
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Oncol. , 25 November 2020
Sec. Head and Neck Cancer
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.524928
 Huanhuan Wang1,2,3†
Huanhuan Wang1,2,3† Yuyu Zhang1,2,3†
Yuyu Zhang1,2,3† Wei Bai4
Wei Bai4 Bin Wang1,2,3
Bin Wang1,2,3 Jinlong Wei1,2,3
Jinlong Wei1,2,3 Rui Ji5
Rui Ji5 Ying Xin6
Ying Xin6 Lihua Dong1,2,3*
Lihua Dong1,2,3* Xin Jiang1,2,3*
Xin Jiang1,2,3*Human papillomavirus (HPV) is a risk factor for squamous cell carcinoma of the head and neck (HNSCC). This study aimed to investigate the feasibility of IHC- p16INK4a (p16) as an alternative modality for diagnosing HPV infection. We searched PubMed, EMBASE, Web of Science, and Cochrane library for studies that evaluated the diagnostic accuracy of IHC-p16 staining. A total of 30 studies involving 2,963 patients were included from 2007 to 2019. The combined sensitivity was 0.94 (95% CI: 0.92–0.95); specificity, 0.90 (95% CI: 0.89–0.91); positive likelihood ratio (LR), 6.80 (95% CI: 5.63–8.21); negative LR, 0.10 (95% CI: 0.07–0.16); diagnostic odds ratio, 85.98 (95% CI: 55.57–133.03); and area under the curve value, 0.9550. Subgroup analysis showed that the IHC-p16 test was more consistent with the in situ hybridization (ISH) test and has greater diagnostic value for oropharyngeal squamous cell carcinoma. The diagnostic efficacy of IHC-p16 varied among countries. In conclusion, IHC-p16 has high sensitivity and specificity for diagnosing HPV infection in HNSCC. The consistency of IHC-p16 findings with those of ISH indicate that their combination can be used to improve the specificity of diagnosis.
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignancy worldwide, with ~830,000 incident cases annually (1). Smoking and drinking are the most established risk factors for HNSCC, but ~20–80% of recent HNSCC cases have been reported to be associated with human papillomavirus (HPV) infection (2–4). The proportion of HPV-related tumors vary by country and tumor site (5, 6). In addition, HPV-associated HNSCC has better disease-free survival and overall survival (7–9) owing to its high radiosensitivity (10–12). Concurrently, the standard treatment modality for HNSCC yields more serious adverse reactions, such as dryness of the mouth, dysphagia, and hearing loss, in patients with HPV-related HNSCC patients (4, 7). Accordingly, de-intensified treatment has become the new standard approach for HPV-positive patients. In general, the treatment for patients with HPV-positive tumors is de-intensified to reduce the adverse reactions and improve the quality of life while ensuring good tumor control. This is achieved by reducing the radiation dose and using radiotherapy alone instead of concurrent chemoradiotherapy (13, 14). Correct diagnosis of HPV infection is the most important step in de-escalating treatment. Only when HPV infection is properly diagnosed can a more suitable population be selected for this new treatment approach. However, there are several diagnostic modalities for HPV infection, and they have varying sensitivity and specificity. Therefore, choosing the appropriate modality for accurate diagnosis of HPV infection will be a key challenge for de-escalating treatment.
Currently, HPVE6/E7mRNA detection is the primary basis for diagnosing HPV infection as it has the advantage of detecting HPV with transcriptional activity (9, 15). Common methods for detecting HPVE6/E7mRNA include polymerase chain reaction (PCR) and in situ hybridization (ISH). PCR-based detection is more sensitive, while ISH-based detection is more specific (16, 17). However, no specific modality has been recommended as the gold standard for diagnosing HPV infection (18, 19). Both PCR and ISH methods have limitations including stringent sampling requirements, long detection time, complicated detection process, and high cost. Therefore, there have been several efforts to develop a novel diagnostic method for HPV infection that is both simple and economical.
Currently, alternative diagnostic methods include PCR or ISH detection of HPV-DNA (20–22). PCR-DNA is a highly sensitive method that can use primers to detect a wide range of HPV types (23). However, its specificity in distinguishing free and integrated DNA is relatively low. This disadvantage is overcome by ISH that can distinguish between the complete and dissociated form of HPV-DNA according to a dot signal and a diffuse signal (24). Even so, the ISH-DNA test for HPV infection in an integrated state is not reliable, and it is impossible to tell whether HPV-DNA is integrated into the host's genome. Although used clinically, the HPV-DNA test can only reflect a transitory infection and cannot identify the HPV driving the carcinogenic process. Further, its accuracy and prognostic relevance are unclear (25, 26).
Increasing studies have used the immunohistochemical (IHC) p16INK4a(p16) staining as an alternative modality for diagnosing HPV infection (27, 28). In HPV infection of squamous epithelial cells, p16 is overexpressed after infection due to inactivation of the Rb protein (23, 29). In general, upregulated p16 expression is believed to be closely associated with HPV infection (30). As such, this test is widely used in oropharyngeal squamous cell carcinoma (OPSCC); however, the relationship between p16 positivity and HPV infection in non-oropharyngeal sites (e.g., paranasal sinuses, mouth, larynx, nasopharynx and hypopharynx) is extremely limited (31, 32). In addition, the diagnostic efficacy of IHC-p16 for HPV infection in all HNSCC patients has not been completely evaluated. Given the advantages of IHC-p16, including its simple operation, short testing time, and being economical, it is essential to better understand its usefulness in the diagnosis of HPV infection. This systematic review and meta-analysis aimed to investigate the feasibility of using IHC-p16 for diagnosing HPV infection in HNSCC and its value for de-escalating treatment. Further, we aimed to assess whether the results varied by tumor site and country.
The protocol for this systematic review was registered on INPLASY (202070068) and is available in full on the inplasy.com (https://doi.org/10.37766/inplasy2020.7.0068).
This study was conducted according to the PRISMA guidelines and the Cochrane diagnostic test manual (33, 34). The search strategy, study selection, methodological quality assessment, data extraction, and data analysis protocols were developed in advance. We searched PubMed, EMBASE, Web of Science, and Cochrane library for relevant articles published from the establishment of the database until October 2019, without language restrictions. The search was assisted by an experienced library staff member. We used a combination of MeSH words and free text words including “Papillomaviridae” and “Head and Neck Neoplasms.” The search strategy and the number of relevant articles identified in each database are shown in the Supplementary Documents. The references of the identified articles were also reviewed to further search for other relevant articles. All articles were searched according to international standards.
