- 1Section of Pathological Anatomy, School of Medicine, Polytechnic University of the Marche Region, United Hospitals, Ancona, Italy
- 2Department of Pathology and Laboratory Medicine, Indiana University School of Medicine, Indianapolis, IN, United States
- 3Department of Pathology and Surgery, Faculty of Medicine, Universidad de Córdoba, Córdoba, Spain
- 4Oncology Unit, Macerata Hospital, Macerata, Italy
Long non-coding RNAs (lncRNAs) are a class of RNA with transcripts longer than 200 nucleotides that lack functional open reading frames. They play various roles in human carcinoma, such as dysregulating gene expression in prostate cancer (PCa), which results in cancer initiation, development, and progression. The non-coding RNA SChLAP1 (second chromosome locus associated with prostate-1) is highly expressed in approximately 25% of PCas with higher prevalence in metastatic compared to localized PCa. Its expression is detectable non-invasively in PCa patient urine samples. Experimental data suggest that targeting SChLAP1 may represent a novel therapeutic application in PCa. This contribution focuses on the role of lncRNAs SChLAP1 expression in PCa diagnosis and prognosis.
Introduction
Prostate cancer (PCa) is the most commonly diagnosed cancer and the second major cause of cancer death in man (1). It is characterized by a wide and heterogeneous spectrum of clinical behaviors, ranging from indolent to aggressive forms. The clinical and morphological methods and features currently used in the routine show a low-predictive value concerning the definition of its level of aggressiveness (2, 3). Predictive and prognostic markers can be determined with clinical and pathological parameters, such as serum prostate-specific antigen (PSA), Gleason score (GS), and TNM stage. Due to PCa heterogeneity, patients with similar TNM stage, GS, and PSA could show opposite outcomes (4).
Additional predictive and prognostic markers are needed to distinguish high-risk from low-risk PCa patients. To this end, genetic and epigenetic investigations have been made to understand the complex genomic landscape of PCa in order to improve its diagnosis and prognosis and to define the potential role of new therapeutical targets (5). Long non-coding RNAs (lncRNAs), a class of RNA with transcripts longer than 200 nucleotides without functional open reading frames, play various roles in human carcinoma (6, 7).
This contribution focuses on the role of lncRNAs second chromosome locus associated with prostate-1 (SChLAP1) expression in PCa diagnosis and prognosis.
Long Non-Coding RNAs
Long non-coding RNAs are a class of RNA with transcripts longer than 200 nucleotides that lack functional open reading frames (6). Based on their locations in the genome relative to protein-coding genes, lncRNAs have been subdivided into intergenic and intragenic. Intragenic lncRNAs can be further subclassified as exonic, intronic, and overlapping lncRNAs (8). Since protein-coding genes have been the focus of most research, the functional role of lncRNAs has been either underestimated or neglected (9).
As shown by Rinn and Chang, “More than 90% of human genome transcripts, including lncRNAs, do not code for proteins” (10). However, accumulating evidence suggests that lncRNAs play a role in the development of various types of cancers, such as PCa, hepatocellular carcinoma, non-small cell lung cancer, leukemia, colon carcinoma, and breast cancer (11–16).
While the mechanism of many lncRNAs remains to be elucidated, it has become clear that lncRNAs contribute to dysregulation of gene expression in PCa, thus resulting in cancer initiation, development, and progression (17).
lncRNAs in Prostate Cancer
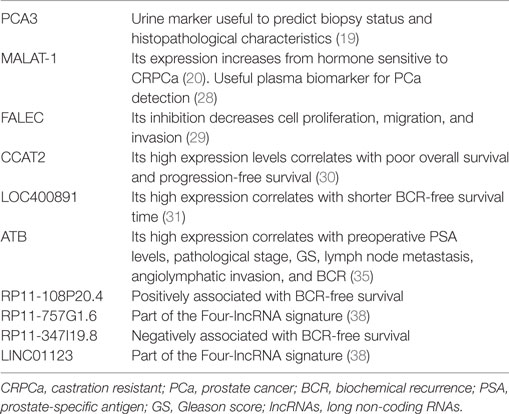
Elucidating the roles of lncRNAs in PCa holds great promise for early detection, prevention, and treatment. A well-known example is prostate cancer antigen3 (PCA3), also known as DD3, initially discovered via expression profiling of prostate sample (18). PCA3 has been extensively studied as a PCa-specific biomarker in body fluids. PCA3 urine RNA assay predicts biopsy status and histopathological characteristics (19). However, it does not predict outcomes such as recurrence and metastasis.
