- 1Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, WA, United States
- 2Department of Pathology, University of Washington, Seattle, WA, United States
Cutaneous T cell lymphomas represent a heterogenous group of lymphoproliferative disorders defined by clonal proliferation of T cells present in the skin. The latest WHO classification in 2016 and WHO-EORTC classification in 2018 has updated the classification of these entities based on the molecular profile. Research in the field of molecular genetics of CTCL has allowed a better understanding of the biology of these tumors and has helped to identify potential targets for therapy that can be tailored to individual patients. In this review, we discuss the latest developments in the molecular profile of CTCLs including biomarkers for diagnosis, prognosis, and potential therapeutic targets. We have also touched upon the utility of various molecular diagnostic modalities. For the purpose of this review, we researched papers in PubMed indexed journals in English literature published in the past 20 years using keywords CTCL, mycosis fungoides, molecular profile, molecular diagnosis, whole genome profile, genomic landscape, TCR clonality.
Introduction
Cutaneous T cell lymphomas (CTCL) are a heterogenous group of lymphoproliferative disorders arising primarily in the skin without the evidence of extracutaneous involvement. For the purposes of this review, we define CTCL according to the WHO 2016 and the 2018 WHO-EORTC classification system. Since, the 2008 WHO classification, molecular advances with gene expression profiling and a better understanding of the genomic landscape of TCL allowed for improved classification. Specific phenotypes such as the T follicular helper cells, are now possible to identify with gene expression profiling. In the 2016 WHO and 2018 WHO-EORTC classification of skin tumors a few provisional entities have been added, like chronic EBV positive mucocutaneous ulcer, primary cutaneous acral CD8+ T cell lymphoma, Primary cutaneous CD4+ small/medium lymphoproliferative disorder. Three distinct types of lymphomatoid papulosis in addition to originally described types A,B,C have also been included (1, 2).
In this review, we discuss the molecular genetic profile of CTCL's along with new molecular biomarkers of diagnostic and prognostic significance with respect to the latest classification.
Molecular Profile of CTCLs
Mycosis Fungoides/Sezary syndrome (MF/SS) comprise the majority of cutaneous T cell lymphomas, most of the genomic studies elucidating the pathogenesis and genetic abnormalities being studied in this group of CTCL. There are no disease specific molecular abnormalities that define these lymphomas, however some changes are more frequent than others.
Whole genomic sequencing studies of MF/SS have revealed somatic mutations in genes involved in TCR/NFκB signaling, Th2 differentiation, cell survival and fate, epigenetic regulation, homologous recombination, and cell-cycle control. These pathways in the pathogenesis of CTCL have opened new avenues for prognostication, disease monitoring, and targeted therapy (3, 4).
Recurrent mutations identified by whole exome/genome sequencing in common oncogenic pathways include- DNA damage repair (i.e., ATM, TP53), cell cycle (i.e., CDKN2A, RB1), apoptosis (i.e., FAS, TNFRSF10A), MAPK pathway (i.e., KRAS, BRAF, MAPK1), and chromatin modifying genes (i.e., ARID1A, DNMT3A, KMT2C). Forty to ninety two percentage of these mutations are single copy number variations (SCNVs) (3, 5–7). Increased burden of somatic single nucleotide variations (SSNVs) have also been identified in various genes like Tp53, RHOA, CD28, DNM3TA. Interestingly, SCNV's are more important mutational drivers for CTCLs than SSNV's when compared to other cancers with significantly higher SCNV/SSNV ratios (5). Increased number of transition (C-T or G-A) mutations are observed in 40–60% of cases along with a higher frequency of dinucleotide variations indicating that ultraviolet (UV) radiation exposure may have a role in the pathogenesis of CTCL (4, 8).
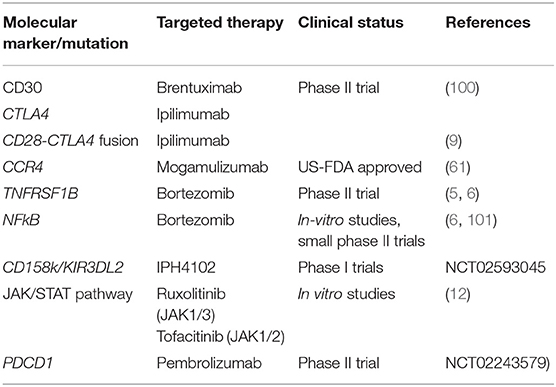
Alterations in T cell signaling pathways have also been described in recent studies which include mutations in co-stimulatory molecules (CD28 and CTLA4), TCR associated enzymes (PLCG1) as well as downstream transcriptional regulators of TCR function (ZEB1) (4, 5, 8). Out of these, a novel CD28-CTLA4 gene fusion (6) resulting in acceleration of TCR signaling has been exploited for anti-CTLA4 therapy (ipilimumab) with promising results (9). Recurrent point mutations and deletions in ZEB1, a zinc-finger transcription repressor, have been found in 56–65% of CTCLs. This functional inactivation of ZEB1 leads to overproduction of TH2 cytokines promoting tumor growth in CTCLs (4, 5, 8). PDCD1, a gene on chromosome 2p expressing PDL1 has also been found to be deleted in 36% of cases. PDL1 is found on activated T cells and gives a negative signal to suppress the T-cell function. This provides a rationale for use of anti PDL1 therapy for CTCL as well.
NFκB signaling pathway has been shown to be affected in cutaneous lymphomas by several mutations leading to its constitutive activation (6, 10) Recurrent point mutation (Thr377Ile) of TNFRSF1B found in 18% MF cases is one such example. NFκB is a nuclear transcription factor regulating gene expression of various growth promoting factors like TNFα, IL-2, IL-6, TGFβ, IFNβ. NFκB is normally sequestered in the cytoplasm by IκB, and it can translocate to the nucleus only when IκB is ubiquitinated or degraded by proteosomes. Recurrent deletions of C-terminus of NFκB leads to proteosomal cleavage of IκB causing constitutive activation (5). These mutations make these tumors amenable to proteosome inhibitors like Bortezomib (11). CARD11 potentiates NFkB signaling in T and B cells has been found to be mutated in a subset of SS cases and has been suggested as a potential therapeutic target as in DLBCL (8).
Activating mutations in JAK/STAT pathway including JAK1, JAK3, STAT3, and STAT5B were found in a subset of cases by many groups. Anti-tumor properties of JAK1/2 inhibitor Ruxolitinib and JAK 1/3 inhibitor Tofacitinib have been tested in CTCL cell lines with promising results (3–5, 12). Two tumor suppressor genes (HNRNPK and SOCS1) which are inhibitors of JAK-STAT signaling have been identified to be recurrently deleted in MF also represent potential therapeutic targets (13).
T-Cell Receptors and Clonality Testing: Diagnostic Utility
T cell development begins in the thymus, where previously uncommitted progenitors migrate from the marrow to become double negative thymocytes and start rearrangement of their T cell receptor (TCR) genes (TCRα, TCRβ, TCRγ, TCRδ). The TCR is a heterodimer of either alpha/beta or gamma/delta type. Each T cell and its progeny have a unique rearrangement of TCR genes formed by diverse recombination of V/J or V/D/J segments during T cell development (14). It is widely accepted that T cell malignancy arise from a single T cell clone, hence testing of malignant cells will demonstrate the same TCR gene sequence, termed as monoclonal. Monoclonality can also be seen in autoimmune and reactive conditions (15, 16) and some malignancies can also have oligoclonal populations. Therefore, assessment of clonality should be performed in appropriate clinico-pathological scenario. Molecular methods used are described below briefly.
