- 1The Hepatosplenic Surgery Center, The First Affiliated Hospital of Harbin Medical University, Harbin, China
- 2Department of Molecular Medicine and Pathology, Faculty of Medical and Health Sciences, The University of Auckland, Auckland, New Zealand
Metastasis, the spread of cancer cells from a primary tumor to a secondary site, represents one of the hallmarks of malignancies and the leading cause of cancer-related death. The process of metastasis is a result of the interaction of genetic heterogeneity, abnormal metabolism, and tumor microenvironments. On the other hand, metabolic reprogramming, another malignancy hallmark, refers to the ability of cancer cells to alter metabolic and nutrient acquisition modes in order to support the energy demands for accomplishing the rapid growth, dissemination, and colonization. Cancer cells remodel metabolic patterns to supplement nutrients for their metastasis and also undergo metabolic adjustments at different stages of metastasis. Genes and signaling pathways involved in tumor metabolic reprogramming crosstalk with those participating in metastasis. Non-coding RNAs are a group of RNA molecules that do not code proteins but have pivotal biological functions. Some of microRNAs and lncRNAs, which are the two most extensively studied non-coding RNAs, have been identified to participate in regulating metabolic remodeling of glucose, lipid, glutamine, oxidative phosphorylation, and mitochondrial respiration, as well as the process of metastasis involving cell motility, transit in the circulation and growth at a new site. This article reviews recent progress on non-coding RNAs operating in the crosstalk between tumor metabolic reprogramming and metastasis, particularly those influencing metastasis through regulating metabolism, and the underlying mechanisms of how they exert their regulatory functions.
Introduction
Metastasis is one of the important cancer hallmarks (1), and a complex multistep process involving intracellular and intercellular signal transduction cascades and comprising the proliferation of primary tumor cells, endovascular intervention, the formation of pre-metastatic niches, and subsequent dissemination of cancer cells or micro-metastases into distant organs (2, 3). Metastasis contributes largely to the mortality for many major cancer types, and exploring the underlying mechanisms is of great significance for seeking effective treatments and improving the prognosis of cancer patients.
Metabolism transforms the absorbed nutrients into small molecule metabolites to sustain the stability of homeostasis, generate bioenergy and regulate cell signaling pathways and physiological activities (4). Metabolic reprogramming is a malignancy hallmark, which refers to the ability of cancer cells to adjust metabolic and nutrient acquisition modes to support their rapid growth, dissemination, and other characteristics (5). The metabolites, such as glucose, amino acids, nucleic acids, and lipids, are remodeled in tumors (Figure 1). Cancer metastasis usually accompanies metabolic reprogramming, and this metabolic change has been recognized at different stages of metastasis (6, 7). Non-coding RNAs (ncRNAs), particularly long-chain non-coding RNA (lncRNA) and microRNA (miRNA), play important roles in regulating metabolic remodeling and the metastasis of cancer cells (8–11). This article aims to discuss the recent progress of how lncRNAs and miRNAs orchestrate in the crosstalk between cancer metabolic reprogramming and metastasis, particularly those influencing metastasis through regulating metabolism-related genes and signaling pathways.
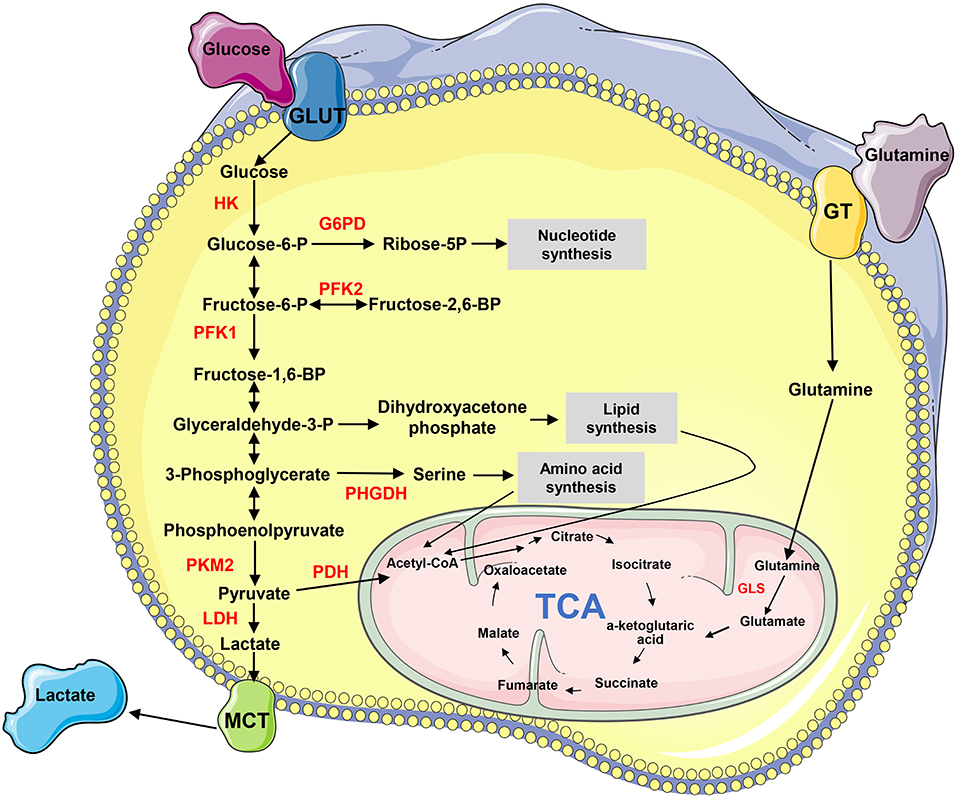
Figure 1. The biochemistry of metabolism in cancer cells. “→” indicates positive regulation or activation. Key enzymes in energy metabolism are marked as red color. G6PD, glucose 6-phosphate dehydrogenase; GLS, glutaminase; GLUT, glucose transporter; GT, glutamine transporter; HK, hexokinase; LDH, lactate dehydrogenase; MCT, Monocarboxylate transporter; PDH, pyruvate dehydrogenase; PFK, phosphofructokinase; PHGHD, D-3-phosphoglycerate dehydrogenase; PKM2, pyruvate kinase M2; TCA, tricarboxylic acid cycle.
Metabolic Reprogramming and Cancer Metastasis
In the 1920s, Warburg observed a special phenomenon that even in aerobic conditions, cancer cells tend to favor metabolism relying on glycolysis rather than the much more efficient oxidative phosphorylation pathway, which is the preference of most normal cells (12, 13). As a result, tumor cells have significantly increased glucose uptake and secretion of lactate, which is converted from pyruvate, the last product of glycolysis (14). This process, now known as the Warburg effect or aerobic glycolysis, marks the start of a new era for studying tumorigenesis and cancer progression in terms of metabolism. Gene mutations, deletions, and translocations have an impact on various signal pathways in cancer cells, and the main oncogenic signal pathways will finally converge to metabolism (15, 16). Metabolites by cancer cells not only supply materials for their proliferation and metastasis but also provide sustaining signals to meet their survival needs in tumor-specific microenvironments (1, 17, 18). In addition, cancer cells affect the metabolism of distant organs, where they metastasize, to facilitate their implantation and growth (19). On the other hand, metastatic cancer cells are also affected by the metabolic alterations of invaded organs and tissues (20, 21).
Tumor cells are characterized by high aerobic glycolysis and high oxidative stress (16). Studies have found that the Warburg effect inhibits anoikis and promotes tumor metastasis (22, 23). Meanwhile, aerobic glycolysis increases glucose consumption, reduces the generation of excessive reactive oxygen species (ROS), and enhances the antioxidant capacity of tumor cells, which obtain the ability to resist anoikis and metastasize (22). Despite the Warburg effect, oxidative metabolism is also a major source of adenosine triphosphate (ATP) in some tumors (24, 25). Mitochondrial oxidative metabolism produces ROS including the hydroxyl (HO−) free radicals, superoxide () and non-radical molecules such as hydrogen peroxide (H2O2) (26). Low or moderate levels of ROS in tumor cells can activate a series of signaling pathways, causing genomic DNA mutations and promoting tumor formation and progression; but high levels of ROS can lead to the death of neoplasm cells (27). The roles of ROS in tumor metastasis remain controversial among different studies. One study shows that antioxidants can promote tumor metastasis (28). Melanoma metastatic cells express higher levels of lactic acid transporter and absorb more lactic acid, enabling them to produce more antioxidants and survive in the blood (29). On the other hand, some studies demonstrate that antioxidants inhibit tumor metastasis, suggesting that ROS may promote the dissemination of tumor cells (30). H2O2 has also been shown to promote tumor metastasis by inhibiting anoikis of detached cancer cells (31).