Another lncRNA investigated in PCa is metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1), originally known to be overexpressed in patients at high risk for non-small cell lung cancer metastasis, as its name implies (20, 21). Its expression is found in many other human solid tumors having close correlation with invasiveness and metastasis (22–26). Ren and colleagues found that MALAT-1 is overexpressed in PCa compared to adjacent normal tissue (20). MALAT-1 expression is significantly higher in castration-resistant PCa (CRPCa) than in primary prostate tumor. Its expression increases from hormone sensitive to CRPCa. The same group of authors showed that this lncRNA could be a promising therapeutic target in patients with CRPCa. The intratumoral administration of therapeutic MALAT-1 siRNA suppressed CRPCa growth and metastasis in vivo, and prolonged the survival of tumor bearing mice (20).
It has also been shown that urine MALAT-1 is an independent predictor of PCa, more accurate than routine PSA. Its use would prevent one-third of unnecessary biopsies in PSA 4–10 ng/ml cohorts, without missing any high-grade PCa (27). Furthermore, in 192 plasma samples, MALAT-1 achieved high diagnostic accuracy in predicting prostate biopsy outcomes and, therefore, it might also be utilized as a plasma-based biomarker for PCa detection (28).
Recently, Zhao et al. investigated the expression profile of FALEC, another lncRNA, in PCa. Like other lncRNAs, its expression is significantly higher in PCa than adjacent normal parenchyma. Its downregulation inhibits cell proliferation, migration, and invasion (29).
Similar results were obtained with CCAT2, a lncRNA involved in proliferation, migration, and invasion of PCa cells. In particular, it was demonstrated that silencing of CCAT2 was able to inhibit N-cadherin, vimentin expression, and improve the expression level of E-cadherin, thus leading to the stimulation of epithelial-mesenchymal transition. High expression level of CCAT2 correlates with poor overall survival and progression-free survival and could be considered an independent prognostic factor in patients with PCa (30). Another promising lncRNA is LOC400891 which showed high expression in patients with an advanced PCa and a shorter biochemical recurrence-free survival time (31).
An interesting feature of lncRNAs is that many of them are not PCa specific. For example, we can observe overexpression of lncRNA-ATB in gastric cancer (32), hepatocellular carcinoma (33), osteosarcoma (34), and other tumors. Its tissue overexpression is directly proportional with the histological grade, high preoperative PSA level, pathological stage, high GS, lymph node metastasis, angiolymphatic invasion, and biochemical recurrence in PCa patients (35).
lncRNA-Based Signature
Signatures, based on microarray lncRNA expression profiling, have been recently developed and widely used in prediction of a series of tumor characteristics and outcomes in various cancer type (36, 37). However, due to its low expression characteristics, a single lncRNA analysis might be associated with false-positive result. To overcome this problem, Huang and colleagues have developed a risk score based on lncRNA expression profile (38). They found four lncRNAs are significantly associated with BCR-free survival. Among the four lncRNAs, two (RP11-108P20.4 and RP11- 757G1.6) were positively associated with BCR-free survival, while the remaining two (RP11-347I19.8 and LINC01123) were negatively associated with BCR-free survival. They estimated a risk score for each patient and then divided patients into a high-risk group and a low-risk group by using the median risk score as the cutoff point. The four-lncRNA signature has been shown to be a powerful prognostic factor, independent of age, tumor and lymph node status, GS, margin status, and adjuvant postoperative radiotherapy (38) in (Table 1).