We reviewed the full text of all observational studies, both retrospective and prospective, and randomized controlled clinical trials that compared the diagnostic efficacy of IHC-p16 positivity with the gold standard modality for HPV diagnosis. The inclusion criteria were: (i) the included patients had HNSCC; (ii) the samples tested were biopsy or puncture specimens; (iii) HPV E6/E7mRNA detection was used as the gold standard for the diagnosis of HPV infection; (iv) p16 expression was detected using IHC; (v) the total sample size was >10. All case reports, preclinical studies, case series, animal studies, and conference summaries were excluded. In addition, papers were also excluded if the specific location of HNSCC was not clearly defined. Further, the included studies must present the specific true positive (TP), false positive (FP), false negative (FN), and true negatives (TN) values or have adequate data so these can be calculated. If data were lacking, we contacted the author by email to ask for the data, and the study was excluded if the author did not respond. Study selection was divided into two parts. First, the authors (JW and BW) screened all the articles independently by browsing the titles and abstracts. Second, the same two authors independently evaluated the full text of the initially included articles. Any disagreements were resolved by the third author (XJ), and the study was finalized for inclusion.
Two authors (JW and BW) independently assessed the methodological quality of the included studies using the QUADAS-2 tool (35). Briefly, the QUADAS-2 tool comprises four domains, namely, patient selection, index test, reference standard, and flow and timing. In addition, the first three sections are evaluated with respect to clinical applicability. Patient selection primarily evaluates whether the selection of patients have introduced bias, including whether the patient selection is random and whether there is inappropriate exclusion. The index test primarily evaluates whether the conduct or interpretation of the test has bias, including whether the process of the experiment is detailed. The reference standard evaluates biases caused by reference criteria and their interpretations. The flow and timing evaluates whether all patients are using the same criteria. Evaluation of the these four parameters helps to assess the risk of bias.
Data extraction was carried out in two parts. First, a researcher (HHW) used a pre-designed data extraction table to extract basic elements from the study, such as author, publication year, and patient source. Then, two other authors (YYZ and WB) independently extracted the specific values of TP, FP, FN, and TN from the text and cross-checked them according to the pre-set standards to ensure the accuracy of the original values extracted. Any differences in the data extraction were resolved through discussion and negotiation. The extracted data were verified by the third author (WB).
Because the head and neck are divided into many regions, and there are two methods for HPVE6/E7mRNA detection, we expected that the data included in the meta-analysis might be uneven. Therefore, we divided the study into several different subgroups based on factors such as tumor location and the detection methods for HPVE6/E7mRNA set as the gold standard. Given that the accuracy of p16 positivity in diagnosing HPV infection is related to the positive threshold, differences in thresholds between studies may have an impact on the sensitivity and specificity. Thus, we further evaluated whether there was a threshold effect using Spearman correlation coefficient. If there was no threshold effect, the sensitivity, specificity, and other indicators were further combined. Sensitivity was defined as the percentage of TP for diagnosing HPV infection in the total number of p16-positive cases (TP+FN). Specificity was defined as the percentage of TN for diagnosis of no-HPV infection in the total number of p16-negative cases (FP+TN). All data were combined using Meta Disc and STATA 15.0 software. We developed a forest map that graphically displays estimates of sensitivity and specificity and visualized heterogeneity between studies. Moreover, heterogeneity was examined using I2 and Cochrane Q-tests. An I2 of >50% indicated heterogeneity, and the source of heterogeneity was further explored. After obtaining the sensitivity and specificity values, we further used the receiver operating characteristic curve (ROC) model to obtain the positive likelihood ratio (LR), negative LR, and the diagnostic odds ratio (OR) and their 95% confidence intervals (CI). Positive LR was defined as the ratio of sensitivity to 1-specificity. Negative LR was defined as the ratio of 1-sensitivity to specificity. The larger the positive LR and the smaller the negative LR, the better the diagnostic experiment. The diagnostic OR was defined as the ratio of positive LR to negative LR. The greater the diagnostic OR, the better the capability of p16 to distinguish between HPV infection and non-HPV infection. The ROC curve was also drawn to obtain the area under the curve (AUC) value to comprehensively evaluate the efficacy of p16 positivity in diagnosing HPV infection. In addition, a funnel plot was used to further evaluate the presence of publication bias.
In total, 2,361 studies were initially identified (Figure 1). After excluding 550 duplicate studies, 1,810 studies were screened, and the full text of 59 studies were reviewed. Of the 59 studies, 29 studies were excluded because the sample size was too small (n = 7) and detailed data were not available (n = 22). Finally, 30 studies (15, 36–64) involving 2,963 patients were included in the meta-analysis.
The characteristics of the 30 studies are shown in Table 1. Some studies used two methods simultaneously. In the 19 (54.3%) studies, HPVE6/E7mRNA was detected using ISH as the gold standard for the diagnosis of HPV infection. Meanwhile, in the 16 (45.7%) studies, HPVE6/E7mRNA was detected via PCR as the gold standard for HPV infection. In total, 5 studies used both ISH and PCR as the gold standard for diagnosis. A total of 2,014 (68.0%) OPSCC cases were included from 25 studies. Other studies included cases with malignant tumors in other subregions of the head and neck, including 752 (25.4%) cases of oral squamous cell carcinoma, 148 (5.0%) cases of hypopharyngeal squamous cell carcinoma, and 49 (1.6%) cases of nasopharyngeal squamous cell carcinoma. With respect to the country of origin, 1,008 (34.0%) cases were from Europe, 1,592 (53.7%) cases from North America, 283 (9.6%) cases from Asia, and 80 (2.7%) cases from Oceania. All the included studies were published after 2007.
The results of quality assessment of the 30 studies are shown in Figure 2. In many studies, not all the factors that might influence the quality assessment were completely reported. With respect to patient selection, 2 studies were assessed to have uncertain risk of bias mainly because patient selection was unclear. For flow and timing, 14 studies were assessed to have uncertain risk of bias mainly because the time interval between tests was not specified. A total of 7 studies were assessed to have high risk of bias in different areas mainly because not all cases were included in the analysis.
The Spearman correlation coefficient was p = 0.081, indicating that there was no threshold effect in this meta-analysis. Therefore, we further combined the sensitivity, specificity, and other indicators of the study. The combined sensitivity of the 30 studies was 0.94 (95% CI: 0.92–0.95) and specificity was 0.90 (95% CI: 0.89–0.91) (Figure 3). We found that p16 positivity had high sensitivity for diagnosing HPV infection. Among the p16-positive patients, 94% had HPV infection; the misdiagnosis rate was only 6%. Among the p-16 negative patients, 90% were not infected with HPV, but there was a 10% of missed diagnosis. The positive LR was 6.80 (95% CI: 5.63–8.21); negative LR, 0.10 (95% CI: 0.07–0.16) (Figure 4); and diagnostic OR, 85.98 (95% CI: 55.57–133.03). These values indicate that p16 positivity was able to distinguish 85.98% of HPV infections from non-infections. The I2-value was 35.1%, indicating good consistency (Figure 5), and the AUC value was 0.9550 (Figure 6), showing that p16 positivity has great diagnostic value.
