- 1Department of Clinical Oncology, Taian City Central Hospital, Tai'an, China
- 2Department of Respiratory Medicine, Taian City Central Hospital, Tai'an, China
- 3Department of Liver Diseases, Central Laboratory, Institute of Clinical Immunology, ShuGuang Hospital Affiliated to Shanghai University of Chinese Traditional Medicine, Shanghai, China
- 4Department of General Surgery, Shuguang Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai, China
- 5Department of Radiation Oncology, The Affiliated Changzhou No.2 People's Hospital of Nanjing Medical University, Changzhou, China
- 6Shanghai University of Medicine & Health Sciences Affiliated Zhoupu Hospital, Shanghai, China
- 7Collaborative Innovation Center for Biomedicine, Shanghai University of Medicine & Health Sciences, Shanghai, China
- 8Shanghai Key Laboratory of Molecular Imaging, Shanghai University of Medicine & Health Sciences, Shanghai, China
CUX1 belongs to the homeodomain transcription factor family and is evolutionarily and functionally conserved from Drosophila to humans. In addition to the involvement in various physiological events including tissue development, cell proliferation, differentiation and migration, and DNA damage response, CUX1 has been implicated in tumorigenesis. Interestingly, CUX1 has been recently recognized as a haploinsufficient tumor suppressor, which is paradoxically overexpressed in tumor cells. While loss of heterozygosity and/or mutations of CUX1 have been frequently detected in many types of cancers, genomic amplification, and overexpression of CUX1 have also been reported in cancer tissues and are correlated with higher tumor grade and poor prognosis. Therefore, deciphering the roles of different CUX1 isoforms and in different tumor stages is required to establish a CUX1-based therapeutic strategy for cancer treatment.
Introduction
CUX1 is previously called CDP (CCAAT displacement protein), Cut-like 1 (CUTL1), or Cut [reviewed in Sansregret and Nepveu (1) and Hulea and Nepveu (2)]. The term “cut” was derived from a Drosophila mutant with the “cut wing” phenotype (3, 4). In 2007, the Human Genome Organization proposed to change the gene root of Cut-like# (CUTL#) to CUT#. Therefore, CUX1 (human gene), Cux1 (mouse gene), and CUX1 (protein) are the simplified nomenclature. CUX1 belongs to the homeodomain (HD) transcription factor family, which was first identified as a sea urchin transcription repressor of the sperm H2B gene by binding to promoter element–CCAAT and competing the binding of other transcriptional activators (5). However, some other studies have shown that CUX1 may also function as either a transcriptional repressor or an activator in a promoter-dependent manner (6–8). CUX1 is present in all metazoans and evolutionarily and functionally conserved from Drosophila to humans, because ectopic expression of human or mouse CUX1 can rescue a wing scalloping mutant phenotype caused by loss of cut (the Drosophila ortholog of CUX1) expression along the prospective wing margins in Drosophila (9). The human CUX1 is at least 340 kb in length and located on the chromosome 7q22 (10). As a transcription factor, CUX1 has been implicated in cell proliferation, differentiation, and migration in various tissues and organs (1, 11–13) [reviewed in Vadnais et al. (8)]. Ectopic overexpression of Cux1 leads to multiorgan hyperplasia in a transgenic mouse model (14, 15). Two distinct Cux1 knockout mouse lines exhibit various phenotypes such as high postnatal lethality, growth retardation, nearly complete hair loss, severely reduced male fertility due to behavioral reasons, cachexia due to muscle wasting and loss of body fat, thin and flaky bones, and abnormal hematopoiesis (16–18) [reviewed in Sansregret and Nepveu (1)]. In addition to its physiological functions, emerging evidence has shown the involvement of CUX1 in tumorigenesis [reviewed in Hulea and Nepveu (2), Liu et al. (19), and Ramdzan and Nepveu (20)], but the exact roles of CUX1 in tumor development are still under debate. In this review, we introduce the protein structures and isoforms of CUX1, describe the various biological processes in which they are involved, summarize the role of CUX1 in tumor development and progression, and discuss the possible explanations related to the paradoxical roles of CUX1 in tumor development.
Structures and Isoforms of CUX1
As a transcriptional factor, CUX1 contains four DNA-binding domains including three Cut repeats (CR1, CR2, and CR3) and one HD (21) (Figure 1). In addition, CUX1 also carries one autoinhibitory domain (ID) at its N-terminus (22) and two active repression domains (R1 and R2) at its C-terminus (23) (Figure 1). CUX1 protein possesses multiple isoforms generated from either proteolysis of full-length CUX1 or alternative transcription initiation of CUX1 gene [reviewed in Sansregret and Nepveu (1) and Hulea and Nepveu (2)]. According to their apparent molecular weight, those CUX1 isoforms were named p200 (full-length CUX1), p150, p110, p90, p80, and p75. Among them, the p150, p110, p90, and p80 are the products of proteolysis of full-length CUX1. The generation of p110 and p90 was mediated by a nuclear cathepsin-L, which removes the N-terminal half of CUX1 (24, 25) (Figure 1), whereas the isoform p80 is a result of two proteolytic events catalyzed by the nuclear cathepsin-L and an unknown caspase at the N- and C-terminal sides, respectively (26), leading to a removal of both the N-terminal half and a region at the C-terminus (Figure 1). Notably, apoptosis onset is not required for such a caspase-mediated p80 processing (26), suggesting the existence of apoptosis-independent role of caspases. The isoform p150 is a proteolytic product of CUX1 at the C-terminal region, but which protease is responsible for p150 processing remains unknown (27, 28). The isoform p75 is encoded by a short CUX1 transcript, which is generated from an alternative transcriptional initiation site within the intron 20 (29). In addition, neutrophil elastase has also been reported to proteolytically process full-length CUX1 to generate short CUX1 isoforms (30, 31).
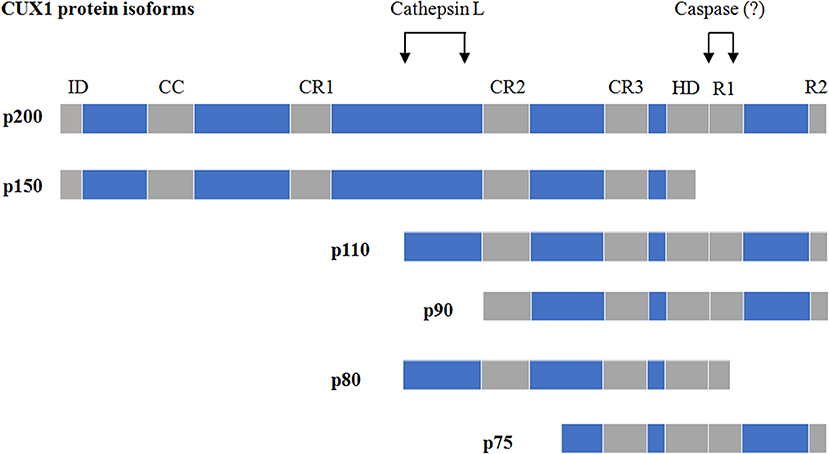
Figure 1. The isoforms of CUX1 and proteinases responsible for their proteolysis. ID, autoinhibitory domain; CR, cut repeats; HD, homeodomain; R1/R2, two active repression domains. The figure is partly modified from Vadnais et al. (8).
