
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 25 February 2020
Sec. Cancer Molecular Targets and Therapeutics
Volume 10 - 2020 | https://doi.org/10.3389/fonc.2020.00218
Sirtuins, class III histone deacetylases, are involved in multiple biological processes in cancer initiation and progression. However, the diverse expression patterns and prognostic values of sirtuins in cancers have yet to be elucidated. In this study, we first evaluated the expression and prognostic values of sirtuins in multiple cancer cohorts using publicly available TCGA pan-cancer datasets. Pan-cancer survival analysis indicated that 6 out of 7 sirtuin family members were significant associated with prognosis of clear cell renal cell carcinoma (KIRC) patients. SIRT1, SIRT3, SIRT4, and SIRT5 were associated with favorable prognosis of KIRC patients, while SIRT6 and SIRT7 were associated with unfavorable prognosis. The expression levels of SIRT4 and SIRT5 in KIRC tissues were lower than that in normal tissues, while SIRT6 and SIRT7 were higher in KIRC tissues. The expression levels of SIRT1, SIRT3, SIRT5, SIRT6, and SIRT7 were significantly correlated with tumor stage and histological grade. DNA methylation may contribute to the dysregulation of sirtuins. Finally, GSEA was conducted to predict the potential functions of sirtuins in KIRC. Our results may provide novel insights for the development of sirtuins-based cancer therapy in KIRC.
Renal cell carcinoma (RCC) is the seventh most common cancer in the developed world, accounting for 2–3% of the total human cancer burden in 2012 (1). RCC can be classified into several histopathological entities, of which clear cell renal cell carcinoma (KIRC) is the most frequent subtype of RCC (75–80%) (2). Surgical resection is an effective method for the treatment of RCC. However, the prognosis of RCC remains unsatisfactory due to late diagnosis and recurrence. Therefore, it is critical to find predictive biomarkers for early diagnosis and prognosis. Epigenetic alterations are an essential event in the development and progression of RCC. They are considered as potential biomarkers for the early diagnosis of RCC and prediction of prognosis (3).
Sirtuins are NAD+ dependent deacetylases that belong to the class III histone deacetylase family. Mammalian sirtuins consist of seven members (SIRT1-SIRT7), which all have a conserved core NAD+ binding domain, but have different N-terminal and C-terminal domains. The seven members have different catalytic activity, target proteins, and functions (4). Sirtuins are involved in the development and progression of aging and various aging-related disease, including cardiovascular disease, diabetes, and neurodegeneration (5). Kidney disease is part of the main aging-related diseases. Aberrant expressions and functions of sirtuins have been reported in acute and chronic kidney disease. SIRT1 exerts renal protective effects through inhibiting fibrosis, cell apoptosis and inflammation, and regulation of blood pressure. SIRT3 attenuates mitochondrial dysfunction and ameliorates oxidative stress to protect against acute kidney injury (6). Recently, a growing number of studies have found that sirtuins are involved in various biological processes related to tumorigenesis, such as cancer-associated metabolic pathway alterations, uncontrolled proliferation, genome instability, tumor microenvironment. Sirtuins are supposed to have complex roles in human cancers, and they act as both oncogene and tumor-suppressor depending on cancer types and experimental conditions (7). Several studies have been conducted to explore the clinical significance of sirtuins in RCC, however, the results are inconsistent. The protein expression of SIRT1, SIRT3, and SIRT6 have been reported to be significantly lower in human RCC tissues compared with that in adjacent normal tissues, and high protein expression levels of SIRT1 and SIRT3 were found to be significantly associated with better survival in RCC patients (8, 9). However, SIRT1 was also reported to be up-regulated in RCC and the expression of SIRT1 was significantly associated with poor prognosis in RCC patients (10). Knockdown of SIRT1 suppressed cell proliferation and promoted apoptosis in RCC cell lines (11). SIRT2 mRNA and protein expression were significantly elevated in RCC tissues compared with normal tissues, and high SIRT2 expression is associated with more advanced tumors and poor prognosis (12). SIRT3 overexpression inhibited the growth of RCC cell lines and enhanced mitochondrial biogenesis (13). However, SIRT3 was reported to be up-regulated in the mitochondrial fraction of human RCC tissues, and knockdown of SIRT3 inhibited RCC cell proliferation and tumor growth in vivo (14). SIRT5 promotes KIRC tumorigenesis through inhibiting SDHA succinylation and silencing SIRT5 inhibited RCC cell proliferation (15).
To date, the expression pattern, prognostic significance, and biological function of sirtuins in KIRC have not yet been fully elucidated. In this study, we extended the knowledge on KIRC by using the gene expression data and clinic data from the public database online. We conducted a comprehensive analysis of the relationship between the expression of sirtuins and clinicopathological parameters of KIRC patients, and conducted a preliminary analysis of their regulation and potential functions.
The RNA-seq data and clinical information were downloaded from TCGA (https://portal.gdc.cancer.gov/). The 16 cancer types selected contained at least 9 samples in the normal group. The genomic alteration of sirtuins was assessed using the cBioPortal (16, 17). Analysis of DNA methylation between normal tissues and KIRC cancer tissues was performed using UALCAN (18).
GSEA was carried out to identify the potential underlying mechanisms of sirtuins expression on the pathogenesis and prognosis of KIRC (19, 20). The gene sets were obtained from the MSigDB database v6.2.Gene set. Permutations were performed 1,000 times for each analysis. The nominal p-value (NOM p < 0.05) and False discovery rate (FDR q < 0.25) were used to select significantly enriched gene sets.
