- 1Discipline of Surgery, School of Medicine, The Queen Elizabeth Hospital, University of Adelaide, Woodville, SA, Australia
- 2The Robinson Research Institute, University of Adelaide, Adelaide, SA, Australia
- 3Department of Medical Oncology, The Queen Elizabeth Hospital, Woodville, SA, Australia
Clinics are increasingly adopting gene-expression profiling to diagnose breast cancer subtype, providing an intrinsic, molecular portrait of the tumor. For example, the PAM50-based Prosigna test quantifies expression of 50 key genes to classify breast cancer subtype, and this method of classification has been demonstrated to be superior over traditional immunohistochemical methods that detect proteins, to predict risk of disease recurrence. However, these tests were largely developed and validated using breast cancer samples from postmenopausal women. Thus, the accuracy of such tests has not been explored in the context of the hormonal fluctuations in estrogen and progesterone that occur during the menstrual cycle in premenopausal women. Concordance between traditional methods of subtyping and the new tests in premenopausal women is likely to depend on the stage of the menstrual cycle at which the tissue sample is taken and the relative effect of hormones on expression of genes versus proteins. The lack of knowledge around the effect of fluctuating estrogen and progesterone on gene expression in breast cancer patients raises serious concerns for intrinsic subtyping in premenopausal women, which comprise about 25% of breast cancer diagnoses. Further research on the impact of the menstrual cycle on intrinsic breast cancer profiling is required if premenopausal women are to benefit from the new technology of intrinsic subtyping.
Introduction
Approximately 25% of breast cancers are diagnosed in women under the age of 50 (1). When breast cancer is diagnosed in young women it carries a high burden, with reduced 5-year survival rates compared to breast cancer in older women (2, 3), and a devastating impact on young families. Breast cancer is considered a chronic disease, with increased mortality extending over the next 40 years, even if the breast cancer is diagnosed at an early stage (3).
In premenopausal women, cyclical production of ovarian hormones estrogen and progesterone occur over the course of the menstrual cycle, causing the mammary gland epithelium to undergo cycles of proliferation, differentiation, and apoptosis (4, 5). Estrogen and progesterone play key roles in the development of breast cancer, with the relative risk of breast cancer related to the breast’s cumulative exposure to these hormones (6, 7).
Breast cancer is not a single disease. There are many mutated genes that can drive tumor development, and biomarkers are essential to classify breast cancer into its different subtypes, each of which responds best to different therapies. Currently, immunohistochemical assays that detect abundance of proteins are used to identify expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) and determine the rate of proliferation of the cancer cells (Ki67). These biomarkers are used collectively to diagnose subtype and thus determine the best treatment option for an individual patient.
Now, clinics are increasingly adopting gene-expression profiling to diagnose breast cancer subtype, providing an intrinsic, molecular portrait of the tumor. For example, the PAM50-based Prosigna test quantifies expression of 50 key genes to classify breast cancer subtype. Tests that diagnose intrinsic breast cancer subtype must be robust, and relatively resistant to fluctuations in gene expression. Generally, good concordance is observed in gene expression between pairs of diagnostic and surgical samples and between classification of subtype by gene expression and traditional immunohistochemical techniques.
However, despite their availability for diagnosing breast cancer subtype in premenopausal women, tests that utilize gene-expression profiling were largely developed and validated using breast cancer samples from postmenopausal women. Fluctuations in ovarian hormones estrogen and progesterone across the menstrual cycle may affect expression of genes currently used in intrinsic subtyping tests. Indeed, studies have found poor concordance between classification of subtype by intrinsic tests and traditional immunohistochemistry in premenopausal breast cancer patients (8, 9).
Here, we outline the role of ovarian hormones in regulation of gene expression in the breast and highlight the deficiencies in knowledge around intrinsic subtyping in premenopausal breast cancer. As different therapeutic strategies are required depending on the tumor type, effective subtyping of breast cancers is necessary to help guide treatment decisions and provide accurate prognostic information for each patient. Intrinsic subtyping offers some significant advantages over traditional subtyping methods; however, the current tests have not been sufficiently validated in the context of premenopausal breast cancers, where there are significant fluctuations in estrogen and progesterone. There is a pressing need for more research into hormonal modulation of breast cancer gene expression in order to provide the optimal subtype diagnosis for premenopausal women.
Hormonally Driven Changes in the Breast During the Menstrual Cycle
During the follicular phase of the menstrual cycle, increasing levels of FSH produced by the pituitary stimulate maturation of estrogen-secreting ovarian follicles. Estrogen acts on the pituitary to further increase the production of FSH and LH. Eventually, the concentration of estrogen peaks, stimulating a peak in LH secretion that triggers ovulation. Following ovulation, LH promotes differentiation of the ovarian follicle into the progesterone producing corpus luteum. The luteal phase is characterized by a high concentration of progesterone and is accompanied by a smaller second rise of estrogen. Progesterone supresses FSH and LH production, resulting in a decrease in estrogen concentration. Levels of progesterone begin to decrease as the corpus luteum ceases to produce progesterone and collapses. Consequently, the end of the menstrual cycle is characterized by low circulating hormones, which, in turn, relieve the negative inhibition of FSH and LH (10, 11) (Figure 1), allowing progression into the next menstrual cycle.
