step TUNEL Apoptosis Assay Kit (Beyotime, Shanghai, China). To evaluate melanoma liver metastasis in vivo, 3.0 × 106 indicated melanoma cells were intrasplenically injected into nude mice to construct liver metastasis model. At the 28th day after injection, the mice were sacrificed and the livers were resected. Hematoxylin-eosin (H&E) staining was performed using the livers. To evaluate melanoma lung metastasis in vivo, 3.0 × 106 indicated melanoma cells were injected into tail vein of nude mice to construct lung metastasis model. At the 28th day after injection, the mice were sacrificed and the lungs were resected. H&E staining was performed using the lungs.
FUT8-AS1 full-length sequences were PCR-amplified with the primers 5’-GGAATTCTCGCTGCGCCGGTGGAGA-3’ (forward) and 5’-GCTCTAGATTTCAGTTGGAAGGAGGTAGG-3’ (reverse). The PCR products were cloned into the EcoR I and Xba I sites of pSPT19 vector (Roche) to construct pSPT19-FUT8-AS1. NF90 binding sites mutated pSPT19-FUT8-AS1 (pSPT19-FUT8-AS1-mut) was constructed using the Fast Mutagenesis System (TransGen Biotech, Beijing, China) with the primers 5’-CTTTATCTTGTAAGAGGACAATCCACATTCAC-3’ (forward) and 5’-TTGTCCTCTTACAAGATAAAGTAGGGCTTCG-3’ (reverse). Wild type and NF90 binding sites mutated FUT8-AS1 was in vitro transcribed and biotinylated from pSPT19-FUT8-AS1 and pSPT19-FUT8-AS1-mut, respectively, using the Biotin RNA Labeling Mix (Roche) and Sp6 RNA polymerase (Roche). After purification, 3 µg of wild type or NF90 binding sites mutated FUT8-AS1 were incubated with 1 mg of whole-cell lysates from CHL-1 cells at 25°C for 1 h. The streptavidin agarose beads (Thermo Fisher Scientific) were used to enrich biotinylated RNA and interacted proteins. The proteins present in the pull-down material were detected using western blot.
Western blot was conducted as previously described (38) with the primary antibodies: for NF90, ab131004, 1:2,000, Abcam; for EZH2, #07-689, 1:2,000, Millipore; for NRAS, ab154291, 1:1,000, Abcam; for GAPDH, ab8245, 1:5,000, Abcam; for phospho-MEK1/2, #9154, 1:1,000, Cell Signaling Technology; for MEK1/2, #8727, 1:1,000, Cell Signaling Technology; for phospho-ERK1/2, #4370, 1:2,000, Cell Signaling Technology; for ERK1/2, #4695, 1:1,000, Cell Signaling Technology.
RIP assays were performed in indicated melanoma cells using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) and an antibody against NF90 (5 µg per reaction; ab131004, Abcam) in accordance with the manufacturer’s manual.
GraphPad Prism v6.0 was employed to perform all statistical analyses. For comparisons, log-rank test, Mann-Whitney test, two-tailed unpaired t test, one-way ANOVA followed by Dunnett’s multiple comparisons test, Spearman correlation analysis, or one-way ANOVA followed by Tukey’s multiple comparisons test was conducted as indicated in figure legends. P < 0.05 was considered as statistically significant.
To search the genes correlated with prognosis of melanoma, we analyzed The Cancer Genome Atlas (TCGA) Skin Cutaneous Melanoma (SKCM) dataset to retrieve the differentially expressed genes between different vital statuses (P-value < 0.01, fold change ≥2, Supplementary Table 1). Among these differentially expressed genes, we noted FUT8-AS1, which has a relative more significant P-value and bigger fold change. Further analysis of the TCGA-SKCM dataset revealed that reduced expression of FUT8-AS1 indicated inferior overall survival (Figure 1A). Two in silico tools, namely the Coding Potential Assessment Tool (CPAT) (http://lilab.research.bcm.edu/cpat/index.php) and the Coding Potential Calculator (CPC) (http://cpc2.cbi.pku.edu.cn/) were employed to calculate the coding potential of FUT8-AS1. CPC and CPAT scores of FUT8-AS1 were equally low as well-known lncRNA HOTAIR (Supplementary Figures 1A, B), which indicated the noncoding nature of FUT8-AS1. Next, we measured FUT8-AS1 expression in our own cohort including 36 benign nevi and 68 melanomas using real-time PCR. The results indicated that FUT8-AS1 was downregulated in melanoma tissues compared with benign nevi (Figure 1B). Correlation analyses between FUT8-AS1 expression and clinicopathological characters indicated that reduced expression of FUT8-AS1 was correlated with thickness, ulceration, and metastasis (Figures 1C–E). Moreover, Kaplan-Meier survival analysis indicated that reduced expression of FUT8-AS1 was correlated with inferior overall survival among these 68 melanomas (Figure 1F). Thus, these findings suggest that FUT8-AS1 is downregulated in melanoma and reduced expression of FUT8-AS1 is correlated with aggressive clinical factors and inferior overall survival.
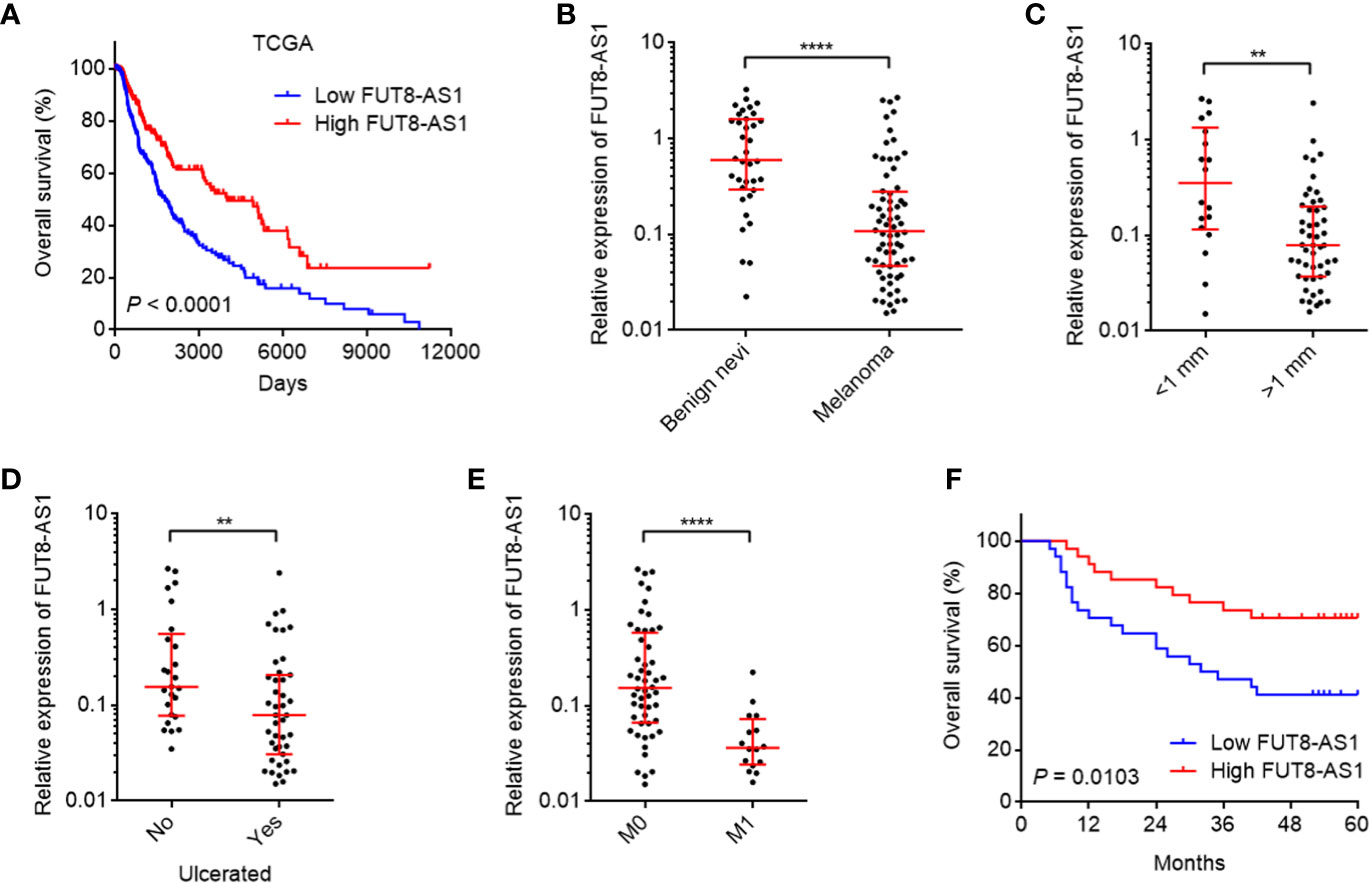
Figure 1 The expression and clinical relevance of FUT8-AS1 in melanoma. (A) Kaplan-Meier survival curves of skin cutaneous melanoma (SKCM) from TCGA dataset stratified by FUT8-AS1 expression (low 50% [n = 229] versus high 50% [n = 229]). P < 0.0001 by log-rank test. (B) FUT8-AS1 expression in 36 benign nevi and 68 melanoma tissues was measured by real-time PCR. (C) FUT8-AS1 expression in 18 melanoma tissues with thickness <1 mm and 50 melanoma tissues with thickness >1 mm. (D) FUT8-AS1 expression in 25 melanoma tissues without ulceration and 43 melanoma tissues with ulceration. (E) FUT8-AS1 expression in 52 melanoma tissues without distant metastasis and 16 melanoma tissues with distant metastasis. For (B–E), data are presented as median with interquartile range. **P < 0.01, ****P < 0.0001 by Mann-Whitney test. (F) Kaplan-Meier survival curves of 68 melanomas stratified by FUT8-AS1 expression (low 50% [n = 34] versus high 50% [n = 34]). P = 0.0103 by log-rank test.
