
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Oncol. , 26 August 2020
Sec. Cancer Metabolism
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.01401
This article is part of the Research Topic Annual Meeting of the International Society of Cancer Metabolism (ISCaM)- Cancer Metabolic Rewiring: Mapping the Road to Clinical Translation View all 12 articles
The International Society of Cancer Metabolism (ISCaM) meeting on Cancer Metabolic Rewiring, held in Braga Portugal in October 2019, provided an outstanding forum for investigators to present current findings and views, and discuss ideas and future directions on fundamental biology as well as clinical translations. The first session on Cancer pH Dynamics was preceded by the opening keynote presentation from our group entitled Intracellular pH Regulation of Protein Dynamics: From Cancer to Stem Cell Behaviors. In this review we introduce a brief background on intracellular pH (pHi) dynamics, including how it is regulated as well as functional consequences, summarize key findings included in our presentation, and conclude with perspectives on how understanding the role of pHi dynamics in stem cells can be relevant for understanding how pHi dynamics enables cancer progression.
Intracellular pH (pHi) was previously thought to be mostly constant for cellular homeostasis and possibly dysregulated in diseases. We now know, however, that pHi is dynamic in normal cells and clearly dysregulated in a number of diseases. In normal cells, pHi changes during cell cycle progression, increasing ~0.3–0.4 pH units at the end of S phase and if this increase is blocked, G2/M is delayed with increased inhibitory phosphorylation of Cdk1-Tyr15 and suppressed cyclin B1 expression (1–3). Additionally, pHi dynamics regulates cell-substrate adhesion remodeling and migration, with increased pHi enabling both behaviors (4–7). Emerging evidence also indicates a critical role for increased pHi in epithelial plasticity, including epithelial to mesenchymal transition (EMT) (8), and stem cell differentiation (9–12). Moreover, it is now well-established that dysregulated pHi is seen with many diseases, most notably cancers, which often have a constitutively increased pHi (13–18), and neurodegenerative disorders, which are associated with a constitutively decreased pHi (19, 20). Our review focuses on dysregulated pHi dynamics in cancer; however, another feature of cancers is a dysregulated extracellular pH that is lower (~7.0) compared with normal tissues (~7.4).
Although many factors contribute to pHi dynamics, the major regulators in most mammalian cells are plasma membrane ion exchangers, including the Na+-H+ exchanger NHE1, the Na+- transporter NBC, and the Na+-dependent Cl−- transporter NDCBE, which are acid-extruders, and Cl−- exchangers of the anion exchanger (AE) family, which are acid loaders (21–23). The BioParadigms Solute Carrier tables1 are an excellent resource on the classification, expression, and transport characteristics of these ion exchangers. Additional plasma membrane ion transport proteins that contribute to pHi dynamics, albeit to less of an extent, include V-ATPases and monocarboxylate transporters of the MCT family. The broad range of ion transport proteins regulate pHi dynamics through changes in their expression and activity, the latter mostly mediated by posttranslational modifications as many are substrates of key signaling kinases, including for NHE1, p90rsk (24), Akt (25, 26), the Rho kinase ROCK (27), and the Ste20 kinase MAP4K4 (28), previously termed NIK. Experimentally, these exchangers can be pharmacologically or genetically targeted to understand how they contribute to pHi dynamics and how pHi dynamics regulates cell behaviors.
We have a relatively strong understanding of how changes in pHi are generated and the effects of pHi changes on myriad cell functions. However, a mechanistic understanding of how pHi changes regulate cell behaviors remains understudied, particularly effects on signaling networks and protein functions. At the ISCaM meeting we presented our work on how changes in pHi regulate protein dynamics to enable cancer and stem cell behaviors, which we summarize in this review. Key to pH-regulated protein structure and function is considering protonation and deprotonation as a protein posttranslational modification, analogous to posttranslational modification by phosphorylation, acetylation, and methylation as we previously described (29). However, studying protonation and deprotonation as a posttranslational modification is more difficult compared with other posttranslational modifications because it is not catalyzed by an enzyme and cannot be detected by mass spectrometry or antibodies. Furthermore, many endogenous “pH sensors” or proteins that are regulated by pH dynamics within the cellular range are coincidence (AND-gate) detectors with their structural conformations, activities, or binding affinities dependent on multiple posttranslational modifications, most commonly phosphorylation or dephosphorylation and protonation or deprotonation.
Most cancer cells have a higher pHi compared with untransformed cells, regardless of the mutational landscape or tissue origin. This higher pHi enables many cancer behaviors, including increased proliferation, directional migration, tumorigenesis, and most recently recognized, the oncogenic and tumor-suppressor functions of proteins with charge-changing mutations (Figure 1). At the ISCaM meeting we presented our findings on pH sensors regulating cell migration and tumorigenesis as well as how pHi dynamics in cancer cells affect the functions of proteins with somatic mutations encoding arginine to histidine substitutions.
