- 1Department of Oncology, The Affiliated Cancer Hospital of Zhengzhou University, Henan Cancer Hospital, Zhengzhou, China
- 2Department of Pathology, Pathology Department of Hebei Medical University, Shijiazhuang, China
Gastric cancer is the fifth most common malignant tumor and second leading cause of cancer-related deaths worldwide. With the improved understanding of gastric cancer, a subset of gastric cancer patients infected with Epstein–Barr virus (EBV) has been identified. EBV-positive gastric cancer is a type of tumor with unique genomic aberrations, significant clinicopathological features, and a good prognosis. After EBV infects the human body, it first enters an incubation period in which the virus integrates its DNA into the host and expresses the latent protein and then affects DNA methylation through miRNA under the action of the latent protein, which leads to the occurrence of EBV-positive gastric cancer. With recent developments in immunotherapy, better treatment of EBV-positive gastric cancer patients appears achievable. Moreover, studies show that treatment with immunotherapy has a high effective rate in patients with EBV-positive gastric cancer. This review summarizes the research status of EBV-positive gastric cancer in recent years and indicates areas for improvement of clinical practice.
Introduction
Epstein–Barr virus (EBV) is the main pathogenic factor for nasopharyngeal carcinoma. However, studies find that EBV infection is also associated with the development of T-cell lymphoma and EBV-associated gastric cancer (1, 2). In 1990, Burke et al. (3) detected EBV in gastric lymphoepithelial carcinoma, and this was the first report on histopathological features of EBV-positive gastric cancer. Furthermore, Shibata and Weiss (4) find EBV to be associated with gastric adenocarcinoma. They demonstrate the presence of the EBV genome specifically in gastric cancer cells and adjacent dysplastic epithelium, but not in surrounding normal cells. Studies find EBV-positive and -negative gastric cancer to have different pathogens and that EBV may play an important role in its pathogenesis (5, 6). In 2004, van Beek et al. (7) analyzed the clinicopathological features of EBV-positive and -negative gastric adenocarcinomas. The results show that EBV-positive gastric cancer has a unique genomic aberration, obvious clinicopathological features, and good prognosis. In 2009, Murphy et al. (8) found EBV-positive gastric cancer to be different from other gastric cancers in terms of patients’ sex, tumor anatomical site, and surgical anatomical structure. In 2014, Liang et al. (9) studied the mechanism of occurrence and development of EBV-positive gastric cancer. The comprehensive epigenomic and transcriptomic analysis identified 216 genes downregulated by EBV-induced hypermethylation; in EBV-positive tumors, the methylation of ACSS1, FAM3B, IHH, and TRABD was significantly increased. Moreover, five signaling pathways (axon guidance, local adhesion formation, interaction between cytokines and receptors, mitogen-activated protein kinase signal transduction, and actin cytoskeleton regulation) were significantly affected by EBV-related genomic and epigenomic changes. However, no specific treatment has been found for EBV-positive gastric cancer, and the EBV-titer is not associated with the risk of gastric cancer (10, 11). Several recent studies find a close relationship between EBV-positive gastric cancer and immune checkpoints (12). In 2018, Panda et al. (13) found EBV-positive gastric cancer with low mutation burden to be a subset of microsatellitestable (MSS) gastric cancer, which may respond to immune checkpoint therapy. Thus, EBV-positive gastric cancer is now considered a unique molecular subtype of gastric cancer (14) and is associated with good prognosis in patients (15). At present, there is no article to summarize and analyze the characteristics, mechanisms, and treatment of EBV-positive gastric cancer, including latency proteins. MicroRNAs (miRNAs) and DNA methylation have important effects on EBV-positive gastric cancer; there is a close relationship between them, and it may reveal potential treatments for EBV-positive gastric cancer. Here, we review recent advances in EBV-positive gastric cancer research to improve the current understanding of this disease and aid in development of newer treatment modalities for this cancer type.
Characteristics of EBV-Positive Gastric Cancer
Gastric cancer is normally classified on the basis of its histomorphological characteristics (16). The Cancer Genome Atlas reports a comprehensive identification of genetic changes associated with gastric cancer and further divides this form of cancer into four subtypes: EBV-positive tumors (9%), microsatellite unstable tumors (22%), genetically stable tumors (20%), and chromosome unstable tumors (50%). Moreover, EBV-positive and MSI gastric cancers have the capability to respond to newer immunotherapy drugs (17). However, as opposed to general gastric cancer, EBV-positive gastric cancer, despite having unique pathological characteristics, has no specific clinical manifestations. A few studies have found higher incidences of EBV-positive gastric cancer in men and patients below the age of 60 years. Camargo et al. (18) find that the average age of EBV-positive gastric cancer patients is 58 years old, and 71% of them are men. EBV-positive gastric cancer often occurs in the proximal stomach (cardia and gastric body), where it forms lumps or ulcers that are accompanied by lymphocyte infiltration. Another noteworthy feature of EBV-positive gastric cancer is the ease of invasion into the submucosa with a low rate of lymph node metastasis. A majority of patients were diagnosed in the advanced stage (52%, stage III and IV), and 2247 (49%) patients died during the median follow-up period of 3 years. An unadjusted Cox regression analysis indicates that the median survival duration of EBV-positive gastric cancer patients is 8.5 years although that of EBV-negative patients is only 5.3 years. It is evident that the prognosis and effective treatment rate of gastric cancer patients with positive EBV is more desirable. We studied the proper treatment to prolong the survival time in EBV-positive gastric cancer patients considering the curable nature of EBV-positive gastric cancer.
How to Test EBV-Positive Gastric Cancer
Immunohistochemistry and In Situ Hybridization
The principles of immunohistochemistry and in situ hybridization (ISH) are different, and detection results vary. The EBER-1 probe used in ISH is a base sequence that can specifically anneal to the small mRNA encoded by the EBV. The probe can detect gastric cancer specimens fixed by formaldehyde and embedded in paraffin, enabling detection of EBV in tumor cells in situ with accurate localization and strong specificity. However, the gastric cancer tissue is often selected to avoid wasting reagents as the EBER-1 probe is expensive. Occasionally the fixation of gastric cancer tissue is poor, the nucleic acid in the tissue is denatured and diffused, the effective binding sites are reduced, and the staining may appear as weak positive or false negative markers. Immunohistochemical detection is based on the LMP-1 membrane protein encoded by EBV, which cannot detect the location or transcriptional quantity of the virus. However, compared with ISH, immunohistochemical methods have the advantage of simple steps, convenient operation, high sensitivity, and low price, making it a reliable primary screening method for EBV. Immunohistochemical positives can be followed up by ISH to exclude the possibility of false positives. The combination of the two methods might improve the accuracy of detection by reducing the chances of false positives and negatives.
Genome Sequencing
EBV-positive gastric cancer is traditionally identified by ISH of viral nucleic acid (19). However, genome sequencing is a potential alternative. Camargo et al. (20) determine the normalized EBV readings in 295 fresh gastric cancer samples by whole genome, whole exome, mRNA, and miRNA sequencing. Formalin-fixed, paraffin-embedded tissue sections were obtained and used for ISH confirmation in 13 cases with high EB viral load and 11 cases with low EBV. In pairwise comparisons, individual samples are uniformly either high or low in all genomic methods for which data are available. The empirical cutoff value of sequencing count confirmed 26 (9%) tumors to be EBV-positive. EBV was either positive or negative based on molecular detection. Conversely, the Epstein–Barr encoding region (EBER)-ISH was either positive or negative in all samples except for one, which was evaluated by the two methods (kappa=0.91). Thus, EBV-positive gastric tumors can be accurately identified by quantifying virus sequences in genomic data. Moreover, simultaneous analysis of human and viral DNA, mRNA, and miRNA can simplify the tumor profile of clinical nursing and research.