The DNA-binding patterns and/or dynamics of CUX1 isoforms are largely determined by which DNA-binding domains are present in them. Although the full-length CUX1 contains all the four DNA-binding domains (three CRs and one HD), it binds to DNA at the –CCAAT motif in a rapid but transient manner and exclusively functions as a transcriptional repressor (32). The isoform p150 with an impaired HD is incapable of binding to DNA and functions as a dominant-negative isoform in the lactating mammary gland (28), whereas the isoforms p110, p90, and p75 with the removal of the N-terminal ID and the CR1 could slowly but stably bind to DNA at the ATCRAT motifs and function as either a repressor or an activator in a promoter-dependent manner (6–8, 33–35).
The Involvement of CUX1 in Various Biological Processes
Physiologically, CUX1 has been reported to play important roles in tissue development, cell migration, proliferation and differentiation, and DNA damage repair.
Roles of CUX1 in the Development of Nervous System
CUX1 expression is detectable in layers II–V in human developing neocortices in the fetal period and disappeared until 3 months of age after birth, suggesting the role of CUX1 in the development of human neocortex (36). Analyses of loss- and gain-of-function of cut mutants in Drosophila have revealed the roles of cut (the Drosophila ortholog of CUX1) in the peripheral nervous system. There are two types of anatomically distinguishable sensory organs, external sensory (es) organs, and internal (chordotonal) sensory organs. In Drosophila, cut is exclusively expressed in cells of the es organ but repressed in cells of the chordotonal organ (37, 38). Therefore, the lethal cut mutants exhibit the transformation of es organs into chordotonal organs (37, 39, 40), whereas forced cut overexpression in Drosophila embryos resulted in the conversion of chordotonal organs into es organs (41). Moreover, the level of Cut is a determinant of the distinct dendrite branching patterns of dendritic arborization (da) sensory in Drosophila (42). Cux1 and Cux2, a homolog protein of Cux1, have been shown to stimulate dendrite branching, spine development, and synapse formation in layers II–III neurons of the cerebral cortex (43, 44). However, an in vitro study showed opposite results indicating that Cux1 suppresses dendritogenesis of neuronal cells (45). Therefore, additional studies are required for clarifying the role of Cux1 in the development of nervous system.
Roles of CUX1 in Cell Proliferation, Differentiation, and Migration
A number of studies have indicated that the expression and/or the DNA-binding dynamics of CUX1 are in a cell cycle–dependent manner (Figure 2). For example, the expression of histone nuclear factor D (HiNF-D), which includes CUX1 as its DNA-binding partner, was upregulated in S-phase in normal cells (46–50); the CUX1-DNA binding was undetectable in G0 and early G1 phase, became detectable in the late G1 phase, and peaked in S phase (11). This dynamic change of CUX1-DNA binding is attributed to at least two posttranslational modifications, Cdc25A-mediated dephosphorylation at the CUX1 HD domain, and cathepsin L–mediated proteolytic cleavage to generate p110 CUX1 (11, 24, 33); in G2 phase, the binding of CUX1-DNA was attenuated due to CyclinA-Cdk1 mediated phosphorylation of CUX1 (51). In addition, Alain Nepveu group also demonstrated that the cyclin B/CDK1–mediated hyperphosphorylation of CUX1 could reset CUX1 DNA-binding activity to the zero level at each cell division (52). On the other hand, a subset of the downstream target genes of CUX1 has been reported to play a role in cell cycle progression (6, 7, 34, 53). These studies together suggest the involvement of CUX1 in cell cycle regulation and cell proliferation. In line with these findings, cells with p110 CUX1 overexpression showed accelerated entry into S phase and cell proliferation, whereas mouse embryo fibroblasts derived from Cux1z/z mutant mice showed an extended G1 phase and retarded cell proliferation (54). Moreover, Cux1 transgenic mice displayed organomegaly and multiorgan hyperplasia (14). All these in vitro and in vivo findings indicate the implication of CUX1 in cell proliferation.
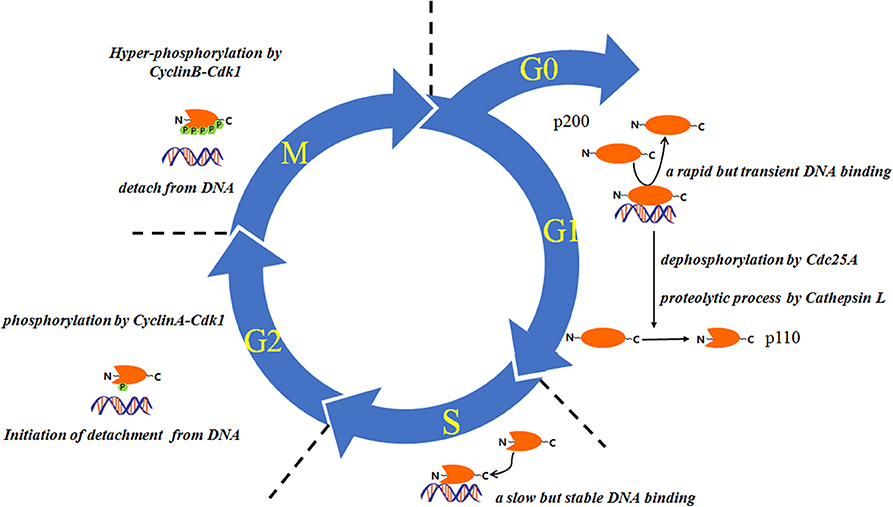
Figure 2. The proteolysis and DNA binding of CUX1 are regulated in a cell cycle–dependent manner. This figure is partly modified from Vanden Heuvel et al. (14).
Early studies have demonstrated that Cux1 is exclusively expressed in undifferentiated cells (21, 55–58), suggesting the role of CUX1 in cell differentiation. In both mice and humans, Cux1 expression is high in long-term hematopoietic stem cell (LT-HSC) but low in short-term hematopoietic stem cell (ST-HSC) and myeloid progenitors, and in vivo Cux1 knockdown led to expansion of myeloid and ST-HSC, suggesting that Cux1 may be essential for maintaining HSC quiescence, suppressing HSC proliferation and self-renewal, and regulating lineage specification and differentiation (59). Cux1 nonfunctional mutant mice on inbred backgrounds die shortly after birth due to retarded differentiation of the lung epithelia, and the survival outbred Cux1 nonfunctional mutant mice exhibit an abnormal pelage because of disrupted hair follicle morphogenesis, suggesting that Cux1 is essential for the differentiation of epithelia in lung and hair follicle (18).