Due to the significant correlation between FUT8-AS1 expression and clinical characteristics of melanoma, we next investigated the potential roles of FUT8-AS1 in melanoma. We constructed CHL-1 and SK-MEL-2 cells stably overexpressing FUT8-AS1 via FUT8-AS1 overexpression lentivirus mediated transfection. The overexpression efficiencies were confirmed by real-time PCR (Figures 2A, B). Glo cell viability assays indicated that both CHL-1 and SK-MEL-2 cells overexpressing FUT8-AS1 had reduced cell viabilities compared with their control cells, respectively (Figures 2C, D). EdU incorporation assays further indicated that both CHL-1 and SK-MEL-2 cells overexpressing FUT8-AS1 had slower cell proliferation rates compared with their control cells, respectively (Figure 2E). Transwell migration assays indicated that both CHL-1 and SK-MEL-2 cells overexpressing FUT8-AS1 had less migrated cell numbers compared with their control cells, respectively (Figure 2F). Transwell invasion assays indicated that both CHL-1 and SK-MEL-2 cells overexpressing FUT8-AS1 had less invasive cell numbers compared with their control cells, respectively (Figure 2G). Thus, these findings suggest that FUT8-AS1 inhibits melanoma cell proliferation, migration, and invasion in vitro.
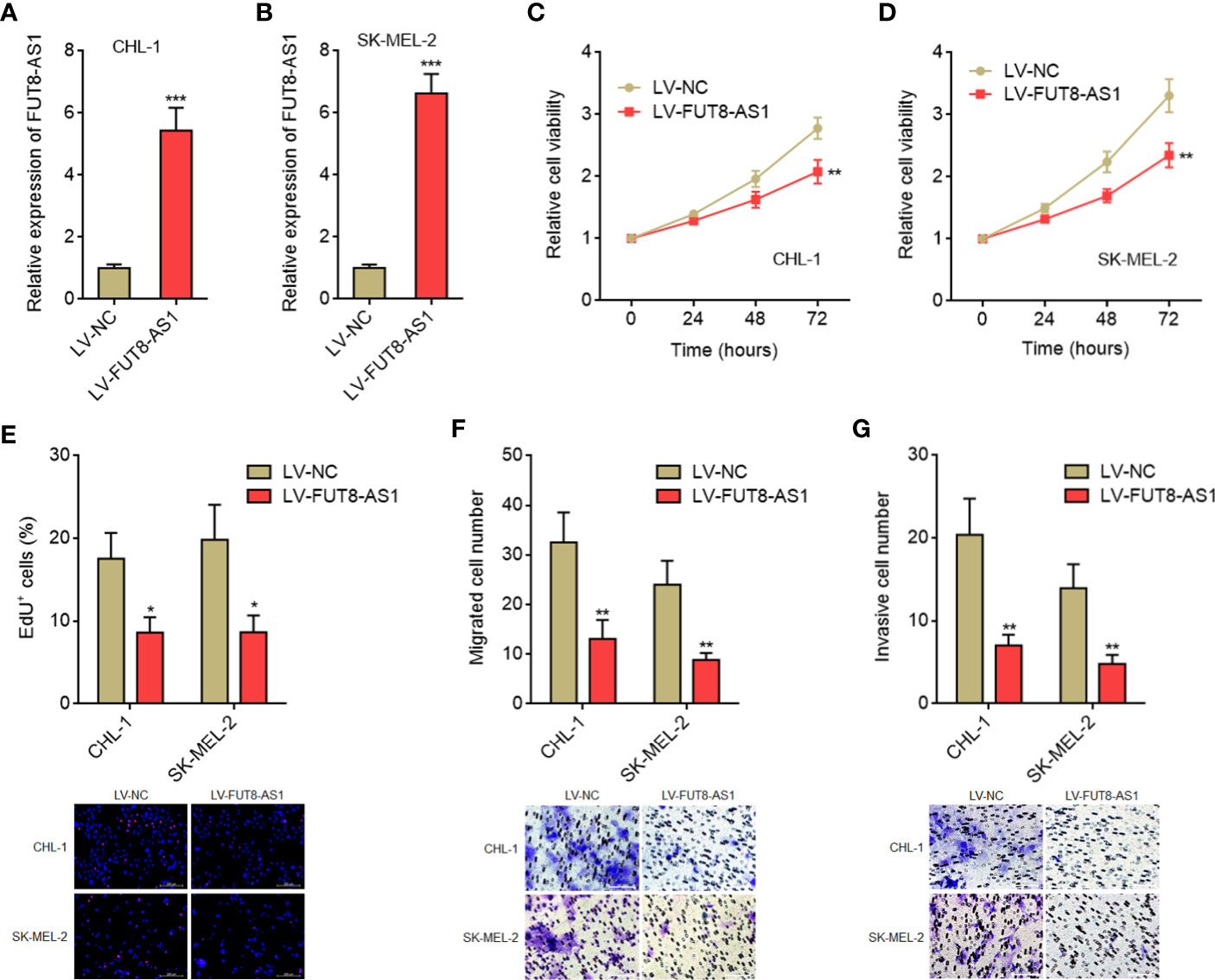
Figure 2 FUT8-AS1 inhibits melanoma cell proliferation, migration, and invasion. (A) FUT8-AS1 expression in CHL-1 cells overexpressing FUT8-AS1or control was measured by real-time PCR. (B) FUT8-AS1 expression in SK-MEL-2 cells overexpressing FUT8-AS1 or control was measured by real-time PCR. (C) Cell viabilities of CHL-1 cells overexpressing FUT8-AS1 or control were determined by the Glo cell viability assay. (D) Cell viabilities of SK-MEL-2 cells overexpressing FUT8-AS1 or control were determined by the Glo cell viability assay. (E) Cell proliferation of CHL-1 and SK-MEL-2 cells overexpressing FUT8-AS1 or control were determined by the EdU incorporation assay. The red color indicates EdU-positive nuclei. Scale bars, 200 µm. (F) Cell migration of CHL-1 and SK-MEL-2 cells overexpressing FUT8-AS1 or control were determined by the transwell migration assay. Scale bars, 100 µm. (G) Cell invasion of CHL-1 and SK-MEL-2 cells overexpressing FUT8-AS1 or control were determined by the transwell invasion assay. Scale bars, 100 µm. Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 by two-tailed unpaired t test.
To further confirm the tumor suppressive roles of FUT8-AS1 in melanoma, we constructed FUT8-AS1 stably silenced CHL-1 and SK-MEL-2 cells via two independent FUT8-AS1 specific shRNAs lentivirus mediated transfection (Figures 3A, B). Glo cell viability assays indicated that FUT8-AS1 silenced CHL-1 and SK-MEL-2 cells both had increased cell viabilities compared with their control cells, respectively (Figures 3C, D). EdU incorporation assays indicated that FUT8-AS1 silenced CHL-1 and SK-MEL-2 cells both had quicker cell proliferation rates compared with their control cells, respectively (Figure 3E). Transwell migration assays indicated that FUT8-AS1 silenced CHL-1 and SK-MEL-2 cells both had more migrated cell numbers compared with their control cells, respectively (Figure 3F). Transwell invasion assays indicated that FUT8-AS1 silenced CHL-1 and SK-MEL-2 cells both had more invasive cell numbers compared with their control cells, respectively (Figure 3G). Collectively, these findings suggest that FUT8-AS1 silencing promotes melanoma cell proliferation, migration, and invasion in vitro, further supporting the tumor suppressive roles of FUT8-A1 in melanoma.
