
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 20 November 2020
Sec. Women's Cancer
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.526850
This article is part of the Research TopicEmerging Diagnostic Approaches for Triple-Negative Breast CancerView all 10 articles
 Simona Ruxandra Volovat1
Simona Ruxandra Volovat1 Constantin Volovat1,2*
Constantin Volovat1,2* Irina Hordila2
Irina Hordila2 Dorin-Alexandru Hordila2
Dorin-Alexandru Hordila2 Ciprian Camil Mirestean2
Ciprian Camil Mirestean2 Oana Tatiana Miron3
Oana Tatiana Miron3 Cristian Lungulescu4
Cristian Lungulescu4 Dragos Viorel Scripcariu5
Dragos Viorel Scripcariu5 Cati Raluca Stolniceanu6
Cati Raluca Stolniceanu6 Assia Adrianova Konsoulova-Kirova7
Assia Adrianova Konsoulova-Kirova7 Cristina Grigorescu5
Cristina Grigorescu5 Cipriana Stefanescu6
Cipriana Stefanescu6 Cristian Constantin Volovat8
Cristian Constantin Volovat8 Iolanda Augustin2
Iolanda Augustin2Noncoding RNAs (ncRNAs) include a diverse range of RNA species, including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs). MiRNAs, ncRNAs of approximately 19–25 nucleotides in length, are involved in gene expression regulation either via degradation or silencing of the messenger RNAs (mRNAs) and have roles in multiple biological processes, including cell proliferation, differentiation, migration, angiogenesis, and apoptosis. LncRNAs, which are longer than 200 nucleotides, comprise one of the largest and most heterogeneous RNA families. LncRNAs can activate or repress gene expression through various mechanisms, acting alone or in combination with miRNAs and other molecules as part of various pathways. Until recently, most research has focused on individual lncRNA and miRNA functions as regulators, and there is limited available data on ncRNA interactions relating to the tumor growth, metastasis, and therapy of cancer, acting either on mRNA alone or as competing endogenous RNA (ceRNA) networks. Triple-negative breast cancer (TNBC) represents approximately 10%–20% of all breast cancers (BCs) and is highly heterogenous and more aggressive than other types of BC, for which current targeted treatment options include hormonotherapy, PARP inhibitors, and immunotherapy; however, no targeted therapies for TNBC are available, partly because of a lack of predictive biomarkers. With advances in proteomics, new evidence has emerged demonstrating the implications of dysregulation of ncRNAs in TNBC etiology. Here, we review the roles of lncRNAs and miRNAs implicated in TNBC, including their interactions and regulatory networks. Our synthesis provides insight into the mechanisms involved in TNBC progression and has potential to aid the discovery of new diagnostic and therapeutic strategies.
Triple negative breast cancer (TNBC) represents a heterogeneous subgroup comprising 12%–17% of all of breast cancer (BC) (1). TNBC is characterized by the absence of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER2), relative to normal tissue or other types of BC, as well as a high proliferative index (determined by mitotic or Ki-67 proliferative indices), high histological grade, and high rates of metastasis (2–4). TNBCs frequently undergo early metastasis to visceral organs and brain (5–7) and are more often diagnosed at an advanced stage. The treatment options for TNBC are limited, and the 5-year survival rate is 77%. Furthermore, this neoplasm is often diagnosed at a relatively young age and is a major cause of female mortality (8–10).
The molecular heterogeneity of TNBC has been recently mapped using messenger RNA (mRNA), microRNA (miRNA), DNA, and epigenetic analyses of data from The Cancer Genome Atlas (TCGA) (11). Six molecular subtypes of TNBC were identified, each displaying a unique biology (12, 13), including basal-like BL1 (with high Ki-67, NRAS, Myc, ATR, and BRCA expression), BL2 (high expression of MET, TP53, EGFR, and EPHA2), immunomodulatory (overexpression of genes involved in immune cell signaling), mesenchymal, mesenchymal stem-like (chemoresistant) (13, 14), and luminal androgen receptor (LAR). Among these molecular subtypes, the BL2 subtype has the worst prognosis, and LAR tumors are associated with the best overall survival (OS) rates.
To improve the survival of patients with TNBC, identification of predictive biomarkers is necessary to assess the risk of metastasis, the response to treatment, or even to develop new therapies. Of novel and potentially useful biomarkers, there is good evidence for close correlation of abnormal expression of noncoding RNAs (ncRNAs) and TNBC development and progression.
Only 2% of the human genome comprises protein-coding gene sequences although more than 75% of the human genome is transcribed. Hence, most transcripts are noncoding genome sequences, including ncRNAs (15, 16). These sequences have regulatory roles in eukaryote cells, in which ncRNA malfunction is important in neoplastic processes (16, 17). Current research focuses on mutations or epigenetic modifications at different levels of gene expression regulation; however, it was recently discovered that this process is also highly modulated post-transcriptionally through repression or enhancement of mRNA. ncRNA expression profiles of tumors can be used to discriminate between different types of cancer (17).
MiRNAs are short, single-stranded, ncRNAs (19–25 nucleotides) that account for 1%–5% of the human genome (28,000 mature miRNAs have been identified) and regulate at least 30% of protein-coding genes (18, 19). They are involved in the regulation of tumor growth, proliferation, differentiation, and apoptosis as oncogenes and tumor suppressors (17, 20).
There are several phases involved in the generation of mature miRNAs. First, a nucleotide sequence is transcribed by RNA polymerase II from intergenic or intron-coding regions, resulting in capped, polyadenylated transcripts named primary-microRNAs (pri-miRNAs). Next, the primary transcript is processed by Drosha and DGCR8, nucleic ribonucleases that are part of a multiprotein complex, the microprocessor complex, which can recognize the junctions between hairpin structures and single-stranded RNA. The resulting product is generated by cleaving the RNA and producing a hairpin-shaped intermediate of 70–100 nucleotides, known as precursor-miRNA (pre-miRNA) (21, 22). The pre-miRNA crosses the nuclear membrane into the cytoplasm through the exportin-5 channel, in which another ribonuclease, Dicer, further processes it into short, double-stranded RNA fragments whose strands separate, and the mature single-stranded molecule adheres to the RNA-induced silencing complex (RISC), which is an effector molecule comprising miRNA and specific proteins.
MiRNA molecules can repress mRNA in two ways. In the first model, the miRNA hybridizes to the 3′ untranslated region (UTR) of their target mRNA. The complex, comprising RISC and the hybrid RNA molecule then mediates the cleavage and degradation of the target mRNA. In the second model, the miRNA does not bind perfectly to complementary sites in the 3′ UTR of its target mRNA, resulting in formation of a complex with RISC that only blocks mRNA translation. Regardless of the mechanism involved, the result is a decreased quantity of the translated product of the gene encoded by the target mRNA (17, 23).
MiRNAs can also promote ribosome biogenesis, resulting in target gene repression (24). Further, miRNAs are implicated in various biological and pathological regulatory processes, including the most malignant phenotype processes (25).
The transcription and function of miRNAs can be altered by slight changes in the activities of proteins involved in this process (26, 27), epigenetic modifications, single nucleotide polymorphisms (SNPs) in miRNA sequences (28, 29), hereditary mutations in the DNA encoding the miRNA (30, 31), or somatic mutation in miRNA genes (32–34). Each specific discrepancy in miRNA processing represents an opportunity for intervening at the post-transcriptional level by regulating miRNA expression. Detection of interactions with other individual precursor molecules is crucial for the discovery of new regulatory mechanisms, and the description of miRNA-protein interactions is indispensable for understanding the complex pathways governing miRNA activity (35).
To date, studies have focused on specific miRNA profiles that intervene at different levels of the malignant process; however, the reasons why miRNAs are over- or under-expressed remain unclear although multiple hypotheses have been proposed (36).
MiRNAs generate unique signatures that reflect the stage of development of cells and tissues of origin. A single miRNA can have multiple mRNA targets, and one mRNA can be the target of several miRNAs. Main miRNAs involved in TNBC are summarized in Table 1. From oncogenic miRNAs involved in TNBC, several are of major interest, including miR-21 and miR-135b. MiR-21 maps to chromosome 17 and is encoded by the MIR21 gene. MiR-21 over-expression is anti-apoptotic and contributes to tumor cell growth. Further, miR-21 can target various mRNA transcripts, and high levels of its expressions are associated with poor patient prognosis in TNBC patients (77). MiR-21 also contributes to EMT and was linked to positive lymph nodes, advanced disease stage, and short OS, resulting in the suggestion that it represents a molecular prognostic and disease progression marker (39). Downregulation of miR-21 in BC stem cells results in reversal of EMT, followed by decreased levels of HIF1α and inhibition of migration and invasion (40). MiR-135b is highly expressed in BC, in which it stimulates cell growth and can promote proliferation, invasion, and migration via large tumor suppressor 2 (LATS2), which it downregulates in TNBC cell lines. The miR-135b-LATS2 axis also represents a potential therapeutic target in TNBC (42). Tumor suppressor miRNAs in TNBC are mainly represented by MiR-101, MiR-125, MiR-145, MiR-200, and MiR-655. MiR-101 is downregulated in TNBC. When its expression is in the normal range, it inhibits cell progression in TNBC and suppresses MCL-1 expression, thereby increasing tumor sensitivity to paclitaxel (53). MiR-125 downregulates MAP2K7 and inhibits EMT when expressed normally. Suppression of miR-125 is linked to poor prognosis and resistance to chemotherapy in patients with TNBC. Further, miR-125 can be considered a prognostic factor in TNBC (54). MiR-145 targets the oncoprotein ROCK1, thereby suppressing BC tumor growth and migration. This miRNA also inhibits apoptosis in response to cIAP1 over-expression in MDA-MB-231 TNBC cells. When miR-145 is downregulated, it has the opposite effect, being correlated with progression and metastasis. Further, miR-145 is a promising diagnostic tool, as it can be easily detected in the serum, as well as a therapeutic target (56, 57). MiR-199a-5p is downregulated in TNBC, in which it has a tumor-suppressor role. High levels of miR-199a-5p repress invasiveness and migration in vitro, and its downregulation has a major impact on tumor growth and invasiveness. MiR-199a-5p is a reliable diagnostic marker and can be processed from blood plasma (60). The miR-200 family comprises MiR-200a, MiR-200b, Mir-200c, MiR-141, and MiR-429 with tumor-suppressor effects on apoptosis and metastasis of cell lines in vitro. In vivo, this family exhibits significantly reduced expression in metastatic TNBC. Levels of miR-200 miRNAs are decreased in mesenchymal subtype TNBC cell lines. Moreover, low expression of this family induces EMT through upregulation of ZEB1 (61, 62) and contributes to TNBC pathogenesis via multiple pathways (63). MiR-200a suppresses the EPH receptor A2 (EPHA2). MiR-200a is downregulated in basal-like BC, leading to elevated EPHA2 expression and consequent poor patient OS (64). MiR-200b-3p and MiR-200b-5p are involved in a dual mechanism of EMT regulation, via targeting of ZEB1/2 and suppression of PKCα (65). MiR-200c downregulation may be involved in invasion and metastasis in TNBC cases with BRCA mutation (66). MiR-655 is also downregulated in TNBC, being correlated with lymph node metastasis. Furthermore, there is a correlation between the level of miR-655 and BC molecular classification. MiR-655 over-expression can downregulate EMT by suppression of PRRX1, inhibiting cell migration and invasion (74).