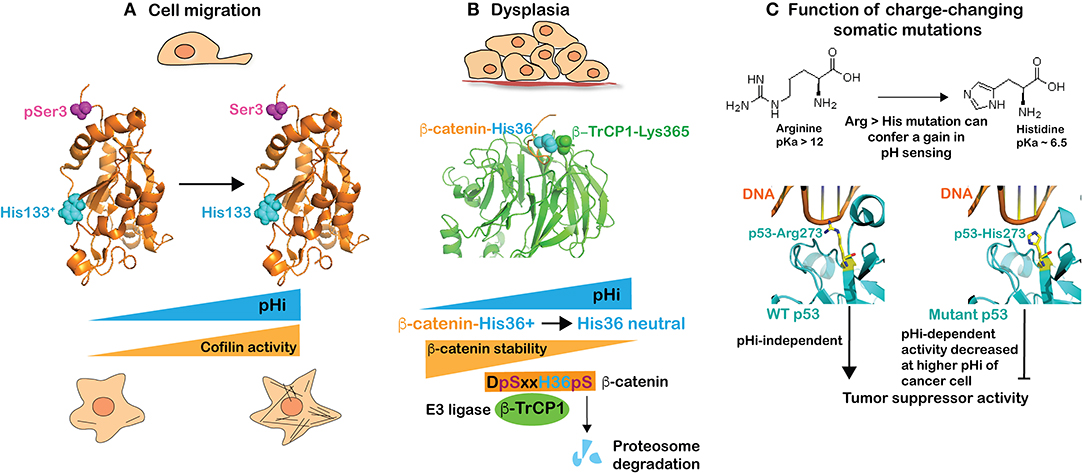
Figure 1. The higher pHi of cancer cells enables many behaviors, including directional migration and tumorigenesis as well as the tumorigenic functions of proteins with charge-changing arginine to histidine mutations. (A) Cell migration is in part dependent on increased activity of cofilin with increased pHi. Cofilin is a coincidence-regulated pH sensor that is activated by deprotonation of His133 (cyan) and dephosphorylation of Ser3 (magenta) for actin polymerization enabling cell migration. (B) Dysplasia is associated with increased pHi, which decreases β-catenin stability. β-catenin is a coincidence-regulated pH sensor with deprotonation of His36 (cyan) and phosphorylation of Ser33/37 by GSK3β enabling binding to the E3 ligase β-TrCP1 for targeting to the proteasome for degradation. Crystal structure data show that β-catenin-His36 is in close proximity to β-TrCP1-Lys365, which suggest that binding would be electrostatically unfavorable with a protonated His36 at lower pHi. (C) Charge changing somatic mutations can confer pH-regulated protein activity. Structure of wild-type p53 (left) and mutant p53-R273H (right) in complex with DNA indicating an electrostatic interaction of Arg273 with the negatively charged phosphate-backbone of DNA that could be partially enabled by protonated, but not neutral, His273.
Cell migration is confirmed to be regulated by pHi in many cell types and species (6, 30–34). An increased pHi of ~0.3–0.4 units is seen in migrating cells and preventing the increased pHi inhibits migratory rate and directionality, and impairs cell polarity. Our presentation described several pH sensors we identified in atomistic detail that collectively regulate different aspects of migration. These include guanine nucleotide exchange factors for the low molecular weight GTPase Cdc42 involved in cell polarity (35), talin binding to actin filaments (36), and focal adhesion kinase (FAK) activity for cell-substrate adhesion dynamics (5) as well as cofilin for actin polymerization (37). The single histidine in cofilin, His133 (human), has an upshifted pKa to ~ 7.2 and must be neutral for increased cofilin activity (Figure 1A). However, cofilin is a coincidence detector and full activity also requires dephosphorylation of Ser3 (Figure 1A) by one of several phosphatases, which releases an autoinhibited interaction between phosphorylated serine and lysine 126 and 127 to allow binding to actin filaments. This AND-gate regulation enables signaling mechanisms to increase cofilin activity in time (with migratory cues) and space (at the leading edge of a migrating cell), and highlights that for many pH sensors a change in protonation state does not function as a binary switch.
