- 1Department of Molecular Hematology, Tohoku University Graduate School of Medicine, Sendai, Japan
- 2Department of Molecular Oncology, Institute of Development, Aging and Cancer, Tohoku University, Sendai, Japan
- 3Medical Mega-Bank Organization, Tohoku University, Sendai, Japan
The transcription factor GATA1 regulates the expression of essential erythroid and megakaryocytic differentiation genes through binding to the DNA consensus sequence WGATAR. The GATA1 protein has four functional domains, including two centrally located zinc-finger domains and two transactivation domains at the N- and C-termini. These functional domains play characteristic roles in the elaborate regulation of diversified GATA1 target genes, each of which exhibits a unique expression profile. Three types of GATA1-related hematological malignancies have been reported. One is a structural mutation in the GATA1 gene, resulting in the production of a short form of GATA1 that lacks the N-terminal transactivation domain and is found in Down syndrome-related acute megakaryocytic leukemia. The other two are cis-acting regulatory mutations affecting expression of the Gata1 gene, which have been shown to cause acute erythroblastic leukemia and myelofibrosis in mice. Therefore, imbalanced gene regulation caused by qualitative and quantitative changes in GATA1 is thought to be involved in specific hematological disease pathogenesis. In the present review, we discuss recent advances in understanding the mechanisms of differential transcriptional regulation by GATA1 during erythroid differentiation, with special reference to the binding kinetics of GATA1 at conformation-specific binding sites.
Introduction
GATA1 is an essential transcription factor (TF) in erythroid and megakaryocyte differentiation that regulates a considerable number of target genes involved in the proliferation, differentiation, and survival of hematopoietic progenitors. GATA1 is a member of GATA factor family, which recognizes the GATA-binding (T/A)GATA(A/G) motif (1). Six members are found in vertebrates and are divided into two subfamilies: GATA1/2/3 belong to the hematopoietic subfamily (2), while GATA4/5/6 are referred to as the endodermal GATA factors (3). GATA1 is expressed in lineage-committed progenitors preprogrammed toward erythrocytes, megakaryocytes, eosinophils, and mast cells (4), whereas GATA2 is abundantly expressed in hematopoietic stem cells, early multipotent progenitors, and monocyte-lineage-committed cells (5, 6). GATA1 and GATA2 display partially overlapping expression patterns during erythroid and megakaryocytic differentiation (7, 8).
Expression of the Gata1 gene in hematopoietic stem cells and early progenitor cells is repressed by an epigenetic mechanism that precludes access of GATA2 (9). Along with differentiation, GATA2 primes Gata1 gene expression and results in Gata1 gene self-activation and Gata2 gene repression by GATA1. Consequently, GATA1 and GATA2 expression levels show dynamic features, referred to as “GATA-factor switching,” during erythroid differentiation (10). GATA1 and GATA2 partially share binding sites at target genes, acting both redundantly and competitively to regulate gene expression (11). Therefore, the balance of GATA1 and GATA2 expression contributes to fine-tuning the transcriptional regulation of target genes.
GATA1 has four functional domains, consisting of two transactivation domains that reside in the amino (N)- and carboxyl (C)-termini and two zinc-finger domains that are referred to as N- and C-terminal zinc fingers in the middle of the protein. The former two function redundantly and cooperatively to transcriptionally regulate individual target genes (12), while the latter two are highly conserved in all GATA factors as DNA-binding and interaction domains for regulatory proteins and other TFs. The C-finger domain (CF) is particularly indispensable for DNA binding by GATA1, while the N-finger domain (NF) is insufficient for DNA binding alone but stabilizes DNA binding by the CF (13, 14). The NF is especially important for Friend of GATA1 (FOG1) interaction (15) and GATA1 homodimerization (16–19). Multiple molecules have been identified to interact with the CF, such as LIM domain only 2 (LMO2) (20) and MED1, a component of the mediator complex (21). Thus, a dual zinc-finger structure appears to modulate DNA-binding and protein–protein interactions to form the characteristic GATA1 complex and leads to the diverse target gene expression regulation.
Several hematopoietic disorders are linked to GATA1 dysfunction. Germ-line and somatic GATA1 gene mutations that produce a short form of GATA that lacks the N-terminal transactivation domain are causal in Diamond–Blackfan anemia (22) and preleukemic disease in Down syndrome patients (23), respectively. Substitution mutations in the NF are associated with X-linked hematopoietic diseases and are accompanied by thrombocytopenia, porphyria, and dyserythropoietic anemia (24). Cell-based complementation approaches have determined how the mutations alter the functions of GATA1 (25). Furthermore, genetically manipulated mouse models that phenocopy human diseases have been established and provide insight into the pathogenesis caused by GATA1 dysfunction (26–29). Furthermore, quantitative reduction of GATA1 has been described as causal in acute erythroblastic leukemia and myelofibrosis in mice (30–32).
GATA-binding motifs are found scattered in a variety of genes that are distributed throughout the genome. However, it is largely unknown how GATA1 properly organizes diversified gene expression to generate distinct expression profiles during erythropoiesis. Recent comprehensive analyses of GATA1 occupancy have shown that diversities in the neighboring sequence of consensus GATA-binding motifs can modify the transcriptional output. Multiple motifs of transcription regulation may act in a synergistic manner when they align in order with the GATA-binding motif in a cis-acting element. In this article, we describe diverse DNA binding and transcriptional regulation mediated by GATA1 during erythropoiesis, focusing on the cis-acting element configuration.