LncRNAs are a category of ncRNA transcripts that regulate gene expression at the transcriptional, translational, and post-translational levels; however, lncRNAs do not encode peptides or proteins although they are vital for the proper functioning of cellular processes (78). LncRNAs exert their functions through various mechanisms, including mediation of interchromosomal interactions, acting as sponges for endogenous RNAs, regulating mRNA decay, and modulating epigenetic components, which are redirected to their targets, among others. Hence, any change in lnRNA expression levels can lead to various illnesses, including malignant disease (15).
The definition and classification of lncRNAs remains somewhat unclear as the exact mechanisms and pathways of action of these molecules have not been fully elucidated although some classes of lncRNA have been intensively studied and no longer represent a mystery.
LncRNAs are >200 nucleotides long with the majority ranging from 1000 to 10,000 nucleotides. In addition, this type of ncRNA generates secondary and three-dimensional structures, allowing it to play dual RNA and protein-like roles (79, 80). LncRNA biogenesis presents some similarities with that of mRNA; it is transcribed by RNA polymerase II, after which the majority of transcripts are spliced. Unlike mRNAs, lncRNAs are predominantly localized in the nucleus and have lower expression levels. Moreover, lncRNA expression patterns are cell type–specific (79, 81).
Most lncRNAs localize to the nucleus and chromatin, where they control DNA sequences and are involved in transcriptional regulation with different functions in the cytoplasm, and a fraction of molecules occur as circulating lncRNAs, which are transmitted via exosomes (78).
LncRNAs can interact with various molecules, including transcription factors, mature mRNAs, chromatin-modifying complexes, RNA binding proteins, DNA, nascent RNA transcripts, microRNA, and chromatin. LncRNA transcripts can bind to active proteins and establish an exact position (cis or trans) (80). Consequently, lncRNAs have important functions in regulating gene expression at the epigenetic, transcription, and post-transcription levels (82).
It is established that dysregulation of lncRNAs contributes to the pathogenesis of cancers (78). Four different archetypes of lncRNA molecular functions have been described, proposing that these molecules can function as decoys, signals, scaffolds, and guides. LncRNAs can act as molecular signals because transcription of an lncRNA occurs at an exact moment and location to integrate developmental signals, interpret cellular context, or respond to diverse stimuli. In this archetype, lncRNAs can act as markers of functionally significant biological events. In the second archetype, lncRNAs function as a molecular decoy; in this context, following their transcription, lncRNAs bind to and titrate their targeted protein. Frequently lncRNAs act as negative regulators of effector molecules. The third lncRNA archetype is the guiding type. LncRNAs can act as gene expression guides in two ways: For genes situated in their immediate vicinity, they act in a cis form, and they are in a trans form for distant genes. LncRNAs bring together components of different complexes and transcription factors that can either repress or activate targeted genes. In the fourth archetype, lncRNAs act as scaffolds. Different lncRNA domains bind distinct effector molecules simultaneously, resulting in either activation or repression of transcription. A better understanding of how the scaffolding complexes are assembled and regulated provides potential new strategies to select and utilize precise signaling constituents to modify cellular activities. These four archetypes of lncRNA molecular functions involve numerous lncRNAs in various pathways specific to TNBC. In a fifth archetype, represented by competing endogenous RNAs (ceRNAs), lncRNAs act as molecular sponges for microRNAs, controlling their bioavailability with functional biological consequences at the cellular or physiological level (82).
Several lncRNAs have important roles in cancer development, some of which have also been identified in TNBC (Table 2). From oncogenic lncRNAs, the main important ones are HOTAIR, MALAT1, HULC, AWPPH and ARNILA. HOTAIR (Hox transcript antisense intergenic RNA) is one of the best-studied lncRNA regulators. It is a spliced, polyadenylated transcript comprising 6 exons; is approximately 2200 nucleotides in length; and maps to chromosome 12q13.13 (83). This sequence has roles in the progression of multiple neoplasms, including pancreatic, colorectal, hepatocellular, gastric, lung, ovarian, and BCs. HOTAIR is oncogenic when upregulated; hence, its expression levels are correlated with patient prognosis, and HOTAIR can be used as a predictive biomarker.
HOTAIR induces migration and invasion of TNBC cell lines and was the first lncRNA to act as a marker of metastasis (83, 84). There are numerous reports of a strong association between lymph node metastasis in TNBC and HOTAIR expression in tissues (84–87) and circulation (88). Moreover, patients with upregulated HOTAIR tend to have poor survival rates. As lncRNAs are highly stable in biological fluids (89–93), detection of HOTAIR in blood samples from patients with TNBC can provide prognostic information and facilitate monitoring of treatment response. LncRNA ARNILA has a key role in TNBC locoregional invasion and metastasis. It can promote EMT by competitively binding to miR-204, which normally inhibits SOX4 expression. Upregulated ARNILA functions as an oncogenic lncRNA because, by sequestering miR-204, it frees SOX4 to induce EMT and metastatic spread. Moreover, silencing of ARNILA effectively inhibits cancer invasion, migration, and metastasis (103). From tumor supressor lncRNAs were described RMST, NEF and Airn. LncRNA RMST was identified as an lncRNA with very low expression levels in TNBC cells compared with healthy tissues. RMST localizes to the cytoplasm and is believed to exert its functions via regulation of mRNA or proteins. When RMST is over-expressed, the number of TNBC cells in the G1/G0 phase increases and the number of migrating cells reduces, indicating that RMST can prevent TNBC cell migration and invasion. In conclusion, RMST warrants further investigation as a potential biomarker and therapeutic tool in patients with TNBC (107).
Fully mature miRNAs interact with complementary regions in the 3’-UTRs of target mRNA transcripts, referred to as miRNA recognition elements (MREs). The consequence is the destruction of targets that are a perfect match or inhibition of translation when the target is a partial match (110).
Every miRNA can target hundreds of mRNAs and contribute to various functions and numerous networks. Individual miRNA mutations alone do not frequently lead to abnormal phenotypes but are strongly related to several network parameters, including cellular milieu, network topology, genomic position, and evolutionary age (111). There is extensive cooperation among miRNAs expressed in various functional contexts. The targeting of a single mRNA by multiple miRNAs leads to selective and specific regulation of its functions. Two co-operating miRNAs can bind to mRNA and form a thermodynamically stable triplex with synergistically increased effectiveness (112, 113). A single miRNA can simultaneously influence several mRNA targets participating in the same process, coordinated with other miRNAs, upstream transcription factors, and downstream mRNA targets. These elements are interconnected and form functional network modules with the general form “transcription factor/miRNA/target gene” (114, 115).
Some examples of lnRNAs-miRNA interactions in TNBC can involve lnRNAs as MALAT1, AWPPH and NEF. MALAT1 maps to chromosome 11q13 and is upregulated in invasive non-small cell lung carcinoma (94). It exhibits an intranuclear location and induces the PI3k/AKT/mTOR and Wnt/β-catenin pathways in multiple neoplasms, including TNBC (95). Zuo et al. suggests that silencing MALAT1 could decrease cell proliferation, migration, and invasion, inducing cell cycle arrest in the G0/G1 phase. Moreover, their study proves that, in TNBC, MALAT1 acts as a sponge for miR-129-5p, thereby promoting disease progression (98). Also, upregulated AWPPH expression is correlated with TNBC (116). A study discovered an association between AWPPH and miR-21, which together promote cell proliferation in TNBC and contribute to the development of chemoresistance. As miR-21 and lncRNA AWPPH are upregulated in TNBC and interact to function in cell proliferation, regulation, and chemoresistance, they have potential for use as blood biomarkers for early diagnosis (102). LncRNA NEF is an lncRNA that has been studied in TNBC cells and patients, demonstrating that NEF downregulates miR-155 and that altered NEF expression leads to high miR-155 levels in plasma. Overexpression of miR-155 combined with low lncRNA NEF levels is a profile that can differentiate patients with TNBC from controls. Downregulated NEF, alongside high levels of circulating miR-155, is correlated with increased migration and invasion of malignant cells. NEF over-expression also downregulates miR-155 expression, and miR-155 over-expression has no effect on lncRNA NEF, suggesting that NEF acts to directly downregulate miR-155 (108).
The arrangement of miRNAs in networks and the number of their interconnections with other molecules has a major influence on their functionality. The connection points of networks comprise miRNAs or mRNAs; those with high numbers of “synapses” are referred to as “hubs” and have major roles in regulating the whole network. mRNAs are situated in hub positions with higher densities of target sites and are evolutionarily selected as major points of direct miRNA-mediated suppression (117).
Several miRNAs associate in cancer networks with p73, a p53 family member. These interactions include inhibition of p73 by miRNAs, downregulation of miRNAs by p73, or modulation of the proteins that stabilize p73 by miRNAs. On the other hand, p73 regulates transcription of crucial miRNA genes having several attractive features in the context of cancer therapy including TNBC: 1) p73, unlike p53, is less often mutated in neoplastic disease; 2) its isoform, Tap73, inhibits all cancer features, developing responsiveness to standard radio- and chemotherapies; and 3) p73 replaces p53 functions, inducing the same axes and regulating stress-response pathways and retaining their function when p53 is dysfunctional (69, 118).
Approximatively 20,000 protein-coding genes have been identified in the human genome, a large number of which are coated with MREs (119). Pseudogenes are genomic loci that are similar to known genes but generally do not encode functional proteins and are defined as “nonfunctional,” “junk,” or “evolutionary relics” (120, 121). Due to their strong resemblance to miRNA sequences, pseudogenes can act as competitors for the same pool of miRNAs through sets of conserved MREs (122).
Salmena et al. were the first to propose the concept of competitive endogenous RNA (ceRNA), which encompasses all types of transcript: mRNA, tRNA, rRNA, lncRNA, pseudogene RNA, and circular RNA. In the ceRNA hypothesis, the traditionally envisaged process is reversed: That is, mRNAs are considered to influence miRNAs, suggesting that mRNAs sharing multiple MREs cross-talk and compete for miRNA binding. The result is reduced miRNA recognition and consequent diminished miRNA activity (102, 123). MREs are “artificial transcripts” comprising repeats of miRNA responsive elements, also termed “miRNA-sponges,” and are considered “the letters” of a “hidden RNA language” represented by ceRNAs. This language represents a complex system of interactions between different RNA species that manages gene expression regulation (124).
Another hypothesis states that ceRNA cross-talk cells center on a simple set of interactions between one miRNA and two target mRNAs (ceRNAs). The most important factors in this model are 1) the rate of transcription and degradation of miRNA and ceRNAs and 2) the rates of association, dissociation, and degradation of the complex. The ideal balance of ceRNA-mediated cross-regulation is obtained when all components of the system are equally represented. Secondary communication in ceRNETs may cause overexpression of the effect of a perturbation at any level of the network. As ceRNAs are key players in gene regulation, they are considered to represent an evolutionary mechanism (125, 126). It is generally accepted that ceRNA networks could be the next candidates to serve as prognostic biomarkers and possibly targets for new therapies in TNBC.
Based on recent topics of research focus, which involve genomic sponges and interactions among them, databases (ceRDB, starBAse, lnCeDB, LncACTdb, HumanViCe) have been created to archive all available information. Relationships are predicted virtually and validated experimentally (125).
