- 1Department of Pharmacy, The Second Xiangya Hospital, Central South University, Changsha, China
- 2Institute of Clinical Pharmacy, Central South University, Changsha, China
Background: Platinum-based agents, including cisplatin, carboplatin, and oxaliplatin, are indispensable for the treatment of lung cancer. The development of toxicity frequently necessitates dose reduction or discontinuation of therapy, despite the clinical response. Pharmacogenomics studies were reviewed to identify the possible genetic variants that underlie individual susceptibility to platinum-related toxicities.
Method: We conducted a systematic search in PubMed and Embase for pharmacogenomics reports that focused on commonly reported platinum-induced toxicities, such as gastrointestinal (GI), hematological, neurological, and other toxicities, in patients diagnosed with lung cancer. Meta-analyses were conducted to determine the association between genetic polymorphisms and platinum-induced toxicity by checking the odds ratio (OR) and 95% confidence interval (CI) using random or fixed-effects models as appropriate.
Results: Twenty eligible studies that met the inclusion criteria with sufficient data were extracted and presented comprehensively. A total of 16 polymorphisms from 11 genes were included in the meta-analysis. MTHFR rs1801131 and MDM2 rs1690924 were significantly correlated with platinum-induced GI toxicity (P = 0.04 and P = 0.02, respectively). Patients with the MTHFR rs1801131AA and MDM2 rs1690924TC/CC genotype tended to have a higher risk of GI toxicity than patients with other genotypes did (OR = 1.73, 95% CI = 0.86–2.18; and OR = 0.51, 95% CI = 0.29–0.88, respectively). Compared to carriers of the MTHFR rs1801133CC genotype, carriers of the CT/TT genotype had a significantly increased risk of hematological toxicity (P = 0.01, OR = 1.68, 95% CI = 1.12–2.52).
Conclusion: In the future, physicians should pay careful attention to MTHFR and MDM2 for personalized chemotherapy treatment among patients with lung cancer.
Background
Lung cancer is the second most commonly diagnosed malignant tumor in men and women and is one of the main causes of mortality worldwide (1). There are two major forms of lung cancer, including small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC); NSCLC accounts for 85% of all cases of lung cancer (2). Because of the difficulty in early diagnosis, most patients are diagnosed with advanced stage disease when surgery is no longer a treatment option. Therefore, chemotherapy is the major treatment choice for these patients (3).
Platinum-based agents, including cisplatin, carboplatin, and oxaliplatin, in combination with other cytotoxic drugs have been recommended as the first-line chemotherapy for lung cancer (4). The antitumor effect of platinum-based agents is by interfering with DNA repair, thereby suppressing and eventually killing cancer cells (5). Unfortunately, platinum can also hamper the growth of normal cells. Although platinum-based agents are effective for cancer therapy, platinum-induced toxicity is very common in clinical settings. The use of platinum-based agents may lead to serious or permanent adverse events, such as hematotoxicity, GI, nephrotoxicity, hearing loss, and other toxicities (6). Severe toxicities can result in dose reduction, treatment delay, or even chemotherapy discontinuation, as well as carry the risk of life-threatening complications (7). Moreover, differences exist among patients considering the severity of platinum-induced toxicities. Therefore, personalized medicine aims to identify patients who have received platinum agent treatment and are more likely to benefit from anticancer agents or more likely to experience adverse events.
Genetic polymorphisms contribute to the differences in platinum-related toxicities, and there is accumulating evidence to support this speculation (8). Therefore, determining the association between polymorphisms and platinum-related toxicities will be beneficial for individualized chemotherapy. Previously, two genome-wide association studies and one whole-exome sequencing study were conducted to identify the genetic markers for platinum-induced toxicities (9–11). In addition, Yin et al. aimed to establish models to explain and predict platinum toxicity interindividual difference by simultaneously incorporating multiple genetic and clinical factors to explore the association of their interactions with platinum-induced toxicities (12, 13).
Although many studies have investigated this issue, there is still no consensus regarding the relationship between genetic polymorphisms and platinum-induced toxicities in patients with lung cancer. For example, Cristina et al. found that ERCC1 C118T was significantly associated with platinum-induced toxicity while other studies presented contradictory results (14–16). Similar associations were found for XRCC1 codon 399, XPD Lys751Gln, and other mutations (17, 18). Hence, quantitative evaluation is needed for determining the association between gene polymorphisms and platinum-induced toxicities.
The aims of this study were as follows: (1) to summarize the pharmacogenomics of platinum-based toxicities in patients with lung cancer and (2) to provide a comprehensive assessment of the association between genetic polymorphisms and platinum-based drug response in patients with lung cancer. We collected all available publications on pharmacogenomics studies that platinum-based toxicities in patients with NSCLC and SCLC and quantitatively studied them using a meta-analysis strategy.
Methods
Search Strategy
For the literature search, two authors (Z. Y. Luo and W. H. Liu) independently performed a systematic literature search in three databases: PubMed database, Cochrane Library, and ISI Web of Knowledge. The search results were reviewed and compared by a third reviewer (Y. Zheng), and discrepancies between searchers were discussed and solved with consensus. The literature was searched from the first available article to June 18, 2019. Publications were retrieved using terms associated with platinum drugs (“platinum” or “cisplatin” or “carboplatin” or “oxaliplatin”) in combination with keywords associated with genetic variation (“polymorphism” or “SNP” or “single nucleotide polymorphism” or “mutation” or “variation”) and “toxicity” or “adverse effect,” and “lung cancer.”
Inclusion Criteria
All identified abstracts were carefully and independently reviewed by two investigators (Z. Y. Luo and W. H. Liu) for eligibility. The inclusion criteria were as follows: (i) clinical studies, regardless of the sample size; (ii) studies that assessed the associations between genetic polymorphisms and platinum-induced severe (grade 3–4) toxicities in patients with lung cancer, and toxicities were evaluated and graded according to the Common Terminology Criteria for Adverse Events; (iii) numbers for each genotype were available or could be calculated in different groups; and (iv) at least two studies that evaluated one polymorphism met the abovementioned three criteria. If the two investigators disagreed about the eligibility of an article, it was resolved by consensus with a third reviewer (J. Q. Luo).
Data Extraction
Data were manually extracted by two reviewers (Z. Y. Luo, W. H. Liu) who were blinded to each other and used the same data recording form. All data were reviewed by the third reviewer (J. Q. Luo) until they reached a consensus on all of the data extraction items. The following information was extracted from each study: name of the first author, publication year, country of the study and ethnicity of the patients, sample size, tumor type, disease stage, chemotherapeutic drugs, platinum-induced toxicities, toxicity evaluation criteria, genes and Single Nucleotide Polymorphism Database number of the investigated polymorphisms, and genotype methods.
