Corrigendum: Knockdown of lncRNA LINC01234 Suppresses the Tumorigenesis of Liver Cancer via Sponging miR-513a-5p
- 1State Key Laboratory of Bioreactor Engineering and Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai, China
- 2Department of Hematology and Oncology, Hwa Mei Hospital, Ningbo Institute of Life and Health Industry, University of Chinese Academy of Sciences, Ningbo, China
- 3Key Laboratory of Diagnosis and Treatment of Digestive System Tumors of Zhejiang Province, Ningbo, China
- 4Department of TCM Oncology, Putuo District Central Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai, China
- 5Department of Hepatobiliary Surgery, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
Background: Liver cancer is a frequent malignancy with poor prognosis and high mortality all over the world. It has been reported many lncRNAs could modulate the tumorigenesis of liver cancer. To identify novel potential targets for liver cancer, the differential expressed lncRNAs between liver cancer and adjacent normal tissues was analyzed with bioinformatics tool.
Methods: The differential expressed lncRNAs between liver cancer and adjacent normal tissues were analyzed with bioinformatics tool. Cell viability and proliferation was tested by CCK8 and Ki67, respectively. Apoptosis of liver cancer cells was tested by flow cytometry. Gene and protein expressions in liver cancer cells were measured by qRT-PCR and western blot, respectively. In vivo model of liver cancer was established to detect the effect of LINC01234 on liver cancer in vivo.
Results: LINC01234 was found to be negatively correlated with the survival rate of patients with liver cancer. Moreover, knockdown of LINC01234 significantly suppressed the proliferation and invasion of liver cancer cells via inducing the apoptosis. Meanwhile, miR-513a-5p was sponged by LINC01234, and USP4 was found to be a direct target of miR-513a-5p. In addition, LINC01234 knockdown inhibited the tumorigenesis of liver cancer via inactivating TGF-β signaling. Furthermore, silencing of LINC01234 notably inhibited the tumor growth of liver cancer in vivo.
Conclusion: Downregulation of LINC01234 could inhibit the tumorigenesis of liver cancer via mediation of miR-513a-5p/USP4/TGF-β axis. Thus, LINC01234 might serve as a new target for the treatment of liver cancer.
Background
Liver cancer is a common malignant tumor, with a 15–17% 5-year survival rate (1, 2). The prognosis of patients with liver cancer is poor because of the high incidence of postoperative recurrence and metastasis (3). Surgery is still the main treatment strategy; however, liver cancer patients are usually diagnosed at advanced stages. Thus, they often miss the optimal opportunity for surgical resection (4). Furthermore, liver cancer is highly resistant to conventional chemotherapy and radiation therapy (5, 6). Currently, clinicopathologic prognostic factors include TNM stage, tumor size, tumor rupture and underlying cirrhosis (7). In addition to these traditional clinical prognostic factors, genetic biomarkers are novel indicators of liver cancer diagnosis and prognosis (8). Molecular biomarkers could predict patient prognosis and served as the target for the treatment of liver cancer (9), while it is still a lack of therapeutic markers for liver cancer. Therefore, it is necessary to adopt a comprehensive approach to identify novel biomarkers for the treatment of liver cancer.
LncRNAs are a class of non-coding RNA transcripts with the length of about 200 nucleotides (10). Intracellular lncRNAs are known to suppress the biological activity of miRNAs and regulate the expression of mRNAs targeted by miRNAs through competing endogenous RNAs (ceRNAs) (11). Therefore, lncRNAs have been considered to be important targets for treating multiple diseases (12, 13). Moreover, lncRNAs play key roles in malignant tumors, including liver cancer (14, 15). However, the effect of lncRNAs on tumorigenesis of liver cancer remains to be further explored.
MicroRNAs (miRNAs) are a class of non-coding small ribonucleic acids which can regulate gene expression by suppression of mRNA translation or degradation of mRNAs (16). Moreover, miRNAs have been considered as important biological regulators for multiple diseases, including liver cancer (1, 17). Meanwhile, in previous studies, lncRNAs have been reported to mediate the tumorigenesis of liver cancer via sponging miRNAs (15, 18). For example, Hong et al. (19) found that lncRNA LINC00460 can contribute to tumor growth and metastasis of hepatocellular carcinoma through miR-342-3p-dependent AGR2 up-regulation. Thus, miRNAs play a key role in liver cancer.
In the current study, we aimed to detect the differentially expressed lncRNAs, which is closely associated with the tumorigenesis of liver cancer. We hope the research could provide a new idea for the development of novel therapeutic strategies against liver cancer.
Materials and Methods
Cell Culture
Liver cancer cell lines (HepG2 and Huh-7), normal liver cell lines (L-02) and 293T cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, United States) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Thermo Fisher Scientific, Waltham, MA, United States), supplemented with 10% FBS, 1% penicillin and streptomycin (Thermo Fisher Scientific) in an incubator (37°C, 5% CO2).
Bioinformatics Analysis
The gene expression data of liver cancer and adjacent normal tissue was obtained from dataset (GSE113850) and the Cancer Genome Atlas (TCGA). The survival curve and the correlation between tumor stage of liver cancer were calculated based on the data from TCGA.
Quantitative Real Time Polymerase Chain Reaction
Total RNA was extracted from liver cancer cell lines using TRIzol reagent (TaKaRa, Tokyo, Japan) according to the manufacturer’s protocol. First-strand cDNA was synthesized using the PrimeScript RT reagent Kit (Takara) according to the manufacturer’s protocol. qRT-PCR was performed in an ABI7500 system using SYBR Green methods. qRT-PCR was performed in triplicate with the following protocol: 2 min at 94°C, followed by 35 cycles (30 s at 94°C and 45 s at 55°C). The primers for all the genes were obtained from GenePharma (Shanghai, China). 2–ΔΔCT method was used to quantify the data. The internal reference gene (U6 or β-actin) was used for normalization. The primers were obtained from GenePharma (Shanghai, China). LINC01234: forward, 5′-TCTCACCTTTCAAACGCTTGTC-3′ and reverse 5′-ACTCTCCTTCCTTTCCTCTGATTC-3′. MiR- 513a-5p: forward, 5′-AGGGAGGTGTCATCTCAACTGA-3′ and reverse 5′-CTCAACTGGTGTCGTGGAGTC-3′. β-actin: forward, 5′-GTCCACCGCAAATGCTTCTA-3′ and reverse 5′-TGCTGTCACCTTCACCGTTC-3′. USP4: forward, 5′-GGA CTATGTATTGGTCCCTACCG-3′ and reverse 5′-TCGATGG TTGCAATGGTGTC-3′. U6: forward, 5′-CTCGCTTCGGCAG CACAT-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′.
Cell Transfection
Lentiviral expressing short-hairpin RNA (shRNA1 or shRNA2) directed target LINC01234 and one non-targeting sequence (negative control) were obtained from Hanbio Biotechnology Co., Ltd (Shanghai, China). Next, LINC01234 shRNA1 or shRNA2 was packaged into lentiviruses. Then the lentiviral vector DNAs were then transfected into 293T cells including lenti-LINC01234 shRNAs and negative control (NC). After transfection, the cells were incubated at 37°C, and then the supernatant was collected. After that, supernatants of two LINC01234 shRNAs and negative control were filtered into particles. Finally, all liver cancer cells were infected with lentiviral particles according to the manufactures’ protocol. After 48 h of incubation, stable liver cancer cells were then selected by puromycin (2.5 μg/mL, Sigma Aldrich, St. Louis, MO, United States). Green fluorescence and qRT-PCR were used to verify the efficiency of transfection.
For miR-513a-5p transfection, liver cancer cells were transfected with miR-513a-5p agomir, miR-513a-5p antagomir or NC by Lipofectamine 2000 according to the previous reference (20). MiR-513a-5p agomir, miR-513a-5p antagomir and negative control RNAs were purchased from GenePharma (Shanghai, China). The efficiency of transfection was detected by q-PCR.
CCK-8 Assay
Liver cancer cells were seeded in 96-well plates (5 × 103 per well) overnight. Then, cells were treated with negative control (NC) or LINC01234 shRNA1 for 0, 24, 48 and 72 h, respectively. Subsequently, cells were treated with 10 μl CCK-8 reagents (Beyotime, Shanghai, China) and further incubated for 2 h at 37°C. Finally, the absorbance of liver cancer cells was measured at 450 nm using a microplate reader (Thermo Fisher Scientific).
