
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 10 March 2020
Sec. Women's Cancer
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.00295
Objective: Despite several nationwide cohort studies of germline BRCA1/2 mutations and several small cohort studies of somatic BRCA1/2 mutations in Chinese epithelial ovarian cancer (EOC) patients, little is known about the impact of these findings on survival outcomes in this population. In this study of 172 retrospectively recruited Chinese EOC patients, germline and somatic BRCA1/2 mutations and their value for predicting survival outcomes were evaluated.
Methods: Unselected patients who visited the study center from January 1, 2011, to January 1, 2015, were recruited and asked to provide peripheral blood samples for this study if they were pathologically confirmed to have primary EOC. All patients received staging surgeries or debulking surgeries involving systemic platinum-based chemotherapy, and the patients were then followed up to December 1, 2017. DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) sections and peripheral blood and sequenced for somatic and germline testing, respectively. The demographic and clinicopathological characteristics of the patients were collected to analyze the distribution of BRCA mutations in subgroups. Survival outcomes were compared among various BRCA mutation statuses using univariate and multivariate models.
Results: In 58 (33.7%) patients, 63 variants were identified, including variants of unknown significance (VUS) in 18 patients (10.5%) and pathogenic or likely pathogenic variants in a partially overlapping set of 41 patients (23.8%). Germline BRCA mutations, somatic BRCA mutations, BRCA1 mutations in general, and BRCA2 mutations in general were found in 35 (20.3%), 7 (4.1%), 28 (16.3%), and 13 (7.6%) patients, respectively. Five recurrent mutations were identified. Personal and family cancer histories as well as hereditary breast and ovarian cancer (HBOC) criteria were associated with deleterious BRCA mutations both overall and in the germline specifically, whereas only age at diagnosis of EOC was associated with somatic BRCA mutations. In univariate and Cox regression analyses, patients with BRCA1/2 mutations in general had significant improvements in progression-free survival (PFS) and overall survival (OS).
Conclusions: In Chinese EOC patients, the distributions and risk factors associated with germline and somatic BRCA1/2 mutations were similar to those previously reported in international studies. Deleterious BRCA mutations in general were associated with improved survival outcomes in this cohort.
Ovarian cancer is the third most common gynecological malignancy and the leading cause of mortality in female cancers (1), representing 1.3% of all new cancer cases in the United States in 2018 (2). In China, the prevalence of ovarian cancer has increased in the past decade, with 52,100 new cases and 22,500 related deaths in 2015 (3). Ovarian cancers are a heterogeneous group of malignancies varying in etiology and molecular biology. Approximately 90% of cases belong to the epithelial type [epithelial ovarian cancer (EOC)], with the most common being high-grade serous carcinoma (HGSC). A majority of EOC patients are diagnosed at advanced stages and have a poor prognosis. As with other malignancies, the tumorigenesis of EOC is a process that drives normal cells toward a malignant state and can involve both somatic (acquired) and germline (inherited) mutations (4, 5). Large-scale cancer sequencing data from cases in The Cancer Genome Atlas (TCGA) revealed that ovarian cancer has the highest prevalence of susceptibility-associated genes (6). In previous reports, ~5 to 10% of invasive EOC cases were hereditary (7–10). In addition, inherited ovarian cancer may present as hereditary breast and ovarian cancer (HBOC) (11, 12). However, in recent reports, ~20% or more of all EOCs have been identified to be associated with germline mutations (13–16). Most cases of inherited susceptibility to EOC are primarily related to germline mutations of BRCA1 and BRCA2, which account for about 80% of hereditary ovarian cancers (17–19). Mutant BRCA is an indispensable founding mutation in EOC (20). Knowledge of the pathogenic molecular mechanism and genetic mutations involved in EOC has promoted genetic counseling and testing as well as potential intervention (21). The emergence of poly(adenosine diphosphate [ADP]-ribose) polymerase inhibitors (PARPis) has necessitated genetic testing (22). However, there is controversy regarding the optimum testing strategy (23–25). Information on BRCA1/2 germline mutations has predictive value for the platinum sensitivity of tumors and the survival outcomes of patients (26–28). In a study by Pennington et al. (28) compared to germline BRCA1/2 mutations, somatic BRCA1/2 mutations had a similar positive impact on overall survival (OS) and platinum responsiveness. On the other hand, in recent reports, homologous recombination deficiency has gained in importance, in addition to the BRCA mutations, on the targeted treatment (23, 29), chemotherapy (30, 31), and prognosis (32, 33) in EOC patients.
Despite several national cohort studies of BRCA germline mutations in China (16, 34, 35) and in Japan (24), no study has attempted to reveal the association between mutations and treatment effects or survival outcomes in Chinese EOC patients. The specific impact of somatic mutations on EOC cohorts is also not well-explored. In our previous report (36), no significant outcomes were discovered due to the limited sample size and short follow-up period.
In the present report, we aimed to determine the frequencies of germline and somatic BRCA1/2 mutations in a single study center based on pathological findings from primary samples or surgical tissues. The mutation status was analyzed in the context of various demographic and clinicopathological characteristics, such as personal and family cancer history as well as histological subtypes. The impact of mutation status on sensitivity to platinum-based chemotherapy and on patient survival outcomes was also described in univariate and multivariate models.
Unselected patients who visited the study center from January 1, 2011, to January 1, 2015, and were diagnosed with EOC were recruited for this study. Once the pathological examination confirmed EOC, the patients were asked to provide samples of peripheral blood for germline testing if they met the following inclusion criteria: (1) age 18 years or older; (2) pathologically confirmed EOC (including EOC, carcinoma of the fallopian tube and primary peritoneal carcinoma); (3) receipt of comprehensive staging surgery or debulking surgery and systemic platinum-based chemotherapy; (4) sufficient formalin-fixed, paraffin-embedded (FFPE) sections for somatic testing, which could be performed between any two systematic chemotherapy treatments; these tissues could be collected by core biopsy or by laparoscopy for patients prepared for neoadjuvant chemotherapy or by laparotomic sampling for patients of primary staging or cytoreductive surgeries; (5) willingness to provide signed consent in advance of the trial; and (6) provision of peripheral blood for germline testing. Patients not meeting all of these inclusion criteria were excluded. Basic patient information regarding age at diagnosis, neoplasm staging and histopathological type was retrieved from medical records. This study was approved by the Institutional Review Board (IRB) of the study center (Registration No. ZS-1245). The Chinese Human Genetic Resources Management Office of the National Ministry of Science and Technology also approved this study (registration No. [2017]1901, http://www.most.gov.cn/bszn/new/rlyc/jgcx/index.htm). As previously mentioned, all patients gave informed consent before enrollment.
