- 1Department of Oncology and Hematology, The Affiliated Hospital of Medical School of Ningbo University, Ningbo, China
- 2Department of Otorhinolaryngology Head and Neck Surgery, Ningbo Medical Center Lihuili Hospital, Ningbo, China
The homeobox A cluster (HOXA) gene family, comprising 11 members, is involved in a wide spectrum of biological functions in human cancers. However, there is little research on the expression profile and prognostic values of HOXA genes in laryngeal squamous cell cancer (LSCC). Based on updated public resources and integrative bioinformatics analysis, we assessed the expression profile and prognostic values of the HOXA family members. Expression and methylation data on HOXA family members were obtained from The Cancer Genome Atlas (TCGA). The prognostic values of HOXA members and clinical features were identified. A gene set enrichment analysis (GSEA) was conducted to explore the mechanism underlying the involvement of HOXA members in LSCC. The associations between tumor immune infiltrating cells (TIICs) and the HOXA family members were evaluated using the Tumor Immune Estimation Resource (TIMER) database. HOXA2 and HOXA4 were downregulated and HOXA7 and HOXA9–13 were upregulated in LSCC. Upregulation of HOXA10, HOXA11, and HOXA13, along with two clinical characteristics (M stage and gender), were associated with a poor LSCC prognosis based on the results of univariate and multivariate Cox proportional hazards regression analyses. Although there were no significant correlations between TIICs and HOXA members, the GSEA results indicated that HOXA members participate in multiple biological processes underlying tumorigenesis. This study comprehensively analyzed the HOXA members, providing insights for further investigation of the HOXA family members as potential targets in LSCC.
Introduction
Laryngeal cancer is one of the most common malignancies in the head and neck region, and laryngeal squamous cell cancer (LSCC) accounts for more than 95% of cases (1). Despite progress regarding comprehensive therapeutic strategies to treat LSCC, the prognosis of LSCC remains unsatisfactory, as 30–40% of patients die within 5 years of diagnosis with advanced LSCC (2). Identification of reliable biomarkers for LSCC prognosis could facilitate individualized treatment.
The HOX gene family is one of the families of homeobox genes that function as developmental regulatory genes (3). In mammals, there are 39 HOX genes in four gene clusters named HOXA, HOXB, HOXC, and HOXD (4). The HOXA cluster comprises 11 genes (including HOXA1, HOXA2, HOXA3, HOXA4, HOXA5, HOXA6, HOXA7, HOXA9, HOXA10, HOXA11, and HOXA13), which encode proteins that contain the DNA-binding homeobox motif (5). The molecular functions of the HOXA family cover a wide spectrum of biological processes, including differentiation, proliferation, migration and cell death. A substantial body of scientific evidence indicates that the expression of particular HOXA genes is dysregulated in certain types of carcinomas, which contributes to carcinogenesis (6–10). For instance, HOXA1 mRNA and protein expression is upregulated in breast cancer, and forced expression of HOXA1 in human breast cancer cells resulted in increased cell proliferation and doxorubicin resistance (11, 12). Aberrantly expressed HOXA6 and HOXA13 were also observed in breast cancer (13). In colorectal cancer, HOXA13 was expressed more in normal colons than in malignant colons, and it was more highly expressed on the left side of the normal colon compared to the right side, indicating that differential HOXA gene expression occurs in an organized manner (10). Additionally, several studies have reported that HOXA9 and HOXA10 can serve as predictive biomarkers of poor survival in glioblastoma multiforme (GBM) (14–16).
Collectively, the differential expression and prognostic values of the HOXA family members have been noticed in various types of cancers. Studying the differential expression of HOXA genes in LSCC provides an opportunity to advance our understanding of LSCC development and to develop new therapeutic agents. In this study, based on updated public resources and integrative bioinformatics analysis, the expression profile and prognostic values of the HOXA family members were comprehensively assessed.
Materials and Methods
The Cancer Genome Atlas (TCGA) mRNA Expression Data of the HOXA Family
The TCGA program was conducted by the National Cancer Institute and National Human Genome Research Institute to molecularly characterize over 20,000 primary cancer samples and matched normal samples spanning 33 cancer types, including 528 cases of primary head and neck squamous carcinoma (HNSC), two cases of metastatic HNSC and 74 adjacent normal control samples. A total of 111 cases of laryngeal squamous cell cancer (LSCC) and 12 normal controls were included in the current study, after matching clinical parameters (including gender, age, smoking history, alcohol consumption, tumor (T) stage, node (N) stage, metastasis (M) stage, clinical stage and primary cancer sites). Subsequently, we used the Genomic Data Commons (GDC) Data Transfer Tool recommended by TCGA to download high-throughput sequencing (HTSeq) Fragments Per Kilobase of transcript per Million mapped reads (FPKM) data on the HOXA family.
Comparison of the mRNA Expression of the HOXA Family in LSCC and Normal Tissues
Using Perl 5.26 software, the mRNA expression levels of the HOXA family were obtained from the HTSeq level 3 data on genome mRNA expression. The differential expression of the HOXA family in LSCC tissues compared to normal tissues was analyzed utilized the limma package in R 3.6.0 software. The results were visualized using the pheatmap package.
Correlation Between mRNA Expression and Methylation of the HOXA Family in LSCC
We used the GDC Data Transfer Tool recommended by TCGA to download data from Illumina HumanMethylation 450K on the methylation levels of cg sites in the gene promoter regions of differentially expressed HOXA members in LSCC tissues. Thereafter, we utilized the corrplot package to further explore the correlation between methylation and HOXA expression in LSCC. The information on cg sites from Illumina HumanMethylation 450K were annotated using the annotation file from the official Illumina website (https://support.illumina.com/downloads/~infinium_humanmethylation450_product_files.html).