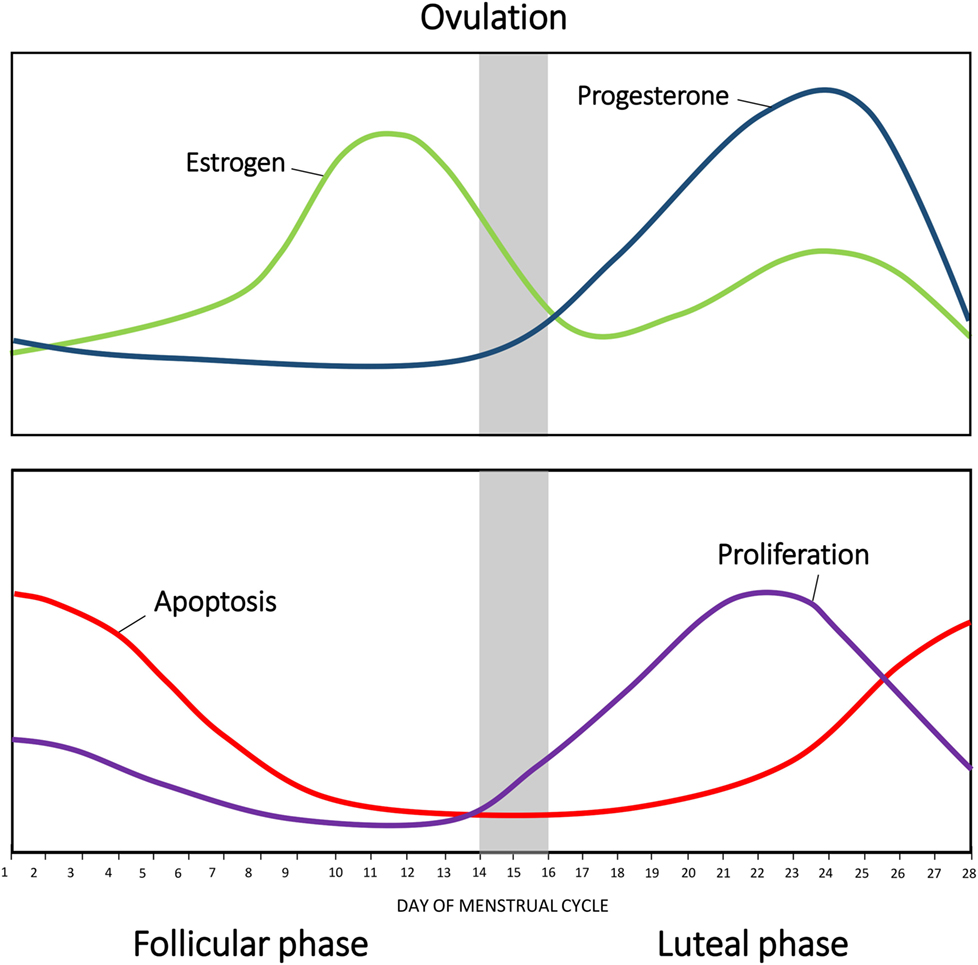
Figure 1. Changes in hormonal levels in accordance with the menstrual cycle. The fluctuations of estrogen (green) and progesterone (blue) during the human menstrual cycle. Net apoptosis (red) and proliferation (purple) in the mammary gland in accordance with the menstrual phase.
Fluctuations in estrogen and progesterone across the menstrual cycle direct the mammary gland epithelium to undergo sequential waves of proliferation, differentiation, and apoptosis (4, 5, 12, 13). Histological analysis of breast tissue by Vogel et al. identified distinct morphological changes in the mammary gland in accordance with different phases of the cycle (14). Five separate phases of breast morphology have been identified, associated with differing concentration of circulating estrogen and progesterone. Each phase has distinct morphological criteria based on the appearances of the luminal cells, myoepithelial vacuolization, intraluminal secretion, stromal edema, and events of cell turnover (5, 14), as summarized in Table 1.
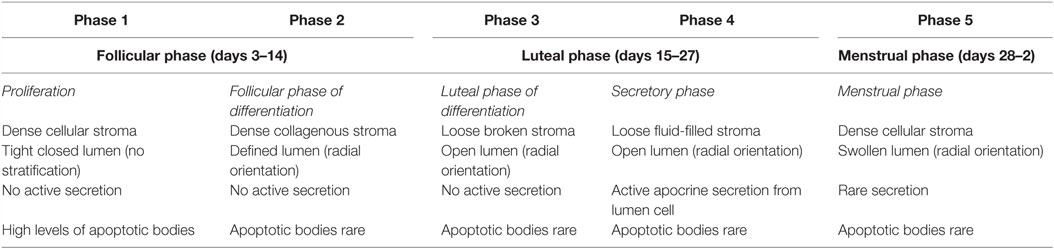
Table 1. Morphological changes in the mammary gland in accordance with the menstrual cycle as described by Vogel et al. (14).
The highest proliferative activity of mammary epithelium is observed in the luteal phase, with rising levels of progesterone (Figure 1). As the concentration of progesterone rises, there is an increase in secondary branching, alveoli budding, and stromal development, accompanied by changes to the extracellular matrix (10, 14). Consistent with this, studies in rodents have shown that administration of exogenous progesterone promotes side-branching and normal secretory alveolar development, whereas estrogen stimulates ductal elongation (15). Ferguson and Anderson showed that epithelial apoptosis increases at the end of the cycle, with decreasing circulating concentration of estrogen and progesterone (16); causing an atrophy of the epithelium, closing of the alveolar lumen, condensation of intralobular stroma, and a variable inflammatory infiltrate (14).
Hormonally driven morphological changes are associated with gene expression changes. Estrogen regulates many genes involved in cell cycle progression; such as Cyclin D (17), and c-MYC (18, 19), and is involved in activation of Cyclin E complexes (20). Estrogen also induces cyclin dependant kinases (Cdk) activation and Rb phosphorylation (20, 21) to promote cell cycle progression. In addition, estrogen treatment inhibits genes responsible for the suppression of cell growth, such as p21 (22). Estrogen is also an inhibitor of apoptosis and increases the expression of antiapoptotic proteins, such as Bcl-2 and Bcl-xL (23). Consistent with this, Bcl-2 is expressed almost exclusively in ER-positive breast cancers and is associated with a good prognosis (24).
Progesterone also plays an important role in cell proliferation and differentiation in the breast, specifically acting during the luteal phase of the menstrual cycle. The proliferative role of progesterone is likely mediated by regulation of cell cycle genes, growth factors, and growth factor receptors. Musgrove et al. illustrated that progesterone treatment of PR-positive breast cancer cells results in an increase in cell cycle progression, which is correlated with an induction of cell cycle genes; including cyclin D1 (25, 26), and c-Myc (25–27). Progesterone also regulates activity of Cdks (28). In addition to stimulating genes associated with cell cycle progression, progesterone has been suggested to inhibit expression of genes responsible for suppression of cell growth, such as tumor suppressor protein p53 (29) and retinoblastoma protein (30). Decline in ovarian hormones also effects gene expression. The fall in estrogen and progesterone at the end of the luteal phase is associated with an increase in apoptotic proteins, such as BAX (10) and FasL (31), and a decrease in antiapoptotic proteins, such as Bcl-2 (10).