CTCL's are clonal disorders arising from skin homing mature T cells showing specific TCR gene rearrangement. CTCL's are routinely diagnosed by clinical presentation, histopathology and immunohistochemistry of the atypical infiltrate with clonality assays serving only as adjuncts to the diagnosis. Comfere et al. reported that only 2.3% dermatologists actually order TCR clonality assays as part of the initial diagnostic evaluation and only 3.7% of pathologists rely on them for diagnosis of cutaneous lymphoproliferative disorders (17).
Early lesions of CTCLs with lesser number of abnormal lymphoid cells typically pose a diagnostic challenge since various benign dermatoses have similar histologic features and immunohistochemistry is also not reliable in such cases (18, 19). Detection of TCR gamma and beta gene rearrangement can be of value in these scenarios as benign dermatitis does not show clonal rearrangement (19). However, the sensitivity of detecting TCR rearrangement by PCR is highly variable ranging from 50–90% in various studies (20–22). The reasons are multifactorial such as stage of disease, method of assay, primer design, PCR products and interobserver variability.
The success of PCR based assays in clonality assessment depends hugely on the choice of oligonucleotide primers and PCR design. This can be partly overcome by using standardized set of primers for e.g., BIOMED 2 (23, 24). Capillary gel electrophoresis was found to be better than conventional PAGE analysis to study the PCR products (25, 26).
Application of Next generation sequencing (NGS) /high throughput sequencing has greatly improved the sensitivity of detection (27–29). Sufficool et al. found a sensitivity of 85% using NGS as opposed to 44% using PCR. Table 1 summarizes various studies showing variable sensitivities of TCR clonality techniques.
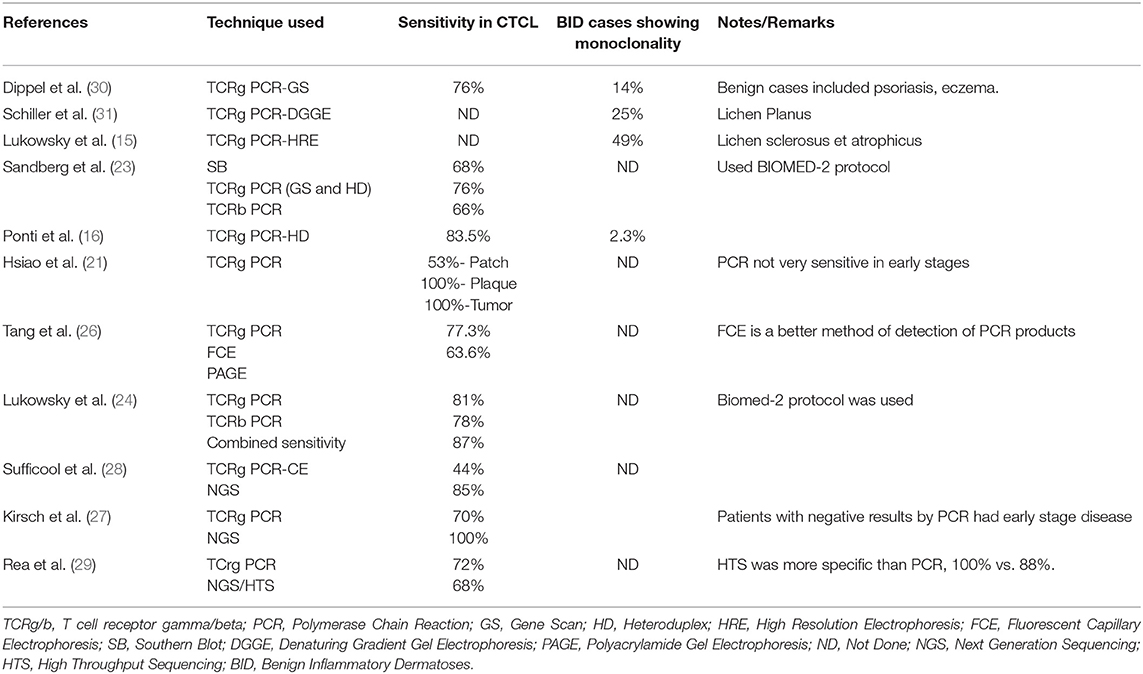
Table 1. Summary of studies showing the sensitivity of various techniques for T cell clonality assessment in CTCL and benign inflammatory skin disorders.
T-cell clonality is itself not diagnostic of a T cell lymphoma as many dermatitis may have dominant T cell clone (16, 18, 30, 31) and 25% of these clonal dermatitis eventually evolve into CTCL (14). Identification of a single clone at two different anatomic sites favors a diagnosis of a malignant process over benign (32). On the contrary, the finding of polyclonality/oligoclonality does not rule out the possibility of CTCL. Therefore, the diagnosis of lymphoma should be made in conjunction with clinical and histopathological features using TCR clonality as an adjunct (23).
TCR gene rearrangement can be utilized for monitoring of residual disease particularly in leukemic involvement by MF/SS. Standard methods used to accomplish this are through flow cytometry using tumor specific TCR Vbeta antibodies (33) or TCR PCR which is more sensitive and specific (34). The monoclonal peak in the original sample is identified and followed up with repeat samplings. This monoclonal peak is however not always tumor-specific as few non-malignant T cell clones may give rise to the same. A large 2015 French study on over 200 patients with MF showed no significant prognostic differences at 4 years of clinical follow-up when MRD was determined by TCR clonality based on PCR studies (35), however a similar study utilizing NGS has not been performed. Weng et al. employed NGS/HTS technique for MRD detection in MF/SS and showed it to be highly sensitive and specific for detecting disease however their study was small (n = 10) and did not have long term outcomes data (<2 years) (36). This is specifically useful in post stem cell transplant recipients where a skin rash could represent drug toxicity, GVHD or recurrence and clinical and pathological distinction can be quite challenging (37, 38). NGS of TCRB can be of value in such critical decision making scenarios (36).
Identification and monitoring of monoclonality, however, seems to have no prognostic relevance even if identified in early lesions (35, 39) However, tumor clone frequency (TCF) obtained by HTS of TCRb gene is a strong and independent prognostic marker for progression free and overall survival in CTCL-MF. TCF > 25% at an early stage of MF has the ability to predict a poorer outcome than any other prognostic marker (40).
Molecular Diagnostic Modalities for TCL
Clinically Used Techniques
PCR Based Assays
PCR of the TCR gamma and/or beta gene is frequently used as an adjunct to asses monoclonality in T cell lymphomas. TCRG PCR is preferred as gamma gene is rearranged earlier and present in most of the T cells and has only 12 segments, hence less primers are to be used (41). Combined use of TCRB and TCRG primers increases the sensitivity of the test than using each of them individually (24, 42). The extracted DNA from fresh tumor, liquid samples, or formalin fixed paraffin embedded tissue specimen can be tested using PCR amplified with commercially available primers. The PCR products are analyzed by capillary electrophoresis or genescan depending upon the size of the amplicon. In straightforward cases, a single dominant peak is seen if the infiltrate is monoclonal whereas multiple peaks when it is polyclonal (43). However, in clinical practice challenging cases and clinical scenarios occur and a good understanding of the starting sample, patient history as well as the specific assay parameters are important for the molecular pathologist in their interpretation of results. This analysis is subjective and liable to inter-observer variability since it is based on qualitative assessment of relative peak heights. Many cases do not have a dominant clone while a dominant peak may represent a mixture of clones of similar size (44). Therefore, it is recommended to run the PCR in duplicate, and only reproducible peaks should be considered positive (41).