Detection of Anti-EBV and Anti-p53 Antibodies
Tumor protein p53, or simply p53, is closely related to the occurrence of gastric cancer, and many studies find EBV infection to be associated with p53 methylation (21–23). In 2019, Camargo et al. found that EBV-positive gastric cancer cases lack the TP53 mutation, suggesting that serological characteristics may provide information for viral carcinogenesis. Consistent with the prevalence of EBV, 99% of patients tested positive for the anti-Epstein–Barr virus nuclear antigen 1 (EBNA) antibody, and 98% of the patients tested positive for the antiviral capsid antigen antibody regardless of the EBV status of the tumor. The levels of p53 antibody and EBV-positivity were negatively correlated. The positive rate of anti-p53 staining was 15% in the literature. However, the results suggest dissimilar correlations between anti-EBV antibody, anti-p53 antibody and tumor EBV-positivity.
Droplet Digital PCR
Droplet digital PCR (ddPCR) is the latest method that can be used in the detection of Plasmodium falciparum (24), multiple viruses (25), nervous system lymphomas (26), etc. A ddPCR-based screening method for detection of EBV–associated gastric carcinoma was established in 2019 (27, 28). This method uses the ddPCR method to calculate EBV-DNA load according to the copy number of EBV BamH1-W fragments and sets the cutoff value of the EBV-DNA load.
What Is the Mechanism of Occurrence and Development of EBV-Positive GC?
Epstein and Barr discovered the EBV in 1964 and identified its original host as the human body; the virus had the capability to infect B-lymphocytes, epithelial cells, and fibroblasts (29, 30). EBV infection in the human body does not immediately lead to gastric cancer. Although the infection rate of EBV in adults is 90%, the incidence rate of gastric cancer remains low; the majority of individuals only carry the virus during the incubation period (31). At present, there are two theories about the mechanisms of EBV infection. The first is that EBV infects B-lymphocytes and oral epithelial cells. As the saliva containing EBV enters the digestive tract, EBV directly infects the epithelial cells. The second theory is that EBV is reactivated in some way in B-lymphocytes in the stomach and then released to infect the epithelial cells (32). Moreover, lymphocytes infected by EBV can encounter epithelial cells through integrin β-1/β-2 and promote cell-to-cell contact by translocating intracellular adhesion molecule-1 to the cell surface. Finally, the virus particles are transmitted through the endocytosis pathway mediated by reticular proteins (33). After phagocytosis, EBV-DNA is transported to the nucleus, where the exposed linear DNA genome is assembled into a functional, small, circular chromosome. After circularization, the viral genome chromatinization can effectively protect it from DNA damage and ensure strict regulation of gene expression (34). The CpG motif of the viral genome is widely methylated, thus successfully establishing latent infection. EBV primarily infects host cells in two ways: lytic and latent infection. However, the virus mostly remains in the latent infection state without replication (19). After entering the incubation period in human bodies, the EBV prompts methylation of the host genome, imbalance of the cellular signaling pathway, abnormal gene expression, generation of a tumor microenvironment of infected gastric epithelial cells, and initiation and development of gastric cancer. Moreover, latent EBV gene products, such as EBERs, BARF-0, EBNA-1, and LMP2A, are involved in the downregulation of the miR-200 family, resulting in reduced E-cadherin expression, which is a key step in the carcinogenesis of EBV-associated gastric cancer (EBVaGC) (35).
Virus Latency Gene Product
EBV has three types of latent phases. As EBV-positive gastric cancer is type I latency, EBERs, EBNA-1, miR-BARTS, and LMP2A are highly expressed and play an important role in viral replication (36, 37). EBER1 upregulates the expression of insulin growth factor-1, thus promoting proliferation of EBVaGC cells (38). EBERs associate with IL-6-STAT3 signaling pathway to induce chemotherapy resistance and promote cell migration (39).
EBNA-1 is an important molecule for EBV latent infection (40, 41). It binds to the viral ORIP sequence in a sequence-dependent manner and aids in EBV attachment to the host cell chromosome (42). EBNA-1 is also a transactivator of viral genes that may induce accumulation of reactive oxygen species (ROS) mediated by miR-34a and NOX2, regulating the activity of tumor cells (43). Additionally, EBNA-1 can lead to the loss of promyelocytic leukemia protein nuclear bodies in the nucleosome of promyelocytic leukemia and weaken the cellular response to DNA damage (44).
BARF-0 and BARF-1 are also involved in the latent state of EBV infection. BARF-0 downregulates the expression of TET2 (45). BARF-1 gene silencing triggers caspase-dependent mitochondrial apoptosis (46). It can also induce alteration in the NF-κB/miR-146a/Smad4 pathway and expression of cyclin-D1 protein in gastric cancer cells (47–49). Moreover, BARF-1 can activate the cell cycle regulator bcl-2 (50). These processes promote proliferation of gastric cancer cells.
LMP2A is the most important molecule in the incubation period of EBV. It can activate the NF-κB-Survivin pathway (51, 52), regulate the expression of cyclin-E and proportion of cells in the S phase (53), mediate Notch signaling, and promote mitochondrial division and cell migration (54). Additionally, it can upregulate miR-155-5p through the NF-κB pathway and inhibit the activation of Smad2 and p-Smad2 (55, 56). LMP2A can also initiate gastric cancer by upregulating or downregulating other genes. For instance, LMP2A downregulates the expression of TET2, COX-2, and HLA (45, 57, 58) and upregulates FOXO1 and FOXO3 (59). LMP2A activates the PI3K/AKT pathway to mediate the transformation process and inhibits apoptosis-induced proliferation by transforming growth factor β1 (60). LMP2A induces STAT3 phosphorylation, resulting in DNMT1 transcriptional activation and PTEN promoter methylation (61). LMP2A also activates CpG island methylation of the AQP3 promoter, induces ERK phosphorylation, and activates DNMT3a transcription, which results in the loss of AQP3 expression (Table 1) (55).
MicroRNAs
EBV is a ubiquitous human carcinogenic virus and is also the first human virus to express miRNAs. The EBV genome contains two regions that encode more than 40 miRNAs that regulate the expression of viral and human genes, such as ebv-miR-BART-1-3p, -2-5p, -3-3p, -4-5p, -5-5p, -7-3p, -9-3p, -10-3p, -17-5p, -10-3p, -18-5p, BART11, etc. (62–67). Studies suggest that EBV miRNAs affect immune response and antigen presentation and recognition, alter the communication between T- and B-cells, drive the production of antibodies during infection, and play a role in apoptosis. Additionally, EBV can induce B-cell transformation and participate in the mechanism of human tumorigenesis. Although EBV infection is related to the occurrence of several diseases, the role of miRNAs remains unclear. Extensive data describes the role of EBV miRNAs in nasopharyngeal carcinoma, and several studies have attempted to evaluate their role in gastric cancer and lymphoma. Song et al. (68) find that the EBV miRNA BART11 downregulates Foxp1 transcription factor, which promotes epithelial-mesenchymal transition by directly affecting gastric tumor cells or indirectly affecting the tumor microenvironment. BART11 also accelerates tumor invasion and metastasis, affecting survival and prognosis in patients. Dong et al. (69) find that BART10-3p and BART22 activate the Wnt signaling pathway by targeting APC and Dkk1, which play an important role in promoting EBVaGC metastasis, thus providing new prognostic biomarkers and potential therapeutic targets in EBVaGC. Wang et al. (70) report that BART3-3p promotes growth and inhibits senescence of gastric cancer cells induced by oncogenes (RAS) or chemotherapy (irinotecan). BART3-3p inhibits the senescence of gastric cancer cells in nude mice by modifying the aging-related SP (SASP) and the infiltration of natural killer cells and macrophages in tumors. BART3-3p directly targets the inhibition of the tumor suppressor gene and leads to the downregulation of p21, the downstream target of p53. The clinical analysis of EBV-positive gastric cancer also displays a negative correlation between the expression of BART3-3p and p21. This study suggests that the expression of BART3-3p is important in the carcinogenesis of EBV-positive gastric cancer. Other miRNAs also play an important role in this process. For instance, miR-BART5 upregulates p53 with PUMA as the target, promoting the survival of host cells (71, 72); miR-BART3-5p targets DICE1 tumor suppressor, and promotes the growth and transformation of cancer cells (73); miR-BART9 specifically inhibits E-cadherin to induce a mesenchymal-like phenotype (74, 75); miR-BART9, -11, and -12 downregulates Bim expression (76); EBV-miR-BART4-5p has an antiapoptotic role that regulates Bid expression in EBV-associated gastric carcinoma (77); EBV-miR-BART20-5p regulates cell proliferation and apoptosis by targeting BAD (78); and miR-BART16 abrogates the production of IFN-stimulated genes in response to IFN-α stimulation and inhibits the antiproliferative effect of IFN-α in latently infected cells (79). Modulation of expression of LMP2A by newly identified EBV-encoded miRNAs, miR-BART22 (80) and miR-BART17-5p, promotes migration and anchorage-independent growth by targeting kruppel-like factor 2 in gastric cancer (81, 82). EBV-miR-BART15-3p targets the anti-apoptotic TAX1BP1 and NLRP3 genes in cancer cells, thus increasing apoptosis (Table 2) (84–86).