In the mammary gland, progesterone elicits its function mainly through a paracrine mechanism. Recently, RANKL has been identified as an important paracrine mediator of progesterone-induced proliferation in the mammary gland (32, 33) and is implicated upstream of Cyclin D (32). Consistent with this, RANKL is required for mammary gland development (34) and was shown to be essential for ductal side branching and alveologenesis in mice (35). In addition, overexpression of its receptor, RANK, in mice resulted in increased proliferation of mammary epithelial cells (36). Wnt-4 has also been identified as a paracrine mediator of progesterone signaling (32, 37), and is important for side-branching of the mammary ductal epithelium (37). To promote optimal proliferation of mammary epithelial cells, estrogen induces expression of the PR. This leads to proliferation of mammary epithelial cells through elevated expression of cell cycle genes, when both estrogen and progesterone are present (38). Conversely, progesterone downregulates its receptor and inhibits synthesis of the ER (39).
Progesterone and estrogen also regulate growth factors and growth factor receptors in the breast, such as epidermal growth factor (EGF) (40) and EGF receptor (EGFR) (41, 42). Many key genes associated with EGFR signaling are upregulated in response to progesterone treatment (41, 43). Furthermore, EGFR signaling has been implicated downstream of estrogen in the mammary gland (44). Estrogen can induce phosphorylation of EGFR, and can directly interact with signal transduction pathways, to activate MAPK, JAK/STAT, SRC, and PI3K signaling pathways downstream of EGFR. In parallel, EGFR can, in turn, phosphorylate and activate ER and PR (45). It has also been shown that EGF family members are induced by estrogen; including EGF (46), TGFα (46, 47), and amphiregulin (Areg) (48, 49), and act as an important mediator of paracrine estrogen-induced proliferation (Figure 2). However, estrogen inhibits EGFR expression in ERα positive cells. In ERα negative cells, secreted amphiregulin activates EGFR signaling to promote cell proliferation (50). Recent studies have also indicated that estrogen treatment induces expression of vascular epithelial growth factor receptor (VEGF), a receptor involved in tumor growth, both in vitro (51) and in vivo (52).
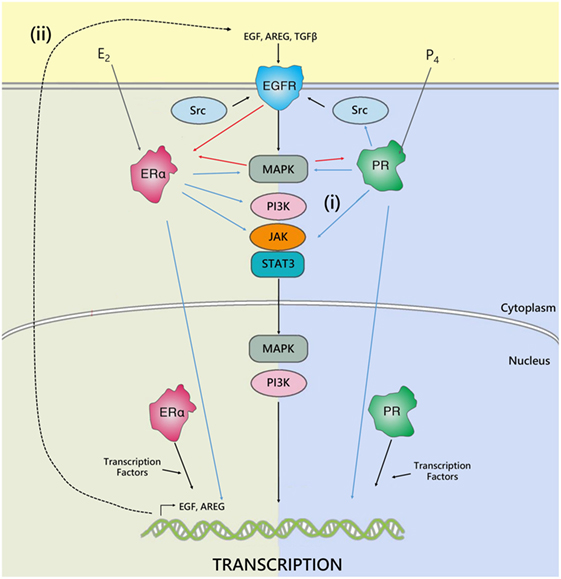
Figure 2. The interplay between ER, PR, and EGFR. Hormone receptors regulate gene transcription either by binding directly to DNA response elements or by recruiting transcription factors and co-regulators. In addition, cross talk occurs between ER, PR, and EGFR to regulate gene expression. The estrogen and progesterone receptor can regulate epidermal growth factor receptor activity by either: (i) directly interfering with their transduction pathways, to activate MAPK, JAK/STAT, SRC, PI3K signaling downstream of EGFR, or (ii) by inducing expression and secretion of paracrine growth factors, such as AREG, TGFβ, or EGF, which act on EGFR to activate pathways involved in cell proliferation, survival, and metastasis. In parallel, EGFR can, in turn, phosphorylate and activate ER and PR. Adapted from Tanos et al. (45).
In breast cancer, increased EGFR signaling is associated with a more aggressive phenotype. Overexpression of growth factor receptors has been associated with increased metastasis and poor survival, together with a lack of response to endocrine therapy (53, 54). As estrogen and progesterone play critical roles in regulation of growth factors, it is possible that the fluctuations of these hormones during the menstrual cycle are sufficient to modulate expression of EGFR and affect downstream signaling. In the luteal phase when progesterone is high and estrogen is present, signaling through growth factor pathways may be increased compared to the follicular phase when progesterone concentration is low. Consistent with this, breast tumors in young women often have significantly higher EGFR expression and worse prognosis (55, 56).
Classification of Breast Cancer Subtypes
Breast cancer is a heterogeneous disease, due to its diverse molecular and cellular features, with different therapeutic strategies required depending on the tumor type and stage. The decision to treat patients with adjuvant therapy has been guided by clinical and pathological features of the tumor. With no adjuvant therapy, 12–58% of women will experience a reoccurrence within 5 years (57–59). Of women diagnosed with breast cancer, the majority (approximately 75–92%) receive adjuvant therapy (57, 60, 61), suggesting that many women receive a treatment that may not provide benefit, exposing them to unnecessary side effects. Ideally, the decision to use adjuvant therapy should be based on the prediction of the degree of benefit, to minimize the number of patients receiving unnecessary treatment. Traditionally, evaluation of ER, PR, HER2, and Ki67 immunoreactivity, together with clinicopathological variables including tumor size, type, and grade, are used to classify breast tumors and guide clinical decisions. Breast cancer can be classified into five major subtypes, i.e., Luminal A, Luminal B, HER2 enriched, Basal-like, and normal breast-like, which show significant differences in incidence, survival, and clinical outcomes (9, 62–64).
Luminal A tumors are the most common, representing 50–60% of all breast cancers (65). Patients with Luminal A breast cancer have a good prognosis; displaying significantly increased overall and disease-free survival compared to other breast cancer subtypes (9, 63, 64). Treatment of early-stage Luminal A breast cancer is based mainly on hormonal therapies, with the addition of adjuvant chemotherapy dependant on the clinical stage. The immunohistochemical profile of Luminal A tumors is characterized by high expression of ER, PR, and luminal cytokeratins 8 and 18, an absence of HER2 expression, and low rate of proliferation measured through Ki67 (65, 66).