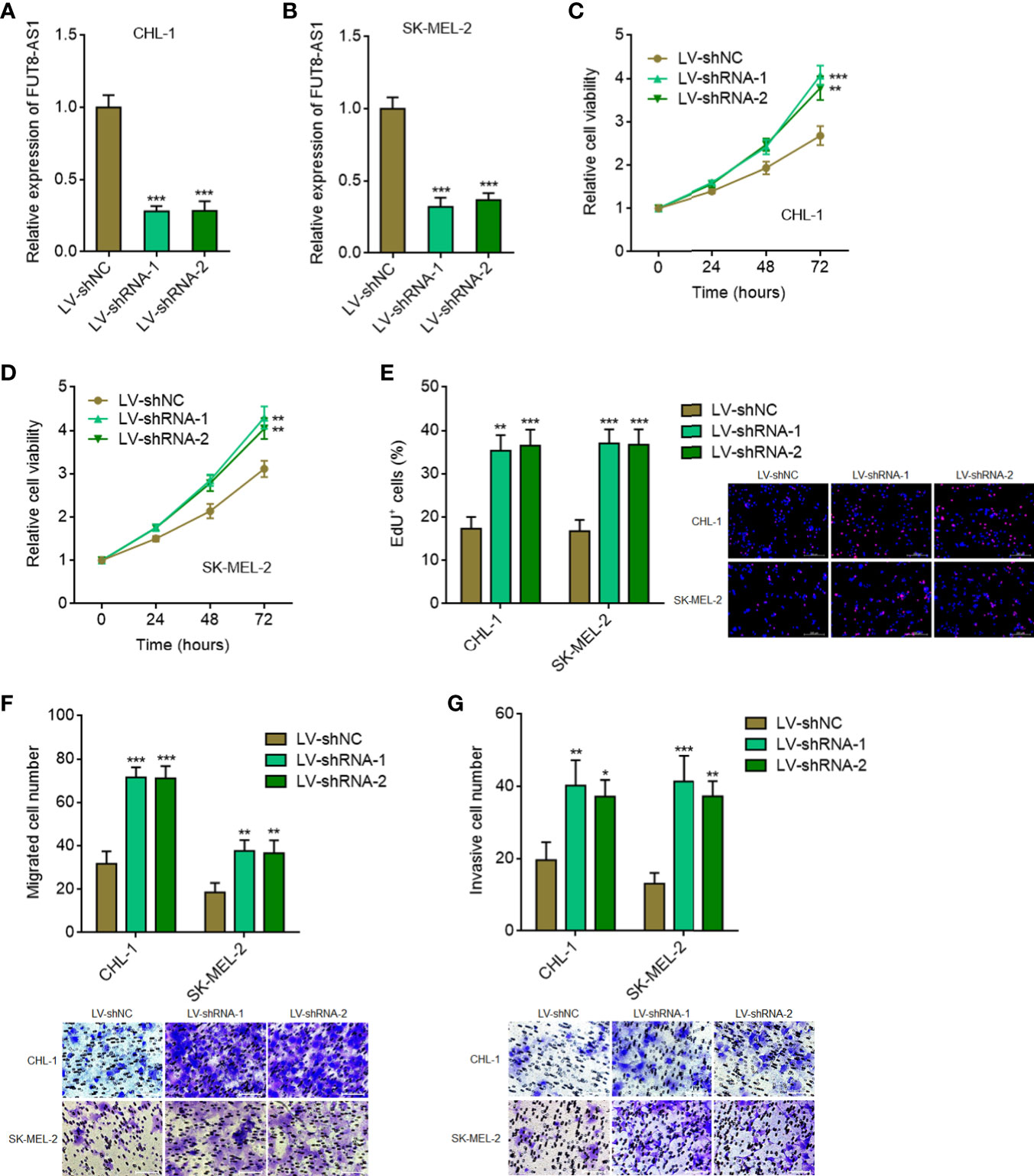
Figure 3 FUT8-AS1 silencing promotes melanoma cell proliferation, migration, and invasion. (A) FUT8-AS1 expression in CHL-1 cells silencing FUT8-AS1 or control was measured by real-time PCR. (B) FUT8-AS1 expression in SK-MEL-2 cells silencing FUT8-AS1 or control was measured by real-time PCR. (C) Cell viabilities of CHL-1 cells silencing FUT8-AS1 or control were determined by the Glo cell viability assay. (D) Cell viabilities of SK-MEL-2 cells silencing FUT8-AS1 or control were determined by the Glo cell viability assay. (E) Cell proliferation of CHL-1 and SK-MEL-2 cells silencing FUT8-AS1 or control were determined by the EdU incorporation assay. The red color indicates EdU-positive nuclei. Scale bars, 200 µm. (F) Cell migration of CHL-1 and SK-MEL-2 cells silencing FUT8-AS1 or control were determined by the transwell migration assay. Scale bars, 100 µm. (G) Cell invasion of CHL-1 and SK-MEL-2 cells silencing FUT8-AS1 or control were determined by the transwell invasion assay. Scale bars, 100 µm. Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA followed by Dunnett’s multiple comparisons test.
To investigate the roles of FUT8-AS1 in melanoma growth in vivo, FUT8-AS1 stably overexpressed and control CHL-1 cells were subcutaneously inoculated into nude mice. Tumor volumes were measured every 7 days and the tumors were excised and weighed at the 28th day after inoculation. The results indicated that the tumors formed by CHL-1 cells overexpressing FUT8-AS1 had a slower growth rate and formed a smaller tumor compared with the tumors formed by control cells (Figures 4A, B). Proliferation marker Ki67 IHC staining indicated that the tumors formed by CHL-1 cells overexpressing FUT8-AS1 had less Ki67 positive cells compared with the tumors formed by control cells (Figure 4C). TUNEL staining indicated that the tumors formed by CHL-1 cells overexpressing FUT8-AS1 had more apoptotic cells compared with the tumors formed by control cells (Figure 4D). To investigate the roles of FUT8-AS1 in melanoma metastasis in vivo, CHL-1 cells overexpressing FUT8-AS1 or control were intrasplenically injected to construct liver metastasis model. The results indicated that CHL-1 cells overexpressing FUT8-AS1 formed less liver metastases compared with control CHL-1 cells (Figure 4E). Furthermore, CHL-1 cells overexpressing FUT8-AS1 or control were injected into the tail veins of nude mice to construct lung metastasis model. The results indicated that CHL-1 cells overexpressing FUT8-AS1 formed less lung metastases compared with control CHL-1 cells (Figure 4F). Thus, these findings suggest that FUT8-AS1 inhibits melanoma growth and metastasis in vivo, further supporting the tumor suppressive roles of FUT8-AS1 in melanoma.
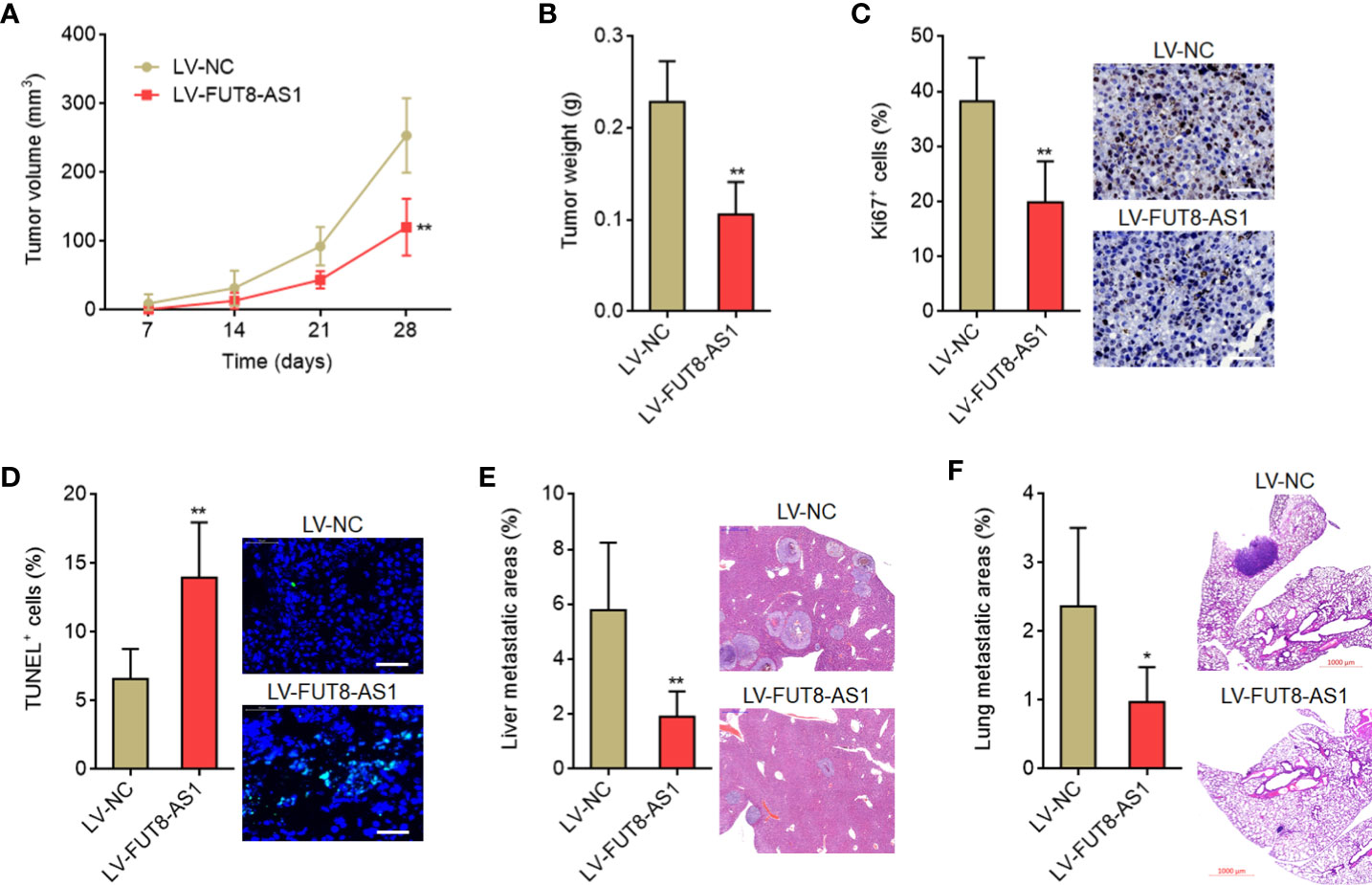
Figure 4 FUT8-AS1 inhibits melanoma growth and metastasis in vivo. (A, B) CHL-1 cells overexpressing FUT8-AS1 or control were subcutaneously inoculated into nude mice. Tumor volumes were measured every 7 days (A). The tumors were excised and weighed at the 28th day after inoculation (B). (C) The tumors formed by CHL-1 cells overexpressing FUT8-AS1 or control were used to perform Ki67 IHC staining. Scale bars, 50 µm. (D) The tumors formed by CHL-1 cells overexpressing FUT8-AS1 or control were used to perform TUNEL staining. Scale bars, 50 µm. (E) CHL-1 cells overexpressing FUT8-AS1 or control were inoculated into spleen of nude mice to construct liver metastasis model. The livers were excised and used to perform H&E staining. Scale bars, 500 µm. (F) CHL-1 cells overexpressing FUT8-AS1 or control were inoculated into tail vein of nude mice to construct lung metastasis model. The lungs were excised and used to perform H&E staining. Scale bars, 1,000 µm. Data are presented as mean ± SD. n = 6 mice in each group. *P < 0.05, **P < 0.01 by Mann-Whitney test.
