
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 05 February 2021
Sec. Head and Neck Cancer
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.522332
This article is part of the Research TopicHead & Neck Cancer and Esophageal Cancer: From Biosignatures to TherapeuticsView all 35 articles
Background: Head and neck squamous cell carcinoma (HNSCC) is a leading cancer with high morbidity and mortality worldwide. The aim is to identify genes with clinical significance by integrated bioinformatics analysis and investigate their function in HNSCC.
Methods: We downloaded and analyzed two gene expression datasets of GSE6631 and GSE107591 to screen differentially expressed genes (DEGs) in HNSCC. Common DEGs were functionally analyzed by Gene ontology and KEGG pathway enrichment analysis. Protein-protein interaction (PPI) network was constructed with STRING database and Cytoscape. ENDOU was overexpressed in FaDu and Cal-27 cell lines, and cell proliferation and migration capability were evaluated with MTT, scratch and transwell assay. The prognostic performance of ENDOU and expression correlation with tumor infiltrates in HNSCC were validated with TCGA HNSCC datasets.
Results: Ninety-eight genes shared common differential expression in both datasets, with core functions like extracellular matrix organization significantly enriched. 15 genes showed prognostic significance, and COBL and ENDOU serve as independent survival markers in HNSCC. In-vitro ENDOU overexpression inhibited FaDu and Cal-27 cells proliferation and migration, indicating its tumor-suppressing role in HNSCC progression. GSEA analysis indicated ENDOU down-stream pathways like DNA replication, mismatch repair, cell cycle and IL-17 signaling pathway. ENDOU showed relative lower expression in HNSCC, especially HPV-positive HNSCC samples. At last, ENDOU showed negative correlation with tumor purity and tumor infiltrating macrophages, especially M2 macrophages.
Conclusion: This study identified ENDOU as a biomarker with prognostic significance in HNSCC progression.
HNSCC is a leading cancer with high morbidity and mortality worldwide (1, 2). It can be categorized by the area of the head or neck in which they begin, like oral cavity, pharynx, oropharynx, larynx, paranasal sinuses and nasal cavity and salivary glands. The most important pathogenic risk factors of HNSCC are tobacco and alcohol consumption and at least 75% of head and neck cancers are attributed to them (3). Molecular pathogenesis is still a comprehensive but largely not understood problem in HNSCC. Due to the phenotypic heterogeneity of each individual patient, it is hard to match suitable and effective treatments. Over the past decades, the development of high-throughput sequencing, increasing researchers have focused on the biological genetic functions of genes involved in the tumor-genesis of HNSCC (4–12).They have illustrated a lot of large-scale gene-expression profiles of HNSCC samples. The first successful therapeutic strategy for HNSCC was to inhibit the epidermal growth factor receptor (EGFR). In-depth understanding of the molecular carcinogenesis of HNSCC could help to develop novel and accurate treatment strategies for individual HNSCC patient.
The Cancer Genome Atlas (TCGA) network group of NIH generated comprehensive genomic characterization of HNSCC in 2015 (5). It profiled 279 HNSCC samples and provided a large-scale landscape of genetic and epigenetic characterizations of HNSCC, pointing the pivotal roles of PIK3CA, TRAF3, E2F1, TP53, NOTCH and other regulators in Wnt signaling pathways in tumor-genesis of HNSCC. Before that, Chen and his colleagues also compared 22-paired samples of HNSCC and normal tissues in 2004 (13). They found critically altered genes in the pathogenic processes of HNSCC through combinatorial analysis of microarray data. However, HNSCC is a cancer with complicated pathogenesis as well as diverse tissue origins. At present, most researches of HNSCC focus on star pathways or molecules, like PI3K signaling pathway or TP53 protein. Therefore, it’s necessary and urgent to dig out more potential targets for HNSCC treatment.
In this study, we downloaded two published and well-generated profiling datasets about HNSCC tumor and normal tissues from the Gene Expression Omnibus (GEO). Then novel potential prognostic markers were dig out to find more avenues to effective clinical treatments of HNSCC via multiple bioinformatics analysis, including biological process functional annotation and pathway enrichment analysis, protein-protein interaction network analysis as well as gene expression profiling interactive survival analysis. Gene ENDOU showed differential expression, correlation with tumor infiltrates and prognostic significance, and were functionally investigated.
The expression profiles of RNAs were screened from the National Center of Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/gds/). The GSE6631 dataset is composed of 22 paired HNSCC tumor and normal samples. The platform is GPL8300 [HG_U95Av2] Affymetrix Human Genome U95 Version 2 Array. The GSE107591 dataset contains 23 normal and 24 tumor tissues. The platform is GPL6244 [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array.
We utilized quartile normalization algorithm to subtract and correct background of these downloaded datasets firstly (14). Then we filtered probes without corresponding gene symbols and calculated the average value of gene symbols with multiple probes. We further used R software Limma package to screen DEGs by filtering p.adj value of student’s t-test and fold change (FC) (15). Finally, with a threshold of p.adj-value <0.05 and absolute value of FC >2 (16, 17), volcano plot was performed by using R software ggplot2 package to identify the DEGs with statistical significance between two groups. Hierarchical clustering and combined analyses were performed for the DEGs.
GO enrichment analyses of differentially expressed mRNAs were implemented with annotation, visualization and integrated discovery (DAVID) (http://david.abcc.ncifcrf.gov/). GO terms (including Molecular Function, Biological Processes and Cellular Components) with p-value less than 0.05 were considered significantly enriched by DEGs. KEGG is a database resource for understanding high-level functions and effects of the biological systems (http://www.genome.jp/kegg/). DAVID was also used to test the significantly statistical enrichment of DEGs in KEGG pathways. Cytoscape (version 3.40) was used to visualize the relationships between biological processes terms and DEGs.
Protein-protein interaction analysis was further conducted in STRING database (https://string-db.org/). The higher or larger score protein-protein interaction pairs were selected to construct PPI networks. Then, the regulatory relationships between genes were visualized via Cytoscape (version 3.4.0) (18). The sub-networks were extracted from the whole PPI network by using MCODE app.
To conduct survival analysis, clinical data and RNA expression data from TCGA dataset were downloaded. Univariate and Multivariate Cox analysis was conducted with SPSS, and forest plot was conducted with R. Then Kaplan-Meier survival plot and LogRank analysis was done. The correlation of ENDOU expression with tumor infiltrating cells were conducted with TIMER (https://cistrome.shinyapps.io/timer/) (19). For macrophage markers, CCL2, CD68, and IL10 were used for tumor associated macrophages (TAM), NOS2, IRF5, and PTGS2 for M1 macrophages and CD163, VSIG4, and MS4A4A for M2 macrophages, referring to previous study (20).
The FaDu and Cal-27 human carcinoma cell lines were propagated and carefully maintained in our laboratory. FaDu and Cal-27 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum at 37°C in a humid atmosphere containing 5% CO2.
CDS of ENDOU (NM_001172440) was inserted into a recombinant lentiviral expression vector (pGSIL) containing green fluorescent proteins (GFP) tag. To generate lentiviral particles, the recombinant expression plasmid was co-transfected with a packaging plasmid system (psPAX2 and pMD2G) into FaDu and Cal-27 cells, and viral particles were collected after 48 h. FaDu and Cal-27 cells were infected with ENDOU lentiviral vector or with a negative control (NC) for 24 h. The infection efficiency was preliminarily assessed in each experiment under a fluorescence microscope and then measured by sorting GFP-positive cells by flow cytometry (Beckman Coulter, USA). The stably infected cells were expanded and harvested for further experiments. The overexpression of ENDOU was examined with western blotting, using monoclonal antibody (Abcam, Cambridge, MA, USA). GAPDH was used as the house-keeping gene.
The cell proliferation activity of FaDu and Cal-27 cells was examined with MTT Cell Proliferation and Cytotoxicity Assay Kit (Dojindo Laboratories, Tokyo, Japan). The cells were incubated for 24, 48, 72, and 96 h. After incubation, the MTT solution was removed and replaced with dimethyl sulfoxide (DMSO; 150 μl, 4%; Sigma). A microplate reader (Bio-Tek, Instruments, Neufahrn, Germany) was used to measure the absorbance at 570 nm.
Cells in logarithmic growth phase were seeded at the upper transwell chamber insert (Corning, USA) at a density of 2 × 104 cells per well. The chamber was placed in a 24- well plate in which the upper chamber contained serum‐free cell culture medium and the lower chamber contained 10% FBS complete medium. The culture was continued for 24 h. The medium was discarded, and stained with a crystal violet solution to observe the number of migrated cells.
FaDu and Cal-27 cells were grown in complete growth medium until 90% confluence after transfection. A 3 mm wound was introduced across the diameter of each plate. Cell migration was observed by microscopy at 24 h.
Student’s t-test or ANOVA were performed to analyze the differential expression. Kaplan-Meier analysis was used to estimate overall survival rate or time; the differences between the survival curves were analyzed using the log-rank test. In all analyses, P<0.05 was considered to indicate a statistically significant result. Continuous data are presented as the mean ± standard deviation.
