- 1First Clinical Medical College, Nanjing Medical University, Nanjing, China
- 2Department of Colorectal Surgery, The First Affiliated Hospital of Nanjing Medical University, Nanjing, China
- 3Department of Radiation Oncology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, China
MicroRNAs (miRNAs) are non-coding small RNA molecules that regulate gene expression at the post-transcriptional/translational level. They act a considerable role not only in the normal progress of development but also in aberrant human diseases, including malignancy. With accumulating proofs of miR-105, the complex role of miR-105 during cancer initiation and progression is gradually emerging. miR-105 acts as a tumor suppressor by inhibiting tumor growth and metastasis or as an oncogene by promoting tumor initiation and invasion, depending on particular tumor contexts and base-pairing genes. In this review, we emphasize the characteristics of miR-105 in cancer to elucidate various deadly tumors and discuss transcriptional regulations that may explain fluctuations in miR-105 expression. This review may provide new ideas for applying miR-105 as a diagnostic and prognostic biomarker.
Introduction
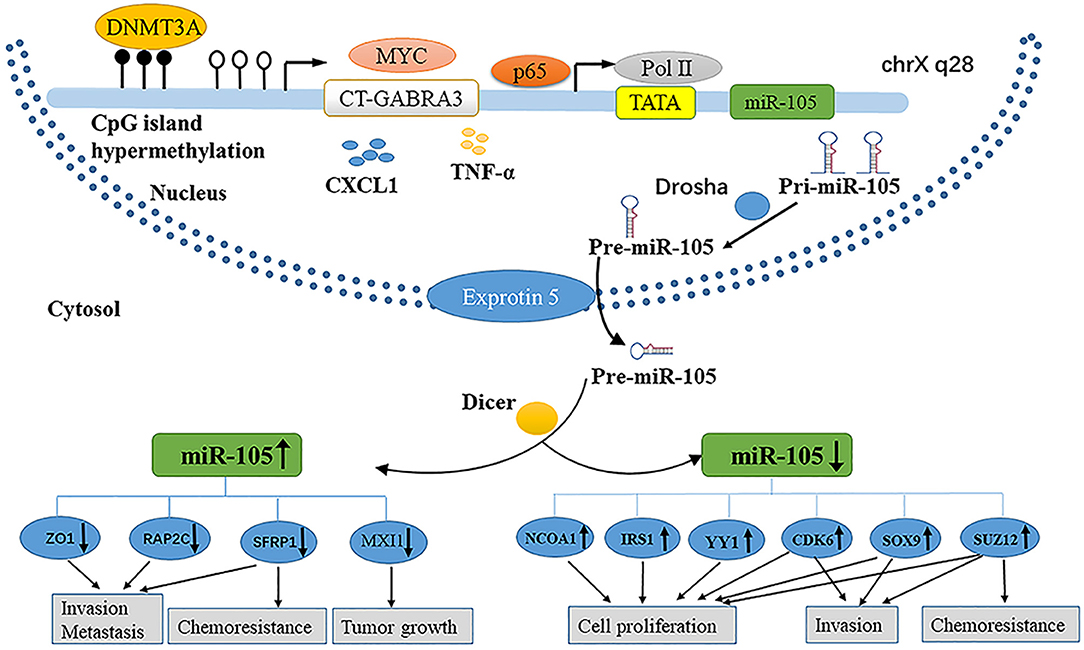
MicroRNAs (miRNAs) are small (~22 nt) non-coding RNAs that regulate gene expression through base-pairing with the 3′ untranslated regions (UTRs) of target messenger RNAs (mRNAs) (1). Similar to protein-coding genes, miRNAs are produced by endogenously transcribed long primary transcripts (pri-miRNAs), which are further cleaved into pre-miRNAs (2) (Figure 1).
Then, the pre-miRNAs are exported from the nuclei by double-stranded RNA-binding proteins (exportin-5) and then cleaved by RNase III family enzymes (Dicer) into incompletely mature miRNAs (3–5). The mature miRNA is incorporated into the RNA-induced silenced complex (RISC) (6, 7). The active RISC complex then binds to a target mRNA with a complementary sequence to trigger the mRNA for subsequent silencing (8). miRNA genes are involved in the regulation of up to 30% of all protein-coding genes (9, 10). Although the scope of miRNA regulation of the human transcriptome is still under investigation, the association of miRNA dysregulation with the occurrence and development of various diseases has been supported by evidence (11).