Roles of ncRNAs in Cancer
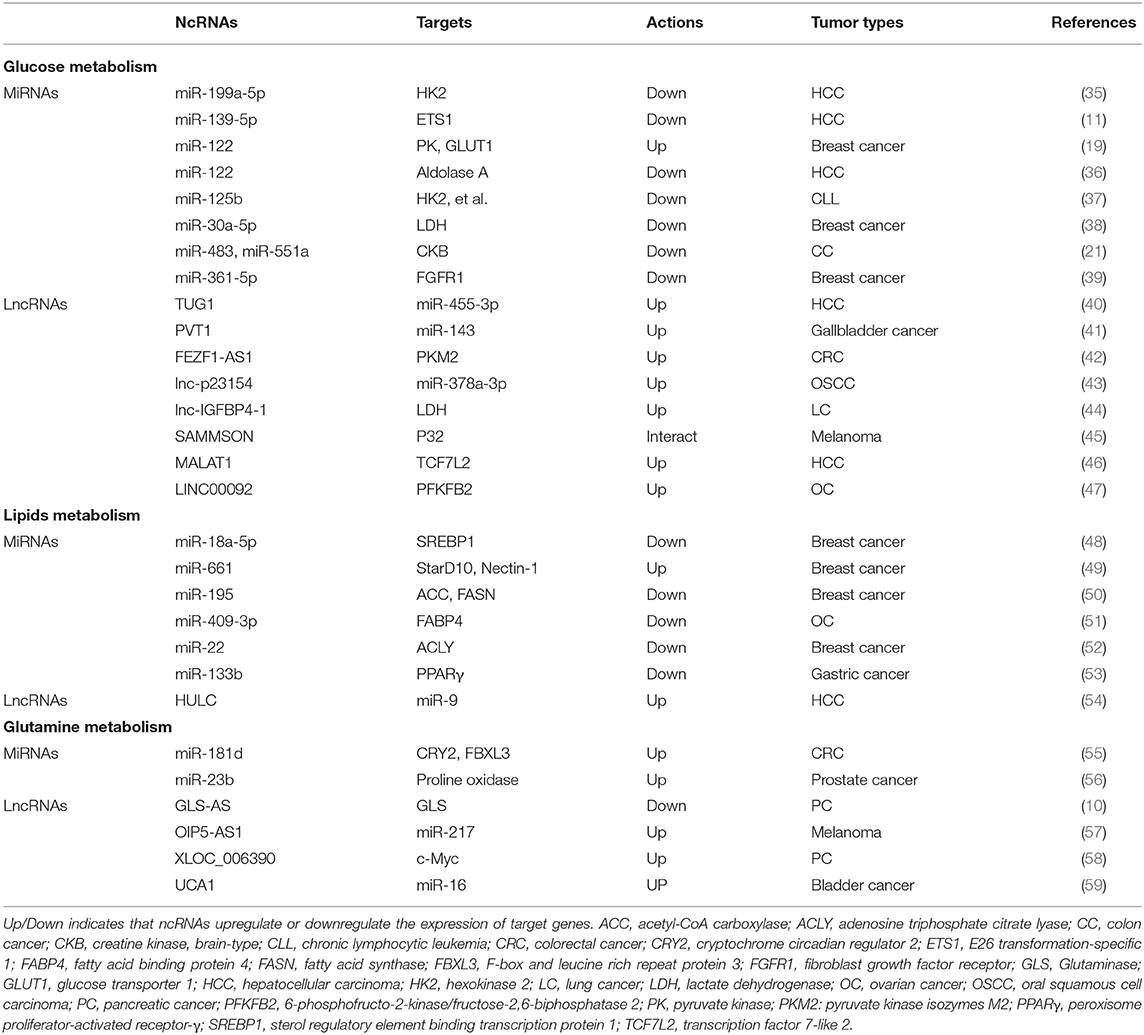
Although the number of ncRNAs in the human genome is unknown, recent studies suggest that thousands of ncRNAs exist in the body. At present, a dozen of abundant and functionally important types of ncRNAs have been discovered, among which miRNA and lncRNA are the two most extensively studied. Because they do not encode proteins, ncRNAs were initially considered to lack biological functions. However, it's now clear and well accepted that ncRNAs affect the development and progression of many malignancies by regulating the proliferation, apoptosis, differentiation, and metastasis of cancer cells (32) (Figure 2). Accumulating evidence also indicates a link between ncRNAs and metabolic changes in cancer (33, 34) (Table 1). NcRNAs are shown to regulate key metabolic enzymes and signaling pathways involved in metabolic reprogramming, resulting in tumorigenesis, cancer progression and metastasis (60, 61).
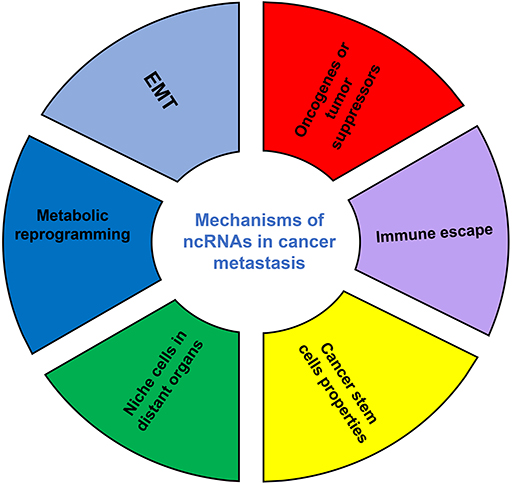
Figure 2. The mechanisms of ncRNAs involved in cancer metastasis. EMT, epithelial-mesenchymal transition.
ncRNAs in Metabolic Reprogramming and Cancer Metastasis
Glucose Metabolism
Cancer cells have significantly enhanced glycolysis during the metastatic process and the glucose supply is dramatically reduced in solid tumors (12, 13). The intermediate metabolites generated in glycolysis are important synthetic materials for tumor growth (62). The acidic and hypoxic microenvironment (TME) inside solid tumors promotes invasion and immune escape (63, 64) by regulating the function and subcellular localization of cytoskeleton proteins, thus promotes the invasion and metastasis of tumor cells through protonation of critical pH-sensitive residues (65, 66). In addition, aerobic glycolysis reduces the oxidative metabolism of glucose, making cancer cells resistant to anoikis and promoting the survival of circulating tumor cells (67).
The first step in glycolysis is the phosphorylation of glucose to form glucose 6-phosphate by a family of enzymes called hexokinases (HKs), which are key glycolytic enzymes to control the rate of glucose metabolism and highly expressed HKs maintain a speedy glycolytic rate in tumor tissues, helping the metastasis of cancer cells (68). HK2 can regulate the expression of matrix metalloproteinase (MMP)-9, SRY-box transcription factor (SOX)-9 and non-processed pseudogene (NANOG) and facilitate the metastasis of ovarian cancer cells (69). Lower expression of miR-139-5p correlates with a worse prognosis of hepatocellular carcinoma (HCC) and overexpressed miR-139-5p restrains aerobic glycolysis, suppressing the metastasis of HCC cells (11). In a mechanism exploration, miR-139-5p regulates the expression of HK1 and 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase (PFKFB) 3 by directly targeting ETS proto-oncogene 1 (ETS1), a transcription factor binding to the promoters of HK1 and PFKFB3 genes, while ETS1 silencing induces the expression of miR-139-5p via a post-transcriptional regulation mode involving Drosha (11). Knockdown of lncRNA-TUG1 induces a marked inhibition of cell migration, invasion, and glycolysis by suppressing miR-455-3p, which is transcriptionally repressed by p21 and directly targets the 3′-UTR of adenosine monophosphate-activated protein kinase subunit β2 (AMPKβ2), thus the lncRNA TUG1/miR-455-3p/AMPKβ2 axis regulates the metastasis and glycolysis of HCC cells through the regulation of HK2 (40). LncRNA PVT1 is upregulated in gallbladder cancer (GBC) tissues and negatively associated with the overall survival of patients. Knockdown of lncRNA PVT1 inhibits the metastasis of GBC cells by regulating aerobic glucose metabolism via modulating HK2 expression by competitively binding to endogenous miR-143 as a competing endogenous RNA (ceRNA) (41).
Pyruvate kinase M2 (PKM2) is an alternatively spliced variant of pyruvate kinase that is preferentially expressed during embryonic development and in many types of cancer cells. PKM2 alters the final rate-limiting step of glycolysis, resulting in a cancer-specific Warburg effect, thus determining the efficiency of glucose utilization and the production of lactic acid (70). The direct interaction of PKM2 in the nucleus with TGF (transforming growth factor)-β-induced factor homeobox 2 (TGIF2), leading to the recruitment of histone deacetylase 3 to the E-cadherin promoter sequence, with subsequent deacetylation of histone H3 and suppression of E-cadherin transcription, thus promoting the process of epithelial-mesenchymal transition (EMT) in colon cancer cells (71). MiR-let-7a inhibits the migration of cervical cancer cells through downregulating PKM2 (72). Breast cancer cells secret miR-122-carrying vesicles into circulation to facilitate their metastasis by increasing nutrient availability in pre-metastatic niches in distant organs including brains and lungs. Mechanistically, the cancer-cell-derived miR-122 suppresses glucose uptake by niche cells via downregulating the glycolytic enzyme pyruvate kinase (19). LncRNA FEZF1-AS1 is one of the most highly expressed lncRNAs in colorectal cancer (CRC) and exerts a promoting function on the metastasis of CRC cells. FEZF1-AS1 binds to and stabilizes the PKM2 protein, thereby activating the STAT3 (signal transducer and activator of transcription 3) signaling pathway and increasing aerobic glycolysis (42).
Glucose transporter (GLUT) is a transmembrane glycoprotein distributed on the cell membrane and mainly carry out transmembrane transport of glucose, thus represents a key factor for cancer cells to take up glucose. High expression of GLUT1 promotes the transport and absorption of glucose, providing abundant materials for glycolysis, thereby enhancing the metastasis of cancer cells (73). GLUT1 gene expression is associated with the invasiveness and MMP-2 activity in pancreatic cancer (74). MiRNAs mediate fine-tuning of genes including GLUTs involved in cancer metabolism to support the biosynthetic and energy requirements for the metastasis of cancer cells (75). MiR-122 secreted by breast cancer cells restricts the expression of GLUT1 in non-cancerous brain astrocytes and lung fibroblasts, allowing cancer cells to obtain sufficient glucose supply during the processes of lung and brain metastases (19). LncRNA lnc-p23154 is shown to be associated with the metastasis of oral squamous cell carcinoma (OSCC) and promotes OSCC cell migration and invasion by suppressing the expression of GLUT1 via its negative effects on miR-378a-3p, which has an inhibitory effect on the expression of GLUT1 (43).
