- 1Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China
- 2College of Life Science, Jiaying University, Meizhou, China
- 3Institute of Cancer Stem Cell, Dalian Medical University, Dalian, China
- 4The First Affiliated Hospital, Zhengzhou University, Zhengzhou, China
- 5The Seventh Affiliated Hospital, Sun Yat-sen University, Shenzhen, China
- 6The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China
Abnormal expression or mutation of RNA splicing proteins are widely observed in human cancers. Here, we identified poly(U) binding splicing factor 60 (PUF60) as one of the most differentially expressed genes out of 97 RNA splicing proteins between normal and bladder cancer tissues by bioinformatics analysis of TCGA bladder cancer expression data. The expression of PUF60 was significantly higher in tumor tissues, while high PUF60 expression was associated with malignant phenotypes of bladder cancer and shorter survival time. Moreover, we identified aurora kinase A (AURKA) as a new downstream target of PUF60 in bladder cancer cells. PUF60 knockdown significantly inhibited cell viability and colony formation capacity in bladder cancer cells, whereas AURKA overexpression reversed this inhibition effect. Overexpression of PUF60 significantly promoted cell viability and colony formation in bladder cancer cells, while treatment with AURKA specific inhibitor reversed this promotive effect. Mechanistically, PUF60 specifically bound to the AURKA promoter, thereby activating its transcription and expression. Furthermore, we showed that there was a significant positive correlation between PUF60 and AURKA expression in bladder cancer tissues, and PUF60 and AURKA expression contributed to tumor progression and malignant phenotypes in the patients with bladder cancer. Collectively, these results indicate that the PUF60/AURKA axis plays a key role in regulating tumorigenesis and progression of bladder cancer, and may be a potential prognostic biomarker and therapeutic target for bladder cancer patients.
Introduction
Bladder cancer is the sixth most common cancer in men and the tenth in both sexes worldwide. There were 549,393 new cases of bladder cancer and 199,922 related deaths in 2018 worldwide (1). Even though diagnosis and treatment of bladder urothelial carcinoma have been improved in the last several decades, it remains an important public health issue worldwide due to poor management of patients (2). The tumor, node and metastasis (TNM) classification system was the most commonly used approach in risk stratification and management of bladder cancer patients. Although it has been updated many times over the last few decades, it still has limitations in predicting therapy response and outcome among bladder cancer patients (3, 4). Therefore, it is urgent to find new biomarkers for predicting the outcomes of bladder cancer, which may lead to a better management of bladder cancer patients. Recently, molecular subtypes based on the gene expression profile in bladder cancer has aroused attention worldwide, and it is promising to identify gene signatures which can better predict survival time and therapy response (5–7).
RNA splicing related proteins are a group of proteins containing RNA binding domains, which are closely involved in pre-mRNA maturation by specifically removing introns (8, 9). There are three protein families which are well-established RNA splicing factors, including serine and arginine-rich (SR) proteins (10), RNA binding motif (RBM) proteins (11) and heterogeneous nuclear ribonucleoproteins (HNRNP) proteins (12). Some other proteins, such as SF3B1 (13), PRMT5 (14), PUF60 (15), U2AF1 (16), and ZRSR2 (17) also participate in the RNA splicing process. It has been reported that abnormal expression or mutations of RNA splicing proteins are widely observed in human diseases, especially in cancers (18–20). However, the exact roles of RNA splicing proteins in bladder cancer development and progression haven’t been systematically elucidated yet. It remains to be further investigated whether there are specific RNA spicing proteins playing an essential role in bladder cancer development.
Poly(U) binding splicing factor 60 (PUF60), also known as FUSE-binding protein-interacting repressor (FIR), is a nucleic acid-binding protein that plays a role in a variety of nuclear processes, including pre-mRNA splicing and transcriptional regulation (21, 22). Overexpression of PUF60 has been reported to be closely associated with the development and progression of multiple cancers, including colon cancer (23–25), hepatocellular carcinoma (26), non-small cell lung cancer (27), breast cancer (28), and esophageal cancer (29, 30). Abnormal expression, mutation or truncation of PUF60 were widely reported in congenital diseases associated with intellectual disability, heart defects and short stature (31–34). In summary, PUF60 plays an important role in human diseases, including cancer and congenital diseases. However, it remains unveiled its role in the initiation, progression and prognosis of bladder cancer.
Aurora kinase A (AURKA) is a member of aurora kinase family, which perform essential functions during cell division (35). AURKA is mainly associated with the spindle poles during mitosis, where it is required for centrosome separation and maturation (36). In the past several decades, emerging studies have proved that AURKA plays an important role in tumor development and progression (37). Specific inhibitors targeting AURKA has been developed and shown promising prospect recently due to the prominent role of AURKA in tumor progression (38–40). In bladder cancer, inhibiting AURKA by its specific inhibitors could decrease cell proliferation (41), induce apoptosis (42, 43) and cause cell cycle arrest (44).
Our TCGA data analysis identified PUF60 as one of the most differentially expressed genes among the 97 well-established RNA splicing factor genes. In this study, we investigated the potential association between PUF60 expression and clinicopathological characteristics in bladder cancer and analyzed the potential of PUF60 to be a new biomarker for malignant phenotypes and poor prognosis in bladder cancer. In addition, we identified AURKA as a new downstream target of PUF60 in bladder cancer cells, and also proved that PUF60 promoted bladder cancer cell growth by transcriptionally upregulating AURKA expression. Collectively, our findings offer new insights into the understanding of the pro-tumorigenic role of PUF60 and its underlying mechanism involved in bladder cancer growth, and suggest that the PUF60/AURKA axis may provide prognostic biomarkers and therapeutic targets for bladder cancer patients.
Materials and Methods
Cell Culture, Antibodies and Chemicals
The human bladder cancer cell lines (5637, UM-UC-3, T24, Biu87, J82) were obtained from American Type Culture Collection (ATCC, Manassas, VA, United States) and cultured in RPMI-1640 (Invitrogen, Carlsbad, CA, United States) supplemented with 10% fetal bovine serum,100 unit/ml penicillin, and 100 μg/ml streptomycin. Cells were maintained in an incubator with a humidified atmosphere of 95% air and 5% CO2 at 37°C.
Anti-PUF60 antibody was purchased from Invitrogen (Carlsbad, CA, United States), anti-AURKA antibody, anti-GAPDH and secondary rabbit antibody from Proteintech (Wuhan, China). AURKA inhibitor was purchased from Selleck (Shanghai, China).
Streptavidin-Agarose Pulldown Assay
The binding of PUF60 to AURKA promoter was confirmed by streptavidin-agarose pulldown assay as previously described. Briefly, 800 ng nuclear proteins from human bladder cancer cell lines were incubated with 8 μg biotin-labeled double-stranded DNA probes of AURKA promoter from −1239 to +43 and 8 μl streptavidin-agarose beads (Sigma-Aldrich) at 4°C overnight. The mixture was then centrifuged at 500 × g to pull down the DNA-protein complex. Precipitated proteins were separated by SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were incubated with primary antibody and horseradish peroxidase-conjugated secondary antibody, and proteins were then detected using the ECL chemiluminescence system (Pierce, Rockford, IL, United States).
Western Blotting
Briefly, cells were collected and lysed by RIPA buffer (150 mM NaCl, 0.5% EDTA, 50 mM Tris, 0.5% NP40) and centrifuged for 15 min at 12,000 rpm and 4°C. 50 μg of harvested total proteins were separated by SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were incubated with primary antibody and horseradish peroxidase-conjugated secondary antibody, and proteins were then detected using the ECL chemiluminescence system (Pierce, Rockford, IL, United States).
Luciferase Reporter Assay
Briefly, 5637 cells were plated in 24-well plates at a density of 1.0 × 105 cells per well then transfected with 483 ng of promoter-luciferase plasmid and 17 ng of pRL-CMV. The luciferase activity was measured using a Dual-Luciferase Assay kit (Promega) 48 h after transfection. Four replicative wells were measured. The primers used for cloning the indicated promoter regions are listed in Supplementary Table 1.
Immunohistochemistry (IHC)
Bladder cancer tissue microarrays with 56 samples were purchased from Outdo Biotech Co., Ltd. (Shanghai, China). Bladder cancer tissue microarrays with 13 pairs of samples were purchased from Alenabio Biotechnology Co., Ltd. (Xi’an, China). The primary antibodies against PUF60 were diluted 1:100, and then incubated at 4°C overnight in a humidified container. After three washes with PBS, the tissue slides were treated with a non-biotin horseradish peroxidase detection system according to manufacturer’s instructions (Dako). IHC scores were evaluated. IHC H-score = Σ(PI × I) = (percentage of cells of weak intensity × 1) + (percentage of cells of moderate intensity × 2) + (percentage of cells of strong intensity × 3).
RNA Extraction and qRT-PCR
Briefly, total RNA was extracted using RaPure Total RNA Micro Kit (Magen, Guangzhou, China). First-strand cDNA was synthesized using HiScript II One Step RT-PCR Kit (Vazyme, Nanjing, China). The primers used to amplify the indicated genes are listed in Supplementary Table 1. Real time q-PCR was performed using ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China) following instructions.
Cell Viability Assay
Cells were seeded in 96-well plates (10,000 cells/well) 24 h after PUF60 siRNA transfection. Cell viability was assessed by the MTS assay (Promega, Madison, WI, United States) 72 h after transfection. Cell viability of stable cell lines with PUF60 overexpression was detected 48 h after plating in 96-well plates (5,000 cells/well).
Colony Formation Assay
Bladder cancer cell lines 5637 or T24 were seeded at a density of 800 cells per well in 6-well plates 24 h after PUF60 siRNA transfection and cultured for 10–14 days. The colonies were then stained with 1% crystal violet and counted. All experiments were performed with three independent trials.
siRNA and Plasmid Construction
The sequences targeting PUF60, 5′-UCAAGAGUGUGCUGGU GAA-3′, 5′-GCUACGGCUUCAUUGAGUA-3′ and negative control siRNA were synthesized by GenePharma Co., Ltd. (Suzhou, China). Transfection was performed according to the manufacturer’s instructions using Lipofectamine RNAiMAX transfection reagent (Invitrogen) and 50 nM siRNA.
For overexpression of PUF60 in bladder cancer cell lines, PUF60 or AURKA was cloned into the pSIN-EF2-puro vector. The PLKO.1-puro vector was used to clone the shRNAs targeting PUF60. The six segments of AURKA promoter region was cloned into the pGL3-basic vector, respectively.
Cell Cycle Analysis
Cells were seeded into six-well plates at a density of 1 × 105 cells per well. After 48 h, the wells were transfected with PUF60 specific siRNA or overexpressing plasmid. At the end of the experiments, adherent cells were trypsinized, counted, washed, and resuspended. The cells were then pelleted and fixed by dropwise addition of 70% ice-cold ethanol at 4°C overnight. The fixed cells were washed with PBS and stained with 50 μg/mL propidium iodide, 50 μg/mL RNase I, and 0.2% Triton X-100 in the dark at 37°C for 30 min and then analyzed with flow cytometry.
Differential Gene Expression Analysis, Functional and Pathway Enrichment Analyses
The cut-off value was ±1.5, false discovery rate (FDR)-adjusted P < 0.05 and fold change (FC) > 1.5 or FC < 1.5 were considered as significantly differentially expressed genes. Median value of PUF60 expression was used to divide GSE13507 patients into PUF60high and PUF60low groups. To investigate functional annotations of the differential genes between PUF60high and PUF60low groups, we employed the Database for Annotation, Visualization and Integrated Discovery (DAVID1) to conduct the gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses.