Tumorigenesis and dysplasia are enabled by increased pHi regulated by NHE1, NBCs, and MCTs, including tumor cell proliferation, growth, and survival (38–40). Our presentation included two of our recent key findings on pHi and tumorigenesis. First, that increased pHi from ~7.30 to ~7.65 in Drosophila eye epithelia by overexpressing Drosophila dnhe2, an ortholog of mammalian NHE1, is sufficient to induce dysplasia in the absence of an activated oncogene (41). Second, that β-catenin, an adherens junction and Wnt pathway protein, is a pH sensor, with pHi not regulating its activity but rather its stability, which decreases at pHi > 7.5 (42). Using a phenotype screen, we found that overexpressing β-catenin suppresses dysplasia in Drosophila eye epithelia with constitutively increased pHi induced by overexpression of dnhe2. These data suggested a lower abundance of β-catenin at higher pHi, which we confirmed in mammalian cells. We also resolved the pH sensing mechanism of His36 (human) in the N-terminus of β-catenin, which when neutral (at higher pHi) increases binding affinity for the E3 ligase β-TrCP1. However, like cofilin described above, β-catenin is a coincidence detector requiring both a neutral His36 and phosphorylated flanking Ser33 and Ser37 for binding β-TrCP1 (Figure 1B). The role of phosphorylated serines in enabling proteasome-mediated degradation of β-catenin has long been recognized (43). The importance of a neutral His36 for binding β-TrCP1 is evident in the crystal structure of β-TrCP1in complex with an N-terminal β-catenin peptide (44) (PDB: 1P22), which shows the proximity of β-catenin-His36 and β-TrCP1-Lys365 (Figure 1B). This suggests that binding would be electrostatically unfavorable with a protonated His36 at lower pHi. Importantly, the DSxxHS motif is conserved in β-catenin across species and occurs in a number of other β-TrCP1 target proteins (45), including the transmembrane protein polycystin 2, the tumor suppressor tensin 2, the centrosomal protein Cep97, the hedgehog pathway protein Gli3, and myosin-XVIIIa, suggesting these substrates may have similar pH sensitive binding to β-TrCP1 and regulated protein stability. We also described that a cancer-associated somatic mutation, β-catenin-H36R, is insensitive to pHi-regulated degradation and, when expressed in Drosophila eye epithelia, enhances Wnt pathway activity, causes tissue overgrowth growth, and induces ectopic tumors. With this mutation, β-catenin stability could be retained at the higher pHi of a cancer cell and enable tumorigenesis. As described in the section below, this is an example of a charge-changing mutation that confers a loss of pH sensing.
Charge-changing somatic mutations can confer a change in pH sensing and enable cancer behaviors specifically at increased pHi. We recently showed that recurrent arginine to histidine mutations in p53 and EGFR can confer a gain in pH sensing to the mutant proteins. Arginine, with a pKa of 12, will be protonated regardless of pHi while histidine, with a pKa near neutral, can titrate with cellular changes in pHi. We found that a highly recurrent arginine to histidine mutation in the tumor suppressor p53 (p53-R273H) could confer pH-dependent DNA binding and transcription of p53 target genes, with decreased transcription at a higher pHi of 7.6 compared with 7.2 (46). The crystal structure of wild-type p53 (47) (PDB: 4HJE) and mutant p53-R273H (48) (PDB: 4IBW) in complex with DNA suggests that wild-type Arg273 forms an electrostatic interaction with the negatively charged phosphate-backbone of DNA (Figure 1C). At the lower pHi of a non-transformed cell, His273 is likely protonated and retains some binding to the negatively-charged DNA but, at the higher pHi of a cancer cell, His273 is likely deprotonated, reducing DNA binding and expression of p53 target genes (Figure 1C). Importantly, lowering pHi in cancer cells expressing p53-R273H recovered p53 transcriptional activity and p53-dependent cell death in response to double-strand breaks (46). We also showed that a cancer-associated arginine to histidine substitution in the epidermal growth factor receptor (EGFR-R776H) that is recurrent in lung cancers confers pH sensing to the mutant protein. Increasing pHi from 7.2 to 7.6 increases activity of EGFR-R776H but not wild-type receptor, and increases cell proliferation and cellular transformation in cells expressing the mutant but not wild-type receptor (46). These results suggest that charge-changing mutations can confer a gain in pH-sensing not seen with the wild-type protein. This work also indicates that charge-changing somatic mutations can confer dynamic function to mutant proteins, specifically inactivating a tumor suppressor and specifically activating an oncogene at the increased pHi of cancer.
Recent findings indicate that pHi dynamics is a key regulator of epithelial plasticity, with increased pHi enabling EMT (8) and epithelial branching morphogenesis (49) as well as differentiation of melanocytes (50), embryonic and adult stem cells (9, 11), and mesenchymal (12) and cardiomyocyte (10) stem cells. These findings raise questions on the role of pHi dynamics in morphogenesis and animal development, which remain largely unresolved. New genetically-encoded tools to measure pHi (51) and genetic and pharmacological approaches to selectively change pHi temporally and spatially will enable new studies necessary to resolve pHi-regulated developmental processes with promise for new approaches to correct impaired morphogenesis.