Cell Apoptosis Analysis
Liver cancer cells were trypsinized, washed with phosphate buffered saline and resuspended in Annexin V Binding Buffer. Then, cells were stained with 5 μl FITC and 5 μl propidium (PI) in the dark for 15 min. Cells were analyzed using flow cytometer (BD, Franklin Lake, NJ, United States) to test the cell apoptosis rate.
Cell Invasion Assay
For cell invasion analysis, transwell assay was performed in this study. The upper chamber is pre-treated with 100 μl of Matrigel. Huh7 cells were seeded into the upper chamber in media with 1% FBS, and the density was adjusted to about 1.0 × 106 cells per chamber. RPMI1640 medium with 10% FBS was added in the lower chamber. After 24 h of incubation at 37°C, the transwell chamber was rinsed twice with PBS (5 min per time), fixed by 5% glutaraldehyde at 4°C and stained with 0.1% crystal violet for 30 minutes. The transwell chamber was washed twice with PBS and then observed under a microscope. The number of invaded cells was regarded to be a reflection of the invasion ability.
Dual Luciferase Reporter Assay
The partial sequences of LINC01234 and 3′-UTR of USP4 containing the putative binding sites of miR-513a-5p were synthetized and obtained from Sangon Biotech (Shanghai, China), then were cloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vectors (Promega, Madison, WI, United States) to construct wild-type reporter vectors LINC01234 (WT/MT) and USP4 (WT/MT), respectively. The LINC01234 (WT/MT) or USP4 (WT/MT) were transfected into 293T cells together with control, vector-control (NC) or miR-513a-5p agomir using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. The relative luciferase activity was analyzed by the Dual-Glo Luciferase Assay System (Promega). The relative luciferase activity was analyzed by the Dual-Glo Luciferase Assay System (Promega).
RNA Pull-Down
For the RNA pulldown assay, the Biotin RNA Labeling Mix (Roche, Basel, Switzerland) was used to transcribe and label probe-control or probe-LINC01234 from LINC01234 shRNA lenti vector in vitro. An RNA structure buffer (Thermo, MA, United States) was used to induce secondary structure formation from the biotin-labeled RNAs. The biotinylated LINC01234 and negative control (bio-NC) were generated via GenePharma and coated to streptavidin-conjugated magnetic beads. Liver cancer cells were lysed and then incubated with the magnetic beads for 6 h. The RNA on the beads was isolated and the enrichment level of miR-513a-5p was detected by PCR.
Wound Healing Assay
Huh-7 cells were plated into a 24-well Cell Culture Cluster and allowed to grow to 80–90% confluence. Then, cells were underlined perpendicular to the cell culture plate with a small pipette head. After washing with PBS 3 times, serum-free medium was used for further culture, and the scratch widths at 0 and 48 h were recorded under an optical microscope.
Immunofluorescence
Liver cancer cells or tumor tissues of mice were prefixed in 4% paraform for 10 min, and fixed in pre-cold methanol for another 10 min. Next, cells were incubated with primary antibodies overnight at 4(C: anti-Ki67 (Abcam; 1:1000), anti-Smad4 (Abcam; 1:1000). Goat anti-rabbit IgG antibody (Abcam; 1:5000) was used as the secondary antibody. The nuclei were stained with DAPI for 15 min at room temperature. The samples were visualized by fluorescence microscope (Olympus CX23, Tokyo, Japan) immediately.
Western Blot Detection
Total protein was isolated from cell lysates or tissues by using RIPA buffer. The concentration of protein was detected with a BCA protein kit (Thermo Fisher Scientific). Then, proteins (40 μg per lane) were separated with 10% SDS-PAGE gel and then transferred into polyvinylidene fluoride (PVDF, Thermo Fisher Scientific) membranes. After blocking with 3% skim milk for 1 h, the membranes were incubated with primary antibodies at 4°C overnight. Subsequently, membranes were incubated with secondary anti-rabbit antibody (Abcam; 1:5000) at room temperature for 1 h. Membranes were scanned by using an Odyssey Imaging System and analyzed with Odyssey v2.0 software (LICOR Biosciences, Lincoln, NE, United States). The primary antibodies used in this study as follows: anti-E-cadherin (Abcam, Cambridge, MA, United States; 1:1000), anti-vimentin (Abcam; 1:1000), anti-α-SMA (Abcam; 1:1000), anti-smad2 (Abcam; 1:1000), anti-smad3 (Abcam; 1:1000), anti-USP4 (Abcam; 1:1000), anti-cleaved caspase 3 (Abcam; 1:1000), anti-Akt (Abcam; 1:1000), anti-ERK (Abcam; 1:1000) and anti-β-actin (Abcam; 1:1000). β-actin was used as an internal control.
In vivo Study
18 BALB/c nude mice (6–8 weeks old) were purchased from Vital River (Beijing, China). The mice were housed within a dedicated SPF facility. Huh7 stable expressed LINC01234 shRNA1 cells were transplanted subcutaneously in each mouse according to the previous reference (21). The tumor volume was measured weekly according to the reference (22). At the end of the experiments, mice were sacrificed and the tumors were collected and weighted. The expression of p-smad2 and p-smad3 were detected by immunohistochemistry (IHC) staining as previously reported (19). All in vivo experiments were performed in accordance with National Institutes of Health guide for the care and use of laboratory animals, following a protocol approved by the Ethics Committees of East China University of Science and Technology.
TUNEL Staining
Apoptosis was also determined by the TUNEL assay according to the manufacturer’s instructions. Briefly, paraffin sections were washed, permeabilized, and then incubated with 50 μl TUNEL reaction mixtures in a wet box for 60 min at 37°C in the dark. For signal conversion, slides were incubated with 50 μl of peroxidase (POD) for 30 min at 37°C, rinsed with PBS, and then incubated with 50 μl diaminobenzidine (DAB) substrate solution for 10 min at 25°C. Finally, the expression of apoptotic cells was observed under an optical microscope.
Statistical Analysis
Each group were performed three independent experiments and all data were expressed as the mean ± standard deviation (SD). Differences were analyzed using Student’s t-test (only 2 groups) or one-way analysis of variance (ANOVA) followed by Tukey’s test (more than 2 groups, Graphpad Prism7). P < 0.05 was considered to indicate a statistically significant difference.
Results
Differentially Expressed lncRNAs Between Liver Cancer and Adjacent Normal Tissues
To detect differentially expressed lncRNAs between liver cancer and normal tissues, a bioinformatics analysis as performed. As indicated in Figure 1A, differentially expressed lncRNAs in GSE113850 data set (DS) were presented using Volcano Plot. In addition, differentially expressed lncRNAs among normal and tumor tissues were also presented in TCGA (Figure 1B). Among these differentially expressed lncRNAs in GSE113850 DS and TCGA, 59 were commonly downregulated, whereas 167 were commonly upregulated (Figure 1C). Moreover, 5 most upregulated overlap linRNAs in liver cancer were presented (Figure 1D). Among these 5 lncRNAs, high expression of LINC01234 was negatively correlated with the survival rate of patient with liver cancer (Figures 1E,F). These data suggested that LINC01234 might play an important role during the tumorigenesis of liver cancer. Thus, LINC01234 was selected for investigation in the following experiments.