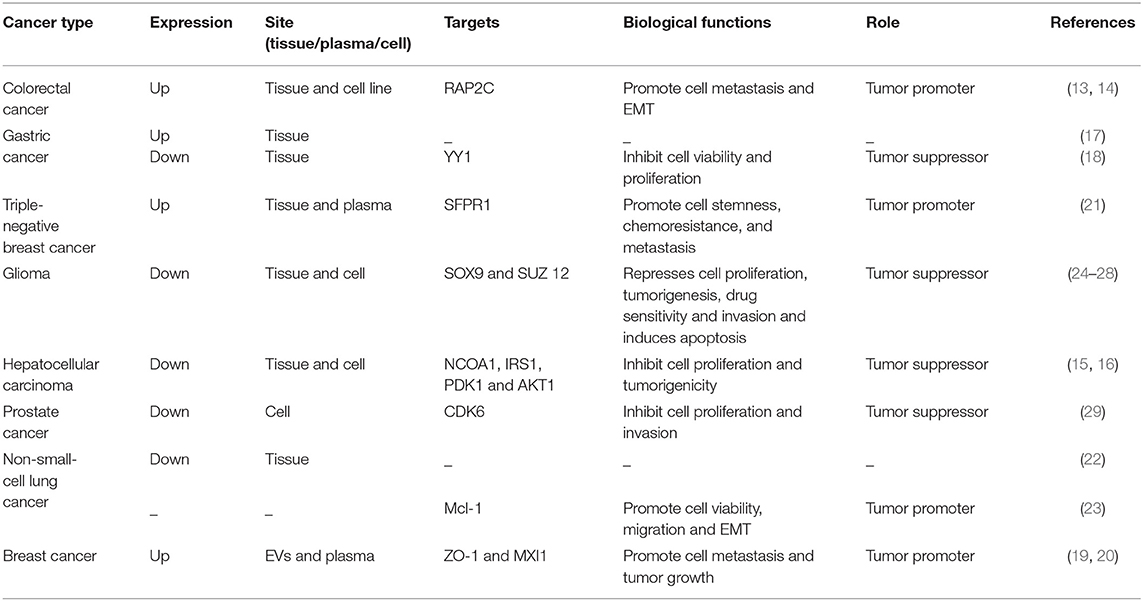
The miRNA-105 family, which consists of three members (miR-105-1, miR-105-2, and miR-767), is located on human chromosome Xq28. The first report showing the relationship between miR-105 and human health demonstrated its downregulation in the primary myelofibrosis granulocytes in comparison with that in normal subjects (12). Afterward, genome-wide miRNA expression profiling studies exhibited that miR-105 is widely expressed in various tissues or organs and is significantly aberrant in malignant cells such as in colorectal cancer (CRC) (13, 14), hepatocellular carcinoma (HCC) (15, 16), gastric cancer (GC) (17, 18), breast cancer (BC) (19–21), lung cancer (22, 23), and glioma (24–28) (Table 1).
However, contradictory results were produced through previous research with regard to whether miR-105 is an oncogene or a tumor suppressor and whether it is a positive or negative prognostic biomarker. In this review, we examine the following according to the properties of miR-105: (1) the oncogenic roles of miR-105, (2) miR-105 as a tumor suppressor, (3) the functions miR-05 in cancer-cell-secreted exosome, (4) the regulation mechanisms explaining the aberrant expression of miR-105, and (5) the diagnostic and prognostic value of miR-105 in cancer. The aim of this review is to emphasize the novel and diverse functions of miR-105 and its sense in cancer therapy.
Oncogenic Roles of miR-105
Colorectal Cancer
The vital roles of miRNAs, especially miR-105, in the pathophysiology of colorectal cancer (CRC) have been demonstrated in various studies (30–32). Through high-throughput sequencing, Hamfjord et al. found that the level of miR-105 expression was upregulated in tumor tissues from eight patients with CRC. Further data confirmed the differences in miRNA expression between adenocarcinomas and neuroendocrine of colon cancer (13). Shen et al. also reported that the miR-105 is overexpressed in CRC tissues and cell lines, and miR-105 expression positively correlates with lymph node invasion, TNM stage, and metastasis. Moreover, miR-105 expression is predominantly stimulated by TNF-α in a time-dependent manner. Upregulated miR-105 expression promotes CRC cell metastasis and epithelial-mesenchymal transition (EMT) in vitro and in vivo by directly targeting the 3′UTRs of a Rap2 subfamily of a small GTP-binding protein (RAP2C) through the activation of the NF-κB signaling pathway (14). Thus, miR-105 has an oncogenic role in human CRC. However, the abovementioned researches are relatively superficial, and future studies are needed to confirm the specific mechanism of miR-105 in CRC.
Gastric Cancer
Although several miRNAs have demonstrated their crucial roles in the occurrence and progression of gastric cancer (GC) (33–35), the exact role of miR-105 in GC remains unclear (17, 18). Liu et al. first reported that miR-105 expression is significantly elevated in GC tissues than in normal tissues (17). However, Zhou et al. recently revealed that a highly methylated miR-105 promoter decreases miR-105 expression in GC tissues (18). Overexpressed miR-105 inhibits cell viability and proliferation by directly targeting the Yin Yang 1 (YY1). The miR-105 role in GC is complex and thus must be investigated further.