Many lncRNAs were shown to exert their biological roles via interacting with proteins (41). To identify whether FUT8-AS1 could also interact with proteins, we used the online in silico tool RNA-Protein Interaction Prediction (RPISeq) (http://pridb.gdcb.iastate.edu/RPISeq/) to predict the potential interaction between FUT8-AS1 and proteins. Notably, we predicted a potential interaction between FUT8-AS1 and NF90 with an interaction probability of 0.95. FUT8-AS1 contains a conserved NF90 binding sequence (5’-CUGUU-3’, 452-456nt of FUT8-AS1, Supplementary Figure 2), supporting the potential interaction between FUT8-AS1 and NF90. NF90 has been revealed to repress miR-145-5p biogenesis via binding pri-miR-145 (42). Our previous report had revealed the tumor suppressive role of miR-145-5p in melanoma (29). Therefore, we next investigated the potential effects of FUT8-AS1 on NF90/miR-145-5p. RNA pull-down assays using in vitro transcribed biotin-labeled FUT8-AS1 showed the specific enrichment of NF90, but not EZH2 (Figure 5A). In addition, the enrichment of NF90 by FUT8-AS1 was significantly reduced by the mutation of NF90 binding sequence in FUT8-AS1 (Figure 5A). RIP assays with NF90 specific antibody showed the enrichment of FUT8-AS1, but not GAPDH mRNA (Figure 5B), which further supports the interaction between FUT8-AS1 and NF90. To investigate whether NF90 regulates miR-145-5p biogenesis in melanoma and whether FUT8-AS1 modulates the effects of NF90 on miR-145-5p, we first detected the effects of NF90 on miR-145-5p. Silencing of NF90 significantly upregulated miR-145-5p expression in both CHL-1 and SK-MEL-2 cells (Figure 5C). RIP assays with NF90 specific antibody revealed the significant enrichment of pri-miR-145 in both CHL-1 and SK-MEL-2 cells (Figures 5D, E), supporting the binding of NF90 to pri-miR-145 in melanoma. Furthermore, overexpression of FUT8-AS1 significantly decreased the binding between pri-miR-145 and NF90 in SK-MEL-2 cells (Figure 5D). Conversely, silencing of FUT8-AS1 increased the binding between pri-miR-145 and NF90 in CHL-1 cells (Figure 5E). These data suggested that FUT8-AS1 reduced the binding of NF90 to pri-miR-145. Next, we measured pri-miR-145 and mature miR-145-5p expression levels in FUT8-AS1 overexpressed CHL-1 and SK-MEL-2 cells. The results indicated that pri-miR-145 was reduced in FUT8-AS1 overexpressed CHL-1 and SK-MEL-2 cells compared with their control cells (Figure 5F), and while mature miR-145-5p was significantly increased in FUT8-AS1 overexpressed CHL-1 and SK-MEL-2 cells (Figure 5G). Furthermore, pri-miR-145 and mature miR-145-5p expression levels in FUT8-AS1 silenced CHL-1 and SK-MEL-2 cells were measured. The results indicated that pri-miR-145 was increased in FUT8-AS1 silenced CHL-1 and SK-MEL-2 cells compared with their control cells (Figure 5H), and while mature miR-145-5p was significantly reduced in FUT8-AS1 silenced CHL-1 and SK-MEL-2 cells (Figure 5I). To elucidate whether the regulation of miR-145-5p by FUT8-AS1 is dependent on NF90, we depleted NF90 in FUT8-AS1 overexpressed and silenced CHL-1 and SK-MEL-2 cells, and then measured mature miR-145-5p expression levels. The results revealed that depletion of NF90 abolished the effects of FUT8-AS1 overexpression and silencing on miR-145-5p (Figures 5J, K). Collectively, these findings suggest that FUT8-AS1 interacted with NF90, relieved the repressive roles of NF90 on miR-145-5p biogenesis, and therefore downregulated pri-miR-145 and upregulated mature miR-145-5p levels in melanoma. The positive correlation between FUT8-AS1 and miR-145-5p expression levels was also found in melanoma tissues (Figure 5L).
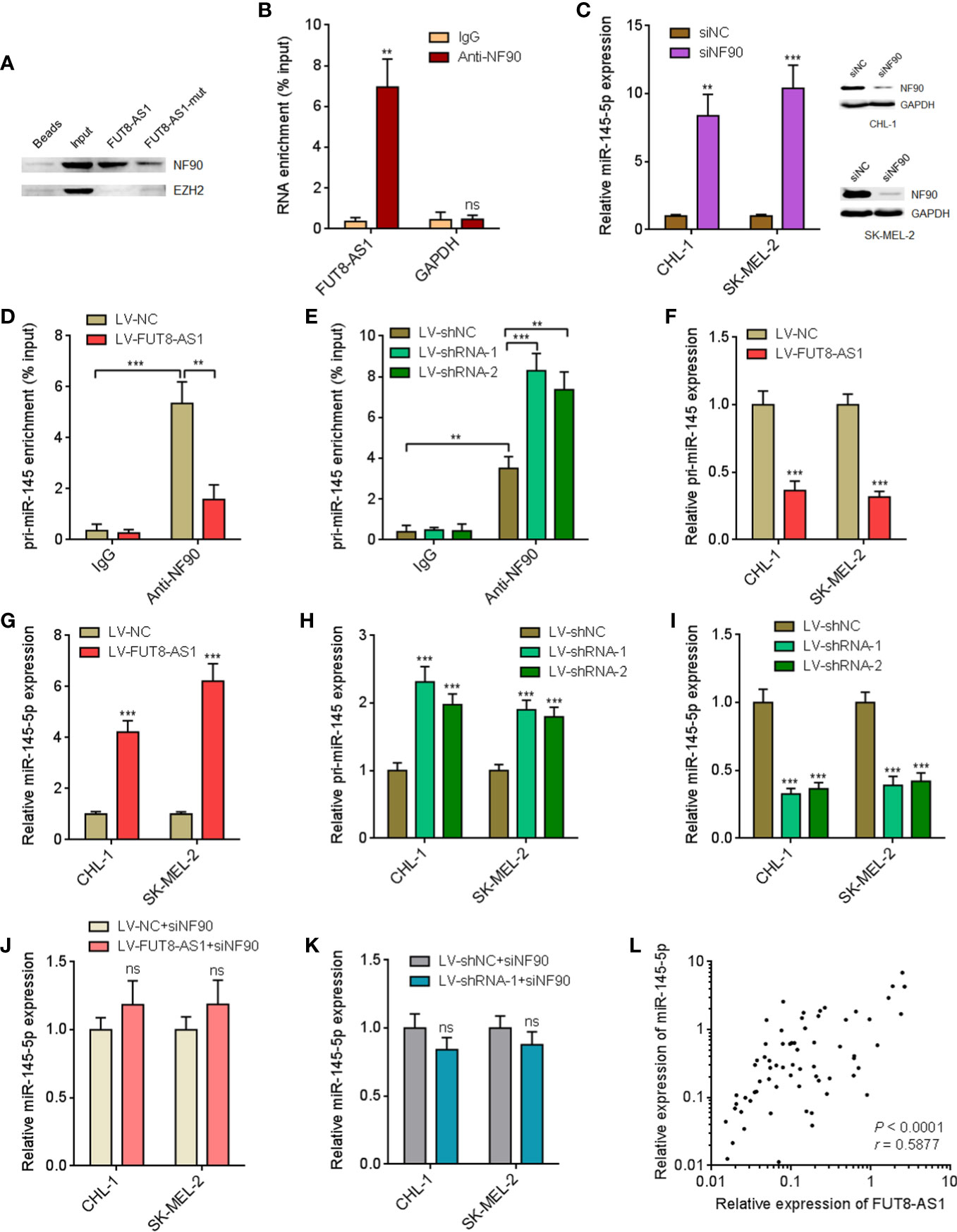
Figure 5 FUT8-AS1 upregulates miR-145-5p via binding NF90. (A) RNA pull-down assays using in vitro transcribed wild-type or NF90 binding site mutant FUT8-AS1. The enriched proteins were measured by western blot. (B) RIP assays were performed in CHL-1 cells using NF90 specific antibody or non-specific IgG. The enriched RNA was detected by real-time PCR. (C) After transfection of NF90 specific siRNAs pool or non-targeting negative control (NC) siRNAs into CHL-1 and SK-MEL-2 cells, miR-145-5p expression levels and NF90 protein expression levels were measured by real-time PCR and western blot, respectively. (D) RIP assays were performed in SK-MEL-2 cells overexpressing FUT8-AS1 or control using NF90 specific antibody or non-specific IgG. The enriched RNA was detected by real-time PCR. (E) RIP assays were performed in CHL-1 cells silencing FUT8-AS1 or control using NF90 specific antibody or non-specific IgG. The enriched RNA was detected by real-time PCR. (F) pri-miR-145 expression levels in CHL-1 and SK-MEL-2 cells overexpressing FUT8-AS1 or control were measured by real-time PCR. (G) Mature miR-145-5p expression levels in CHL-1 and SK-MEL-2 cells overexpressing FUT8-AS1 or control were measured by real-time PCR. (H) pri-miR-145 expression levels in CHL-1 and SK-MEL-2 cells silencing FUT8-AS1 or control were measured by real-time PCR. (I) Mature miR-145-5p expression levels in CHL-1 and SK-MEL-2 cells silencing FUT8-AS1 or control were measured by real-time PCR. (J) After transfection of NF90 specific siRNAs pool into CHL-1 and SK-MEL-2 cells overexpressing FUT8-AS1 or control, mature miR-145-5p expression levels were measured by real-time PCR. (K) After transfection of NF90 specific siRNAs pool into CHL-1 and SK-MEL-2 cells silencing FUT8-AS1 or control, mature miR-145-5p expression levels were measured by real-time PCR. (L) miR-145-5p expression levels in the same 68 melanoma tissues used in Figure 1 were measured by real-time PCR. The correlation between miR-145-5p and FUT8-AS1 expression levels in these 68 melanoma tissues was calculated by Spearman correlation analysis. r = 0.5877, P < 0.0001. For (B–K), data are presented as mean ± SD. **P < 0.01, ***P < 0.001, ns, not significant, by two-tailed unpaired t test [B, C, D, E (the comparison between IgG and Anti-NF90 for LV-shNC), F, G, J, K] or one-way ANOVA followed by Dunnett’s multiple comparisons test [E (the comparison between LV-shRNA-1, LV-shRNA-2, and LV-shNC for Anti-NF90), H, I].
