- 1Department of Cancer Biology, Institute of Cancer Research, London, United Kingdom
- 2Division of Computational and Systems Medicine, Department of Surgery and Cancer, Imperial College London, London, United Kingdom
Although there has been a renewed interest in the field of cancer metabolism in the last decade, the link between metabolism and DNA damage/DNA repair in cancer has yet to be appreciably explored. In this review, we examine the evidence connecting DNA damage and repair mechanisms with cell metabolism through three principal links. (1) Regulation of methyl- and acetyl-group donors through different metabolic pathways can impact DNA folding and remodeling, an essential part of accurate double strand break repair. (2) Glutamine, aspartate, and other nutrients are essential for de novo nucleotide synthesis, which dictates the availability of the nucleotide pool, and thereby influences DNA repair and replication. (3) Reactive oxygen species, which can increase oxidative DNA damage and hence the load of the DNA-repair machinery, are regulated through different metabolic pathways. Interestingly, while metabolism affects DNA repair, DNA damage can also induce metabolic rewiring. Activation of the DNA damage response (DDR) triggers an increase in nucleotide synthesis and anabolic glucose metabolism, while also reducing glutamine anaplerosis. Furthermore, mutations in genes involved in the DDR and DNA repair also lead to metabolic rewiring. Links between cancer metabolism and DNA damage/DNA repair are increasingly apparent, yielding opportunities to investigate the mechanistic basis behind potential metabolic vulnerabilities of a substantial fraction of tumors.
Introduction
Over the past decade, a renewed interest in cancer metabolism has emerged. The idea originated from the work of Otto Warburg and colleagues—first published in the 1920s—who noticed the propensity for cancer cells to consume increased quantities of glucose (1, 2). We now understand that there is extensive metabolic rewiring in cancer cells, and we are starting to decipher how cancer cells reprogram their metabolism to adapt to changes in their microenvironment and support their high metabolic needs (3–5). Cancer metabolism is a broad topic and our current understanding of how metabolic reprogramming is linked to malignant transformation has been extensively reviewed elsewhere (6–8). Therefore, in this review, we decided to focus on the connections between cell metabolism and another major aspect of cancer biology, DNA-repair/DNA-damage pathways.
The major hallmarks, as described by Pavlova and Thompson (7), include: deregulated glucose and amino acid uptake, opportunistic ways of acquiring nutrients, use of metabolic intermediates for biomass and nicotinamide adenine dinucleotide phosphate (NADPH) synthesis, increased demand for nitrogen, and alterations in metabolite-driven gene expression and interaction with the microenvironment (7, 9). Importantly, the wide differences in metabolic dependencies across cancer types, or stage of progression, as well as the dynamic shifts between metabolic pathways, make the study of cancer metabolism and the development of new therapies targeting these pathways very challenging.