Molecular testing for infectious agents that are seen in some CTCLs are mostly PCR based. Input samples may include testing of peripheral blood or tumor tissues. However, understanding the pathobiology of how the infectious agent leads to tumorigenesis is important in diagnosis. One example include Adult T-cell leukemia/lymphoma, a monoclonal integration site of HTLV1 virus genome into the host cell leads to upregulation of HBZ in most cases impacting cell growth, immune response and differentiation.
Next Generation Sequencing Based Assays
NGS has vastly improved TCR clonality assessment mainly through improved specificity via sequencing of rearranged TCR. Published evidenced is increasing for the use of this technology in determining T cell clonality status (27–29, 44) In this technique, TCRG and TCRB loci are PCR amplified using consensus primers, PCR products are purified and made into libraries and ligated to adapters which are then sequenced by synthesis using labeled nucleotides. The malignant clone is identified by sequence abundance. If most frequent two sequences consist of the predominant sequence, the population is called clonal (28). This technique has advantage of being quantitative, robust and reproducible. NGS has successfully proven to be useful for early detection of CTCL as well in differentiating malignant from benign lesions especially where PCR is negative (27, 44). A clonal sequence can be identified in as low as 1 in 50,000 malignant cells when flowcytometry is not able to reveal residual disease (36). Patients in molecular remission can thus be reliably identified using NGS. Minimal residual disease (MRD) detection by NGS have been described to be reliable with some clinically validated platforms for use in T cell lymphomas such as sezary syndrome (36) and other CTCLs (27).
Molecular Techniques Being Used for Research Domain
Single Cell Sequencing
Although this technology is still relatively contained within the research space, clinical applications using this technology will likely advance patient care in the next decade. Single cell sequencing studies have shown evidence of tumor heterogeneity in CTCLs and there is also preliminary evidence that there may be a prognostic impact (45–47). Demonstration of specific gene expression profiles enabling examination of the tumor microenvironment along with the tumor cells in the same assay provides useful insights to tumor biology and immune response, which ultimately helps in tailoring personalized treatments, understanding prognosis, response to therapy, and reasons for drug resistance (48, 49).
Gaydosik et al. performed single cell RNA sequencing (scRNA) using droplet based sequencing to study thousands of individual cells in advanced CTCL's along with skin samples from healthy controls. The results not only revealed significant inter-tumor but also intra-tumor heterogeneity. Many CTCL's had differential expression of genes that was unique to that tumor signifying its importance in personalizing patient specific therapy. A common gene expression profile of highly proliferating lymphocytes was identified which was present in all tumors indicating its potential diagnostic role (47).
Buus et al. demonstrated distinct subpopulations of tumor cells in Sezary syndrome (SS) through ScRNA sequencing and flow cytometry while studying drug resistance to histone deacetylator inhibitor. Some cell populations were responsive to the drug while others were resistant. This study sheds light on the utility of this technique in its therapeutic implications (45).
Similarly, Borcherding et al. described the transcritpomic diversity in Sezary cells using ScRNA sequencing can be used to predict disease stage and guide therapy (46).
Gene Expression Profiling
In the past decade, gene expression profiling have been used to identify unique T cell lymphoma subtypes leading to new provisional diagnostic entities. The molecular platforms include mostly research use only, however increasingly clinical grade assays are becoming available. The earliest and possibly most widely available are gene expression microarrays (50). These assays are robust with well-established analysis tools and pipelines but time consuming and require highly skilled staff. Newer platforms that enable direct capture and counting of specific RNA molecules via a barcode or color-coded bead combinations have improved on prior generations of DNA probe based gene expression arrays (51). Gene expression by RNA sequencing is yet another platform that is based on NGS technology, where instead of sequencing input sample of DNA, RNA is used and therefore an expression profile is generated based on the relative numbers of cDNA molecules that are created in the NGS library preparation. A recent study compared RNA sequencing results from 181 fresh and FFPE CTCL or inflammatory dermatoses skin tissues and showed that specific genes were upregulated in benign skin conditions or low grade CTCL and that a gene signature could be applied to differentiate patients with more aggressive disease (52). In addition to RNA gene expression profiling, microRNA profiling is also possible with the technologies described in this section. More detailed description of specific expression profiles are described in the disease entities below.
Disease Entities: Molecular Markers of Diagnostic, Prognostic, and Therapeutic Significance
Mycosis Fungoides/Sezary Syndrome (MF/SS)
Mycosis fungoides is the most common form of CTCL encompassing 50–60% of all CTCLs (53). Sezary syndrome (SS) is the leukemic variant presenting with erythrodermic lesions along with lymph node and peripheral blood involvement at presentation. MF is skin limited and leukemic involvement occurs only in few cases progressing to advanced disease. MF/SS are considered distinct entities but included in the same staging system (34, 53). MF and SS arise from different types of CD4+ memory T cell. MF T-cells are T resident memory (Trm) cells exhibiting CCR4+/CLA+/ L-selectin-/CCR7– (TRM), whereas SS malignant T-cell are central memory cells (Tcm) (CCR4+/L-selectin+/CCR7+) (54). Trm cells are skin tropic and stay within the epithelial barriers while Tcm cells have the ability to shuffle between skin, lymph nodes and blood (55), which provides an understanding of some clinical differences between MF and SS.
MF passes through various clinical stages and early lesions (eMF) posing a significant diagnostic challenge when differentiating from benign dermatoses (18). The utility of T cell clonality studies, discussed above, can be useful but must be interpreted with caution with full understanding of the clinical and pathologic picture. High throughput sequencing of TCR CDR3 region can identify a smaller clone of malignant cells in comparison to PCR which may miss a low level clonal process (27).
Gene expression profiling of MF/SS has elucidated a diagnostic panel of genes that can reliably characterize them. Litvinov et al. describes a 17 gene signature including IL2RA, CCR4, STAT5A, and TOX that is able to identify patients who are at risk of progression and differentiate MF/SS from benign dermatoses (56). Nebozhyn et al. proposed a panel of five genes (STAT4, GATA3, PLS3, CD1D, and TRAIL), Michel et al. proposed a panel of 4 genes (PLS3, Twist1, CD158k/KIR3DL2, and NKp46) using qRTPCR showing the ability to diagnose 91 and 100% cases of SS, respectively (57, 58). Amongst these Twist1 alone has the power to diagnose 91% SS cases. CD158k/KIR3DL2 has emerged as a therapeutic target with phase 1 clinical trials currently underway (52). CD158k/KIR3DL2 is highly expressed in SS and advanced MF confers resistance to activation induced cell death in SS and has been found efficacious in reducing tumor size and improved survival in vivo and in vitro (59, 60).
Expression of CCR4 by the malignant T cells in CTCL has been exploited for targeted therapy by the use of anti-CCR4 monoclonal antibody mogamulizumab. This drug has recently been approved by US-FDA for treatment of relapsed or refractory MF and SS after phase 3 multicentric clinical trial (MAVORIC study) showed a significant improvement in progression free survival as compared to conventional treatment by acetone deacetylase inhibitor vorinostat (61). Granulomatous drug eruption is a frequent side effect of mogamulizumab which can be confused with disease progression but additional studies showed this side effect might represent adequate treatment response (62).
TOX, a gene that encodes a member of homeobox family and regulate T cell development in the thymus, is upregulated in early as well as advanced MF. Presence of TOX mRNA can help differentiate eMF from benign cases (63). Moreover, high expression of TOX mRNA in eMF is also strongly correlated with disease progression potentially allowing for intensifying treatment at an early stage for patients (64, 65). Some of the molecular markers and aberrations have already been discussed in the above paragraphs and hence, will not be discussed again.