Survival Analysis of HOXA Members in LSCC
The prognostic values of the HOXA members were investigated using the following two steps: (1) the associations between HOXA members, as well as each clinical parameter, and overall survival among LSCC patients were assessed using univariate Cox proportional hazards regression analyses and (2) using multivariate Cox proportional hazards regression analysis, the independent prognostic values of the HOXA members were then obtained by controlling for the significant clinical parameters from step 1. All the analyses were performed using the survival package in R 3.6.0 software.
Associations Between Tumor Immune Infiltrating Cells (TIICs) and the HOXA Family Using the Tumor Immune Estimation Resource (TIMER) Database
Tumor cells and TIICs interact through multiple genes and pathways during cancer progression. To explore the correlations between TIICs and HOXA members, we utilized the TIMER platform (https://cistrome.shinyapps.io/timer/), which is an online tool for assessing the specific gene(s) associated with TIICs (17). In TIMER, the TIICs include B-cells, CD4+ T-cells, CD8+ T-cells, dendritic cells, macrophages and neutrophils.
Gene Set Enrichment Analysis (GSEA)
To evaluate the potential mechanism underlying the involvement of HOXA members in the carcinogenesis of LSCC, we performed GSEA (version 4.0.1; http://software.broadinstitute.org/gsea/index.jsp) to identify the to identify the pathways related to the differential HOXA expression in the TCGA LSCC tissues (18). The annotated gene set file c2.cp.kegg.v7.0.symbols.gmt (from the Msig database) was used as the reference. GSEA was performed using a random combination number of 1,000 permutations and a false discovery rate (FDR) < 0.05 to identify the significantly enriched pathways.
Statistical Analysis
The HTSeq FPKM mRNA data from the TCGA database was handled using Perl 5.26 software. The limma package was applied to analyze the expression of HOXA members in LSCC tissues, the corrplot package was used for the correlation between methylation and expression of HOXA members, the survival package was used for the analysis of prognostic values, the ggplot package was used to plot forest plots related to the multivariate Cox proportional hazards regression analysis.
Results
Expression Status of HOXA Members in LSCC Tissues
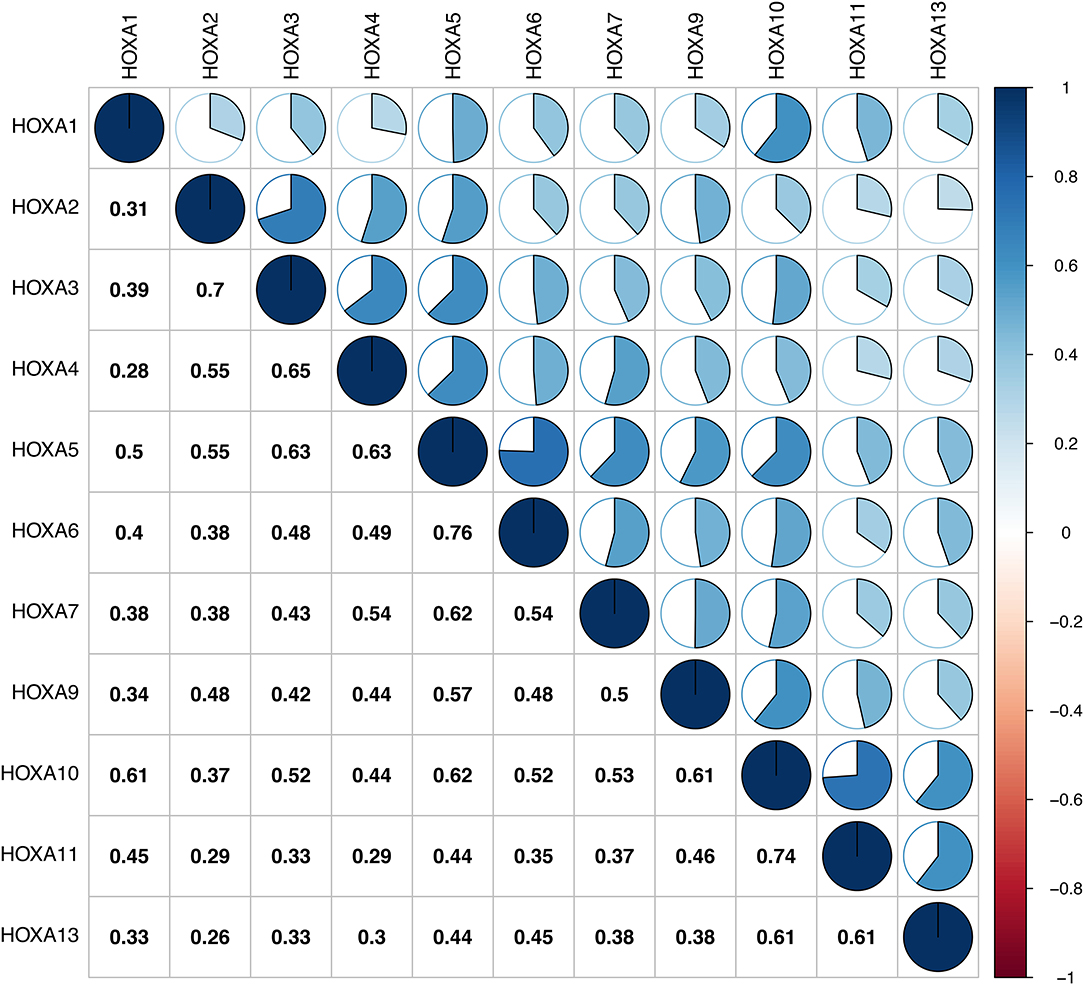
First of all, the mRNA expression data on HOXA members (HOXA1–13) from 111 LSCC samples and 12 normal control samples, which originated from TCGA, were obtained using Perl software. Pearson's correlation of HOXA family genes were calculated and used to assess whether these genes were correlated with each other using the corrplot package. As shown in Figure 1, the HOXA family genes were correlated to a significant degree.