Lactate dehydrogenase A (LDHA) is another important rate-limiting enzyme in glucose metabolism by catalyzing the interconversion of pyruvate and lactate. The phosphorylation of LDHA at Y10 is positively correlated with the progression of metastatic breast cancer (76). LDHA can promote the process of EMT by activating EMT-related proteins and facilitate the metastasis of lung adenocarcinoma (77). In breast cancer cells, miR-30a-5p suppresses LDHA expression by directly targeting its 3′-UTR, thus inhibits glycolysis by decreasing glucose uptake, lactate production, ATP generation, and extracellular acidification rate, and increasing oxygen consumption. As a result, glycolysis regulated by miR-30a-5p plays a critical role in the metastasis of breast cancer cells (38). Lnc-IGFBP4-1 promotes the metastasis of lung cancer cells through a possible mechanism of metabolic reprogramming by enhancing the expression of LDH and ATP production (44).
In addition to the above glycolytic enzymes, other kinases in glycolysis are also regulated by ncRNAs. The high level of glucose uptake and aerobic glycolysis stimulates the hexosamine biosynthetic flux, leading to an increased level of UDP-β-DN-acetylglucosamine (UDP-GlcNAc), which is the final product of hexosamine biosynthesis (78, 79) and catalyzed by O-GlcNAcylation transferase (OGT) (80). OGT is post-transcriptionally inhibited by miRNA-101 and upregulated OGT increases O-GlcNAcylation level and promotes the metastasis of CRC cells (81). MiR-551a and miR-483 suppress hepatic colonization and metastasis of CRC cells by convergently dysregulating creatine kinase brain-type (CKB), which is released into the extracellular microenvironments by hypoxic metastatic cells and catalyzes the production of phosphocreatine that helps to generate ATP and fuel metastatic cells (21). In ovarian cancer, the chemokine CXCL14 (C-X-C motif ligand 14)-high expressed cancer-associated fibroblasts mediate the upregulation of lncRNA LINC00092, which downregulates PFKFB2 (6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2), a glycolytic enzyme involved in synthesis and degradation of fructose-2,6-bisphosphate, thereby promoting metastasis by altering glycolysis (47). MiR-361-5p inhibits the glycolysis and invasion of breast cancer cells by respectively targeting MMP-1 and fibroblast growth factor receptor 1 (FGFR1), which is a promoter of glycolytic enzyme and a suppresser of OXPHOS (oxidative phosphorylation) (39).
Mitochondrial Respiration and Oxidative Phosphorylation
Tumor cells employ glycolysis as the main energy supply method even in oxygen-rich environments due to their impaired mitochondrial function, along with the reductive carboxylation of glutamine to malate (12). Hypoxia and reduced mitochondrial capacity promote cancer cell dependence on glycolysis for ATP production that is supported by cytosolic reductive metabolism, while preventing this metabolic adaptation can increase the accumulation of ROS and a reduction of metastatic capacity of cancer cells (82). In addition, the metabolic interaction between cancer cell mitochondrial respiration and catabolism in carcinoma-associated fibroblasts enhances the growth and metastasis of cancer cells (83). The expression of miR-485 is downregulated in breast cancer tissues and the overexpression of its both mature forms, miR-485-3p and miR-485-5p, restrains mitochondrial respiration and suppresses the metastasis of breast cancer cells by downregulating peroxisome proliferator-activated receptor-gamma coactivator (PGC)−1α (84).
It is well known that OXPHOS produces more ATP than glycolysis under the same condition (13). Studies used to suggest that OXPHOS plays a minor role in the energy metabolism of tumor cells due to their deficient mitochondrial functions. However, a study shows that 80% of the energy generated by MCF-7 breast cancer cells in vitro is derived from mitochondrial OXPHOS and only 20% from glycolysis (25). Another study also finds that genes involved in OXPHOS are significantly upregulated in breast, leukemia, lung, lymphoma and ovarian cancers (85). Therefore, tumor cells may use aerobic glycolysis for energy and glycolytic metabolites are transferred into anabolic pathways to support malignancy, but this process may be suspended during cancer cell metastasis. In support, it is shown that aggressive cancer cells favor OXPHOS for energy supply (86). Although the dispute exists, it seems that OXPHOS is also an important metabolic pattern of tumor cells (87).
It is unclear how primary and metastatic tumors select different metabolic pathways, glycolysis or OXPHOS, for their energy supply. However, it is acceptable that glycolysis is more remarkable in aggressive and fast-growing tumors, and different types of cancer may have different metabolism pathways. A subset of glioma cells relies on glycolysis while the others in the same tumor depend on OXPHOS for energy supply, indicating a characteristic of metabolic heterogeneity (88). A genetic analysis shows that breast cancer cells utilizing glycolysis as the main energy metabolic method prefer to metastasize to the liver, but cells adopting OXPHOS as the principal metabolic pathway are more likely to metastasize to bone and lungs, and pyruvate dehydrogenase kinase-1 (PDK1) is required for liver metastasis (89). Despite metabolic heterogeneity that exists in the same tumor and metabolic types affect the organs where tumor cells metastasize, little information is available regarding the influence of ncRNAs on OXPHOS. A study suggests that miR-155 and miR-210 derived from melanoma exosomes promote glycolysis and inhibit OXPHOS in tumor cells and contribute to the creation of a pre-metastatic niche (90).
Lipid Metabolism
Lipid anabolism is an important indicator of abnormal tumor metabolism because lipids provide components of biofilms and regulate fluidity and lipid molecule signal transduction of cytomembranes, and thus participates in the metastasis of cancer cells (91, 92). Under metabolic stress, tumor cells strengthen the coupling of fatty acid anabolism and catabolism to establish a fatty acid circulation network to promote their growth and metastasis (93).
In a mouse model with subcutaneous osteosarcoma, the serum metabolic profiling reveals an increase of key metabolites in glycolysis and tricarboxylic acid cycle (TCA); while in mice with lung metastasis, serum metabolic profile shows a decrease of most metabolites, except for cholesterol and free fatty acids, suggesting that elevated lipid metabolism may be associated with tumor metastasis (94). Sterol regulatory element-binding transcription protein 1 (SREBP1), a candidate target of miR-18a-5p, is the master transcription factor that controls lipid metabolism; and miR-18a-5p can suppress the invasion and migration of breast cancer cells by regulating SREBP1 (48). MiR-661 is required for the efficient invasion of breast cancer cells by destabilizing StAR-related lipid transfer protein 10 and the cell-cell adhesion protein Nectin-1, leading to the downregulation of epithelial markers (49). Acetyl-CoA carboxylase (ACC), fatty acid synthase (FASN) and 3-hydroxy-3-methyl-glutaryl-CoA reductase (HMGR) are potential targets of miR-195, whose ectopic expression regulates cellular triglyceride and cholesterol levels, leading to decreased proliferation and metastasis of breast cancer cells (50). Hypoxia decreases the expression of miR-409-3p, a regulator of FABP4 (fatty acid binding protein 4), which increases the metastatic potential of ovarian cancer cells (51).
Fatty acid anabolism requires the activation of ACC, adenosine triphosphate citrate lyase (ACLY) and FASN (93), which are upregulated in aggressive tumors and associated with a poor prognosis (95, 96). ACLY participates in the de novo synthesis of fatty acids by converting citrate into oxaloacetate and cytosolic acetyl-CoA. In gastric cancer, highly expressed ACLY is closely related to advanced stages and lymph metastasis (97). ACLY regulates low molecular weight isoform of cyclin E (LMW-E), which can also enhance the enzyme activity of ACLY in a positive feedback way, promoting the formation of lipid droplets and the metastasis of breast cancer cells (98). ACLY stabilizes CTNNB1 (beta-catenin 1) protein and enhances its transcriptional activity, thus promoting the migration and invasion of colon cancer cells (99). MiR-22 restricts the metastasis of breast cancer cells by inhibiting the expression of ACLY (52). Overexpression of miR-133b in gastric cancer increases the levels of nuclear PPAR-γ, which decreases the transcriptional activity of ACLY, and then represses the invasion of cells (53). ACC is a rate-limiting enzyme for fatty acid synthesis, which catalyzes acetyl-CoA to form malonyl-CoA (100). ACC1 is highly modulated by phosphorylation and allosteric regulation and plays a key role in promoting a speedy adaptation to novel microenvironments (101). In HCC, highly expressed ACC1 is closely related to poor differentiation, vascular invasion, and poor prognosis, and has been regarded as a biomarker for early diagnosis (102). Contradictorily, ACC1 inhibition is shown to promote the metastasis of breast cancer cells (103), indicating the roles of ACC1 may be cancer type-dependent. Acyl-CoA oxidase 1 (ACOX1) is a key enzyme of the fatty acid oxidation pathway and its overexpression alleviates the migration and invasion of colorectal cancer (104). The class III deacetylase sirtuin 1 (SIRT1) prevents the trans-activation effect of activator protein (AP-1) on miR-15b-5p by deacetylation of AP-1, then upregulates the expression of ACOX1, which act as a direct target for miR-15b-5p (104).