In addition to cell proliferation and differentiation, some recent studies have disclosed the involvement of Cux1 in cell migration and invasion. A high-throughput RNAi screening demonstrated that Cux1 knockdown led to impaired cell migration and invasion in NIH-3T3 and a series of human cancer cell lines (12). Correspondingly, MEF cells (MEFs) derived from Cux1 knockout mice are defective in migration and invasion compared to MEFs derived from wild-type mice (13). Interestingly, the migration defect in Cux1 knockout MEFs can be completely rescued by p110 CUX1 but partially rescued by p200 CUX1 (full-length CUX1) (35). Moreover, stimulated or inhibited proteolytic processing of p200 CUX1 can, respectively, enhance or decrease cell migration, suggesting that CUX1-mediated cell migration may be attributed to its proteolytic products (13, 35).
Relationship Between CUX1 and DNA Damage Repair
Genomic integrity is critical for proper cellular function and faithful transmission of genetic information to progeny. In addition to implicating in cell proliferation, differentiation, and migration, accumulating evidence has indicated the involvement of CUX1 in DNA damage response (DDR). According to a genome-wide location analysis of p110 CUX1, 18 DDR-related genes, including ATM (ATM serine/threonine kinase) and ATR (ATR serine/threonine kinase), were suggested to be the putative targets of p110 CUX1 (6, 7). A subsequent study further confirmed the direct transcriptional regulation of CUX1 on those DDR genes and suggested that CUX1 is required for ATM- and/or ATR-mediated DNA repair in response to DNA damages induced by ionizing radiation (IR) and/or ultraviolet (UV), respectively (6). In addition to direct transcriptional regulation of DDR-related genes, CUX1 was reported to function as an accessory factor to promote DNA damage repair independent of its transcriptional activity (60–64). By directly interacting with 8-oxoguanine DNA glycosylase 1 (OGG1), CUX1 can stimulate the DNA binding, Schiff-base formation, glycosylase, and apurinic/apyrimidinic (AP)-lyase activities of OGG1 to enhance the removal of ROS-induced DNA adducts, 7,8-dihydro-8-oxoguanine (8-oxoG) (63). The direct activity of DNA damage repair of CUX1 is mainly attributed to its CUT domains because a CUX1 recombinant protein containing only CUT domains 1 and 2 is sufficient to accelerate DNA damage repair (62). More interestingly, in line with this finding, some other CUT domain proteins have also shown to be directly involved in base excision repair (65, 66), suggesting that the CUT domain may serve as a therapeutic target of tumor in response to DNA damage.
Roles of CUX1 in Tumor Development
In addition to its physiological functions, CUX1 has been implicated in tumor development in many species including Drosophila, mouse, and humans. But whether CUX1 functions as an oncogene or tumor suppressor is still under debate, because the results of CUX1 studies on tumor development are controversial.
CUX1 Serves as an Oncogene
There are several lines of evidence indicating the oncogenic role of CUX1. First, elevated expression of CUX1 has been observed in many types of cancers, including colorectal cancer (67), multiple myeloma (68), uterine leiomyomas (69), high-grade breast cancer (12), pancreatic cancer (70), melanoma (71), and glioma (72); second, CUX1 expression is positively associated with poor prognosis in glioma, glioblastoma, colorectal cancer, breast cancer, and pancreatic cancer (12, 61, 67, 72); third, mouse mammary tumor virus (MMTV) p200, p110, and p75 CUX1 transgenic mice develop late-onset mammary carcinoma (64, 73); fourth, active Kras mutations, such as KRASG12D and KRASQ61L, have been observed in mammary carcinomas from MMTV-p200 CUX1 transgenic mice, and CUX1 can cooperate with KRASG12V, an active Kras mutant, to promote lung tumor formation in vivo (64); fifth, CUX1 is a transcriptional target downstream of the transforming growth factor β and/or PI3K-AKT signalings and contributes to enhanced proliferation, migration/invasion, and reduced apoptosis in tumor cells (12, 70). More interestingly, a very recent study has demonstrated that CUX1 can generate a circular RNA (circ-CUX1) to promote tumor progression in neuroblastoma (NB) (74). This circ-CUX1 carries exon 2 and partial intron 2 of CUX1 and is up-regulated in NB tissues and cell lines. The levels of circ-CUX1 negatively associate with the survival probability in NB patients. Circ-CUX1 can directly interact with EWSR1 and facilitate EWSR1-MAZ interaction, resulting in transactivation of MAZ and transcriptional alteration of CUX1 and other genes associated with tumor progression (74).
The mechanisms by which CUX1 promotes tumor development have been investigated in many types of cancers especially in breast cancer and pancreatic cancer. For example, by cooperating with GLIS1, CUX1 can stimulate autocrine activation of the Wnt/β-catenin pathway to enhance cell migration and invasion in breast cancer (75). CUX1 stimulates migration and invasion by transcriptionally activating or repressing a series of target genes related to cell motility, including activating snail and slug and repressing E-cadherin, in breast cancer cells (13). CUX1 stabilizes Src and in turn activates its downstream signaling molecules such as RhoA, Rac1, Cdc42, and ROCK by transcriptionally upregulating C-terminal Src kinase (Csk) in pancreatic cancer (76). CUX1 can transcriptionally upregulate WNT5A and GRIA3 to reduce apoptosis and promote proliferation, migration, and invasiveness in pancreatic cancer (77, 78). The p110 CUX1 activates a transcriptional program that reinforces the spindle assembly checkpoint and delays mitosis until extranumerary centrosomes have clustered to two poles, thereby enabling bipolar mitosis and survival of tetraploid cells (53). CUX1 was reported to promote the aggressiveness of pancreatic neuroendocrine tumor partly through modulating MMP9 expression (79). In addition, CUX1 was enriched in tumor-associated macrophages (TAMs) and interacted with nuclear factor κB (NF-κB) p65 to attenuate the activation of NF-κB signaling, leading to a decrease in T-cell attraction and an increase in angiogenesis in pancreatic cancer (80).
CUX1 Acts as a Tumor Suppressor
Although some previous studies have indicated the oncogenic role of CUX1 in tumor progression, other studies also exhibit substantial evidence to support CUX1 as an important tumor suppressor in many types of cancers.
The evidence of CUX1 as a tumor suppressor first emerged from cytogenetic studies showing 7q− (deletions within the long arm of chromosome 7) in many types of cancers, including myeloid leukemia, pancreatic carcinoma, kidney carcinoma, colon carcinoma, ovarian carcinoma, lung carcinoma, head-and-neck carcinoma, cholangiocarcinoma, uterine leiomyoma, and breast cancer (81–88). Next, loss of heterozygosity (LOH) analyses confirmed LOH at 7q22 in a subset of breast cancer, uterine leiomyoma, and ovarian cancer (88–91), suggesting that genes in this region including CUX1 may function as tumor suppressors. Furthermore, CUX1 was late identified as a haploinsufficient tumor suppressor in acute myeloid leukemia, because RNAi-mediated cut (the Drosophila ortholog of CUX1) knockdown led to the development of melanotic pseudotumors in a Drosophila tumor model (92). More importantly, a comprehensive study by interrogating total 7,651 genome sequences derived from 28 tumor types revealed nonsense and frameshift mutations in CUX1 in 1–5% of tumors and found that CUX1 deficiency can lead to activation of the pro-oncogenic PI3K-AKT signaling (93). In addition, CUX1 has been shown to negatively regulate invasion in castrate-resistant prostate cancer (94) and multidrug resistance in gastric cancer (95). It is worth mentioning that, although loss and/or inactivation of a CUX1 allele have been documented in many studies, there is so far no case of a tumor where both alleles have been lost or inactivated, suggesting the coexistence of an inactivated and an activated CUX1 alleles in tumor cells.