Triple-Negative Breast Cancer
miR-105 functions as a tumor promoter by targeting corresponding genes in BC (19–21). Li et al. reported that miR-105 is upregulated specifically in the cancer tissues of patients with triple-negative breast cancer (TNBC). Currently, the treatment of TNBC is very disappointing (36). High miR-105 expression correlated with poor survival in patients with TNBC, and miR-105 promoted the stemness, chemoresistance, and metastasis of the TNBC cells by activating Wnt/β-catenin signaling by targeting SFPR1. Moreover, circulating miR-105 levels were significantly elevated in the plasmas of 74 patients with TNBC relative to the miR-105 levels of 44 patients without TNBC or 12 healthy controls, indicating that the circulating miR-105 may serve as a powerful biomarker for TNBC (21).
miR-105 as a Tumor Suppressor
Glioma
Yan et al. investigated that miR-105 is significantly lower in anaplastic gliomas and secondary glioblastomas than in low-grade gliomas. High miR-105 levels are commonly related to favorable prognosis in patients with glioma (25), and miR-105 level is reduced in glioblastoma unlike in low-grade gliomas (28). Guan et al. found that gliomas show larger decrease in miR-105 expression than non-neoplastic brains. Furthermore, low miR-105 expression is statistically associated with advanced tumor grade and unfavorable clinical outcome (24). Liu et al. reported that miR-105 is commonly downregulated in glioma tissues and cells and the level of miR-105 expression decreases with increasing tumor stage. Gain and loss-of-function experiments indicated that miR-105 upregulation represses proliferation and invasion and induces apoptosis by directly targeting the mRNA of SOX9 and inhibiting the downstream TCF4, c-MYC, Cyclin D1, and AXIN2 protein expression (27). In another study, Zhang et al. confirmed that miR-105 level is remarkably reduced in glioma tissues and cell lines and ectopic miR-105 inhibits cell proliferation, tumorigenesis, metastasis, and drug sensitivity by downregulating the suppressor of Zeste 12 in glioma (26). The abovementioned researches suggest that targeting miR-105 and its downstream factors is a promising therapeutic strategy for glioma treatment.
Hepatocellular Carcinoma
To date, human endogenous miRNA dysregulation has been involved in the carcinogenesis and progression of HCC (37–39). Shen et al. found that HCC cell lines and clinical HCC tissues show significant decrease in their miR-105 expression levels in contrast to normal hepatocytes and adjacent non-cancerous tissues. Decreased miR-105 expression promotes the proliferation and tumorigenicity of HCC cells in vitro and in vivo and activates the phosphoinositide 3-kinase (PI3K)/AKT signaling pathway by directly upregulating insulin receptor substrate-1, 3-phosphoinositide-dependent protein kinase-1, and AKT1, leading to decreased cyclin-dependent kinase inhibitors of 1A and 1B (p21Cip1 and p27Kip1) and increased cyclin D1 expression in HCC (16). Another study showed that miR-105 expression was reduced in HCC tissues (15). The upregulated expression of miR-105 suppresses the proliferation of HCC cells in vitro by targeting the nuclear receptor coactivator 1 (NCOA1). Furthermore, decreased miR-105 correlates with reduced median OS and PFS in patients with HCC. The research on the mechanism of miR-105 loss will help future scholars better understand the tumor transformation in HCC and design new treatment strategies.
Other Tumors
Apart from the aforementioned tumors, the miR-105 as a tumor suppressor has been widely studied in other sort of cancer, including prostate cancer and non-small-cell lung cancer (NSCLC). Honeywell et al. reported that prostate cancer cell lines have downregulated miR-105 expression compared with normal prostate epithelial cells. miR-105 upregulation inhibits tumor cell proliferation and invasion in vitro and tumor growth in vivo, and CDK6 is deemed as a new target of miR-105 (29). Consistently, Lu et al. also reported that the level of miR-105-1 is decreased in NSCLC tissues and that a decreased miR-105-1 level is associated with a large tumor size and poor overall survival (OS) and disease-free survival of patients with NSCLC (22). Interestingly, Jin et al. found that an elevated miR-105 significantly enhances the viability and migration of NSCLC cells in vitro through activating the mTOR and p38MAPK pathways by targeting Mcl1 (23). However, further researches are needed to fully characterize the biological function of miR-105 in these cancers.