Thereafter, the differentially expressed HOXA members were analyzed using the limma package and visualized using the pheatmap package, as shown in Figure 2A. As shown in Figure 2B, HOXA2 and HOXA4 were significantly downregulated in LSCC tissues compared to control tissues, while HOXA7, HOXA9, HOXA10, HOXA11, and HOXA13 were significantly upregulated in LSCC tissues. There were no significant differences in HOXA1, HOXA3, HOXA5, and HOXA6 expression between LSCC and control tissues.
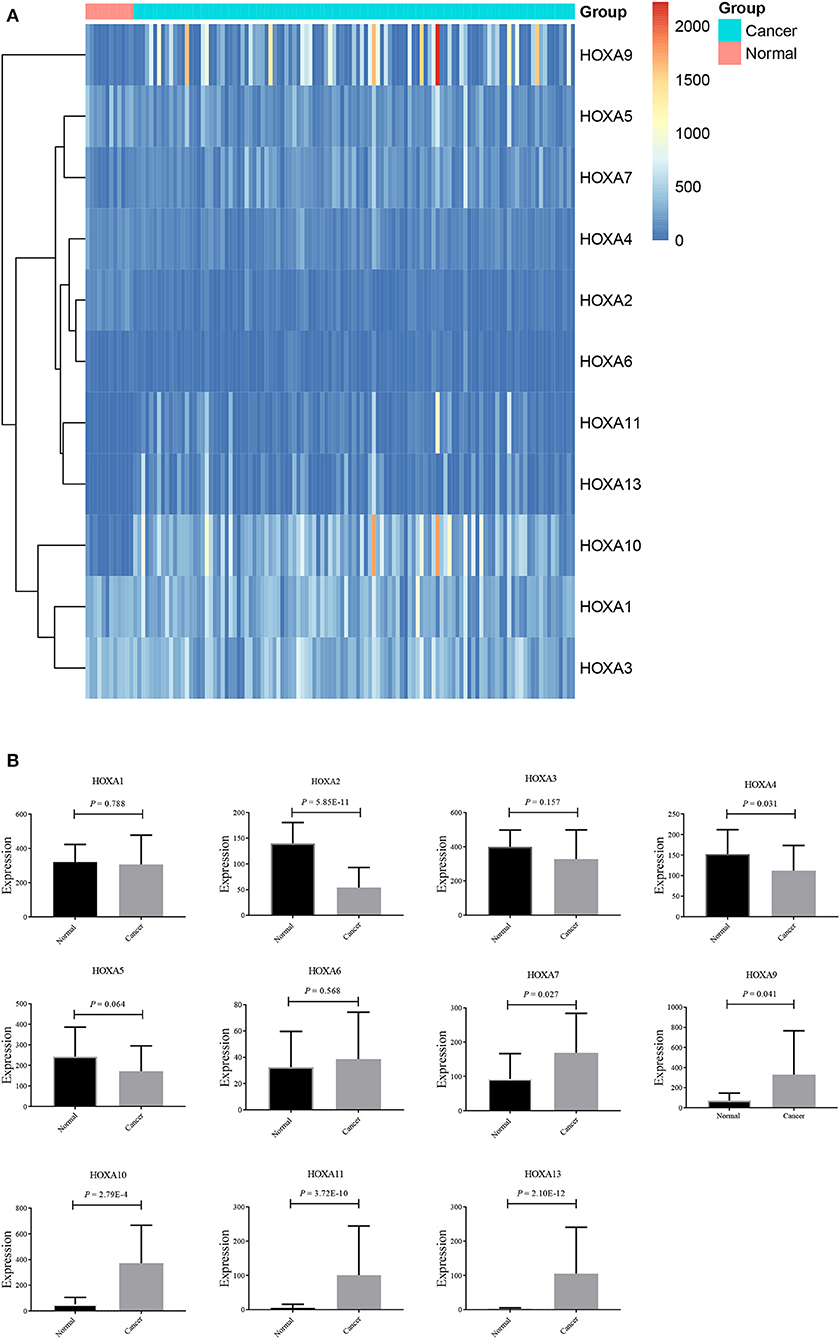
Figure 2. Expression profile of HOXA members in LSCC represented by a heatmap (A), and histograms (B).