In our previous report, we have found that miR-145-5p activated MAPK signaling via directly targeting NRAS (29). Therefore, we further investigated the effects of FUT8-AS1 on NRAS/MAPK signaling. Our findings revealed that NRAS mRNA levels were decreased in FUT8-AS1 overexpressed cells and increased in FUT8-AS1 silenced cells (Figures 6A, B). Consistently, NRAS protein levels were decreased in FUT8-AS1 overexpressed cells and increased in FUT8-AS1 silenced cells (Figures 6C, D). Next, the effects of FUT8-AS1 on MAPK signaling were investigated. As shown in Figures 6E, F, the phosphorylation levels of MEK1/2 and ERK1/2 were decreased in FUT8-AS1 overexpressed cells and increased in FUT8-AS1 silenced cells (Figures 6E, F). Thus, these data suggest that FUT8-AS1 represses NRAS/MAPK signaling in melanoma.
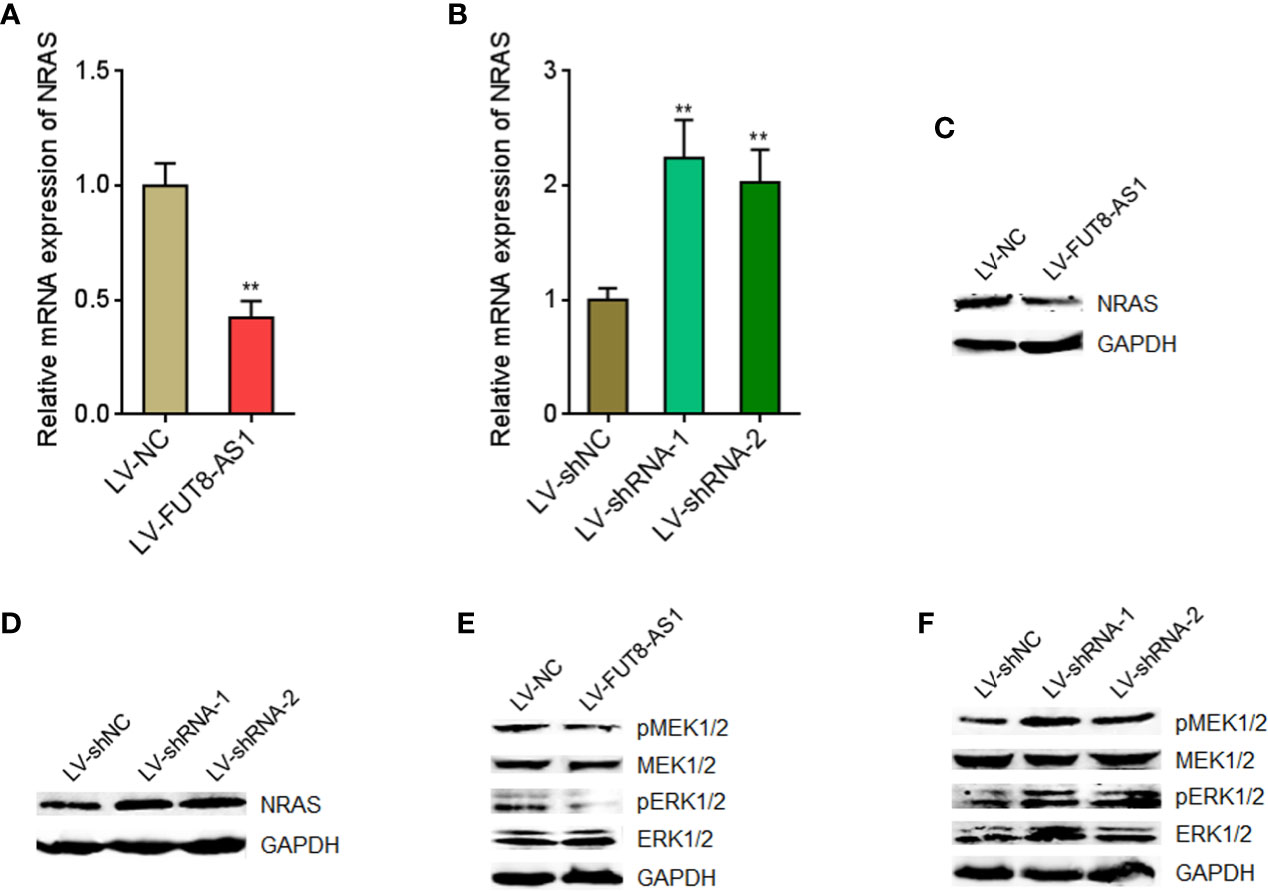
Figure 6 FUT8-AS1 represses NRAS/MAPK signaling. (A) NRAS mRNA expression levels in SK-MEL-2 cells overexpressing FUT8-AS1 or control were measured by real-time PCR. (B) NRAS mRNA expression levels in CHL-1 cells silencing FUT8-AS1 or control were measured by real-time PCR. (C) NRAS protein levels in SK-MEL-2 cells overexpressing FUT8-AS1 or control were measured by western blot. (D) NRAS protein levels in CHL-1 cells silencing FUT8-AS1 or control were measured by western blot. (E) Phosphorylation levels of MEK1/2 and ERK1/2 in SK-MEL-2 cells overexpressing FUT8-AS1 or control were measured by western blot. (F) Phosphorylation levels of MEK1/2 and ERK1/2 in CHL-1 cells silencing FUT8-AS1 or control were measured by western blot. Data are presented as mean ± SD. **P < 0.01 by two-tailed unpaired t test (A) or one-way ANOVA followed by Dunnett’s multiple comparisons test (B).
To elucidate whether the tumor suppressive roles of FUT8-AS1 in melanoma were dependent on the regulation of miR-145-5p/NRAS/MAPK signaling axis, we inhibited miR-145-5p in FUT8-AS1 overexpressed CHL-1 cells (Figure 7A). Glo cell viability assays indicated that the reduced cell viabilities caused by FUT8-AS1 overexpression were reversed by miR-145-5p inhibition (Figure 7B). EdU incorporation assays indicated that the slower cell proliferation rates caused by FUT8-AS1 overexpression were reversed by miR-145-5p inhibition (Figure 7C). Transwell migration assays indicated that the reduced migrated cell number caused by FUT8-AS1 overexpression were reversed by miR-145-5p inhibition (Figure 7D). Transwell invasion assays indicated that the reduced invasive cell number caused by FUT8-AS1 overexpression was reversed by miR-145-5p inhibition (Figure 7E). Collectively, these findings showed that the suppressive roles of FUT8-AS1 in melanoma cell proliferation, migration, and invasion were reversed by miR-145-5p inhibition.