Possible Explanations on the Opposite Roles of CUX1 in Tumor Progression
It seems paradoxical that CUX1 possesses both oncogenic and tumor-suppressive features. One possible explanation comes from the protective role of CUX1 in DNA damage repair, because the machinery of DNA damage repair is a double-edge sword in tumor initiation and progression. On the one hand, an effective DNA-repair machinery is required to prevent accumulation of DNA lesions, genomic instability, and subsequent malignant transformation in normal cells, suggesting the suppressive roles of DNA repair in tumor initiation (96). On the other hand, a basal repair activity is also essential for tumor cells to avoid DNA damage–induced cell death (97). Given the involvement of CUX1 in both exogenous DNA damage (induced by temozolomide, H2O2, UV, and IR) and endogenous DNA damage (induced by intracellular ROS) (6, 7, 61, 63, 64), it is conceivable that CUX1 may, respectively, function as a tumor suppressor or an oncogene in the stages of tumor initiation or progression.
The second explanation rises from the existence of various CUX1 isoforms and their divergent transcriptional activities. By alternative transcriptional initiation and/or proteolytic processing, CUX1 generates several short isoforms that possess distinct DNA-binding capacity and transcriptional activities compared to the full-length CUX1 (p200 CUX1). p200 CUX1 binds to DNA in a rapid but transient manner and exclusively functions as a transcriptional repressor through either competition occupancy for CCAAT or Sp1 binding sites or active repression by the recruitment of histone deacetylases (HDACs) (23, 98). However, in contrast to full-length CUX1, the short isoforms can bind to DNA stably and function as either transcriptional repressors or activators in a promote-specific manner (33–35). Because most of the short isoforms are derived from the full-length CUX1 transcript, RNAi approach is not able to specifically knock down indicated isoforms, which makes RNAi not suitable to determine the roles of full-length CUX1 and its isoforms in tumor progression. So far, from the results of overexpression studies, it is clear that the CUX1 short isoforms may mainly function as oncogenes. For example, MMTV-p110, p75 transgenic mice developed mammary tumors after a long latency period, and genes involved in Wnt/β-catenin signaling were directly regulated by those short CUX1 isoforms (73). Stimulation or inhibition of the proteolytic processing of p200 CUX1 toward p110 CUX1 can, respectively, enhance or attenuate cell migration, suggesting that the p110 CUX1 but not the p200 CUX1 plays a major role in promoting cell migration (13). p200 CUX1 is proteolytically processed into p110 CUX1 by a nuclear cathepsin L at the G1/S transition, and forced overexpression of p110 CUX1 stimulates cell proliferation (24, 54). Moreover, the nuclear accumulation and activity of cathepsin L were increased in many transformed cells in parallel with augmented CUX1 processing, and the cell-permeable but not the non–cell-permeable inhibitors of cathepsin L delay the entry into S phase and proliferation in transformed cells (99). These findings suggest that the short CUX1 isoforms contribute to its oncogenic role in tumor progression. However, the exact role of p200 CUX1 in tumor progression is still under debate. A previous study had shown that MMTV-p200 mice developed mammary tumors with a slightly higher penetrance than the MMTV-p75 or p110 CUX1 mice by promoting faster DNA repair, thereby allowing transformed cells to avoid senescence and continue to proliferate (64), suggesting the oncogenic role of p200 CUX1. But recent findings showing that many types of tumors possess nonsense or frameshift mutations of CUX1 paradoxically suggest that p200 CUX1 may function as a tumor suppressor (93). Very recently, by employing K562 cells, which predominantly express the p200 CUX1, Arthur et al. (100) have demonstrated that p200 CUX1 binds distal cis-regulatory elements associated with gene activation, but the coexistence of p200 CUX1 and its short isoforms in many other cancer cells is still a bottleneck in the process to disclose the exact roles of them in tumor development. Therefore, more precise and sophisticated studies, for example, by employing CRISPR/Cas9 genome editing approach to establish either p200 CUX1 unprocessable or p200 CUX1 null mutants, are required to define the roles of p200 CUX1 and its isoforms in tumor development.
The third explanation derives from the presence of CUX1 in both tumor cells and TAMs and the inhibitory effects of CUX1 on the NF-κB signaling. The Michl P group has demonstrated the overexpression of CUX1 in both tumor cells and TAMs in pancreatic cancer (70, 80). They demonstrated that the full-length CUX1 interacts with NF-κB p65 and HDAC1 to form a protein trimer to repress NF-κB signaling in TAMs, resulting in inhibition of M1 polarization and enhanced angiogenesis and tumor progression (80). While inhibition on NF-κB signaling in TAMs promotes tumor progression, aberrant and constitutive activation NF-κB signaling is frequent in tumoral cells and shows positive effects on tumor progression (101, 102). Therefore, if CUX1 can inhibit NF-κB signaling in both tumor cells and TAMs, CUX1-mediated inhibition on NF-κB signaling in tumor cells or TAMs may, respectively, suppress or promote tumor development, which may contribute to the paradoxical roles of CUX1 to tumor development.
Summary
As an evolutionarily conserved transcription factor, CUX1 is expressed in almost all metazoans. Because of the proteolytic processing or alternative transcriptional initiation, CUX1 possesses multiple isoforms with differential DNA-binding capacity and transcriptional activity. The full-length CUX1 binds to DNA in a rapid but transient manner to exclusively repress gene expression by either passively occupying the binding sites of transcriptional activators or actively recruiting HDACs to achieve epigenetic silencing. CUX1 is physiologically implicated in tissue development, cell proliferation, differentiation and migration, and DNA damage repair. The inbred Cux1−/− or inactive mice are postnatal lethal due to retarded differentiation of the lung epithelia, and the survival outbred Cux1−/− mice exhibit an abnormal pelage because of disrupted hair follicle morphogenesis. The pathological involvement of CUX1 in tumorigenesis is complicated. Both sides of evidence, respectively, support the tumor suppressive or oncogenic roles of CUX1 in tumor development and progression. So far, the short isoforms of CUX1, such as p110 and p75 CUX1, seem to carry oncogenic features, while the exact role of full-length CUX1 in tumor progression remains elusive. Therefore, further studies specifically targeting full-length CUX1 or short isoforms are required to decipher the role of CUX1 in tumor progression.
