- 1Department of Urology, Osaka University Graduate School of Medicine, Suita, Japan
- 2Department Pathology, University of Alabama at Birmingham, Birmingham, AL, United States
- 3Department Pathology, University Hospital Cologne, Cologne, Germany
- 4Department of Therapeutic Urologic Oncology, Osaka University Graduate School of Medicine, Suita, Japan
Recent studies showed the clinical utility of next-generation sequencing of urinary cell-free DNA (cfDNA) from patients with urothelial bladder cancer (UBC). In this study, we aimed to develop urinary cfDNA analysis by droplet digital PCR (ddPCR) as a high-throughput and rapid assay for UBC detection and prognosis. We analyzed urinary cfDNA of 202 samples from 2 cohorts. Test cohort was designed for investigating clinical utility of urinary cfDNA, and was composed of 74 samples from patients with UBC, and 52 samples of benign hematuria patients. Validation cohort was designed for validation and assessment of clinical utility comparing urinary cfDNA with UroVysion (Abbott, Illinois, USA), and was composed of 40 samples from patients with UBC, and 36 prospectively collected samples from patients under surveillance after surgery for urothelial carcinoma. We performed ddPCR analysis of hotspot gene mutations (TERT promoter and FGFR3). In the test cohort, the sensitivity of urinary cfDNA diagnosis was 68.9% (51/74) and the specificity was 100% in patients with UBC. The sensitivity increased to 85.9% when used in conjunction with urine cytology. In addition, patients with high TERT C228T allele frequency (≥14%) had significantly worse prognosis in bladder tumor recurrence than patients with low TERT C228T allele frequency or negative TERT C228T (p = 0.0322). In the validation cohort, the sensitivity of urinary cfDNA was 57.5% (23/40) and the specificity was 100% in UBC patients. The sensitivity of the combination of urine cytology with our hotspot analysis (77.5%) was higher than that of urine cytology with UroVysion (68.9%). In the post-surgical surveillance group, patients positive for the TERT C228T mutation had significantly worse prognosis for bladder tumor recurrence than mutation negative patients (p < 0.001). In conclusion, ddPCR analysis of urinary cfDNA is a simple and promising assay for the clinical setting, surpassing UroVysion for detection and prognosis determination in UBC.
Introduction
Urothelial bladder cancer (UBC) is one of the most common cancers in the world (1). Approximately 70% of UBC patients are diagnosed with non-muscle invasive bladder cancer at initial presentation (2). Non-muscle invasive bladder cancer is treated by transurethral resection of the bladder tumor (TURBT) and intravesical instillation therapy. Fifty to seventy percent of patients experience bladder tumor recurrence and 10–15% experience disease progression to muscle-invasive bladder cancer or distant metastasis (3, 4). For these reasons non-muscle invasive bladder cancer patients require cystoscopy, urine cytology, and computed tomography scans for a long period at regular intervals, but current follow-up methods are suboptimal due to their low sensitivity or high invasiveness (5).
Several urine-based diagnostic tools (UroVysion, NMP22, and etc) are approved for clinical use by The Food and Drug Administration. UroVysion is designed to detect aneuploidy for chromosome 3, 7, 17, and loss of 9p21 locus via fluorescence in situ hybridization (FISH) of cells in urinary sediments (6). Importantly, UroVysion can predict disease recurrence earlier than cystoscopy examination, suggesting that analysis of chromosomal changes by urinary analysis could predict disease recurrence in patients without visible tumors (7). However, none of these currently available tests are recommended for routine use due to their low sensitivity and high cost (2, 8).
There is an urgent need to develop useful and non-invasive assays for detection and surveillance of UBC. Several researchers have reported the utility of genomic analysis of urinary DNA from urothelial carcinoma patients by next-generation sequencing (NGS) (9–11).
NGS methods can produce a large amount of DNA data simultaneously, but a major hurdle in these methods is the data processing steps, or bioinformatics. Analysis by ddPCR can detect mutations with high sensitivity and is easy to interpret. In this study, we analyzed hotspot mutations in UBC using ddPCR analysis of urinary cfDNA focusing on TERT promoter and FGFR3 mutations. Furthermore, we compared the clinical utility of cfDNA analysis with that of UroVysion.
Materials and Methods
Patients and Samples
To select genes suitable for ddPCR analysis, 66 UBC tissues of formalin-fixed paraffin-embedded (FFPE) samples from Japanese patients were obtained by transurethral resection of bladder tumor (TURBT) performed at Osaka University Hospital. The samples were then analyzed by massively parallel sequencing as previously described (Table S1) (12–16).
