- 1Department of Oncology, Renmin Hospital of Wuhan University, Wuhan, China
- 2Department of Thoracic Surgery, National Cancer Center, National Clinical Research Center for Cancer, Cancer Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing, China
As extracellular vesicles, exosomes are released from most cells to perform cell–cell communication. Recent studies have shown that exosomes could be released into tumor microenvironment and blood to promote tumor progression through packaging and transmitting various bioactive molecules, such as cholesterol, proteins, lipids, miRNAs, mRNAs, and long non-coding RNAs (lncRNAs) to distant cells. LncRNAs have emerged as a major class of non-coding transcripts. A lot of LncRNAs have been discovered during the past few years of research on genomics. They have been proven to participate in various biological functions and disease processes through multiple mechanisms. In this review, we analyzed the role of exosome-derived lncRNAs in lung carcinogenesis and metastasis. We also highlight opportunities for the clinical potential of exosomes with specific lncRNAs as biomarkers and therapeutic intervention in lung cancer.
Introduction
Lung cancer is still the most common malignancy with the highest morbidity and mortality. In China, it has been estimated that 733,300 new cases of lung cancer were diagnosed and 610,200 cases died in 2015 (1). In America, approximately 228,150 people were firstly diagnosed with lung cancer and 142,670 patients with lung cancer died in 2019 (2). High mortality rate occurred in lung cancer patients mainly due to late-stage diagnosis of the disease (3). Patients often go to the hospital with symptoms of chest distress, hemoptysis, or systemic involvement. The 5-year survival rate of lung cancer is as low as 4% for patient with distant metastases (4), and the overall 5-year survival rate is only 24%. In view of this, it is of great importance to clarify the mechanism of lung cancer metastasis, find relevant biomarkers for early diagnosis, and treat patients with precision therapy.
Recent studies have shown that exosomes play an important role in the metastasis of lung cancer cells (5–8). Characterized by inflammation, angiogenesis, immunosuppression, and organotropism, pre-metastatic niche is used to spread tumor cells (9–11). It has been confirmed that exosomes secreted by primary tumor site possibly contribute to the establishment of premetastatic niche (12–14). Various bioactive molecules, such as proteins, RNAs, cholesterol, etc., are encapsulated by exosomes and transported to adjacent cells or distant tissues to optimize tumor microenvironment (15–17). LncRNAs are emerging as epigenetic regulators affecting transcription and playing a key role in human health (18–21). In addition to directly regulating intracellular biological activities, lncRNAs can also be detached from the surface of primary cells in the form of exosomes, which were subsequently transmitted to adjacent cells or distant organs through circulatory system (22–24). Exosome-transmitted lncRNAs have been detected in both blood and tumor tissues in lung cancer (25–27). In this review, we highlight the exosomal lncRNAs in lung cancer and summarize the potential biomarkers and mechanisms of exosome-derived lncRNAs.
Exosome Biology and Function
As one of three types of extracellular vesicles, exosomes have a diameter of 40–150 nm, which can be secreted by a vast majority of cells (28–30). The formation mechanism of exosomes is different from other types of vesicles. For the first step, the plasma membrane buds inward to form early endosomes (membrane-bound vacuoles) (31, 32). After undergoing several changes, the late endosomes named multivesicular bodies (MVBs) form with the membrane-bound vacuoles budding inward and pinching off to shape intraluminal vesicles (ILVs) (33, 34). Subsequently, the ILV-loaded MVBs release into extracellular space to act as exosomes or fuse with lysosomes to degrade the ILV contents (35–37). Vesicles with exosome-like structures were discovered by (38), while the true definition of exosomes was in 1983 (39). During the past decade, with the in-depth study of exosomes, it has been found that exosomes are involved in the development and prognosis of various diseases (40–44), and especially, play an irreplaceable role in tumor invasion (45), metastasis (46), and progression (47). Exosomes contain a variety of bioactive molecules, including proteins, RNAs, cholesterol, and lipids (24, 48–52). Figure 1 shows the structure and contents of exosomes. The contents of exosomes are the key to the biological function of exosomes in the pathophysiological process. For example, plasma exosomal proteins derived from endothelial cells play an important role in small cerebral vascular disease caused by Alzheimer’s disease (53). Exosomes shuttling high levels of microRNA-221-3p promote the resistance of breast cancer cells to adriamycin by targeting PIK3R1 (54).
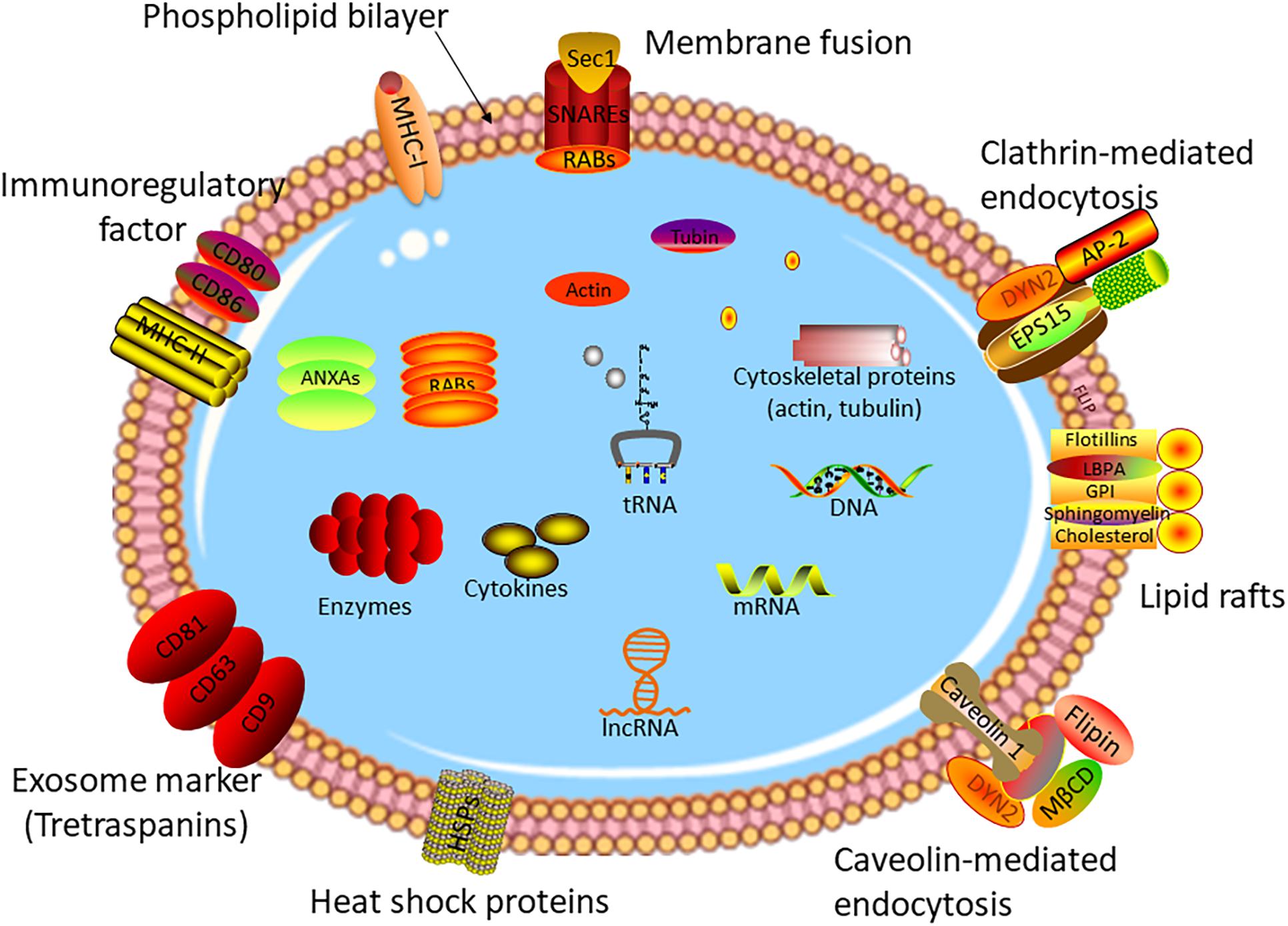
Figure 1. Structure of exosome. Exosomes are small vesicles with a double-membrane structure. The phospholipid bilayer is embedded with many transmembrane proteins, marker proteins, and receptors, such as heat chock proteins, lipid rafts, immune-regulatory molecules, cytoskeleton proteins, membrane fusion proteins, etc. Exosomes consisted of DNA, RNAs, cytokines, enzymes, and other bioactive molecules, which are transmitted from original cells to target cells.