Composite Elements with an E-Box and a GATA-Binding Motif
A number of molecules reportedly interact with GATA1 and modulate its function. TFs recruited with GATA1 diversify the transcriptional output based on how the molecules complex with GATA1. Accretion of TFs to the neighboring regions of the GATA-binding motif appears to modify the DNA binding, kinetics, and stoichiometry of GATA1, and subsequently modifies the formation of GATA1-centered transcriptional complexes on the cis-acting elements (Figure 1).
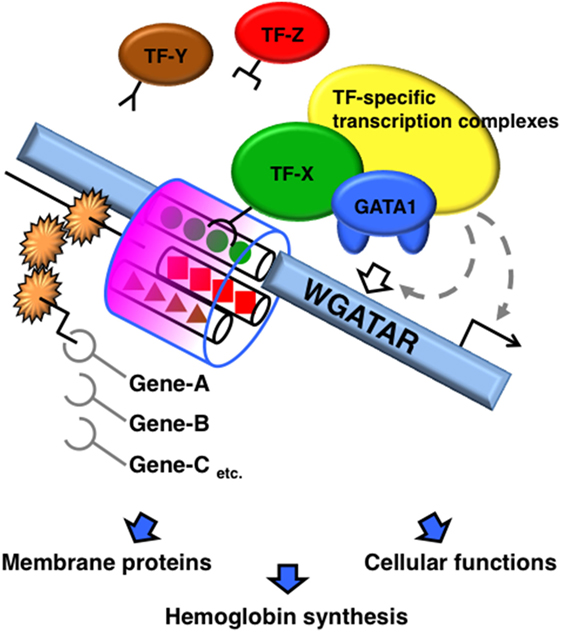
Figure 1. Schematic diagram of GATA1 functional modifications mediated by sequences adjacent to the GATA-binding site. A transcription factor (TF) recruited to the region adjacent to the GATA-binding motifs modifies the DNA binding, kinetics, and stoichiometry of GATA1. Consequently, the GATA1-centered transcriptional complex formation on the cis-acting elements is altered, which alters the transcriptional activity of GATA1.
Transcription factors that interact with GATA1 have specific motifs that are enriched in repetitive regions of the genome. Chromatin immunoprecipitation (ChIP)-sequencing analyses identified GATA1 (20, 33–35) and GATA2 (36) interactions and co-localization with SCL on the E-box–GATA combined motif, thus forming a large protein complex with E2A, LMO2, and LIM domain-binding 1. SCL is a member of the basic helix-loop-helix family of TFs, which recognize DNA-binding sites containing the E-box motif (CANNTG) as a heterodimer with E2A (37). The E-box sequence is enriched 7–12 bp upstream of GATA motifs in erythroid-specific genes. The DNA-binding affinities of GATA1 and SCL to the combined E-box–GATA motif are increased by co-localization with each other, thereby modifying the transcriptional activity of GATA1 (37–39). SCL occupancy is often found at GATA-occupied loci lacking an E-box (40). By contrast, SCL occupancy is abrogated by the lack of GATA1 on the E-box–GATA motif in certain genes, such as genes encoding erythroid-specific 5-aminolevulinate synthase, uroporphyrinogen III synthase, and pyruvate kinase, in which single-nucleotide mutations of the GATA motif have been found in patients with erythroid disorders (41). Thus, the combination of GATA-binding and E-box motifs contributes to specific transcriptional regulation by varying the DNA-binding characteristics and structure of the complex containing GATA1 and SCL, and possibly by varying other components of the complex (Figure 1).
The large complex binding to the E-box–GATA motif usually acts as a transcription activator (40). Interestingly, in the genes activated by GATA2 and repressed by GATA1, SCL is co-localized with GATA2 when the gene is activated, while the SCL occupancy is decreased when GATA1 takes the place of GATA2 (40). One plausible explanation for the difference in SCL occupancy is that differences in undesignated sequences in the composite E-box–GATA motif may influence complex formation, depending on whether GATA1 or GATA2 is present. Binding analyses utilizing naked DNA have indicated that SCL preferentially binds to CAGGTG (33) or CAGATG (42) motifs in essential hematopoiesis genes, whereas CATCTG sequences are enriched in GATA2-occupied loci in lineage-negative hematopoietic progenitors (43). Furthermore, the spacer lengths and sequences between E-box and GATA-binding motif vary at individual loci. We envision delicate differences in sequence alignment among E-box–GATA composite elements conferring changes in complex structure, DNA-binding affinity, and protein–protein interactions, consequently diversifying target gene expression profiles.
CACC-Box Adjacent to GATA-Binding Motif
Genome-wide ChIP-sequencing analyses have revealed that the NCNCNCCCN (extended CACC) motif is frequently found in the region of GATA1 ChIP peaks in erythroid-committed cells (44, 45). The CACC motif is a known consensus sequence recognized by Krüppel-like transcription factors (KLFs). Among the 17 mammalian KLFs, KLF1 (EKLF), KLF2, and KLF13 are involved in erythrocyte maturation and differentiation (46–48). In particular, the quantitative ratio of GATA1 and KLF1 in the nucleus is tuned by their acetylation states, which are mediated by HDACs, to control erythroid-specific gene expression (49–51). Many KLF1 variants have been found in recent years as the causes of different types of red cell disorders (52, 53).