DNA Methylation
DNA methylation is arguably the most important mechanism in EBV-positive gastric cancer (87). Liang et al. (9) find that 216 genes were downregulated by EBV-related hypermethylation. It was also found that five signaling pathways—axon guidance, local adhesion formation, interaction between cytokines and receptors, mitogen-activated protein kinase signal transduction, and actin cytoskeleton regulation—were jointly affected by EBV-related genomic and epigenomic changes. Thus, with advances in high-throughput sequencing technology, it is possible to fully describe the mechanism of EBV-induced DNA methylation. Zhao et al. (88) displayed that the promoters of 886 genes involved in cancer-related pathways were abnormally hypermethylated in EBV-positive AGS cells, including p14ARF, AQP3, p15, p16INK4A, DLC-1, p73, Rec8, ACSS1, WWOX, FAM3B, BCL7A, IHH, BLU, TRABD, TFF1, TIMP3, FHIT, DAPK, FSD1, GSTP1, APC, SSTR1, CRBP1, Mark1, SCRN1, etc. (55, 89–95). Two of these genes, PIK3CA and ARID1A, presented with the highest methylation rate (96–100). Yu et al. (92) find that methylation levels of the promoter of the meiosis-specific gene, Rec8, were significantly higher in EBV-positive than EBV-negative gastric cancer tissues, and methylation levels in both these subtypes were significantly higher than that in, e.g., tissue uninfected by EBV. It is a newer tumor suppressor that is downregulated by promoter methylation in gastric cancer, especially in the EBV-related subtypes. The antitumor effect of Rec8 can be partially explained by the downregulation of cell growth–related genes (G6PD, SLC2A1, NOL3, MCM2, SNAI1, and SNAI2) and upregulation of apoptosis or migration inhibitors (Gadd45G and LDHA) and tumor inhibitors (PinX1, IGFBP3, and ETS2). The results suggest that methylation of the Rec8 gene promoter is an independent risk factor for reducing the survival in patients with gastric cancer. Further, Zhao et al. (91) indicate that SSTR1 is a newer methylated gene in gastric cancer cells in response to EBV infection, which may act as a potential tumor suppressor. Additionally, proteins expressed during the incubation period can directly affect the methylation of multiple gene promoters. For example, Wang et al. (55) find that LMP2A induces ERK phosphorylation. LMP2A also increases transcription of DNMT3a by activating CpG island methylation of AQP3 promoter in EBVaGC; resulting in the loss of AQP3 expression. Qi et al. (57) find that the overexpression of LMP1 and LMP2A inhibits COX-2 expression, mediated through the reduction of TRAF2; p-ERK aids LMP1-inhibition of COX-2 in gastric cancer. Hino et al. (61) find that LMP2A induces phosphorylation of STAT3 and activates transcription of DNMT1, leading to the CpG island methylation of PTEN promoter and loss of PTEN expression in EBV-related gastric cancer. Additionally, LMP2A plays an important role in epigenetic abnormalities in host gastric cells and occurrence and maintenance of EBV-related cancers (Table 3).
Helicobacter pylori
Simultaneous infection with EBV and Helicobacter pylori can occur (102). Helicobacter pylori and EBV infection are associated with IL-10 and IL-1RN polymorphisms (103). However, evidence for the possible interaction or antagonism of these infectious factors in carcinogenesis of gastric cancer is limited. Camargo et al. (18) compare the serological characteristics of Helicobacter pylori in 58 EBV-positive and 111 EBV-negative gastric cancer patients at the National Cancer Institute’s International EBV-Gastric Cancer Consortium, United States. The results suggest that the overall serum positive rate for an individual’s five immunogenic proteins is as high as 90%. Moreover, catalase antibodies were marginally associated with EBV-positive tumors. Taken together, these results suggest that infection with Helicobacter pylori is related to the occurrence and development of EBV-positive gastric cancer.
What Is the Relationship Between EBV-Positive Gastric Cancer and Immunotherapy?
Statistical analysis reveals that EBV-positive gastric cancer with a low mutation burden is a subset of MSS gastric cancer and may respond to immune checkpoint therapy. In 2019, Roh et al. (104) comprehensively analyzed the status of SPC (specificity), MSI, and EBV. The results validate that the combined application of SPC, MSI, and EBV statuses could predict the efficacy and prognosis of adjuvant chemotherapy in stage II or III gastric cancer.
Gene expression profile analysis of EBVaGC patients indicates significant changes in immune response genes, which may allow recruitment of reactive immune cells to better survival outcomes in patients (105). Moreover, EBVaGC is characterized by high and low density of CD8+ T-cells and CD204+ macrophages, respectively (106, 107). Both the infiltrating immune cells and specific immune microenvironment contribute to antitumor immunity (108). However, tumor cells in EBVaGC evade immune responses via a variety of strategies. It is reported that indoleamine-pyrrole 2,3-dioxygenase (IDO1) is an effective immunosuppressive enzyme, which is upregulated in EBVaGC to resist tumor immune responses (109).
EBVaGC was found to express high levels of programmed death ligand 1 (PD-L1) in cancer and infiltrating immune cells (110). As tumor cells recruit PD-L1 to interact with programmed cell death protein 1 (PD-1) on the surface of T-cells to escape from antitumor immunity, the high expression of PD-L1 in EBVaGC can be considered to be related to tumor progression (111). Additionally, several studies have also found PD-L1 expression to be increased in patients with EBV-positive gastric cancer; patients with MSI gastric cancer showed better prognosis (112, 113).
In 2018, Kim et al. (114) reported on the molecular characterization of tissue and circulatory tumor DNA (ctDNA) in 61 patients with metastatic gastric cancer who received rescue treatment with perbrolizumab in a prospective phase 2 clinical trial. The results indicate that the objective effective rate (ORR) of perbrolizumab in the treatment of EBV-positive metastatic gastric cancer was 100%, significantly higher than the ORR of 85.7% in the treatment of metastatic microsatellite instability gastric cancer. There is a high correlation between PD-L1 positive and EBV-positive/MSI-H, suggesting that immunotherapy may be as effective in EBV-positive gastric cancer patients as it is in MSI-H patients.
In 2020, Kim et al. verified the effectiveness of immunotherapy in the treatment of EBV-positive gastric cancer (115). A total of 300 gastric cancer patients (Asian) were included in this study, of which PD-L1Cps ≥1 was positive in 178 cases (59.3%) and PD-L1Cps <1 in 122 cases (40.7%). PD-L1Cps ≥1 was significantly associated with stage I tumor (P=0.022), high microsatellite instability (MSI-H) (P<0.001), positive EBV status (P=0.008), and positive Helicobacter pylori status (P=0.001). In the gene expression profile, PD-L1CPs were highly positively correlated with mutation load (P<0.001), EBV (P<0.001), and microsatellite subtype (P<0.001). PD-L1 was expressed in 59.3% of gastric cancer patients and was associated with positive MSI and EBV status. These results suggest that patients with EBV-positive gastric cancer can benefit from immunotherapy. Research regarding the treatment of EBV-positive gastric cancer is currently under way, and we eagerly await the results.
