- Department of Gastroenterology, Affiliated Hospital of Qingdao University, Qingdao, China
Non-coding RNAs (ncRNAs) are reported to be expressed in human cancers, including pancreatic ductal adenocarcinoma (PDAC). These ncRNAs affect the growth, migration and invasion of tumor cells by regulating cell cycle and apoptosis, as well as playing important roles in epigenetic processes, transcription and post-transcriptional regulation. It is still unclear whether alterations in ncRNAs influence PDAC development and progression. Because of this, analysis based on existing data on ncRNAs, which are crucial for modulating pancreatic tumorigenesis, will be important for future research on PDAC. Here, we summarize ncRNAs with tumor-promoting functions: HOTAIR, HOTTIP, MALAT1, lncRNA H19, lncRNA PVT1, circ-RNA ciRS-7, circ-0030235, circ-RNA_100782, circ-LDLRAD3, circ-0007534, circRHOT1, circZMYM2, circ-IARS, circ-RNA PDE8A, miR-21, miR-155, miR-221/222, miR-196b, miR-10a. While others including GAS5, MEG3, and lncRNA ENST00000480739, has_circ_0001649, miR-34a, miR-100, miR-217, miR-143 inhibit the proliferation and invasion of PDAC. Hence, we summarize the functions of ncRNAs in the occurrence, development and metastasis of PDAC, with the goal to provide guidance in the clinical diagnosis and treatment of PDAC.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a malignant tumor with a high incidence, malignancy and mortality rate. PDAC is the seventh leading cause of cancer-related death throughout the world (1). According to 2019 statistics from the American Cancer Society, PDAC mortality rate ranks 4th in both men and women (2). Due to a lack of effective treatments, the 5-years survival rate for PDAC remains below 8% (3). This high mortality rate is largely due to late presentation and detection of the disease, when patients become non-candidates for surgical resection. In addition, the mechanisms behind PDAC tumorigenesis and progression are still unclear.
Mutations in KRAS, TP53, SMAD4, CDKN2A commonly contribute to PDAC progression (4–7). In addition, PDAC development requires the involvement of various signal transduction pathways including the Hippo, Hedgehog, Wnt/Notch, JNK, PI3K, K-ras, and transforming growth factor (TGF) -β signaling pathways. Moreover, genome-wide association studies (GWAS) have identified a large number of pathways and gene sets involved in the development of PDAC (8, 9).
Non-coding RNAs (ncRNAs) have widely been identified in mammals as unique RNA transcripts. Nc-RNAs are classified as small RNAs (<200 bp) and long RNAs (>200 bp) based on nucleotide length, and include microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), small nucleolar RNAs (snoRNAs), tRNA-derived stress-induced RNAs (tiRNAs), enhancer non-coding RNAs (eRNAs), circular RNAs (circRNAs), and long non-coding RNAs (lncRNAs) (10, 11). In addition, ncRNAs are also categorized based on their localization into cytoplasmic and nuclear ncRNAs. Even though ncRNAs are not translated into proteins, they are critical for DNA replication, translation, RNA splicing and epigenetic regulation. NcRNAs also participate in the cellular processes including differentiation, proliferation, apoptosis and metabolism. Subsequent studies have shown that ncRNAs play a vital role as either oncogenes or tumor suppressors in tumorigenesis. Herein, we summarize the roles and functions of ncRNAs in the diagnosis and treatment of PDAC.
LncRNAs
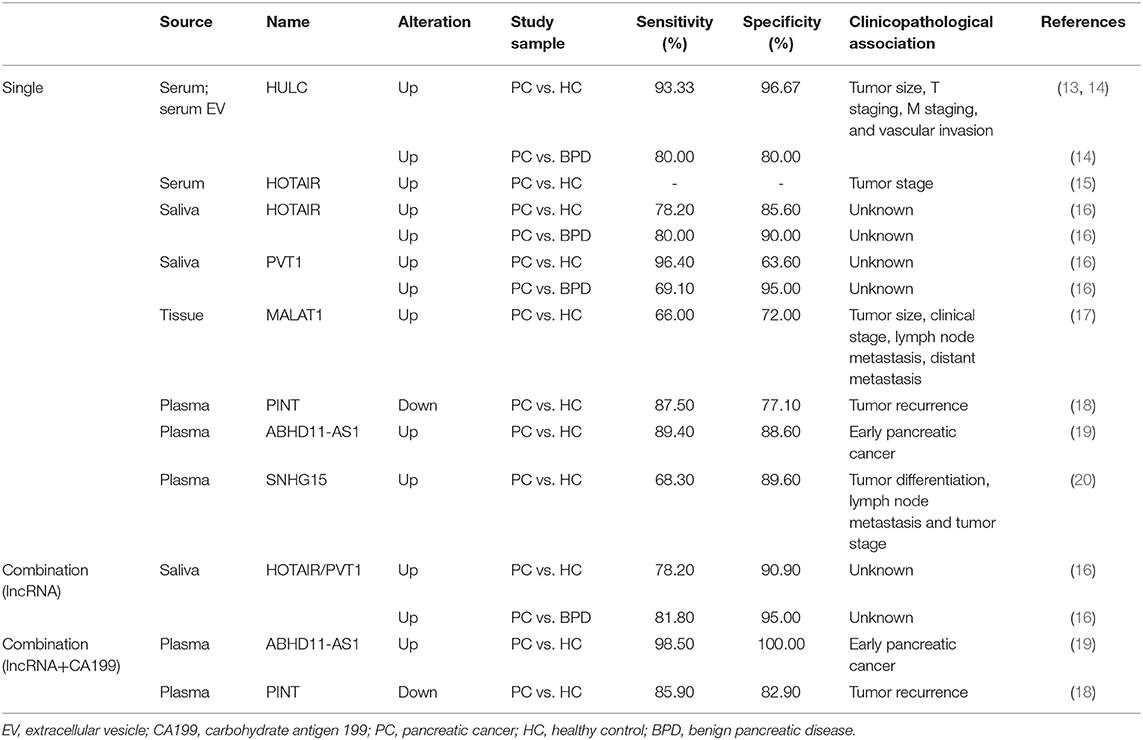
LncRNAs, which contain a length of more than 200 nucleotides, are transcribed by RNA polymerase II and contain a 5′ cap and 3′ poly A tail (12). These ncRNAs are widely distributed throughout the genome but have zero protein-coding capacity. They are involved in many biological processes, including transcriptional regulation in cis or trans, chromatin remolding, nuclear transport, genomic imprinting and oncogenic progression. Most lncRNAs are expressed in specific different tumor types, making them potential targets for cancer diagnosis and treatment. LncRNAs act as candidate diagnostic biomarkers for PDAC as summarized in Table 1. The functions and regulatory mechanisms of lnRNAs and other ncRNAs are depicted in Figure 1. Therefore, lncRNAs are of interest in the exploration of novel diagnostic and therapeutic approaches.
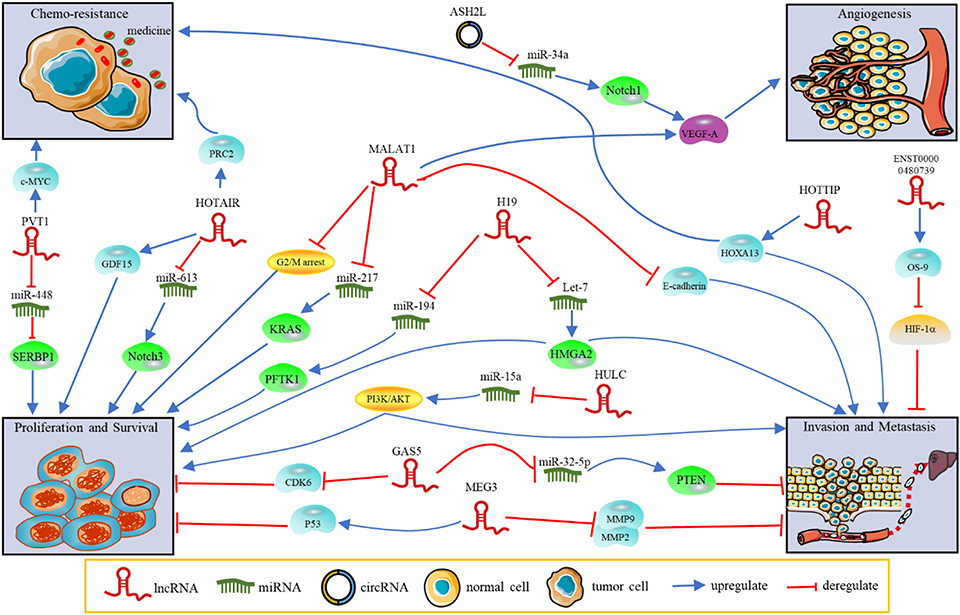
Figure 1. The functions and regulatory mechanisms of ncRNAs in PDAC. NcRNAs regulate tumor progression, such as proliferation, invasion, metastasis, angiogenesis and chemoresistance.
