
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 30 October 2020
Sec. Hematologic Malignancies
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.584766
This article is part of the Research Topic Innovative Multi-Disciplinary Approaches for Precision Studies in Leukemia View all 11 articles
 Cong Lu1,2,3
Cong Lu1,2,3 Jiang Zhu1,2,3
Jiang Zhu1,2,3 Xiangjun Chen1,2,3
Xiangjun Chen1,2,3 Yanjie Hu1,2,3
Yanjie Hu1,2,3 Wei Xie1,2,3
Wei Xie1,2,3 Junxia Yao1,2,3*
Junxia Yao1,2,3* Shiang Huang1,2,3*
Shiang Huang1,2,3*The role of CXC chemokine receptors in tumors has been an increasingly researched focus in recent years. However, significant prognostic values of CXCR members in acute myeloid leukemia are yet to be explored profoundly. In this study, we firstly made an analysis of the relationship of CXCR family members and AML using samples from TCGA. Our results suggested that transcriptional expressions of CXCRs serve an important role in AML. CXCR transcript expressions, except CXCR1 expression, were significantly increased in AML. It displayed the expression pattern of CXCR members in different AML subtypes according to FAB classification. The correlations of CXCR transcript expression with different genotypes and karyotypes were also present. High CXCR2 expression was found to have a significantly worse prognosis compared with that of low CXCR2 expression, and CXCR2 was also found to be an independent prognostic factor. We also established a CXCR signature to identify high-risk subgroups of patients with AML. It was an independent prognostic factor and could become a powerful method to predict the survival rate of patients.
Acute myelocytic leukemia (AML) is a malignant clonal disease of hematopoietic stem cells, which is characterized by a block of leukemic blasts in the bone marrow and other tissues (1). Adult patients could attain complete remission (CR) after standardized chemotherapy treatment; however, the short duration of CR is still an urgent problem for clinician due to a high relapse rate (2). Recent studies suggest that the interaction of the CXC chemokine receptor (CXCR) members and their ligands as well as the complex regulatory network of them take an effect on certain tumor-related processes (3) including activation, proliferation and invasion of leukemic cells (4).
The CXCR family consists of proteins CXCR1–7 (5). CXCR1 and CXCR2 exhibit a high affinity toward a common ligand IL-8. This receptor-ligand interaction induces leukocyte chemotaxis, cell proliferation, and migration and is critical for inflammation and metastasis of tumors (6–8). The CXC chemokine ligand (CXCL) 9, 10, 11/CXCR3 axis regulates tumor differentiation and activation and the paracrine signal transduction for immune cell development (9, 10). CXCR4, expressed on the surface of hematopoietic stem cells and leukemia blast cells, is activated by CXCL12, and it participates in leukemia cell proliferation and infiltration, as well as in conferring resistance to chemotherapy drugs (11–13). CXCL13 is the ligand for CXCR5, also known as the Burkitt lymphoma receptor 1 (BLR1), and the CXCL13/CXCR5 axis is necessary for B cell homing to lymph node follicles and for the production of immunoglobulin, which coordinates the humoral immunity of the body (14). CXCL16 selectively binds to its sole receptor, CXCR6, and is mainly expressed in natural killer, CD8+ T, and CD4+ T cells. CXCL16/CXCR6 binding plays an essential role in cell adhesion and activation of the immune response (15). CXCR7 is a receptor of CXCL12; however, it is unable to mediate G-protein activation to directly induce cell migration and is considered to be an atypical chemokine receptor (ACKR3). Ligand CXCL-11 or CXCL-12, when bound to CXCR7, can rapidly mediate ligand internalization and degradation (16).
Original expression and clinical data of CXCR family members in AML were down from The Cancer Genome Atlas (TCGA) database (http://Cancegenome.nih.gov/). They were divided into eight subtypes (M0-M7) according to the French-American and British (FAB) classification of AML. The CXCR family mRNA expression level, gender, age, survival status, survival time, risk stratification, chromosome karyotype analysis, gene detection, and other clinical data were extracted from AML patients in TCGA. This study was approved by the Ethics Committee at hospital. Validation of mRNA prognostic power in this study used expression data of TARGET database.
Differences of the expression levels of CXCRs between AML patients and normal tissues were obtained from the GEPIA website (http://gepia.cancer-pku.cn/). the Gene Expression Profiling Interactive Analysis server (GEPIA) is a newly developed interactive web server (17) for analyzing the RNA sequencing expression data of thousands of tumor and normal samples from the TCGA and the GTEx projects using a standard processing pipeline (18). Since AML samples from TCGA database were all tumor samples, GEPIA matches normal samples from the GTEs database.
cBioPortal (http://www.cbioportal.org) is a multi-functional open network platform, which is a set of tools that propose data mining, data integration, and visualization based on the TCGA database. The intuitive web interface enables the complex cancer genome profiles to be integrated and explored by clinicians (19).
STRING (https://string-db.org/) is a database of known and predicted protein-protein interactions networks designed for protein functional enrichment analysis. The interactions include direct (physical) and indirect (functional) associations. They stem from computational prediction, from knowledge transfer between organisms, and from interactions aggregated from other (primary) databases. The STRING database currently covers 24,584,628 proteins from 5,090 organisms (20, 21). The Search Tool by choosing Multiple Proteins was used to construct the PPI networks of CXCR member
SPSS 19.0 software (SPSS, Chicago, IL) and R language (3.6.3) were used for the statistical analysis. Non-parametric tests were used to compare two or more independent sample sets of data. The patient samples were divided into high- and low- expression groups based on their median expression value. Kaplan-Meier survival analysis and log-rank tests were conducted to analyze the overall survival (OS) using survival R package. Univariate and multivariate analyses were performed on the categorized data to identify independent predictors of outcome. P values below 0.05 were considered significant. We chose the candidate genes and they were subjected to multiple proportional risk regression to construct a gene signature as a risk score model. The risk score model included the expression level of mRNA for each optimal prognosis, with weights determined by the estimated regression coefficients of their multivariate Cox regression model, as shown below
We compared the transcript expressions of the CXCRs in patients with AML with those of normal control subjects, using server GEPIA (22). 173 patients with AML from TCGA and 70 normal subjects from the Genotype-Tissue Expression (GTEx) portal were displayed on the website. Remarkably, all CXCR transcripts, except for CXCR1, showed a higher expression of mRNA in patients with AML, in comparison to that in normal subjects (Figure 1A). Of note, the expression of the CXCR4 transcript was much higher in the AML group. The decreasing order of expression levels was as follows: CXCR4, CXCR7, CXCR2, CXCR6, CXCR5, CXCR3, and CXCR1. Although, CXCR2 expression is down-regulated in samples from most types of cancers compared to samples from normal subjects, it was found to be upregulated in AML (Figure 1B).