Author Contributions
NL, QS, LW, XW, and YF performed extensive literature search and discussion. NL drafted the manuscript. JL and HW edited the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31671453, 31870905 to HW, 81773224 to JL), the Scientific Program of Shanghai Municipal Health Commission (SHWJ2019211 to HW) and the Scientific Program of Jiangsu Province (BK20191157, BE2018643 to JL).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Sansregret L, Nepveu A. The multiple roles of CUX1: insights from mouse models and cell-based assays. Gene. (2008) 412:84–94. doi: 10.1016/j.gene.2008.01.017
2. Hulea L, Nepveu A. CUX1 transcription factors: from biochemical activities and cell-based assays to mouse models and human diseases. Gene. (2012) 497:18–26. doi: 10.1016/j.gene.2012.01.039
3. Jack J, Dorsett D, Delotto Y, Liu S. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development. (1991) 113:735–47.
4. Liu S, McLeod E, Jack J. Four distinct regulatory regions of the cut locus and their effect on cell type specification in Drosophila. Genetics. (1991) 127:151–9.
5. Barberis A, Superti-Furga G, Busslinger M. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell. (1987) 50:347–59. doi: 10.1016/0092-8674(87)90489-2
6. Vadnais C, Davoudi S, Afshin M, Harada R, Dudley R, Clermont PL, et al. CUX1 transcription factor is required for optimal ATM/ATR-mediated responses to DNA damage. Nucleic Acids Res. (2012) 40:4483–95. doi: 10.1093/nar/gks041
7. Harada R, Vadnais C, Sansregret L, Leduy L, Berube G, Robert F, et al. Genome-wide location analysis and expression studies reveal a role for p110 CUX1 in the activation of DNA replication genes. Nucleic Acids Res. (2008) 36:189–202. doi: 10.1093/nar/gkm970
8. Vadnais C, Awan AA, Harada R, Clermont PL, Leduy L, Berube G, et al. Long-range transcriptional regulation by the p110 CUX1 homeodomain protein on the ENCODE array. BMC Genomics. (2013) 14:258. doi: 10.1186/1471-2164-14-258
9. Ludlow C, Choy R, Blochlinger K. Functional analysis of Drosophila and mammalian cut proteins in flies. Dev Biol. (1996) 178:149–59. doi: 10.1006/dbio.1996.0205
10. Thoennissen NH, Lasho T, Thoennissen GB, Ogawa S, Tefferi A, Koeffler HP. Novel CUX1 missense mutation in association with 7q- at leukemic transformation of MPN. Am J Hematol. (2011) 86:703–5. doi: 10.1002/ajh.22069
11. Coqueret O, Berube G, Nepveu A. The mammalian cut homeodomain protein functions as a cell-cycle-dependent transcriptional repressor which downmodulates p21WAF1/CIP1/SDI1 in S phase. EMBO J. (1998) 17:4680–94. doi: 10.1093/emboj/17.16.4680
12. Michl P, Ramjaun AR, Pardo OE, Warne PH, Wagner M, Poulsom R, et al. CUTL1 is a target of TGF(beta) signaling that enhances cancer cell motility and invasiveness. Cancer Cell. (2005) 7:521–32. doi: 10.1016/j.ccr.2005.05.018
13. Kedinger V, Sansregret L, Harada R, Vadnais C, Cadieux C, Fathers K, et al. p110 CUX1 homeodomain protein stimulates cell migration and invasion in part through a regulatory cascade culminating in the repression of E-cadherin and occludin. J Biol Chem. (2009) 284:27701–11. doi: 10.1074/jbc.M109.031849
14. Vanden Heuvel GB, Brantley JG, Alcalay NI, Sharma M, Kemeny G, Warolin J, et al. Hepatomegaly in transgenic mice expressing the homeobox gene Cux-1. Mol Carcinog. (2005) 43:18–30. doi: 10.1002/mc.20091
15. Ledford AW, Brantley JG, Kemeny G, Foreman TL, Quaggin SE, Igarashi P, et al. Deregulated expression of the homeobox gene Cux-1 in transgenic mice results in downregulation of p27(kip1) expression during nephrogenesis, glomerular abnormalities, and multiorgan hyperplasia. Dev Biol. (2002) 245:157–71. doi: 10.1006/dbio.2002.0636
16. Luong MX, Van Der Meijden CM, Xing D, Hesselton R, Monuki ES, Jones SN, et al. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol Cell Biol. (2002) 22:1424–37. doi: 10.1128/MCB.22.5.1424-1437.2002
17. Sinclair AM, Lee JA, Goldstein A, Xing D, Liu S, Ju R, et al. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood. (2001) 98:3658–67. doi: 10.1182/blood.V98.13.3658
18. Ellis T, Gambardella L, Horcher M, Tschanz S, Capol J, Bertram P, et al. The transcriptional repressor CDP. (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. (2001) 15:2307–19. doi: 10.1101/gad.200101
19. Liu KC, Lin BS, Zhao M, Wang KY, Lan XP. Cutl1: a potential target for cancer therapy. Cell Signal. (2013) 25:349–54. doi: 10.1016/j.cellsig.2012.10.008
20. Ramdzan ZM, Nepveu A. CUX1, a haploinsufficient tumour suppressor gene overexpressed in advanced cancers. Nat Rev Cancer. (2014) 14:673–82. doi: 10.1038/nrc3805
21. Neufeld EJ, Skalnik DG, Lievens PM, Orkin SH. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat Genet. (1992) 1:50–5. doi: 10.1038/ng0492-50
22. Truscott M, Raynal L, Wang Y, Berube G, Leduy L, Nepveu A. The N-terminal region of the CCAAT displacement protein. (CDP)/Cux transcription factor functions as an autoinhibitory domain that modulates DNA binding. J Biol Chem. (2004) 279:49787–94. doi: 10.1074/jbc.M409484200
23. Mailly F, Berube G, Harada R, Mao PL, Phillips S, Nepveu A. The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol Cell Biol. (1996) 16:5346–57. doi: 10.1128/MCB.16.10.5346
24. Goulet B, Baruch A, Moon NS, Poirier M, Sansregret LL, Erickson A, et al. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol Cell. (2004) 14:207–19. doi: 10.1016/S1097-2765(04)00209-6
25. Goulet B, Truscott M, Nepveu A. A novel proteolytically processed CDP/Cux isoform of 90 kDa is generated by cathepsin L. Biol Chem. (2006) 387:1285–93. doi: 10.1515/BC.2006.159
26. Truscott M, Denault JB, Goulet B, Leduy L, Salvesen GS, Nepveu A. Carboxyl-terminal proteolytic processing of CUX1 by a caspase enables transcriptional activation in proliferating cells. J Biol Chem. (2007) 282:30216–26. doi: 10.1074/jbc.M702328200
27. Zhu Q, Gregg K, Lozano M, Liu J, Dudley JP. CDP is a repressor of mouse mammary tumor virus expression in the mammary gland. J Virol. (2000) 74:6348–57. doi: 10.1128/JVI.74.14.6348-6357.2000
28. Maitra U, Seo J, Lozano MM, Dudley JP. Differentiation-induced cleavage of Cutl1/CDP generates a novel dominant-negative isoform that regulates mammary gene expression. Mol Cell Biol. (2006) 26:7466–78. doi: 10.1128/MCB.01083-06
29. Goulet B, Watson P, Poirier M, Leduy L, Berube G, Meterissian S, et al. Characterization of a tissue-specific CDP/Cux isoform, p75, activated in breast tumor cells. Cancer Res. (2002) 62:6625–33.