All patients received staging surgeries or primary or interval debulking surgeries along with systematic chemotherapy, which was initiated with platinum-based chemotherapy. Demographic data and medical history, including the date of diagnosis, were retrieved from the patients' medical records. Data regarding histological classification and grading were obtained from the electronic pathology database at the study center. Staging was determined by the International Federation of Gynecology and Obstetrics (FIGO) 2014 staging system (37). Personal and family cancer histories were addressed, especially personal breast cancer history and the aspects of cancer history specified by the “Criteria for further genetic risk evaluation” from the National Comprehensive Cancer Network (NCCN) HBOC guideline of “Genetic/Familial High-Risk Assessment: Breast and Ovarian” (38).
Tumors were sorted into the following categories depending on their response to platinum-based chemotherapy: (1) platinum sensitive, if the time between administration and first relapse of the disease was more than 6 months; (2) platinum resistant, if the patient had a relapse 4 weeks to 6 months after administration; or (3) refractory, if the disease relapsed within 4 weeks under platinum-based agents. Follow-ups were conducted for all enrolled patients until December 1, 2017, or death. Disease relapse or progression was determined by medical imaging, serology, or histology. Progression-free survival (PFS) was defined as the time span between initial diagnosis and disease relapse or progression, while OS was defined as the time span between initial diagnosis and patient death.
For germline BRCA1/2 analyses, DNA was extracted from 200 μl of peripheral blood with the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany). For somatic BRCA1/2 analysis, 10 FFPE sections measuring five micrometers in thickness and taken from representative blocks were made for each case. The cancerous region was then marked by two experienced pathologists (YY and HW) and macro-dissected following deparaffinization using xylene. DNA was then extracted from the tumor tissue using the QIAamp DNA FFPE Tissue Kit (QIAGEN, Hilden, Germany).
Sequencing libraries were prepared using an Oncomine BRCA research assay (Thermo Fisher, Waltham, Massachusetts, USA); the libraries were then subjected to emulsion PCR using the Ion OneTouch 2 System and sequenced on an Ion Torrent Personal Genome Machine sequencer to a median of 500X depth for germline mutations and 1000X for somatic mutations according to the manufacturer's instructions. Upon completion, the raw data were sent through a pipeline customized by Life Technologies. The parameters for variation calling are available (in JSON format) upon request. The sequences covered all exons of the BRCA1 and BRCA2 genes as well as ±20 bp flanking regions to enable the detection of variations affecting potential splice sites. However, due to its high cost, we did not include multiplex ligation-dependent probe amplification (MLPA) in the tests for BRCA1/2.
The identified BRCA1/2 variations were classified according to the 2015 American College of Medical Genetics and Genomics (ACMG) guidelines (39). Pathogenic and likely pathogenic mutations were regarded as deleterious mutations.
Comparisons of continuous variables were conducted with parametric methods if the assumption of normality was met. Non-normally distributed variables and categorical data were compared between patients with and without specific deleterious mutations by using non-parametric tests. Kaplan–Meier survival curves were generated, and proportional hazards models were used to estimate the hazard ratios (HRs) and 95% confidence intervals (95% CIs) for the effects of deleterious mutations on PFS and OS. A multivariable analysis of disease-free survival was performed with adjustment for the important baseline risk factors of major histological subtype (HGSC vs. others), age at diagnosis, and stage (stage I–II vs. III–IV). Unless otherwise stated, all analyses used a two-sided significance level of 0.05 and were conducted with Statistical Product and Service Solutions (SPSS) Statistics 20.0 software (IBM Corporation, Armonk, NY, USA).
The demographic and clinicopathological characteristics of the patients are listed in Table 1. In total, 172 patients were recruited and had final genetic testing outcomes. The median age at diagnosis in the cohort was 52.5 years (range 18–81). Most cases were ovarian carcinomas (168 cases, 97.7%), stage III-IV (140 cases, 81.4%), and HGSC (138 cases, 80.2%). Based on personal and family cancer history, 31 patients met the HBOC criteria. No patients had ever received PARPi treatment. Progression and mortality occurred in 106 (61.6%) and 50 (29.1%) patients, and the median PFS and OS were 23 months (range 0–68) and 39 (12–93) months, respectively.
In 58 (33.7%) patients, 63 variants were identified, including variants of unknown significance (VUS) in 18 patients (10.5%) and pathogenic or likely pathogenic variants in a partially overlapping set of 41 patients (23.8%) (Figure 1). Germline BRCA mutations, somatic BRCA mutations, BRCA1 mutations in general, and BRCA2 mutations in general were found in 35 (20.3%), 7 (4.1%), 28 (16.3%), and 13 (7.6%) patients, respectively. The mutation details are listed in Supplement Table 1. Likely pathogenic or pathogenic mutations consisted of 19 (46.3%) frameshift deletions, 4 (9.8%) frameshift insertions, 12 (29.3%) non-sense mutations, 4 (9.8%) splice-site mutations, and 2 (4.9%) missense mutations.
As shown in Table 2 and Supplement Table 1, 35 (19.7%) and seven (4.1%) patients had at least one germline and somatic deleterious mutation, respectively; 28 (16.3%) and 13 (7.6%) patients harbored at least one deleterious mutation of BRCA1 and BRCA2, respectively. Two (1.2%), 5 (2.9%), and 2 (1.2%) patients carried dual germline/somatic mutations; dual germline and dual somatic mutations classified as VUS; and dual likely pathogenic and pathogenic mutations, respectively. In cases No. 063 and No. 068, the distance between the co-occurring mutations exceeded the amplicon length; therefore, we were unable to determine whether these mutations were in cis or in trans. The criteria for reclassification were unmet for these mutations. In case No. 032, the patient harbored one likely pathogenic germline variant and one somatic pathogenic variant (Supplement Table 1).