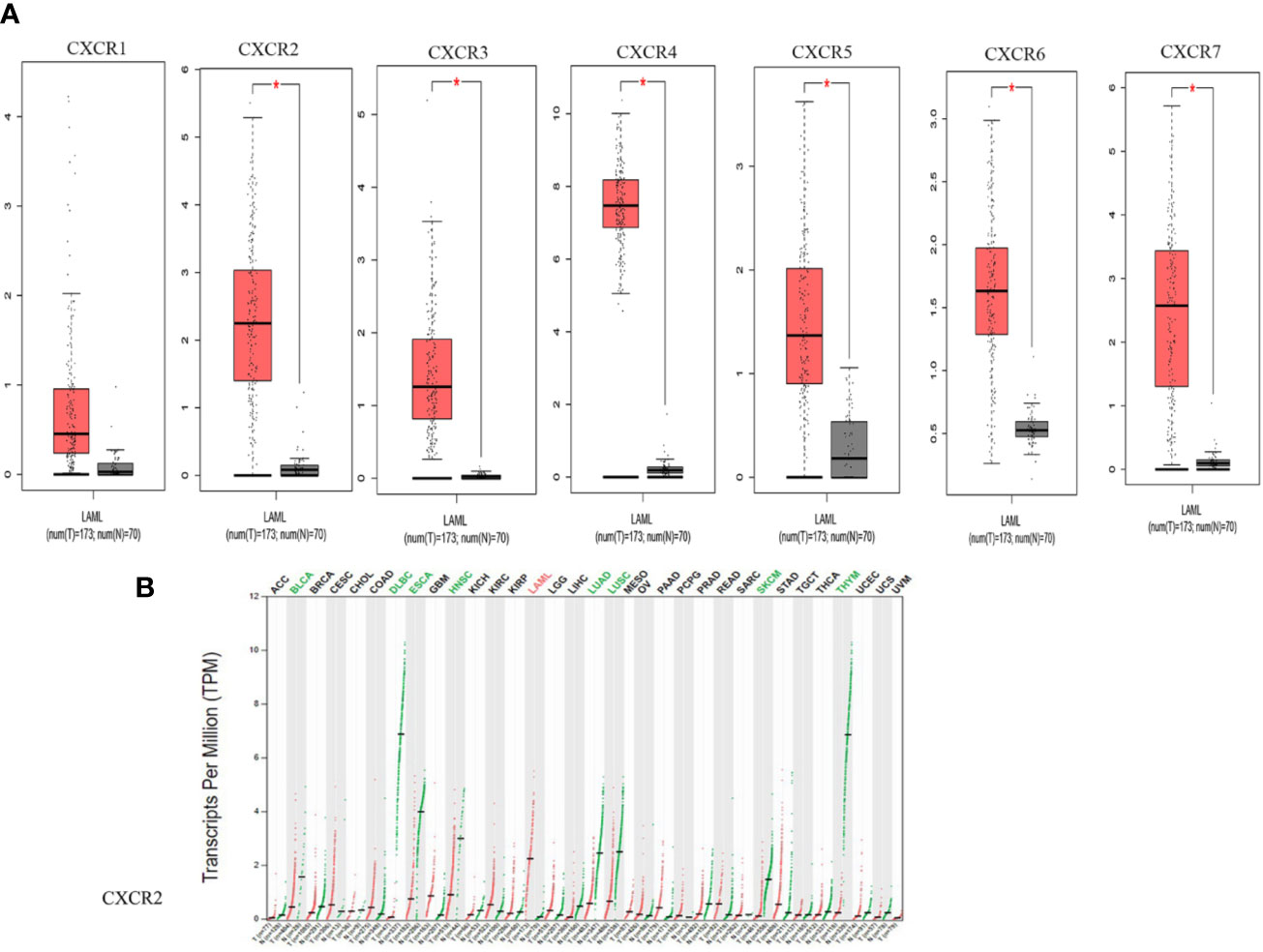
Figure 1 The mRNA expression from GEPIA. (A) CXCRs between AML and normal samples. (*P < 0.05). (B) CXCR2 expression profile across all tumor samples and paired normal tissues.
We analyzed transcript expression of each CXCR in the M0 to M5 FAB subtypes of 151 patients with AML; two cases of AML-M6 and one case of AML-M7 were not analyzed. Higher expression of CXCR1 transcripts was found more frequently in the AML-M5 subtype than in other subtypes (P = 0.0038). CXCR2 showed the highest expression in AML-M5, and the lowest expression was seen in AML-M3 (P < 0.0001). CXCR3 transcripts showed a higher expression in AML-M3, as compared to that in the other subtypes (P = 0.013). AML-M4/M5 patients showed an increased expression of CXCR4 transcripts compared to that in other subtypes, with the highest expression found in AML-M5 (P = 0.003). Moreover, no significant difference was found between the expressions of CXCR5 and CXCR6 among the different AML subtypes. CXCR7 showed the highest expression in AML-M0, while the lowest expression was observed in AML-M5 patients (P = 0.027) (Figure 2).
To assess the relationship between CXCR transcript expression and the mutation status of traditional prognostic genes, we compared transcript expression of CXCRs in patients having mutations in FLT3, IDH1, and NPM1, with that in the mutation-negative control group. Patients with a FLT3 mutation showed a significant decrease in the expression of CXCR3, CXCR5, and CXCR6 transcripts compared to that in mutation-negative patients (Figure 3A). Similarly, patients with a NPM1 mutation showed a significant decrease in the expression CXCR3 and CXCR6 transcripts compared to that in mutation-negative patients (Figure 3B). As for the mutant IDH1, the CXCR4 transcript showed a significant increase in its expression in mutant IDH1 patients compared to that in the control group (P = 0.032, Figure 3C)
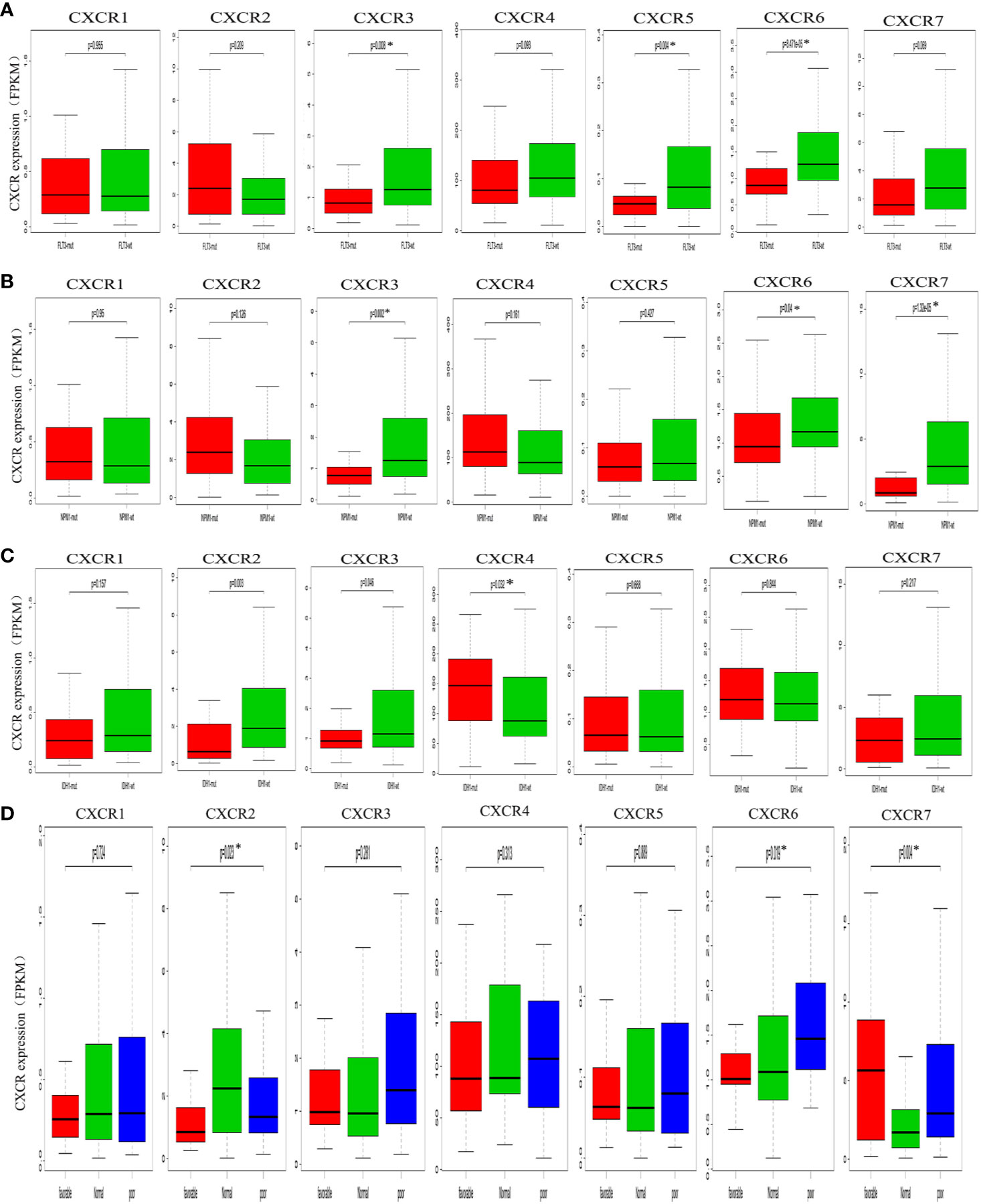
Figure 3 Analysis of prognostic gene expressions between the mutant group and the wild-type group. (A) FLT3 mutation. (B) NPM1 mutation. (C) IDH1 mutation. (D) cytogenetic karyotype.