30. Goulet B, Markovic Y, Leduy L, Nepveu A. Proteolytic processing of cut homeobox 1 by neutrophil elastase in the MV4;11 myeloid leukemia cell line. Mol Cancer Res. (2008) 6:644–53. doi: 10.1158/1541-7786.MCR-07-0268
31. Yu L, Zhong L, Xiong L, Dan W, Li J, Ye J, et al. Neutrophil elastase-mediated proteolysis of the tumor suppressor p200 CUX1 promotes cell proliferation and inhibits cell differentiation in APL. Life Sci. (2020) 242:117229. doi: 10.1016/j.lfs.2019.117229
32. Moon NS, Berube G, Nepveu A. CCAAT displacement activity involves CUT repeats 1 and 2, not the CUT homeodomain. J Biol Chem. (2000) 275:31325–34. doi: 10.1074/jbc.M002912200
33. Moon NS, Premdas P, Truscott M, Leduy L, Berube G, Nepveu A. S phase-specific proteolytic cleavage is required to activate stable DNA binding by the CDP/Cut homeodomain protein. Mol Cell Biol. (2001) 21:6332–45. doi: 10.1128/MCB.21.18.6332-6345.2001
34. Truscott M, Harada R, Vadnais C, Robert F, Nepveu A. p110 CUX1 cooperates with E2F transcription factors in the transcriptional activation of cell cycle-regulated genes. Mol Cell Biol. (2008) 28:3127–38. doi: 10.1128/MCB.02089-07
35. Kedinger V, Nepveu A. The roles of CUX1 homeodomain proteins in the establishment of a transcriptional program required for cell migration and invasion. Cell Adh Migr. (2010) 4:348–52. doi: 10.4161/cam.4.3.11407
36. Saito T, Hanai S, Takashima S, Nakagawa E, Okazaki S, Inoue T, et al. Neocortical layer formation of human developing brains and lissencephalies: consideration of layer-specific marker expression. Cereb Cortex. (2011) 21:588–96. doi: 10.1093/cercor/bhq125
37. Blochlinger K, Bodmer R, Jan LY, Jan YN. Patterns of expression of cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. (1990) 4:1322–31. doi: 10.1101/gad.4.8.1322
38. Jarman AP, Ahmed I. The specificity of proneural genes in determining Drosophila sense organ identity. Mech Dev. (1998) 76:117–25. doi: 10.1016/S0925-4773(98)00116-6
39. Bodmer R, Barbel S, Sheperd S, Jack JW, Jan LY, Jan YN. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell. (1987) 51:293–307. doi: 10.1016/0092-8674(87)90156-5
40. Merritt DJ, Hawken A, Whitington PM. The role of the cut gene in the specification of central projections by sensory axons in Drosophila. Neuron. (1993) 10:741–52. doi: 10.1016/0896-6273(93)90174-P
41. Blochlinger K, Jan LY, Jan YN. Transformation of sensory organ identity by ectopic expression of cut in Drosophila. Genes Dev. (1991) 5:1124–35. doi: 10.1101/gad.5.7.1124
42. Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. (2003) 112:805–18. doi: 10.1016/S0092-8674(03)00160-0
43. Cubelos B, Nieto M. Intrinsic programs regulating dendrites and synapses in the upper layer neurons of the cortex. Commun Integr Biol. (2010) 3:483–6. doi: 10.4161/cib.3.6.12755
44. Cubelos B, Sebastian-Serrano A, Beccari L, Calcagnotto ME, Cisneros E, Kim S, et al. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron. (2010) 66:523–35. doi: 10.1016/j.neuron.2010.04.038
45. Li N, Zhao CT, Wang Y, Yuan XB. The transcription factor Cux1 regulates dendritic morphology of cortical pyramidal neurons. PLoS ONE. (2010) 5:e10596. doi: 10.1371/journal.pone.0010596
46. van Wijnen AJ, Wright KL, Lian JB, Stein JL, Stein GS. Human H4 histone gene transcription requires the proliferation-specific nuclear factor HiNF-D. Auxiliary roles for HiNF-C. (Sp1-like) and HiNF-A. (high mobility group-like). J Biol Chem. (1989) 264:15034–42.
47. Holthuis J, Owen TA, Van Wijnen AJ, Wright KL, Ramsey-Ewing A, Kennedy MB, et al. Tumor cells exhibit deregulation of the cell cycle histone gene promoter factor HiNF-D. Science. (1990) 247:1454–7. doi: 10.1126/science.2321007
48. Van Wijnen AJ, Owen TA, Holthuis J, Lian JB, Stein JL, Stein GS. Coordination of protein-DNA interactions in the promoters of human H4, H3, and H1 histone genes during the cell cycle, tumorigenesis, and development. J Cell Physiol. (1991) 148:174–89. doi: 10.1002/jcp.1041480120
49. Wright KL, Dell'Orco RT, van Wijnen AJ, Stein JL, Stein GS. Multiple mechanisms regulate the proliferation-specific histone gene transcription factor HiNF-D in normal human diploid fibroblasts. Biochemistry. (1992) 31:2812–8. doi: 10.1021/bi00125a023
50. Van Wijnen AJ, Van Gurp MF, De Ridder MC, Tufarelli C, Last TJ, Birnbaum M, et al. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc Natl Acad Sci USA. (1996) 93:11516–21. doi: 10.1073/pnas.93.21.11516
51. Santaguida M, Ding Q, Berube G, Truscott M, Whyte P, Nepveu A. Phosphorylation of the CCAAT displacement protein. (CDP)/Cux transcription factor by cyclin A-Cdk1 modulates its DNA binding activity in G(2). J Biol Chem. (2001) 276:45780–90. doi: 10.1074/jbc.M107978200
52. Sansregret L, Gallo D, Santaguida M, Leduy L, Harada R, Nepveu A. Hyperphosphorylation by cyclin B/CDK1 in mitosis resets CUX1 DNA binding clock at each cell cycle. J Biol Chem. (2010) 285:32834–43. doi: 10.1074/jbc.M110.156406
53. Sansregret L, Vadnais C, Livingstone J, Kwiatkowski N, Awan A, Cadieux C, et al. Cut homeobox 1 causes chromosomal instability by promoting bipolar division after cytokinesis failure. Proc Natl Acad Sci USA. (2011) 108:1949–54. doi: 10.1073/pnas.1008403108
54. Sansregret L, Goulet B, Harada R, Wilson B, Leduy L, Bertoglio J, et al. The p110 isoform of the CDP/Cux transcription factor accelerates entry into S phase. Mol Cell Biol. (2006) 26:2441–55. doi: 10.1128/MCB.26.6.2441-2455.2006
55. Skalnik DG, Strauss EC, Orkin SH. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J Biol Chem. (1991) 266:16736–44.