Discussion
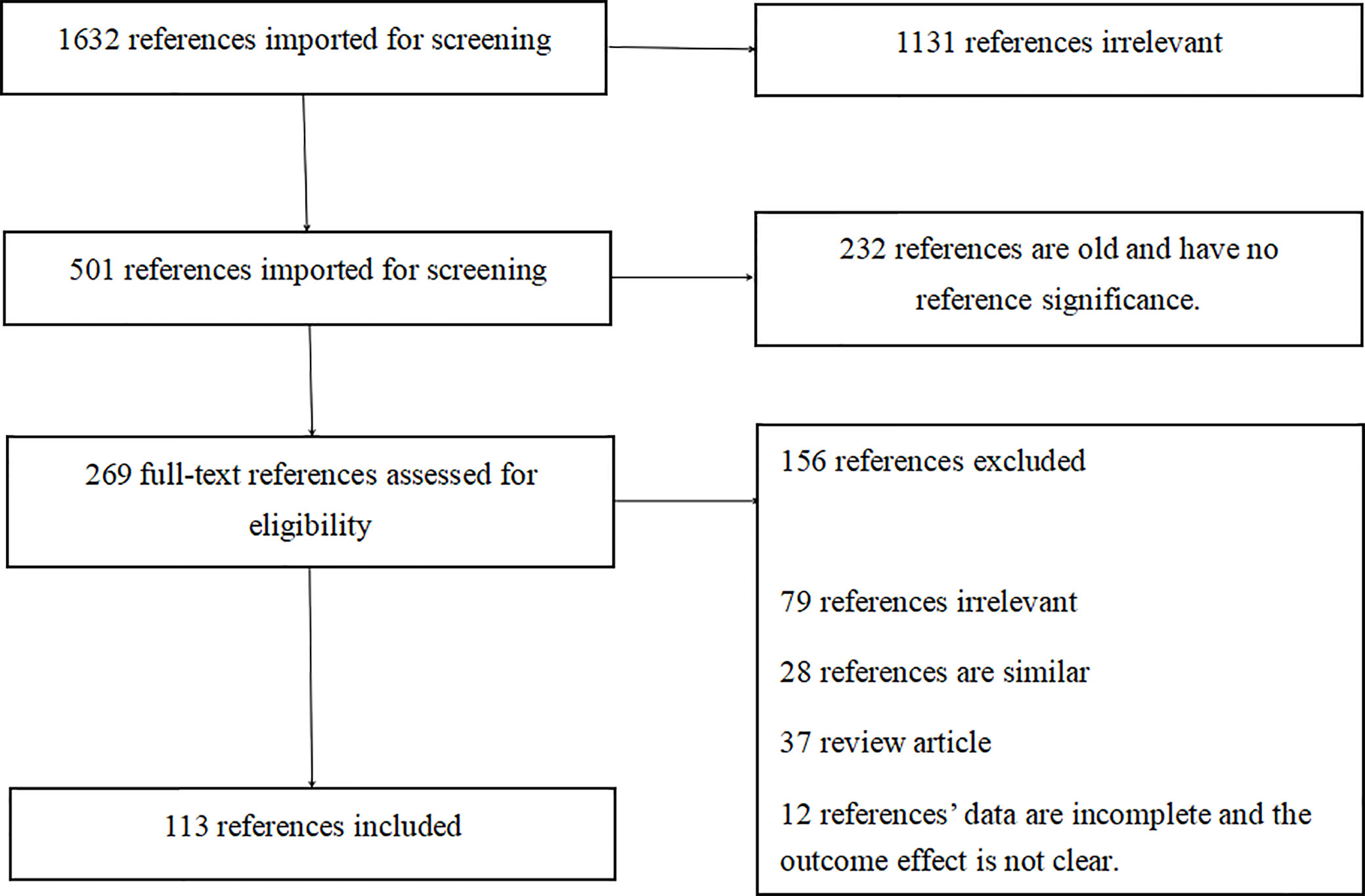
This paper reviews articles dating back to the discovery of EBV-positive gastric cancer through more recent studies. We collected and read 1632 articles on Pubmed and finally selected 113 representative articles for collection (Figures 1 and 2). We comprehensively and systematically review EBV-positive gastric cancer, its pathological features, detection methods, pathogenesis, and potential treatment. We also describe how EBV infects the human body and affects the host’s miRNA through the expression of proteins in the latent period until it results in DNA methylation and the onset of gastric cancer. With the developments in scientific research and improvements in detection technology, our understanding of EBV-positive gastric cancer has improved. We are gradually beginning to comprehend how DNA methylation contributes to the occurrence and development of gastric cancer due to EBV infection. Moreover, recent studies have helped to understand the occurrence and development of EBV-positive gastric cancer from the perspective of gene mutations, miRNA expression, and biology. However, these details are still insufficient for improved treatment of EBV-positive gastric cancer. Although the prognosis in patients with EBV-positive gastric cancer is significantly better than other types of cancer, there is still no unified treatment regimen. On the basis of several pathogenic mechanisms, we anticipate the use of therapies that target miRNAs, DNA methylation, or immunotherapy to manage this cancer type. Furthermore, recent studies indicate that immunotherapy can help achieve complete remission in EBV-positive gastric cancer. Thus, it would be possible to decide whether to administer surgical treatment or immunotherapy in patients with early EBV-positive gastric cancer, whether anti-Helicobacter pylori therapy imparts significant therapeutic effects in EBV-positive gastric cancer patients, and whether these treatment modalities can also be administered in EBV-negative gastric cancer patients. Additionally, we may be able to address the issue regarding treatment of patients infected with EBV, but whose gastric cancer is not caused by EBV. In summary, we have reviewed the detection and pathogenesis of EBV-positive gastric cancer and its correlation with immunotherapy. We are in the initial stages of understanding the pathogenesis of EBV-positive gastric cancer; several unknown challenges and treatment options remain to be explored and discovered. We believe that further research and a better understanding of EBV will play a vital role in the treatment and prognosis of patients with gastric cancer.
Search Strategy and Selection Criteria
Data for this review were identified by searching the PubMed and references from relevant articles using the search terms “EBV,” “EBV Gastric cancer,” “miRNA EBV,” “DNA methylation,” “immunotherapy,” and “Helicobacter pylori.” Abstracts and reports from meetings were included only when they related directly to previously published work. Only articles published in English between 1987 and 2020 were included (Figures 1 and 2).
Author Contributions
XC and HLv conceived the article and decided the theme of the manuscript. YW, HLei, and S-QW reviewed published articles and materials and integrated them. KS and KJ analyzed the materials and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study received funding from the National Natural Science Foundation of China (81472714); and Joint Co-construction Project of Medical Science and Technique Foundation Plan of Henan Province (2018020486) 1000 Talents Program of Central Plains of Henan Province (204200510023).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to editor Laura Lorenzon and experts reviewing manuscripts, for their hard work and guidance. Thanks to the National Science and Nature Fund Committee and the affiliated Cancer Hospital of Zhengzhou University.
References
1. Takayama T, Sakabe T, Fujii M, Yamada E, Uno M, Ono Y. In vitro production of human antibodies specifically reactive with human gastric cancer cells of established lines and autologous tissues. J Surg Oncol (1987) 36(3):215–24. doi: 10.1002/jso.2930360313
2. Bocian J, Januszkiewicz-Lewandowska D. Epstein-Barr virus infection - life cycle, methods of diagnosis, associated diseases. Postepy Hig Med Dosw (Online) (2011) 65:286–98. doi: 10.5604/17322693.943104
3. Burke AP, Yen TS, Shekitka KM, Sobin LH. Lymphoepithelial carcinoma of the stomach with Epstein-Barr virus demonstrated by polymerase chain reaction. Mod Pathol (1990) 3(3):377–80.