Fatty acid catabolism also plays a role in the process of cancer metastasis. Monoacylglycerol lipase, functioning with hormone-sensitive lipase (LIPE) to hydrolyze intracellular triglyceride stores to fatty acids and glycerol, is highly expressed in prostate cancer and is related to the EMT process (105). Phospholipase D (PLD), an enzyme that hydrolyzes phosphatidylcholine to produce the signal molecule phosphatidic acid and soluble choline, promotes the metastasis of cancer cells (106). Adipocytes are the carrier of energy, sources of hormones and cytokines, and a crucial component of tumor microenvironments, and also facilitate tumor metastasis. For instance, melanoma cells have a higher level of fatty acid oxidation after they absorb fatty acid oxidase enzymes in exosomes secreted by adipose cells (107). However, the roles of ncRNAs in fatty acid catabolism and adipocytes remain to be clarified.
Glutamine Metabolism
Cancer cells have shown increased glutamine uptake and catabolism. Glutaminase (GLS) catalyzes glutamine into glutamate, which is subsequently catalyzed by glutamate dehydrogenase (GDH) to form α-ketoglutarate, finally entering the TCA as an important energy source. Although glutamine is a non-essential amino acid, it is indispensable in specific conditions and its metabolism has important biological significance for cancer cells. Glutamine can increase the expression of hypoxia-inducible factor (HIF)-1α, enhance the pro-autophagic effect of its target BNIP3 (BCL2/adenovirus E1B 19kDa interacting protein 3), and promote the metastasis of melanoma cells (108). Metabolite fumarate, an intermediate product of glutamine metabolism, reduces the level of ROS and maintains the balance of redox by activating glutathione peroxidase, and thus promoting the metastasis of cancer cells (109). C-Myc is an oncogenic transcription factor and promotes glutamine catabolism to fuel the growth of cancer cells by upregulating GLS and suppresses proline oxidase primarily through upregulating miR-23b (56). LncRNA GLS-AS regulates a feedback loop of glutaminase and c-Myc, thus being involved in the metastasis and representing a therapeutic target for the metabolic reprogramming of pancreatic cancer cells (10). MicroRNA-133a-3p targets GABARAPL1 (gamma-aminobutyric acid receptor-associated protein-like 1) to inhibit autophagy-mediated glutaminolysis, thereby inhibiting metastasis of gastric cancer (110).
ncRNAs and Metabolic Signaling Pathways
As discussed above, ncRNAs participate in the metastasis-related metabolic reprogramming by regulating individual genes. In the following paragraphs, we discuss how dysregulated ncRNAs modulate the key metabolic signaling pathways to affect the metastasis of cancer cells (Figure 3).
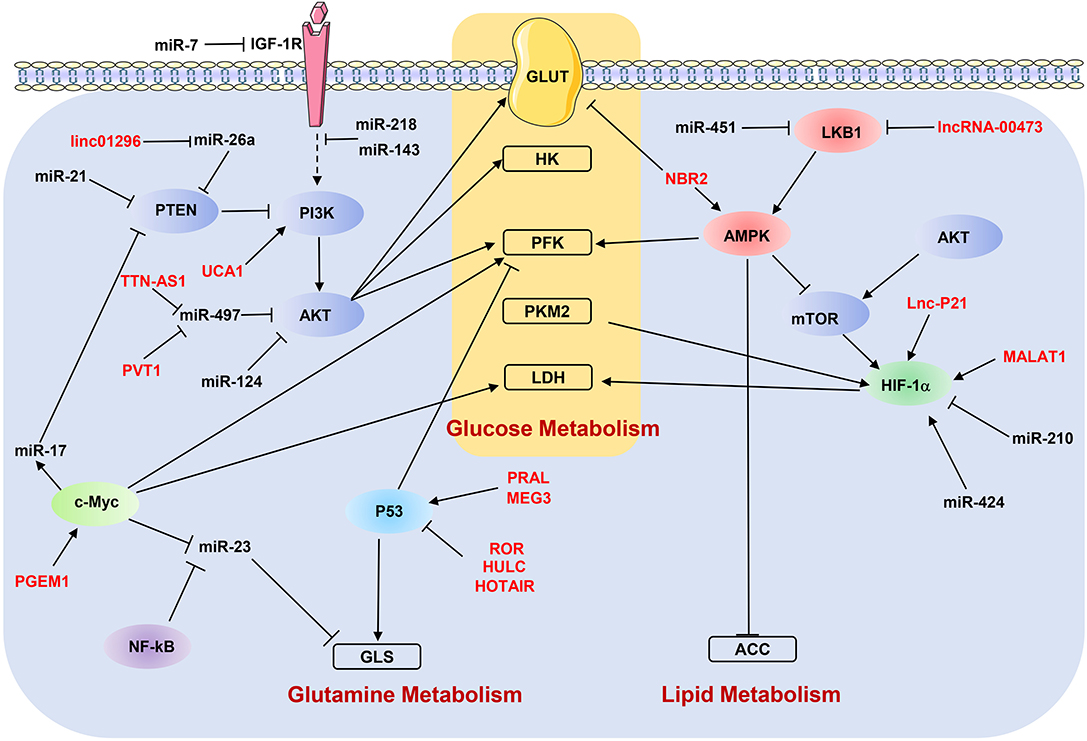
Figure 3. NcRNAs regulate metabolic reprogramming by targeting signaling pathways. “→” indicates positive regulation or activation; “⊥”, negative regulation or blockade; dotted lines “—–”, indirect regulation. LncRNAs are marked as red color. ACC, acetyl-CoA carboxylase; AMPK, AMP-activated protein kinase; GLS, glutaminase; GLUT, glucose transporter; HK, hexokinase; IGF-1R, insulin-like growth factor-1 receptor; LDH, lactate dehydrogenase; LKB1, liver kinase B1; PFK, phosphofructokinase; PKM2, pyruvate kinase M2; PTEN, phosphatase and tensin homolog.
Hypoxia-Inducible Pathways
Hypoxic microenvironments are very frequently observed in almost all the solid tumors, have an impact on cell biological behaviors and extracellular matrix remodeling, increase metastatic capacities and contribute greatly to therapy resistance (111). Under aerobic conditions, tumor cells have a high rate of glucose-dependent metabolism (112), while in hypoxic microenvironments they have to adjust themselves to lower oxygen situations by changing the metabolic pattern, resulting in the suppression of anti-cancer immunity and high potential capacities of invasion and migration (113). HIFs are the master driving forces of the cellular adaption to hypoxia and well-known transcription factors by regulating a vast array of genes involved in angiogenesis and metastasis, and in particular, GLUTs and glycolytic enzymes including HK2, 6-phosphofructokinase, and LDHA are regulated by HIFs (111). The expression and stabilization of HIFs are controlled by mRNAs. For example, miR-365 directly targets homeobox A9 (HOXA9) by binding to 3′-UTR region, and the downregulation of HOXA9 increases the expression of HIF-1α and its downstream glycolytic genes HK2, GLUT1, and PDK1, promoting glycolysis and metastasis of cutaneous squamous cell carcinoma (114). The knockdown of miR-592 in HCC cells strengthens glycolysis by enhancing WSB1-induced HIF-1α stability and promotes HCC cell migration in vitro (115).
LKB1-AMP Activated Protein Kinase Pathway
AMPK (5′ adenosine monophosphate-activated protein kinase) is a critical sensor to maintain cellular energy homeostasis. AMPK phosphorylates ACC1 and SREBP1 to inhibit the synthesis of fatty acids, cholesterol, and triglycerides, and activate fatty acid uptake. It also stimulates glycolysis by activating phosphorylation of PFKFB3 and glycogen phosphorylase (116). Downregulating AMPK shows a promoting effect on the growth and biosynthesis of cancer cells (117), and many types of cancer cells have a shortage of AMPK to maintain their glycolytic phenotypes (16). Activating AMPK is required for an increased AMP/ATP ratio and switching the oxidative metabolic to glycolytic phenotypes (116). Liver kinase B1 (LKB1) is a tumor suppressor that locates at the upstream of AMPK and can repress ATP depletion by phosphorylating and activating AMPK when cellular ATP levels are limited (118). Under tumor microenvironments, miR-7 inhibits autophagy by upregulating the LKB1-AMPK signaling pathway, leading to a reduced intracellular glucose supply, and as a result, the proliferation and metastasis of pancreatic cancer cells are inhibited (119). MiR-451 regulates the proliferation, migration and responsiveness to glucose deprivation of glioma cells by targeting the LKB1/AMPK pathway, and depresses the LKB-1-associated protein CAB39 that promotes glioma cells adapting to metabolic stress (120). Taurine up-regulated gene 1 (TUG1) is highly expressed in HCC cells and upregulates miR-455-3p at the transcriptional level, thus the TUG1/miR-455-3p/AMPKβ2 axis promotes glycolysis and metastasis by upregulating HK2 (40). Higher expression of lncRNA MACC1-AS1 correlates with the lung metastasis of gastric cancer cells, and MACC1-AS1 is elevated under metabolic stress and facilitates metabolic plasticity by increasing MACC1 mRNA expression and strengthening glycolysis and anti-oxidative abilities via the AMPK/Lin28 pathway (121). LncRNA MACC1-AS1 is also upregulated in pancreatic cancer and related to poor prognosis, and its knockdown inhibits the metastasis of pancreatic cancer cells by upregulating the expression of PAX8 (paired-box gene 8), which plays a role in activating NOTCH1 signaling and promoting cell aerobic glycolysis (122).