LncRNAs as Potential Oncogenes and Biomarkers in PDAC
MALAT1
Metastasis-associated lung adenocarcinoma transcript-1 (MALAT1, also known as NEAT2) was initially discovered in lung cancer and has been subsequently detected to be overexpressed in multiple tumors as a negative prognosis factor. MALAT1 is highly expressed in PDAC tissues and positively correlates with tumor size, clinical stage, lymph node metastasis, distant metastasis and prognosis (21, 22). Its expression is also up-regulated in cancer stem cells (CSCs), which is closely related to drug resistance. In addition, it can interact with RNA-binding protein human antigen R (HuR) to regulate T-cell intracellular antigen-1 (TIA-1) mediated autophagic activation at the post-transcriptional level. Furthermore, it can also regulate KRAS expression through competitive inhibition to promote PDAC cell proliferation (21, 23). Recently, human enhancer of zeste homology 2 (EZH2) was shown to be recruited to the E-cadherin promoter through MALAT1, which repressed the expression of E-cadherin and facilitated the invasion and metastasis of PDAC cells (24). Knockout of MALAT1 induced G2/M cell cycle arrest, inhibition of epithelial-mesenchymal transition (EMT), decreased cancer stem cell-like properties, repressed N-myc downregulated gene-1 (NORG-1) and hindered the growth and invasion of cancer cells (25, 26). Other work has used different databases to identify the top three key target genes of MALAT1, which include CCND1, RAF-mitogen-activated kinase 8 (MAPK8) and VEGFA (17). This suggests that these may participate in the mTOR signaling pathway, pathways in cancer, and the MAPK signaling pathway in PDAC. Diminished expression of MALAT1 decreases the expression of Yes-associated protein 1 (YAP1) and elevates large tumor suppressor 1 (LATS1) levels (27). It has been suggested that MALAT1 regulates PDAC via the Hippo-YAP pathway. It also has been reported to regulate multiple signaling pathway, including phosphoinositide-3-kinase-AKT (PI3K-AKT), NF-κb, mTOR, MAPK and WNT pathways in multiple cancer types. Thus, the complex mechanisms and roles that MALAT1 plays in PDAC are worth further exploration.
HOTAIR
HOX antisense transcript intergenic RNA (HOTAIR) transcribed from the HOXC locus. Its overexpression has been linked to the poor prognosis of different cancers, including breast, gastric, colorectal, bladder and esophageal squamous cell carcinoma (28–30). Increased expression of HOTAIR has been observed in PDAC tissues and is negatively correlated with overall survival. HOTAIR inhibits the expression of cell cycle interferon related genes, targets and binds to the tumor suppressor gene GDF15, and accelerates the proliferation of pancreatic cancer cells. Furthermore, knockout of HOTAIR in the pancreatic cancer cell lines Panc-1 and L3.6Pl significantly decreases the progression of cells and interacts with the Polycomb Repressive Complex 2 (PRC2) (28, 31). HOTAIR promotes the proliferation of pancreatic cancer cells by acting as a competing endogenous RNA via sponging miR-613 to regulate the expression of NOTCH3 (32). Overexpressing HOTAIR regulates the trimethylation of histone H3 at lysine 27 to inhibit the expression of TRAIL receptor death receptor 5 (DR5) through EZH2. HOTAIR overexpression also improves the resistance of pancreatic cancer cells to TRAIL induced apoptosis (33). In addition, knockout of HOTAIR can enhance the radio-sensitivity of PDAC cells by increasing the expression of Wnt inhibitory factor 1 (WIF-1) (34). The HOTAIR-WIF-1 axis can be used as a potential target for PDAC radiotherapy, which needs to be further evaluated. The salivary HOTAIR of pancreatic cancer patients was significantly higher than expression levels observed in healthy individuals. The expression of HOTAIR in patients' saliva was significantly reduced after the PDAC tumor was surgically removed (16). This indicates that HOTAIR can be evaluated in patients undergoing resection and that it may be a promising novel diagnostic marker and therapeutic target.
HOTTIP
The lincRNA HOXA distal transcript antisense RNA (HOTTIP) is another HOX-related lncRNA. Studies have found that the expression levels of HOTTIP are significantly increased in multiple PDAC cell lines and PDAC specimens (35). HOTTIP interacts with the WD repeat containing protein 5 (WDR5)/mixed lineage leukemia (MLL) complex to enhance the methylation of histone 3 on lysine 4 (H3K4) in order to modulate the proliferation and differentiation of PDAC cells (36, 37). Up-regulation of HOTTIP promotes the secretion of IL-6 and expression of PD-L1 in neutrophils, thereby inhibiting the activity of T cells and promoting the immune escape of ovarian cancer cells (38). This may be used for reference in immunotherapy of PDAC. Decreased expression of HOTTIP in pancreatic cancer cells leads to increased G0/G1 phase cells, decreased Vimentin and Snai1 expression, and increased E-cadherin expression. Furthermore, HOTTIP knockout can reduce the expression level of HOXA13 and enhance the sensitivity of human pancreatic cancer cells to gemcitabine (35). In turn, others have shown that HOTTIP is not involved in the regulation of HOXA13, but regulates several other HOX genes (39). Further studies will be required to fully understand these relationships.
PVT1
Plasmacytoma variant translocation 1(PVT1) was the first lncRNA gene identified in human Burkitt's lymphoma as a recurrent breakpoint. PVT1 and MYC have correlated one another and co-amplified. It was confirmed in a variety of solid tumors, including colon and breast cancers, that increased expression of PVT1 could increase MYC protein (40). A GWAS study identified a risk locus at 8q24.21, which interacts with MYC promoters, that reached genome-wide significance located PVT1 (41). It was also identified that in human PDAC cells, PVT1 acts as an oncogene promoting EMT via TGF-β/Smad signaling (42). PVT1 also acts as a sponge for miRNAs to regulate the development of PDAC. PVT1 could promote the proliferation and metastasis of PDAC cells by acting as a miR-448 sponge to inhibit SERBP1 (43). It has also been reported that PVT1 acts as a sponge to modulate cytoprotective autophagy and promote the development of PDAC via the PVT1/miR-20a-5p/ULK1/autophagy pathway (44). Of note, PVT1 also participates in drug resistance. Research indicates that PVT1 regulates gemcitabine chemosensitivity in PDAC through miR1207 (45). Curcumin can inhibit the PRC2-PVT1-c-Myc axis by inhibiting the PRC2 subunit Enhancer of EZH2 to enhance the sensitivity of PDAC cells to gemcitabine (46). PVT1, along with MALAT1 and HOTTIP, could act as a prospective biomarker to predict the efficacy of gemcitabine in PDAC patients (47), and as such, future assessment is warranted.
H19
LncRNA H19 is a maternally imprinted gene that is highly expressed in PDAC tissues and is involved in tumor progression. It increases high-mobility group AT-hook 2 (HMGA2) mediated EMT by antagonizing let-7 and promotes both tumor cell metastasis and invasion (48, 49). MiR-675 reduces the activation of H19 by binding to the 3′ untranslated region (UTR) on E2F-1 mRNA and altering the expression of E2F-1 protein (50). In addition, the Wnt-signaling pathway is involved in regulating PDAC cell proliferation and migration via the H19/miR-194/PFFTK1 axis (51). H19 is also believed to play a role in cancer therapy and is the earliest lncRNA used in the treatment of PDAC. BC-819 (DTA-H19) carries a diphtheria toxin-A chain (DTA), which can be applied to the treatment of tumors expressing high levels of H19 (52). DTA-H19 combined with gemcitabine can reduce the tumor size and delay the progression of PDAC in vivo. Altogether, these data suggest that H19 is a promising therapeutic marker for PDAC.
HULC
Highly up-regulated in liver cancer (HULC) is another lncRNA that modulates the proliferation of PDAC. HULC can regulate the viability, proliferation, migration and invasion of PDAC cells. Up-regulation of HULC activates the PI3K/AKT pathway via negative regulation of miR-15a expression (53). HULC levels are significantly increased in PDAC compared to the non-tumor tissues. Higher expression of HULC in PDAC is correlated with poor clinical outcomes in patients. It is thus suggested that HULC is a promising prognostic biomarker candidate (54). Recently, a report suggested that serum extracellular vesicle (EV) HULC expression is increased in PDAC patients in comparison to intraductal papillary mucinous patients and healthy individuals (13). Thus, HULC may be a new potential diagnosis maker for PDAC and may merit further investigation.
LncRNAs as Potential Suppressors and Biomarkers in PDAC
GAS5
Growth arrest-specific transcript 5 (GAS5) was originally identified by screening potential tumor suppressor genes expressed at high levels during growth using a functional cDNA library (55). GAS5 is one of the few lncRNAs that are negatively correlated with tumor development in breast cancer, malignant pleural mesothelioma and hepatocellular carcinoma (56–58). GAS5 negatively regulates miR-32-5p to promote the expression of pleiotrophin (PTEN), which can block the activation of the PI3K/Akt signaling pathway, inhibiting the proliferation and survival of PDAC cells (59). Studies have found that GAS5 overexpression can significantly inhibit both the proliferation and invasion of PANC-l and BxPC-3 cells in vitro (60). After inhibiting of GAS5 expression by RNA interference, a larger number of cells were found to be arrested in the S phase of the cell cycle. This suggested that GAS5 regulates the cell cycle of PDAC. Furthermore, GAS5 regulates the cell cycle of PDAC cells by inhibiting cyclin-dependent kinase 6 (CDK6) and blocking proliferation and differentiation. Studies have shown that GAS5 inhibits drug resistance in PDAC by negative regulation of miR-181c-5p and reducing the inactivation of the Hippo signal transduction pathway (61). Overexpression of GAS5 inhibits PDAC cell proliferation, migration and gemcitabine resistance through miR-221/suppressor of cytokine signaling 3 (SOCS3) mediated EMT and tumor CSCs (62). Overall, GAS5 could be as a novel target for PDAC drug resistance therapy.