We also analyzed the relationship between the CXCR transcript expression and cytogenetic karyotype, which is an important parameter for AML prognosis. Patients with the karyocyte t(15;17), t(8;21), and inv(16) were designated the favorable group; patients with normal karyocytes were designated the normal group; patients with the karyocyte 5q-/7q- or complex karyotype were designated the poor group. The results showed that patients of the favorable group had the highest CXCR7 expression (P = 0.004), those of the normal group had the highest CXCR2 transcript expression (P = 0.023), and those of the poor group had the highest CXCR6 transcript expression (P = 0.019, Figure 3D).
To assess the diagnostic and prognostic significance of CXCRs in AML, the correlation between expression of each CXCR transcript with AML risk stratification was estimated. The patients were divided into two groups according to their risk stratification. The intermediate-risk and high-risk patients were combined into one medium/high-risk group, and the expression of each CXCR transcript in the medium/high-risk group was compared to that of the low-risk group. The expression of CXCR1, CXCR2, and CXCR6 transcripts was significantly higher in patients of the medium/high-risk group than that in patients of the low risk group (P < 0.05) (Figure 4).
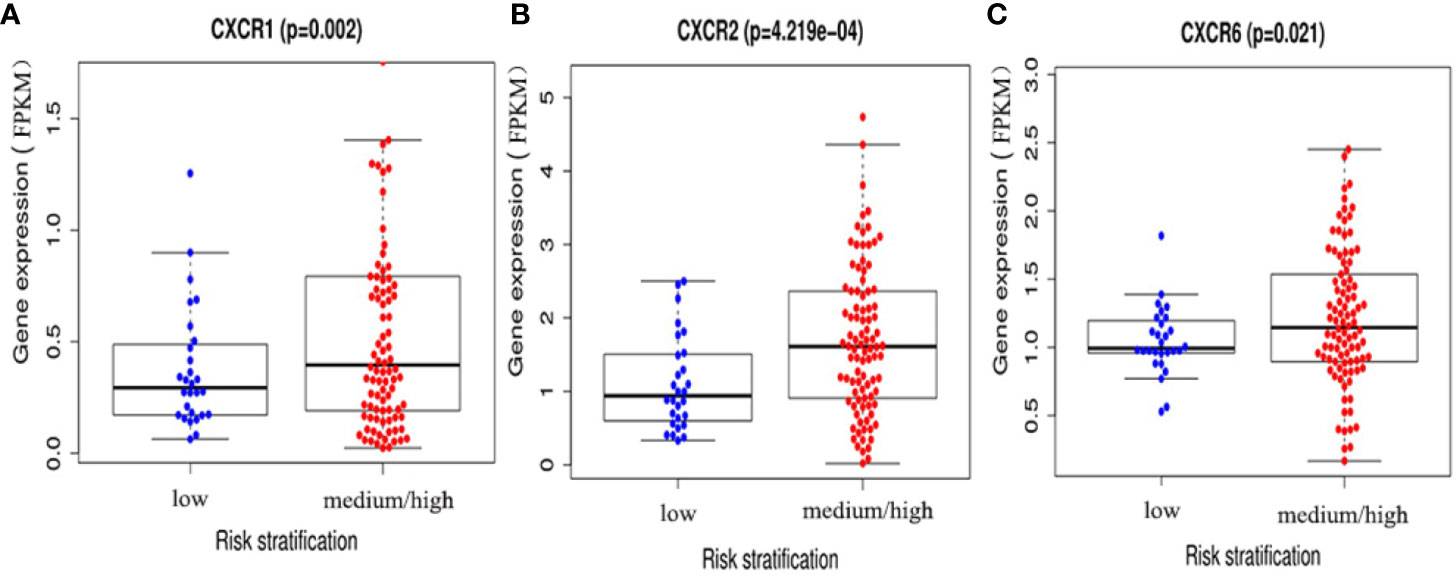
Figure 4 The expressions of (A) CXCR1, (B) CXCR2, and (C) CXCR6 between AML low and medium-high risk group.
The information of 122 patients with AML who are eligible for survival analysis was retrieved from TCGA database. The screening criteria for these patients as follows: 1) complete survival data present; 2) survival time longer than 30 days; 3) RNA-sequencing expression data present. The relevant clinical characteristics are shown in Table 1.
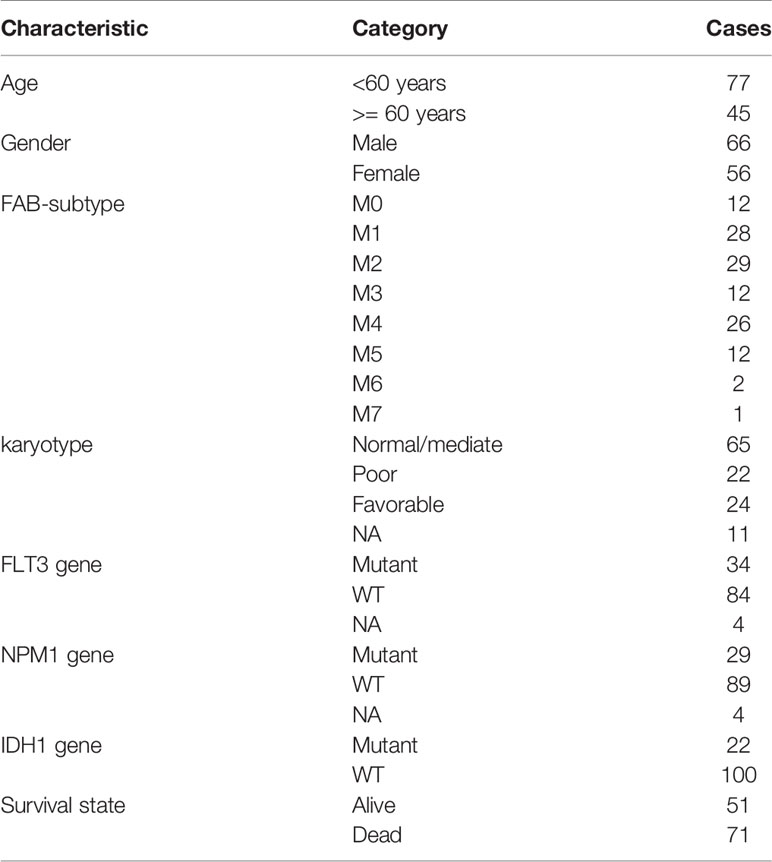
Table 1 Clinical characteristics of 122 AML patients enrolled from the TCGA database for survival analysis.
The prognostic value of CXCRs was estimated by analyzing the survival data and CXCR transcript levels in 122 patients from TCGA database. For each CXCR transcript, patients were divided into either the high- or low-expression group according to the median value of CXCR transcript expression. The median values of each CXCR mRNA expression (FPKM) were shown as follows: CXCR1 0.287, CXCR2 1.772, CXCR3 1.052, CXCR4 92.207, CXCR5 0.065, CXCR6 1.137, and CXCR7 2.400. Using Kaplan-Meier survival analysis, it was found that the survival time of the CXCR2 high-expression group was significantly shorter than that of the low-expression group (P = 0.029). However, no difference in survival times was observed between the high- and low-expression groups for other CXCR transcripts (Figure 5).
Furthermore, univariate and multivariate COX analyses were used to determine whether CXCR2 was an independent prognostic factor. Gender, age, white blood cell (WBC) counts, blast cell percentage, risk stratification, and FLT3 and IDH1 gene mutation were also analyzed to identify the independent factors affecting patient survival. Age, risk stratification, and CXCR2 expression were found to be the independent factors affecting patient survival (Table 2).
Given the increasing focus on the prognostic value of gene signatures, and the prognostic significance of CXCR transcripts in AML, the potential of CXCR signatures as a risk score model for AML was explored.
We chose CXCR1–7 as the candidate genes; these were subjected to multiple proportional risk regression analysis to construct a risk score model based on gene signatures. The risk score model was constructed as follows:
where exp. represents expression levels.
The forest plot for the gene signature model and its concordance index (C-index = 0.66, P = 4.216e-4) are shown in Figure 6. The patients were divided into high- and low-risk groups according to the median risk score, and it was found that the high-risk group displayed a significant reduction in OS compared to that of the low-risk group (P = 2.28e−04, Figure 7A). The heatmap of the core genes and risk-score of patients with AML in the two groups are shown in Figures 7B, C. Additionally, receiver operating characteristic (ROC) curves were built to evaluate the performance of the CXCR signature risk model at three time points (Figure 8). The AUC corresponding to 1, 2, and 3 years was 0.719, 0.705, and 0.684, using the Kaplan-Meier method.
