
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Oncol. , 22 November 2019
Sec. Hematologic Malignancies
Volume 9 - 2019 | https://doi.org/10.3389/fonc.2019.01304
 Xiaoxue Wang1,2
Xiaoxue Wang1,2 Xinyue Huang1
Xinyue Huang1 Hui Pang2
Hui Pang2 Sheng Xiao3
Sheng Xiao3 Hongcang Gu2,4
Hongcang Gu2,4 Heyang Zhang1
Heyang Zhang1 Baixun Wang1
Baixun Wang1 Lijun Zhang1
Lijun Zhang1 Xiaojing Yan1*
Xiaojing Yan1*Background: Myeloid/Lymphoid Neoplasm with FGFR1 Rearrangement is a rare kind of hematological malignant disease.
Case presentation: A 37-year-old male patient experienced three distinct disease stages from myeloproliferative neoplasm (MPN), T-cell lymphoblastic lymphoma (T-LBL) to a much more complexed phage of a mixed phenotype acute leukemia (MPAL). Both genetic and genomic alternations were detected including chromosomal abnormality and genic mutations.
Result: Karyotyping and fluorescence in situ hybridization (FISH) analysis of either bone marrow or lymph node sample confirmed the presence of the FGFR1 rearrangement. Amplifications of RUNX1, ERG, and U2AF1 genes were identified by next generation sequencing. Furthermore, a frame-shift variant of F330fs*>149 in the RUNX1 gene and a missense mutation of R2263Q in NOTCH1 were also detected.
Conclusion: The FGFR1 rearrangement functions as a trigging oncogenic event. Then other genetic events such as RUNX1 and/or NOTCH1 alternations further lead to progression of disease with trilineage blasts assignment.
Myeloid/lymphoid neoplasms with fibroblast growth factor receptor-1 (FGFR1) rearrangement has been presented as an independent disease classification in the 2016 World Health Organization classification of tumors of hematopoietic and lymphoid tissues (1). Rearrangement of the FGFR1 gene locus leads to more than 10 fused genes, which code for fusion proteins containing the N-terminal derived from a partner gene and the C-terminal of tyrosine kinase domain from the FGFR1 gene, which causes constitutive activation of FGFR1 (2). The patients with FGFR1 rearrangement usually initiate with myeloproliferative neoplasm (MPN) and undergo through diverse clinical courses such as lymphoblastic lymphoma (LBL), acute lymphoblastic leukemia (ALL) or acute myeloid leukemia (AML) (3). Here, we reported an interesting and very rare case which presented sequentially as MPN, T-LBL in lymph nodes and mixed phenotype acute leukemia
(MPAL) in bone marrow. To the best of our knowledge, our case represents the first report of this disease with three distinct developmental phases.
A 37-year-old male patient was admitted to our center in August 2017 due to abnormal complete blood count (CBC) with WBC 34.1*109/L and a hemoglobin (HGB) level of 205 g/L. Further bone marrow (BM) evaluation showed hypercellularity with trilineage growth including prominent granulocytic, erythroid and megakaryocytic proliferation (Figure 1A). Myelofibrosis was absent, also JAK2 V617F/JAK2 exon 12/MPL/CALR gene mutations or BCR/ABL fusion gene were not identified. The EPO level decreased to 0.82 mIU/ml, thus the clinical diagnosis of polycythemia vera (PV) was made by combining with BM biopsy findings and increased hemoglobin. After treatment of phlebotomy and hydroxyurea, patient's CBC dropped back normal.