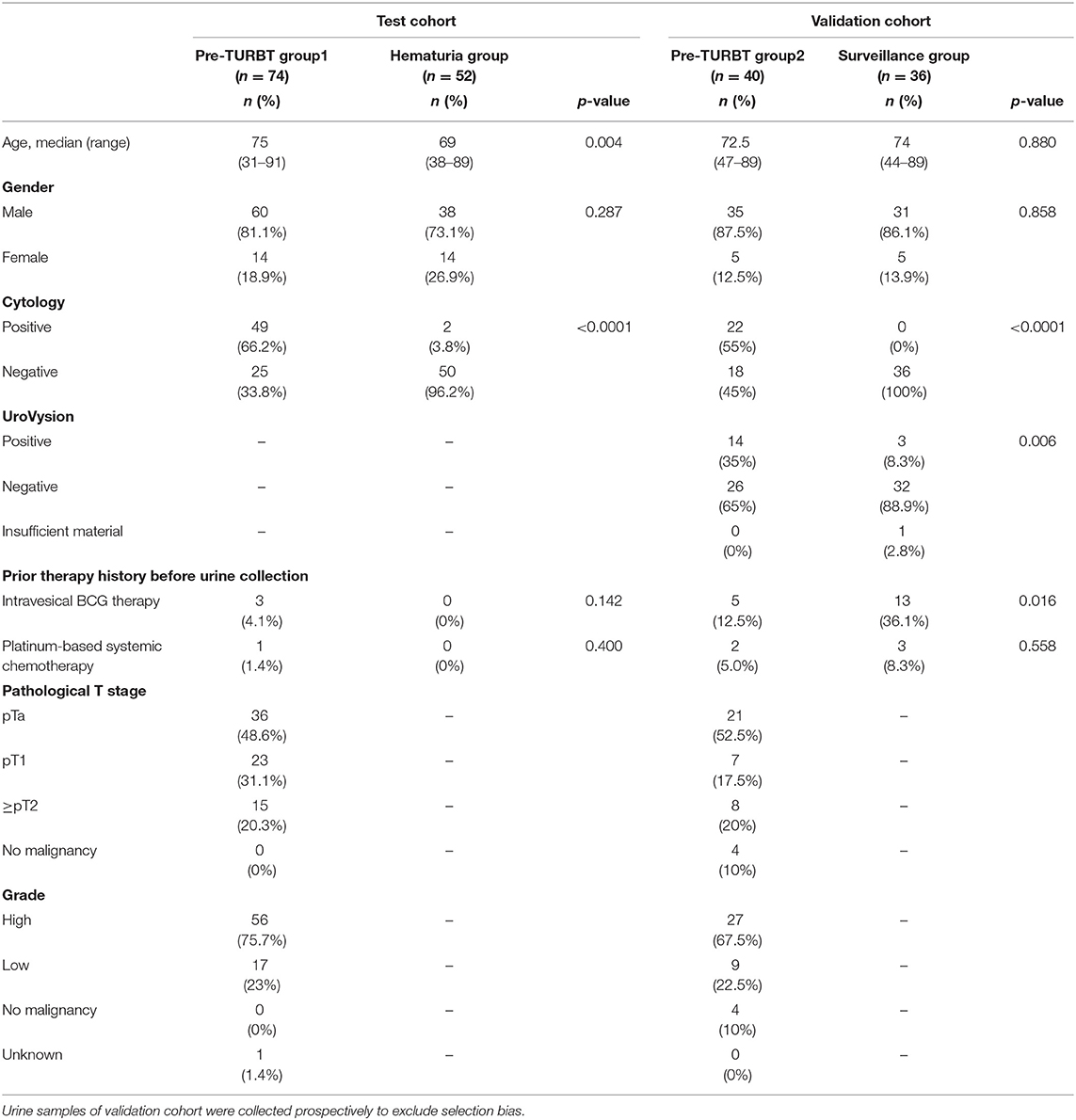
We analyzed 202 urine samples from 2 distinct and independent cohorts: test and validation cohorts. The test cohort consisted of 74 urine samples collected before TURBT (pre-TURBT group 1), and 52 samples collected from patients with microscopic or macroscopic hematuria with no malignant findings in the urinary tract, confirmed by a urologist (hematuria group). In this group, two patients with hematuria and positive urine cytology were also included as they revealed no malignant findings in the lower and upper urinary tract after a follow-up of more than 1 year, confirmed by a detailed examination by a urologist. All the samples of the validation cohort were collected prospectively to exclude the selection bias. The validation cohort was composed of 40 urine samples collected before TURBT (pre-TURBT group 2), and 36 samples collected from patients receiving the standard surveillance protocol after TURBT or radical nephroureterectomy with negative urine cytology (surveillance group) (Table 1). For the validation cohort, we performed both UroVysion assay and urinary cfDNA analysis. All patients were treated at Osaka University Hospital during 2013–2019 and provided written informed consent. This study was approved by the Institutional Review Board of Osaka University (IRB #668-3).
Pathological Diagnosis
Histological diagnosis was performed by experienced pathologists according to the 8th edition of the AJCC stage classification (17), and the World Health Organization 2016 criteria (18). The urine cytology was also evaluated by pathologists according to our institutional criteria. Positive urine cytology was defined to be class IV and V. We used the highest urine cytology class for data analysis of patients receiving several cytology tests.