Krüppel-like transcription factors have three conserved C2H2-type zinc fingers at the C-terminus that are involved in DNA binding and directly interact with GATA1 (54). When the GATA and CACC motifs are located close together in the regulatory region, GATA1 and KLF1 improve the DNA-binding affinity of each other, synergistically increasing gene expression in hematopoietic and non-hematopoietic cells (54, 55). Thus, a composite element composed of GATA- and CACC-binding motifs works as a cis-regulatory region distinct from each individual motif’s function (Figure 1). ChIP-sequencing peaks of GATA1 and KLF1 overlap in several gene loci. However, the number of co-occupied loci is lower than expected based on the significant cooperative function of GATA1 and KLF1 in the erythroid differentiation (56, 57). One plausible explanation is that other KLFs share CACC motif binding with KLF1 when adjacent to GATA motif. In addition, TFs belonging to specificity protein (SP) family bind to the CACC motif and have a triple-C2H2-type DNA-binding domain highly conserved with KLFs. SP1, which has relatively ubiquitous expression pattern and plays key roles in critical biological process through regulating metabolic genes (58), is reported to interact and cooperatively work with GATA1 to control gene expression (54, 59). Furthermore, ZBP-89, a Krüppel-type zinc finger TF, has a potential to bind CACC motifs and complex with GATA1 (60). KLF2 regulates embryonic erythropoiesis through redundantly and cooperatively working with KLF1 (47, 61). Therefore, other TFs that share CACC motif binding with KLF1 may influence KLF1 function and consequently influence GATA1 function (Figure 1). Questions regarding how the undesignated nucleotides in the consensus CACC motif contribute to the preferential binding of KLFs and SPs remain to be answered, and GATA1 function may be modified by alternate factors recruited to the CACC motif.
Binding Motifs for Collaborative TFs
In addition to the E-box and CACC motifs, a variety of cis-regulatory sequences found adjacent to the GATA-binding motif have been reported to mediate the transcriptional output of GATA1 during erythroid differentiation. Consensus NFE2-binding (C/T)GCTGA(C/G)TCA(C/T) motifs are found close to the GATA-binding motif in genes encoding β-globin and erythroid-specific membrane protein (62, 63). CP2 was originally identified as a regulator for human α-globin gene expression (64) and regulates erythroid differentiation through the binding at the CNRG-N5-6-CNR(G/C) motif. A CP2-binding motif adjacent to the GATA motif is required in the regulation of erythroid-specific genes, such as mouse Gata1, mouse Klf1, mouse Nfe2, mouse Epor, human/mouse UROS, the human/mouse globin gene clusters, and human CDC6, in vivo (65–72).
We assume that the sequence adjacent to the GATA motif may alter the DNA-binding mode of GATA1, the formation of transcription complexes, and organization of chromatin structures if a TF recognizes and binds the sequence. Given these associations, we propose that sequence alignments of regions neighboring the GATA-binding motifs influence the transcriptional activity of GATA1 (Figure 1).
Composite Elements with Dual GATA1-Binding Motifs
We noticed that one or more GATA-binding motifs are found in scattered regions throughout the genes involved in erythropoiesis. Given that the sequence adjacent to the GATA-binding motif is varied in each region, individual GATA1 is predicted to access cis-regulatory regions differentially to contribute to gene expression and to allow sophisticated gene regulation. cis-Targeting experiments have found that each GATA-binding motif independently and redundantly plays a role in the expression of certain genes in mice (65, 66, 73, 74). However, the regulation of the spatiotemporal control of gene expression is poorly understood.
Focusing on the GATA-binding motif, one significant observation is that two or more GATA-binding motifs are sometimes oriented closely in line, generating composite GATA elements in which GATA-binding motifs are aligned side-by-side in either tandem or palindromic orientation. Considering that GATA1 forms a homodimer through NF and CF interactions (19), composite GATA elements composed of two GATA-binding motifs may have a more significant function than two single GATA-binding motifs. Furthermore, the DNA-binding structure of GATA3 revealed that the NF binds the opposite face of DNA that is bound by the CF and interacts with the C-terminal basic-tail of the CF that is inserted into the minor groove (75). Therefore, when two GATA-binding motifs are aligned side-by-side, the direction of the two GATA-binding motifs seems to be important.
Palindromic Dual GATA-Binding Motifs
When two GATA-binding motifs are aligned in a palindromic orientation, two types of composite GATA elements are generated: either a head-to-head (YTATCW–WGATAR) or tail-to-tail (WGATAR–YTATCW) orientation. The difference in orientation may influence GATA1 binding modes. For simplicity’s sake, we refer to the GATA-binding motifs aligned in head-to-head and tail-to-tail palindromic orientations as Pal-GATA or rPal-GATA motifs, respectively.
GATA1 binds to DNA with the CF while the NF scarcely functions during DNA binding (76). Indeed, the association and dissociation kinetics of GATA1 on a single-GATA motif on naked DNA, as measured in DNA-binding surface plasmon resonance (SPR) studies, do not change regardless of the NF function (77). By contrast, an electrophoretic mobility shift assay showed that the NF can associate with DNA on GAT(N) sequences, although the binding affinity is too weak to support GATA1–DNA binding independently (14). Therefore, in cases where a GAT(N) sequence is aligned adjacent to a GATA-binding motif that is bound by the CF, the NF may contribute to DNA binding. Particularly, the NF increases GATA–DNA-binding affinity in the Pal-GATA motif on both naked and chromatin DNA (14, 78).