Second Chromosome Locus Associated with Prostate-1
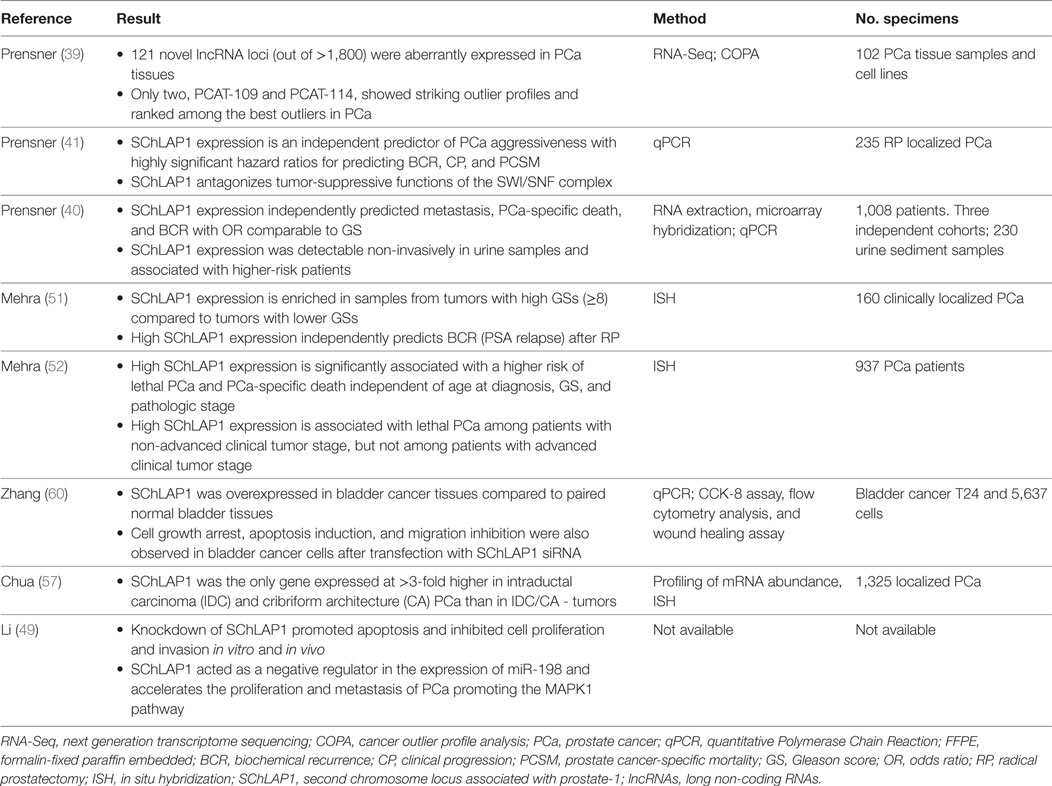
About 1,800 lncRNAs were identified by Presner et al. through the application of RNA sequencing techniques (i.e., transcriptome sequencing) on a consistent number of tissue samples. Of these 1,800 lncRNAs, 121 resulted transcriptionally dysregulated in PCa (39). Such 121 Prostate Cancer-Associated Transcripts represent an unbiased list of potentially functional lncRNAs associated with PCa. By performing a cancer outlier profile analysis to identify intergenic lncRNAs selectively upregulated in a subset of cancers, they found two lncRNAs, PCAT-109 and PCAT-114, both located on Chromosome 2q31.3 in a “gene desert,” a region of the genome that are lacking of protein-coding genes. Both genes showed “outlier profiles and ranked among the best outliers in PCa” (40). In particular, PCAT-114 was found to be overexpressed in prostate cell lines. It was named SChLAP1 after its genomic location (40). Schlap1 gene has a transcript length of 24,484 Kb. The complete gene is composed of 7 exons and 1,675 nucleotides. The primary transcript (isoform 1) is composed of 5 exons with a length of 1,436 nucleotides. As a result of a spicing process, a total of 8 isoforms were found, with isoform 1, isoform 2, and 3 accounting for >90% of transcripts (41). RNA-seq, performed on 27 different tissue samples from 95 human individuals, showed that SChLAP1 expression was highly specific for prostate tissue, being present at minor levels in bladder, kidney, and testis samples (42).
SChLAP1’s Working Mechanisms: Interaction with SWI/SNF Complex and miR-198
In vitro and in vivo gain-of-function and loss-of-function experiments have shown that SChLAP1 plays a crucial role in cancer cell invasiveness and metastasis, antagonizing the activity of the SWI/SNF chromatin-modifying complex, a multiprotein system able to move nucleosomes at gene promoters. In particular, such experiments showed that the inactivation of SWI/SNF complex promoted cancer progression and that multiple SWI/SNF components were somatically inactivated in cancer (41, 43). As shown by Prensner, even though “other lncRNAs, such as HOTAIR and HOTTIP, are known to assist epigenetic complexes such as PRC2 and MLL by facilitating their genomic binding and enhancing their functions, SChLAP1 is the first lncRNA that impairs a major epigenetic complex with well-documented tumor suppressor function” (41, 44).