Sample Processing
The urine samples were processed to obtain the cfDNA following the method described in our earlier study (19). Briefly, post collection, the urine samples were centrifuged at 2,000 × g for 30 min, and the supernatants were stored at −80°C until use. Subsequently, the supernatants were thawed in a water bath at 27°C, and 12 mL of each supernatant was used for cfDNA purification after centrifugation at 16,000 × g for 10 min. The supernatant was processed by QIAamp Circulating Nucleic Acid Kit (QIAGEN, Hilden, Germany) as previously reported (19). The cell pellet was analyzed by UroVysion analysis.
Massively Parallel Sequencing
Mutation and data analysis were performed by massively parallel sequencing that assign a unique identifier to each template molecule for increasing the sensitivity as previously reported. In brief, the TERTSeqS (12–16) assay which targets the promoter region of TERT, and the UroSeqS (12–16) assay which targets 10 genes (CDKN2A, ERBB2, FGFR3, HRAS, KRAS, MET, MLL, PIK3CA, TP53, and VHL) were performed on the 66 FFPE UBC tissue samples. Multiplex PCR was used to detect these mutations. The 59 primer pairs for this PCR were listed in Table S2. PCR products were purified AMPure XP beads (Beckman Coulter, PA, USA) and sequenced on an Illumina Miseq (Illumina, Inc., San Diego, CA, USA). The average unique coverage depth was 27,688 × (range 667–108,370 ×) for TERTSeqs and 3,356 × (range 51–21,650 ×) for UroSeqs. These data of Japanese patients were derived from the results published before (13, 14).
Droplet Digital PCR (ddPCR)
Analysis of urine supernatants by ddPCR was performed on the QX100 Droplet Digital PCR System (Bio-Rad Laboratories, Hercules, CA, USA), including primers and probes (FAM, mutant type; HEX, wild type), and ddPCR Supermix for Probes (No dUTP) according to the manufacturer's protocol. Primers and probes (TERT promoters (g.1295228 C>T:C228T and g.1295250 C>T:C250T), and FGFR3 S249C) for ddPCR, and PCR protocols were used as previously reported (19).
Statistical Analysis
Statistical analysis was performed using JMP Pro 14.0.0 (SAS Institute Inc., Cary, NC, USA). The patient characteristics were compared using the Mann-Whitney U test and chi-square test. A log rank test was performed for analysis of the difference between the two groups. The Cox proportional hazard model was used for univariate and multivariate analysis. The best cutoff value was determined by receiver-operating characteristics curve analysis. Differences were considered statistically significant when the p < 0.05.
Results
Selection of Targeted Genes for Urinary cfDNA Analysis