There are three important observations about the N-finger function regarding its DNA binding. First, the DNA-binding kinetics at Pal-GATA motifs with R216 substitution mutations are similar to those of GATA1 lacking an entire NF, indicating the R216 residue is essential for NF-DNA association. The R216 residue is located on the opposite face to the FOG1 association face (20). Therefore, NF-DNA binding appears to be independent of FOG1 association. To date, multiple substitution mutations in the NF have been found in inherited human diseases (24, 78). The disease pathogenesis caused by the R216 mutation likely differs from that due to impaired FOG1 association, although the disease phenotypes partially overlap (79–82).
Second, the NF prefers to bind to a specific configuration of two GATA-binding motifs. SPR analysis showed that GATA1 lacking an NF binds to rPal-GATA motif similarly to wild-type GATA1 (77). GATA1 with a substitution mutation at R216 binds to tandem-oriented GATA motifs (Tandem-GATA motif) similarly to wild-type GATA1 (77), suggesting that the NF-DNA association is not critical for binding to the rPal-GATA motif and Tandem-GATA motif. By contrast, the SPR parameters of GATA1 binding at Pal-GATA motifs do not fit the 1:1 binding model found at single-GATA motifs (77). GATA1 binds to single-GATA motifs monovalently through the CF, while Pal-GATA motif binding is bivalent and uses both the CF and NF (Figure 2A). Thus, GATA1 differentially binds single-GATA and Pal-GATA motifs.
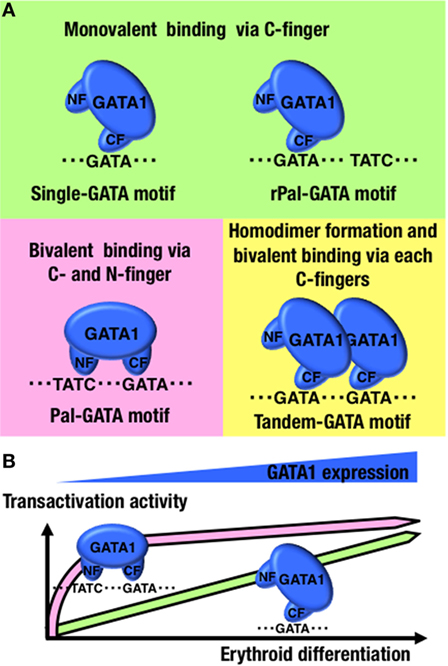
Figure 2. GATA1-binding kinetics at conformation-specific binding sites. (A) Schematic diagrams of the GATA1-binding modes at the indicated configurations of GATA-binding motifs. (B) Differences in transactivation dynamics of GATA1 between single- and Pal-GATA motifs. NF, N-finger domain; CF, C-finger domain.
Lastly and importantly, if the NF loses its DNA-binding capacity, the disabled GATA1 binds to Pal-GATA motifs monovalently, as it would at single-GATA motifs. Regardless of the NF function in DNA binding, GATA1 is able to bind to any configuration of GATA-binding motifs, although the GATA1 occupancy levels at Pal-GATA motifs depend on the binding capacity of the NF (77). Therefore, monovalent binding of GATA1 to the Pal-GATA motif is proposed to retain some activity, though not comparable to the full activity from bivalent GATA1 binding. Importantly, time-course reporter analyses in which GATA1 was introduced into non-hematopoietic cells have revealed that bivalent binding of GATA1 at Pal-GATA motifs is required for transactivation during early phases of GATA1 induction or at low GATA1 expression levels (Figure 2B) (77). GATA1 shows a dynamic expression profile during erythroid differentiation (10). The bivalent binding of GATA1 to Pal-GATA motifs may induce allosteric binding effects and enable GATA1 to precipitously induce target genes during the early erythroid differentiation phases (Figure 1).
Double GATA Motifs Oriented in Tandem
Similar to GATA1 binding at Pal-GATA motifs, SPR parameters of GATA1 binding at Tandem-GATA motifs does not fit the 1:1 binding model, and the SPR values are not altered by substitution mutations at R216 (77). Furthermore, if GATA1 lacks homodimerization capacities, then the disabled GATA1 binds to Tandem-GATA motifs similarly to GATA1 binding at single-GATA motifs (77). This finding generates an image of two GATA1 molecules in a dimer formation bivalently binding to two tandem-oriented GATA-binding motifs through the individual CFs (Figure 2A). Considering that GATA1 recognizes and binds to single-GATA motifs through the CF, there might be cases in which two GATA1 molecule monomers bind to two GATA-binding motifs aligned in any configuration if GATA1 is abundant, as described in previous studies of transgenic zebrafish (18). The secondary GATA1 molecule more efficiently binds to Tandem-GATA motifs than other configurations of dual GATA-binding motifs by forming a GATA1 homodimer.
In mice and zebrafish, GATA1 dimerization is important for erythropoiesis because it regulates specific genes, including the Gata1 gene (18, 19). Similar to the Pal-GATA motifs, the GATA1 monomer binds to Tandem-GATA motifs monovalently if GATA1 fails to form a homodimer (77). Taking into consideration the above-mentioned issues, bivalent binding of GATA1 homodimers to Tandem-GATA motifs might have a specific function that differs from the monovalent GATA1 monomers binding at Tandem-GATA motifs (Figure 1).