Luminal B tumors account for 15–20% of all breast cancers (65). Patients with Luminal B breast cancer have poorer outcomes from endocrine therapy, however, have a better response to chemotherapy, achieving pathological complete response (pCR) to neoadjuvant chemotherapy in 16% of tumors compared to 6% in Luminal A tumors (67). From the immunohistochemical point of view, Luminal B tumors are characterized by a lower expression of ER and PR, and higher Ki67 index, and display a higher histological grade, compared to Luminal A tumors (66). Like Luminal A tumors, they express luminal cytokeratins 8 and 18 (65, 66).
Human epidermal growth factor receptor 2-enriched tumors represent 15–20% of breast cancer subtypes (65). Patients with HER2-enriched tumors have poor prognosis and overall survival (9, 63, 64). The immunohistochemical profile of HER2-enriched tumors is characterized by variable ER or PR expression and overexpression of HER2 (66). Consequently, treatment of HER2-enriched tumors includes monoclonal antibodies which directly target the HER2 receptor given in conjunction with chemotherapy (68).
Basal-like tumors comprise 15–20% of all breast cancers (66), and are associated with an aggressive clinical behavior and a high rate of metastasis (69). Patients with Basal-like tumors have a poor prognosis, displaying lower overall and disease free survival compared to other subtypes (9, 63, 64). Treatment of basal-like tumors involves systemic chemotherapy. The immunohistochemical profile of basal-like tumors is characterized by a triple-negative phenotype; ER, PR, and HER2 negative.
Normal breast-like tumors account for 5–10% of all breast cancers (65). They lack the expression of ER, PR, and HER2, however, are not considered basal-like tumors as expression of basal cytokeratin 5 and EGFR is absent (66). However, normal breast-like tumors are poorly defined, and it is argued that they are an artifact of having a high percentage of normal cells in the tumor specimen (70, 71). It has been suggested that these tumors could be grouped into the recently discovered claudin-low subtype, which also displays basal-like characteristics, while also sharing biomarkers in common with normal-like breast epithelial cells. Similar to the basal-like subtype, claudin-low tumors have been associated with therapeutic resistance and poor survival outcomes (72), due to their highly migratory nature.
In clinical practice, identifying triple-negative and HER-2-positive breast cancers can be achieved with standard pathological testing, and recommendations for appropriate adjuvant therapy in early-stage disease are well defined. However, for patients with ER-positive and HER-2-negative disease, distinguishing between those with Luminal A disease and those with Luminal B disease is more challenging and has implications for treatment recommendations (73). Identifying those patients with good prognosis Luminal A disease who will have a small absolute benefit from adjuvant chemotherapy can avoid unnecessary chemotherapy and its associated side effects, while identifying those with Luminal B disease and a higher risk of relapse can prevent under treatment in this group (73, 74).
Gene-Expression Profiling in Breast Cancer
In 2000, Perou et al. (62) proposed a new classification system for breast cancer subtypes, separating them into distinct subgroups based on gene-expression profiles, as opposed to protein expression signatures used in traditional subtyping methods. Intrinsic subtyping by gene-expression profiling is predictive of overall and relapse-free survival (8, 9, 63, 64, 75) and can predict the relative risk of relapse and the patient benefit from hormonal therapy and chemotherapy (71). Therefore, gene-expression profiling can be used to inform risk prediction and help guide treatment decisions, to decrease the number of patients receiving unnecessary treatment. The main genes associated with each subtype, together with pathological characteristics and prognosis, are summarized in Table 2.
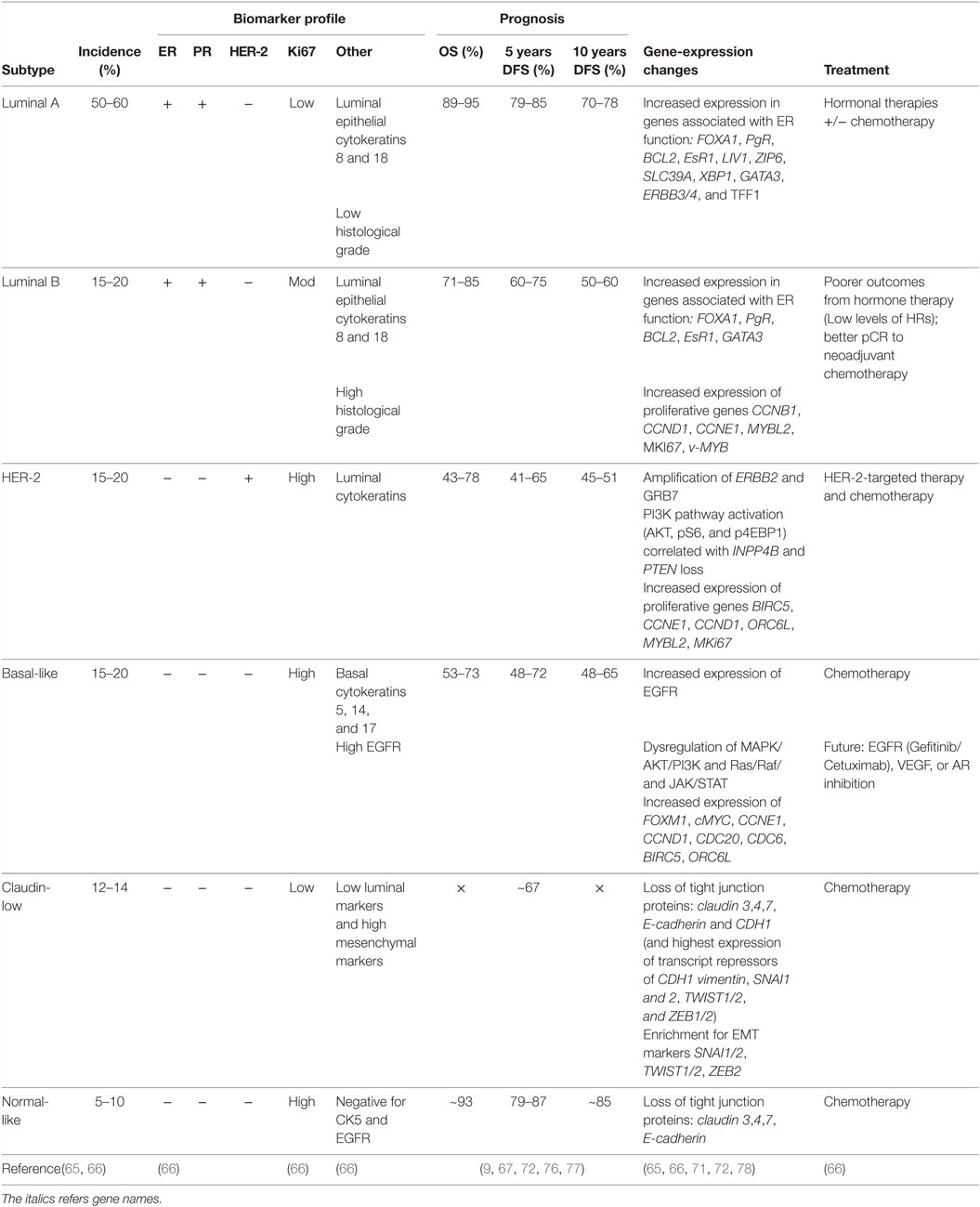
Table 2. Summary of clinical and pathological characteristics, prognosis, and gene-expression changes of breast cancer subtypes.