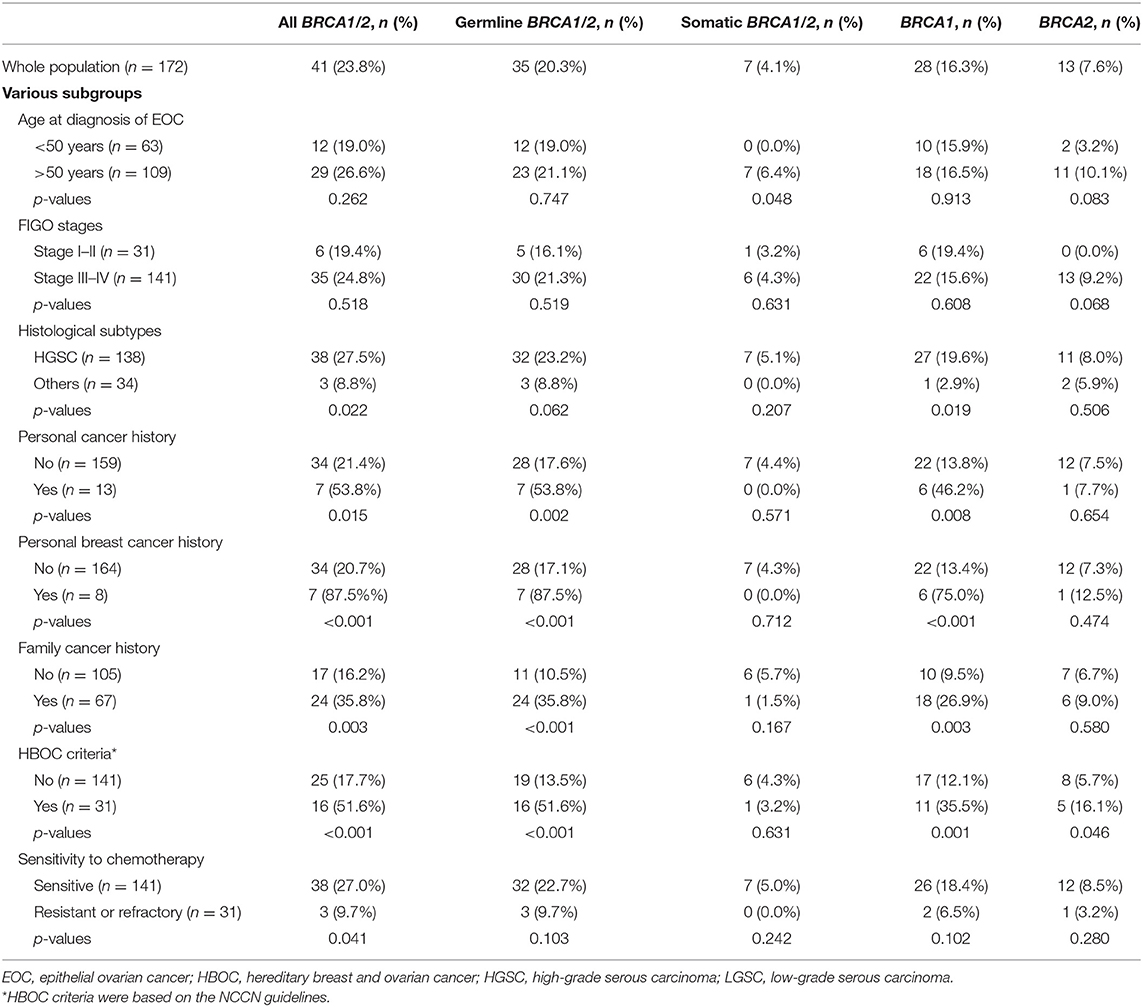
Table 2. Distributions of pathogenic or likely pathogenic mutations in the overall and subgroup populations.
Five recurrent mutations were identified in the cohort. Three pathogenic mutations occurred in two patients (BRCA1 c.5470_5477delATTGGGCA, BRCA1 c.66dup, and BRCA2 c.1963delC). Two VUS missense mutations (BRCA2 c.1568A>G and BRCA2 c.6325G>A) appeared in three and four patients, respectively. All recurrent mutations were in the germline except for BRCA2 c.1964delC.
As shown in Table 2, FIGO stages had no significant association with deleterious mutations of BRCA genes in general or of BRCA1 or BRCA2 in particular. Personal and family cancer history and HBOC criteria were associated with overall and germline BRCA mutations. HBOC criteria were also associated with overall BRCA1 and BRCA2 mutations. For somatic BRCA mutations, only age at diagnosis had a significant impact, while for germline BRCA mutations, histological subtype also had a significant impact. However, only overall deleterious BRCA mutations had a significant impact on sensitivity to platinum-based chemotherapy (Table 2).
The survival outcomes associated with various deleterious BRCA mutations are shown in Figures 2, 3 and Supplement Table 2. In univariate and Cox regression analyses, patients with overall BRCA1/2 mutations had significant improvements in PFS and OS (Figure 2, Supplement Table 2). Overall, germline BRCA1/2 mutations were associated with significant improvement in OS but not PFS. Overall, BRCA2 mutations were associated with significantly improved OS in the Cox regression model. Somatic BRCA1/2 and overall BRCA1 had no significant impact on survival outcomes. However, in the subgroup analysis of HGSC patients (n = 138), various mutation statuses had no significant impact on survival outcomes, except that overall BRCA mutations affected PFS (Supplement Table 2).