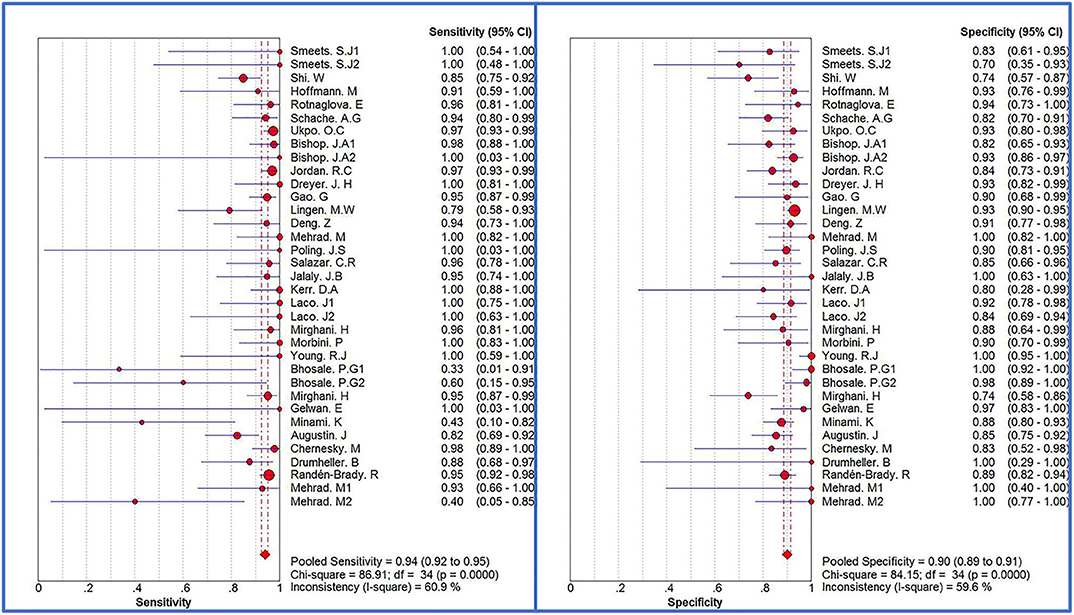
Figure 3. Forest plots of sensitivity and specificity of 30 original studies combined for diagnosis of HPV infection compared with p16 positive. Among them, 5 studies used different detection methods or included patients with different tumors, so they were divided into two records, named 1.2, respectively.
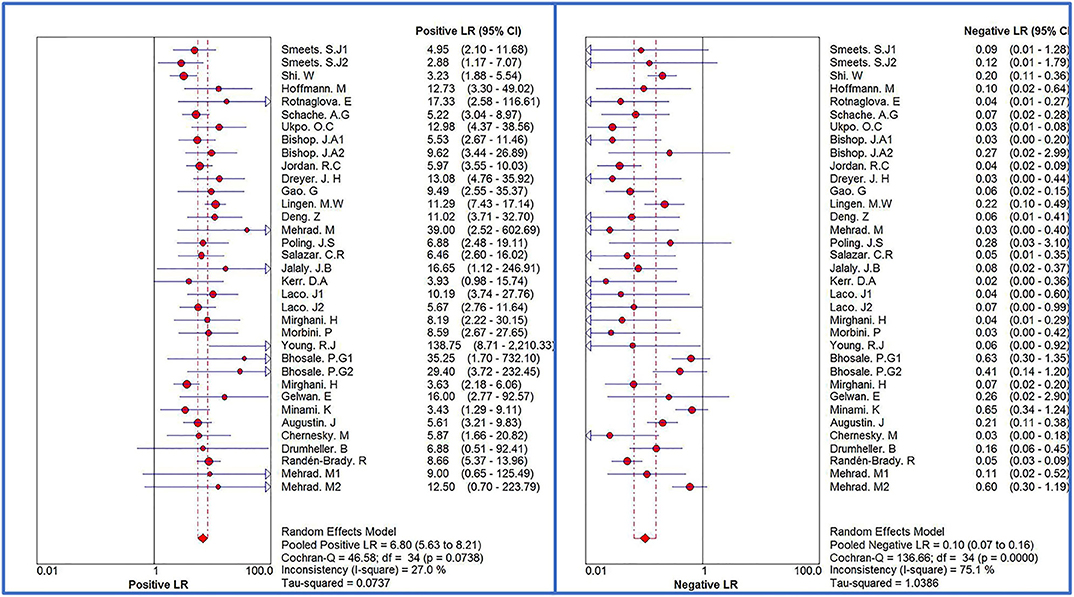
Figure 4. Forest plots of positive LR and negative LR of 30 original studies combined for diagnosis of HPV infection compared with p16 positive. Among them, 5 studies used different detection methods or included patients with different tumors, so they were divided into two records, named 1.2, respectively.
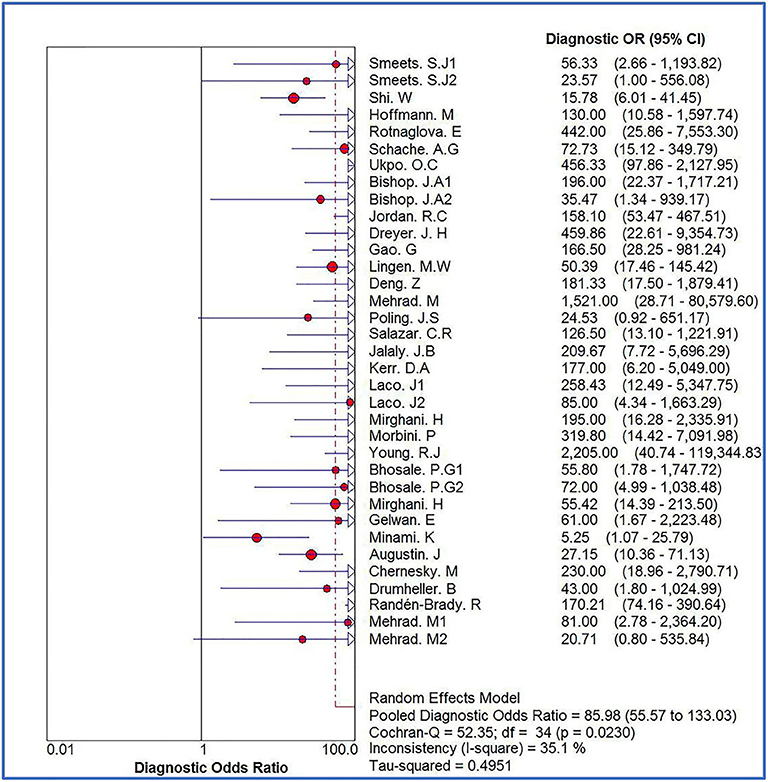
Figure 5. Forest plot of diagnostic OR of 30 original studies combined for diagnosis of HPV infection compared with p16 positive. Heterogeneity test I2 = 35.1%.