Statistical Analysis
The patients were divided into two groups: those with grade 3–4 (severe) toxicities and those with grade 0–2 (no or mild) toxicities. The pooled odds ratio (OR) and associated 95% CI were calculated and used to evaluate the strength of association in different genotype groups. The significance of the pooled estimates of the OR was determined using the Z-test. The Cochran's Q-test and I2 metric were performed to determine the possibility of between-study heterogeneity. The heterogeneity of publications in each meta-analysis was considered to be significant at P < 0.05 for the Q statistics and I2 > 50% for the I2 metric. Sensitivity analysis was conducted if publication bias existed; one study was excluded at a time, and the others were analyzed to estimate whether the results were affected markedly by a single study. Subgroup analysis based on toxicity [categorized as overall, gastrointestinal (GI), and hematological toxicities] was conducted to assess the sources of heterogeneity across the studies. Potential publication bias was assessed using the Begg and Egger tests, with P < 0.05 considered to indicate a significant publication bias. All statistical analyses were performed with by the Cochrane Collaboration software (Review Manager 5, the Cochrane Collaboration, Oxford, United Kingdom).
Results
Literature Review and Characteristics of the Included Studies
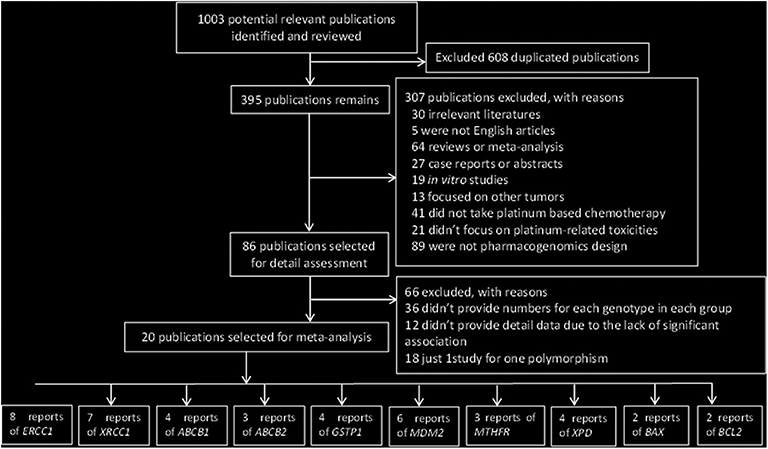