Toward a goal of resolving the role of pHi dynamics in cell fate decisions, at the ISCaM meeting we discussed our findings on pHi-regulated embryonic and adult stem cell differentiation. As we previously described (11), with differentiation of naïve clonal mouse embryonic stem cells (mESC) to primed epiblast-like cells there is an NHE1-dependent transient increase in pHi of ~ 0.3 units (Figure 2A). Preventing this increase in pHi blocks differentiation, as indicated by sustained expression of the mESC markers Rex1, Stra8, and Nanog, and attenuated expression of the epiblast markers Brachyury, fibroblast growth factor 5, and Pax6. An increase in pHi is also necessary for differentiation of adult follicle stem cells in the Drosophila ovary to prefollicle cells and follicle cells (9, 11) (Figure 2B), the later necessary for germ cell maturation. Consistent with germ cells requiring enrichment from differentiated follicle cells, preventing the increase in pHi along the follicle stem cell lineage impairs ovary morphology and adult oogenesis and substantially decreases fertility (9). These findings were obtained by genetically silencing Drosophila dnhe2, an acid extruder, or overexpressing a newly identified Drosophila ae2, an ortholog of the mammalian acid loader AE2.
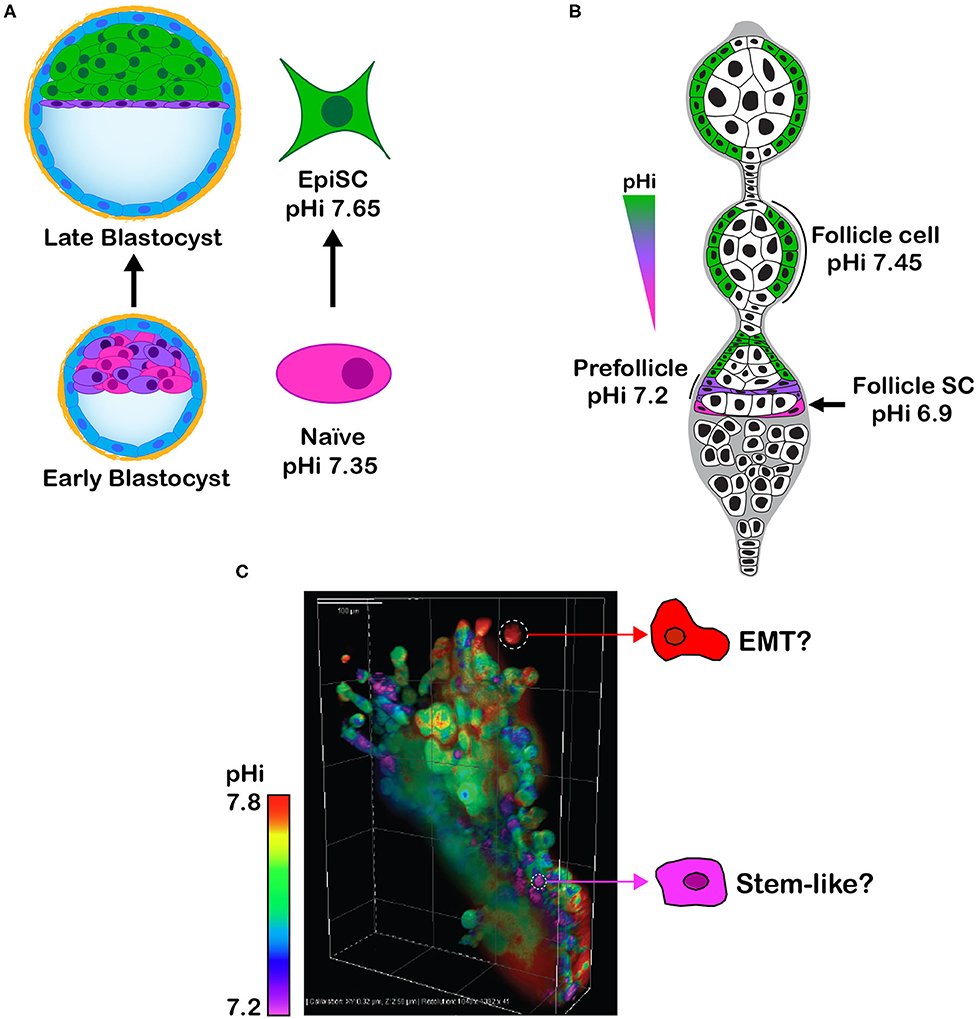
Figure 2. (A) Schematic showing that clonal self-renewing mESC (Naïve), derived from the inner cell mass of the early blastocyst, have a lower pHi than differentiated primed epiblast-like stem cells (EpiSC), which are analogous to cells in the late epiblast stage. (B) Schematic of Drosophila germarium showing an increase in pHi from self-renewing follicle stem cell (Follicle SC) to differentiated prefollicle and follicle cell. (C) Image of lung cancer H1299 cells expressing the pHi biosensor mCherry-pHluorin and grown in Matrigel as 3D spheroids shows intracellular pHi heterogeneity that might reflect phenotypic heterogeneity, such as cells with a higher pHi undergoing EMT and cells with a lower pHi being self-renewing tumor initiating stem-like cells.