Conclusion
Accession to DNA via the consensus GATA-binding motif is a fundamental issue in the role of GATA1 as a TF. However, simple GATA1–DNA interactions only explain certain aspects of transcriptional regulation and fail to address comprehensive GATA1 regulation of various target genes, each of which has its own expression profile. Recent findings have made great advances in our knowledge of how the molecules recruited to regions adjacent to GATA-binding motifs modify the binding kinetics, reaction stoichiometry, and complex formations centered with GATA1, which may allosterically regulate GATA1 transcriptional activity (Figure 1). Intriguingly, a feature of GATA1 being a dual zinc-finger structure allows bivalent GATA1 binding to two GATA-binding motifs aligned side-by-side in both palindromic and tandem orientations. This variety may allow further functional diversity in GATA1 binding above monovalent binding for simple GATA-binding motifs. The molecular mechanisms of GATA1 transcriptional activity have been dissected in recent decades, and the contributions of GATA1 mutations to disease pathogenesis have been vigorously investigated. Nonetheless, how the configuration of cis-acting elements in the sequence surrounding GATA-binding motifs can generate diversified GATA1 transcriptional activity remains to be explored.
Author Contributions
All the authors listed have made substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported in part by JSPS KAKENHI (15H04759; RS), Grant-in-Aid for JSPS Fellow (15J04864; AH), Platform for Drug Discovery, Informatics, and Structural Life Science from Japan Agency for Medical Research and Development (AMED) (RS), AMED-Core Research for Evolutional Science and Technology (AMED-CREST) (RS), Friends of Leukemia Research Fund (RS), and Children’s Cancer Association of Japan (RS).
References
1. Evans T, Reitman M, Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci U S A (1988) 85:5976–80. doi:10.1073/pnas.85.16.5976
2. Shimizu R, Yamamoto M. Gene expression regulation and domain function of hematopoietic GATA factors. Semin Cell Dev Biol (2005) 16:129–36. doi:10.1016/j.semcdb.2004.11.001
3. Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem (2000) 275:38949–52. doi:10.1074/jbc.R000029200
4. Yamamoto M, Ko LJ, Leonard MW, Beug H, Orkin SH, Engel JD. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev (1990) 4:1650–62. doi:10.1101/gad.4.10.1650
5. Bresnick EH, Martowicz ML, Pal S, Johnson KD. Developmental control via GATA factor interplay at chromatin domains. J Cell Physiol (2005) 205:1–9. doi:10.1002/jcp.20393
6. Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene (2002) 21:3368–76. doi:10.1038/sj.onc.1205326
7. Ohneda K, Yamamoto M. Roles of hematopoietic transcription factors GATA-1 and GATA-2 in the development of red blood cell lineage. Acta Haematol (2002) 108:237–45. doi:10.1159/000065660
8. Elefanty AG, Antoniu M, Custodio M, Carmo-Fonseca M, Grosveld FG. GATA transcription factors associate with a novel class of nuclear bodies in erythroblasts and megakaryocytes. EMBO J (1996) 15:319–33.
9. Takai J, Moriguchi T, Suzuki M, Yu L, Ohneda K, Yamamoto M. The Gata1 5′ region harbors distinct cis-regulatory modules that direct gene activation in erythroid cells and gene inactivation in HSCs. Blood (2013) 122:3450–60. doi:10.1182/blood-2013-01-476911
10. Kaneko H, Shimizu R, Yamamoto M. GATA factor switching during erythroid differentiation. Curr Opin Hematol (2010) 17:163–8. doi:10.1097/MOH.0b013e32833800b8
11. Suzuki M, Kobayashi-Osaki M, Tsutsumi S, Pan X, Ohmori S, Takai J, et al. GATA factor switching from GATA2 to GATA1 contributes to erythroid differentiation. Genes Cells (2013) 18:921–33. doi:10.1111/gtc.12086
12. Kaneko H, Kobayashi E, Yamamoto M, Shimizu R. N- and C-terminal transactivation domains of GATA1 protein coordinate hematopoietic program. J Biol Chem (2012) 287:21439–49. doi:10.1074/jbc.M112.370437
13. Trainor CD, Ghirlando R, Simpson MA. GATA zinc finger interactions modulate DNA binding and transactivation. J Biol Chem (2000) 275:28157–66. doi:10.1074/jbc.M000020200
14. Trainor CD, Omichinski JD, Vandergon TL, Gronenborn AM, Clore GM, Felsenfeld G. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol Cell Biol (1996) 16:2238–47. doi:10.1128/MCB.16.5.2238
15. Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, et al. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell (1997) 90:109–19. doi:10.1016/S0092-8674(00)80318-9
16. Crossley M, Merika M, Orkin SH. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol (1995) 15:2448–56. doi:10.1128/MCB.15.5.2448
17. Mackay JP, Kowalski K, Fox AH, Czolij R, King GF, Crossley M. Involvement of the N-finger in the self-association of GATA-1. J Biol Chem (1998) 273:30506–7. doi:10.1074/jbc.273.46.30560