The initial research yielded 1,003 publications. A total of 608 articles were excluded owing to duplicate publication; in addition, 306 studies were excluded in the first round of review, among which 30 were irrelevant literature articles, 5 were not English articles, 64 were reviews or meta-analysis, 27 were case reports or abstracts, 19 were non-clinical-related studies, 13 focused on other tumors, 41 did not evaluate platinum-based chemotherapy, 20 did not focus on platinum-related toxicities, and 89 were not pharmacogenomic studies. After reading the full text of these articles, we found that 12 studies did not provide detailed data owing to the lack of significant association between gene polymorphisms and platinum-based toxicities.
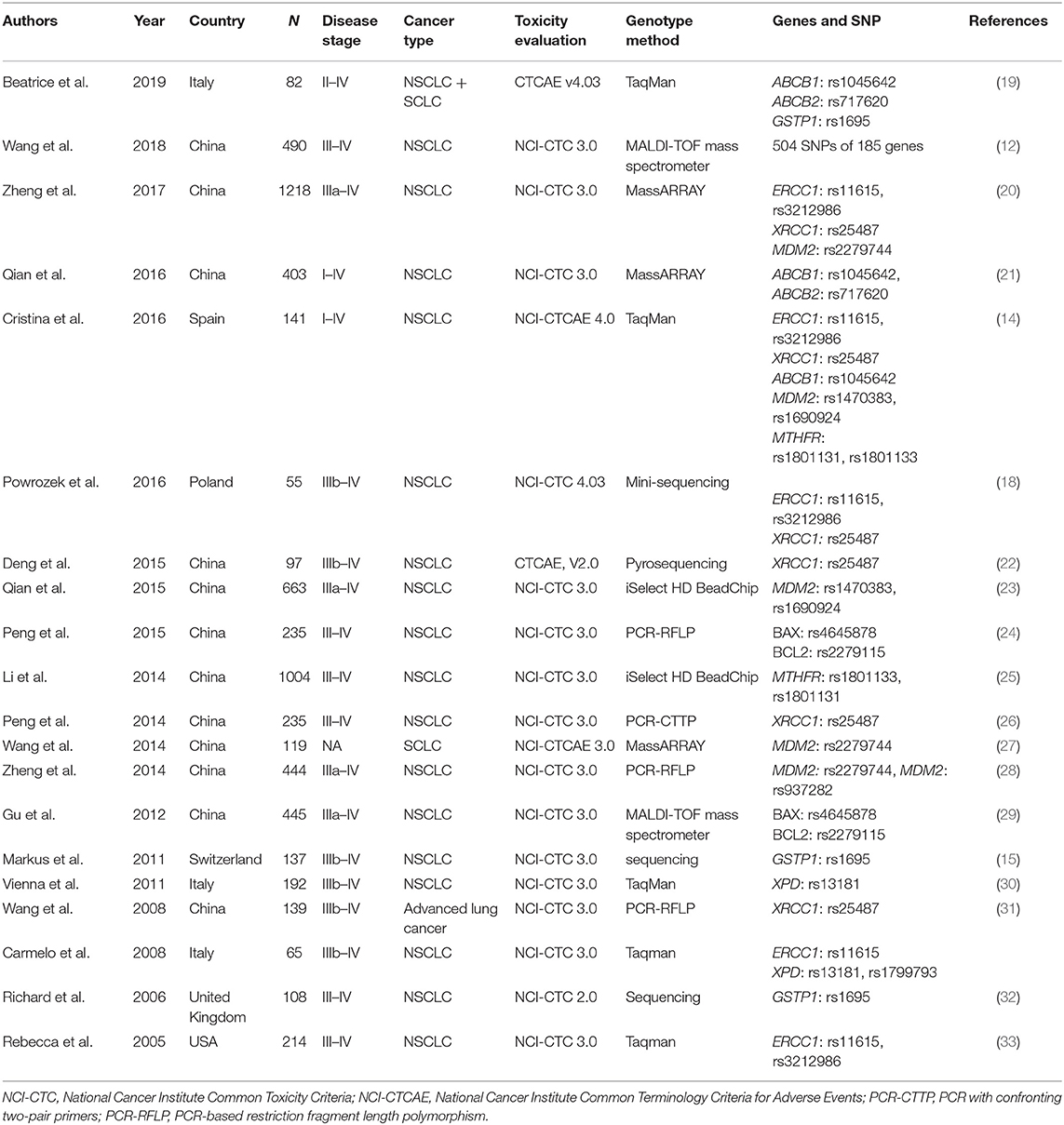
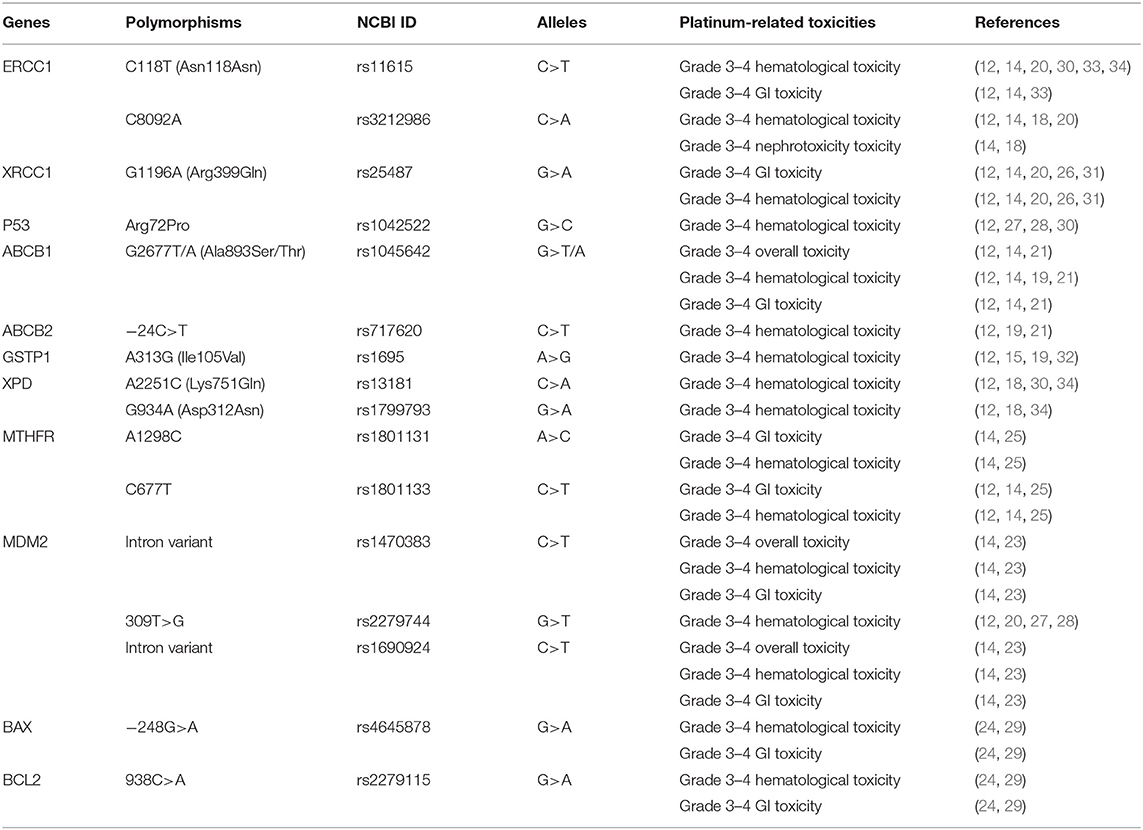
Finally, 20 publications with sufficient data met the inclusion criteria and were extracted (Figure 1). The general characteristics of the studies included in the meta-analysis are presented in Table 1. The included publications included 16 polymorphisms in 11 genes, and the detailed information of these mutation are listed in Table 2. A candidate gene approach was employed for the identification of single nucleotide polymorphisms (SNPs) that conferred susceptibility to platinum-based toxicities, and this methodology directly evaluated the relationship between one variant and a particular toxicity or several toxicities.
Quantitative Synthesis of the Association Between Polymorphisms and Platinum-Related Toxicities
The main results of this meta-analysis showing the association between polymorphisms and risk of platinum-based grade 3–4 toxicities are shown in Table 3 and Supplemental Figures.
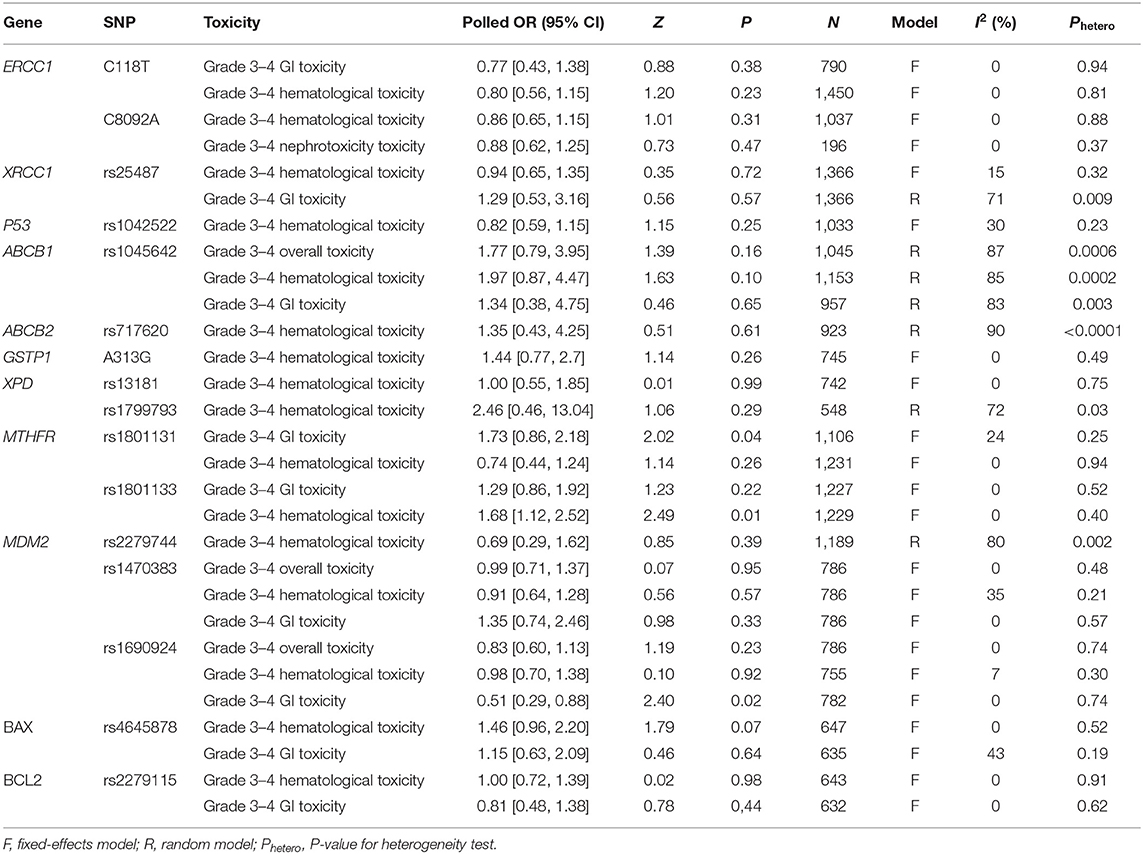
Table 3. Summary of meta-analysis of the association of genetic polymorphisms with platinum induced toxicities.