LncRNA ENST00000480739
The lncRNA ENST00000480739 is a relatively rare tumor suppressor that was recently uncovered. ENST00000480739 expression in pancreatic tumor specimens is significantly lower than that which is observed in adjacent non-tumor tissues (63). It is also negatively correlated to tumor stage and could be used as an independent prognostic factor in PDAC patients who underwent surgery. ENST00000480739 inhibits tumor invasion through the regulation of osteosarcoma amplified-9 (OS-9), modulates hypoxia-inducible factor-1α (HIF-1α) and inhibits EMT (63). It not only has the potential to inhibit metastasis but can also be used as a biomarker for both risk prediction and treatment screening in PDAC.
MEG3
Maternally expressed gene 3 (MEG3) acts as a tumor suppressor and shows to be down-regulated in several tumors, such as hepatocellular, prostate, gastric and lung cancers (64–67). Hu et al. (68) found that MEG3 inhibits the proliferation, induces apoptosis via p53 activation and is upregulated along with p53 by fenofibrate to restrain the proliferation of PDAC cells. Other studies found that the MEG3 expression levels in human pancreatic cancer tissues are lower than corresponding non-cancerous tissues (69). These were also found to be negatively correlated to patients' clinicopathological features. In vitro studies have shown that MEG3 plays an anticancer role in the regulation of cell proliferation, migration, invasion, induction of EMT, and cancer stem cell (CSC) properties. Furthermore, study has shown that MEG3 overexpression plays an anticancer role through the in vitro modulation of the PI3K/AKT/ B cell lymphoma-2 (Bcl-2)/Bax/Cyclin D1/P53 and P13K/AKT/matrix metalloproteinases-2(MMP-2) /MMP-9 signaling pathways (70).
Numerous lncRNAs participate in pancreatic cancer tumorigenesis. Further, many of the PDAC susceptibility loci that were previously identified in GWAS are located in lncRNAs such as 7q32.3 (LINC-PINT) and 17q25.1 (LINC00673) (41, 71). Interestingly, LINC-PINT, through the TGF -β pathway, inhibits PDAC growth in early stages (72). This may be a potential target for early treatment for patients but it requires further testing to prove. Additionally, LINC00673 is also correlated with good outcomes in PDAC patients. It can negatively regulate miR-504 to inhibit the progression of PDAC (73). There are many potential lncRNAs biomarkers that remain to be explored and translated to clinical practices.
CircRNAs
Circular RNAs (circRNAs) are a new type of endogenous ncRNA that used to be considered as miRNA sponges. CircRNAs are stable since they lack 5′ cap or 3′ Poly A tail terminal ends that block traditional RNA degradation pathways, existing as a closed loop structure. Heterogeneous circRNAs may contribute to the development of many different tumors.
CircRNAs in PDAC
Numerous studies have demonstrated that circRNAs are aberrantly expressed in PDAC. One study identified circRNAs in six pairs of PDAC and para-cancerous tissues using a microarray (74). Additional microarray data revealed that there were 115 upregulated and 141 downregulated circRNAs in PDACs (75). Another study uncovered that 453 circRNAs were differentially expressed and were significantly different in extracellular vesicles isolated from the plasma of 8 PDAC patients or healthy controls (76). It has been suggested that aberrant expression of circRNAs in PDAC are related to proliferation and development. These aberrantly expressed circRNAs may be involved in the regulation of PDAC and are expected to be diagnostic markers, though this remains to be tested.
CircRNAs Regulate PDAC Progression
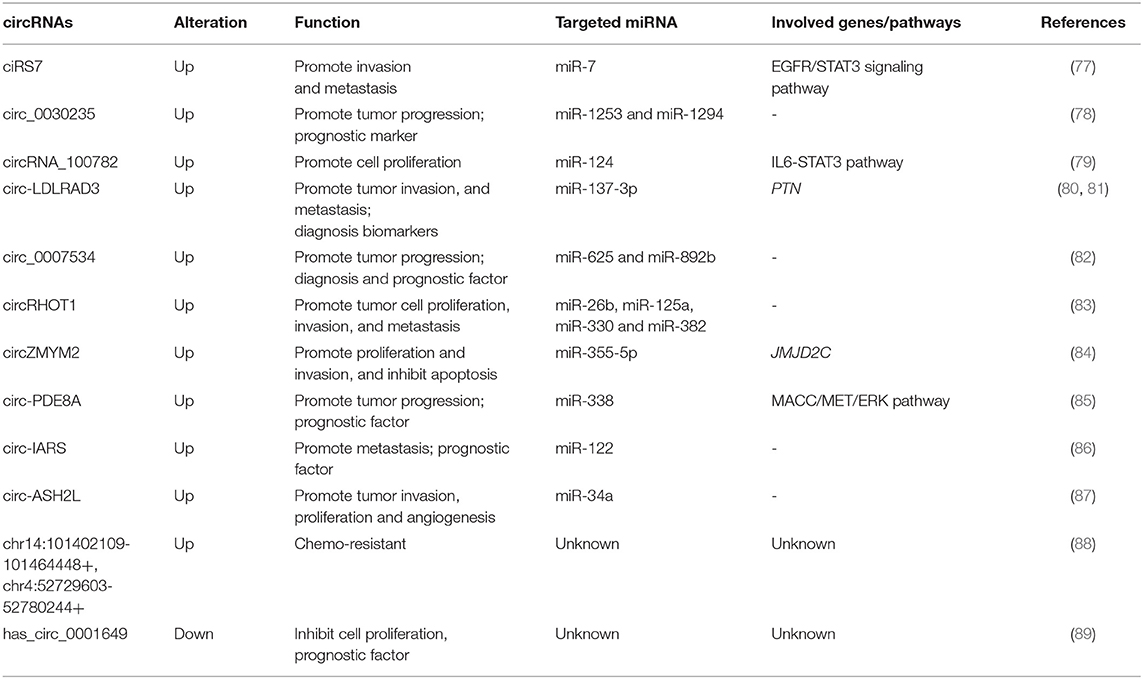
CircRNAs can regulate a variety of different pathways (Table 2). The circRNA ciRS7 is expressed in PDAC tissues compared to para-cancerous tissues and can negatively regulate miR-7, a cancer suppressor. It can also affect the proliferation and invasion of PDAC cells through epidermal growth factor receptor (EGFR), as well as signal transducer and activator of transcription 3 (STAT3) signaling pathways (77). In addition, overexpression of ciRS7 promotes lymph node metastasis and venous invasion in PDAC cases. Down-regulation of circ-LDLRAD3 can inhibit PDAC cell proliferation and metastasis through the up-regulation of miR-137-3p/ PTEN (80). Others have proven that the circRHOT1 regulates the PDAC cells proliferation, invasion and metastasis by binding to miRNAs, including miR-26b, miR-125a, miR-330, and miR-382 (83). There is mounting evidence underlining that circRNAs act as miRNA sponges that regulate the progression of PDAC. Chen et al. (79) elucidated that silencing of circRNA_100782 down-regulates the expression levels of interleukin-6 receptor (IL6R) and STAT3 by acting as a miR-124 sponge that inhibits BxPC3 cell proliferation. Another study has shown that circ_0030235 promotes cell proliferation by directly sponging miR-1253 and miR-1294 in vitro (78). An additional study also revealed that circ_0007534 functions as a sponge for miR-625 and miR-892b to facilitate the malignant behavior of PDAC cells (82). Studies have identified that circZMYM2 (hsa_circ_0099999) is up-regulated in both PDAC cells and tissues, promoting tumor progression by influencing JMJD2C expression levels via acting as miR-355-5p sponges (84).