Two GEO datasets of HNSCC, GSE6631 and GSE107591 were first normalized (Figure S1) and analyzed to acquire significantly differential expressed genes (DEGs), respectively. Both datasets contains more than twenty tumor and normal samples, and with the same microarray platform of Affymetrix gene expression microarray. The GSE107591 dataset contains 23 normal and 24 tumor tissues. As GSE6631 comprises 22 paired HNSCC tumor and normal samples, we conducted paired comparison. With a threshold of p.adj-value <0.05 and absolute value of FC >2, the expression of DEGs in HNSCC tumor and normal tissues were shown in Figure 1. We found 91 genes significantly up-regulated and 121 genes down-regulated in GSE6631 dataset (Figures 1A, C). For GSE107591, we found 194 up-regulated and 278 down-regulated genes in HNSCC tumor samples compared with normal tissues (Figures 1B, D).
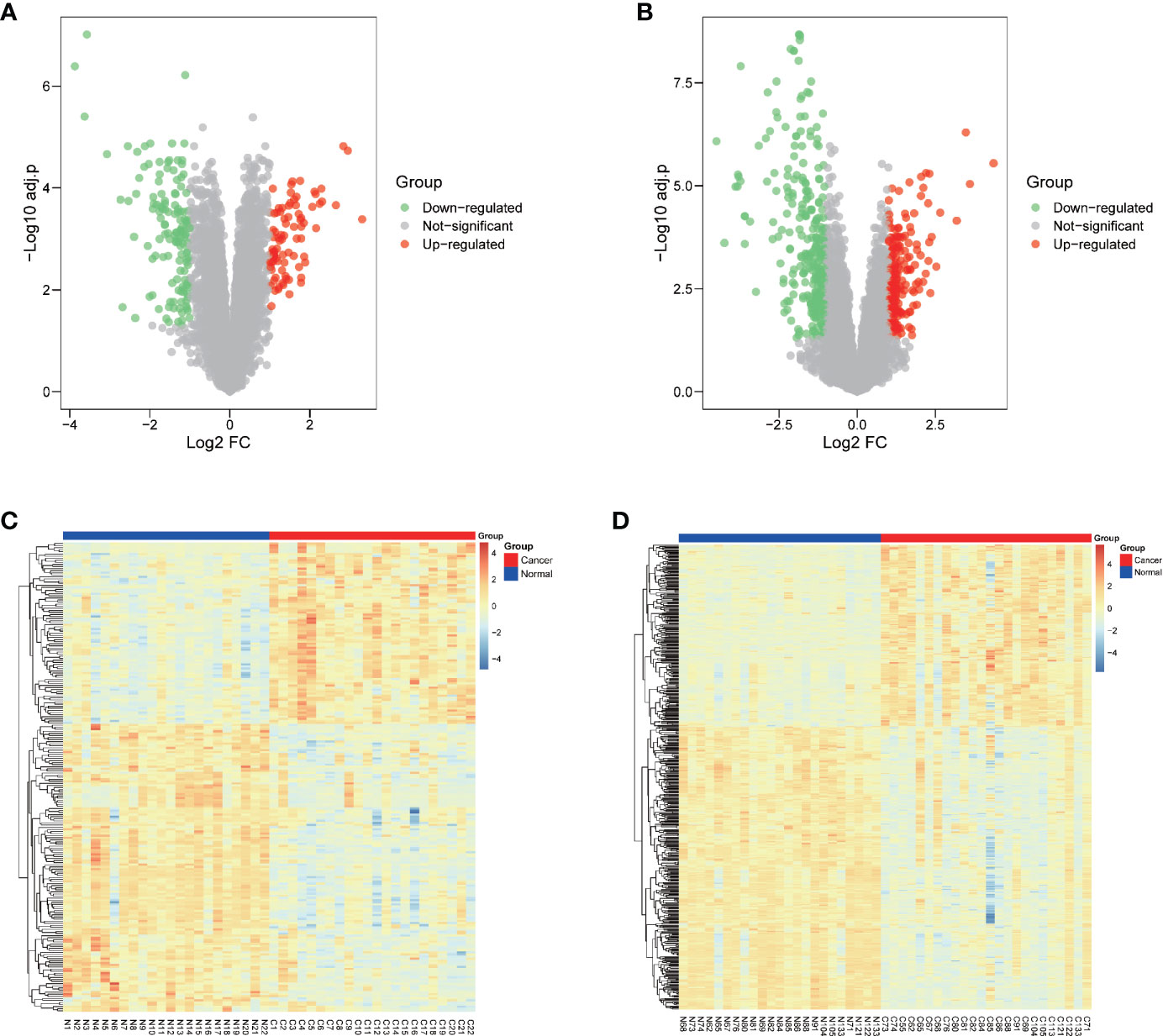
Figure 1 Differentially expressed genes (DEGs) of normal and Head and neck squamous cell carcinoma (HNSCC) tissue from GSE6631 and GSE107591. (A) Volcano plot showing significantly deregulated genes in GSE6631; (B) Volcano plot showing significantly deregulated genes in GSE107591; (C) Heatmap clustering of all DEGs in normal and HNSCC tissue from GSE6631; (D) Heatmap clustering of all DEGs in normal and HNSCC tissue from GSE107591; UP, genes with significant increased expression in HNSCC group were labeled in red. DW, genes with significant decreased expression in HNSCC group were labeled in green, genes with no different were in HNSCC group were labelled in grey.
To further figure out the roles of these DEGs in HNSCC, we searched the common DEGs both altered in these two datasets. A total of 98 DEGs (37 up-regulated and 61 down-regulated DEGs) showed dysregulation in both datasets (Figure 2A). To further detailed unravel the function of these common DEGs in HNSCC, we constructed protein-protein interaction (PPI) network via STRING database (https://string-db.org/). The genes with larger scores were selected to construct PPI networks (Figure 2B), and their function was uploaded to DAVID database for KEGG pathway and GO enrichment analysis. As shown in Figure 2C, biological processes with the largest number of common DEGs and smallest p-value includes extracellular matrix disassemble, collagen catabolic process, extracellular matrix organization, cell adhesion, aging and angiogenesis. As for the KEGG enrichment analysis in Figure 2C, not surprisingly, items like Pathways in cancer, ECM-receptor interaction, Focal adhesion and PI3K-Akt signaling pathways are significantly enriched and showed the largest number of common DEGs. The PI3K-Akt signaling was inappropriately activated in many cancers, including head and neck cancer (21). Somatic mutations of PIK3CA had been also described before and are found in about 15% of HNSCC (22–24). These findings might help us find more new therapy targets for HNSCC.
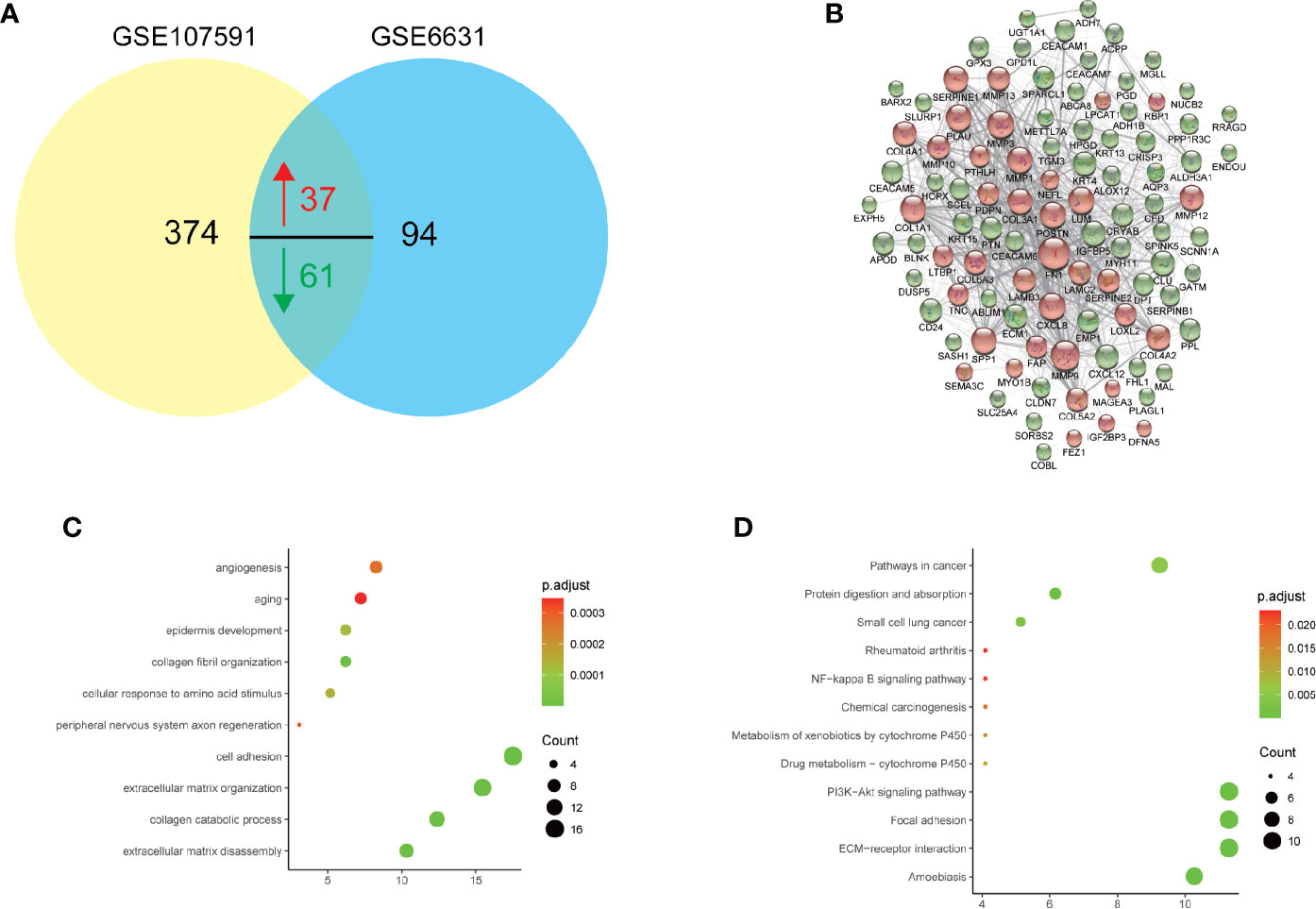
Figure 2 Gene Ontology (GO) and KEGG pathway analysis of 98 commonly deregulated genes in GSE6631 and GSE107591. P value, count and ratio of top 10 enriched functional items were shown. (A) Venn plot of 98 commonly deregulated genes in GSE6631 and GSE107591; (B) Protein-protein interaction (PPI) network of 98 commonly deregulated genes; (C) Top 10 enriched Gene Ontology biological processes, including extracellular matrix disassembly were significantly identified; (D) Top 10 enriched KEGG pathways including ECM-receptor interaction were shown.
