- 1Department of Gastrointestinal Cancer Biology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, China
- 2Key Laboratory of Cancer Immunology and Biotherapy, Tianjin, China
- 3National Clinical Research Center for Cancer, Tianjin, China
Circular RNAs (circRNAs), which are generated mainly from back-splicing of exons in precursor mRNAs (pre-mRNAs), are a novel class of endogenous covalently closed RNA molecules. Their functions as microRNA sponges, protein scaffolds, and modulators of transcription and splicing, as well as occasional templates for polypeptide production, are beginning to be recognized, though the investigation of circRNAs is in its infancy. circRNAs play critical roles in diverse cellular processes. Aberrant expression of circRNAs in malignancies sustains cellular growth and proliferation, promotes cellular invasiveness, and circumvents cellular senescence and death, suggesting their potential for exploitation as clinical biomarkers and therapeutic targets. In this review, we highlight recent progress in research on circRNAs in cancer, emphasizing the molecular mechanisms and potential clinical value of circRNAs.
Introduction
Circular RNAs (circRNAs), a novel class of endogenous covalently closed RNA molecules, have attracted great attention in the past few years. They are generally derived from back-splicing of precursor mRNA (pre-mRNA), during which the 3′ splice donor sequence is joined to the downstream 5′ splice acceptor sequence (1, 2). In 1976, Sanger et al. (3) first identified the single-stranded viroid RNA in circular formats with high thermal stability via electron microscopy. Subsequently, this kind of circRNA transcript was found in hepatitis delta virus (4), yeast (5), archaea (6), fruit flies (7), and mammals (8). In 1991, Nigro et al. (9) discovered the presence of circRNA transcripts derived from the Deleted in Colon Cancer (DCC) gene in humans for the first time; subsequently, other genes, including ETS-1 gene (10), the human cytochrome P450 gene (11), the human dystrophin gene (12), and the antisense noncoding RNA in the INK4 locus (13), were identified to produce circular transcripts. However, these circRNAs were long considered as aberrant splicing by-products with low abundance and limited biological function (14).
With the development of high-throughput RNA sequencing (RNA-seq) and bioinformatics tools, numerous circRNAs have been identified. Salzman et al. (15) explored the circRNA map in pediatric acute lymphoblastic leukemia samples and revealed that large portions of spliced gene transcripts are circRNAs. Jeck et al. (16) identified >25,000 circRNAs that are not degraded by exonucleases in human fibroblasts. Circular splicing of RNA is accepted to be a general feature of gene expression, but whether these circRNAs are functional is a primary concern of researchers.
Studies to date have reported that circRNAs are dysregulated in the pathophysiologic processes of several diseases, including cardiovascular disease (17), neurodegenerative disease such as Alzheimer's disease (18), metabolic disorder (19), diseases caused by viral infection (20), and cancer, which is the focus of the following sections (21). Abnormal expressed circRNAs can modulate gene transcription via indirect interactions with other transcription factors, such as microRNAs (miRNAs) and RNA binding proteins (RBPs) (22, 23). Sporadic studies have also pointed out that some circRNAs containing translation initiation elements can be translated into functional proteins and peptides (24, 25). Moreover, the competitive splicing mechanism of circRNAs also affects the expression of their parent genes, thus producing biological effects (26). In addition to functioning as regulators of gene expression, circRNAs are also novel promising biomarkers for disease diagnosis and prognosis assessment due to their stable closed circular structure and tissue- and developmental stage-specific expression patterns (27, 28).
However, the understanding of circRNAs is in its infancy, and knowledge of the biological characteristics of these molecules requires further supplementation. More work is needed to explore the emerging roles of circRNAs in cancer. In this review, we introduce the biogenetic model, expression profile, and functional mechanism of circRNAs and summarize recent progress in circRNA research and their application in cancer.
Mechanisms for circRNA Generation
circRNAs are divided into three main types according to their sources, namely, exonic circular RNAs (ecircRNAs), circular intronic RNAs (ciRNAs), and exon–intron circular RNAs (elciRNAs), among which ecircRNAs are the most common (29). Several circularization mechanisms produce circRNAs. These elements shorten the spatial distance between the two ends of the loop sequence and provide possibilities for back-splicing.
Intron pairing-driven circularization is mediated by 30–40 nucleotide (nt) reverse complementary sequences in the flanking regions of circularized exons; these sequences form double-stranded RNA structures and therefore promote circRNA production as cis-acting elements (2, 16). Alu is the most common reverse complementary element in mammals. In addition, some specific RBPs can bind to both sides of flanking intron sequences and bring splice donors and splice acceptors sufficiently close through protein–protein interactions (30), such as Quaking (QKI) (31), NF90/NF110 (32), and FUS (33). In contrast, other RBPs may inhibit circRNA production. For example, adenosine deaminase acting on RNA 1 (ADAR1), a common Alu editing element, was identified to deaminate adenosine nucleosides to inosine (A-to-I editing) in regions that complementary and proximal to the splice sites of circularized exons and destabilize intron pairing interactions, thereby antagonizing circRNA biogenesis (34). Lariat-driven circularization occurs in the process of exon skipping. This process produces a lariat intermediate containing introns and exons, which then undergo back-splicing to form circRNAs. The production of ciRNAs is a special situation in the lariat-driven model (35). CircRNA biogenesis in vivo is indeed very complicated. The expression level of a circRNA may be influenced by various circularization mechanisms. In addition, alternative splicing is a key component of circRNA production and gene expression regulation (36, 37).
circRNA Expression and Characteristics
circRNAs have been identified to be expressed widely in most organisms. The size of circRNAs ranges from <100 to several thousand nucleotides (30), and the common size reported in human cells is a few hundred nucleotides comprising 2–3 exons. The expression of most circRNAs is generally lower than that of their linear transcripts (38), but a few circular transcripts exhibit an expression level slightly or much higher than those of the their linear transcripts, such as the products of the CDR1 and Sry genes (8, 15, 16, 22). In addition, due to their unique closed circular structure, circRNAs are protected from degradation by exonucleases and are more stable than linear RNAs (16). Multiple studies have reported that circRNAs have longer half-lives than linear transcripts both in vitro and in vivo (39, 40). Moreover, circRNA expression is generally tissue- and developmental stage-specific. Several studies have shown that many circRNAs are upregulated in the nervous system (41–44), a characteristic that may be related to their posttranscriptional accumulation in neurons (45–47). Therefore, scientists hypothesized that the intracellular level of circRNAs was negatively correlated with the cell proliferation index (48). This hypothesis may explain why circRNA expression in tumor cells is generally lower than that in normal cells. Moreover, circRNAs may have higher sequence conservation than other types of RNA in mammals (41). Taken together, the high conservation, stability, and specificity of circRNAs imply that circRNAs may have multiple biological functions and clinical applications and are unlikely to be simply splicing by-products.
Techniques for Studying circRNAs
Although circRNAs were discovered decades ago, they could not be detected by early routine poly(A)-enriched RNA sequencing technology. Panda et al. (49) proposed a novel method, RNase R treatment followed by polyadenylation and poly (A)+ RNA depletion (RPAD), to enrich highly pure circRNAs. In this method, total RNAs are first treated with RNase R to deplete linear RNAs. The remaining RNAs with 3'-OH ends are polyadenylated and removed by poly (A)+ RNA depletion via oligo (dT) beads. The RPAD method eliminated the interference of linear RNAs and significantly improved the reliability of the data. However, RPAD is unsuitable for joint analysis of circRNAs with other molecules such as miRNAs and mRNAs. Researchers should adopt different methods to optimize RNA sequencing libraries for the specific experimental purpose.
