
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 09 July 2019
Sec. Molecular and Cellular Oncology
Volume 9 - 2019 | https://doi.org/10.3389/fonc.2019.00587
This article is part of the Research Topic Role of RNA in Molecular Diagnostics of Cancer View all 12 articles
Small nucleolar RNAs (SnoRNAs) are a class of non-coding RNAs divided into two classes: C/D box snoRNAs and H/ACA box snoRNAs. The canonical function of C/D box and H/ACA box snoRNAs are 2'-O-ribose methylation and pseudouridylation of ribosomal RNAs (rRNAs), respectively. Emerging evidence has demonstrated that snoRNAs are involved in various physiological and pathological cellular processes. Mutations and aberrant expression of snoRNAs have been reported in cell transformation, tumorigenesis, and metastasis, indicating that snoRNAs may serve as biomarkers and/or therapeutic targets of cancer. Hence, further study of the functions and underlying mechanism of snoRNAs is valuable. In this review, we summarize the biogenesis and functions of snoRNAs, as well as the association of snoRNAs in different types of cancers and their potential roles in cancer diagnosis and therapy.
Small nucleolar RNAs (snoRNAs) are an extensively studied non-coding RNAs that primarily accumulate in the nucleoli and consist of 60–300 nucleotides (NTs). SnoRNAs are mostly responsible for the posttranscriptional modification and maturation of ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), and other cellular RNAs. SnoRNAs are divided into two classes: C/D box snoRNAs and H/ACA box snoRNAs. C/D box snoRNAs guide−2′-O-ribose methylation, and H/ACA box snoRNAs direct the pseudouridylation of NTs (1–4).
The box C/D family of snoRNAs is characterized by a kink-turn (k-turn) (stem–bulge–stem) structure and contains two conserved sequence elements: box C (RUGAUGA) and box D (CUGA) located at the 5′ and 3′ ends of the RNA molecule, respectively (Figure 1) (1). The k-turn motif is essential to the assembly of a small nucleolar ribonucleoprotein (snoRNP) including fibrillarin, Nop56, Nop58, and 15.5kD (3, 5, 6). Fibrillarin is an enzyme that is responsible for substrate methylation. Nop56, Nop58, and 15.5kD contribute to maturation, stability, and localization of snoRNAs. There are a few C/D box snoRNAs that do not form canonical snoRNPs to function in RNA methylation (7, 8). Most C/D box snoRNAs contain less conserved copies of box C and box D in the central region of the snoRNA, identified as box C' and box D', respectively, which generally carry one or two base modifications. The upstream elements of the box D/D′ motifs are complementary to target RNAs, allowing for alignment and methylation of appropriate NTs (3, 9, 10).
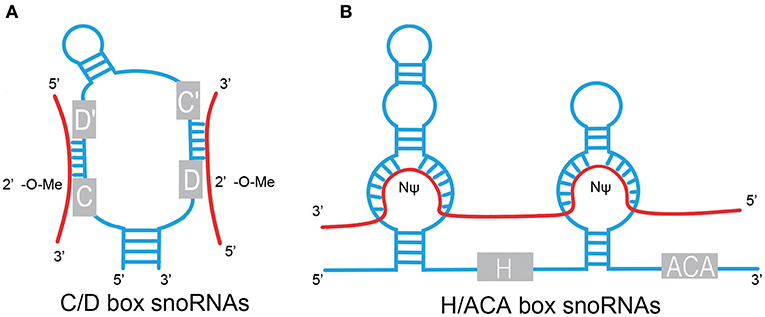
Figure 1. Structural features of C/D box and H/ACA box snoRNAs. (A) C/D box. snoRNAs contain two conserved sequence elements, named box C (RUGAUGA), and box D (CUGA). Box C and box D are close to each other by the base pairing of the 5′ and 3′ termini and fold into a k-turn motif. The antisense elements, upstream of the box D/D′ motifs, are complementary to target RNAs and catalyze site-specific 2'-O-methylation (2′-O-Me) of the NT in target RNAs. (B) H/ACA box snoRNAs contain evolutionarily conserved structural elements, including box H (ANANNA), box ACA motif, and two pseudouridylation pockets. Pseudouridylation pockets are complementary to the substrate RNAs and responsible for pseudouridylation (NΨ). Red line, target RNAs.
H/ACA box snoRNAs are composed of conserved box H and box ACA motifs (Figure 1). Box H is an ANANNA sequence (N represents any NT), and ACA box is a trinucleotide (ACA). H/ACA box snoRNAs are marked by the “hairpin–hinge–hairpin–tail” secondary structure with box H in the hinge region and box ACA on the 3′ end of the last hairpin. An internal loop is located in the hairpin of H/ACA box snoRNAs with a 9–13 NT sequence on each strand complementary to the substrate RNAs, which forms a so-called pseudouridylation pocket. The distance between the site responsible for pseudouridylation and the H/ACA box is 14–16 NTs (3, 6, 9, 11). Similar to the C/D box snoRNAs, a set of core proteins are correlated with the H/ACA box snoRNAs to form stable and functional snoRNPs, including NHP2, NOP10, GAR1, and the pseudouridine synthase dyskerin (8).
In vertebrates, most snoRNAs are encoded in the introns of protein-coding or non-coding genes, except for a small number of snoRNAs transcribed autonomously by RNA polymerase II (3). The biogenesis of most intronic snoRNAs includes cotranscription with the host gene, splicing, debranching of the intron lariat, and exonucleolytic digestion in the nucleoplasm. Recruitment of ribonucleoproteins to the nascent intronic snoRNAs is essential to the maturation of snoRNAs; this process is cotranscriptionally initiated. These proteins are critical to both processing stability and nucleolar localization (3, 12). In addition, many auxiliary factors are involved in snoRNP assembly and maturation, such as Shq1, Naf1, and NUFLP. SnoRNPs are transported to Cajal bodies, where they perform additional processing and maturation tasks (13). Thereafter, they are delivered to the nucleolus.