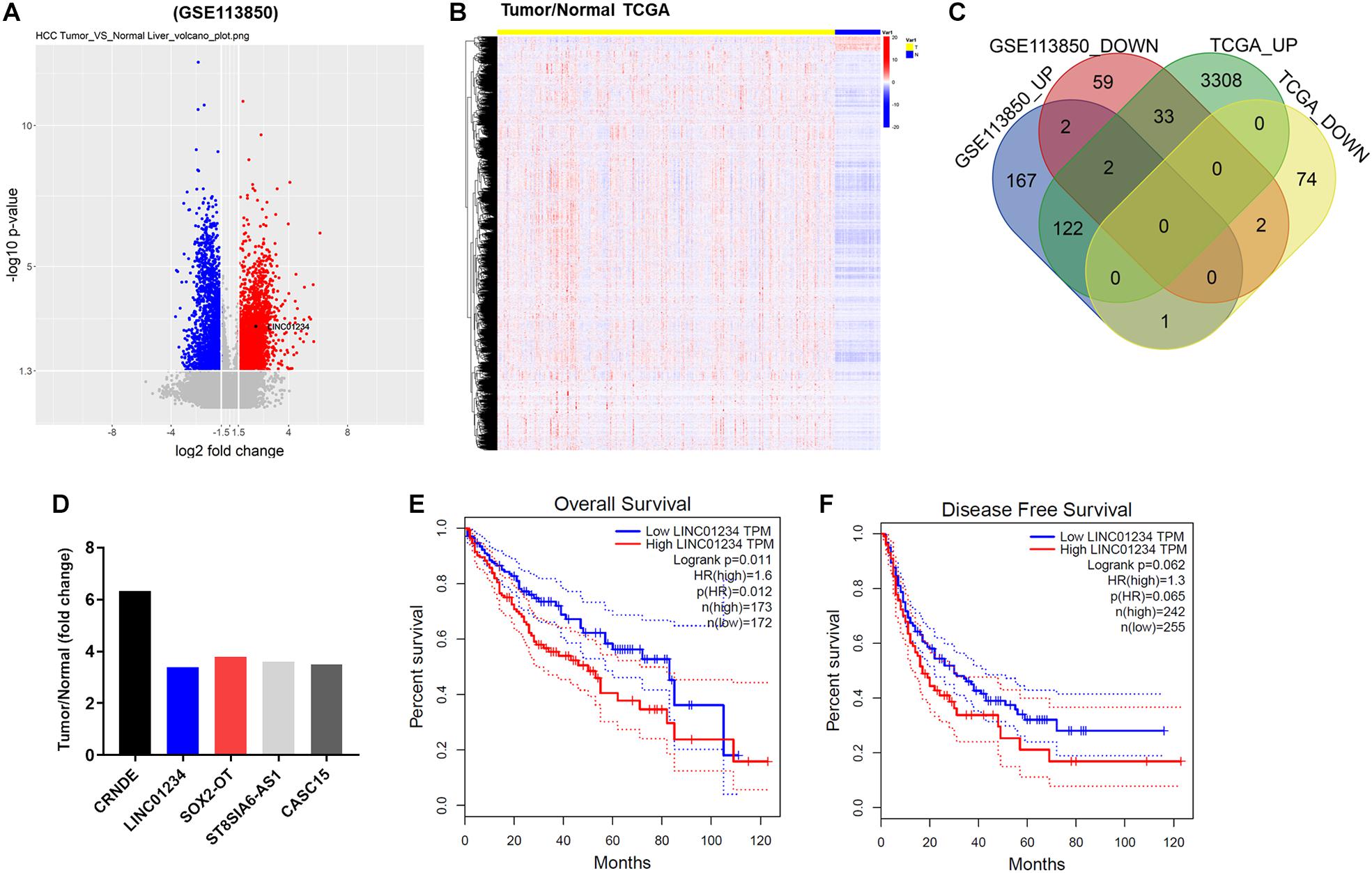
Figure 1. The differentially expressed lncRNAs between liver cancer and adjacent normal tissues. (A) Volcano plots illustrating the lncRNAs differentially expressed in liver cancer detected in the GSE113850 dataset. Red indicates a higher expression level, while blue indicates a lower expression level. (B) Differentially expressed lncRNAs among normal and liver cancer tissues were presented by TCGA. (C) Venn diagram showing the overlap among the differentially expressed lncRNAs in the GSE113850 dataset. (D) Dataset indicated overlap 5 upregulated lncRNAs in liver cancer tissues. (E,F) Overall and disease free survival of liver cancer were analyzed by TCGA.
LINC01234 Knockdown Significantly Decreased the Proliferation of Liver Cancer Cells
In order to test the gene expression, qRT-PCR was used. As indicated in Figure 2A, the expression of LINC01234 in liver cancer cells was much higher than that in normal liver cells. Moreover, LINC01234 shRNA1 and shRNA2 were stably transfected into liver cancer cells (Figures 2B,C), and the expression of LINC01234 in liver cancers was significantly downregulated when transfected with LINC01234 shRNA (Figure 2D). Since LINC01234 shRNA1 exhibited better transfection efficiency, it was therefore used in subsequent experiments. Furthermore, the viability and proliferation of Huh-7 and HepG2 cells were significantly inhibited by LINC01234 knockdown (Figures 2E–H). Meanwhile, Huh-7 cells were more sensitive to LINC01234 shRNA1 than HepG2 cells. Thus, Huh-7 cells were used in the following experiments. Taken together, LINC01234 silencing significantly decreased the proliferation of liver cancer cells.
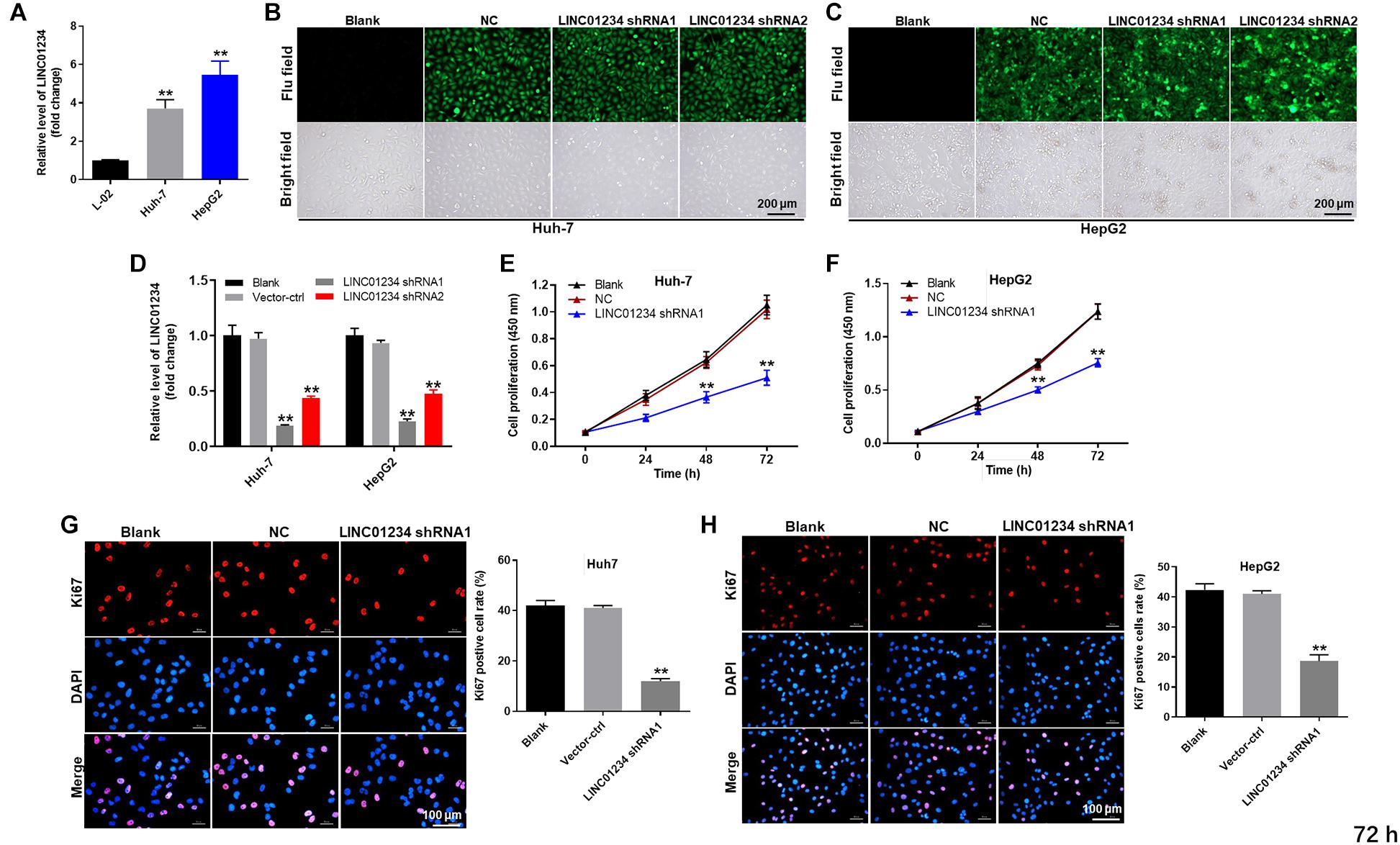
Figure 2. LINC01234 knockdown significantly decreased the proliferation of liver cancer cells. (A) The expression of LINC01234 in Huh-7, HepG2 and L-02 cells was detected by qRT-PCR. (B,C) Huh-7 cells were transfected with LINC01234 shRNA1 or shRNA2 for 24 h. Then, the efficiency of cell transfection was detected by green fluorescence staining. (D) The expression of LINC01234 in liver cancer cells was detected by qRT-PCR. The proliferation of (E) Huh-7 or (F) HepG2 cells was detected by CCK-8 assay. (G,H) Ki67 staining was performed to test the proliferation of Huh-7 and HepG2 cells. Red indicates the Ki67 staining. Blue indicates DAPI staining. **P < 0.01 compared to control.