All data analysis was conducted using SPSS Version 20 software or Graphpad Prism 7.0. In Figures 2A,C, we compared the expression of each sirtuin between tumor tissues and corresponding paired normal tissues, using the paired t-test or Wilcoxon matched-pairs signed test. In Figures 1A, 2B, 3, when two groups were with normal distribution, we used the standard Student's test for equal variance or Welch t-test for unequal variance. Otherwise, we used the Mann-Whitney U-test (non-normal distribution). In Table 1, patients were divided into low and high groups based on the median value of sirtuins expression. The chi-square and Fisher exact tests were applied to identify the correlations between the sirtuins expression and clinical features of KIRC patients. The Kaplan-Meier curve was used to compare the influence of sirtuins expression on overall survival and disease-free survival, and a log-rank test was used to estimate the difference in survival. A value of P < 0.05 was considered statistically significant.
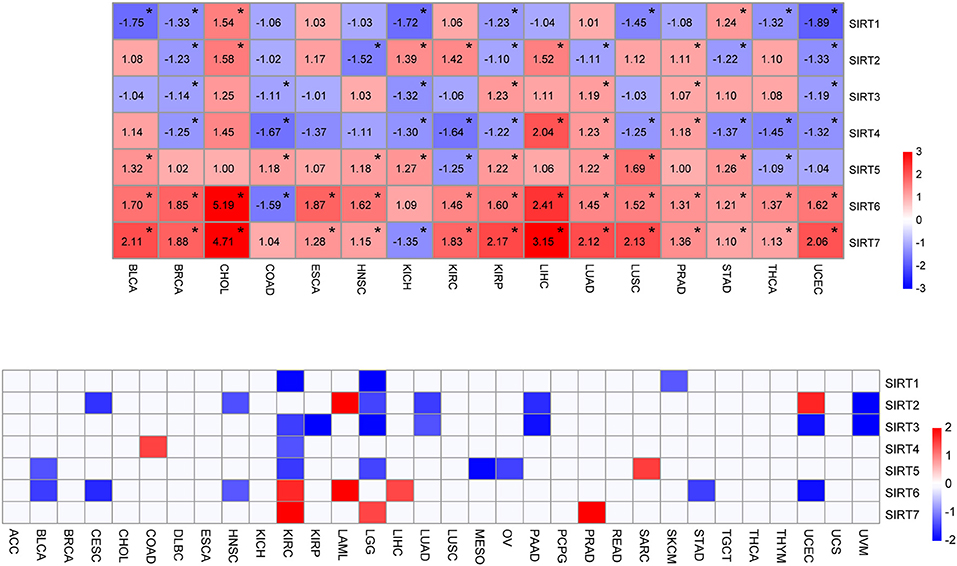
Figure 1. Expression level and survival analysis of sirtuins across TCGA cancer types. (A) Heatmap of sirtuins with changed expressions in 16 cancer cohorts compared to normal tissues (*P < 0.05). (B) Summary of hazard ratios (HR) illustrating cancer-sirtuin pairs with altered prognosis. ACC, Adrenocortical Carcinoma; BLCA, Bladder Urothelial Carcinoma; BRCA, Breast Invasive Carcinoma; CESC, Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma; CHOL, Cholangiocarcinoma; COAD, Colon Adenocarcinoma; DLBC, Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; ESCA, Esophageal Carcinoma; HNSC, Head and Neck Squamous Cell Carcinoma; KICH, Kidney Chromophobe; KIRC, Kidney Renal Clear Cell Carcinoma; KIRP, Kidney Renal Papillary Cell Carcinoma; LAML, Acute Myeloid Leukemia; LGG, Brain Lower Grade Glioma; LIHC, Liver Hepatocellular Carcinoma; LUAD, Lung Adenocarcinoma; LUSC, Lung Squamous Cell Carcinoma; MESO, Mesothelioma; OV, Ovarian Serous Cystadenocarcinoma; PAAD, Pancreatic Adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, Prostate Adenocarcinoma; READ, Rectum Adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous Melanoma; STAD, Stomach Adenocarcinoma; TGCT, Testicular Germ Cell Tumors; THCA, Thyroid carcinoma; THYM, Thymoma; UCEC, Uterine Corpus Endometrial Carcinoma; UCS, Uterine Carcinosarcoma; UVM, Uveal Melanoma.
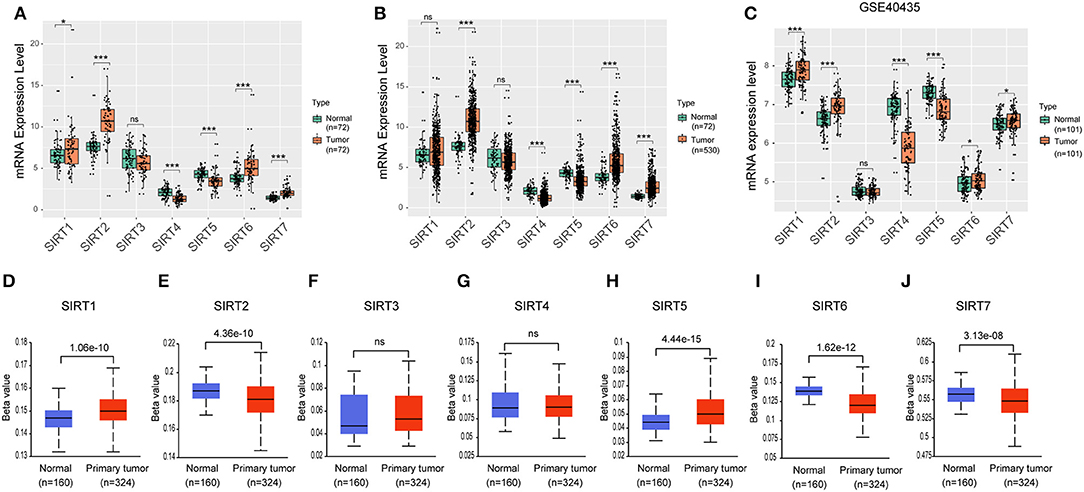
Figure 2. Expression level of sirtuins in KIRC. Boxplots show sirtuins expression in normal tissues compared with paired cancer tissues (A), or unpaired KIRC tissues (B). (C) Boxplots show sirtuins expression in normal and KIRC tissues from GSE40435. (D–J) Promoter methylation status of sirtuins in KIRC (*P < 0.05, ***P < 0.001).