CircRNAs as Diagnostic, Prognostic and Therapeutic Biomarkers for PDAC
CircRNAs have the ability to be strong biomarkers for PDAC since they exhibit stable expression and have high serum concentrations. Yang et al. (81) investigated that circ-LDLRAD3 up-regulation in PDAC tissues, plasmas and PDAC cell lines, and identified its association with venous and lymphatic invasion and metastasis. Furthermore, this study confirmed that circ-LDLRAD3 holds potential to be a new diagnosis biomarker, as it has higher sensitivity and specificity when combined with CA19-9 than using CA19-9 alone (81). Clinical studies have shown that the overexpression of circ_0030235, circ_0007534 in PDAC are associated with tumor stage, lymph node invasion and poor overall survival (78, 82). Exosomes play an important role in the development and prognosis of tumors. Owing to their stability, they can be detected in blood plasma and equipped higher level than existing checks. Studies have identified that exosomal circ-PDE8A promotes the progression of PDAC by binding to miR-338 to activate the MACC/MET/ERK pathway (85). The same group also found that high expression levels of circ-IARS in plasma exosomes positively correlated with tumor metastasis including vessel invasion, liver metastasis and tumor-node-metastasis (TNM) stage. circ-IARS is also negatively associated with survival of PDAC patients (86). Shao et al. (88) developed Gemcitabine resistant cell lines (PANC-1-GR) and verified that the expression levels of two circRNAs (chr14:101402109-101464448+, chr4:52729603-52780244+) were significantly correlated to drug resistance observed in PANC-1-GR, as well as the plasma of gemcitabine non-responsive PDAC patients. CircRNAs can be used not only as biomarkers, but also to provide information about tumor stage and classification. Conversely, expression levels of has_circ_0001649 were decreased in PDAC tissues and cells when compared to normal control, and were associated with tumor stage and differentiation grade (89). Has_circ_0001649 may be regarded as a novel prognostic biomarker for PDAC patients who had undergone surgery.
Currently, the roles of circRNAs in tumor progression and clinical application have gained attention. However, the studies of circRNAs in PDAC are still at infancy stage. Further studies on circRNAs in PDAC should not only be based on the databases established, but also require a great deal of work to fully understand their functions.
MiRNAs
MicroRNAs (miRNAs) are small non-coding RNAs ~19–25 nucleotides in length, that regulate gene expression at the post transcriptional level through RNA interference (90). MiRNAs play a significant role in the initiation and progression of a tumor by regulating tumor growth, anti-apoptotic, metastasis and invasion (91, 92). These have potential as cancer diagnosis markers, prognosis predictors, and for the monitoring of therapy.
MiRNAs as Diagnosis and Prognostic Biomarkers for PDAC
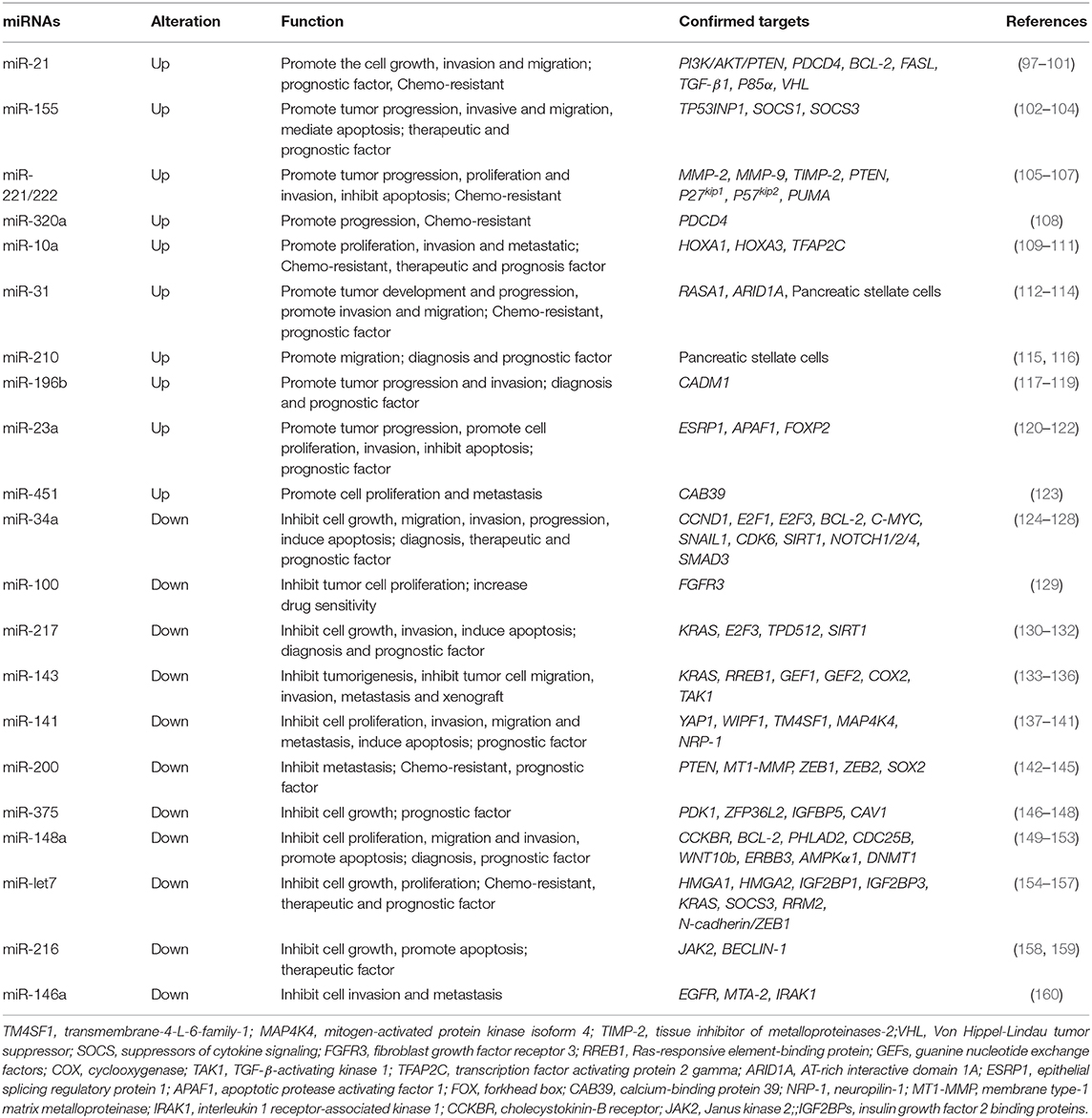
MiRNAs are conservative, generalized, testable and keep stable when existing outside cells. Ongoing research has revealed that miRNAs exhibit fair sensitivity and specificity as diagnosis biomarkers. The potential for miRNAs as diagnostic and therapeutic markers leads to the analyses of deregulated miRNAs in PDAC cases (93). Abnormal expression levels of miRNAs in PDAC tissues, blood and saliva have shown to be closely related to the initiation and development of PDAC (94, 95). Studies have found that plasma miRNAs (miR-16, miR-196a) combined with serum CA19-9 can increase the sensitivity and specificity for diagnosis in comparison with using CA19-9 alone (96). MiRNAs can also be identified in the saliva and act as diagnosis biomarkers for unresectable pancreatic tumors (95). This can lead to the creation of specific miRNA profiles for diagnosis and treatment. In addition, published studies have demonstrated that miRNAs can interact with lncRNAs and circRNAs (Figure 1). They can also regulate key signaling molecules and pathways in disease development and progression (Tables 2, 3). At the same time, it is challenging to fully understand the interaction network between different miRNAs and other ncRNAs. Further differentiation of the roles that miRNAs play in the development and progression of PDAC are necessary to explore their specificity and sensitivity as biomarkers for PDAC.
Conclusion
With the rocket development of next-generation sequencing and bioinformatic analyses, ncRNAs and their prominent roles in oncogenesis, specifically the progression of PDAC reveal enormous potential for ncRNAs in the diagnosis and treatment of cancers. Although ncRNAs have gradually become a research hotspot, the limitations in detection approaches and inclusion in larger databases make their roles in PDAC difficult to fully understand. Understanding the relationship between the function and mechanism of ncRNAs in PDAC will help classify ncRNAs and their roles in the clinic. A great deal of work remains to be completed to uncover the complex mechanisms of ncRNAs, which lead to tumorigenesis and progression, to ultimately select the most effective diagnostic and therapeutic targets.
Author Contributions
RG and YJ designed the study. RG collected data and wrote the manuscript. YJ revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. (2019) 69:7–34. doi: 10.3322/caac.21551
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. (2016) 66:7–30. doi: 10.3322/caac.21332
4. Bailey JM, Hendley AM, Lafaro KJ, Pruski MA, Jones NC, Alsina J, et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene. (2016) 35:4282–8. doi: 10.1038/onc.2015.441
5. Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. (2008) 321:1801–6. doi: 10.1126/science.1164368
6. Roberts NJ, Norris AL, Petersen GM, Bondy ML, Brand R, Gallinger S, et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov. (2016) 6:166–75. doi: 10.1158/2159-8290.CD-15-0402
7. Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. (2015) 518:495–501. doi: 10.1038/nature14169
8. Walsh N, Zhang H, Hyland PL, Yang Q, Mocci E, Zhang M, et al. Agnostic pathway/gene set analysis of genome-wide association data identifies associations for pancreatic cancer. J Natl Cancer Inst. (2019) 111:557–567. doi: 10.1093/jnci/djy155
9. Klein AP, Wolpin BM, Risch HA, Stolzenberg-Solomon RZ, Mocci E, Zhang M, et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nature communications. (2018) 9:556. doi: 10.1038/s41467-018-02942-5
10. Adams BD, Parsons C, Walker L, Zhang WC, Slack F J. Targeting noncoding RNAs in disease. J Clin Invest. (2017) 127:761–71. doi: 10.1172/JCI84424
11. Rigoutsos I, Lee SK, Nam SY, Anfossi S, Pasculli B, Pichler M, et al. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol. (2017) 18:98. doi: 10.1186/s13059-017-1224-0
12. Batista PJ, Chang H Y. Long non-coding RNAs: cellular address codes in development and disease. Cell. (2013) 152:1298–307. doi: 10.1016/j.cell.2013.02.012
13. Takahashi K, Ota Y, Kogure T, Suzuki Y, Iwamoto H, Yamakita K, et al. Circulating extracellular vesicle-encapsulated HULC is a potential biomarker for human pancreatic cancer. Cancer Sci. (2020) 111:98–111. doi: 10.1111/cas.14232
14. Ou ZL, Luo Z, Lu YB. Long non-coding RNA HULC as a diagnostic and prognostic marker of pancreatic cancer. World J Gastroenterol. (2019) 25:6728–42. doi: 10.3748/wjg.v25.i46.6728
15. Ma Y, Hu M, Zhou L, Ling S, Li Y, Kong B, et al. Long non-coding RNA HOTAIR promotes cancer cell energy metabolism in pancreatic adenocarcinoma by upregulating hexokinase-2. Oncol Lett. (2019) 18:2212–9.