18. Nishikawa K, Kobayashi M, Masumi A, Lyons SE, Weinstein BM, Liu PP, et al. Self-association of Gata1 enhances transcriptional activity in vivo in zebra fish embryos. Mol Cell Biol (2003) 23:8295–305. doi:10.1128/MCB.23.22.8295-8305.2003
19. Shimizu R, Trainor CD, Nishikawa K, Kobayashi M, Ohneda K, Yamamoto M. GATA-1 self-association controls erythroid development in vivo. J Biol Chem (2007) 282:15862–71. doi:10.1074/jbc.M701936200
20. Wilkinson-White L, Gamsjaeger R, Dastmalchi S, Wienert B, Stokes PH, Crossley M, et al. Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1. Proc Natl Acad Sci U S A (2011) 108:14443–8. doi:10.1073/pnas.1105898108
21. Stumpf M, Waskow C, Krötschel M, van Essen D, Rodriguez P, Zhang X, et al. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc Natl Acad Sci U S A (2006) 103:18504–9. doi:10.1073/pnas.0604494103
22. Sankaran VG, Ghazvinian R, Do R, Thiru P, Vergilio JA, Beggs AH, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest (2012) 122:2439–43. doi:10.1172/JCI63597
23. Ahmed M, Sternberg A, Hall G, Thomas A, Smith O, O’Marcaigh A, et al. Natural history of GATA1 mutation in Down syndrome. Blood (2004) 103:2480–9. doi:10.1182/blood-2003-10-3383
24. Singleton BK, Frayne J, Anstee DJ. Blood group phenotypes resulting from mutations in erythroid transcription factors. Curr Opin Hematol (2012) 19:486–93. doi:10.1097/MOH.0b013e328358f92e
25. Campbell AE, Wilkinson-White L, Mackay JP, Matthews JM, Blobel GA. Analysis of disease-causing GATA1 mutations in murine gene complementation systems. Blood (2013) 121:5218–27. doi:10.1182/blood-2013-03-488080
26. Shimizu R, Ohneda K, Engel JD, Trainor CD, Yamamoto M. Transgenic rescue of GATA-1-deficient mice with GATA-1 lacking a FOG-1 association site phenocopies patients with X-linked thrombocytopenia. Blood (2004) 103:2560–7. doi:10.1182/blood-2003-07-2514
27. Shimizu R, Kobayashi E, Engel JD, Yamamoto M. Induction of hyperproliferative fetal megakaryopoiesis by an N-terminally truncated GATA1 mutant. Genes Cells (2009) 14:1119–31. doi:10.1111/j.1365-2443.2009.01338.x
28. Hasegawa A, Shimizu R, Mohandas N, Yamamoto M. Mature erythrocyte membrane homeostasis is compromised by loss of the GATA1-FOG1 interaction. Blood (2012) 119:2615–23. doi:10.1182/blood-2011-09-382473
29. Shimizu R, Yamamoto M. GATA-related hematopoietic disorders. Exp Hematol (2016) 44:696–705. doi:10.1016/j.exphem.2016.05.010
30. Shimizu R, Kuroha T, Ohneda O, Pan X, Ohneda K, Takahashi S, et al. Leukemogenesis caused by incapacitated GATA-1 function. Mol Cell Biol (2004) 24:10814–25. doi:10.1128/MCB.24.24.10814-10825.2004
31. Centurione L, Di Baldassarre A, Zingariello M, Bosco D, Gatta V, Rana RA, et al. Increased and pathologic emperipolesis of neutrophils within megakaryocytes associated with marrow fibrosis in GATA-1(low) mice. Blood (2004) 104:3573–80. doi:10.1182/blood-2004-01-0193
32. Vannucchi AM, Bianchi L, Cellai C, Paoletti F, Rana RA, Lorenzini R, et al. Development of myelofibrosis in mice genetically impaired for GATA-1 expression (GATA-1(low) mice). Blood (2002) 100:1123–32. doi:10.1182/blood-2002-06-1913
33. Wadman IA, Osada H, Grütz GG, Agulnick AD, Westphal H, Forster A, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J (1997) 16:3145–57. doi:10.1093/emboj/16.11.3145
34. Osada H, Grutz G, Axelson H, Forster A, Rabbitts TH. Association of erythroid transcription factors: complexes involving the LIM protein RBTN2 and the zinc-finger protein GATA1. Proc Natl Acad Sci U S A (1995) 92:9585–9. doi:10.1073/pnas.92.21.9585
35. Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature (1996) 384:270–2. doi:10.1038/384270a0
36. Wozniak RJ, Keles S, Lugus JJ, Young KH, Boyer ME, Tran TM, et al. Molecular hallmarks of endogenous chromatin complexes containing master regulators of hematopoiesis. Mol Cell Biol (2008) 28:6681–94. doi:10.1128/MCB.01061-08
37. Lécuyer E, Hoang TSCL. From the origin of hematopoeisis to stem cells and leukemia. Exp Hematol (2004) 32:11–24. doi:10.1016/j.exphem.2003.10.010
38. Lahlil R, Lécuyer E, Herblot S, Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol Cell Biol (2004) 24:1439–52. doi:10.1128/MCB.24.4.1439-1452.2004
39. Yun WJ, Kim YW, Kang Y, Lee J, Dean A, Kim A. The hematopoietic regulator TAL1 is required for chromatin looping between the β-globin LCR and human γ-globin genes to activate transcription. Nucleic Acids Res (2014) 42:4283–93. doi:10.1093/nar/gku072
40. Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, et al. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood (2009) 113:2191–201. doi:10.1182/blood-2008-07-169417