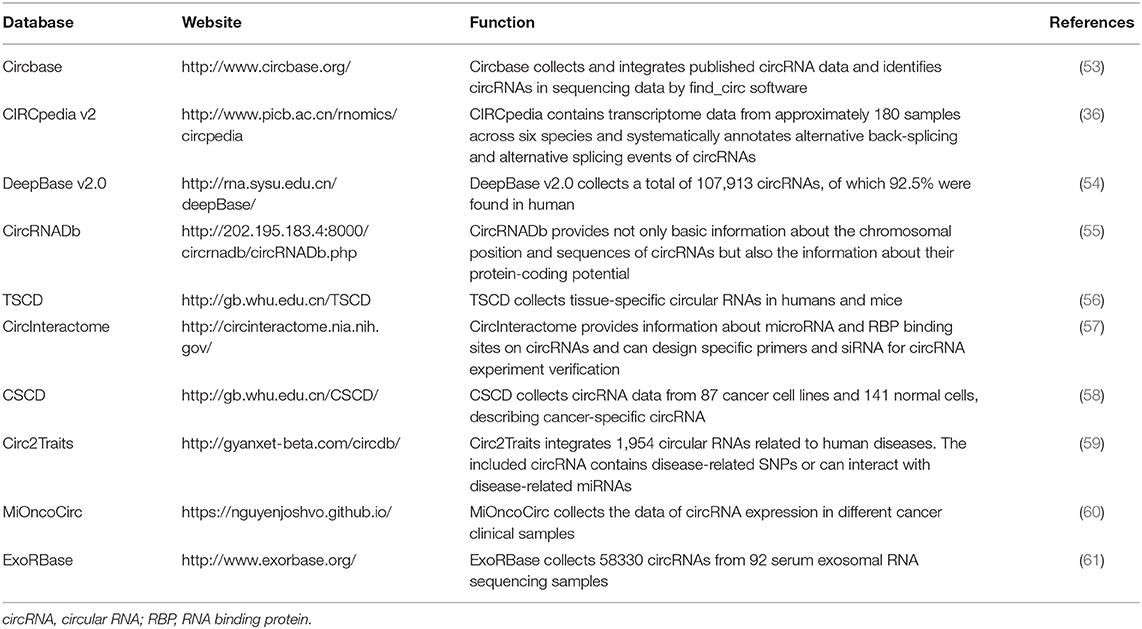
To date, a variety of algorithms have been developed to identify circRNAs. CircRNAs are generated primarily from pre-mRNA back-splicing, not canonical splicing; thus, the mapping algorithms used in early transcriptome analysis cannot directly match the fragments to the genome. Therefore, sequencing reads that span back-splicing sites require further genomic alignment and correction (50, 51). Various bioinformatics algorithms have been developed for circRNA annotation and quantification (51, 52). The use of two or more tools simultaneously is recommended to meet specific research demands for circRNA identification (52). In addition, multiple databases have been developed for circRNA analysis (Table 1). Through these databases, researchers can search basic information about circRNAs, predict interactions of circRNAs with target molecules and their translation potential, and evaluate their relationships with diseases.
Functions of circRNAs
Numerous reports have confirmed that circRNAs can regulate gene expression directly or, more commonly, through binding with miRNAs, RBPs, and other gene expression regulators, thereby regulating various biological processes.
miRNA Sponging
miRNAs are a large class of small (~22 nt) noncoding single-stranded RNAs that can bind to their target mRNAs to inhibit their translation or promote their degradation. Recent studies have reported that many circRNAs can act as competitive endogenous RNAs (ceRNAs), binding with miRNAs through miRNA response elements (MREs) to downregulate the function of the target miRNA. The most representative example is CDR1as (antisense to the cerebellar degeneration-related protein 1 transcript), also known as ciRS-7 (22, 62). CDR1as contains 73 binding sites for miR-7, and the interaction of these two RNAs can inhibit the function of miR-7. In addition, their interaction provides a novel mechanism for miR-7 transport. Similarly, circSry, derived from the mouse sex-determining gene Sry, contains 16 binding sites for miR-138 and can act as a miRNA sponge (29). Notably, most ecircRNAs are localized mainly in the cytoplasm, indicating their availability to bind with miRNAs and regulate their function (39). However, most circRNAs do not contain multiple miRNA binding sites (29). Although this mechanism is the most widely studied mechanism of circRNAs, its importance remains to be confirmed. The abundance of most circRNAs is generally low, which also limits the universality of the miRNA sponge hypothesis.
Protein Scaffolding
CircRNAs contain many RBP binding sites in addition to MREs. For example, human antigen R (HuR) has been reported to bind with numerous circRNAs in human cervical carcinoma HeLa cells (63). CircPABPN1, derived from the PABPN1 gene, can compete with PABPN1 mRNA for binding to HuR, thereby inhibiting the translation of PABPN1. In addition, elciRNAs and ciRNAs, predominantly localized in the nucleus, were shown to be able to interact with small nuclear ribonucleoprotein U1 (snRNP U1) and enhance RNA polymerase II (Pol II) transcriptional activity on their parental gene as cis regulators (35, 64). Additionally, the binding of circRNAs to some functional proteins may affect multiple signaling pathways leading to homeostasis changes. Du et al. (65) found that the circRNA circ-Foxo3 interacted with the anti-senescence protein ID-1, the transcription factor E2F1, and the anti-stress proteins FAK and HIF1α and retained them in the cytoplasm to hinder their corresponding functions.
Translational Templates
Although circRNAs have historically been considered noncoding RNAs, several recent studies have indicated that some circRNAs contain translation initiation sites and have translational potential (39). For example, some circRNAs with internal ribosome entry sites (IRESs) can be translated into proteins in vitro and in vivo (24). FBXW7-185aa is a novel protein encoded by circ-FBXW7 and contains an IRES (66). N6-methyladenosine(m6A), the most common RNA base modification, is abundant in circRNA sequences and can facilitate translation initiation by recruiting eIF4G2 and YTHDF3 (67). Another study in 2017 found that circ-ZNF609 contained an open reading frame and could be translated in human skeletal muscle. To date, only a few circRNAs have been reported to be involved in the translation process. Although some bioinformatics tools have been developed to predict the translation potential of circRNAs, their accuracy needs to be further improved and experimentally verified. In addition, whether these proteins or peptides formed from circRNAs have important functions needs exploration.
Other Functions
Moreover, the biosynthesis of circRNAs can compete with the splicing of their linear transcripts, thus affecting their expression and corresponding functions (68). Because of their stability, circRNAs can also accumulate over time and may thus act as memory molecules for the transcriptional history of a cell (69). The presence of circRNAs in vesicles also suggests their function as signaling molecules (70).
The Functions of circRNA and Cancer
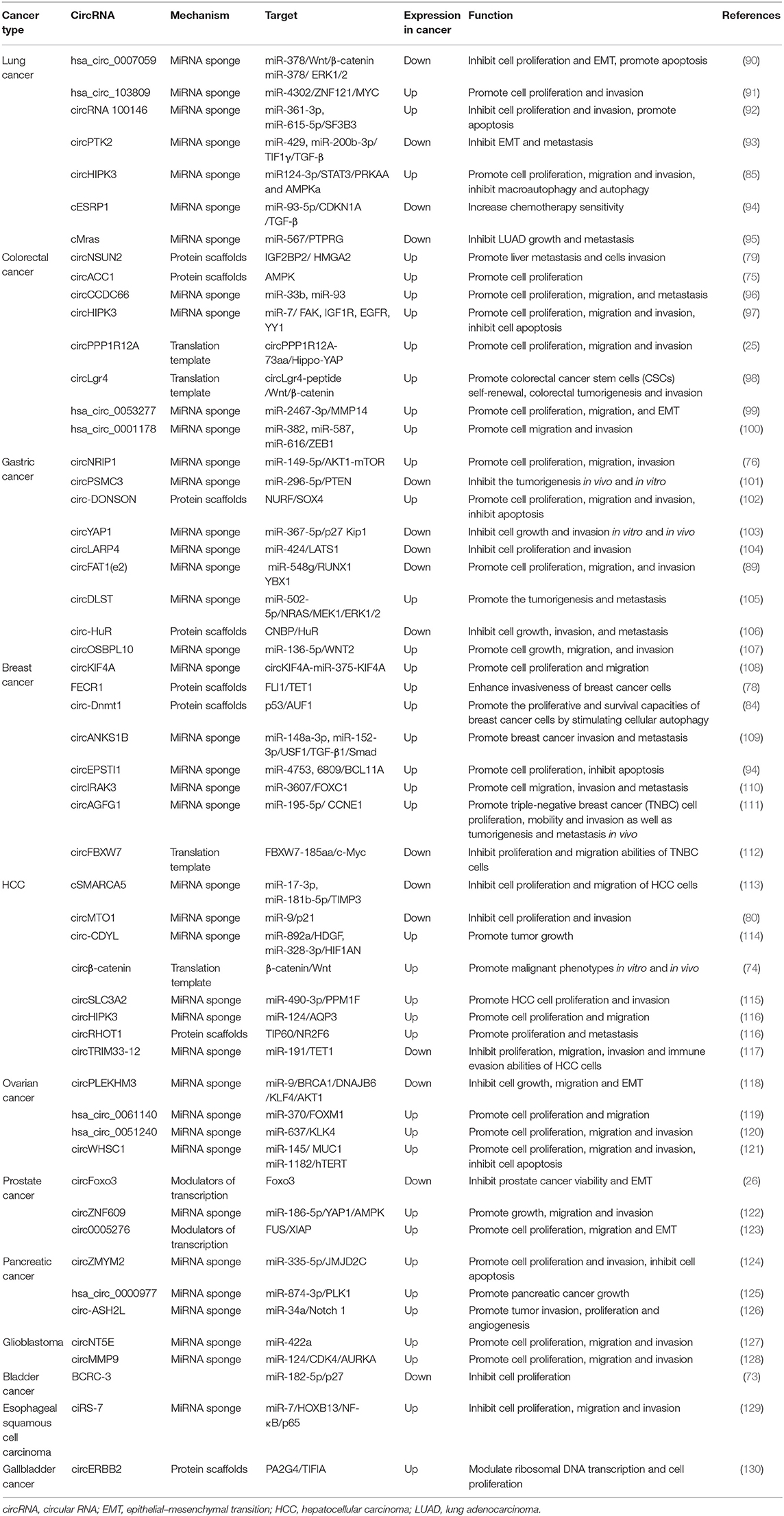
CircRNAs are abnormally expressed in many cancers, such as lung cancer, breast cancer, digestive system cancers, ovarian cancer, and glioblastoma. Hang et al. (71) identified 185 differentially expressed circRNAs between non-small-cell lung cancer (NSCLC) tissues and adjacent normal tissues through RNA sequencing. In addition, Zeng et al. (72) detected 192 upregulated and 239 downregulated circRNAs in colorectal cancer (CRC) tissues from patients with or without pulmonary metastasis. Whether this differential expression is related to cancer development has been a focus of circRNA research.