Exosomes Loaded With Cargos and Secreted by Donor Cells
The current mainstream view is that cell membrane invaginates and internalizes extracellular ligands and particles to form nascent exosomes, which originate from ILVs. ILVs fused with the serous membrane to form multivesicular endosomes (MVEs) with packaging various bioactive molecules. Endosomal sorting complex required for transport (ESCRT) is the key molecular mechanism in MVE membrane shaping and scissing, and as a primary driver, ESCRT enables cargo selection and ILV budding (55–57). Mediated by Rab family and soluble N-ethyl-maleimide-sensitive factor attachment protein receptor (SNARE) family, mature exosomes are released by donor cells (58, 59). The Rab families are small GTPases that control the transport and secretion of intracellular secretory MVBs by moving the cytoskeleton and positioning the vesicles in the cytoplasmic membrane (60, 61). More than 70 subtypes have been found with the in-depth study of Rab GTPases, which were located on different cell membrane surfaces. MVE traffic regulated by Rab GTPases affects physiology and disease. For example, Rab5 is an endosome organizer to transfer dipeptidyl peptidase 4 (DPPIV) and bile salt export pump (BSEP) to the bile canaliculi (62). SNARE was originally discovered in bovine brain extract, which was verified to be a key molecule in the release of neurotransmitters (63). SNARE complex assembly was initiated by the docking of GTPases and membrane (64). SNARE complex drives membrane fusion through Sec3 interacting with Sso2 (target membrane protein of SNARE) (65). Located on the surface of secretory vesicles, activated RAL-1 recruits SYX-5 at the surface of plasma membrane to promote fusion of MVBs, and further to promote secretion of exosomes (66).
Exosome Uptake and Function
Exosome uptake is a very complex biological process. Exosomes containing bioactive molecules bind to acceptor cells directly by cell membrane fusion, endocytosis, and cell-specific uptake (67). Recipient cells identify and capture exosomes probably depending on their size or surface biomolecules (67). Eguchi and Yang have demonstrated that clathrin-mediated endocytosis and macropinocytosis are effective means to promote exosomes to be ingested by target cells (68, 69). Caveola-related endocytosis was also detected in EB virus-infected cells (70). Lipid is the main component of cell membrane. As microdomains interspersed in cell membrane, lipid rafts play a key role in the passage of molecules across cell membranes. It has been proven that some lipid rafts exist in caveolin-1-mediated invaginations or planar regions within cell membrane, and these regions are rich in flotillin proteins (67, 71, 72). So, lipid rafts may be involved in the process of internalizing exosomes through flotillin-mediated endocytosis.
The function of exosomes is complex and worth exploring. It is interesting to note that the target cell phenotype may be changed by exosomes (73). Exosomal miR-21 triggered phenotypic changes of hypoxic oral squamous cell carcinoma to promote cancer metastasis in a hypoxia-inducible factor-1-dependent manner (74). Melanoma-derived exosomes, composed of tumor microenvironment oncoproteins, induced bone marrow progenitor cells to a prometastatic phenotype (75). In addition, exosomes have been emerging as ideal biomarkers for containing active molecules of special significance. For example, Zhu and colleagues found that exosome-transmitted transfer ribonucleic acid (tRNA)-derived small RNAs, such as tRNA-ValTAC-3 and tRNA-ValAAC-5, were dramatically elevated in exosomes separated from the liver cancer patient’s blood, which indicated that exosomal tsRNA could function as a potential biomarker for cancer diagnosis (76). LncRNA growth arrest-specific 5 (GAS5) mediated by exosomes was also identified as a promising serum-based biomarker for early non-small-cell lung cancer (NSCLC) diagnosis (77). It is widely proven that some exosomal DNAs, RNAs, proteins, and other bioactive molecules are emerging as critical and ideal non-invasive biomarkers for various benign and malignant diseases (78–82).
As mentioned before, exosomes can encapsulate bioactive molecules and then intelligently identify the recipient cells to achieve information transmission. Exosomes loaded with special drugs to treat diseases gradually attracted the attention of scholars. The main advantages of exosomes in drug delivery are the cell-targeted therapy, non-toxicity, and non-immunogenicity. Zou and colleagues synthesized Apt-Exos, which combined the natural delivery advantages of exosomes and superiority of aptamers of molecular recognition to provide an ideal delivery platform for cancer theranostics (83). It has been confirmed that miRNA cargo in exosomes derived from administered cells has therapeutic effects in brain repair and recovery after neurological injury (84). As a novel therapeutic approach, drugs transmitted by exosomes has entered the preclinical research phase (85). In recent years, exosome-related nanovesicles have been used to the precise treatment of diseases (86–88).
Characteristics of LncRNAs
Gene transcription in organisms is a complex and orderly process. Most genome sequences are transcribed into coding RNAs or non-coding RNAs (89). As a class of non-coding RNAs, lncRNAs are recently identified as a group of long RNA transcripts with no apparent protein-coding function (90), which are divided into five categories, as follows: intronic, sense, antisense, intergenic, and bidirectional. With the implementation of the human genome project and the development of molecular biology, lncRNAs have been increasingly discovered (91–93). They are evolutionarily conservative and contain relatively few exons (94). It has been demonstrated that lncRNAs are by-products of RNA polymerase II transcription, which can mediate chromosome remodeling and prevent the recruitment of chromatin modifiers to affect gene expression (95–100). In cytoplasm, lncRNAs are largely reported to sponge miRNAs to regulate gene expression (101–105). For example, FOXF1 adjacent non-coding developmental regulatory RNA (FENDRR) was identified as downregulated in lung cancer, and high expression of which could suppress the progression of NSCLC by regulating miRNA-761/TIM2 axis (106). CDKN2B antisense RNA 1 (CDKN2B-AS1) was proven to be highly expressed in NSCLC (107), which promoted cancer cell proliferation and migration (108). LncRNAs also form complementary double chains with transcripts of protein-coding genes and produce endogenous siRNAs under the action of dicer enzyme to regulate gene expression level (109). In addition, lncRNAs can bind with proteins, regulate protein functions, and participate in protein degradation (110–112). Peng and colleagues found that lncRNA rhabdomyosarcoma 2-associated transcript (RMST) increased the DNA methyltransferases (DNMT3B) stability by promoting the interaction between RNA-binding protein HuR and DNMT3B 3’ UTR (113). Recent studies have shown that lncRNAs are also involved in autophagy regulation and m(6)A methylation (111, 114–117). In general, lncRNAs are involved in all aspects of life for their complex functions of chromatin modification, transcriptional regulation, pre-mRNA processing and splicing, RNA stability modulation, etc (118–121). Figure 2 shows the structure and function of lncRNA. Furthermore, as bioactive molecules, lncRNAs can be transmitted by exosomes to target cells or organs to exhibit their biological functions (23, 24).
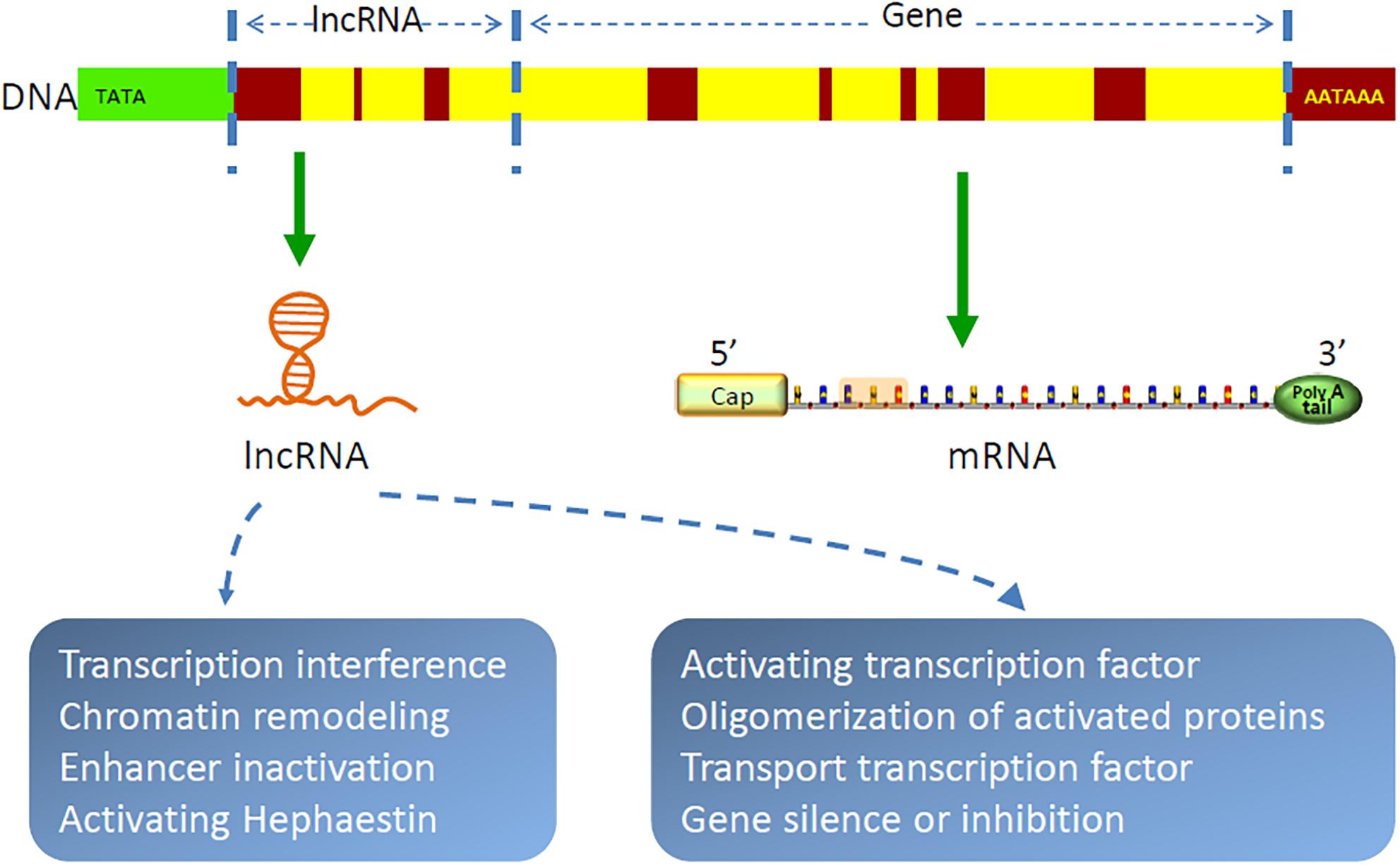
Figure 2. Structure and function of lncRNAs. LncRNAs are by-products of RNA polymerase II transcription, which function as chromatin remodeling, regulating gene transcription and translation, etc.