In vivo, SChLAP1 has been shown to be implicated in tumor cell proliferation and metastasization, as evidenced by both the reduction of tumor growth kinetics and the decreased number and dimensions of metastatic sites as a consequence of the intracardiac injection of 22Rv1 cells with SChLAP1 knockdown in CB-17 SCID mice (41).
Recent studies have shown interaction between SChLAP1 and miR-198. MiR-198 is downregulated in many cancers, such as gastric cancer, lung cancer, and hepatocellular carcinoma (45–47). miR-198 suppress the proliferation and invasion of colorectal carcinoma (48). miR-198 might exert its anticancer effect through inhibition of MAPKs signaling pathway (49). In PCa tissue, a low expression of miR-198 was found. As shown by Li et al., “knockdown of SChLAP1 significantly increased the expression of miR-198 and SChLAP1 overexpression markedly decreased it. Thus, SChLAP1 acted as a negative regulator in the expression of miR-198” and subsequently modulated the MAPK1 signaling pathway in PCa (49) (Figure 1). Transfecting PCa cells with a designed-specific siRNA to knockdown SChLAP1 expression, investigators have obtained, as expected, a significantly reduction in cell proliferation together with an increase in apoptosis-associated proteins. Furthermore, SChLAP1 knockdown determined a decrease of MMP-9, MMP-14, and VEGF expressions both in vitro and in vivo, confirming its involvement in cancer invasiveness and metastasis (49). All such findings show multiple interactions between SChLAP1 and factors involved in oncogenesis and cancer progression and explain the mechanism through which SChLAP1 promotes migration and invasion of PCa (50). Understanding this molecular pathway is essential for exploring new potential strategies for early diagnosis and therapy.
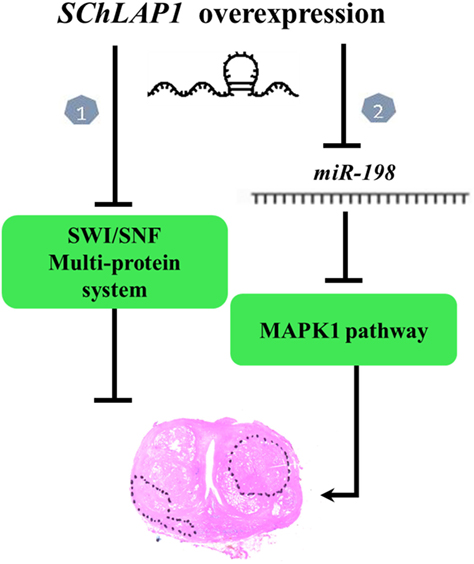
Figure 1. Working model of second chromosome locus associated with prostate-1 (SChLAP1) in prostate cancer (PCa). There are two pathways. In Pathway 1, loss of SWI/SNF functionality promotes cancer progression (41). In Pathway 2, miR-198 might exert its anticancer effect through inhibition of MAPKs signaling pathway. SChLAP1 acts as a negative regulator in the expression of miR-198 and subsequently modulates the MAPK1 signaling pathway in PCa (49).
SChLAP1: Predictor of Aggressive PCa
Second chromosome locus associated with prostate-1 is highly expressed in approximately 25% of PCa, expression being higher in metastatic compared to localized prostate cancers. It was associated with ETS gene fusions (41). Multivariate and univariate regression analyses have demonstrated that SChLAP1 expression is an independent predictor of PCa aggressiveness with highly significant hazard ratios for predicting BCR, clinical progression to systemic disease, and PCa-specific mortality, compared to other clinical factors such as advanced clinical stage and the GS (41). Moreover, SChLAP1 expression was significantly associated with extracapsular extension, seminal vesicle invasion, and positive surgical margin status (40).
Validation in three-independent cohorts has confirmed the prognostic value of SChLAP1 for metastasis. On multivariate modeling, SChLAP1 expression independently predicted metastasis within 10 years, death within 10 years, and biochemical recurrence within 5 years with odds ratios, comparable to GS. Among all known genes, the lncRNA SChLAP1 ranked first for elevated expression in patients with metastatic progression by receiver-operator-curve area-under-the-curve analyses and was the only prostate-specific gene, ideal for development as a non-invasive biomarker (40).
Similar results have been obtained by Mehra and colleagues using a novel RNA in situ hybridization (ISH) assay for detection of SChLAP1 in formalin-fixed, paraffin-embedded tissue (51). They found that high SChLAP1 expression independently predicts biochemical PCa recurrence after radical prostatectomy in patients with clinically localized PCa and that it is associated with the development of lethal PCa.