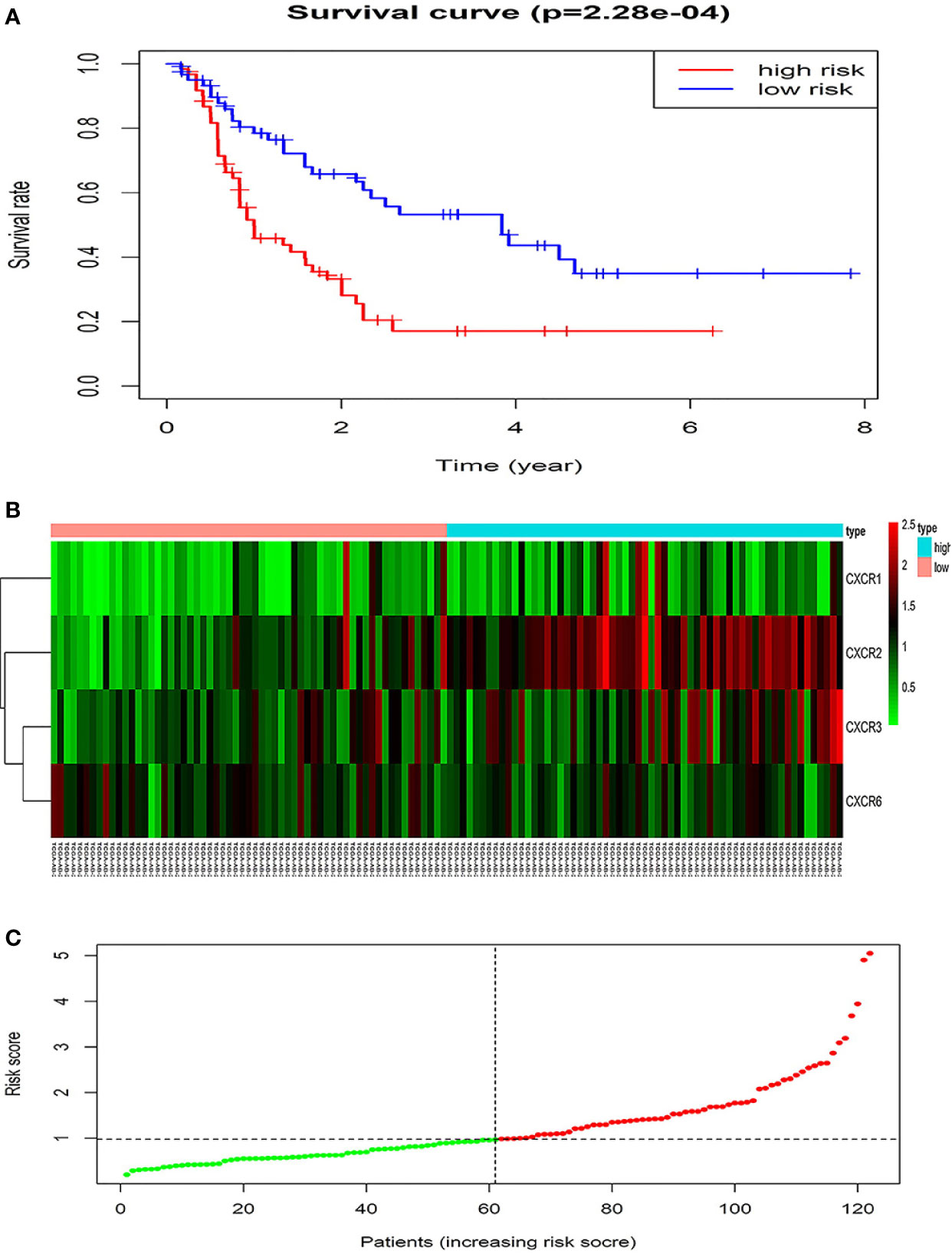
Figure 7 The prognostic values of CXCR signature. (A) The survival curves of low- and high- risk group. (B) The heatmap of gene signature. (C) The risk score of AML patients in two groups.
To determine if CXCR signature risk score could be used as an independent prognostic predictor for OS, we performed univariate and multivariate Cox regression analysis. The factors included were similar to those used in the previous regression analysis. The results showed that CXCR signature risk-score was an independent prognostic predictor of OS. Additionally, we found that age greater than 60 years and high-risk of AML (in the traditional risk stratification) were also independent predictors of a shortened OS (Table 3).
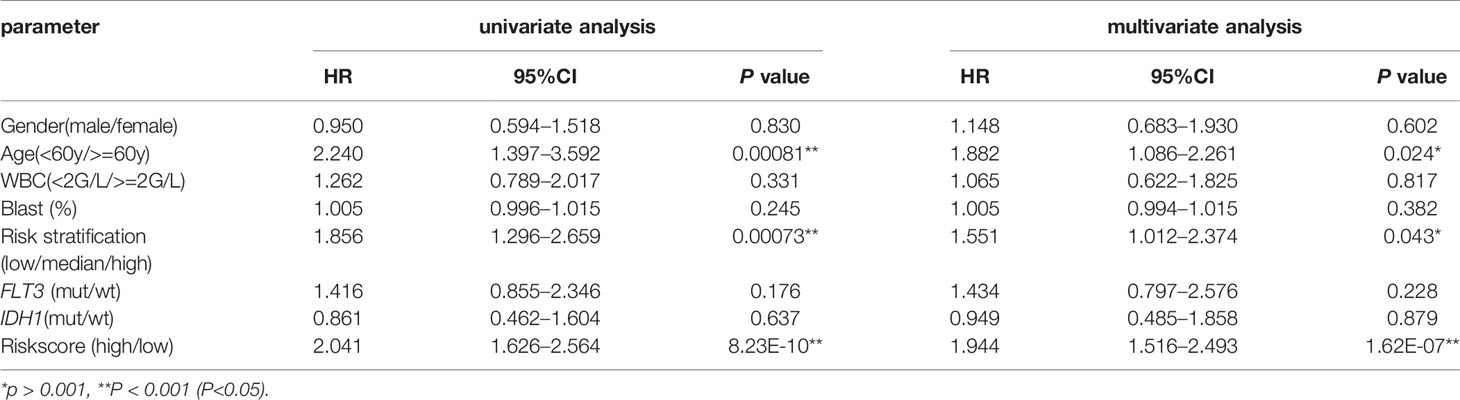
Table 3 Univariate and multivariate overall survival (OS) analysis of CXCR signature in AML patients.
We used the c-Bioportal online tool to estimate the correlation between the CXCRs (RNA Seq V2 RSEM); the Pearson’s correlations are listed in Table 4. The data showed that CXCR1 expression was associated with CXCR2 (r = 0.523). The expression of CXCR3 was found to be closely correlated to that of CXCR5 (r = 0.538) and CXCR6 (r = 0.412).
The STRING database was used to cluster and construct a network of CXCRs and the 47 most frequently altered neighboring genes (Figure 9A). Cytoscape software was used to screen the hub genes from the constructed network. CXCR4 and IL-10 were found to be important hub genes of the network (Figure 9B).