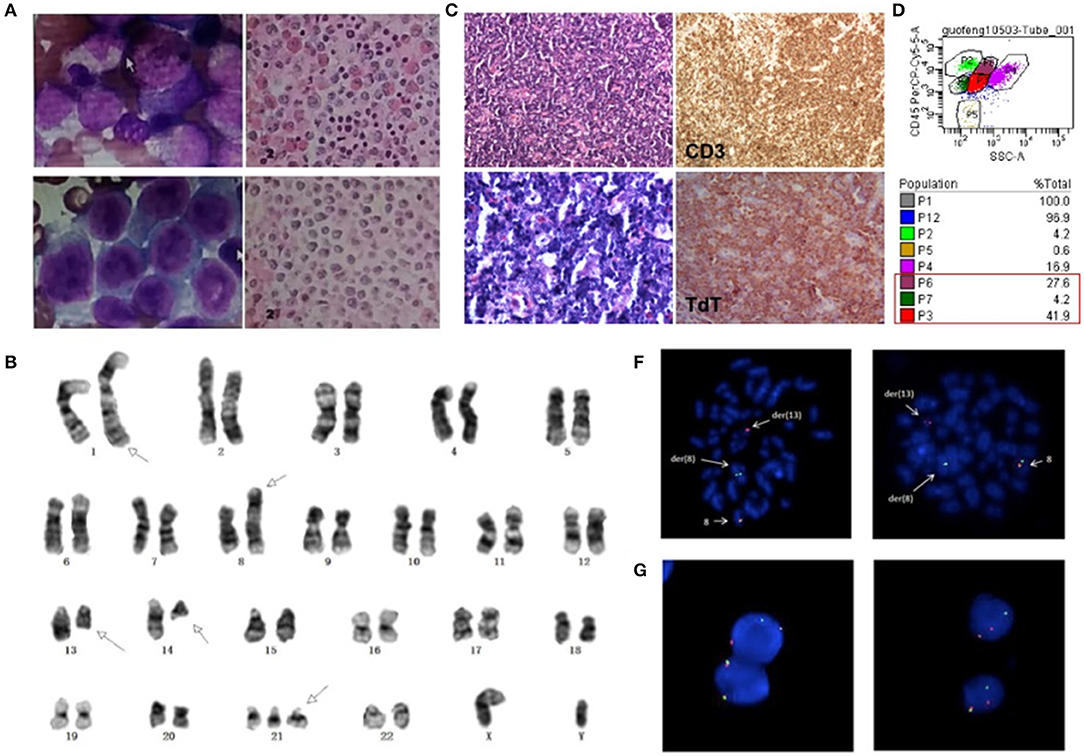
Figure 1. (A) Morphologic changes of BM smear and biopsy showed trilineage hypercellularity at initial onset. (B) Cervical lymph node biopsy with infiltration of T lymphoblasts. (C) BM smear showed markedly increased abnormal blasts and BM biopsy showed diffuse hyperplasia of immature cells at leukemic stage. (D) Flow cytometry analysis showed three separate clonal populations in BM sample. (E) G-banding karyotype of the bone marrow (BM) cells demonstrated the t(8;13)(p11.2;ql2.l); t(l;l4)(q42;q12) and trisomy 21. Arrows show the abnormal chromosomes. FISH analysis of BM cells (F) and lymph nodes (G) with an FGFR1 break-apart DNA probe revealing FGFR1 rearrangement (the 5'FGFR1 is labeled green, the 3'FGFR1 is labeled red).
Three months later, the patient came back to the hospital for bilateral cervical lymphadenopathy with fever and fatigue. Ultrasonographic findings suggest splenomegaly and generalized lymphadenopathy on both sides of the diaphragm. Immunohistochemical staining for cervical lymph node biopsy displayed as diffused abnormal proliferative lymphoblastic cells with CD3(+) TdT(+) CD99(+) CD4(+) Ki67(70%), while few scattered cells were positive for MPO, CD117 and CD8 (Figure 1B). Then, the patient was diagnosed as T-lymphoblastic lymphoma (LBL). The CBC of the patient showed a WBC count of 27.16*109/L and a HGB level of 200 g/L. Intriguingly, bone marrow aspirate revealed hypercellularity with predominant blasts (Figure 1C) and flow cytometry showed three separate clonal populations (Figure 1D): (1) The majority of blasts were cells with ambiguous lineage (P3 group, 41.9%), mainly expressed CD33, CD34, CD13, HLA-DR, CD123; partially expressed CD117, CD22, CD19, CD10, cCD79a, and MPO. (2) The myeloid subset (P6 group, 27.6%) mainly expressed CD33, CD13, HLA-DR, CD123, CD4, CD11c and partially expressed CD14 and CD22. (3) The B-lymphoblast subset (P7 group, 4.2%) mainly expressed CD34, CD19, CD22, CD10, cCD79a, HLA-DR and partially expressed CD33 and CD13. Accordingly, the patient was diagnosed as MPAL. Karyotyping analysis of BM sample illustrated as 47, XY, t(1;14)(q42;q12), t(8;13)(p11.2;q12), +21 [20] (Figure 1E) and fluorescence in situ hybridization (FISH) analysis using the FGFR1 break-apart probe confirmed the presence of the FGFR1 rearrangement in all metaphase cells scored (Figure 1F). Then, next generation sequencing (NGS) assay of 118 commonly mutated genes in hematological disorders (listed in Supplementary Table 1) identified the amplifications of RUNX1, ERG, and U2AF1 genes on chromosome 21p. Remarkably, a frame-shift variant of F330fs*>149 in the RUNX1 gene and a missense mutation of R2263Q in NOTCH1 were also detected. However, the NGS assay didn't identify any gene mutation using the BM samples collected during the patient's first visit. Interestingly, FGFR1 rearrangement was also detected in 36% (18/50) of cells in lymph node biopsy sample (Figure 1G). Based on the clinical course and laboratory findings, the patient was finally diagnosed as myeloid/lymphoid neoplasm with FGFR1 rearrangement.