Lung Cancer-Related Exosomal LncRNAs
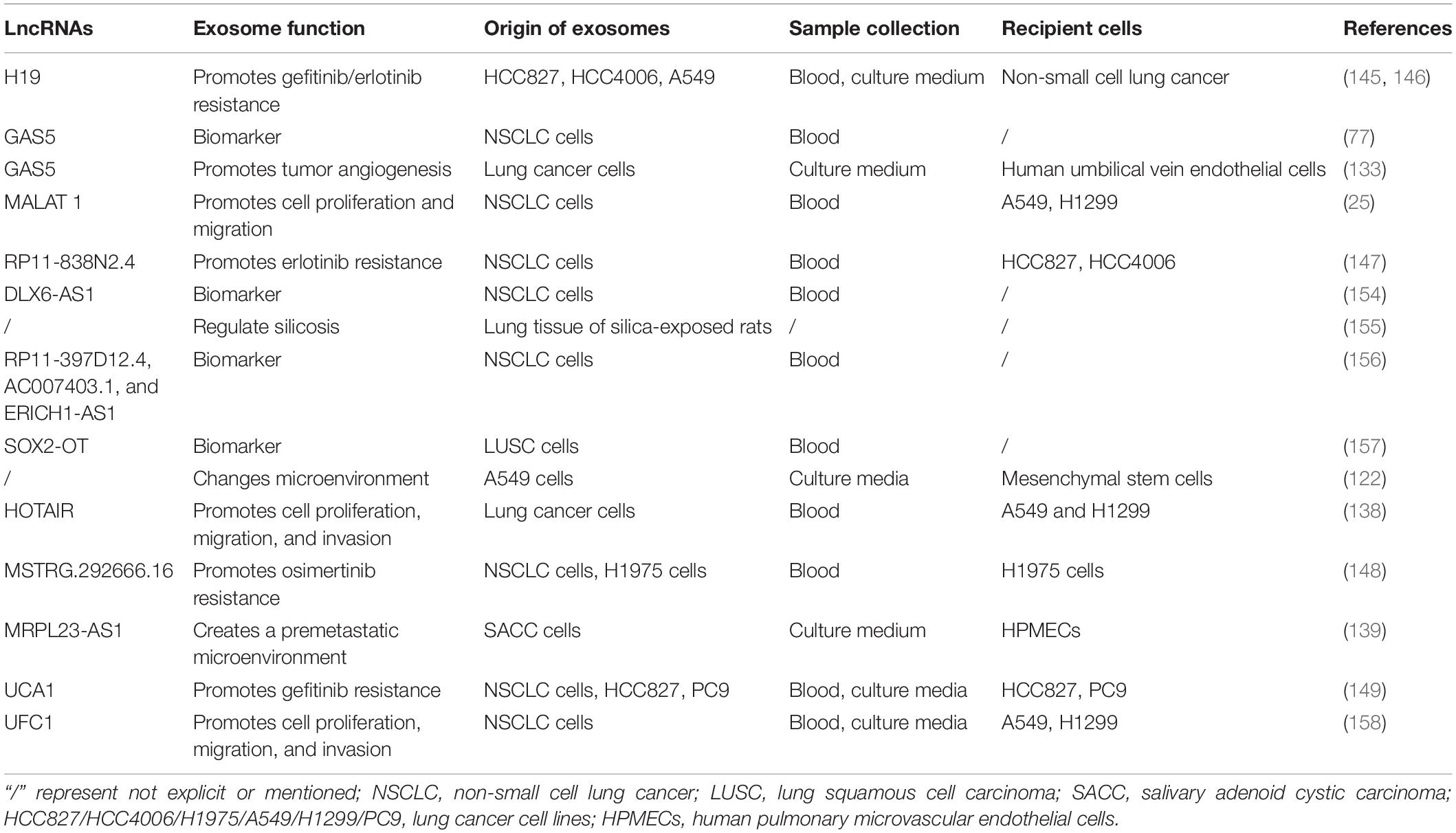
Although the free exosome database (ExoCarta) was launched in 2009, the first exosomal lncRNA in lung cancer was reported by Wang and colleagues in 2016. Wang and colleagues verified the new mechanisms that lung cancer cell-derived exosomes could regulate mesenchymal stem cells by transmitting lncRNAs (122). In recent years, some definite lncRNAs are discovered in exosomes secreted from lung cancer cells, which affect tumor progression, metastasis, and invasion. The latest researches concerning exosomal lncRNAs and lung cancer are summarized in Table 1.
Exosome-Derived LncRNAs in Lung Cancer Progress
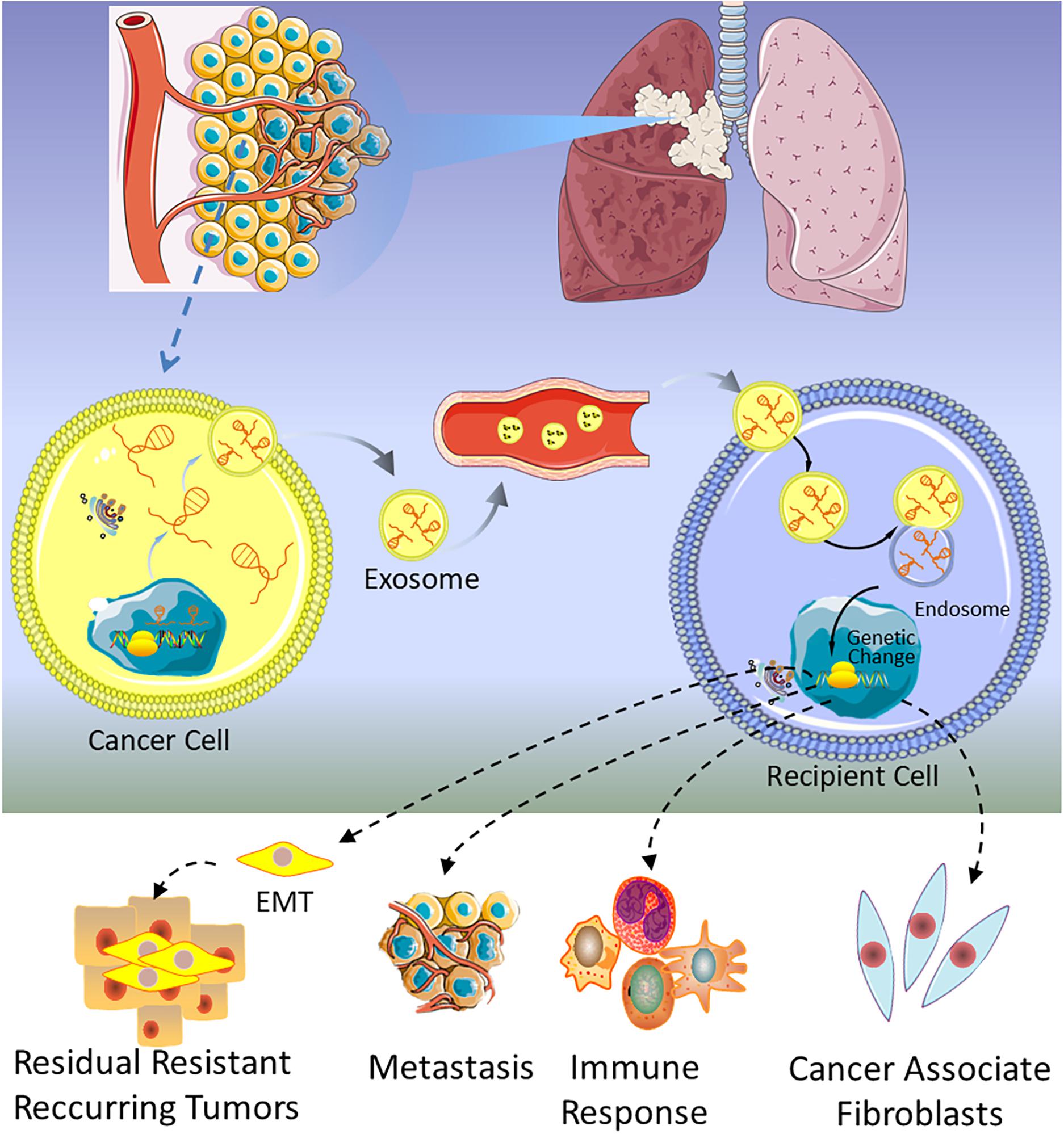
Cargos carried by exosomes are involved in all stages of tumor progression. In the process of tumor progression, the most important molecular mechanism is that tumor cells adhere to the stroma, further migrate to blood, reach premetastatic niches, establish new tumor lesions, and achieve tumor metastasis. It is reported that exosomal lncRNAs promote tumor progression by facilitating tumor premetastatic niche formation (73, 123, 124). Furthermore, epithelial to mesenchymal transition (EMT) is also an important mechanism of tumorigenesis. EMT promotes tumor cells to escape from the original tumor site to form new metastases (125). Several studies have shown that tumor-derived exosomal lncRNAs play vital roles in regulating EMT (126–128).