16. Xie Z, Chen X, Li J, Guo Y, Li H, Pan X, et al. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget. (2016) 7:25408–19. doi: 10.18632/oncotarget.8323
17. Xie ZC, Dang YW, Wei DM, Chen P, Tang RX, Huang Q, et al. Clinical significance and prospective molecular mechanism of MALAT1 in pancreatic cancer exploration: a comprehensive study based on the GeneChip, GEO, Oncomine, and TCGA databases. Onco Targets Ther. (2017) 10:3991–4005. doi: 10.2147/OTT.S136878
18. Li L, Zhang G-Q, Chen H, Zhao Z-J, Chen H-Z, Liu H, et al. Plasma and tumor levels of Linc-pint are diagnostic and prognostic biomarkers for pancreatic cancer. Oncotarget. (2016) 7:71773–81. doi: 10.18632/oncotarget.12365
19. Liu Y, Feng W, Liu W, Kong X, Li L, He J, et al. Circulating lncRNA ABHD11-AS1 serves as a biomarker for early pancreatic cancer diagnosis. J Cancer. (2019) 10:3746–56. doi: 10.7150/jca.32052
20. Guo XB, Yin HS, Wang JY. Evaluating the diagnostic and prognostic value of long non-coding RNA SNHG15 in pancreatic ductal adenocarcinoma. Eur Rev Med Pharmacol Sci. (2018) 22:5892–8. doi: 10.26355/eurrev_201809_15917
21. Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH, et al. Long non-coding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther. (2016) 15:2232–43. doi: 10.1158/1535-7163.MCT-16-0008
22. Pang EJ, Yang R, Fu XB, Liu Y F. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. (2015) 36:2403–7. doi: 10.1007/s13277-014-2850-8
23. Liu P, Yang H, Zhang J, Peng X, Lu Z, Tong W, et al. The lncRNA MALAT1 acts as a competing endogenous RNA to regulate KRAS expression by sponging miR-217 in pancreatic ductal adenocarcinoma. Sci Rep. (2017) 7:5186. doi: 10.1038/s41598-017-05274-4
24. Han T, Jiao F, Hu H, Yuan C, Wang L, Jin ZL, et al. EZH2 promotes cell migration and invasion but not alters cell proliferation by suppressing E-cadherin, partly through association with MALAT-1 in pancreatic cancer. Oncotarget. (2016) 7:11194–207. doi: 10.18632/oncotarget.7156
25. Jiao F, Hu H, Han T, Yuan C, Wang L, Jin Z, et al. Long non-coding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int J Mol Sci. (2015) 16:6677–93. doi: 10.3390/ijms16046677
26. Cheng Y, Imanirad P, Jutooru I, Hedrick E, Jin UH, Rodrigues Hoffman A, et al. Role of metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1) in pancreatic cancer. PLoS ONE. (2018) 13:e0192264. doi: 10.1371/journal.pone.0192264
27. Zhou Y, Shan T, Ding W, Hua Z, Shen Y, Lu Z, et al. Study on mechanism about long noncoding RNA MALAT1 affecting pancreatic cancer by regulating Hippo-YAP signaling. J Cell Physiol. (2018) 233:5805–14. doi: 10.1002/jcp.26357
28. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. (2010) 464:1071–6. doi: 10.1038/nature08975
29. Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY, Xue WQ, et al. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. (2013) 104:1675–82. doi: 10.1111/cas.12296
30. Hajjari M, Behmanesh M, Sadeghizadeh M and Zeinoddini M. Up-regulation of HOTAIR long non-coding RNA in human gastric adenocarcinoma tissues. Med Oncol. (2013) 30:670. doi: 10.1007/s12032-013-0670-0
31. Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. (2013) 32:1616–25. doi: 10.1038/onc.2012.193
32. Cai H, Yao J, An Y, Chen X, Chen W, Wu D, et al. LncRNA HOTAIR acts a competing endogenous RNA to control the expression of notch3 via sponging miR-613 in pancreatic cancer. Oncotarget. (2017) 8:32905–17. doi: 10.18632/oncotarget.16462
33. Yang SZ, Xu F, Zhou T, Zhao X, McDonald JM, Chen Y. The long non-coding RNA HOTAIR enhances pancreatic cancer resistance to TNF-related apoptosis-inducing ligand. J Biol Chem. (2017) 292:10390–7. doi: 10.1074/jbc.M117.786830
34. Jiang Y, Li Z, Zheng S, Chen H, Zhao X, Gao W, et al. The long non-coding RNA HOTAIR affects the radiosensitivity of pancreatic ductal adenocarcinoma by regulating the expression of Wnt inhibitory factor 1. Tumour Biol. (2016) 37:3957–67. doi: 10.1007/s13277-015-4234-0
35. Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H, et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med. (2015) 13:84. doi: 10.1186/s12967-015-0442-z
36. Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long non-coding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. (2011) 472:120–4. doi: 10.1038/nature09819
37. Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao X, et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. (2017) 410:68–81. doi: 10.1016/j.canlet.2017.09.019
38. Shang A, Wang W, Gu C, Chen C, Zeng B, Yang Y, et al. Long non-coding RNA HOTTIP enhances IL-6 expression to potentiate immune escape of ovarian cancer cells by upregulating the expression of PD-L1 in neutrophils. J Exp Clin Cancer Res. (2019) 38:411. doi: 10.1186/s13046-019-1394-6
39. Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget. (2015) 6:10840–52. doi: 10.18632/oncotarget.3450
40. Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. (2014) 512:82–6. doi: 10.1038/nature13311
41. Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet. (2014) 46:994–1000. doi: 10.1038/ng.3052
42. Zhang X, Feng W, Zhang J, Ge L, Zhang Y, Jiang X, et al. Long non-coding RNA PVT1 promotes epithelialmesenchymal transition via the TGFbeta/Smad pathway in pancreatic cancer cells. Oncol Rep. (2018) 40:1093–102. doi: 10.3892/or.2018.6462
43. Zhao L, Kong H, Sun H, Chen Z, Chen B, Zhou M. LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J Cell Physiol. (2018) 233:4044–55. doi: 10.1002/jcp.26072
44. Huang F, Chen W, Peng J, Li Y, Zhuang Y, Zhu Z, et al. LncRNA PVT1 triggers Cyto-protective autophagy and promotes pancreatic ductal adenocarcinoma development via the miR-20a-5p/ULK1 Axis. Mol Cancer. (2018) 17:98. doi: 10.1186/s12943-018-0845-6
45. You L, Wang H, Yang G, Zhao F, Zhang J, Liu Z, et al. Gemcitabine exhibits a suppressive effect on pancreatic cancer cell growth by regulating processing of PVT1 to miR1207. Mol Oncol. (2018) 12:2147–64. doi: 10.1002/1878-0261.12393
46. Yoshida K, Toden S, Ravindranathan P, Han H and Goel A. Curcumin sensitizes pancreatic cancer cells to gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1 expression. Carcinogenesis. (2017) 38:1036–46. doi: 10.1093/carcin/bgx065
47. Wang CJ, Shi SB, Tian J, Xu J, Niu ZX. lncRNA MALAT1, HOTTIP, and PVT1 as predictors for predicting the efficacy of GEM based chemotherapy in first-line treatment of pancreatic cancer patients. Oncotarget. (2017) 8:95108–15. doi: 10.18632/oncotarget.19345
48. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. (2013) 52:101–12. doi: 10.1016/j.molcel.2013.08.027
49. Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, et al. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biol. (2014) 35:9163–9. doi: 10.1007/s13277-014-2185-5
50. Ma L, Tian X, Guo H, Zhang Z, Du C, Wang F, et al. Long non-coding RNA H19 derived miR-675 regulates cell proliferation by down-regulating E2F-1 in human pancreatic ductal adenocarcinoma. J Cancer. (2018) 9:389–99. doi: 10.7150/jca.21347
51. Sun Y, Zhu Q, Yang W, Shan Y, Yu Z, Zhang Q, et al. LncRNA H19/miR-194/PFTK1 axis modulates the cell proliferation and migration of pancreatic cancer. J Cell Biochem. (2019) 120:3874–86. doi: 10.1002/jcb.27669
52. Hanna N, Ohana P, Konikoff FM, Leichtmann G, Hubert A, Appelbaum L, et al. Phase 1/2a, dose-escalation, safety, pharmacokinetic and preliminary efficacy study of intratumoral administration of BC-819 in patients with unresectable pancreatic cancer. Cancer Gene Ther. (2012) 19:374–81. doi: 10.1038/cgt.2012.10
53. Feng H, Wei B, Zhang Y. Long non-coding RNA HULC promotes proliferation, migration and invasion of pancreatic cancer cells by down-regulating microRNA-15a. Int J Biol Macromol. (2019) 126:891–8. doi: 10.1016/j.ijbiomac.2018.12.238
54. Peng W, Gao W, Feng J. Long non-coding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. (2014) 31:346. doi: 10.1007/s12032-014-0346-4
55. Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. (1988) 54:787–93. doi: 10.1016/S0092-8674(88)91065-3
56. Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. (2009) 28:195–208. doi: 10.1038/onc.2008.373
57. Renganathan A, Kresoja-Rakic J, Echeverry N, Ziltener G, Vrugt B, Opitz I, et al. GAS5 long non-coding RNA in malignant pleural mesothelioma. Mol Cancer. (2014) 13:119. doi: 10.1186/1476-4598-13-119
58. Yang L and Jiang J. GAS5 Regulates RECK expression and inhibits invasion potential of HCC cells by sponging miR-135b. Biomed Res Int. (2019) 2019:2973289. doi: 10.1155/2019/2973289
59. Gao ZQ, Wang JF, Chen DH, Ma XS, Wu Y, Tang Z, et al. Long non-coding RNA GAS5 suppresses pancreatic cancer metastasis through modulating miR-32–5p/PTEN axis. Cell Biosci. (2017) 7:66. doi: 10.1186/s13578-017-0192-0
60. Lu X, Fang Y, Wang Z, Xie J, Zhan Q, Deng X, et al. Downregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6. Cell Tissue Res. (2013) 354:891–6. doi: 10.1007/s00441-013-1711-x
61. Gao ZQ, Wang JF, Chen DH, Ma XS, Yang W, Zhe T, et al. Long non-coding RNA GAS5 antagonizes the chemoresistance of pancreatic cancer cells through down-regulation of miR-181c-5p. Biomed Pharmacother. (2018) 97:809–17. doi: 10.1016/j.biopha.2017.10.157
62. Liu B, Wu S, Ma J, Yan S, Xiao Z, Wan L, et al. lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Mol Ther Nucleic Acids. (2018) 13:472–82. doi: 10.1016/j.omtn.2018.09.026
63. Sun YW, Chen YF, Li J, Huo YM, Liu DJ, Hua R, et al. A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1alpha in pancreatic ductal adenocarcinoma. Br J Cancer. (2014) 111:2131–41. doi: 10.1038/bjc.2014.520
64. Zhuo H, Tang J, Lin Z, Jiang R, Zhang X, Ji J, et al. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol Carcinog. (2016) 55:209–19. doi: 10.1002/mc.22270
65. Ribarska T, Goering W, Droop J, Bastian KM, Ingenwerth M and Schulz W A. Deregulation of an imprinted gene network in prostate cancer. Epigenetics. (2014) 9:704–17. doi: 10.4161/epi.28006
66. Yan J, Guo X, Xia J, Shan T, Gu C, Liang Z, et al. MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Med Oncol. (2014) 31:879. doi: 10.1007/s12032-014-0879-6
67. Terashima M, Tange S, Ishimura A, Suzuki T. MEG3 Long non-coding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J Biol Chem. (2017) 292:82–99. doi: 10.1074/jbc.M116.750950
68. Hu D, Su C, Jiang M, Shen Y, Shi A, Zhao F, et al. Fenofibrate inhibited pancreatic cancer cells proliferation via activation of p53 mediated by upregulation of LncRNA MEG3. Biochem Biophys Res Commun. (2016) 471:290–5. doi: 10.1016/j.bbrc.2016.01.169
69. Ma L, Wang F, Du C, Zhang Z, Guo H, Xie X, et al. Long non-coding RNA MEG3 functions as a tumour suppressor and has prognostic predictive value in human pancreatic cancer. Oncol Rep. (2018) 39:1132–40. doi: 10.3892/or.2018.6178
70. Gu L, Zhang J, Shi M, Zhan Q, Shen B, Peng C. lncRNA MEG3 had anti-cancer effects to suppress pancreatic cancer activity. Biomed Pharmacother. (2017) 89:1269–76. doi: 10.1016/j.biopha.2017.02.041
71. Childs EJ, Mocci E, Campa D, Bracci PM, Gallinger S, Goggins M, et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet. (2015) 47:911–6. doi: 10.1038/ng.3341
72. Lu H, Yang D, Zhang L, Lu S, Ye J, Li M, et al. Linc-pint inhibits early stage pancreatic ductal adenocarcinoma growth through TGF-β pathway activation. Oncol Lett. (2019) 17:4633–9. doi: 10.3892/ol.2019.10111
73. Gong Y, Dai HS, Shu JJ, Liu W, Bie P, Zhang LD. LNC00673 suppresses proliferation and metastasis of pancreatic cancer via target miR-504/ HNF1A. J Cancer. (2020) 11:940–8. doi: 10.7150/jca.32855
74. Li H, Hao X, Wang H, Liu Z, He Y, Pu M, et al. Circular RNA expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cell Physiol Biochem. (2016) 40:1334–44. doi: 10.1159/000453186
75. Zhang Q, Wang JY, Zhou SY, Yang SJ, Zhong S L. Circular RNA expression in pancreatic ductal adenocarcinoma. Oncol Lett. (2019) 18:2923–30. doi: 10.3892/ol.2019.10624
76. Li Q, Geng S, Yuan H, Li Y, Zhang S, Pu L, et al. Circular RNA expression profiles in extracellular vesicles from the plasma of patients with pancreatic ductal adenocarcinoma. FEBS Open Bio. (2019) 9:2052–62. doi: 10.1002/2211-5463.12741
77. Liu L, Liu FB, Huang M, Xie K, Xie QS, Liu CH, et al. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int. (2019) 18:580–6. doi: 10.1016/j.hbpd.2019.03.003
78. Xu Y, Yao Y, Gao P, Cui Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun. (2019) 509:138–42. doi: 10.1016/j.bbrc.2018.12.088
79. Chen G, Shi Y, Zhang Y, Sun J. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco Targets Ther. (2017) 10:5783–94. doi: 10.2147/OTT.S150678
80. Yao J, Zhang C, Chen Y, Gao S. Downregulation of circular RNA circ-LDLRAD3 suppresses pancreatic cancer progression through miR-137–3p/PTN axis. Life Sci. (2019) 239:116871. doi: 10.1016/j.lfs.2019.116871
81. Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. (2017) 23:8345–54. doi: 10.3748/wjg.v23.i47.8345
82. Hao L, Rong W, Bai L, Cui H, Zhang S, Li Y, et al. Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b. J Cell Biochem. (2019) 120:3780–9. doi: 10.1002/jcb.27658
83. Qu S, Hao X, Song W, Niu K, Yang X, Zhang X, et al. Circular RNA circRHOT1 is upregulated and promotes cell proliferation and invasion in pancreatic cancer. Epigenomics. (2019) 11:53–63. doi: 10.2217/epi-2018-0051
84. An Y, Cai H, Zhang Y, Liu S, Duan Y, Sun D, et al. circZMYM2 competed endogenously with miR-335–5p to regulate JMJD2C in pancreatic cancer. Cell Physiol Biochem. (2018) 51:2224–36. doi: 10.1159/000495868
85. Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. (2018) 432:237–50. doi: 10.1016/j.canlet.2018.04.035
86. Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. (2018) 37:177. doi: 10.1186/s13046-018-0822-3
87. Chen Y, Li Z, Zhang M, Wang B, Ye J, Zhang Y, et al. Circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. (2019) 38:466. doi: 10.1186/s13046-019-1436-0
88. Shao F, Huang M, Meng F, Huang Q. Circular RNA signature predicts gemcitabine resistance of pancreatic ductal adenocarcinoma. Front Pharmacol. (2018) 9:584. doi: 10.3389/fphar.2018.00584
89. Jiang Y, Wang T, Yan L, Qu L. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: hsa_circ_0001649. Gene. (2018) 675:88–93. doi: 10.1016/j.gene.2018.06.099
90. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
91. Liu H. MicroRNAs in breast cancer initiation and progression. Cell Mol Life Sci. (2012) 69:3587–99. doi: 10.1007/s00018-012-1128-9
92. Li SD, Zhang JR, Wang YQ, Wan X P. The role of microRNAs in ovarian cancer initiation and progression. J Cell Mol Med. (2010) 14:2240–9. doi: 10.1111/j.1582-4934.2010.01058.x
93. Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. (2007) 26:4442–52. doi: 10.1038/sj.onc.1210228
94. Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res. (2009) 2:807–13. doi: 10.1158/1940-6207.CAPR-09-0094
95. Humeau M, Vignolle-Vidoni A, Sicard F, Martins F, Bournet B, Buscail L, et al. Salivary microRNA in pancreatic cancer patients. PLoS ONE. (2015) 10:e0130996. doi: 10.1371/journal.pone.0130996
96. Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, et al. Combination of plasma microRNAs with serum CA19–9 for early detection of pancreatic cancer. Int J Cancer. (2012) 131:683–91. doi: 10.1002/ijc.26422
97. Wang P, Zhuang L, Zhang J, Fan J, Luo J, Chen H, et al. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol Oncol. (2013) 7:334–45. doi: 10.1016/j.molonc.2012.10.011
98. Ying H, Elpek KG, Vinjamoori A, Zimmerman SM, Chu GC, Yan H, et al. PTEN is a major tumor suppressor in pancreatic ductal adenocarcinoma and regulates an NF-kappaB-cytokine network. Cancer Discov. (2011) 1:158–69. doi: 10.1158/2159-8290.CD-11-0031
99. Paik WH, Kim HR, Park JK, Song BJ, Lee SH, Hwang J H. Chemosensitivity induced by down-regulation of microRNA-21 in gemcitabine-resistant pancreatic cancer cells by indole-3-carbinol. Anticancer Res. (2013) 33:1473–81.