Each subtype displays a distinct gene-expression profile. Luminal A tumors are characterized by a high level of ER, and as such display an increased expression in genes associated with ER function, such as Bcl2, EsR1, PgR, and FOXA1 (62, 63). Compared to Luminal A, Luminal B tumors display an increase in expression of proliferative genes, and consequently possess a more aggressive phenotype, higher proliferative index, and worse prognosis (9, 63, 64, 67, 79). Like Luminal A tumors, Luminal B tumors also express genes associated with ER activation, including Bcl2, FOXA1, CCND1, and GATA3 (79); however, increased expression of proliferative genes, such as CCNB1, CCND1, CCNE1, MYBL2, and MKi67, appears to be the hallmark of luminal B tumors (63, 64, 79).
HER-2-enriched tumors are characterized by a high expression of human epidermal growth factor receptor-2, ErbB2, and other genes associated with the HER2 pathway (65, 80). HER2 functions as a receptor tyrosine kinase and signals through PI3K/AKT/mTOR, JAK/STAT, MAPK and Ras/Raf pathways to promote cell survival, proliferation, and migration (66). Consequently, a dysregulation of signaling in HER-2-enriched tumors can lead to sustained proliferative signaling, a hallmark of cancer. HER-2-enriched tumors also overexpress GRB7, an adaptor protein involved in receptor tyrosine kinase signaling, which can also promote activation of PI3K/AKT/mTOR, JAK/STAT, and MAPK signaling pathways to allow for sustained proliferative signaling (66). HER-2-enriched tumors have increased expression of proliferative genes, including BIRC5, CCND1, CCNE1, ORC6L, and MKi67, and are often associated with a more aggressive and highly proliferative tumor (80).
Basal-like tumors express high levels of basal cytokeratins 5, 14, and 17, and do not express ER, PR, and HER2. Consequently, basal-like tumors cannot be treated with many conventional therapies, however, have a better response to chemotherapy compared to other subtypes. The EGFR is often overexpressed in basal-like tumors, where increased EGFR expression correlates with poor patient survival. Basal-like tumors display a dysregulation in PI3K/AKT, JAK/STAT, and ERK/MAP signaling pathways and a high expression of proliferative genes, such as FOXM1, c-MYC, CCNE1 BIRC5, and CCND1 (66). In addition, basal-like tumors overexpress genes involved in the progression through the cell cycle (CDC20, CDC6) (66) and genes associated with the EGFR pathway (43). Absence of ER expression results in low expression of estrogen-related genes, EsR1, Bcl2, and PgR (66).
Claudin-low tumors are enriched for epithelial-to-mesenchymal transition markers, such as SNAI1/2, TWIST1/2, and ZEB2 (72), with low expression of tight junction proteins, such as claudin 3, 4, and 7, E-cadherin, and CDH1 (72). The claudin-low subtype is highly migratory and therefore has a poor prognosis (76).
PAM50
PAM50 is a list of 50 genes that classify breast cancers into one of five intrinsic subtypes from formalin-fixed, paraffin-embedded tissues by real time polymerase chain reaction (RT-PCR) (62). These 50 genes identified were refined from a list of 1,906 genes, which were found in four previous microarray studies. The list was minimized to genes that have passed previously established formalin-fixed paraffin-embedded (FFPE) performance criteria, and were further refined through statistical analyzes, allowing for identification of genes which showed a highest correlation to each intrinsic subtype. Differential gene expression between subtypes is shown by microarray in Figure 3. Subsequent studies have shown that classification of intrinsic subtypes using the PAM50 test retains the prognostic and predictive significance characteristic to breast cancer subtypes (9, 70, 74, 81, 82). Furthermore, several studies have shown that the PAM50 classification method provides better information on prognosis than immunohistochemistry-based surrogates (8, 9, 64). This suggests that subtyping by immunohistochemistry is inferior to genomic profiling, identifying a requirement for gene-expression profiling in a clinical setting.
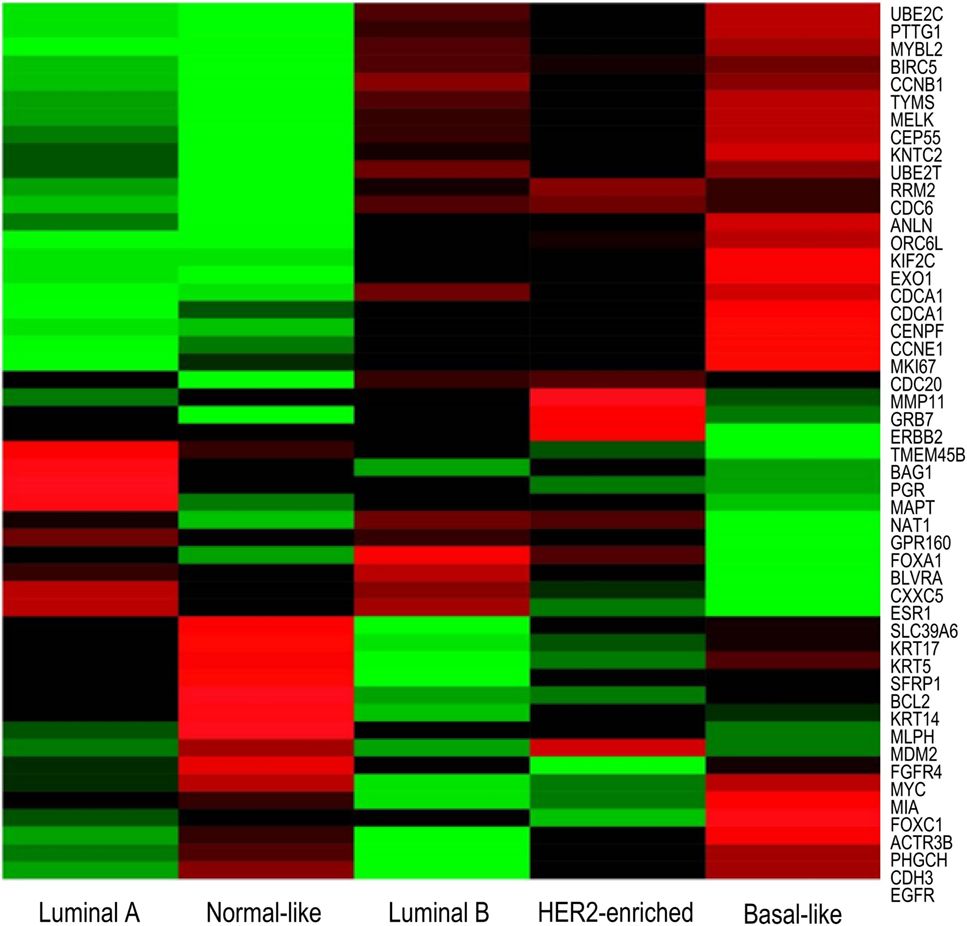
Figure 3. Microarray heatmap of PAM50 genes expression in “intrinsic” breast cancer subtypes. Molecular profiles have distinct gene expression. Expression values of genes included in the PAM50 signature are shown as red/green according to their relative expression level for each subtype. Highest gene expression (red), lowest (green), and average (black) (71).