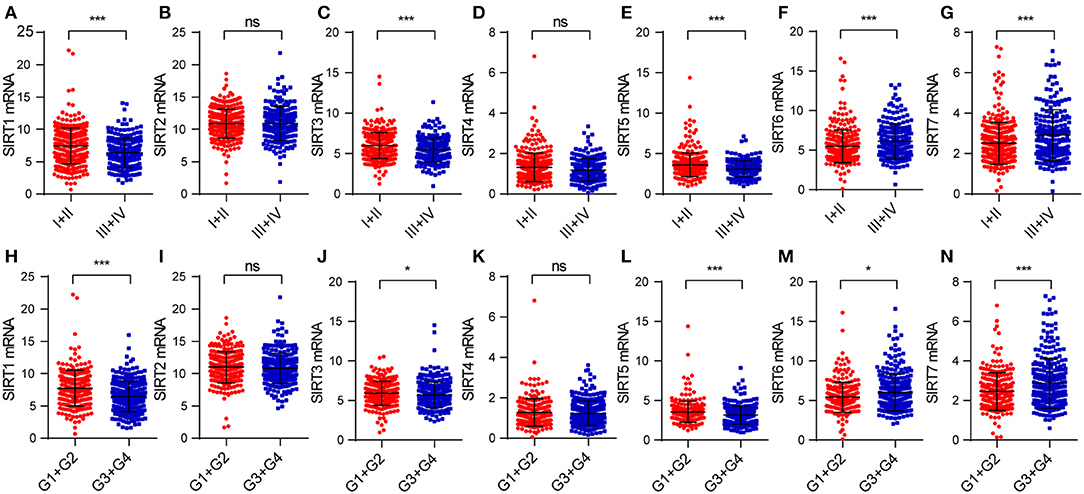
Figure 3. The relationship between sirtuins expression and clinicopathological features in KIRC patients: (A–G) pathological stage, (H–N) histological grade (*P < 0.05, ***P < 0.001).
To determine the expression pattern and prognostic value of sirtuins across cancer types, we downloaded the clinical information of 32 cancer cohorts and corresponding cancer tissue RNA-seq datasets from the TCGA database. We removed cancer types with <9 samples in the normal tissue group to ensure that there were sufficient samples in each group for statistical analysis. We analyzed the expression of sirtuins in 16 cancer types, including BLCA, BRCA, CHOL, COAD, ESCA, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, LUSC, PRAD, STAD, THCA, and UCEC (Figure 1A). Of all the 112 comparisons between cancers and control tissues, 79 were statistically significant (70.5%). SIRT5, SIRT6, and SIRT7 exhibited up-regulation in multiple cancer types. SIRT6 and SIRT7 were up-regulated in 14 of the 15 significantly altered cancer types. SIRT1 and SIRT4 significantly decreased in 7 cancer types and 9 cancer types, respectively. According to the median value, sirtuins were divided into high-expression and low-expression groups. Kaplan-Meier curve and log-rank test analysis were used for overall survival analysis. An overview of these results was shown in Figure 1B and Supplementary Table 1, there was an apparent heterogeneity between different cancer types. We observed a unique role of sirtuins in KIRC comparing to other cancer types. Six members of sirtuins were significantly associated with overall survival in patients with KIRC, the subsequent studies will focus on the role of sirtuins in KIRC.
We further investigated the expression of sirtuins in KIRC in more detail. In 72 paired KIRC tissues and corresponding adjacent normal tissues, expressions of SIRT4 and SIRT5 were down-regulated in cancer tissues than in normal tissues, while expressions of SIRT6 and SIRT7 were up-regulated in cancer tissues (Figure 2A). Similar results were shown between 530 KIRC tissues and 72 normal tissues (Figure 2B). We further confirmed these results using an independent KIRC cohort from the GEO database (GSE40435). The expression of SIRT4 and SIRT5 were down-regulated in cancer tissues, while SIRT6 and SIRT7 were up-regulated (Figure 2C). This was consistent with our result that SIRT4 and SIRT5 were favorable prognostic factors for KIRC patients, SIRT6 and SIRT7 were unfavorable. It is clear that promoter methylation plays an important role in renal tumorigenesis by silencing tumor suppressor genes (21). There was a relatively high frequency of promoter methylation as compared to the number of somatic alterations in RCC (22). To determine the methylation status of sirtuins in KIRC tissues, we applied UALCAN to explore the level of methylation in the promoter region and its relationship with sirtuins mRNA expression. The promoter of SIRT1 and SIRT5 were hypermethylated in KIRC tissues than that in normal tissues, while SIRT2, SIRT6, and SIRT7 were hypomethylated in KIRC tissues (Figures 2D–J).
We explored the correlation between sirtuins expression and clinicopathological parameters of patients with KIRC. In total, we identified 530 KIRC samples with mRNA expression data and clinical information. We applied the chi-square test to identify the correlation between the sirtuins expression and clinicopathological parameters of KIRC patients. SIRT3 and SIRT5 were highly expressed in female patients. Low expressions of SIRT1, SIRT3, and SIRT5 were significantly correlated with advanced TNM stage and poor histological grade. High SIRT6 expression was associated with advanced TNM stage and poor histological grade stage. High SIRT7 expression was associated with poor histological grade (Table 1). We further analyzed the relationship between the expression of sirtuins and tumor stage, histological grade. The result showed that SIRT1, SIRT3, and SIRT5 were lower in advanced TNM stage and poor histological grade, while SIRT6 and SIRT7 were higher (Figure 3). This was consistent with that SIRT1, SIRT3, and SIRT5 might be a favorable factor for KIRC patients, whereas SIRT6 and SIRT7 might be unfavorable.