To further explore the clinical significance of DEGs in HNSCC, we first validated the expression of 98 common DEGs with TCGA HNSCC dataset. A total of 2230 DEGs were acquired in HNSCC datasets and 92 genes (in 98 common DEGs, Table S1) showed significant dysregulation in all datasets (Figures 3A, B). Then the prognostic significance of the 92 DEGs were analyzed with univariate analysis. As shown in Figure 3C, 15 genes showed promising performance with HNSCC overall survival. Nine genes of LAMC2 (HR=1.5, P=0.00319), SEMA3C (HR=1.35, P=0.029), FAP (HR=1.42, P=0.0112), COBL (HR=1.38, P=0.0184), SERPINE1 (HR=1.61, P=0.000616),PLAU (HR=1.49, P=0.00373), MYO1B (HR=1.35, P=0.028), DUSP5 (HR=1.31, P=0.0498), LAMB3 (HR=1.49, P=0.00407) were hazardous, and 6 genes of CEACAM1 (HR=0.709, P=0.00049), CEACAM6 (HR=0.751, P=0.0362), ALDH3A1(HR=0.763, P=0.0478), ENDOU (HR=0.711, P=0.0135), SPINK5 (HR=0.736, P=0.0249) and METTL7A (HR=0.765, P=0.0494) were potential protective markers. At last, the multivariate analysis was conducted and only COBL and ENDOU showed significance (Figure 3D), indicating COBL (HR=1.55, P=0.00233) and ENDOU (HR=0.712, P=0.0361) as independent prognostic overall markers in HNSCC, and the Kaplan-Meier overall survival plot of COBL and ENDOU was shown in Figures 3E, F, and other 13 genes were shown in Figure S2.
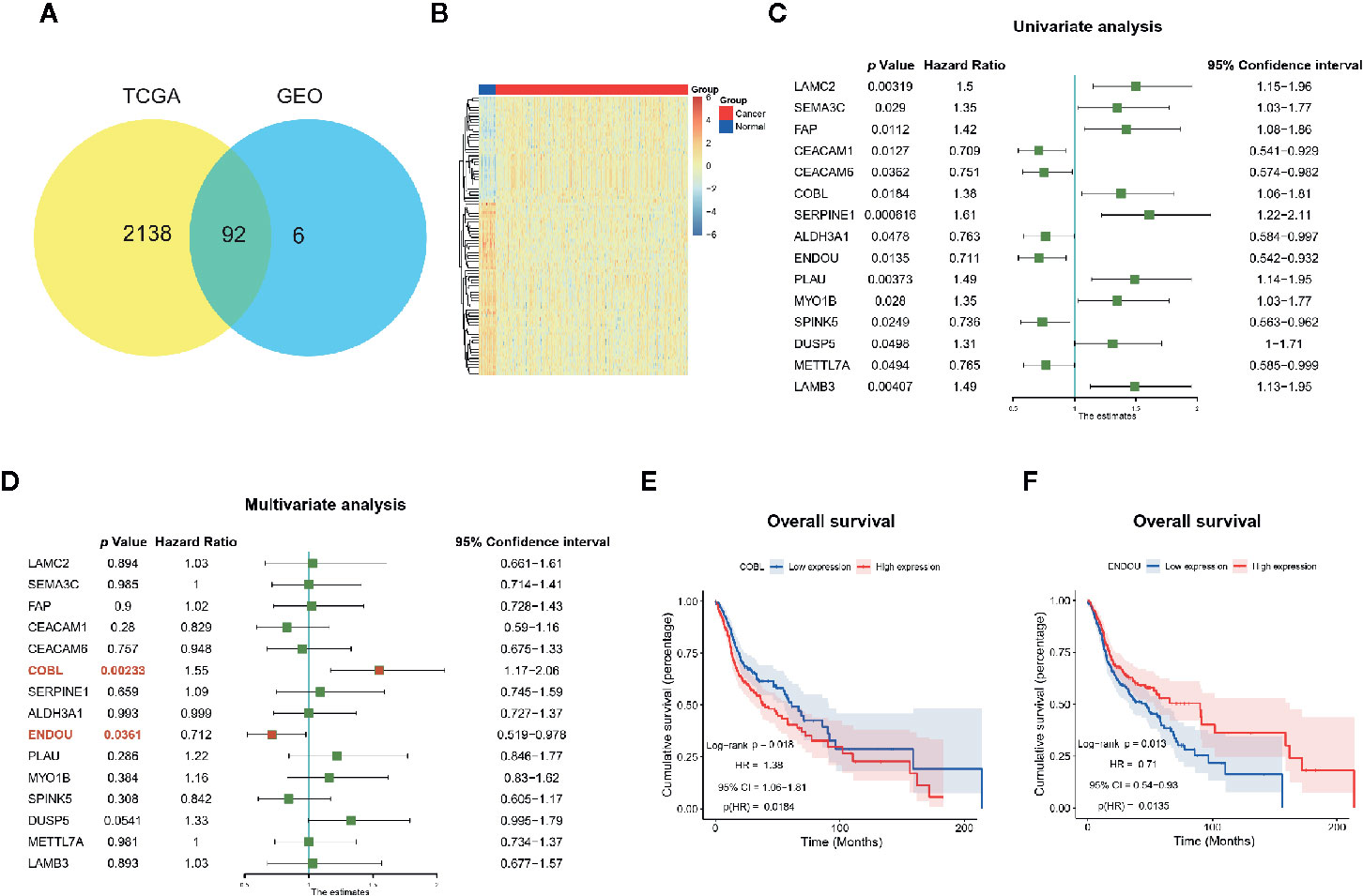
Figure 3 Prognostic significance of genes in The Cancer Genome Atlas (TCGA) HNSCC dataset. (A) Ninety-two genes showed consistent dysregulation in TCGA dataset, and were shown with Venn plot; (B) Heatmap of the dysregulated 92 genes in TCGA HNSCC cancer and normal samples; (C) Univariate analysis identified 15 genes with prognostic significance in TCGA HNSCC datasets; (D) Multivariate analysis further identified COBL and ENDOU as independent prognostic markers in HNSCC; (E, F) Kaplan-Meier overall survival plot of COBL (E) and ENDOU (F) in HNSCC.
Cordon-Bleu WH2 Repeat Protein (COBL) contains WH2 domains (WASP, Wiskott-Aldrich syndrome protein, homology domain-2) and has been reported to play an important role in the reorganization of the actin cytoskeleton (25–27). In acute lymphoblastic leukemia, COBL is a hotspot for IKZF1 deletions (28), and we will study the function of COBL in HNSCC in the future. ENDOU, also known as PP11 (Placental Protein 11), encodes uridylate-specific endoribonuclease expressed in the placenta. ENDOU was reported to displays RNA binding capability and cleaves single stranded RNA in a Mn (2+)-dependent manner at uridylates (29). Studies of ENDOU in cancer dates back to 1980s (30, 31). ENDOU was annotated with the function of in proteolysis (Gene Ontology term GO:0006508), and members like matrix metalloproteinases (MMPs) play an important function in cancer progression, by depredating extracellular matrix. Considering the significant enrichment of functional items of extracellular matrix in Figures 2C, D, we chose ENDOU for the following study.
At last, the clinical association between ENDOU expression and clinicopathological variables in TCGA HNSCC patients was analyzed. As shown in Table 1, ENDOU expression showed significant correlation with survival status (P=0.03), gender (P=0.011), alcohol history (P=0.017), clinical N stage (P=0.001), lymphovascular invasion status (P=0.001), neoplasm histologic grade (P<0.001) and pathologic N stage (P<0.001). In sum, ENDOU may serve as an independent prognostic marker in HNSCC.