Lastly, microRNA profiling has been extensively performed to evaluate its role in the diagnosis and management of MF/SS. Invaluable diagnostic potential with 95% sensitivity and specificity in differentiating eMF from benign lesions of a panel of three microRNAs namely miR-155, miR-203, and miR-205 has been elucidated (66). Measuring their levels in plasma can be used to monitor tumor burden and response to therapy (67). eMF and benign lesions display different miR profiles and as the disease progresses, different miRNAs become deregulated implicating their role in disease progression (68). 3-miRNA classifier, based on miR-106b-5p, miR-148a-3p, and miR-338-3p, can successfully segregate patients into high-risk and low-risk groups of disease progression (69).
All these studies confer that there is a wide variety of genomic alterations in MF/SS and the genetic signature of tumors can differ between patients. Hence, choosing personalized therapy for each patient is worthwhile according to the individual tumor's molecular pattern (70).
Primary Cutaneous CD30+ T Cell Lymphoproliferative Disorders
The cell membrane protein, CD30 (aka TNFRSF8), is a member of tumor necrosis factor receptor family and is expressed on activated B and T cells as well as a number of lymphoid malignancies. These include a spectrum of disorders comprising of various types of lymphomatoid papulosis (LyP) and cutaneous Anaplastic large cell lymphoma (pcALCL). They are the second most common CTCL about 25% of all CTCLs (53). Clinical presentation is different in both LyP and pcALCL but they have overlapping pathological features (71).
The latest WHO-EORTC classification (2) has described new subtypes of LyP described types D- resembling primary cutaneous aggressive epidermotropic cytotoxic T-cell lymphoma (72); type E- angiocentric and angiodestructive and clinically characterized by large necrotic eschar-like lesions (73); LyP with presence of rearrangements of DUSP-IRF4 locus on 6p25.3 (74). These are histological subtypes with little difference in the prognosis. There differentiation from other aggressive lymphomas is important.
LyP
Presence of detectable monoclonal T cell population varies in LyP and largely depend upon the number of T cells present in the lesion which is higher in type C than type A (75). Various studies have proved that MF or other lymphomas arising in patients with LyP have the same TCR rearrangement as the original lesion suggesting a clonal relationship between them (76). A recent study showed a high proportion of LyP have increased expression of SATB1 (91.7%) and that these cases were characterized by prominent epidermal hyperplasia and infiltration by polymorphs, with better response to methotrexate and interferon therapy (77). Although rare, rearrangement in the DUSP22-IRF4 locus has led to a newly recognized subtype of LyP harboring a biphasic histological pattern with small-medium sized T cells exhibiting epidermotropism along with large T cells in the dermis, both endorsing CD30 positivity (78).
pcALCL
This is an indolent CD30+ lymphoma and involvement of extracutaneous sites needs to be ruled out because systemic ALCL portends a poor prognosis. There are various differences between pc ALCL and systemic ALCL on molecular level. Most cases of pcALCL are negative for classic chromosomal rearrangements seen in systemic ALCL—ALK, DUSP22/IRF4, and TP63 (79). DUSP/IRF4 rearrangement is found in 20% pcALCLs (78) and show a biphasic histological pattern similar to LyP with this alteration (80). There is no clinical or prognostic significance of this genetic change in pcALCL (81). DUSP22/IRF4 is also not diagnostically specific for pcALCL as it has also been identified in few systemic ALCLs where is associated with better prognosis than TP63 positive cases (82).
A novel NPM1-TYK2 gene fusion has been identified in 2 cases (1LyP, 1pcALCL) of CD30+ LPDs by whole transcriptome sequencing. This chimeric protein activates STAT5 pathway and hence has been implicated as a therapeutic target (83). TP63 rearrangement has also been described in two cases of pcALCL and both these cases had worse clinical progression like systemic ALCL (84).
ALK positivity by immunohistochemistry has been found in only a handful of cases of pcALCL and these cases showed a favorable prognosis (85) unlike their systemic counterpart. ALK translocation has not been found in pcALCL to date (79).
Most widely used targeted therapy is anti CD30 brentuximab in these neoplasms. Other potential therapeutic targets being developed are KIR3DL2 (86), NPM-TYK2, and miR155 (87).
Cuatneous Lymphomas With T Follicular Helper (TFH) Phenotype
Recent revision of WHO classification of hematolymphoid neoplasms has proposed a category of nodal T cell lymphomas wcell lymphoma with TFH phenotype and includes Angioimmunoblastic T cell lymphoma (AITL), Follicular T cell lymphoma (FTCL), and nodal peripheral T cell lymphoma (PTCL) with TFH phenotype. Diagnosis requires presence of atleast two markers of TFH lineage- CD279/PD-1, CD10, BCL6, CXCL13, ICOS, SLAM-associated protein (SAP), and CXCR5 (1).
Amongst these, AITL frequently involves skin in about 50% cases (88). TCR rearrangement was found in 82% cases (89).
RHOA p.G17v mutation has been found in 60–70% cases of AITL with equal frequencies in lymph nodes as well as skin (90, 91). This mutation is diagnostic of AITL and not present in other PTCL-NOS. Patients harboring this change also tend to have classic AITL histological features and an increased expression of TFH markers making it an important molecular marker of this lineage (92, 93).
Other mutations found in these cases by whole exome sequencing were TET2, DNMT3A, IDH2 in various studies (94–96). Mutations in TCR related genes (PLCG1, CD28, PI3K elements, CTNNB1, and GTF2I) correlated with early disease progression (97). AITL is frequently infiltrated by atypical B cells which sometimes resemble Reed-Sternberg cells posing a diagnostic difficulty. Rearrangement of both Ig genes and TCR has been identified in some cases of AITL (98). When differential expression of genes was studied, TET2 and DNMT3A mutations were found in B and T cells while RHOA and IDH2 mutations were found only in tumor cells (99).
Conclusion
Novel molecular discoveries have vastly advanced the field of CTCLs providing not only a deeper understanding of tumor biology, immune response, but also have introduced new targeted therapeutic strategies. Table 2 summarizes various molecular biomarkers being studied for targeted therapy in CTCL.
Expert opinion: T cell clonality detection by NGS helps identify primary clonal sequences that is helpful for diagnosis of CTCL as well as follow up minimal residual analysis. TCR gamma PCR is the standard of care in most of the labs but it has a variable sensitivity and a lower specificity. In our opinion, NGS should be included in the diagnostic work up as well as management of MRD.
Author Contributions
RW performed the literature search and wrote most parts of the manuscript. CY conceived the design and framework of the paper, wrote some parts, and edited the whole manuscript.