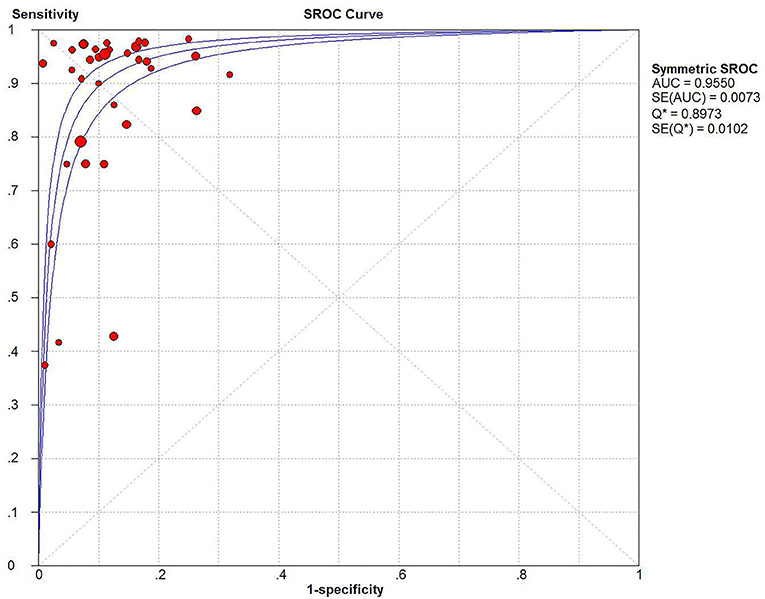
Figure 6. SROC curve and 95% CI of 30 studies combined for diagnosis of HPV infection compared with p16 positive, AUC value = 0.9550.
In addition, we investigated whether the diagnostic efficacy of p16 positivity is consistent when different testing methods are used as the gold standard. PCR and ISH were analyzed separately. For ISH HPVE6/E7mRNA, the combined sensitivity was 0.94 (95% CI: 0.92–0.96); specificity, 0.91 (95% CI: 0.89–0.93); positive LR, 7.53 (95% CI: 5.77–9.83); and negative LR, 0.12 (95% CI: 0.06–0.22). The diagnostic OR was 101.09 (95% CI: 59.93–170.54); I2-value, 12.1%; and AUC value, 0.9627. For PCR HPVE6/E7mRNA, the combined sensitivity was 0.93 (95% CI: 0.91–0.95); specificity, 0.89 (95% CI: 0.87–0.91); positive LR, 6.17 (95% CI: 4.70–8.09); negative LR, 0.09 (95% CI: 0.05–0.17); diagnostic OR, 75.25 (95% CI: 38.28–147.91); I2-value, 48.9%; and AUC value, 0.9424. After all the studies were grouped according to the test methods, the possibility of heterogeneity in each group was reduced. When ISH was used as the gold standard, the combined sensitivity, specificity, and diagnostic OR were higher, and the AUC value was larger than those in the PCR. This indicates that when ISH is used as the gold standard, p16 has higher diagnostic efficacy for HPV infection. The combined results are shown in Table 2.
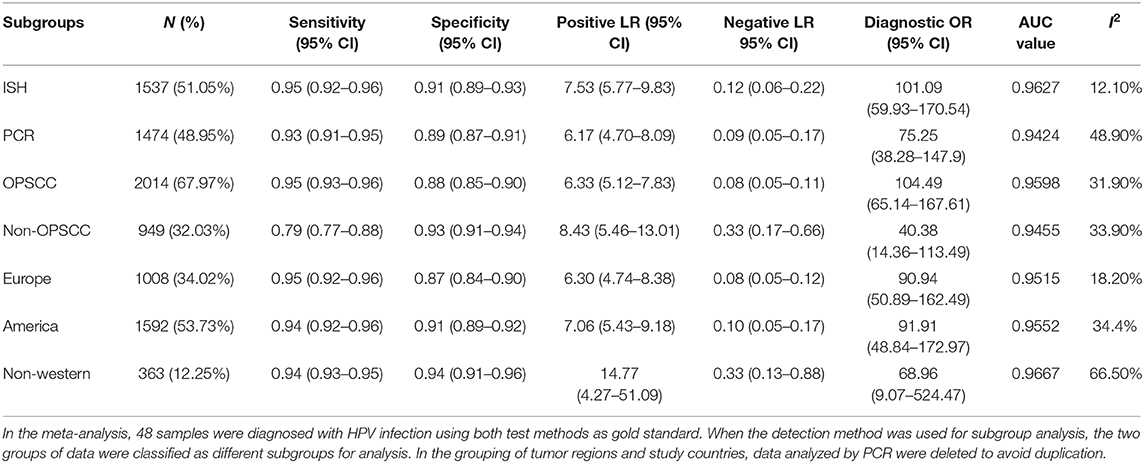
Table 2. Summary of combined effect values after grouping according to gold standard detection method, tumor location and study country.
To further explore whether the diagnostic efficacy of p16 positivity for HPV infection is consistent across all sites of squamous cell carcinomas, we conducted subgroup analysis according to tumor site. In patients with OPSCC, the combined sensitivity was 0.95 (95% CI: 0.93–0.96); specificity, 0.88 (95% CI: 0.85–0.90); positive LR, 6.33 (95% CI: 5.12–7.83); negative LR, 0.08 (95% CI: 0.05–0.11); diagnostic OR, 104.49 (95% CI: 65.14–167.61); I2-value, 31.9%; and AUC value, 0.9598. In non-OPSCC patients, the combined sensitivity was 0.79 (95% CI: 0.77–0.88); specificity, 0.93 (95% CI: 0.91–0.94); positive LR, 8.43 (95% CI: 5.46–13.01); negative LR, 0.33 (95% CI: 0.17–0.66); diagnostic OR, 40.38 (95% CI: 14.36–113.49); I2-value, 33.9%; and AUC value, 0.9455. Compared with non-OPSCC patients, the sensitivity, positive LR, diagnostic OR, and AUC value of HPV infection diagnosed according to p16 positivity is higher in OPSCC patients, indicating that p16 positivity has higher diagnostic efficacy for HPV infection in OPSCC (Table 2).
In subgroup analysis by country to investigate whether the diagnostic efficacy of p16 positivity for HPV infection is consistent across countries, we grouped studies according to their origin: European, north American, and non-Western. The results showed higher diagnostic efficacy of p16 positivity in European and American countries. In contrast, in non-western countries, the combined diagnostic OR was only 68.96, and heterogeneity was observed, indicating that p16 positivity had no significant diagnostic value. The above results are shown in Table 2. The forest plots for all subgroups are shown in the Supplementary Document.
The funnel plot for publication bias showed no statistically significant difference (p = 0.61), indicating that there was no publication bias (Figure 7).