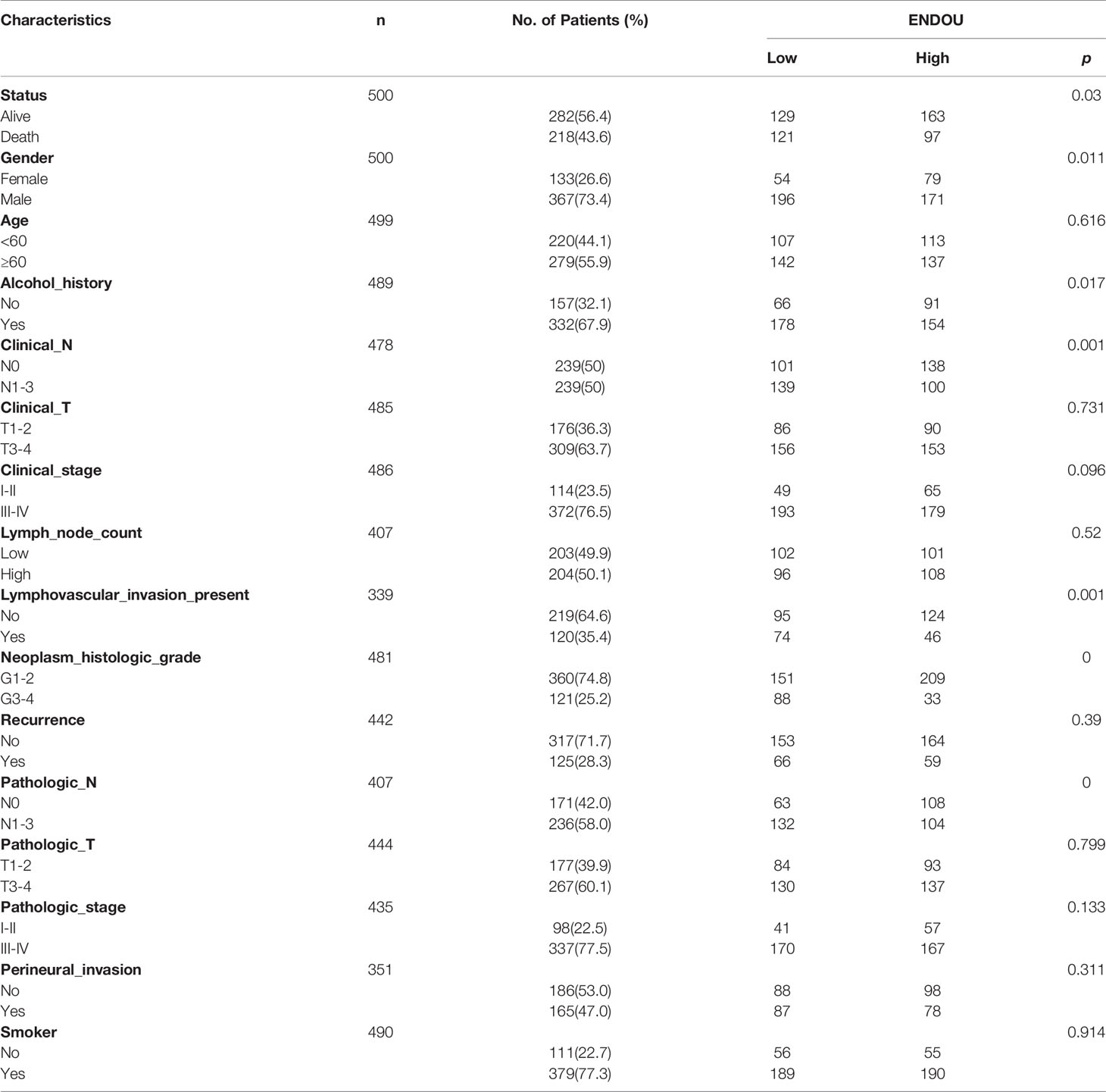
Table 1 Clinical association between ENDOU expression and clinicopathological variables in HNSCC patients.
As ENDOU shows consistent lower expression and hazardous prognostic significance in HNSCC, to explore the function of ENDOU in HNSCC, we conducted in-vitro over-expression (OE) studies in two cell lines of Fadu and Cal-27. The proliferation and migration capability was examined. As shown in Figure 4A, the proliferation capability of Fadu and Cal-27cells in ENDOU overexpression group was significantly decreased. Then wound healing assay and transwell assay was applied to study the effect of ENDOU overexpression on cell migration and invasion. In Figures 4B, C, the migration distance of ENDOU overexpression group was significantly larger than vector groups, and cells numbers was significantly decreased, implying that ENDOU may serve as a tumor suppressor in HNSCC.
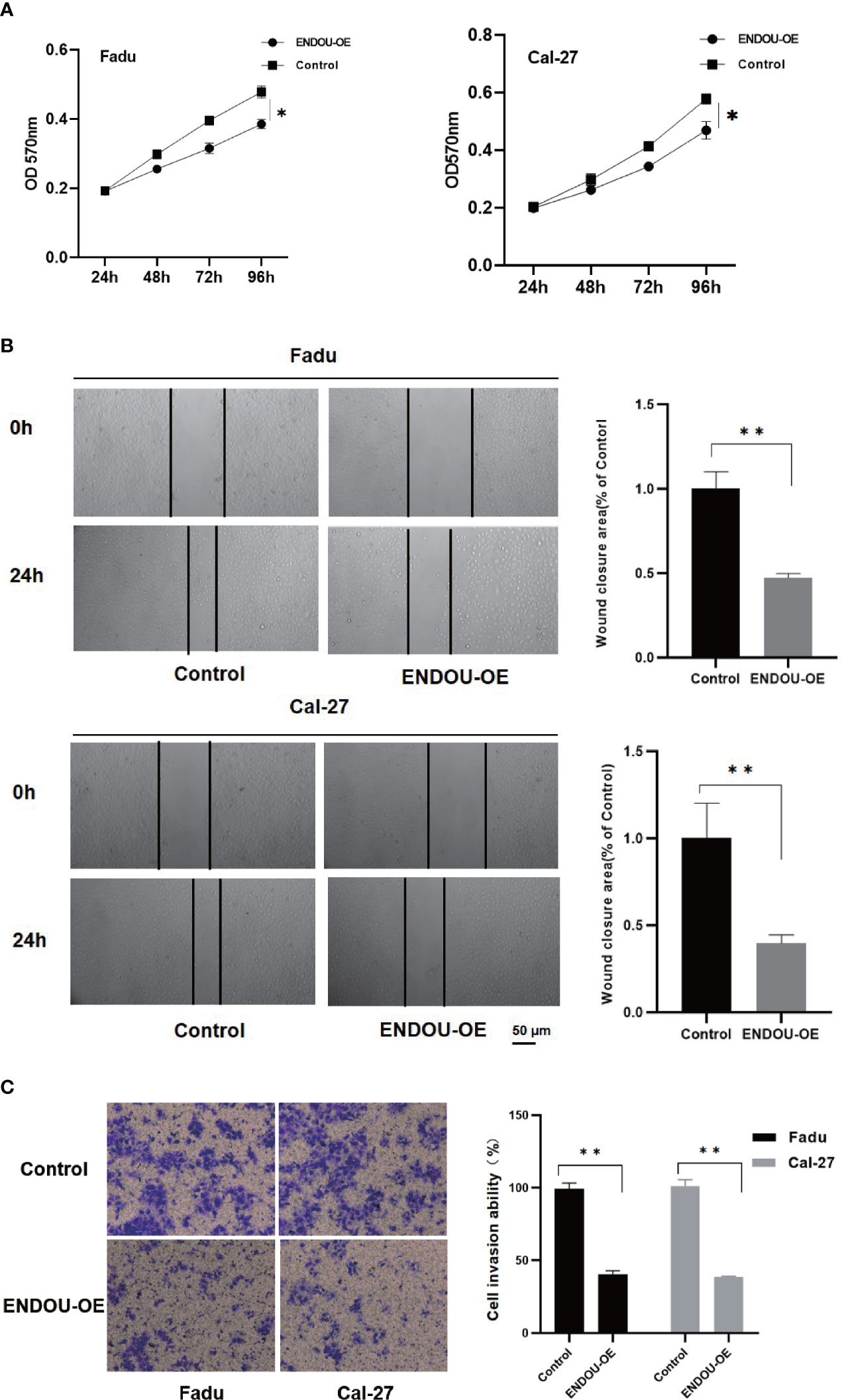
Figure 4 ENDOU overexpression inhibits Fadu and Cal-27 cells proliferation and migration in vitro. (A) MTT assay results indicated ENDOU overexpression (OE) inhibited Fadu and Cal-27 cells proliferation. Each assay was replicated three times. *P < 0.05. (B) Scratch healing assay showed decreased migration capability in ENDOU overexpressing Fadu and Cal-27 cells. Each assay was replicated three times. *P < 0.05. (C) Transwell assay showed that ENDOU overexpression (OE) attenuated the invasion capability of Fadu and Cal-27 cells. Each assay was replicated three times. *P < 0.05, **P < 0.01.