We further explored the prognostic value of sirtuins in patients with KIRC. We assessed the relationship between individual sirtuin and the survival of KIRC patients. Decreased SIRT1, SIRT3, SIRT4, and SIRT5 expressions and increased SIRT6 and SIRT7 expressions were strongly associated with poor overall survival (Figures 4A–G). Further, Kaplan-Meier curves for disease-free survival showed that low SIRT1, SIRT3 and SIRT5, and high SIRT6 were significantly correlated with poor disease-free survival (Figures 4H–N).
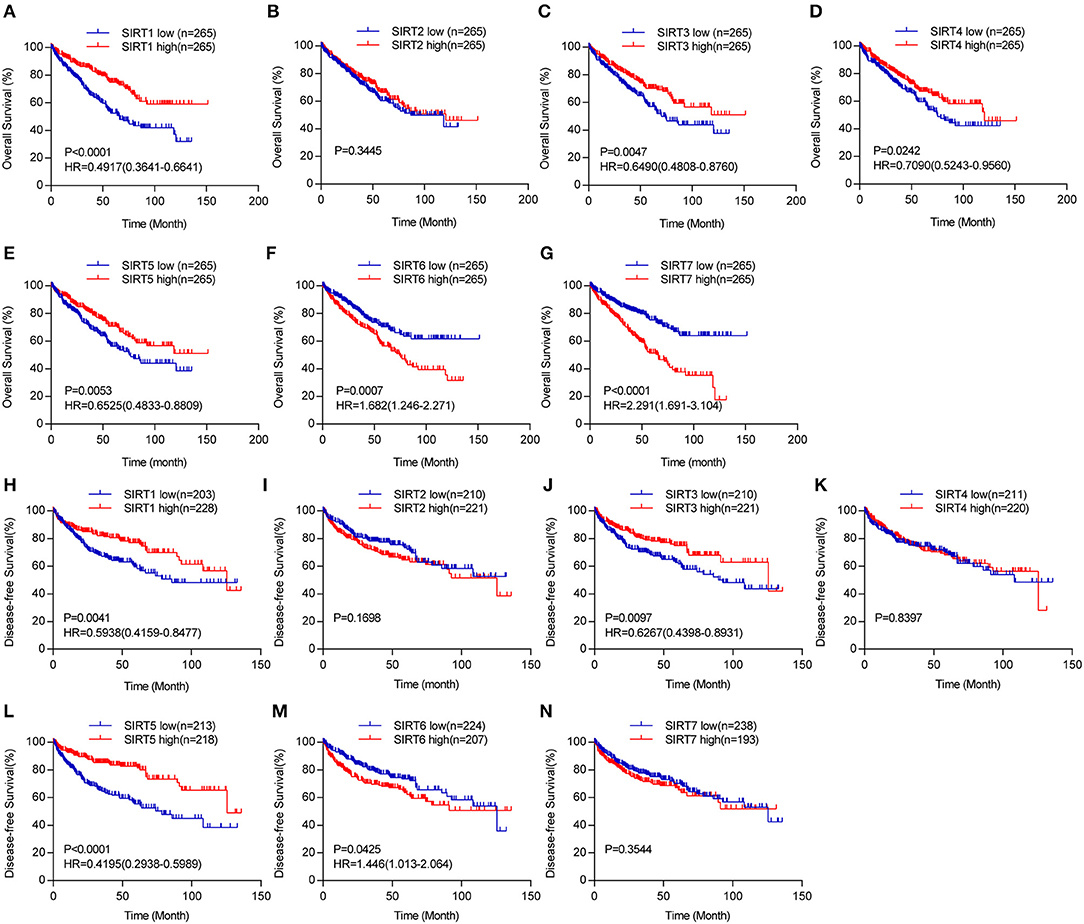
Figure 4. The prognostic value of sirtuins in KIRC. Kaplan-Meier curves show the correlation between sirtuins expression and overall survival (A–G), and disease-free survival (H–N) of KIRC patients.
We next investigated the potential mechanisms that dysregulate sirtuins expression in KIRC. We applied the cBioPortal online tool for KIRC (The Cancer Genome Atlas, Provisional) to analyze the genetic alteration frequency of sirtuins. A total of 448 patients were analyzed. The oncoprints include missense mutation, deletion, and amplification. The percentage of genetic alterations of sirtuins in KIRC varied from 0.2 to 1.1% (Figures 5A,B). The percentages of genetic mutation in SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, and SIRT7 were 0.67% (mutation), 0.22% (amplification), 0.22% (amplification), 1.12% (mutation), 0.22% (mutation), 0.67% (0.22% mutation, 0.45% deep deletion), 0.89% (0.22% mutation, 0.45% amplification, 0.22 multiple alterations). Missense mutation was identified in SIRT1, SIRT4, SIRT5, SIRT6, and SIRT7, and the mutation in the SIR2 domain was verified in SIRT1 and SIRT4. Besides, frameshift insertion was found in SIRT1 and SIRT4 (Figure 5C).