56. Luo W, Skalnik DG. CCAAT displacement protein competes with multiple transcriptional activators for binding to four sites in the proximal gp91phox promoter. J Biol Chem. (1996) 271:18203–10. doi: 10.1074/jbc.271.30.18203
57. Pattison S, Skalnik DG, Roman A. CCAAT displacement protein, a regulator of differentiation-specific gene expression, binds a negative regulatory element within the 5' end of the human papillomavirus type 6 long control region. J Virol. (1997) 71:2013–22. doi: 10.1128/JVI.71.3.2013-2022.1997
58. Skalnik DG. Transcriptional mechanisms regulating myeloid-specific genes. Gene. (2002) 284:1–21. doi: 10.1016/S0378-1119(02)00387-6
59. An N, Khan S, Imgruet MK, Gurbuxani SK, Konecki SN, Burgess MR, et al. Gene dosage effect of CUX1 in a murine model disrupts HSC homeostasis and controls the severity and mortality of MDS. Blood. (2018) 131:2682–97. doi: 10.1182/blood-2017-10-810028
60. Aly M, Ramdzan ZM, Nagata Y, Balasubramanian SK, Hosono N, Makishima H, et al. Distinct clinical and biological implications of CUX1 in myeloid neoplasms. Blood Adv. (2019) 3:2164–78. doi: 10.1182/bloodadvances.2018028423
61. Kaur S, Ramdzan ZM, Guiot MC, Li L, Leduy L, Ramotar D, et al. CUX1 stimulates APE1 enzymatic activity and increases the resistance of glioblastoma cells to the mono-alkylating agent temozolomide. Neuro Oncol. (2018) 20:484–93. doi: 10.1093/neuonc/nox178
62. Ramdzan ZM, Ginjala V, Pinder JB, Chung D, Donovan CM, Kaur S, et al. The DNA repair function of CUX1 contributes to radioresistance. Oncotarget. (2017) 8:19021–38. doi: 10.18632/oncotarget.14875
63. Ramdzan ZM, Pal R, Kaur S, Leduy L, Berube G, Davoudi S, et al. The function of CUX1 in oxidative DNA damage repair is needed to prevent premature senescence of mouse embryo fibroblasts. Oncotarget. (2015) 6:3613–26. doi: 10.18632/oncotarget.2919
64. Ramdzan ZM, Vadnais C, Pal R, Vandal G, Cadieux C, Leduy L, et al. RAS transformation requires CUX1-dependent repair of oxidative DNA damage. PLoS Biol. (2014) 12:e1001807. doi: 10.1371/journal.pbio.1001807
65. Kaur S, Coulombe Y, Ramdzan ZM, Leduy L, Masson JY, Nepveu A. Special AT-rich Sequence-binding Protein 1. (SATB1) Functions as an accessory factor in base excision repair. J Biol Chem. (2016) 291:22769–80. doi: 10.1074/jbc.M116.735696
66. Pal R, Ramdzan ZM, Kaur S, Duquette PM, Marcotte R, Leduy L, et al. CUX2 protein functions as an accessory factor in the repair of oxidative DNA damage. J Biol Chem. (2015) 290:22520–31. doi: 10.1074/jbc.M115.651042
67. Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. (2012) 487:330–7. doi: 10.1038/nature11252
68. De Vos J, Thykjaer T, Tarte K, Ensslen M, Raynaud P, Requirand G, et al. Comparison of gene expression profiling between malignant and normal plasma cells with oligonucleotide arrays. Oncogene. (2002) 21:6848–57. doi: 10.1038/sj.onc.1205868
69. Moon NS, Rong Zeng W, Premdas P, Santaguida M, Berube G, Nepveu A. Expression of N-terminally truncated isoforms of CDP/CUX is increased in human uterine leiomyomas. Int J Cancer. (2002) 100:429–32. doi: 10.1002/ijc.10510
70. Ripka S, Neesse A, Riedel J, Bug E, Aigner A, Poulsom R, et al. CUX1: target of Akt signalling and mediator of resistance to apoptosis in pancreatic cancer. Gut. (2010) 59:1101–10. doi: 10.1136/gut.2009.189720
71. Fan X, Wang H, Zhou J, Wang S, Zhang X, Li T, et al. The transcription factor CUTL1 is associated with proliferation and prognosis in malignant melanoma. Melanoma Res. (2014) 24:198–206. doi: 10.1097/CMR.0000000000000064
72. Wu X, Feng F, Yang C, Zhang M, Cheng Y, Zhao Y, et al. Upregulated expression of CUX1 correlates with poor prognosis in glioma patients: a bioinformatic analysis. J Mol Neurosci. (2019) 69, 527–37. doi: 10.1007/s12031-019-01355-3
73. Cadieux C, Kedinger V, Yao L, Vadnais C, Drossos M, Paquet M, et al. Mouse mammary tumor virus p75 and p110 CUX1 transgenic mice develop mammary tumors of various histologic types. Cancer Res. (2009) 69:7188–97. doi: 10.1158/0008-5472.CAN-08-4899
74. Li H, Yang F, Hu A, Wang X, Fang E, Chen Y, et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol Med. (2019) 11:e10835. doi: 10.15252/emmm.201910835
75. Vadnais C, Shooshtarizadeh P, Rajadurai CV, Lesurf R, Hulea L, Davoudi S, et al. Autocrine activation of the Wnt/beta-catenin pathway by CUX1 and GLIS1 in breast cancers. Biol Open. (2014) 3:937–46. doi: 10.1242/bio.20148193
76. Aleksic T, Bechtel M, Krndija D, Von Wichert G, Knobel B, Giehl K, et al. CUTL1 promotes tumor cell migration by decreasing proteasome-mediated Src degradation. Oncogene. (2007) 26:5939–49. doi: 10.1038/sj.onc.1210398
77. Ripka S, Riedel J, Neesse A, Griesmann H, Buchholz M, Ellenrieder V, et al. Glutamate receptor GRIA3–target of CUX1 and mediator of tumor progression in pancreatic cancer. Neoplasia. (2010) 12:659–67. doi: 10.1593/neo.10486
78. Ripka S, Konig A, Buchholz M, Wagner M, Sipos B, Kloppel G, et al. WNT5A–target of CUTL1 and potent modulator of tumor cell migration and invasion in pancreatic cancer. Carcinogenesis. (2007) 28:1178–87. doi: 10.1093/carcin/bgl255
79. Jiao H, Zeng L, Zhang J, Yang S, Lou W. THBS2, a microRNA-744-5p target, modulates MMP9 expression through CUX1 in pancreatic neuroendocrine tumors. Oncol Lett. (2020) 19:1683–92. doi: 10.3892/ol.2020.11273
80. Kuhnemuth B, Muhlberg L, Schipper M, Griesmann H, Neesse A, Milosevic N, et al. CUX1 modulates polarization of tumor-associated macrophages by antagonizing NF-kappaB signaling. Oncogene. (2015) 34:177–87. doi: 10.1038/onc.2013.530
81. Fischer K, Brown J, Scherer SW, Schramm P, Stewart J, Fugazza G, et al. Delineation of genomic regions in chromosome band 7q22 commonly deleted in myeloid leukemias. Recent Results Cancer Res. (1998) 144:46–52. doi: 10.1007/978-3-642-46836-0_6
82. Berker-Karauzum S, Luleci G, Ozbilim G, Erdogan A, Kuzucu A, Demircan A. Cytogenetic findings in thirty lung carcinoma patients. Cancer Genet Cytogenet. (1998) 100:114–23. doi: 10.1016/S0165-4608(96)00422-0
83. Dave BJ, Hopwood VL, King TM, Jiang H, Spitz MR, Pathak S. Genetic susceptibility to lung cancer as determined by lymphocytic chromosome analysis. Cancer Epidemiol Biomarkers Prev. (1995) 4:743–9.