In addition to the accepted role for cell metabolism in cancer, it is well established that DNA-repair/DNA-damage pathways are important in cancer progression because dysregulation leads to higher levels of genomic instability, increased mutation rate, and enhanced intra-tumor heterogeneity (10–13). There are currently three principal mechanisms through which changes in cell metabolic status are thought to have an influence on DNA-damage/DNA-repair pathways: chromatin remodeling, double-strand break (DSB) repair, and redox homeostasis (Figure 1). Uncovering new links between these important aspects of cancer biology might lead to the development of new targeted therapies in DNA-repair deficient cancers or even improving the efficacy of existing therapies, such as PARP inhibitors, anthracyclines, and platinum salts. In this review, we examine work seeking to uncover the links between DNA-repair/DNA-damage and cell metabolism.
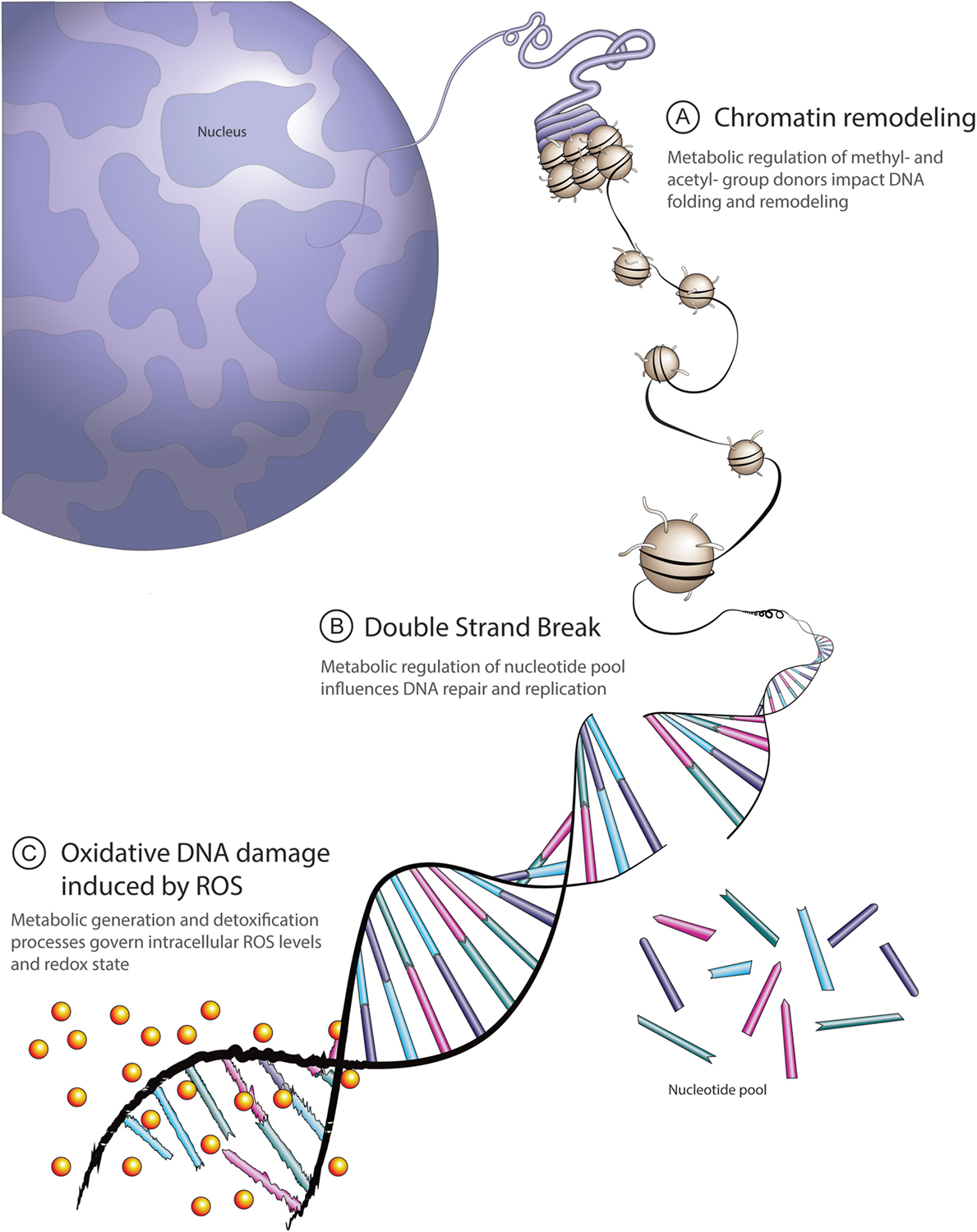
Figure 1. Overview of principal links between cell metabolism and DNA repair. (A) Methyl-group donors from the S-adenosylmethionine pathway and acetyl-donors from citrate cycle-derived acetyl coenzyme A contribute to dynamic chromatin packaging and remodeling essential to DNA double-strand repair. (B) Metabolic intermediates derived from glucose, glutamine, and aspartate are required for de novo nucleotide synthesis. The ready availability of a pool of nucleotides facilitates appropriate DNA repair and replication. (C) Intracellular reactive oxygen species (ROS) levels reflect a balance between generation and detoxification. A principal ROS detoxification mechanism involves reduced glutathione (GSH), determined by glutamine and cysteine availability, as well as NADPH levels. High ROS-induced DNA damage leads to excessive burden on the DNA repair machinery.
Metabolic Status Affects DNA Folding and Repair Pathways
The first link between DNA repair and cell metabolism involves DNA folding and organization. Chromatin packaging and remodeling, through different histone posttranslational modifications—including acetylation, methylation, phosphorylation, and ubiquitination, as well as through DNA modifications such as methylation—can regulate gene expression levels by modulating the access to DNA of different protein complexes (14, 15). Interestingly, similar mechanisms can also regulate access of DNA-repair proteins to the DNA double-helix (16). When repairing DSBs, the first step involves unfolding DNA to allow access of the repair complexes to the DSB (17). There is a growing body of evidence suggesting that these mechanisms can in fact regulate the choice of DNA-repair pathway (i.e., homologous recombination), or non-homologous end-joining used to repair the DSB (16). Substrates added to histones or DNA are derived from metabolic intermediates (18–20) (Figure 2). For example, methyl-group donors mostly come from the S-adenosylmethionine (SAM) pathway (18), while the sole acetyl-group donor is acetyl coenzyme A (acetyl-CoA), known for the transfer of its acetyl group to oxaloacetate to form citrate and start the tricarboxylic acid (TCA) cycle (21). It has been shown that expression levels of the enzyme, ATP citrate lyase, can regulate the availability of acetyl-CoA in the cell (22). Restricting the amount of acetyl-group donors can disrupt proper DNA organization and have an impact on DNA folding and DNA remodeling essential to successful DNA DSB repair (22, 23). Acetyl-CoA can also be generated from acetate and CoA from the acetyl-CoA synthetase 2 enzyme. In fact, under metabolic stress conditions in which oxygen and lipids are deprived, acetate-generated acetyl-CoA is enhanced and acetate-derived carbons are incorporated in lipid synthesis (24, 25). Acetate-derived acetyl-CoA has also been shown to be involved in histone acetylation, suggesting that acetate availability could influence histone acetylation in cancer cells (26).