Correlation of HOXA Expression and Methylation in LSCC
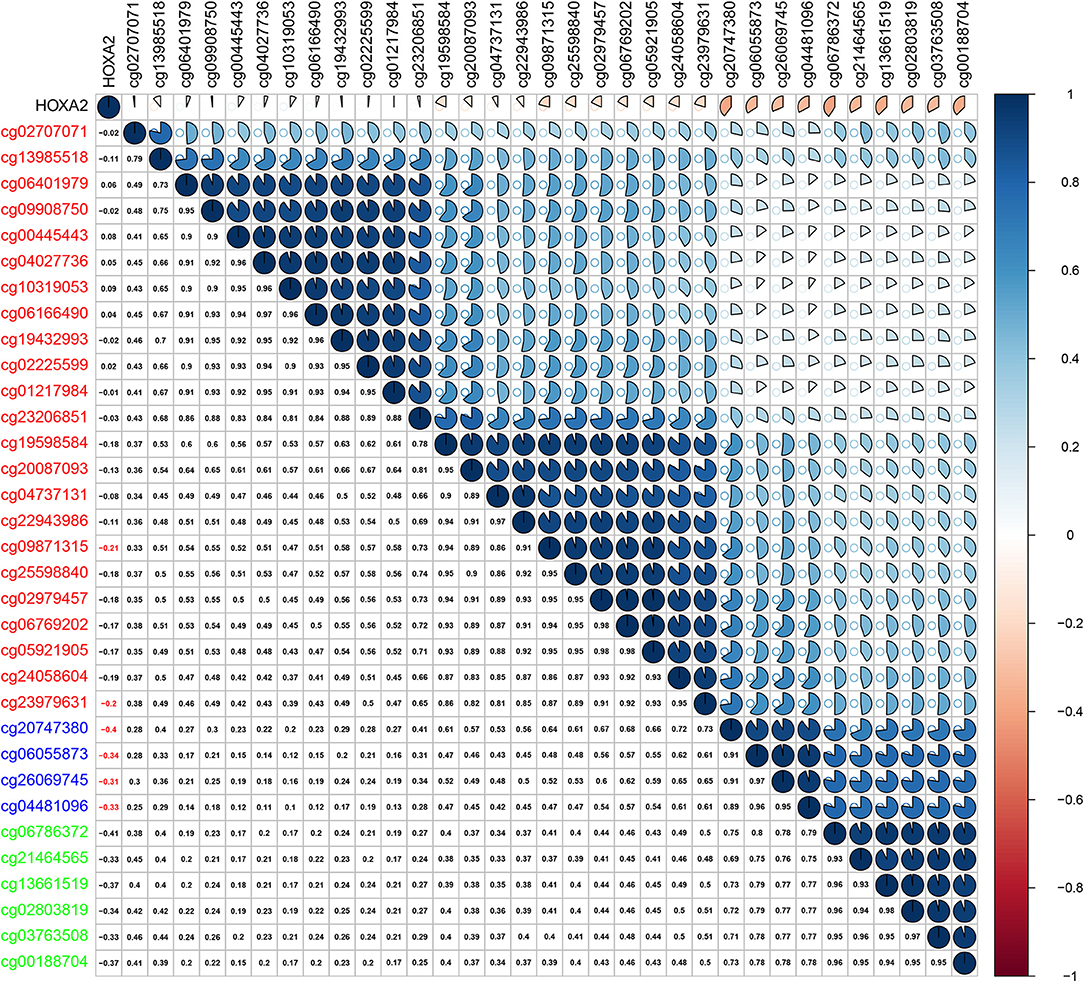
Methylation of gene promoter regions is one of the most common mechanisms that influences gene expression during the progression of human cancer. We identified seven differentially expressed HOXA members in LSCC (downregulated HOXA2 and HOXA4 and upregulated HOXA7, HOXA9, HOXA10, HOXA11, and HOXA13). The Pearson's correlation results showed that six of seven differentially expressed HOXA members (including HOXA4, HOXA7, HOXA9, HOXA10, HOXA11, and HOXA13) was negative associated with methylation level (Figure S1), and only five of the 32 assessed CG sites in the promoter region of HOXA2 exhibited negative correlation with HOXA2 expression in LSCC (Figure 3). These results indicated the inverse correlation between expression and methylation level of HOXA members in LSCC.
Prognostic Values of HOXA Members in LSCC
Subsequently, the prognostic values of HOXA members were analyzed. First, the predictive capabilities of differentially expressed HOXA members (HOXA2, HOXA4, HOXA7, HOXA9, HOXA10, HOXA11, and HOXA13) and clinical features were assessed by univariate Cox proportional hazards regression analyses. The results showed that the expression of three HOXA members (HOXA10, HOXA11, and HOXA13) and two clinical features (M stage and male) were associated with poor outcome of LSCC patients (hazard ratio [HR] for HOXA10: 1.379 (1.081–1.759); HR for HOXA11: 1.179 (1.000–1.391); HR for HOXA13: 1.129 (0.999–1.277); HR for M stage: 8.225 (1.901–35.594); and HR for male: 3.367 [1.708–6.639]) (Table 1). Second, the independent prognostic values of HOXA10, HOXA11, and HOXA13 were assessed using multivariate Cox proportional hazards regression analysis to control for the prognostic effects of the clinical features. The results showed that the expression of HOXA10, HOXA11, and HOXA13 and two clinical parameters (M stage and gender) were independent prognostic biomarkers of LSCC outcome. The results of the multivariate Cox proportional hazards regression analysis are exhibited in forest plots in Figure 4.
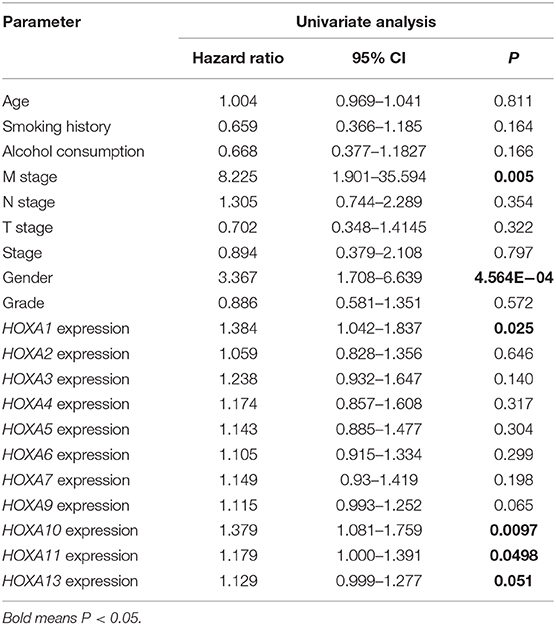
Table 1. Univariate Cox proportional hazards regression analyses of HOXA members and clinical features in LSCC.