To explore the potential mechanism of ENDOU as a tumor suppressor in HNSCC, we applied Gene Set Enrichment Analysis (GSEA) analysis to enrich ENDOU related KEGG pathways and biological processes. As shown in Figure 5, DNA replication, mismatch repair, cell cycle and IL-17 showed significant enrichment, which supported the cellular phenotype of ENDOU in HNSCC.
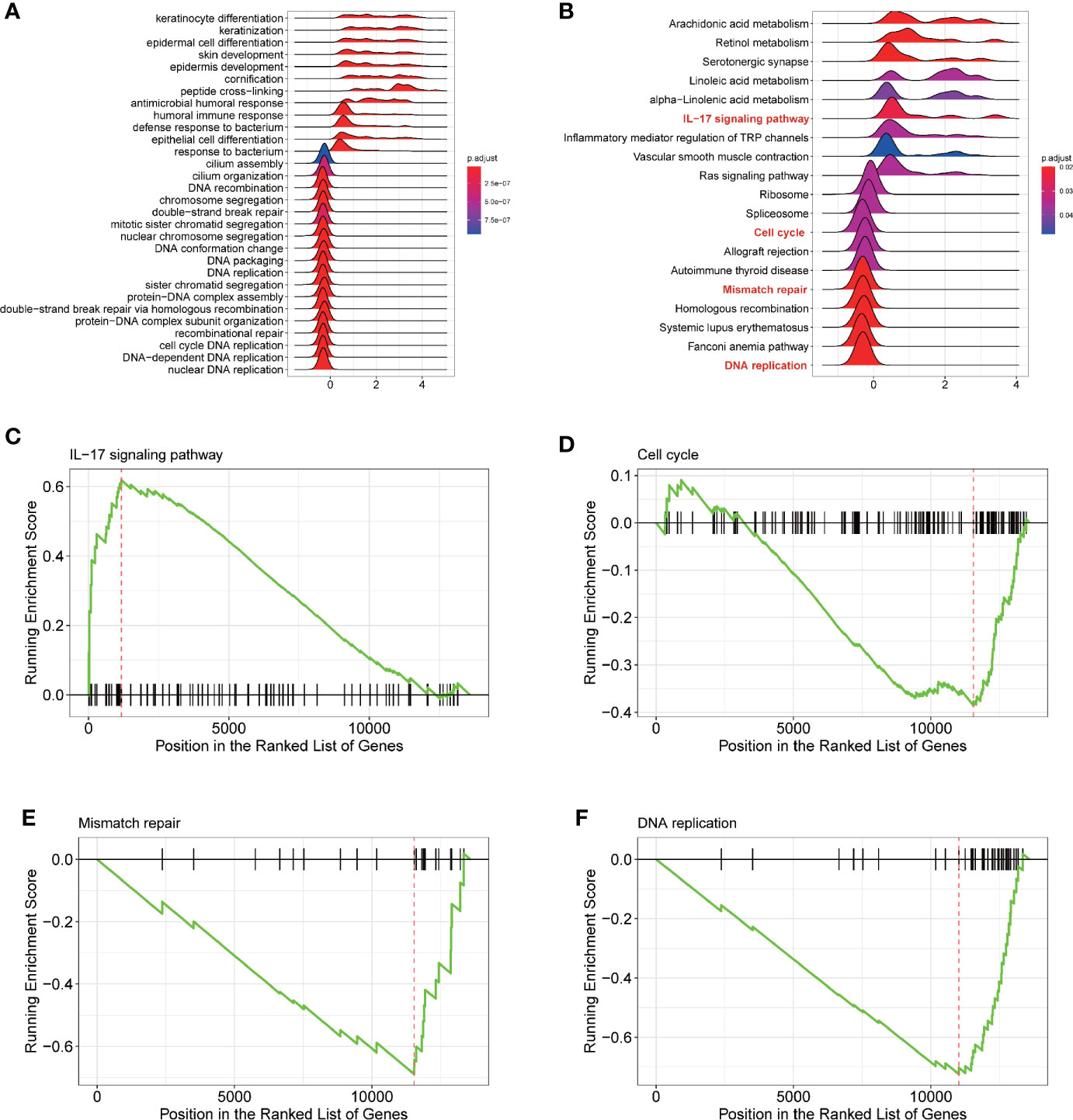
Figure 5 Gene Set Enrichment Analysis (GSEA) analysis reveals DNA replication, Mismatch repair, Cell cycle and IL-17 signaling pathway as potential ENDOU functional mechanism. (A) GSEA analysis of Gene ontology enrichment items of ENDOU; (B) GSEA analysis of KEGG pathway enrichment items of ENDOU; (C) GSEA Enrichment plot of IL-17 signaling pathway in head and neck squamous cell carcinoma (HNSCC); (D) GSEA Enrichment plot of Cell cycle in HNSCC; (E) GSEA Enrichment plot of Mismatch repair in HNSCC; (F) GSEA Enrichment plot of DNA replication in HNSCC.
In addition to HNSCC, we analyzed the expression of ENDOU in other cancer types, as shown in Figure 6A, decreased expression was also observed in bladder cancer (BLCA), breast cancer (BRCA), liver cancer (LIHC) and other cancer samples, indicating a universal role of ENDOU in cancer. For HNSCC, Human papillomavirus (HPV) positive HNSCC has a far better prognosis than HPV negative HNSCC, and HPV infection has been linked with intratumoral immune cell infiltrates (e.g. IL-17+CD8+T lymphocytes) in HNSCC (32, 33). Then we compared the expression of ENDOU in HPV-negative and HPV-positive HNSCC samples, and ENDOU showed significant lower expression in HPV-positive HNSCC samples (Figure 6A). At last, we analyzed the expression of ENDOU with tumor immune infiltrates, in HPV-negative and HPV-positive HNSCC samples. As shown in Figure 6B, ENDOU showed significant negative correlation with neutrophils, dendritic cells and especially macrophages. As the correlation was most significant in tumor infiltrating macrophages, correlation of TAMs, M1 and M2 macrophages markers with ENDOU expression were analyzed. As shown in Figure 6C, the markers of M2 macrophages (CD163, VSIG4, and MS4A4A) all showed significant negative correlation coefficient with ENDOU, implicating potential role of ENDOU in tumor infiltrating M2 macrophages.
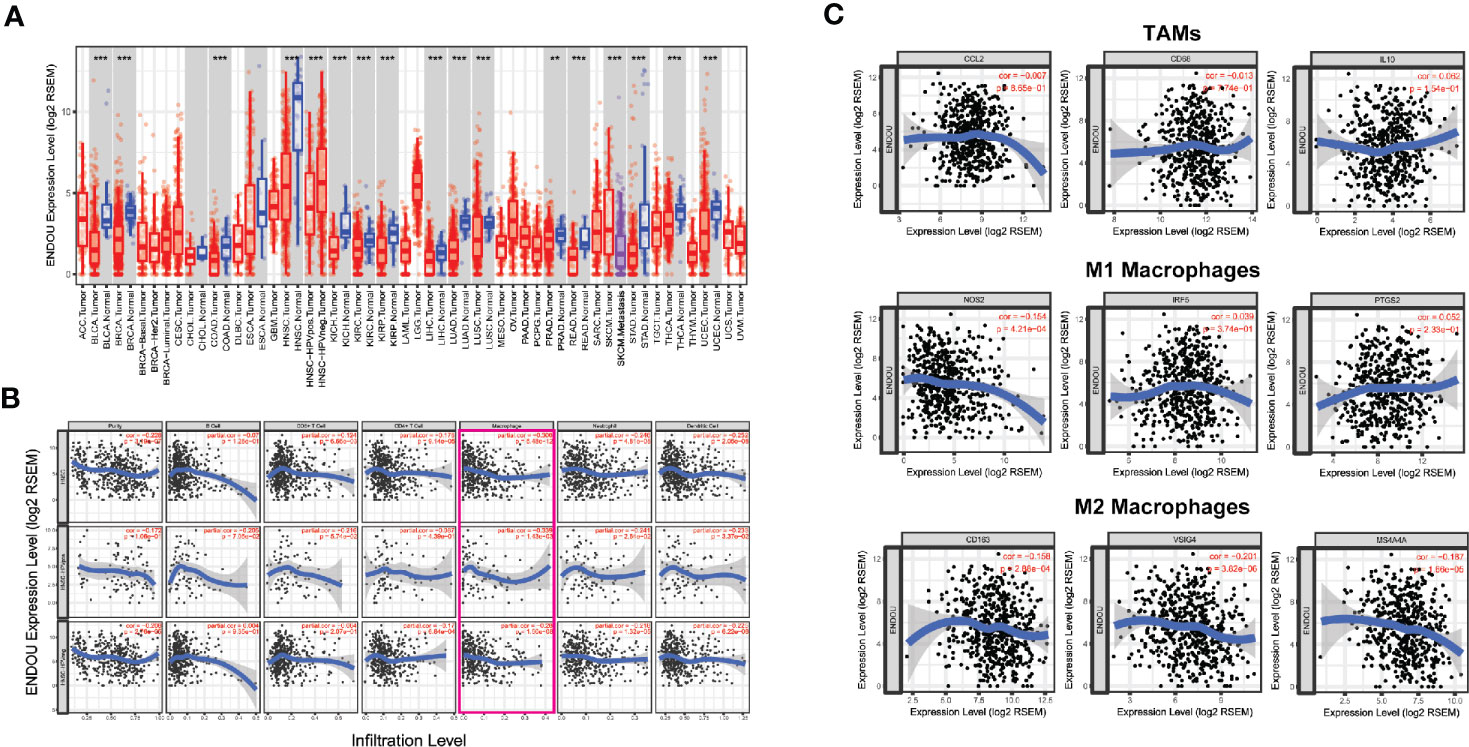
Figure 6 ENDOU showed significant down-regulation in head and neck squamous cell carcinoma (HNSCC) development and correlates with tumor infiltrating immune cells. (A) ENDOU was significantly downregulated in HNSCC samples, compared with normal tissue. ENDOU showed significant lower expression in HPV-positive HNSCC samples. Decreased expression was also observed in bladder cancer (BLCA), breast cancer (BRCA), liver cancer (LIHC) and other cancer samples. **P < 0.01; ***P < 0.001; (B) The correlation of ENDOU expression with tumor infiltrates were analyzed, and ENDOU expression showed significant negative correlation with tumor purity, macrophages, neutrophils and dendritic cells; (C) ENDOU expression showed negative correlation with M2 macrophages markers, including CD163, VSIG4, and MS4A4A.