Statistical Analysis
Data of Sanchez-Carbayo and Blaveri studies were obtained from Oncomine database. Gene expression and clinical information data of other datasets used in our analysis were obtained from TCGA database or GEO database.
Statistical analyses were performed using GraphPad Prism (version 8). Chi-square test and t-test or Mann Whitney test were applied for variance analysis, Spearman rank correlation method was for correlation analysis, and Kaplan–Meier analysis was for survival analysis. The cut-off value for Kaplan–Meier analysis was determined by X-tile software. Each of the statistical tests was two- tailed, and P < 0.05 was regarded as statistically significant.
Results
PUF60 Was Highly Expressed in Bladder Cancer
We first downloaded the mRNA expression data of normal and bladder cancer tissues from TCGA database and analyzed the mRNA expression of a total of 97 RNA splicing proteins (including SRSF family, RBM family, HNRNP family, SF3B1, PRMT5, PUF60, U2AF1, and ZRSR2) between normal and carcinoma tissues. 25 genes were significantly differentially expressed (Figure 1A) (FC > 1.5, P < 0.05), and HNRNPF, HNRNPA2B, RBM42, and PUF60 were the four most differentially expressed genes. Since the roles of HNRNPF and HNRNPA2B have been investigated in bladder cancer before, while the roles of PUF60 in the development and progression of bladder cancer remains unclear, we set off to investigate the roles and functions of PUF60 in bladder cancer.
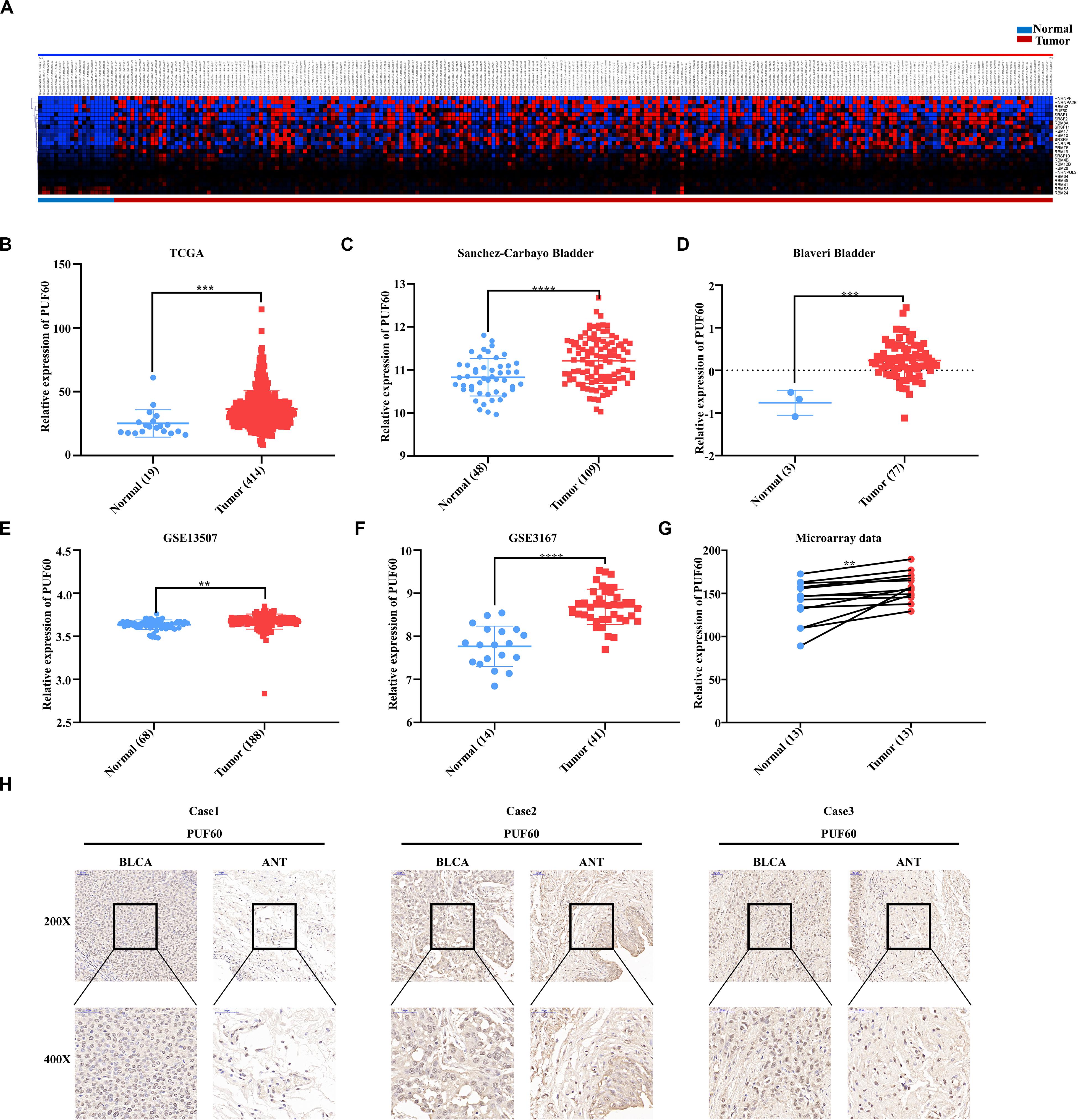
Figure 1. PUF60 was highly expressed in bladder cancer. (A) Heat map of differentially expressed RNA splicing-related genes between normal bladder tissues and bladder cancer tissues of TCGA data. (B) Relative PUF60 mRNA expression in normal and tumor tissues from TCGA (P = 0.0007). (C,D) Relative PUF60 mRNA expression in normal and tumor tissues from two bladder cancer cohorts from Oncomine database (Sanchez-Carbayo Bladder: P < 0.0001; Blaveri Bladder: P = 0.0001). (E,F) Relative PUF60 mRNA expression in normal and tumor tissues of GSE13507 and GSE3167 from GEO database (GSE13507: P = 0.016; GSE3167: P < 0.0001). (G) Relative expression of PUF60 in paired normal and tumor tissues from tissue microarray data (P = 0.0051). (H) Three cases of representative IHC images of PUF60 in bladder cancer tissues (BLCA) and adjacent normal tissues (ANT). Data was analyzed by t-test; **P < 0.01, ***P < 0.001, ****P < 0.0001.
Next, we confirmed the expression of PUF60 in TCGA datasets, and found significantly higher expression of PUF60 mRNA in carcinoma tissues compared to normal bladder tissues (Figure 1B). To further verify the expression of PUF60 in bladder cancer, we collected the published RNA-sequencing or microarray data of bladder cancer from Oncomine database and GEO database, which include four datasets containing both normal and carcinoma tissues. We found that PUF60 mRNA was significantly highly expressed in tumor tissues compared to normal bladder tissues in all four datasets (Figures 1C–F). To further evaluate the PUF60 protein expression in bladder cancer, 13 bladder cancer tissues and paired adjacent normal tissues were analyzed by immunohistochemistry (IHC), confirming that bladder cancer tissues have significantly higher PUF60 protein expression compared to paired normal tissues (Figures 1G,H). This result was consistent with our analysis of PUF60 mRNA expression in public databases.
High PUF60 Expression Was Associated With Malignant Phenotypes in Bladder Cancer
Our previous work proved that PUF60 was highly expressed in bladder cancer, but it was unclear the association between PUF60 mRNA expression and bladder cancer phenotypes, such as histopathological type, T stage, grade and molecular subtypes. Hence, we analyzed the association between PUF60 mRNA expression and molecular subtypes, grade, N stage or T stage of TCGA bladder cancer expression data. We found that the mRNA expression of PUF60 was significantly higher in basal bladder cancer tissues, which was a malignant molecular subtype with poor survival (Figure 2A). In contrast, we didn’t find any association between PUF60 mRNA expression and other pathological phenotypes (Figures 2B–D). To further investigate the mRNA expression profile of PUF60 in different phenotypes of bladder cancer, we searched the GEO database for bladder cancer datasets that have over 90 samples as well as clinical and histopathological information of patients. Seven datasets from different studies (GSE86411, GSE128192, GSE120736, GSE128959, GSE48276, GSE124305, and GSE31684) were selected. Our analysis of GSE86411 and GSE128192 revealed that PUF60 mRNA was significantly highly expressed in micropapillary bladder cancer and Sarcomatoid urothelial bladder cancer (SARC), which displayed a high propensity for distant metastasis and were associated with short survival (Figures 2E,F), compared to conventional urothelial carcinoma. We also analyzed the PUF60 mRNA expression in superficial and infiltrating bladder urothelial carcinoma of GSE120736 dataset, finding that infiltrating tissues tended to have higher PUF60 mRNA expression (Figure 2G). Intriguingly, when we investigated the PUF60 mRNA expression in primary, progressive and recurrent bladder cancer tissues of GSE128959 dataset, we found that primary or progressive bladder cancer had higher PUF60 mRNA expression than recurrent cancer (Figure 2H). This suggests that PUF60 may play an important role in bladder cancer initiation and progression. Moreover, we found PUF60 mRNA was highly expressed in advanced T stage bladder cancer in most of datasets (Figures 2I–L) and high grade bladder cancer also tended to have higher PUF60 mRNA expression (Figure 2M). Unfortunately, there weren’t datasets with enough patient information regarding N stage and M stage. Finally, our analysis of PUF60 mRNA expression in different molecular subtypes of bladder cancer showed that basal types had higher PUF60 mRNA expression (Figure 2N), which was consistent with our previous analysis in TCGA dataset. These results indicated that PUF60 are closely associated with most of malignant phenotypes in bladder cancer, and it was a potential molecular marker for malignant behavior in bladder cancer.