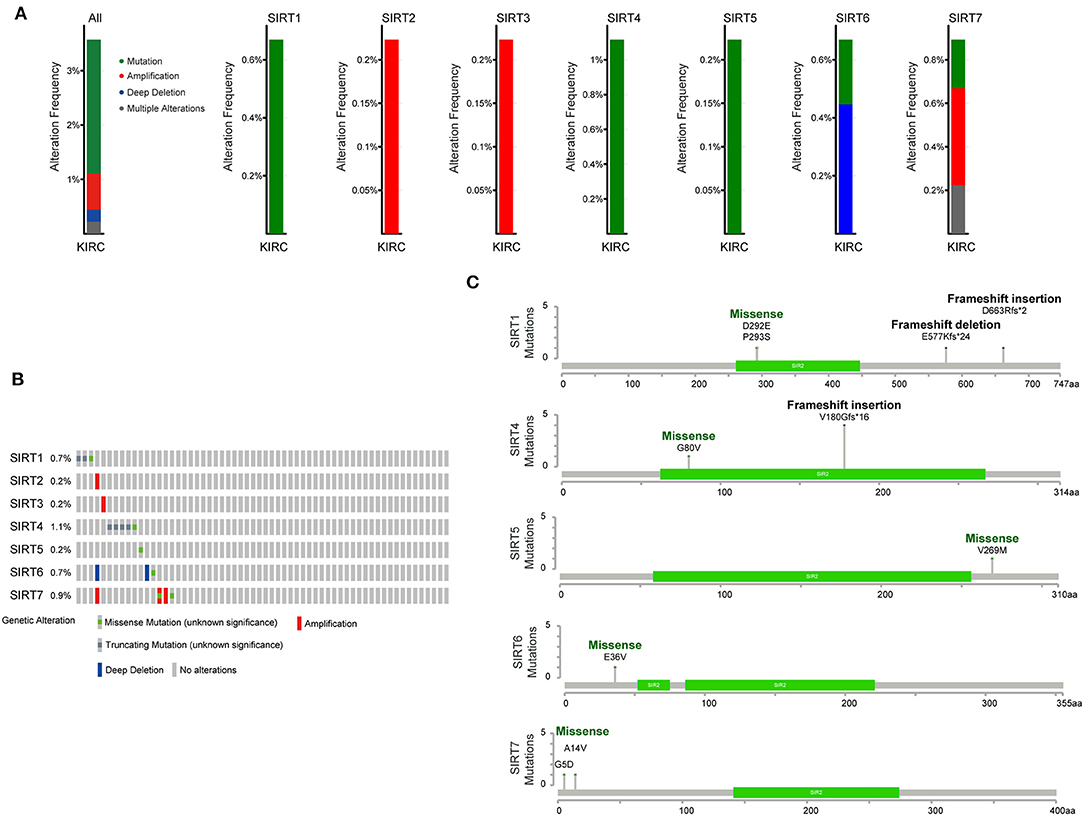
Figure 5. Genomic analysis of sirtuins in KIRC. (A) Summary of genomic alteration of sirtuins in KIRC. (B) Oncoprint visual summary of alteration on a query of sirtuins. (C) The mutations of SIRT1, SIRT4, SIRT5, SIRT6, and SIRT7 were plotted. The SIR2 domain is displayed in green.
GSEA was performed to investigate the potential mechanisms of sirtuins in KIRC. Samples were divided into high group and low group according to the median value.
We analyzed KEGG gene sets using the GSEA. Pathways involving the TCA cycle and oxidative phosphorylation were significantly enriched and positively correlated with SIRT3, SIRT4, and SIRT5, whereas negatively correlated with SIRT7. The well-known pathways associated with cell cycle were significantly regulated by sirtuins alteration in KIRC (Figure 6).
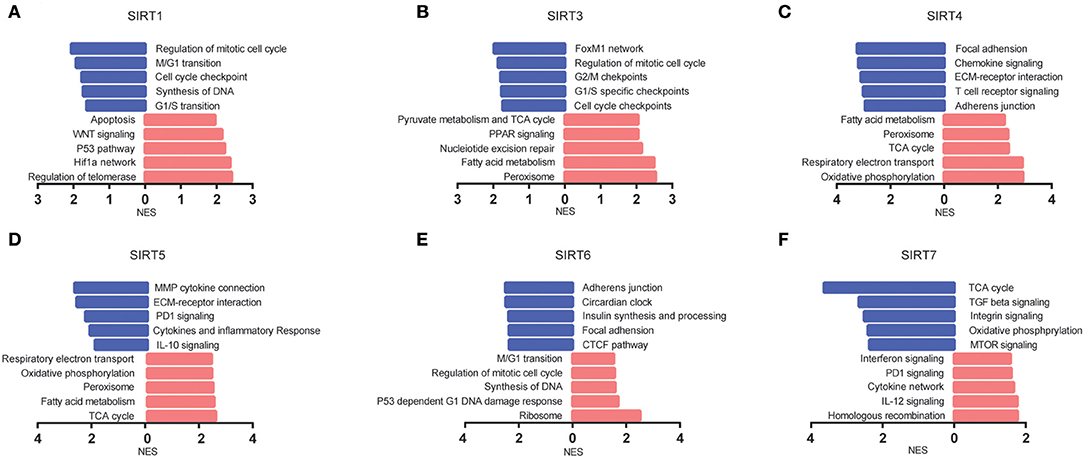
Figure 6. The pathways associated with sirtuins alterations were predicted by GSEA. (A–F) Pathway alterations in the high-expression group vs. low-expression group. Pathways enriched in phenotype high were shown in red (right), and pathways enriched in phenotype low were shown in blue (left). All gene sets were significantly enriched at nominal p < 5% and FDR < 25%.
Sirtuins are involved in a few cancer-related biological processes, including cancer metabolism, genome stability, and tumor microenvironment. Sirtuins act as tumor suppressors or tumor promoters depending on the cellular context and experimental conditions. Due to the complex role of sirtuins in cancer, a better understanding of each sirtuin will be important for the future development of sirtuins-based therapy. Our results showed that sirtuins are heterogeneous in different cancers. According to the pan-cancer survival analysis and expression profiling analysis of TCGA, 6 out of 7 sirtuin family members were significantly correlated with the survival of KIRC patients. Next, we found that the expression of sirtuins was related to the histological grade and pathological stage of KIRC. We found that promoter methylation status may contribute to the dysregulation of sirtuins in KIRC.