PI3K/AKT/mTOR Pathway
In tumor cells, the classical PI3K/AKT/mTOR signaling pathway is often highly expressed and is involved in regulating cancer growth, proliferation, invasion, and metastasis (123, 124). Recent studies also show that PI3K/AKT/mTOR pathway also functions in tumor cell metabolisms such as glycolysis (125), lipid metabolism (126) and amino acid metabolism (127). Meanwhile, ncRNAs also participate in the regulation of cell metabolism through the PI3K/AKT/mTOR pathway, thus affecting tumor cell metastasis. MiR-204-5p expressed in breast cancer cells affects the mTOR pathway, reduces the oxygen consumption and extracellular acidification rates, and inhibits metastasis (128). Circular RNA circNRIP1 acts as ceRNA to bind to miR-149-5p, activates the AKT/mTOR signaling pathway, thus increases glucose uptake, lactate contents and ATP production, regulates the Warburg effect and promotes gastric cancer cell metastasis (129). LINC00963 promotes non-small cell lung cancer metastasis by preventing ubiquitination of glycolytic kinase PGK1 (phosphoglycerate kinase 1), which activates the AKT/mTOR pathway (130). MiR-384 regulates the expression of pleiotrophin, which can upregulate lipogenic genes, mediate de novo lipid synthesis and promote the metastasis of HCC cells via the N-syndecan/PI3K/Akt/mTOR pathway (131).
Conclusion and Expectation
Metastasis is a major obstacle for successful treatments of cancer and represents the leading cause of cancer-related death. Metabolic reprogramming is another hallmark of cancer because cancer cells obtain their energy supply mainly depending on glycolysis rather than mitochondrial oxidative phosphorylation through altered oncogenic metabolic pathways. The changes in metabolisms of glucose, lipid, and glutamine and mitochondrial respiration and oxidative phosphorylation involve the hypoxia-inducible, LKB1-AMP activated protein kinase and other signaling pathways in cancer cells. These metabolic alterations render cancer cells obtaining energy and metabolites for fast bioenergetics and metabolic fluxes, and being prone to metastasize to distant organs, where normal cells also undergo metabolic changes to form pre-metastatic niches for the implantation and growth of cancer cells. Many ncRNAs, particularly miRNAs and lncRNAs, have been identified as major participants in the metabolic gene regulatory networks and the multi-staged process of metastasis. Some of them either inhibit or promote cancer metastasis involving cell motility, transit in the circulation, and growth at a new site by regulating the metabolism of glycolysis, lipid, and glutamine (Figure 4).

Figure 4. NcRNAs participate in tumor metastasis by regulating metabolic reprogramming. Red and green pellets represent ncRNAs that inhibit and promote, respectively, the metastasis through regulating glycolysis, lipid metabolism or glutamine metabolism.
Despite cumulative studies showing the altered expression profiles of ncRNAs during metabolic rearrangement in cancer, their roles and molecular characteristics remain largely unexplored. Without a doubt, the dysregulation of ncRNAs influences multiple metabolic processes and plays a critical role in tumor metastasis. More ncRNAs are being identified as potential diagnostic biomarkers or therapeutic targets for cancers. For example, a meta-analysis of a large cohort of cancer patients reveals that lncRNA SNHG12 serves as a diagnostic and prognostic biomarker and a druggable therapeutic target with promising clinical potential in multiple types of cancer (132). MiR-34, miR-16, and miR-155 have been regarded as cancer therapeutic targets and are being evaluated in clinical trials (133). Furthermore, therapeutic small RNA drugs are becoming novel promising therapeutics since the first small-interfering RNA (siRNA) drug, Patisiran, which acts by binding and degrading transthyretin mRNA, was approved for the treatment of a rare polyneuropathy by FDA in 2018. However, it seems that ncRNAs are likely to play “fine-tuning” rather than “definite” roles. NcRNAs interact with each other and with other types of factors to form complicated networks, which regulate metabolic reprogramming and metastasis of cancer cells. Therefore, a deeper and more comprehensive understanding of the complicated networks of interactions that ncRNAs coordinate in the metabolic and metastatic processes may help translate the discoveries into a strategy for the diagnosis and treatment of cancer (32).
In addition, thousands of ncRNA sequences exist within cells and more types of ncRNAs have been recently discovered, such as circular RNA (circRNA), piwi-interacting RNA (piRNA), small interfering RNAs (siRNA), enhancer RNAs (eRNA) and promoter-associated RNA (PAR), which engage in cellular processes including chromatin remodeling, transcription, post-transcriptional modifications and signal transduction (134, 135). Further investigation on the new ncRNAs and their interactions with other ncRNAs and the associated networks may provide a unique opportunity to elucidate underlying mechanisms of how ncRNAs operate in the crosstalk between cancer metabolic reprogramming and metastasis.
Author Contributions
XS designed the study and finalized the manuscript. ZL drafted the manuscript. Both authors approved the final manuscript.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2017YFC1308602) and a Supportive Fund by Heilongjiang Provincial Department of Science and Technology (GX18C010).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ACC, acetyl-CoA carboxylase; ACLY, adenosine triphosphate citrate lyase; ACOX1, Acyl-CoA oxidase 1; AMPK, AMP-activated protein kinase; AMPKβ2, adenosine monophosphate-activated protein kinase subunit β2; ATP, adenosine triphosphate; BNIP3, BCL2/adenovirus E1B 19kDa interacting protein 3; ceRNA, competing endogenous RNA; CKB, creatine kinase brain-type; CTNNB1, beta-catenin 1; CRC, colorectal cancer; EMT, epithelial-mesenchymal transition; FABP4, fatty acid binding protein 4; FASN, fatty acid synthase; FBXL3, F-box/LRR-repeat protein 3; FDA, Food and Drug Administration; PFKFB2, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2; FGFR1, fibroblast growth factor receptor 1; GABARAPL1, gamma-aminobutyric acid receptor-associated protein-like 1; GBC, gallbladder cancer; GDH, glutamate dehydrogenase; GLS, glutaminase; GLUT, glucose transporter; HCC, hepatocellular carcinoma; HIF, hypoxia-inducible factor; HK, hexokinases; HMGR, 3-hydroxy-3-methyl-glutaryl-CoA reductase; H2O2, hydrogen peroxide; HOXA9, Homeobox A9; siRNA, small-interfering RNA; LDH, lactate dehydrogenase; LKB1, liver kinase B1; LIPE, hormone-sensitive lipase; LMW-E, low molecular weight isoform of cyclin E; lncRNA, long-chain non-coding RNA; MMP, metalloproteinase; miRNA, microRNA; ncRNA, non-coding RNA; NOTCH1, Notch homolog 1; OGT, O-GlcNAcylation transferase; OXPHOS, oxidative phosphorylation; PAX8, paired-box gene 8; PDK1, pyruvate dehydrogenase kinase 1; PFKFB, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase; PGC, peroxisome proliferator-activated receptor gamma coactivator; PGK1, phosphoglycerate kinase 1; PLD, Phospholipase D; PKM2, pyruvate kinase M2; PPAR, peroxisome proliferator activated receptor; ROS, oxygen species; SREBP1, sterol regulatory element binding transcription protein 1; TCA, tricarboxylic acid cycle; TGF-β, transforming growth factor-β; TGIF2, TGF-β-induced factor homeobox 2; TME, tumor micro-environment; UDP-GlcNAc, UDP-β-DN-acetylglucosamine; 3′-UTR, three prime untranslated region.