At least one mutation was detected of 69.7% (46/66) of UBC tissues tested by massively parallel sequencing. TERT promoter mutations were detected in 48.5% (32/66) of tissues, TP53 mutations were found in 27.3% (18/66) of tissues, and FGFR3 mutations were found in 24.2% (16/66) of tissues with high frequency. Although TP53 mutations are frequently found in UBC tissues, TP53 mutations are not suitable for ddPCR analysis because TP53 does not have hotspot mutations (Figure S1). However, TERT promoter mutations C228T and C250T, and FGFR3 S249C are hotspot mutations with high frequency in UBC (Figure S1). We selected these 3 gene regions for urinary cfDNA hotspot mutation analysis by ddPCR.
Urinary cfDNA Analysis of the Test Cohort
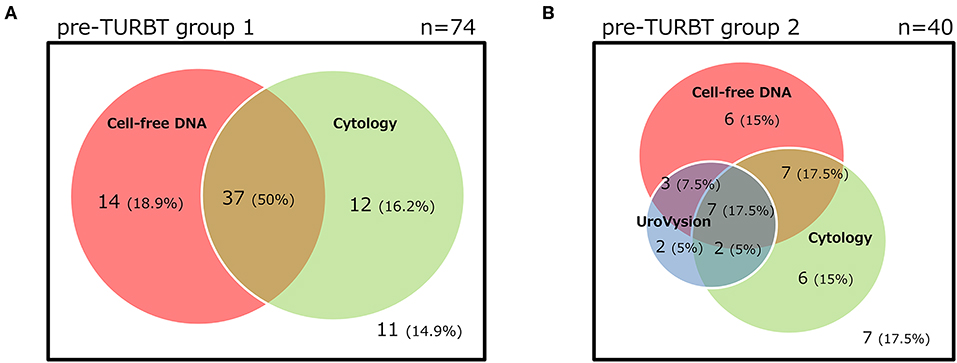
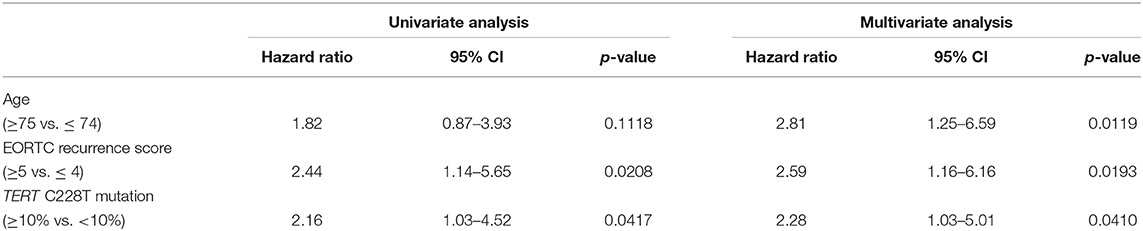
The positive rate of urine cytology was 66.2% (49/74) in pre-TURBT group 1, and 3.8% (2/52) in hematuria group (Table 1). Clinical-pathological features and mutant allele frequency (MAF) of each mutation are shown in Figure 1. Among the patients in the pre-TURBT group 1 before urine samples were collected, three (4.1%) patients had received at least one intravesical BCG therapy, and one (1.4%) patient had been treated with platinum-based regimens as adjuvant systemic chemotherapy for radical nephroureterectomy (Table 1, Figure 1A). The TERT C228T mutation was detected in both ≤ pT1 tumor and ≥pT2 tumor in pre-TURBT group 1 at high frequency (Table 2, Figure 1A). The FGFR3 S249C mutation was mainly detected in ≤ pT1 tumor in pre-TURBT group 1 (Table 2, Figure 1A). There was no association between the positive rate of the three urinary cfDNA mutations and prior BCG instillation therapy history (p = 0.235) (Figure 1A). The sensitivity of urinary cfDNA analysis (TERT C228T, TERT C250T, and FGFR3 S249C) was 68.9% in pre-TURBT 1, and the specificity was 96.2% using the hematuria group as the control cohort. When used in conjunction with urine cytology, the sensitivity increased to 85.1% in pre-TURBT group 1 (Figure 2A). Next, we analyzed the prognostic potential of urinary cfDNA. Although there was no association between presence of the TERT C228T mutation and bladder tumor recurrence, by stratifying the MAF of the TERT C228T at the best cut off point, the MAF of the TERT C228T mutation (≥14%) before TURBT was significantly associated with bladder tumor recurrence (p = 0.0322) (Figure 3). MAF of the TERT C228T mutation in urinary cfDNA before TURBT was an independent factor associated with bladder tumor recurrence after adjustment for European Organization for Research and Treatment of Cancer (EORTC) recurrence score and age (Table 3).
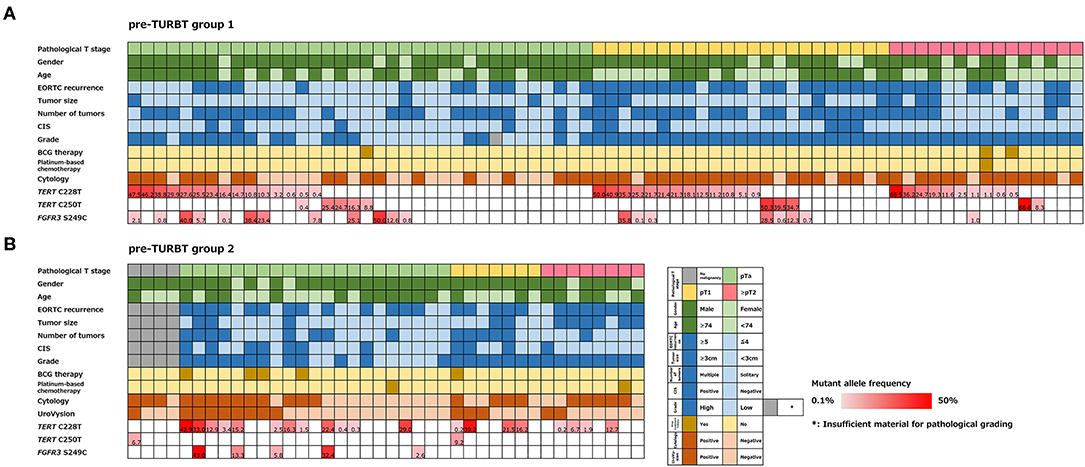
Figure 1. Alteration landscape of pre-TURBT group 1 (A) and 2 (B), combined with tumor stage, gender, age, EORTC recurrence score, tumor size, concurrent CIS, grade, prior treatment history of BCG instillation and platinum-based chemotherapy, urine cytology, UroVysion, and mutant allele frequency (MAF) of each mutation.
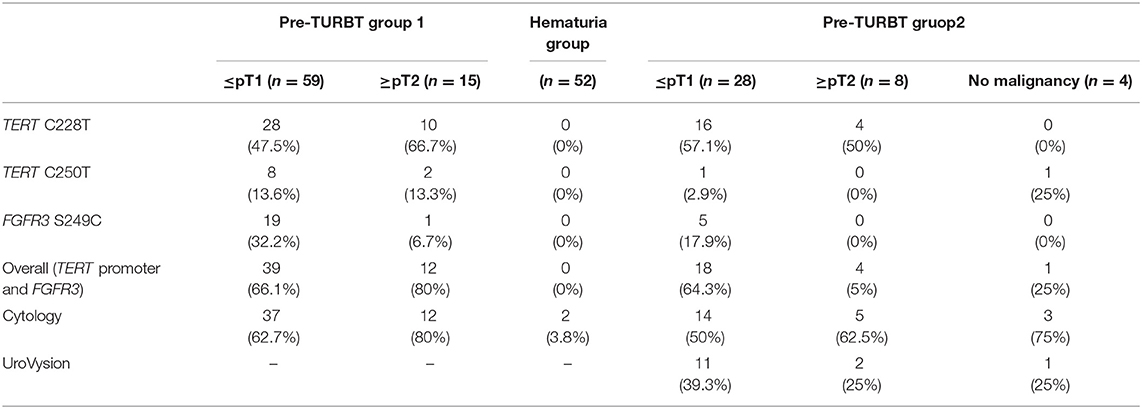
Table 2. Positive rate of urinary cell-free DNA, urine cytology, and UroVysion for pre-TURBT group 1,2 and hematuria group.