At present, head and neck cancer is the sixth most common cancer with a poor prognosis and high mortality over the world accounting for approximately 4% of all cancers in the United States (34). HNSCC is a kind of highly heterogeneous malignancy and appropriate clinical treatments remain a major challenge for HNSCC because of heterogeneity. Hence, personalized care for HNSCC patients requires a better understanding of the molecular mechanism of HNSCC. Though some biomarkers in head and neck squamous cell carcinoma progression have been reported yet (35, 36), accepted biomarkers for HNSCC prognosis in the clinic is still raw. Effective prognostic care for HNSCC patients requires a better understanding of the molecular mechanism. With the recent development of next generation sequencing and other “omics” profiling methods, we began to focus on genomic analysis of HNSCC to illustrate the detailed or new causes of HNSCC pathogenesis and to try to develop new markers for treatments of this cancer.
In the present study, gene expression data of HNSCC were downloaded from GEO, which revealed significant differences between survival status and clinical treatments of HNSCC patients. Hence, we reanalyzed two published microarray datasets, GSE6631 and GSE107591 in this study. Finally, we found 98 common DEGs between 45 normal and 46 HNSCC tumor samples. Gene functional annotation and pathway enrichment analysis of these DEGs showed that extracellular matrix disassemble collagen catabolic process, extracellular matrix organization, cell adhesion, aging and angiogenesis were enriched in HNSCC. These results revealed that our bioinformatics analysis may help a better understanding of the regulation of these genes in HNSCC. There are also many researchers focusing on extracellular matrix organization play an important role in HNSCC (37, 38). They pointed out that extracellular matrix organization was one of the most frequently altered pathways in HNSCC, consistent with our results.
By integrating the clinical information in TCGA, we screened 9 hazardous and 6 protective prognostic markers. Of the 15 genes, LAMC2 (39–41), SERPINE1 (41–43), PLAU (44), MYO1B (45), LAMB3 (46), CEACAM1 (41, 47), CEACAM6 (48, 49), ALDH3A1 (50) and SPINK5 (51) has been reported as HNSCC prognostic markers, and in this study, we first uncovered the clinical significance of SEMA3C, FAP, COBL, DUSP5, ENDOU and METTL7A with HNSCC survival. Semaphorin 3C (SEMA3C) has been reported to play a pivotal role in tumor microenvironment driven neuroblastoma metastasis and progression (52). Fibroblast Activation Protein Alpha (FAP) was proven to serve as a marker in metastatic prostate cancer (53), pancreatic cancer (54) and ovarian cancer (55). Cordon-Bleu WH2 Repeat Protein (COBL) has been reported to play an important role in the reorganization of the actin cytoskeleton (25–27), and as a hotspot for IKZF1 deletions (28) in acute lymphoblastic leukemia. Dual Specificity Phosphatase 5 (DUSP5) is tumor suppressor in ovarian cancer (56). Methyltransferase Like 7A (METTL7A) promoted cell viability and reduced apoptosis following methotrexate in choriocarcinoma (57). Further functional and mechanism studies of SEMA3C, FAP, COBL, DUSP5, ENDOU, and METTL7A may provide more detailed information to reveal their potential role as therapeutic targets in HNSCC.
Then, ENDOU not only showed significant lower expression in HNSCC cancer samples (compared with normal samples), but also decreased level in higher stage cancer samples, implicating its tumor suppressing function in HNSCC occurrence and progression. ENDOU, also known as PP11 (Placental Protein 11), encodes uridylate-specific endoribonuclease expressed in the placenta. ENDOU was reported to displays RNA binding capability and cleaves single stranded RNA in a Mn (2+)-dependent manner at uridylates (29). Studies of ENDOU in cancer dates back to 1980s (30, 31), and this is the first time investigating the role of ENDOU in HNSCC. In this study, ENDOU was negatively correlated with tumor purity and then were functionally investigated in cancer cell lines. MTT, scratch healing and transwell assay demonstrated that ENDOU indeed inhibited the proliferation and migration capability, supporting it tumor suppressing role in HNSCC cancer cells. Meanwhile, there are several limits in this part. First, the effect of ENDOU overexpression can be validated in other HNSCC cell lines, and if possible, the impact of ENDOU silencing or mutation on HNSCC cell lines to support the conclusion. Secondly, an in vivo study of ENDOU would provide more solid evidence, as the function of genes in vitro and in vivo are not always consistent. Lastly, the underlying mechanism of ENDOU inhibiting FaDu cells proliferation and migration may be explored.
At last, tumor infiltrating immune cells in tumor microenvironment plays a pivotal role in HNSCC progression and prognosis prediction (58, 59). In this study, we found ENDOU negative correlates with tumor infiltrating neutrophils, dendritic cells and macrophages, especially M2 macrophages. Macrophages were reported to be involved in HNSCC metastasis (60–62), chemotherapy (63) and prognosis (64). Macrophages were polarized into M1 and M2 subtypes depending on their environment (65), and were reported to be functionally distinct in cancer (66). M2 polarized macrophages were found to be correlated with poor prognosis in nasopharyngeal carcinoma patients (67). HNSCC cancer cells were found to contribute to M2 polarization in several ways, like secreting microRNA-21 abundant exosomes (68) and lactic acid (69). Based on the founding that tumor suppressor gene ENDOU was negatively correlated with M2 markers of CD163, VSIG4, and MS4A4A, we speculated that ENDOU may also inhibit macrophages M2 polarization in tumor microenvironment.
In sum, our multiple bioinformatics analysis identified 98 genes with core functions like extracellular matrix organization significantly enriched. 15 genes showed prognostic significance, and COBL and ENDOU serve as independent survival markers in HNSCC. In-vitro ENDOU overexpression inhibited FaDu and Cal-27 cells proliferation and migration, indicating its tumor-suppressing role in HNSCC progression. GSEA analysis indicated ENDOU down-stream pathways like DNA replication, mismatch repair, cell cycle and IL-17 signaling pathway. ENDOU showed relative lower expression in HNSCC, especially HPV-positive HNSCC samples. At last, ENDOU showed negative correlation with tumor purity and tumor infiltrating macrophages, especially M2 macrophages. It was the first time that ENDOU was identified as a tumor suppressor, with prognostic significance and negative correlation with tumor infiltrating M2 macrophages. However, further biological and basic studies are needed to validate our findings.
All datasets generated for this study are included in the article/Supplementary Material.
The experiment was approved the Ethics Committee of Eye Nose and Throat Hospital, Fudan University.
CX, YZ, and LZ conceived and designed the study. YZ analyzed the data. YPS, YS, and MZ performed the experiments. YZ and LZ wrote the paper. CX and YPS reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by Natural Science Foundation ofShanghai (16ZR1419300), the National Natural Science Foundation (81972529) and Science and Technology Commission of Shanghai Municipality (9411961300).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.522332/full#supplementary-material
Supplementary Figure 1 | Normalization and MDS plot of GSE6631 and GSE107591; A, B) Normalization boxplot of GSE6631 and GSE107591; C, D) MDS plot of GSE6631 and GSE107591.
Supplementary Figure 2 | Kaplan-Meier survival plot of (A) PLAU; (B) ALDH3A1; (C) SERPINE1; (D) CEACAM6; (E) CEACAM61; (F) LAMB3; (G) METTL7A; (H) DUSP5; (I) SPINK5; (J) MYO1B; (K) FAP; (L) SEMA3C; (M) LAMC2 in HNSCC.