4. Shibata D, Weiss LM. Epstein-Barr virus-associated gastric adenocarcinoma. Am J Pathol (1992) 140(4):769–74.
5. zur Hausen A, van Grieken NC, Meijer GA, Hermsen MA, Bloemena E, Meuwissen SG, et al. Distinct chromosomal aberrations in Epstein-Barr virus-carrying gastric carcinomas tested by comparative genomic hybridization. Gastroenterology (2001) 121(3):612–8. doi: 10.1053/gast.2001.27200
6. Wu MS, Shun CT, Wu CC, Hsu TY, Lin MT, Chang MC, et al. Epstein-Barr virus-associated gastric carcinomas: relation to H. pylori infection and genetic alterations. Gastroenterology (2000) 118(6):1031–8. doi: 10.1016/S0016-5085(00)70355-6
7. van Beek J, zur Hausen A, Klein Kranenbarg E, van de Velde CJ, Middeldorp JM, van den Brule AJ, et al. EBV-positive gastric adenocarcinomas: a distinct clinicopathologic entity with a low frequency of lymph node involvement. J Clin Oncol (2004) 22(4):664–70. doi: 10.1200/JCO.2004.08.061
8. Murphy G, Pfeiffer R, Camargo MC, Rabkin CS. Meta-analysis shows that prevalence of Epstein-Barr virus-positive gastric cancer differs based on sex and anatomic location. Gastroenterology (2009) 137(3):824–33. doi: 10.1053/j.gastro.2009.05.001
9. Liang Q, Yao X, Tang S, Zhang J, Yau TO, Li X, et al. Integrative identification of Epstein-Barr virus-associated mutations and epigenetic alterations in gastric cancer. Gastroenterology (2014) 147(6):1350–62 e4. doi: 10.1053/j.gastro.2014.08.036
10. Figueiredo C, Camargo MC, Leite M, Fuentes-Panana EM, Rabkin CS, Machado JC. Pathogenesis of Gastric Cancer: Genetics and Molecular Classification. Curr Top Microbiol Immunol (2017) 400:277–304. doi: 10.1007/978-3-319-50520-6_12
11. Varga MG, Cai H, Waterboer T, Murphy G, Shimazu T, Taylor PR, et al. Epstein-Barr Virus Antibody Titers Are Not Associated with Gastric Cancer Risk in East Asia. Dig Dis Sci (2018) 63(10):2765–72. doi: 10.1007/s10620-018-5154-9
12. Dai C, Geng R, Wang C, Wong A, Qing M, Hu J, et al. Concordance of immune checkpoints within tumor immune contexture and their prognostic significance in gastric cancer. Mol Oncol (2016) 10(10):1551–8. doi: 10.1016/j.molonc.2016.09.004
13. Panda A, Mehnert JM, Hirshfield KM, Riedlinger G, Damare S, Saunders T, et al. Immune Activation and Benefit From Avelumab in EBV-Positive Gastric Cancer. J Natl Cancer Inst (2018) 110(3):316–20. doi: 10.1093/jnci/djx213
14. Jacome AA, Lima EM, Kazzi AI, Chaves GF, Mendonca DC, Maciel MM, et al. Epstein-Barr virus-positive gastric cancer: a distinct molecular subtype of the disease? Rev Soc Bras Med Trop (2016) 49(2):150–7. doi: 10.1590/0037-8682-0270-2015
15. Gasenko E, Isajevs S, Camargo MC, Offerhaus GJA, Polaka I, Gulley ML, et al. Clinicopathological characteristics of Epstein-Barr virus-positive gastric cancer in Latvia. Eur J Gastroenterol Hepatol (2019) 31(11):1328–33. doi: 10.1097/MEG.0000000000001521
16. Chia NY, Tan P. Molecular classification of gastric cancer. Ann Oncol (2016) 27(5):763–9. doi: 10.1093/annonc/mdw040
17. Rodriquenz MG, Roviello G, D’Angelo A, Lavacchi D, Roviello F, Polom K. MSI and EBV Positive Gastric Cancer’s Subgroups and Their Link With Novel Immunotherapy. J Clin Med (2020) 9(5):1427. doi: 10.3390/jcm9051427
18. Camargo MC, Kim KM, Matsuo K, Torres J, Liao LM, Morgan DR, et al. Anti-Helicobacter pylori Antibody Profiles in Epstein-Barr virus (EBV)-Positive and EBV-Negative Gastric Cancer. Helicobacter (2016) 21(2):153–7. doi: 10.1111/hel.12249
19. Rowlands DC, Ito M, Mangham DC, Reynolds G, Herbst H, Hallissey MT, et al. Epstein-Barr virus and carcinomas: rare association of the virus with gastric adenocarcinomas. Br J Cancer (1993) 68(5):1014–9. doi: 10.1038/bjc.1993.472
20. Camargo MC, Bowlby R, Chu A, Pedamallu CS, Thorsson V, Elmore S, et al. Validation and calibration of next-generation sequencing to identify Epstein-Barr virus-positive gastric cancer in The Cancer Genome Atlas. Gastric Cancer (2016) 19(2):676–81. doi: 10.1007/s10120-015-0508-x
21. Chatterjee K, Das P, Chattopadhyay NR, Mal S, Choudhuri T. The interplay between Epstein-Bar virus (EBV) with the p53 and its homologs during EBV associated malignancies. Heliyon (2019) 5(11):e02624. doi: 10.1016/j.heliyon.2019.e02624
22. Xu DM, Kong YL, Wang L, Zhu HY, Wu JZ, Xia Y, et al. EBV-miR-BHRF1-1 Targets p53 Gene: Potential Role in Epstein-Barr Virus Associated Chronic Lymphocytic Leukemia. Cancer Res Treat (2020) 52(2):492–504. doi: 10.4143/crt.2019.457
23. Wang Q, Lingel A, Geiser V, Kwapnoski Z, Zhang L. Tumor Suppressor p53 Stimulates the Expression of Epstein-Barr Virus Latent Membrane Protein 1. J Virol (2017) 91(20):e00312-17. doi: 10.1128/JVI.00312-17
24. Pomari E, Silva R, Moro L, La Marca G, Perandin F, Verra F, et al. Droplet Digital PCR for the Detection of Plasmodium falciparum DNA in Whole Blood and Serum: A Comparative Analysis with Other Molecular Methods. Pathogens (2020) 9(6):478. doi: 10.3390/pathogens9060478
25. Cao WW, He DS, Chen ZJ, Zuo YZ, Chen X, Chang YL, et al. Development of a droplet digital PCR for detection and quantification of porcine epidemic diarrhea virus. J Vet Diagn Invest (2020) 32(4):572–6. doi: 10.1177/1040638720924753
26. Chen K, Ma Y, Ding T, Zhang X, Chen B, Guan M. Effectiveness of digital PCR for MYD88(L265P) detection in vitreous fluid for primary central nervous system lymphoma diagnosis. Exp Ther Med (2020) 20(1):301–8. doi: 10.3892/etm.2020.8695
27. Shuto T, Nishikawa J, Shimokuri K, Yanagi A, Takagi T, Takagi F, et al. Establishment of a Screening Method for Epstein-Barr Virus-Associated Gastric Carcinoma by Droplet Digital PCR. Microorganisms (2019) 7(12):628. doi: 10.3390/microorganisms7120628
28. Koh J, Lee KW, Nam SK, Seo AN, Kim JW, Kim JW, et al. Development and Validation of an Easy-to-Implement, Practical Algorithm for the Identification of Molecular Subtypes of Gastric Cancer: Prognostic and Therapeutic Implications. Oncologist (2019) 24(12):e1321–e30. doi: 10.1634/theoncologist.2019-0058
29. Epstein A. Why and How Epstein-Barr Virus Was Discovered 50 Years Ago. Curr Top Microbiol Immunol (2015) 390(Pt 1):3–15. doi: 10.1007/978-3-319-22822-8_1
30. Daskalogianni C, Pyndiah S, Apcher S, Mazars A, Manoury B, Ammari N, et al. Epstein-Barr virus-encoded EBNA1 and ZEBRA: targets for therapeutic strategies against EBV-carrying cancers. J Pathol (2015) 235(2):334–41. doi: 10.1002/path.4431
31. Lieberman PM. Virology. Epstein-Barr virus turns 50. Science (2014) 343(6177):1323–5. doi: 10.1126/science.1252786
32. Hutt-Fletcher LM. The Long and Complicated Relationship between Epstein-Barr Virus and Epithelial Cells. J Virol (2017) 91(1):e01677-16. doi: 10.1128/JVI.01677-16
33. Nanbo A, Kachi K, Yoshiyama H, Ohba Y. Epstein-Barr virus exploits host endocytic machinery for cell-to-cell viral transmission rather than a virological synapse. J Gen Virol (2016) 97(11):2989–3006. doi: 10.1099/jgv.0.000605
34. Lieberman PM. Keeping it quiet: chromatin control of gammaherpesvirus latency. Nat Rev Microbiol (2013) 11(12):863–75. doi: 10.1038/nrmicro3135
35. Shinozaki A, Sakatani T, Ushiku T, Hino R, Isogai M, Ishikawa S, et al. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res (2010) 70(11):4719–27. doi: 10.1158/0008-5472.CAN-09-4620
36. Luo B, Wang Y, Wang XF, Liang H, Yan LP, Huang BH, et al. Expression of Epstein-Barr virus genes in EBV-associated gastric carcinomas. World J Gastroenterol (2005) 11(5):629–33. doi: 10.3748/wjg.v11.i5.629
37. Sugiura M, Imai S, Tokunaga M, Koizumi S, Uchizawa M, Okamoto K, et al. Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: unique viral latency in the tumour cells. Br J Cancer (1996) 74(4):625–31. doi: 10.1038/bjc.1996.412
38. Iwakiri D, Eizuru Y, Tokunaga M, Takada K. Autocrine growth of Epstein-Barr virus-positive gastric carcinoma cells mediated by an Epstein-Barr virus-encoded small RNA. Cancer Res (2003) 63(21):7062–7.