There are several important questions to resolve on the role of pHi dynamics in stem cell differentiation. First is whether pHi is a conserved regulator of stem cell differentiation in different tissues, perhaps using established and well-characterized models for intestinal epithelial (52) and skin epidermal (53) stem cell lineages. Second is how pHi dynamics regulates activity of pathways and functions of proteins with established roles in stem cell behaviors. One possibility is a role for pH sensing by β-catenin (as described above) in Wnt signaling, because high Wnt pathway activity (54) at low pHi may retain self-renewal of stem cells and inhibit differentiation. Third is whether pHi-regulated stem cell differentiation can inform regenerative medicine approaches to correct or restore impaired cell and tissue functions.
To consider how pHi dynamics in stem cells and cancer might be linked, we concluded our presentation by showing new data on pHi heterogeneity in spheroids of clonal human lung cancer cells (Figure 2C). Using H1299 cells expressing the previously described (41) genetically encoded and ratiometric pH biosensor mCherry-pHluorin, we observe distinct intercellular differences in pHi when grown in 3D (Figure 2C). Distinct pH heterogeneity (including intracellular and extracellular pH) is seen in cancer spheroids (55–58) and a mouse model of breast ductal carcinoma (59); however, whether this heterogeneity reflects differences in mutational signatures, cell identity, phenotypes, or epithelial or metabolic plasticity remains unresolved. For example, might cells with a lower pHi be stem-like tumor initiating cells? Could cells with a higher pHi have increased glycolysis to fuel rapid proliferation or be undergoing EMT for metastasis? The possibility that a lower pHi could enable tumor initiating cells raises caution on the idea of lowering pHi to limit cancer progression. Tumor heterogeneity, whether genetic, epigenetic, or phenotypic, is increasingly being recognized as a challenge for cancer therapies (60, 61), and improved understanding of the determinants and consequences of pHi heterogeneity could contribute to resolving these therapeutic challenges.
The field has taken a first important step in identifying a number of normal and pathological cell behaviors regulated by pHi dynamics. A second step in understanding how pHi regulates the signaling pathways mediating these behaviors is now emerging. A third step of improved mechanistic understanding is an important future direction to resolve design principles and functions of pH sensitive proteins mediating pHi-regulated cell behaviors. This third step is experimentally challenging and remains largely unexplored, but holds promise for identifying new therapeutic targets and informing the design of therapeutics for regenerative medicine and treating diseases with dysregulated pHi dynamics, including cancer.
All authors contributed to obtaining data included in the figures, including data on pHi and cancer (KW and DB), pHi and stem cell differentiation (YL and DB), and contributed to writing and editing the manuscript.
This work was supported by National Institutes of Health grants F32CA177055 (KW) and R01CA197855 and R01GM11634 (DB).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank members of the Barber laboratory for their contributions and suggestions. We apologize for not being able to include all relevant publications on the topics we present because of space limitations.
1. Flinck M, Kramer SH, Schnipper J, Andersen AP, Pedersen SF. The acid-base transport proteins NHE1 and NBCn1 regulate cell cycle progression in human breast cancer cells. Cell Cycle. (2018) 17:1056–67. doi: 10.1080/15384101.2018.1464850
2. Ochi H, Aoto S, Tachibana K, Hara M, Chiba K. Block of CDK1-dependent polyadenosine elongation of Cyclin B mRNA in metaphase-i-arrested starfish oocytes is released by intracellular pH elevation upon spawning. Mol Reprod Dev. (2016) 83:79–87. doi: 10.1002/mrd.22599
3. Putney LK, Barber DL. Na-H exchange-dependent increase in intracellular pH times G2/M entry and transition. J Biol Chem. (2003) 278:44645–9. doi: 10.1074/jbc.M308099200