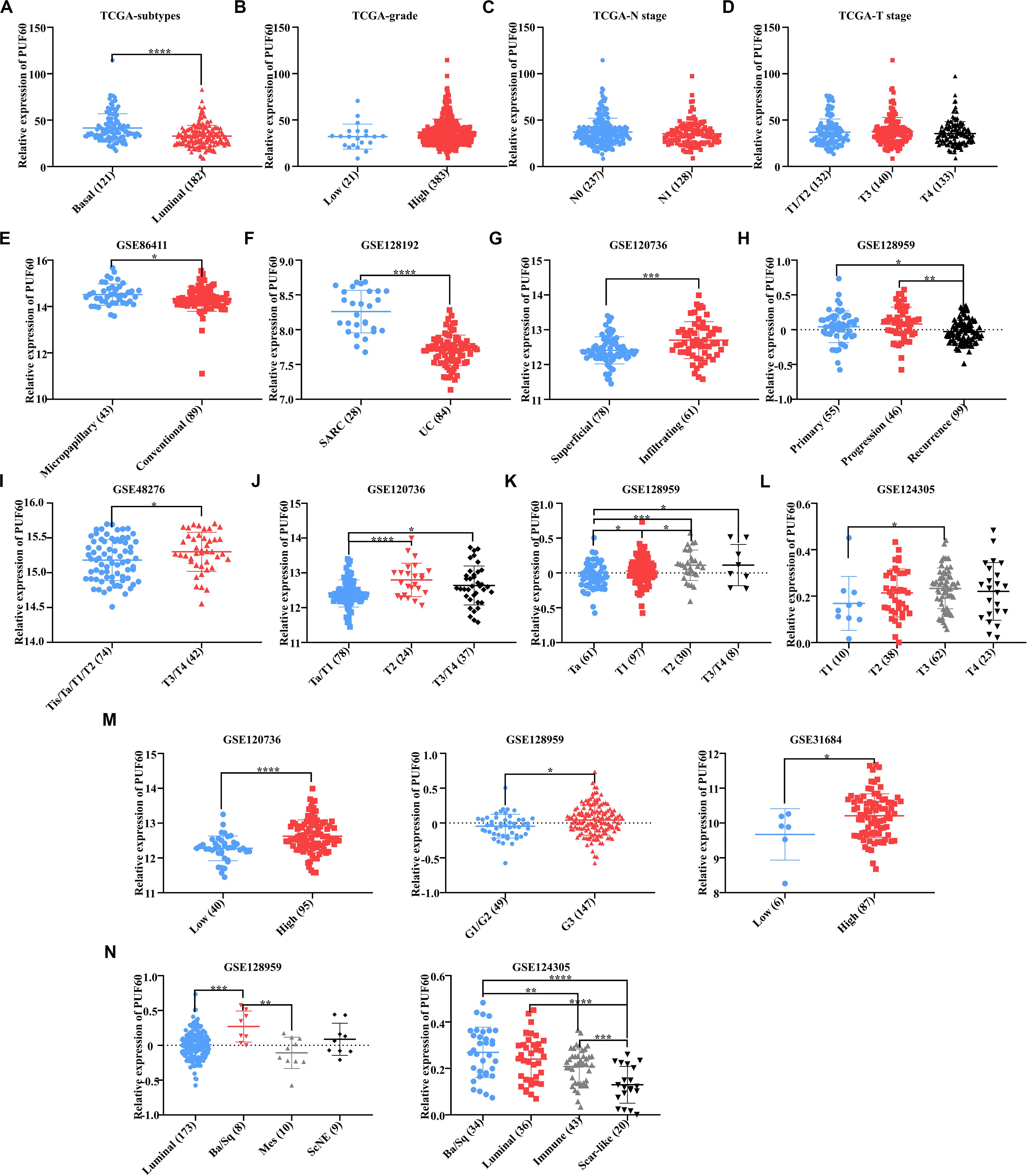
Figure 2. High PUF60 expression was associated with malignant phenotypes in bladder cancer. (A) Relative PUF60 mRNA expression in basal-like and luminal-like bladder cancer tissues from TCGA data (P < 0.0001). (B) Relative PUF60 mRNA expression in low and high grade bladder cancer tissues from TCGA data (P = 0.1219). (C) Relative PUF60 mRNA expression in lymph node negative (N0) and positive (N1) tissues from TCGA data (P = 0.1821). (D) Relative PUF60 mRNA expression of different T stage tissues from TCGA data (P = 0.3815). (E) Relative PUF60 mRNA expression in bladder micropapillary urothelial carcinoma and conventional urothelial carcinoma of GSE86411 data (P = 0.0381). (F) Relative PUF60 mRNA expression in sarcomatoid urothelial bladder cancer (SARC) and conventional bladder urothelial carcinoma (UC) of GSE128192 data (P < 0.0001). (G) Relative PUF60 mRNA expression in superficial and infiltrating bladder cancer of GSE120736 (P = 0.0002). (H) Relative PUF60 mRNA expression in primary, progressive and recurrent bladder cancer of GSE128959 data (Recurrence vs. Primary: P = 0.0323; Recurrence vs. Progressive: P = 0.0019). (I–L) Relative PUF60 mRNA expression in different T stage tissues of four independent GEO datasets: GSE48276 (I), GSE120736 (J), GSE128959 (K), GSE124305 (L) (GSE48276: P = 0.0359; GSE120736: T2 vs. Ta/T1: P < 0.0001, T3/T4 vs. Ta/T1: P = 0.0115; GSE128959: T1 vs. Ta: P = 0.0246, T2 vs. Ta: P = 0.0007, T3/T4 vs. Ta: P = 0.0416, T2 vs. T1: P = 0.0472; GSE124305: P = 0.0425). (M) Relative PUF60 mRNA expression in low and high grade bladder cancer from three independent GEO datasets (left to right: GSE120736, GSE128959, and GSE31684) (GSE120736: P < 0.0001; GSE128959: P = 0.0108; GSE31684: P = 0.0490). (N) Relative PUF60 mRNA expression in different molecular subtypes of GSE128959 (left) and GSE124305 (right); luminal, Luminal-like subtype bladder cancer; Ba/Sq, Basal/Squamous-like subtype bladder cancer; Mes, Mesenchymal-like subtype bladder cancer; ScNE, Small-cell/Neuroendocrine-like subtype bladder cancer; Immune, Immune subtype bladder cancer; Scar-like, Scar-like subtype bladder cancer (GSE128959: luminal vs. Ba/Sq: P = 0.0004, Ba/Sq vs. Mes: P = 0.0027; GSE124305: Ba/Sq vs. Scar-like: P < 0.0001, Ba/Sq vs. Immune: P = 0.0049, Luminal vs. Scar-like: P < 0.0001, Immune vs. Scar-like: P = 0.0003). Data was analyzed by t-test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
PUF60 Predicted Unfavorable Outcomes in Bladder Cancer Patients
We have proved above that PUF60 mRNA was highly expressed and associated with malignant phenotypes in bladders. To examine whether PUF60 could serve as a prognostic biomarker for bladder cancer patients. We conducted Kaplan–Meier survival analysis according to the PUF60 mRNA expression of TCGA dataset in bladder cancer patients. We found that patients with high mRNA expression of PUF60 had significant shorter overall survival time (Figure 3A). To confirm the prognostic value of PUF60 protein expression for bladder cancer patients, we next conducted Kaplan–Meier survival analysis of our tissue microarray data according to the PUF60 protein expression score. The result showed that patients with higher PUF60 protein expression had a poorer overall survival (Figure 3B), consistent with our analysis of TCGA data. The difference between low and high PUF60 protein expression is not significant though, probably due to our limited number of samples. We also analyzed the association between PUF60 mRNA expression and clinical pathological characteristics of patients, but no significant difference was found (Table 1). To further prove our conclusion, we conducted the Kaplan–Meier survival analysis in several GEO datasets with the survival information. Patients with high PUF60 mRNA expression are prone to have shorter overall survival time in three out of the four datasets (Figures 3C, E, G, I). In addition, we analyzed the disease free survival according to the PUF60 mRNA expression in three independent GEO datasets. The result revealed that patients with high PUF60 mRNA expression had significantly shorter survival time in two out of the three cohorts (Figures 3D,F,H), which was consistent with the analysis of overall survival. These results supported that PUF60 was a potential prognostic biomarker for bladder cancer patients.
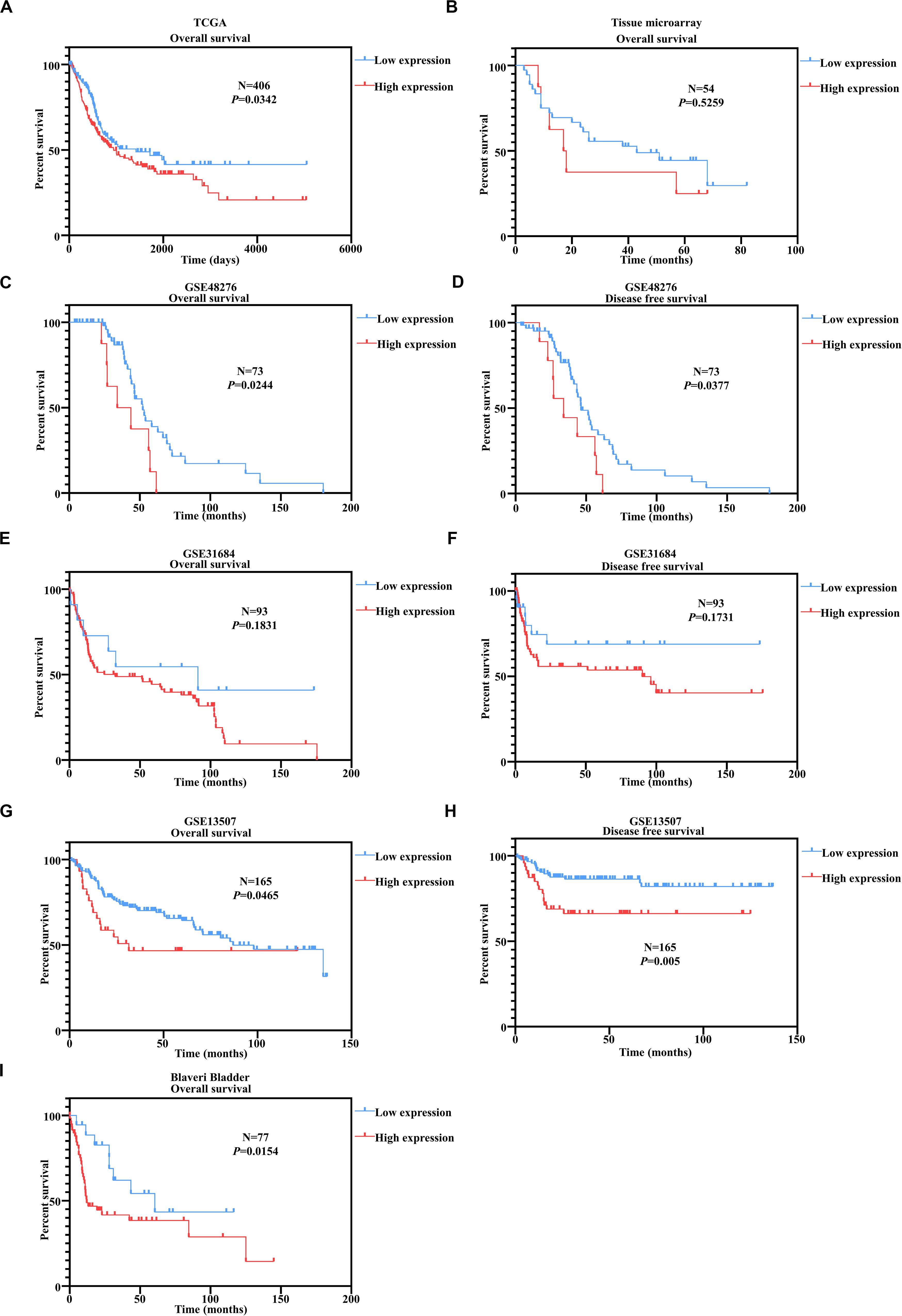
Figure 3. PUF60 predicted unfavorable outcomes in bladder cancer patients. (A) Kaplan–Meier analysis of overall survival according to the PUF60 mRNA expression from TCGA data. (B) Kaplan–Meier analysis of overall survival according to the PUF60 protein expression from tissue microarray data. (C,D) Kaplan–Meier analysis of overall (left) and disease free survival (right) according to the PUF60 mRNA expression from GSE48276 data. (E,F) Kaplan–Meier analysis of overall (left) and disease free survival (right) according to the PUF60 mRNA expression from GSE31684 data. (G,H) Kaplan–Meier analysis of overall (left) and disease free survival (right) according to the PUF60 mRNA expression from GSE13507 data. (I) Kaplan–Meier analysis of overall (left) and disease free survival (right) according to the PUF60 mRNA expression from Blaveri bladder studies.