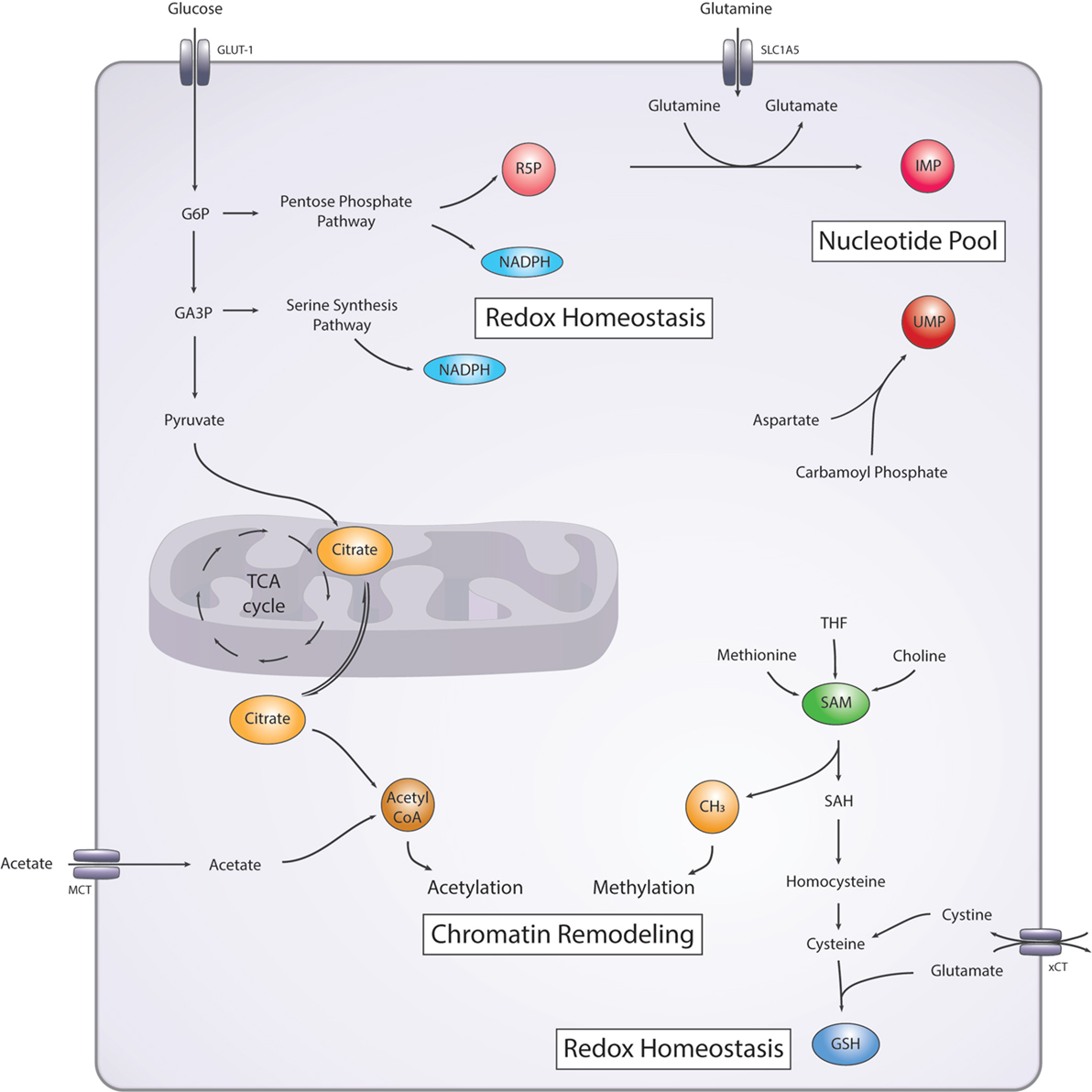
Figure 2. Simplified diagram of the main metabolic pathways involved in DNA damage/repair. Nucleotide pool: nucleotide precursor ribose-5-phosphate (R5P) is generated from the pentose-phosphate pathway (PPP). Purines and pyrimidines precursors, inosine monophosphate (IMP), and uridine monophosphate (UMP), respectively are synthesized from glutamine, and aspartate and carbamoyl phosphate, respectively. Redox homeostasis: nicotinamide adenine dinucleotide phosphate (NADPH) is generated from the PPP and the serine synthesis pathway. Glutathione (GSH) is generated from cysteine and glutamate. Chromatin remodeling: methyl-group donors (CH3) are generated from the S-adenosyl methionine (SAM) pathway and acetyl-group donors (acetyl coenzyme A) are generated from the citrate cycle or from acetate. GLUT-1, glucose transporter 1; SCL1A5, alanine, serine, cysteine-preferring transporter 2 (ASCT2); G6P, glucose 6-phosphate; GA3P, glyceraldehyde 3-phosphate; SAH, S-adenosyl homocysteine; xCT, cystine-glutamate antiporter; MCT, monocarboxylate transporter family.
Additionally, changes in the activity of the SAM pathway due to availability of necessary nutrients or substrates also influence DNA or histone methylation by regulating the pool of methyl-donors (27). The SAM pathway is interconnected with methionine, tetrahydrofolate (THF), one-carbon metabolism, and choline pathways. These pathways tend to compensate for one another; however, depletion of choline, folate, or methionine can still have an impact on the final SAM concentration (18). Such changes in the SAM pathway not only influence gene expression of cancer-associated genes through epigenetic modifications but can also impact DNA folding during DNA repair processes. The latter can also be influenced directly by metabolic intermediates such as fumarate. DNA-PK dependent activation of fumarase contributes to enhanced local generation of fumarate, which in turn promotes DNA repair via inhibition of KDM2B-mediated histone demethylase activity (28). Accumulation of fumarate can also lead to epithelial-to-mesenchymal transition and poor clinical outcome (29). Since modifications in histone and DNA are important for proper repair of DNA DSBs (30), changes in nutrient and substrate levels that disrupt these modifications are bound to affect DNA-repair pathways.