Exosomal miR-105 by Cancer-Cell Secretion
Exosomes are small (50–100 nm) extracellular vesicles (EVs) with lipid bilayer membrane, whose release are conducted by fusion with the cell membrane (40–42). Exosomes have been identified as key factors mediating interactions between tumor cells and other stromal cells in tumor microenvironment. Exosomal miRNAs secreted by tumor cells can enter other non-cancer cells by endocytosis (40, 42–45). Cancer-secreted miRNAs act as an essential role in facilitating the cancer initiation and progression (46–49). Zhou et al. first found that metastatic BC (MBC) cells, which secrete exosomal RNAs, significantly stimulate the migration efficiency of human microvascular endothelial cells (HMVECs). The level of miR-105 expression is markedly higher in exosomes derived from MBC cells than in non-cancerous mammary epithelial cells. Meanwhile, the ectopic cancer-derived miR-105 is transferred via exosomes, thereby dramatically reducing ZO-1 expression and breaking the barrier function of HMVECs that promote metastasis both in vitro and in vivo (19). Yan et al. found that the uptake of EVs from BC cells by patient-derived cancer-associated fibroblasts (CAFs) induces the enrichment of a gene signature related to MYC activation in CAFs compared with non-cancerous mammary epithelial cell EVs or phosphate buffer saline by gene set enrichment analysis. In search of the potential upstream event leading to MYC activation, they observed MAX-interacting protein 1 (MXI1), which is a well-known antagonist of MYC transcriptional activity, is downregulated by >50% (50, 51). Moreover, miR-105 directly targets MXI1 in which its expression level is greatly higher in EVs derived from BC cells than in non-cancerous mammary epithelial cells. The abnormal expression of miR-105 in EV is the result of the MYC oncoprotein in BC cells and in turn, acts on MXI1 in CAFs and activates MYC signaling to induce a metabolic program that is suitable for tumor growth. Thus, the MYC-miR-105-MXI1-MYC-signaling axis-mediated metabolic reprogramming of stromal cells contributes to a sustained tumor growth by conditioning the shared metabolic environment (20) (Figure 2).
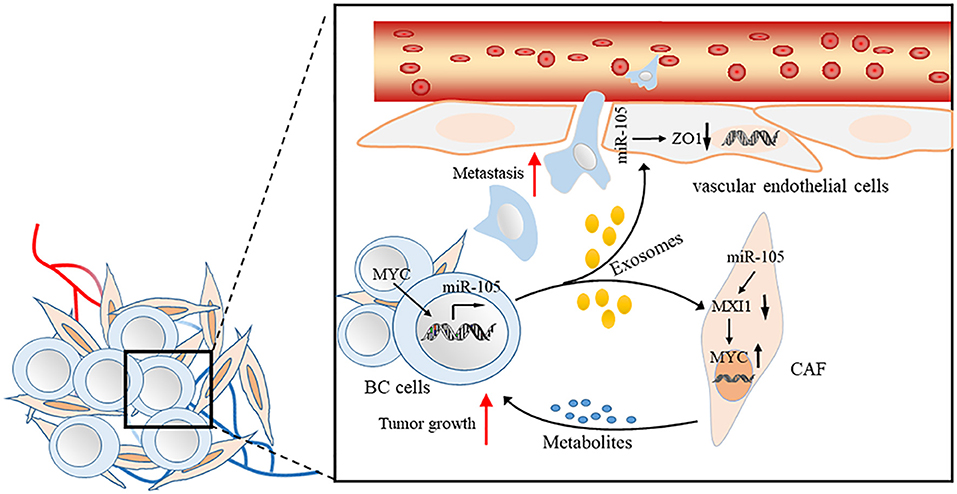
Figure 2. Proposed model of promoting BC cell metastasis and tumor growth by cancer-secreted EVs through a miR-105-mediated mechanism identified herein.
miR-105 is present in serum-derived exosomes, and the level of miR-105 expression in patients with distant metastasis is higher than in patients who did not develop metastasis (19). These studies provide insights into the regulation of gene expression and cellular components of the tumor microenvironment by exosomally derived miRNAs and indicate that miR-105 has potential clinical applications as a prognostic biomarker and a therapeutic target for BC.
Regulation Mechanisms of miR-105 Expression
Parallel to protein-encoding genes, the expression of miRNA is regulated not only by different mechanisms at a genetic or epigenetic level but also by the dysregulation of specific transcription factors (52). Similarly, the dysregulation of miR-105 in cancer is involved in some of the aforementioned mechanisms (Figure 1).