ERCC1 C118T and C8092A
The most extensively studied polymorphism was ERCC1 C118T. The association between C118T polymorphism and platinum-based grade 3–4 hematological and GI toxicity was found and replicated by six and three studies, respectively. We first performed a meta-analysis to determine the association between C118T polymorphism and grade 3–4 hematological toxicity by including 1,450 subjects. No significant relationship was detected between C118T polymorphism and grade 3–4 hematological toxicity (OR = 0.80, 95% CI: 0.56–1.15, P = 0.23) using a fixed-effect model. On subgroup analyses based on ethnicity, the combined OR for risk and the I2 were consistent and did not show any apparent fluctuation (Supplemental Table 1).
We further analyzed the relationship between C118T polymorphism and grade 3–4 GI toxicity by including 790 patients. Pooled data from these investigations showed grade 3–4 GI toxicity rates of 13.49 and 14.15% in the CC + CT genotype and the TT genotype groups, respectively. No significant relationship was detected between C118T polymorphism and grade 3–4 GI toxicity (OR = 0.77, 95% CI: 0.43–1.38, P = 0.38) using a fixed-effect model. This association did not significantly change on subgroup analyses based on ethnicity.
A total of four and two studies examined the association between C8092A mutation and platinum-based grade 3–4 hematological toxicity and nephrotoxicity, respectively. No significant association was detected between C8092A polymorphism and grade 3–4 hematological toxicity (OR = 0.86, 95% CI: 0.65–1.15, P = 0.31) using a fixed-effect model. This association had no significant change on subgroup analyses based on ethnicity. Among 196 patients who were included to determine the association between C8092A polymorphism and grade 3–4 nephrotoxicity, no significant association was observed (OR = 0.88, 95% CI: 0.62–1.25, P = 0.58) using a fixed-effect model.
XRCC1 G1196A
Five studies were eligible to determine the association between G1196A polymorphism and grade 3–4 GI and hematological toxicities. Pooled analysis showed that there was no significant association between G1196A polymorphism and grade 3–4 GI toxicity (OR = 1.29, 95% CI: 0.53–3.16, P = 0.56) using a random-effect model (Table 3 and Supplemental Figure 2). A sensitivity analysis was conducted because of publication bias (I2 = 71%). After removing the study by Peng et al., the publication bias disappeared (I2 = 10%). We further examined the raw data of this study and found that the G1196A mutation was not considered in the Hardy–Weinberg equation (P = 0.002). No significant association was detected between G1196A polymorphism and grade 3–4 hematological toxicity (OR = 0.94, 95% CI: 0.65–1.35, P = 0.35) using a fixed-effect model. On subgroup analyses based on ethnicity, the combined risk of G1196A mutation on platinum-based grade 3–4 GI toxicities was consistent and did not show any apparent fluctuation (Supplemental Table 1).
P53 Arg72Pro
A total of four studies with 1,033 patients were included to determine the association between P53 Arg72Pro polymorphism and grade 3–4 hematological toxicity. However, the pooled analysis showed no significant association between Arg72Pro polymorphism and grade 3–4 hematological toxicity (OR = 0.82, 95% CI: 0.59–1.15, P = 0.25) using a fixed-effect model. This association did not significantly change on subgroup analyses based on ethnicity (Supplemental Table 1).
ABCB1 G2677T/A
A total of three, four, and three studies were available to determine the association between G2677T/A polymorphism and platinum-induced grade 3–4 overall, hematological, and GI toxicities, respectively. On overall analysis, no significant association was detected between G2677T/A polymorphism and grade 3–4 overall toxicity (OR = 1.77, 95% CI: 0.79–3.95, P = 0.16) using a random-effect model. In addition, publication bias disappeared (I2 = 0%) after the study by Wang et al. was removed. The possible reason is that the call rate for G2677T/A genotypes was <90% in their study, which could have led to inaccuracy in their results. On subgroup analyses based on ethnicity, different ethnic populations showed distinct effects for the G2677T/A polymorphism. There were significant protective effects of the T/A allele on the risk of platinum-based overall toxicity in the non-Chinese subgroup (Supplemental Table 1).
Moreover, no significant association was detected between the G2677T/A polymorphism and grade 3–4 hematological toxicity (OR = 1.97, 95% CI: 0.87–4.47, P = 0.10) using a random-effect model. After removing the study by Qian et al., publication bias disappeared (I2 = 17%). Furthermore, the association between the G2677T/A polymorphism and grade 3–4 GI toxicity was analyzed. No significant association was detected between the G2677T/A polymorphism and grade 3–4 GI toxicity (OR = 1.34, 95% CI: 0.38–4.75, P = 0.46) using a random-effect model. In addition, the publication bias vanished (I2 = 0%) when the study by Qian et al. was removed. The study by Qian et al. was a high-quality well-designed study, with the appropriate sample size. The association between the G2677T/A polymorphism and grade 3–4 hematological and GI toxicities was still non-significant after the study by Qian et al. was removed.