39. Banerjee AS, Pal AD, Banerjee S. Epstein-Barr virus-encoded small non-coding RNAs induce cancer cell chemoresistance and migration. Virology (2013) 443(2):294–305. doi: 10.1016/j.virol.2013.05.020
40. Imai S, Koizumi S, Sugiura M, Tokunaga M, Uemura Y, Yamamoto N, et al. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc Natl Acad Sci USA (1994) 91(19):9131–5. doi: 10.1073/pnas.91.19.9131
41. Murray PG, Niedobitek G, Kremmer E, Grasser F, Reynolds GM, Cruchley A, et al. In situ detection of the Epstein-Barr virus-encoded nuclear antigen 1 in oral hairy leukoplakia and virus-associated carcinomas. J Pathol (1996) 178(1):44–7. doi: 10.1002/(SICI)1096-9896(199601)178:1<44::AID-PATH471>3.0.CO;2-0
42. Cheng TC, Hsieh SS, Hsu WL, Chen YF, Ho HH, Sheu LF. Expression of Epstein-Barr nuclear antigen 1 in gastric carcinoma cells is associated with enhanced tumorigenicity and reduced cisplatin sensitivity. Int J Oncol (2010) 36(1):151–60. doi: 10.3892/ijo_00000486
43. Kim SM, Hur DY, Hong SW, Kim JH. EBV-encoded EBNA1 regulates cell viability by modulating miR34a-NOX2-ROS signaling in gastric cancer cells. Biochem Biophys Res Commun (2017) 494(3-4):550–5. doi: 10.1016/j.bbrc.2017.10.095
44. Sivachandran N, Dawson CW, Young LS, Liu FF, Middeldorp J, Frappier L. Contributions of the Epstein-Barr virus EBNA1 protein to gastric carcinoma. J Virol (2012) 86(1):60–8. doi: 10.1128/JVI.05623-11
45. Namba-Fukuyo H, Funata S, Matsusaka K, Fukuyo M, Rahmutulla B, Mano Y, et al. TET2 functions as a resistance factor against DNA methylation acquisition during Epstein-Barr virus infection. Oncotarget (2016) 7(49):81512–26. doi: 10.18632/oncotarget.13130
46. Mohidin TB, Ng CC. BARF1 gene silencing triggers caspase-dependent mitochondrial apoptosis in Epstein-Barr virus-positive malignant cells. J Biosci (2015) 40(1):41–51. doi: 10.1007/s12038-015-9502-z
47. Kim DH, Chang MS, Yoon CJ, Middeldorp JM, Martinez OM, Byeon SJ, et al. Epstein-Barr virus BARF1-induced NFkappaB/miR-146a/SMAD4 alterations in stomach cancer cells. Oncotarget (2016) 7(50):82213–27. doi: 10.18632/oncotarget.10511
48. Chang MS, Kim DH, Roh JK, Middeldorp JM, Kim YS, Kim S, et al. Epstein-Barr virus-encoded BARF1 promotes proliferation of gastric carcinoma cells through regulation of NF-kappaB. J Virol (2013) 87(19):10515–23. doi: 10.1128/JVI.00955-13
49. Wiech T, Nikolopoulos E, Lassman S, Heidt T, Schopflin A, Sarbia M, et al. Cyclin D1 expression is induced by viral BARF1 and is overexpressed in EBV-associated gastric cancer. Virchows Arch (2008) 452(6):621–7. doi: 10.1007/s00428-008-0594-9
50. Chang MS, Lee HS, Jung EJ, Kim CW, Lee BL, Kim WH. Cell-cycle regulators, bcl-2 and NF-kappaB in Epstein-Barr virus-positive gastric carcinomas. Int J Oncol (2005) 27(5):1265–72. doi: 10.3892/ijo.27.5.1265
51. Hino R, Uozaki H, Inoue Y, Shintani Y, Ushiku T, Sakatani T, et al. Survival advantage of EBV-associated gastric carcinoma: survivin up-regulation by viral latent membrane protein 2A. Cancer Res (2008) 68(5):1427–35. doi: 10.1158/0008-5472.CAN-07-3027
52. Yasui M, Kunita A, Numakura S, Uozaki H, Ushiku T, Fukayama M. Cancer stem cells in Epstein-Barr virus-associated gastric carcinoma. Cancer Sci (2020) 111(7):2598–607. doi: 10.1111/cas.14435
53. Liu X, Gao Y, Luo B, Zhao Y. Construction and Antiapoptosis Activities of Recombinant Adenoviral Expression Vector Carrying EBV Latent Membrane Protein 2A. Gastroenterol Res Pract (2011) 2011:182832. doi: 10.1155/2011/182832
54. Pal AD, Basak NP, Banerjee AS, Banerjee S. Epstein-Barr virus latent membrane protein-2A alters mitochondrial dynamics promoting cellular migration mediated by Notch signaling pathway. Carcinogenesis (2014) 35(7):1592–601. doi: 10.1093/carcin/bgu069
55. Wang J, Liu W, Zhang X, Zhang Y, Xiao H, Luo B. LMP2A induces DNA methylation and expression repression of AQP3 in EBV-associated gastric carcinoma. Virology (2019) 534:87–95. doi: 10.1016/j.virol.2019.06.006
56. Shi Q, Zhang Y, Liu W, Xiao H, Qi Y, Li J, et al. Latent membrane protein 2A inhibits expression level of Smad2 through regulating miR-155-5p in EBV-associated gastric cancer cell lines. J Med Virol (2020) 92(1):96–106. doi: 10.1002/jmv.25579
57. Qi YF, Liu M, Zhang Y, Liu W, Xiao H, Luo B. EBV down-regulates COX-2 expression via TRAF2 and ERK signal pathway in EBV-associated gastric cancer. Virus Res (2019) 272:197735. doi: 10.1016/j.virusres.2019.197735
58. Deb Pal A, Banerjee S. Epstein-Barr virus latent membrane protein 2A mediated activation of Sonic Hedgehog pathway induces HLA class Ia downregulation in gastric cancer cells. Virology (2015) 484:22–32. doi: 10.1016/j.virol.2015.05.007
59. Liu W, Song YY, Wang JY, Xiao H, Zhang Y, Luo B. Dysregulation of FOXO transcription factors in Epstein-Barr virus-associated gastric carcinoma. Virus Res (2020) 276:197808. doi: 10.1016/j.virusres.2019.197808