LINC01234 Sponged miR-513a-5p
For the purpose of exploring the mechanism by which LINC01234 mediated the progression of liver cancer, Starbase1 was performed. As shown in Figure 3A, LINC01234 had a putative miR-513a-5p targeting site. In addition, miR-513a-5p agomir/antagomir was stably transfected into Huh-7 cells (Figure 3B). Meanwhile, co-transfection of the wild-type LINC01234 vector (WT-LINC01234) with miR-513a-5p agomir, significantly reduced luciferase activities compared with mutant LINC01234 vector (MT-LINC01234) (Figure 3C). Additionally, LINC01234 bound to miR-513a-5p (Figure 3D) was verified with pull down assay. Moreover, USP4 was found to be the direct target of miR-513a-5p (Figures 3E,F). However, the difference of USP4 level between liver cancer and adjacent normal tissues is not significant (Figure 3G). Besides, the difference of USP4 expression among different stages of liver cancer is not obvious (Figure 3H). Additionally, the expression of USP4 in liver cancer cells was notably upregulated by miR-513a-5p antagomir but inhibited by miR-513a-5p agomir (Figure 3I). Altogether, LINC01234 knockdown inhibited the tumorigenesis of liver cancer via mediation of miR-513a-5p/USP4 axis.
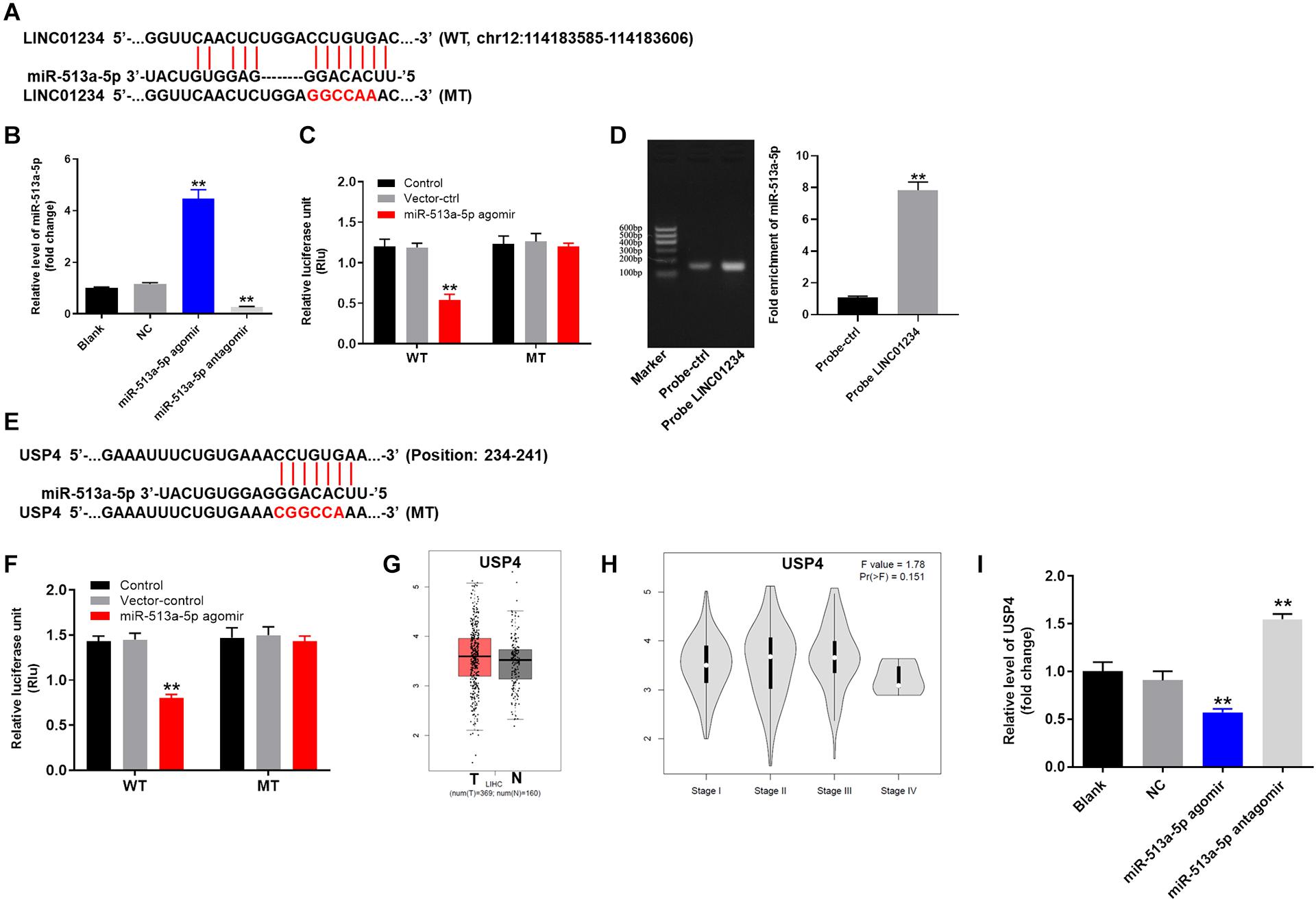
Figure 3. LINC01234 sponged miR-513a-5p. (A) Gene structure of LINC01234 indicating the predicted miR-513a-5p binding site in its 3′UTR. (B) Huh-7 cells were transfected with miR-513a-5p agomir/antagomir for 24 h. Then, cell transfection was verified by qRT-PCR. (C) The luciferase activity in Huh-7 cells after co-transfecting a plasmid encoding the wild-type (WT) or mutant (MT) LINC01234 3′-UTR and miR-513a-5p. (D) Co-localization of LINC01234 and miR-513a-5p detected using RNA pull-down. (E) Gene structure of USP4 indicating the predicted miR-513a-5p binding site in its 3′UTR. (F) The luciferase activity in Huh-7 cells after co-transfecting a plasmid encoding the wild-type (WT) or mutant (MT) USP4 3′-UTR and miR-513a-5p. (G) The differential expression of USP4 in normal and tumor tissues was analyzed from the data of TCGA. (H) The correlation between USP4 and different stages of liver cancer was analyzed. (I) The expression of USP4 in liver cancer cells was measured by qRT-PCR. **P < 0.01 compared to control.
Knockdown of LINC01234 Significantly Suppressed the Progression of Liver Cancer in vitro
Next, flow cytometry was performed to detect the cell apoptosis. As revealed in Figure 4A and Supplementary Figure S1, downregulation of LINC01234 notably induced the apoptosis of liver cancer cells. In addition, knockdown of LINC01234 greatly inhibited the migration and invasion of liver cancer cells (Figures 4B,C). Furthermore, the expression of E-cadherin in Huh-7 cells was significantly upregulated by LINC01234 shRNA1 (Figure 4D). In contrast, knockdown of LINC01234 greatly decreased the expression of vimentin and α-SMA in liver cancer cells (Figure 4D). To sum up, knockdown of LINC01234 significantly suppressed the progression of liver cancer in vitro.