Many lncRNAs have been proposed as ceRNAs for miRNAs. In TNBC, ARF6 overexpression causes loss of MiR-145, resulting in promotion of cell invasion. LincRNA-RoR, a regulator of reprogramming, is a ceRNA for miR-145, leading to competitive inhibition with ARF6, and loss of mature miR-145 expression (126–130).
Another important discovery is circular RNAs (circRNAs), which are ceRNAs created by the direct ligation of the 5′ and 3′ ends of linear RNA. Circular forms are transitional in the course of RNA splicing, exhibit superior stability relative to linear RNAs, and could consequently serve as effective miRNA sponges (131).
Direct regulatory relationships in these networks manifest as correlations among genes regulated by the same transcription factors, transcription factors with their individual correlated targets, or incidental correlations between gene expression levels. It has become clear that co-expression networks created in this way do not accurately represent the underlying regulatory processes and do not retain many of the properties associated with biological networks (132).
Recently, weighted gene co-expression network analysis (WGCNA) has been developed as a data mining method for studying ceRNA networks in TNBC. WGCNA was performed using mRNAs and lncRNAs extracted from mammary tissue of patients with triple-negative disease and the information subsequently correlated with Ki-67 status, using TCGA database. The RAD51AP1 and TYMS mRNAs were discovered to play a major role in overall survival of patients with TNBC based on analysis of this ceRNA network. The results of that study demonstrate that specific mRNAs and lncRNAs are part of a ceRNA network in TNBC, which could serve as target for treatment of patients with TNBC in the future (133).
A study of 116 TNBC and 11 normal tissues from TCGA analyzed integrated expression profiles, including data on miRNAs, lncRNAs, and mRNAs in a WGCNA to identify the features of dysregulated genes. Seven key molecules (AKAP12, FOS, EMX2OS, MYCNOS, RP11-542B15.1, hsa-miR-9, and hsa-miR-183) were overexpressed and correlated with poor patient outcomes; however, overexpression of hsa-miR-145, LINC00461, RP11-576D8.4, and RP11-496D24.2 was correlated with better OS (134).
An lncRNA-miRNA-mRNA regulatory axis was also hypothesized as positively correlated with TNBC in a ceRNA, in which five molecules (TERT, TRIML2, PHBP4, miR-1-3p, and miR-133a-3p) were significantly associated with prognosis in patients with TNBC (135).
Study of lncRNA-mRNA-miRNA axis interactions in a cohort of 1441 patients with TNBC from TGCA database showed that nine lncRNAs, including HOTTIP, ST7-AS2, LINC00491, ST7-OT4, NLGN1-AS1, ELFN2, C1orf143, C10orf126, and C8orf31, were associated with TNBC TNM stage, suggesting that they have oncogenic roles in TNBC. Further, differentially expressed lncRNAs (DElncRNAs) were found to be involved in progression of TNBC with four lncRNAs, RBMS3-AS3, C8orf31, LINC00452, and LINC00200, showing great potential as prognostic predictors in TNBC (136).
In a study of 155 DElncRNAs, DEmRNAs, and DEmiRNAs, OSTN-AS1 was identified as related to immunologic function and immune-related marker co-expression and proposed as a novel, possibly immune-related, prognostic marker (137).
A biomarker is a molecule that, ideally, is easy to detect and offers credible information on diagnosis, prognosis, or other disease parameters (138). MiRNAs are considered possible strong biomarkers in TNBC and can be evaluated in tissue specimens and as circulating miRNAs, where the latter present new opportunities for identification of powerful biomarkers in TNBC (139, 140).
MiRNAs useful as biomarkers can be identified singly or as part of a group of miRNAs all implicated in TNBC, referred to as miR-signatures. Examples of miRNAs useful as independent markers and also included in various miR-signatures include miR-10b, miR-155, and miR-21. Further, these molecules are dysregulated in numerous neoplasms in addition to TNBC (71, 141).
An miR-signature comprising 11 miRNAs (miR-21, miR-125b, miR-155, miR-181a, miR-181b, miR-10b, miR-195, miR-31, miR-183, miR-130a-3p, miR-451a) can distinguish between TNBC tumor and normal breast tissue (139). Further, in a cohort of 11 patients with TNBC, tissue biopsies were obtained prior to systemic therapy. A signature comprising three miRs (miR-200b-3p, miR-190a, and miR-512-5p) was demonstrated as being associated with complete pathological response to different treatment protocols (140). Another study in a cohort of 173 TNBC cases (up to 50 years old) discovered an additional miR-signature (miR-16, miR-655, miR-421, miR-374a, miR-374b, miR-497, miR-125b, miR-374a, miR-155, miR-16, miR-125b) that could function as a predictive biomarker for OS and disease-free survival (116). Further, miR-148a and miR-629-3p are associated with lung metastases, and miR-141 was found to promote brain metastasis (67, 142, 143). The miR-10 family is also implicated in TNBC progression and metastasis (144).
High expression of miR-95-3p is significantly correlated with decreased OS and relapse-free survival in patients treated with anthracycline-based chemotherapy, and another five-miRNA signature (including let-7d-3p, miR-30a-3p, miR-30c-5p, miR-128-3p, and miR-95-3p) was validated as a novel prognostic and predictive biomarker in TNBC, predicting patient response to anthracycline-based chemotherapy (145). Further, two miR-signatures were detected in a study of 173 TNBC cases. The first four-miR signature (miR-155, miR-16, miR-374a, and miR-125b) is associated with poor OS rate. The second also comprises four miRs (miR-155, miR-27a, miR-30e, and miR-493) and is linked to BC classification based on ER/PR/HER2/basal cytokeratin/EGFR status as well as separation of cases into low- and high-risk categories (50) with the ability to predict patient response to therapy with the two most common systemic protocols used to treat TNBC (anthracycline or anthracycline plus taxanes) (146).
Many recent studies have described circulating miRNAs as promising diagnostic, prognostic, or predictive biomarkers for BC. The processes of miRNA biogenesis and maturation occur in the cell nucleus and cytoplasm, and miRNAs can be discharged out of cells from the cytoplasm to become extracellular circulating miRNAs. There are multiple mechanisms for miR transportation within organisms, including 1) wrapped in large apoptotic bodies, 2) in HDL or LDL lipoprotein complexes, 3) in microvesicles or smaller exosomes, and 4) in an AGO protein complex (147). Exosomes and lipid vesicles have critical roles in cell–cell communication. Cell-free circulating miRNAs can be detected and isolated from the microenvironment as well as from different body fluids, such as plasma, serum, saliva, urine, seminal fluid, breast milk, and cerebrospinal fluid (148) (Figure 1).
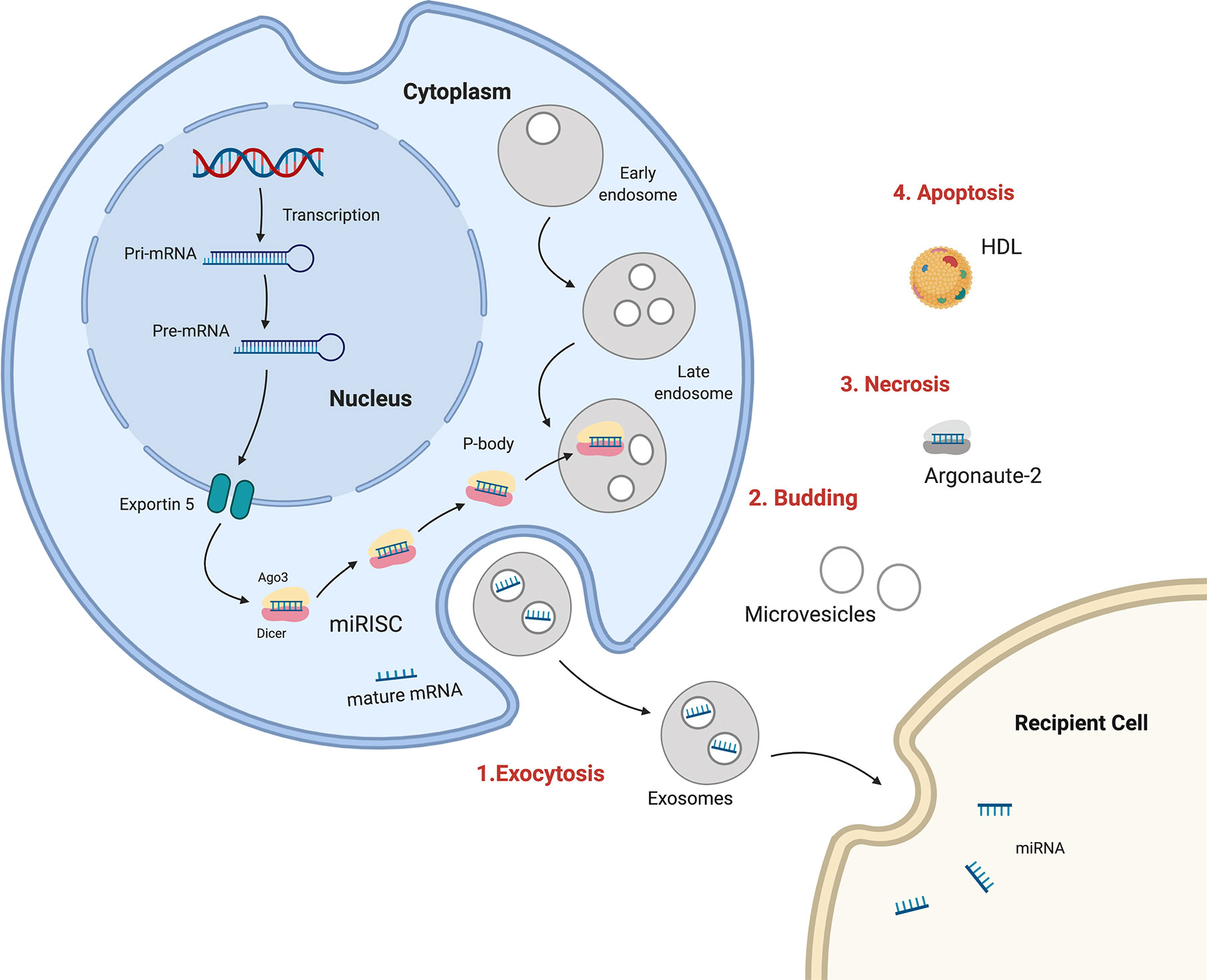
Figure 1 Sources and routes of circulating mRNAs. 1. Exosomal secretion- Pri-miRNA is transcribed , processed by Drosha and transported in cytoplasm by Exportin5, where is integrated in RISC complex and target mRNA in exosomes which are released in human fluids. 2. Budding from plasma membrane and forming microvesicles. 3. Necrosis, with the releasing of AGO-miRNA complexes. 4. Apoptosis, bounding high-density lipoproteins (HDL).