The first lncRNA found to be involved in lung cancer metastases was metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) (129), and subsequently, Zhang and colleagues found that exosomal MALAT1 was highly expressed in NSCLC patients’ serum, which accelerated tumor migration and proliferation by suppressing cell apoptosis and shorting cell cycle (25). These studies suggest that exosomal MALAT1 may act as a non-invasive biomarker for diagnosis of NSCLC or be a promising therapeutic target for NSCLC. At the same time, a similar mechanism of exosomal MALAT1 in many other malignancies is also verified (130–132). As shown in Table 1, several lncRNAs have been discovered in lung cancer, which are involved in the progression, metastasis, invasion, and proliferation of tumor. For example, GAS5 has been proven to be a potential therapeutic target for lung cancer by inhibiting angiogenesis (133). Lung cancer-derived exosomal GAS5 regulates the apoptosis, proliferation, and tube formation of human umbilical vein endothelial cells (HUVECs) (133). Located on the cytogenetic band 12q13.13, homeobox transcript antisense intergenic RNA (HOTAIR) gene has five transcripts, which are all identified as lncRNAs. The expression level of HOTAIR was significantly increased in patients with (chronic obstructive pulmonary disease) COPD or lung cancer, especially in patients with advanced stage of the tumor (134, 135). Exosomal HOTAIR was first identified from serum of patients with glioblastoma multiforme (136), and then it was found to be highly expressed in bronchoalveolar lavage (BAL) of smokers, NSCLC, and healthy patients (137). NSCLC cell-derived exosomal HOTAIR promoted cancer cell proliferation, migration, and invasion by sequestrating miRNA-203 (138). Unlike the above lncRNAs, exosomal MRPL23 antisense RNA 1 (MRPL23-AS1) derived from salivary adenoid cystic carcinoma (SACC), which increased microvascular permeability and promoted the metastasis of SACC to the lungs (139). Figure 3 shows that exosomal lncRNAs secreted by lung cancer cells are transmitted to target cells, which could regulate metastasis, immune response, EMT, and cancer-associated fibroblasts.
Exosome-Derived LncRNAs in Lung Cancer Drug Resistance
Despite a variety of new antitumor drugs appearing every year, the cure rate and 5-year survival rate for lung cancer patients are still far from ideal. One important reason is that tumor cells quickly develop resistance once they are repeatedly exposed to the drugs. The latest research demonstrated that lncRNAs play an important role in lung cancer drug resistance. Colon cancer-associated transcript-1 (CCAT1) effectively sponged for miR-130a-3p and contributed to cisplatin resistance in NSCLC cells by targeting sex-determining region Y-box 4 (SOX4) (140). The deletion of lncRNA X inactivate-specific transcript (XIST) increased cisplatin chemosensitivity of NSCLC cells by inhibition of ATG7-mediated autophagy (141). Nuclear paraspeckle assembly transcript 1 (NEAT1) promoted paclitaxel resistance of NSCLC cells through enhancing caspase-3 expression and activating Akt/mTOR pathway (142).
The recent emerging exosome-derived lncRNAs are also important reasons for tumor cells acquiring drug resistance. Exosomal lncRNA activated in RCC with sunitinib resistance (lncARSR) was delivered to renal cell carcinoma to promote sunitinib resistance through sponging miR-34/miR-449, which further enhanced AXL and c-MET expression (15). A similar research was conducted that androgen-regulated transcript 1 (PART1) derived from exosomes facilitated gefitinib resistance via increasing Bcl-2 expression by competitively binding to miR-129 in ESCC cells (143). Exosomes separated from temozolomide-resistant glioblastoma (GBM) cells were full of SET-binding factor 2 (SBF2) antisense RNA1 (SBF2-AS1), which was the key factor to lead sensitive GBM cells turning to temozolomide (TMZ) resistance (144).
So far, four exosome-derived lncRNAs are confirmed to mediate drug resistance in lung cancer. One study finds that NSCLC cells exposed to gefitinib increases the expression of H19, which is delivered to other cells through exosomes secreted by primary tumor. Furthermore, exosomal H19 disseminates gefitinib resistance by targeting gefitinib-sensitive tumor cells (145). A similar study shows that H19 facilitates erlotinib resistance in NSCLC via miR-615-3p/ATG7 axis (146). Another exosome-mediated lncRNA in lung cancer is RP11-838N2.4, which facilitates erlotinib resistance in NSCLC (147). A series of experiments proved that exosomal RP11-838N2.4 is secreted from NSCLC cells of erlotinib resistance, and then transmitted to sensitive cells to promote erlotinib resistance. The third lung cancer-derived exosomal lncRNA is MSTRG.292666.16, which contributes to the acquired osimertinib resistance of lung cancer cells through regulating expression levels of miR-21, miR-125b, TGFβ, and ARF6 (148). The fourth exosomal lncRNA associated with drug resistance in lung cancer is urothelial carcinoma-associated 1 (UCA1), which facilitates gefitinib resistance in NSCLC by repressing miRNA-143 expression (149). These findings verify the mechanisms of exosome-derived lncRNAs regulating target therapy resistance and providing new insights for us to study drug resistance in lung cancer.
Prospects of Exosome-Derived LncRNAs
Through the above literature review, we find that exosome-derived lncRNAs combine the advantages of exosomes and lncRNA characteristics, enabling lncRNAs to target distant cells or organs through the circulatory system. On the one hand, although many exosomal lncRNAs have been proven to be diagnostic markers for cancer, tumor-specific lncRNAs are still to be explored at present. Especially in the field of lung cancer research, the mechanism of exosome-derived lncRNAs in tumor progression is still in its infancy. On the other hand, the modification of exosomes and lncRNAs through biotechnology provides new ideas and insights for accurate evaluation of tumorigenesis and effective intervention in tumor progression. In addition, current studies have shown that lncRNA functions are far more complex than we know, and they may encode proteins (150, 151). Ahadi and colleagues found that there were miRNA seed sequences in lncRNAs, such as miR-18a, miR-93, and miR-106b (152). These exosome-derived lncRNAs also contained RNA and protein binding sites. This new finds may verify that exosomal lncRNAs are involved in tumorigenesis. The emerging exosome-wrapped lncRNAs would be the expectable method to cure lung cancer or at least detect lung cancer at its early stage to improve survival rate.
Considering the targeted delivery function of exosomes, cell biology, pharmaceutical science, and nanotechnology are attracted to the new era of human-derived nanovesicles. Aptamer-mediated drug delivery system is considered to be a prominent therapeutic for nanodelivery (87). For example, after fusing with anti-epithelial cell adhesion molecule cancer (EpCAM), aptamer could carry transferrin/aptamer across blood–brain barrier to cure brain diseases (153). Aptamer-targeted exosomal delivery is the embodiment of the combination of nanomedicine and exosome, which provide a much more optimized targeted drug delivery systems compared with traditional nanoparticle-based systems. With the help of nanoengineering technology, exosome-derived lncRNAs could be modified to carry a specific sequence or molecule that inhibits cancer cells, which will achieve a real sense of precision treatment for lung cancer.