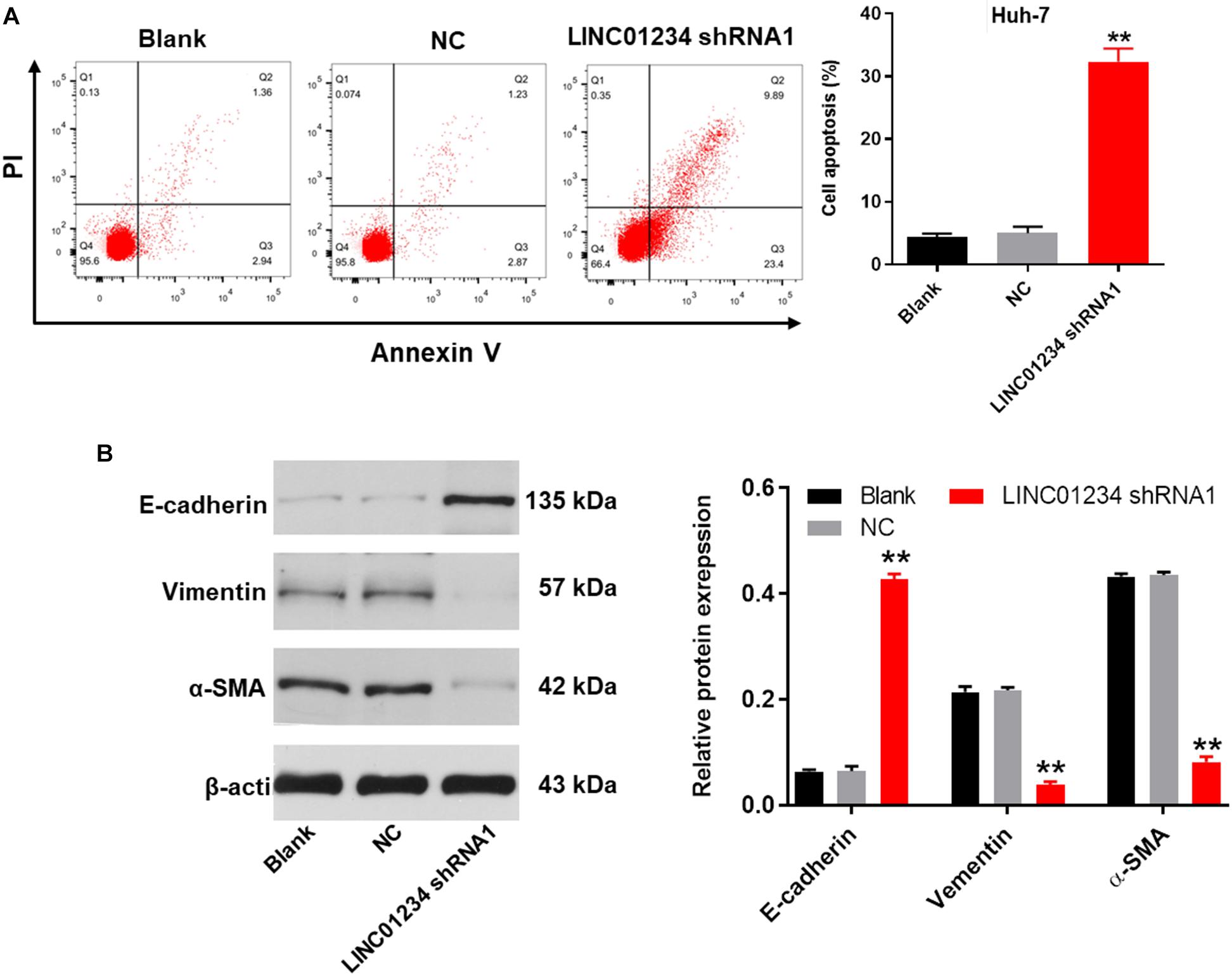
Figure 4. Knockdown of LINC01234 significantly suppressed the progression of liver cancer in vitro. (A) The apoptotic Huh-7 cells were examined by flow cytometry. (B) Cell migration was measured by wound healing assay. (C) Cell invasion was detected by transwell assay. (D) The protein expressions of E-cadherin, vimentin and α-SMA in Huh-7 cells were detected by western blot. The relative protein expressions were quantified by normalizing to β-actin. **P < 0.01 compared to control.
Downregulation of LINC01234 Notably Inhibited the Tumorigenesis of Liver Cancer via Mediation of miR-513a-5p/USP4/TGF-β1 Axis
In order to investigate the protein expressions in liver cancer cells, western blot was used. As indicated in Figures 5A,B, the expressions of USP4, p-Smad2 and p-Smad3 in Huh-7 cells were significantly downregulated in the presence of LINC01234 shRNA1, while this phenomenon was partially reversed by miR-513a-5p antagomir. In addition, the expression of Smad4 in cytoplasm of Huh-7 cells was significantly decreased by LINC01234 knockdown, which was partially reversed in the presence of miR-513a-5p antagomir (Figure 5C). This data suggested that Smad4 transferred from the cytoplasm to the nucleus in the presence of LINC01234 shRNA1. Moreover, LINC01234 shRNA1 greatly suppressed the expressions of p-Akt and p-ERK in Huh-7 cells. In contrast, the expression of cleaved caspase 3 in Huh-7 cells was notably upregulated in the presence of LINC01234 knockdown (Figures 5D,E). However, downregulation of miR-513a-5p partially reversed the effect of LINC01234 shRNA1 on these proteins (Figures 5D,E). In summary, silencing of LINC01234 notably inhibited the tumorigenesis of liver cancer via mediation of miR-513a-5p/USP4/TGF-β1 axis.
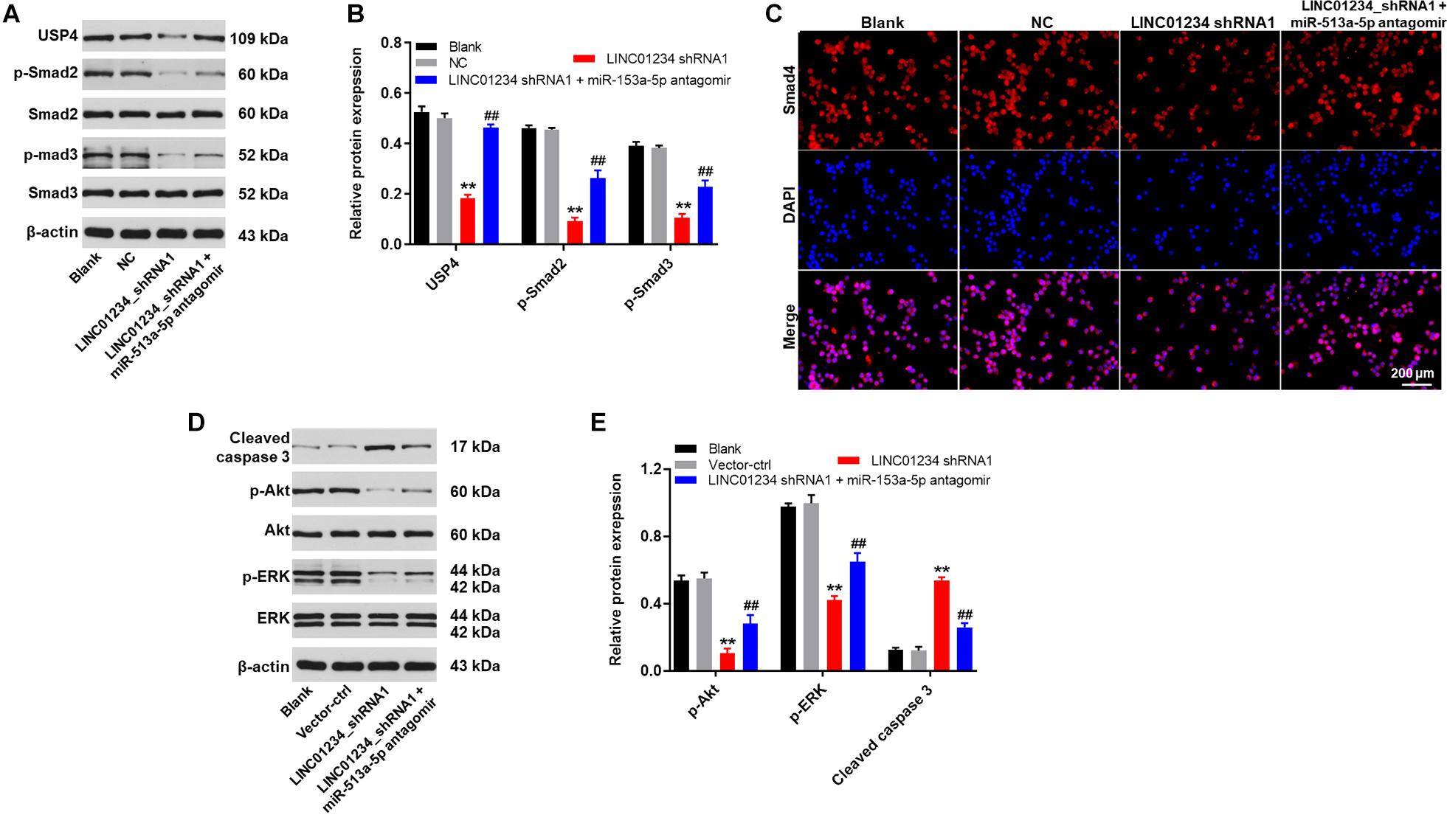
Figure 5. Downregulation of LINC01234 notably inhibited the tumorigenesis of liver cancer via mediation of miR-513a-5p/USP4/TGF-β1 axis. (A) The protein expressions of USP4, Smad2, Smad3, p-Smad2 and p-Smad3 in Huh-7 cells were detected by western blot. (B) The relative protein expressions were quantified by normalizing to β-actin. (C) The location of Smad4 was explored by immunofluorescence staining. Red indicates Smad4 fluorescence. Blue indicates DAPI fluorescence. (D) The protein expressions of Akt, p-Akt, cleaved caspase 3, ERK and p-ERK in Huh-7 cells were measured by western blot. (E) The relative protein expressions were quantified by normalizing to β-actin. **P < 0.01 compared to control. ##P < 0.01 compared to LINC01234 shRNA1.