One well-studied function of snoRNAs is their role in the modification, maturation, and stabilization of rRNA (3). As the research intensifies, hundreds of 2′-O-methylation or pseudouridylation residues located within conserved and functional regions of rRNAs have been found. A sequence upstream of box D and/or box D' recognizes target RNAs and results in the 2′-O-methylation of the fifth NT by the methylase fibrillarin (7, 14, 15). The conversion of uridines to pseudouridine is carried out by dyskerin, and the site destined for pseudouridylation is located 14–16 NTs upstream of box H and/or box ACA (9, 16, 17).
In addition to the rRNA modification function, some snoRNAs act in on pre-rRNA cleavage (18, 19). For example, SNORD3 promotes the proper pre-rRNA formation for subsequent endonucleotic processing (20–23). Furthermore, SNORD118, SNORD14, SNORD22, SNORA71, and probably, SNORD13 are involved in pre-rRNA cleavage (24–27).
Recent evidence indicates that snoRNAs play an important role in the regulation of gene expression. SNORD115 is encoded in the imprinted SNURF–SNRPN locus on human chromosome 15, which is frequently deleted in Prader–Willi syndrome (PWS) (28–30). SNORD115 contains an 18-NT sequence complementary to the alternative exon Vb of the serotonin receptor subtype 2C (5-HT2cR) and regulates the alternative splicing of 5-HT2cR (31). Besides the regulation of alternative splicing, SNORD115 influences RNA editing. The site-specific adenosine-to-inosine (A-to-I) base conversion is essential to the generation of 5-HT2cR (32). Vitali et al. demonstrated that SNORD115 inhibited the efficiency of the ADAR2-mediated RNA editing of 5-HT2cR by forming a bona fide snoRNP particle (33). Moreover, using bioinformatic prediction and experimental verification, Rishore et al. identified that SNORD115 regulated the alternative splicing of five pre-mRNAs (DPM2, TAF1, RALGPS1, PBRM1, and CRHR1). They demonstrated that SNORD115 generated shorter RNAs, called processed small nucleolar RNAs (psnoRNAs), that directed the regulation of alternative splicing (34). Two gene clusters encoding SNORD115 and SNORD116 are located in the imprinted locus 15q11-q13 containing 47 repeats of SNORD115 along with 27 copies of SNORD116. Using genome-wide array analysis after overexpressing SNORD115 and SNORD116 in human embryonic kidney (HEK) 293T cells, Falaleeva et al. found that SNORD115 and SNORD116 influenced the expression levels of over 200 genes and modified each other's activity (35). Bazeley et al. found that energetically favorable putative targets of SNORD116 were correlated with exons that were capable of alternative splicing and speculated that SNORD116 was involved in the regulation of alternative splicing (36). Wu et al. identified that 5′ snoRNA-capped and 3′ polyadenylated lncRNA (SPA) required snoRNP complexes to protect them from trimming by 5′-3′ exoribonuclease 2 (XRN2). The most well-understood function of SNORD27 is to guide the methylation of rRNA. Recent research demonstrated that SNORD27 regulated the alternative splicing of the transcription factor E2F7 pre-mRNA by directing RNA–RNA interaction (7). A similar function was reported for SNORD88C, which produced small RNAs derived from snoRNAs (sdRNAs) containing the box C' that was complementary to several pre-mRNAs including FGFR3 and regulated the alternative splicing of FGFR3 pre-mRNA (37). In another study, Huang et al. discovered that SNORA50A inhibited mRNA 3' processing by blocking the Fib1-poly(A) site interaction, which was the first report that snoRNA regulated mRNA 3' processing (38).
Michel et al. found that three snoRNAs (SNORD32A, 33, and 35A) encoded in the ribosomal protein L13a (Rpl13a) locus were increased significantly under oxidative stress induced by palmitate and hydrogen peroxide treatment. Knockdown of these three snoRNAs simultaneously increased resistance against palmitate in vivo (39). Furthermore, snoRNA ACA11 has been found to suppress oxidative stress by downregulating ribosomal protein genes and other snoRNAs (40). Brandis et al. identified that the loss of snoRNA U60 reduced plasma membrane-to-endoplasmic reticulum cholesterol trafficking and increased de novo cholesterol synthesis. This finding suggested that U60 played a function in regulating intracellular cholesterol trafficking (41). Other reports demonstrated that snoRNAs regulate cellular metabolic homeostasis. For example, snoRNA U17 regulated cellular cholesterol trafficking via the encoding of hypoxia-upregulated mitochondrial movement regulator (HUMMR) by its target mRNA. Four snoRNAs encoded in Rpl13a, snoRNAs U32A, U33, U34A, and U35A, regulated systemic glucose metabolism (42, 43).
SnoRNAs are widely involved in many physiological and pathological processes; indeed, emerging evidence suggests that snoRNAs have tumor-suppressive or oncogenic functions in various cancer types (Table 1). SnoRNAs are reported to participate in many biological cancer processes, including the invasion of growth suppressors and cell death, activation of invasion and metastasis, and angiogenesis and sustained proliferative signaling. The underlying molecular mechanisms are diverse.