SIRT1, the best known and most studied member of the sirtuins family, was reported to play a dual role in tumorigenesis. SIRT1 plays a tumor-suppressor role in various human cancers, including breast cancer, bladder cancer, and glioblastoma (23, 24). Meanwhile, overexpression of SIRT1 was also reported in several cancers and associated with an unfavorable prognosis (25, 26). The expression of SIRT1 was reported to predict shorter overall survival, disease-free survival, and cancer-specific survival in KIRC (10). SIRT1 act as a direct target of miRNA-200a and miRNA-22, miRNA-200a and miR-22 could inhibit cell proliferation, arrested cell cycle progression, and promoted cell apoptosis in RCC cell lines (11, 27). However, it was also reported that SIRT1 expression was significantly lower in RCC than in normal tissues and high expression of SIRT1 was correlated with a better prognosis for RCC patients (8). SIRT1 overexpression inhibited RCC cell line proliferation by repressing FGB expression (9). In our study, the mRNA expression of SIRT1 was up-regulated in KIRC than in normal tissues. Low SIRT1 was associated with high TNM stage and advanced histologic grade. Besides, Kaplan-Meier curves showed that low expression of SIRT1 was significantly correlated with poor overall survival and disease-free survival.
SIRT2 is mainly located in the cytoplasm. Recent studies found that SIRT2 may mainly act tumor suppressor by maintaining genome stability (28, 29). The previous study reported that the higher mRNA and protein expression levels of SIRT2 were found in RCC samples than normal tissues. High SIRT2 levels predicted poor survival of RCC patients (12). In our study, we found that the mRNA level of SIRT2 was higher in KIRC tissues than control tissues. However, SIRT2 did not show a significant association with advanced tumor characters or survival.
SIRT3, SIRT4, and SIRT5 are known as mitochondrial sirtuins. They orchestrate numerous aspects of mitochondrial biology, including redox balance, metabolism homeostasis, and mitochondrial dynamics (30). In agreement with SIRT3 functioning as a tumor suppressor, SIRT3 was a favorable prognostic indicator for multiple cancer in our study, including KIRC, KIRP, LGG, LUAD, PAAD, UCEC, and UVM. SIRT3 was deleted or down-regulated in many types of human cancers (31). In renal cancer, SIRT3 was reported to be down-regulated in KIRC tissues and predicted a favorable survival of KIRC patients (8, 13, 32). Overexpression of SIRT3 in RCC cell lines inhibited cell proliferation and reversed the Warburg effect (13). In our study, although SIRT3 did not show a significant difference between KIRC and normal tissues, SIRT3 expression was inversely correlated with high TNM stage and advanced histological grade in KIRC patients. In addition, low SIRT3 expression was significantly associated with poor overall survival and disease-free survival of KIRC patients, which is consistent with the previous view that SIRT3 act as a tumor suppressor. SIRT4 exerts tumor suppressor activity in many cancers (33). Until now, little was known about the role of SIRT4 in RCC. In this study, SIRT4 expression was lower in KIRC tissues than in normal tissues. SIRT4 expression was significantly and positively correlated with overall survival of KIRC patients. Currently, the role of SIRT5 in cancer has not been widely reported. SIRT5 was overexpressed in non-small cell lung cancer and colorectal cancer, high SIRT5 expression was an unfavorable predictor of survival. Overexpression of SIRT5 promoted cancer cell growth and drug resistance (34, 35). SIRT5 was recently reported to be down-regulated in glioblastoma and its down-regulation was associated with poor prognosis (35, 36). SIRT5 was reported to be up-regulated in KIRC, SIRT5 promote RCC tumorigenesis by inhibiting SDHA succinylation. In our study, SIRT5 expression decreased in KIRC tissues, and its expression level was negatively correlated with the TNM stage and histological grade in KIRC patients. Lower SIRT5 expression was significantly correlated with poor overall survival and disease-free survival.
SIRT6 has been reported to regulate inflammation, genome instability, glucose metabolism which are associated with tumor suppressor (37). SIRT6 act as a tumor suppressor in several cancers (38), including pancreatic cancer (39), breast cancer (40), and hepatocellular carcinoma (41, 42). While in other cancer types, such as lung cancer (43, 44) and melanoma (45), SIRT6 was up-regulated and act as a tumor promoter. In this study. SIRT6 expression was found to be higher in KIRC tissues than in normal tissues and was significantly and positively correlated with tumor stage and histologic grade. Furthermore, an elevated level of SIRT6 was significantly associated with worse overall survival and disease-free survival in patients with KIRC. Accumulative studies showed that SIRT7 was a biomarker for poor prognosis (46). In agreement with a tumor-promoting role for SIRT7, our study found that SIRT7 was significantly up-regulated in multiple cancers. SIRT7 may be an unfavorable prognostic factor for KIRC and high SIRT7 expression was positively correlated with tumor pathologic stage and histological grade.
It was reported that profound changes in cellular metabolism take place during KIRC tumorigenesis. Because of the frequent alterations of signaling networks that regulate metabolic behavior, KIRC was described as a “metabolic disease” (47). Cellular metabolism in KIRC involved impaired Krebs cycle and oxidative phosphorylation, and a subsequent Warburg metabolic shift to aerobic glycolysis, and all of those features were associated with poor prognosis (48, 49). However, the molecular mechanisms of these metabolic changes are not fully elucidated. In this study, GSEA analyses showed that sirtuins were closely related to metabolic pathways, such as oxidative phosphorylation, Krebs cycle, fatty acid metabolism. Therefore, the analysis above suggested the functions of sirtuins in cellular metabolism, providing insight into the promising therapeutic target in KIRC. Gender has been considered as an independent risk factor for survival in KIRC patients, with a benefit for women (50). The mechanism of this difference is unclear. According to our results, SIRT3 and SIRT5 were highly expressed in female patients. Further experiments may be helpful to understand the relationship between SIRT3/SIRT5 and gender in KIRC. In general, our study investigated the expression pattern and prognosis role of sirtuins in KIRC by using TCGA data, however, the results are still preliminary, further in vivo and in vitro experiments are needed to validate the role of sirtuins in KIRC. Despite these limitations, this study might be helpful to guide further investigation of sirtuins in KIRC.