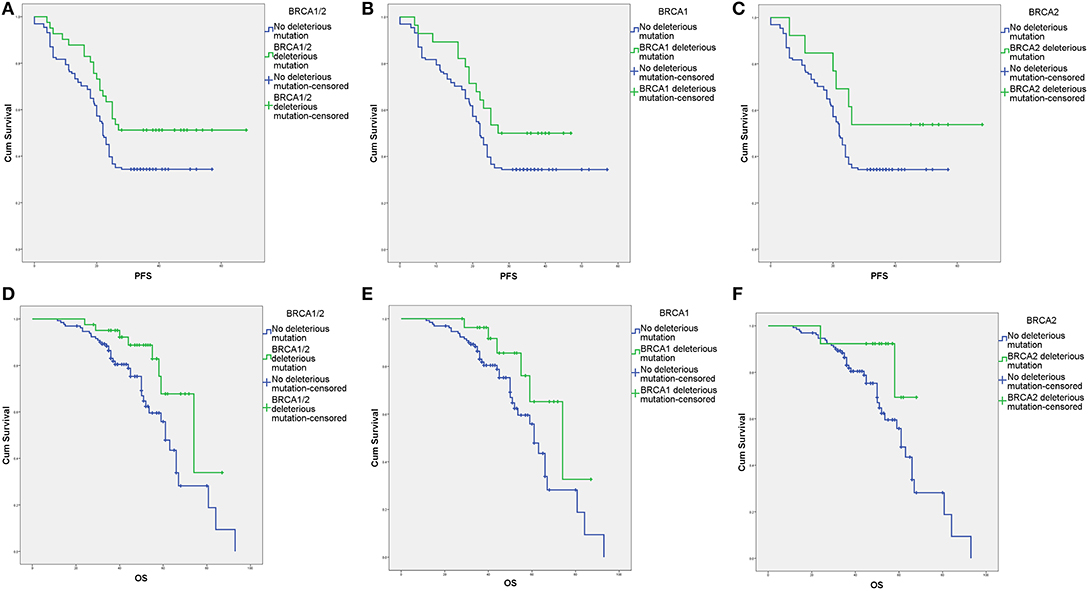
Figure 2. Survival outcomes of various BRCA aberrations according to Kaplan–Meier analysis. (A) Progression-free survival (PFS) with and without deleterious BRCA1/2 mutations. (B) PFS with and without deleterious BRCA1 mutations. (C) PFS with and without deleterious BRCA2 mutations. (D) Overall survival (OS) with and without deleterious BRCA1/2 mutations. (E) OS with and without deleterious BRCA1 mutations. (F) OS with and without deleterious BRCA2 mutations.
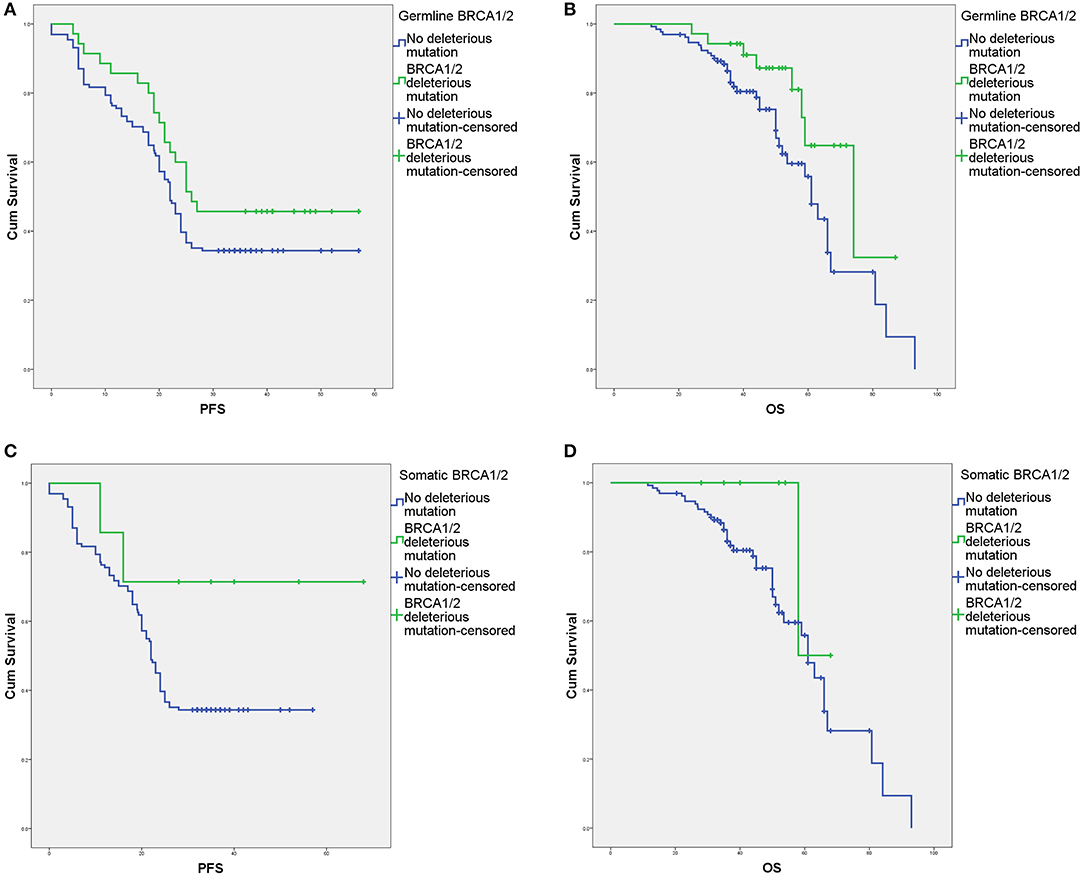
Figure 3. Survival outcomes of germline and somatic BRCA aberrations according to Kaplan–Meier analysis. (A) Progression-free survival (PFS) with and without deleterious germline BRCA1/2 mutations. (B) Overall survival (OS) with and without deleterious germline BRCA1/2 mutations. (C) PFS with and without deleterious somatic BRCA1/2 mutations. (D) OS with and without deleterious somatic BRCA1/2 mutations.