P53 is a well-known tumor suppressor that responds to diverse cellular stresses to regulate the expression of target genes involved in cell cycle arrest, apoptosis, and DNA repair (73). Increasing evidence suggests that snoRNAs are closely associated with the p53 regulatory pathway. For instance, a recent study identified that snoRNAs and FBL were usually overexpressed in human breast and prostate cancers and that this overexpression promoted tumorigenicity in vitro and in vivo (74). Further research found that oncogene Myc upregulated the expression level of FBL and led to elevated snoRNA biogenesis, inducing p53 suppression. Cellular stress induced by knockdown of snoRNA pathway genes increased the accumulation of p53 by promoting the binding of ribosomal protein L5 (RPL5) and ribosomal protein L11 (RPL11) with MDM2 and led to p53 stabilization. Furthermore, impaired snoRNA biogenesis induced by FBL depletion promoted PTB binding to the p53 internal ribosome entering site (IRES) and regulated the initiation of p53 translation (74). A similar finding has been reported by Langhendries et al. U3 and U8 were upregulated in breast cancer, and depletion of U3 and U8 inhibited tumorigenicity of breast cancer cells in vivo and in vitro (50). Depletion of U3 or U8 led to ribosome biogenesis dysfunction by inhibiting pre-rRNA processing and reduced production of mature rRNAs (50). SnoRNA42 was overexpressed in non-small cell lung cancer (NSCLC) and played an oncogenic role by regulating the expression of p53. Doxorubicin-induced DNA damage induced the expression of the GAS5-derived snoRNAs (U44 and U77) in a p53-dependent manner. Chromatin immunoprecipitation sequencing (ChIP-seq) experiments confirmed that p53 directly controlled GAS5 transcription. GAS5-derived snoRNAs are closely correlated with p53 levels in colorectal tissue, suggesting that GAS5-derived snoRNAs have a critical role in p53-associated signaling pathways in colorectal cancer (CRC) (75).
There are several other snoRNAs associated with the p53 regulatory pathway. In a multistage germline copy number variation (CNV)-based genome-wide association studies (GWAS), a low-frequency duplication at 15q13.3 containing SNORA18L5 was strongly correlated to risk of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Further studies showed that SNORA18L5 was overexpressed in HCC tissues compared with adjacent normal tissues and played an oncogenic role in HCC (57). Overexpression of snoRNA18L5 kept RPL5 and RPL11 from binding to MDM2, resulting in increased MDM2-mediated ubiquitination degradation of p53 (57). It is well known that p53 mutations in cancers often exert oncogenic gain-of-function properties that contribute to tumorigenicity. Increasing evidence shows that in addition to wild-type p53, mutant p53 also interacts with snoRNAs in cancers (76). For instance, Pourebrahim et al. identified that p53-mutant mice developed osteosarcomas with increased metastasis as compared with p53-null mice. Using comprehensive transcriptome RNA sequencing analysis of 16 tumors, they also found that a cluster of snoRNAs were upregulated in p53 mutant tumors. An analysis of regulatory elements showed that the Est2-binding motif was highly enriched in these deregulated snoRNAs. Genetic deletion of the Esr2 transcription factor reduced the expression of these snoRNAs in p53 mutant mice and abrogated the mutant p53 prometastatic phenotype (69).
There are many other signaling pathways associated with snoRNAs, including the phosphoinositide 3-kinase (PI3K)–AKT and Wnt/β-catenin pathways discussed here. The PI3K–AKT pathway is a highly conserved cell signaling system in most multicellular organisms, which is critical to diverse cellular processes such as stemness, cell proliferation, differentiation, and cell death (77). The role of the PI3K–AKT signaling pathway in cancer is well-documented (77). A number of snoRNAs have been implicated in PI3K–AKT signaling, either directly or indirectly. ACA11 knockdown significantly decreased the phosphorylation of PI3K and AKT, whereas ACA11 overexpression promoted HCC cell growth and metastasis by activating the PI3K–AKT pathway (59). In another study, SNORD126 activated PI3K–AKT signaling by upregulating FGFR2 and promoted HCC and CRC cell growth (58). PH domain leucine-rich repeat protein phosphatase (PHLPP) is an endogenous inhibitor of the AKT pathway (77). In gallbladder cancer, SNORA74B knockdown suppressed activation of the AKT/mechanistic target of rapamycin (mTOR) pathway via inducing PHLPP expression (67). In the Wnt/β-catenin pathway, the major pathway regulating the development and progression of HCC, SNORD76 knockdown significantly decreased the level of β-catenin, c-Myc, and cyclinD1. Conversely, overexpression of SNORD76 promoted HCC tumorigenicity through activation of the Wnt/β-catenin pathway (60).
In 2008, Kawaji et al. performed an unbiased sequencing of human small RNAs (19–40 NTs) and found independent classes of small RNAs originating from noncoding RNAs (78). Since then, a wealth of sdRNAs have been identified. In 2009, Taft et al. performed a systematic analysis of small RNA by deep sequencing libraries from various eukaryotic tissues and found that small RNAs with evolutionarily conserved size and position were derived from a large proportion of snoRNA loci in animals (human, mouse, chicken, and fruit fly), Arabidopsis, and fission yeast (79). In recent years, the processing patterns of snoRNAs have raised substantial attention, and increasing evidence shows that these small RNAs have a role in cancers. SnoRNA–miR-28, an miRNA-like non-coding RNA derived from snoRNAs, was significantly upregulated in breast tumors and promoted proliferation of tumor cells (80). SNORD28, a p53-repressed snoRNA located in SNHG1, can be processed into smaller miRNA-like molecules that are capable of binding argonaute (AGO) and exerting miRNA-like effects. This procession can be repressed by p53, the underlying mechanism of which is unclear (80). At the same time, snoRNA–miR-28 acts as an miRNA and directly interacts with TAF9B's 3'-untranslated region (3'-UTR), leading to the reduction of TAF9B mRNA and protein expression levels. The reduced expression impairs the stability of p53 by promoting the binding of MDM2 to p53. Collectively, a regulatory loop exists between p53, SNHG1, snoRNA-miR-28, and TAF9B (80). miR-768-5p, another miRNA-like non-coding RNA derived from SNORD17, was reported to bind to YB-1 and may play a role in the development of cancers (52).