Cell Proliferation
Via high-throughput sequencing, Xie et al. (73) screened a circRNA related to bladder cancer (BC), BCRC-3. BCRC-3 is poorly expressed in BC tissues and cell lines, and it can bind to miR-182-5p to act as a ceRNA, thereby upregulating the expression of p27 and inhibiting BC cell proliferation. Liang et al. (74) found that circβ-catenin was upregulated in liver cancer tissues compared to adjacent tissues. Silencing circβ-catenin significantly inhibited malignant phenotypes. Mechanistically, circβ-catenin can be translated into a 370-amino acid (aa) β-catenin isoform. This β-catenin−370aa construct competed with GSK3β to inhibit its degradation of β-catenin, thereby activating the Wnt/β-catenin pathway in liver cancer to promote tumor growth. Moreover, circACC1 has been reported to bind the regulatory β and γ subunits of AMP-activated protein kinase (AMPK), resulting in a ternary complex, which in turn activates AMPK enzyme activity and then promotes fatty acid b-oxidation and glycolysis (75). Overexpression of circACC1 can promote tumorigenesis. A positive correlation between AMPK activation and elevated circACC1 expression was identified in CRC tissues. Zhang et al. (76) revealed that circNRIP1 can sponge miR-149-5p and thus affect the AKT1/mammalian target of rapamycin (mTOR) axis, acting as a tumor promoter in gastric cancer (GC). This study also suggested that circNRIP1 can assemble into exosomes and participate in exosomal communication among GC cells (76).
Invasion and Metastasis
Recently, Hu et al. (77) identified a circRNA, circASAP1, that is associated with pulmonary metastasis after curative resection in hepatocellular carcinoma (HCC) patients. Studies have shown that circASAP1 acts as a sponge of miR-326 and miR-532-5p, which have mitogen-activated protein kinase (MAPK)1 and colony stimulating factor (CSF-1) in common as target genes, thereby promoting HCC cell invasion and macrophage infiltration. In addition, Chen et al. (78) discovered a circRNA, FECR1, derived from Friend leukemia virus integration 1 (FLI1). Overexpression of FECR1 can increase the invasiveness of breast cancer cells. Mechanism studies have shown that FECR1 can recruit TET1 and act in trans to downregulate DNMT1, causing DNA demethylation of FLI1 and promoting cell metastasis. FECR1 may be a potential therapeutic target for metastatic breast cancer. m6A-modified circNSUN2 can form a ternary complex with insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2) and high-mobility group A2 (HMGA2) to enhance HMGA2 mRNA stability and subsequently promote liver metastasis in CRC (79). CircFoxo3 expression is low in high-grade prostate cancer, and overexpression of circFoxo3 in DU145 cells can inhibit the epithelial–mesenchymal transition (EMT) and reduce cell viability by enhancing Foxo3 expression (26). CircMTO1 (hsa_circRNA_104135) is significantly downregulated in HCC and can affect p21 expression by targeting miR-9 and, in turn, promote cell proliferation and invasion (80).
Cell Cycle
Circ-Foxo3 has been shown to bind with CDK2 and p21, leading to formation of the circ-Foxo3–p21–CDK2 ternary complex (81). CDK2 generally interacts with cyclin A/E to promote cell cycle progression, while p21 inhibits these effects. Formation of this ternary complex enhanced the inhibitory effect of p21 on CDK2, which prevented cell cycle transition from G1 to S phase and thus inhibited cell proliferation. Cheng et al. (82) found significant upregulation of circTP63 in lung squamous cell carcinoma (LUSC) tissues. CircTP63 competitively binds to miR-873-3p, thereby abolishing the inhibitory effect of miR-873-3p on its target gene Forkhead Box M1 (FOXM1). Elevated FOXM1 expression further enhances the expression of centromere protein (CENP)A and CENPB, ultimately promoting cell cycle progression and cell proliferation.
Cell Death
Circ-Foxo3 was reported to be downregulated in tumor tissues and cells (83). On the one hand, transfection of circ-Foxo3 can induce apoptosis as a stress stimulus. On the other hand, highly expressed circ-Foxo3 interacts with p53 and murine double minute 2 (MDM2) and inhibits ubiquitination-mediated degradation of Foxo3 by MDM2. Overexpression of Foxo3 promotes apoptosis via its target Puma. Du et al. (84) detected abnormally increased circ-Dnmt1 expression in breast cancer tissues and cells and found that circ-Dnmt1 interacted with p53 and AUF1, leading to their nuclear translocation, thus mediating cell autophagy. Transfection of circ-Dnmt1 into breast cancer cells can induce autophagy to maintain cell homeostasis and ultimately promote cell proliferation and tumorigenesis. Furthermore, Chen et al. (85) found that circHIPK3 expression was upregulated in STK11 mutant lung cancer cell lines. Silencing circHIPK3 can induce autophagy through the miR124-3p–STAT3–PRKAA/AMPKa axis. Thus, circHIPK3 is a key upstream regulator of autophagy in lung cancer.
Others
CircRNAs can also regulate the drug sensitivity of tumors. By high-throughput sequencing of circRNAs of five pairs of cisplatin-sensitive and cisplatin-resistant ovarian cancer tissues, Zhao et al. (86) found that CDR1as was significantly downregulated in cisplatin-resistant tissues. Mechanism studies confirmed that CDR1as can regulate the sensitivity of ovarian cancer to cisplatin through the miR-1270/SCAI signaling pathway and promote ovarian cancer development. In addition, circRNAs derived from fusion genes have been shown to play a special role in cancers. Recent studies have demonstrated that chromosomal translocations in cancer could not only encode oncogenic fusion proteins but also generate circRNAs. This kind of fusion circRNA (f-circRNA) is unique to cancer and may play an important role in tumorigenesis. F-circRNAs were first identified in a study on leukemia, in which F-circPR and F-circM9 (derived from PML-RARα and MLL-AF9 fusion genes, respectively) were found to promote cellular transformation, cell viability, and drug resistance (87). In addition, F-circEA-2a, derived from the EML4-ALK fusion gene in NSCLC, is located mainly in the cytoplasm and can promote cell migration and invasion (88). The tumor-suppressive effect of circFAT1 (e2) in GC can simultaneously arise through the cytoplasmic circFAT1 (e2)/miR-548g/RUNX1 axis and the nuclear circFAT1 (e2)/YBN1 regulatory network (89). Thus, the mechanism of circRNAs in vivo is very complicated, and our current knowledge of this process is only nascent. Additional studies are listed in Table 2.