Conflict of Interest
CY has research funding with OBI pharmaceutical and Pfizer for unrelated projects.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. (2016) 127:2375–90. doi: 10.1182/blood-2016-01-643569
2. Willemze R, Cerroni L, Kempf W, Berti E, Facchetti F, Swerdlow S, et al. The 2018 update of the WHO-EORTC classification for primary cutaneous lymphomas. Blood. (2019) 133:1703–14. doi: 10.1182/blood-2018-11-881268
3. da Silva Almeida AC, Abate F, Khiabanian H, Martinez-Escala E, Guitart J, Tensen C, et al. The mutational landscape of cutaneous T cell lymphoma and Sézary syndrome. Nat Genet. (2015) 47:1465–70. doi: 10.1038/ng.3442
4. McGirt LY, Jia P, Baerenwald DA, Duszynski RJ, Dahlman KB, Zic JA, et al. Whole-genome sequencing reveals oncogenic mutations in mycosis fungoides. Blood. (2015) 126:508–19. doi: 10.1182/blood-2014-11-611194
5. Choi J, Goh G, Walradt T, Hong BS, Bunick C, Chen K, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. (2015) 47:1011–9. doi: 10.1038/ng.3356
6. Ungewickell A, Bhaduri A, Rios E, Reuter J, Lee CS, Mah A, et al. Genomic analysis of mycosis fungoides and Sézary syndrome identifies recurrent alterations in TNFR2. Nat Genet. (2015) 47:1056–60. doi: 10.1038/ng.3370
7. Damsky WE, Choi J. Genetics of cutaneous T cell lymphoma: from bench to bedside. Curr Treat Options Oncol. (2016) 17:33. doi: 10.1007/s11864-016-0410-8
8. Wang L, Ni X, Covington KR, Yang BY, Shiu J, Zhang X, et al. Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet. (2015) 47:1426–34. doi: 10.1038/ng.3444
9. Sekulic A, Liang WS, Tembe W, Izatt T, Kruglyak S, Kiefer JA, et al. Personalized treatment of Sézary syndrome by targeting a novel CTLA4:CD28 fusion. Mol Genet Genomic Med. (2015) 3:130–6. doi: 10.1002/mgg3.121
10. Izban KF, Ergin M, Qin JZ, Martinez RL, Pooley RJ Jr, Saeed S, et al. Constitutive expression of NF-kappa B is a characteristic feature of mycosis fungoides: implications for apoptosis resistance and pathogenesis. Hum Pathol. (2000) 31:1482–90. doi: 10.1053/hupa.2000.20370
11. Chang TP, Poltoratsky V, Vancurova I. Bortezomib inhibits expression of TGF-β1, IL-10, and CXCR4, resulting in decreased survival and migration of cutaneous T cell lymphoma cells. J Immunol. (2015) 194:2942–53. doi: 10.4049/jimmunol.1402610
12. Pérez C, González-Rincón J, Onaindia A, Almaráz C, García-Díaz N, Pisonero H, et al. Mutated JAK kinases and deregulated STAT activity are potential therapeutic targets in cutaneous T-cell lymphoma. Haematologica. (2015) 100:e450–3. doi: 10.3324/haematol.2015.132837
13. Bastidas Torres AN, Cats D, Mei H, Szuhai K, Willemze R, Vermeer M, et al. Genomic analysis reveals recurrent deletion of JAK-STAT signaling inhibitors HNRNPK and SOCS1 in mycosis fungoides. Genes Chromosomes Cancer. (2018) 57:653–64. doi: 10.1002/gcc.22679
14. Wood GS. Analysis of clonality in cutaneous T cell lymphoma and associated diseases. Ann N Y Acad Sci. (2001) 941:26–30. doi: 10.1111/j.1749-6632.2001.tb03707.x
15. Lukowsky A, Muche JM, Sterry W, Audring H. Detection of expanded T cell clones in skin biopsy samples of patients with lichen sclerosus et atrophicus by T cell receptor-gamma polymerase chain reaction assays. J Invest Dermatol. (2000) 115:254–9. doi: 10.1046/j.1523-1747.2000.00040.x
16. Ponti R, Quaglino P, Novelli M, Fierro MT, Comessatti A, Peroni A, et al. T-cell receptor gamma gene rearrangement by multiplex polymerase chain reaction/heteroduplex analysis in patients with cutaneous T-cell lymphoma (mycosis fungoides/Sézary syndrome) and benign inflammatory disease: correlation with clinical, histological and immunophenotypical findings. Br J Dermatol. (2005) 153:565–73. doi: 10.1111/j.1365-2133.2005.06649.x
17. Comfere N, Sundram U, Hurley MY, Swick B. Views of dermatopathologists about clonality assays in the diagnosis of cutaneous T-cell and B-cell lymphoproliferative disorders. J Cutan Pathol. (2018) 45:39–47. doi: 10.1111/cup.13072
18. Jawed SI, Myskowski PL, Horwitz S, Moskowitz A, Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sézary syndrome): part I. Diagnosis: clinical and histopathologic features and new molecular and biologic markers. J Am Acad Dermatol. (2014) 70:205.e201–216; quiz 221–202. doi: 10.1016/j.jaad.2013.07.049
19. Xu C, Wan C, Wang L, Yang HJ, Tang Y, Liu WP. Diagnostic significance of TCR gene clonal rearrangement analysis in early mycosis fungoides. Chin J Cancer. (2011) 30:264–72. doi: 10.5732/cjc.010.10344
20. Hodges E, Krishna MT, Pickard C, Smith JL. Diagnostic role of tests for T cell receptor (TCR) genes. J Clin Pathol. (2003) 56:1–11. doi: 10.1136/jcp.56.1.1
21. Hsiao PF, Hsiao CH, Lin YC, Tseng M, Tsai TF, Jee SH. Histopathologic-molecular correlation in early mycosis fungoides using T-cell receptor gamma gene rearrangement by polymerase chain reaction with laser capture microdissection. J Formos Med Assoc. (2007) 106:265–72. doi: 10.1016/S0929-6646(09)60251-5
22. Ponti R, Fierro MT, Quaglino P, Lisa B, Paola F, Michela O, et al. TCRgamma-chain gene rearrangement by PCR-based GeneScan: diagnostic accuracy improvement and clonal heterogeneity analysis in multiple cutaneous T-cell lymphoma samples. J Invest Dermatol. (2008) 128:1030–8. doi: 10.1038/sj.jid.5701109
23. Sandberg Y, Heule F, Lam K, Lugtenburg PJ, Wolvers-Tettero I, van Dongen JJ, et al. Molecular immunoglobulin/T- cell receptor clonality analysis in cutaneous lymphoproliferations. Experience with the BIOMED-2 standardized polymerase chain reaction protocol. Haematologica. (2003) 88:659–70. Available online at: http://www.haematologica.org/content/88/6/659.long
24. Lukowsky A, Muche JM, Mobs M, Assaf C, Humme D, Hummel M, et al. Evaluation of T-cell clonality in archival skin biopsy samples of cutaneous T-cell lymphomas using the biomed-2 PCR protocol. Diagn Mol Pathol. (2010) 19:70–7. doi: 10.1097/PDM.0b013e3181b2a1b7
25. Lee SC, Berg KD, Racke FK, Griffin C, Eshleman JR. Pseudo-spikes are common in histologically benign lymphoid tissues. J Mol Diagn. (2000) 2:145–52. doi: 10.1016/S1525-1578(10)60630-7
26. Tang MB, Chong TK, Tan ES, Sun Y, Tan SH. A comparative study of polymerase chain reaction detection of clonal T-cell receptor gamma chain gene rearrangements using polyacrylamide gel electrophoresis versus fluorescence capillary electrophoresis Ann Acad Med Singapore. (2008) 37:27–31. Available online at: http://www.annals.edu.sg/pdf/37VolNo1Jan2008/V37N1p27.pdf