To our knowledge, this study is the first to describe the positive impact of BRCA mutation status on survival outcomes in a Chinese EOC cohort. In our study, overall BRCA mutations, germline BRCA mutations, and overall BRCA2 mutations were all associated with improved survival outcomes, as in previous reports (27, 28, 40–42). We did not find a significant impact of germline mutations on PFS or a significant impact of somatic mutations on PFS or OS. The limited sample size was probably the main reason. As shown in Table 2, HGSC patients harbored more deleterious BRCA1/2 and BRCA1 mutations than patients with other subtypes, and the BRCAness phenotype was independently associated with improved OS in HGSC patients (43). However, in our study, the effects of sample size were so obvious that HGSC patients, the majority of the cohort, had no significant associations between mutation status and survival outcome (Supplement Table 2). A larger cohort of Chinese EOC patients (NCT03015376) would provide more details and profound perspectives on such issues. The improved survival outcomes were likely caused by a higher sensitivity to platinum in patients with deleterious mutations than in those without such mutations, as revealed in the Pennington et al. (28) study.
Overall, we found that germline and somatic BRCA1/2 mutations had prevalence rates of 20.3 and 4.1%, respectively. Disparities in both germline and somatic testing exist (44, 45). Regarding other Chinese studies, our germline mutation prevalence approximated the values reported by Li et al. (1331 cases, 22.4%) (16). Wang et al. (481 cases, 19.6%) (46), Shi et al. (916 cases, 17.0%), and a meta-analysis (21.8%) (47), but it was significantly lower than the value reported by Wu et al. (826 cases, 28.4%) (35). Our somatic BRCA1/2 mutation prevalence was similar to those in two small cohort studies, which reported prevalence values of 4.0% (2/50) (48) and 6.4% (4/62) (36). These differences may originate from the selection of subjects and the database used for analysis. As addressed in a recently published meta-analysis (47), the variant profile in ethnically Chinese people was distinctive from those in other ethnic groups with no distinct founder pathogenic variants. However, this conclusion should be treated cautiously. China needs a uniform platform such as TCGA to further promote genomic testing. Chinese healthcare providers, including gynecologists, still have greatly divergent opinions and suggestions about genetic testing for gynecologic oncology, and these positions are also significantly different from the requirements and recommendations in the guidelines and consensus statements (49–51).
There are several issues worth considering in relation to the selection bias in this report. In our study, HGSC had a significantly higher frequency of overall BRCA1/2 and BRCA1 mutations than other subtypes. These tumors displayed defective homologous recombination due to germline and somatic BRCA mutations, epigenetic inactivation of BRCA and abnormalities in DNA repair genes (9, 13, 14). The quality of the data that can be obtained from FFPE tissues as opposed to fresh tissues needs clarification. The fact that the distribution of somatic mutations was consistent with other Chinese studies (36, 48) and with a Polish study using FFPE tissues from HGSC patients (52) could guarantee the quality of our sample preparation. It was suggested that an adjusted targeted capture-based enrichment protocol was superior to commonly applied multiplex PCR-based protocols for reliable BRCA1/2 variant detection, including CNV detection, using FFPE tumor samples (53). As previously reported, the prevalence of BRCA1/2 mutations in patients with peritoneal carcinoma or fallopian tube carcinoma is comparable to that in EOC patients (54). Therefore, we did not perform a subgroup analysis of these three site-specific cancers. We also did not evaluate the impact of neoadjuvant therapy on prognosis by BRCA status since primary debulking surgery seems to ensure a longer PFS than neoadjuvant chemotherapy alone in women with the BRCA wild-type genotype (55). However, patients with pathogenic mutations in BRCA may benefit from intraperitoneal therapy (56). Finally, no patients in our study had ever utilized PARPis. The high cost of genetic testing is an important barrier to universal performance. However, somatic and germline mutations and expression loss of BRCA1/2 are sufficiently common in ovarian cancer to warrant assessment for the prediction of treatment benefit in clinical trials of PARPis (57).
We reported the previously unknown association between BRCA mutation and survival outcomes in Chinese EOC patients, which was the most important strength of our study. However, the present study has several limitations. We did not exploit all mechanisms of BRCA1/2 regulation, such as large gene rearrangements and epigenetic modifications. Second, the present study population did not include a sufficient sample of histological subtypes other than HGSC; this limitation impairs the ability to capture the mutational landscape of these tumors. Third, as reported previously (58–61). a multigene panel for germline and/or somatic testing would probably be cost effective, providing a substantial rate of clinically actionable pathogenic variants (62). There is a potential benefit to be gained from rescreening with a multigene panel in patients who previously underwent non-informative genetic screening (63). On the other hand, up to 50% of ovarian HGSC patients may exhibit homologous recombination defects (64), which are associated with the indication of PARPis as maintenance therapy, even in newly diagnosed EOC patients (23, 25, 29). These advancements in genetic and genomic testing in EOC will promote further exploration in the future. Importantly, we did not include the MLPA analysis in our study, which is also very important for the evaluation of BRCAness (43, 65). The lack of MLPA obviously decreases the deleterious mutation frequency in patients with EOC or breast cancer and causes bias regarding the association between BRCAness and survival prognosis (66). We did not include BRCA methylation in the study. However, unlike BRCA mutation, BRCA1 methylation was not associated with improved survival or greater sensitivity to platinum chemotherapy (67). Last but not least, a discussion of cosegregation analysis and potential interventions (such as NCT03294343 and NCT04190667) should be provided to all patients with deleterious mutations.
In conclusion, we reported the prevalence of germline and somatic BRCA1/2 mutations in an unselected Chinese EOC cohort comprising 172 patients. Germline mutations were associated with cancer history, especially the aspects relevant to the HBOC criteria. Somatic mutations were associated only with age at diagnosis of EOC. In our study, overall deleterious BRCA mutations predicted increased sensitivity to platinum-based chemotherapy and significantly improved survival outcomes. A larger cohort must be examined with multigene panel testing and even the homologous recombination deficiency model to clarify the indications for PARPis.
All datasets generated for this study are included in the article/Supplementary Material.