References
1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
2. Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. (2017) 168:670–91. doi: 10.1016/j.cell.2016.11.037
3. Lu M, Zhu WW, Wang X, Tang JJ, Zhang KL, Yu GY, et al. ACOT12-Dependent alteration of acetyl-CoA drives hepatocellular carcinoma metastasis by epigenetic induction of epithelial-mesenchymal transition. Cell Metab. (2019) 29:886–900.e5. doi: 10.1016/j.cmet.2018.12.019
4. Elia I, Doglioni G, Fendt SM. Metabolic hallmarks of metastasis formation. Trends Cell Biol. (2018) 28:673–84. doi: 10.1016/j.tcb.2018.04.002
5. Luo M, Brooks M, Wicha MS. Asparagine and glutamine: co-conspirators fueling metastasis. Cell Metab. (2018) 27:947–9. doi: 10.1016/j.cmet.2018.04.012
6. Schild T, Low V, Blenis J, Gomes AP. Unique metabolic adaptations dictate distal organ-specific metastatic colonization. Cancer Cell. (2018) 33:347–54. doi: 10.1016/j.ccell.2018.02.001
7. Lehuede C, Dupuy F, Rabinovitch R, Jones RG, Siegel PM. Metabolic plasticity as a determinant of tumor growth and metastasis. Cancer Res. (2016) 76:5201–8. doi: 10.1158/0008-5472.CAN-16-0266
8. Beltran-Anaya FO, Cedro-Tanda A, Hidalgo-Miranda A, Romero-Cordoba SL. Insights into the regulatory role of non-coding RNAs in cancer metabolism. Front Physiol. (2016) 7:342. doi: 10.3389/fphys.2016.00342
9. Yu XH, Wang HF, Wu JB, Wang SS, Tang YJ, Tang YL, et al. Non-coding RNAs derailed: the many influences on the fatty acid reprogramming of cancer. Life Sci. (2019) 231:116509. doi: 10.1016/j.lfs.2019.05.065
10. Deng SJ, Chen HY, Zeng Z, Deng S, Zhu S, Ye Z, et al. Nutrient stress-dysregulated antisense lncRNA GLS-AS impairs GLS-mediated metabolism and represses pancreatic cancer progression. Cancer Res. (2019) 79:1398–412. doi: 10.1158/0008-5472.CAN-18-0419
11. Hua S, Lei L, Deng L, Weng X, Liu C, Qi X, et al. miR-139-5p inhibits aerobic glycolysis, cell proliferation, migration, and invasion in hepatocellular carcinoma via a reciprocal regulatory interaction with ETS1. Oncogene. (2018) 37:1624–36. doi: 10.1038/s41388-017-0057-3
12. Warburg O. On the origin of cancer cells. Science. (1956) 123:309–14. doi: 10.1126/science.123.3191.309
13. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. (2009) 324:1029–33. doi: 10.1126/science.1160809
14. Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. (2010) 70:859–62. doi: 10.1158/0008-5472.CAN-09-3556
15. Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. (2016) 5:e189. doi: 10.1038/oncsis.2015.49
16. Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. (2011) 11:85–95. doi: 10.1038/nrc2981
17. Weber GF. Time and circumstances: cancer cell metabolism at various stages of disease progression. Front Oncol. (2016) 6:257. doi: 10.3389/fonc.2016.00257
18. Alderton GK. Metastasis: metabolic reprogramming in disseminated cells. Nat Rev Cancer. (2014) 14:703. doi: 10.1038/nrc3842
19. Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. (2015) 17:183–94. doi: 10.1038/ncb3094
20. Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. (2014) 159:1603–14. doi: 10.1016/j.cell.2014.11.025
21. Loo JM, Scherl A, Nguyen A, Man FY, Weinberg E, Zeng Z, et al. Extracellular metabolic energetics can promote cancer progression. Cell. (2015) 160:393–406. doi: 10.1016/j.cell.2014.12.018
22. Lu J, Tan M, Cai Q. The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. (2015) 356:156–64. doi: 10.1016/j.canlet.2014.04.001
23. Lu J. The Warburg metabolism fuels tumor metastasis. Cancer Metastasis Rev. (2019) 38:157–64. doi: 10.1007/s10555-019-09794-5
24. Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. (2012) 15:827–37. doi: 10.1016/j.cmet.2012.05.001
25. Guppy M, Leedman P, Zu X, Russell V. Contribution by different fuels and metabolic pathways to the total ATP turnover of proliferating MCF-7 breast cancer cells. Biochem J. (2002) 364:309–15. doi: 10.1042/bj3640309
26. Stowe DF, Camara AK. Mitochondrial reactive oxygen species production in excitable cells: modulators of mitochondrial and cell function. Antioxidants Redox Signal. (2009) 11:1373–414. doi: 10.1089/ars.2008.2331
27. Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. (2013) 12:931–47. doi: 10.1038/nrd4002
28. Wang H, Liu X, Long M, Huang Y, Zhang L, Zhang R, et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci Transl Med. (2016) 8:334ra51. doi: 10.1126/scitranslmed.aad6095
29. Tasdogan A, Faubert B, Ramesh V, Ubellacker JM, Shen B, Solmonson A, et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature. (2020) 577:115–20. doi: 10.1038/s41586-019-1847-2
30. Porporato PE, Payen VL, Perez-Escuredo J, De Saedeleer CJ, Danhier P, Copetti T, et al. A mitochondrial switch promotes tumor metastasis. Cell Rep. (2014) 8:754–66. doi: 10.1016/j.celrep.2014.06.043
31. Moloney JN, Cotter TG. ROS signalling in the biology of cancer. Semin Cell Dev Biol. (2018) 80:50–64. doi: 10.1016/j.semcdb.2017.05.023
32. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. (2018) 18:5–18. doi: 10.1038/nrc.2017.99
33. Shankaraiah RC, Veronese A, Sabbioni S, Negrini M. Non-coding RNAs in the reprogramming of glucose metabolism in cancer. Cancer Lett. (2018) 419:167–74. doi: 10.1016/j.canlet.2018.01.048
34. Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. (2012) 13:239–50. doi: 10.1038/nrm3313
35. Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen T, et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology. (2015) 62:1132–44. doi: 10.1002/hep.27929
36. Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. (2009) 28:3526–36. doi: 10.1038/onc.2009.211
37. Tili E, Michaille JJ, Luo Z, Volinia S, Rassenti LZ, Kipps TJ, et al. The down-regulation of miR-125b in chronic lymphocytic leukemias leads to metabolic adaptation of cells to a transformed state. Blood. (2012) 120:2631–8. doi: 10.1182/blood-2012-03-415737
38. Li L, Kang L, Zhao W, Feng Y, Liu W, Wang T, et al. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Lett. (2017) 400:89–98. doi: 10.1016/j.canlet.2017.04.034
39. Ma F, Zhang L, Ma L, Zhang Y, Zhang J, Guo B. MiR-361-5p inhibits glycolytic metabolism, proliferation and invasion of breast cancer by targeting FGFR1 and MMP-1. J Exp Clin Cancer Res. (2017) 36:158. doi: 10.1186/s13046-017-0630-1
40. Lin YH, Wu MH, Huang YH, Yeh CT, Cheng ML, Chi HC, et al. Taurine up-regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology. (2018) 67:188–203. doi: 10.1002/hep.29462
41. Chen J, Yu Y, Li H, Hu Q, Chen X, He Y, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. (2019) 18:33. doi: 10.1186/s12943-019-0947-9
42. Bian Z, Zhang J, Li M, Feng Y, Wang X, Yao S, et al. LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res. (2018) 24:4808–19. doi: 10.1158/1078-0432.CCR-17-2967
43. Wang Y, Zhang X, Wang Z, Hu Q, Wu J, Li Y, et al. LncRNA-p23154 promotes the invasion-metastasis potential of oral squamous cell carcinoma by regulating Glut1-mediated glycolysis. Cancer Lett. (2018) 434:172–83. doi: 10.1016/j.canlet.2018.07.016
44. Yang B, Zhang L, Cao Y, Chen S, Cao J, Wu D, et al. Overexpression of lncRNA IGFBP4-1 reprograms energy metabolism to promote lung cancer progression. Mol Cancer. (2017) 16:154. doi: 10.1186/s12943-017-0722-8
45. Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. (2016) 531:518–22. doi: 10.1038/nature17161
46. Malakar P, Stein I, Saragovi A, Winkler R, Stern-Ginossar N, Berger M, et al. Long noncoding RNA MALAT1 regulates cancer glucose metabolism by enhancing mTOR-mediated translation of TCF7L2. Cancer Res. (2019) 79:2480–93. doi: 10.1158/0008-5472.CAN-18-1432
47. Zhao L, Ji G, Le X, Wang C, Xu L, Feng M, et al. Long noncoding RNA LINC00092 acts in cancer-associated fibroblasts to drive glycolysis and progression of ovarian cancer. Cancer Res. (2017) 77:1369–82. doi: 10.1158/0008-5472.CAN-16-1615
48. Zhang N, Zhang H, Liu Y, Su P, Zhang J, Wang X, et al. SREBP1, targeted by miR-18a-5p, modulates epithelial-mesenchymal transition in breast cancer via forming a co-repressor complex with Snail and HDAC1/2. Cell Death Differ. (2019) 26:843–59. doi: 10.1038/s41418-018-0158-8
49. Vetter G, Saumet A, Moes M, Vallar L, Le Bechec A, Laurini C, et al. miR-661 expression in SNAI1-induced epithelial to mesenchymal transition contributes to breast cancer cell invasion by targeting Nectin-1 and StarD10 messengers. Oncogene. (2010) 29:4436–48. doi: 10.1038/onc.2010.181
50. Singh R, Yadav V, Kumar S, Saini N. MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Sci Rep. (2015) 5:17454. doi: 10.1038/srep17454
51. Gharpure KM, Pradeep S, Sans M, Rupaimoole R, Ivan C, Wu SY, et al. FABP4 as a key determinant of metastatic potential of ovarian cancer. Nat Commun. (2018) 9:2923. doi: 10.1038/s41467-018-04987-y
52. Liu H, Huang X, Ye T. MiR-22 down-regulates the proto-oncogene ATP citrate lyase to inhibit the growth and metastasis of breast cancer. Am J Transl Res. (2018) 10:659–69.