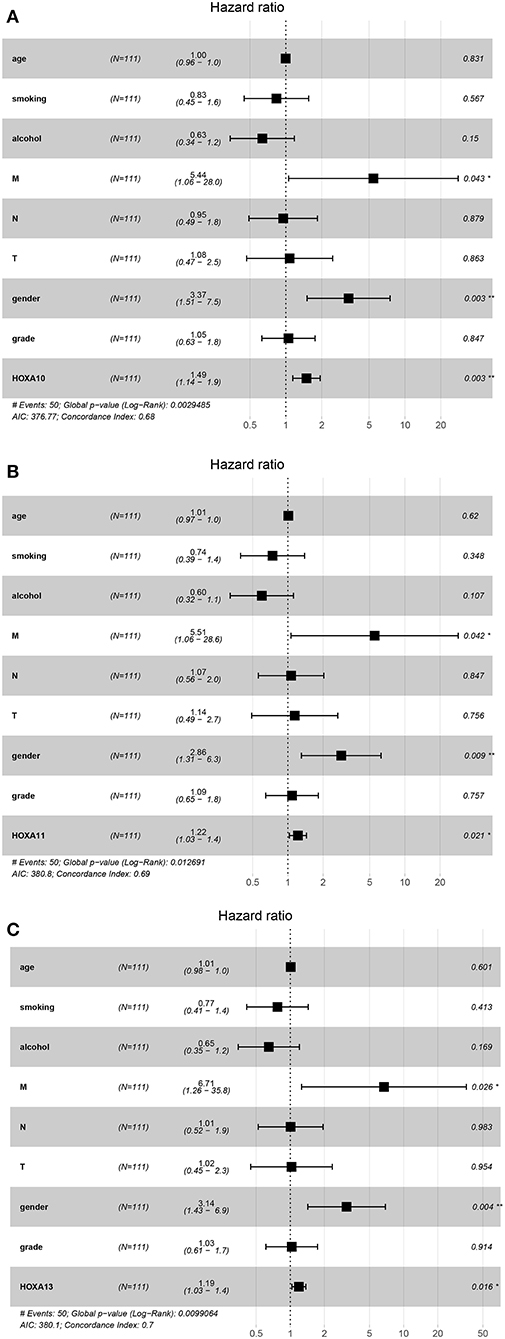
Figure 4. Forest plots of the results of multivariate Cox regression analyses of significant prognostic factors: HOXA10 (A), HOXA11 (B), and HOXA13 (C). *stands for P < 0.05; **stands for P < 0.01.
Correlations Between TIICs and HOXA Members
Considering the increasing evidence on the associations between immunological features and prognosis in cancer, we further explored the correlations between TIICs and HOXA members. The TIMER database is a public resource used to explore the associations between certain gene products and immune cells around tumor cells. The first column in Figure 5 shows scatterplots of the expression of HOXA members against tumor purity. HOXA members with high expression in the microenvironment cells are expected to have a negative association with tumor purity, while HOXA members with high expression in tumor cells are expect to have a positive association with tumor purity (17). In accordance with our aforementioned findings, HOXA7, HOXA10, and HOXA13 were highly expressed in LSCC tissues, with positive associations with tumor purity (Figure 5). However, there were no significant correlations between TIICs and HOXA members (Figure S2).
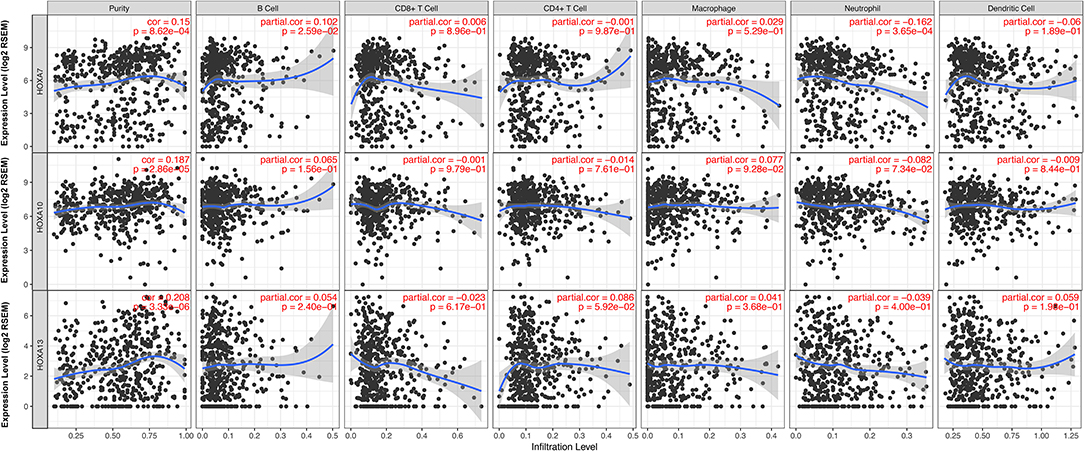
Figure 5. Correlations between tumor infiltrating immune cells (TIICs; B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) and HOXA members (including HOXA7, HOXA10, and HOXA13) in LSCC. Tumor purity is shown in the panels on the left.
Potential Mechanism Underlying the Effects of Prognostic HOXA Members on LSCC Carcinogenesis
A GSEA of differentially expressed HOXA members with statistical prognostic value was conducted to evaluate the potential biological mechanism by which differential expression of HOXA10, HOXA11, and HOXA13 affects the carcinogenesis of LSCC. The GSEA indicated that high expression of HOXA10 was related to “WNT signaling pathway,” “pathway in cancer,” “basal cell carcinoma,” “cell cycle,” “mismatch repair,” and “DNA replication” (Figure 6A), high expression of HOXA11 was related to “DNA replication,” “mismatch repair,” and “nucleotide excision repair” (Figure 6B) and high expression of HOXA13 was related to “colorectal cancer” and “WNT signaling pathway” (Figure 6C).
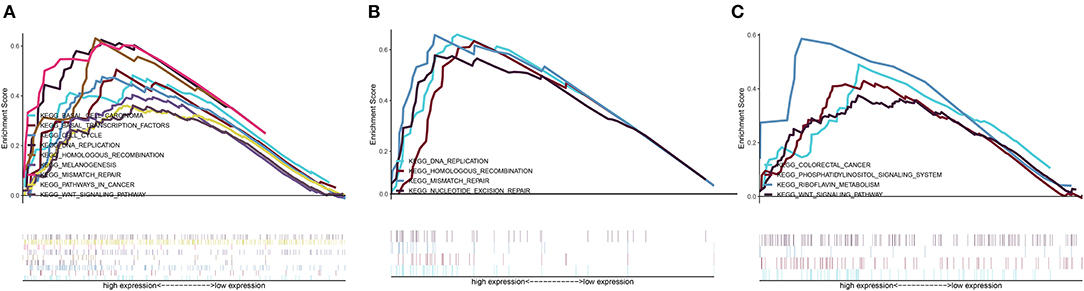
Figure 6. Cancer-related Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with HOXA10 (A), HOXA11 (B), and HOXA13 (C) based on a gene set enrichment analysis (GSEA).