Conclusion
In this review, we briefly summarize the functional characteristics of exosomes and lncRNAs, expound the role of exosome-derived lncRNAs in tumors, especially in lung cancer, and look forward to the advantages of the combination of exosome-derived lncRNAs with nanomedicine in achieving targeted drug delivery and tumor precision treatment. At present, there are relatively few studies about exosome-derived lncRNAs in lung cancer, but more studies that can give us an in-depth understanding of their regulatory mechanism are under way. Although the functions of exosome-derived lncRNAs in tumors are similar, the tumorigenesis and progression of any cancer has its own unique rules and mechanisms. Currently, the mainstream research direction of exosome-derived lncRNAs is that they can be used as non-invasive tumor markers to diagnose tumors and as major gene targets for antitumor therapy. However, the pathophysiological mechanism of exosomal lncRNA in tumors still needs to be further studied. Meanwhile, under the current technical conditions, exosomal lncRNAs are still difficult to use as biomarkers due to their small number in body fluids and heterogeneity, which may cause false negatives or positives in cancer diagnosis. In future studies, more specific exosomal lncRNAs in different cancer should be discovered. With the combination of nanoengineering and molecular biology, exosome-mediated lncRNAs for precision nanomedicine will provide novel methods for cancer diagnosis and therapy.
Author Contributions
TF wrote the manuscript. JH and NS revised and approved the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (No. 2018YFC1312100/2018YFC1312102), National Natural Science Foundation of China (81802299 and 81502514), the CAMS Innovation Fund for Medical Sciences (2016-I2M-1-001 and 2017-I2M-1-005), the Fundamental Research Funds for the Central Universities (3332018070), the National Key Basic Research Development Plan (2018YFC1312105) and the National Key Research and Development Program of China (2016YFC0901401).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. (2016) 66:115–32. doi: 10.3322/caac.21338
2. Siegel RL, Miller KD. Cancer statistics, 2019. CA Cancer J Clin. (2019) 69:7–34. doi: 10.3322/caac.21551
3. Blandin Knight S, Crosbie PA, Balata H, Chudziak J, Hussell T, Dive C. Progress and prospects of early detection in lung cancer. Open Biol. (2017) 7:170070. doi: 10.1098/rsob.170070
4. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr., Wu YL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. (2017) 389:299–311. doi: 10.1016/s0140-6736(16)30958-8
5. Zhang X, Sai B, Wang F, Wang L, Wang Y, Zheng L, et al. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol Cancer. (2019) 18:40. doi: 10.1186/s12943-019-0959-5
6. Huang WT, Chong IW, Chen HL, Li CY, Hsieh CC, Kuo HF, et al. Pigment epithelium-derived factor inhibits lung cancer migration and invasion by upregulating exosomal thrombospondin 1. Cancer Lett. (2019) 442:287–98. doi: 10.1016/j.canlet.2018.10.031
7. Lukic A, Wahlund CJE, Gomez C, Brodin D, Samuelsson B, Wheelock CE, et al. Exosomes and cells from lung cancer pleural exudates transform LTC4 to LTD4, promoting cell migration and survival via CysLT1. Cancer Lett. (2019) 444:1–8. doi: 10.1016/j.canlet.2018.11.033
8. Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. (2018) 9:5395. doi: 10.1038/s41467-018-07810-w
9. Carpenter RA, Kwak JG. Implantable pre-metastatic niches for the study of the microenvironmental regulation of disseminated human tumour cells. Nat Biomed Eng. (2018) 2:915–29. doi: 10.1038/s41551-018-0307-x
10. Liu Y, Cao X. Characteristics and significance of the pre-metastatic niche. Cancer Cell. (2016) 30:668–81. doi: 10.1016/j.ccell.2016.09.011
11. Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer. (2017) 17:302–17. doi: 10.1038/nrc.2017.6
12. Ortiz A, Gui J, Zahedi F, Yu P, Cho C, Bhattacharya S, et al. An interferon-driven oxysterol-based defense against tumor-derived extracellular vesicles. Cancer Cell. (2019) 35:33–45.e36. doi: 10.1016/j.ccell.2018.12.001
13. Hsu YL, Huang MS, Hung JY, Chang WA, Tsai YM, Pan YC, et al. Bone-marrow-derived cell-released extracellular vesicle miR-92a regulates hepatic pre-metastatic niche in lung cancer. Oncogene. (2019) 39:739–53. doi: 10.1038/s41388-019-1024-y
14. Guo Y, Ji X, Liu J, Fan D, Zhou Q, Chen C, et al. Effects of exosomes on pre-metastatic niche formation in tumors. Mol Cancer. (2019) 18:39. doi: 10.1186/s12943-019-0995-1
15. Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. (2016) 29:653–68. doi: 10.1016/j.ccell.2016.03.004
16. Zhang Y, Li M, Hu C. Exosomal transfer of miR-214 mediates gefitinib resistance in non-small cell lung cancer. Biochem Biophys Res Commun. (2018) 507:457–64. doi: 10.1016/j.bbrc.2018.11.061
17. van Balkom BW, de Jong OG, Smits M, Brummelman J, den Ouden K, de Bree PM, et al. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood. (2013) 121:3997–4006, S1–15. doi: 10.1182/blood-2013-02-478925
18. Kulkarni S, Lied A, Kulkarni V, Rucevic M, Martin MP, Walker-Sperling V, et al. CCR5AS lncRNA variation differentially regulates CCR5, influencing HIV disease outcome. Nat Immunol. (2019) 20:824–34. doi: 10.1038/s41590-019-0406-1
19. Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. (2014) 15:423–37. doi: 10.1038/nrg3722
20. James AR, Schroeder MP, Neumann M, Bastian L, Eckert C, Gokbuget N, et al. Long non-coding RNAs defining major subtypes of B cell precursor acute lymphoblastic leukemia. J Hematol Oncol. (2019) 12:8. doi: 10.1186/s13045-018-0692-3
21. Li Y, Zeng C, Hu J, Pan Y, Shan Y, Liu B, et al. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J Hematol Oncol. (2018) 11:89. doi: 10.1186/s13045-018-0632-2
22. Liu Y, Lin L, Zou R, Wen C, Wang Z, Lin F. MSC-derived exosomes promote proliferation and inhibit apoptosis of chondrocytes via lncRNA-KLF3-AS1/miR-206/GIT1 axis in osteoarthritis. Cell Cycle. (2018) 17:2411–22. doi: 10.1080/15384101.2018.1526603
23. Li X, Liu R, Huang Z, Gurley EC, Wang X, Wang J, et al. Cholangiocyte-derived exosomal long noncoding RNA H19 promotes cholestatic liver injury in mouse and humans. Hepatology. (2018) 68:599–615. doi: 10.1002/hep.29838
24. Li B, Xu H, Han H, Song S, Zhang X, Ouyang L, et al. Exosome-mediated transfer of lncRUNX2-AS1 from multiple myeloma cells to MSCs contributes to osteogenesis. Oncogene. (2018) 37:5508–19. doi: 10.1038/s41388-018-0359-0
25. Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. (2017) 490:406–14. doi: 10.1016/j.bbrc.2017.06.055
26. Li J, Chen J, Wang S, Li P, Zheng C, Zhou X, et al. Blockage of transferred exosome-shuttled miR-494 inhibits melanoma growth and metastasis. J Cell Physiol. (2019) 234:15763–74. doi: 10.1002/jcp.28234
27. Liu R, Tang A, Wang X, Chen X, Zhao L, Xiao Z, et al. Inhibition of lncRNA NEAT1 suppresses the inflammatory response in IBD by modulating the intestinal epithelial barrier and by exosome-mediated polarization of macrophages. Int J Mol Med. (2018) 42:2903–13. doi: 10.3892/ijmm.2018.3829
28. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. (2013) 200:373–83. doi: 10.1083/jcb.201211138
29. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. (2002) 2:569–79. doi: 10.1038/nri855
30. Kalluri R. The biology and function of exosomes in cancer. J Clin Investig. (2016) 126:1208–15. doi: 10.1172/jci81135
31. Kalluri R, LeBleu VS. The biology function and biomedical applications of exosomes. Science. (2020) 367:eaau6977. doi: 10.1126/science.aau6977
32. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. (2019) 88:487–514. doi: 10.1146/annurev-biochem-013118-111902
33. Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. (2007) 23:519–47. doi: 10.1146/annurev.cellbio.23.090506.123319