Accurate testing of predictive biomarkers is important, as discrepancies between IHC and intrinsic classification of breast tumors may lead to differences in treatment decisions and patient outcomes. To address this, ER, PR, and HER2 concordance between immunohistochemistry and gene profiling has been investigated. A number of studies have reported a discordance of below 10% for ER and PR status (8, 9, 70, 83, 84). Cheang et al. evaluated concordance in subtype classification between PAM50 and immunohistochemistry, finding that of Luminal tumors as defined by PAM50, 8% did not stain positive for ER through immunohistochemistry (8). Consistent with this, Chia et al. identified 8% of Luminal tumors instead being classified as either HER-2-positive, or triple-negative through immunohistochemistry (9). As patients with ER negative or HER-2-positive tumors will receive chemotherapy, any discordances in classification of luminal tumors can have critical implications on treatment options, where women may receive unnecessary chemotherapy with no benefit.
Although several studies have reported high overall concordance in HER-2 expression between gene profiling and immunohistochemistry (85–88), others report low concordance (70, 81). Chia et al. (9) found that of HER-2-enriched tumors identified by PAM50, only 66% of these stained positive for HER2 expression by immunohistochemistry. Instead, 31% of HER2-enriched tumors were classified as Luminal A or B tumors, and 4% classified as basal-like through immunohistochemistry. Cheang et al. (8) also identified a low concordance of 69% in HER2 status between PAM50 and immunohistochemistry, with 6% of tumors instead classified as Luminal A tumors, and 16% as triple-negative.
As different subtypes each respond best to different treatments and the absolute benefit of adjuvant therapies depends on the risk of relapse, this poses the question; in discordant cases which result should be used to guide treatment decisions, and is it appropriate to deny a patient treatment that would otherwise be indicated by a different subtyping method? Studies by Chia et al. (9) and Cheang et al. (8) included only premenopausal women and described low concordance between immunohistochemistry and gene profiling. This low concordance may be due to the fluctuations in circulating hormones during the menstrual cycle and the relative effect of hormonal stimulation on gene versus protein expression. While it is believed that PAM50 is more reflective of the true biology of the tumor than protein-based immunohistochemistry, the paucity of data on premenopausal women makes it difficult to determine the efficacy of the PAM50 test compared to the traditional gold standard for tumor subtyping.
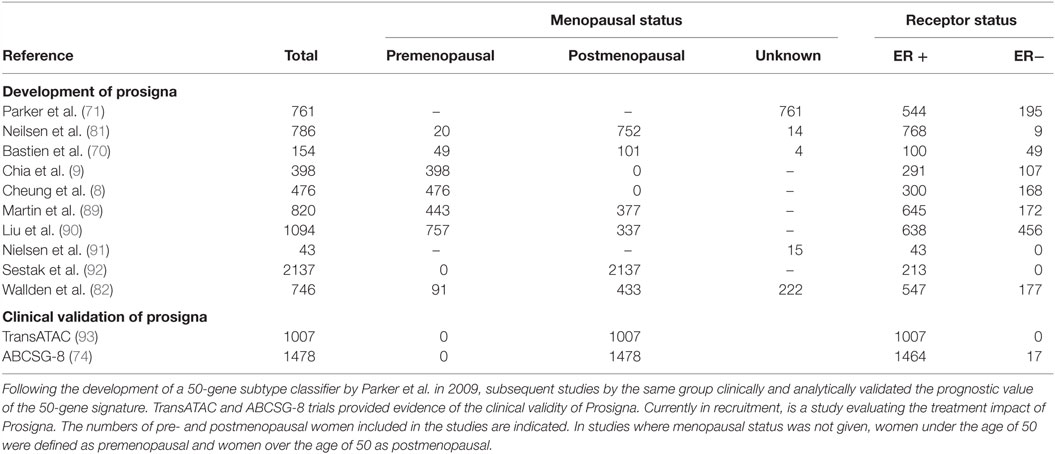
Prosigna is an in vitro diagnostic assay which is based on the PAM50 gene signature assay. The Prosigna test is performed on FFPE tissue and identifies the patient’s risk of distant reoccurrences of disease; it aims to aid clinicians and patients in treatment decisions. The development of Prosigna from PAM50 is summarized in Table 3. Of note, the validation of the Prosigna test – necessary for FDA approval – was based on two clinical trials (the TransATAC and ABCSG-8 clinical trials) incorporating data from over 2,400 postmenopausal women enrolled in adjuvant aromatase inhibitor trials. Although studies have examined the use of PAM50 in managing adjuvant therapy in premenopausal breast cancer (8, 9), there have been no large-scale clinical trials into the efficacy of the Prosigna test for premenopausal women. Thus, the accuracy of the Prosigna test has never been properly validated in the context of the hormonal fluctuations that occur during the menstrual cycle in premenopausal women.