PIWI-interacting RNAs are identified as inhibitor of transposable elements (TEs) in the germline and play an important role in regulating target RNA to silence its expression via base-pairing recognition (81). In addition to miRNA-like functions, these sdRNAs also have the ability to play piRNA-like functions. Pi-sno-75, a piRNA derived from SNORD75 located in GAS5, can specifically bind to PIWIL1 and PIWIL4 (piRNA binding proteins in Homo sapiens) and bearing modification of 2′-O-methylation at the 3′ terminus (82). Microarray results showed that TRAIL, a tumor-specific suppressor, was upregulated by pi-sno-75 synthesized in breast cancer cells. ChIP results showed that overexpression of pi-sno-75 increased the H3K4me3 level and decreased the H3K27me3 level within −169 base pair (bp) of TRAIL promoter in a PIWIL1/PIWIL4-dependent manner. Further research identified the molecular mechanism of this regulation: the pi-sno-75/PIWIL complex can interact with WDR5 and recruit the entire hCOMPASS-like complex along with SMYD3 to the promoter region of the TRAIL gene (82). Pi-sno-75 exhibits antitumor activity in vitro and in vivo by upregulating the expression of TRAIL (82). Uzunova et al. identified that sdRNAs derived from SNORD44, SNORD74, SNORD78, and SNORD81 were upregulated in prostate cancer. In particular, the levels of, SNORD78 and its sdRNAs were obviously higher in patients with metastatic diseases (65).
Cancer stem cells have the capacities of self-renewal, differentiation, and tumorigenicity. The presence of cancer stem cells has been reported in various cancers, which may explain why current chemotherapies cannot consistently eradicate cancers (83). A growing body of research suggests that snoRNAs play an important role in cancer stem cells. Self-renewal activity is essential in leukemogenesis. Zhou et al. identified that amino-terminal enhancer of split (AES), C/D box snoRNAs, and rRNA 2′-O-methylation are essential for the AML1–ETO–induced self-renewal of leukemia cells in vitro and in vivo (54). Knockdown of AES leads to reduction of snoRNAs (primarily C/D box snoRNAs) and suppression of translation efficiency in AE9a-transduced leukemia cells. DDX21 is an RNA helicase that binds to snoRNA/RNP to facilitate rRNA modification. The underlying mechanism involves the knockdown of AES, which reduces the association of DDX21 with the C/D box snoRNP complex including FBL, NOP598, NOP56, and NCL and followed by suppression of snoRNAs. C/D box snoRNAs are highly expressed in acute myeloid leukemia (AML) and correlated closely with the in vivo frequency of leukemic stem cells. Knockdown of SNORD14D or SNORD35A suppressed the clonogenic potential of leukemia cells in vitro and delayed leukemogenesis in vivo (54). In previous studies, Mannoor et al. demonstrated that (aldehyde dehydrogenase 1) was a cancer stem cell marker, as ALDH1+ cancer cells have extensive self-renewal, proliferation, and in vivo tumorigenic potentials. There are 22 snoRNAs that display differential expression in ALDH+ cancer cells compared with ALDH− cancer cells (44). SNORA3 and SNORA43 overexpressed in lung cancer stem cells and inversely associated with survival in NSCLC patients. Along with CD133, another important maker for lung cancer stem cells, SNORA42 was confirmed to be especially dysregulated in lung cancer stem cells (44). Knockdown of SNORA42 inhibited self-renewal capacity and in vitro tumorigenesis by inducing apoptosis and reducing the transcript level of stem cell-associated genes including OCT4, Nanog, Sox2, Notch1, Smo, and ABCS2, suggesting SNORA42 may associate with the expression of core stem cell transcription factors in lung cancer stem cells (44). SNORD78 is also reported to be upregulated in cancer stem cells in NSCLC and is required for the self-renewal of cancer stem cells in NSCLC (46).
As mentioned above, aberration in the expression of snoRNAs was found to be prevalent in many cancers; some of these anomalies are cancer type specific. Substantial research has shown that many snoRNAs are stably expressed and detectable in body fluids including the blood plasma, serum, and urine of cancer patients. Their expression levels are closely associated with diagnosis, prognosis, and classification of subtypes. Given these specifics, snoRNAs have the potential to become cancer biomarkers (84, 85).
The clinical value of snoRNA expression analysis in the diagnosis of certain subtypes of peripheral T-cell lymphoma (PTCL) has been demonstrated. Moreover, Valleron et al. found that the overexpression of snoRNA HBI-239 and HBI-239-processed miRNA predicted good prognosis in angio-immunoblastic T-cell lymphoma (AITL) and PTCL not otherwise specified (PTCL-NOS) (86). Berquet et al. investigated the snoRNA expression profiles in B-cell chronic lymphocytic leukemia (CLL) patients and identified that immunoglobulin heavy chain variable region gene (IGHV)-mutant patients exhibiting overexpression of 20 snoRNAs had a shorter treatment-free survival (TFS) (87). SNORA70F and SNORD116-118 were developed into a 2-snoRNA model that appeared to distinguish two different prognostic CLL groups (88). Furthermore, a research group identified that snoRNA expression profiles can be used for the classification of leukemia subgroups due to the differential expression among these various groups (89). SNORD33, SNORD66, and SNORD76 were overexpressed in NSCLC patients and were detectable in plasma, yielding 81.1% sensitivity and 95.8% specificity in distinguishing NSCLC patients. SNORA42 was overexpressed in NSCLC patients; the levels were inversely correlated with the survival of patients, providing a potential marker for diagnosis and prognosis in NSCLC. SNORA47, SNORA68, and SNORA78 were reported to accurately predict overall survival of NSCLC patients in a training set of 77 cases, which may become an snoRNA-based model for predicting overall survival in lung cancer patients (44, 90, 91). SNORD42 and SNORD21 were also reported to be promising predictive biomarkers for prognosis in CRC patients (55, 92). The expression level of SNORA18L5 in HCC tissues was correlated with the survival time of patients; patients with high SNORA23 expression had a shorter disease-free survival time (57, 70). RUN44, RUN43, and RUN48 were downregulated in breast cancer and were associated with a poor prognosis. In a recent study, Krishnan et al. found a large number of snoRNAs that were promising prognostic markers for breast cancer (51, 93). SNORD114-10 was downregulated in metastatic omentum tissues, suggesting that this snoRNA may provide a prediagnostic biomarker for ovarian cancer metastasis (72). These studies strongly indicate that snoRNAs offer promising novel diagnostic and prognostic markers across a range of cancer types. However, further investigation is necessary, and many challenges must be overcome prior to their application in clinical settings.