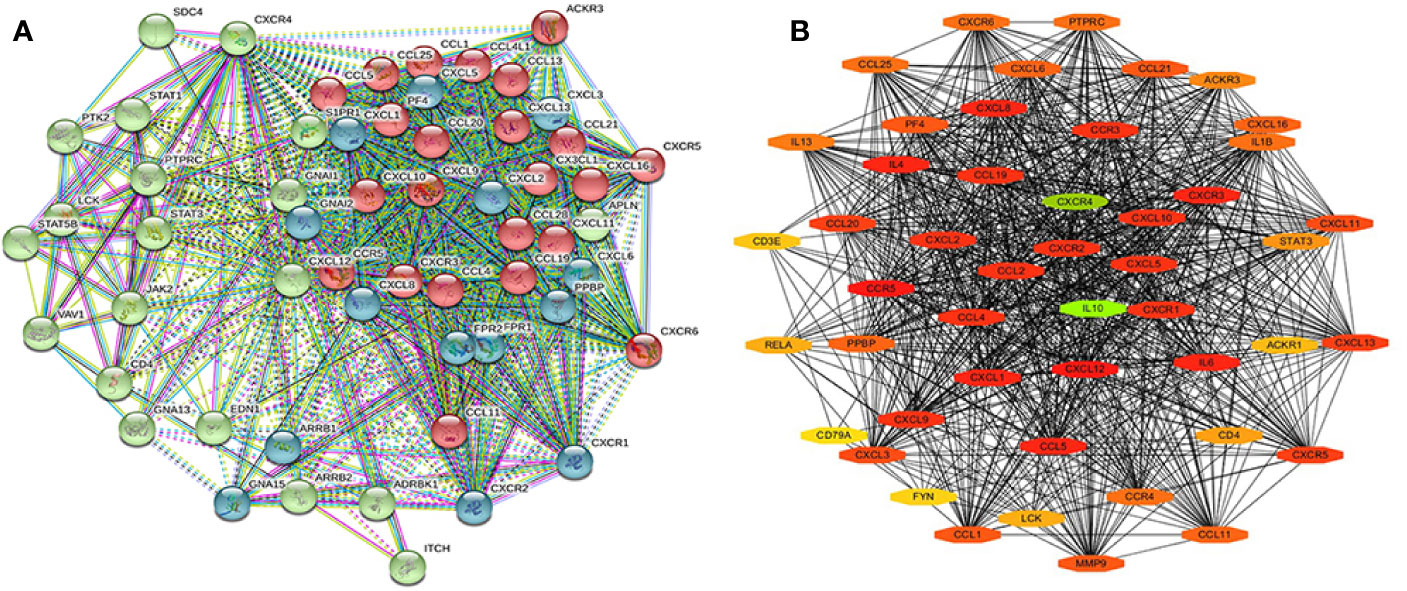
Figure 9 The PPI of CXCR family members and their neighbor genes (A) the PPI network analyzed by String. (B) The hub genes analyzed by Cytoscape.
Gene ontology (GO) enrichment analysis was used to predict the functional role of target host genes, and classify them into functional categories, including biological process (BP), cellular component (CC), and molecular function (MF). GO:0070098 (chemokine-mediated signaling pathway), GO:0030595 (leukocyte chemotaxis), GO:0060326 (cell chemotaxis), GO:0019221 (cytokine-mediated signaling pathway), and GO:0007166 (cell surface receptor signal pathway) were found to be significantly regulated by CXCRs and were classified under BP (Figure 10A) (23). These are well-known signal pathways involving cytokines and their receptors. Several important GO enriched BP pathways are associated with anti-inflammatory and immune responses. The top 20 GO terms for CCs and MF are shown in Figures 10B, C.
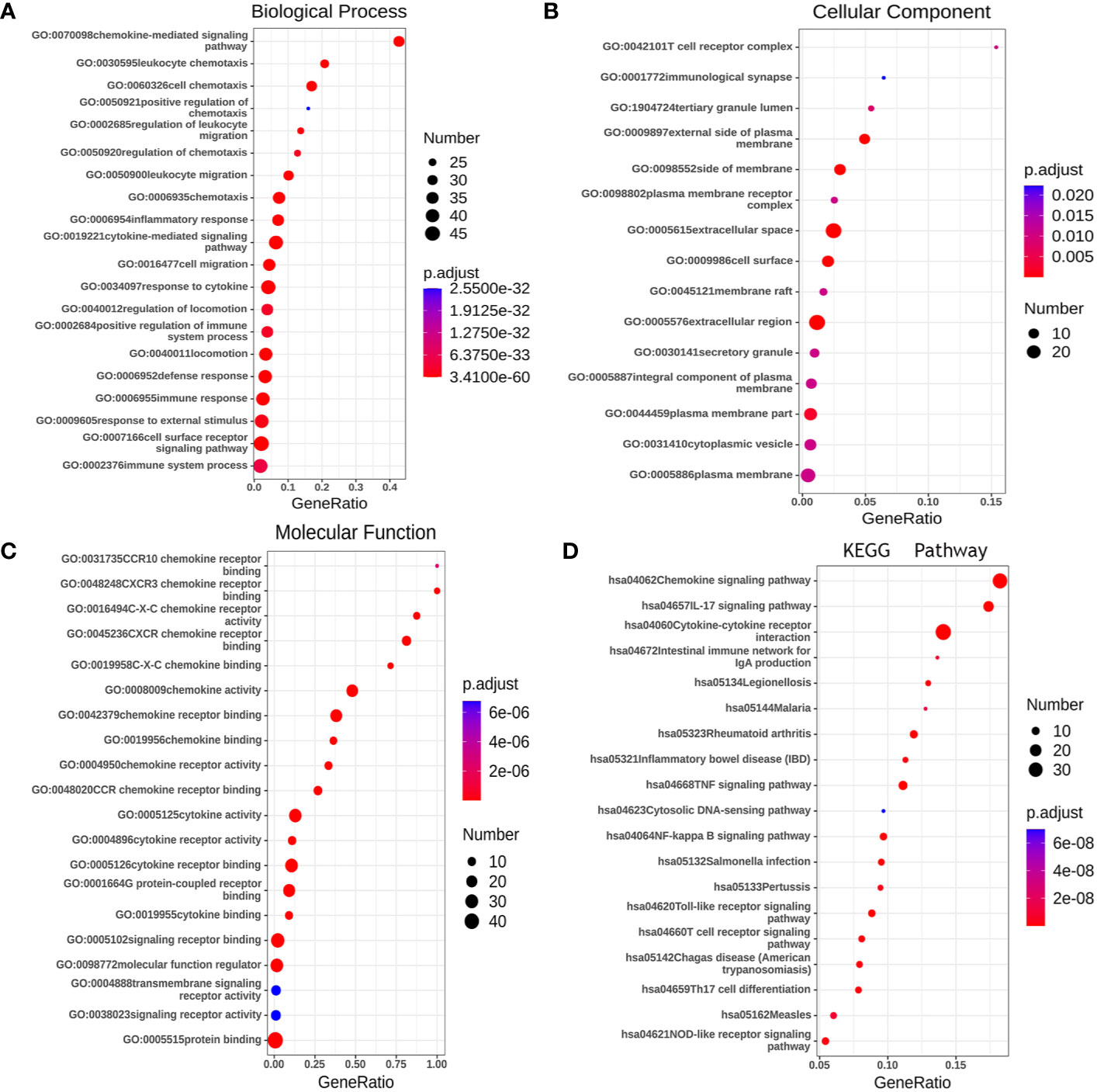
Figure 10 The functions of CXCRs and neighborhood genes predicted by GO and KEGG analysis by String. (A) Biological Process. (B) Cellular Component. (C) Molecular Function. (D) KEGG enrichment.
We used the Kyoto Encyclopedia of Genes and Genomes (KEGG) to functionally analyze the CXCRs, as well as frequently altered neighboring genes. The 20 most significantly CXCR-enriched pathways, using KEGG analysis in patients with AML, are shown in Figure 10D (P < 0.0001). KEGG pathway analysis showed that these CXCR genes are most significantly enriched in cytokine-cytokine receptor interaction, chemokine signaling pathway, IL-17 signaling pathway, TNF signaling pathway and NF-kappa B signaling, and Toll-like receptor signaling pathway, which is related to the process of tumor invasion and metastasis.
The prognostic value of CXCR2 and the CXCR signature risk score model were verified using the TARGET database. We selected 295 patients with AML from the TARGET database to analyze the prognostic value of CXCR genes and evaluate the risk score model. The median value of CXCR2 expression (counts) in this cohort was 65. It was observed that CXCR2 could act as an independent prognostic factor (P = 0.030, Figure 11A). We used the risk score model [Risk score = 0.65×(CXCR1exp.) + 2.01 × (CXCR2exp.) + 1.89 × (CXCR3exp.) + 0.65 × (CXCR6exp.)] established to calculate the risk score of each patient. The median value of CXCR signature risk score was 940.872. The risk score model could accurately categorize the patients in the TARGET database into high- and low-risk groups according to the median value (P = 4.079e-02, Figure 11B).