4. Cardone RA, Casavola V, Reshkin SJ. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat Rev Cancer. (2005) 5:786–95. doi: 10.1038/nrc1713
5. Choi CH, Webb BA, Chimenti MS, Jacobson MP, Barber DL. pH sensing by FAK-His58 regulates focal adhesion remodeling. J Cell Biol. (2013) 202:849–59. doi: 10.1083/jcb.201302131
6. Denker SP, Barber DL. Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J Cell Biol. (2002) 159:1087–96. doi: 10.1083/jcb.200208050
7. Stock C, Schwab A. Ion channels and transporters in metastasis. Biochim Biophys Acta. (2015) 1848:2638–46. doi: 10.1016/j.bbamem.2014.11.012
8. Amith SR, Wilkinson JM, Fliegel L. Na+/H+ exchanger NHE1 regulation modulates metastatic potential and epithelial-mesenchymal transition of triple-negative breast cancer cells. Oncotarget. (2016) 7:21091–113. doi: 10.18632/oncotarget.8520
9. Benitez M, Tatapudy S, Liu Y, Barber DL, Nystul TG. Drosophila anion exchanger 2 is required for proper ovary development and oogenesis. Dev Biol. (2019) 452:127–33. doi: 10.1016/j.ydbio.2019.04.018
10. Li X, Karki P, Lei L, Wang H, Fliegel L. Na+/H+ exchanger isoform 1 facilitates cardiomyocyte embryonic stem cell differentiation. Am J Physiol Heart Circ Physiol. (2009) 296:H159–70. doi: 10.1152/ajpheart.00375.2008
11. Ulmschneider B, Grillo-Hill BK, Benitez M, Azimova DR, Barber DL, Nystul TG. Increased intracellular pH is necessary for adult epithelial and embryonic stem cell differentiation. J Cell Biol. (2016) 215:345–55. doi: 10.1083/jcb.201606042
12. Gao W, Zhang H, Chang G, Xie Z, Wang H, Ma L, et al. Decreased intracellular pH induced by cariporide differentially contributes to human umbilical cord-derived mesenchymal stem cells differentiation. Cell Physiol Biochem. (2014) 33:185–94. doi: 10.1159/000356661
13. Cardone RA, Alfarouk KO, Elliott RL, Alqahtani SS, Ahmed SBM, Aljarbou AN, et al. The role of sodium hydrogen exchanger 1 in dysregulation of proton dynamics and reprogramming of cancer metabolism as a sequela. Int J Mol Sci. (2019) 20:3694. doi: 10.3390/ijms20153694
14. Stock C, Pedersen SF. Roles of pH and the Na(+)/H(+) exchanger NHE1 in cancer: from cell biology and animal models to an emerging translational perspective? Semin Cancer Biol. (2017) 43:5–16. doi: 10.1016/j.semcancer.2016.12.001
15. Swietach P. What is pH regulation, and why do cancer cells need it? Cancer Metastasis Rev. (2019) 38:5–15. doi: 10.1007/s10555-018-09778-x
16. Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer. (2011) 11:671–7. doi: 10.1038/nrc3110
17. White KA, Grillo-Hill BK, Barber DL. Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. J Cell Sci. (2017) 130:663–69. doi: 10.1242/jcs.195297
18. Parks SK, Chiche J, Pouyssegur J. Disrupting proton dynamics and energy metabolism for cancer therapy. Nat Rev Cancer. (2013) 13:611–23. doi: 10.1038/nrc3579
19. Harguindey S, Reshkin SJ, Orive G, Arranz JL, Anitua E. Growth and trophic factors, pH and the Na+/H+ exchanger in Alzheimer's disease, other neurodegenerative diseases and cancer: new therapeutic possibilities and potential dangers. Curr Alzheimer Res. (2007) 4:53–65. doi: 10.2174/156720507779939841
20. Majdi A, Mahmoudi J, Sadigh-Eteghad S, Golzari SE, Sabermarouf B, Reyhani-Rad S. Permissive role of cytosolic pH acidification in neurodegeneration: a closer look at its causes and consequences. J Neurosci Res. (2016) 94:879–87. doi: 10.1002/jnr.23757
21. Boedtkjer E, Bunch L, Pedersen SF. Physiology, pharmacology and pathophysiology of the pH regulatory transport proteins NHE1 and NBCn1: similarities, differences, and implications for cancer therapy. Curr Pharm Des. (2012) 18:1345–71. doi: 10.2174/138161212799504830
22. Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. (2010) 11:50–61. doi: 10.1038/nrm2820
23. Parker MD, Boron WF. The divergence, actions, roles, and relatives of sodium-coupled bicarbonate transporters. Physiol Rev. (2013) 93:803–959. doi: 10.1152/physrev.00023.2012
24. Takahashi E, Abe J, Gallis B, Aebersold R, Spring DJ, Krebs EG, et al. p90(RSK) is a serum-stimulated Na+/H+ exchanger isoform-1 kinase. Regulatory phosphorylation of serine 703 of Na+/H+ exchanger isoform-1. J Biol Chem. (1999) 274:20206–14. doi: 10.1074/jbc.274.29.20206