Epigenetic regulation is an important mechanism governing miRNA expression (53, 54). Zhou et al. reported that the expression of miR-105 is downregulated concomitantly with the DNA hypermethylation of CpGs in the upstream region of the miR-105 promoter in GC. Furthermore, the knockdown of DNA methyltransferase (DNMT3A) could regain the level of miR-105 expression, which is suppressed in GC (18). Meanwhile, Loriot et al. demonstrated that the hypomethylation of CpGs in the upstream region of the miR-105 promoter activates the cancer-germline transcript (CT-GABRA3) that inducess the high expression of miR-105 and miR-767 in melanoma, immortalized fibroblast, and embryonal carcinoma cell lines (55). Through chromatin immunoprecipitation (ChIP) assays, Ji et al. confirmed that the NF-κB transcription complex p65 binds to the upstream promoter regions of miR-105 genes to silence transcription; p65 is recruited beforehand by the fibroblast growth factor 2 (FGF2), and miR-105 expression is restored by suppressing p65 in human osteoarthritis chondrocytes (56). Yan et al. found that the abnormal expression of MYC upregulates miR-105 expression both in the cells and in EVs, while MYC knockdown downregulates intracellularly and secretes miR-105 in BC cells expressing high miR-105 levels. Then, a thorough search of the promoter of GABRA3, the hosting gene harboring hsa-mir-105-1/2, revealed the presence of an E-box which responded to MYC to activate reporter gene expression (20). The alteration of the tumor microenvironment has a novel mechanism of the miRNA expression (57). Shen et al. found that the level of miR-105 expression is markedly stimulated by TNF-α in a time-dependent manner through employing RT-qPCR analysis in CRC cells (14). By next-generation sequencing and RT-qPCR, Hsu et al. found that miR-105 expression is upregulated in cells that is treated with CXCL1 secreted from tumor-associated dendritic cells in colon cancer (58).
Clinical Implications
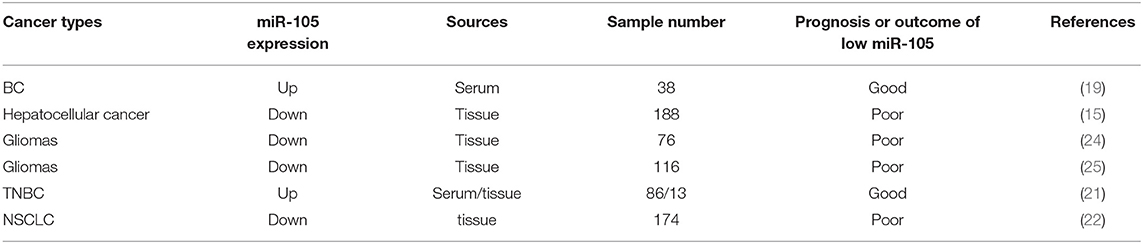
Improving the therapeutic result with the early diagnosis and accurate prognosis evaluation of cancer is critical. Cancer cells or tissues may present aberrant miRNA expression profiles, and specific miRNA features can be used not only for diagnosis but also for classifying patients with cancer as subgroups with different prognosis for individualized treatment (59–63). The role of miR-105 in cancer diagnosis and prognosis is extensively investigated, but it exhibits apparently conflicting outcomes (Table 2).
Zhou et al. demonstrated that patients who later developed distant metastases have higher levels of circulating (exosomal) and tumor miR-105 than patients who did not and had normal mammary tissues, suggesting that cancer-derived miR-105 can serve as a blood-based marker for the early diagnosis of BC metastasis (19). High NCOA1 and low miR-105-1 levels significantly decrease OS (P < 0.001) and PFS (P = 0.002), implying that NCOA1 and miR-105-1 might have a potential prognostic value for patients with HCC (15). Guan et al. found that a decreased miR-105 expression is statistically associated with advanced clinical features and poor OS (P < 0.001) of patients with glioma, suggesting that miR-105 downregulation may be used as a malignant prognostic marker in gliomas (24). Similarly, Yan et al. indicated that low miR-105 expression can be used for identifying patients with a high risk of unfavorable outcomes, particularly as a prognostic marker for patient risk stratification in anaplastic gliomas and secondary and proneural glioblastomas (25). A high level of miR-105 is associated with advanced clinical stages and the increased rates of the distant metastasis of CRC (14). Li et al. found that circulating miR-105/93-3p can act as an early diagnostic biomarker for TNBC, and the levels of miR-105/93-3p are significantly elevated in the plasmas of patients with TNBC relative to the levels in the plasmas of patients without TNBC or healthy controls. As expected, the patients with high miR-105/93-3p levels in TNBC tissues showed a positive correlation with chemoresistance and poor survival (21). Reduced miR-105-1 expression is associated with a larger tumor size, as well as the poor OS and disease-free survival of patients with NSCLC, demonstrating that downregulated miR-105-1 can be used as an independent malignant predictor in patients with NSCLC (22).