This systematic review and meta-analysis investigated the diagnostic accuracy of p16 for HPV infection. We found that p16 expression has high sensitivity and moderate specificity as an alternative biomarker for the diagnosis of HPV infection. The findings of this meta-analysis are consistent with those of previous studies where p16 expression for diagnosing HPV infection had 90% sensitivity and >80% specificity (27, 28, 65). Concurrently, the misdiagnosis rate was 5–20%. This suggests that p16 alone has inadequate diagnostic efficacy for HPV infection (41, 66, 67). In some cases of HPVE6/E7mRNA-negative HNSCC, p16 staining was still diffuse, indicating that p16 expression was not specific to HPV infection (36, 68). High expression of p16 was also found in cervical adenocarcinoma, suggesting that the high expression of p16 can be carried out in a non-HPV dependent manner (69). Some researchers also highlighted that p16 overexpression may be related to Rb dysfunction, but Rb dysfunction may not be related to HPV infection (70). Rb protein is the upstream protein of p16, and its mutation can lead to up-regulation of p16 expression (71), and the false-positive rate is ~25% (72). Therefore, the overexpression of upstream protein and gene mutation of p16 may also be important causes of p16 upregulation. In addition, IHC-p16 was performed in only one section of the tumor tissue. The staining results may vary between sections, leading to incorrect results. In addition, the cut-off value for p16 positivity also widely varied between studies, ranging from 5 to 75%. There also many terms used for its definition, such as diffusion and powerful staining, which are unspecific (36, 73). Therefore, different diagnostic cut-off value, different staining levels, and the subjectivity of the diagnoser may lead to partial negative results, which may be the important reasons for false negative errors. It is also important that mutations and deletions in the p16 gene itself prevent it from being overexpressed in a HPV-dependent manner. The correlation between p16 and HPV infection may differ according to different patterns. In general, p16 positivity is not completely indicative of HPV infection. Aside from IHC-p16 being a simple and more readily available method, it also costs 2–6 times lower than other detection methods and has high sensitivity. However, its specificity is relatively moderate (73). Therefore, the clinical use of p16 in the diagnosis of HPV infection should be fully considered. When considering de-escalating treatment, the diagnosis of HPV infection should be more specific. Therefore, p16 alone may not be the optimal biomarker. A recent meta-analysis showed that the combination of IHC-p16 with HPV-DNA testing significantly improved the specificity of the diagnosis of HPV infection (74). To overcome the limitations of a single detection method, a novel strategy of using a combination of different detection methods for HPV is proposed (75). The combination of IHC-p16 with other HPV-specific tests may be more appropriate for selecting patients eligible for de-escalated treatment (75).
The studies in this meta-analysis used different modalities as the gold standard for diagnosing HPV infection. The results showed that the diagnostic efficacy of IHC-p16 for detecting HPVE6/E7mRNA differed between PCR and ISH, with IHC-p16 being more consistent with ISH for diagnosing HPV infection. PCR is widely used because of its high sensitivity and specificity (19), but PCR detection usually requires a higher level of skills and special experimental conditions to avoid contamination. Further, it is more difficult to replicate clinically. Meanwhile, ISH has higher specificity (19), but it has lower sensitivity in cases of low viral load. Thus, the selection of the appropriate diagnostic modality should be individualized, placing high importance on reducing the misdiagnosis when considering de-escalating treatment for HPV-positive HNSCC patients. Lower misdiagnosis rates can prevent the wrongful treatment de-escalation for HPV-negative patients, which can lead to poor local control of the tumor. Compared with missed diagnosis of HPV infection, the consequences of misdiagnosis are more fatal. Considering the current incidence of HPV-related HNSCC and the socio-economic cost of various test methods, the Supplementary Diagnostic modality should be a more practical strategy based on p16 positivity. Accordingly, ISH-HPVE6/E7mRNA should be evaluated in p16-positive tumors to improve the specificity of detection and prevent unreasonable de-escalation of treatment.
In subgroup analysis according to tumor location, p16 positivity for HPV diagnosis had higher sensitivity and specificity in OPSCC. The diagnostic OR was also two times higher than that of non-OPSCC. This may be related to the different infection rates of HPV in different tumor sites (5). Accordingly, p16 expression has been reported to have diagnostic value in OPSCC. Previous studies have also reported that the positive predictive value of p16 expression is lower for tumors outside the oropharynx, suggesting that IHC-p16 should not be used as an alternative biomarker for non-OPSCC (76, 77). Collectively, these findings indicate that IHC-p16 should be used cautiously in the diagnosis of HPV infection in non-OPSCC.
In subgroup analysis according to country, we found that the diagnostic efficacy of p16 expression varied between countries. This difference may be related to the different infection rates of HPV, which is influenced by alcohol and tobacco smoking and sexual behavior. Collectively, these results suggested the optimal diagnostic biomarker for HPV infection may different by country or region.
This meta-analysis was conducted according to stringent guidelines. Relevant studies were identified from four databases using a pre-defined search strategy, and data were extracted according to pre-set tables. Further, the risk of bias for each study was analyzed, and the data were analyzed statistically using two software. However, this study also has some limitations, including the lack of prospective data and multivariate analysis.
IHC-p16 staining is a highly effective modality for diagnosing HPV infection, particularly for OPSCC patients. However, the diagnostic efficacy varies between countries, and misdiagnosis could not be eliminated. When selecting patients for treatment de-escalation, HPVE6/E7mRNA should be detected using ISH based on p16 positivity to ensure accurate treatment.
All datasets generated for this study are included in the article/Supplementary Material.
XJ and LD: conceptualization and validation. BW: software. WB: formal analysis. HW: investigation. YZ: resources. HW, BW, and YZ: writing-original draft preparation. RJ, YX, and XJ: writing-review and editing. XJ: funding acquisition. All authors read and approved the manuscript.