The Institutional Review Board of Peking Union Medical College Hospital approved this study (No. HS-1245). In addition, the Chinese Human Genetic Resources Management Office of the National Ministry of Science and Technology approved this study (registration no. [2017]1901, http://www.most.gov.cn/bszn/new/rlyc/jgcx/index.htm).
MW and ZL conceived of the original idea for the study, interpreted the results, carried out the statistical analysis, edited the paper, and acted as overall guarantors. YY, LL, and JL obtained ethical approval, contributed to the preparation of the data set, interpreted the results, and contributed to drafts of the paper. HW, JW, and JG contributed to the study design and interpretation of the results and commented on drafts of the paper. Specifically, colleagues from the Department of Obstetrics and Gynecology were in charge of the diagnosis, treatment, and follow-up of the patients, while colleagues from the Department of Pathology were in charge of the pathological evaluation and molecular diagnosis.
This study was supported by the Chinese Academy of Medical Sciences Initiative for Innovative Medicine (CAMS-2017-I2M-1-002 and CAMS-2016-I2M-1-002), by the National Science-Technology Support Plan Projects (2015BAI13B04), and by the National Natural Science Foundation of China (81602162). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00295/full#supplementary-material
1. Tangjitgamol S, Manusirivithaya S, Laopaiboon M, Lumbiganon P. Interval debulking surgery for advanced epithelial ovarian cancer: a Cochrane systematic review. Gynecol Oncol. (2009) 112:257–64. doi: 10.1016/j.ygyno.2008.09.041
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. (2017) 67:7–30. doi: 10.3322/caac.21387
3. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. (2016) 66:115–32. doi: 10.3322/caac.21338
4. Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. (2009) 458:719–24. doi: 10.1038/nature07943
5. Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. (2011) 331:1553–8. doi: 10.1126/science.1204040
6. Lu C, Xie M, Wendl MC, Wang J, McLellan MD, Leiserson MD, et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun. (2015) 6:10086. doi: 10.1038/ncomms10086
7. Russo A, Calo V, Bruno L, Rizzo S, Bazan V, Di Fede G. Hereditary ovarian cancer. Crit Rev Oncol Hematol. (2009) 69:28–44. doi: 10.1016/j.critrevonc.2008.06.003
8. Narod SA, Madlensky L, Bradley L, Cole D, Tonin P, Rosen B, et al. Hereditary and familial ovarian cancer in southern Ontario. Cancer. (1994) 74:2341–6.
9. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. (2011) 474:609–15. doi: 10.1038/nature10166
10. Kanchi KL, Johnson KJ, Lu C, McLellan MD, Leiserson MD, Wendl MC, et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun. (2014) 5:3156. doi: 10.1038/ncomms4156
11. Easton DF, Bishop DT, Ford D, Crockford GP. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. The Breast Cancer Linkage Consortium. Am J Hum Genet. (1993) 52:678–701.
12. Lynch HT, Albano WA, Lynch JF, Lynch PM, Campbell A. Surveillance and management of patients at high genetic risk for ovarian carcinoma. Obstet Gynecol. (1982) 59:589–96.
13. Ding L, Bailey MH, Porta-Pardo E, Thorsson V, Colaprico A, Bertrand D, et al. Perspective on oncogenic processes at the end of the beginning of cancer genomics. Cell. (2018) 173:305–20 e10. doi: 10.1016/j.cell.2018.03.033
14. Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, et al. A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell. (2018) 33:690–705 e9. doi: 10.1016/j.ccell.2018.03.014
15. Norquist BM, Harrell MI, Brady MF, Walsh T, Lee MK, Gulsuner S, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. (2016) 2:482–90. doi: 10.1001/jamaoncol.2015.5495
16. Li A, Xie R, Zhi Q, Deng Y, Wu Y, Li W, et al. BRCA germline mutations in an unselected nationwide cohort of Chinese patients with ovarian cancer and healthy controls. Gynecol Oncol. (2018) 151:145–52. doi: 10.1016/j.ygyno.2018.07.024
17. Boyd J. Molecular genetics of hereditary ovarian cancer. Oncology. (1998) 12:399–406; discussion 9–10, 13.
18. Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Kwan E, et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet. (2001) 68:700–10. doi: 10.1086/318787
19. Matloff ET. The breast surgeon's role in BRCA1 and BRCA2 testing. Am J Surg. (2000) 180:294–8. doi: 10.1016/s0002-9610(00)00460-8
20. Jonsson P, Bandlamudi C, Cheng ML, Srinivasan P, Chavan SS, Friedman ND, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature. (2019) 571:576–9. doi: 10.1038/s41586-019-1382-1
21. Hartmann LC, Lindor NM. The role of risk-reducing surgery in hereditary breast and ovarian cancer. N Engl J Med. (2016) 374:454–68. doi: 10.1056/NEJMra1503523
22. Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. (2018) 379:2495–505. doi: 10.1056/NEJMoa1810858
23. Coleman RL, Fleming GF, Brady MF, Swisher EM, Steffensen KD, Friedlander M, et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N Engl J Med. (2019) 381:2403–15. doi: 10.1056/NEJMoa1909707
24. Enomoto T, Aoki D, Hattori K, Jinushi M, Kigawa J, Takeshima N, et al. The first Japanese nationwide multicenter study of BRCA mutation testing in ovarian cancer: CHARacterizing the cross-sectionaL approach to Ovarian cancer geneTic TEsting of BRCA (CHARLOTTE). Int J Gynecol Cancer. (2019) 29:1043–9. doi: 10.1136/ijgc-2019-000384