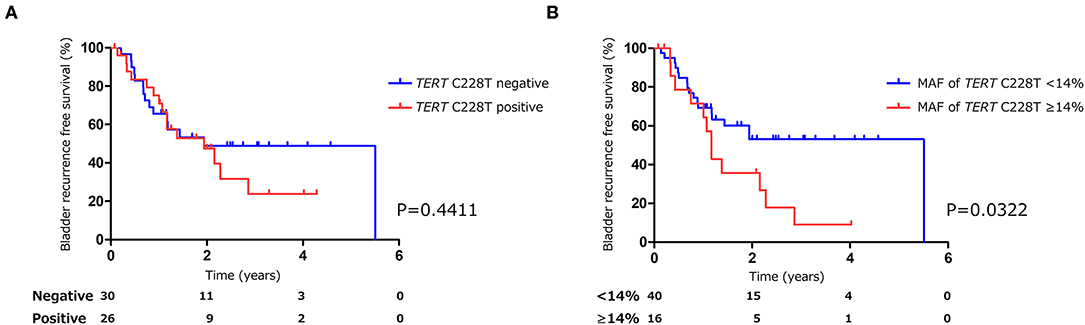
Figure 3. Kaplan-Meier analysis of bladder tumor recurrence free survival of pre-TURBT group1 stratified by TERT C228T mutation (A), and by mutant allele frequency (14%) of TERT C228T (B).
Urinary cfDNA Analysis of the Validation Cohort
The positive rate of urine cytology was 55% (22/40) in pre-TURBT group 2, and 0% in the surveillance group (Table 1). Among the patients in the pre-TURBT group 2 before urine samples were collected at the time of TURBT, five (12.5%) patients had received at least one intravesical BCG therapy, and two (5.0%) had been treated with platinum-based adjuvant chemotherapy for radical nephroureterectomy of upper tract urothelial carcinoma. Among the patients in the surveillance group, 13 (36.1%) patients had received at least one intravesical BCG therapy, and 3 (8.3%) had received platinum-based systemic chemotherapy before urine samples were collected (Table 1). Four patients were determined to have no evidence of tumor malignancy by pathological assessment of TURBT tissues in pre-TURBT group 2. Of these 4, one had the TERT C250T mutation, three had positive urine cytology, and 1 had a positive result by UroVysion (Table 2, Figure 1B). The positive rate of the 3 urinary cfDNA mutations was 57.5% in pre-TURBT group 2. There was no association between the positive rate of the three urinary cfDNA mutations and prior BCG instillation therapy or prior systemic chemotherapy in the pre-TURBT group 2 (p = 0.477), and surveillance group (p = 0.677) (Figure 1B). In addition, the sensitivity of urine cytology in conjunction with urinary cfDNA analysis (77.5%) was higher than when in conjunction with UroVysion (67.5%) (Figure 2B). Of the 36 patients in the surveillance group, 6 patients experienced bladder tumor recurrence during the follow-up period after sample collection (median: 364 days) (Figure 4). Of these 6 patients, 4 were positive for the TERT C228T mutation, but 1 patient tested positive by UroVysion assay. Patients in the surveillance group with the TERT C228T mutation had significantly worse prognosis for bladder tumor recurrence (p = 0.0006). This association was not detected by UroVysion assay (p = 0.6228) (Figure 4).