1. Jou A, Hess J. Epidemiology and Molecular Biology of Head and Neck Cancer. Oncol Res Treat (2017) 40(6):328–32. doi: 10.1159/000477127
2. Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol (2006) 24(14):2137–50. doi: 10.1200/JCO.2005.05.2308
3. Moskovitz JM, Moy J, Seiwert TY, Ferris RL. Immunotherapy for Head and Neck Squamous Cell Carcinoma: A Review of Current and Emerging Therapeutic Options. Oncologist (2017) 22(6):680–93. doi: 10.1634/theoncologist.2016-0318
4. Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science (2011) 333(6046):1154–7. doi: 10.1126/science.1206923
5. Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature (2015) 517(7536):576–82. doi: 10.1038/nature14129
6. Diehn M, Eisen MB, Botstein D, Brown PO. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat Genet (2000) 25(1):58–62. doi: 10.1038/75603
7. El-Naggar AK, Kim HW, Clayman GL, Coombes MM, Le B, Lai S. Differential expression profiling of head and neck squamous carcinoma: significance in their phenotypic and biological classification. Oncogene (2002) 21(53):8206–19. doi: 10.1038/sj.onc.1206021
8. Lemaire F, Millon R, Young J, Cromer A, Wasylyk C, Schultz I, et al. Differential expression profiling of head and neck squamous cell carcinoma (HNSCC). Br J Cancer (2003) 89(10):1940–9. doi: 10.1038/sj.bjc.6601373
9. Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science (2011) 333(6046):1157–60. doi: 10.1126/science.1208130
10. Emmert-Buck MR, Strausberg RL, Krizman DB, Bonaldo MF, Bonner RF, Bostwick DG, et al. Molecular profiling of clinical tissues specimens: feasibility and applications. J Mol Diagn (2000) 2(2):60–6. doi: 10.1016/s1525-1578(10)60617-4
11. Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet (1999) 23(1):41–6. doi: 10.1038/12640
12. Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, et al. Gene expression profiles in normal and cancer cells. Science (1997) 276(5316):1268–72. doi: 10.1126/science.276.5316.1268
13. Kuriakose MA, Chen WT, He ZM, Sikora AG, Zhang P, Zhang ZY, et al. Selection and validation of differentially expressed genes in head and neck cancer. Cell Mol Life Sci (2004) 61(11):1372–83. doi: 10.1007/s00018-004-4069-0
14. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res (2003) 31(4):e15. doi: 10.1093/nar/gng015
15. Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics (2006) 7:252. doi: 10.1186/1471-2164-7-252
16. Zou Y, Zhang M, Zeng D, Ruan Y, Shen L, Mu Z, et al. Periplaneta americana Extracts Accelerate Liver Regeneration a Complex Network of Pathways. Front Pharmacol (2020) 11:1174. doi: 10.3389/fphar.2020.01174
17. Babačić H, Lehtiö J, Pico de Coaña Y, Pernemalm M, Eriksson H. In-depth plasma proteomics reveals increase in circulating PD-1 during anti-PD-1 immunotherapy in patients with metastatic cutaneous melanoma. J Immunother Cancer (2020) 8(1). doi: 10.1136/jitc-2019-000204
18. Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics (2009) 25(21):2857–9. doi: 10.1093/bioinformatics/btp517
19. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res (2017) 77(21):e108–10. doi: 10.1158/0008-5472.CAN-17-0307
20. Pan JH, Zhou H, Cooper L, Huang JL, Zhu SB, Zhao XX, et al. LAYN Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Gastric and Colon Cancers. Front Immunol (2019) 10:6. doi: 10.3389/fimmu.2019.00006
21. Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer (2009) 9(8):550–62. doi: 10.1038/nrc2664
22. Kozaki K, Imoto I, Pimkhaokham A, Hasegawa S, Tsuda H, Omura K, et al. PIK3CA mutation is an oncogenic aberration at advanced stages of oral squamous cell carcinoma. Cancer Sci (2006) 97(12):1351–8. doi: 10.1111/j.1349-7006.2006.00343.x
23. Qiu W, Schonleben F, Li X, Ho DJ, Close LG, Manolidis S, et al. PIK3CA mutations in head and neck squamous cell carcinoma. Clin Cancer Res (2006) 12(5):1441–6. doi: 10.1158/1078-0432.CCR-05-2173
24. Murugan AK, Hong NT, Fukui Y, Munirajan AK, Tsuchida N. Oncogenic mutations of the PIK3CA gene in head and neck squamous cell carcinomas. Int J Oncol (2008) 32(1):101–11. doi: 10.3892/ijo.32.1.101
25. Hou W, Nemitz S, Schopper S, Nielsen ML, Kessels MM, Qualmann B. Arginine Methylation by PRMT2 Controls the Functions of the Actin Nucleator Cobl. Dev Cell (2018) 45(2):262–275 e8. doi: 10.1016/j.devcel.2018.03.007
26. Hou W, Izadi M, Nemitz S, Haag N, Kessels MM, Qualmann B. The Actin Nucleator Cobl Is Controlled by Calcium and Calmodulin. PloS Biol (2015) 13(9):e1002233. doi: 10.1371/journal.pbio.1002233
27. Schwintzer L, Koch N, Ahuja R, Grimm J, Kessels MM, Qualmann B. The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J (2011) 30(15):3147–59. doi: 10.1038/emboj.2011.207
28. Lopes BA, Meyer C, Barbosa TC, Poubel CP, Mansur MB, Duployez N, et al. IKZF1 Deletions with COBL Breakpoints Are Not Driven by RAG-Mediated Recombination Events in Acute Lymphoblastic Leukemia. Transl Oncol (2019) 12(5):726–32. doi: 10.1016/j.tranon.2019.02.002
29. Laneve P, Gioia U, Ragno R, Altieri F, Di Franco C, Santini T, et al. The tumor marker human placental protein 11 is an endoribonuclease. J Biol Chem (2008) 283(50):34712–9. doi: 10.1074/jbc.M805759200
30. Inaba N, Renk T, Daume E, Bohn H. Ectopic production of placenta-”specific” tissue proteins (PP5 and PP11) by malignant breast tumors. Arch Gynecol (1981) 231(1):87–90. doi: 10.1007/BF02110028
31. Inaba N, Renk T, Wurster K, Rapp W, Bohn H. Ectopic synthesis of pregnancy specific beta 1-glycoprotein (SP1) and placental specific tissue proteins (PP5, PP10, PP11, PP12) in nontrophoblastic malignant tumours. Possible markers in oncology. Klin Wochenschr (1980) 58(15):789–91. doi: 10.1007/BF01478287
32. Song L, Xie H, Tong F, Yan B, Zhang S, Fu E, et al. Association of lnc-IL17RA-11 with increased radiation sensitivity and improved prognosis of HPV-positive HNSCC. J Cell Biochem (2019) 120(10):17438–48. doi: 10.1002/jcb.29008
33. Partlova S, Boucek J, Kloudova K, Lukesova E, Zabrodsky M, Grega M, et al. Distinct patterns of intratumoral immune cell infiltrates in patients with HPV-associated compared to non-virally induced head and neck squamous cell carcinoma. Oncoimmunology (2015) 4(1):e965570. doi: 10.4161/21624011.2014.965570
34. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin (2017) 67(1):7–30. doi: 10.3322/caac.21387
35. Wicker C, Takiar V, Suganya R, Arnold S, Brill Y, Chen L, et al. Evaluation of antioxidant network proteins as novel prognostic biomarkers for head and neck cancer patients. Oral Oncol (2020) 111:104949. doi: 10.1016/j.oraloncology.2020.104949
36. Li W, Chen Y, Nie X. Regulatory Mechanisms of lncRNAs and Their Target Gene Signaling Pathways in Laryngeal Squamous Cell Carcinoma. Front Pharmacol (2020) 11:1140. doi: 10.3389/fphar.2020.01140
37. Koontongkaew S, Amornphimoltham P, Monthanpisut P, Saensuk T, Leelakriangsak M. Fibroblasts and extracellular matrix differently modulate MMP activation by primary and metastatic head and neck cancer cells. Med Oncol (2012) 29(2):690–703. doi: 10.1007/s12032-011-9871-6
38. Taitz A, Petruzzelli GJ, Lozano Y, Shankar R, Young MR. Bi-directional stimulation of adherence to extracellular matrix components by human head and neck squamous carcinoma cells and endothelial cells. Cancer Lett (1995) 96(2):253–60. doi: 10.1016/0304-3835(95)03939-T
39. Ge Y, Li W, Ni Q, He Y, Chu J, Wei P. Weighted Gene Co-Expression Network Analysis Identifies Hub Genes Associated with Occurrence and Prognosis of Oral Squamous Cell Carcinoma. Med Sci Monit (2019) 25:7272–88. doi: 10.12659/MSM.916025
40. Mendez E, Houck JR, Doody DR, Fan W, Lohavanichbutr P, Rue TC, et al. A genetic expression profile associated with oral cancer identifies a group of patients at high risk of poor survival. Clin Cancer Res (2009) 15(4):1353–61. doi: 10.1158/1078-0432.CCR-08-1816
41. Zhao L, Chi W, Cao H, Cui W, Meng W, Guo W, et al. Screening and clinical significance of tumor markers in head and neck squamous cell carcinoma through bioinformatics analysis. Mol Med Rep (2019) 19(1):143–54. doi: 10.3892/mmr.2018.9639
42. Arroyo-Solera I, Pavon MA, Leon X, Lopez M, Gallardo A, Cespedes MV, et al. Effect of serpinE1 overexpression on the primary tumor and lymph node, and lung metastases in head and neck squamous cell carcinoma. Head Neck (2019) 41(2):429–39. doi: 10.1002/hed.25437
43. Saleh AD, Cheng H, Martin SE, Si H, Ormanoglu P, Carlson S, et al. Integrated Genomic and Functional microRNA Analysis Identifies miR-30-5p as a Tumor Suppressor and Potential Therapeutic Nanomedicine in Head and Neck Cancer. Clin Cancer Res (2019) 25(9):2860–73. doi: 10.1158/1078-0432.CCR-18-0716
44. Yang K, Zhang S, Zhang D, Tao Q, Zhang T, Liu G, et al. Identification of SERPINE1, PLAU and ACTA1 as biomarkers of head and neck squamous cell carcinoma based on integrated bioinformatics analysis. Int J Clin Oncol (2019) 24(9):1030–41. doi: 10.1007/s10147-019-01435-9
45. Yamada Y, Koshizuka K, Hanazawa T, Kikkawa N, Okato A, Idichi T, et al. Passenger strand of miR-145-3p acts as a tumor-suppressor by targeting MYO1B in head and neck squamous cell carcinoma. Int J Oncol (2018) 52(1):166–78. doi: 10.3892/ijo.2017.4190
46. Liu L, Jung SN, Oh C, Lee K, Won HR, Chang JW, et al. LAMB3 is associated with disease progression and cisplatin cytotoxic sensitivity in head and neck squamous cell carcinoma. Eur J Surg Oncol (2019) 45(3):359–65. doi: 10.1016/j.ejso.2018.10.543
47. Tam K, Schoppy DW, Shin JH, Tay JK, Moreno-Nieves U, Mundy DC, et al. Assessing the Impact of Targeting CEACAM1 in Head and Neck Squamous Cell Carcinoma. Otolaryngol Head Neck Surg (2018) 159(1):76–84. doi: 10.1177/0194599818756627
48. Bednarek K, Kostrzewska-Poczekaj M, Szaumkessel M, Kiwerska K, Paczkowska J, Byzia E, et al. Downregulation of CEACAM6 gene expression in laryngeal squamous cell carcinoma is an effect of DNA hypermethylation and correlates with disease progression. Am J Cancer Res (2018) 8(7):1249–61.