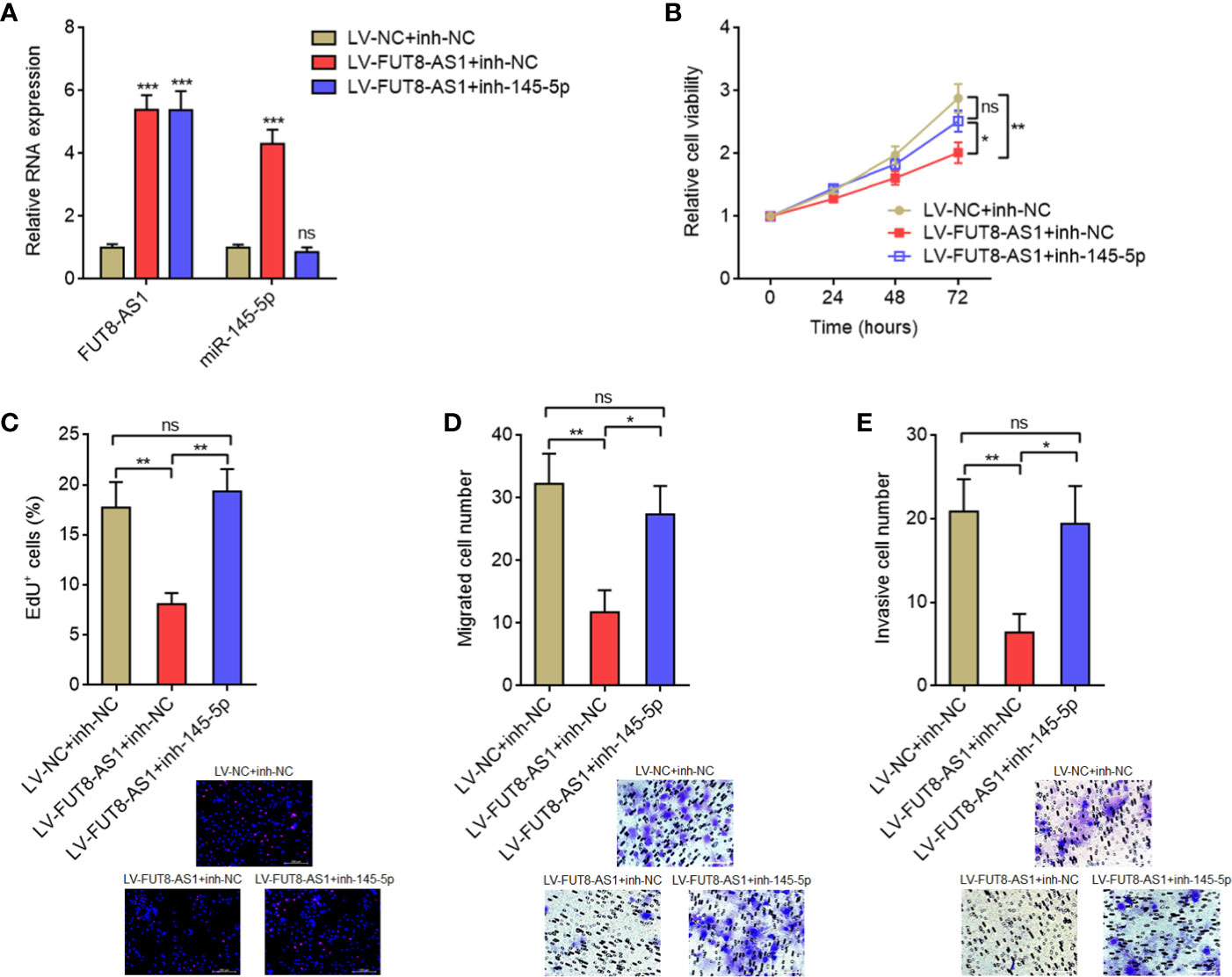
Figure 7 miR-145-5p inhibition reversed the tumor suppressive roles of FUT8-AS1 in melanoma. (A) FUT8-AS1 and miR-145-5p expression levels in CHL-1 cells overexpressing FUT8-AS1 and concurrently silencing miR-145-5p were measured by real-time PCR. (B) Cell viabilities of CHL-1 cells overexpressing FUT8-AS1 and concurrently silencing miR-145-5p were determined by the Glo cell viability assay. (C) Cell proliferation of CHL-1 cells overexpressing FUT8-AS1 and concurrently silencing miR-145-5p were determined by the EdU incorporation assay. The red color indicates EdU-positive nuclei. Scale bars, 200 µm. (D) Cell migration of CHL-1 cells overexpressing FUT8-AS1 and concurrently silencing miR-145-5p were determined by the transwell migration assay. Scale bars, 100 µm. (E) Cell invasion of CHL-1 cells overexpressing FUT8-AS1 and concurrently silencing miR-145-5p were determined by the transwell invasion assay. Scale bars, 100 µm. Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant, by one-way ANOVA followed by Tukey’s multiple comparisons test.
To further elucidate whether the roles of FUT8-AS1 in melanoma were dependent on the regulation of MAPK signaling, we treated FUT8-AS1 silenced and control CHL-1 cells with MEK1/2 inhibitor MEK162. Glo cell viability assays indicated that MEK162 treatment abolished the increased cell viabilities caused by FUT8-AS1 silencing (Figure 8A, compared with Figure 3C). EdU incorporation assays indicated that MEK162 treatment abolished the accelerated cell proliferation rate caused by FUT8-AS1 silencing (Figure 8B, compared with Figure 3E). Transwell migration assays indicated that MEK162 treatment abolished the increased migrated cell number caused by FUT8-AS1 silencing (Figure 8C, compared with Figure 3F). Transwell invasion assays indicated that MEK162 treatment abolished the increased invasive cell number caused by FUT8-AS1 silencing (Figure 8D, compared with Figure 3G). Thus, these findings suggested that the roles of FUT8-AS1 in melanoma were dependent on the regulation of miR-145-5p/NRAS/MAPK signaling.
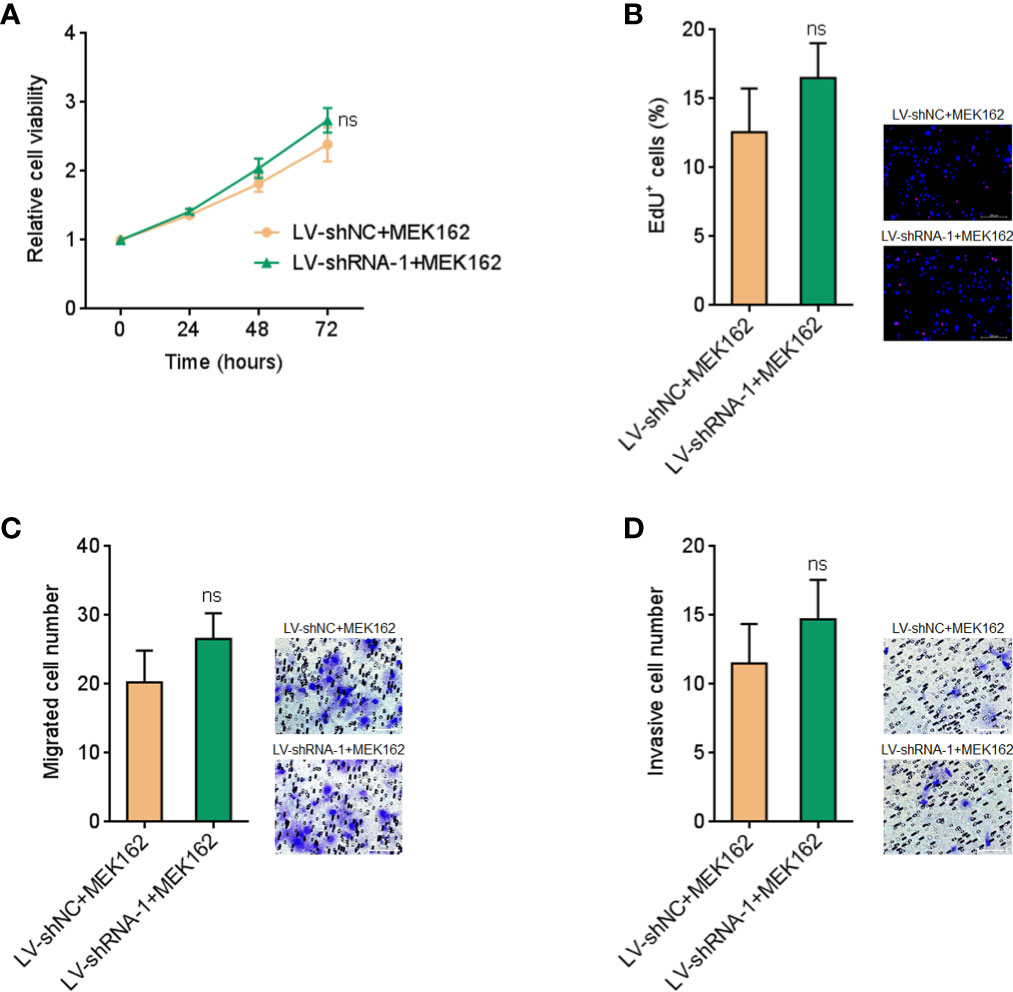
Figure 8 MEK1/2 inhibitor MEK162 abolished the oncogenic roles of FUT8-AS1 silencing in melanoma. (A) CHL-1 cells silencing FUT8-AS1 or control were treated with 1 µM MEK162. Then, cell viabilities of treated cells were determined by the Glo cell viability assay. (B) CHL-1 cells silencing FUT8-AS1 or control were treated with 1 µM MEK162. Then, cell proliferation of treated cells was determined by the EdU incorporation assay. The red color indicates EdU-positive nuclei. Scale bars, 200 µm. (C) CHL-1 cells silencing FUT8-AS1 or control were treated with 1 µM MEK162. Then, cell migration of treated cells was determined by the transwell migration assay. Scale bars, 100 µm. (D) CHL-1 cells silencing FUT8-AS1 or control were treated with 1 µM MEK162. Then, cell invasion of treated cells was determined by the transwell invasion assay. Scale bars, 100 µm. Data are presented as mean ± SD. ns, not significant, by two-tailed unpaired t test.