Oncotype DX
Oncotype DX evaluates the expression of 21 genes associated with tumor proliferation, invasion, and estrogen signaling (94) (Table 4). In 2004, Paik et al. selected 250 candidate genes from published literature and genomic databases that have been shown to be correlated with disease outcome. The list of genes was reduced to 16 genes, which showed the highest correlation to distant recurrence after 10 years. Relative expression of these genes, in relation to expression of five reference genes, provide a reoccurrence score which is significantly correlated with likelihood of breast cancer reoccurrence in 10 years (94–98). Oncotype DX, therefore, impacts adjuvant treatment decisions and influences treatment recommendations. The development of Oncotype DX is summarized in Table 5. Like the PAM50 gene set, the genes used in the Oncotype DX test rely heavily on genes related to ER and growth factor signaling and proliferation, which are differentially expressed by normal breast epithelial across the menstrual cycle, as discussed above.
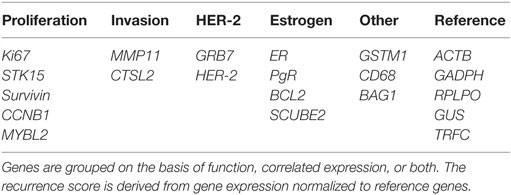
Table 4. Panel of 21 genes used in the Oncotype DX assay to determine the risk of distant recurrence.
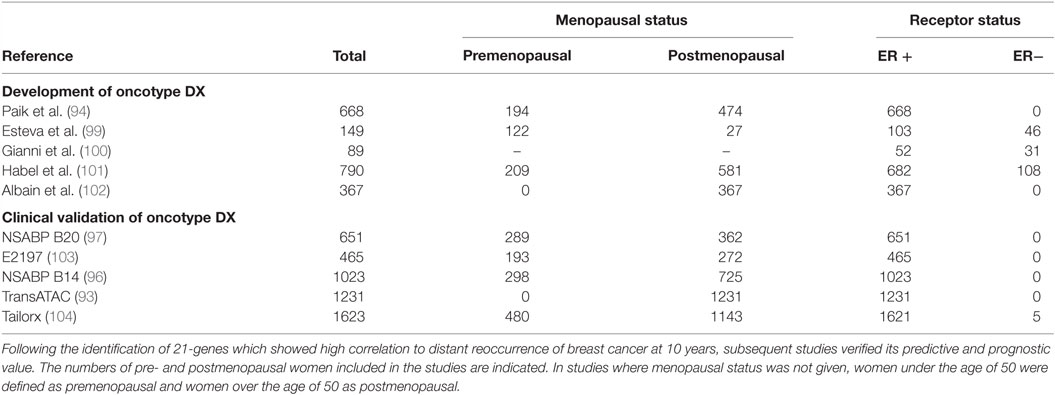
Table 5. The development of Oncotype DX, a 21-gene assay which identifies patient benefit from chemotherapy.
EndoPredict
EndoPredict is an RT-PCR-based diagnostic test which evaluates the expression of eight proliferative and hormone receptor-associated genes. In conjunction with the tumors clinicopathological factors, it identifies the risk of distant metastasis within 10 years (105). EndoPredict is used to guide treatment decisions for both chemotherapy and hormonal therapy in ER-positive, HER-2-negative breast cancer.
The EndoPredict gene signature was identified from gene expression profiles of breast cancer samples taken predominantly from postmenopausal women (105). Similar to Prosigna, initial clinical validation of EndoPredict was based on two clinical trials (ABCSG-6 and ABCSG-8 clinical trials), which incorporated data exclusively from postmenopausal women, who were enrolled in aromatase inhibitor trials (105). In 2014, results of EndoPredict prognostic validity from a third clinical study were published. This study included both pre- and postmenopausal women, 54 and 46% of patients, respectively, and suggested that EndoPredict is prognostic in both pre- and postmenopausal women with breast cancer (106). The development of EndoPredict is summarized in Table 6. There are a lack of studies which validate the efficacy of EndoPredict in premenopausal women, and whether EndoPredict is an appropriate tool for guiding treatment decisions in premenopausal women has not yet been sufficiently investigated.
MammaPrint
MammaPrint is a diagnostic test which evaluates the expression of 70 genes associated with metastasis, proliferation, invasion, survival, and angiogenesis (113). The list of 70 genes was identified from whole-genome expression arrays, and selected for on the basis of those which were significantly correlated with disease outcome (113). Interestingly, MammaPrint does not measure expression of commonly used diagnostic markers ER, PR, or HER2.
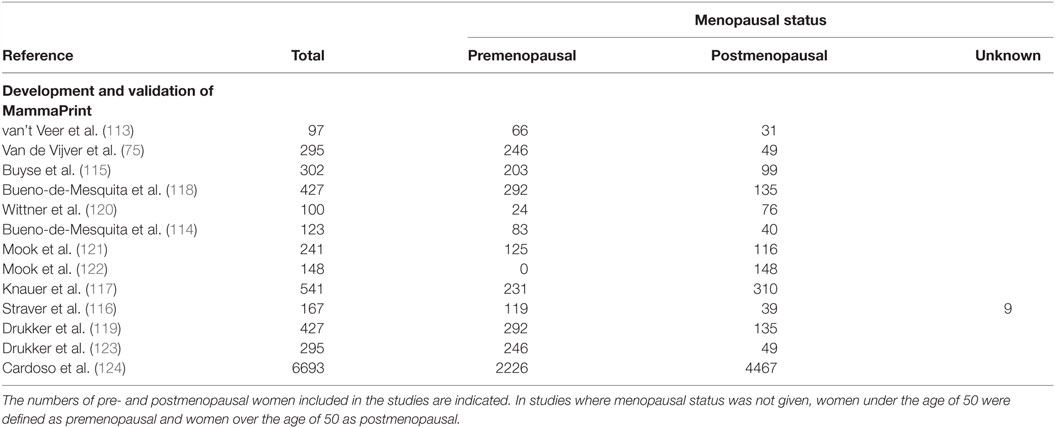
Through the relative expression of these 70 genes, MammaPrint classifies tumors into high or low risk groups, which corresponds with patient’s clinical outcome. Studies have shown that risk groups identified by MammaPrint correspond with patients overall survival (114), disease-free metastasis (75, 115), and the benefit from adjuvant chemotherapy (116, 117). Most studies validating the diagnostic capabilities of MammaPrint were small-scale retrospective studies, which included both pre and postmenopausal women, as summarized in Table 7. MammaPrint had initially been developed and validated in patients under the age of 55, suggesting that MammaPrint is targeted toward the younger population. Furthermore, the first prospective study which evaluated the impact of MammaPrint in assisting with treatment decisions included patients under the age of 55 (118, 119). As MammaPrint was developed in a younger population, and does not measure the gene expression of ER or PR, it is likely that MammaPrint is appropriate for diagnosing breast cancer in premenopausal women.