Interestingly, high SChLAP1 expression is associated with lethal PCa among patients with non-advanced clinical tumor stage. Similarly, high SChLAP1 expression is associated with lethal PCa among patients with low grade tumors (GS ≤7). However, using SChLAP1 as prognostic test, investigators have obtained a sensitivity around 24% and a specificity of 94% in the group of non-advanced clinical tumor stage and in the group with a 6–7 GS (52). Considering the low sensitivity of the test in the identification of an aggressive disease in contrast with low-risk morphological features, the evaluation of SChLAP1expression alone does not seem to improve treatment decision. In conjunction with other prognostic tools, SChLAP1 has been shown to improve upon established clinical algorithms for the risk stratification of PCa patients, specifically the CAPRA-S score (53, 54), i.e., one of the best clinic-pathological models to date. SChLAP1 further improves prediction upon both the Decipher test 50 and the CCP gene signature (55, 56).
Recently, Chua et al. have investigated SChLAP1 expression in subsets of PCa characterized by cribriform architecture (CA) and intraductal carcinoma (IDC), features both associated with increased risks of biochemical relapse and metastasis. Besides the histological presentation, IDC/CA+ cancers harbor an increased percentage of genome aberration (PGA). This is in agreement with the observation that tumors with IDC or genomic instability have a greater metastatic potential. Using mRNA abundance analyses and assessing >25,000 genes, they found that SChLAP1 was surprisingly the only gene with more than threefold higher expression in IDC/CA+ compared to IDC/CA− cancers (57).
The association of SChLAP1 expression within IDC/CA+ tumors has also been further demonstrated by SChLAP1 RNA-ISH in prostatectomy TMA-cores. The SChLAP1+, IDC/CA+ subgroup has shown a significant increase of biochemical relapse, independent of PGA. Combining histology features of cribriform architecture and intraductal carcinoma with genomic instability or SChLAP1 expression can stratify patients for recurrence more accurately than any parameter alone. Interestingly, SChLAP1 RNA-ISH diffuse expression has been observed in the cribriform architecture and intraductal carcinoma and in the adjacent invasive adenocarcinoma. This further supports a field defect and a common clonal ancestor to both histopathologies (58).
SChLAP1 in Urine Sediments
Second chromosome locus associated with prostate-1 expression is detectable non-invasively in PCa patient in urine samples. Its expression is both more frequent and more highly elevated in GS 7 compared to GS 6 patients even if it is less sensitive than PCA3 and TMPRSS-ERG gene fusion (40). “SChLAP1 expression may complement existing urine diagnostic assays, including PCA3 and TMPRSS2-ERG, and that clinical application of a SChLAP1 urine test would be most effective in conjunction with these, and potentially other, urine assays” (59).
SChLAP1 As a Potential Drug Target
RNA interference (RNAi) technology using short interfering RNA (si-RNA) has shown great potential in the treatment of cancers through silencing of specific genes. In vitro and in vivo experiments have demonstrated that SChLAP1-knockdown promoted apoptosis and inhibited cell proliferation and invasion (41, 49). SChLAP1 is also overexpressed in bladder cancer compared to paired normal bladder tissues. Cell transfected with SChLAP1 siRNA showed growth arrest, apoptosis, and migration inhibition, suggesting oncogenic roles in bladder cancer and a potential therapeutic target (Table 2) (60). Such results might be the starting point to investigate the therapeutical potential of antagonizing SChLAP1 oncogenic functions (60).
lncRNA in Diagnosis, Prognosis, and Treatment: Pros and Cons
There are Pros and Cons with the use of lncRNA in the diagnosis and treatment of PCa patients. Indeed, lncRNA may represent a useful biomarker that can give to clinicians fundamental information on tumor biological behavior and aggressiveness, leading to the possibility of designing personalized and tailored strategies for a single PCa patient. This may also allow an optimization of patients’ outcome and to avoid useful costs and consequences of not effective therapies for PCa patients.
As far as the Cons, tumor aggressiveness is the result of a complex process that involves lncRNA and a variety of driver genes, leading to the necessity for uropathologists to test and validate not a single driver gene, but a panel of genes with direct consequences on the relative costs of these procedures.
Conclusion
In conclusion, research on lncRNAs in PCa is at its onset. However, as shown in this review:
• The first set of data has revealed central roles with clinical significance for lncRNAs in different stages of the disease.