Great progressions in molecule targeted therapy and immunotherapy have highlighted the therapeutic strategies based on molecular aberrations in melanoma (43). Current targeted therapies mainly focus on aberrant proteins. Recently, increasing evidences have revealed that except for proteins, ncRNAs also demonstrate important roles in cancers and would be developed as therapeutic targets (4, 44, 45). In this study, via analyzing public TCGA dataset, we identified a novel melanoma correlated lncRNA FUT8-AS1. FUT8-AS1 is locating at chromosome 14q23.3 and has only one exon. The knowledge of FUT8-AS1 is lacking with only one report showing that FUT8-AS1 is correlated with outcome of a subpopulation of glioblastoma multiforme patients (46). In this study, we further investigated the expression, clinical significance, roles, and mechanism of action of FUT8-AS1 in melanoma.
FUT8-AS1 is reduced in melanoma tissues compared with benign nevi. Reduced expression of FUT8-AS1 is correlated with thickness, ulceration, metastasis, and inferior overall survival of melanomas. Gain and loss-of-function assays demonstrated that FUT8-AS1 suppresses melanoma cell proliferation, migration, and invasion in vitro. Furthermore, FUT8-AS1 represses melanoma growth and metastasis in vivo. Thus, FUT8-AS1 is identified as a tumor suppressive lncRNA in melanoma. FUT8-AS1 would also be a potential prognostic biomarker for melanoma. Enhancing FUT8-AS1 and/or its effects in downstream signaling would be potential therapeutic strategies for melanoma.
In addition, we identified a relative novel molecular mechanism of FUT8-AS1, which is the promotion of miR-145-5p biogenesis via competitively binding NF90. Previous reports about the effects of lncRNAs on miRNAs are mainly the competitive binding of miRNAs by lncRNAs, which further relieve the repressive roles of miRNAs on their mRNA targets (47). lncRNA-ATB is a classic example, which competitively binds miR-200s and upregulates miR-200s targets ZEB1 and ZEB2 in hepatocellular carcinoma (48). In our previous report, we have demonstrated that lncRNA MHENCR competitively binds miR-425 and miR-489, and therefore upregulates their targets IGF1 and SPIN1 in melanoma (39). In this study, we found that FUT8-AS1 does not bind miR-145-5p, but promotes miR-145-5p biogenesis.
Combining bioinformatic prediction and experimental verification, we found that FUT8-AS1 directly binds NF90. NF90 is a well-characterized dual strand RNA binding protein. Intriguingly, our previously identified tumor suppressive miR-145-5p was recently reported to be downstream target of NF90 by Zhuang et al. (42). NF90 directly binds pri-miR-145 and represses the biogenesis of mature miR-145-5p (42). In this study, we found that via competitively binding NF90, FUT8-AS1 represses the binding of NF90 to pri-miR-145. Therefore, FUT8-AS1 represses the effects of NF90 on miR-145-5p biogenesis. Lastly, FUT8-AS1 downregulates pri-miR-145 and upregulates mature miR-145-5p. The significant positive correlation between FUT8-AS1 and miR-145-5p expression in melanoma tissues supports the positive modulation of miR-145-5p by FUT8-AS1 in human melanoma. Furthermore, functional rescue assays showed that miR-145-5p inhibitors reverse the tumor suppressive roles of FUT8-AS1 in melanoma, supporting miR-145-5p as an important downstream target of FUT8-AS1. Several reports, including our own, have identified NRAS as a critical downstream target of miR-145-5p (29, 49, 50). Via targeting NRAS, miR-145-5p further represses MAPK signaling. MAPK signaling has been frequently shown to be aberrantly activated in melanoma (3). Therefore, in this study we further investigated the effects of FUT8-AS1 on NRAS/MAPK signaling via regulating miR-145-5p. In line with expectation, our results showed that FUT8-AS1 represses the expression of NRAS and further represses MAPK signaling in melanoma. MAPK signaling inhibitor MEK162 abolished the oncogenic roles of FUT8-AS1 silencing in melanoma, which further supports that miR-145-5p/NRAS/MAPK signaling is the critical mediator of the roles of FUT8-AS1 in melanoma. Therefore, we identified relative novel mechanism of action of lncRNA and also the detailed downstream molecular signaling cascade of FUT8-AS1 in melanoma.
In summary, our findings suggested FUT8-AS1 as an important tumor suppressive lncRNA in melanoma, which represses melanoma cell proliferation, migration, and invasion in vitro, and melanoma tumor growth and metastasis in vivo. FUT8-AS1 exerts its tumor suppressive roles via binding NF90, relieving the repressing roles of NF90 on miR-145-5p biogenesis, upregulating miR-145-5p, repressing NRAS, and lastly repressing MAPK signaling in melanoma. Furthermore, our data and TCGA data both suggested that FUT8-AS1 is correlated with inferior outcome of melanoma patients. Overall, these data provide novel molecular driver of melanoma progression and potential target for melanoma outcome prediction and therapy.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by the Review Board of the 969th Hospital of PLA. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Review Board of the 969th Hospital of PLA.
Study concept and design: X-JC, SL, LY. Acquisition of data: SL, D-MH, D-ZH, W-JS, X-CZ. Analysis and interpretation of data: X-JC, SL, D-MH, J-QL, LY. Drafting and editing of the manuscript: X-JC, SL. All authors contributed to the article and approved the submitted version.
This work was supported by the National Nature Science Foundation of China (81971854).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.586085/full#supplementary-material
Supplementary Figure 1 | FUT8-AS1 is a noncoding RNA. (A) The coding potential of FUT8-AS1 was calculated by the Coding Potential Calculator (CPC). GAPDH and HOTAIR were used as coding and noncoding RNA controls, respectively. (B) The coding potential of FUT8-AS1 was calculated by the Coding Potential Assessment Tool (CPAT). GAPDH and HOTAIR were used as coding and noncoding RNA controls, respectively.
Supplementary Figure 2 | FUT8-AS1 has a conserved NF90 binding sequence. The structure of FUT8-AS1 was predicted by the mfold web server (http://unafold.rna.albany.edu/?q=mfold). Red line indicates the conserved NF90 binding sequence (5′-CUGUU-3′, 452-456nt of FUT8-AS1).