ABCB2 −24C>T
Only three studies were qualified for analyzing the association between the −24C>T polymorphism and platinum-induced grade 3–4 hematological toxicity. No significant association was observed between the −24C>T polymorphism and grade 3–4 hematological toxicity (OR = 1.35, 95% CI: 0.43–4.25, P = 0.51) using a random-effect model. In addition, the publication bias vanished (I2 = 0%) when the study by Wang et al. was removed. The call rate for the −24C>T genotype was 83% in this study, which could have led to inaccuracy in the result. On subgroup analyses based on ethnicity, the association between the −24C>T polymorphism and grade 3–4 hematological toxicity was still non-significant (Supplemental Table 1).
GSTP1 A313G
Data from 745 patients included in 4 studies were used for analyzing the association between the A313G polymorphism and platinum-induced grade 3–4 hematological toxicity. There was no significant relationship between the A313G polymorphism and grade 3–4 hematological toxicity (OR = 1.44, 95% CI: 0.77–2.70, P = 0.26) using a fixed-effect model. On subgroup analyses based on ethnicity, the association between the A313G polymorphism and grade 3–4 hematological toxicity was still non-significant (Supplemental Table 1).
XPD A2251C and G934A
A total of four studies determined the association between the A2251C polymorphism and grade 3–4 hematological toxicity. No significant correlation was detected between the A2251C polymorphism and grade 3–4 hematological toxicity (OR = 1.00, 95% CI: 0.55–1.85, P = 0.99) using a fixed-effect model. Three studies examined the association between the G934A polymorphism and grade 3–4 hematological toxicity. No significant association was detected between the G934A polymorphism and grade 3–4 hematological toxicity (OR = 2.46, 95% CI: 0.46–13.04, P = 0.29) using a random-effect model. The publication bias disappeared (I2 = 0%) when the study by Tibaldi et al. or that by Powrozek et al. was removed. The main reason for this phenomenon was the small sample size of these two studies.
On subgroup analyses based on ethnicity, a non-significant association was found between these two polymorphisms and grade 3–4 hematological toxicity (Supplemental Table 1).
MTHFR A1298C and C677T
In total, three included studies evaluated the association between the A1298C polymorphism and grade 3–4 GI and hematological toxicities. Carriers of the AA genotype had more severe GI toxicity than carriers of the AC + CC genotype did (OR = 1.73, 95% CI: 0.86–2.18, P = 0.04) on a fixed-effect model. However, no significant association was found between the A1298C polymorphism and grade 3–4 hematological toxicities (OR = 0.74, 95% CI: 0.44–1.24, P = 0.26) using a fixed-effect model.
Two studies evaluated the association between the C677T polymorphism and grade 3–4 GI and hematological toxicities. No significant association was detected between the C677T polymorphism and grade 3–4 GI toxicity (OR = 1.29, 95% CI: 0.86–1.92, P = 0.22) using a fixed-effect model. The pooled data showed that patients with the 677CC genotype had an increased risk of severe hematological toxicity than the carriers of the CT + TT genotype did (OR = 1.86, 95% CI: 1.12–2.52, P = 0.01) using a fixed-effect model. The association between these two polymorphisms and grade 3–4 hematological toxicity was unchanged on subgroup analyses based on ethnicity (Supplemental Table 1).
MDM2 rs1470383, rs2279744, and rs1690924
Four studies with 1,189 Chinese patients evaluated the correlation between the rs2279744 polymorphism and grade 3–4 hematologic toxicity. There was no significant correlation between the rs2279744 polymorphism and grade 3–4 hematological toxicity (OR = 0.69, 95% CI: 0.29–1.62, P = 0.39) using a random-effect model. Similarly, we investigated the influence of a single study on the overall risk by excluding one study at a time. However, the combined overall risk and I2 were consistent and did not show any apparent fluctuation.
Data from 786 subjects in 2 studies were used for analyzing the association between the rs1470383 polymorphism and platinum-based toxicities, including grade 3–4 overall, hematological, and GI toxicities. The pooled results showed no significant correlations between the rs1470383 polymorphism and grade 3–4 overall, hematological, and GI toxicities.
Two studies were used for analyzing the association between the rs1690924 polymorphism and platinum-based toxicities, including grade 3–4 overall, hematological, and GI toxicities. The pooled results showed no significant correlations between the rs1470383 polymorphism and grade 3–4 overall and hematological toxicities. The pooled results from all patients indicated that the carriers of the TT + TC genotype had a markedly increased risk of grade 3–4 GI toxicity than carriers of the CC genotype did (OR = 0.51, 95% CI: 0.29–2.43, P = 0.02) using a fixed-effect model.
BAX rs4645878
Two studies were included to analyze the association between the rs4645878 polymorphism and grade 3–4 hematological and GI toxicities. The pooled analysis showed no significant difference between the rs4645878 mutation and grade 3–4 hematological and GI toxicities using a fixed-effect model.
BCL2 rs2279115
The two studies that were included to the determine association between the rs4645878 polymorphism and grade 3–4 hematological and GI toxicities were included to evaluate the association between the rs2279115 polymorphism and grade 3–4 hematological and GI toxicities. The pooled analysis showed no significant differences between the rs4645878 mutation and grade 3–4 hematological and GI toxicities using a fixed-effect model.
Discussion
Researchers had intense interest in the influence of genetic factors on the platinum-based adverse events considering interindividual differences. In the present meta-analysis, we included 20 studies with 6,287 patients with lung cancer who were treated with platinum-based regiments, and we systematically evaluated the impact of genetic polymorphisms on platinum-based grade 3–4 toxicities. A total of 16 polymorphisms in 11 genes were analyzed, and our results provided evidence that MTHFR A1298C and C677T and MDM2 rs1470383 polymorphisms were significantly associated with platinum-based grade 3–4 toxicities.
To date, platinum-based chemotherapy is widely used as first-line therapy for the treatment of lung cancer and is highly cost-effective in Chinese patients (35). As the cytotoxic effects of platinum are not specific, the mechanism of toxicity appears to involve multiple systems during chemotherapy. In the current study, we found that A1298C and C677T mutations of MTHFR were significantly associated with platinum-based grade 3–4 GI and hematological toxicities, respectively. MTHFR encodes the methylenetetrahydrofolate reductase enzyme that is involved in the folate metabolism pathway. The folate metabolism pathway plays an essential role in platinum cytotoxicity, and MTHFR is essential for transmethylation reactions including DNA methylation and DNA synthesis, thereby contributing to cancer prognosis (36). Both A1298C and C677T polymorphisms were associated with reduced enzyme activity and correlated with DNA hypomethylation, both of which alter the sensitivity of tumor cell to platinum compounds (37).