27. Kirsch IR, Watanabe R, O'Malley JT, Williamson DW, Scott LL, Elco CP, et al. TCR sequencing facilitates diagnosis and identifies mature T cells as the cell of origin in CTCL. Sci Transl Med. (2015) 7:308ra158. doi: 10.1126/scitranslmed.aaa9122
28. Sufficool KE, Lockwood CM, Abel HJ, Hagemann I, Schumacher JA, Kelley TW, et al. T-cell clonality assessment by next-generation sequencing improves detection sensitivity in mycosis fungoides. J Am Acad Dermatol. (2015) 73:228–36.e222. doi: 10.1016/j.jaad.2015.04.030
29. Rea B, Haun P, Emerson R, Vignali M, Farooqi M, Samimi S, et al. Role of high-throughput sequencing in the diagnosis of cutaneous T-cell lymphoma. J Clin Pathol. (2018) 71:814–20. doi: 10.1136/jclinpath-2018-205004
30. Dippel E, Assaf C, Hummel M, Schrag HJ, Stein H, Goerdt S, et al. Clonal T-cell receptor gamma-chain gene rearrangement by PCR-based GeneScan analysis in advanced cutaneous T-cell lymphoma: a critical evaluation. J Pathol. (1999) 188:146–54. doi: 10.1002/(SICI)1096-9896(199906)188:2<146::AID-PATH334>3.0.CO;2-7
31. Schiller PI, Flaig MJ, Puchta U, Kind P, Sander C. Detection of clonal T cells in lichen planus Arch Dermatol Res. (2000) 292:568–9. doi: 10.1007/s004030000178
32. Thurber SE, Zhang B, Kim YH, Schrijver I, Zehnder J, Kohler S. T-cell clonality analysis in biopsy specimens from two different skin sites shows high specificity in the diagnosis of patients with suggested mycosis fungoides. J Am Acad Dermatol. (2007) 57:782–90. doi: 10.1016/j.jaad.2007.06.004
33. Gibson JF, Huang J, Liu KJ, Carlson KR, Foss F, Choi J, et al. Cutaneous T-cell lymphoma (CTCL): current practices in blood assessment and the utility of T-cell receptor (TCR)-Vβ chain restriction. J Am Acad Dermatol. (2016) 74:870–7. doi: 10.1016/j.jaad.2015.12.018
34. Olsen E, Vonderheid E, Pimpinelli N, Willemze R, Kim Y, Knobler R, et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood. (2007) 110:1713–22. doi: 10.1182/blood-2007-03-055749
35. Hurabielle C, Ingen-Housz-Oro S, Ortonne N, Cornillet-Lefebvre P, Merah A, D'Incan M, et al. Frequency and prognostic value of cutaneous molecular residual disease in mycosis fungoides: a prospective multicentre trial of the Cutaneous Lymphoma French Study Group. Br J Dermatol. (2015) 173:1015–23. doi: 10.1111/bjd.14017
36. Weng WK, Armstrong R, Arai S, Desmarais C, Hoppe R, Kim Y, et al. Minimal residual disease monitoring with high-throughput sequencing of T cell receptors in cutaneous T cell lymphoma. Sci Transl Med. (2013) 5:214ra171. doi: 10.1126/scitranslmed.3007420
37. Kanakry CG, Coffey DG, Towlerton AM, Vulic A, Storer BE, Chou J, et al. Origin and evolution of the T cell repertoire after posttransplantation cyclophosphamide. JCI Insight. (2016) 1:e86252. doi: 10.1172/jci.insight.86252
38. Xu L, You X, Zheng P, Zhang BM, Gupta P, Lavori P, et al. Methodologic considerations in the application of next-generation sequencing of human TRB repertoires for clinical use. J Mol Diagn. (2017) 19:72–83. doi: 10.1016/j.jmoldx.2016.07.009
39. Massone C, Crisman G, Kerl H, Cerroni L. The prognosis of early mycosis fungoides is not influenced by phenotype and T-cell clonality. Br J Dermatol. (2008) 159:881–6. doi: 10.1111/j.1365-2133.2008.08761.x
40. de Masson A, O'Malley JT, Elco CP, Garcia S, Divito SJ, Lowry EL, et al. High-throughput sequencing of the T cell receptor β gene identifies aggressive early-stage mycosis fungoides. Sci Transl Med. (2018) 10:eaar5894. doi: 10.1126/scitranslmed.aar5894
41. Raess PW, Bagg A. The role of molecular pathology in the diagnosis of cutaneous lymphomas. Patholog Res Int. (2012) 2012:913523. doi: 10.1155/2012/913523
42. Zhang B, Beck AH, Taube JM, Kohler S, Seo K, Zwerner J, et al. Combined use of PCR-based TCRG and TCRB clonality tests on paraffin-embedded skin tissue in the differential diagnosis of mycosis fungoides and inflammatory dermatoses. J Mol Diagn. (2010) 12:320–7. doi: 10.2353/jmoldx.2010.090123
43. Langerak AW, Groenen PJ, Brüggemann M, Beldjord K, Bellan C, Bonello L, et al. EuroClonality/BIOMED-2 guidelines for interpretation and reporting of Ig/TCR clonality testing in suspected lymphoproliferations. Leukemia. (2012) 26:2159–71. doi: 10.1038/leu.2012.246
44. Schumacher JA, Duncavage EJ, Mosbruger TL, Szankasi P, Kelley TW. A comparison of deep sequencing of TCRG rearrangements vs traditional capillary electrophoresis for assessment of clonality in T-Cell lymphoproliferative disorders. Am J Clin Pathol. (2014) 141:348–59. doi: 10.1309/AJCP5TYGBVW4ZITR
45. Buus TB, Willerslev-Olsen A, Fredholm S, Blümel E, Nastasi C, Gluud M, et al. Single-cell heterogeneity in Sézary syndrome. Blood Adv. (2018) 2:2115–26. doi: 10.1182/bloodadvances.2018022608
46. Borcherding N, Voigt AP, Liu V, Link BK, Zhang W, Jabbari A. Single-cell profiling of cutaneous T-cell lymphoma reveals underlying heterogeneity associated with disease progression. Clin Cancer Res. (2019) 25:2996–3005. doi: 10.1158/1078-0432.CCR-18-3309
47. Gaydosik AM, Tabib T, Gerskin LJ, Bayan CY, Conway JF, Lafyatis R, et al. Single-cell lymphocyte heterogeneity in advanced Cutaneous T-Cell Lymphoma skin tumors. Clin Cancer Res. (2019) 25:4443–54. doi: 10.1158/1078-0432.CCR-19-0148
48. Baslan T, Hicks J. Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat Rev Cancer. (2017) 17:557–69. doi: 10.1038/nrc.2017.58
49. Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N, Werb Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat Cell Biol. (2018) 20:1349–60. doi: 10.1038/s41556-018-0236-7
50. Segundo-Val IS, Sanz-Lozano CS. Introduction to the gene expression analysis. Methods Mol Biol. (2016) 1434:29–43. doi: 10.1007/978-1-4939-3652-6_3
51. Tsang HF, Xue VW, Koh SP, Chiu Y, Ng LP, Wong SC. NanoString, a novel digital color-coded barcode technology: current and future applications in molecular diagnostics. Expert Rev Mol Diagn. (2017) 17:95–103. doi: 10.1080/14737159.2017.1268533
52. Litvinov IV, Tetzlaff MT, Thibault P, Gangar P, Moreau L, Watters AK, et al. Gene expression analysis in Cutaneous T-Cell Lymphomas (CTCL) highlights disease heterogeneity and potential diagnostic and prognostic indicators. Oncoimmunology. (2017) 6:e1306618. doi: 10.1080/2162402X.2017.1306618