Knockdown of LINC01234 Notably Attenuated the Tumor Growth of Liver Cancer in vivo Through Inactivation of TGF-β Signaling
Finally, to detect the effect of LINC01234 on liver cancer in vivo, xenograft mice model was established. As shown in Figures 6A,B, tumor sizes of mice were significantly decreased by LINC01234 knockdown. Consistently, knockdown of LINC01234 notably downregulated the tumor weights of mice (Figure 6C). Meanwhile, the expression of LINC01234 in tissues of mice was greatly inhibited by LINC01234 knockdown (Figure 6D). Furthermore, Ki67 and Tunnel assays indicated tumor cell proliferation was obviously suppressed by LINC01234 shRNA1 via inducing apoptosis (Figures 6E,F). The expressions of p-Smad2 and p-Smad3 in tumor tissues were notably decreased by LINC01234 knockdown (Figures 6G,H). Altogether, knockdown of LINC01234 notably attenuated the tumor growth of liver cancer through inactivation of TGF-β signaling in vivo.
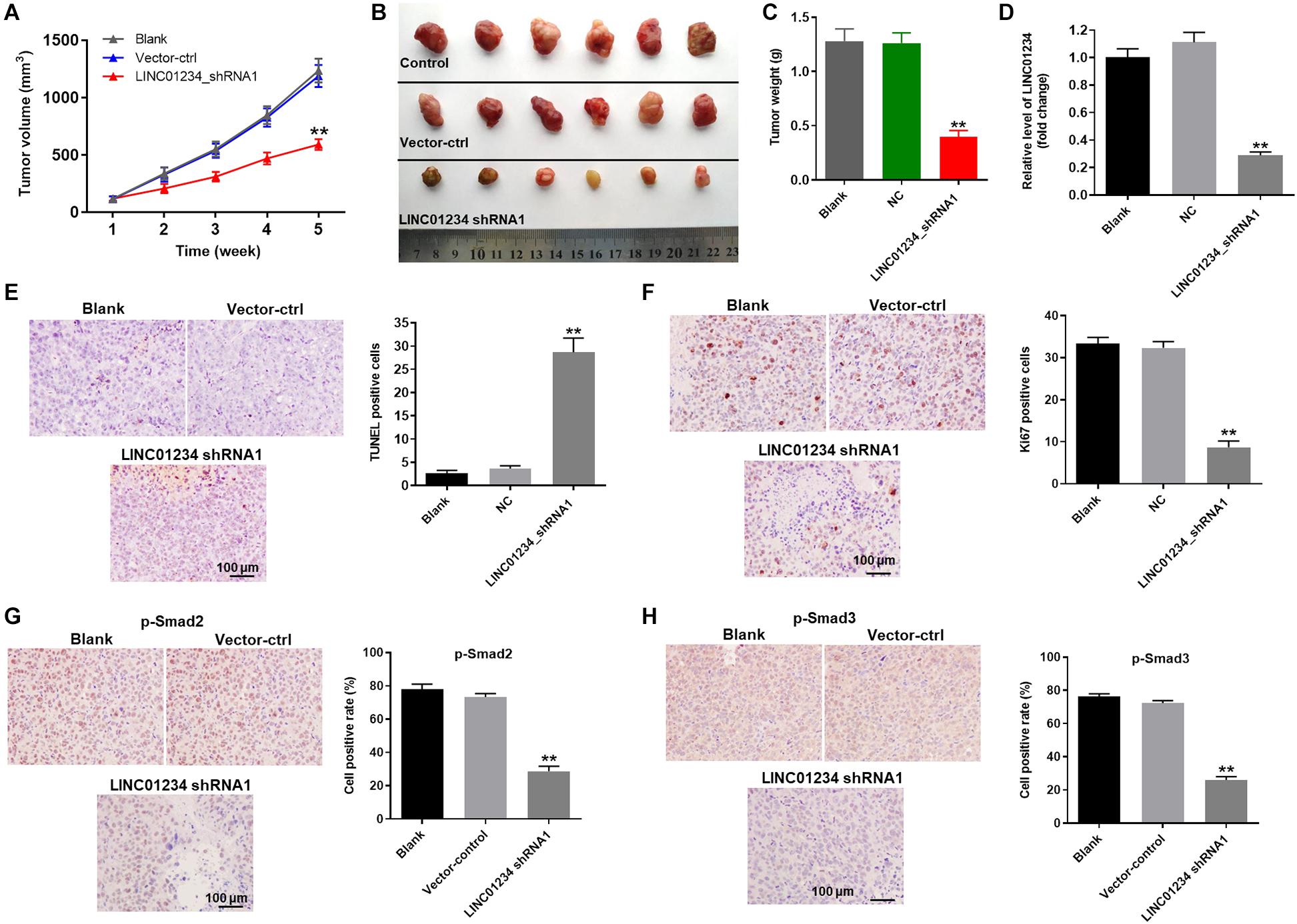
Figure 6. Knockdown of LINC01234 notably inhibited the tumor growth of liver cancer in vivo through inactivation of TGF-β signaling. Mice were subcutaneously injected Huh-7 cells transfected with vector-control or LINC01234 shRNA1 after tumors in mice were allowed to grow for 5 weeks. (A) Volumes of tumors were measured at the indicated times after transplantation. At the end of study, (B) tumor tissues were collected and imaged. (C) Tumors of each mouse were weighted. (D) The expression of LINC01234 in tumor tissues of mice was detected by qRT-PCR. (E) The apoptotic cells in tumor tissues of mice were detected by TUNEL staining. (F) Ki67 staining was performed to test the proliferation of liver cancer cells. (G,H) The expressions of p-Smad2 and p-Smad3 in tumor tissues of mice were detected by IHC staining. **P < 0.01 compared to control.
Discussion
LncRNAs are a group of non-coding RNAs widely distributed in humans (23), which are different from linear non-coding RNAs such as miRNAs. It has been reported that lncRNAs may mediate upregulation or downregulation of gene expression (24), and lncRNAs are stable and widely expressed in many tumor tissues (25, 26). It suggests the possibility that lncRNAs may be involved in paracrine signaling or cell-to-cell crosstalk. The present findings indicate that LINC01234 knockdown could suppress liver cancer cell proliferation via inducing apoptosis. To the best knowledge, this study firstly explored the function of LINC01234 in liver cancer. Current study suggested that LINC01234 likely promote tumorigenesis of liver cancer, particularly during the advanced stages of the disease; LINC01234 might serve as an important biomarker for diagnosis and treatment of liver cancer.
MiRNAs are known to play important roles in the progression of multiple diseases, including liver cancer (17, 27). Our research found miR-513a-5p antagomir partially reversed the inhibitory effect of LINC01234 knockdown on liver cancer, further verified the function of miR-513a-5p in liver cancer progression. In addition, Zhu et al. (28) demonstrated that MRVI1-AS1 enhances nasopharyngeal cancer malignancy by sponging miR-513a-5p. Our present findings are consistent with these results, confirming that miR-513a-5p could act as a key mediator in malignant tumors. However, a previous report indicated that LINC01234 silencing could exert an anti-oncogenic effect in esophageal cancer cells through sponging miR-193a-5p (29). This difference may result from the different tumor type.
It was recently suggested that USP4 plays a key role in multiple malignant tumors (30, 31). In addition, USP4 reportedly mediates cell proliferation, survival and metastasis (32, 33). The present study indicated that USP4 is a direct target of miR-513a-5p. It has been previously confirmed that USP4 could act as a tumor promoter in liver cancer (34). These results further confirm the role of USP4 during the development of liver cancer. Moreover, Jiang et al. (35) indicated that miR-148a dysregulation could discriminate poor prognosis of hepatocellular carcinoma in association with USP4 overexpression. Our result was consistent to this previous study, confirming that LINC01234 could mediate the progression of liver cancer via indirectly targeting USP4.