60. Fukuda M, Longnecker R. Epstein-Barr virus latent membrane protein 2A mediates transformation through constitutive activation of the Ras/PI3-K/Akt Pathway. J Virol (2007) 81(17):9299–306. doi: 10.1128/JVI.00537-07
61. Hino R, Uozaki H, Murakami N, Ushiku T, Shinozaki A, Ishikawa S, et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res (2009) 69(7):2766–74. doi: 10.1158/0008-5472.CAN-08-3070
62. Zheng X, Wang J, Wei L, Peng Q, Gao Y, Fu Y, et al. Epstein-Barr Virus MicroRNA miR-BART5-3p Inhibits p53 Expression. J Virol (2018) 92(23):e01677-16. doi: 10.1128/JVI.01022-18
63. Lung RW, Hau PM, Yu KH, Yip KY, Tong JH, Chak WP, et al. EBV-encoded miRNAs target ATM-mediated response in nasopharyngeal carcinoma. J Pathol (2018) 244(4):394–407. doi: 10.1002/path.5018
64. Cai L, Ye Y, Jiang Q, Chen Y, Lyu X, Li J, et al. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat Commun (2015) 6:7353. doi: 10.1038/ncomms8353
65. Cai LM, Lyu XM, Luo WR, Cui XF, Ye YF, Yuan CC, et al. EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. Oncogene (2015) 34(17):2156–66. doi: 10.1038/onc.2014.341
66. Min K, Lee SK. EBV miR-BART10-3p Promotes Cell Proliferation and Migration by Targeting DKK1. Int J Biol Sci (2019) 15(3):657–67. doi: 10.7150/ijbs.30099
67. Song Y, Li X, Zeng Z, Li Q, Gong Z, Liao Q, et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget (2016) 7(24):36783–99. doi: 10.18632/oncotarget.9170
68. Song Y, Li Q, Liao S, Zhong K, Jin Y, Zeng T. Epstein-Barr virus-encoded miR-BART11 promotes tumor-associated macrophage-induced epithelial-mesenchymal transition via targeting FOXP1 in gastric cancer. Virology (2020) 548:6–16. doi: 10.1016/j.virol.2020.05.011
69. Dong M, Gong LP, Chen JN, Zhang XF, Zhang YW, Hui DY, et al. EBV-miR-BART10-3p and EBV-miR-BART22 promote metastasis of EBV-associated gastric carcinoma by activating the canonical Wnt signaling pathway. Cell Oncol (Dordr) (2020). doi: 10.1007/s13402-020-00538-0
70. Wang J, Zheng X, Qin Z, Wei L, Lu Y, Peng Q, et al. Epstein-Barr virus miR-BART3-3p promotes tumorigenesis by regulating the senescence pathway in gastric cancer. J Biol Chem (2019) 294(13):4854–66. doi: 10.1074/jbc.RA118.006853
71. Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med (2008) 205(11):2551–60. doi: 10.1084/jem.20072581
72. Babu SG, Ponia SS, Kumar D, Saxena S. Cellular oncomiR orthologue in EBV oncogenesis. Comput Biol Med (2011) 41(10):891–8. doi: 10.1016/j.compbiomed.2011.07.007
73. Lei T, Yuen KS, Xu R, Tsao SW, Chen H, Li M, et al. Targeting of DICE1 tumor suppressor by Epstein-Barr virus-encoded miR-BART3* microRNA in nasopharyngeal carcinoma. Int J Cancer (2013) 133(1):79–87. doi: 10.1002/ijc.28007
74. Hsu CY, Yi YH, Chang KP, Chang YS, Chen SJ, Chen HC. The Epstein-Barr virus-encoded microRNA MiR-BART9 promotes tumor metastasis by targeting E-cadherin in nasopharyngeal carcinoma. PloS Pathog (2014) 10(2):e1003974. doi: 10.1371/journal.ppat.1003974
75. Tsai CY, Liu YY, Liu KH, Hsu JT, Chen TC, Chiu CT, et al. Comprehensive profiling of virus microRNAs of Epstein-Barr virus-associated gastric carcinoma: highlighting the interactions of ebv-Bart9 and host tumor cells. J Gastroenterol Hepatol (2017) 32(1):82–91. doi: 10.1111/jgh.13432
76. Marquitz AR, Mathur A, Nam CS, Raab-Traub N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology (2011) 412(2):392–400. doi: 10.1016/j.virol.2011.01.028
77. Shinozaki-Ushiku A, Kunita A, Isogai M, Hibiya T, Ushiku T, Takada K, et al. Profiling of Virus-Encoded MicroRNAs in Epstein-Barr Virus-Associated Gastric Carcinoma and Their Roles in Gastric Carcinogenesis. J Virol (2015) 89(10):5581–91. doi: 10.1128/JVI.03639-14
78. Kim H, Choi H, Lee SK. Epstein-Barr virus miR-BART20-5p regulates cell proliferation and apoptosis by targeting BAD. Cancer Lett (2015) 356(2 Pt B):733–42. doi: 10.1016/j.canlet.2014.10.023
79. Hooykaas MJG, van Gent M, Soppe JA, Kruse E, Boer IGJ, van Leenen D, et al. EBV MicroRNA BART16 Suppresses Type I IFN Signaling. J Immunol (2017) 198(10):4062–73. doi: 10.4049/jimmunol.1501605
80. Lung RW, Tong JH, Sung YM, Leung PS, Ng DC, Chau SL, et al. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia (2009) 11(11):1174–84. doi: 10.1593/neo.09888
81. Yoon JH, Min K, Lee SK. Epstein-Barr Virus miR-BART17-5p Promotes Migration and Anchorage-Independent Growth by Targeting Kruppel-Like Factor 2 in Gastric Cancer. Microorganisms (2020) 8(2):58. doi: 10.3390/microorganisms8020258
82. Treece AL, Duncan DL, Tang W, Elmore S, Morgan DR, Dominguez RL, et al. Gastric adenocarcinoma microRNA profiles in fixed tissue and in plasma reveal cancer-associated and Epstein-Barr virus-related expression patterns. Lab Invest (2016) 96(6):661–71. doi: 10.1038/labinvest.2016.33
83. Cai L, Long Y, Chong T, Cai W, Tsang CM, Zhou X, et al. EBV-miR-BART7-3p Imposes Stemness in Nasopharyngeal Carcinoma Cells by Suppressing SMAD7. Front Genet (2019) 10:939.