Nucleotide Levels Affect DNA-Repair Potential of Cancer Cells
Another major mechanism upon which cell metabolism can regulate DNA-repair/DNA-damage is through the regulation of the pool of nucleotides used for DNA replication and repair (31, 32). Many different metabolic pathways are involved in de novo nucleotide synthesis and can have an impact on the levels of intracellular nucleotides available (Figure 2) (31, 33, 34). An important precursor in the synthesis of the ribose backbone, essential for both purines and pyrimidines, comes from ribose-5-phosphate, an intermediate of the pentose-phosphate pathway (PPP) (31). Briefly, the PPP utilizes glucose-6-phosphate (G6P), an intermediate of glycolysis, which can be redirected to generate metabolic intermediates necessary for nucleotide and protein synthesis, as well as generation of NADPH. The PPP is a major focus in cancer metabolism, and it has been carefully reviewed elsewhere (31, 35, 36). The increased glucose consumption observed in cancer cells has been shown, at least in some cancers, to be used to fuel the PPP, where it can potentially be used to generate reducing power in the form of NADPH and nucleotide precursors (32). Additionally, other metabolic pathways are involved more specifically in purine or pyrimidine ring synthesis. The amide group of glutamine is essential in two steps of inosine monophosphate synthesis, an intermediate in de novo purine synthesis, and in one step of uridine monophosphate synthesis, an intermediate in de novo pyrimidine synthesis (37, 38). In glioblastoma, it has been shown that α-ketoglutarate is diverted out of the TCA cycle to synthesize more glutamine through the glutamine synthetase enzyme. The resulting glutamine is then used for de novo purine synthesis (39). Furthermore, aspartate is essential for the synthesis of pyrimidines, and glycine is also essential for purine synthesis (38). Loss-of-function mutations in arginosuccinate synthase 1, which leads to reduced arginine production in the urea cycle, promotes the use of aspartate for pyrimidine synthesis (33). Aspartate is mostly synthesized from glutamate and oxaloacetate. Therefore, the availability of glutamine and/or other amino acids/metabolic substrates can influence the amount and ratio of nucleotides produced in a cell and might provide a regulatory mechanism for DNA-repair.
Deregulated Redox Homeostasis Promotes DNA Damage
The metabolic regulation of reactive oxygen species (ROS) levels is the third and final link that we address here between DNA-repair/DNA-damage and cell metabolism. High ROS levels affect many aspects of tumor biology and, here, we focus on their role in inducing DNA damage and genomic instability. Most of the DNA lesions that are formed by ROS-induced damage are single strand breaks (SSBs) that can be repaired through nucleotide or base excision repair (NER/BER) (40). However, these SSBs can lead to stalling of the replication fork or error in replication, ultimately leading to DSBs (40). The accumulation of oxidative DNA damage increases the burden on the DNA-repair machinery. Tight regulation of cellular redox stress is essential, since high ROS levels can lead to oxidative stress and oxidative damage of proteins, DNA, and lipids, while a certain level of ROS is essential for activating signaling pathways involved in multiple biological processes (41–43). Cells have evolved a number of ways to balance ROS levels (Figure 2). Glutathione (GSH) is one of the major ROS-scavenging molecules (44), and it occurs in two versions; the reduced form, sulfhydryl GSH, and the oxidized, glutathione disulfide (GSSG). The enzyme GSH peroxidase catalyzes the reduction of H2O2 to water and lipid hydroperoxides to their corresponding lipid alcohols and in turn oxidizes GSH to GSSG. The reverse reaction is catalyzed by the enzyme glutathione reductase, also known as glutathione-disulfide reductase using the reducing potential of NADPH (45).
Glutathione is synthesized in the cytoplasm from cysteine and glutamate, and cysteine availability is the rate-limiting step of the synthesis reaction (46, 47). Cysteine can be synthesized from serine through the transsulfuration pathway (47), or it can be transported into the cells as cystine in exchange for glutamate using the cystine-glutamate antiporter xCT (39). Interestingly, mTORC2 can phosphorylate and inhibit the xCT antiporter, therefore providing a mechanism to regulate intracellular cysteine levels (48). Furthermore, it has been shown that, in some cancer types, growing cells in DMEM containing cystine leads to xCT dependent glutamine dependency in vitro, providing evidence for context dependent changes in metabolism (5). The main source of NADPH comes from the PPP and a significant amount also comes from the serine synthesis pathway through the THF pathway. Both harness the energy from the catabolism of metabolic intermediates to generate NADPH (31, 47). Interestingly, an important regulator of ROS levels is the transcription factor nuclear factor E2-related factor 2 (NRF2) that has been shown to directly regulate the key serine synthesis enzymes (PHGDH, PSAT1, ATF4) (49). This provides a potential mechanism through which NRF2 can regulate ROS levels by influencing NADPH and GSH levels. Importantly, the DNA-repair protein, BRCA1, regulates NRF2, providing an additional link between DNA repair pathways and ROS levels (50). Overall, there are still many open questions with regards to how cancer cells can rewire their metabolism based on different nutrient availability, but there is growing evidence to suggest that maintaining cellular redox homeostasis plays a major role on DNA-damage/DNA-repair pathways.
DNA Damage Response (DDR) Triggers Metabolic Rewiring
While DNA-repair pathways can be influenced by cellular metabolic status and nutrient availability in the tumor microenvironment, accumulation of DNA damage due to extrinsic and intrinsic genotoxic stress, or deficient DNA repair can also cause abrupt rewiring of cell metabolism. Cells have evolved the DDR pathway (DDR) to monitor this genotoxic stress and maintain accurate transmission of genetic information to subsequent generations. The DDR can, therefore, halt cell-cycle progression, induce DNA-repair mechanisms, or trigger programmed cell death when DNA damage is irreparable (51). Ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) kinases are two key enzymes in the recognition of DNA damage and implementation of the DDR (51, 52). Upon activation by DNA-damage, ATM and ATR generate a second wave of phosphorylation that impacts many downstream effector proteins. An important aspect of DDR, driven by ATM and ATR activation, is the induction of metabolic rewiring to promote the resolution of genotoxic stress. ATM has been shown to activate the PPP through induction of the rate-limiting enzyme glucose-6-phosphate dehydrogenase (G6PD), to support the synthesis of reducing power in the form of NADPH, and generate ribose-5-phosphate for nucleotide synthesis (53, 54).