Conclusion
miRNAs play a vital role in the initiation and progression of human malignancies. On the one hand, miR-105 facilitates cell proliferation, promotes metastasis and chemoresistance, and initiates EMT in tumorigenesis, as illustrated in Figure 1. On the other hand, miR-105 inhibits proliferation, tumorigenesis, migration, invasion, and drug sensitivity. The role of this miRNA depends on the cellular and histological features of tumors and target mRNAs. The fluctuation of miR-105 expression is correlated with epigenetic alterations, especially the regression of miR-105 in cancer. Nevertheless, further researches are needed to provide sufficient evidence to explain the activation of miR-105 in cancer.
Author Contributions
JL, ZZ, and FC contributed to conception and manuscript writing. TH, WP, and QG searched the literature. YS participated in its coordination and modification. All the authors have read and approved the final manuscript.
Funding
The work is funded by Jiangsu Key Medical Discipline (General Surgery) (ZDXKA2016005).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Ambros V. microRNAs: tiny regulators with great potential. Cell. (2001) 107:823–6. doi: 10.1016/S0092-8674(01)00616-X
2. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. (2003) 425:415–9. doi: 10.1038/nature01957
3. Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. (2001) 409:363–6. doi: 10.1038/35053110
4. Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. (2004) 10:185–91. doi: 10.1261/rna.5167604
5. Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. (2001) 293:834–8. doi: 10.1126/science.1062961
6. Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. (2002) 16:720–8. doi: 10.1101/gad.974702
7. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. (2009) 11:228–34. doi: 10.1038/ncb0309-228
8. Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. (2005) 123:631–40. doi: 10.1016/j.cell.2005.10.022
9. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. (2005) 120:15–20. doi: 10.1016/j.cell.2004.12.035
10. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. (2003) 115:787–98. doi: 10.1016/S0092-8674(03)01018-3
11. Srinivasan S, Selvan ST, Archunan G, Gulyas B, Padmanabhan P. MicroRNAs -the next generation therapeutic targets in human diseases. Theranostics. (2013) 3:930–42. doi: 10.7150/thno.7026
12. Guglielmelli P, Tozzi L, Pancrazzi A, Bogani C, Antonioli E, Ponziani V, et al. MicroRNA expression profile in granulocytes from primary myelofibrosis patients. Exp Hematol. (2007) 35:1708–18. doi: 10.1016/j.exphem.2007.08.020
13. Hamfjord J, Stangeland AM, Hughes T, Skrede ML, Tveit KM, Ikdahl T, et al. Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS ONE. (2012) 7:e34150. doi: 10.1371/journal.pone.0034150
14. Shen Z, Zhou R, Liu C, Wang Y, Zhan W, Shao Z, et al. MicroRNA-105 is involved in TNF-alpha-related tumor microenvironment enhanced colorectal cancer progression. Cell Death Dis. (2017) 8:3213. doi: 10.1038/s41419-017-0048-x
15. Ma YS, Wu TM, Lv ZW, Lu GX, Cong XL, Xie RT, et al. High expression of miR-105–1 positively correlates with clinical prognosis of hepatocellular carcinoma by targeting oncogene NCOA1. Oncotarget. (2017) 8:11896–905. doi: 10.18632/oncotarget.14435
16. Shen G, Rong X, Zhao J, Yang X, Li H, Jiang H, et al. MicroRNA-105 suppresses cell proliferation and inhibits PI3K/AKT signaling in human hepatocellular carcinoma. Carcinogenesis. (2014) 35:2748–55. doi: 10.1093/carcin/bgu208
17. Liu D, Hu X, Zhou H, Shi G, Wu J. Identification of aberrantly expressed miRNAs in gastric cancer. Gastroenterol Res Prac. (2014) 2014:473817. doi: 10.1155/2014/473817
18. Zhou GQ, Han F, Shi ZL, Yu L, Li XF, Yu C, et al. DNMT3A-mediated down-regulation of microRNA-105 promotes gastric cancer cell proliferation. Eur Rev Med Pharmacol Sci. (2017) 21:3377–83.
19. Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. (2014) 25:501–15. doi: 10.1016/j.ccr.2014.03.007
20. Yan W, Wu X, Zhou W, Fong MY, Cao M, Liu J, et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol. (2018) 20:597–609. doi: 10.1038/s41556-018-0083-6
21. Li HY, Liang JL, Kuo YL, Lee HH, Calkins MJ, Chang HT, et al. miR-105/93-3p promotes chemoresistance and circulating miR-105/93-3p acts as a diagnostic biomarker for triple negative breast cancer. Breast Cancer Res. (2017) 19:133. doi: 10.1186/s13058-017-0918-2
22. Lu G, Fu D, Jia C, Chai L, Han Y, Liu J, et al. Reduced miR-105-1 levels are associated with poor survival of patients with non-small cell lung cancer. Oncol Lett. (2017) 14:7842–7848. doi: 10.3892/ol.2017.7228
23. Jin X, Yu Y, Zou Q, Wang M, Cui Y, Xie J, et al. MicroRNA-105 promotes epithelial-mesenchymal transition of nonsmall lung cancer cells through upregulating Mcl-1. J Cell Biochem. (2018) 120:5880–8. doi: 10.1002/jcb.27873
24. Guan Y, Chen L, Bao Y, Li Z, Cui R, Li G, et al. Identification of low miR-105 expression as a novel poor prognostic predictor for human glioma. Int J Clin Exp Med. (2015) 8:10855–64.