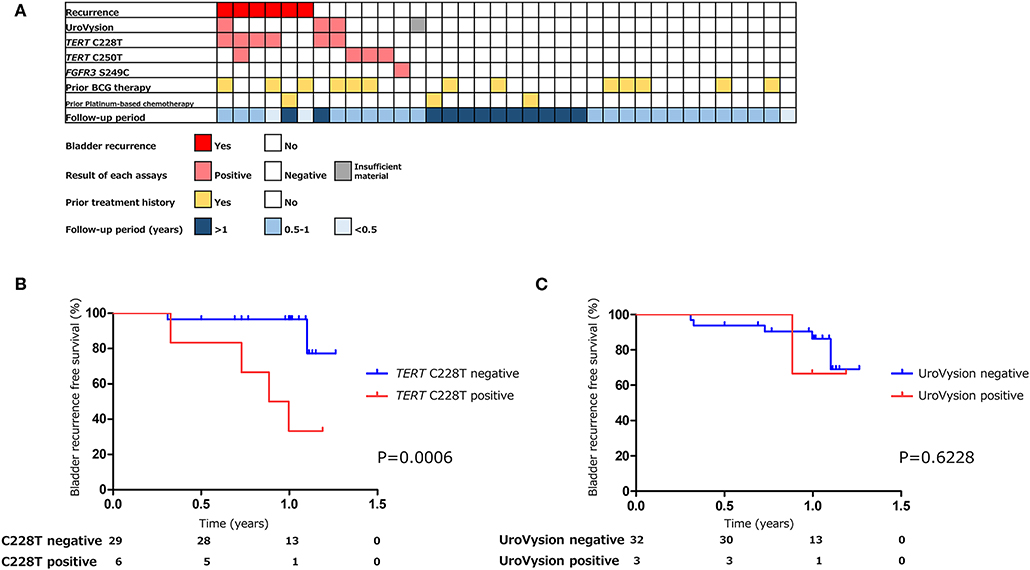
Figure 4. The association of urinary markers and bladder tumor recurrence in surveillance group (n = 36) (A). Kaplan–Meier analysis of bladder tumor recurrence free survival of surveillance group stratified by TERT C228T mutation (B), and UroVysion (C).
Discussion
In this study, we showed the clinical utility of hotspot mutation analysis of urinary cell-free DNA by ddPCR for patients with UBC. This method aids in detection and surveillance of UBC in a non-invasive and simple manner. The sensitivity of urinary cfDNA analysis by ddPCR was high in both pre-TURBT groups. In combination with urine cytology, the sensitivity of urinary cfDNA assay was higher enough for clinical use than UroVysion. The mutant allele frequency of TERT C228T in urinary cfDNA before TURBT was significantly associated with bladder tumor recurrence. Furthermore, the utility of prognostic prediction of urinary cfDNA analysis of the TERT C228T was confirmed by samples prospectively collected in surveillance group. The TERT C228T mutation could predict disease recurrence more accurately than UroVysion.
Because urothelial carcinoma is directly and constantly in contact with urine, urinary cfDNA have clinical potential in cancer screening and disease monitoring of urothelial carcinoma (20, 21). There have been several studies on urinary cfDNA from UBC patients analyzed by NGS (9–11). Dudley et al. reported the usefulness of urinary cfDNA analysis by hybrid capture-based cancer personalized profiling by targeted deep sequencing (9). They reported that the sensitivity for early-stage bladder cancer was 84% (45/54), and urinary cfDNA mutations detected by NGS were significantly associated with bladder tumor recurrence (p < 0.0001). The sensitivity of urinary cfDNA analysis targeting TERT promoter and FGFR3 mutations by ddPCR was lower than the NGS method, but the sensitivity was comparable to the NGS method when combined with urine cytology. Hotspot mutation analysis by ddPCR is simple enough for clinical use. We also showed that hotspot mutations of TERT promoter and FGFR3 are frequently identified in tumor tissues of Japanese UBC patients consistent with those in previous report in Western countries (22). This confirms that our ddPCR method can be utilized for various ethnicities. Mutations in the upstream of TERT gene mainly affect two hotspots (C228T and C250T) (12, 23). This region recruits transcription factor and engages in long-range chromatin interactions (24, 25). The C228T mutation in the TERT promoter region has been shown to increase TERT expression more than the C250T mutation (26). We demonstrated that the TERT C228T mutation was significantly associated with bladder tumor recurrence in the surveillance group, and that ≥14% MAF of TERT C228T, and not just a positive for TERT C228T, was associated with bladder tumor recurrence in pre-TURBT group 1. This observation could be due to the volume of the TERT C228T reflect in bladder cancer activity, playing an important biological role in recurrence of UBC.
UroVysion is a FISH assay that detects aneuploidy of urothelial cells in urine. The overall sensitivity of UroVysion in previous studies varied from 36 to 86% (27, 28). This wide range of clinical performance might be due to bias in the selection of patients. In this study, the sensitivity of UroVysion in pre-TURBT group 2, in which samples were collected prospectively to exclude selection bias, was determined to be 35%. Kim et al. reported that UroVysion can predict disease recurrence and progression in patients with non-muscle invasive bladder cancer present as negative by cystoscopy (7). Of the 6 patients who experienced bladder tumor recurrence during the follow-up period, one had tested positive by UroVysion, and four patients tested positive for TERT C228T. TERT C228T analysis has been shown to predict disease recurrence more precisely UroVysion.
In this study, the MAF of urinary cfDNA was observed to be more than 50% in some cases (Figure 1), and as the occurrence of SNPs in the three gene regions evaluated in this study was rare (29), the mutated genes in urinary cfDNA were thought to be mainly derived from a urothelial tumor, not from normal cells.
This study has several limitations. First, the size of each patient group was small; especially, the number of patients who experienced bladder tumor recurrence was only six in the surveillance group. Larger size and multi-institutional study are needed to warrant current method. Second, the design of the primers needs cost-benefit optimization. The analysis of three gene regions by ddPCR presented in this study is useful in a clinical setting due to its simplicity. Addition of other mutated genes analyzed could increase the sensitivity but would increase the cost per test. Currently, our TERT promoter and FGFR3 mutation analysis is sufficient for clinical use in combination with urine cytology. Finally, there could be possibilities of contamination by blood cells or epithelium though we performed centrifugation to remove cellular components in processing urine samples, which might result in a lower MAF than the actual value. Therefore, careful interpretation should be made for the cases with lower MAF in urinary cfDNA.