Increased carbon flux to the PPP from glucose derivatives is driven not only by increased glucose consumption often observed in cancer cells (55, 56) but also through ROS-mediated inactivation of many glycolytic enzymes, notably PKM2 that promotes flux into the oxidative arm of the PPP as well as serine biosynthesis (57). Many studies have shown increased glucose dependency following DDR (58), and these data demonstrate the challenge posed to cancer cells under genotoxic stress. They need to contain ROS levels, enhance nucleotide synthesis to repair DNA damage, and inhibit cell-cycle progression until resolution. To this end, SIRT4 activation upon DDR has been shown to inhibit glutamine consumption in HepG2, HeLa, HEK293T cells, and in lung tissue (58). The reduced glutamine consumption or, more specifically, reduced anaplerosis of glutamine into the TCA cycle, confers improved cell survival compared to SIRT4 knockout cells that cannot activate this response (58). While this study suggests that DDR undoubtedly causes changes in metabolism, conflict in the literature indicates that the nature of these changes is context- and cell type-dependent. For example, the concept that cells undergoing genotoxic stress need to synthesize more nucleotides to repair the DNA damage contradicts the reduced glutamine consumption. As mentioned previously, the amide group of glutamine is essential for purine and pyrimidine de novo synthesis. Other interesting points to note are that in non-cancerous tissues/cells, there are increases in fatty acid oxidation (FAO) and oxidative phosphorylation (OXPHOS) in response to acute or chronic genotoxic stress (59). Increased FAO/OXPHOS is driven by AMP kinase (AMPK) activation resulting from the depletion of ATP. In the context of that study, cells are depleted of ATP because of the utilization of NAD+ by the DNA repair enzyme PARP-1. PARP-1 generates PAR chains from NAD+ and PARylates DNA to induce repair. This leads to depletion of NAD+, reduction of ATP synthesis, and activation of AMPK (59). This study further supports that DNA damage impacts cell metabolism. However, given the current literature, it is still unclear how a cell’s nutrient availability can dictate the nature of its metabolic rewiring.
Metabolic Changes Driven by DDR Gene Mutations
The p53 protein encoded by the TP53 gene is another key player in DDR through its role in DNA repair and cell-cycle regulation (60–62). While p53 levels are kept low under normal conditions, upon DNA damage or other cellular stress, p53 is activated by phosphorylation events that prevent its degradation (60). Wild-type p53 activation regulates cell-cycle entry by transcriptionally repressing cyclin B and CDC25B, and it also plays an important role in DNA-repair. More specifically, it determines whether or not to repair the DNA or trigger senescence or apoptosis. p53 also plays many modulatory roles in the metabolic rewiring of cells. Among these, is its regulation of glycolysis through transcriptional activation of the TP53-induced glycolysis and apoptosis regulator (TIGAR) protein (63). TIGAR inhibits glycolysis by dephosphorylating fructose-2,6-biphosphate (F2,6B) leading to degradation of F2,6P and inhibition of phosphofructo kinase 1, an enzyme integral to glycolysis. p53 has also been shown to inhibit other enzymes of glycolysis as well as directly repress the expression of glucose transporters, GLUT1 and GLUT4 (61, 64). One might surmise that this impediment in glycolysis potentially leads to a surplus in glycolytic intermediates, leading to redirection toward the PPP. However, p53 has also been shown to inhibit the flux of glucose to the PPP by directly binding to G6PD and preventing its active dimer formation (61).
In addition, p53 has been shown to transcriptionally regulate proteins involved in the electron transport chain and mitochondrial stability, therefore, providing a mechanism to regulate OXPHOS (61, 65). Together, these data suggest that the role of p53 in metabolic regulation is context and tissue dependent. Importantly, TP53 is one of the most mutated genes in cancer and many gain-of-function mutations have been described, leading to different changes in p53 depending on the mutation and the context. For example, mutant p533KR cannot induce cell-cycle-arrest, senescence, or apoptosis, but it can still regulate metabolic target genes resulting in decreased ROS levels and reduced glycolytic flux (61, 66). Therefore, mutations in DDR or DNA-repair genes may result from the application of selective pressures established by the metabolic niches of cancer cells.
Other genes involved in DNA-repair are also commonly mutated in cancer (13). Some of the most well-known DNA-repair genes associated with cancer are BRCA1 and BRCA2 but mutations in other genes such as ATM, ATR, and CDK12 [for a more extensive list, see Ref. (13)] also lead to DNA-repair deficient cancers (13, 67–70). Some cancers with a similar phenotype to DNA-repair mutant cancer do not harbor any mutations in those pathways. Together, these DNA-repair deficient cancers have been described as having a “BRCAness” phenotype (13, 71). Some metabolic changes have been described in the context of BRCA1 or BRCA2 mutations (50, 72–74); however, it is still unclear whether these are due to their DNA-repair function or some alternative role. Since connections have been drawn between metabolism and DNA-repair, it seems likely that the “BRCAness” phenotype and BRCA1/2 mutations would be associated with changes in metabolism. Again, as with most other mutations found in cancer, the metabolic changes associated with these DNA repair defects might reflect the context and tissue of origin.