circRNAs as Biomarkers in Cancer
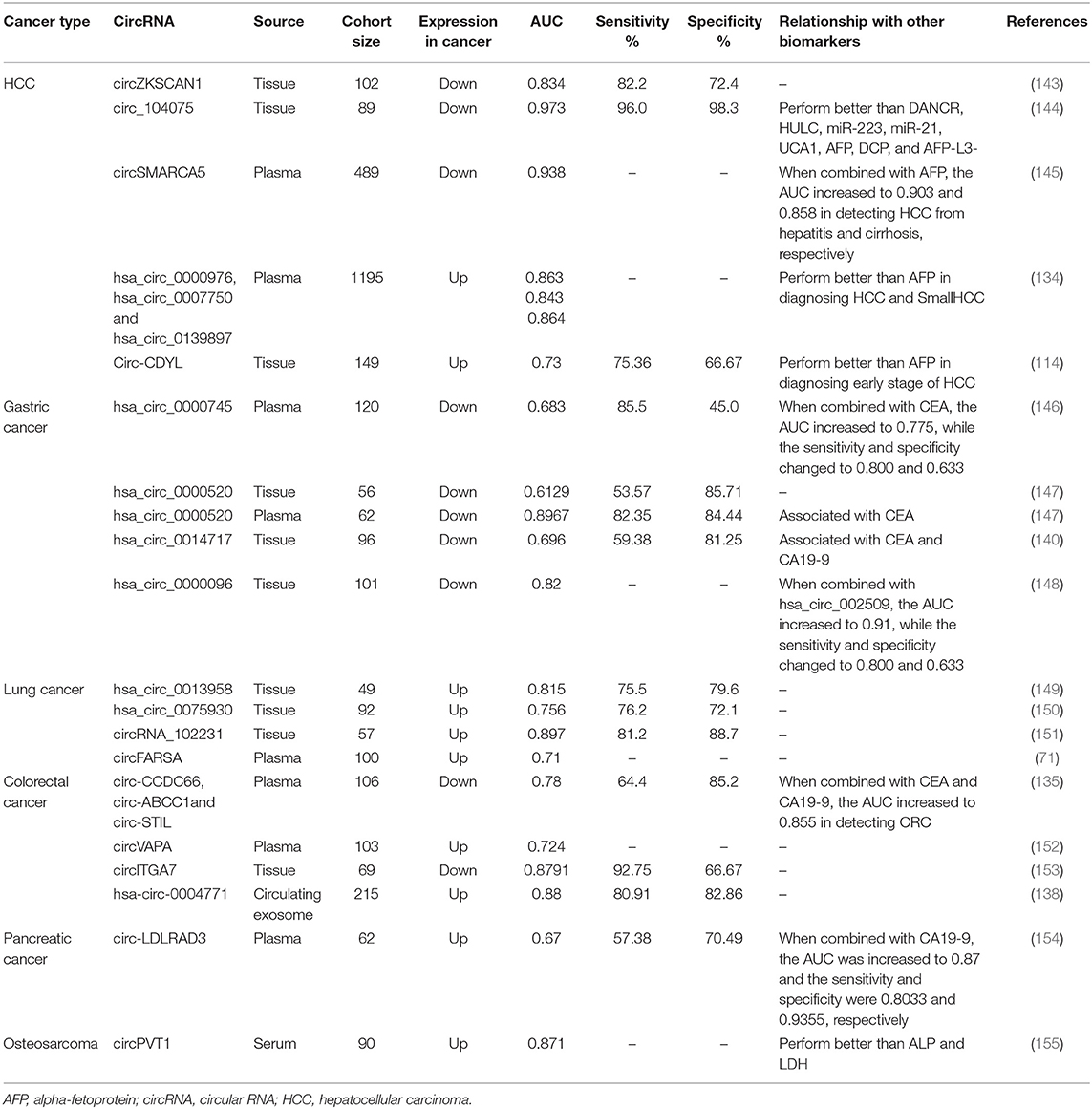
CircRNAs have a covalently closed circular structure, which increases their resistance to exonuclease digestion and their accumulation in body fluids and tissues (131, 132). In addition, circRNAs are often expressed in a tissue- and developmental stage-specific manner. The properties of circRNAs have inspired numerous studies on their application as promising biomarkers in cancer.
Diagnostic Biomarkers
circRNAs in Tissues
Wei et al. (114) found that circ-CDYL and its target genes (HDGF and HIF1AN) were highly expressed in Barcelona Clinic Liver Cancer (BCLC) stages 0 and A of HCC. Their sensitivity and specificity as combined biomarkers were higher than those of alpha-fetoprotein (AFP) in the early stage of HCC. These results indicated that a panel combining circ-CDYL with HDGF and HIF1AN could be used as a monitoring indicator for early HCC or high-risk populations. In addition, Zhong et al. (133) detected highly expressed CDR1as in nasopharyngeal carcinoma (NPC) biopsy samples. However, unfortunately, current methods for detecting circRNAs in tissue are complex and invasive, which greatly limit their roles in the early diagnosis and screening of cancer.
circRNAs in Peripheral Blood
Numerous studies have confirmed that circRNAs can be stably enriched in peripheral blood, urine, and saliva. Therefore, circRNAs could become suitable biomarkers for liquid biopsy. Recently, a study on the use of plasma circRNAs in the diagnosis of hepatitis B virus (HBV)-associated HCC was published (134). This study included 1,195 plasma samples, which were divided into a training set and two validation sets. The researchers found that the plasma expression levels of hsa_circ_0000976, hsa_circ_0007750, and hsa_circ_0139897 in HCC patients were significantly higher than those in healthy controls and patients with chronic hepatitis B or HBV-related liver cirrhosis. They also designed an HCC prediction model named CircPanel through binary logistic regression analysis. CircPanel showed higher sensitivity and specificity (both higher than 80%) than AFP level for distinguishing HCC patients from controls. Notably, CircPanel can also diagnose AFP-negative HCC and AFP-negative small HCC (solitary, diameter ≤3 cm) with high diagnostic accuracy. In another study, Lin et al. (135) found that the plasma levels of circ-CCDC66, circ-ABCC1, and circ-STIL were significantly decreased in CRC patients compared with controls. The combination of these three circRNAs had a sensitivity and specificity of 64.4% and 85.2%, respectively, for diagnosing CRC. Notably, f-circRNAs derived from fusion genes are generally cancer-specific and thus have high specificity in diagnosing cancer, which is a unique advantage for tumor biomarkers. Tan et al. (136) reported a fusion circRNA named F-circEA derived from an EML4-ALK fusion gene that was positively expressed in five of six NSCLC patients with EML4-ALK translocation. F-circEA also exists specifically in the plasma of EMLA4-ALK-positive NSCLC patients.
circRNAs in Exosomes
Exosomal circRNAs have received gradually increasing attention in recent years. Li et al. (137) identified more than 1,000 circRNAs in human serum exosomes for the first time and suggested that these circRNAs could distinguish between colon cancer patients and healthy controls. Pan et al. (138) used qRT-PCR to detect hsa-circ-0004771 in circulating exosomes from 170 patients and 45 healthy controls and confirmed that hsa-circ-0004771 could clearly distinguish between patients with stage I/II CRC and those with benign intestinal diseases. Li et al. (139) detected high expression of circ-PDE8A in liver-metastatic pancreatic ductal adenocarcinoma (PDAC) tissues, and this characteristic was confirmed to be closely related to lymphatic infiltration, T status, and TNM stage. Circ-PDE8A is an independent risk factor for the survival of PDAC patients. Further research confirmed the presence of circ-PDE8A-rich exosomes secreted from tumor cells in the plasma of PDAC patients. Similarly, circ-PDE8A in plasma exosomes may also be a diagnostic and prognostic marker for PDAC.
circRNAs in Other Body Fluids
The use of circRNAs in other body fluid samples has also been studied. For example, gastric juice examination is a highly organ-specific test for the diagnosis of gastric diseases. Shao et al. (140) explored the feasibility of hsa_circ_0014717 in gastric juice as a biomarker for screening patients with GC. The expression of hsa_circ_0014717 in gastric juice from 38 healthy people, 30 patients with gastric ulcers, 15 patients with chronic atrophic gastritis, and 39 patients with GC was measured by qRT-PCR. Hsa_circ_0014717 was significantly downregulated in patients with chronic atrophic gastritis compared with healthy controls, suggesting the potential utility of hsa_circ_0014717 in gastric juice as a biomarker for screening high-risk populations for GC. In addition, saliva is a body fluid used for disease research due to its convenient and noninvasive sampling method. Bahn et al. (141) detected and verified the presence of circRNAs in cell-free saliva, which suggested a new direction for the application of circRNAs as biomarkers. Moreover, Chen et al. (142) detected highly expressed circPRMT5 in exosomes isolated from urine samples from patients with urothelial carcinoma of the bladder (UCB). Additional studies are listed in Table 3.