Since snoRNAs participate in tumorigenesis, tumor aggressiveness, and staging, they are ideal candidates for cancer therapy. The regulation of snoRNA expression may contribute to the goal of cancer treatment. For example, antisense oligonucleotide (ASO)-mediated downregulation of SNORA23 expression reduced tumor growth, dissemination, and liver metastasis in pancreatic ductal adenocarcinoma (70). Our studies demonstrated that SNORD44 was downregulated in CRC and that overexpression of SNORD44 by an oncolytic adenovirus inhibited CRC growth (94). With the expansion of research and the further development of applicable technologies, snoRNAs may become major cancer therapeutic targets in the near future.
Although the landscape of snoRNA function in its entirety remains still unclear, previous studies have confirmed the critical roles of snoRNAs in rRNA processing, gene transcription, RNA splicing, and other cellular processes. As sequencing and microarray analysis technologies evolve, more and more tumor-related snoRNAs are being discovered. Recent studies have demonstrated that aberrant expression and mutations in specific snoRNAs were associated with tumorigenesis and metastasis. The tissue- and cancer-specific expression of snoRNAs can be used as reliable prognostic markers for cancer diagnosis and novel therapeutic targets. However, the field is still in its infancy, and our understanding of the role of snoRNAs in tumors remains incomplete. Currently, the effect of snoRNA on cellular signaling pathways is also poorly understood. Functional screening of RNA interference libraries and/or clustered regularly interspaced short palindromic repeats (CRISPR)-based libraries is necessary to explore snoRNA functions in signaling pathways. SnoRNAs do not always act directly; indeed, most of their actions are indirect binding to other molecules. The identification of more molecules that interact with snoRNAs using RNA precipitation combined with high-throughput mass spectrometry and RNA immunoprecipitation (RIP) technologies is promising. Moreover, animal models are important for understanding the function of non-coding RNA in tumorigenesis and development. However, animal models of snoRNAs in cancer remain insufficient to date. Furthermore, emerging evidence suggests that some non-coding RNAs contain translated open reading frames that are able to encode proteins. This hints at whether snoRNAs also have the ability to encode proteins and provides a whole new direction for investigating snoRNA function. A better grasp of the role of snoRNAs in tumors will help us to understand tumors more comprehensively and may offer not only novel diagnostic biomarkers but also effective therapeutic targets in the near future.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by the National Natural Science Fund (31671348 and 31301064).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. (1995) 64:897–934. doi: 10.1146/annurev.bi.64.070195.004341
2. Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. (2002) 84:775–90. doi: 10.1016/S0300-9084(02)01402-5
3. Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. (2007) 35:1452–64. doi: 10.1093/nar/gkl1172
4. Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. (2002) 109:145–8. doi: 10.1016/S0092-8674(02)00718-3
5. Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. (2001) 20:4214–21. doi: 10.1093/emboj/20.15.4214
6. Weinstein LB, Steitz JA. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol. (1999) 11:378–84. doi: 10.1016/S0955-0674(99)80053-2
7. Falaleeva M, Pages A, Matuszek Z, Hidmi S, Agranat-Tamir L, Korotkov K, et al. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc Natl Acad Sci USA. (2016) 113:E1625–34. doi: 10.1073/pnas.1519292113
8. Massenet S, Bertrand E, Verheggen C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. (2017) 14:680–92. doi: 10.1080/15476286.2016.1243646
9. Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. (1996) 86:823–34. doi: 10.1016/S0092-8674(00)80156-7
10. Falaleeva M, Welden JR, Duncan MJ, Stamm S. C/D-box snoRNAs form methylating and non-methylating ribonucleoprotein complexes: old dogs show new tricks. Bioessays. (2017) 39. doi: 10.1002/bies.201600264
11. Bratkovic T, Rogelj B. Biology and applications of small nucleolar RNAs. Cell Mol Life Sci. (2011) 68:3843–51. doi: 10.1007/s00018-011-0762-y
12. Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. (2007) 8:209–20. doi: 10.1038/nrm2124
13. Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. (2003) 22:4283–93. doi: 10.1093/emboj/cdg394
14. Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol. (2002) 14:319–27. doi: 10.1016/S0955-0674(02)00334-4
15. Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. (2006) 34:D158–62. doi: 10.1093/nar/gkj002
16. Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. (1997) 89:799–809. doi: 10.1016/S0092-8674(00)80263-9
17. Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. (1997) 11:941–56. doi: 10.1101/gad.11.7.941
18. Atzorn V, Fragapane P, Kiss T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol Cell Biol. (2004) 24:1769–78. doi: 10.1128/MCB.24.4.1769-1778.2004
19. Eliceiri GL. Small nucleolar RNAs. Cell Mol Life Sci. (1999) 56:22–31. doi: 10.1007/s000180050003
20. Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. (1990) 60:897–908. doi: 10.1016/0092-8674(90)90338-F
21. Savino R, Gerbi SA. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. (1990) 9:2299–308. doi: 10.1002/j.1460-2075.1990.tb07401.x
22. Hughes JM, Ares M Jr. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5' external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. (1991) 10:4231–9. doi: 10.1002/j.1460-2075.1991.tb05001.x
23. Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. (2012) 3:397–414. doi: 10.1002/wrna.117
24. Granneman S, Vogelzangs J, Luhrmann R, van Venrooij WJ, Pruijn GJ, Watkins NJ. Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP. Mol Cell Biol. (2004) 24:8600–10. doi: 10.1128/MCB.24.19.8600-8610.2004
25. Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. (1993) 73:1233–45. doi: 10.1016/0092-8674(93)90651-6
26. Liang WQ, Fournier MJ. U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes Dev. (1995) 9:2433–43. doi: 10.1101/gad.9.19.2433
27. Tycowski KT, Shu MD, Steitz JA. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science. (1994) 266:1558–61. doi: 10.1126/science.7985025
28. Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet. (2001) 10:2687–700. doi: 10.1093/hmg/10.23.2687
29. Butler MG, Hanchett JM, Thompson T. Clinical findings and natural history of prader–willi syndrome. In: Management of Prader–Willi Syndrome. 3rd ed. (2006). p. 3–48.
30. Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. (2006) 311:230–2. doi: 10.1126/science.1118265
31. Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA. (2000) 97:14311–6. doi: 10.1073/pnas.250426397
32. Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. (1997) 387:303–8. doi: 10.1038/387303a0
33. Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaille J, et al. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol. (2005) 169:745–53. doi: 10.1083/jcb.200411129
34. Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, et al. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet. (2010) 19:1153–64. doi: 10.1093/hmg/ddp585
35. Falaleeva M, Surface J, Shen M, de la Grange P, Stamm S. SNORD116 and SNORD115 change expression of multiple genes and modify each other's activity. Gene. (2015) 572:266–73. doi: 10.1016/j.gene.2015.07.023
36. Bazeley PS, Shepelev V, Talebizadeh Z, Butler MG, Fedorova L, Filatov V, et al. snoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions. Gene. (2008) 408:172–9. doi: 10.1016/j.gene.2007.10.037
37. Ono M, Yamada K, Avolio F, Scott MS, van Koningsbruggen S, Barton GJ, et al. Analysis of human small nucleolar RNAs (snoRNA) and the development of snoRNA modulator of gene expression vectors. Mol Biol Cell. (2010) 21:1569–84. doi: 10.1091/mbc.e10-01-0078
38. Huang C, Shi J, Guo Y, Huang W, Huang S, Ming S, et al. A snoRNA modulates mRNA 3' end processing and regulates the expression of a subset of mRNAs. Nucleic Acids Res. (2017) 45:8647–60. doi: 10.1093/nar/gkx651
39. Michel CI, Holley CL, Scruggs BS, Sidhu R, Brookheart RT, Listenberger LL, et al. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. (2011) 14:33–44. doi: 10.1016/j.cmet.2011.04.009
40. Chu L, Su MY, Maggi LB Jr, Lu L, Mullins C, Crosby S, et al. Multiple myeloma-associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J Clin Invest. (2012) 122:2793–806. doi: 10.1172/JCI63051
41. Brandis KA, Gale S, Jinn S, Langmade SJ, Dudley-Rucker N, Jiang H, et al. Box C/D small nucleolar RNA (snoRNA) U60 regulates intracellular cholesterol trafficking. J Biol Chem. (2013) 288:35703–13. doi: 10.1074/jbc.M113.488577
42. Jinn S, Brandis KA, Ren A, Chacko A, Dudley-Rucker N, Gale SE, et al. snoRNA U17 regulates cellular cholesterol trafficking. Cell Metab. (2015) 21:855–67. doi: 10.1016/j.cmet.2015.04.010
43. Lee J, Harris AN, Holley CL, Mahadevan J, Pyles KD, Lavagnino Z, et al. Rpl13a small nucleolar RNAs regulate systemic glucose metabolism. J Clin Invest. (2016) 126:4616–25. doi: 10.1172/JCI88069
44. Mannoor K, Shen J, Liao J, Liu Z, Jiang F. Small nucleolar RNA signatures of lung tumor-initiating cells. Mol Cancer. (2014) 13:104. doi: 10.1186/1476-4598-13-104
45. Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu L, et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. (2012) 31:2794–804. doi: 10.1038/onc.2011.449
46. Zheng D, Zhang J, Ni J, Luo J, Wang J, Tang L, et al. Small nucleolar RNA 78 promotes the tumorigenesis in non-small cell lung cancer. J Exp Clin Cancer Res. (2015) 34:49. doi: 10.1186/s13046-015-0170-5
47. Dong XY, Rodriguez C, Guo P, Sun X, Talbot JT, Zhou W, et al. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum Mol Genet. (2008) 17:1031–42. doi: 10.1093/hmg/ddm375
48. Dong XY, Guo P, Boyd J, Sun X, Li Q, Zhou W, et al. Implication of snoRNA U50 in human breast cancer. J Genet Genom. (2009) 36:447–54. doi: 10.1016/S1673-8527(08)60134-4
49. Siprashvili Z, Webster DE, Johnston D, Shenoy RM, Ungewickell AJ, Bhaduri A, et al. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet. (2016) 48:53–8. doi: 10.1038/ng.3452
50. Langhendries JL, Nicolas E, Doumont G, Goldman S, Lafontaine DL. The human box C/D snoRNAs U3 and U8 are required for pre-rRNA processing and tumorigenesis. Oncotarget. (2016) 7:59519–34. doi: 10.18632/oncotarget.11148
51. Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, et al. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. (2011) 104:1168–77. doi: 10.1038/sj.bjc.6606076
52. Blenkiron C, Hurley DG, Fitzgerald S, Print CG, Lasham A. Links between the oncoprotein YB-1 and small non-coding RNAs in breast cancer. PLoS ONE. (2013) 8:e80171. doi: 10.1371/journal.pone.0080171
53. Valleron W, Laprevotte E, Gautier EF, Quelen C, Demur C, Delabesse E, et al. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia. (2012) 26:2052–60. doi: 10.1038/leu.2012.111
54. Zhou F, Liu Y, Rohde C, Pauli C, Gerloff D, Kohn M, et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat Cell Biol. (2017) 19:844–55. doi: 10.1038/ncb3563