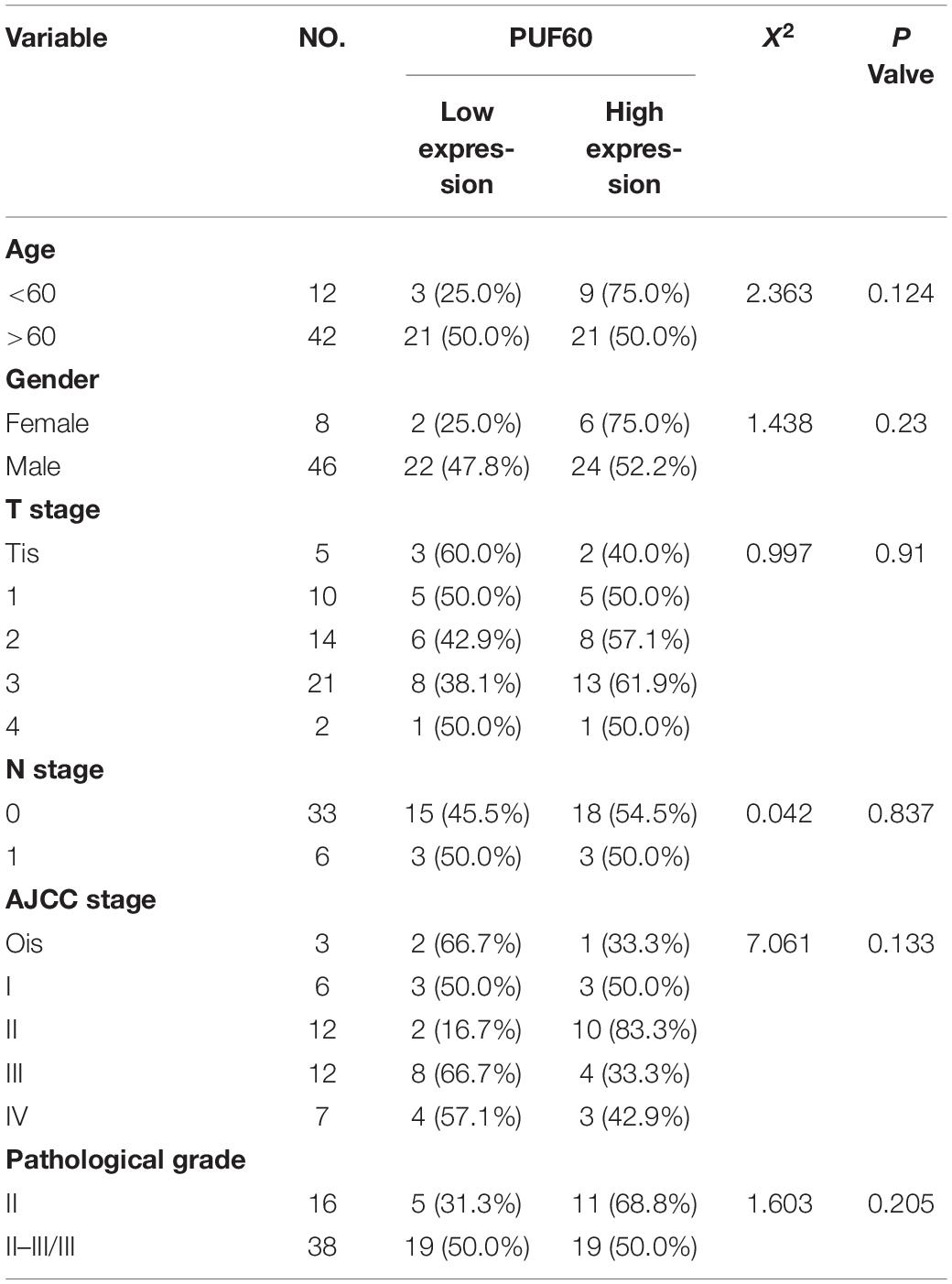
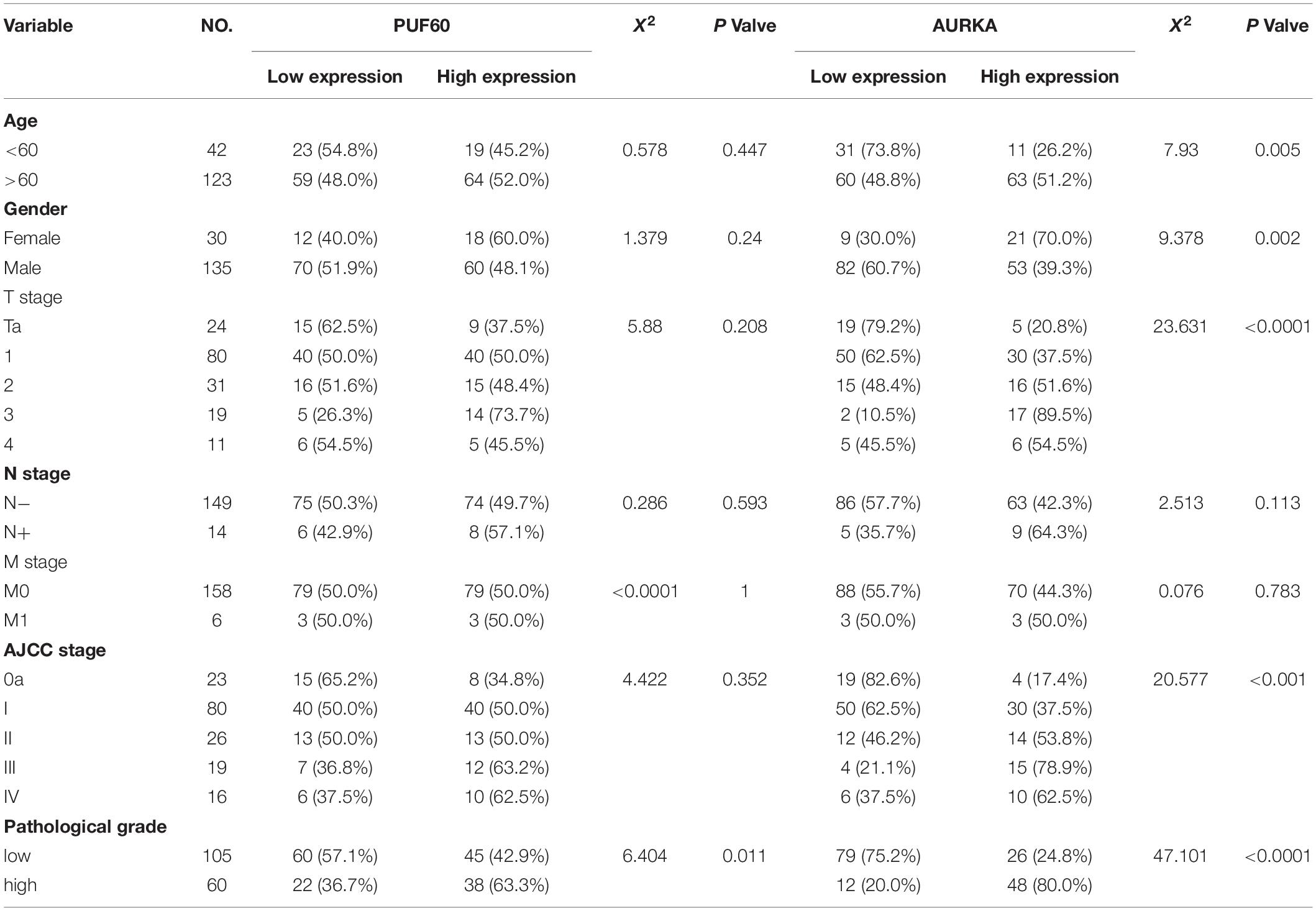
Table 1. Correlation between PUF60 and clinical pathology characteristics in bladder cancer of tissue microarray data.
PUF60 Promotes Bladder Cancer Cell Growth and Cell Cycle Progression
To investigate the biological functions of PUF60 in bladder cancer, we detected the protein expression of PUF60 in five bladder cancer cell lines (5637, UM-UC-3, T24, Biu87, J82) by western blot (Figure 4A). We then chose 5637 cells with high PUF60 expression to knock down its expression (Figure 4B), and T24 cells with low PUF60 expression to overexpress its expression (Figure 4C). We found that knockdown of PUF60 significantly inhibited the proliferation and clonogenicity of 5637 cells (Figures 4D,E), whereas overexpression of PUF60 significantly increased the proliferation and clonogenicity in T24 cells (Figures 4F,G). Furthermore, we found that knockdown of PUF60 caused G1/S arrest in 5637 cells (Figure 4H), while overexpression of PUF60 promoted cell cycle progression in T24 cells (Figure 4I).
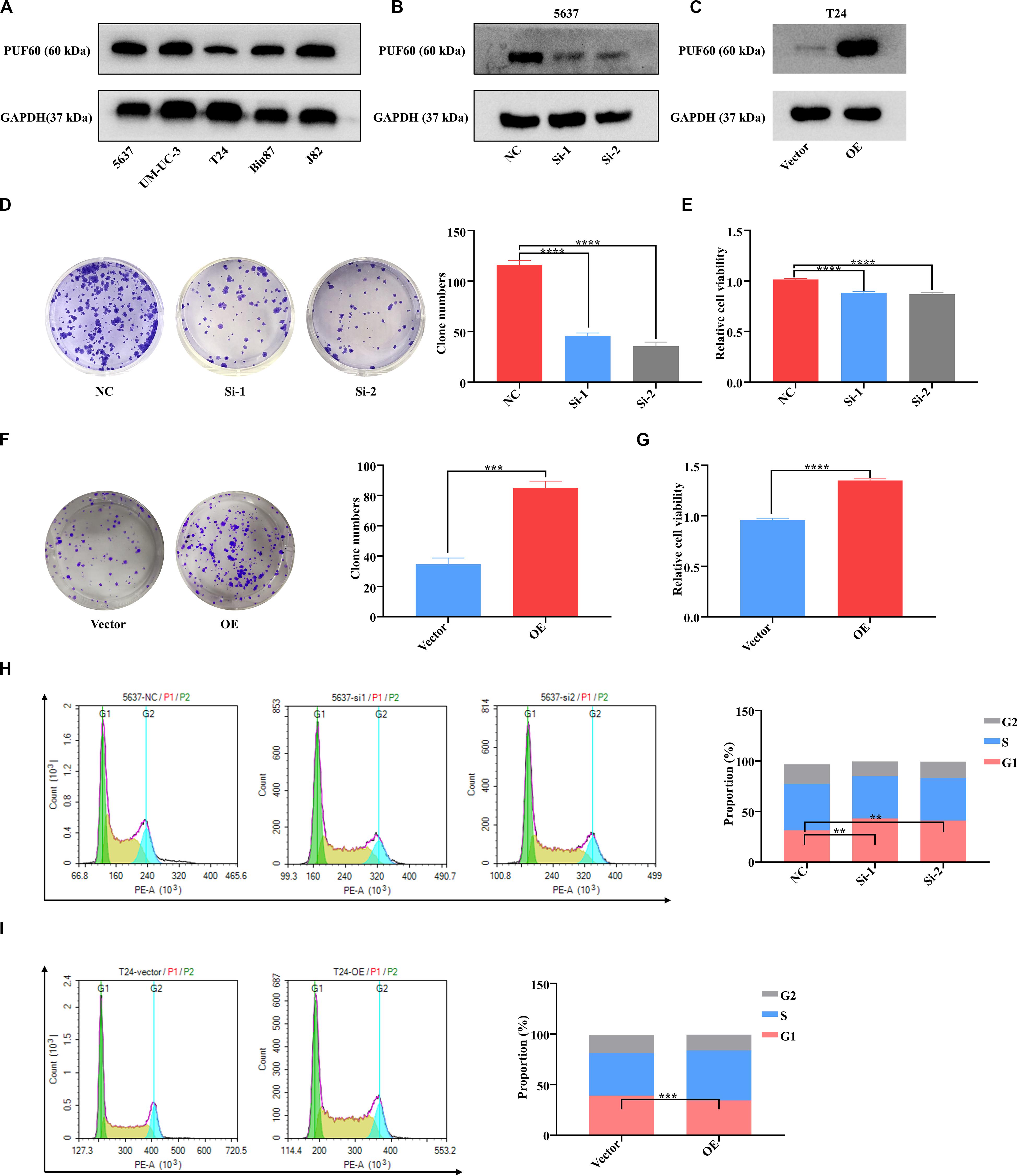
Figure 4. PUF60 promoted bladder cancer cell growth and cell cycle progression. (A) Endogenous expression of PUF60 was detected by western blot in different bladder cancer cell lines. (B) PUF60 expression was knocked down by its specific siRNAs in 5637 cells and detected by western blot. (C) PUF60 was overexpressed in T24 cells and detected by western blot. (D,E) Knockdown of PUF60 inhibited the clonogenicity (D) and cell viability (E) of 5637 cells (D: NC vs. Si-1: P < 0.0001, NC vs. Si-2: P < 0.0001; E: NC vs. Si-1: P < 0.0001, NC vs. Si-2: P < 0.0001). (F,G) Overexpression of PUF60 promoted the clonogenicity (F) and cell viability (G) of T24 cells (F: OE vs. vector: P = 0.0001; G: OE vs. vector: P < 0.0001). (H) Knockdown of PUF60 inhibited cell cycle progression in 5637 cells (NC vs. Si-1: P = 0.0018, NC vs. Si-2: P < 0.0027). (I) Overexpression of PUF60 promoted cell cycle progression in T24 cells (OE vs. vector: P = 0.0005). Data was analyzed by t-test; **P < 0.01, ***P < 0.001, ****P < 0.0001.