41. Wakabayashi A, Ulirsch JC, Ludwig LS, Fiorini C, Yasuda M, Choudhuri A, et al. Insight into GATA1 transcriptional activity through interrocation of cis element disrupted in human erythroid disorders. Proc Natl Acad Sci U S A (2016) 113:4434–9. doi:10.1073/pnas.1521754113
42. Hsu HY, Lin SY, Huang CJ, Lian SL, Ho YH. Changes of serum copper and zinc levels in patients with nasopharyngeal carcinoma by radiotherapy. Biol Trace Elem Res (1994) 46:1–16. doi:10.1007/BF02790064
43. Hewitt KJ, Kim DH, Devadas P, Prathibha R, Zuo C, Sanalkumar R, et al. Hematopoietic signaling mechanism revealed from a stem/progenitor cell cistrome. Mol Cell (2015) 59:62–74. doi:10.1016/j.molcel.2015.05.020
44. Zhang Y, Wu W, Cheng Y, King DC, Harris RS, Taylor J, et al. Primary sequence and epigenetic determinants of in vivo occupancy of genomic DNA by GATA1. Nucleic Acids Res (2009) 37:7024–38. doi:10.1093/nar/gkp747
45. Tallack MR, Whitington T, Yuen WS, Wainwright EN, Keys JR, Gardiner BB, et al. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res (2010) 20:1052–63. doi:10.1101/gr.106575.110
46. Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol Cell Biol (1993) 13:2776–86. doi:10.1128/MCB.13.5.2776
47. Basu P, Morris PE, Haar JL, Wani MA, Lingrel JB, Gaensler KM, et al. KLF2 is essential for primitive erythropoiesis and regulates the human and murine embryonic beta-like globin genes in vivo. Blood (2005) 106:2566–71. doi:10.1182/blood-2005-02-0674
48. Gordon AR, Outram SV, Keramatipour M, Goddard CA, Colledge WH, Metcalfe JC, et al. Splenomegaly and modified erythropoiesis in KLF13-/- mice. J Biol Chem (2008) 283:11897–904. doi:10.1074/jbc.M709569200
49. Varricchio L, Dell’Aversana C, Nebbioso A, Migliaccio G, Altucci L, Mai A, et al. Identification of NuRSERY, a new functional HDAC complex composed by HDAC5, GATA1, EKLF and pERK present in human erythroid cells. Int J Biochem Cell Biol (2014) 50:112–22. doi:10.1016/j.biocel.2014.02.019
50. Thein SL. The molecular basis of β-thalassemia. Cold Spring Harb Perspect Med (2013) 3:a011700. doi:10.1101/cshperspect.a011700
51. Crispino JD, Weiss MJ. Erythro-megakaryocytic transcription factors associated with hereditary anemia. Blood (2014) 123:3080–8. doi:10.1182/blood-2014-01-453167
52. Siatecka M, Bieker JJ. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood (2011) 118:2044–55. doi:10.1182/blood-2011-03-331371
53. Perkins A, Xu X, Higgs DR, Patrinos GP, Amaud L, Bieker JJ, et al. Krüppeling erythropoiesis: an unexpected broad spectrum of human red blood cell disorders due to KLF1 variants. Blood (2016) 127:1856–62. doi:10.1182/blood-2016-01-694331
54. Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol Cell Biol (1995) 15:2437–47. doi:10.1128/MCB.15.5.2437
55. Kang Y, Kim YW, Yun J, Shin J, Kim A. KLF1 stabilizes GATA-1 and TAL1 occupancy in the human β-globin locus. Biochim Biophys Acta (2015) 1849:282–9. doi:10.1016/j.bbagrm.2014.12.010
56. Wontakal SN, Guo X, Smith C, MacCarthy T, Bresnick EH, Bergman A, et al. A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proc Natl Acad Sci U S A (2012) 109:3832–7. doi:10.1073/pnas.1121019109
57. Pilon AM, Ajay SS, Kumar SA, Steiner LA, Cherukuri PF, Wincovitch S, et al. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood (2011) 118:e139–48. doi:10.1182/blood-2011-05-355107
58. Archer MC. Role of sp transcription factors in the regulation of cancer cell metabolism. Genes Cancer (2011) 2:712–9. doi:10.1177/1947601911423029
59. Imanishi M, Imamura C, Higashi C, Yan W, Negi S, Futaki S, et al. Zinc finger–zinc finger interaction between the transcription factors, GATA-1 and Sp1. Biochem Biophys Res Commun (2010) 400:625–30. doi:10.1016/j.bbrc.2010.08.116
60. Woo AJ, Moran TB, Schindler YL, Choe SK, Langer NB, Sullivan MR, et al. Identification of ZBP-89 as a novel GATA-1-associated transcription factor involved in megakaryocytic and erythroid development. Mol Cell Biol (2008) 28:2675–89. doi:10.1128/MCB.01945-07
61. Alhashem YN, Vinjamur DS, Basu M, Klingmüller U, Gaensler KM, Lloyd JA. Transcription factors KLF1 and KLF2 positively regulate embryonic and fetal beta-globin genes through direct promoter binding. J Biol Chem (2011) 286:24819–27. doi:10.1074/jbc.M111.247536
62. Johnson KD, Grass JA, Boyer ME, Kiekhaefer CM, Blobel GA, Weiss MJ, et al. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc Natl Acad Sci U S A (2002) 99:11760–5. doi:10.1073/pnas.192285999
63. Steiner LA, Maksimova Y, Schulz V, Wong C, Raha D, Mahajan MC, et al. Chromatin architecture and transcription factor binding regulate expression of erythrocyte membrane protein genes. Mol Cell Biol (2009) 29:5399–412. doi:10.1128/MCB.00777-09
64. Lim LC, Fang L, Swendeman SL, Sheffery M. Characterization of the molecularly cloned murine alpha-globin transcription factor CP2. J Biol Chem (1993) 268:18008–17.