49. Cameron S, de Long LM, Hazar-Rethinam M, Topkas E, Endo-Munoz L, Cumming A, et al. Focal overexpression of CEACAM6 contributes to enhanced tumourigenesis in head and neck cancer via suppression of apoptosis. Mol Cancer (2012) 11:74. doi: 10.1186/1476-4598-11-74
50. Okazaki S, Shintani S, Hirata Y, Suina K, Semba T, Yamasaki J, et al. Synthetic lethality of the ALDH3A1 inhibitor dyclonine and xCT inhibitors in glutathione deficiency-resistant cancer cells. Oncotarget (2018) 9(73):33832–43. doi: 10.18632/oncotarget.26112
51. Lv Z, Wu K, Qin X, Yuan J, Yan M, Zhang J, et al. A Novel Tumor Suppressor SPINK5 Serves as an Independent Prognostic Predictor for Patients with Head and Neck Squamous Cell Carcinoma. Cancer Manag Res (2020) 12:4855–69. doi: 10.2147/CMAR.S236266
52. Delloye-Bourgeois C, Bertin L, Thoinet K, Jarrosson L, Kindbeiter K, Buffet T, et al. Microenvironment-Driven Shift of Cohesion/Detachment Balance within Tumors Induces a Switch toward Metastasis in Neuroblastoma. Cancer Cell (2017) 32(4):427–443 e8. doi: 10.1016/j.ccell.2017.09.006
53. Hintz HM, Gallant JP, Vander Griend DJ, Coleman IM, Nelson PS, LeBeau AM. Imaging Fibroblast Activation Protein Alpha Improves Diagnosis of Metastatic Prostate Cancer with Positron Emission Tomography. Clin Cancer Res (2020) 26(18):4882–91. doi: 10.1158/1078-0432.CCR-20-1358
54. Ogawa Y, Masugi Y, Abe T, Yamazaki K, Ueno A, Fujii-Nishimura Y, et al. Three distinct stroma types in human pancreatic cancer identified by image analysis of fibroblast subpopulations and collagen. Clin Cancer Res (2020). doi: 10.1158/1078-0432.CCR-20-2298
55. Hussain A, Voisin V, Poon S, Karamboulas C, Bui NHB, Meens J, et al. Distinct fibroblast functional states drive clinical outcomes in ovarian cancer and are regulated by TCF21. J Exp Med (2020) 217(8). doi: 10.1084/jem.20191094
56. Hua KT, Wang MY, Chen MW, Wei LH, Chen CK, Ko CH, et al. The H3K9 methyltransferase G9a is a marker of aggressive ovarian cancer that promotes peritoneal metastasis. Mol Cancer (2014) 13:189. doi: 10.1186/1476-4598-13-189
57. Jun F, Peng Z, Zhang Y, Shi D. Quantitative proteomic analysis identifies novel regulators of methotrexate resistance in choriocarcinoma. Gynecol Oncol (2020) 157(1):268–79. doi: 10.1016/j.ygyno.2020.01.013
58. Badr M, Johrens K, Allgauer M, Boxberg M, Weichert W, Tinhofer I, et al. Morphomolecular analysis of the immune tumor microenvironment in human head and neck cancer. Cancer Immunol Immunother (2019) 68(9):1443–54. doi: 10.1007/s00262-019-02378-w
59. Chimote AA, Hajdu P, Sfyris AM, Gleich BN, Wise-Draper T, Casper KA, et al. Kv1.3 Channels Mark Functionally Competent CD8+ Tumor-Infiltrating Lymphocytes in Head and Neck Cancer. Cancer Res (2017) 77(1):53–61. doi: 10.1158/0008-5472.CAN-16-2372
60. Tsujikawa T, Yaguchi T, Ohmura G, Ohta S, Kobayashi A, Kawamura N, et al. Autocrine and paracrine loops between cancer cells and macrophages promote lymph node metastasis via CCR4/CCL22 in head and neck squamous cell carcinoma. Int J Cancer (2013) 132(12):2755–66. doi: 10.1002/ijc.27966
61. Topf MC, Harshyne L, Tuluc M, Mardekian S, Vimawala S, Cognetti DM, et al. Loss of CD169(+) Subcapsular Macrophages during Metastatic Spread of Head and Neck Squamous Cell Carcinoma. Otolaryngol Head Neck Surg (2019) 161(1):67–73. doi: 10.1177/0194599819829741
62. She L, Qin Y, Wang J, Liu C, Zhu G, Li G, et al. Tumor-associated macrophages derived CCL18 promotes metastasis in squamous cell carcinoma of the head and neck. Cancer Cell Int (2018) 18:120. doi: 10.1186/s12935-018-0620-1
63. Balermpas P, Rodel F, Liberz R, Oppermann J, Wagenblast J, Ghanaati S, et al. Head and neck cancer relapse after chemoradiotherapy correlates with CD163+ macrophages in primary tumour and CD11b+ myeloid cells in recurrences. Br J Cancer (2014) 111(8):1509–18. doi: 10.1038/bjc.2014.446
64. Ooft ML, van Ipenburg JA, Sanders ME, Kranendonk M, Hofland I, de Bree R, et al. Prognostic role of tumour-associated macrophages and regulatory T cells in EBV-positive and EBV-negative nasopharyngeal carcinoma. J Clin Pathol (2018) 71(3):267–74. doi: 10.1136/jclinpath-2017-204664
65. Wang Y, Smith W, Hao D, He B, Kong L. M1 and M2 macrophage polarization and potentially therapeutic naturally occurring compounds. Int Immunopharmacol (2019) 70:459–66. doi: 10.1016/j.intimp.2019.02.050
66. Yuan A, Hsiao YJ, Chen HY, Chen HW, Ho CC, Chen YY, et al. Opposite Effects of M1 and M2 Macrophage Subtypes on Lung Cancer Progression. Sci Rep (2015) 5:14273. doi: 10.1038/srep14273
67. Huang H, Liu X, Zhao F, Lu J, Zhang B, Peng X, et al. M2-polarized tumour-associated macrophages in stroma correlate with poor prognosis and Epstein-Barr viral infection in nasopharyngeal carcinoma. Acta Otolaryngol (2017) 137(8):888–94. doi: 10.1080/00016489.2017.1296585
68. Hsieh CH, Tai SK, Yang MH. Snail-overexpressing Cancer Cells Promote M2-Like Polarization of Tumor-Associated Macrophages by Delivering MiR-21-Abundant Exosomes. Neoplasia (2018) 20(8):775–88. doi: 10.1016/j.neo.2018.06.004
Keywords: head and neck squamous cell carcinoma, ENDOU, prognosis, tumor suppressor, bioinformatics
Citation: Xu C, Zhang Y, Shen Y, Shi Y, Zhang M and Zhou L (2021) Integrated Analysis Reveals ENDOU as a Biomarker in Head and Neck Squamous Cell Carcinoma Progression. Front. Oncol. 10:522332. doi: 10.3389/fonc.2020.522332
Received: 22 December 2019; Accepted: 07 December 2020;
Published: 05 February 2021.
Edited by:
Jorge A. R. Salvador, University of Coimbra, PortugalReviewed by:
Wenyue Sun, National Cancer Institute, United StatesCopyright © 2021 Xu, Zhang, Shen, Shi, Zhang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Zhou, Wmw3Njg1NkAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.