The patient was treated with CHOP and hyper-CVAD chemotherapy. However, complete remission (CR) was not achieved. The patient could not find suitable HLA-matched donor for allogeneic hematopoietic stem cell transplantation (HSCT), and no further treatment were performed. Unfortunately, the patient died 10 months after onset.
Myeloid/lymphoid neoplasms with FGFR1 rearrangement, also known as the 8p11 myeloproliferative syndrome (EMS) or stem cell leukemia/lymphoma syndrome (SCLL), is extremely rare with around 100 cases reported until now which represented as MPN or more complicated situations (4, 5). It has been reported that FGFR1 could activate multiple signaling pathways as a receptor tyrosine kinase and constitutive activation of FGFR1 by gene fusion is a critical event in oncogenesis (2, 4). The prognosis of patients were generally poor and HSCT is probably the best choice for a long-term survival (5, 6). The presence of multiple lineage disorders suggests the involvement of stem cells for patients harboring FGFR1 rearrangement (3). Gain of an additional copy of chromosome 21 is associated with progression of EMS, but its exact role remains unclear (2). Here we detected the amplifications of RUNX1, ERG, and U2AF1 genes on chromosome 21p, which have been reported playing important roles in the pathogenesis of hematological malignancies (7, 8). A high frequency of RUNX1 mutation can be seen in patients with FGFR1 rearrangement (9), which have also been detected in a variety of hematological malignancies. Here, we also identified a missense variant of NOTCH1 and this gene is recurrently found mutated in T-ALL but firstly reported in EMS. While, RUNX1 and NOTCH1 mutations were identified in the late stage of the disease (MPAL), but not in the initial stage of the disease (MPN). These findings indicated that FGFR1 rearrangement might be the initial oncogenic event in the hematopoietic pluripotent stem cells, which promote the trilineage hematopoiesis. Then other genetic events such as RUNX1 and/or NOTCH1 abnormalities further accelerated the progress of the disease, resulting in acute leukemia with trilineage blasts assignment (Figure 2). The abnormal hematopoietic stem cells present various phenotypes because of distinct microenvironments, such as lymphoma in lymph nodes or leukemia/MPN in bone marrow.
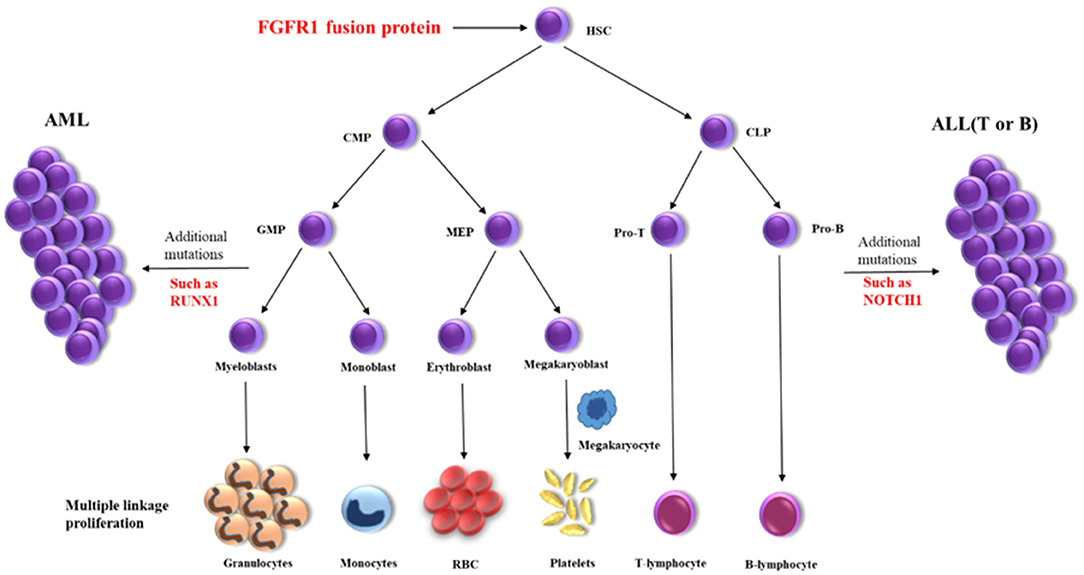
Figure 2. Schematic diagram of the disease progression mechanism. HSC, hematopoietic stem cells; CMP, conu11on myeloid progenitor; CLP, common lymphoid progenitor; GMP, granulocyte macrophage progenitor; MEP, megakaryocyte erythroid progenitor; Pro-T, T progenitor cells; Pro-B, B progenitor cells.