55. Yoshida K, Toden S, Weng W, Shigeyasu K, Miyoshi J, Turner J, et al. SNORA21—an oncogenic small nucleolar RNA, with a prognostic biomarker potential in human colorectal cancer. EBioMed. (2017) 22:68–77. doi: 10.1016/j.ebiom.2017.07.009
56. Yang X, Li Y, Li L, Liu J, Wu M, Ye M. SnoRNAs are involved in the progression of ulcerative colitis and colorectal cancer. Dig Liver Dis. (2017) 49:545–51. doi: 10.1016/j.dld.2016.12.029
57. Cao P, Yang A, Wang R, Xia X, Zhai Y, Li Y, et al. Germline duplication of SNORA18L5 increases risk for HBV-related hepatocellular carcinoma by altering localization of ribosomal proteins and decreasing levels of p53. Gastroenterology. (2018) 155:542–56. doi: 10.1053/j.gastro.2018.04.020
58. Fang X, Yang D, Luo H, Wu S, Dong W, Xiao J, et al. SNORD126 promotes HCC and CRC cell growth by activating the PI3K–AKT pathway through FGFR2. J Mol Cell Biol. (2017) 9:243–55. doi: 10.1093/jmcb/mjw048
59. Wu L, Zheng J, Chen P, Liu Q, Yuan Y. Small nucleolar RNA ACA11 promotes proliferation, migration and invasion in hepatocellular carcinoma by targeting the PI3K/AKT signaling pathway. Biomed Pharmacother. (2017) 90:705–12. doi: 10.1016/j.biopha.2017.04.014
60. Chen L, Han L, Wei J, Zhang K, Shi Z, Duan R, et al. SNORD76, a box C/D snoRNA, acts as a tumor suppressor in glioblastoma. Sci Rep. (2015) 5:8588. doi: 10.1038/srep08588
61. Wu L, Chang L, Wang H, Ma W, Peng Q, Yuan Y. Clinical significance of C/D box small nucleolar RNA U76 as an oncogene and a prognostic biomarker in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. (2018) 42:82–91. doi: 10.1016/j.clinre.2017.04.018
62. Li G, He Y, Liu X, Zheng Z, Zhang M, Qin F, et al. Small nucleolar RNA 47 promotes tumorigenesis by regulating EMT markers in hepatocellular carcinoma. Minerva Med. (2017) 108:396–404. doi: 10.23736/S0026-4806.17.05132-1
63. Xu G, Yang F, Ding CL, Zhao LJ, Ren H, Zhao P, et al. Small nucleolar RNA 113-1 suppresses tumorigenesis in hepatocellular carcinoma. Mol Cancer. (2014) 13:216. doi: 10.1186/1476-4598-13-216
64. Crea F, Quagliata L, Michael A, Liu HH, Frumento P, Azad AA, et al. Integrated analysis of the prostate cancer small-nucleolar transcriptome reveals SNORA55 as a driver of prostate cancer progression. Mol Oncol. (2016) 10:693–703. doi: 10.1016/j.molonc.2015.12.010
65. Martens-Uzunova ES, Hoogstrate Y, Kalsbeek A, Pigmans B, Vredenbregt-van den Berg M, Dits N, et al. C/D-box snoRNA-derived RNA production is associated with malignant transformation and metastatic progression in prostate cancer. Oncotarget. (2015) 6:17430–44. doi: 10.18632/oncotarget.4172
66. Stepanov GA, Filippova JA, Nushtaeva AA, Kuligina EV, Koval OA, Richter VA, et al. Artificial analogues of circulating box C/D RNAs induce strong innate immune response and microRNA activation in human adenocarcinoma cells. Adv Exp Med Biol. (2016) 924:121–5. doi: 10.1007/978-3-319-42044-8_24
67. Qin Y, Meng L, Fu Y, Quan Z, Ma M, Weng M, et al. SNORA74B gene silencing inhibits gallbladder cancer cells by inducing PHLPP and suppressing Akt/mTOR signaling. Oncotarget. (2017) 8:19980–96. doi: 10.18632/oncotarget.15301
68. Xu B, Ye MH, Lv SG, Wang QX, Wu MJ, Xiao B, et al. SNORD47, a box C/D snoRNA, suppresses tumorigenesis in glioblastoma. Oncotarget. (2017) 8:43953–66. doi: 10.18632/oncotarget.16693
69. Pourebrahim R, Zhang Y, Liu B, Gao R, Xiong S, Lin PP, et al. Integrative genome analysis of somatic p53 mutant osteosarcomas identifies Ets2-dependent regulation of small nucleolar RNAs by mutant p53 protein. Genes Dev. (2017) 31:1847–57. doi: 10.1101/gad.304972.117
70. Cui L, Nakano K, Obchoei S, Setoguchi K, Matsumoto M, Yamamoto T, et al. Small nucleolar noncoding RNA SNORA23, up-regulated in human pancreatic ductal adenocarcinoma, regulates expression of spectrin repeat-containing nuclear envelope 2 to promote growth and metastasis of xenograft tumors in mice. Gastroenterology. (2017) 153:292–306.e2. doi: 10.1053/j.gastro.2017.03.050
71. Zou AE, Ku J, Honda TK, Yu V, Kuo SZ, Zheng H, et al. Transcriptome sequencing uncovers novel long noncoding and small nucleolar RNAs dysregulated in head and neck squamous cell carcinoma. RNA. (2015) 21:1122–34. doi: 10.1261/rna.049262.114
72. Yang Y, Zhang H, Xie Y, Zhang S, Zhu J, Yin G, et al. Preliminary screening and identification of differentially expressed metastasis-related ncRNAs in ovarian cancer. Oncol Lett. (2018) 15:368–74. doi: 10.3892/ol.2017.7338