The processes used to validate miRNAs as biomarkers in TNBC have strengths and weaknesses. The great advantage of miRNAs in the pre-analytical phase is that they can be detected in biological fluids, require a minimally invasive procedure for collection, and are stable under various conditions (repetitive freeze-thaw, extreme pH values, up to 24 h at room temperature, or for long periods at 70°C) (149). Problems with validation of miRNA biomarkers include several biases related to the daily habits of patients (physical activity, smoking, diet, kidney pathology, and circadian rhythm), sample collection, and handling (149–153). Real-time quantitative polymerase chain reaction (RT-qPCR) is the standard analytical technique used to assess circulating miRNAs; however, some alternative platforms, including other PCR-based assays, microarray, and next-generation sequencing (NGS) can also be used. Validation can be affected in the analytical phase by noncirculating miRNA contamination (blood cells, skin, and activated platelets) and by hemolysis (154–157). Finally, issues of concern in the postanalytical phase include a lack of a standardized protocols and procedures (158).
IVarious studies have confirmed the importance of analysis of miRNAs as a noninvasive approach for screening and management of BC molecular subtypes (Table 3). Downregulation of miR-195-5p and miR-495 has value as a prospective circulating surrogate molecular marker for early detection of luminal or TNBC (171). Another seven-serum miRNA panel (miR-489-3p + miR-199a-3p + miR-195-5p + miR-15a-5p + miR-7-5p + let-7c-5p + let-7i-5p) can serve as a diagnostic marker for patients with TNBC (172). Further, a panel of nine miRNAs (miR-15a, miR-18a, miR-107, miR-133a, miR-139-5p, miR-143, miR-145, miR-365, and miR-425) is a potential blood-based multimarker test for BC detection (173). Moreover, another five circulating miRNA combination (miR-1246, miR-1307-3p, miR-4634, miR-6861-5p, and miR-6875-5p) can detect TNBC with a sensitivity of 97.3%, specificity of 82.9%, and accuracy of 89.7% for BC in the test cohort. Furthermore, this combination can detect early stage BC (sensitivity 98.0% for stage 0) (174).
In stage II and III TNBC patients, the overexpression of many circulating miRNAs (hsa-miRNA-188-5p, hsa-miRNA-1202, hsa-miRNA-4281, hsa-miRNA-1207-5p, hsa-miRNA-4270, hsa-miRNA-1225-5p, hsa-miRNA-642b-3p, hsa-miRNA-1290, hsa-miRNA-3141, miRNA-127-3p, miRNA-148b, miRNA-409-3p, miRNA-652, and miRNA-801) has been identified and validated (175). Serum miR-21 expression level is associated with lymph node metastasis and high Ki-67 expression in TNBC (p < 0.01) (176).
Some circulating plasma miRNAs can accurately discriminate TNBC from non-TNBC. In TNBC, miR-199a-5p, miR-21, and miR-16 are downregulated (AUC: 0.88, 0.80, and 0.87, respectively), and their levels are restored to normal after surgery. MiR-489, miR-200b, miR-193b, miR-125b, miR-105, miR-93 3p are upregulated in TNBC (AUC: 0.99, 0.88, 0.91, 0.97, 0.93, and 0.66, respectively), which has been validated in diagnostic settings. In addition, circulating miR-34a is downregulated in patients with both TNBC and HR+/HER2- BC tumors unresponsive to neoadjuvant chemotherapy compared to those with responsive tumors (AUC = 0.59) (177).
In a meta-analysis of 21 relevant studies (2510 patients) exploring the prognostic value of miRNAs in TNBC by measuring miR expression levels in tumor or blood samples, six miRNAs were assessed: miR-155, miR-21, miR-27a/b, miR-374a/b, miR-210, and miR-454. The results indicated that decreased expression of miR-155 is associated with reduced OS (adjusted hazard ratio (HR) = 0.58, 95% confidence interval (CI): 0.34–0.99; crude HR = 0.67, 95% CI: 0.58–0.79). High miR21 expression was also predictive of reduced OS (crude HR = 2.50, 95% CI: 1.56–4.01), and elevated levels of miR-27a/b, miR-210, and miR-454 expression were associated with shorter OS, and levels of miR-454 and miR-374a/b expression were associated with DFS (178).
In a prospective observational study using frozen tissue sections, 165 TNBC samples and 33 paired normal breast tissues were analyzed using transcriptome microarrays. An integrated mRNA-lncRNA signature, based on eight mRNAs and two lncRNAs (HIST2H2BC and SNRPEP4), was evaluated. This signature is more feasible for predicting 2-year recurrence-free survival than classic prognostic factors (AUC 0.714) and can accurately predict clinical outcomes and benefit from of taxane chemotherapy in patients with TNBC (179). Expression levels of Linc00339 in several BC cell lines were compared with those in a normal mammary gland epithelial cell line, and higher expression of miR-377-3p indicates longer OS in patients with TNBC. Hyperexpression of miR-377-3p produces a delay in TNBC cell growth by regulating cell cycle distribution and apoptosis and miR-377-3p regulates HOXC6 expression, influencing Linc00339-mediated TNBC proliferation. Hence, a Linc00339/miR-377-3p/HOXC6 axis influences TNBC progression and may be a promising therapeutic target for TNBC treatment (180).
Measurement of HIF1A-AS2 expression in 86 TNBC specimens, 30 non-TNBC specimens, and 30 adjacent mammary specimens demonstrated that it is upregulated in TNBC tissues compared with non-TNBC tissues, leading to the conclusion that HIF1A-AS2 expression is associated with OS in patients with TNBC (181).
Another study analyzed the expression of the lncRNA, HOTAIR, in 163 cases of TNBC and found that its expression in tumor tissues is strongly correlated with lymph node metastasis and has a direct strong association with androgen receptor (AR) expression. These data suggest the involvement of HOTAIR in regulation of AR-mediated pathways, leading to its proposal as a prognostic marker correlated with new therapeutic strategies for patients with the LAR type of TNBC (93). Furthermore, plasma urothelial carcinoma associated 1 (UCA1) levels are significantly elevated in patients with TNBC, indicating that this molecule may act as a specific biomarker for TNBC diagnosis (182).
A recently discovered lncRNA, hepatocellular carcinoma upregulated EZH2-associated lncRNA (HEIH), is overexpressed in TNBC tissues and cell lines compared with adjacent normal mammary tissues and a normal mammary epithelial cell line. Downregulation of HEIH inhibits TNBC cell proliferation and promotes apoptosis by regulating the miR-4458/SOCS1 axis. HEIH is also involved in clinical progression (183).
Comparison between the genome-wide methylation profiles in peripheral blood DNA from 233 patients with TNBC and 231 controls led to the identification and validation of increased methylation at cg06588802 in the long intergenic noncoding RNA, LINC00299 in patients with TNBC compared with controls, suggesting that hypermethylation of LINC00299 in peripheral blood may constitute a useful circulating biomarker for TNBC (184).
The paradigm proposed for the origin of cancer related to its hallmarks involves the highly dynamic accumulation of mutations and chromosomal genetic changes occurring at different times and influenced by epigenetic mechanisms.
Translating current knowledge of TNBC into oncological practice involves identification of new biomarkers/molecules/assays, which reveal new pathways or novel ways to understand these diseases. EMT is a current research focus with the metastatic process involving multiple genes/pathways: for example, ETS, RAS, integrins, WNT/beta-catenin, SRC, and Notch (185–191).
Cancer stem cells express phenotypic plasticity, which is a major factor in tumor malignancy, and miRNAs are involved in the maintenance of stemness in TNBC (192–196). Specifically, miRNA-31 promotes mammary stem cell expansion and tumorigenesis by suppressing WNT signaling antagonists (197).
LncRNAs are also central players in the battle against malignant diseases. LncRNAs contribute to regulatory networks by various mechanisms. Nuclear lncRNAs may be considered to have epigenetic functions of regulation or to act as guides by recruiting chromatin modification factors to cytogenetic loci.
Studies of miRNAs and lncRNAs can also improve information regarding the molecular classification of TNBC. Recently a novel TNBC classification system, the FUSCC classification, was established by integrating the expression profiles of mRNAs and lncRNAs (198). Therefore, it is essential to explore the mechanisms and interactions involved in the regulatory networks, including lncRNA, miRNA, mRNA, genetic mutations, and epigenetic changes in TNBC. A new cross-talk pattern, represented by the ceRNA hypothesis, in which lncRNAs can serve as a ceRNAs involved in regulation of mRNA expression and also including miRNAs, represents a new perspective regarding ncRNA interactions.
Possible tissue biomarkers have been identified in numerous studies, particularly miRNAs, singly or associated in specific signatures, relevant for TNBC subtypes, prognosis, and prediction of treatment efficacy.
The importance of circulating lncRNAs and miRNAs as robust noninvasive biomarkers, particularly for the diagnosis of TNBC, is demonstrated by emerging evidence. A major issue for biomarkers can arise if circulating RNA profiles do not resemble miRNAs within the tumor. An analysis of 210 plasma miRNAs suggested that they may match those in tumors although other studies report differences in expression between serum and tumor miRNAs, attributable to only a small proportion of miRNAs being released from the tumor into the circulation. Differentially expressed miRNA and target genes are suggested to form complex interaction networks, affecting many biological processes (199).
LncRNA, miRNA, and mRNA interactions in cancer and the molecular mechanisms involved in these interactions correlated with tumorigenesis and cancer progression remain undefined. Future biomarkers in TNBC may be represented by genomic signatures developed into clinically accepted tests for the early detection and clinical management of patients with TNBC.
Circulating miRNAs and lncRNAs are also promising tests for early detection, prognosis, and monitoring treatment effects. LncRNA-miRNA-mRNA interaction networks provide opportunities for further experimental studies and improvement of biomarker predictions for developing novel therapeutic approaches in TNBC.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Foulkes W, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med (2010) 363:1938–48. doi: 10.1056/NEJMra1001389
2. Irvin W, Carey LA. What is triple-negative breast cancer? Eur J Cancer (2008) 44:2799–805. doi: 10.1016/j.ejca.2008.09.034
3. Stockmans G, Deraedt K, Wildiers H, Moerman P, Paridaens P. Triple-negative breast cancer. Curr Opin Oncol (2008) 20:614–20. doi: 10.1097/CCO.0b013e328312efba
4. Thike A, Cheok PY, Jara-Lazaro AR, Than B, Than P, Than PH, et al. Triple-negative breast cancer: Clinicopathological characteristics and relationship with basal-like breast cancer. Mod Pathol (2010) 23:123–33. doi: 10.1038/modpathol.2009.145
5. Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res (2008) 68:3018–114. doi: 10.1158/0008-5472.CAN-07-5644
6. Lin N, Claus E, Sohl J, Razzak AR, Arnaout A, Winer EP, et al. Sites of distant recurrence and clinical outcomes in patients with metastatic triple-negative breast cancer: High incidence of central nervous system metastases. Cancer (2008) 113:2638–45. doi: 10.1002/cncr.23930
7. Heitz F, Harter P, Lueck HJ, Fissler-Eckhoff A, Lorenz-Salehi F, Scheil-Bertram S, et al. Triple-negative and HER2-overexpressing breast cancers exhibit an elevated risk and an earlier occurrence of cerebral metastases. Eur J Cancer (2009) 45:2792. doi: 10.1016/j.ejca.2009.06.027
8. Liedtke C, Rody A, Gluz O, Baumann K, Beyer D, Kohls EB, et al. The prognostic impact of age in different molecular subtypes of breast cancer. Breast Cancer Res Treat (2015) 152:667–73. doi: 10.1007/s10549-015-3491-3
9. Skandan SP. 5-year Overall survival of triple negative breast cancer: A single institution experience. J Clin Oncol (2016) 34:e12580. doi: 10.1200/JCO.2016.34.15_suppl.e12580
10. Howlader N, Noone AM, Krapcho M, et al. eds. SEER Cancer Statistics Review, 1975-2016. Bethesda, MD: National Cancer Institute (2019). Availble from seer.cancer.gov/csr/1975_2016/, based on November 2018 SEER data submission, posted to the SEER web site April 2019.
11. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature (2012) 490(7428):61–70. doi: 10.1038/nature11412
12. Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shy Y, et al. TNBC type: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform (2012) 11:147–56. doi: 10.4137/CIN.S9983
13. Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest (2011) 121:2750–67. doi: 10.1172/JCI45014
14. Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Cancer Res (2010) 12:R68. doi: 10.1186/bcr2635
15. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature (2012) 489:101–8. doi: 10.1038/nature11233
16. Nagini S. Breast Cancer: Current Molecular Therapeutic Targets and New Players. Anti-Cancer Agents Medicinal Chem (2017) 17:152–63. doi: 10.2174/1871520616666160502122724
17. Pecorino L. Molecular Biology of Cancer: Mechanisms, Targets, and Therapeutics. 3rd edition. New York: Oxford University Press (2012), ISBN: ISBN 978–0–19–957717–0.
18. Liu W, Mao SY, Zhu WY. Impact of tiny miRNAs on cancers. World J Gastroenterol (2007) 13:497–502. doi: 10.3748/wjg.v13.i4.497
19. Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell (2005) 120:21–4. doi: 10.1016/j.cell.2004.12.031
20. Avery-Kiejda KA, Braye SG, Mathe A, Forbes JF, Scott RJ. Decreased expression of key tumour suppressor microRNAs is associated with lymph node metastases in triple negative breast cancer. BMC Cancer (2014) 14:51. doi: 10.1186/1471-2407-14-51
21. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature (2003) 425(6956):415–9. doi: 10.1038/nature01957
22. Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev (2004) 18:3016–27. doi: 10.1101/gad.1262504
23. Wu W. MicroRNA and Cancer Methods and Protocols. 2nd ed. New York:Humana Press (2018)ISSN 1940-6029; ISBN 978-1-4939-7435-1. doi: 10.1007/978-1-4939-7435-1
24. Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up regulate translation. Science; (2007) 318:1931–4. doi: 10.1126/science.1149460
25. Tufekci KU, Meuwissen RL, Genc S. The role of microRNAs in biological processes. Methods Mol Biol (2014) 1107:15–31. doi: 10.1007/978-1-62703-748-8_2
26. Lujambio A, Esteller M. How epigenetics can explain human metastasis: A new role for microRNAs. Cell Cycle (2009) 8:377–82. doi: 10.4161/cc.8.3.7526
27. Davis-Dusenbery BN, Hata A. Mechanisms of control of microRNA biogenesis. J Biochem (2010) (2010) 148:381–92. doi: 10.1158/0008-5472.CAN-05-3632
28. Sun G, Yan J, Noltner K, Feng J, Sarkis DA. SNPs in human miRNA genes affect biogenesis and function. RNA (2009) 15:1640–51. doi: 10.1261/rna.1560209
29. Saunders MA, Liang H, Li W-H. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA (2007) 104:3300–5. doi: 10.1073/pnas.0611347104
30. Hughes AE, Bradley DT, Campbell M, Lechner J, Dash DP, Simpson DA, et al. Mutation Altering the miR-184 Seed Region Causes Familial Keratoconus with Cataract. Am J Hum Genet (2011) 89:628–33. doi: 10.1016/j.ajhg.2011.09.014
31. Conte I, Hadfield KD, Barbato S, et al. MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proc Natl Acad Sci USA (2015) (2015) 112:E3236–45. doi: 10.1073/pnas.1401464112
32. Czubak K, Lewandowska MA, Klonowska K, Roszkowski K, Kowalewski J, Figlerowicz M, et al. High copy number variation of cancer-related microRNA genes and frequent amplification of DICER1 and DROSHA in lung cancer. Oncotarget (2015) 6:23399–416. doi: 10.18632/oncotarget.4351
33. Oak N, Ghosh R, Huang KL, Wheeler DA, Ding L, Plon SE, et al. Framework for microRNA variant annotation and prioritization using human population and disease datasets. Hum Mutat (2019) 40:73–89. doi: 10.1002/humu.23668
34. Galka-Marciniak P, Urbanek-Trzeciak MO, Nawrocka PM, Dutkiewicz A, Giefing M, Lewandowska MA, et al. Somatic Mutations in miRNA Genes in Lung Cancer-Potential Functional Consequences of Non-Coding Sequence Variants. Cancers (2019) 11:793. doi: 10.3390/cancers11060793
35. Sohel MH. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achievements Life Sci (2016) 10(2):175–86. doi: 10.1016/j.als.2016.11.007
36. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol (2009) 11:228–34. doi: 10.1038/ncb0309-228
37. D’Ippolito E, Plantamura I, Bongiovanni L, Casalini P, Baroni S, Piovan C, et al. miR-9 and miR-200 regulate PDGFRβ-mediated endothelial differentiation of tumor cells in triple-negative breast cancer. Cancer Res (2016) 76:5562–72. doi: 10.1158/0008-5472.CAN-16-0140
38. Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol (2010) 12:247–56. doi: 10.1038/ncb2024
39. Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metas¬tasis and patient poor prognosis. RNA; (2008) 14:2348–60. doi: 10.1261/rna.1034808
40. Han X, Yan S, Weijie Z, Feng W, Liuxing W, Mengquan L, et al. Critical role of miR-10b in transforming growth factor β1-induced epithelial–mesenchymal transition in breast cancer. Cancer Gene Ther (2014) 21:60–7. doi: 10.1038/cgt.2013.82
41. Fong MY, Zhou W, Liu L, Chin AR, Somlo G, Palomares M, et al. Breast-cancer-secreted miR-122 reprograms glucose me¬tabolism in premetastatic niche to promote metastasis. Nat Cell Biol (2015) 17:183–94. doi: 10.1038/ncb3094
42. Hua K, Jin J, Zhao J, Song J, Song H, Li D, et al. MiR-135b, upregulated in breast cancer, promotes cell growth and disrupts the cell cycle by regulating LATS2. Int J Oncol (2016) 48:1997– 2006. doi: 10.3892/ijo.2016.3405
43. Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med (2011) 3:279–90. doi: 10.1002/emmm.201100136
44. Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene; (2014) 33:679–89. doi: 10.1038/onc.2012.636
45. Neel JC, Lebrun JJ. Activin and TGFβ regulate expression of the microRNA-181 family to promote cell migration and invasion in breast cancer cells. Cell Signal (2013) 25:1556–66. doi: 10.1016/j.cellsig.2013.03.013
46. Liu H, Wang Y, Li X, Zhang YJ, Li J, Zheng YQ, et al. Expression and regulatory function of miRNA-182 in triple-negative breast cancer cells through its targeting of profilin 1. Tumour Biol (2013) 34:1713–22. doi: 10.1007/s13277-013-0708-0
47. Falkenberg N, Anastasov N, Schaub A, Radulovic V, Schmitt M, Magdolen V, et al. Secreted uPAR isoform 2 (uPAR7b) is a novel direct target of miR-221. Oncotarget (2015) 6:8103– 8114. doi: 10.18632/oncotarget.3516
48. Nassirpour R, Mehta PP, Baxi SM, Yin J. miR-221 promotes tumorigenesis in human triple negative breast cancer cells. PLoSOne (2013) 8:e62170. doi: 10.1371/journal.pone.0062170
49. Jin C, Yan B, Lu Q, Lin Y, Ma L. Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumour Biol (2016) 37:7383–94. doi: 10.1007/s13277-015-4605-6
50. Usmani A, Shoro AA, Memon Z, Hussain M, Rehman R. Diagnostic, prognostic and predictive value of MicroR- NA-21 in breast cancer patients, their daughters and healthy individuals in breast cancer patients, their daughters and healthy individuals. Am J Cancer Res (2015) 5:2484–90.