Associations Between Genome-Wide Expression Profiles and PUF60 Expression
To further clarify the potential underlying mechanism of PUF60-regulated bladder cancer cell growth, we selected GSE1357 dataset, which included the most samples of primary bladder cancer and intact clinical information of patients in all the datasets, to conduct gene expression profile analysis. The median value of PUF60 mRNA expression was used to divide patients into PUF60high and PUF60low groups, and a total of 40 up-regulated and 125 down-regulated genes were identified to be significantly associated with PUF60 mRNA expression (FDR-adjusted P < 0.05 and FC > 1.5 or FC < 1.5, Figure 5A). Aberrantly expressed genes were displayed/identified in an expression heat map (Figure 5B). Among the up-regulated genes in the PUF60high group, 12 out 40 were those promoting the proliferation and progression of bladder cancer or serving as independent unfavorable biomarkers for patients, including MCM2 (45), CKS1B (46), CDCA8 (47), TK1 (48), RAD21 (49), AURKA (42, 44), CDCA5 (50), TRIP13 (51), TACC3 (52), IQGAP3 (53–55), CDC20 (56, 57), RECQL4 (58). On the other hand, some of the down-regulated genes included the well-established tumor-suppressing genes in cancers, such as IFI16 (59, 60), ERGIC2 (61–63), SLC5A8 (64–72) (Figure 5C). These results indicated that PUF60 may play an important role in the initiation and progression of bladder cancer through up-regulating some oncogenes while down-regulating certain tumor-suppressing genes.
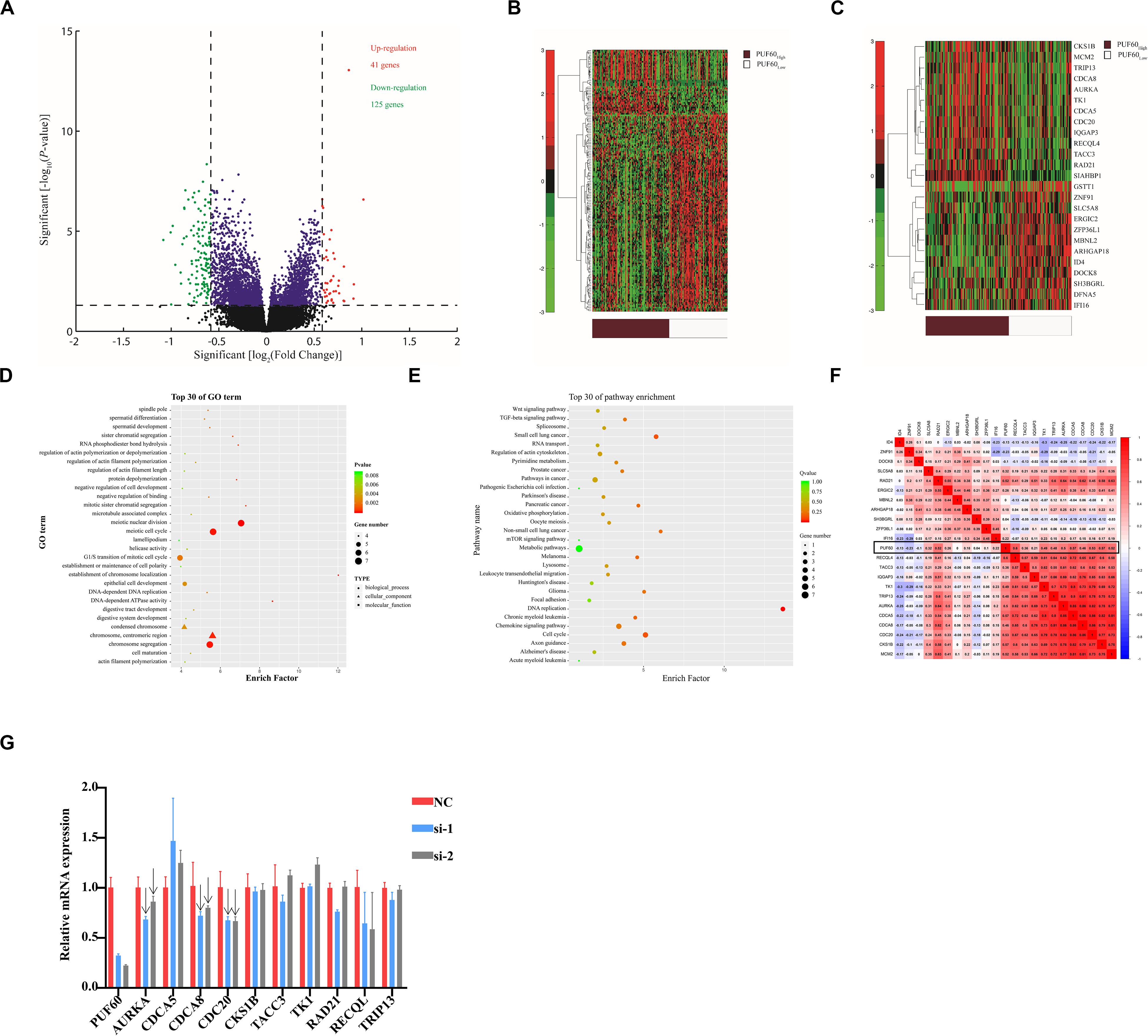
Figure 5. Associations between genome-wide expression profiles and PUF60 expression. (A) Volcano plot of differentially expressed gene profiles between PUF60high and PUF60low. (B) Expression heat map of PUF60-associated genes between PUF60high and PUF60low. (C) Expression heat map of differentially expressed genes closely related to bladder cancer development. (D,E) GO and KEGG analyses of differentially expressed genes between PUF60high and PUF60low. (F) Correlation between PUF60 and genes identified relevant to bladder cancer development from TCGA data. (G) mRNA levels of PUF60 and its possible downstream genes were detected by real time q-PCR in cells with PUF60 knockdown.
To elucidate the potential molecular pathways that involve PUF60 in the development and progression of bladder cancer, we conducted GO terms and KEGG enrichments analyses based on differential genes in the two groups. The results show that biological processes relevant to cell division, including meiotic nuclear division, meiotic cell cycle and chromosome segregation, were among the top 10 in GO terms analysis (Figure 5D). KEGG analysis also showed that DNA replication and cell cycle were among the top differential pathways (Figure 5E). These results were consistent with our cellular experiments in vitro. All these evidences indicated that PUF60 might be one of the dominant factors that mediate the initiation and progression of various cancers by influencing some key biological processes or pathways. To further validate the correlation between PUF60 and the differentially expressed genes identified above in the GSE13507 dataset, we analyzed the correlation between PUF60 and genes identified above using TCGA expression data, and found that 22 out of 25 genes were also significantly correlated with PUF60 (Figure 5F). To test whether PUF60 can regulate the expression of genes identified above, we knocked down PUF60 expression by its specific siRNAs in bladder cancer cell line 5637, and then detected the mRNA levels of 10 genes that were significantly associated with PUF60 in both TCGA and GES13507 datasets. We found mRNA expression of AURKA, CDCA8, CDC20 were decreased upon knockdown of PUF60 by its specific siRNA, indicating that they are the potential downstream targets of PUF60 in bladder cancer (Figure 5G, indicated by the arrows).
PUF60 Promoted Bladder Cancer Cell Growth via Transcriptionally Upregulating AURKA Expression
Considering the well-acknowledged vital roles of AURKA in bladder cancer development and progression, we conjectured that AURKA was the most potential target of PUF60. To confirm our hypothesis, we knocked down PUF60 expression by its specific siRNAs in 5637 cells, which significantly decreased the mRNA and protein expression of AURKA (Figure 6A). In contrast, overexpression of PUF60 in T24 cells significantly increased the expression of AURKA (Figure 6B). Next, we analyzed the association between mRNA expression of PUF60 or AURKA and the clinical pathological characteristics in GSE13507 datasets (Table 2), showing that higher AURKA mRNA expression was significantly associated with age, gender, AJCC stage and grade of patients.