34. Appelqvist H, Wäster P, Kågedal K, Öllinger K. The lysosome: from waste bag to potential therapeutic target. J Mol Cell Biol. (2013) 5:214–26. doi: 10.1093/jmcb/mjt022
35. Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. (2019) 21:9–17. doi: 10.1038/s41556-018-0250-9
36. Guix FX, Sannerud R, Berditchevski F, Arranz AM, Horré K, Snellinx A, et al. Tetraspanin 6: a pivotal protein of the multiple vesicular body determining exosome release and lysosomal degradation of amyloid precursor protein fragments. Mol Neurodegener. (2017) 12:25. doi: 10.1186/s13024-017-0165-0
37. Villarroya-Beltri C, Baixauli F, Mittelbrunn M, Fernández-Delgado I, Torralba D, Moreno-Gonzalo O, et al. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun. (2016) 7:13588. doi: 10.1038/ncomms13588
38. Trams EG, Lauter CJ, Salem N Jr., Heine U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta. (1981) 645:63–70. doi: 10.1016/0005-2736(81)90512-5
39. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. (1983) 33:967–78. doi: 10.1016/0092-8674(83)90040-5
40. Sindi HA, Russomanno G, Satta S, Abdul-Salam VB, Jo KB, Chaudhry BQ, et al. Therapeutic potential of KLF2-induced exosomal microRNAs in pulmonary hypertension. Nat Commun. (2020) 11:1185. doi: 10.1038/s41467-020-14966-x
41. Magayr TA, Song X, Streets AJ, Vergoz L, Chang L, Valluru MK, et al. Global microRNA profiling in human urinary exosomes reveals novel disease biomarkers and cellular pathways for autosomal dominant polycystic kidney disease. Kidney Int. (2020) 98:420–35. doi: 10.1016/j.kint.2020.02.008
42. Jiao Y, Li W, Wang W, Tong X, Xia R, Fan J, et al. Platelet-derived exosomes promote neutrophil extracellular trap formation during septic shock. Crit Care. (2020) 24:380. doi: 10.1186/s13054-020-03082-3
43. Xie Y, Li J, Li P, Li N, Zhang Y, Binang H, et al. RNA-seq profiling of serum exosomal circular RNAs reveals Circ-PNN as a potential biomarker for human colorectal cancer. Front Oncol. (2020) 10:982. doi: 10.3389/fonc.2020.00982
44. Kita S, Maeda N, Shimomura I. Interorgan communication by exosomes, adipose tissue, and adiponectin in metabolic syndrome. J Clin Investig. (2019) 129:4041–9. doi: 10.1172/jci129193
45. Kwon Y, Kim M, Kim Y, Jung HS, Jeoung D. Exosomal MicroRNAs as mediators of cellular interactions between cancer cells and macrophages. Front Immunol. (2020) 11:1167. doi: 10.3389/fimmu.2020.01167
46. Zhang C, Fan Y, Che X, Zhang M, Li Z, Li C, et al. Anti-PD-1 therapy response predicted by the combination of exosomal PD-L1 and CD28. Front Oncol. (2020) 10:760. doi: 10.3389/fonc.2020.00760
47. Lovisa F, Di Battista P, Gaffo E, Damanti CC, Garbin A, Gallingani I, et al. RNY4 in circulating exosomes of patients with pediatric anaplastic large cell lymphoma: an active player? Front Oncol. (2020) 10:238. doi: 10.3389/fonc.2020.00238
48. Joyce DP, Kerin MJ, Dwyer RM. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int J Cancer. (2016) 139:1443–8. doi: 10.1002/ijc.30179
49. Weick EM, Puno MR, Januszyk K, Zinder JC, DiMattia MA, Lima CD. Helicase-dependent RNA decay illuminated by a Cryo-EM structure of a human nuclear RNA exosome-MTR4 complex. Cell. (2018) 173:1663–77.e1621. doi: 10.1016/j.cell.2018.05.041
50. Wang F, Li L, Piontek K, Sakaguchi M, Selaru FM. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology. (2018) 67:940–54. doi: 10.1002/hep.29586
51. Mukhamedova N, Hoang A. Exosomes containing HIV protein Nef reorganize lipid rafts potentiating inflammatory response in bystander cells. PLoS Pathog. (2019) 15:e1007907. doi: 10.1371/journal.ppat.1007907
52. Higuchi H, Yamakawa N, Imadome KI. Role of exosomes as a proinflammatory mediator in the development of EBV-associated lymphoma. Blood. (2018) 131:2552–67. doi: 10.1182/blood-2017-07-794529
53. Abner EL, Elahi FM, Jicha GA, Mustapic M, Al-Janabi O, Kramer JH, et al. Endothelial-derived plasma exosome proteins in Alzheimer’s disease angiopathy. FASEB J. (2020) 34:5967–74. doi: 10.1096/fj.202000034R
54. Pan X, Hong X, Lai J, Cheng L, Cheng Y, Yao M, et al. Exosomal MicroRNA-221-3p confers adriamycin resistance in breast cancer cells by targeting PIK3R1. Front Oncol. (2020) 10:441. doi: 10.3389/fonc.2020.00441
55. Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. (2010) 11:556–66. doi: 10.1038/nrm2937
56. Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol. (2013) 5:a016766. doi: 10.1101/cshperspect.a016766
57. Quinney KB, Frankel EB, Shankar R, Kasberg W, Luong P, Audhya A. Growth factor stimulation promotes multivesicular endosome biogenesis by prolonging recruitment of the late-acting ESCRT machinery. Proc Natl Acad Sci USA. (2019) 116:6858–67. doi: 10.1073/pnas.1817898116
58. Blanc L, Vidal M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases. (2018) 9:95–106. doi: 10.1080/21541248.2016.1264352
59. Gross JC, Chaudhary V, Bartscherer K, Boutros M. Active Wnt proteins are secreted on exosomes. Nat Cell Biol. (2012) 14:1036–45. doi: 10.1038/ncb2574
60. Raiborg C, Wenzel EM, Pedersen NM, Olsvik H, Schink KO, Schultz SW, et al. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. (2015) 520:234–8. doi: 10.1038/nature14359
61. Zhu X, Yin J, Liang S, Liang R, Zhou X, Chen Z, et al. The multivesicular bodies (MVBs)-localized AAA ATPase LRD6-6 inhibits immunity and cell death likely through regulating MVBs-mediated vesicular trafficking in rice. PLoS Genet. (2016) 12:e1006311. doi: 10.1371/journal.pgen.1006311
62. Hammerling BC, Najor RH, Cortez MQ, Shires SE, Leon LJ, Gonzalez ER, et al. A Rab5 endosomal pathway mediates Parkin-dependent mitochondrial clearance. Nat Commun. (2017) 8:14050. doi: 10.1038/ncomms14050
63. Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. (1993) 362:318–24. doi: 10.1038/362318a0
64. Heider MR, Munson M. Exorcising the exocyst complex. Traffic. (2012) 13:898–907. doi: 10.1111/j.1600-0854.2012.01353.x
65. Yue P, Zhang Y, Mei K, Wang S, Lesigang J, Zhu Y, et al. Sec3 promotes the initial binary t-SNARE complex assembly and membrane fusion. Nat Commun. (2017) 8:14236. doi: 10.1038/ncomms14236
66. Hyenne V, Apaydin A, Rodriguez D, Spiegelhalter C, Hoff-Yoessle S, Diem M, et al. RAL-1 controls multivesicular body biogenesis and exosome secretion. J Cell Biol. (2015) 211:27–37. doi: 10.1083/jcb.201504136
67. Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. (2014) 3:24641. doi: 10.3402/jev.v3.24641
68. Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, et al. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem. (2014) 289:22258–67. doi: 10.1074/jbc.M114.588046
69. Yang Y, Shen G, Wang H, Li H, Zhang T, Tao N, et al. Interferometric plasmonic imaging and detection of single exosomes. Proc Natl Acad Sci USA. (2018) 115:10275–80. doi: 10.1073/pnas.1804548115
70. Nanbo A, Kawanishi E, Yoshida R, Yoshiyama H. Exosomes derived from Epstein-Barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. J Virol. (2013) 87:10334–47. doi: 10.1128/jvi.01310-13
71. Glebov OO, Bright NA, Nichols BJ. Flotillin-1 defines a clathrin-independent endocytic pathway in mammalian cells. Nat Cell Biol. (2006) 8:46–54. doi: 10.1038/ncb1342
72. Kobayashi J, Hasegawa T, Sugeno N, Yoshida S, Akiyama T, Fujimori K, et al. Extracellular alpha-synuclein enters dopaminergic cells by modulating flotillin-1-assisted dopamine transporter endocytosis. FASEB J. (2019) 33:10240–56. doi: 10.1096/fj.201802051R
73. Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer – implications for future improvements in cancer care. Nat Rev Clin Oncol. (2018) 15:617–38. doi: 10.1038/s41571-018-0036-9
74. Li L, Li C, Wang S, Wang Z, Jiang J, Wang W, et al. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. (2016) 76:1770–80. doi: 10.1158/0008-5472.can-15-1625
75. Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. (2012) 18:883–91. doi: 10.1038/nm.2753
76. Zhu L, Li J, Gong Y, Wu Q, Tan S, Sun D, et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer. (2019) 18:74. doi: 10.1186/s12943-019-1000-8