53. Cheng Y, Jia B, Wang Y, Wan S. miR-133b acts as a tumor suppressor and negatively regulates ATP citrate lyase via PPARgamma in gastric cancer. Oncol Rep. (2017) 38:3220–6. doi: 10.3892/or.2017.5944
54. Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. (2015) 75:846–57. doi: 10.1158/0008-5472.CAN-14-1192
55. Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. (2009) 458:762–5. doi: 10.1038/nature07823
56. Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci USA. (2012) 109:8983–8. doi: 10.1073/pnas.1203244109
57. Luan W, Zhang X, Ruan H, Wang J, Bu X. Long noncoding RNA OIP5-AS1 acts as a competing endogenous RNA to promote glutamine catabolism and malignant melanoma growth by sponging miR-217. J Cell Physiol. (2019) 234:16609–18. doi: 10.1002/jcp.28335
58. He J, Li F, Zhou Y, Hou X, Liu S, Li X, et al. LncRNA XLOC_006390 promotes pancreatic carcinogenesis and glutamate metabolism by stabilizing c-Myc. Cancer Lett. (2020) 469:419–28. doi: 10.1016/j.canlet.2019.11.021
59. Li HJ, Li X, Pang H, Pan JJ, Xie XJ, Chen W. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jap J Clin Oncol. (2015) 45:1055–63. doi: 10.1093/jjco/hyv132
60. Sun H, Huang Z, Sheng W, Xu MD. Emerging roles of long non-coding RNAs in tumor metabolism. J Hematol Oncol. (2018) 11:106. doi: 10.1186/s13045-018-0648-7
61. Singh PK, Brand RE, Mehla K. MicroRNAs in pancreatic cancer metabolism. Nat Rev Gastroenterol Hepatol. (2012) 9:334–44. doi: 10.1038/nrgastro.2012.63
62. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. (2008) 7:11–20. doi: 10.1016/j.cmet.2007.10.002
63. Huber V, Camisaschi C, Berzi A, Ferro S, Lugini L, Triulzi T, et al. Cancer acidity: an ultimate frontier of tumor immune escape and a novel target of immunomodulation. Semin Cancer Biol. (2017) 43:74–89. doi: 10.1016/j.semcancer.2017.03.001
64. Rohani N, Hao L, Alexis MS, Joughin BA, Krismer K, Moufarrej MN, et al. Acidification of tumor at stromal boundaries drives transcriptome alterations associated with aggressive phenotypes. Cancer Res. (2019) 79:1952–66. doi: 10.1158/0008-5472.CAN-18-1604
65. Magalhaes MA, Larson DR, Mader CC, Bravo-Cordero JJ, Gil-Henn H, Oser M, et al. Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J Cell Biol. (2011) 195:903–20. doi: 10.1083/jcb.201103045
66. Choi CH, Webb BA, Chimenti MS, Jacobson MP, Barber DL. pH sensing by FAK-His58 regulates focal adhesion remodeling. J Cell Biol. (2013) 202:849–59. doi: 10.1083/jcb.201302131
67. Kamarajugadda S, Stemboroski L, Cai Q, Simpson NE, Nayak S, Tan M, et al. Glucose oxidation modulates anoikis and tumor metastasis. Mol Cell Biol. (2012) 32:1893–907. doi: 10.1128/MCB.06248-11
68. Zhang J, Wang S, Jiang B, Huang L, Ji Z, Li X, et al. c-Src phosphorylation and activation of hexokinase promotes tumorigenesis and metastasis. Nat Commun. (2017) 8:13732. doi: 10.1038/ncomms13732
69. Siu MKY, Jiang YX, Wang JJ, Leung THY, Han CY, Tsang BK, et al. Hexokinase 2 regulates ovarian cancer cell migration, invasion and stemness via FAK/ERK1/2/MMP9/NANOG/SOX9 signaling cascades. Cancers. (2019) 11:813. doi: 10.3390/cancers11060813
70. Chaneton B, Gottlieb E. Rocking cell metabolism: revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem Sci. (2012) 37:309–16. doi: 10.1016/j.tibs.2012.04.003
71. Hamabe A, Konno M, Tanuma N, Shima H, Tsunekuni K, Kawamoto K, et al. Role of pyruvate kinase M2 in transcriptional regulation leading to epithelial-mesenchymal transition. Proc Natl Acad Sci USA. (2014) 111:15526–31. doi: 10.1073/pnas.1407717111
72. Guo M, Zhao X, Yuan X, Jiang J, Li P. MiR-let-7a inhibits cell proliferation, migration, and invasion by down-regulating PKM2 in cervical cancer. Oncotarget. (2017) 8:28226–236. doi: 10.18632/oncotarget.15999
73. Nagarajan A, Dogra SK, Sun L, Gandotra N, Ho T, Cai G, et al. Paraoxonase 2 facilitates pancreatic cancer growth and metastasis by stimulating GLUT1-mediated glucose transport. Mol Cell. (2017) 67:685–701 e6. doi: 10.1016/j.molcel.2017.07.014
74. Ito H, Duxbury M, Zinner MJ, Ashley SW, Whang EE. Glucose transporter-1 gene expression is associated with pancreatic cancer invasiveness and MMP-2 activity. Surgery. (2004) 136:548–56. doi: 10.1016/j.surg.2004.05.032
75. Hatziapostolou M, Polytarchou C, Iliopoulos D. miRNAs link metabolic reprogramming to oncogenesis. Trends Endocrinol Metab. (2013) 24:361–73. doi: 10.1016/j.tem.2013.03.002
76. Jin L, Chun J, Pan C, Alesi GN, Li D, Magliocca KR, et al. Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene. (2017) 36:3797–806. doi: 10.1038/onc.2017.6
77. Hou XM, Yuan SQ, Zhao D, Liu XJ, Wu XA. LDH-A promotes malignant behavior via activation of epithelial-to-mesenchymal transition in lung adenocarcinoma. Biosci Rep. (2019) 39:BSR20181476. doi: 10.1042/BSR20181476
78. Ruan HB, Singh JP, Li MD, Wu J, Yang X. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol Metab. (2013) 24:301–9. doi: 10.1016/j.tem.2013.02.002
79. Ma Z, Vosseller K. O-GlcNAc in cancer biology. Amino Acids. (2013) 45:719–33. doi: 10.1007/s00726-013-1543-8
80. Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S, et al. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. Cell Metab. (2012) 16:226–37. doi: 10.1016/j.cmet.2012.07.006
81. Jiang M, Xu B, Li X, Shang Y, Chu Y, Wang W, et al. O-GlcNAcylation promotes colorectal cancer metastasis via the miR-101-O-GlcNAc/EZH2 regulatory feedback circuit. Oncogene. (2019) 38:301–16. doi: 10.1038/s41388-018-0435-5
82. Labuschagne CF, Cheung EC, Blagih J, Domart MC, Vousden KH. Cell clustering promotes a metabolic switch that supports metastatic colonization. Cell Metab. (2019) 30:720–34.e5. doi: 10.1016/j.cmet.2019.07.014
83. Sotgia F, Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe RC, Witkiewicz AK, et al. Mitochondrial metabolism in cancer metastasis: visualizing tumor cell mitochondria and the “reverse Warburg effect” in positive lymph node tissue. Cell Cycle. (2012) 11:1445–54. doi: 10.4161/cc.19841
84. Lou C, Xiao M, Cheng S, Lu X, Jia S, Ren Y, et al. MiR-485-3p and miR-485-5p suppress breast cancer cell metastasis by inhibiting PGC-1alpha expression. Cell Death Dis. (2016) 7:e2159. doi: 10.1038/cddis.2016.27
85. Hu J, Locasale JW, Bielas JH, O'Sullivan J, Sheahan K, Cantley LC, et al. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat Biotechnol. (2013) 31:522–9. doi: 10.1038/nbt.2530
86. Choudhury AR, Singh KK. Mitochondrial determinants of cancer health disparities. Semin Cancer Biol. (2017) 47:125–46. doi: 10.1016/j.semcancer.2017.05.001
87. Ho J, de Moura MB, Lin Y, Vincent G, Thorne S, Duncan LM, et al. Importance of glycolysis and oxidative phosphorylation in advanced melanoma. Mol Cancer. (2012) 11:76. doi: 10.1186/1476-4598-11-76
88. Griguer CE, Oliva CR, Gillespie GY. Glucose metabolism heterogeneity in human and mouse malignant glioma cell lines. J Neuro Oncol. (2005) 74:123–33. doi: 10.1007/s11060-004-6404-6
89. Dupuy F, Tabaries S, Andrzejewski S, Dong Z, Blagih J, Annis MG, et al. PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab. (2015) 22:577–89. doi: 10.1016/j.cmet.2015.08.007
90. Shu S, Yang Y, Allen CL, Maguire O, Minderman H, Sen A, et al. Metabolic reprogramming of stromal fibroblasts by melanoma exosome microRNA favours a pre-metastatic microenvironment. Sci Rep. (2018) 8:12905. doi: 10.1038/s41598-018-31323-7
91. Menendez JA. Fine-tuning the lipogenic/lipolytic balance to optimize the metabolic requirements of cancer cell growth: molecular mechanisms and therapeutic perspectives. Biochim Biophys Acta. (2010) 1801:381–91. doi: 10.1016/j.bbalip.2009.09.005
92. Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, et al. FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab. (2012) 15:279–91. doi: 10.1016/j.cmet.2011.12.018
93. Luo X, Cheng C, Tan Z, Li N, Tang M, Yang L, et al. Emerging roles of lipid metabolism in cancer metastasis. Mol Cancer. (2017) 16:76. doi: 10.1186/s12943-017-0646-3
94. Hua Y, Qiu Y, Zhao A, Wang X, Chen T, Zhang Z, et al. Dynamic metabolic transformation in tumor invasion and metastasis in mice with LM-8 osteosarcoma cell transplantation. J Proteome Res. (2011) 10:3513–21. doi: 10.1021/pr200147g
95. Hao Q, Li T, Zhang X, Gao P, Qiao P, Li S, et al. Expression and roles of fatty acid synthase in hepatocellular carcinoma. Oncol Rep. (2014) 32:2471–6. doi: 10.3892/or.2014.3484
96. Kuhajda FP. Fatty acid synthase and cancer: new application of an old pathway. Cancer Res. (2006) 66:5977–80. doi: 10.1158/0008-5472.CAN-05-4673
97. Qian X, Hu J, Zhao J, Chen H. ATP citrate lyase expression is associated with advanced stage and prognosis in gastric adenocarcinoma. Int J Clin Exp Med. (2015) 8:7855–60.