53. Willemze R, Jaffe ES, Burg G, Cerroni L, Berti E, Swerdlow SH, et al. WHO-EORTC classification for cutaneous lymphomas. Blood. (2005) 105:3768–85. doi: 10.1182/blood-2004-09-3502
54. Campbell JJ, Clark RA, Watanabe R, Kupper TS. Sezary syndrome and mycosis fungoides arise from distinct T-cell subsets: a biologic rationale for their distinct clinical behaviors. Blood. (2010) 116:767–71. doi: 10.1182/blood-2009-11-251926
55. Clark RA. Resident memory T cells in human health and disease. Sci Transl Med. (2015) 7:269rv261. doi: 10.1126/scitranslmed.3010641
56. Litvinov IV, Netchiporouk E, Cordeiro B, Doré MA, Moreau L, Pehr K, et al. The use of transcriptional profiling to improve personalized diagnosis and management of cutaneous T-cell lymphoma (CTCL). Clin Cancer Res. (2015) 21:2820–9. doi: 10.1158/1078-0432.CCR-14-3322
57. Nebozhyn M, Loboda A, Kari L, Rook AH, Vonderheid E, Lessin S, et al. Quantitative PCR on 5 genes reliably identifies CTCL patients with 5% to 99% circulating tumor cells with 90% accuracy. Blood. (2006) 107:3189–96. doi: 10.1182/blood-2005-07-2813
58. Michel L, Jean-Louis F, Begue E, Bensussan A, Bagot M. Use of PLS3, Twist, CD158k/KIR3DL2, and NKp46 gene expression combination for reliable Sézary syndrome diagnosis. Blood. (2013) 121:1477–8. doi: 10.1182/blood-2012-10-460535
59. Bagot M. New targeted treatments for cutaneous T-cell lymphomas. Indian J Dermatol. (2017) 62:142–5. doi: 10.4103/ijd.IJD_73_17
60. Van Der Weyden C, Bagot M, Neeson P, Darcy PK, Prince H. IPH4102, a monoclonal antibody directed against the immune receptor molecule KIR3DL2, for the treatment of cutaneous T-cell lymphoma. Expert Opin Investig Drugs. (2018) 27:691–7. doi: 10.1080/13543784.2018.1498081
61. Kim YH, Bagot M, Pinter-Brown L, Rook AH, Porcu P, Horwitz SM, et al. Mogamulizumab versus vorinostat in previously treated cutaneous T-cell lymphoma (MAVORIC): an international, open-label, randomised, controlled phase 3 trial. Lancet Oncol. (2018) 19:1192–204. doi: 10.1016/S1470-2045(18)30379-6
62. Chen L, Carson KR, Staser KW, Mehta-Shah N, Schaffer A, Rosman IS, et al. Mogamulizumab-associated cutaneous granulomatous drug eruption mimicking mycosis fungoides but possibly indicating durable clinical response. JAMA Dermatol. (2019) 155: 968–71. doi: 10.1001/jamadermatol.2019.0369
63. Zhang Y, Wang Y, Yu R, Huang Y, Su M, Xiao C, et al. Molecular markers of early-stage mycosis fungoides. J Invest Dermatol. (2012) 132:1698–706. doi: 10.1038/jid.2012.13
64. Huang Y, Litvinov IV, Wang Y, Su MW, Tu P, Jiang X, et al. Thymocyte selection-associated high mobility group box gene (TOX) is aberrantly over-expressed in mycosis fungoides and correlates with poor prognosis. Oncotarget. (2014) 5:4418–25. doi: 10.18632/oncotarget.2031
65. Yu X, Li Z. TOX gene: a novel target for human cancer gene therapy. Am J Cancer Res. (2015) 5:3516–24. Available online at: http://www.ajcr.us/AJCR_V5N12.html
66. Ralfkiaer U, Hagedorn PH, Bangsgaard N, Løvendorf MB, Ahler CB, Svensson L, et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL). Blood. (2011) 118:5891–900. doi: 10.1182/blood-2011-06-358382
67. Dusílková N, Bašová P, Polívka J, Kodet O, Kulvait V, Pešta M, et al. Plasma miR-155, miR-203, and miR-205 are biomarkers for monitoring of primary cutaneous T-cell lymphomas. Int J Mol Sci. (2017) 18:2136. doi: 10.3390/ijms18102136
68. Ralfkiaer U, Lindahl LM, Litman T, Gjerdrum L, Ahler CB, Gniadecki R, et al. MicroRNA expression in early mycosis fungoides is distinctly different from atopic dermatitis and advanced cutaneous T-cell lymphoma. Anticancer Res. (2014) 34:7207–17. Available online at: http://ar.iiarjournals.org/content/34/12/7207.long
69. Lindahl LM, Besenbacher S, Rittig AH, Celis P, Willerslev-Olsen A, Gjerdrum LMR, et al. Prognostic miRNA classifier in early-stage mycosis fungoides: development and validation in a Danish nationwide study. Blood. (2018) 131:759–70. doi: 10.1182/blood-2017-06-788950
70. Izykowska K, Przybylski GK, Gand C, Braun FC, Grabarczyk P, Kuss AW, et al. Genetic rearrangements result in altered gene expression and novel fusion transcripts in Sézary syndrome. Oncotarget. (2017) 8:39627–39. doi: 10.18632/oncotarget.17383
71. Bekkenk MW, Geelen FA, van Voorst Vader PC, Heule F, Geerts M, van Vloten WA, et al. Primary and secondary cutaneous CD30(+) lymphoproliferative disorders: a report from the Dutch Cutaneous Lymphoma Group on the long-term follow-up data of 219 patients and guidelines for diagnosis and treatment. Blood. (2000) 95:3653–61. doi: 10.1182/blood.V95.12.3653.012k23_3653_3661
72. Saggini A, Gulia A, Argenyi Z, Fink-Puches R, Lissia A, Magaña M, et al. A variant of lymphomatoid papulosis simulating primary cutaneous aggressive epidermotropic CD8+ cytotoxic T-cell lymphoma. Description of 9 cases. Am J Surg Pathol. (2010) 34:1168–75. doi: 10.1097/PAS.0b013e3181e75356
73. Kempf W, Kazakov DV, Paredes BE, Laeng H, Palmedo G, Kutzner H. Primary cutaneous anaplastic large cell lymphoma with angioinvasive features and cytotoxic phenotype: a rare lymphoma variant within the spectrum of CD30+ lymphoproliferative disorders. Dermatology. (2013) 227:346–52. doi: 10.1159/000355479
74. Karai LJ, Kadin ME, Hsi ED, Sluzevich J, Ketterling RP, Knudson RA, et al. Chromosomal rearrangements of 6p25.3 define a new subtype of lymphomatoid papulosis. Am J Surg Pathol. (2013) 37:1173–81. doi: 10.1097/PAS.0b013e318282d01e
75. Greisser J, Palmedo G, Sander C, Kutzner H, Kazakov DV, Roos M, et al. Detection of clonal rearrangement of T-cell receptor genes in the diagnosis of primary cutaneous CD30 lymphoproliferative disorders. J Cutan Pathol. (2006) 33:711–5. doi: 10.1111/j.1600-0560.2006.00560.x
76. de la Garza Bravo MM, Patel KP, Loghavi S, Curry JL, Torres Cabala CA, Cason RC, et al. Shared clonality in distinctive lesions of lymphomatoid papulosis and mycosis fungoides occurring in the same patients suggests a common origin. Hum Pathol. (2015) 46:558–69. doi: 10.1016/j.humpath.2014.12.008
77. Sun J, Yi S, Qiu L, Fu W, Wang A, Liu F, et al. SATB1 defines a subtype of cutaneous CD30. J Invest Dermatol. (2018) 138:1795–804. doi: 10.1016/j.jid.2018.02.028