84. Choi H, Lee SK. TAX1BP1 downregulation by EBV-miR-BART15-3p enhances chemosensitivity of gastric cancer cells to 5-FU. Arch Virol (2017) 162(2):369–77. doi: 10.1007/s00705-016-3109-z
85. Schroder K, Tschopp J. The inflammasomes. Cell (2010) 140(6):821–32. doi: 10.1016/j.cell.2010.01.040
86. Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol (2019) 19(8):477–89. doi: 10.1038/s41577-019-0165-0
87. Matsusaka K, Kaneda A, Nagae G, Ushiku T, Kikuchi Y, Hino R, et al. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res (2011) 71(23):7187–97. doi: 10.1158/0008-5472.CAN-11-1349
88. Zhao J, Liang Q, Cheung KF, Kang W, Lung RW, Tong JH, et al. Genome-wide identification of Epstein-Barr virus-driven promoter methylation profiles of human genes in gastric cancer cells. Cancer (2013) 119(2):304–12. doi: 10.1002/cncr.27724
89. Farrell PJ. Epstein-Barr Virus and Cancer. Annu Rev Pathol (2019) 14:29–53. doi: 10.1146/annurev-pathmechdis-012418-013023
90. Ushiku T, Chong JM, Uozaki H, Hino R, Chang MS, Sudo M, et al. p73 gene promoter methylation in Epstein-Barr virus-associated gastric carcinoma. Int J Cancer (2007) 120(1):60–6. doi: 10.1002/ijc.22275
91. Zhao J, Liang Q, Cheung KF, Kang W, Dong Y, Lung RW, et al. Somatostatin receptor 1, a novel EBV-associated CpG hypermethylated gene, contributes to the pathogenesis of EBV-associated gastric cancer. Br J Cancer (2013) 108(12):2557–64. doi: 10.1038/bjc.2013.263
92. Yu J, Liang Q, Wang J, Wang K, Gao J, Zhang J, et al. REC8 functions as a tumor suppressor and is epigenetically downregulated in gastric cancer, especially in EBV-positive subtype. Oncogene (2017) 36(2):182–93. doi: 10.1038/onc.2016.187
93. Okada T, Nakamura M, Nishikawa J, Sakai K, Zhang Y, Saito M, et al. Identification of genes specifically methylated in Epstein-Barr virus-associated gastric carcinomas. Cancer Sci (2013) 104(10):1309–14. doi: 10.1111/cas.12228
94. He D, Zhang YW, Zhang NN, Zhou L, Chen JN, Jiang Y, et al. Aberrant gene promoter methylation of p16, FHIT, CRBP1, WWOX, and DLC-1 in Epstein-Barr virus-associated gastric carcinomas. Med Oncol (2015) 32(4):92. doi: 10.1007/s12032-015-0525-y
95. Zhao Z, Liu W, Liu J, Wang J, Luo B. The effect of EBV on WIF1, NLK, and APC gene methylation and expression in gastric carcinoma and nasopharyngeal cancer. J Med Virol (2017) 89(10):1844–51. doi: 10.1002/jmv.24863
96. Boger C, Kruger S, Behrens HM, Bock S, Haag J, Kalthoff H, et al. Epstein-Barr virus-associated gastric cancer reveals intratumoral heterogeneity of PIK3CA mutations. Ann Oncol (2017) 28(5):1005–14. doi: 10.1093/annonc/mdx047
97. Ignatova E, Seriak D, Fedyanin M, Tryakin A, Pokataev I, Menshikova S, et al. Epstein-Barr virus-associated gastric cancer: disease that requires special approach. Gastric Cancer (2020) 236(6):951–60. doi: 10.1007/s10120-020-01095-z
98. He CY, Qiu MZ, Yang XH, Zhou DL, Ma JJ, Long YK, et al. Classification of gastric cancer by EBV status combined with molecular profiling predicts patient prognosis. Clin Transl Med (2020) 10(1):353–62. doi: 10.1002/ctm2.32
99. Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet (2011) 43(12):1219–23. doi: 10.1038/ng.982
100. Gulley ML. Genomic assays for Epstein-Barr virus-positive gastric adenocarcinoma. Exp Mol Med (2015) 47:e134. doi: 10.1038/emm.2014.93
101. Wang J, Liu W, Zhao Z, Zhang Y, Luo B. Association of the Aquaporin 3 Gene Polymorphism (rs2231231) with Epstein-Barr Virus-Associated Cancers in China. Intervirology (2018) 61(2):72–8.
102. Su X, Ye Z, Wang Z, Long Y, Qiu M, He C. Epstein-Barr virus infection associated with pepsinogens and Helicobacter pylori infection in patients with gastric cancer. Virus Res (2018) 256:1–5. doi: 10.1016/j.virusres.2018.07.017
103. Teresa F, Serra N, Capra G, Mascarella C, Gagliardi C, Di Carlo P, et al. Helicobacter pylori and Epstein-Barr Virus Infection in Gastric Diseases: Correlation with IL-10 and IL1RN Polymorphism. J Oncol (2019) 2019:1785132. doi: 10.1155/2019/1785132
104. Roh CK, Choi YY, Choi S, Seo WJ, Cho M, Jang E, et al. Single Patient Classifier Assay, Microsatellite Instability, and Epstein-Barr Virus Status Predict Clinical Outcomes in Stage II/III Gastric Cancer: Results from CLASSIC Trial. Yonsei Med J (2019) 60(2):132–9. doi: 10.3349/ymj.2019.60.2.132
105. Kim SY, Park C, Kim HJ, Park J, Hwang J, Kim JI, et al. Deregulation of immune response genes in patients with Epstein-Barr virus-associated gastric cancer and outcomes. Gastroenterology (2015) 148(1):137–47 e9. doi: 10.1053/j.gastro.2014.09.020
106. van Beek J, zur Hausen A, Snel SN, Berkhof J, Kranenbarg EK, van de Velde CJ, et al. Morphological evidence of an activated cytotoxic T-cell infiltrate in EBV-positive gastric carcinoma preventing lymph node metastases. Am J Surg Pathol (2006) 30(1):59–65. doi: 10.1097/01.pas.0000176428.06629.1e
107. Ichimura T, Abe H, Morikawa T, Yamashita H, Ishikawa S, Ushiku T, et al. Low density of CD204-positive M2-type tumor-associated macrophages in Epstein-Barr virus-associated gastric cancer: a clinicopathologic study with digital image analysis. Hum Pathol (2016) 56:74–80. doi: 10.1371/journal.ppat.1003341
108. Song HJ, Srivastava A, Lee J, Kim YS, Kim KM, Ki Kang W, et al. Host inflammatory response predicts survival of patients with Epstein-Barr virus-associated gastric carcinoma. Gastroenterology (2010) 139(1):84–92 e2. doi: 10.18632/oncotarget.9076
109. Strong MJ, Xu G, Coco J, Baribault C, Vinay DS, Lacey MR, et al. Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy. PloS Pathog (2013) 9(5):e1003341. doi: 10.5230/jgc.2016.16.1.1
110. Derks S, Liao X, Chiaravalli AM, Xu X, Camargo MC, Solcia E, et al. Abundant PD-L1 expression in Epstein-Barr Virus-infected gastric cancers. Oncotarget (2016) 7(22):32925–32. doi: 10.1002/jso.25022
111. Cho J, Kang MS, Kim KM. Epstein-Barr Virus-Associated Gastric Carcinoma and Specific Features of the Accompanying Immune Response. J Gastric Cancer (2016) 16(1):1–7. doi: 10.1093/jnci/djx215
112. Pereira MA, Ramos M, Faraj SF, Dias AR, Yagi OK, Zilberstein B, et al. Clinicopathological and prognostic features of Epstein-Barr virus infection, microsatellite instability, and PD-L1 expression in gastric cancer. J Surg Oncol (2018) 117(5):829–39. doi: 10.1038/s41591-018-0101-z
113. Almhanna K, Antonia S. PD-L1 Antibodies for EBV-Positive Gastric Cancer, Going Beyond PD-L1 Expression and Microsatellite Instability. J Natl Cancer Inst (2018) 110(3):221–2. doi: 10.1016/j.prp.2020.152881
114. Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med (2018) 24(9):1449–58. doi: 10.3389/fgene.2019.00939
Keywords: Epstein–Barr virus, gastric cancer, miRNA, DNA methylation, immune checkpoint
Citation: Sun K, Jia K, Lv H, Wang S-Q, Wu Y, Lei H and Chen X (2020) EBV-Positive Gastric Cancer: Current Knowledge and Future Perspectives. Front. Oncol. 10:583463. doi: 10.3389/fonc.2020.583463
Received: 15 July 2020; Accepted: 09 September 2020;
Published: 14 December 2020.
Edited by:
Laura Lorenzon, Catholic University of the Sacred Heart, ItalyCopyright © 2020 Sun, Jia, Lv, Wang, Wu, Lei and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaobing Chen, emx5eWNoZW54YjA4MDdAenp1LmVkdS5jbg==
 Keran Sun
Keran Sun Keqi Jia
Keqi Jia Huifang Lv1
Huifang Lv1 Xiaobing Chen
Xiaobing Chen