This research was funded in part by grants from the National Natural Science Foundation of China (81570344 to YX), the Norman Bethune Program of Jilin University (2015225 to YX and 2015203 to XJ), National Key R&D Program of China (2017YFC0112100 to XJ), and the Jilin Provincial Science and Technology Foundations (20180414039GH to YX and 20190201200JC to XJ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Editage (www.editage.cn) for English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.524928/full#supplementary-material
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Zaravinos A. An updated overview of HPV-associated head and neck carcinomas. Oncotarget. (2014) 5:3956–69. doi: 10.18632/oncotarget.1934
3. Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. (2011) 29:4294–301. doi: 10.1200/JCO.2011.36.4596
4. Gillison ML, Broutian T, Pickard RK, Tong ZY, Xiao W, Kahle L, et al. Prevalence of oral HPV infection in the United States2009-2010. Jama. (2012) 307:693–703. doi: 10.1001/jama.2012.101
5. Betiol J, Villa LL, Sichero L. Impact of HPV infection on the development of head and neck cancer. Braz J Med Biol Res. (2013) 46:217–26. doi: 10.1590/1414-431X20132703
6. Castellsagué X, Alemany L, Quer M, Halec G, Quirós B, Tous S, et al. HPV involvement in head and neck cancers: comprehensive assessment of biomarkers in 3680 patients. J Natl Cancer Inst. (2016) 108:djv403. doi: 10.1093/jnci/djv403
7. Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. (2010) 363:24–35. doi: 10.1056/NEJMoa0912217
8. O'Sullivan B, Huang SH, Siu LL, Waldron J, Zhao H, Perez-Ordonez B, et al. Deintensification candidate subgroups in human papillomavirus-related oropharyngeal cancer according to minimal risk of distant metastasis. J Clin Oncol. (2013) 31:543–50. doi: 10.1200/JCO.2012.44.0164
9. Weinberger PM, Yu Z, Haffty BG, Kowalski D, Harigopal M, Brandsma J, et al. Molecular classification identifies a subset of human papillomavirus–associated oropharyngeal cancers with favorable prognosis. J Clin Oncol. (2006) 24:736–47. doi: 10.1200/JCO.2004.00.3335
10. Petrelli F, Sarti E, Barni S. Predictive value of human papillomavirus in oropharyngeal carcinoma treated with radiotherapy: an updated systematic review and meta-analysis of 30 trials. Head Neck. (2014) 36:750–9. doi: 10.1002/hed.23351
11. Lindel K, Beer KT, Laissue J, Greiner RH, Aebersold MD. Human papillomavirus positive squamous cell carcinoma of the oropharynx: a radiosensitive subgroup of head and neck carcinoma. Cancer. (2001) 92:805–13. doi: 10.1002/1097-0142(20010815)92:4<805::AID-CNCR1386>3.0.CO;2-9
12. Rieckmann T, Tribius S, Grob TJ, Meyer F, Busch CJ, Petersen C, et al. HNSCC cell lines positive for HPV and p16 possess higher cellular radiosensitivity due to an impaired DSB repair capacity. Radiother Oncol. (2013) 107:242–6. doi: 10.1016/j.radonc.2013.03.013
13. Mesia R, Taberna M. HPV-related oropharyngeal carcinoma de-escalation protocols. Lancet Oncol. (2017) 18:704–705. doi: 10.1016/S1470-2045(17)30250-4
14. Chen AM, Felix C, Wang PC, Hsu S, Basehart V, Garst J, et al. Reduced-dose radiotherapy for human papillomavirus-associated squamous-cell carcinoma of the oropharynx: a single-arm, phase 2 study. Lancet Oncol. (2017) 18:803–11. doi: 10.1016/S1470-2045(17)30246-2
15. Jordan RC, Lingen MW, Perez-Ordonez B, He X, Pickard R, Koluder M, et al. Validation of methods for oropharyngeal cancer HPV status determination in US cooperative group trials. Am J Surg Pathol. (2012) 36:945–54. doi: 10.1097/PAS.0b013e318253a2d1
16. Cantley RL, Gabrielli E, Montebelli F, Cimbaluk D, Gattuso P, Petruzzelli G. Ancillary studies in determining human papillomavirus status of squamous cell carcinoma of the oropharynx: a review. Patholog Res Int. (2011) 2011:138469. doi: 10.4061/2011/138469
17. The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. (2015) 517:576–82. doi: 10.1038/nature14129
18. Braakhuis BJ, Brakenhoff RH, Meijer CJ, Snijders PJ, Leemans RC. Human papilloma virus in head and neck cancer: the need for a standardised assay to assess the full clinical importance. Eur J Cancer. (2009) 45:2935–9. doi: 10.1016/j.ejca.2009.08.018
19. Robinson M, Sloan P, Shaw R. Refining the diagnosis of oropharyngeal squamous cell carcinoma using human papillomavirus testing. Oral Oncol. (2010) 46:492–6. doi: 10.1016/j.oraloncology.2010.02.013
20. Kumar S, Chera BS, Beaty B, Marron D, Jefferys S, Amdur R, et al. Multiplexed digital PCR for detection of HPV in tissue samples from oropharyngeal cancer patients. J Clin Oncol. (2019) 37:e17552. doi: 10.1200/JCO.2019.37.15_suppl.e17552
21. Sekee TR, Burt FJ, Goedhals D, Goedhals J, Munsamy Y, Seedat YR. Human papillomavirus in head and neck squamous cell carcinomas in a South African cohort. Papillomavirus Res. (2018) 6:58–62. doi: 10.1016/j.pvr.2018.10.006
22. Lin T, Mohamed ASR, Elhalawani H, Jethanandani A, Rock CD, Williams B, et al. P16 and HPV-DNA tests discordance in human papilloma virus (HPV)-associated oropharyngeal cancer: results from a case-matched study. Int J Radiation Oncol Biol Phys. (2018) 100:1330–1. doi: 10.1016/j.ijrobp.2017.12.065
23. Zhang HS, Postigo AA, Dean CD. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a TGFbeta, contact inhibition. Cell. (1999) 97:53–61. doi: 10.1016/S0092-8674(00)80714-X
24. Venuti A, Paolini F. HPV detection methods in head and neck cancer. Head Neck Pathol. (2012) 6(Suppl. 1):S63–74. doi: 10.1007/s12105-012-0372-5
25. Jung AC, Briolat J, Millon R, de Reynies A, Rickman D, Thomas E, et al. Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int J Cancer. (2010) 126:1882–94. doi: 10.1002/ijc.24911
26. Holzinger D, Schmitt M, Dyckhoff G, Benner A, Pawlita M, Bosch XF. Viral RNA patterns and high viral load reliably define oropharynx carcinomas with active HPV16 involvement. Cancer Res. (2012) 72:4993–5003. doi: 10.1158/0008-5472.CAN-11-3934
27. Lassen P, Eriksen JG, Hamilton-Dutoit S, Tramm T, Alsner J, Overgaard J. Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol. (2009) 27:1992–8. doi: 10.1200/JCO.2008.20.2853
28. Nauta IH, Rietbergen MM, van Bokhoven AAJD, Bloemena E, Lissenberg-Witte BI, Heideman DAM, et al. Evaluation of the eighth TNM classification on p16-positive oropharyngeal squamous cell carcinomas in the Netherlands and the importance of additional HPV DNA testing. Annals Oncol. (2018) 29:1273–9. doi: 10.1093/annonc/mdy060