98. Lucenay KS, Doostan I, Karakas C, Bui T, Ding Z, Mills GB, et al. Cyclin E associates with the lipogenic enzyme ATP-citrate lyase to enable malignant growth of breast cancer cells. Cancer Res. (2016) 76:2406–18. doi: 10.1158/0008-5472.CAN-15-1646
99. Wen J, Min X, Shen M, Hua Q, Han Y, Zhao L, et al. ACLY facilitates colon cancer cell metastasis by CTNNB1. J Exp Clin Cancer Res. (2019) 38:401. doi: 10.1186/s13046-019-1391-9
100. Wakil SJ, Abu-Elheiga LA. Fatty acid metabolism: target for metabolic syndrome. J Lipid Res. (2009) 50 (Suppl.) S138–43. doi: 10.1194/jlr.R800079-JLR200
101. Brownsey RW, Boone AN, Elliott JE, Kulpa JE, Lee WM. Regulation of acetyl-CoA carboxylase. Biochem Soc Transac. (2006) 34:223–7. doi: 10.1042/BST0340223
102. Wang MD, Wu H, Fu GB, Zhang HL, Zhou X, Tang L, et al. Acetyl-coenzyme A carboxylase alpha promotion of glucose-mediated fatty acid synthesis enhances survival of hepatocellular carcinoma in mice and patients. Hepatology. (2016) 63:1272–86. doi: 10.1002/hep.28415
103. Rios Garcia M, Steinbauer B, Srivastava K, Singhal M, Mattijssen F, Maida A, et al. Acetyl-CoA carboxylase 1-dependent protein acetylation controls breast cancer metastasis and recurrence. Cell Metab. (2017) 26:842–55.e5. doi: 10.1016/j.cmet.2017.09.018
104. Sun LN, Zhi Z, Chen LY, Zhou Q, Li XM, Gan WJ, et al. SIRT1 suppresses colorectal cancer metastasis by transcriptional repression of miR-15b-5p. Cancer Lett. (2017) 409:104–15. doi: 10.1016/j.canlet.2017.09.001
105. Nomura DK, Lombardi DP, Chang JW, Niessen S, Ward AM, Long JZ, et al. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chem Biol. (2011) 18:846–56. doi: 10.1016/j.chembiol.2011.05.009
106. Henkels KM, Boivin GP, Dudley ES, Berberich SJ, Gomez-Cambronero J. Phospholipase D (PLD) drives cell invasion, tumor growth and metastasis in a human breast cancer xenograph model. Oncogene. (2013) 32:5551–62. doi: 10.1038/onc.2013.207
107. Lazar I, Clement E, Dauvillier S, Milhas D, Ducoux-Petit M, LeGonidec S, et al. Adipocyte exosomes promote melanoma aggressiveness through fatty acid oxidation: a novel mechanism linking obesity and cancer. Cancer Res. (2016) 76:4051–7. doi: 10.1158/0008-5472.CAN-16-0651
108. Vara-Perez M, Maes H, Van Dingenen S, Agostinis P. BNIP3 contributes to the glutamine-driven aggressive behavior of melanoma cells. Biol Chem. (2019) 400:187–93. doi: 10.1515/hsz-2018-0208
109. Jin L, Li D, Alesi GN, Fan J, Kang HB, Lu Z, et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. (2015) 27:257–70. doi: 10.1016/j.ccell.2014.12.006
110. Zhang X, Li Z, Xuan Z, Xu P, Wang W, Chen Z, et al. Novel role of miR-133a-3p in repressing gastric cancer growth and metastasis via blocking autophagy-mediated glutaminolysis. J Exp Clin Cancer Res. (2018) 37:320. doi: 10.1186/s13046-018-0993-y
111. Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. (2014) 14:430–9. doi: 10.1038/nrc3726
112. Iwamoto H, Abe M, Yang Y, Cui D, Seki T, Nakamura M, et al. Cancer lipid metabolism confers antiangiogenic drug resistance. Cell Metab. (2018) 28:104–17.e5. doi: 10.1016/j.cmet.2018.05.005
113. Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, Tanaka H, et al. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. (2001) 414:550–4. doi: 10.1038/35107085
114. Zhou L, Wang Y, Zhou M, Zhang Y, Wang P, Li X, et al. HOXA9 inhibits HIF-1alpha-mediated glycolysis through interacting with CRIP2 to repress cutaneous squamous cell carcinoma development. Nat Commun. (2018) 9:1480. doi: 10.1038/s41467-018-03914-5
115. Jia YY, Zhao JY, Li BL, Gao K, Song Y, Liu MY, et al. miR-592/WSB1/HIF-1alpha axis inhibits glycolytic metabolism to decrease hepatocellular carcinoma growth. Oncotarget. (2016) 7:35257–69. doi: 10.18632/oncotarget.9135
116. Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. (2012) 13:251–62. doi: 10.1038/nrm3311
117. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. (2011) 13:132–41. doi: 10.1038/ncb2152
118. Hezel AF, Bardeesy N. LKB1; linking cell structure and tumor suppression. Oncogene. (2008) 27:6908–19. doi: 10.1038/onc.2008.342
119. Gu DN, Jiang MJ, Mei Z, Dai JJ, Dai CY, Fang C, et al. microRNA-7 impairs autophagy-derived pools of glucose to suppress pancreatic cancer progression. Cancer Lett. (2017) 400:69–78. doi: 10.1016/j.canlet.2017.04.020
120. Godlewski J, Nowicki MO, Bronisz A, Nuovo G, Palatini J, De Lay M, et al. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell. (2010) 37:620–32. doi: 10.1016/j.molcel.2010.02.018
121. Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang S, et al. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. (2018) 17:69. doi: 10.1186/s12943-018-0820-2
122. Qi C, Xiaofeng C, Dongen L, Liang Y, Liping X, Yue H, et al. Long non-coding RNA MACC1-AS1 promoted pancreatic carcinoma progression through activation of PAX8/NOTCH1 signaling pathway. J Exp Clin Cancer Res. (2019) 38:344. doi: 10.1186/s13046-019-1332-7
123. Hua H, Zhu Y, Song YH. Ruscogenin suppressed the hepatocellular carcinoma metastasis via PI3K/Akt/mTOR signaling pathway. Biomed Pharmacother. (2018) 101:115–22. doi: 10.1016/j.biopha.2018.02.031
124. Jeong YJ, Choi Y, Shin JM, Cho HJ, Kang JH, Park KK, et al. Melittin suppresses EGF-induced cell motility and invasion by inhibiting PI3K/Akt/mTOR signaling pathway in breast cancer cells. Food Chem Toxicol. (2014) 68:218–25. doi: 10.1016/j.fct.2014.03.022
125. Makinoshima H, Takita M, Saruwatari K, Umemura S, Obata Y, Ishii G, et al. Signaling through the phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) axis is responsible for aerobic glycolysis mediated by glucose transporter in epidermal growth factor receptor (EGFR)-mutated lung adenocarcinoma. J Biol Chem. (2015) 290:17495–504. doi: 10.1074/jbc.M115.660498
126. Yamauchi Y, Furukawa K, Hamamura K. Positive feedback loop between PI3K-Akt-mTORC1 signaling and the lipogenic pathway boosts Akt signaling: induction of the lipogenic pathway by a melanoma antigen. Cancer Res. (2011) 71:4989–97. doi: 10.1158/0008-5472.CAN-10-4108
127. Toda K, Kawada K, Iwamoto M, Inamoto S, Sasazuki T, Shirasawa S, et al. Metabolic alterations caused by KRAS mutations in colorectal cancer contribute to cell adaptation to glutamine depletion by upregulation of asparagine synthetase. Neoplasia. (2016) 18:654–65. doi: 10.1016/j.neo.2016.09.004
128. Hong BS, Ryu HS, Kim N, Kim J, Lee E, Moon H, et al. Tumor suppressor miRNA-204-5p regulates growth, metastasis, and immune microenvironment remodeling in breast cancer. Cancer Res. (2019) 79:1520–34. doi: 10.1158/0008-5472.CAN-18-0891
129. Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. (2019) 18:20. doi: 10.1186/s12943-018-0935-5
130. Yu T, Zhao Y, Hu Z, Li J, Chu D, Zhang J, et al. MetaLnc9 facilitates lung cancer metastasis via a PGK1-activated AKT/mTOR pathway. Cancer Res. (2017) 77:5782–94. doi: 10.1158/0008-5472.CAN-17-0671
131. Bai PS, Xia N, Sun H, Kong Y. Pleiotrophin, a target of miR-384, promotes proliferation, metastasis and lipogenesis in HBV-related hepatocellular carcinoma. J Cell Mol Med. (2017) 21:3023–43. doi: 10.1111/jcmm.13213
132. Tamang S, Acharya V, Roy D, Sharma R, Aryaa A, Sharma U, et al. SNHG12: An LncRNA as a potential therapeutic target and biomarker for human cancer. Front Oncol. (2019) 9:901. doi: 10.3389/fonc.2019.00901
133. Hanna J, Hossain GS, Kocerha J. The potential for microRNA therapeutics and clinical research. Front Genet. (2019) 10:478. doi: 10.3389/fgene.2019.00478
134. Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. (2014) 157:77–94. doi: 10.1016/j.cell.2014.03.008
Keywords: non-coding RNA, metabolic reprogramming, cancer metastasis, microRNA, long non-coding RNA
Citation: Li Z and Sun X (2020) Non-Coding RNAs Operate in the Crosstalk Between Cancer Metabolic Reprogramming and Metastasis. Front. Oncol. 10:810. doi: 10.3389/fonc.2020.00810
Received: 17 February 2020; Accepted: 24 April 2020;
Published: 29 May 2020.
Edited by:
Yue Zhao, University of Cologne, GermanyReviewed by:
Yingjun Zhao, Fudan University Shanghai Cancer Center, ChinaGeorg F. Weber, University of Cincinnati, United States
Copyright © 2020 Li and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueying Sun, c3VueHVleWluZ0BocmJtdS5lZHUuY24=; ay5zdW5AYXVja2xhbmQuYWMubno=
 Ziyi Li
Ziyi Li Xueying Sun
Xueying Sun