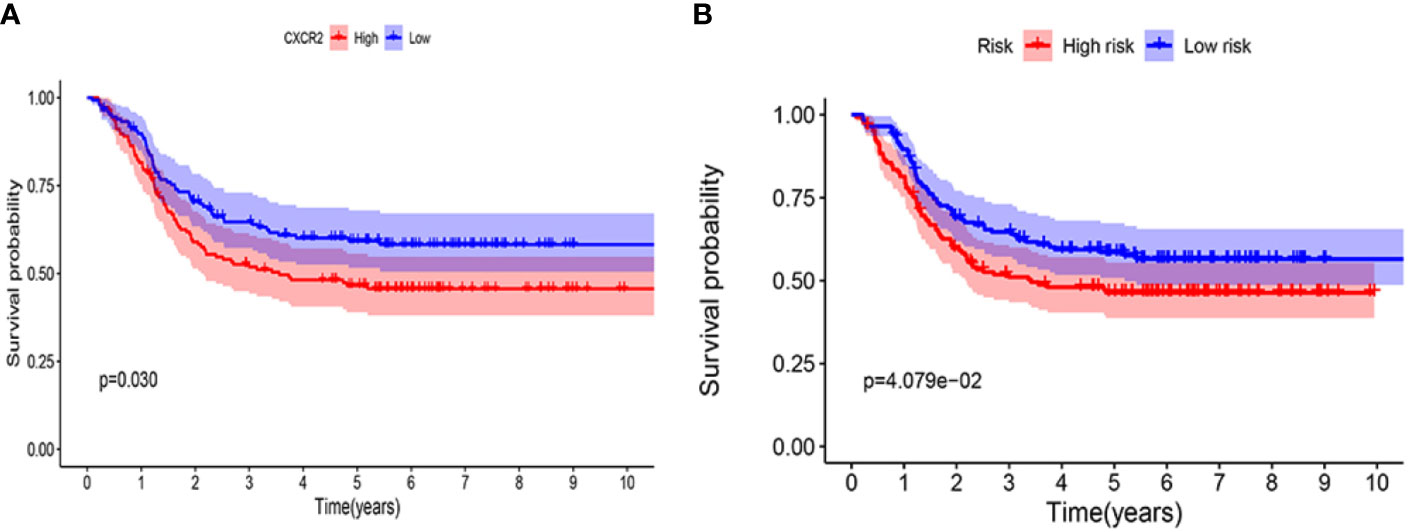
Figure 11 Kaplan-Meier survival curves in validation cohort using TARGET database. (A) CXCR2. (B) CXCR gene signature.
The CXCR family members have been reported to play important roles in different types of cancers (24, 25). Although CXCRs are known to be involved in tumorigenesis and prognosis of numerous cancers, there is a lack of detailed bioinformatic analyses of CXCRs in AML. This is the first study to investigate the transcript levels and prognostic value of CXCRs in AML. Additionally, we evaluated CXCR signatures as predictors of the risk of developing AML.
Our analysis revealed that all CXCRs, except CXCR1, were up-regulated in patients with AML compared to normal subjects. Notably, CXCR2 expression was found to significantly increase in AML but is known to be no apparent increase or even decreased in most of other tumors, indicating a critical role of CXCR2 in AML.
We also showed that CXCR4 expression was increased AML-M4 and AML-M5. This is consistent with the findings of our previous study, wherein we demonstrated that CXCR4 expression is higher in AML-M4 and AML-M5 than in subtypes M2 and M3 (26). Further, we found that the transcript expression of other CXCRs was significantly different in different AML subtypes. Very few studies have reported the correlation between the FAB subtypes and transcript levels of CXCRs; however, this correlation needs to be investigated further.
We assessed the correlation between CXCR transcript expression and the cytogenetic and molecular profile of patients with AML. Previous studies have reported a correlation between CXCR4 and FLT3 in AML. Rombouts et al. showed that CXCR4 showed a higher expression in FLT3-ITD mutant group than in the FLT3-wild-type group (27). This finding is consistent with our previous study as well (26). These results showed that patients with a FLT3 mutation had lower expression of CXCR3, CXCR5, and CXCR6 transcripts than that in mutation-negative patients, suggesting a possible anti-cancer effect of CXCR3/5/6 in AML. However, we have not been able to demonstrate this.
Konoplev et al. evaluated the relationship between CXCR4 and NPM1 in a group of 117 untreated adults with AML and found that a mutation in NPM1 is not correlated with CXCR4 or pCXCR4 protein levels, suggesting that CXCR4 and NPM pathways play independent roles in adult AML (28). In contrast, Mannelli et al. demonstrated that the NPM1-mutated cases of AML displayed a significantly higher expression of CXCR4 compared to NPM1-wild-type cases (29). With respect to studies investigating the relationship between CXCR4 and other prognostic genes, Kuo et al. found that CXCR4 expression was significantly higher in CEBPA wild-type patients than in CEBPA mutant patients; thus, CEBPA has been speculated to affect the CXCR4 expression (30). In vitro studies first detected the increased expression of endogenous CXCR4 in AML cell lines and demonstrated that CEBPA mutants modulated CXCR4 activation. Currently, the correlation between CXCR4/CXCR7 expression and IDH1 has been reported in human glioma (31, 32) but has not been reported in AML yet. In this study, we found that CXCR4 transcript expression in mutant IDH1 patients was significantly higher than that in IDH1 wild-type patients. This helps to better understand the relationship between CXCR4 and IDH1 in AML. Despite these results, there are very few reports on the correlation between other CXCRs and AML prognostic genes; hence, this further investigation is required.
Risk stratification for AML the molecular and genetic characteristics takes into account, as well as age, WBC count, and several other factors. Correlation analysis of CXCRs with AML risk stratification revealed that CXCR1, CXCR2, and CXCR6 transcript expressions were higher in the medium/high-risk group than those in the low-risk group. To determine the prognostic value of CXCRs, we analyzed the OS of patients with AML. OS analysis showed that CXCR2 was a predictor of shorter OS and was independent of other classical factors such as age, gender, WBC counts, blast cell percentage, risk stratification, and FLT3/IDH1 gene mutation.
The CXCR1/CXCR2 pathway is the most widely studied pathway in tumors. CXCL1, 2, 3, and 8 are angiogenic chemokines that bind to receptor CXCR2, with the highest affinity exhibited by CXCL1 (33). While studying the effects and ligands of CXCR1/CXCR2, Cheng et al. found that CXCL8 is up-regulated in co-cultures of bone marrow mesenchymal cells and leukemia cell lines compared with CXCL8 expression in single cultures (34). Inhibition of CXCL8/CXCR2 binding can lead to cell cycle arrest in G0/G1 phase, inhibition of AML cell proliferation, inhibition of AKT phosphorylation, and cell apoptosis. Elevated expression of IL-8 and CXCR2 was found in stem and progenitor cells isolated from AML and myelodysplastic syndrome (MDS) patients. Schinke et al. demonstrated that an increased CXCR2 expression was a poor prognostic factor for AML and MDS, further reinforcing the prominent role of the IL-8/CXCR2 axis in AML and MDS (35). Hao et al. investigated the relationship between CXCL1/CXCL2, clinical characteristics, and prognosis in patients with AML (36). Expression of CXCL1 and CXCL2 was detected using quantitative-PCR in bone marrow samples from 160 patients with de novo AML. Furthermore, CR was assessed and event-free survival (EFS) and overall survival were calculated. An increased expression of CXCL2 was found to correlate with the monochromosomal karyotype (P = 0.001). However, CXCL2 was negatively correlated with EFS (P = 0.069) and overall survival (P = 0.055), although this was not statistically significant.
The role of CXCR4 in the development of AML has become an attractive subject of investigation in the recent years. CXCL12 binds and activates its homologous receptor CXCR4 in the microenvironment of the bone marrow to mediate the transport of leukemia cells, while keeping in close contact with stromal cells and the extracellular matrix to generate growth-promoting and anti-apoptotic signals. Increased CXCR4 expression in AML cells is associated with poor prognosis (37). Rombouts et al. found that patients with increased CXCR4 expression in the CD34+ subset of cells had significantly reduced chances of survival and a higher probability of relapse, suggesting that the Stromal Cell-Derived Factor-1(SDF-1)/CXCR4 axis may influence responsiveness to therapy and contribute to an unfavorable prognosis of AML (27). Additionally, we have previously suggested that CXCR4 is an independent prognostic factor for AML (26). The results of a previous report suggested that CXCR4 is expressed in a subset of patients with AML and is associated with poor prognosis, and CXCR4 expression appears to be an independent prognostic factor for reduced survival in a heterogeneous group of patients with AML (38). However, we did not find any obvious prognostic value of CXCR4. This may be due to insufficient samples in the dataset or erroneous methods of detection for CXCR4 expression.