78. Wada DA, Law ME, Hsi ED, Dicaudo D, Ma L, Lim MS, et al. Specificity of IRF4 translocations for primary cutaneous anaplastic large cell lymphoma: a multicenter study of 204 skin biopsies. Mod Pathol. (2011) 24:596–605. doi: 10.1038/modpathol.2010.225
79. Prieto-Torres L, Rodriguez-Pinilla SM, Onaindia A, Ara M, Requena L, Piris MÁ. CD30-positive primary cutaneous lymphoproliferative disorders: molecular alterations and targeted therapies. Haematologica. (2019) 104:226–35. doi: 10.3324/haematol.2018.197152
80. Onaindia A, Montes-Moreno S, Rodríguez-Pinilla SM, Batlle A, González de Villambrosía S, Rodríguez AM, et al. Primary cutaneous anaplastic large cell lymphomas with 6p25.3 rearrangement exhibit particular histological features. Histopathology. (2015) 66:846–55. doi: 10.1111/his.12529
81. Fauconneau A, Pham-Ledard A, Cappellen D, Frison E, Prochazkova-Carlotti M, Parrens M, et al. Assessment of diagnostic criteria between primary cutaneous anaplastic large-cell lymphoma and CD30-rich transformed mycosis fungoides; a study of 66 cases. Br J Dermatol. (2015) 172:1547–54. doi: 10.1111/bjd.13690
82. Feldman AL, Dogan A, Smith DI, Law ME, Ansell SM, Johnson SH, et al. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively parallel genomic sequencing. Blood. (2011) 117:915–9. doi: 10.1182/blood-2010-08-303305
83. Velusamy T, Kiel MJ, Sahasrabuddhe AA, Rolland D, Dixon CA, Bailey NG, et al. A novel recurrent NPM1-TYK2 gene fusion in cutaneous CD30-positive lymphoproliferative disorders. Blood. (2014) 124:3768–71. doi: 10.1182/blood-2014-07-588434
84. Vasmatzis G, Johnson SH, Knudson RA, Ketterling R, Braggio E, Fonseca R, et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood. (2012) 120:2280–9. doi: 10.1182/blood-2012-03-419937
85. Oschlies I, Lisfeld J, Lamant L, Nakazawa A, d'Amore ES, Hansson U, et al. ALK-positive anaplastic large cell lymphoma limited to the skin: clinical, histopathological and molecular analysis of 6 pediatric cases. A report from the ALCL99 study. Haematologica. (2013) 98:50–6. doi: 10.3324/haematol.2012.065664
86. Battistella M, Janin A, Jean-Louis F, Collomb C, Leboeuf C, Sicard H, et al. KIR3DL2 (CD158k) is a potential therapeutic target in primary cutaneous anaplastic large-cell lymphoma. Br J Dermatol. (2016) 175:325–33. doi: 10.1111/bjd.14626
87. Benner MF, Ballabio E, van Kester MS, Saunders NJ, Vermeer MH, Willemze R, et al. Primary cutaneous anaplastic large cell lymphoma shows a distinct miRNA expression profile and reveals differences from tumor-stage mycosis fungoides. Exp Dermatol. (2012) 21:632–4. doi: 10.1111/j.1600-0625.2012.01548.x
88. Dogan A, Attygalle AD, Kyriakou C. Angioimmunoblastic T-cell lymphoma. Br J Haematol. (2003) 121:681–91. doi: 10.1046/j.1365-2141.2003.04335.x
89. Oishi N, Sartori-Valinotti JC, Bennani NN, Wada D, He R, Cappel MA, et al. Cutaneous lesions of angioimmunoblastic T-cell lymphoma: clinical, pathological, and immunophenotypic features. J Cutan Pathol. (2019) 46:637–44. doi: 10.1111/cup.13475
90. Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. (2014) 46:171–5. doi: 10.1038/ng.2872
91. Leclaire Alirkilicarslan A, Dupuy A, Pujals A, Parrens M, Vergier B, Robson A, et al. Expression of TFH markers and detection of RHOA p.G17V and IDH2 p.R172K/S mutations in cutaneous localizations of angioimmunoblastic T-cell lymphomaS. Am J Surg Pathol. (2017) 41:1581–92. doi: 10.1097/PAS.0000000000000956
92. Nagao R, Kikuti YY, Carreras J, Kikuchi T, Miyaoka M, Matsushita H, et al. Clinicopathologic analysis of angioimmunoblastic T-cell lymphoma with or without RHOA G17V mutation using formalin-fixed paraffin-embedded sections. Am J Surg Pathol. (2016) 40:1041–50. doi: 10.1097/PAS.0000000000000651
93. Ondrejka SL, Grzywacz B, Bodo J, Makishima H, Polprasert C, Said J, et al. Angioimmunoblastic T-cell lymphomas with the RHOA p.Gly17Val mutation have classic clinical and pathologic features. Am J Surg Pathol. (2016) 40:335–41. doi: 10.1097/PAS.0000000000000555
94. Odejide O, Weigert O, Lane AA, Toscano D, Lunning M, Kopp N, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. (2014) 123:1293–6. doi: 10.1182/blood-2013-10-531509
95. Palomero T, Couronné L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. (2014) 46:166–70. doi: 10.1038/ng.2873
96. Wang M, Zhang S, Chuang SS, Ashton-Key M, Ochoa E, Bolli N, et al. Angioimmunoblastic T cell lymphoma: novel molecular insights by mutation profiling. Oncotarget. (2017) 8:17763–70. doi: 10.18632/oncotarget.14846
97. Vallois D, Dobay MP, Morin RD, Lemonnier F, Missiaglia E, Juilland M, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. (2016) 128:1490–502. doi: 10.1182/blood-2016-02-698977
98. de Leval L, Gisselbrecht C, Gaulard P. Advances in the understanding and management of angioimmunoblastic T-cell lymphoma. Br J Haematol. (2010) 148:673–89. doi: 10.1111/j.1365-2141.2009.08003.x
99. Nguyen TB, Sakata-Yanagimoto M, Asabe Y, Matsubara D, Kano J, Yoshida K, et al. Identification of cell-type-specific mutations in nodal T-cell lymphomas. Blood Cancer J. (2017) 7:e516. doi: 10.1038/bcj.2016.122
100. Duvic M, Tetzlaff MT, Gangar P, Clos AL, Sui D, Talpur R. Results of a phase II trial of brentuximab vedotin for CD30+ cutaneous T-cell lymphoma and lymphomatoid papulosis. J Clin Oncol. (2015) 33:3759–65. doi: 10.1200/JCO.2014.60.3787
Keywords: cutaneous T cell lymphoma (CTCL), molecular biology, TCR clonality, mycosis fungoides, NGS, single cell sequencing
Citation: Walia R and Yeung CCS (2020) An Update on Molecular Biology of Cutaneous T Cell Lymphoma. Front. Oncol. 9:1558. doi: 10.3389/fonc.2019.01558
Received: 16 October 2019; Accepted: 23 December 2019;
Published: 22 January 2020.
Edited by:
Basem M. William, The Ohio State University, United StatesReviewed by:
Pier Paolo Piccaluga, University of Bologna, ItalyKelly Quek, University of Texas MD Anderson Cancer Center, United States
Copyright © 2020 Walia and Yeung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cecilia C. S. Yeung, Y3lldW5nJiN4MDAwNDA7ZnJlZGh1dGNoLm9yZw==
†These authors have contributed equally to this work
 Ritika Walia
Ritika Walia Cecilia C. S. Yeung
Cecilia C. S. Yeung