• There is evidence that lncRNAs, including SChLAP1, are critical in PCa development and progression.
• Concerning future perspective, mainly based on experimental data, targeting SChLAP1 may become a novel therapeutic application in PCa.
Author Contributions
RM: conception and design. AC and SG: drafting the manuscript. AD and RMa: acquisition of data. LC and AL-B: critical revision of the manuscript. MS: supervision.
Conflict of Interest Statement
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties. No writing assistance was utilized in the production of this manuscript.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin (2016) 66:7–30. doi:10.3322/caac.21332
2. Klotz L. Prostate cancer overdiagnosis and overtreatment. Curr Opin Endocrinol Diabetes Obes (2013) 20:204–9. doi:10.1097/MED.0b013e328360332a
3. Alcover J, Filella X. Identification of candidates for active surveillance: should we change the current paradigm? Clin Genitourin Cancer (2015) 13:499–504. doi:10.1016/j.clgc.2015.06.001
4. Fraser M, Berlin A, Bristow RG, van der Kwast T. Genomic, pathological, and clinical heterogeneity as drivers of personalized medicine in prostate cancer. Urol Oncol (2015) 33:85–94. doi:10.1016/j.urolonc.2013.10.020
5. Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM. Beyond PSA: the next generation of prostate cancer biomarkers. Sci Transl Med (2012) 4:127rv3. doi:10.1126/scitranslmed.3003180
6. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature (2012) 482:339–46. doi:10.1038/nature10887
7. Diederichs S, Bartsch L, Berkmann JC, Fröse K, Heitmann J, Hoppe C, et al. The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol Med (2016) 8:442–57. doi:10.15252/emmm.201506055
8. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res (2012) 22:1775–89. doi:10.1101/gr.132159.111
9. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer (2011) 10:38. doi:10.1186/1476-4598-10-38
10. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem (2012) 81:145–66. doi:10.1146/annurev-biochem-051410-092902
11. Smolle MA, Bauernhofer T, Pummer K, Calin GA, Pichler M. Current insights into Long Non-Coding RNAs (lncRNAs) in prostate cancer. Int J Mol Sci (2017) 18:E473. doi:10.3390/ijms18020473
12. Heery R, Finn SP, Cuffe S, Gray SG. Long non-coding RNAs: key regulators of epithelial-mesenchymal transition, tumour drug resistance and cancer stem cells. Cancers (2017) 9:E38. doi:10.3390/cancers9040038
13. Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene (2007) 26:851–8. doi:10.1038/sj.onc.1209846
14. Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell (2007) 12:215–29. doi:10.1016/j.ccr.2007.07.027
15. Pibouin L, Villaudy J, Ferbus D, Muleris M, Prospéri MT, Remvikos Y, et al. Cloning of the mRNA of overexpression in colon carcinoma-1: a sequence overexpressed in a subset of colon carcinomas. Cancer Genet Cytogenet (2002) 133:55–60. doi:10.1016/S0165-4608(01)00634-3
16. Zhang C, Wang X, Li X, Zhao N, Wang Y, Han X, et al. The landscape of DNA methylation-mediated regulation of long non-coding RNAs in breast cancer. Oncotarget (2017) 8(31):51134–50. doi:10.18632/oncotarget.17705
17. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov (2011) 1:391–407. doi:10.1158/2159-8290.CD-11-0209
18. Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res (1999) 59:5975–9.