77. Li C, Lv Y, Shao C, Chen C, Zhang T, Wei Y, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. (2019) 234:20721–7. doi: 10.1002/jcp.28678
78. Sharma A, Johnson A. Exosome DNA: critical regulator of tumor immunity and a diagnostic biomarker. J Cell Physiol. (2019) 235:1921–32. doi: 10.1002/jcp.29153
79. In ’t Veld S, Wurdinger T. Tumor-educated platelets. Blood. (2019) 133:2359–64. doi: 10.1182/blood-2018-12-852830
80. Pulliam L, Sun B, Mustapic M, Chawla S, Kapogiannis D. Plasma neuronal exosomes serve as biomarkers of cognitive impairment in HIV infection and Alzheimer’s disease. J Neurovirol. (2019) 25:702–9. doi: 10.1007/s13365-018-0695-4
81. Chen T, Wang C, Yu H, Ding M, Zhang C, Lu X, et al. Increased urinary exosomal microRNAs in children with idiopathic nephrotic syndrome. EBioMedicine. (2019) 39:552–61. doi: 10.1016/j.ebiom.2018.11.018
82. Mori MA, Ludwig RG, Garcia-Martin R, Brandao BB, Kahn CR. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. (2019) 30:656–73. doi: 10.1016/j.cmet.2019.07.011
83. Zou J, Shi M, Liu X, Jin C, Xing X, Qiu L. Aptamer-functionalized exosomes: elucidating the cellular uptake mechanism and the potential for cancer-targeted chemotherapy. Anal Chem. (2019) 91:2425–30. doi: 10.1021/acs.analchem.8b05204
84. Zhang ZG, Buller B, Chopp M. Exosomes – beyond stem cells for restorative therapy in stroke and neurological injury. Nat Rev Neurol. (2019) 15:193–203. doi: 10.1038/s41582-018-0126-4
85. Abbaszadeh H, Ghorbani F, Derakhshani M, Movassaghpour A, Yousefi M. Human umbilical cord mesenchymal stem cell-derived extracellular vesicles: a novel therapeutic paradigm. J Cell Physiol. (2019) 235:706–17. doi: 10.1002/jcp.29004
86. Yang B, Chen Y, Shi J. Exosome biochemistry and advanced nanotechnology for next-generation theranostic platforms. Adv Mater. (2019) 31:e1802896. doi: 10.1002/adma.201802896
87. Tran PHL, Xiang D, Tran TTD, Yin W, Zhang Y, Kong L, et al. Exosomes and nanoengineering: a match made for precision therapeutics. Adv Mater. (2019) 32:e1904040. doi: 10.1002/adma.201904040
88. Rehman FU, Du T, Shaikh S, Jiang X, Chen Y, Li X, et al. Nano in nano: biosynthesized gold and iron nanoclusters cargo neoplastic exosomes for cancer status biomarking. Nanomedicine. (2018) 14:2619–31. doi: 10.1016/j.nano.2018.07.014
89. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. (2012) 489:101–8. doi: 10.1038/nature11233
90. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. (2012) 22:1775–89. doi: 10.1101/gr.132159.111
91. Sarropoulos I, Marin R, Cardoso-Moreira M, Kaessmann H. Developmental dynamics of lncRNAs across mammalian organs and species. Nature. (2019) 571:510–4. doi: 10.1038/s41586-019-1341-x
92. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. (2015) 47:199–208. doi: 10.1038/ng.3192
93. Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. (2015) 11:1110–22. doi: 10.1016/j.celrep.2015.04.023
94. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. (2011) 25:1915–27. doi: 10.1101/gad.17446611
95. McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. (2015) 521:232–6. doi: 10.1038/nature14443
96. Nojima T, Tellier M, Foxwell J, Ribeiro de Almeida C, Tan-Wong SM, Dhir S, et al. Deregulated expression of mammalian lncRNA through loss of SPT6 induces R-loop formation, replication stress, and cellular senescence. Mol Cell. (2018) 72:970–84.e977. doi: 10.1016/j.molcel.2018.10.011
97. Kim DH, Sung S. Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev Cell. (2017) 40:302–12.e304. doi: 10.1016/j.devcel.2016.12.021
98. Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. (2019) 21:542–51. doi: 10.1038/s41556-019-0311-8
99. Yu F, Zhang G, Shi A, Hu J, Li F, Zhang X, et al. LnChrom: a resource of experimentally validated lncRNA-chromatin interactions in human and mouse. Database (2018) 2018:1–7. doi: 10.1093/database/bay039
100. Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. (2018) 561:132–6. doi: 10.1038/s41586-018-0453-z
101. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
102. Zhuang M, Zhao S, Jiang Z, Wang S, Sun P, Quan J, et al. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBioMedicine. (2019) 41:286–98. doi: 10.1016/j.ebiom.2018.12.049
103. Wang YG, Wang T, Shi M, Zhai B. Long noncoding RNA EPB41L4A-AS2 inhibits hepatocellular carcinoma development by sponging miR-301a-5p and targeting FOXL1. J Exp Clin Cancer Res. (2019) 38:153. doi: 10.1186/s13046-019-1128-9
104. Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. (2018) 17:141. doi: 10.1186/s12943-018-0894-x
105. Sun Z, Ou C, Liu J, Chen C, Zhou Q, Yang S, et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. (2019) 38:2627–44. doi: 10.1038/s41388-018-0628-y
106. Zhang G, Wang Q, Zhang X, Ding Z, Liu R. LncRNA FENDRR suppresses the progression of NSCLC via regulating miR-761/TIMP2 axis. Biomed Pharmacother. (2019) 118:109309. doi: 10.1016/j.biopha.2019.109309
107. Liu Z, Zhu Y, Xu L, Zhang J, Xie H, Fu H, et al. Tumor stroma-infiltrating mast cells predict prognosis and adjuvant chemotherapeutic benefits in patients with muscle invasive bladder cancer. Oncoimmunology. (2018) 7:e1474317. doi: 10.1080/2162402x.2018.1474317
108. Zhao J, Hao S, Wang L, Hu C, Zhang S, Guo L, et al. Long non-coding RNA ANRIL promotes the invasion and metastasis of thyroid cancer cells through TGF-β/Smad signaling pathway. Oncotarget. (2016) 7:57903–18. doi: 10.18632/oncotarget.11087
109. Zhuo W, Liu Y, Li S, Guo D, Sun Q, Jin J, et al. Long noncoding RNA GMAN, Up-regulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of Ephrin A1 by competitively binding GMAN-AS. Gastroenterology. (2019) 156:676–91.e611. doi: 10.1053/j.gastro.2018.10.054
110. Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. (2011) 470:284–8. doi: 10.1038/nature09701
111. Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins. Trends Cell Biol. (2018) 28:113–27. doi: 10.1016/j.tcb.2017.10.001
112. Li S, Mi L, Yu L, Yu Q, Liu T, Wang GX, et al. Zbtb7b engages the long noncoding RNA Blnc1 to drive brown and beige fat development and thermogenesis. Proc Natl Acad Sci USA. (2017) 114:E7111–20. doi: 10.1073/pnas.1703494114
113. Peng WX, Koirala P, Zhang W, Ni C, Wang Z, Yang L, et al. lncRNA RMST enhances DNMT3 expression through interaction with HuR. Mol Ther. (2020) 28:9–18. doi: 10.1016/j.ymthe.2019.09.024
114. Cai Q, Wang S, Jin L, Weng M, Zhou D, Wang J, et al. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol Cancer. (2019) 18:82. doi: 10.1186/s12943-019-1016-0
115. Zhang J, Wang P, Wan L, Xu S, Pang D. The emergence of noncoding RNAs as Heracles in autophagy. Autophagy. (2017) 13:1004–24. doi: 10.1080/15548627.2017.1312041
116. Xu S, Wang P, Zhang J, Wu H, Sui S, Zhang J, et al. Ai-lncRNA EGOT enhancing autophagy sensitizes paclitaxel cytotoxicity via upregulation of ITPR1 expression by RNA-RNA and RNA-protein interactions in human cancer. Mol Cancer. (2019) 18:89. doi: 10.1186/s12943-019-1017-z
117. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. (2016) 537:369–73. doi: 10.1038/nature19342
118. Brockdorff N, Bowness JS, Wei G. Progress toward understanding chromosome silencing by Xist RNA. Genes Dev. (2020) 34:733–44. doi: 10.1101/gad.337196.120
119. Dong X, Yang Z, Yang H, Li D, Qiu X. Long Non-coding RNA MIR4435-2HG promotes colorectal cancer proliferation and metastasis through miR-206/YAP1 Axis. Front Oncol. (2020) 10:160. doi: 10.3389/fonc.2020.00160
120. Calixto CPG, Tzioutziou NA, James AB, Hornyik C, Guo W, Zhang R, et al. Cold-dependent expression and alternative splicing of Arabidopsis long non-coding RNAs. Front Plant Sci. (2019) 10:235. doi: 10.3389/fpls.2019.00235
121. Chen R, Zhang X, Wang C. LncRNA HOXB-AS1 promotes cell growth in multiple myeloma via FUT4 mRNA stability by ELAVL1. J Cell Biochem. (2019) 121:4043–51. doi: 10.1002/jcb.29573