In conclusion, we demonstrated the clinical utility of TERT promoter and FGFR3 hotspot mutation analysis in urinary cfDNA from UBC patients by ddPCR. The combination of urine cytology with urinary cfDNA analysis showed a higher sensitivity than when combined with UroVysion, enough to justify utilization of ddPCR analysis in the clinic. Liquid biopsy analysis of TERT promoter and FGFR3 mutations in urinary cfDNA could be a novel diagnostic and prognostic biomarker for UBC.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The studies involving human participants were reviewed and approved by Osaka University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YH: conceptualization, formal analysis, methodology, investigation, and writing-original draft. KF: conceptualization, supervision, and writing-review and editing. KM: sample collection. M-LE: data analysis. ET: investigation and sample collection. MM, YK, KN, CW, and YI: investigation. TK, KH, AK, TU, MU, RI, and NN: supervision. GN: conceptualization and supervision.
Funding
This study was supported by Osaka University Grant.
Conflict of Interest
GN equity or royalty from the licensed technologies from Johns Hopkins that are related to the work described in this paper.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all other researchers in our laboratory.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00755/full#supplementary-material
References
1. Spiess PE, Agarwal N, Bangs R, Boorjian SA, Buyyounouski MK, Clark PE, et al. Bladder cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. (2017) 15:1240–67. doi: 10.6004/jnccn.2017.0156
2. Babjuk M, Burger M, Compérat EM, Gontero P, Mostafid AH, Palou J, et al. European association of urology guidelines on non-muscle-invasive bladder cancer (TaT1 and carcinoma in situ) - (2019). update. Euro Urol. (2019) 76:639–57 doi: 10.1016/j.eururo.2019.08.016
3. Prout GR Jr, Barton BA, Griffin PP, Friedell GH. Treated history of noninvasive grade 1 transitional cell carcinoma. The national bladder cancer group. J Urol. (1992) 148:1413–9. doi: 10.1016/S0022-5347(17)36924-0
4. Sylvester RJ, Van Der Meijden AP, Oosterlinck W, Witjes JA, Bouffioux C, Denis L, et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. (2006) 49:466–5. doi: 10.1016/j.eururo.2005.12.031
5. Lotan Y, Roehrborn CG. Sensitivity and specificity of commonly available bladder tumor markers versus cytology: results of a comprehensive literature review and meta-analyses. Urology. (2003) 61:109–18. doi: 10.1016/S0090-4295(02)02136-2
6. Sokolova IA, Halling KC, Jenkins RB, Burkhardt HM, Meyer RG, Seelig SA, et al. The development of a multitarget, multicolor fluorescence in situ hybridization assay for the detection of urothelial carcinoma in urine. J Mol Diagn. (2000) 2:116–23. doi: 10.1016/S1525-1578(10)60625-3
7. Kim PH, Sukhu R, Cordon BH, Sfakianos JP, Sjoberg DD, Hakimi AA, et al. Reflex fluorescence in situ hybridization assay for suspicious urinary cytology in patients with bladder cancer with negative surveillance cystoscopy. BJU Int. (2014) 114:354–9. doi: 10.1111/bju.12516
8. Chang SS, Boorjian SA, Chou R, Clark PE, Daneshmand S, Konety BR, et al. Diagnosis and treatment of non-muscle invasive bladder cancer: AUA/SUO guideline. J Urol. (2016) 196:1021–9. doi: 10.1016/j.juro.2016.06.049
9. Dudley JC, Schroers-martin J, Lazzareschi DV, Shi WY, Chen SB, Esfahani MS, et al. Detection and surveillance of bladder cancer using urine tumor DNA. Cancer Discov. (2019) 9:500–9. doi: 10.1158/2159-8290.CD-18-0825
10. Satyal U, Srivastava A, Abbosh PH. Urine biopsy-liquid gold for molecular detection and surveillance of bladder cancer. Front Oncol. (2019) 9:1266. doi: 10.3389/fonc.2019.01266
11. Ward DG, Gordon NS, Boucher RH, Pirrie SJ, Baxter L, Ott S, et al. Targeted deep sequencing of urothelial bladder cancers and associated urinary DNA: a 23-gene panel with utility for non-invasive diagnosis and risk stratification. BJU Int. (2019) 124:532–44. doi: 10.1111/bju.14808
12. Kinde I, Munari E, Faraj SF, Hruban RH, Schoenberg M, Bivalacqua, et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. (2013) 73:7162–7. doi: 10.1158/0008-5472.CAN-13-2498
13. Springer SU, Chen CH, Rodriguez Pena MDC, Li L, Douville C, Wang Y, et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. Elife. (2018) 7:e43237. doi: 10.7554/eLife.32143
14. Eich ML, Rodriguez Pena MDC, Springer SU, Taheri D, Tregnago AC, Salles DC, et al. Incidence and distribution of UroSEEK gene panel in a multi-institutional cohort of bladder urothelial carcinoma. Mod Pathol. (2019) 32:1544–50. doi: 10.1038/s41379-019-0276-y
15. Rodriguez Pena MDC, Tregnago AC, Eich ML, Springer S, Wang Y, Taheri D, et al. Spectrum of genetic mutations in de novo PUNLMP of the urinary bladder. Virchows Arch. (2017) 471:761–7. doi: 10.1007/s00428-017-2164-5
16. Nguyen D, Taheri D, Springer S, Cowan M, Guner G, Mendoza Rodriguez MA, et al. High prevalence of TERT promoter mutations in micropapillary urothelial carcinoma. Virchows Arch. (2016) 469:427–34. doi: 10.1007/s00428-016-2001-2