Conclusion
Historically, cancer has been thought of as a genetic disease driven by the accumulation of multiple mutation “hits” (75–77). However, in recent years, this paradigm has begun to shift, and cancer is often regarded as a “metabolic disease” (78, 79), which is influenced by complex interactions between the tumor and its microenvironment. Therefore, when examining data suggesting that mutations lead to the rewiring of cancer metabolism, in fact, it may be more pertinent to question whether these mutations arise in the first place from selective pressures applied by extrinsic metabolic factors in the tumor microenvironment. In a noteworthy example, while KRAS-mutant cancers exhibit resistance to low-glucose growth conditions, glucose deprivation has been shown to drive the accumulation of novel KRAS mutations in KRAS wild-type cancers (80). This indicates that extrinsic factors, such as the availability of essential nutrients, directly influence the fate of resulting genetic alterations that confer growth and survival advantages of cancer cells in their given microenvironment.
With regards to mutations in DNA-repair genes, it is possible that the highly dynamic and fluctuating conditions encountered in the tumor microenvironment provide an evolutionary pressure to select for cancer cells that have heightened genomic instability and are more proficient at adapting to these changes as a result. Consistently, it has been shown that tumors with increased heterogeneity are less responsive to therapy and behave more aggressively than tumors arising from single clonal populations (81–83). As we have discussed, a multitude of extrinsic metabolic factors affects DNA repair and, subsequently, genomic stability (Figures 1 and 2). Therefore, when considering future strategies to refine existing or develop novel cancer therapies, it will be essential at all stages of research and drug design to take account of the dynamic interplay between microenvironment and metabolic factors that are proving to influence treatment efficacy so substantially.
Author Contributions
MOT and GP wrote the manuscript. MOT and NP designed the figure. NP drew the figure and contributed to the revision of the manuscript, and bibliography.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
MOT and NP are supported by Cancer Research UK Ph.D. studentships. Work in the GP lab is supported by the Institute of Cancer Research and a Cancer Research UK Grand Challenge award (C59824/A25044).
References
1. Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol (1927) 8(6):519–30. doi:10.1085/jgp.8.6.519
2. Warburg O. On the origin of cancer cells. Science (1956) 123(3191):309–14. doi:10.1126/science.123.3191.309
3. Wellen KE, Thompson CB. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol Cell (2010) 40(2):323–32. doi:10.1016/j.molcel.2010.10.004
4. Anastasiou D. Tumour microenvironment factors shaping the cancer metabolism landscape. Br J Cancer (2017) 116(3):277–86. doi:10.1038/bjc.2016.412
5. Muir A, Danai LV, Gui DY, Waingarten CY, Lewis CA, Vander Heiden MG. Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. Elife (2017) 6:e27713. doi:10.7554/eLife.27713
6. Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer (2011) 11(2):85–95. doi:10.1038/nrc2981
7. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab (2016) 23(1):27–47. doi:10.1016/j.cmet.2015.12.006
8. Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol (2017) 14(2):113. doi:10.1038/nrclinonc.2017.1
9. DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv (2016) 2(5):e1600200. doi:10.1126/sciadv.1600200
10. Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature (2013) 501(7467):338–45. doi:10.1038/nature12625
11. Chae YK, Anker JF, Carneiro BA, Chandra S, Kaplan J, Kalyan A, et al. Genomic landscape of DNA repair genes in cancer. Oncotarget (2016) 7(17):23312–21. doi:10.18632/oncotarget.8196
12. Gavande NS, VanderVere-Carozza PS, Hinshaw HD, Jalal SI, Sears CR, Pawelczak KS, et al. DNA repair targeted therapy: the past or future of cancer treatment? Pharmacol Ther (2016) 160:65–83. doi:10.1016/j.pharmthera.2016.02.003
13. Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer (2016) 16(2):110–20. doi:10.1038/nrc.2015.21
14. Attwood JT, Yung RL, Richardson BC. DNA methylation and the regulation of gene transcription. Cell Mol Life Sci (2002) 59(2):241–57. doi:10.1007/s00018-002-8420-z
15. Miller JL, Grant PA. The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell Biochem (2013) 61:289–317. doi:10.1007/978-94-007-4525-4_13
16. Jeggo PA, Downs JA. Roles of chromatin remodellers in DNA double strand break repair. Exp Cell Res (2014) 329(1):69–77. doi:10.1016/j.yexcr.2014.09.023
17. Sinha M, Peterson CL. Chromatin dynamics during repair of chromosomal DNA double-strand breaks. Epigenomics (2009) 1(2):371–85. doi:10.2217/epi.09.22
18. Niculescu MD, Zeisel SH. Diet, methyl donors and DNA methylation: interactions between dietary folate, methionine and choline. J Nutr (2002) 132(8 Suppl):2333S–5S.
19. Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab (2015) 21(6):805–21. doi:10.1016/j.cmet.2015.05.014
20. Reid MA, Dai Z, Locasale JW. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat Cell Biol (2017) 19(11):1298–306. doi:10.1038/ncb3629
21. Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol (2014) 15(8):536–50. doi:10.1038/nrm3841
22. Sivanand S, Rhoades S, Jiang Q, Lee JV, Benci J, Zhang J, et al. Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Mol Cell (2017) 67(2):252–65.e256. doi:10.1016/j.molcel.2017.06.008
23. Zaidi N, Swinnen JV, Smans K. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res (2012) 72(15):3709–14. doi:10.1158/0008-5472.CAN-11-4112
24. Kamphorst JJ, Chung MK, Fan J, Rabinowitz JD. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab (2014) 2:23. doi:10.1186/2049-3002-2-23
25. Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell (2015) 27(1):57–71. doi:10.1016/j.ccell.2014.12.002
26. Mews P, Donahue G, Drake AM, Luczak V, Abel T, Berger SL. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature (2017) 546(7658):381–6. doi:10.1038/nature22405
27. Mehrmohamadi M, Mentch LK, Clark AG, Locasale JW. Integrative modelling of tumour DNA methylation quantifies the contribution of metabolism. Nat Commun (2016) 7:13666. doi:10.1038/ncomms13666
28. Jiang Y, Qian X, Shen J, Wang Y, Li X, Liu R, et al. Local generation of fumarate promotes DNA repair through inhibition of histone H3 demethylation. Nat Cell Biol (2015) 17(9):1158–68. doi:10.1038/ncb3209
29. Sciacovelli M, Goncalves E, Johnson TI, Zecchini VR, da Costa AS, Gaude E, et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature (2016) 537(7621):544–7. doi:10.1038/nature19353
30. Russo G, Landi R, Pezone A, Morano A, Zuchegna C, Romano A, et al. DNA damage and repair modify DNA methylation and chromatin domain of the targeted locus: mechanism of allele methylation polymorphism. Sci Rep (2016) 6:33222. doi:10.1038/srep33222
31. Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci (2014) 39(8):347–54. doi:10.1016/j.tibs.2014.06.005
32. Rao X, Duan X, Mao W, Li X, Li Z, Li Q, et al. O-GlcNAcylation of G6PD promotes the pentose phosphate pathway and tumor growth. Nat Commun (2015) 6:8468. doi:10.1038/ncomms9468
33. Rabinovich S, Adler L, Yizhak K, Sarver A, Silberman A, Agron S, et al. Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature (2015) 527(7578):379–83. doi:10.1038/nature15529
34. Kim J, Hu Z, Cai L, Li K, Choi E, Faubert B, et al. CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nature (2017) 546(7656):168–72. doi:10.1038/nature22359
35. Jiang P, Du W, Wu M. Regulation of the pentose phosphate pathway in cancer. Protein Cell (2014) 5(8):592–602. doi:10.1007/s13238-014-0082-8
36. Kowalik MA, Columbano A, Perra A. Emerging role of the pentose phosphate pathway in hepatocellular carcinoma. Front Oncol (2017) 7:87. doi:10.3389/fonc.2017.00087
37. Cory JG, Cory AH. Critical roles of glutamine as nitrogen donors in purine and pyrimidine nucleotide synthesis: asparaginase treatment in childhood acute lymphoblastic leukemia. In Vivo (2006) 20(5):587–9.
38. Lane AN, Fan TW. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res (2015) 43(4):2466–85. doi:10.1093/nar/gkv047
39. Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol (2015) 17(12):1556–68. doi:10.1038/ncb3272
40. Lindahl T. Instability and decay of the primary structure of DNA. Nature (1993) 362(6422):709–15. doi:10.1038/362709a0
41. Alleman RJ, Katunga LA, Nelson MA, Brown DA, Anderson EJ. The “Goldilocks Zone” from a redox perspective-adaptive vs. deleterious responses to oxidative stress in striated muscle. Front Physiol (2014) 5:358. doi:10.3389/fphys.2014.00358
42. Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol (2014) 24(10):R453–62. doi:10.1016/j.cub.2014.03.034
43. Panieri E, Santoro MM. ROS homeostasis and metabolism: a dangerous liason in cancer cells. Cell Death Dis (2016) 7(6):e2253. doi:10.1038/cddis.2016.105
44. Shanware NP, Mullen AR, DeBerardinis RJ, Abraham RT. Glutamine: pleiotropic roles in tumor growth and stress resistance. J Mol Med (Berl) (2011) 89(3):229–36. doi:10.1007/s00109-011-0731-9
45. Lu SC. Regulation of glutathione synthesis. Mol Aspects Med (2009) 30(1–2):42–59. doi:10.1016/j.mam.2008.05.005
46. Hultberg B, Hultberg M. High glutathione turnover in human cell lines revealed by acivicin inhibition of gamma-glutamyltranspeptidase and the effects of thiol-reactive metals during acivicin inhibition. Clin Chim Acta (2004) 349(1–2):45–52. doi:10.1016/j.cccn.2004.05.024
47. Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat Rev Cancer (2016) 16(10):650–62. doi:10.1038/nrc.2016.81
48. Gu Y, Albuquerque CP, Braas D, Zhang W, Villa GR, Bi J, et al. mTORC2 regulates amino acid metabolism in cancer by phosphorylation of the cystine-glutamate antiporter xCT. Mol Cell (2017) 67(1):128–38.e127. doi:10.1016/j.molcel.2017.05.030
49. DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet (2015) 47(12):1475–81. doi:10.1038/ng.3421
50. Gorrini C, Baniasadi PS, Harris IS, Silvester J, Inoue S, Snow B, et al. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J Exp Med (2013) 210(8):1529–44. doi:10.1084/jem.20121337
51. Marechal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol (2013) 5(9):a012716. doi:10.1101/cshperspect.a012716
52. Mirzayans R, Severin D, Murray D. Relationship between DNA double-strand break rejoining and cell survival after exposure to ionizing radiation in human fibroblast strains with differing ATM/p53 status: implications for evaluation of clinical radiosensitivity. Int J Radiat Oncol Biol Phys (2006) 66(5):1498–505. doi:10.1016/j.ijrobp.2006.08.064
53. Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J (2011) 30(3):546–55. doi:10.1038/emboj.2010.330
54. Aird KM, Worth AJ, Snyder NW, Lee JV, Sivanand S, Liu Q, et al. ATM couples replication stress and metabolic reprogramming during cellular senescence. Cell Rep (2015) 11(6):893–901. doi:10.1016/j.celrep.2015.04.014
55. Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev (2009) 23(5):537–48. doi:10.1101/gad.1756509
56. Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer (2011) 11(5):325–37. doi:10.1038/nrc3038
57. Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science (2011) 334(6060):1278–83. doi:10.1126/science.1211485
58. Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell (2013) 23(4):450–63. doi:10.1016/j.ccr.2013.02.024
59. Brace LE, Vose SC, Stanya K, Gathungu RM, Marur VR, Longchamp A, et al. Increased oxidative phosphorylation in response to acute and chronic DNA damage. NPJ Aging Mech Dis (2016) 2:16022. doi:10.1038/npjamd.2016.22
60. Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene (1999) 18(53):7644–55. doi:10.1038/sj.onc.1203015
61. Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. Metabolic regulation by p53 family members. Cell Metab (2013) 18(5):617–33. doi:10.1016/j.cmet.2013.06.019
62. Broustas CG, Lieberman HB. DNA damage response genes and the development of cancer metastasis. Radiat Res (2014) 181(2):111–30. doi:10.1667/RR13515.1
63. Li H, Jogl G. Structural and biochemical studies of TIGAR (TP53-induced glycolysis and apoptosis regulator). J Biol Chem (2009) 284(3):1748–54. doi:10.1074/jbc.M807821200
64. Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res (2004) 64(7):2627–33. doi:10.1158/0008-5472.CAN-03-0846
65. Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science (2006) 312(5780):1650–3. doi:10.1126/science.1126863
66. Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell (2012) 149(6):1269–83. doi:10.1016/j.cell.2012.04.026
67. Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst (2003) 95(19):1482–5. doi:10.1093/jnci/djg050
68. Thull DL, Vogel VG. Recognition and management of hereditary breast cancer syndromes. Oncologist (2004) 9(1):13–24. doi:10.1634/theoncologist.9-1-13
69. Atchley DP, Albarracin CT, Lopez A, Valero V, Amos CI, Gonzalez-Angulo AM, et al. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol (2008) 26(26):4282–8. doi:10.1200/JCO.2008.16.6231
70. O’Donovan PJ, Livingston DM. BRCA1 and BRCA2: breast/ovarian cancer susceptibility gene products and participants in DNA double-strand break repair. Carcinogenesis (2010) 31(6):961–7. doi:10.1093/carcin/bgq069
71. Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer (2004) 4(10):814–9. doi:10.1038/nrc1457
72. Privat M, Radosevic-Robin N, Aubel C, Cayre A, Penault-Llorca F, Marceau G, et al. BRCA1 induces major energetic metabolism reprogramming in breast cancer cells. PLoS One (2014) 9(7):e102438. doi:10.1371/journal.pone.0102438
73. Rytelewski M, Tong JG, Buensuceso A, Leong HS, Maleki Vareki S, Figueredo R, et al. BRCA2 inhibition enhances cisplatin-mediated alterations in tumor cell proliferation, metabolism, and metastasis. Mol Oncol (2014) 8(8):1429–40. doi:10.1016/j.molonc.2014.05.017
74. Cuyas E, Fernandez-Arroyo S, Alarcon T, Lupu R, Joven J, Menendez JA. Germline BRCA1 mutation reprograms breast epithelial cell metabolism towards mitochondrial-dependent biosynthesis: evidence for metformin-based “starvation” strategies in BRCA1 carriers. Oncotarget (2016) 7(33):52974–92. doi:10.18632/oncotarget.9732
75. Nordling CO. A new theory on cancer-inducing mechanism. Br J Cancer (1953) 7(1):68–72. doi:10.1038/bjc.1953.8
76. Ashley DJ. The two “hit” and multiple “hit” theories of carcinogenesis. Br J Cancer (1969) 23(2):313–28. doi:10.1038/bjc.1969.41
77. Knudson AG Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A (1971) 68(4):820–3. doi:10.1073/pnas.68.4.820
78. Coller HA. Is cancer a metabolic disease? Am J Pathol (2014) 184(1):4–17. doi:10.1016/j.ajpath.2013.07.035
79. Seyfried TN, Flores RE, Poff AM, D’Agostino DP. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis (2014) 35(3):515–27. doi:10.1093/carcin/bgt480
80. Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science (2009) 325(5947):1555–9. doi:10.1126/science.1174229
81. Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol (1999) 9(12):M57–60. doi:10.1016/S0962-8924(99)01661-X
82. Jogi A, Vaapil M, Johansson M, Pahlman S. Cancer cell differentiation heterogeneity and aggressive behavior in solid tumors. Ups J Med Sci (2012) 117(2):217–24. doi:10.3109/03009734.2012.659294
Keywords: metabolism, DNA repair, DNA damage, cancer, reactive oxygen species
Citation: Turgeon MO, Perry NJ and Poulogiannis G (2018) DNA Damage, Repair, and Cancer Metabolism. Front. Oncol. 8:15. doi: 10.3389/fonc.2018.00015
Received: 15 December 2017; Accepted: 17 January 2018;
Published: 05 February 2018
Edited by:
Leonardo Freire-de-Lima, Universidade Federal do Rio de Janeiro, BrazilReviewed by:
Daniele Vergara, University of Salento, ItalyBernd Groner, Georg Speyer Haus, Germany
Vandna Kukshal, Washington University in St. Louis, United States
Copyright: © 2018 Turgeon, Perry and Poulogiannis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George Poulogiannis, Z2VvcmdlLnBvdWxvZ2lhbm5pc0BpY3IuYWMudWs=
 Marc-Olivier Turgeon
Marc-Olivier Turgeon Nicholas J. S. Perry
Nicholas J. S. Perry George Poulogiannis
George Poulogiannis