In conclusion, we systematically analyzed the prognostic value and expression of sirtuins in multiple cancer types and provided a thorough understanding of the heterogeneity and complexity of the roles of sirtuins in cancer. Our results indicated that SIRT1, SIRT3, SIRT4, and SIRT5 may play a beneficial role in KIRC, while SIRT6 and SIRT7 may act as unfavorable factors in KIRC. Further studies are required to validate our findings and promote the clinical utility of sirtuins serving as prognostic indicators or therapeutic targets for cancer.
Publicly available datasets were analyzed in this study. This data can be found here: TCGA (https://portal.gdc.cancer.gov/).
YT and NL developed the idea and designed the research. YT analyzed the data. YT and FP drafted the manuscript. BL and GG revised the writing. All authors read and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (81873574, 81602572).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00218/full#supplementary-material
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. (2015) 136:E359–86. doi: 10.1002/ijc.29210
2. Moch H. An overview of renal cell cancer: pathology and genetics. Semin Cancer Biol. (2013) 23:3–9. doi: 10.1016/j.semcancer.2012.06.006
3. Joosten SC, Smits KM, Aarts MJ, Melotte V, Koch A, Tjan-Heijnen VC, et al. Epigenetics in renal cell cancer: mechanisms and clinical applications. Nat Rev Urol. (2018) 15:430–51. doi: 10.1038/s41585-018-0023-z
4. Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. (2000) 403:795–800. doi: 10.1038/35001622
5. Guarente L. Franklin H. Epstein lecture: sirtuins, aging, and medicine. N Engl J Med. (2011) 364:2235–44. doi: 10.1056/NEJMra1100831
6. Morigi M, Perico L, Benigni A. Sirtuins in renal health and disease. J Am Soc Nephrol. (2018) 29:1799–809. doi: 10.1681/ASN.2017111218
7. Carafa V, Altucci L, Nebbioso A. Dual tumor suppressor and tumor promoter action of sirtuins in determining malignant phenotype. Front Pharmacol. (2019) 10:38. doi: 10.3389/fphar.2019.00038
8. Jeh SU, Park JJ, Lee JS, Kim DC, Do J, Lee SW, et al. Differential expression of the sirtuin family in renal cell carcinoma: aspects of carcinogenesis and prognostic significance. Urol Oncol. (2017) 35:675 e9–15. doi: 10.1016/j.urolonc.2017.08.016
9. Chen Y, Zhu Y, Sheng Y, Xiao J, Xiao Y, Cheng N, et al. SIRT1 downregulated FGB expression to inhibit RCC tumorigenesis by destabilizing STAT3. Exp Cell Res. (2019) 382:111466. doi: 10.1016/j.yexcr.2019.06.011
10. Noh SJ, Kang MJ, Kim KM, Bae JS, Park HS, Moon WS, et al. Acetylation status of P53 and the expression of DBC1, SIRT1, and androgen receptor are associated with survival in clear cell renal cell carcinoma patients. Pathology. (2013) 45:574–80. doi: 10.1097/PAT.0b013e3283652c7a
11. Fu H, Song W, Chen X, Guo T, Duan B, Wang X, et al. MiRNA-200a induce cell apoptosis in renal cell carcinoma by directly targeting SIRT1. Mol Cell Biochem. (2018) 437:143–52. doi: 10.1007/s11010-017-3102-1
12. Wei R, He D, Zhang X. Role of SIRT2 in regulation of stemness of cancer stem-like cells in renal cell carcinoma. Cell Physiol Biochem. (2018) 49:2348–57. doi: 10.1159/000493835
13. Liu H, Li S, Liu X, Chen Y, Deng H. SIRT3 overexpression inhibits growth of kidney tumor cells and enhances mitochondrial biogenesis. J Proteome Res. (2018) 17:3143–52. doi: 10.1021/acs.jproteome.8b00260
14. Choi J, Koh E, Lee YS, Lee HW, Kang HG, Yoon YE, et al. Mitochondrial Sirt3 supports cell proliferation by regulating glutamine-dependent oxidation in renal cell carcinoma. Biochem Biophys Res Commun. (2016) 474:547–53. doi: 10.1016/j.bbrc.2016.04.117
15. Ma Y, Qi Y, Wang L, Zheng Z, Zhang Y, Zheng J. SIRT5-mediated SDHA desuccinylation promotes clear cell renal cell carcinoma tumorigenesis. Free Radic Biol Med. (2019) 134:458–67. doi: 10.1016/j.freeradbiomed.2019.01.030
16. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. (2013) 6:pl1. doi: 10.1126/scisignal.2004088
17. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. (2012) 2:401–4. doi: 10.1158/2159-8290.CD-12-0095
18. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. (2017) 19:649–58. doi: 10.1016/j.neo.2017.05.002
19. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
20. Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. (2003) 34:267–73. doi: 10.1038/ng1180
21. Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. (2005) 2(Suppl 1):S4–11. doi: 10.1038/ncponc0354
22. Morris MR, Maher ER. Epigenetics of renal cell carcinoma: the path towards new diagnostics and therapeutics. Genome Med. (2010) 2:59. doi: 10.1186/gm180
23. Hou Z, Xu X, Zhou L, Fu X, Tao S, Zhou J, et al. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumour Biol. (2017) 39:1010428317718135. doi: 10.1177/1010428317718135
24. Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. (2009) 185:203–11. doi: 10.1083/jcb.200809167
25. Li C, Liu Z, Yang K, Chen X, Zeng Y, Liu J, et al. miR-133b inhibits glioma cell proliferation and invasion by targeting Sirt1. Oncotarget. (2016) 7:36247–54. doi: 10.18632/oncotarget.9198
26. Bu P, Luo C, He Q, Yang P, Li X, Xu D. MicroRNA-9 inhibits the proliferation and migration of malignant melanoma cells via targeting sirituin 1. Exp Ther Med. (2017) 14:931–8. doi: 10.3892/etm.2017.4595