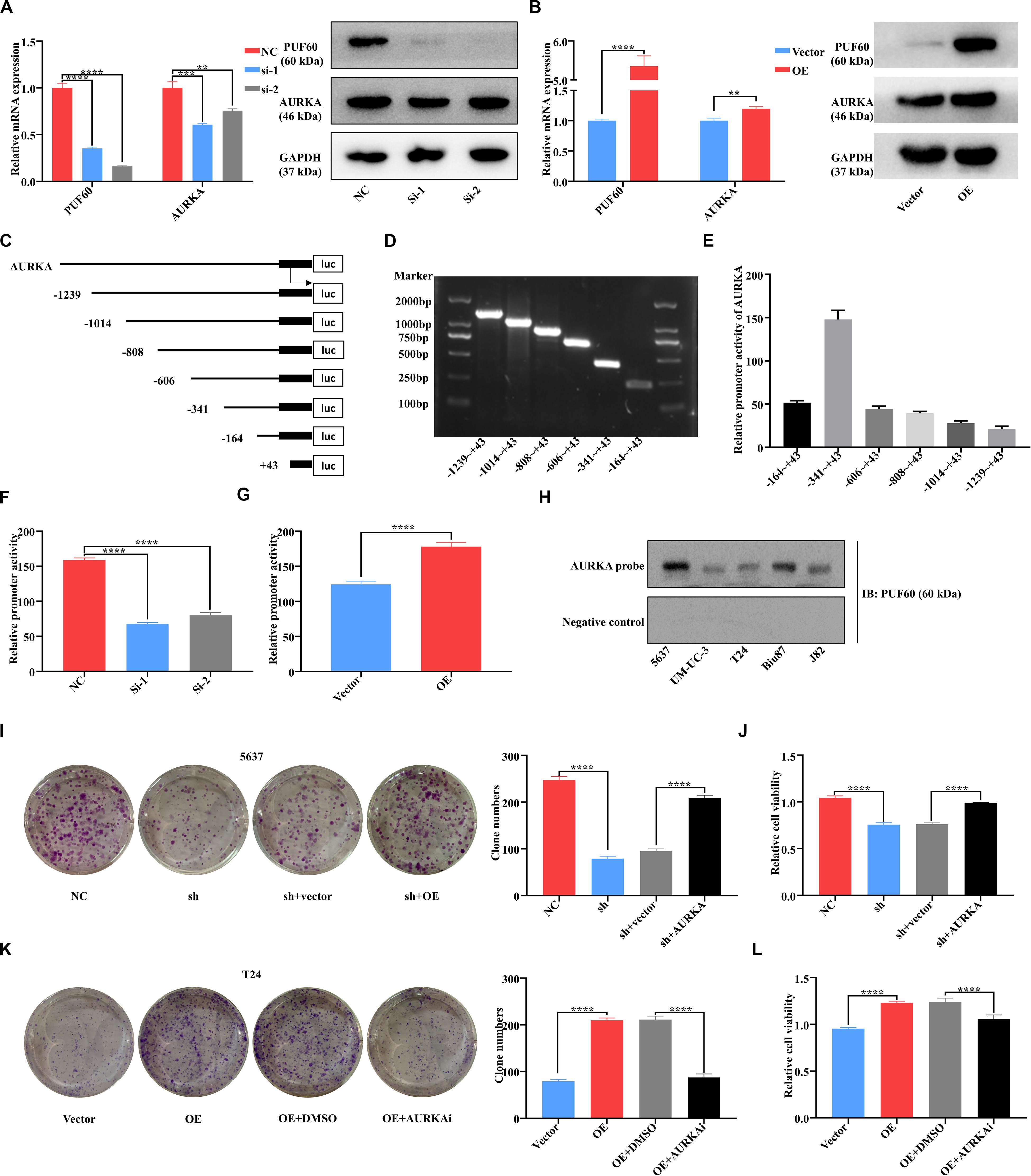
Figure 6. PUF60 promoted bladder cancer cell growth via transcriptionally upregulating AURKA expression. (A) PUF60 and AURKA expression was detected by RT-qPCR and western blot in 5637 cells with PUF60 knocked down (PUF60: NC vs. Si-1: P < 0.0001, NC vs. Si-2: P < 0.0001; AURKA: NC vs. Si-1: P = 0.0005, NC vs. Si-2: P < 0.0032). (B) PUF60 and AURKA expression was detected by RT-qPCR and western blot in T24 cells overexpressing PUF60 (PUF60: OE vs. vector: P < 0.0001; AURKA: OE vs. vector: P = 0.0033). (C) Ideograph of different segments of AURKA promoter. (D) Different segments of AURKA promoter were amplified by PCR using specific primers, and PCR products were detected by agarose gel electrophoresis. (E) Relative promoter activity of different segments of AURKA promoter measured by dual luciferase assay. (F) Relative activity of AURKA promoter was measured after knockdown of PUF60 in 5637 cells (NC vs. Si-1: P < 0.0001, NC vs. Si-2: P < 0.0001). (G) Relative activity of AURKA promoter was measured after overexpression of PUF60 in T24 cells (OE vs. vector: P < 0.0001). (H) Binding of PUF60 on the 5′-biotin labeled AURKA promoter probe or a control non-specific probe was detected by Western blot using anti-PUF60 antibody. (I,J) Knockdown of PUF60 inhibited the clonogenicity (I) and viability (J) of 5637 cells, which was reversed by AURKA overexpression (I: NC vs. sh: P < 0.0001, sh + vector vs. sh + AURKA: P < 0.0001; J: NC vs. sh: P < 0.0001, sh + vector vs. sh + AURKA: P < 0.0001). (K,L) Overexpression of PUF60 promoted the clonogenicity (K) and viability (L) of T24 cells, which was reversed by AURKA inhibitor (K: OE vs. vector: P < 0.0001, OE + DMSO vs. OE + AURKAi: P < 0.0001; L: OE vs. vector: P < 0.0001, OE + DMSO vs. OE + AURKAi: P < 0.0001). Clonogenicity was determined by colony formation assay. Viability was measured by MTS assay. Data was analyzed by t-test; **P < 0.01, ***P < 0.001, ****P < 0.0001.
Recent studies reported that RNA binding proteins were widely associated with gene transcriptional regulation, leading us to examine whether PUF60 upregulated AURKA expression by transcriptional activation. We constructed the luciferase reporter plasmids that have six different segments of the AURKA promoter (Figures 6C,D). The luciferase reporter assay showed that −341 ∼+43 was the core promoter of AURKA (Figure 6E), so we chose this plasmid for following validation assays. We then knocked down PUF60 expression in 5637 cells, and found that it significantly decreased the AURKA promoter activity (Figure 6F), while overexpression of PUF60 in T24 cells significantly increased the AURKA promoter activity (Figure 6G). To further validate that PUF60 transcriptionally activated AURKA expression, we conducted streptavidin-agarose pulldown assay, which showed that there was obvious binding of PUF60 at the AURKA promoter (Figure 6H). The results proved that PUF60 regulated AURKA expression by specifically binding to its promoter, thus activating its transcription.
To corroborate that PUF60 indeed regulated bladder cancer cell growth by mediating AURKA expression, we conducted the expression rescue experiments. We found that knockdown of PUF60 significantly inhibited 5637 cells growth, while overexpression of AURKA could partially reverse such an effect (Figures 6I,J). Consistently, overexpression of PUF60 significantly increased T24 cells growth, which was partially reversed by AURKA specific inhibitor (Figures 6K,L). These results suggested that PUF60 regulated bladder cancer cell growth by transcriptionally activating AURKA expression.
AURKA Was Highly Expressed and Positively Correlated With PUF60 Expression in Bladder Cancer
We analyzed the AURKA mRNA expression between normal and tumor tissues in our TCGA and GEO data. Four out of five datasets showed that AURKA mRNA was significantly highly expressed in tumor tissues (Figures 7A–E). Next, we analyzed the mRNA expression correlation between PUF60 and AURKA in all datasets we used, and AURKA mRNA expression had significant positive correlation with PUF60 mRNA expression in 9 out of 11 datasets (Figures 7F–P). These results are in line with the regulation relationship between PUF60 and AURKA.
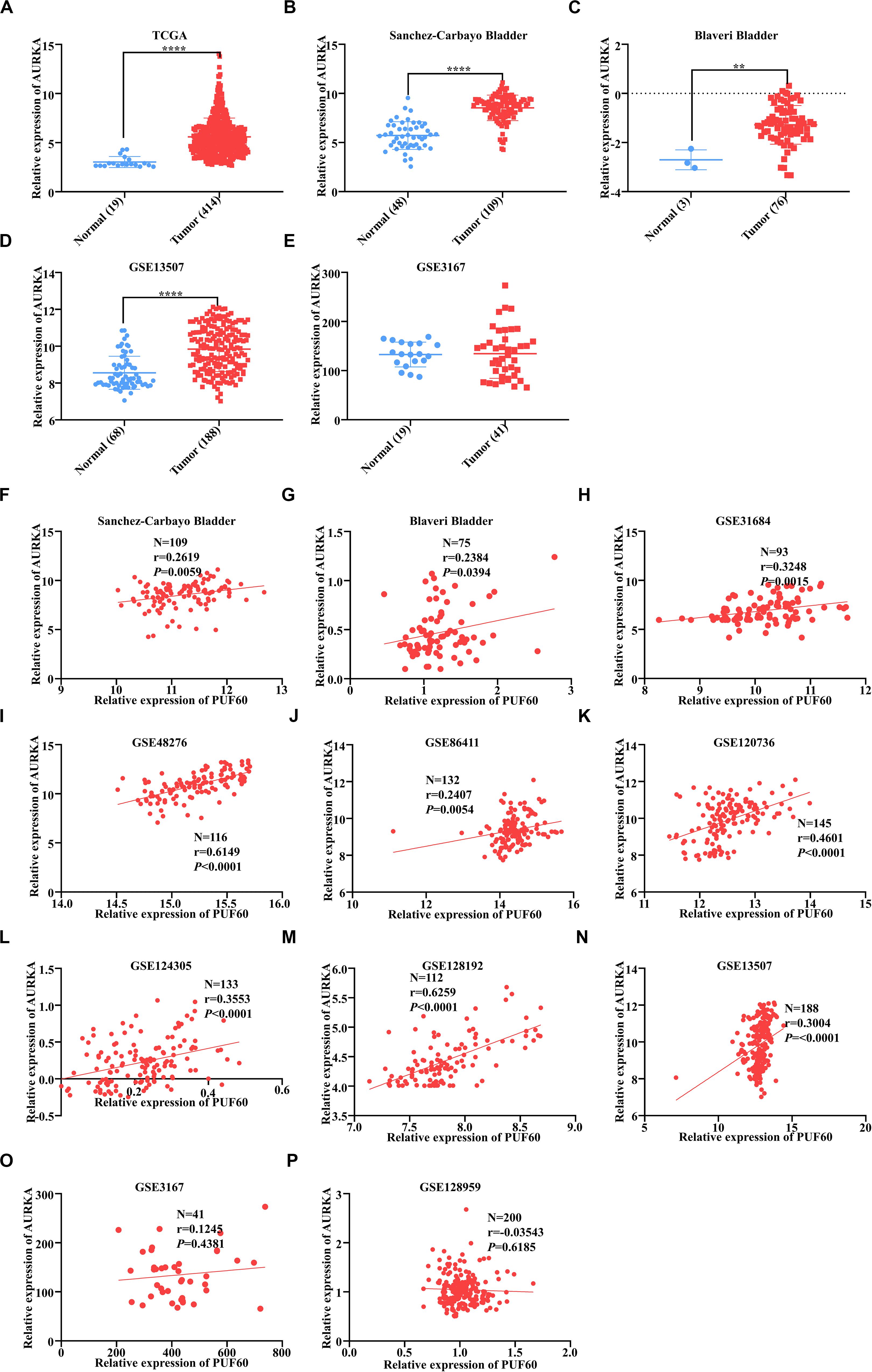
Figure 7. AURKA was highly expressed and positively correlated with PUF60 expression in bladder cancer. (A–E) Relative AURKA mRNA expression between normal and tumor tissues in TCGA data (A), Sanchez-Carbayo bladder study (B), Blaveri bladder study (C), GEO13507 (D), and GEO3167 data (E) (A: P < 0.0001; B: P < 0.0001; C: P = 0.0026; D: P < 0.0001; E: P = 0.8837). (F–P) The correlation between AURKA and PUF60 mRNA expression in all datasets used before. Data was analyzed by t-test; **P < 0.01, ****P < 0.0001.
Discussion
Over the past three decades, scientists have acquired a better understanding of human cancer initiation and progression with the rapid development of genome sequencing technique (73). Molecular subtypes of cancers rather than conventional clinical pathological subtypes have gained increasing attention in prognosis and treatment response prediction for patients (74). Molecular subtypes of bladder cancer have come into our sight because of its promising roles in predicting prognosis and guiding clinical treatment (5–7). It is of importance to identify genes associated with different molecular subtypes in bladder cancer.
RNA splicing proteins are a group of proteins not only involved in pre-mRNA splicing but also RNA export and transcriptional regulation. It has been reported that these proteins were widely associated with human diseases (8, 9, 18–20, 75). In the present study, we identified PUF60 as one of the most differentially expressed genes between normal and tumor bladder tissues among the 97 RNA splicing proteins. Next, we confirmed that the protein and mRNA expression of PUF60 were overexpressed in bladder cancer by analyzing our tissue microarray data and expression data from Oncomine database and GEO database. Furthermore, we found high PUF60 mRNA expression was associated with more malignant histological subtypes, higher pathological grade, advanced T stage and malignant molecular subtypes in bladder cancer. Kaplan–Meier survival analysis also indicated that patients with higher PUF60 mRNA expression are prone to have shorter survival time. To investigate its biological functions in bladder cancer cells, we conducted in vitro cell experiment, demonstrating that knockdown of PUF60 inhibited bladder cancer cell growth and cell cycle progression, while overexpression had opposite effects. These results showed that PUF60 served as an oncogene in bladder cancer. In our future study, we will perform animal experiments to verify the oncogenic role of PUF60/AURKA in bladder cancer in vivo. Our analysis also showed that PUF60 was associated with the malignant phenotypes of bladder cancer, and our current data indicated that PUF60 promoted cell cycle progression and growth in bladder cancer cells. It is worth investigating whether PUF60 is involved in other biological and functional processes of bladder cancer progression, such as migration, invasion and maintenance of stemness.