51. Sossey-Alaoui K, Safina A, Li X, Vaughan MM, Hicks DG, Bakin AV, et al. Down-Regulation of WAVE3, a Metastasis Promoter Gene, Inhibits Invasion and Metastasis of Breast Cancer Cells. Am J Pathol (2007) 170:2112–21. doi: 10.2353/ajpath.2007.060975
52. Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle ; (2009) 8:712–5. doi: 10.4161/cc.8.5.7753
53. Liu X, Tang H, Chen J, Song C, Yang L, Liu P, et al. MicroR-NA-101 inhibits cell progression and increases paclitaxel sen- sitivity by suppressing MCL-1 expression in human triple- negative breast cancer. Oncotarget (2015) 6:20070–83. doi: 10.18632/oncotarget.4039
54. Hong L, Pan F, Jiang H, Zhang L, Liu Y, Cai C, et al. MiR-125b inhibited epithelial-mesenchymal transition of triple- negative breast cancer by targeting MAP2K7. Onco Tar- gets Ther (2016) 9:2639–48. doi: 10.2147/OTT.S102713
55. Yan M, Li X, Tong D, Han C, Zhao R, He Y, et al. MiR-136 sup-presses tumor invasion and metastasis by targeting RASAL2 in triple-negative breast cancer. Oncol Rep (2016) 36:65–71. doi: 10.3892/or.2016.4767
56. Zheng M, Sun X, Li Y, Zuo W. MicroRNA-145 inhibits growth and migration of breast cancer cells through targeting oncoprotein ROCK1. Tumour Biol (2016) 37:8189–96. doi: 10.1007/s13277-015-4722-2
57. Zheng M, Wu Z, Wu A, Huang Z, He N. MiR-145 promotes TNF-α-induced apoptosis by facilitating the formation of RIP1-FADDcaspase-8 complex in triple-negative breast cancer. Tumour Biol (2016) 37:8599–607. doi: 10.1007/s13277-015-4631-4
58. Fkih M, Privat I, Trimeche M, Penault-Llorca F, Bignon YJ, Kenani A. miR-10b, miR-26a, miR-146a And miR-153 expression in triple negative vs non triple negative breast cancer: potential biomarkers. Pathol Oncol Res (2017) 23:815–27. doi: 10.1007/s12253-017-0188-4
59. Furuya K, Sasaki A, Tsunoda Y, Tsuji M, Udaka Y, Oyamada H, et al. Eribulin upregulates miR-195 expression and downregulates Wnt3a expression in non-basal-like type of triple-negative breast cancer cell MDA-MB-231. Hum Cell (2016) 29:76–82. doi: 10.1007/s13577-015-0126-2
60. Chen J, Shin VY, Siu MT, Ho JC, Cheuk I, Kwong A. miR-199a-5p confers tumorsuppressive role in triple-negative breast cancer. BMC Cancer (2016) 16:887. doi: 10.1186/s12885-016-2916-7
61. Ramaiah MJ, Naushad MS, Lavanya PON, Gayatri A, Vainshnave S, Pal-Bhadra M. Epigenetic regulation of miR-200 as the potential strategy for the therapy against triple-negative breast cancer. Gene (2018) 641:248–58. doi: 10.1016/J.GENE.2017.10.018
62. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol (2008) 10:593–601. doi: 10.1038/ncb1722
63. Korpal MA, Khew-Goodall Y, Goodal GJ, Krpal M, Lee ES, Hu G, et al. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem (2008) 283:14910–4. doi: 10.1074/jbc.C800074200
64. Tsouko E, Wang J, Frigo DE, Aydogdu E, Williams C. MiR-200a inhibits migration of triple-negative breast can¬cer cells through direct repression of the EPHA2 oncogene. Carcinogenesis; (2015) 36:1051–60. doi: 10.1093/carcin/bgv087
65. Rhodes LV, Martin EC, Segar HC, Miller DF, Buechlein A, Rusch DB, et al. Dual regulation by microRNA-200b-3p and microRNA-200b-5p in the inhibition of epithelial-to-mesenchymal transition in triple-negative breast cancer. Oncotarget (2015) 6:16638–52. doi: 10.18632/oncotarget.3184
66. Erturk E, Cecener G, Tezcan G, Egeli U, Tunca B, Gokgoz S, et al. BRCA mutations cause reduction in miR-200c expres- sion in triple negative breast cancer. Gene (2015) 556:163–9. doi: 10.1016/j.gene.2014.11.047
67. Wang C, Zheng X, Shen C, Shi Y. MicroRNA-203 suppresses cell proliferation and migration by targeting BIRC5 and LASP1 in human triple-negative breast cancer cells. J Exp Clin Cancer Res (2012) 31:58. doi: 10.1186/1756-9966-31-58
68. DeCastro AJ, Dunphy KA, Hutchinson J, Balboni AL, Cherukuri P, Jerry DJ, et al. MiR203 mediates subversion of stem cell properties during mammary epithelial differentiation via repression of ΔNP63α and promotes mesenchymal-to-epithelial transition. Cell Death Dis (2013) 4:e514. doi: 10.1038/cddis.2013.37
69. Piovan C, Palmieri D, Di Leva G, Braccioli L, Casalini P, Nuovo G, et al. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol (2012) 6:458–72. doi: 10.1016/j.molonc.2012.03.003
70. Chao CH, Chang CC, Wu MJ, Ko HW, Wang D, Hung MC, et al. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J Clin Invest (2014) 124:3093–106. doi: 10.1172/JCI73351
71. Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath C, Schwartz G, et al. Altered Micro-RNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res (2017) 67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019
72. Wang J, Tsouko E, Jonsson P, Bergh J, Hartman J, Aydogdu E, et al. MiR-206 inhibits cell migration through direct target- ing of the actin-binding protein coronin 1C in triple-negative breast cancer. Mol Oncol (2014) 8:1690–702. doi: 10.1016/j.molonc.2014.07.006
73. Zavala V, Perez-Moreno E, Tapia T, Camus M, Carvallo P. MiR-146a and miR-638 in BRCA1-deficient triple negative breast cancer tumors, as potential biomarkers for improved overall survival. Cancer Biomark (2016) 16:99–107. doi: 10.3233/CBM-150545
74. Lv ZD, Kong B, Liu XP, Jin LY, Dong Q, Li FN, et al. MiR-655 suppresses epithelial-to-mesenchymal transition by targeting Prrx1 in triple-negative breast cancer. J Cell Mol Med (2016) 20:864–73. doi: 10.1111/jcmm.12770
75. Phan B, Majid S, Ursu S, deSemir D, Nosrati M, Bezrookove V, et al. Tumor suppressor role of microRNA-1296 in triple-negative breast cancer. Oncotarget (2016) 7(15):19519–30. doi: 10.18632/oncotarget.6961
76. Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. En Relat Cancer (2010) 17:F19–36. doi: 10.1677/ERC-09-0184
77. Dong G, Liang X, Wang D, Gao H, Wang L, Liu J. High expression of miR-21 in triple negative breast cancers was cor- related with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol (2014) 31:57. doi: 10.1007/s12032-014-0057-x
78. Chi Y, Wang D, Wang J, Yu W, Yang J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells (2019) 8:1015. doi: 10.3390/cells8091015
79. Novikova IV, Hennelly SP, Sanbonmatsu KY. Tackling Structures of Long Noncoding RNAs. Int J Mol Sci (2013) 14:23672–84. doi: 10.3390/ijms141223672
80. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs, Mol. Cell (2011) 43:904–14. doi: 10.1016/j.molcel.2011.08.018
81. Rutenberg-Schoenberg M, Sexton AN, Simon MD. The properties of long noncoding RNAs that regulate chromatin, Annu. Rev Genom Hum Genet (2016) 17:69–94. doi: 10.1146/annurev-genom-090314-024939
82. Akhade VS, Pal D, Kanduri C. Long Noncoding RNA: Genome Organization and Mechanism of Action. Adv Exp Med Biol (2017) 1008:47–74. doi: 10.1007/978-981-10-5203-3_2
83. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature (2010) 464:1071–6. doi: 10.1038/nature08975
84. Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep (2014) 32:395–402. doi: 10.3892/or.2014.3186
85. Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW, Jin HY, et al. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol (2014) 134:121–8. doi: 10.1016/j.ygyno.2014.03.556
86. Sun Z, Wu XY, Wu CL. The association between LncRNA HOTAIR and cancer lymph node metastasis and distant metastasis: a meta-analysis. Neoplasma ; (2018) 65:178–84. doi: 10.4149/neo_2018_170114N34
87. Cai B, Wu Z, Liao K, Zhang S. Long noncoding RNA HOTAIR can serve as a common molecular marker for lymph node metastasis: a meta-analysis. Tumour Biol (2014) 35:8445–50. doi: 10.1007/s13277-014-2311-4
88. Lu R, Zhang J, Zhang W, Huang Y, Wang N, Zhang Q, et al. Circulating HOTAIR expression predicts the clinical response to neoadjuvant chemotherapy in patients with breast cancer. Cancer Biomark (2018) 22(2):249–56. doi: 10.3233/CBM-170874
89. Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta (2010) 1799:597–615. doi: 10.1016/j.bbagrm.2010.10.001
90. Shi T, Gao G, Cao Y. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis Markers (2016) 2016:9085195. doi: 10.1155/2016/9085195
91. Jiao ZY, Tian Q, Li N, Wang HB, Li KZ. Plasma long non-coding RNAs (lncRNAs) serve as potential biomarkers for predicting breast cancer. Eur Rev Med Pharmacol Sci (2018) 22:1994–9. doi: 10.26355/eurrev_201804_14727
92. Avazpour N, Hajjari M, Tahmasebi Birgani M. HOTAIR: A Promising Long Non-coding RNA with Potential Role in Breast Invasive Carcinoma. Front Genet (2017) 8:170:170. doi: 10.3389/fgene.2017.00170
93. Collina F, Aquino G, Brogna M, Cipolletta S, Buonfanti G, DeLaurentiis M, et al. LncRNA HOTAIR up-regulation is strongly related with lymph nodes metastasis and LAR subtype of Triple Negative Breast Cancer. J Cancer (2019) 10(9):2018–24. doi: 10.7150/jca.29670
94. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res (2013) 73:(3) 1180–1189. doi: 10.1158/0008-5472.CAN-12-2850
95. Dong Y, Liang G, Yuan B, Yuan C, Yang C, Gao R, et al. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol (2015) 36:(3) 1477–1486. doi: 10.1007/s13277-014-2631-4
96. Li L, Xu Y, Liu S, Fu S, Tu J, Hu Y, et al. LncRNA MALAT1 promotes relapse of breast cancer patients with postoperative fever. Am J Transl Res (2018) 10:(10) 3186–3197.
97. Huang XJ, Xia Y, He GF, Zheng LL, Cai YP, Yin Y, et al. MALAT1 promotes angiogenesis of breast cancer. Oncol Rep (2018) 40:(5) 2683–2689. doi: 10.3892/or.2018.6705
98. Zuo Y, Li Y, Zhou Z, Ma M, Fu K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed Pharmacother (2017) 95:922–8. doi: 10.1016/j.biopha.2017.09.005
99. Shi F, Xiao F, Ding P, Qin H, Huang R. Long noncoding RNA highly up-regulated in liver cancer predicts unfavorable outcome and regulates metastasis by MMPs in triple-negative breast cancer, Arch. Med Res (2016) 47:(6) 446–453. doi: 10.1016/j.arcmed.2016.11.001
100. Ding Y, Sun C, Li J, Liu J, Pu L, Xiong S. The Significance of Long Non-coding RNA HULC in Predicting Prognosis and Metastasis of Cancers: a Meta-Analysis Pathol. Oncol Res (2019) 25:311. doi: 10.1007/s12253-017-0351-y
101. Wang K, Li X, Song C, Li M. LncRNA AWPPH promotes the growth of triple-negative breast cancer byup-regulating frizzled homolog 7 (FZD7). Biosci Rep (2018) 38(6):BSR20181223. doi: 10.1042/BSR20181223
102. Liu A, Qu H, Gong W, Xiang JY, Yang MM, Zhang W. LncRNA AWPPH and miRNA-21 regulates cancer cell proliferation and chemosensitivity in triple-negative breast cancer by interacting with each other. J Cell Biochem (2019) 120(9):14860–6. doi: 10.1002/jcb.28747