100. Dong J, Zhao YP, Zhou L, Zhang TP, Chen G. Bcl-2 upregulation induced by miR-21 via a direct interaction is associated with apoptosis and chemoresistance in MIA PaCa-2 pancreatic cancer cells. Arch Med Res. (2011) 42:8–14. doi: 10.1016/j.arcmed.2011.01.006
101. Sun J, Jiang Z, Li Y, Wang K, Chen X, Liu G. Downregulation of miR-21 inhibits the malignant phenotype of pancreatic cancer cells by targeting VHL. Onco Targets Ther. (2019) 12:7215–26. doi: 10.2147/OTT.S211535
102. Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. (2007) 104:16170–5. doi: 10.1073/pnas.0703942104
103. Huang C, Li H, Wu W, Jiang T, Qiu Z. Regulation of miR-155 affects pancreatic cancer cell invasiveness and migration by modulating the STAT3 signaling pathway through SOCS1. Oncol Rep. (2013) 30:1223–30. doi: 10.3892/or.2013.2576
104. Wang J, Guo J, Fan H. MiR-155 regulates the proliferation and apoptosis of pancreatic cancer cells through targeting SOCS3. Eur Rev Med Pharmacol Sci. (2019) 23:5168–75. doi: 10.26355/eurrev_201906_18181
105. Xu Q, Li P, Chen X, Zong L, Jiang Z, Nan L, et al. miR-221/222 induces pancreatic cancer progression through the regulation of matrix metalloproteinases. Oncotarget. (2015) 6:14153–64. doi: 10.18632/oncotarget.3686
106. Park JK, Lee EJ, Esau C, Schmittgen T D. Antisense inhibition of microRNA-21 or−221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. (2009) 38: e190–9. doi: 10.1097/MPA.0b013e3181ba82e1
107. Sarkar S, Dubaybo H, Ali S, Goncalves P, Kollepara SL, Sethi S, et al. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer Res. (2013) 3:465–77.
108. Wang W, Zhao L, Wei X, Wang L, Liu S, Yang Y, et al. MicroRNA-320a promotes 5-FU resistance in human pancreatic cancer cells. Sci Rep. (2016) 6:27641. doi: 10.1038/srep27641
109. Ohuchida K, Mizumoto K, Lin C, Yamaguchi H, Ohtsuka T, Sato N, et al. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann Surg Oncol. (2012) 19:2394–402. doi: 10.1245/s10434-012-2252-3
110. Xiong G, Huang H, Feng M, Yang G, Zheng S, You L, et al. MiR-10a-5p targets TFAP2C to promote gemcitabine resistance in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. (2018) 37:76. doi: 10.1186/s13046-018-0739-x
111. Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI, et al. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. (2009) 137:2136–45.e1–7. doi: 10.1053/j.gastro.2009.08.065
112. Masamune A, Nakano E, Hamada S, Takikawa T, Yoshida N, Shimosegawa T. Alteration of the microRNA expression profile during the activation of pancreatic stellate cells. Scand J Gastroenterol. (2014) 49:323–31. doi: 10.3109/00365521.2013.876447
113. Kent OA, Mendell JT, Rottapel R. Transcriptional regulation of miR-31 by oncogenic KRAS mediates metastatic phenotypes by repressing RASA1. Mol Cancer Res. (2016) 14:267–77. doi: 10.1158/1541-7786.MCR-15-0456
114. Zhang L, Wang C, Yu S, Jia C, Yan J, Lu Z, et al. Loss of ARID1A expression correlates with tumor differentiation and tumor progression stage in pancreatic ductal adenocarcinoma. Technol Cancer Res Treat. (2018) 17:1533034618754475. doi: 10.1177/1533034618754475
115. Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le QT, et al. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol. (2010) 3:109–13. doi: 10.1593/tlo.09256
116. Takikawa T, Masamune A, Hamada S, Nakano E, Yoshida N, Shimosegawa T. miR-210 regulates the interaction between pancreatic cancer cells and stellate cells. Biochem Biophys Res Commun. (2013) 437:433–9. doi: 10.1016/j.bbrc.2013.06.097
117. Wang HL, Zhou R, Liu J, Chang Y, Liu S, Wang XB, et al. MicroRNA-196b inhibits late apoptosis of pancreatic cancer cells by targeting CADM1. Sci Rep. (2017) 7:11467. doi: 10.1038/s41598-017-11248-3
118. Kanno S, Nosho K, Ishigami K, Yamamoto I, Koide H, Kurihara H, et al. MicroRNA-196b is an independent prognostic biomarker in patients with pancreatic cancer. Carcinogenesis. (2017) 38:425–31. doi: 10.1093/carcin/bgx013
119. Bartsch DK, Gercke N, Strauch K, Wieboldt R, Matthäi E, Wagner V, et al. The combination of MiRNA-196b, LCN2, and TIMP1 is a potential set of circulating biomarkers for screening individuals at risk for familial pancreatic cancer. J Clin Med. (2018) 7:295. doi: 10.3390/jcm7100295
120. Wu G, Li Z, Jiang P, Zhang X, Xu Y, Chen K, et al. MicroRNA-23a promotes pancreatic cancer metastasis by targeting epithelial splicing regulator protein 1. Oncotarget. (2017) 8:82854–71. doi: 10.18632/oncotarget.20692
121. Liu N, Sun YY, Zhang XW, Chen S, Wang Y, Zhang ZX, et al. Oncogenic miR-23a in pancreatic ductal adenocarcinogenesis via inhibiting APAF1. Dig Dis Sci. (2015) 60:2000–8. doi: 10.1007/s10620-015-3588-x
122. Diao H, Ye Z, Qin R. miR-23a acts as an oncogene in pancreatic carcinoma by targeting FOXP2. J Investig Med. (2018) 66:676–83. doi: 10.1136/jim-2017-000598
123. Guo R, Gu J, Zhang Z, Wang Y, Gu C. MiR-451 promotes cell proliferation and metastasis in pancreatic cancer through targeting CAB39. Biomed Res Int. (2017) 2017:2381482. doi: 10.1155/2017/2381482
124. Long LM, Zhan JK, Wang HQ, Li S, Chen YY, Liu YS. The clinical significance of miR-34a in pancreatic ductal carcinoma and associated molecular and cellular mechanisms. Pathobiology. (2017) 84:38–48. doi: 10.1159/000447302
125. Xia J, Duan Q, Ahmad A, Bao B, Banerjee S, Shi Y, et al. Genistein inhibits cell growth and induces apoptosis through up-regulation of miR-34a in pancreatic cancer cells. Curr Drug Targets. (2012) 13:1750–6. doi: 10.2174/138945012804545597
126. Feng SD, Mao Z, Liu C, Nie YS, Sun B, Guo M, et al. Simultaneous overexpression of miR-126 and miR-34a induces a superior antitumor efficacy in pancreatic adenocarcinoma. Onco Targets Ther. (2017) 10:5591–604. doi: 10.2147/OTT.S149632
127. Tang Y, Tang Y, Cheng YS. miR-34a inhibits pancreatic cancer progression through Snail1-mediated epithelial-mesenchymal transition and the Notch signaling pathway. Sci Rep. (2017) 7:38232. doi: 10.1038/srep38232
128. Hu QL, Jiang QY, Jin X, Shen J, Wang K, Li YB, et al. Cationic microRNA-delivering nanovectors with bifunctional peptides for efficient treatment of PANC-1 xenograft model. Biomaterials. (2013) 34:2265–76. doi: 10.1016/j.biomaterials.2012.12.016
129. Li Z, Li X, Yu C, Wang M, Peng F, Xiao J, et al. MicroRNA-100 regulates pancreatic cancer cells growth and sensitivity to chemotherapy through targeting FGFR3. Tumour Biol. (2014) 35:11751–9. doi: 10.1007/s13277-014-2271-8
130. Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. (2010) 31:1726–33. doi: 10.1093/carcin/bgq160
131. Yang J, Zhang HF, Qin C F. MicroRNA-217 functions as a prognosis predictor and inhibits pancreatic cancer cell proliferation and invasion via targeting E2F3. Eur Rev Med Pharmacol Sci. (2017) 21:4050–57.