29. Klussmann JP, Gultekin E, Weissenborn SJ, Wieland U, Dries V, Dienes HP, et al. Expression of p16 protein identifies a distinct entity of tonsillar carcinomas associated with human papillomavirus. Am J Pathol. (2003) 162:747–53. doi: 10.1016/S0002-9440(10)63871-0
30. Kumar B, Cordell KG, Lee JS, Worden FP, Prince ME, Tran HH, et al. EGFR p16, HPV Titer, Bcl-xL and p53, sex, and smoking as indicators of response to therapy and survival in oropharyngeal cancer. J Clin Oncol. (2008) 26:3128–37. doi: 10.1200/JCO.2007.12.7662
31. Combes JD, Franceschi S. Role of human papillomavirus in non-oropharyngeal head and neck cancers. Oral Oncology. (2014) 50:370–9. doi: 10.1016/j.oraloncology.2013.11.004
32. Leidy J, Chen K, Stockl T, Ye W, Wu Q, Liang J, et al. P16 expression and prognostic value in laryngeal squamous cell carcinomas - a large cohort study from chinese patients. Lab Invest. (2012) 92:311A−2A. doi: 10.1038/labinvest.2012.37
33. McInnes MDF, Moher D, Thombs BD, McGrath TA, Bossuyt PM, Clifford T, et al. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies: the PRISMA-DTA statement. Jama. (2018) 319:388–96. doi: 10.1001/jama.2017.19163
34. Moher D, Liberati A, Tetzlaff J, Altman GD. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. (2009) 339:b2535. doi: 10.1136/bmj.b2535
35. Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. (2011) 155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009
36. Smeets SJ, Hesselink AT, Speel EJM, Haesevoets A, Snijders PJF, Pawlita M, et al. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer. (2007) 121:2465–72. doi: 10.1002/ijc.22980
37. Shi W, Kato H, Perez-Ordonez B, Pintilie M, Huang S, Hui A, et al. Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J Clin Oncol. (2009) 27:6213–21. doi: 10.1200/JCO.2009.23.1670
38. Hoffmann M, Ihloff AS, Görögh T, Weise JB, Fazel A, Krams M, et al. p16INK4a overexpression predicts translational active human papillomavirus infection in tonsillar cancer. Int J Cancer. (2010) 127:1595–602. doi: 10.1002/ijc.25174
39. Rotnaglova E, Tachezy R, Salakova M, Prochazka B, Kosl'abova E, Vesela E, et al. HPV involvement in tonsillar cancer: prognostic significance and clinically relevant markers. Int J Cancer. (2011) 129:101–10. doi: 10.1002/ijc.25889
40. Schache AG, Liloglou T, Risk JM, Filia A, Jones TM, Sheard J, et al. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity specificity, prognostic discrimination. Clin Cancer Res. (2011) 17:6262–71. doi: 10.1158/1078-0432.CCR-11-0388
41. Ukpo OC, Flanagan JJ, Ma XJ, Luo Y, Thorstad WL, Lewis JS Jr. High-risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol. (2011) 35:1343–50. doi: 10.1097/PAS.0b013e318220e59d
42. Bishop JA, Ma XJ, Wang H, Luo Y, Illei PB, Begum S, et al. Detection of transcriptionally active high-risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol. (2012) 36:1874–82. doi: 10.1097/PAS.0b013e318265fb2b
43. Mehrad M, Carpenter DH, Chernock RD, Wang H, Ma XJ, Luo Y, et al. Papillary squamous cell carcinoma of the head and neck clinicopathologic and molecular features with special reference to human papillomavirus. Am J Surg Pathol. (2013) 37:1349–56. doi: 10.1097/PAS.0b013e318290427d
44. Dreyer JH, Hauck F, Oliveira-Silva M, Barros MHM, Niedobitek G. Detection of HPV infection in head and neck squamous cell carcinoma: a practical proposal. Virchows Archiv. (2013) 462:381–9. doi: 10.1007/s00428-013-1393-5
45. Gao G, Chernock RD, Gay HA, Thorstad WL, Zhang TR, Wang H, et al. A novel RT-PCR method for quantification of human papillomavirus transcripts in archived tissues and its application in oropharyngeal cancer prognosis. Int J Cancer. (2013) 132:882–90. doi: 10.1002/ijc.27739
46. Lingen MW, Xiao W, Schmitt A, Jiang B, Pickard R, Kreinbrink P, et al. Low etiologic fraction for high-risk human papillomavirus in oral cavity squamous cell carcinomas. Oral Oncol. (2013) 49:1–8. doi: 10.1016/j.oraloncology.2012.07.002
47. Deng Z, Hasegawa M, Aoki K, Matayoshi S, Kiyuna A, Yamashita Y, et al. A comprehensive evaluation of human papillomavirus positive status and p16INK4a overexpression as a prognostic biomarker in head and neck squamous cell carcinoma. Int J Oncol. (2014) 45:67–76. doi: 10.3892/ijo.2014.2440
48. Mehrad M, Zhao H, Gao G, Wang X, Lewis JS Jr. Transcriptionally-active human papillomavirus is consistently retained in the distant metastases of primary oropharyngeal carcinomas. Head Neck Pathol. (2014) 8:157–63. doi: 10.1007/s12105-013-0509-1
49. Poling JS, Ma XJ, Bui S, Luo Y, Li R, Koch WM, et al. Human papillomavirus (HPV) status of non-tobacco related squamous cell carcinomas of the lateral tongue. Oral Oncol. (2014) 50:306–10. doi: 10.1016/j.oraloncology.2014.01.006
50. Salazar CR, Anayannis N, Smith RV, Wang Y, Haigentz M Jr, Garg M, et al. Combined P16 and human papillomavirus testing predicts head and neck cancer survival. Int J Cancer. (2014) 135:2404–12. doi: 10.1002/ijc.28876
51. Jalaly JB, Lewis JS Jr, Collins BT, Wu X, Ma XJ, Luo Y, et al. Correlation of p16 immunohistochemistry in FNA biopsies with corresponding tissue specimens in HPV-related squamous cell carcinomas of the oropharynx. Cancer Cytopathol. (2015) 123:723–31. doi: 10.1002/cncy.21600
52. Kerr DA, Arora KS, Mahadevan KK, Hornick JL, Krane JF, Rivera MN, et al. Performance of a branch chain RNA in situ hybridization assay for the detection of high-risk human papillomavirus in head and neck squamous cell carcinoma. Am J Surg Pathol. (2015) 39:1643–52. doi: 10.1097/PAS.0000000000000516
53. Laco J, Sieglova K, Vosmikova H, Dundr P, Nemejcova K, Michalek J, et al. The presence of high-risk human papillomavirus (HPV) E6/E7 mRNA transcripts in a subset of sinonasal carcinomas is evidence of involvement of HPV in its etiopathogenesis. Virchows Arch. (2015) 467:405–15. doi: 10.1007/s00428-015-1812-x
54. Mirghani H, Casiraghi O, Amen F, He M, Ma XJ, Saulnier P, et al. Diagnosis of HPV-driven head and neck cancer with a single test in routine clinical practice. Modern Pathol. (2015) 28:1518–27. doi: 10.1038/modpathol.2015.113
55. Morbini P, Alberizzi P, Tinelli C, Paglino C, Bertino G, Comoli P, et al. Identification of transcriptionally active HPV infection in formalin-fixed paraffin-embedded biopsies of oropharyngeal carcinoma. Hum Pathol. (2015) 46:681–9. doi: 10.1016/j.humpath.2014.12.014