CXCR7 is a newly discovered receptor of CXCL12 that co-exists with CXCR4, and CXCR4/CXCR7 has been declared to play a role in AML (39). Kim et al. investigated the expression levels and function of CXCR7 in AML cells in vitro, and showed that CXCR7 was involved in the regulation of autocrine CXCL12 in AML cells (40). Faaij et al. analyzed the expression of chemokine receptors in children with skin involvement of AML and showed that skin-residing AML cells intracellularly expressed CXCR4 and CXCR7 in 90.9% of evaluated cases (41). These results suggested that chemokine receptor interactions are involved in the homing and retention of AML blast cells in the skin.
The role of the CXCR3, CXCR5, and CXCR6 axes in AML is not well known. The therapeutic potential of CXCR3-CXCL9/10/11 and CXCR5/CXCL13 signaling pathways in tumors has been investigated previously (42, 43). Abnormally high levels of CXCR5 and CXCL13 in the serum of lymphoma patients are significantly associated with poor prognosis (44). Similarly, abnormally high expression levels of CXCR6 and CXCL16 are found to be closely related to tumor proliferation and metastasis, and have been reported to be associated with human ovarian cancer (45) and the metastasis of liver cancer cells (46).
The traditional approach of predicting the prognosis of a disease using one single gene cannot compete with the predictive value of several different potential biomarkers; thus, there is a growing concern over the prognostic value of gene signatures. Yu et al. had reported a CXCR signature for gastric cancer, and a Kaplan-Meier survival analysis revealed that the OS significantly reduced in the high-risk CXCR signature group compared with that of the low-risk CXCR signature group (47). In this study, we established a CXCR signature for AML, which comprehensively determined the patient’s prognosis based on the expression of each CXCR transcript and its prognosis coefficient. Using the Kaplan-Meier survival analysis for the CXCR signature, median OS in the low risk-group was shown to be significantly higher than that of the high-risk group. Additionally, it is known AML prognosis increasingly relies on detection of multigene Panel Testing. We did a bioinformatics analysis and verified it in different database. Experimental validation need to be further studied.
In conclusion, we analyzed the expression, clinical features, and prognostic value of CXCRs in AML. Our results suggested that transcriptional expressions of CXCRs serve an important role in AML. CXCR transcript expressions, except CXCR1 expression, were significantly increased in AML. High CXCR2 expression was found to have a significantly worse prognosis compared with that of low CXCR2 expression, and CXCR2 was also found to be an independent prognostic factor. We established a CXCR signature to identify high-risk subgroups of patients with AML. These results might help improve the treatment and prognosis of patients with AML.
Publicly available datasets were analyzed in this study. This data can be found here: TCGA database (http://cancergenome.nih.gov/).
The studies involving human participants were reviewed and approved by the Academic Committee of Wuhan Union Hospital, Huazhong University of Science and technology. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
The concept of the study was designed by CL, SH, and JY. WX, YH, and CL made the literature research. JZ provided data acquisition. XC and CL made the statistical analysis. CL and JY wrote the manuscript with inputs from all co-authors. The first version was prepared by CL. WX, YH, and JZ made a revision. JY and SH conceived and coordinated the project. SH was the guarantor of integrity of entire study. All authors contributed to the article and approved the submitted version.
This study was funded by Hubei Provincial Natural Science Foundation of China (grant no. 2018CFB485) and National Natural Science Foundation of China (grant no. 81200348)
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Thanks to all the authors for their contributions to this research. Everyone has made an important effort for the work. We would like to thank Editage (www.editage.com) for English language editing. We also would thank Shiyi Cao for his consultation on statistical analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.584766/full#supplementary-material
1. Pollyea DA, Jordan CT. Therapeutic targeting of acute myeloid leukemia stem cells. Blood (2017) 129(12):1627–35. doi: 10.1182/blood-2016-10-696039
2. Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med (2015) 373(12):1136–52. doi: 10.1056/NEJMra1406184
3. Balkwill FR. The chemokine system and cancer. J Pathol (2012) 226(2):148–57. doi: 10.1002/path.3029
4. Mollica Poeta V, Massara M, Capucetti A, Bonecchi R. Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy. Front Immunol (2019) 10:379. doi: 10.3389/fimmu.2019.00379
5. Vandercappellen J, Van Damme J, Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett (2008) 267(2):226–44. doi: 10.1016/j.canlet.2008.04.050
6. Ha H, Debnath B, Neamati N. Role of the CXCL8-CXCR1/2 axis in cancer and inflammatory diseases. Theranostics (2017) 7(6):1543–88. doi: 10.7150/thno.15625
7. Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T, et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev (2016) 31:61–71. doi: 10.1016/j.cytogfr.2016.08.002
8. Bi H, Zhang Y, Wang S, Fang W, He W, Yin L, et al. Interleukin-8 promotes cell migration via CXCR1 and CXCR2 in liver cancer. Oncol Lett (2019) 18(4):4176–84. doi: 10.3892/ol.2019.10735
9. Neo SY, Lundqvist A. The Multifaceted Roles of CXCL9 within the Tumor Microenvironment. Adv Exp Med Biol (2020) 1231:45–51. doi: 10.1007/978-3-030-36667-4_5
10. Kuo PT, Zeng Z, Salim N, Mattarollo S, Wells JW, Leggatt GR. The Role of CXCR3 and Its Chemokine Ligands in Skin Disease and Cancer. Front Med (Lausanne) (2018) 5:271. doi: 10.3389/fmed.2018.00271
11. Burger JA, Peled A. CXCR4 antagonists: targeting the microenvironment in leukemia and other cancers. Leukemia (2009) 23(1):43–52. doi: 10.1038/leu.2008.299
12. Möhle R, Schittenhelm M, Failenschmid C, Bautz F, Kratz-Albers K, Serve H, et al. Functional response of leukaemic blasts to stromal cell-derived factor-1 correlates with preferential expression of the chemokine receptor CXCR4 in acute myelomonocytic and lymphoblastic leukaemia. Br J Haematol (2000) 110(3):563–72. doi: 10.1046/j.1365-2141.2000.02157.x
13. Möhle R, Bautz F, Rafii S, Moore MA, Brugger W, Kanz L. The chemokine receptor CXCR-4 is expressed on CD34+ hematopoietic progenitors and leukemic cells and mediates transendothelial migration induced by stromal cell-derived factor-1. Blood (1998) 91(12):4523–30.