25. Yan W, Li R, Liu Y, Yang P, Wang Z, Zhang C, et al. MicroRNA expression patterns in the malignant progression of gliomas and a 5-microRNA signature for prognosis. Oncotarget. (2014) 5:12908–15. doi: 10.18632/oncotarget.2679
26. Zhang J, Wu W, Xu S, Zhang J, Zhang J, Yu Q, et al. MicroRNA-105 inhibits human glioma cell malignancy by directly targeting SUZ12. Tumour Biol. (2017) 39: 5766. doi: 10.1177/1010428317705766
27. Liu X, Wang H, Zhu Z, Ye Y, Mao H, Zhang S. MicroRNA-105 targets SOX9 and inhibits human glioma cell progression. FEBS Lett. (2016) 590:4329–42. doi: 10.1002/1873-3468.12458
28. Ma X, Yoshimoto K, Guan Y, Hata N, Mizoguchi M, Sagata N, et al. Associations between microRNA expression and mesenchymal marker gene expression in glioblastoma. Neuro Oncol. (2012) 14:1153–62. doi: 10.1093/neuonc/nos145
29. Honeywell DR, Cabrita MA, Zhao H, Dimitroulakos J, Addison CL. miR-105 inhibits prostate tumour growth by suppressing CDK6 levels. PLoS ONE. (2013) 8:e70515. doi: 10.1371/journal.pone.0070515
30. Jahid S, Sun J, Edwards RA, Dizon D, Panarelli NC, Milsom JW, et al. miR-23a promotes the transition from indolent to invasive colorectal cancer. Cancer Disc. (2012) 2:540–53. doi: 10.1158/2159-8290.CD-11-0267
31. Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J, et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. (2012) 72:3631–41. doi: 10.1158/0008-5472.CAN-12-0667
32. Gao J, Li N, Dong Y, Li S, Xu L, Li X, et al. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene. (2015) 34:4142–52. doi: 10.1038/onc.2014.348
33. Tang H, Deng M, Tang Y, Xie X, Guo J, Kong Y, et al. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res. (2013) 19:5602–12. doi: 10.1158/1078-0432.CCR-13-1326
34. Xia J, Wu Z, Yu C, He W, Zheng H, He Y, et al. miR-124 inhibits cell proliferation in gastric cancer through down-regulation of SPHK1. J Pathol. (2012) 227:470–80. doi: 10.1002/path.4030
35. Duan Y, Hu L, Liu B, Yu B, Li J, Yan M, et al. Tumor suppressor miR-24 restrains gastric cancer progression by downregulating RegIV. Mol Cancer. (2014) 13:127. doi: 10.1186/1476-4598-13-127
36. Palma G, Frasci G, Chirico A, Esposito E, Siani C, Saturnino C, et al. Triple negative breast cancer: looking for the missing link between biology and treatments. Oncotarget. (2015) 6:26560–74. doi: 10.18632/oncotarget.5306
37. Liu H, Liu Y, Liu W, Zhang W, Xu J. EZH2-mediated loss of miR-622 determines CXCR4 activation in hepatocellular carcinoma. Nat Commun. (2015) 6:8494. doi: 10.1038/ncomms949
38. Tao ZH, Wan JL, Zeng LY, Xie L, Sun HC, Qin LX, et al. miR-612 suppresses the invasive-metastatic cascade in hepatocellular carcinoma. J Exp Med. (2013) 210:789–803. doi: 10.1084/jem.20120153
39. Chang Y, Yan W, He X, Zhang L, Li C, Huang H, et al. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. (2012) 143:177–87.e178. doi: 10.1053/j.gastro.2012.04.009
40. Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. (2008) 10:1470–6. doi: 10.1038/ncb1800
41. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. (2007) 9:654–9. doi: 10.1038/ncb1596
42. Yuan A, Farber EL, Rapoport AL, Tejada D, Deniskin R, Akhmedov NB, et al. Transfer of microRNAs by embryonic stem cell microvesicles. PloS ONE. (2009) 4:e4722. doi: 10.1371/journal.pone.0004722
43. Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. (2011) 71:3792–801. doi: 10.1158/0008-5472.CAN-10-4455
44. Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. (2010) 39:133–44. doi: 10.1016/j.molcel.2010.06.010
45. Zhuang G, Wu X, Jiang Z, Kasman I, Yao J, Guan Y, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. (2012) 31:3513–23. doi: 10.1038/emboj.2012.183
46. Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC, Tsai PH, et al. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. (2017) 36:4929–42. doi: 10.1038/onc.2017.105