17. American Joint Committee on Cancer. AJCC Cancer Staging Manual, 8th edition. Chicago, IL (2017).
18. WHO Classification of Tumours of Urinary System & Male Genital Organs, 4th edition. Geneva: World Health Organization (2016).
19. Hayashi Y, Fujita K, Matsuzaki K, Matsushita M, Kawamura K, Koh Y, et al. Diagnostic potential of TERT promoter and FGFR3 mutations in urinary cell-free DNA in upper tract urothelial carcinoma. Cancer Sci. (2019) 110:1771–9. doi: 10.1111/cas.14000
20. Hayashi Y, Fujita K. A new era in the detection of urothelial carcinoma by sequencing cell-free DNA. Transl Androl Urol. (2019) 8:S497–501. doi: 10.21037/tau.2019.08.26
21. Yoshida T, Kates M, Fujita K, Bivalacqua TJ, McConkey DJ. Predictive biomarkers for drug response in bladder cancer. Int J Urol. (2019) 26:1044–53. doi: 10.1111/iju.14082
22. Pietzak EJ, Bagrodia A, Cha EK, Drill EN, Iyer G, Isharwal S, et al. ext-generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur Urol:. (2017) 72:952–9. doi: 10.1016/j.eururo.2017.05.032
23. Vinagre J, Almeida A, Pópulo H, Batista R, Lyra J, Pinto V, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. (2013) 4:2185. doi: 10.1038/ncomms3185
24. Stern JL, Theodorescu D, Vogelstein B, Papadopoulos N, Cech TR. Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. (2015) 29:2219–24. doi: 10.1101/gad.269498.115
25. Min J, Shay JW. TERT promoter mutations enhance telomerase activation by long-range chromatin interactions. Cancer Discov. (2016) 6:1212–4. doi: 10.1158/2159-8290.CD-16-1050
26. Chiba K, Lorbeer FK, Shain AH, McSwiggen DT, Schruf E, Oh A, et al. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science. (2017) 357:1416–20. doi: 10.1126/science.aao0535
27. Lokeshwar VB, Habuchi T, Grossman HB, Murphy WM, Hautmann SH, Hemstreet GP III, et al. Bladder tumor markers beyond cytology: international consensus panel on bladder tumor markers. Urology. (2005) 66:35–63. doi: 10.1016/j.urology.2005.08.064
28. van Rhijn BW, van der Poel HG, van der Kwast TH. Urine markers for bladder cancer surveillance: a systematic review. Eur Urol. (2005) 47:736–48. doi: 10.1016/j.eururo.2005.03.014
Keywords: bladder cancer, TERT promoter, FGFR3, cell-free DNA, liquid biopsy, UroVysion, PCR, prognosis
Citation: Hayashi Y, Fujita K, Matsuzaki K, Eich M-L, Tomiyama E, Matsushita M, Koh Y, Nakano K, Wang C, Ishizuya Y, Kato T, Hatano K, Kawashima A, Ujike T, Uemura M, Imamura R, Netto GJ and Nonomura N (2020) Clinical Significance of Hotspot Mutation Analysis of Urinary Cell-Free DNA in Urothelial Bladder Cancer. Front. Oncol. 10:755. doi: 10.3389/fonc.2020.00755
Received: 05 March 2020; Accepted: 20 April 2020;
Published: 19 May 2020.
Edited by:
Masaki Shiota, Kyushu University, JapanReviewed by:
Shigehiro Tsukahara, Kyushu University, JapanTakashi Matsumoto, Kyushu University, Japan
Copyright © 2020 Hayashi, Fujita, Matsuzaki, Eich, Tomiyama, Matsushita, Koh, Nakano, Wang, Ishizuya, Kato, Hatano, Kawashima, Ujike, Uemura, Imamura, Netto and Nonomura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazutoshi Fujita, a2F6dWZ1aml0YTJAZ21haWwuY29t
 Yujiro Hayashi
Yujiro Hayashi Kazutoshi Fujita
Kazutoshi Fujita Kyosuke Matsuzaki1
Kyosuke Matsuzaki1 Kosuke Nakano
Kosuke Nakano Motohide Uemura
Motohide Uemura Ryoichi Imamura
Ryoichi Imamura