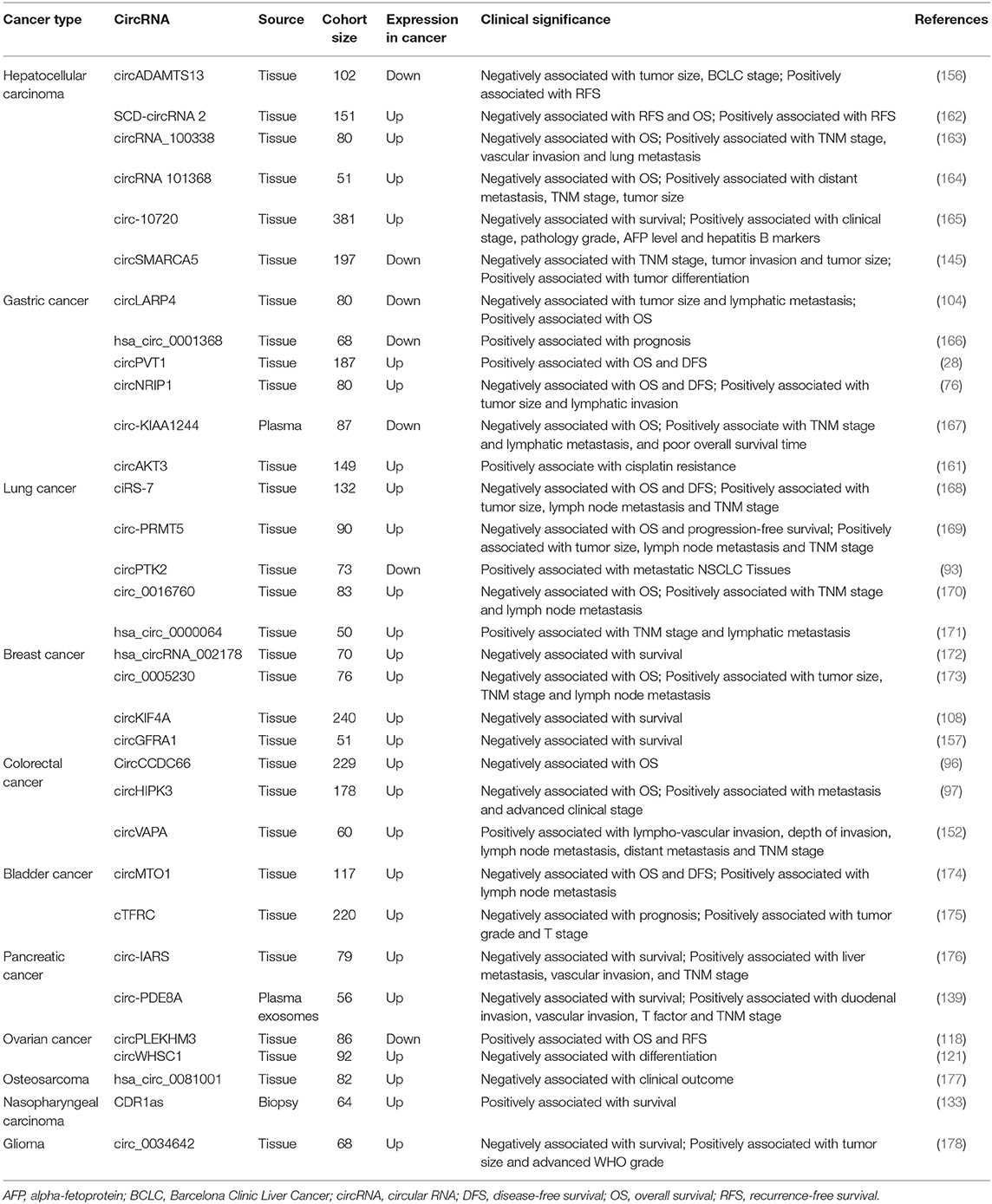
Prognostic Biomarkers
circRNAs Predict Patient Survival
CircRNAs have wide application prospects in monitoring the treatment efficacy and assessing the prognosis of cancer. By using RNA-seq to identify circRNAs related to HCC progression, Qiu et al. (156) determined that circADAMTS13 was significantly downregulated in HCC tissues compared with matched nontumor tissues. Moreover, its expression was positively associated with recurrence-free survival (RFS). Receiver operating characteristic (ROC) curve analysis was used to evaluate the diagnostic value of circADAMTS13 in HCC patients, and the area under ROC curve (AUC) was 0.987. He et al. (157) determined that circGFRA1 was significantly upregulated in triple-negative breast cancer via circRNA microarray analysis. In addition, Kaplan–Meier survival analysis showed that upregulation of circGFRA1 was associated with poorer clinical survival. CircHIPK3 is significantly upregulated in CRC tissues and cell lines and is associated with the T status of tumors, lymph node and distant metastasis, and advanced clinical stage (97). High expression of circHIPK3 in CRC is an independent prognostic factor for poor overall survival (OS). Compared with paired noncancerous tissues, GC tissues exhibit significant downregulation of hsa_circ_0000096; moreover, the AUC increased from 0.82 to 0.91 when hsa_circ_0000096 was combined with hsa_circ_002509 (148). Analysis of 90 paired samples of NSCLC and adjacent normal tissues showed that hsa_circ_0014130 was significantly upregulated in NSCLC and was found to be related to tumor size, TNM stage, and lymphatic metastasis (158). ROC analysis indicated that hsa_circ_0014130 might be a prognostic biomarker in NSCLC.
In addition to identifying the differential expression of hsa_circ_002059 in GC and adjacent tissues, Li et al. (159) also found that the expression levels of hsa_circ_002059 in 36 pairs of matched plasma samples from preoperative and postoperative GC patients were significantly different. Considering that circRNAs can be secreted into the tumor microenvironment and the circulatory system by tumor cells, it is reasonable to speculate that circRNAs can be used as biomarkers to evaluate the tumor-bearing status of patients. As mentioned above, circHIPK3 can regulate the autophagy process in lung cancer through the miR124-3p–STAT3–PRKAA/AMPKa axis (85). In addition, researchers found that the ratio (C:L ratio) of circHIPK3 to linear HIPK3 (linHIPK3) can reflect the level of autophagy in cells—a higher C:L ratio indicates a lower level of autophagy and poorer prognosis.
circRNAs Predict Not Only Tumor Metastasis but Also Drug Resistance
Recent studies have also reported that circRNAs can predict tumor metastasis and drug resistance. Xu et al. (160) analyzed the circRNA expression profile of three pairs of CRC patients with or without liver metastasis. Further verification demonstrated that circRNA_0001178 and circRNA_0000826 were significantly upregulated in metastatic CRC (CRC-m) tissues. The AUCs were 0.945 for circRNA_0001178 and 0.816 for circRNA_0000826, and both two circRNAs could differentiate liver metastases from CRC. CircASAP1 is highly expressed in the tissues of HCC patients with pulmonary metastases after curative resection (77), and its expression is positively correlated with the levels of CSF-1, MAPK1, and CD68+ tumor-associated macrophage. Patients with high circASAP1 expression tend to have relatively low OS and high recurrence rates. Therefore, circASAP1 can be used as a prognostic indicator for HCC.
In addition, cisplatin is one of the main chemotherapeutic drugs used to treat GC. Huang et al. (161) analyzed the correlation between the clinical efficacy of cisplatin and circRNA expression in 105 cisplatin-treated patients, and they found that the expression of circAKT3 in cisplatin-resistant patients was higher than that in cisplatin-sensitive patients. CircAKT3 is an effective predictive biomarker for cisplatin resistance in GC patients (AUC = 0.91). Confirmation of the relationships between circRNA expression and drug sensitivity would provide guidance for reasonable clinical medication and thus improve patient prognosis. Additional studies are listed in Table 4.
Conclusions and Perspectives
As the understanding of circRNAs has increased, their perception by the scientific community has changed dramatically. CircRNAs are not “splicing noise” but rather a class of structurally stable RNA molecules with multiple biological functions. circRNAs are generally derived from back-splicing of pre-mRNA and are widely expressed across biological systems.
The relationship between circRNAs and cancer has recently become an area of research interest. Numerous circRNAs are dysregulated and play regulatory roles in the development of cancer. Several examples of research on the roles of circRNAs in cancer are given above. In summary, circRNAs, through various signaling pathways, can participate in and affect processes related to cell proliferation, migration and invasion, apoptosis, autophagy, and drug resistance, as well as others. This capacity has inspired researchers to consider the therapeutic possibilities of targeting circRNAs and their associated pathways. In addition, researchers have also tried to synthesize artificial circRNAs for disease treatment. For example, an artificial circRNA with eight miRNA-122 binding sites was used to competitively bind to miRNA-122, which is required for the hepatitis C virus (HCV) life cycle, thereby inhibiting the propagation of HCV, with an efficiency comparable to that of the anti-miR drug miravirsen (179). This study proposed the idea that engineered circRNAs could be used for disease-specific treatments, similar to targeted drugs; however, this idea is far from clinical translation. Due to their stable structure, conservative sequence, and specific expression pattern, circRNAs have the potential to be used as biomarkers for cancer. circRNAs have wide clinical application prospects—from diagnostic assessments to predicting patient prognosis and treatment response. However, most related studies have been single-center and retrospective works.
The study of circRNAs is just beginning, and the mechanism of circRNA biogenesis is not well understood. Even though a few circRNAs have been functionally characterized, our understanding of circRNAs remains incomplete. In addition to the identified functions as miRNA sponges, protein scaffolds, translation templates, and transcriptional regulators, other mechanisms are awaiting discovery, and the cooperation and the relative importance of these mechanisms cannot be evaluated at present. In addition, the transport and degradation mechanisms of circRNAs are poorly understood. Therefore, much work remains to be done. The discovery of circRNAs has undoubtedly enriched the content of RNA regulatory networks and has offered new approaches for the development of clinically translatable diagnostic/prognostic biomarkers and therapeutic targets for cancer.