25. Meima ME, Webb BA, Witkowska HE, Barber DL. The sodium-hydrogen exchanger NHE1 is an Akt substrate necessary for actin filament reorganization by growth factors. J Biol Chem. (2009) 284:26666–75. doi: 10.1074/jbc.M109.019448
26. Snabaitis AK, Cuello F, Avkiran M. Protein kinase B/Akt phosphorylates and inhibits the cardiac Na+/H+ exchanger NHE1. Circ Res. (2008) 103:881–90. doi: 10.1161/CIRCRESAHA.108.175877
27. Tominaga T, Ishizaki T, Narumiya S, Barber DL. p160ROCK mediates RhoA activation of Na-H exchange. EMBO J. (1998) 17:4712–22. doi: 10.1093/emboj/17.16.4712
28. Yan W, Nehrke K, Choi J, Barber DL. The Nck-interacting kinase (NIK) phosphorylates the Na+-H+ exchanger NHE1 and regulates NHE1 activation by platelet-derived growth factor. J Biol Chem. (2001) 276:31349–56. doi: 10.1074/jbc.M102679200
29. Schonichen A, Webb BA, Jacobson MP, Barber DL. Considering protonation as a posttranslational modification regulating protein structure and function. Annu Rev Biophys. (2013) 42:289–314. doi: 10.1146/annurev-biophys-050511-102349
30. Jensen HH, Pedersen GA, Morgen JJ, Parsons M, Pedersen SF, Nejsum LN. The Na(+) /H(+) exchanger NHE1 localizes as clusters to cryptic lamellipodia and accelerates collective epithelial cell migration. J Physiol. (2019) 597:849–67. doi: 10.1113/JP277383
31. Magalhaes MA, Larson DR, Mader CC, Bravo-Cordero JJ, Gil-Henn H, Oser M, et al. Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J Cell Biol. (2011) 195:903–20. doi: 10.1083/jcb.201103045
32. Parks SK, Pouyssegur J. The Na(+)/HCO3(-) co-transporter SLC4A4 plays a role in growth and migration of colon and breast cancer cells. J Cell Physiol. (2015) 230:1954–63. doi: 10.1002/jcp.24930
33. Patel H, Barber DL. A developmentally regulated Na-H exchanger in Dictyostelium discoideum is necessary for cell polarity during chemotaxis. J Cell Biol. (2005) 169:321–9. doi: 10.1083/jcb.200412145
34. Schwab A, Stock C. Ion channels and transporters in tumour cell migration and invasion. Philos Trans R Soc Lond B Biol Sci. (2014) 369:20130102. doi: 10.1098/rstb.2013.0102
35. Frantz C, Karydis A, Nalbant P, Hahn KM, Barber DL. Positive feedback between Cdc42 activity and H+ efflux by the Na-H exchanger NHE1 for polarity of migrating cells. J Cell Biol. (2007) 179:403–10. doi: 10.1083/jcb.200704169
36. Srivastava J, Barreiro G, Groscurth S, Gingras AR, Goult BT, Critchley DR, et al. Structural model and functional significance of pH-dependent talin-actin binding for focal adhesion remodeling. Proc Natl Acad Sci USA. (2008) 105:14436–41. doi: 10.1073/pnas.0805163105
37. Frantz C, Barreiro G, Dominguez L, Chen X, Eddy R, Condeelis J, et al. Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J Cell Biol. (2008) 183:865–79. doi: 10.1083/jcb.200804161
38. Andersen AP, Samsoe-Petersen J, Oernbo EK, Boedtkjer E, Moreira JMA, Kveiborg M, et al. The net acid extruders NHE1, NBCn1 and MCT4 promote mammary tumor growth through distinct but overlapping mechanisms. Int J Cancer. (2018) 142:2529–42. doi: 10.1002/ijc.31276
39. Chiche J, Le Fur Y, Vilmen C, Frassineti F, Daniel L, Halestrap AP, et al. In vivo pH in metabolic-defective Ras-transformed fibroblast tumors: key role of the monocarboxylate transporter, MCT4, for inducing an alkaline intracellular pH. Int J Cancer. (2012) 130:1511–20. doi: 10.1002/ijc.26125
40. Pouyssegur J, Franchi A, Pages G. pHi, aerobic glycolysis and vascular endothelial growth factor in tumour growth. Novartis Found Symp. (2001) 240:186–96; discussion 196–8. doi: 10.1002/0470868716.ch13
41. Grillo-Hill BK, Choi C, Jimenez-Vidal M, Barber DL. Increased H(+) efflux is sufficient to induce dysplasia and necessary for viability with oncogene expression. Elife. (2015) 4:e03270. doi: 10.7554/eLife.03270
42. White KA, Grillo-Hill BK, Esquivel M, Peralta J, Bui VN, Chire I, et al. β-Catenin is a pH sensor with decreased stability at higher intracellular pH. J Cell Biol. (2018) 217:3965–76. doi: 10.1083/jcb.201712041
43. Stamos JL, Weis WI. The beta-catenin destruction complex. Cold Spring Harb Perspect Biol. (2013) 5:a007898. doi: 10.1101/cshperspect.a007898
44. Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(β-TrCP1) ubiquitin ligase. Mol Cell. (2003) 11:1445–56. doi: 10.1016/S1097-2765(03)00234-X
45. Shafique S, Younis S, Niaz H, Rashid S. Elucidation, functional clustering and structural characterization of betaTrCP1 substrates through a molecular dynamics study. Mol Biosyst. (2016) 12:2233–46. doi: 10.1039/C6MB00189K
46. White KA, Ruiz DG, Szpiech ZA, Strauli NB, Hernandez RD, Jacobson MP, et al. Cancer-associated arginine-to-histidine mutations confer a gain in pH sensing to mutant proteins. Sci Signal. (2017) 10:eaam9331. doi: 10.1126/scisignal.aam9931
47. Chen G, Hou Z, Gulbranson DR, Thomson JA. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell. (2010) 7:240–8. doi: 10.1016/j.stem.2010.06.017
48. Eldar A, Rozenberg H, Diskin-Posner Y, Rohs R, Shakked Z. Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions. Nucleic Acids Res. (2013) 41:8748–59. doi: 10.1093/nar/gkt630
49. Jenkins EC Jr, Debnath S, Gundry S, Gundry S, Uyar U, Fata JE, et al. Intracellular pH regulation by Na(+)/H(+) exchanger-1 (NHE1) is required for growth factor-induced mammary branching morphogenesis. Dev Biol. (2012) 365:71–81. doi: 10.1016/j.ydbio.2012.02.010
50. Raja DA, Gotherwal V, Burse SA, Subramaniam YJ, Sultan F, Vats A, et al. pH-controlled histone acetylation amplifies melanocyte differentiation downstream of MITF. EMBO Rep. (2020). 21:e48333. doi: 10.15252/embr.201948333
51. Grillo-Hill BK, Webb BA, Barber DL. Ratiometric imaging of pH probes. Methods Cell Biol. (2014) 123:429–48. doi: 10.1016/B978-0-12-420138-5.00023-9
52. Gehart, H. Clevers H. Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol. (2019) 16:19–34. doi: 10.1038/s41575-018-0081-y
53. Gonzales KAU, Fuchs E. Skin and its regenerative powers: an alliance between stem cells and their niche. Dev Cell. (2017) 43:387–401. doi: 10.1016/j.devcel.2017.10.001
54. Kretzschmar K, Clevers H. Wnt/β-catenin signaling in adult mammalian epithelial stem cells. Dev Biol. (2017) 428:273–82. doi: 10.1016/j.ydbio.2017.05.015
55. Alvarez-Perez J, Ballesteros P, Cerdan S. Microscopic images of intraspheroidal pH by 1H magnetic resonance chemical shift imaging of pH sensitive indicators. MAGMA. (2005) 18:293–301. doi: 10.1007/s10334-005-0013-z
56. Lee AH, Tannock IF. Heterogeneity of intracellular pH and of mechanisms that regulate intracellular pH in populations of cultured cells. Cancer Res. (1998) 58:1901–8.
57. McIntyre A, Hulikova A, Ledaki I, Snell C, Singleton D, Steers G, et al. Disrupting hypoxia-induced bicarbonate transport acidifies tumor cells and suppresses tumor growth. Cancer Res. (2016) 76:3744–55. doi: 10.1158/0008-5472.CAN-15-1862
58. Swietach P, Wigfield S, Cobden P, Supuran CT, Harris AL, Vaughan-Jones RD. Tumor-associated carbonic anhydrase 9 spatially coordinates intracellular pH in three-dimensional multicellular growths. J Biol Chem. (2008) 283:20473–83. doi: 10.1074/jbc.M801330200
59. Lobo RC, Hubbard NE, Damonte P, Mori H, Penzvalto Z, Pham C, et al. Glucose uptake and intracellular pH in a mouse model of ductal carcinoma in situ (DCIS) suggests metabolic heterogeneity. Front Cell Dev Biol. (2016) 4:93. doi: 10.3389/fcell.2016.00093
60. Hausser J, Alon U. Tumour heterogeneity and the evolutionary trade-offs of cancer. Nat Rev Cancer. (2020) 20:247–57. doi: 10.1038/s41568-020-0241-6
Keywords: intracellular pH, cancer, stem cell, protein structure, protonation
Citation: Liu Y, White KA and Barber DL (2020) Intracellular pH Regulates Cancer and Stem Cell Behaviors: A Protein Dynamics Perspective. Front. Oncol. 10:1401. doi: 10.3389/fonc.2020.01401
Received: 12 May 2020; Accepted: 02 July 2020;
Published: 26 August 2020.
Edited by:
Silvia Pastorekova, Slovak Academy of Sciences, SlovakiaReviewed by:
Ana P. Gomes, Moffitt Cancer Center, United StatesCopyright © 2020 Liu, White and Barber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diane L. Barber, ZGlhbmUuYmFyYmVyQHVjc2YuZWR1
†Present address: Katharine A. White, Department of Chemistry and Biochemistry, Harper Cancer Research Institute, University of Notre Dame, South Bend, IN, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.