122. Wang S, Li X, Zhu R, Han Q, Zhao RC. Lung cancer exosomes initiate global long non-coding RNA changes in mesenchymal stem cells. Int J Oncol. (2016) 48:681–9. doi: 10.3892/ijo.2015.3272
123. Feng T, Zhang P, Sun Y, Wang Y, Tong J, Dai H, et al. High throughput sequencing identifies breast cancer-secreted exosomal LncRNAs initiating pulmonary pre-metastatic niche formation. Gene. (2019) 710:258–64. doi: 10.1016/j.gene.2019.06.004
124. Kahroba H, Hejazi MS, Samadi N. Exosomes: from carcinogenesis and metastasis to diagnosis and treatment of gastric cancer. Cell Mol Life Sci. (2019) 76:1747–58. doi: 10.1007/s00018-019-03035-2
125. Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. (2013) 27:2192–206. doi: 10.1101/gad.225334.113
126. Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W, et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol. (2017) 143:991–1004. doi: 10.1007/s00432-017-2361-2
127. Li Z, Jiang P, Li J, Peng M, Zhao X, Zhang X, et al. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene. (2018) 37:3822–38. doi: 10.1038/s41388-018-0237-9
128. Wang B, Tan Z, Guan F. Tumor-derived exosomes mediate the instability of cadherins and promote tumor progression. Int J Mol Sci. (2019) 20:3652. doi: 10.3390/ijms20153652
129. Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med. (2013) 91:791–801. doi: 10.1007/s00109-013-1028-y
130. Zhang P, Zhou H, Lu K, Lu Y, Wang Y, Feng T. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. OncoTargets Ther. (2018) 11:291–9. doi: 10.2147/ott.s155134
131. Cui Y, Fu S, Sun D, Xing J, Hou T, Wu X. EPC-derived exosomes promote osteoclastogenesis through LncRNA-MALAT1. J Cell Mol Med. (2019) 23:3843–54. doi: 10.1111/jcmm.14228
132. Cooper DR, Wang C, Patel R, Trujillo A, Patel NA, Prather J, et al. Human adipose-derived stem cell conditioned media and exosomes containing MALAT1 promote human dermal fibroblast migration and ischemic wound healing. Adv Wound Care. (2018) 7:299–308. doi: 10.1089/wound.2017.0775
133. Cheng Y, Dai X, Yang T, Zhang N, Liu Z, Jiang Y. Low long noncoding RNA growth arrest-specific transcript 5 expression in the exosomes of lung cancer cells promotes tumor angiogenesis. J Oncol. (2019) 2019:2476175. doi: 10.1155/2019/2476175
134. Xia H, Xue J, Xu H, Lin M, Shi M, Sun Q, et al. Andrographolide antagonizes the cigarette smoke-induced epithelial-mesenchymal transition and pulmonary dysfunction through anti-inflammatory inhibiting HOTAIR. Toxicology. (2019) 42:284–94. doi: 10.1016/j.tox.2019.05.009
135. Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. (2013) 436:319–24. doi: 10.1016/j.bbrc.2013.05.101
136. Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C, et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer. (2018) 17:74. doi: 10.1186/s12943-018-0822-0
137. Wu F, Yin Z, Yang L, Fan J, Xu J, Jin Y, et al. Smoking induced extracellular vesicles release and their distinct properties in non-small cell lung cancer. J Cancer. (2019) 10:3435–43. doi: 10.7150/jca.30425
138. Zhang C, Xu L, Deng G, Ding Y, Bi K, Jin H, et al. Exosomal HOTAIR promotes proliferation, migration and invasion of lung cancer by sponging miR-203. Sci China Life Sci. (2020) 63:1265–8. doi: 10.1007/s11427-019-1579-x
139. Chen CW, Fu M, Du ZH, Zhao F, Yang WW, Xu LH, et al. Long noncoding RNA MRPL23-AS1 promoteoid cystic carcinoma lung metastasis. Cancer Res. (2020) 80:2273–85. doi: 10.1158/0008-5472.Can-19-0819
140. Hu B, Zhang H, Wang Z, Zhang F, Wei H, Li L. LncRNA CCAT1/miR-130a-3p axis increases cisplatin resistance in non-small-cell lung cancer cell line by targeting SOX4. Cancer Biol Ther. (2017) 18:974–83. doi: 10.1080/15384047.2017.1385679
141. Sun W, Zu Y, Fu X, Deng Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol Rep. (2017) 38:3347–54. doi: 10.3892/or.2017.6056
142. Li B, Gu W, Zhu X. NEAT1 mediates paclitaxel-resistance of non-small cell of lung cancer through activation of Akt/mTOR signalling pathway. J Drug Target. (2019) 27:1061–7. doi: 10.1080/1061186x.2019.1585437
143. Kang M, Ren M, Li Y, Fu Y, Deng M, Li C. Exosome-mediated transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell carcinoma via functioning as a competing endogenous RNA. J Exp Clin Cancer Res. (2018) 37:171. doi: 10.1186/s13046-018-0845-9
144. Zhang Z, Yin J, Lu C, Wei Y, Zeng A, You Y. Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. J Exp Clin Cancer Res. (2019) 38:166. doi: 10.1186/s13046-019-1139-6
145. Lei Y, Guo W, Chen B, Chen L, Gong J, Li W. Tumorreleased lncRNA H19 promotes gefitinib resistance via packaging into exosomes in nonsmall cell lung cancer. Oncol Rep. (2018) 40:3438–46. doi: 10.3892/or.2018.6762
146. Pan R, Zhou H. Exosomal transfer of lncRNA H19 promotes erlotinib resistance in non-small cell lung cancer via miR-615-3p/ATG7 Axis. Cancer Manage Res. (2020) 12:4283–97. doi: 10.2147/cmar.S241095
147. Zhang W, Cai X, Yu J, Lu X, Qian Q, Qian W. Exosome-mediated transfer of lncRNA RP11838N2.4 promotes erlotinib resistance in non-small cell lung cancer. Int J Oncol. (2018) 53:527–38. doi: 10.3892/ijo.2018.4412
148. Deng Q, Fang Q, Xie B, Sun H, Bao Y, Zhou S. Exosomal long non-coding RNA MSTRG.292666.16 is associated with osimertinib (AZD9291) resistance in non-small cell lung cancer. Aging. (2020) 12:8001–15. doi: 10.18632/aging.103119
149. Chen X, Wang Z, Tong F, Dong X, Wu G, Zhang R. lncRNA UCA1 promotes gefitinib resistance as a ceRNA to target FOSL2 by sponging miR-143 in non-small cell lung cancer. Mol Ther Nucleic Acids. (2020) 19:643–53. doi: 10.1016/j.omtn.2019.10.047
150. Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S, et al. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science. (2014) 343:1248636. doi: 10.1126/science.1248636
151. Huang JZ, Chen M, Chen D, Gao XC, Zhu S, Huang H, et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell. (2017) 68:171–84.e176. doi: 10.1016/j.molcel.2017.09.015
152. Ahadi A, Brennan S, Kennedy PJ, Hutvagner G, Tran N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci Rep. (2016) 6:24922. doi: 10.1038/srep24922
153. Macdonald J, Henri J, Goodman L, Xiang D, Duan W, Shigdar S. Development of a bifunctional aptamer targeting the transferrin receptor and epithelial cell adhesion molecule (EpCAM) for the treatment of brain cancer metastases. ACS Chem Neurosci. (2017) 8:777–84. doi: 10.1021/acschemneuro.6b00369
154. Zhang X, Guo H, Bao Y, Yu H, Xie D, Wang X. Exosomal long non-coding RNA DLX6-AS1 as a potential diagnostic biomarker for non-small cell lung cancer. Oncol Lett. (2019) 18:5197–204. doi: 10.3892/ol.2019.10892
155. Sai L, Yu G, Bo C, Zhang Y, Du Z, Li C, et al. Profiling long non-coding RNA changes in silica-induced pulmonary fibrosis in rat. Toxicol Lett. (2019) 310:7–13. doi: 10.1016/j.toxlet.2019.04.003
156. Tang Q, Ni Z, Cheng Z, Xu J, Yu H, Yin P. Three circulating long non-coding RNAs act as biomarkers for predicting NSCLC. Cell Physiol Biochem. (2015) 37:1002–9. doi: 10.1159/000430226
157. Teng Y, Kang H, Chu Y. Identification of an exosomal long noncoding RNA SOX2-OT in plasma as a promising biomarker for lung squamous cell carcinoma. Genet Test Mol Biomarkers. (2019) 23:235–40. doi: 10.1089/gtmb.2018.0103
Keywords: exosome, lncRNA, lung cancer, biomarkers, intercellular communication
Citation: Fan T, Sun N and He J (2020) Exosome-Derived LncRNAs in Lung Cancer. Front. Oncol. 10:1728. doi: 10.3389/fonc.2020.01728
Received: 16 May 2020; Accepted: 03 August 2020;
Published: 23 September 2020.
Edited by:
Jessy Deshane, University of Alabama at Birmingham, United StatesReviewed by:
Runsheng Chen, Institute of Biophysics (CAS), ChinaCristina Teixido, Hospital Clínic de Barcelona, Spain
Copyright © 2020 Fan, Sun and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Sun, c3VubmFuQHZpcC4xMjYuY29t; Jie He, cHJvZi5qaWVoZUBnbWFpbC5jb20=
 Tao Fan
Tao Fan Nan Sun2*
Nan Sun2* Jie He
Jie He