The gene encoding murine double minute 2 (MDM2) is a proto-oncogene and a key negative regulator of p53. MDM2 plays a role in P53-independent antitumor activity through directing binding, ubiquitination, and degradation of the p53 gene (38). A previous meta-analysis found that the MDM2 gene polymorphism (rs2279744) was associated with the risk of lung cancer and the clinical outcomes (39). A previous study verified that unnatural change in the expression of MDM2, mediated by polymorphisms, contributed to subsequent attenuation of the p53 pathway, thereby accelerating the spread of NSCLC (40). In the current study, the MDM2 rs1690924 mutation was significantly associated with grade 3–4 GI toxicity, and the carriers of the TT + TC genotype had a markedly increased risk of grade 3–4 GI toxicity than the carriers of the CC genotype did. To date, only one study found an association between this polymorphism and overall survival (41). However, the function of MDM2 rs1690924 polymorphism is unknown.
Findings on genetic polymorphisms that affect platinum-induced toxicities were inconclusive in most studies, and the sample size of most studies was generally small. Polymorphisms in genes encoding the nucleotide excision repair (NER) pathway are among the most clinically relevant genetic determinants with susceptibility to platinum-based toxicities (42). The NER pathway is responsible for repairing DNA intrastrand cross-links induced by platinum-based chemotherapy, and the most commonly reported candidate gene associated with platinum-based toxicities include ERCC1, XRCC1, and XPD (43). In fact, genes involved in the NER pathway were most commonly evaluated in this meta-analysis. However, results from our meta-analysis showed no significant association between the gene polymorphisms of the NER pathway and platinum-induced severe toxicities.
Although previous studies validated that polymorphisms in genes act as potential risk factors for platinum-induced toxicities considering individual differences, the results of our research indicated that some polymorphisms correlated with platinum-induced toxicities had limited contribution to the interindividual differences in platinum-induced toxicities. The reasons for negative or conflicting results may be complicated. (i) The incidence of toxicities between cisplatin, carboplatin, and oxaliplatin varied (44). (ii) Although all patients in these studies were receiving treatment with platinum-based drugs, the use of non-platinum drugs, such as antimicrotubule agents, antifolate agents, or pyrimidine antagonists, as part of the chemotherapy regimens can affect toxicities profiles (45). (iii) The evaluation of toxicities was based on a standard handbook, while bias may have occurred when certain toxicities such as GI toxicity mainly depend on the subjective assessment by physicians. (iv) Platinum-induced toxicities were aggravated by the cumulative dosage; therefore, different chemotherapy cycles may also affect the results. (v) Platinum-induced toxicities may be affected by other factors, such as the demographic characteristics, molecular features of tumors, comorbidity, ethnicity, and intestinal bacteria.
Till date, the candidate gene approach is the most widely used strategy to identify platinum-induced toxicities and their associated polymorphisms. Novel variants will not be identified using this method, although the likelihood of a positive or negative association with a particular adverse event may be considerable in robust studies. Nevertheless, although previous GWAS and whole-exon sequencing had found novel genetic factors associated with platinum-induced toxicities, all these identified interactions need further verification to determine the mechanism.
The current meta-analysis has several limitations. First, studies without detailed data were excluded because of the lack of information regarding a significant association between gene polymorphisms and platinum-induced toxicities, and we were unable to contact these authors to provide us with the detailed data. This may have caused publication bias. Second, some of the included studies had small sample sizes, which may result in a decreased power to detect significant differences in the distribution of genotypes between grade 3–4 toxicities and grade 0–2 toxicities. Third, the occurrence of platinum-induced toxicities is affected by various factors, and hence, the pooled OR in this meta-analysis was based on the crude OR from the original studies. Because we could not obtain the raw data from individual studies, the pooled OR in this study was not adjusted for potential confounding factors such as sex, age, smoking status, comedications, gene–gene interactions, and gene–environment interactions, among other factors.
The results of our study indicated that single grade 3–4 toxicities that were associated with genetic polymorphisms might partly be the cause for inter-individual differences. Hence, further pharmacogenomics research is needed to determine the novel mutations and their associations. The combined effect of the genetic and clinical factors via gene–gene and gene–environment interactions should be considered in future studies, which may help to predict the risk of lung cancer. Nevertheless, more number of prospective, high-quality, multicenter clinical trials are urgently needed to explore the impact of gene polymorphisms on platinum-based toxicities in patients with lung cancer.
Conclusion
Our study found that MTHFR A1298C and C677T polymorphisms and MDM2 rs1470383 polymorphisms were significantly correlated with platinum-induced severe toxicities in patients with lung cancer. These polymorphisms should be considered for personalized chemotherapy treatment for lung cancer in the future.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
ZL and WL conceived and designed the study. ZL and YW performed research. ZL and JL conducted data analysis. ZL, HY, and WL accomplishment the manuscript, reference collection, data management, statistical analyses, paper writing, and study design.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer J-YY declared a shared affiliation, with no collaboration, with the authors to the handling editor at the time of review.