Discussion
Homeobox genes were first identified in the fruit fly Drosophila (19). A total of 39 HOX genes are located on various chromosomes, which are clustered into four clusters, namely HOXA, HOXB, HOXC and HOXD (4). The genes in these four cluster each encode a 61-amino acid homeodomain, and these genes are key components of master regulatory pathways during normal embryonic development (3). A typical characteristic of the homeodomain is its DNA-binding nature; the proteins function as transcription factors by binding to the promoters of various target genes (20). Increasing evidence has shown that the protein products of HOXA genes not only act as transcriptional factors promoting carcinogenesis but also serve as tumor-suppressor factors, based on their aberrant expression patterns in certain organs. Increasing published or public genomic data and multiple online platforms provide us the opportunity for exploring the expression profiles of families of genes in human cancers and their clinical practice value. This study demonstrated the distinct expression profile and methylation profile, prognostic values and biological processes related to HOXA members in LSCC.
Previous research has shown that, according to expression data, HOXA genes contribute to the development of human cancers. Reverse transcriptase-polymerase chain reaction (RT-PCR) showed that HOXA7 and HOXA9 mRNAs were significantly overexpressed in esophageal squamous cell carcinoma tissues compared to non-cancerous surrounding tissues (21), while HOXA9 was epigenetically downregulated in lung cancer (22). HOXA13 expression increased in breast cancer (13), whereas it was downregulated in colorectal cancer (10). However, the expression of the entire HOXA family in LSCC was not previously comprehensively investigated. This in silico study demonstrated the expression profile of HOXA members in LSCC and showed that HOXA2 and HOXA4 were downregulated in LSCC tissues compared to normal control tissues. In contrast, HOXA7, HOXA9, HOXA10, HOXA11, and HOXA13 were upregulated in LSCC tissues compared to normal control tissues. Unfortunately, no significant differences in the mRNA expression of HOXA1, HOXA3, HOXA5, and HOXA6 were identified in LSCC tissues compared to normal control tissues.
According to the Pearson's correlation between HOXA mRNA expression and the methylation level of cg sites in the promoter regions in LSCC, among the seven differentially expressed HOXA members (HOXA2, HOXA4, HOXA7, HOXA9, HOXA10, HOXA11, and HOXA13), most expression levels, particularly regarding HOXA4 and HOXA9, are affected by the methylation level. These results are in accordance with previous findings showing a negative correlation between HOXA4 methylation and expression in patients with acute myeloid leukemia (23).
Several reports have identified HOXA gene signatures in GBM, and high expression of HOXA9 and HOXA10 were reported to be predictors of poor outcome in patients with GBM (14, 15). Moreover, it was reported that novel methylation markers in HOXA9 also served as an independent indicator of prognosis in invasive bladder cancer (24). Additionally, multiple highly expressed HOXA members were reported to be significantly correlated with poor overall survival in patients with acute myeloid leukemia (25). In this study, univariate Cox proportional hazards regression analyses were performed to analyze the prognostic values of HOXA members in LSCC. In fact, four HOXA members were significantly associated with poor clinical outcomes in LSCC (HOXA1, HOXA10, HOXA11, and HOXA13). Thus, although no significant differential expression of HOXA1 was found in LSCC, the univariate Cox proportional hazards regression showed that HOXA1 expression was significantly associated with prognosis. The predictive potential of HOXA has also been reported in breast cancer (12). In breast cancer, HOXA1 knockdown inhibited cell proliferation and increased apoptosis and cell cycle arrest by influencing the aberrant expression of several cell cycle and apoptosis-associated proteins, comprising cyclin D1, B-cell lymphoma 2 (Bcl-2) and Bcl-2-like protein 4 (12). Thus, although HOXA1 was not differentially expressed in LSCC, the prognostic value of HOXA1 has been highlighted in various human cancers, including in LSCC. Exploration of the HOXA1-related mechanisms is still required.
In hepatocellular carcinoma cells, HOXA10 knockdown induced cell cycle arrest at the G0/G1 phase and apoptosis by reducing the expression of Cyclin D1 and Survivin (26). Decreased expression of HOXA10 accelerated the acetylation of p53 (Lys382) and suppressed the transcription of histone deacetylase 1 (HDAC1; a potential deacetylase for p53) to activate p53 transcription (26). Additionally, HOXA10 might promote cell proliferation by elevating Bcl-2 expression and inhibiting apoptosis in gastric cancer, and high expression of HOXA10 predicted poor overall survival in gastric cancer patients (27). In this study, we found high expression of HOXA10 in LSCC tissues. Both univariate and multivariate Cox proportional hazards regression analyses affirmed the prognostic value of HOXA10 in the prediction of poor outcome in LSCC patients.
Overexpression of HOXA11 has been observed in ovarian cancer (28), bladder cancer (29), renal cell carcinoma (29) and lung cancer (30), while downregulation of HOXA11 has been observed in gastric cancer (31) and glioblastoma (32). In glioblastoma, overexpression of HOXA11 confers a tumor suppressive effect, reduces treatment resistance and contributes to a favorable prognosis (32). However, overexpression of HOXA11 showed a poor association with overall survival in lung cancer (33). HOXA11 was significantly downregulated in cisplatin-resistant lung adenocarcinoma cell lines compared with parent cell lines, and in vitro experiments showed that overexpression of HOXA11 increased cisplatin sensitivity by inhibiting Akt/β-catenin signaling (34). Our results showed high expression of HOXA11 in LSCC, which was associated with unfavorable outcomes in LSCC patients. However, given that there is little relevant research on the topic, the biological and prognostic values of HOXA11 warrant further intensive investigation. It may be useful to systematically explore the prognostic value of HOXA11 using meta-analysis.