Author Contributions
JL, XZ, and MY collected literatures and wrote the manuscript. HL contributed to writing design and revised the manuscript. All authors read and approved the final manuscript.
Funding
This review was funded by the National Natural Science Foundation of China (81772620), National Key R&D Program of China (2018ZX09201015), and Tianjin Science and Technology Major Project of Chronic Diseases Prevention and Control (17ZXMFSY00130).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. (2015) 12:381–8. doi: 10.1080/15476286.2015.1020271
2. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. (2014) 159:134–47. doi: 10.1016/j.cell.2014.09.001
3. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. (1976) 73:3852–6. doi: 10.1073/pnas.73.11.3852
4. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. (1986) 323:558–60. doi: 10.1038/323558a0
5. Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. Some yeast mitochondrial RNAs are circular. Cell. (1980) 19:313–9. doi: 10.1016/0092-8674(80)90505-X
6. Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. (2012) 40:3131–42. doi: 10.1093/nar/gkr1009
7. Houseley JM, Garcia-Casado Z, Pascual M, Paricio N, O'Dell KM, Monckton DG, et al. Noncanonical RNAs from transcripts of the Drosophila muscleblind gene. J Heredity. (2006) 97:253–60. doi: 10.1093/jhered/esj037
8. Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. (1993) 73:1019–30. doi: 10.1016/0092-8674(93)90279-Y
9. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. Scrambled exons. Cell. (1991) 64:607–13. doi: 10.1016/0092-8674(91)90244-S
10. Cocquerelle C, Daubersies P, Majerus MA, Kerckaert JP, Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. (1992) 11:1095–8. doi: 10.1002/j.1460-2075.1992.tb05148.x
11. Zaphiropoulos PG. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol. (1997) 17:2985–93. doi: 10.1128/MCB.17.6.2985
12. Surono A, Takeshima Y, Wibawa T, Ikezawa M, Nonaka I, Matsuo M. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Human Mol Genet. (1999) 8:493–500. doi: 10.1093/hmg/8.3.493
13. Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. (2010) 6:e1001233. doi: 10.1371/journal.pgen.1001233
14. Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. (1993) 7:155–60. doi: 10.1096/fasebj.7.1.7678559
15. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. (2012) 7:e30733. doi: 10.1371/journal.pone.0030733
16. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19:141–57. doi: 10.1261/rna.035667.112
17. Condorelli G, Latronico MV, Cavarretta E. microRNAs in cardiovascular diseases: current knowledge and the road ahead. J Am Coll Cardiol. (2014) 63:2177–87. doi: 10.1016/j.jacc.2014.01.050
18. Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet. (2013) 4:307. doi: 10.3389/fgene.2013.00307
19. Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. (2017) 136:1629–42. doi: 10.1161/CIRCULATIONAHA.117.029004
20. Wong DK, Yuen MF, Poon RT, Yuen JC, Fung J, Lai CL. Quantification of hepatitis B virus covalently closed circular DNA in patients with hepatocellular carcinoma. J Hepatol. (2006) 45:553–9. doi: 10.1016/j.jhep.2006.05.014
21. He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. (2017) 396:138–44. doi: 10.1016/j.canlet.2017.03.027
22. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
23. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. (2013) 340:440–1. doi: 10.1126/science.1238522
24. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. (1995) 268:415–7. doi: 10.1126/science.7536344
25. Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. (2019) 18:47. doi: 10.1186/s12943-019-1010-6
26. Shen Z, Zhou L, Zhang C, Xu J. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. (2020) 468:88–101. doi: 10.1016/j.canlet.2019.10.006
27. Wang C, Tan S, Liu WR, Lei Q, Qiao W, Wu Y, et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer. (2019) 18:134. doi: 10.1186/s12943-019-1061-8
28. Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. (2017) 388:208–19. doi: 10.1016/j.canlet.2016.12.006
29. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. (2014) 15:409. doi: 10.1186/s13059-014-0409-z
30. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. (2014) 20:1829–42. doi: 10.1261/rna.047126.114
31. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. (2015) 160:1125–34. doi: 10.1016/j.cell.2015.02.014
32. Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. (2017) 67:214–27.e7. doi: 10.1016/j.molcel.2017.05.023
33. Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. (2017) 8:14741. doi: 10.1038/ncomms14741
34. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. (2015) 10:170–7. doi: 10.1016/j.celrep.2014.12.019
35. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
36. Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. (2016) 26:1277–87. doi: 10.1101/gr.202895.115
37. Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun. (2016) 7:12060. doi: 10.1038/ncomms12060
38. Maass PG, Glazar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med. (2017) 95:1179–89. doi: 10.1007/s00109-017-1582-9
39. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. (2014) 32:453–61. doi: 10.1038/nbt.2890
40. Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. (2016) 44:1370–83. doi: 10.1093/nar/gkv1367
41. Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. (2015) 58:870–85. doi: 10.1016/j.molcel.2015.03.027
42. Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. (2014) 9:1966–80. doi: 10.1016/j.celrep.2014.10.062
43. You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. (2015) 18:603–10. doi: 10.1038/nn.3975
44. Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. (2015) 16:126. doi: 10.1186/s13059-015-0690-5
45. Hanan M, Soreq H, Kadener S. CircRNAs in the brain. RNA Biol. (2017) 14:1028–34. doi: 10.1080/15476286.2016.1255398
46. Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. (2016) 6:38907. doi: 10.1038/srep38907
47. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. (2008) 40:1413–5. doi: 10.1038/ng.259
48. Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. (2015) 5:8057. doi: 10.1038/srep08057
49. Panda AC, De S, Grammatikakis I, Munk R, Yang X, Piao Y, et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. (2017) 45:e116. doi: 10.1093/nar/gkx297
50. Jakobi T, Dieterich C. Computational approaches for circular RNA analysis. Wiley Interdiscip Rev RNA. (2019) 10:e1528. doi: 10.1002/wrna.1528
51. Zeng X, Lin W, Guo M, Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol. (2017) 13:e1005420. doi: 10.1371/journal.pcbi.1005420
52. Hansen TB. Improved circRNA Identification by combining prediction algorithms. Front Cell Dev Biol. (2018) 6:20. doi: 10.3389/fcell.2018.00020
53. Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. (2014) 20:1666–70. doi: 10.1261/rna.043687.113
54. Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang ZL, et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. (2016) 44:D196–202. doi: 10.1093/nar/gkv1273
55. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. (2016) 6:34985. doi: 10.1038/srep34985
56. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Briefings Bioinformatics. (2017) 18:984–92.
57. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. (2016) 13:34–42. doi: 10.1080/15476286.2015.1128065
58. Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. (2018) 46:D925–9. doi: 10.1093/nar/gkx863
59. Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. (2013) 4:283. doi: 10.3389/fgene.2013.00283
60. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. (2019) 176:869–81.e13. doi: 10.1016/j.cell.2018.12.021
61. Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. (2018) 46:D106–12. doi: 10.1093/nar/gkx891
62. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. (2013) 73:5609–12. doi: 10.1158/0008-5472.CAN-13-1568
63. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. (2017) 14:361–9. doi: 10.1080/15476286.2017.1279788
64. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. (2015) 22:256–64. doi: 10.1038/nsmb.2959
65. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. (2017) 38:1402–12. doi: 10.1093/eurheartj/ehw001
66. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. (2018) 110:304–15. doi: 10.1093/jnci/djx166
67. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. (2017) 27:626–41. doi: 10.1038/cr.2017.31
68. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. (2014) 56:55–66. doi: 10.1016/j.molcel.2014.08.019
69. Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. (2019) 38:e100836. doi: 10.15252/embj.2018100836
70. Lasda E, Parker R. Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS ONE. (2016) 11:e0148407. doi: 10.1371/journal.pone.0148407
71. Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin G, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. (2018) 7:2783–91. doi: 10.1002/cam4.1514
72. Zeng Y, Xu Y, Shu R, Sun L, Tian Y, Shi C, et al. Altered expression profiles of circular RNA in colorectal cancer tissues from patients with lung metastasis. Int J Mol Med. (2017) 40:1818–28. doi: 10.3892/ijmm.2017.3189
73. Xie F, Li Y, Wang M, Huang C, Tao D, Zheng F, et al. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol Cancer. (2018) 17:144. doi: 10.1186/s12943-018-0892-z
74. Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. (2019) 20:84. doi: 10.1186/s13059-019-1685-4
75. Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R, et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. (2019) 30:157–73.e7. doi: 10.1016/j.cmet.2019.05.009
76. Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. (2019) 18:20. doi: 10.1186/s12943-018-0935-5
77. Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, et al. Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. (2019). doi: 10.1002/hep.31068. [Epub ahead of print].
78. Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. (2018) 19:218. doi: 10.1186/s13059-018-1594-y
79. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. (2019) 10:4695. doi: 10.1038/s41467-019-12651-2
80. Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. (2017) 66:1151–64. doi: 10.1002/hep.29270
81. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. (2016) 44:2846–58. doi: 10.1093/nar/gkw027
82. Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. (2019) 10:3200. doi: 10.1038/s41467-019-11162-4
83. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. (2017) 24:357–70. doi: 10.1038/cdd.2016.133
84. Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. (2018) 37:5829–42. doi: 10.1038/s41388-018-0369-y
85. Chen X, Mao R, Su W, Yang X, Geng Q, Guo C, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy. (2019) :1-13. doi: 10.1080/15548627.2019.1634945
86. Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol Therapy Nucleic Acids. (2019) 18:24–33. doi: 10.1016/j.omtn.2019.07.012
87. Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. (2016) 165:289–302. doi: 10.1016/j.cell.2016.03.020
88. Tan SY, Sun D, Pu WC, Gou QH, Guo CL, Gong YL, et al. Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol Cancer. (2018) 17:5. doi: 10.1186/s12943-018-0887-9
89. Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin M, et al. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. (2019) 442:222–32. doi: 10.1016/j.canlet.2018.10.040
90. Gao S, Yu Y, Liu L, Meng J, Li G. Circular RNA hsa_circ_0007059 restrains proliferation and epithelial-mesenchymal transition in lung cancer cells via inhibiting microRNA-378. Life Sci. (2019) 233:116692. doi: 10.1016/j.lfs.2019.116692
91. Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. (2018) 500:846–51. doi: 10.1016/j.bbrc.2018.04.172
92. Chen L, Nan A, Zhang N, Jia Y, Li X, Ling Y, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. (2019) 18:13. doi: 10.1186/s12943-019-0943-0
93. Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer. (2018) 17:140. doi: 10.1186/s12943-018-0889-7
94. Huang W, Yang Y, Wu J, Niu Y, Yao Y, Zhang J, et al. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-beta signalling. Cell Death Differ. (2019) 27:1709–27. doi: 10.1038/s41418-019-0455-x
95. Yu C, Tian F, Liu J, Su M, Wu M, Zhu X, et al. Circular RNA cMras inhibits lung adenocarcinoma progression via modulating miR-567/PTPRG regulatory pathway. Cell Prolif. (2019) 52:e12610. doi: 10.1111/cpr.12610
96. Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. (2017) 77:2339–50. doi: 10.1158/0008-5472.CAN-16-1883
97. Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. (2018) 9:417. doi: 10.1038/s41419-018-0454-8
98. Zhi X, Zhang J, Cheng Z, Bian L, Qin J. circLgr4 drives colorectal tumorigenesis and invasion through Lgr4-targeting peptide. Int J Cancer. (2019). doi: 10.1002/ijc.32549. [Epub ahead of print].
99. Xiao H, Liu M. Circular RNA hsa_circ_0053277 promotes the development of colorectal cancer by upregulating matrix metallopeptidase 14 via miR-2467-3p sequestration. J Cell Physiol. (2019) 235:2881–90. doi: 10.1002/jcp.29193
100. Ren C, Zhang Z, Wang S, Zhu W, Zheng P, Wang W. Circular RNA hsa_circ_0001178 facilitates the invasion and metastasis of colorectal cancer through upregulating ZEB1 via sponging multiple miRNAs. Biol Chem. (2019) 401:487–96. doi: 10.1515/hsz-2019-0350
101. Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang W, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. (2019) 18:25. doi: 10.1186/s12943-019-0958-6
102. Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu X, et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. (2019) 18:45. doi: 10.1186/s12943-019-1006-2
103. Liu H, Liu Y, Bian Z, Zhang J, Zhang R, Chen X, et al. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 (Kip1) axis. Mol Cancer. (2018) 17:151. doi: 10.1186/s12943-018-0902-1
104. Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. (2017) 16:151. doi: 10.1186/s12943-017-0719-3
105. Zhang J, Hou L, Liang R, Chen X, Zhang R, Chen W, et al. CircDLST promotes the tumorigenesis and metastasis of gastric cancer by sponging miR-502-5p and activating the NRAS/MEK1/ERK1/2 signaling. Mol Cancer. (2019) 18:80. doi: 10.1186/s12943-019-1015-1
106. Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y, et al. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer. (2019) 18:158. doi: 10.1186/s12943-019-1094-z
107. Wang S, Zhang X, Li Z, Wang W, Li B, Huang X, et al. Circular RNA profile identifies circOSBPL10 as an oncogenic factor and prognostic marker in gastric cancer. Oncogene. (2019) 38:6985–7001. doi: 10.1038/s41388-019-0933-0
108. Tang H, Huang X, Wang J, Yang L, Kong Y, Gao G, et al. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol Cancer. (2019) 18:23. doi: 10.1186/s12943-019-0946-x
109. Zeng K, He B, Yang BB, Xu T, Chen X, Xu M, et al. The pro-metastasis effect of circANKS1B in breast cancer. Mol Cancer. (2018) 17:160. doi: 10.1186/s12943-018-0914-x
110. Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J, et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. (2018) 430:179–92. doi: 10.1016/j.canlet.2018.05.033
111. Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. (2019) 18:4. doi: 10.1186/s12943-018-0933-7
112. Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou X, et al. circFBXW7 inhibits malignant progression by sponging miR-197-3p and encoding a 185-aa protein in triple-negative breast cancer. Mol Therapy Nucleic acids. (2019) 18:88–98. doi: 10.1016/j.omtn.2019.07.023
113. Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. (2018) 68:1214–27. doi: 10.1016/j.jhep.2018.01.012
114. Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, et al. A noncoding regulatory RNAs network driven by circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. (2019) 71:130–47. doi: 10.1002/hep.30795
115. Wang H, Chen W, Jin M, Hou L, Chen X, Zhang R, et al. CircSLC3A2 functions as an oncogenic factor in hepatocellular carcinoma by sponging miR-490-3p and regulating PPM1F expression. Mol Cancer. (2018) 17:165. doi: 10.1186/s12943-018-0909-7
116. Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. (2018) 9:175. doi: 10.1038/s41419-017-0204-3
117. Zhang PF, Wei CY, Huang XY, Peng R, Yang X, Lu JC, et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer. (2019) 18:105. doi: 10.1186/s12943-019-1031-1
118. Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J, et al. CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer. (2019) 18:144. doi: 10.1186/s12943-019-1080-5
119. Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Therapy Nucleic Acids. (2018) 13:55–63. doi: 10.1016/j.omtn.2018.08.010
120. Zhang M, Xia B, Xu Y, Zhang Y, Xu J, Lou G. Circular RNA (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif Cells Nanomed Biotechnol. (2019) 47:1224–33. doi: 10.1080/21691401.2019.1593999
121. Zong ZH, Du YP, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. (2019) 38:437. doi: 10.1186/s13046-019-1437-z
122. Jin C, Zhao W, Zhang Z, Liu W. Silencing circular RNA circZNF609 restrains growth, migration and invasion by up-regulating microRNA-186-5p in prostate cancer. Artif Cells Nanomed And Biotechnol. (2019) 47:3350–8. doi: 10.1080/21691401.2019.1648281
123. Feng Y, Yang Y, Zhao X, Fan Y, Zhou L, Rong J, et al. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis. (2019) 10:792. doi: 10.1038/s41419-019-2028-9
124. An Y, Cai H, Zhang Y, Liu S, Duan Y, Sun D, et al. circZMYM2 competed endogenously with miR-335-5p to regulate JMJD2C in pancreatic cancer. Cell Physiol Biochem. (2018) 51:2224–36. doi: 10.1159/000495868
125. Huang WJ, Wang Y, Liu S, Yang J, Guo SX, Wang L, et al. Silencing circular RNA hsa_circ_0000977 suppresses pancreatic ductal adenocarcinoma progression by stimulating miR-874-3p and inhibiting PLK1 expression. Cancer Lett. (2018) 422:70–80. doi: 10.1016/j.canlet.2018.02.014
126. Chen Y, Li Z, Zhang M, Wang B, Ye J, Zhang Y, et al. Circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. (2019) 38:466. doi: 10.1186/s13046-019-1436-0
127. Wang R, Zhang S, Chen X, Li N, Li J, Jia R, et al. CircNT5E Acts as a Sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. (2018) 78:4812–25. doi: 10.1158/0008-5472.CAN-18-0532
128. Wang R, Zhang S, Chen X, Li N, Li J, Jia R, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. (2018) 17:166. doi: 10.1186/s12943-018-0911-0