Acknowledgments
We would like to thank the researchers and study participants for their contributions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.01573/full#supplementary-material
References
2. Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. (2008) 359:1367–80. doi: 10.1056/NEJMra0802714
3. Lebwohl D, Canetta R. Clinical development of platinum complexes in cancer therapy: an historical perspective and an update. Eur J Cancer. (1998) 34:1522–34. doi: 10.1016/s0959-8049(98)00224-x
4. Novello S, Barlesi F, Califano R, Cufer T, Ekman S, Levra MG, et al. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2016) 27:v1–27. doi: 10.1093/annonc/mdw326
5. Shi Y, Sun Y. Medical management of lung cancer: experience in China. Thorac Cancer. (2015) 6:10–6. doi: 10.1111/1759-7714.12168
6. Pilkington G, Boland A, Brown T, Oyee J, Bagust A, Dickson R. A systematic review of the clinical effectiveness of first-line chemotherapy for adult patients with locally advanced or metastatic non-small cell lung cancer. Thorax. (2015) 70:359–67. doi: 10.1136/thoraxjnl-2014-205914
7. Jiang N, Chen XC, Zhao Y. Analysis of the risk factors for myelosuppression after concurrent chemoradiotherapy for patients with advanced non-small cell lung cancer. Support Care Cancer. (2013) 21:785–91. doi: 10.1007/s00520-012-1580-y
8. Xiong Y, Huang BY, Yin JY. Pharmacogenomics of platinum-based chemotherapy in non-small cell lung cancer: focusing on DNA repair systems. Med Oncol. (2017) 34:48. doi: 10.1007/s12032-017-0905-6
9. Cao S, Wang S, Ma H, Tang S, Sun C, Dai J, et al. Genome-wide association study of myelosuppression in non-small-cell lung cancer patients with platinum-based chemotherapy. Pharmacogenomics J. (2016) 16:41–6. doi: 10.1038/tpj.2015.22
10. Green H, Hasmats J, Kupershmidt I, Edsgard D, de Petris L, Lewensohn R, et al. Using whole-exome sequencing to identify genetic markers for carboplatin and gemcitabine-induced toxicities. Clin Cancer Res. (2016) 22:366–73. doi: 10.1158/1078-0432.CCR-15-0964
11. Cao S, Wang C, Ma H, Yin R, Zhu M, Shen W, et al. Genome-wide association study on platinum-induced hepatotoxicity in non-small cell lung cancer patients. Sci Rep. (2015) 5:11556. doi: 10.1038/srep11556
12. Wang LY, Cui JJ, Liu JY, Guo AX, Zhao ZY, Liu YZ, et al. Gene-gene and gene-environment interaction data for platinum-based chemotherapy in non-small cell lung cancer. Sci Data. (2018) 5:180284. doi: 10.1038/sdata.2018.284
13. Cui JJ, Wang LY, Zhu T, Gong WJ, Zhou HH, Liu ZQ, et al. Gene-gene and gene-environment interactions influence platinum-based chemotherapy response and toxicity in non-small cell lung cancer patients. Sci Rep. (2017) 7:5082. doi: 10.1038/s41598-017-05246-8
14. Perez-Ramirez C, Canadas-Garre M, Alnatsha A, Villar E, Delgado JR, Faus-Dader MJ, et al. Pharmacogenetic predictors of toxicity to platinum based chemotherapy in non-small cell lung cancer patients. Pharmacol Res. (2016) 111:877–84. doi: 10.1016/j.phrs.2016.08.002
15. Joerger M, Burgers SA, Baas P, Smit EF, Haitjema TJ, Bard MP, et al. Germline polymorphisms in patients with advanced nonsmall cell lung cancer receiving first-line platinum-gemcitabine chemotherapy: a prospective clinical study. Cancer. (2012) 118:2466–75. doi: 10.1002/cncr.26562
16. Gandara DR, Kawaguchi T, Crowley J, Moon J, Furuse K, Kawahara M, et al. Japanese-US common-arm analysis of paclitaxel plus carboplatin in advanced non-small-cell lung cancer: a model for assessing population-related pharmacogenomics. J Clin Oncol. (2009) 27:3540–6. doi: 10.1200/jco.2008.20.8793
17. Li Y, Huang XE, Jin GF, Shen HB, Xu L. Lack of any relationship between chemotherapy toxicity in non-small cell lung cancer cases and polymorphisms in XRCC1 codon 399 or XPD codon 751. Asian Pac J Cancer Prev. (2011) 12:739–42.
18. Powrozek T, Mlak R, Krawczyk P, Homa I, Ciesielka M, Koziol P, et al. The relationship between polymorphisms of genes regulating DNA repair or cell division and the toxicity of platinum and vinorelbine chemotherapy in advanced NSCLC patients. Clin Transl Oncol. (2016) 18:125–31. doi: 10.1007/s12094-015-1343-6
19. De Troia B, Dalu D, Filipazzi V, Isabella L, Tosca N, Ferrario S, et al. ABCB1 c.3435C>T polymorphism is associated with platinum toxicity: a preliminary study. Cancer Chemother Pharmacol. (2019) 83:803–8. doi: 10.1007/s00280-019-03794-6
20. Zheng Y, Deng Z, Yin J, Wang S, Lu D, Wen X, et al. The association of genetic variations in DNA repair pathways with severe toxicities in NSCLC patients undergoing platinum-based chemotherapy. Int J Cancer. (2017) 141:2336–47. doi: 10.1002/ijc.30921
21. Qian CY, Zheng Y, Wang Y, Chen J, Liu JY, Zhou HH, et al. Associations of genetic polymorphisms of the transporters organic cation transporter 2 (OCT2), multidrug and toxin extrusion 1 (MATE1), and ATP-binding cassette subfamily C member 2 (ABCC2) with platinum-based chemotherapy response and toxicity in non-small cell lung cancer patients. Chin J Cancer. (2016) 35:85. doi: 10.1186/s40880-016-0145-8
22. Deng JH, Deng J, Shi DH, Ouyang XN, Niu PG. Clinical outcome of cisplatin-based chemotherapy is associated with the polymorphisms of GSTP1 and XRCC1 in advanced non-small cell lung cancer patients. Clin Transl Oncol. (2015) 17:720–6. doi: 10.1007/s12094-015-1299-6
23. Qian J, Liu H, Gu S, Wu Q, Zhao X, Wu W, et al. Genetic variants of the MDM2 gene are predictive of treatment-related toxicities and overall survival in patients with advanced NSCLC. Clin Lung Cancer. (2015) 16:e37–53. doi: 10.1016/j.cllc.2015.02.001
24. Peng Y, Wang L, Qing Y, Li C, Ren T, Li Q, et al. Polymorphisms of BCL2 and BAX genes associate with outcomes in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Sci Rep. (2015) 5:17766. doi: 10.1038/srep17766
25. Li X, Shao M, Wang S, Zhao X, Chen H, Qian J, et al. Heterozygote advantage of methylenetetrahydrofolate reductase polymorphisms on clinical outcomes in advanced non-small cell lung cancer (NSCLC) patients treated with platinum-based chemotherapy. Tumour Biol. (2014) 35:11159–70. doi: 10.1007/s13277-014-2427-6