65. Ohneda K, Shimizu R, Nishimura S, Muraosa Y, Takahashi S, Engel JD, et al. A minigene containing four discrete cis elements recapitulates GATA-1 gene expression in vivo. Genes Cells (2002) 7:1243–54. doi:10.1046/j.1365-2443.2002.00595.x
66. Shimizu R, Hasegawa A, Ottolenghi S, Ronchi A, Yamamoto M. Verification of the in vivo activity of three distinct cis-acting elements within the Gata1 gene promoter-proximal enhancer in mice. Genes Cells (2013) 18:1032–41. doi:10.1111/gtc.12096
67. Bosè F, Fugazza C, Casalgrandi M, Capelli A, Cunningham JM, Zhao Q, et al. Functional interaction of CP2 with GATA-1 in the regulation of erythroid promoters. Mol Cell Biol (2006) 26:3942–54. doi:10.1128/MCB.26.10.3942-3954.2006
68. Solis C, Aizencang GI, Astrin KH, Bishop DF, Desnick RJ. Uroporphyrinogen III synthase erythroid promoter mutations in adjacent GATA1 and CP2 elements cause congenital erythropoietic porphyria. J Clin Invest (2001) 107:753–62. doi:10.1172/JCI10642
69. Kim CG, Swendeman SL, Barnhart KM, Sheffery M. Promoter elements and erythroid cell nuclear factors that regulate alpha-globin gene transcription in vitro. Mol Cell Biol (1990) 10:5958–66. doi:10.1128/MCB.10.11.5958
70. Sabath DE, Koehler KM, Yang WQ, Phan V, Wilson J. DNA-protein interactions in the proximal zeta-globin promoter: identification of novel CCACCC- and CCAAT-binding proteins. Blood Cells Mol Dis (1998) 24:183–98. doi:10.1006/bcmd.1998.0185
71. Chae JH, Kim CG. CP2 binding to the promoter is essential for the enhanced transcription of globin genes in erythroid cells. Mol Cells (2003) 15:40–7.
72. Fernández-Morales B, Pavón L, Calés C. CDC6 expression is regulated by lineage specific transcription factor GATA1. Cell Cycle (2012) 11:3055–66. doi:10.4161/cc.21471
73. Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, et al. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med (2002) 195:1387–95. doi:10.1084/jem.20020656
74. Snow JW, Trowbridge JJ, Fujiwara T, Emambokus NE, Grass JA, Orkin SH, et al. A single cis element maintains repression of the key developmental regulator Gata2. PLoS Genet (2010) 6:e1001103. doi:10.1371/journal.pgen.1001103
75. Bates DL, Chen Y, Kim G, Guo L, Chen L. Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA. J Mol Biol (2008) 381:1292–306. doi:10.1016/j.jmb.2008.06.072
76. Martin DI, Orkin SH. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf1. Genes Dev (1990) 4:1886–98. doi:10.1101/gad.4.11.1886
77. Hasegawa A, Kaneko H, Ishihara D, Nakamura M, Watanabe A, Yamamoto M, et al. GATA1 binding kinetics on conformation-specific binding sites elicit differential transcriptional regulation. Mol Cell Biol (2016) 36:2151–67. doi:10.1128/MCB.00017-16
78. Ciovacco WA, Raskind WH, Kacena MA. Human phenotypes associated with GATA-1 mutations. Gene (2008) 427:1–6. doi:10.1016/j.gene.2008.09.018
79. Yu C, Niakan KK, Matsushita M, Stamatoyannopoulos G, Orkin SH, Raskind WH. X-linked thrombocytopenia with thalassemia from a mutation in the amino finger of GATA-1 affecting DNA binding rather than FOG-1 interaction. Blood (2002) 100:2040–5. doi:10.1182/blood-2002-02-0387
80. Tubman VN, Levine JE, Campagna DR, Monahan-Earley R, Dvorak AM, Neufeld EJ, et al. X-linked gray platelet syndrome due to a GATA1 Arg216Gln mutation. Blood (2007) 109:3297–9. doi:10.1182/blood-2006-02-004101
81. Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, et al. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet (2000) 24:266–70. doi:10.1038/73480
82. Freson K, Matthijs G, Thys C, Mariën P, Hoylaerts MF, Vermylen J, et al. Different substitutions at residue D218 of the X-linked transcription factor GATA1 lead to altered clinical severity of macrothrombocytopenia and anemia and are associated with variable skewed X inactivation. Hum Mol Genet (2002) 11:147–52. doi:10.1093/hmg/11.2.147
Keywords: cis-acting elements, GATA1 transcription factor, DNA-binding domain, protein–protein interactions, erythropoiesis
Citation: Hasegawa A and Shimizu R (2017) GATA1 Activity Governed by Configurations of cis-Acting Elements. Front. Oncol. 6:269. doi: 10.3389/fonc.2016.00269
Received: 13 September 2016; Accepted: 19 December 2016;
Published: 09 January 2017
Edited by:
Laura Gutiérrez, Hospital Clínico San Carlos, SpainReviewed by:
Anna Rita Migliaccio, Icahn School of Medicine at Mount Sinai, USAIoannis S. Vizirianakis, Aristotle University of Thessaloniki, Greece
Eric Milot, Université de Montréal, Canada
Copyright: © 2017 Hasegawa and Shimizu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ritsuko Shimizu, cnNoaW1penUmI3gwMDA0MDttZWQudG9ob2t1LmFjLmpw
 Atsushi Hasegawa
Atsushi Hasegawa Ritsuko Shimizu
Ritsuko Shimizu