14. Hussain M, Adah D, Tariq M, Lu Y, Zhang J, Liu J. CXCL13/CXCR5 signaling axis in cancer. Life Sci (2019) 227:175–86. doi: 10.1016/j.lfs.2019.04.053
15. Lepore F, D’Alessandro G, Antonangeli F, Santoro A, Esposito V, Limatola C, et al. CXCL16/CXCR6 Axis Drives Microglia/Macrophages Phenotype in Physiological Conditions and Plays a Crucial Role in Glioma. Front Immunol (2018) 9:2750. doi: 10.3389/fimmu.2018.02750
16. Naumann U, Cameroni E, Pruenster M, Mahabaleshwar H, Raz E, Zerwes HG, et al. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One (2010) 5(2):e9175. doi: 10.1371/journal.pone.0009175
17. Wang K, Chen R, Feng Z, Zhu YM, Sun XX, Huang W, et al. Identification of differentially expressed genes in non-small cell lung cancer. Aging (Albany NY) (2019) 11(23):11170–85. doi: 10.18632/aging.102521
18. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res (2019) 47(W1):W556–60. doi: 10.1093/nar/gkz430
19. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal (2013) 6(269):pl1. doi: 10.1126/scisignal.2004088
20. von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res (2003) 31(1):258–61. doi: 10.1093/nar/gkg034
21. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res (2019) 47(D1):D607–13. doi: 10.1093/nar/gky1131
22. Zhou Q, Zhang F, He Z, Zuo MZ. E2F2/5/8 Serve as Potential Prognostic Biomarkers and Targets for Human Ovarian Cancer. Front Oncol (2019) 9:161. doi: 10.3389/fonc.2019.00161
23. Eslamloo K, Xue X, Hall JR, Smith NC, Caballero-Solares A, Parrish CC, et al. Transcriptome profiling of antiviral immune and dietary fatty acid dependent responses of Atlantic salmon macrophage-like cells. BMC Genomics (2017) 18(1):706. doi: 10.1186/s12864-017-4099-2
24. Guo K, Feng G, Yan Q, Sun L, Zhang K, Shen F, et al. CXCR4 and CXCR3 are two distinct prognostic biomarkers in breast cancer: Database mining for CXCR family members. Mol Med Rep (2019) 20(6):4791–802. doi: 10.3892/mmr.2019.10784
25. Cabrero-de Las Heras S, Martínez-Balibrea E. CXC family of chemokines as prognostic or predictive biomarkers and possible drug targets in colorectal cancer. World J Gastroenterol (2018) 24(42):4738–49. doi: 10.3748/wjg.v24.i42.4738
26. Du W, Lu C, Zhu X, Hu D, Chen X, Li J, et al. Prognostic significance of CXCR4 expression in acute myeloid leukemia. Cancer Med (2019) 8(15):6595–603. doi: 10.1002/cam4.2535
27. Rombouts EJ, Pavic B, Löwenberg B, Ploemacher RE. Relation between CXCR-4 expression, Flt3 mutations, and unfavorable prognosis of adult acute myeloid leukemia. Blood (2004) 104(2):550–7. doi: 10.1182/blood-2004-02-0566
28. Konoplev S, Lin P, Yin CC, Lin E, Nogueras González GM, Kantarjian HM, et al. CXC chemokine receptor 4 expression, CXC chemokine receptor 4 activation, and wild-type nucleophosmin are independently associated with unfavorable prognosis in patients with acute myeloid leukemia. Clin Lymphoma Myeloma Leuk (2013) 13(6):686–92. doi: 10.1016/j.clml.2013.05.013
29. Mannelli F, Cutini I, Gianfaldoni G, Bencini S, Scappini B, Pancani F, et al. CXCR4 expression accounts for clinical phenotype and outcome in acute myeloid leukemia. Cytometry B Clin Cytom (2014) 86(5):340–9. doi: 10.1002/cytob.21156
30. Kuo YY, Hou HA, Chen YK, Li LY, Chen PH, Tseng MH, et al. The N-terminal CEBPA mutant in acute myeloid leukemia impairs CXCR4 expression. Haematologica (2014) 99(12):1799–807. doi: 10.3324/haematol.2014.107821
31. Bianco AM, Uno M, Oba-Shinjo SM, Clara CA, de Almeida Galatro TF, Rosemberg S, et al. CXCR7 and CXCR4 Expressions in Infiltrative Astrocytomas and Their Interactions with HIF1α Expression and IDH1 Mutation. Pathol Oncol Res (2015) 21(2):229–40. doi: 10.1007/s12253-014-9813-7
32. Birner P, Tchorbanov A, Natchev S, Tuettenberg J, Guentchev M. The chemokine receptor CXCR7 influences prognosis in human glioma in an IDH1-dependent manner. J Clin Pathol (2015) 68(10):830–4. doi: 10.1136/jclinpath-2015-202886
33. Susek KH, Karvouni M, Alici E, Lundqvist A. The Role of CXC Chemokine Receptors 1-4 on Immune Cells in the Tumor Microenvironment. Front Immunol (2018) 9:2159. doi: 10.3389/fimmu.2018.02159
34. Cheng J, Li Y, Liu S, Jiang Y, Ma J, Wan L, et al. CXCL8 derived from mesenchymal stromal cells supports survival and proliferation of acute myeloid leukemia cells through the PI3K/AKT pathway. FASEB J (2019) 33(4):4755–64. doi: 10.1096/fj.201801931R
35. Schinke C, Giricz O, Li W, Shastri A, Gordon S, Barreyro L, et al. IL8-CXCR2 pathway inhibition as a therapeutic strategy against MDS and AML stem cells. Blood (2015) 125(20):3144–52. doi: 10.1182/blood-2015-01-621631
36. Hao X, Gu M, Sun J, Cong L. A-kinase interacting protein 1 might serve as a novel biomarker for worse prognosis through the interaction of chemokine (C-X-C motif) ligand 1/chemokine (C-X-C motif) ligand 2 in acute myeloid leukemia. J Clin Lab Anal (2020) 34(2):e23052. doi: 10.1002/jcla.23052
37. Cho BS, Kim HJ, Konopleva M. Targeting the CXCL12/CXCR4 axis in acute myeloid leukemia: from bench to bedside. Korean J Intern Med (2017) 32(2):248–57. doi: 10.3904/kjim.2016.244
38. Rady AS, Badawy RH, Gamal BME, Darwish AD, Aziz RSA, Gammal ME, et al. Association of CXCR4 Expression and Clinical Outcome in Different Subsets of De Novo Acute Myeloid Leukemia Patients. Clin Lab (2020) 66(3). doi: 10.7754/Clin.Lab.2019.190725
39. Sartina E, Suguihara C, Ramchandran S, Nwajei P, Rodriguez M, Torres E, et al. Antagonism of CXCR7 attenuates chronic hypoxia-induced pulmonary hypertension. Pediatr Res (2012) 71(6):682–8. doi: 10.1038/pr.2012.30
40. Kim HY, Lee SY, Kim DY, Moon JY, Choi YS, Song IC, et al. Expression and functional roles of the chemokine receptor CXCR7 in acute myeloid leukemia cells. Blood Res (2015) 50(4):218–26. doi: 10.5045/br.2015.50.4.218
41. Faaij CM, Willemze AJ, Révész T, Balzarolo M, Tensen CP, Hoogeboom M, et al. Chemokine/chemokine receptor interactions in extramedullary leukaemia of the skin in childhood AML: differential roles for CCR2, CCR5, CXCR4 and CXCR7. Pediatr Blood Cancer (2010) 55(2):344–8. doi: 10.1002/pbc.22500
42. Chow MT, Ozga AJ, Servis RL, Frederick DT, Lo JA, Fisher DE, et al. Intratumoral Activity of the CXCR3 Chemokine System Is Required for the Efficacy of Anti-PD-1 Therapy. Immunity (2019) 50(6):1498–512.e5. doi: 10.1016/j.immuni.2019.04.010
43. De Roos AJ, Mirick DK, Edlefsen KL, LaCroix AZ, Kopecky KJ, Madeleine MM, et al. Markers of B-cell activation in relation to risk of non-Hodgkin lymphoma. Cancer Res (2012) 72(18):4733–43. doi: 10.1158/0008-5472.CAN-12-1639
44. Cha Z, Qian G, Zang Y, Gu H, Huang Y, Zhu L, et al. Circulating CXCR5+CD4+ T cells assist in the survival and growth of primary diffuse large B cell lymphoma cells through interleukin 10 pathway. Exp Cell Res (2017) 350(1):154–60. doi: 10.1016/j.yexcr.2016.11.017
45. Chandrasekar B, Bysani S, Mummidi S. CXCL16 signals via Gi, phosphatidylinositol 3-kinase, Akt, I kappa B kinase,and nuclear factor-kappa B and induces cell-cell adhesion and aortic smooth muscle cell proliferation. J Biol Chem (2004) 279(5):3188–96. doi: 10.1074/jbc.M311660200
46. Yang Y, Sheng L, Jiani H, Xiangyu D, Bo Z. CD133+ ovarian cancer stem cells maintain high abilities of invasion and migration via CXCL16/CXCR6 axis. J Third Mil Med Univ (2015) 37(8):754–61. doi: 10.16016/j.1000-5404.201411128
Keywords: acute myeloid leukemia, CXC chemokine receptor, FAB subtypes, risk stratification, gene signature, prognostic role
Citation: Lu C, Zhu J, Chen X, Hu Y, Xie W, Yao J and Huang S (2020) Risk Stratification in Acute Myeloid Leukemia Using CXCR Gene Signatures: A Bioinformatics Analysis. Front. Oncol. 10:584766. doi: 10.3389/fonc.2020.584766
Received: 18 July 2020; Accepted: 09 October 2020;
Published: 30 October 2020.
Edited by:
Sandra Marmiroli, University of Modena and Reggio Emilia, ItalyReviewed by:
Francesca Chiarini, Italian National Research Council, ItalyCopyright © 2020 Lu, Zhu, Chen, Hu, Xie, Yao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junxia Yao, eWFvanVueGlhQHlhaG9vLmNvbQ==; Shiang Huang, c2EyaHVhbmdAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.