26. Peng Y, Li Z, Zhang S, Xiong Y, Cun Y, Qian C, et al. Association of DNA base excision repair genes (OGG1, APE1 and XRCC1) polymorphisms with outcome to platinum-based chemotherapy in advanced nonsmall-cell lung cancer patients. Int J Cancer. (2014) 135:2687–96. doi: 10.1002/ijc.28892
27. Wang X, Wang YZ, Ma KW, Chen X, Li W. MDM2 rs2279744 and TP53 rs1042522 polymorphisms associated with etoposide- and cisplatin-induced grade III/IV neutropenia in Chinese extensive-stage small-cell lung cancer patients. Oncol Res Treat. (2014) 37:176–80. doi: 10.1159/000360785
28. Zheng D, Chen Y, Gao C, Wei Y, Cao G, Lu N, et al. Polymorphisms of p53 and MDM2 genes are associated with severe toxicities in patients with non-small cell lung cancer. Cancer Biol Ther. (2014) 15:1542–51. doi: 10.4161/15384047.2014.956599
29. Gu S, Wu Q, Zhao X, Wu W, Gao Z, Tan X, et al. Association of CASP3 polymorphism with hematologic toxicity in patients with advanced non-small-cell lung carcinoma treated with platinum-based chemotherapy. Cancer Sci. (2012) 103:1451–9. doi: 10.1111/j.1349-7006.2012.02323.x
30. Ludovini V, Floriani I, Pistola L, Minotti V, Meacci M, Chiari R, et al. Association of cytidine deaminase and xeroderma pigmentosum group D polymorphisms with response, toxicity, and survival in cisplatin/gemcitabine-treated advanced non-small cell lung cancer patients. J Thorac Oncol. (2011) 6:2018–26. doi: 10.1097/JTO.0b013e3182307e1f
31. Wang Z, Xu B, Lin D, Tan W, Leaw S, Hong X, et al. XRCC1 polymorphisms and severe toxicity in lung cancer patients treated with cisplatin-based chemotherapy in Chinese population. Lung Cancer. (2008) 62:99–104. doi: 10.1016/j.lungcan.2008.02.019
32. Booton R, Ward T, Heighway J, Ashcroft L, Morris J, Thatcher N. Glutathione-S-transferase P1 isoenzyme polymorphisms, platinum-based chemotherapy, and non-small cell lung cancer. J Thorac Oncol. (2006) 1:679–83. doi: 10.1097/01243894-200609000-00013
33. Suk R, Gurubhagavatula S, Park S, Zhou W, Su L, Lynch TJ, et al. Polymorphisms in ERCC1 and grade 3 or 4 toxicity in non-small cell lung cancer patients. Clin Cancer Res. (2005) 11:1534–8. doi: 10.1158/1078-0432.CCR-04-1953
34. Tibaldi C, Giovannetti E, Vasile E, Mey V, Laan AC, Nannizzi S, et al. Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. (2008) 14:1797–803. doi: 10.1158/1078-0432.CCR-07-1364
35. Peng Y, Ma F, Tan C, Wan X, Yi L, Peng L, et al. Model-based economic evaluation of ceritinib and platinum-based chemotherapy as first-line treatments for advanced non-small cell lung cancer in China. Adv Ther. (2019) 36:3047–58. doi: 10.1007/s12325-019-01103-4
36. Zhang X, Zhang D, Huang L, Li G, Chen L, Ma J, et al. Discovery of novel biomarkers of therapeutic responses in Han Chinese pemetrexed-based treated advanced NSCLC patients. Front Pharmacol. (2019) 10:944. doi: 10.3389/fphar.2019.00944
37. Weisberg IS, Jacques PF, Selhub J, Bostom AG, Chen Z, Curtis Ellison R, et al. The 1298A–>C polymorphism in methylenetetrahydrofolate reductase (MTHFR): in vitro expression and association with homocysteine. Atherosclerosis. (2001) 156:409–15. doi: 10.1016/s0021-9150(00)00671-7
38. Hashemi M, Omrani M, Eskandari-Nasab E, Hasani SS, Mashhadi MA, Taheri M. A 40-bp insertion/deletion polymorphism of Murine Double Minute2 (MDM2) increased the risk of breast cancer in Zahedan, Southeast Iran. Iran Biomed J. (2014) 18:245–9. doi: 10.6091/ibj.13332.2014
39. Luan L, Wang H, Zhao B, Wang F, Shi J, Xu X. Association of MDM2 gene SNP 309 polymorphism and human non-small cell lung cancer susceptibility: a meta-analysis. Pathol Res Pract. (2019) 215:152538. doi: 10.1016/j.prp.2019.152538
40. Zhuo W, Zhang L, Zhu B, Ling J, Chen Z. Association of MDM2 SNP309 variation with lung cancer risk: evidence from 7196 cases and 8456 controls. PLoS ONE. (2012) 7:e41546. doi: 10.1371/journal.pone.0041546
41. Perez-Ramirez C, Canadas-Garre M. Pharmacogenetics of platinum-based chemotherapy: impact of DNA repair and folate metabolism gene polymorphisms on prognosis of non-small cell lung cancer patients. Pharmacogenomics J. (2019) 19:164–77. doi: 10.1038/s41397-018-0014-8
42. Perez-Ramirez C, Canadas-Garre M, Molina MA, Robles AI, Faus-Dader MJ, Calleja-Hernandez MA. Contribution of genetic factors to platinum-based chemotherapy sensitivity and prognosis of non-small cell lung cancer. Mutat Res. (2017) 771:32–58. doi: 10.1016/j.mrrev.2016.11.003
43. Zhang R, Zhou F, Cheng L, Yu A, Zhu M, Wang M, et al. Genetic variants in nucleotide excision repair pathway predict survival of esophageal squamous cell cancer patients receiving platinum-based chemotherapy. Mol Carcinog. (2018) 57:1553–65. doi: 10.1002/mc.22877
44. Yu J, Xiao J, Yang Y, Cao B. Oxaliplatin-based doublets versus cisplatin or carboplatin-based doublets in the first-line treatment of advanced nonsmall cell lung cancer. Medicine. (2015) 94:e1072. doi: 10.1097/md.0000000000001072
Keywords: platinum, pharmacogenomics, toxicity, individual difference, meta-analysis
Citation: Liu W, Wang Y, Luo J, Yuan H and Luo Z (2020) Genetic Polymorphisms and Platinum-Based Chemotherapy-Induced Toxicities in Patients With Lung Cancer: A Systematic Review and Meta-Analysis. Front. Oncol. 9:1573. doi: 10.3389/fonc.2019.01573
Received: 16 October 2019; Accepted: 30 December 2019;
Published: 17 March 2020.
Edited by:
Mukerrem Betul Yerer Aycan, Erciyes University, TurkeyReviewed by:
Ji-Ye Yin, Xiangya Hospital, Central South University, ChinaMassimiliano Berretta, Centro di Riferimento Oncologico di Aviano (IRCCS), Italy
Copyright © 2020 Liu, Wang, Luo, Yuan and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiying Luo, bHpoeTE5OTA4OUBjc3UuZWR1LmNu
 Wenhui Liu
Wenhui Liu Ying Wang1,2
Ying Wang1,2 Jianquan Luo
Jianquan Luo Zhiying Luo
Zhiying Luo