27. Zhang S, Zhang D, Yi C, Wang Y, Wang H, Wang J. MicroRNA-22 functions as a tumor suppressor by targeting SIRT1 in renal cell carcinoma. Oncol Rep. (2016) 35:559–67. doi: 10.3892/or.2015.4333
28. Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. (2011) 20:487–99. doi: 10.1016/j.ccr.2011.09.004
29. Serrano L, Martinez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. (2013) 27:639–53. doi: 10.1101/gad.211342.112
30. Tang X, Chen XF, Chen HZ, Liu DP. Mitochondrial Sirtuins in cardiometabolic diseases. Clin Sci. (2017) 131:2063–78. doi: 10.1042/CS20160685
31. Torrens-Mas M, Oliver J, Roca P, Sastre-Serra J. SIRT3: oncogene and tumor suppressor in cancer. Cancers. (2017) 9:90. doi: 10.3390/cancers9070090
32. Zhou Y, Cheng S, Chen S, Zhao Y. Prognostic and clinicopathological value of SIRT3 expression in various cancers: a systematic review and meta-analysis. Onco Targets Ther. (2018) 11:2157–67. doi: 10.2147/OTT.S157836
33. Huang G, Zhu G. Sirtuin-4 (SIRT4), a therapeutic target with oncogenic and tumor-suppressive activity in cancer. Onco Targets Ther. (2018) 11:3395–400. doi: 10.2147/OTT.S157724
34. Wang YQ, Wang HL, Xu J, Tan J, Fu LN, Wang JL, et al. Sirtuin5 contributes to colorectal carcinogenesis by enhancing glutaminolysis in a deglutarylation-dependent manner. Nat Commun. (2018) 9:545. doi: 10.1038/s41467-018-02951-4
35. Lu W, Zuo Y, Feng Y, Zhang M. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol. (2014) 35:10699–705. doi: 10.1007/s13277-014-2372-4
36. Chen X, Xu Z, Zeng S, Wang X, Liu W, Qian L, et al. SIRT5 downregulation is associated with poor prognosis in glioblastoma. Cancer Biomark. (2019) 24:449–59. doi: 10.3233/CBM-182197
37. Lerrer B, Gertler AA, Cohen HY. The complex role of SIRT6 in carcinogenesis. Carcinogenesis. (2016) 37:108–18. doi: 10.1093/carcin/bgv167
38. Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. (2012) 151:1185–99. doi: 10.1016/j.cell.2012.10.047
39. Kugel S, Sebastian C, Fitamant J, Ross KN, Saha SK, Jain E, et al. SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell. (2016) 165:1401–15. doi: 10.1016/j.cell.2016.04.033
40. Khongkow M, Olmos Y, Gong C, Gomes AR, Monteiro LJ, Yague E, et al. SIRT6 modulates paclitaxel and epirubicin resistance and survival in breast cancer. Carcinogenesis. (2013) 34:1476–86. doi: 10.1093/carcin/bgt098
41. Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen X, et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat Cell Biol. (2012) 14:1203–11. doi: 10.1038/ncb2590
42. Marquardt JU, Fischer K, Baus K, Kashyap A, Ma S, Krupp M, et al. Sirtuin-6-dependent genetic and epigenetic alterations are associated with poor clinical outcome in hepatocellular carcinoma patients. Hepatology. (2013) 58:1054–64. doi: 10.1002/hep.26413
43. Azuma Y, Yokobori T, Mogi A, Altan B, Yajima T, Kosaka T, et al. SIRT6 expression is associated with poor prognosis and chemosensitivity in patients with non-small cell lung cancer. J Surg Oncol. (2015) 112:231–7. doi: 10.1002/jso.23975
44. Bai L, Lin G, Sun L, Liu Y, Huang X, Cao C, et al. Upregulation of SIRT6 predicts poor prognosis and promotes metastasis of non-small cell lung cancer via the ERK1/2/MMP9 pathway. Oncotarget. (2016) 7:40377–86. doi: 10.18632/oncotarget.9750
45. Garcia-Peterson LM, Ndiaye MA, Singh CK, Chhabra G, Huang W, Ahmad N. SIRT6 histone deacetylase functions as a potential oncogene in human melanoma. Genes Cancer. (2017) 8:701–12. doi: 10.18632/genesandcancer.153
46. Paredes S, Villanova L, Chua KF. Molecular pathways: emerging roles of mammalian Sirtuin SIRT7 in cancer. Clin Cancer Res. (2014) 20:1741–6. doi: 10.1158/1078-0432.CCR-13-1547
47. Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol. (2010) 7:277–85. doi: 10.1038/nrurol.2010.47
48. Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. (2013) 499:43–9. doi: 10.1038/nature12222
49. Ricketts CJ, De Cubas AA, Fan H, Smith CC, Lang M, Reznik E, et al. The cancer genome atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep. (2018) 23:313–26 e5. doi: 10.1016/j.celrep.2018.03.075
50. May M, Aziz A, Zigeuner R, Chromecki T, Cindolo L, Schips L, et al. Gender differences in clinicopathological features and survival in surgically treated patients with renal cell carcinoma: an analysis of the multicenter CORONA database. World J Urol. (2013) 31:1073–80. doi: 10.1007/s00345-013-1071-x
Keywords: sirtuins, epigenetics, TCGA, cancer, clear cell renal cell carcinoma
Citation: Tan Y, Li B, Peng F, Gong G and Li N (2020) Integrative Analysis of Sirtuins and Their Prognostic Significance in Clear Cell Renal Cell Carcinoma. Front. Oncol. 10:218. doi: 10.3389/fonc.2020.00218
Received: 06 October 2019; Accepted: 07 February 2020;
Published: 25 February 2020.
Edited by:
Sofia Diana Merajver, Michigan Medicine, University of Michigan, United StatesReviewed by:
Maria Munoz Caffarel, Biodonostia Health Research Institute (IIS Biodonostia), SpainCopyright © 2020 Tan, Li, Peng, Gong and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ning Li, bGluaW5neHlAY3N1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.