TGF-β signaling plays a key role in various kinds of diseases, including malignant tumors (36, 37). It has been reported that TGF-β can activate Smad2 and Smad3 (38). Moreover, Smad4 is known to be a core of TGF-β signaling (39). In this study, we found that LINC01234 knockdown downregulated the expression of p-Smad2, p-Smad3 in liver cancer cells and increased the level of Smad4 in Cytoplasm of Huh-7 cells. Based on these data, the mechanism underlying the anti-tumor effects of LINC01234 knockdown was associated with the inactivation of TGF-β signaling pathway. According to Zhang et al. (33), USP4 could positively regulate TGF-β signaling pathway. Current study was consistent to this data, suggesting that LINC01234 could modulate the tumorigenesis of liver cancer via mediation of USP4/TGF-β axis.
Besides, we also found that the expression of E-cadherin, α-SMA and vimentin were notably regulated by LINC01234 shRNA1 in Huh-7 cells. E-cadherin, α-SMA and vimentin played important roles in EMT process (9). Miettinen et al. (40) revealed that activation of TGF-β signaling could enhance the EMT process. Consistent to these findings, we suggested that LINC01234 could promote liver cancer through activation of TGF-β1/EMT signaling.
Components of the PI3K/Akt pathway it is commonly activated in more types of cancer (15). In the current research, LINC01234 knockdown significantly inactivated PI3K/Akt signaling. An earlier report similarly found that inactivation of PI3K/Akt signaling contributes to cancer cell apoptosis (41). Moreover, our findings suggested that knockdown of LINC01234 inactivated EMT process in liver cancer cells. Xiao et al. (42) revealed that USP4 could activate PI3K/Akt to regulate the progression of peritoneal dialysis. Activation of PI3K/Akt could cause the upregulation of EMT process (43). Together with these reports, we suggested that USP4 may promote EMT process in liver cancer through activation of PI3K/Akt. Frankly speaking, the interaction between TGF-β signaling and PI3K/Akt in the condition of LINC01234 knockdown was still unclear and more investigation are needed.
Conclusion
To sum up, LINC01234 was upregulated in liver cancer. In addition, LINC01234 knockdown could inhibit the growth, migration and invasion of liver cancer cells via mediating the miR-513a-5p/USP4 axis. Thus, LINC01234 may act as a key biomarker for diagnosis and treatment of liver cancer.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
WX, KL, CS, XW, WD, JW, and JL conceived and supervised the study. YL, BX, XL, and JL designed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (81803082) and China postdoctoral Science Foundation (2018M632046).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript has been released as a pre-print at Researchsquare, [Xu et al. (44)].
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.571565/full#supplementary-material
FIGURE S1 | Knockdown of LINC01234 significantly induced the apoptosis of HepG2 cells. The apoptotic HepG2 cells were examined by flow cytometry.
FIGURE S2 | The melt curve plots of primers.
Abbreviations
lncRNAs, long non-coding RNAs; USP4, ubiquitin specific protease 4.
Footnotes
References
1. Chen W, Bian H, Xie X, Yang X, Bi B, Li C, et al. Negative feedback loop of ERK/CREB/miR-212-3p inhibits HBeAg-induced macrophage activation. J Cell Mol Med. (2020) 20:1–11. doi: 10.1155/2018/2461363
2. Ma X, Mo M, Tan HJJ, Tan C, Zeng X, Zhang G, et al. LINC02499, a novel liver-specific long non-coding RNA with potential diagnostic and prognostic value, inhibits hepatocellular carcinoma cell proliferation migration, and invasion. Hepatol Res. (2020) 50:726–40. doi: 10.1111/hepr.13491
3. Xiong XR, Tian XL, Huo RJ, Dong YT, Liu D, Bai JC, et al. 1alpha, 25-dihydroxyvitamin D3 inhibits transforming growth factor beta1-induced epithelial-mesenchymal transition via beta-catenin pathway. Chin Med J (Engl). (2020) 133:1298–303. doi: 10.1097/cm9.0000000000000830
4. Hu J, Zhou ZY, Ran HL, Yuan XC, Zeng X, Zhang ZY. Diagnosis of liver tumors by multimodal ultrasound imaging. Medicine (Baltimore). (2020) 99:e21652. doi: 10.1097/md.0000000000021652
5. Xiao Y, Gong Q, Wang W, Liu F, Kong Q, Pan F, et al. The combination of Biochanin A and SB590885 potentiates the inhibition of tumour progression in hepatocellular carcinoma. Cancer Cell Int. (2020) 20:371. doi: 10.1186/s12935-020-01463-w
6. Li S, Ji J, Zhang Z, Peng Q, Hao L, Guo Y, et al. Cisplatin promotes the expression level of PD-L1 in the microenvironment of hepatocellular carcinoma through YAP1. Mol Cell Biochem. (2020). 10:37–39. doi: 10.1007/s11010-020-03861-0
7. Jiao Y, Li Y, Jia B, Chen Q, Pan G, Hua F, et al. The prognostic value of lncRNA SNHG4 and its potential mechanism in liver cancer. Biosci Rep. (2020) 40:BSR20190729. doi: 10.1042/BSR20190729
8. Guan YF, Huang QL, Ai YL, Chen QT, Zhao WX, Wang XM, et al. Nur77-activated lncRNA WFDC21P attenuates hepatocarcinogenesis via modulating glycolysis. Oncogene. (2020) 39:2408–23. doi: 10.1038/s41388-020-1158-y
9. Shi C, Yang J, Hu L, Liao B, Qiao L, Shen W, et al. Glycochenodeoxycholic acid induces stemness and chemoresistance via the STAT3 signaling pathway in hepatocellular carcinoma cells. Aging (Albany N Y). (2020) 12:103751. doi: 10.18632/aging.103751
10. Xu W, Yu S, Xiong J, Long J, Zheng Y, Sang X. CeRNA regulatory network-based analysis to study the roles of noncoding RNAs in the pathogenesis of intrahepatic cholangiocellular carcinoma. Aging (Albany N Y). (2020) 12:1047–86. doi: 10.18632/aging.102634
11. Ji H, Niu C, Zhan X, Xu J, Lian S, Xu B, et al. Identification, functional prediction, and key lncRNA verification of cold stress-related lncRNAs in rats liver. Sci Rep. (2020) 10:521. doi: 10.1038/s41598-020-57451-7
12. Ren F, Wang J, Aniagu S, Li J, Jiang Y, Chen T. Effects of trichloroethylene on the expression of long intergenic noncoding RNAs in B6C3F1 mouse liver. Chem Res Toxicol. (2020). 9:152–59. doi: 10.1021/acs.chemrestox.9b00382
13. Gupta M, Chandan K, Sarwat M. Role of miRNA and Long non-coding RNA in hepatocellular carcinoma. Curr Pharm Des. (2020) 26:415–28. doi: 10.2174/1381612826666200115093835
14. Sadeghi H, Nazemalhosseini-Mojarad E, Sahebi U, Fazeli E, Azizi-Tabesh G, Yassaee VR, et al. Novel long noncoding RNAs upregulation may have synergistic effects on the CYP24A1 and PFDN4 biomarker role in human colorectal cancer. J Cell Physiol. (2020). 12:156–67. doi: 10.1002/jcp.29992
15. Sun C, Huang S, Hou Y, Li Z, Xia D, Zhang L, et al. Long noncoding RNA AC092171.4 promotes hepatocellular carcinoma progression by sponging microRNA-1271 and upregulating GRB2. Aging (Albany N Y). (2020) 12:14141–56. doi: 10.18632/aging.103419
16. Ross AK, Coutinho de Almeida R, Ramos YFM, Li J, Meulenbelt I, Guilak F. The miRNA-mRNA interactome of murine induced pluripotent stem cell-derived chondrocytes in response to inflammatory cytokines. FASEB J. (2020). 7:636–39. doi: 10.1096/fj.202000889R
17. Huang Z, Wen J, Yu J, Liao J, Liu S, Cai N, et al. MicroRNA-148a-3p inhibits progression of hepatocelluar carcimoma by repressing SMAD2 expression in an Ago2 dependent manner. J Exp Clin Cancer Res. (2020) 39:150. doi: 10.1186/s13046-020-01649-0