103. Yang F, Shen Y, Zhang W, Jin J, Huang D, Fang H, et al. An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell Death Differ (2018) 25(12):2209–20. doi: 10.1038/s41418-018-0123-6
104. Wang O, Yang F, Liu Y, Lv L, Ma R, Chen C, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am J Trans Res (2017) 9(2):533–45. doi: 10.1042/BSR20181223
105. Han J, Han B, Wu X, Hao J, Dong X, Shen Q, et al. Knockdown of lncRNA H19 restores chemo-sensitivity inpaclitaxel-resistant triple-negative breast cancer through triggering apoptosis and regulating Aktsignaling pathway. Toxicol Appl Pharmacol (2018) 359:55–61. doi: 10.1016/j.taap.2018.09.018
106. Yang J, Meng X, Yu Y, Pan L, Zheng Q, Lin W. LncRNA POU3F3 promotes proliferation and inhibits apoptosis of cancer cells in triple-negative breast cancer by inactivating caspase 9. Biosci Biotechnol Biochem (2019) 83:6, 1117–23. doi: 10.1080/09168451.2019.1588097
107. Wang L, Liu D, Wu X, Zeng Y, Li L, Hou Y, et al. Long non-coding RNA (LncRNA) RMST in triple-negative breast cancer (TNBC): Expression analysis and biological roles research. J Cell Physiol (2018) 233(10):6603–12. doi: 10.1002/jcp.26311
108. Xiang S, Zhaoyun L, Zhiyong Y. LncRNA NEF is downregulated in triple negative breast cancer and correlated with poor prognosis. Acta Biochim Biophys Sin (2019) 51:386–92. doi: 10.1093/abbs/gmz021
109. Liu L, Yu D, Shi H, Li J, Meng L. Reduced lncRNA Aim enhances the malignant invasion of triple-negative breast cancer cells mainly by activating Wnt/β-catenin/mTOR/PI3K signaling. Die Pharmazie (2017) 72(10):599–603(5). doi: 10.1691/ph.2017.7547
110. Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol (2010) 17:1169–74. doi: 10.1038/nsmb.1921
111. Xiao Y, Xu C, Guan J, Ping Y, Fan H, Li Y, et al. Discovering dysfunction of multiple microRNAs cooperation in disease by a conserved microRNA co-expression network. PLoSOne (2012) 7(2):e32201. doi: 10.1371/journal.pone.0032201
112. Schmitz U, Lai X, Winter F, Wolkenhauer O, Vera J, Gupta SK. Cooperative gene regulation by microRNA pairs and their identification using a computational workflow. Nucleic Acids Res (2014) 42(12):7539–52. doi: 10.1093/nar/gku465
113. Lai X, Gupta SK, Schmitz U, Marquardt S, Knoll S, Spitschak A, et al. MiR-205-5p and miR-342- 3p cooperate in the repression of the E2F1 transcription factor in the context of anticancer chemotherapy resistance. Theranostics (2018) 8(4):1106–20. doi: 10.7150/thno.19904
114. Nazarov PV, Reinsbach SE, Muller A, Nicot N, Philippidou D, Vallar L, et al. Interplay of microRNAs, transcription factors and target genes: linking dynamic expression changes to function. Nucleic Acids Res (2013) 41(5):2817–31. doi: 10.1093/nar/gks1471
115. Sengupta D, Bandyopadhyay S. Topological patterns in microRNA-gene regulatory network: studies in colorectal and breast cancer. Mol Biosyst (2013) 9(6):1360–71. doi: 10.1039/c3mb25518b
116. Cascione L, Gasparini P, Lovat F, Carasi S, Pulvirenti A, Ferro A, et al. Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PloS One (2013) 8(2):e55910. doi: 10.1371/journal.pone.0055910
117. Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet (2016) 17(12):719–32. doi: 10.1038/nrg.2016.134
118. Logotheti S, Marquardt S, Putzer B. “p73-Governed miRNA Networks: Translating BioinformaticsApproaches to Therapeutic Solutions for Cancer Metastasis”. In: Computational Biology of Non-Coding RNA-Methods and Protocols. New York: Springer Nature, Humana Press (2019). p. 33–55. doi: 10.1007/978-1-4939-8982-9_2
119. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res (2009) 19:92–105. doi: 10.1101/gr.082701.108
120. D’Errico I, Gadaleta G, Saccone C. Pseudogenes in metazoa: origin and features. Brief Funct Genomic Proteomic (2004) 3:157–67. doi: 10.1093/bfgp/3.2.157
121. Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Carter DR. Pseudogenes: Pseudofunctional or key regulators in health and disease? RNA (2011) 17(5):792–8. doi: 10.1261/rna.2658311
122. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature (2010) 465:1033–8. doi: 10.1038/nature09144
123. Chan WL, Chang JG. Pseudogene-derived endogenous siRNAs and their function. Methods Mol Biol (2014) 1167:227–39. doi: 10.1007/978-1-4939-0835-6_15
124. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell (2011) 146(3):353–8. doi: 10.1016/j.cell.2011.07.014
125. Ala U, Karreth FA, Bosia C, Pagnani A, Taulli R, Leopold V, et al. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci USA (2013) 110:7154–9. doi: 10.1073/pnas.1222509110
126. Cacchiarelli D, Santoni D, Bozzoni I. MicroRNAs as prime players in a combinatorial view of evolution. RNA Biol (2008) 5(3):120–2. doi: 10.4161/rna.5.3.6569
127. Shen X, Xie B, Ma Z, Yu W, Wang W, Xu D. Identification of novel long non-coding RNAs in triple negative breast cancer. Oncotarget (2015) 6:21730–9. doi: 10.18632/oncotarget.4419
128. Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, et al. Endogenous miRNA sponge lincRNA-RoR regulates Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell (2013) 25:69–80. doi: 10.1016/j.devcel.2013.03.002
129. Hashimoto S, Onodera Y, Hashimoto A, Tanaka M, Hamaguchi M, Yamada A, et al. Requirement for Arf6 in breast cancer invasive activities. Proc Natl Acad Sci U S A (2004) 101:6647–52. doi: 10.1073/pnas.0401753101
130. Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res (2015) 13:330–8. doi: 10.1158/1541-7786.MCR-14-0251
131. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA (2014) 20:1829–42. doi: 10.1261/rna.047126.114
132. Quackenbush J, Glass K. Transcriptomics and Network Medicine, pp.251. In: Loscalzo J, Barabasi A-L, Silveman EK, editors. Network Medicine, Complex Systems in Human Disease and Therapeutics. London: Harvard University Press (2017). p. 251.
133. Le K, Guo H, Zhang Q, Huang X, Xu M, Huang Z, et al. Gene and lncRNA co-expression network analysis reveals novel ceRNA network for triple-negative breast cancer. Sci Rep (2019) 9(1):15122. doi: 10.1038/s41598-019-51626-7
134. Yuan N, Zhang G, Bie F, Ma M, Ma Y, Jiang X, et al. Integrative analysis of lncRNAs and miRNAs with coding RNAs associated with ceRNAcrosstalk network in triple negative breast cancer. Onco Targets Ther (2017) 10:5883–97. doi: 10.2147/OTT.S149308
135. Song X, Zhang C, Liu Z, Liu Q, He K, Yu Z. Characterization of ceRNA network to reveal potential prognostic biomarkers in triple-negative breast cancer. Peer J (2019) 7:e7522. doi: 10.7717/peerj.7522 eCollection 2019.
136. Yang R, Xing L, Wang M, Chi H, Zhang L, Chen J, et al. Comprehensive Analysis of Differentially Expressed Profiles of lncRNAs/mRNAs and miRNAs with Associated ceRNA Networks in Triple-Negative Breast Cancer. Cell Physiol Biochem (2018) 50(2):473–88. doi: 10.1159/000494162
137. Liu Z, Mi M, Li X, Zheng X, Wu G, Zhang L. lncRNA OSTN-AS1 May Represent a Novel Immune-Related Prognostic Marker for Triple-Negative Breast Cancer Based on Integrated Analysis of a ceRNA Network. Front Genet (2019) 10:850:850. doi: 10.3389/fgene.2019.00850
138. Morrow DA, De Lemos JA. Benchmarks for the assessment of novel cardiovascular biomarkers. Circulation (2007) 115:949–52. doi: 10.1161/CIRCULATIONAHA.106.683110
139. Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W, et al. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PloS One (2014) 9(5):e96228. doi: 10.1371/journal.pone.0096228
140. Kolacinska A, Morawiec J, Pawlowska Z, Szemraj J, Szymanska B, Malachowska B, et al. Association of microRNA-93, 190, 200b and receptor status in core biopsies from stage III breast cancer patients. DNA Cell Biol (2014) 33(9):624–9. doi: 10.1089/dna.2014.2419
141. Sempere LF, Preis M, Yezefski T, Ouyang H, Suriawinata AA, Silahtaroglu A, et al. Fluorescence-based code-detection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clin Cancer Res (2010) 16(16):4246–55. doi: 10.1158/1078-0432.CCR-10-1152
142. Song X, Zhang C, Liu Z, Liu Q, He K, Yu Z. miR-629-3p may serve as a novel biomarker and potential therapeutic target for lung metastases of triple-negative breast cancer. Breast Cancer Res BCR (2017) 19:72. doi: 10.1186/s13058-017-0865-y
143. Debeb BG, Lacerda L, Anfossi S, Diagaradjane P, Chu K, Bambhrolia A, et al. miR-141-Mediated Regulation of Brain Metastasis From Breast Cancer. JNCI J Natl Cancer Inst (2016) 108(8):diw026. doi: 10.1093/jnci/djw026
144. Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA (2006) 103:9136–41. doi: 10.1073/pnas.0508889103
145. Turashvili G, Lightbody ED, Tyryshkin K, SenGupta SK, Elliott BE, Madarnas Y, et al. Novel prognostic and predictive microRNA targets for triple-negativebreast cancer. FASEB J (2018) 32(11):5937–54. doi: 10.1096/fj.201800120R
146. Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, et al. microRNA expression pro- filing identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget (2014) 5(5):1174–84. doi: 10.18632/oncotarget.1682
147. Matamala N, Vargas MT, Gonzalez-Campora R, Minambres R, Arias JI, et al. Tumor MicroRNA Expression Profiling Identifies Circulating MicroRNAs for Early Breast Cancer Detection. Clin Chem (2015) 61:1098–106. doi: 10.1373/clinchem.2015.238691
148. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The MicroRNA Spectrum in 12 Body Fluids. Clin Chem (2010) 56:1733–41. doi: 10.1373/clinchem.2010.147405
149. Takahashi K, Yokota S-I, Tatsumi N, Fukami T, Yokoi T, Nakajima M. Cigarette smoking substantially alters plasma microRNA profiles in healthy subjects. Toxicol Appl Pharmacol (2013) 272:154–60. doi: 10.1016/j.taap.2013.05.018
150. Baggish AL, Hale AM, Weiner RB, Lewis GD, Systrom D, Wang F, et al. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J Physiol (2011) 589:3983–94. doi: 10.1113/jphysiol.2011.213363
151. Witwer KW. XenomiRs and miRNA homeostasis in health and disease: Evidence that diet and dietary miRNAs directly and indirectly influence circulating miRNA profiles. RNABoil (2012) 9:1147–54. doi: 10.4161/rna.21619
152. Lima-Oliveira G, Guidi GC, Salvagno GL, Brocco G, Danese E, Lippi G. Estimation of the imprecision on clinical chemistry testing due tofist clenching and maintenance during venipuncture. Clin Biochem (2016) 49(18):1364–7. doi: 10.1016/j.clinbiochem.2016.07.007
153. Cheng HH, Yi HS, Kim Y, Kroh EM, Chien JW, Eaton KD, et al. Plasma Processing Conditions Substantially Influence Circulating microRNA Biomarker Levels. PloS One (2013) 8(6):e64795. doi: 10.1371/journal.pone.0064795
154. Bustin SA, Nolan T. Pitfalls of Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction. J Biomol Tech (2004) 15(3):155–66.
155. Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Valley MP, van Zandwijk N, et al. Haemolysis during Sample Preparation Alters microRNA Content of Plasma. PloS One (2011) 6(9):e24145. doi: 10.1371/journal.pone.0024145
156. Willeit P, Zampetaki A, Dudek K, Kaudewitz D, King A, Kirkby NS, et al. Circulating MicroRNAs as Novel Biomarkers for Platelet Activation. Circ Res (2013) 112:595–600. doi: 10.1161/CIRCRESAHA.111.300539
157. Boeckel J-N, Thome CE, Leistner D, Zeiher AM, Fichtlscherer S, Dimmeler S. Heparin Selectively A_ects the Quantification of MicroRNAs in Human Blood Samples. Clin Chem (2013) 59:1125–7. doi: 10.1373/clinchem.2012.199505
158. Faraldi M, Gomarasca M, Banfi G, Lombardi G. Free Circulating miRNAs Measurement in Clinical Settings: The Still Unsolved Issue of the Normalization. Adv Virus Res (2018) 87:113–39. doi: 10.1016/bs.acc.2018.07.003
159. Chen W, Cai F, Zhang B, Barekati Z, Zhong XY. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: potential biomarkers. Tumor Biol (2013) 34:455–62. doi: 10.1007/s13277-012-0570-5
160. Niedźwiecki S, Piekarski J, Szymańska B, Pawlowska Z, Jeziorski A. Serum levels of circulating miRNA-21, miRNA-10b and miRNA-200c in triple-negative breast cancer patients. Ginekologia Polska (2018) 89:415–20. doi: 10.5603/GP.a2018.0071
161. Bottani M, Banfi G, Lombardi G. Circulating miRNAs as Diagnostic and Prognostic Biomarkers in Common Solid Tumors: Focus on Lung, Breast, Prostate Cancers, and Osteosarcoma. J Clin Med (2019) 8:1661. doi: 10.3390/jcm8101661
162. Shin VY, Siu JM, Cheuk I, Ng EK, Kwong A. Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. Br J Cancer (2015) 112(11):1751–9. doi: 10.1038/bjc.2015.143
163. Li HY, Liang JL, Kuo YL, Lee HH, Calkins MJ, Chang HT, et al. miR-105/93-3p promotes chemoresistance and circulating miR-105/93-3p acts as a diagnostic biomarker for triple negative breast cancer. Breast Cancer Res: BCR (2017) 19(1):133. doi: 10.1186/s13058-017-0918-2
164. Jiang L, He D, Yang D, Chen Z, Pan Q, Mao A, et al. MiR-489 Regulates Chemoresistance in Breast Cancer via EpithelialMesenchymal Transition Pathway. FEBS Lett (2014) 588(11). doi: 10.1016/j.febslet.2014.04.024
165. Ding L, Gu H, Xiong X, Ao H, Cao J, Lin W, et al. MicroRNAs involved in carcinogenesis, prognosis, therapeuticresistance and applications in human triple-negative breast cancer. Cells (2019) 8(12):1492. doi: 10.3390/cells8121492
166. Zubor P, Kubatka P, Kajo K, Dankova Z, Polacek H, Bielik T, et al. Why the Gold Standard Approach by Mammography Demands Extension by Multiomics? Application of Liquid Biopsy miRNA Profiles to Breast Cancer Disease Management. Int J Mol Sci (2019) 20:2878. doi: 10.3390/ijms20122878
167. Eichelser C, Flesch-Janys D, Chang-Claude J, Pantel K, Schwarzenbach H. Deregulated Serum Concentrations of Circulating Cell–Free MicroRNAs miR-17, miR-34a, miR-155, and miR-373 in Human Breast Cancer Development and Progression. Clin Chem (2013) 59(10):1489–96. doi: 10.1373/clinchem.2013.205161
168. Ritter A, Hirschfeld M, Berner K, Rucker G, Jager M, Weiss D, et al. Circulating non coding RNA biomarker potential in neoadjuvant chemotherapy of triple negative breast cancer? Int J Oncol (2019) 56(1):47–68. doi: 10.3892/ijo.2019.4920 Advance online publication.
169. Mutlu M, Raza U, Saatci Ö, Eyupoglu E, Yurdusev E, Sahin O. miR-200c: a versatile watchdog in cancer progression, EMT, and drug resistance. J Mol Med (2016) 94:629–44. doi: 10.1007/s00109-016-1420-5
170. Zeng Z, Chen X, Zhu D, Luo Z, Yang M. Low Expression of Circulating MicroRNA-34c is Associated with Poor Prognosis in Triple-Negative Breast Cancer. Yonsei Med J (2017) 58(4):697–702. doi: 10.3349/ymj.2017.58.4.697
171. Mishra S, Srivastava AK, Suman S, Kumar V, Shukla Y. Circulating miRNAs revealed as surrogate molecular signatures for the early detection of breast cancer. Cancer Lett (2015) 369:67–75. doi: 10.1016/j.canlet.2015.07.045
172. Qattan A, Intabli H, Alkhayal W, Eltabache C, Tweigieri T, Amer SB. Robust expression of tumor suppressor miRNA’s let-7 and miR-195 detected in plasma of Saudi female breast cancer patients. BMC Cancer (2017) 17:799. doi: 10.1186/s12885-017-3776-5
173. Kodahl AR, Lyng MB, Binder H, Cold S, Gravgaard K, Knoop AS, et al. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: a case control study. Mol Oncol (2014) 8(5):874–83. doi: 10.1016/j.molonc.2014.03.002
174. Shimomura A, Shiino S, Kawauchi J, Takizawa S, Sakamoto H, Matsuzaki J, et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci (2016) 107:326–34. doi: 10.1111/cas.12880
175. Cuk K, Zucknick M, Madhavan D, Schott S, Golatta M, Heil J, et al. Plasma MicroRNA panel for minimally invasive detection of breastcancer. PloS One (2013) 8(10):e76729. doi: 10.1371/journal.pone.0076729
176. Song N, Liang B, Wang D. The function of MiR-21 expression differences and pathogenesis on familial and triple negative breast Cancer serum. Pak J Pharm Sci (2016) 29:679–84.
177. Zhu W, Liu M, Fan Y, Ma F, Xu N, Xu B. Dynamics of circulating microRNAs as a novel indicator of clinical response to neoadjuvant chemotherapy in breast cancer. Cancer Med (2018) 7:4420–33. doi: 10.1002/cam4.1723
178. Lü L, Mao X, Shi P, He B, Xu K, Zhang P. MicroRNAs in the prognosis of triple-negative breast cancer: A systematic review and meta-analysis. Med (Baltimore) (2017) 96(22):e7085. doi: 10.1097/MD.0000000000007085
179. Fan CN, Ma L, Liu N. Comprehensive analysis of novel three-long noncoding RNA signatures as a diagnostic and prognostic biomarkers of human triple-negative breast cancer. J Cell Biochem (2019) 120(3):3185–96. doi: 10.1002/jcb.27584
180. Wang X, Chen T, Zhang Y, Zhang N, Li C, Li Y, et al. Long noncoding RNA Linc00339 promotes triple-negative breast cancer progression through miR-37-3p/HOXC6 signaling pathway. J Cell Physiol (2019) 234(8):13303–17. doi: 10.1002/jcp.28007
181. Wang Y, Zhang G, Han J. HIF1A-AS2 predicts poor prognosis and regulates cell migration and invasion in triple-negative breast cancer. J Cell Biochem (2019) 120(6):10513–8. doi: 10.1002/jcb.28337
182. Liu M, Xing LQ, Liu YJ. A three-long noncoding RNA signature as a diagnostic biomarker for differentiating between triple-negative and non-triple-negative breast cancers. Med (Baltimore) (2017) 96(9):e6222. doi: 10.1097/MD.0000000000006222
183. Li P, Zhou B, Lv Y, Qian Q, Mesa KG, Schick M. LncRNA HEIH regulates cell proliferation and apoptosis through miR-4458/SOCS1 axis in triple-negative breast cancer. Hum Cell (2019) 32(4):522–8. doi: 10.1007/s13577-019-00273-1
184. Bermejo JL, Huang G, Manoochehri M, Mesa KG, Schick M, Silos RG. Long intergenic noncoding RNA 299 methylation in peripheral blood isa biomarker for triple-negative breast cancer. Epigenomics (2019) 11(1):81–93. doi: 10.2217/epi-2018-0121
185. Galliher AJ, Schiemann WP. Beta3 integrin and Src facilitate transforming growth factor-beta mediated induction of epithelial-mesenchymal transition in mammary epithelial cells. Breast Cancer Res BCR (2006) 8(4):R42. doi: 10.1186/bcr1524
186. Watanabe S, Ueda Y, Akaboshi S, Hino Y, Sekita Y, Nakao M. HMGA2 maintains oncogenic RAS-induced epithelial-mesenchymal transition in human pancreatic cancer cells. Am J Pathol (2009) (2009) 174:854–68. doi: 10.2353/ajpath.2009.080523
187. Taki M, Verschueren K, Yokoyama K, Nagayama M, Kamata N, et al. Involvement of ETS-1 transcription factor in inducing matrix metalloproteinase-2 expression by epithelial-mesenchymal transition in human squamous carcinoma cells. Int J Oncol (2006) 28:487–96. doi: 10.3892/ijo.28.2.487
188. Mamuya FA, Duncan MK. aV integrins and TGF-beta-induced EMT: A circle of regulation. J Cell Mol Med (2012) 16(3):445–55. doi: 10.1111/j.1582-4934.2011.01419.x
189. Borok Z. Role for _3 integrin in EMT and pulmonary fibrosis. J Clin Investig (2009) 119:7–10. doi: 10.1172/JCI38084
190. Van der Velden JL, Guala AS, Leggett SE, Sluimer J, Badura EC, Janssen-Heininger YM. Induction of a mesenchymal expression program in lung epithelial cells by Wnt/_-catenin requires the presence of c-Jun N-terminal kinase 1. Am J Respir Cell Mol Biol (2012) 47:306–14. doi: 10.1165/rcmb.2011-0297OC
191. Bao B, Wang Z, Ali S, Banerjee S, Azmi AS, Miele L, et al. Notch-1 induces epithelial-mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells. Cancer Lett (2011) 307:26–36. doi: 10.1016/j.canlet.2011.03.012
192. Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res (2004) 14:1902–10. doi: 10.1101/gr.2722704
193. Lehmann U, Hasemeier B, Christgen M, Muller M, Romermann D, Langer F, et al. Epigenetic inactivation of microRNA gene hsa-miR-9-1 in human breast cancer. J Pathol (2008) 214:17–24. doi: 10.1101/gr.2722704
194. Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene LncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer (2012) 11:5. doi: 10.1186/1476-4598-11-5
195. Fabbri M, Calin GA. Epigenetics and miRNAs in human cancer. Adv Genet (2010) 70:87–99. doi: 10.1016/B978-0-12-380866-0.60004-6
196. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci (2003) 100:3983–8. doi: 10.1073/pnas.0530291100
197. Davar D, Tarhini A, Kirkwood JM. Adjuvant therapy for melanoma. Cancer J (2012) 18:192–202. doi: 10.1097/PPO.0b013e31824f118b
198. Liu YR, Jiang YZ, Xu XE, Yu KD, Jin X, Hu X, et al. Comprehensive transcriptome analysis identifies novel molecular subtypes and subtype-specific RNAs of triple-negative breast cancer. Breast Cancer Res (2016) 18(1):33. doi: 10.1186/s13058-016-0690-8
Keywords: miRNA, LncRNA, biomarkers, ceRNA, circulating miRNA, triple negative breast cancer
Citation: Volovat SR, Volovat C, Hordila I, Hordila D-A, Mirestean CC, Miron OT, Lungulescu C, Scripcariu DV, Stolniceanu CR, Konsoulova-Kirova AA, Grigorescu C, Stefanescu C, Volovat CC and Augustin I (2020) MiRNA and LncRNA as Potential Biomarkers in Triple-Negative Breast Cancer: A Review. Front. Oncol. 10:526850. doi: 10.3389/fonc.2020.526850
Received: 14 January 2020; Accepted: 13 October 2020;
Published: 20 November 2020.
Edited by:
Marilena Valeria Iorio, Istituto Nazionale dei Tumori (IRCCS), ItalyReviewed by:
Praveen Vikas, University of Iowa Hospitals and Clinics, United StatesCopyright © 2020 Volovat, Volovat, Hordila, Hordila, Mirestean, Miron, Lungulescu, Scripcariu, Stolniceanu, Konsoulova-Kirova, Grigorescu, Stefanescu, Volovat and Augustin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Constantin Volovat, dm9sb3ZhdGNvbnN0YW50aW5AZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.