19. Nakanishi H, Groskopf J, Fritsche HA, Bhadkamkar V, Blase A, Kumar SV, et al. PCA3 molecular urine assay correlates with prostate cancer tumor volume: implication in selecting candidates for active surveillance. J Urol (2008) 179:1804–9. doi:10.1016/j.juro.2008.01.013
20. Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol (2013) 190:2278–87. doi:10.1016/j.juro.2013.07.001
21. Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage nonsmall cell lung cancer. Oncogene (2003) 22:8031. doi:10.1038/sj.onc.1206928
22. Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol (2012) 29:1810. doi:10.1007/s12032-011-0004-z
23. Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett (2010) 584:4575. doi:10.1016/j.febslet.2010.10.008
24. Yamada K, Kano J, Tsunoda H, Yoshikawa H, Okubo C, Ishiyama T, et al. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci (2006) 97:106. doi:10.1111/j.1349-7006.2006.00147.x
25. Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long noncoding RNA and its important 30 end functional motif in colorectal cancer metastasis. Int J Oncol (2011) 39:169. doi:10.3892/ijo.2011.1007
26. Chen D, Liu L, Wang K, Yu H, Wang Y, Liu J, et al. The role of MALAT-1 in the invasion and metastasis of gastric cancer. Scand J Gastroenterol (2017) 52:790–6. doi:10.1080/00365521.2017.1280531
27. Wang F, Ren S, Chen R, Lu J, Shi X, Zhu Y, et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget (2014) 5:11091–102. doi:10.18632/oncotarget.2691
28. Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer (2013) 49:2949–59. doi:10.1016/j.ejca.2013.04.026
29. Zhao R, Sun F, Bei X, Wang X, Zhu Y, Jiang C, et al. Upregulation of the long non-coding RNA FALEC promotes proliferation and migration of prostate cancer cell lines and predicts prognosis of PCa patients. Prostate (2017) 77(10):1107–17. doi:10.1002/pros.23367
30. Zheng J, Zhao S, He X, Zheng Z, Bai W, Duan Y, et al. The up-regulation of long non-coding RNA CCAT2 indicates a poor prognosis for prostate cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Biochem Biophys Res Commun (2016) 480(4):508–14. doi:10.1016/j.bbrc.2016.08.120
31. Wang J, Cheng G, Li X, Pan Y, Qin C, Yang H, et al. Overexpression of long non-coding RNA LOC400891 promotes tumor progression and poor prognosis in prostate cancer. Tumour Biol (2016) 37:9603–13. doi:10.1007/s13277-016-4847-y
32. Saito T, Kurashige J, Nambara S, Komatsu H, Hirata H, Ueda M, et al. A Long non-coding RNA activated by transforming growth Factor-β is an independent prognostic marker of gastric cancer. Ann Surg Oncol (2015) 22(Suppl 3):S915–22. doi:10.1245/s10434-015-4554-8
33. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell (2014) 25:666–81. doi:10.1016/j.ccr.2014.03.010
34. Han F, Wang C, Wang Y, Zhang L. Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s. Am J Cancer Res (2017) 7:770–83.
35. Xu S, Yi XM, Tang CP, Ge JP, Zhang ZY, Zhou WQ. Long non-coding RNA ATB promotes growth and epithelial-mesenchymal transition and predicts poor prognosis in human prostate carcinoma. Oncol Rep (2016) 36:10–22. doi:10.3892/or.2016.4791
36. Zhu X, Tian X, Yu C, Shen C, Yan T, Hong J, et al. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol Cancer (2016) 15:60. doi:10.1186/s12943-016-0544-0
37. Sun J, Chen X, Wang Z, Guo M, Shi H, Wang X, et al. A potential prognostic long non-coding RNA signature to predict metastasis-free survival of breast cancer patients. Sci Rep (2015) 5:16553. doi:10.1038/srep16553
38. Huang TB, Dong CP, Zhou GC, Lu SM, Luan Y, Gu X, et al. A potential panel of four-long noncoding RNA signature in prostate cancer predicts biochemical recurrence-free survival and disease-free survival. Int Urol Nephrol (2017) 49:825–35. doi:10.1007/s11255-017-1536-8
39. Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol (2011) 29:742–9. doi:10.1038/nbt.1914
40. Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM, et al. Nomination and validation of the long noncoding RNA SChLAP1 as a risk factor for metastatic prostate cancer progression: a multi-institutional high-throughput analysis. Lancet Oncol (2014) 15:1469–80. doi:10.1016/S1470-2045(14)71113-1
41. Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet (2013) 45:1392–8. doi:10.1038/ng.2771
42. Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics (2014) 13:397–406. doi:10.1074/mcp.M113.035600