HOXA13 is expressed more in normal colons than in malignant colons. Additionally, HOXA13 was differentially expressed based on location, with higher expression on the left side of the normal colon compared to the right side (10). Differential expression of HOXA13 was also reported in breast cancer (13), gastric cancer (35), prostate carcinoma (36) and thyroid cancer (37). HOXA13 knockdown significantly restored the epithelial characteristics and reduced the mesenchymal characteristics of the cancer cells via the transforming growth factor (TGF)-β signaling pathway (35). Moreover, HOXA13 expression negatively affects cisplatin sensitivity in human esophageal squamous cells and overall survival in patients with esophageal squamous cell carcinoma (38). Our results showed that multiple cancer-associated pathways were identified in LSCC tissues with high expression of HOXA13, and high expression of HOXA13 in LSCC predicted poor overall survival.
Conclusion
This in silico study demonstrated the expression profile of HOXA family members in LSCC and the biological and prognostic values of the HOXA family in LSCC, providing insights for further investigation of HOXA members as potential targets in LSCC.
Data Availability Statement
The data that support the findings of this study are openly available in The Cancer Genome Atlas (TCGA) program at https://portal.gdc.cancer.gov/.
Author Contributions
JL and CZ designed the research study and analyzed the data from public database. JL, MY, and CZ were involved in data analysis. CZ was responsible for writing of manuscript. JL and MY contributed to the revised manuscript. All authors reviewed the manuscript.
Funding
This work was supported by the Ningbo Health Branding Subject Fund (No. PPXK2018-02).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00368/full#supplementary-material
Figure S1. Pearson's correlation between methylation levels and expression of all differentially expressed HOXA members in LSCC [including HOXA2 (A), HOXA4 (B), HOXA7 (C), HOXA9 (D), HOXA10 (E), HOXA11 (F), and HOXA13 (G)].
Figure S2. Correlations between tumor infiltrating immune cells (TIICs; B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) and all HOXA members in LSCC.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. (2019) 69:7–34. doi: 10.3322/caac.21551
2. Thompson LD. Laryngeal dysplasia, squamous cell carcinoma, and variants. Surg Pathol Clin. (2017) 10:15–33. doi: 10.1016/j.path.2016.10.003
3. Gehring WJ, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. (1986) 20:147–73. doi: 10.1146/annurev.ge.20.120186.001051
4. Ruddle FH, Bartels JL, Bentley KL, Kappen C, Murtha MT, Pendleton JW. Evolution of hox genes. Annu Rev Genet. (1994) 28:423–42. doi: 10.1146/annurev.ge.28.120194.002231
5. Sanchez-Herrero E. Hox targets and cellular functions. Scientifica. (2013) 2013:738257. doi: 10.1155/2013/738257
6. Maeda K, Hamada J, Takahashi Y, Tada M, Yamamoto Y, Sugihara T, et al. Altered expressions of HOX genes in human cutaneous malignant melanoma. Int J Cancer. (2005) 114:436–41. doi: 10.1002/ijc.20706
7. Makiyama K, Hamada J, Takada M, Murakawa K, Takahashi Y, Tada M, et al. Aberrant expression of HOX genes in human invasive breast carcinoma. Oncol Rep. (2005) 13:673–9. doi: 10.3892/or.13.4.673
8. Abe M, Hamada J, Takahashi O, Takahashi Y, Tada M, Miyamoto M, et al. Disordered expression of HOX genes in human non-small cell lung cancer. Oncol Rep. (2006) 15:797–802. doi: 10.3892/or.15.4.797
9. Hassan NM, Hamada J, Murai T, Seino A, Takahashi Y, Tada M, et al. Aberrant expression of HOX genes in oral dysplasia and squamous cell carcinoma tissues. Oncol Res. (2006) 16:217–24. doi: 10.3727/000000006783981080
10. Kanai M, Hamada J, Takada M, Asano T, Murakawa K, Takahashi Y, et al. Aberrant expressions of HOX genes in colorectal and hepatocellular carcinomas. Oncol Rep. (2010) 23:843–51. doi: 10.3892/or_00000706
11. Zhang X, Zhu T, Chen Y, Mertani HC, Lee KO, Lobie PE. Human growth hormone-regulated HOXA1 is a human mammary epithelial oncogene. J Biol Chem. (2003) 278:7580–90. doi: 10.1074/jbc.M212050200
12. Liu J, Liu J, Lu X. HOXA1 upregulation is associated with poor prognosis and tumor progression in breast cancer. Exp Ther Med. (2019) 17:1896–902. doi: 10.3892/etm.2018.7145
13. Hur H, Lee JY, Yun HJ, Park BW, Kim MH. Analysis of HOX gene expression patterns in human breast cancer. Mol Biotechnol. (2014) 56:64–71. doi: 10.1007/s12033-013-9682-4
14. Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. (2008) 26:3015–24. doi: 10.1200/JCO.2007.15.7164
15. Costa BM, Smith JS, Chen Y, Chen J, Phillips HS, Aldape KD, et al. Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res. (2010) 70:453–62. doi: 10.1158/0008-5472.CAN-09-2189
16. Gaspar N, Marshall L, Perryman L, Bax DA, Little SE, Viana-Pereira M, et al. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Res. (2010) 70:9243–52. doi: 10.1158/0008-5472.CAN-10-1250
17. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. (2017) 77:e108–10. doi: 10.1158/0008-5472.CAN-17-0307
18. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
19. Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. (1978) 276:565–70. doi: 10.1038/276565a0
20. Errico MC, Felicetti F, Bottero L, Mattia G, Boe A, Felli N, et al. The abrogation of the HOXB7/PBX2 complex induces apoptosis in melanoma through the miR-221&222-c-FOS pathway. Int J Cancer. (2013) 133:879–92. doi: 10.1002/ijc.28097
21. Chen KN, Gu ZD, Ke Y, Li JY, Shi XT, Xu GW. Expression of 11 HOX genes is deregulated in esophageal squamous cell carcinoma. Clin Cancer Res. (2005) 11:1044–9. Available online at: https://clincancerres.aacrjournals.org/content/clincanres/11/3/1044.full.pdf
22. Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci USA. (2007) 104:5527–32. doi: 10.1073/pnas.0701059104
23. Musialik E, Bujko M, Kober P, Grygorowicz MA, Libura M, Przestrzelska M, et al. Promoter DNA methylation and expression levels of HOXA4, HOXA5 and MEIS1 in acute myeloid leukemia. Mol Med Rep. (2015) 11:3948–54. doi: 10.3892/mmr.2015.3196
24. Kim YJ, Yoon HY, Kim JS, Kang HW, Min BD, Kim SK, et al. HOXA9, ISL1 and ALDH1A3 methylation patterns as prognostic markers for nonmuscle invasive bladder cancer: array-based DNA methylation and expression profiling. Int J Cancer. (2013) 133:1135–42. doi: 10.1002/ijc.28121
25. Chen SL, Qin ZY, Hu F, Wang Y, Dai YJ, Liang Y. The role of the HOXA gene family in acute myeloid leukemia. Genes. (2019) 10:621. doi: 10.3390/genes10080621
26. Zhang Y, Chen J, Wu SS, Lv MJ, Yu YS, Tang ZH, et al. HOXA10 knockdown inhibits proliferation, induces cell cycle arrest and apoptosis in hepatocellular carcinoma cells through HDAC1. Cancer Manag Res. (2019) 11:7065–76. doi: 10.2147/CMAR.S199239
27. Song C, Han Y, Luo H, Qin Z, Chen Z, Liu Y, et al. HOXA10 induces BCL2 expression, inhibits apoptosis, and promotes cell proliferation in gastric cancer. Cancer Med. (2019) 8:5651–61. doi: 10.1002/cam4.2440
28. Cheng W, Liu J, Yoshida H, Rosen D, Naora H. Lineage infidelity of epithelial ovarian cancers is controlled by HOX genes that specify regional identity in the reproductive tract. Nat Med. (2005) 11:531–7. doi: 10.1038/nm1230
29. Cantile M, Cindolo L, Napodano G, Altieri V, Cillo C. Hyperexpression of locus C genes in the HOX network is strongly associated in vivo with human bladder transitional cell carcinomas. Oncogene. (2003) 22:6462–8. doi: 10.1038/sj.onc.1206808
30. Zhang R, Zhang TT, Zhai GQ, Guo XY, Qin Y, Gan TQ, et al. Evaluation of the HOXA11 level in patients with lung squamous cancer and insights into potential molecular pathways via bioinformatics analysis. World J Surg Oncol. (2018) 16:109. doi: 10.1186/s12957-018-1375-9
31. Cui Y, Gao D, Linghu E, Zhan Q, Chen R, Brock MV, et al. Epigenetic changes and functional study of HOXA11 in human gastric cancer. Epigenomics. (2015) 7:201–13. doi: 10.2217/epi.14.92
32. Se YB, Kim SH, Kim JY, Kim JE, Dho YS, Kim JW, et al. Underexpression of HOXA11 is associated with treatment resistance and poor prognosis in glioblastoma. Cancer Res Treat. (2017) 49:387–98. doi: 10.4143/crt.2016.106
33. Yang X, Deng Y, He RQ, Li XJ, Ma J, Chen G, et al. Upregulation of HOXA11 during the progression of lung adenocarcinoma detected via multiple approaches. Int J Mol Med. (2018) 42:2650–64. doi: 10.3892/ijmm.2018.3826
34. Zhang Y, Yuan Y, Li Y, Zhang P, Chen P, Sun S. An inverse interaction between HOXA11 and HOXA11-AS is associated with cisplatin resistance in lung adenocarcinoma. Epigenetics. (2019) 14:949–60. doi: 10.1080/15592294.2019.1625673
35. He YX, Song XH, Zhao ZY, Zhao H. HOXA13 upregulation in gastric cancer is associated with enhanced cancer cell invasion and epithelial-to-mesenchymal transition. Eur Rev Med Pharmacol Sci. (2017) 21:258–65. Available online at: https://www.europeanreview.org/article/12092
36. Javed S, Langley SE. Importance of HOX genes in normal prostate gland formation, prostate cancer development and its early detection. BJU Int. (2014) 113:535–40. doi: 10.1111/bju.12269
37. Cantile M, Scognamiglio G, La Sala L, La Mantia E, Scaramuzza V, Valentino E, et al. Aberrant expression of posterior HOX genes in well differentiated histotypes of thyroid cancers. Int J Mol Sci. (2013) 14:21727–40. doi: 10.3390/ijms141121727
Keywords: HOXA family, TCGA, prognosis, GSEA, LSCC
Citation: Li J, Ye M and Zhou C (2020) Expression Profile and Prognostic Values of HOXA Family Members in Laryngeal Squamous Cell Cancer. Front. Oncol. 10:368. doi: 10.3389/fonc.2020.00368
Received: 29 November 2019; Accepted: 02 March 2020;
Published: 31 March 2020.
Edited by:
Jorge A. R. Salvador, University of Coimbra, PortugalReviewed by:
Hong-Quan Duong, Hanoi University of Public Health, VietnamMaria Cossu Rocca, European Institute of Oncology (IEO), Italy
Copyright © 2020 Li, Ye and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chongchang Zhou, emhvdTkwMDcwOTkwMDcwOUAxNjMuY29t
 Jinyun Li1
Jinyun Li1 Chongchang Zhou
Chongchang Zhou