129. Li RC, Ke S, Meng FK, Lu J, Zou XJ, He ZG, et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. (2018) 9:838. doi: 10.1038/s41419-018-0852-y
130. Huang X, He M, Huang S, Lin R, Zhan M, Yang D, et al. Circular RNA circERBB2 promotes gallbladder cancer progression by regulating PA2G4-dependent rDNA transcription. Mol Cancer. (2019) 18:166. doi: 10.1186/s12943-019-1098-8
131. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. (2013) 9:e1003777. doi: 10.1371/journal.pgen.1003777
132. Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE. (2015) 10:e0141214. doi: 10.1371/journal.pone.0141214
133. Zhong Q, Huang J, Wei J, Wu R. Circular RNA CDR1as sponges miR-7-5p to enhance E2F3 stability and promote the growth of nasopharyngeal carcinoma. Cancer Cell Int. (2019) 19:252. doi: 10.1186/s12935-019-0959-y
134. Yu J, Ding WB, Wang MC, Guo XG, Xu J, Xu QG, et al. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: A large-scale, multicenter study. Int J Cancer. (2020) 146:1754–63. doi: 10.1002/ijc.32647
135. Lin J, Cai D, Li W, Yu T, Mao H, Jiang S, et al. Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer. Clin Biochem. (2019) 74:60–8. doi: 10.1016/j.clinbiochem.2019.10.012
136. Tan S, Gou Q, Pu W, Guo C, Yang Y, Wu K, et al. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. (2018) 28:693–5. doi: 10.1038/s41422-018-0033-7
137. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. (2015) 25:981–4. doi: 10.1038/cr.2015.82
138. Pan B, Qin J, Liu X, He B, Wang X, Pan Y, et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. (2019) 10:1096. doi: 10.3389/fgene.2019.01096
139. Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. (2018) 432:237–50. doi: 10.1016/j.canlet.2018.04.035
140. Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. (2017) 6:1173–80. doi: 10.1002/cam4.1055
141. Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. (2015) 61:221–30. doi: 10.1373/clinchem.2014.230433
142. Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. (2018) 24:6319–30. doi: 10.1158/1078-0432.CCR-18-1270
143. Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. (2017) 11:422–37. doi: 10.1002/1878-0261.12045
144. Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. (2018) 9:1091. doi: 10.1038/s41419-018-1132-6
145. Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang L, et al. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin Chim Acta Int J Clin Chem. (2019) 492:37–44. doi: 10.1016/j.cca.2019.02.001
146. Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. (2017) 23:6330–8. doi: 10.3748/wjg.v23.i34.6330
147. Sun H, Tang W, Rong D, Jin H, Fu K, Zhang W, et al. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. (2018) 21:299–306. doi: 10.3233/CBM-170379
148. Li P, Chen H, Chen S, Mo X, Li T, Xiao B, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. (2017) 116:626–33. doi: 10.1038/bjc.2016.451
149. Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. (2017) 284:2170–82. doi: 10.1111/febs.14132
150. Li J, Wang J, Chen Z, Chen Y, Jin M. Hsa_circ_0079530 promotes cell proliferation and invasion in non-small cell lung cancer. Gene. (2018) 665:1–5. doi: 10.1016/j.gene.2018.04.059
151. Zong L, Sun Q, Zhang H, Chen Z, Deng Y, Li D, et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacotherapy. (2018) 102:639–44. doi: 10.1016/j.biopha.2018.03.084
152. Li XN, Wang ZJ, Ye CX, Zhao BC, Huang XX, Yang L. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed Pharmacotherapy. (2019) 112:108611. doi: 10.1016/j.biopha.2019.108611
153. Li X, Wang J, Zhang C, Lin C, Zhang J, Zhang W, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. (2018) 246:166–79. doi: 10.1002/path.5125
154. Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. (2017) 23:8345–54. doi: 10.3748/wjg.v23.i47.8345
155. Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. (2018) 14:321–30. doi: 10.7150/ijbs.24360
156. Qiu L, Huang Y, Li Z, Dong X, Chen G, Xu H, et al. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol Oncol. (2019) 13:441–55. doi: 10.1002/1878-0261.12424
157. He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. (2017) 36:145. doi: 10.1186/s13046-017-0614-1
158. Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S, et al. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. (2018) 8:2878. doi: 10.1038/s41598-018-21300-5
159. Li P, Chen S, Chen H, Mo X, Li T, Shao Y, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta Int J Clin Chem. (2015) 444:132–6. doi: 10.1016/j.cca.2015.02.018
160. Xu H, Wang C, Song H, Xu Y, Ji G. RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol Cancer. (2019) 18:8. doi: 10.1186/s12943-018-0932-8
161. Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. (2019) 18:71. doi: 10.1186/s12943-019-0969-3
162. Dong W, Dai ZH, Liu FC, Guo XG, Ge CM, Ding J, et al. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine. (2019) 45:155–67. doi: 10.1016/j.ebiom.2019.06.030
163. Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. (2017) 7:5428. doi: 10.1038/s41598-017-05432-8
164. Li S, Gu H, Huang Y, Peng Q, Zhou R, Yi P, et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle. (2018) 17:2349–59. doi: 10.1080/15384101.2018.1526599
165. Meng J, Chen S, Han JX, Qian B, Wang XR, Zhong WL, et al. Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. (2018) 78:4150–62. doi: 10.1158/0008-5472.CAN-17-3009
166. Lu J, Zhang PY, Li P, Xie JW, Wang JB, Lin JX, et al. Circular RNA hsa_circ_0001368 suppresses the progression of gastric cancer by regulating miR-6506-5p/FOXO3 axis. Biochem Biophys Res Commun. (2019) 512:29–33. doi: 10.1016/j.bbrc.2019.02.111
167. Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. (2018) 17:137. doi: 10.1186/s12943-018-0888-8
168. Yan B, Zhang W, Mao XW, Jiang LY. Circular RNA ciRS-7 correlates with advance disease and poor prognosis, and its down-regulation inhibits cells proliferation while induces cells apoptosis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. (2018) 22:8712–21. doi: 10.26355/eurrev_201812_16636
169. Wang Y, Li Y, He H, Wang F. Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene. (2019) 720:144099. doi: 10.1016/j.gene.2019.144099
170. Li Y, Hu J, Li L, Cai S, Zhang H, Zhu X, et al. Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem Biophys Res Commun. (2018) 503:2089–94. doi: 10.1016/j.bbrc.2018.07.164
171. Luo YH, Zhu XZ, Huang KW, Zhang Q, Fan YX, Yan PW, et al. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacotherapy. (2017) 96:892–8. doi: 10.1016/j.biopha.2017.12.015
172. Liu T, Ye P, Ye Y, Lu S, Han B. Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA-328-3p-mediated inhibition of COL1A1. J Cell Mol Med. (2020) 24:2189–201. doi: 10.1111/jcmm.14875
173. Xu Y, Yao Y, Leng K, Ji D, Qu L, Liu Y, et al. Increased expression of circular RNA circ_0005230 indicates dismal prognosis in breast cancer and regulates cell proliferation and invasion via miR-618/ CBX8 signal pathway. Cell Physiol Biochem. (2018) 51:1710–22. doi: 10.1159/000495675
174. Li Y, Wan B, Liu L, Zhou L, Zeng Q. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun. (2019) 508:991–6. doi: 10.1016/j.bbrc.2018.12.046
175. Su H, Tao T, Yang Z, Kang X, Zhang X, Kang D, et al. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. (2019) 18:27. doi: 10.1186/s12943-019-0951-0
176. Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. (2018) 37:177. doi: 10.1186/s13046-018-0822-3
177. Kun-Peng Z, Chun-Lin Z, Jian-Ping H, Lei Z. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int J Biol Sci. (2018) 14:1513–20. doi: 10.7150/ijbs.27523
178. Yang M, Li G, Fan L, Zhang G, Xu J, Zhang J. Circular RNA circ_0034642 elevates BATF3 expression and promotes cell proliferation and invasion through miR-1205 in glioma. Biochem Biophys Res Commun. (2019) 508:980–5. doi: 10.1016/j.bbrc.2018.12.052
Keywords: circular RNAs, molecular mechanism, cancer, clinical application, biomarker
Citation: Liu J, Zhang X, Yan M and Li H (2020) Emerging Role of Circular RNAs in Cancer. Front. Oncol. 10:663. doi: 10.3389/fonc.2020.00663
Received: 02 December 2019; Accepted: 09 April 2020;
Published: 20 May 2020.
Edited by:
Alfons Navarro, University of Barcelona, SpainReviewed by:
Shengfang Ge, Shanghai Jiao Tong University, ChinaYun Dai, First Affiliated Hospital of Jilin University, China
Copyright © 2020 Liu, Zhang, Yan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Li, bGlodWlAdGptdWNoLmNvbQ==
 Jing Liu
Jing Liu Xin Zhang
Xin Zhang Meinan Yan1,2,3
Meinan Yan1,2,3