84. Mertens F, Johansson B, Hoglund M, Mitelman F. Chromosomal imbalance maps of malignant solid tumors: a cytogenetic survey of 3185 neoplasms. Cancer Res. (1997) 57:2765–80.
85. Solinas-Toldo S, Wallrapp C, Muller-Pillasch F, Bentz M, Gress T, Lichter P. Mapping of chromosomal imbalances in pancreatic carcinoma by comparative genomic hybridization. Cancer Res. (1996) 56:3803–7.
86. Storto PD, Saidman SL, Demetris AJ, Letessier E, Whiteside TL, Gollin SM. Chromosomal breakpoints in cholangiocarcinoma cell lines. Genes Chromosomes Cancer. (1990) 2:300–10. doi: 10.1002/gcc.2870020408
87. Xing YP, Powell WL, Morton CC. The del(7q) subgroup in uterine leiomyomata: genetic and biologic characteristics. Further evidence for the secondary nature of cytogenetic abnormalities in the pathobiology of uterine leiomyomata. Cancer Genet Cytogenet. (1997) 98:69–74. doi: 10.1016/S0165-4608(96)00406-2
88. Zeng WR, Watson P, Lin J, Jothy S, Lidereau R, Park M, et al. Refined mapping of the region of loss of heterozygosity on the long arm of chromosome 7 in human breast cancer defines the location of a second tumor suppressor gene at 7q22 in the region of the CUTL1 gene. Oncogene. (1999) 18:2015–21. doi: 10.1038/sj.onc.1202519
89. Zeng WR, Scherer SW, Koutsilieris M, Huizenga JJ, Filteau F, Tsui LC, et al. Loss of heterozygosity and reduced expression of the CUTL1 gene in uterine leiomyomas. Oncogene. (1997) 14:2355–65. doi: 10.1038/sj.onc.1201076
90. Ishwad CS, Ferrell RE, Hanley K, Davare J, Meloni AM, Sandberg AA, et al. Two discrete regions of deletion at 7q in uterine leiomyomas. Genes Chromosomes Cancer. (1997) 19:156–60. doi: 10.1002/(SICI)1098-2264(199707)19:3andlt;156::AID-GCC4andgt;3.0.CO;2-X
91. Neville PJ, Thomas N, Campbell IG. Loss of heterozygosity at 7q22 and mutation analysis of the CDP gene in human epithelial ovarian tumors. Int J Cancer. (2001) 91:345–9. doi: 10.1002/1097-0215(200002)9999:9999andlt;::AID-IJC1050andgt;3.0.CO;2-1
92. Mcnerney ME, Brown CD, Wang X, Bartom ET, Karmakar S, Bandlamudi C, et al. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood. (2013) 121:975–83. doi: 10.1182/blood-2012-04-426965
93. Wong CC, Martincorena I, Rust AG, Rashid M, Alifrangis C, Alexandrov LB, et al. Inactivating CUX1 mutations promote tumorigenesis. Nat Genet. (2014) 46:33–8. doi: 10.1038/ng.2846
94. Dorris ER, O'neill A, Treacy A, Klocker H, Teltsh O, Kay E, et al. The transcription factor CUX1 negatively regulates invasion in castrate resistant prostate cancer. Oncotarget. (2020) 11:846–57. doi: 10.18632/oncotarget.27494
95. Li T, Wang H, Sun Y, Zhao L, Gang Y, Guo X, et al. Transcription factor CUTL1 is a negative regulator of drug resistance in gastric cancer. J Biol Chem. (2013) 288:4135–47. doi: 10.1074/jbc.M112.345942
96. Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. (2001) 411:366–74. doi: 10.1038/35077232
97. Farmer H, Mccabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. (2005) 434:917–21. doi: 10.1038/nature03445
98. Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld EJ, et al. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J Biol Chem. (1999) 274:7803–15. doi: 10.1074/jbc.274.12.7803
99. Goulet B, Sansregret L, Leduy L, Bogyo M, Weber E, Chauhan SS, et al. Increased expression and activity of nuclear cathepsin L in cancer cells suggests a novel mechanism of cell transformation. Mol Cancer Res. (2007) 5:899–907. doi: 10.1158/1541-7786.MCR-07-0160
100. Arthur RK, An N, Khan S, McNerney ME. The haploinsufficient tumor suppressor, CUX1, acts as an analog transcriptional regulator that controls target genes through distal enhancers that loop to target promoters. Nucleic Acids Res. (2017) 45:6350–61. doi: 10.1093/nar/gkx218
101. Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. (2009) 8:33–40. doi: 10.1038/nrd2781
Keywords: CUX1, haploinsufficient tumor suppressor, tumor progression, DNA damage, KRAS mutation
Citation: Liu N, Sun Q, Wan L, Wang X, Feng Y, Luo J and Wu H (2020) CUX1, A Controversial Player in Tumor Development. Front. Oncol. 10:738. doi: 10.3389/fonc.2020.00738
Received: 11 February 2020; Accepted: 17 April 2020;
Published: 29 May 2020.
Edited by:
Zhigang Zhang, Shanghai Cancer Institute, ChinaReviewed by:
Hua Wang, Anhui Medical University, ChinaLiwei Dong, National Center for Liver Cancer, China
Dechun Feng, National Institute on Alcohol Abuse and Alcoholism (NIAAA), United States
Yinying Lu, Beijing 302 Hospital, China
Copyright © 2020 Liu, Sun, Wan, Wang, Feng, Luo and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hailong Wu, d3VobEBzdW1ocy5lZHUuY24=; Judong Luo, anVkb25nbHVvQDE2My5jb20=
 Ning Liu
Ning Liu Qiliang Sun2
Qiliang Sun2 Long Wan
Long Wan Hailong Wu
Hailong Wu