43. Roberts CW, Orkin SH. The SWI/SNF complex – chromatin and cancer. Nat Rev Cancer (2004) 4:133–42. doi:10.1038/nrc1273
44. Atala A. Re: The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. J Urol (2014) 192:613. doi:10.1016/j.juro.2014.05.031
45. Huang WT, Wang HL, Yang H, Ren FH, Luo YH, Huang CQ, et al. Lower expressed miR-198 and its potential targets in hepatocellular carcinoma: a clinicopathological and in silico study. Onco Targets Ther (2016) 22:5163–80. doi:10.2147/OTT.S108828
46. Wu S, Zhang G, Li P, Chen S, Zhang F, Li J, et al. miR-198 targets SHMT1 to inhibit cell proliferation and enhance cell apoptosis in lung adenocarcinoma. Tumour Biol (2016) 37:5193–202. doi:10.1007/s13277-015-4369-z
47. Cui Z, Zheng X, Kong D. Decreased miR-198 expression and its prognostic significance in human gastric cancer. World J Surg Oncol (2016) 6:33. doi:10.1186/s12957-016-0784-x
48. Wang M, Wang J, Kong X, Chen H, Wang Y, Qin M, et al. MiR-198 represses tumor growth and metastasis in colorectal cancer by targeting fucosyl transferase 8. Sci Rep (2014) 1:6145. doi:10.1038/srep06145
49. Li Y, Luo H, Xiao N, Duan J, Wang Z, Wang S. Long noncoding RNA SChLAP1 accelerates the proliferation and metastasis of prostate cancer via targeting miR-198 and promoting the MAPK1 pathway. Oncol Res (2017). doi:10.3727/096504017X14944585873631
50. Ma W, Chen X, Ding L, Ma J, Jing W, Lan T, et al. The prognostic value of long noncoding RNAs in prostate cancer: a systematic review and meta-analysis. Oncotarget (2017) 8(34):57755–65. doi:10.18632/oncotarget.17645
51. Mehra R, Shi Y, Udager AM, Prensner JR, Sahu A, Iyer MK, et al. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia (2014) 16:1121–7. doi:10.1016/j.neo.2014.11.006
52. Mehra R, Udager AM, Ahearn TU, Cao X, Feng FY, Loda M, et al. Overexpression of the long non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur Urol (2016) 70:549–52. doi:10.1016/j.eururo.2015.12.003
53. Cooperberg MR, Hilton JF, Carroll PR. The CAPRA-S score: a straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer (2011) 117:5039–46. doi:10.1002/cncr.26169
54. Cooperberg MR, Davicioni E, Crisan A, Jenkins RB, Ghadessi M, Karnes RJ. Combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high-risk prostatectomy cohort. Eur Urol (2015) 67:326–33. doi:10.1016/j.eururo.2014.05.039
55. Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest (2004) 113:913–23. doi:10.1172/JCI20032
56. Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol (2011) 12:245–55. doi:10.1016/S1470-2045(10)70295-3
57. Chua MLK, Lo W, Pintilie M, Murgic J, Lalonde E, Bhandari V, et al. A prostate cancer “nimbosus”: genomic instability and SChLAP1 dysregulation underpin aggression of intraductal and cribriform subpathologies. Eur Urol (2017) 72(5):665–74. doi:10.1016/j.eururo.2017.04.034
58. Spratt DE. Convergence of genomic instability and SChLAP1: weathering the storm of intraductal carcinoma of the prostate. Eur Urol (2017) 72(5):675–6. doi:10.1016/j.eururo.2017.05.015
59. Whitman EJ, Groskopf J, Ali A, Chen Y, Blase A, Furusato B, et al. PCA3 score before radical prostatectomy predicts extracapsular extension and tumor volume. J Urol (2008) 180:1975–8. doi:10.1016/j.juro.2008.07.060
Keywords: second chromosome locus associated with prostate-1, metastatic prostate cancer, long non-coding RNA, lethal prostate cancer, prognostic biomarker, marker of aggressiveness
Citation: Cimadamore A, Gasparrini S, Mazzucchelli R, Doria A, Cheng L, Lopez-Beltran A, Santoni M, Scarpelli M and Montironi R (2017) Long Non-coding RNAs in Prostate Cancer with Emphasis on Second Chromosome Locus Associated with Prostate-1 Expression. Front. Oncol. 7:305. doi: 10.3389/fonc.2017.00305
Received: 06 July 2017; Accepted: 27 November 2017;
Published: 12 December 2017
Edited by:
Caterina Nardella, University of Trento, ItalyReviewed by:
Jun Yan, Nanjing University, ChinaChandi C. Mandal, Central University of Rajasthan, India
Copyright: © 2017 Cimadamore, Gasparrini, Mazzucchelli, Doria, Cheng, Lopez-Beltran, Santoni, Scarpelli and Montironi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rodolfo Montironi, ci5tb250aXJvbmlAdW5pdnBtLml0
†These authors have contributed equally to this work.
 Alessia Cimadamore
Alessia Cimadamore Silvia Gasparrini1†
Silvia Gasparrini1† Antonio Lopez-Beltran
Antonio Lopez-Beltran Rodolfo Montironi
Rodolfo Montironi