132. Chen Q, Wang P, Fu Y, Liu X, Xu W, Wei J, et al. MicroRNA-217 inhibits cell proliferation, invasion and migration by targeting Tpd52l2 in human pancreatic adenocarcinoma. Oncol Rep. (2017) 38:3567–73. doi: 10.3892/or.2017.6036
133. Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, et al. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. (2010) 24:2754–9. doi: 10.1101/gad.1950610
134. Hu Y, Ou Y, Wu K, Chen Y and Sun W. miR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway. Tumour Biol. (2012) 33:1863–70. doi: 10.1007/s13277-012-0446-8
135. Pham H, Rodriguez CE, Donald GW, Hertzer KM, Jung XS, Chang HH, et al. miR-143 decreases COX-2 mRNA stability and expression in pancreatic cancer cells. Biochem Biophys Res Commun. (2013) 439:6–11. doi: 10.1016/j.bbrc.2013.08.042
136. Huang FT, Peng JF, Cheng WJ, Zhuang YY, Wang LY, Li CQ, et al. MiR-143 targeting TAK1 attenuates pancreatic ductal adenocarcinoma progression via MAPK and NF-kappaB pathway in vitro. Dig Dis Sci. (2017) 62:944–57. doi: 10.1007/s10620-017-4472-7
137. Zhu ZM, Xu YF, Su QJ, Du JD, Tan XL, Tu YL, et al. Prognostic significance of microRNA-141 expression and its tumor suppressor function in human pancreatic ductal adenocarcinoma. Mol Cell Biochem. (2014) 388:39–49. doi: 10.1007/s11010-013-1897-y
138. Pan Y, Lu F, Xiong P, Pan M, Zhang Z, Lin X, et al. WIPF1 antagonizes the tumor suppressive effect of miR-141/200c and is associated with poor survival in patients with PDAC. J Exp Clin Cancer Res. (2018) 37:167. doi: 10.1186/s13046-018-0848-6
139. Zhao G, Wang B, Liu Y, Zhang JG, Deng SC, Qin Q, et al. miRNA-141, downregulated in pancreatic cancer, inhibits cell proliferation and invasion by directly targeting MAP4K4. Mol Cancer Ther. (2013) 12:2569–80. doi: 10.1158/1535-7163.MCT-13-0296
140. Xu L, Li Q, Xu D, Wang Q, An Y, Du Q, et al. hsa-miR-141 downregulates TM4SF1 to inhibit pancreatic cancer cell invasion and migration. Int J Oncol. (2014) 44:459–66. doi: 10.3892/ijo.2013.2189
141. Ma L, Zhai B, Zhu H, Li W, Jiang W, Lei L, et al. The miR-141/neuropilin-1 axis is associated with the clinicopathology and contributes to the growth and metastasis of pancreatic cancer. Cancer Cell Int. (2019) 19:248. doi: 10.1186/s12935-019-0963-2
142. Soubani O, Ali AS, Logna F, Ali S, Philip PA, Sarkar F H. Re-expression of miR-200 by novel approaches regulates the expression of PTEN and MT1-MMP in pancreatic cancer. Carcinogenesis. (2012) 33:1563–71. doi: 10.1093/carcin/bgs189
143. Diaz-Riascos ZV, Ginesta MM, Fabregat J, Serrano T, Busquets J, Buscail L, et al. Expression and role of microRNAs from the miR-200 family in the tumor formation and metastatic propensity of pancreatic cancer. Mol Ther Nucleic Acids. (2019) 17:491–503. doi: 10.1016/j.omtn.2019.06.015
144. Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. (2009) 11:1487–95. doi: 10.1038/ncb1998
145. Lu Y, Lu J, Li X, Zhu H, Fan X, Zhu S, et al. MiR-200a inhibits epithelial-mesenchymal transition of pancreatic cancer stem cell. BMC Cancer. (2014) 14:85. doi: 10.1186/1471-2407-14-85
146. Zhou J, Song S, He S, Zhu X, Zhang Y, Yi B, et al. microRNA-375 targets PDK1 in pancreatic carcinoma and suppresses cell growth through the Akt signaling pathway. Int J Mol Med. (2014) 33:950–6. doi: 10.3892/ijmm.2014.1638
147. Yonemori K, Seki N, Kurahara H, Osako Y, Idichi T, Arai T, et al. ZFP36L2 promotes cancer cell aggressiveness and is regulated by antitumor microRNA-375 in pancreatic ductal adenocarcinoma. Cancer Sci. (2017) 108:124–135. doi: 10.1111/cas.13119
148. Basu A, Alder H, Khiyami A, Leahy P, Croce CM, Haldar S. microRNA-375 and microRNA-221: potential non-coding RNAs associated with antiproliferative activity of benzyl isothiocyanate in pancreatic cancer. Genes Cancer. (2011) 2:108–19. doi: 10.1177/1947601911409212
149. Zhang R, Li M, Zang W, Chen X, Wang Y, Li P, et al. MiR-148a regulates the growth and apoptosis in pancreatic cancer by targeting CCKBR and Bcl-2. Tumour Biol. (2014) 35:837–44. doi: 10.1007/s13277-013-1115-2
150. Idichi T, Seki N, Kurahara H, Fukuhisa H, Toda H, Shimonosono M, et al. Molecular pathogenesis of pancreatic ductal adenocarcinoma: impact of passenger strand of pre-miR-148a on gene regulation. Cancer Sci. (2018) 109:2013–2026. doi: 10.1111/cas.13610
151. Liffers ST, Munding JB, Vogt M, Kuhlmann JD, Verdoodt B, Nambiar S, et al. MicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25B. Lab Invest. (2011) 91:1472–9. doi: 10.1038/labinvest.2011.99
152. Peng L, Liu Z, Xiao J, Tu Y, Wan Z, Xiong H, et al. MicroRNA-148a suppresses epithelial-mesenchymal transition and invasion of pancreatic cancer cells by targeting Wnt10b and inhibiting the Wnt/beta-catenin signaling pathway. Oncol Rep. (2017) 38:301–308. doi: 10.3892/or.2017.5705
153. Feng H, Wang Y, Su J, Liang H, Zhang CY, Chen X, et al. microRNA-148a suppresses the proliferation and migration of pancreatic cancer cells by down-regulating ErbB3. Pancreas. (2016) 45:1263–71. doi: 10.1097/MPA.0000000000000677
154. Kugel S, Sebastian C, Fitamant J, Ross KN, Saha SK, Jain E, et al. SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell. (2016) 165:1401–1415. doi: 10.1016/j.cell.2016.04.033
155. Karmakar S, Kaushik G, Nimmakayala R, Rachagani S, Ponnusamy MP, Batra SK. MicroRNA regulation of K-Ras in pancreatic cancer and opportunities for therapeutic intervention. Semin Cancer Biol. (2019) 54:63–71. doi: 10.1016/j.semcancer.2017.11.020
156. Patel K, Kollory A, Takashima A, Sarkar S, Faller DV, Ghosh SK. MicroRNA let-7 downregulates STAT3 phosphorylation in pancreatic cancer cells by increasing SOCS3 expression. Cancer Lett. (2014) 347:54–64. doi: 10.1016/j.canlet.2014.01.020
157. Bhutia YD, Hung SW, Krentz M, Patel D, Lovin D, Manoharan R, et al. Differential processing of let-7a precursors influences RRM2 expression and chemosensitivity in pancreatic cancer: role of LIN-28 and SET oncoprotein. PLoS ONE. (2013) 8:e53436. doi: 10.1371/journal.pone.0053436
158. Wang S, Chen X, Tang M. MicroRNA-216a inhibits pancreatic cancer by directly targeting Janus kinase 2. Oncol Rep. (2014) 32:2824–30. doi: 10.3892/or.2014.3478
159. Zhang X, Shi H, Lin S, Ba M, Cui S. MicroRNA-216a enhances the radiosensitivity of pancreatic cancer cells by inhibiting beclin-1-mediated autophagy. Oncol Rep. (2015) 34:1557–64. doi: 10.3892/or.2015.4078
Keywords: non-coding RNAs, pancreatic ductal adenocarcinoma, miRNA, lncRNA, circ-RNA
Citation: Gong R and Jiang Y (2020) Non-coding RNAs in Pancreatic Ductal Adenocarcinoma. Front. Oncol. 10:309. doi: 10.3389/fonc.2020.00309
Received: 02 December 2019; Accepted: 20 February 2020;
Published: 17 March 2020.
Edited by:
Ondrej Slaby, Brno University of Technology, CzechiaReviewed by:
Maria Paola Paronetto, Foro Italico University of Rome, ItalyFengbiao Mao, University of Michigan, United States
Copyright © 2020 Gong and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yueping Jiang, eXVlcGluZ21kQGhvdG1haWwuY29t
 Ruining Gong
Ruining Gong Yueping Jiang
Yueping Jiang