56. Young RJ, Urban D, Angel C, Corry J, Lyons B, Vallance N, et al. Frequency and prognostic significance of p16 INK4A protein overexpression and transcriptionally active human papillomavirus infection in laryngeal squamous cell carcinoma. British J Cancer. (2015) 112:1098–104. doi: 10.1038/bjc.2015.59
57. Bhosale PG, Pandey M, Desai RS, Patil A, Kane S, Prabhash K, et al. Low prevalence of transcriptionally active human papilloma virus in Indian patients with HNSCC and leukoplakia. Oral Surg Oral Med Oral Pathol Oral Radiol. (2016) 122:609–18.e7. doi: 10.1016/j.oooo.2016.06.006
58. Mirghani H, Casiraghi O, Guerlain J, Amen F, He MX, Ma XJ, et al. Diagnosis of HPV driven oropharyngeal cancers: comparing p16 based algorithms with the RNAscope HPV-test. Oral Oncol. (2016) 62:101–8. doi: 10.1016/j.oraloncology.2016.10.009
59. Gelwan E, Malm IJ, Khararjian A, Fakhry C, Bishop JA, Westra HW. Nonuniform distribution of high-risk human papillomavirus in squamous cell carcinomas of the oropharynx: rethinking the anatomic boundaries of oral and oropharyngeal carcinoma from an oncologic HPV perspective. Am J Surg Pathol. (2017) 41:1722–8. doi: 10.1097/PAS.0000000000000929
60. Minami K, Kogashiwa Y, Ebihara Y, Nakahira M, Sugasawa M, Fujino T, et al. Human papillomavirus and p16 protein expression as prognostic biomarkers in mobile tongue cancer. Acta Oto-Laryngol. (2017) 137:1121–6. doi: 10.1080/00016489.2017.1339327
61. Augustin J, Outh-Gauer S, Mandavit M, Gasne C, Grard O, Denize T, et al. Evaluation of the efficacy of the 4 tests (p16 immunochemistry polymerase chain reaction, DNA. and RNA in situ hybridization) to evaluate a human papillomavirus infection in head and neck cancers: a cohort of 348 French squamous cell carcinomas. Hum Pathol. (2018) 78:63–71. doi: 10.1016/j.humpath.2018.04.006
62. Chernesky M, Jang D, Schweizer J, Arias M, Doerwald-Munoz L, Gupta M, et al. HPV E6 oncoproteins and nucleic acids in neck lymph node fine needle aspirates and oral samples from patients with oropharyngeal squamous cell carcinoma. Papillomavirus Res. (2018) 6:1–5. doi: 10.1016/j.pvr.2018.05.003
63. Drumheller B, Cohen C, Lawson D, Siddiqui TM. Automated RNA in situ hybridization for 18 high risk human papilloma viruses in squamous cell carcinoma of the head and neck: comparison with p16 immunohistochemistry. Appl Immunohistochem Mol Morphol. (2019) 27:160–4. doi: 10.1097/PAI.0000000000000550
64. Randén-Brady R, Carpén T, Jouhi L, Syrjänen S, Haglund C, Tarkkanen J, et al. In situ hybridization for high-risk HPV E6/E7 mRNA is a superior method for detecting transcriptionally active HPV in oropharyngeal cancer. Hum Pathol. (2019) 90:97–105. doi: 10.1016/j.humpath.2019.05.006
65. Thavaraj S, Stokes A, Guerra E, Bible J, Halligan E, Long A, et al. Evaluation of human papillomavirus testing for squamous cell carcinoma of the tonsil in clinical practice. J Clin Pathol. (2011) 64:308–12. doi: 10.1136/jcp.2010.088450
66. Rietbergen MM, Brakenhoff RH, Bloemena E, Witte BI, Snijders PJ, Heideman DA, et al. Human papillomavirus detection and comorbidity: critical issues in selection of patients with oropharyngeal cancer for treatment De-escalation trials. Ann Oncol. (2013) 24:2740–5. doi: 10.1093/annonc/mdt319
67. Wasylyk B, Abecassis J, Jung CA. identification of clinically relevant HPV-related HNSCC: in p16 should we trust? Oral Oncol. (2013) 49:e33–7. doi: 10.1016/j.oraloncology.2013.07.014
68. Begum S, Cao D, Gillison M, Zahurak M, Westra HW. Tissue distribution of human papillomavirus 16 DNA integration in patients with tonsillar carcinoma. Clin Cancer Res. (2005) 11:5694–9. doi: 10.1158/1078-0432.CCR-05-0587
69. Murphy N, Ring M, Killalea AG, Uhlmann V, O'Donovan M, Mulcahy F, et al. p16INK4A as a marker for cervical dyskaryosis: CIN and cGIN in cervical biopsies and ThinPrep smears. J Clin Pathol. (2003) 56:56–63. doi: 10.1136/jcp.56.1.56
70. Marur S, D'Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. (2010) 11:781–9. doi: 10.1016/S1470-2045(10)70017-6
71. Hayes DN, Van Waes C, Seiwert YT. Genetic landscape of human papillomavirus-associated head and neck cancer and comparison to tobacco-related tumors. J Clin Oncol. (2015) 33:3227–34. doi: 10.1200/JCO.2015.62.1086
72. St Guily JL, Jacquard AC, Pretet JL, Haesebaert J, Beby-Defaux A, Clavel C, et al. Human papillomavirus genotype distribution in oropharynx and oral cavity cancer in France–The EDiTH VI study. J Clin Virol. (2011) 51:100–4. doi: 10.1016/j.jcv.2011.03.003
73. Lewis JS Jr. p16 Immunohistochemistry as a standalone test for risk stratification in oropharyngeal squamous cell carcinoma. Head Neck Pathol. (2012) 6(Suppl. 1):S75–82. doi: 10.1007/s12105-012-0369-0
74. Prigge ES, Arbyn M, von Knebel Doeberitz M, Reuschenbach M. Diagnostic accuracy of p16(INK4a) immunohistochemistry in oropharyngeal squamous cell carcinomas: a systematic review and meta-analysis. Int J Cancer. (2017) 140:1186–98. doi: 10.1002/ijc.30516
75. Westra WH. The changing face of head and neck cancer in the 21st century: the impact of HPV on the epidemiology and pathology of oral cancer. Head Neck Pathol. (2009) 3:78–81. doi: 10.1007/s12105-009-0100-y
76. Taberna M, Resteghini C, Swanson B, Pickard RK, Jiang B, Xiao W, et al. Low etiologic fraction for human papillomavirus in larynx squamous cell carcinoma. Oral Oncol. (2016) 61:55–61. doi: 10.1016/j.oraloncology.2016.08.009
Keywords: human papillomavirus, squamous cell cancers of the head and neck, immunohistochemical staining, p16, meta-analysis
Citation: Wang H, Zhang Y, Bai W, Wang B, Wei J, Ji R, Xin Y, Dong L and Jiang X (2020) Feasibility of Immunohistochemical p16 Staining in the Diagnosis of Human Papillomavirus Infection in Patients With Squamous Cell Carcinoma of the Head and Neck: A Systematic Review and Meta-Analysis. Front. Oncol. 10:524928. doi: 10.3389/fonc.2020.524928
Received: 07 January 2020; Accepted: 10 September 2020;
Published: 25 November 2020.
Edited by:
Wojciech Golusiński, Poznan University of Medical Sciences, PolandReviewed by:
Amanda Psyrri, University General Hospital Attikon, GreeceCopyright © 2020 Wang, Zhang, Bai, Wang, Wei, Ji, Xin, Dong and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihua Dong, NDE5NTIyNjM3QHFxLmNvbQ==; Xin Jiang, amlhbmd4QGpsdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.