25. Gonzalez-Martin A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. (2019) 381:2391–402. doi: 10.1056/NEJMoa1910962
26. Yang D, Khan S, Sun Y, Hess K, Shmulevich I, Sood AK, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. (2011) 306:1557–65. doi: 10.1001/jama.2011.1456
27. Bolton KL, Chenevix-Trench G, Goh C, Sadetzki S, Ramus SJ, Karlan BY, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. (2012) 307:382–90. doi: 10.1001/jama.2012.20
28. Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, Rendi MH, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. (2014) 20:764–75. doi: 10.1158/1078-0432.CCR-13-2287
29. Ray-Coquard I, Pautier P, Pignata S, Perol D, Gonzalez-Martin A, Berger R, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. (2019) 381:2416–28. doi: 10.1056/NEJMoa1911361
30. Lai CH, Vallikad E, Lin H, Yang LY, Jung SM, Liu HE, et al. Maintenance of pegylated liposomal doxorubicin/carboplatin in patients with advanced ovarian cancer: randomized study of an Asian Gynecologic Oncology Group. J Gynecol Oncol. (2020) 31:e5. doi: 10.3802/jgo.2020.31.e5
31. Lee YJ, Kim D, Shim JE, Bae SJ, Jung YJ, Kim S, et al. Genomic profiling of the residual disease of advanced high-grade serous ovarian cancer after neoadjuvant chemotherapy. Int J Cancer. (2020) 146:1851–61. doi: 10.1002/ijc.32729
32. Lu R, Xie S, Wang Y, Zheng H, Zhang H, Deng M, et al. MUS81 Participates in the progression of serous ovarian cancer associated with dysfunctional DNA repair system. Front Oncol. (2019) 9:1189. doi: 10.3389/fonc.2019.01189
33. Takaya H, Nakai H, Sakai K, Nishio K, Murakami K, Mandai M, et al. Intratumor heterogeneity and homologous recombination deficiency of high-grade serous ovarian cancer are associated with prognosis and molecular subtype and change in treatment course. Gynecol Oncol. (2020) 156:415–22. doi: 10.1016/j.ygyno.2019.11.013
34. Shi T, Wang P, Xie C, Yin S, Shi D, Wei C, et al. BRCA1 and BRCA2 mutations in ovarian cancer patients from China: ethnic-related mutations in BRCA1 associated with an increased risk of ovarian cancer. Int J Cancer. (2017) 140:2051–9. doi: 10.1002/ijc.30633
35. Wu X, Wu L, Kong B, Liu J, Yin R, Wen H, et al. The first nationwide multicenter prevalence study of germline BRCA1 and BRCA2 mutations in Chinese ovarian cancer patients. Int J Gynecol Cancer. (2017) 27:1650–7. doi: 10.1097/IGC.0000000000001065
36. Li W, Shao D, Li L, Wu M, Ma S, Tan X, et al. Germline and somatic mutations of multi-gene panel in Chinese patients with epithelial ovarian cancer: a prospective cohort study. J Ovarian Res. (2019) 12:80. doi: 10.1186/s13048-019-0560-y
37. Mutch DG, Prat J. 2014 FIGO staging for ovarian, fallopian tube and peritoneal cancer. Gynecol Oncol. (2014) 133:401–4. doi: 10.1016/j.ygyno.2014.04.013
38. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®). Genetic/Familial High-Risk Assessment: Breast and Ovarian. Version 2.2019 - July 30, 2018. Available online at: https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf
39. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. (2015) 17:405–24. doi: 10.1038/gim.2015.30
40. Alsop K, Fereday S, Meldrum C, Defazio A, Emmanuel C, George J, et al. BRCA Mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol. (2012) 30:2654–63. doi: 10.1200/JCO.2011.39.8545
41. Chetrit A, Hirsh-Yechezkel G, Ben-David Y, Lubin F, Friedman E, Sadetzki S. Effect of BRCA1/2 mutations on long-term survival of patients with invasive ovarian cancer: the National Israeli Study of Ovarian Cancer. J Clin Oncol. (2008) 26:20–5. doi: 10.1200/jco.2007.11.6905
42. Norquist BM, Brady MF, Harrell MI, Walsh T, Lee MK, Gulsuner S, et al. Mutations in homologous recombination genes and outcomes in ovarian carcinoma patients in GOG 218: an NRG Oncology/Gynecologic Oncology Group Study. Clin Cancer Res. (2018) 24:777–83. doi: 10.1158/1078-0432.ccr-17-1327
43. Hjortkjaer M, Malik Aagaard Jorgensen M, Waldstrom M, Ornskov D, Sogaard-Andersen E, Jakobsen A, et al. The clinical importance of BRCAness in a population-based cohort of Danish epithelial ovarian cancer. Int J Gynecol Cancer. (2019) 29:166–73. doi: 10.1136/ijgc-2018-000017
44. Huang M, Kamath P, Schlumbrecht M, Miao F, Driscoll D, Oldak S, et al. Identifying disparities in germline and somatic testing for ovarian cancer. Gynecol Oncol. (2019) 153:297–303. doi: 10.1016/j.ygyno.2019.03.007
45. Hinchcliff EM, Bednar EM, Lu KH, Rauh-Hain JA. Disparities in gynecologic cancer genetics evaluation. Gynecol Oncol. (2019) 153:184–91. doi: 10.1016/j.ygyno.2019.01.024
46. Wang J, Li W, Shi Y, Huang Y, Sun T, Tang L, et al. Germline mutation landscape of Chinese patients with familial breast/ovarian cancer in a panel of 22 susceptibility genes. Cancer Med. (2019) 8:2074–84. doi: 10.1002/cam4.2093
47. Gao X, Nan X, Liu Y, Liu R, Zang W, Shan G, et al. Comprehensive profiling of BRCA1 and BRCA2 variants in breast and ovarian cancer in Chinese patients. Hum Mutat. (2020) 41:696–708. doi: 10.1002/humu.23965
48. Zhao Q, Yang J, Li L, Cao D, Yu M, Shen K. Germline and somatic mutations in homologous recombination genes among Chinese ovarian cancer patients detected using next-generation sequencing. J Gynecol Oncol. (2017) 28:e39. doi: 10.3802/jgo.2017.28.e39