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin (2020) 70:7–30. doi: 10.3322/caac.21590
2. Gide TN, Quek C, Menzies AM, Tasker AT, Shang P, Holst J, et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell (2019) 35:238–55.e6. doi: 10.1016/j.ccell.2019.01.003
3. Coppe JP, Mori M, Pan B, Yau C, Wolf DM, Ruiz-Saenz A, et al. Mapping phospho-catalytic dependencies of therapy-resistant tumours reveals actionable vulnerabilities. Nat Cell Biol (2019) 21:778–90. doi: 10.1038/s41556-019-0328-z
4. Esposito R, Bosch N, Lanzos A, Polidori T, Pulido-Quetglas C, Johnson R. Hacking the Cancer Genome: Profiling Therapeutically Actionable Long Non-coding RNAs Using CRISPR-Cas9 Screening. Cancer Cell (2019) 35:545–57. doi: 10.1016/j.ccell.2019.01.019
5. Hanniford D, Ulloa-Morales A, Karz A, Berzoti-Coelho MG, Moubarak RS, Sanchez-Sendra B, et al. Epigenetic Silencing of CDR1as Drives IGF2BP3-Mediated Melanoma Invasion and Metastasis. Cancer Cell (2020) 37:55–70.e15. doi: 10.1016/j.ccell.2019.12.007
6. Li JK, Chen C, Liu JY, Shi JZ, Liu SP, Liu B, et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer (2017) 16:111. doi: 10.1186/s12943-017-0681-0
7. Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y, et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell (2018) 33:706–20.e9. doi: 10.1016/j.ccell.2018.03.006
8. Mondal T, Juvvuna PK, Kirkeby A, Mitra S, Kosalai ST, Traxler L, et al. Sense-Antisense lncRNA Pair Encoded by Locus 6p22.3 Determines Neuroblastoma Susceptibility via the USP36-CHD7-SOX9 Regulatory Axis. Cancer Cell (2018) 33:417–34.e7. doi: 10.1016/j.ccell.2018.01.020
9. Hu WL, Jin L, Xu A, Wang YF, Thorne RF, Zhang XD, et al. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol (2018) 20:492–502. doi: 10.1038/s41556-018-0066-7
10. Yuan JH, Liu XN, Wang TT, Pan W, Tao QF, Zhou WP, et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol (2017) 19:820–32. doi: 10.1038/ncb3538
11. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet (2014) 15:7–21. doi: 10.1038/nrg3606
12. Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigo R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet (2018) 19:535–48. doi: 10.1038/s41576-018-0017-y
13. Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov (2013) 12:847–65. doi: 10.1038/nrd4140
14. Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol (2019) 20:5–20. doi: 10.1038/s41580-018-0059-1
15. Mo S, Zhang L, Dai W, Han L, Wang R, Xiang W, et al. Antisense lncRNA LDLRAD4-AS1 promotes metastasis by decreasing the expression of LDLRAD4 and predicts a poor prognosis in colorectal cancer. Cell Death Dis (2020) 11:155. doi: 10.1038/s41419-020-2338-y
16. Wu C, Wei Y, Zhu Y, Li K, Zhu Y, Zhao Y, et al. Identification of cancer-related potential biomarkers based on lncRNA-pseudogene-mRNA competitive networks. FEBS Lett (2018) 592:973–86. doi: 10.1002/1873-3468.13011
17. Dika E, Riefolo M, Porcellini E, Broseghini E, Ribero S, Senetta R, et al. Defining the Prognostic Role of MicroRNAs in Cutaneous Melanoma. J Invest Dermatol (2020) 140:2260–67. doi: 10.1016/j.jid.2020.03.949
18. Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J (2016) 283:3739–54. doi: 10.1111/febs.13839
19. Huang Z, Zhou JK, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer (2020) 19:77. doi: 10.1186/s12943-020-01188-4
20. Keshavarz M, Asadi MH. Long non-coding RNA ES1 controls the proliferation of breast cancer cells by regulating the Oct4/Sox2/miR-302 axis. FEBS J (2019) 286:2611–23. doi: 10.1111/febs.14825
21. Grzywa TM, Klicka K, Paskal W, Dudkiewicz J, Wejman J, Pyzlak M, et al. miR-410-3p is induced by vemurafenib via ER stress and contributes to resistance to BRAF inhibitor in melanoma. PLoS One (2020) 15:e0234707. doi: 10.1371/journal.pone.0234707
22. Yang Z, Liao B, Xiang X, Ke S. miR-21-5p promotes cell proliferation and G1/S transition in melanoma by targeting CDKN2C. FEBS Open Bio (2020) 10:752–60. doi: 10.1002/2211-5463.12819
23. Gerloff D, Lutzkendorf J, Moritz RKC, Wersig T, Mader K, Muller LP, et al. Melanoma-Derived Exosomal miR-125b-5p Educates Tumor Associated Macrophages (TAMs) by Targeting Lysosomal Acid Lipase A (LIPA). Cancers (Basel) (2020) 12:464. doi: 10.3390/cancers12020464
24. Tupone MG, D’Aguanno S, Di Martile M, Valentini E, Desideri M, Trisciuoglio D, et al. microRNA-378a-5p iS a novel positive regulator of melanoma progression. Oncogenesis (2020) 9:22. doi: 10.1038/s41389-020-0203-6
25. Zhao G, Wei Z, Guo Y. MicroRNA-107 is a novel tumor suppressor targeting POU3F2 in melanoma. Biol Res (2020) 53:11. doi: 10.1186/s40659-020-00278-3
26. Zhao G, Yin Y, Zhao B. miR-140-5p is negatively correlated with proliferation, invasion, and tumorigenesis in malignant melanoma by targeting SOX4 via the Wnt/beta-catenin and NF-kappaB cascades. J Cell Physiol (2020) 235:2161–70. doi: 10.1002/jcp.29122
27. Luan W, Qian Y, Ni X, Bu X, Xia Y, Wang J, et al. miR-204-5p acts as a tumor suppressor by targeting matrix metalloproteinases-9 and B-cell lymphoma-2 in malignant melanoma. Onco Targets Ther (2017) 10:1237–46. doi: 10.2147/OTT.S128819
28. Meng P, Ma S, Liu Y, Zhao N, Xia X, Bi Y. MiR-128-3p targets JAG1 to inhibit cell proliferation and invasion in melanoma. G Ital Dermatol Venereol (2020). doi: 10.23736/S0392-0488.19.06504-0
29. Liu S, Gao G, Yan D, Chen X, Yao X, Guo S, et al. Effects of miR-145-5p through NRAS on the cell proliferation, apoptosis, migration, and invasion in melanoma by inhibiting MAPK and PI3K/AKT pathways. Cancer Med (2017) 6:819–33. doi: 10.1002/cam4.1030
30. Kobayashi H, Tomari Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim Biophys Acta (2016) 1859:71–81. doi: 10.1016/j.bbagrm.2015.08.007
31. Liu N, Liu Z, Liu X, Chen H. Comprehensive Analysis of a Competing Endogenous RNA Network Identifies Seven-lncRNA Signature as a Prognostic Biomarker for Melanoma. Front Oncol (2019) 9:935. doi: 10.3389/fonc.2019.00935
32. Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature (2016) 531:518–22. doi: 10.1038/nature17161
33. Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ, et al. The lncRNA SLNCR1 Mediates Melanoma Invasion through a Conserved SRA1-like Region. Cell Rep (2016) 15:2025–37. doi: 10.1016/j.celrep.2016.04.018
34. Sang B, Zhang YY, Guo ST, Kong LF, Cheng Q, Liu GZ, et al. Dual functions for OVAAL in initiation of RAF/MEK/ERK prosurvival signals and evasion of p27-mediated cellular senescence. Proc Natl Acad Sci USA (2018) 115:E11661–70. doi: 10.1073/pnas.1805950115
35. Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature (2017) 548:343–6. doi: 10.1038/nature23451
36. Hosono Y, Niknafs YS, Prensner JR, Iyer MK, Dhanasekaran SM, Mehra R, et al. Oncogenic Role of THOR, a Conserved Cancer/Testis Long Non-coding RNA. Cell (2017) 171:1559–72.e20. doi: 10.1016/j.cell.2017.11.040
37. Chen X, Gao G, Liu S, Yu L, Yan D, Yao X, et al. Long Noncoding RNA PVT1 as a Novel Diagnostic Biomarker and Therapeutic Target for Melanoma. BioMed Res Int (2017) 2017:7038579. doi: 10.1155/2017/7038579
38. Chen X, Liu S, Zhao X, Ma X, Gao G, Yu L, et al. Long noncoding RNA ILF3-AS1 promotes cell proliferation, migration, and invasion via negatively regulating miR-200b/a/429 in melanoma. Biosci Rep (2017) 37:BSR20171031. doi: 10.1042/BSR20171031
39. Chen X, Dong H, Liu S, Yu L, Yan D, Yao X, et al. Long noncoding RNA MHENCR promotes melanoma progression via regulating miR-425/489-mediated PI3K-Akt pathway. Am J Transl Res (2017) 9:90–102.
40. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet (2015) 47:199–208. doi: 10.1038/ng.3192
41. Luo K, Geng J, Zhang Q, Xu Y, Zhou X, Huang Z, et al. LncRNA CASC9 interacts with CPSF3 to regulate TGF-beta signaling in colorectal cancer. J Exp Clin Cancer Res (2019) 38:249. doi: 10.1186/s13046-019-1263-3
42. Zhuang J, Shen L, Yang L, Huang X, Lu Q, Cui Y, et al. TGFbeta1 Promotes Gemcitabine Resistance through Regulating the LncRNA-LET/NF90/miR-145 Signaling Axis in Bladder Cancer. Theranostics (2017) 7:3053–67. doi: 10.7150/thno.19542
43. Etxeberria I, Bolanos E, Quetglas J II, Gros A, Villanueva A, Palomero J, et al. Intratumor Adoptive Transfer of IL-12 mRNA Transiently Engineered Antitumor CD8(+) T Cells. Cancer Cell (2019) 36:613–29.e7. doi: 10.1016/j.ccell.2019.10.006
44. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell (2016) 29:452–63. doi: 10.1016/j.ccell.2016.03.010
45. Yuan JH, Yang F, Chen BF, Lu Z, Huo XS, Zhou WP, et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology (2011) 54:2025–35. doi: 10.1002/hep.24606
46. Shergalis A, Bankhead A,3, Luesakul U, Muangsin N, Neamati N. Current Challenges and Opportunities in Treating Glioblastoma. Pharmacol Rev (2018) 70:412–45. doi: 10.1124/pr.117.014944
47. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell (2011) 146:353–8. doi: 10.1016/j.cell.2011.07.014
48. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell (2014) 25:666–81. doi: 10.1016/j.ccr.2014.03.010
49. Yin Y, Yan ZP, Lu NN, Xu Q, He J, Qian X, et al. Downregulation of miR-145 associated with cancer progression and VEGF transcriptional activation by targeting N-RAS and IRS1. Biochim Biophys Acta (2013) 1829:239–47. doi: 10.1016/j.bbagrm.2012.11.006
Keywords: FUT8-AS1, melanoma, malignancy, NF90, microRNA biogenesis, MAPK signaling
Citation: Chen X-j, Liu S, Han D-m, Han D-z, Sun W-j, Zhao X-c, Liang J-q and Yu L (2021) FUT8-AS1 Inhibits the Malignancy of Melanoma Through Promoting miR-145-5p Biogenesis and Suppressing NRAS/MAPK Signaling. Front. Oncol. 10:586085. doi: 10.3389/fonc.2020.586085
Received: 22 July 2020; Accepted: 20 November 2020;
Published: 19 May 2021.
Edited by:
Luisa Lanfrancone, European Institute of Oncology (IEO), ItalyReviewed by:
Mattia Riefolo, University of Bologna, ItalyCopyright © 2021 Chen, Liu, Han, Han, Sun, Zhao, Liang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang-jun Chen, Y3hqXzI1M0AxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.