To clarify the underlying molecular mechanism by which PUF60 promoted bladder cancer growth, we analyzed the association between genome-wide expression profiles and PUF60 mRNA expression based on the data from GSE13507. AURKA, a vital gene involved in bladder cancer progression (41–44), was identified as a possible target of PUF60. A recent study indicated that pervasive chromatin-RNA binding protein interactions played an important role in the gene transcriptional regulation process beyond our expectation (76). Hence, we explored whether PUF60 upregulated AURKA expression transcriptionally by binding to its promoter, which was proved by our luciferase reporter assay and streptavidin-agarose pulldown assay. We will explore the potential downstream targets of the PUF60/AURKA axis in our follow-up study, which will help explain why the knockdown of PUF60 was only partially rescued by the overexpression of AURKA. Currently, it is unclear whether the RNA splicing functions of PUF60 were involved in the regulation of AURKA expression. Moreover, though our work proved that PUF60 regulated the promoter activity of AURKA, our present data cannot distinguish whether PUF60 directly or indirectly bound to the AURKA promoter. Considering that PUF60 is not a transcription factor, we speculate that PUF60 might interact with other components of the transcription machinery to regulate AURKA expression. Noteworthily, we cannot rule out the possibility that PUF60 could influence the expression of certain transcription factors that can directly bind to the AURKA promoter to regulate its expression. Lastly, we analyzed the correlation between PUF60 and AURKA expression in all datasets we used, finding significant correlation between the two genes, which further supported our conclusions.
In summary, we found that PUF60 was highly expressed in bladder cancer cells and associated with malignant phenotypes of bladder cancer. PUF60 promoted bladder cancer cell growth by activating AURKA signaling. High expression of PUF60 and AURKA predicted poor prognosis in bladder cancer patients. Our findings have demonstrated that PUF60 plays an important role in bladder cancer growth and provided new insights into the understanding of the pro-tumorigenic role of PUF60, indicating the PUF60/AURKA axis may serve as a potential clinical prognostic biomarker or a possible therapeutic target for bladder cancer.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
QL, WD, FZ, and LH conceived the study, participated in the design of the study, and interpretation of data. QL, XA, MC, NW, SS, YL, CZ, KL, XW, TT, YP, and HQ performed the experiments. QL, FX, WD, FZ, and LH analyzed and prepared data for publication. QL, MC, WD, and LH drafted and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the funds from the National Natural Science Foundation of China (81772483, 81902681, 81672665, 81702761, 81972623, 81972569, and 81772925), and the 5010 Program of Sun Yat-sen University (for XA).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the Deng’s Laboratory for their advice and technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.568015/full#supplementary-material
TABLE S1 | Primers for PCR and real-time PCR.
Footnotes
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmström P-U, Choi W, et al. Bladder cancer. Lancet. (2016) 388:2796–810. doi: 10.1016/s0140-6736(16)30512-8
3. Paner GP, Stadler WM, Hansel DE, Montironi R, Lin DW, Amin MB. Updates in the Eighth Edition of the tumor-node-metastasis staging classification for urologic cancers. Eur Urol. (2018) 73:560–9. doi: 10.1016/j.eururo.2017.12.018
4. Gallan AJ, Choy B, Paner GP. Contemporary grading and staging of urothelial neoplasms of the urinary bladder: new concepts and approaches to challenging scenarios. Surg Pathol Clin. (2018) 11:775–95. doi: 10.1016/j.path.2018.07.006
5. Lerner SP, McConkey DJ, Hoadley KA, Chan KS, Kim WY, Radvanyi F, et al. Bladder cancer molecular taxonomy: summary from a consensus meeting. Bladder Cancer. (2016) 2:37–47. doi: 10.3233/BLC-150037
6. Sanli O, Dobruch J, Knowles MA, Burger M, Alemozaffar M, Nielsen ME, et al. Bladder cancer. Nat Rev Dis Primers. (2017) 3:17022. doi: 10.1038/nrdp.2017.22
7. Aine M, Eriksson P, Liedberg F, Hoglund M, Sjodahl G. On molecular classification of bladder cancer: out of one. Many. Eur Urol. (2015) 68:921–3. doi: 10.1016/j.eururo.2015.07.021
8. Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. (1996) 65:367–409. doi: 10.1146/annurev.bi.65.070196.002055
10. Valcarcel J, Green MR. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. (1996) 21:296–301. doi: 10.1016/0968-0004(96)10039-6
11. Elliott DJ. The role of potential splicing factors including RBMY, RBMX, hnRNPG-T and STAR proteins in spermatogenesis. Int J Androl. (2004) 27:328–34. doi: 10.1111/j.1365-2605.2004.00496.x
12. Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. (2012) 3:1–12. doi: 10.1002/wrna.100
13. Obeng EA, Chappell RJ, Seiler M, Chen MC, Campagna DR, Schmidt PJ, et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. (2016) 30:404–17. doi: 10.1016/j.ccell.2016.08.006
14. Bezzi M, Teo SX, Muller J, Mok WC, Sahu SK, Vardy LA, et al. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. (2013) 27:1903–16. doi: 10.1101/gad.219899.113
15. Kralovicova J, Sevcikova I, Stejskalova E, Obuca M, Hiller M, Stanek D, et al. PUF60-activated exons uncover altered 3’ splice-site selection by germline missense mutations in a single RRM. Nucleic Acids Res. (2018) 46:6166–87. doi: 10.1093/nar/gky389
16. Shirai CL, Ley JN, White BS, Kim S, Tibbitts J, Shao J, et al. Mutant U2AF1 expression alters hematopoiesis and Pre-mRNA splicing in vivo. Cancer Cell. (2015) 27:631–43. doi: 10.1016/j.ccell.2015.04.008
17. Madan V, Kanojia D, Li J, Okamoto R, Sato-Otsubo A, Kohlmann A, et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun. (2015) 6:6042. doi: 10.1038/ncomms7042
18. Gruber AJ, Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet. (2019) 20:599–614. doi: 10.1038/s41576-019-0145-z
19. Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. (2016) 16:413–30. doi: 10.1038/nrc.2016.51
20. Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. (2016) 35:2413–27. doi: 10.1038/onc.2015.318
21. Page-McCaw PS, Amonlirdviman K, Sharp PA. PUF60: a novel U2AF65-related splicing activity. RNA. (1999) 5:1548–60. doi: 10.1017/s1355838299991938
22. Liu J, He L, Collins I, Ge H, Libutti D, Li J, et al. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol Cell. (2000) 5:331–41. doi: 10.1016/s1097-2765(00)80428-1
23. Kobayashi S, Hoshino T, Hiwasa T, Satoh M, Rahmutulla B, Tsuchida S, et al. Anti-FIRs (PUF60) auto-antibodies are detected in the sera of early-stage colon cancer patients. Oncotarget. (2016) 7:82493–503. doi: 10.18632/oncotarget.12696
24. Kajiwara T, Matsushita K, Itoga S, Tamura M, Tanaka N, Tomonaga T, et al. SAP155-mediated c-myc suppressor far-upstream element-binding protein-interacting repressor splicing variants are activated in colon cancer tissues. Cancer Sci. (2013) 104:149–56. doi: 10.1111/cas.12058
25. Matsushita K, Tomonaga T, Shimada H, Shioya A, Higashi M, Matsubara H, et al. An essential role of alternative splicing of c-myc suppressor FUSE-binding protein-interacting repressor in carcinogenesis. Cancer Res. (2006) 66:1409–17. doi: 10.1158/0008-5472.CAN-04-4459
26. Malz M, Bovet M, Samarin J, Rabenhorst U, Sticht C, Bissinger M, et al. Overexpression of far upstream element (FUSE) binding protein (FBP)-interacting repressor (FIR) supports growth of hepatocellular carcinoma. Hepatology. (2014) 60:1241–50. doi: 10.1002/hep.27218
27. Muller B, Bovet M, Yin Y, Stichel D, Malz M, Gonzalez-Vallinas M, et al. Concomitant expression of far upstream element (FUSE) binding protein (FBP) interacting repressor (FIR) and its splice variants induce migration and invasion of non-small cell lung cancer (NSCLC) cells. J Pathol. (2015) 237:390–401. doi: 10.1002/path.4588
28. Sun D, Lei W, Hou X, Li H, Ni W. PUF60 accelerates the progression of breast cancer through downregulation of PTEN expression. Cancer Manag Res. (2019) 11:821–30. doi: 10.2147/CMAR.S180242
29. Ogura Y, Hoshino T, Tanaka N, Ailiken G, Kobayashi S, Kitamura K, et al. Disturbed alternative splicing of FIR (PUF60) directed cyclin E overexpression in esophageal cancers. Oncotarget. (2018) 9:22929–44. doi: 10.18632/oncotarget.25149
30. Kobayashi S, Hiwasa T, Ishige T, Rahmutulla B, Kano M, Hoshino T, et al. Anti-FIRDeltaexon2, a splicing variant form of PUF60, autoantibody is detected in the sera of esophageal squamous cell carcinoma. Cancer Sci. (2019) 110:2004–13. doi: 10.1111/cas.14024
31. El Chehadeh S, Kerstjens-Frederikse WS, Thevenon J, Kuentz P, Bruel AL, Thauvin-Robinet C, et al. Dominant variants in the splicing factor PUF60 cause a recognizable syndrome with intellectual disability, heart defects and short stature. Eur J Hum Genet. (2016) 25:43–51. doi: 10.1038/ejhg.2016.133
32. Zhao JJ, Halvardson J, Zander CS, Zaghlool A, Georgii-Hemming P, Mansson E, et al. Exome sequencing reveals NAA15 and PUF60 as candidate genes associated with intellectual disability. Am J Med Genet Part B Neuropsychiatric Genet. (2018) 177:10–20. doi: 10.1002/ajmg.b.32574
33. Zhang YM, Yang HB, Shi JL, Chen H, Shu XM, Lu X, et al. The prevalence and clinical significance of anti-PUF60 antibodies in patients with idiopathic inflammatory myopathy. Clin Rheumatol. (2018) 37:1573–80. doi: 10.1007/s10067-018-4031-4
34. Low KJ, Ansari M, Abou Jamra R, Clarke A, El Chehadeh S, FitzPatrick DR, et al. PUF60 variants cause a syndrome of ID, short stature, microcephaly, coloboma, craniofacial, cardiac, renal and spinal features. Eur J Hum Genet. (2017) 25:552–9. doi: 10.1038/ejhg.2017.27
35. Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. (2003) 4:842–54. doi: 10.1038/nrm1245
36. Marumoto T, Zhang D, Saya H. Aurora-A–a guardian of poles. Nat Rev Cancer. (2005) 5:42–50. doi: 10.1038/nrc1526
37. Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. (2010) 10:825–41. doi: 10.1038/nrc2964
38. Yan M, Wang C, He B, Yang M, Tong M, Long Z, et al. Aurora-A Kinase: a potent oncogene and target for cancer therapy. Med Res Rev. (2016) 36:1036–79. doi: 10.1002/med.21399
39. Damodaran AP, Vaufrey L, Gavard O, Prigent C. Aurora A Kinase is a priority pharmaceutical target for the treatment of cancers. Trends Pharmacol Sci. (2017) 38:687–700. doi: 10.1016/j.tips.2017.05.003
40. Tao X, Chon HS, Fu S, Kavanagh JJ, Hu W. Update on aurora kinase inhibitors in gynecologic malignancies. Recent Pat Anticancer Drug Discov. (2008) 3:162–77. doi: 10.2174/157489208786242322
41. Guo M, Lu S, Huang H, Wang Y, Yang MQ, Yang Y, et al. Increased AURKA promotes cell proliferation and predicts poor prognosis in bladder cancer. BMC Syst Biol. (2018) 12(Suppl. 7):118. doi: 10.1186/s12918-018-0634-2
42. Mobley A, Zhang S, Bondaruk J, Wang Y, Majewski T, Caraway NP, et al. Aurora Kinase A is a biomarker for bladder cancer detection and contributes to its aggressive behavior. Sci Rep. (2017) 7:40714. doi: 10.1038/srep40714
43. Yuan Q, Sun T, Ye F, Kong W, Jin H. MicroRNA-124-3p affects proliferation, migration and apoptosis of bladder cancer cells through targeting AURKA. Cancer Biomark. (2017) 19:93–101. doi: 10.3233/CBM-160427
44. Zhou N, Singh K, Mir MC, Parker Y, Lindner D, Dreicer R, et al. The investigational Aurora kinase A inhibitor MLN8237 induces defects in cell viability and cell-cycle progression in malignant bladder cancer cells in vitro and in vivo. Clin Cancer Res. (2013) 19:1717–28. doi: 10.1158/1078-0432.CCR-12-2383
45. Kruger S, Thorns C, Stocker W, Muller-Kunert E, Bohle A, Feller AC. Prognostic value of MCM2 immunoreactivity in stage T1 transitional cell carcinoma of the bladder. Eur Urol. (2003) 43:138–45. doi: 10.1016/s0302-2838(02)00580-8
46. Kawakami K, Enokida H, Tachiwada T, Nishiyama K, Seki N, Nakagawa M. Increased SKP2 and CKS1 gene expression contributes to the progression of human urothelial carcinoma. J Urol. (2007) 178:301–7. doi: 10.1016/j.juro.2007.03.002
47. Li S, Liu X, Liu T, Meng X, Yin X, Fang C, et al. Identification of biomarkers correlated with the TNM staging and overall survival of patients with bladder cancer. Front Physiol. (2017) 8:947. doi: 10.3389/fphys.2017.00947
48. Rausch S, Hennenlotter J, Teepe K, Kuehs U, Aufderklamm S, Bier S, et al. Muscle-invasive bladder cancer is characterized by overexpression of thymidine kinase 1. Urol Oncol. (2015) 33:e21–9. doi: 10.1016/j.urolonc.2015.06.007
49. Yu Z, Xu Q, Wang G, Rowe M, Driskell C, Xie Q, et al. DNA topoisomerase IIalpha and RAD21 cohesin complex component are predicted as potential therapeutic targets in bladder cancer. Oncol Lett. (2019) 18:518–28. doi: 10.3892/ol.2019.10365
50. Chang IW, Lin VC, He HL, Hsu CT, Li CC, Wu WJ, et al. CDCA5 overexpression is an indicator of poor prognosis in patients with urothelial carcinomas of the upper urinary tract and urinary bladder. Am J Transl Res. (2015) 7:710–22.