49. Li L, Qiu L, Wu M. A survey of willingness about genetic counseling and tests in patients of epithelial ovarian cancer. Zhonghua Yi Xue Za Zhi. (2017) 97:3412–5. doi: 10.3760/cma.j.issn.0376-2491.2017.43.011
50. Liu GY, Zhang W. Will Chinese ovarian cancer patients benefit from knowing the BRCA2 mutation status? Chin J Cancer. (2012) 31:1–4. doi: 10.5732/cjc.011.10432
51. Shipman H, Flynn S, MacDonald-Smith CF, Brenton J, Crawford R, Tischkowitz M, et al. Universal BRCA1/BRCA2 testing for ovarian cancer patients is welcomed, but with care: how women and staff contextualize experiences of expanded access. J Genet Couns. (2017) 26:1280–91. doi: 10.1007/s10897-017-0108-5
52. Koczkowska M, Zuk M, Gorczynski A, Ratajska M, Lewandowska M, Biernat W, et al. Detection of somatic BRCA1/2 mutations in ovarian cancer - next-generation sequencing analysis of 100 cases. Cancer Med. (2016) 5:1640–6. doi: 10.1002/cam4.748
53. Zakrzewski F, Gieldon L, Rump A, Seifert M, Grutzmann K, Kruger A, et al. Targeted capture-based NGS is superior to multiplex PCR-based NGS for hereditary BRCA1 and BRCA2 gene analysis in FFPE tumor samples. BMC Cancer. (2019) 19:396. doi: 10.1186/s12885-019-5584-6
54. Choi MC, Bae JS, Jung SG, Park H, Joo WD, Song SH, et al. Prevalence of germline BRCA mutations among women with carcinoma of the peritoneum or fallopian tube. J Gynecol Oncol. (2018) 29:e43. doi: 10.3802/jgo.2018.29.e43
55. Petrillo M, Marchetti C, De Leo R, Musella A, Capoluongo E, Paris I, et al. BRCA mutational status, initial disease presentation, and clinical outcome in high-grade serous advanced ovarian cancer: a multicenter study. Am J Obstet Gynecol. (2017) 217:334 e1–9. doi: 10.1016/j.ajog.2017.05.036
56. Naumann RW, Morris JC, Tait DL, Higgins RV, Crane EK, Drury LK, et al. Patients with BRCA mutations have superior outcomes after intraperitoneal chemotherapy in optimally resected high grade ovarian cancer. Gynecol Oncol. (2018) 151:477–80. doi: 10.1016/j.ygyno.2018.10.003
57. Hennessy BT, Timms KM, Carey MS, Gutin A, Meyer LA, Flake DD, 2nd, et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J Clin Oncol. (2010) 28:3570–6. doi: 10.1200/jco.2009.27.2997
58. Minion LE, Dolinsky JS, Chase DM, Dunlop CL, Chao EC, Monk BJ. Hereditary predisposition to ovarian cancer, looking beyond BRCA1/BRCA2. Gynecol Oncol. (2015) 137:86–92. doi: 10.1016/j.ygyno.2015.01.537
59. Singh J, Thota N, Singh S, Padhi S, Mohan P, Deshwal S, et al. Screening of over 1000 Indian patients with breast and/or ovarian cancer with a multi-gene panel: prevalence of BRCA1/2 and non-BRCA mutations. Breast Cancer Res Treat. (2018) 170:189–96. doi: 10.1007/s10549-018-4726-x
60. Eoh KJ, Kim JE, Park HS, Lee ST, Park JS, Han JW, et al. Detection of germline mutations in patients with epithelial ovarian cancer using multi-gene panels: beyond BRCA1/2. Cancer Res Treat. (2018) 50:917–25. doi: 10.4143/crt.2017.220
61. Graffeo R, Livraghi L, Pagani O, Goldhirsch A, Partridge AH, Garber JE. Time to incorporate germline multigene panel testing into breast and ovarian cancer patient care. Breast Cancer Res Treat. (2016) 160:393–410. doi: 10.1007/s10549-016-4003-9
62. Feliubadalo L, Lopez-Fernandez A, Pineda M, Diez O, Del Valle J, Gutierrez-Enriquez S, et al. Opportunistic testing of BRCA1, BRCA2 and mismatch repair genes improves the yield of phenotype driven hereditary cancer gene panels. Int J Cancer. (2019) 145:2682–91. doi: 10.1002/ijc.32304
63. Frey MK, Kim SH, Bassett RY, Martineau J, Dalton E, Chern JY, et al. Rescreening for genetic mutations using multi-gene panel testing in patients who previously underwent non-informative genetic screening. Gynecol Oncol. (2015) 139:211–5. doi: 10.1016/j.ygyno.2015.08.006
64. Moschetta M, George A, Kaye SB, Banerjee S. BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer. Ann Oncol. (2016) 27:1449–55. doi: 10.1093/annonc/mdw142
65. Weren RD, Mensenkamp AR, Simons M, Eijkelenboom A, Sie AS, Ouchene H, et al. Novel BRCA1 and BRCA2 tumor test as basis for treatment decisions and referral for genetic counselling of patients with ovarian carcinomas. Hum Mutat. (2017) 38:226–35. doi: 10.1002/humu.23137
66. Riahi A, Chabouni-Bouhamed H, Kharrat M. Prevalence of BRCA1 and BRCA2 large genomic rearrangements in Tunisian high risk breast/ovarian cancer families: implications for genetic testing. Cancer Genet. (2017) 210:22–7. doi: 10.1016/j.cancergen.2016.11.002
Keywords: epithelial ovarian cancer, BRCA mutations, germline mutations, somatic mutations, progression-free survival, overall survival
Citation: You Y, Li L, Lu J, Wu H, Wang J, Gao J, Wu M and Liang Z (2020) Germline and Somatic BRCA1/2 Mutations in 172 Chinese Women With Epithelial Ovarian Cancer. Front. Oncol. 10:295. doi: 10.3389/fonc.2020.00295
Received: 13 January 2020; Accepted: 20 February 2020;
Published: 10 March 2020.
Edited by:
Giuseppe Vizzielli, Agostino Gemelli University Polyclinic, ItalyReviewed by:
Michael Dean, National Cancer Institute at Frederick, United StatesCopyright © 2020 You, Li, Lu, Wu, Wang, Gao, Wu and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Wu, d3VtaW5nQHB1bWNoLmNu; Zhiyong Liang, bGlhbmd6aGl5b25nMTIyMEB5YWhvby5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.