47. Baroni S, Romero-Cordoba S, Plantamura I, Dugo M, D'Ippolito E, Cataldo A, et al. Exosome-mediated delivery of miR-9 inducescancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. (2016) 7:e2312. doi: 10.1038/cddis.2016.224
48. Singh R, Pochampally R, Watabe K, Lu Z, Mo YY. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol Cancer. (2014) 13:256. doi: 10.1186/1476-4598-13-256
49. Fang T, Lv H, Lv G, Li T, Wang C, Han Q, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. (2018) 9:191. doi: 10.1038/s41467-017-02583-0
50. Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. (1993) 72:223–32. doi: 10.1016/0092-8674(93)90662-A
51. Lee TC, Ziff EB. Mxi1 is a repressor of the c-Myc promoter and reverses activation by USF. J Biol Chem. (1999) 274:595–606. doi: 10.1074/jbc.274.2.595
52. Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J. (2012) 18:215–22. doi: 10.1097/PPO.0b013e318250c001
53. Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. (2007) 67:1424–9. doi: 10.1158/0008-5472.CAN-06-4218
54. Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. (2012) 31:1609–22. doi: 10.1038/onc.2011.354
55. Loriot A, Van Tongelen A, Blanco J, Klaessens S, Cannuyer J, van Baren N, et al. A novel cancer-germline transcript carrying pro-metastatic miR-105 and TET-targeting miR-767 induced by DNA hypomethylation in tumors. Epigenetics. (2014) 9:1163–71. doi: 10.4161/epi.29628
56. Ji Q, Xu X, Xu Y, Fan Z, Kang L, Li L, et al. miR-105/Runx2 axis mediates FGF2-induced ADAMTS expression in osteoarthritis cartilage. J Mol Med. (2016) 94:681–94. doi: 10.1007/s00109-016-1380-9
57. Qin Q, Furong W, Baosheng L. Multiple functions of hypoxia-regulated miR-210 in cancer. J Exp Clin Cancer Res. (2014) 33:50. doi: 10.1186/1756-9966-33-50
58. Hsu YL, Chen YJ, Chang WA, Jian SF, Fan HL, Wang JY, et al. Interaction between tumor-associated dendritic cells and colon cancer cells contributes to tumor progression via CXCL1. Int J Mol Sci. (2018) 19:2427. doi: 10.3390/ijms19082427
59. Zheng W, Zhao J, Tao Y, Guo M, Ya Z, Chen C, et al. MicroRNA-21: a promising biomarker for the prognosis and diagnosis of non-small cell lung cancer. Oncol Let. (2018) 16:2777–82. doi: 10.3892/ol.2018.8972
60. Ghodousi ES, Rahgozar S. MicroRNA-326 and microRNA-200c: two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J Cell Biochem. (2018) 119:6024–32. doi: 10.1002/jcb.26800
61. Li P, Dong J, Zhou X, Sun W, Huang H, Chen T, et al. Expression patterns of microRNA-329 and its clinical performance in diagnosis and prognosis of breast cancer. OncoTarget Ther. (2017) 10:5711–8. doi: 10.2147/OTT.S147974
62. Bai Y, Lin H, Fang Z, Luo Q, Fang Y, Su Y, et al. Plasma microRNA-19a as a potential biomarker for esophageal squamous cell carcinoma diagnosis and prognosis. Biomarks Med. (2017) 11:431–41. doi: 10.2217/bmm-2016-0286
Keywords: miR-105, cancer, oncogene, tumor suppressor, exosome, clinical implication
Citation: Li J, Zhang Z, Chen F, Hu T, Peng W, Gu Q and Sun Y (2019) The Diverse Oncogenic and Tumor Suppressor Roles of microRNA-105 in Cancer. Front. Oncol. 9:518. doi: 10.3389/fonc.2019.00518
Received: 15 January 2019; Accepted: 29 May 2019;
Published: 20 June 2019.
Edited by:
Luisa Lanfrancone, Istituto Europeo di Oncologia s.r.l., ItalyReviewed by:
Giuseppe Palma, National Cancer Institute G. Pascale Foundation (IRCCS), ItalyGiovanni Blandino, Istituti Fisioterapici Ospitalieri (IRCCS), Italy
Copyright © 2019 Li, Zhang, Chen, Hu, Peng, Gu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yueming Sun, c3VueXVlbWluZ0Buam11LmVkdS5jbg==
†These authors have contributed equally to this work
 Jie Li
Jie Li Zhiyuan Zhang
Zhiyuan Zhang Fangyu Chen
Fangyu Chen Tao Hu1,2
Tao Hu1,2