73. Joerger AC, Fersht AR. The p53 pathway: origins, inactivation in cancer, and emerging therapeutic approaches. Annu Rev Biochem. (2016) 85:375–404. doi: 10.1146/annurev-biochem-060815-014710
74. Su H, Xu T, Ganapathy S, Shadfan M, Long M, Huang TH, et al. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene. (2014) 33:1348–58. doi: 10.1038/onc.2013.89
75. Krell J, Frampton AE, Mirnezami R, Harding V, De Giorgio A, Roca Alonso L, et al. Growth arrest-specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer. PLoS ONE. (2014) 9:e98561. doi: 10.1371/journal.pone.0098561
76. Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. (2014) 25:304–17. doi: 10.1016/j.ccr.2014.01.021
77. LoRusso PM. Inhibition of the PI3K/AKT/mTOR pathway in solid tumors. J Clin Oncol. (2016) 34:3803–15. doi: 10.1200/JCO.2014.59.0018
78. Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, et al. Hidden layers of human small RNAs. BMC Genomics. (2008) 9:157. doi: 10.1186/1471-2164-9-157
79. Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. (2009) 15:1233–40. doi: 10.1261/rna.1528909
80. Yu F, Bracken CP, Pillman KA, Lawrence DM, Goodall GJ, Callen DF, et al. p53 represses the oncogenic Sno-MiR-28 derived from a SnoRNA. PLoS ONE. (2015) 10:e0129190. doi: 10.1371/journal.pone.0129190
81. Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: its biogenesis and functions. Annu Rev Biochem. (2015) 84:405–33. doi: 10.1146/annurev-biochem-060614-034258
82. He X, Chen X, Zhang X, Duan X, Pan T, Hu Q, et al. (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. (2015) 43:3712–25. doi: 10.1093/nar/gkv214
83. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. (2017) 23:1124–34. doi: 10.1038/nm.4409
84. Thorenoor N, Slaby O. Small nucleolar RNAs functioning and potential roles in cancer. Tumour Biol. (2015) 36:41–53. doi: 10.1007/s13277-014-2818-8
85. Mannoor K, Liao J, Jiang F. Small nucleolar RNAs in cancer. Biochim Biophys Acta. (2012) 1826:121–8. doi: 10.1016/j.bbcan.2012.03.005
86. Valleron W, Ysebaert L, Berquet L, Fataccioli V, Quelen C, Martin A, et al. Small nucleolar RNA expression profiling identifies potential prognostic markers in peripheral T-cell lymphoma. Blood. (2012) 120:3997–4005. doi: 10.1182/blood-2012-06-438135
87. Berquet L, Valleron W, Grgurevic S, Quelen C, Zaki O, Quillet-Mary A, et al. Small nucleolar RNA expression profiles refine the prognostic impact of IGHV mutational status on treatment-free survival in chronic lymphocytic leukaemia. Br J Haematol. (2016) 172:819–23. doi: 10.1111/bjh.13544
88. Ronchetti D, Mosca L, Cutrona G, Tuana G, Gentile M, Fabris S, et al. Small nucleolar RNAs as new biomarkers in chronic lymphocytic leukemia. BMC Med Genomics. (2013) 6:27. doi: 10.1186/1755-8794-6-27
89. Teittinen KJ, Laiho A, Uusimaki A, Pursiheimo JP, Gyenesei A, Lohi O. Expression of small nucleolar RNAs in leukemic cells. Cell Oncol. (2013) 36:55–63. doi: 10.1007/s13402-012-0113-5
90. Liao J, Yu L, Mei Y, Guarnera M, Shen J, Li R, et al. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer. (2010) 9:198. doi: 10.1186/1476-4598-9-198
91. Gao L, Ma J, Mannoor K, Guarnera MA, Shetty A, Zhan M, et al. Genome-wide small nucleolar RNA expression analysis of lung cancer by next-generation deep sequencing. Int J Cancer. (2015) 136:E623–9. doi: 10.1002/ijc.29169
92. Okugawa Y, Toiyama Y, Toden S, Mitoma H, Nagasaka T, Tanaka K, et al. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut. (2017) 66:107–17. doi: 10.1136/gutjnl-2015-309359
93. Krishnan P, Ghosh S, Wang B, Heyns M, Graham K, Mackey JR, et al. Profiling of small nucleolar RNAs by next generation sequencing: potential new players for breast cancer prognosis. PLoS ONE. (2016) 11:e0162622. doi: 10.1371/journal.pone.0162622
Keywords: small nucleolar RNA, cancer, rRNA processing, mRNA splicing, sdRNA, biomarker, therapy
Citation: Liang J, Wen J, Huang Z, Chen X, Zhang B and Chu L (2019) Small Nucleolar RNAs: Insight Into Their Function in Cancer. Front. Oncol. 9:587. doi: 10.3389/fonc.2019.00587
Received: 23 February 2019; Accepted: 17 June 2019;
Published: 09 July 2019.
Edited by:
Cesar Wong, Hong Kong Polytechnic University, Hong KongReviewed by:
Zexian Liu, Sun Yat-sen University Cancer Center (SYSUCC), ChinaCopyright © 2019 Liang, Wen, Huang, Chen, Zhang and Chu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Chu, bGlhbmdjaHVAdGpoLnRqbXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.