51. Yan X, Guo ZX, Liu XP, Feng YJ, Zhao YJ, Liu TZ, et al. Four novel biomarkers for bladder cancer identified by weighted gene coexpression network analysis. J Cell Physiol. (2019) 234:19073–87. doi: 10.1002/jcp.28546
52. Lin ZR, Wang MY, He SY, Cai ZM, Huang WR. TACC3 transcriptionally upregulates E2F1 to promote cell growth and confer sensitivity to cisplatin in bladder cancer. Cell Death Dis. (2018) 9:72. doi: 10.1038/s41419-017-0112-6
53. Xu Y, Kim YH, Jeong P, Piao XM, Byun YJ, Kang HW, et al. Diagnostic value of combined IQGAP3/BMP4 and IQGAP3/FAM107A expression ratios in urinary cell-free DNA for discriminating bladder cancer from hematuria. Urol Oncol. (2019) 37:86–96. doi: 10.1016/j.urolonc.2018.10.023
54. Kim WT, Kim YH, Jeong P, Seo SP, Kang HW, Kim YJ, et al. Urinary cell-free nucleic acid IQGAP3: a new non-invasive diagnostic marker for bladder cancer. Oncotarget. (2018) 9:14354–65. doi: 10.18632/oncotarget.24436
55. Xu Y, Kim YH, Jeong P, Piao XM, Byun YJ, Seo SP, et al. Urinary cell-free DNA IQGAP3/BMP4 ratio as a prognostic marker for non-muscle-invasive bladder cancer. Clin Genitourinary Cancer. (2019) 17:e704–11. doi: 10.1016/j.clgc.2019.04.001
56. Choi JW, Kim Y, Lee JH, Kim YS. High expression of spindle assembly checkpoint proteins CDC20 and MAD2 is associated with poor prognosis in urothelial bladder cancer. Virchows Arch. (2013) 463:681–7. doi: 10.1007/s00428-013-1473-6
57. Kidokoro T, Tanikawa C, Furukawa Y, Katagiri T, Nakamura Y, Matsuda K. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene. (2008) 27:1562–71. doi: 10.1038/sj.onc.1210799
58. Patterson K, Arya L, Bottomley S, Morgan S, Cox A, Catto J, et al. Altered RECQL5 expression in urothelial bladder carcinoma increases cellular proliferation and makes RECQL5 helicase activity a novel target for chemotherapy. Oncotarget. (2016) 7:76140–50. doi: 10.18632/oncotarget.12683
59. Lin W, Zhao Z, Ni Z, Zhao Y, Du W, Chen S. IFI16 restoration in hepatocellular carcinoma induces tumour inhibition via activation of p53 signals and inflammasome. Cell Prolif. (2017) 50:e12392. doi: 10.1111/cpr.12392
60. Choubey D, Panchanathan R. IFI16, an amplifier of DNA-damage response: role in cellular senescence and aging-associated inflammatory diseases. Ageing Res Rev. (2016) 28:27–36. doi: 10.1016/j.arr.2016.04.002
61. Kwok SC, Liu X, Mangel P, Daskal I. PTX1(ERGIC2)-VP22 fusion protein upregulates interferon-beta in prostate cancer cell line PC-3. DNA Cell Biol. (2006) 25:523–9. doi: 10.1089/dna.2006.25.523
62. Liu X, Daskal I, Kwok SC. Effects of PTX1 expression on growth and tumorigenicity of the prostate cancer cell line PC-3. DNA Cell Biol. (2003) 22:469–74. doi: 10.1089/104454903322247343
63. Kwok SC, Liu X, Daskal I. Molecular cloning, expression, localization, and gene organization of PTX1, a human nuclear protein that is downregulated in prostate cancer. DNA Cell Biol. (2001) 20:349–57. doi: 10.1089/10445490152122460
64. Orellana-Manzano A, O’Ryan MG, Lagomarcino AJ, George S, Munoz MS, Mamani N, et al. Helicobacter pylori infection is associated with decreased expression of SLC5A8, a cancer suppressor gene, in young children. Front Cell Infect Microbiol. (2016) 6:121. doi: 10.3389/fcimb.2016.00121
65. Gurav A, Sivaprakasam S, Bhutia YD, Boettger T, Singh N, Ganapathy V. Slc5a8, a Na+-coupled high-affinity transporter for short-chain fatty acids, is a conditional tumour suppressor in colon that protects against colitis and colon cancer under low-fibre dietary conditions. Biochem J. (2015) 469:267–78. doi: 10.1042/BJ20150242
66. Elangovan S, Pathania R, Ramachandran S, Ananth S, Padia RN, Srinivas SR, et al. Molecular mechanism of SLC5A8 inactivation in breast cancer. Mol Cell Biol. (2013) 33:3920–35. doi: 10.1128/MCB.01702-12
67. Coothankandaswamy V, Elangovan S, Singh N, Prasad PD, Thangaraju M, Ganapathy V. The plasma membrane transporter SLC5A8 suppresses tumour progression through depletion of survivin without involving its transport function. Biochem J. (2013) 450:169–78. doi: 10.1042/BJ20121248
68. Helm J, Coppola D, Ganapathy V, Lloyd M, Centeno BA, Chen DT, et al. SLC5A8 nuclear translocation and loss of expression are associated with poor outcome in pancreatic ductal adenocarcinoma. Pancreas. (2012) 41:904–9. doi: 10.1097/MPA.0b013e31823f429f
69. Thangaraju M, Gopal E, Martin PM, Ananth S, Smith SB, Prasad PD, et al. SLC5A8 triggers tumor cell apoptosis through pyruvate-dependent inhibition of histone deacetylases. Cancer Res. (2006) 66:11560–4. doi: 10.1158/0008-5472.CAN-06-1950
70. Whitman SP, Hackanson B, Liyanarachchi S, Liu S, Rush LJ, Maharry K, et al. DNA hypermethylation and epigenetic silencing of the tumor suppressor gene, SLC5A8, in acute myeloid leukemia with the MLL partial tandem duplication. Blood. (2008) 112:2013–6. doi: 10.1182/blood-2008-01-128595
71. Babu E, Ramachandran S, CoothanKandaswamy V, Elangovan S, Prasad PD, Ganapathy V, et al. Role of SLC5A8, a plasma membrane transporter and a tumor suppressor, in the antitumor activity of dichloroacetate. Oncogene. (2011) 30:4026–37. doi: 10.1038/onc.2011.113
72. Li H, Myeroff L, Smiraglia D, Romero MF, Pretlow TP, Kasturi L, et al. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc Natl Acad Sci USA. (2003) 100:8412–7. doi: 10.1073/pnas.1430846100
73. Lappalainen T, Scott AJ, Brandt M, Hall IM. Genomic analysis in the age of human genome sequencing. Cell. (2019) 177:70–84. doi: 10.1016/j.cell.2019.02.032
74. Zhao L, Lee VHF, Ng MK, Yan H, Bijlsma MF. Molecular subtyping of cancer: current status and moving toward clinical applications. Brief Bioinform. (2019) 20:572–84. doi: 10.1093/bib/bby026
75. Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. (2011) 27:89–97. doi: 10.1016/j.tig.2010.12.001
Keywords: PUF60, AURKA, bladder cancer, transcriptional regulation, biomarker
Citation: Long Q, An X, Chen M, Wang N, Sui S, Li Y, Zhang C, Lee K, Wang X, Tian T, Pan Y, Qiu H, Xie F, Deng W, Zheng F and He L (2020) PUF60/AURKA Axis Contributes to Tumor Progression and Malignant Phenotypes in Bladder Cancer. Front. Oncol. 10:568015. doi: 10.3389/fonc.2020.568015
Received: 31 May 2020; Accepted: 04 September 2020;
Published: 07 October 2020.
Edited by:
Shunbin Xiong, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Yun Zhang, Texas Southern University, United StatesYixin Yao, University of Texas MD Anderson Cancer Center, United States
Copyright © 2020 Long, An, Chen, Wang, Sui, Li, Zhang, Lee, Wang, Tian, Pan, Qiu, Xie, Deng, Zheng and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wuguo Deng, ZGVuZ3dnQHN5c3VjYy5vcmcuY24=; Fufu Zheng, emZmY25AMTYzLmNvbQ==; Liru He, aGVsckBzeXN1Y2Mub3JnLmNu
†These authors have contributed equally to this work
 Qian Long
Qian Long Xin An
Xin An Miao Chen
Miao Chen Nan Wang2
Nan Wang2 Yangxun Pan
Yangxun Pan Fangyun Xie
Fangyun Xie Wuguo Deng
Wuguo Deng