Our case provides a typical clinico-pathogenic model of myeloid/lymphoid neoplasm with FGFR1 rearrangement, which has not been reported previously. More clinical data are needed to identify the pathogenesis and interaction mechanism as well as optimal therapeutic approach to improve clinical outcomes.
All datasets generated for this study are included in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by China Medical University. The patients/participants provided their written informed consent to participate in this study.
Written informed consent was obtained from the patient for publication of this case report and the accompanying images.
XW collected and analyzed all data, wrote the draft. HP and HG responsible for the revision. XH, HZ, BW, and LZ cared for the patient and provided clinical history. SX performed the Karyotype, FISH, and NGS. XY designed the study and responsible for the revisions.
This study received financial support from the National Youth Top-notch Talent of Ten Thousand Talent Program, the National Natural Science Foundation of China (NSFC, 81170519; NSFC, 81900153), and New Century Excellent Talents in University (NCET-13-1037).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.01304/full#supplementary-material
1. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. (2016) 127:2391–405. doi: 10.1182/blood-2016-03-643544.
2. Jackson CC, Medeiros LJ, Miranda RN. 8p11 myeloproliferative syndrome: a review. Hum Pathol. (2010). 41:461–76. doi: 10.1016/j.humpath.2009.11.003
3. Kumar KR, Chen W, Koduru PR, Luu HS. Myeloid and lymphoid neoplasm with abnormalities of FGFR1 presenting with trilineage blasts and RUNX1 rearrangement: a case report and review of literature. Am J Clin Pathol. (2015) 143:738–48. doi: 10.1309/AJCPUD6W1JLQQMNA
4. Macdonald D, Reiter A, Cross NC. The 8p11 myeloproliferative syndrome: a distinct clinical entity caused by constitutive activation of FGFR1. Acta Haematol. (2002) 107:101–7. doi: 10.1159/000046639
5. Strati P, Tang G, Duose DY, Mallampati S, Luthra R, Patel KP, et al., Myeloid/lymphoid neoplasms with FGFR1 rearrangement. Leuk Lymphoma. (2018) 59:1672–6. doi: 10.1080/10428194.2017.1397663
6. Umino K, Fujiwara SI, Ikeda T, Toda Y, Ito S, Mashima K, et al. Clinical outcomes of myeloid/lymphoid neoplasms with fibroblast growth factor receptor-1 (FGFR1) rearrangement. Hematology. (2018) 23:470–7. doi: 10.1080/10245332.2018.1446279
7. Ilagan JO, Ramakrishnan A, Hayes B, Murphy ME, Zebari AS, Bradley P, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. (2015) 25:4–26. doi: 10.1101/gr.181016.114
8. Mandoli A, Singh AA, Prange KHM, Tijchon E, Oerlemans M, Dirks R, et al. The hematopoietic transcription factors RUNX1 and ERG prevent AML1-ETO oncogene overexpression and onset of the apoptosis program in t(8;21) AMLs. Cell Rep. (2016) 17:2087–100. doi: 10.1016/j.celrep.2016.08.082
Keywords: FGFR1, MPN, MPAL, Runx1, Notch
Citation: Wang X, Huang X, Pang H, Xiao S, Gu H, Zhang H, Wang B, Zhang L and Yan X (2019) Myeloid/Lymphoid Neoplasm With FGFR1 Rearrangement Accompanying RUNX1 and NOTCH1 Gene Mutations. Front. Oncol. 9:1304. doi: 10.3389/fonc.2019.01304
Received: 23 August 2019; Accepted: 11 November 2019;
Published: 22 November 2019.
Edited by:
Adam Finn Binder, Jefferson University Hospitals, Thomas Jefferson University, United StatesReviewed by:
Husheng Ding, Mayo Clinic, United StatesCopyright © 2019 Wang, Huang, Pang, Xiao, Gu, Zhang, Wang, Zhang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojing Yan, eWFueGlhb2ppbmdfcHBAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.