18. Luo T, Gao Y, Zhangyuan G, Xu X, Xue C, Jin L, et al. lncRNA PCBP1-AS1 aggravates the progression of hepatocellular carcinoma via regulating PCBP1/PRL-3/AKT pathway. Cancer Manag Res. (2020) 12:5395–408. doi: 10.2147/cmar.s249657
19. Hong H, Sui C, Qian T, Xu X, Zhu X, Fei Q, et al. Long noncoding RNA LINC00460 conduces to tumor growth and metastasis of hepatocellular carcinoma through miR-342-3p-dependent AGR2 up-regulation. Aging (Albany N Y). (2020) 12:10544–55. doi: 10.18632/aging.103278
20. Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J, et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. (2020) 13:5. doi: 10.1186/s13045-019-0839-x
21. Wang C, Jiang X, Li X, Song S, Meng Q, Wang L, et al. Long noncoding RNA HULC accelerates the growth of human liver cancer stem cells by upregulating CyclinD1 through miR675-PKM2 pathway via autophagy. Stem Cell Res Ther. (2020) 11:8. doi: 10.1186/s13287-019-1528-y
22. Jiang B, Yang B, Wang Q, Zheng X, Guo Y, Lu W. lncRNA PVT1 promotes hepatitis B viruspositive liver cancer progression by disturbing histone methylation on the cMyc promoter. Oncol Rep. (2020) 43:718–26. doi: 10.3892/or.2019.7444
23. Qiu S, Chen G, Peng J, Liu J, Chen J, Wang J, et al. LncRNA EGOT decreases breast cancer cell viability and migration via inactivation of the Hedgehog pathway. FEBS Open Bio. (2020) 10:817–26. doi: 10.1002/2211-5463.12833
24. Xu Y, Xia X, Jiang Y, Wu D, Wang S, Fu S, et al. Down-regulated lncRNA AGAP2-AS1 contributes to pre-eclampsia as a competing endogenous RNA for JDP2 by impairing trophoblastic phenotype. J Cell Mol Med. (2020) 24:4557–68. doi: 10.1111/jcmm.15113
25. Zhang L, Zhang D, Qin ZY, Li J, Shen ZY. The role and possible mechanism of long noncoding RNA PVT1 in modulating 3T3-L1 preadipocyte proliferation and differentiation. IUBMB Life. (2020) 72:1460–7. doi: 10.1002/iub.2269
26. Liu C, Lu Z, Liu H, Zhuang S, Guo P. LncRNA XIST promotes the progression of laryngeal squamous cell carcinoma via sponging miR-125b-5p to modulate TRIB2. Biosci Rep. (2020). 5:15–19. doi: 10.1042/BSR20193172
27. Komoll RM, Hu Q, Olarewaju O, von Dohlen L, Yuan Q, Xie Y, et al. MicroRNA-342-3p is a potent tumour suppressor in hepatocellular carcinoma. J Hepatol. (2020) 40:BSR20193172. doi: 10.1016/j.jhep.2020.07.039
28. Zhu Y, He D, Bo H, Liu Z, Xiao M, Xiang L, et al. The MRVI1-AS1/ATF3 signaling loop sensitizes nasopharyngeal cancer cells to paclitaxel by regulating the Hippo-TAZ pathway. Oncogene. (2019) 38:6065–81. doi: 10.1038/s41388-019-0858-7
29. Ma J, Han LN, Song JR, Bai XM, Wang JZ, Meng LF, et al. Long noncoding RNA LINC01234 silencing exerts an anti-oncogenic effect in esophageal cancer cells through microRNA-193a-5p-mediated CCNE1 downregulation. Cell Oncol (Dordr). (2020) 43:377–94. doi: 10.1007/s13402-019-00493-5
30. Liang Y, Song X, Li Y, Ma T, Su P, Guo R, et al. Targeting the circBMPR2/miR-553/USP4 axis as a potent therapeutic approach for breast cancer. Mol Ther Nucleic Acids. (2019) 17:347–61. doi: 10.1016/j.omtn.2019.05.005
31. Nguyen HH, Kim T, Nguyen T, Hahn MJ, Yun SI, Kim KKA. Selective inhibitor of ubiquitin-specific protease 4 suppresses colorectal cancer progression by regulating beta-catenin signaling. Cell Physiol Biochem. (2019) 53:157–71. doi: 10.33594/000000127
32. Geng N, Li Y, Zhang W, Wang F, Wang X, Jin Z, et al. A PAK5-DNPEP-USP4 axis dictates breast cancer growth and metastasis. Int J Cancer. (2020) 146:1139–51. doi: 10.1002/ijc.32523
33. Zhang J, Na S, Pan S, Long S, Xin Y, Jiang Q, et al. Inhibition of USP4 attenuates pathological scarring by downregulation of the TGFbeta/Smad signaling pathway. Mol Med Rep. (2019) 20:1429–35. doi: 10.3892/mmr.2019.10370
34. Qiu C, Liu Y, Mei Y, Zou M, Zhao Z, Ye M, et al. Ubiquitin-speci fi c protease 4 promotes metastasis of hepatocellular carcinoma by increasing TGF-beta signaling-induced epithelial-mesenchymal transition. Aging (Albany N Y). (2018) 10:2783–99. doi: 10.18632/aging.101587
35. Heo MJ, Kim YM, Koo JH, Yang YM, An J, Lee SK, et al. microRNA-148a dysregulation discriminates poor prognosis of hepatocellular carcinoma in association with USP4 overexpression. Oncotarget. (2014) 5:2792–806. doi: 10.18632/oncotarget.1920
36. Shi X, Shao X, Liu B, Lv M, Pandey P, Guo C, et al. Genome-wide screening of functional long noncoding RNAs in the epicardial adipose tissues of atrial fibrillation. Biochim Biophys Acta Mol Basis Dis. (2020) 1866:165757. doi: 10.1016/j.bbadis.2020.165757
37. Wang C, Gao Y, Zhang Z, Chen C, Chi Q, Xu K, et al. Ursolic acid protects chondrocytes, exhibits anti-inflammatory properties via regulation of the NF-kappaB/NLRP3 inflammasome pathway and ameliorates osteoarthritis. Biomed Pharmacother. (2020) 130:110568. doi: 10.1016/j.biopha.2020.110568
38. Nyati S, Gregg BS, Xu J, Young G, Kimmel L, Nyati MK, et al. TGFBR2 mediated phosphorylation of BUB1 at Ser-318 is required for transforming growth factor-beta signaling. Neoplasia. (2020) 22:163–78. doi: 10.1016/j.neo.2020.02.001
39. Sakamoto R, Kajihara I, Miyauchi H, Maeda-Otsuka S, Yamada-Kanazawa S, Sawamura S, et al. Inhibition of endoglin exerts anti-tumor effects through the regulation of non-Smad TGF-beta signaling in angiosarcoma. J Invest Dermatol. (2020). 17:234–42. doi: 10.1016/j.jid.2020.01.031
40. Miettinen PJ, Ebner R, Lopez AR, Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol. (1994) 127:2021–36. doi: 10.1083/jcb.127.6.2021
41. Pan S, Ren F, Li L, Liu D, Li Y, Wang A, et al. MiR-328-3p inhibits cell proliferation and metastasis in colorectal cancer by targeting Girdin and inhibiting the PI3K/Akt signaling pathway. Exp Cell Res. (2020) 390:111939. doi: 10.1016/j.yexcr.2020.111939
42. Xiao L, Peng X, Liu F, Tang C, Hu C, Xu X, et al. AKT regulation of mesothelial-to-mesenchymal transition in peritoneal dialysis is modulated by Smurf2 and deubiquitinating enzyme USP4. BMC Cell Biol. (2015) 16:7. doi: 10.1186/s12860-015-0055-7
43. Peng Y, Li F, Zhang P, Wang X, Shen Y, Feng Y, et al. IGF-1 promotes multiple myeloma progression through PI3K/Akt-mediated epithelial-mesenchymal transition. Life Sci. (2020) 249:117503. doi: 10.1016/j.lfs.2020.117503
Keywords: liver cancer, LINC01234, USP4, miR-513a-5p, TGF-β
Citation: Xu W, Li K, Song C, Wang X, Li Y, Xu B, Liang X, Deng W, Wang J and Liu J (2020) Knockdown of lncRNA LINC01234 Suppresses the Tumorigenesis of Liver Cancer via Sponging miR-513a-5p. Front. Oncol. 10:571565. doi: 10.3389/fonc.2020.571565
Received: 11 June 2020; Accepted: 21 August 2020;
Published: 16 October 2020.
Edited by:
Erica Golemis, Fox Chase Cancer Center, United StatesReviewed by:
Hanqing Liu, Jiangsu University, ChinaBojan Bujisic, Massachusetts General Hospital and Harvard Medical School, United States
Copyright © 2020 Xu, Li, Song, Wang, Li, Xu, Liang, Deng, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanli Deng, tcldwl@163.com; Junqing Wang, wangjunqingmd@hotmail.com; Jianwen Liu, jianwen_ecust@126.com
 Wen Xu1
Wen Xu1 Jianwen Liu
Jianwen Liu