In 2016, results from a second prospective study were published, comparing MammaPrint to clinicopathological tools for selecting patients for adjuvant chemotherapy (124). The median age of the patients was 55 years. The study found that approximately 46% of patients, who were classified as high risk by clinicopathological features, were also classified as a low risk of metastasis by MammaPrint. Although these tumors presented with a high clinical risk, the results from the study suggested that these patients received no significant benefit from chemotherapy. Therefore, the authors concluded that using MammaPrint to guide treatment can reduce the number of patients receiving unnecessary chemotherapy.
Breast Cancer Index
The Breast Cancer Index is an RT-PCR based assay which classifies patients into risk groups to predict the likelihood of benefit from endocrine therapy, and the risk of early or late recurrence (125–128). The Breast Cancer Index evaluates two independent biomarkers; the HOXB13:IL17BR gene ratio, which is associated with endocrine therapy response (129), and the molecular grade index, which is determined by the expression of five proliferative-related genes (130). Classification of breast cancer through the expression of these seven genes aims to identify patients which are most likely to benefit from adjuvant therapy. The development of the Breast Cancer Index is summarized in Table 8. Similar to Prosigna, the clinical validation of the Breast Cancer Index was based on retrospective studies which used samples exclusively from postmenopausal women and, therefore, results cannot be generalized to premenopausal women.
Hormonal Modulation of Breast Cancer Biomarkers
The abundance of ER fluctuates in normal breast tissue during the menstrual cycle, with ER positivity higher during the follicular phase, compared to the luteal phase (136, 137). Similarly, ER positivity in breast cancers is significantly higher in the follicular phase, when progesterone is absent, compared to the luteal phase (138, 139). This suggests that hormonal fluctuations during the menstrual cycle alter expression of hormone receptors and are highly likely to affect expression of genes associated with hormone signaling.
If menstrual cycle stage affects hormonal receptor signaling and the expression of genes and proteins used in subtype diagnosis, it is expected that there would be discrepancies between diagnostic and surgical breast cancer samples, which would be taken from the woman at different times and, therefore, different stages of the menstrual cycle. However, studies on concordance between biopsy and surgical samples have not specifically investigated premenopausal women. In 2013, Dekker et al. assessed the concordance of ER and HER2 expression between core needle biopsy and surgical resections (140). A concordance of ER was found in 99.1% of patients, and in 96.4% of cases for HER2. The menopausal status of women was not specified in the study; however, the mean age of women enrolled was 63 years old, which suggests that a majority of patients were postmenopausal. In a pooled study of 2507 invasive breast tumors, predominantly from postmenopausal women, concordance of ER was found for 93.4% of patients, and 97.8% for HER2 (140). This high concordance in receptor status led the authors to conclude that ER and HER2 status can be reliably determined from the core needle biopsy. However, these studies focused primarily on postmenopausal women, and the effect of premenopausal factors was not investigated.
In addition to studies investigating concordance in traditional biomarkers, several studies have evaluated the gene expression changes between core biopsies and surgical excisions (141–143). In 2012, Riis et al. compared whole gene-expression profiles of 13 women (1/13 premenopausal; 12/13 postmenopausal) and identified 228 genes differentially expressed, a majority of which are immunoregulatory or stress related (142). Two genes from the PAM50 signature gene list had differential expression between samples; GRB7 and ACTR3B. From the Oncotype DX test, only one gene showed differential expression, GRB7. Additionally, Jeselsohn et al. compared gene expression between core biopsies and surgical excisions in postmenopausal women with ER-positive breast cancer (141). The authors identified significant changes in the expression level of 14 genes, a majority of which are immunoregulatory. Two genes involved in the Oncotype DX test and PAM50 intrinsic classification, MYC and CCNB1, showed differential expression between core biopsies and surgical excisions. A recent study utilized a genome-wide approach to determine gene expression changes between core biopsies and surgical excisions. The authors collected 56 paired core-cuts from postmenopausal breast cancer patients, and classified tumors into one of the five intrinsic subtypes based on the PAM50 gene signature (143). No systematic differences in categorization of the tumors into intrinsic subtypes were identified; however, discordances were identified between the Luminal A versus Luminal B subtype. While these studies have generally found good concordance between diagnostic and surgical samples, concordance in premenopausal women has not been specifically investigated.
A change in biomarker status can have important clinical consequences for adjuvant treatment. Studies evaluating the concordance of hormone receptors and gene expression profiles between core biopsies and surgical excisions suggest that biopsies taken at diagnosis are representative of the whole tumor. However, these studies were performed predominantly again in postmenopausal women, and it is possible that hormonal fluctuations in premenopausal women may alter the expression of these biomarkers. As such, the biopsy taken at diagnosis or during surgery may be influenced by fluctuating concentrations of estrogen and progesterone and, therefore, not represent the true tumor profile. This can lead to an incorrect diagnosis and risk prediction and sub-optimal treatment of premenopausal women.
A recent study compared the expression of estrogen-related genes between pre- and postmenopausal women with ER-positive breast cancer (144). It was suggested that the different hormonal environments of pre- and postmenopausal women may affect the biological characteristics of the breast tumor. The authors found that expression of estrogen-related genes PgR, TFF1, and GATA3, were significantly higher in premenopausal women compared to in postmenopausal women. Consistent with this, studies have also shown that expression of estrogen-regulated genes is significantly associated with the level of estrogen in the blood (145, 146). It is likely that the fluctuating concentration of estrogen during the menstrual cycle affects the expression of these estrogen-related genes. In 2013, Haynes et al. compared the expression of estrogen-related genes between women at different stages of the menstrual cycle (145). They found that the expression of key estrogen-related genes was highest during the follicular phase of the menstrual cycle when estrogen concentration peaks. However, it remains unknown how menstrual cycling can affect the expression of these genes within the same tumor. As Oncotype DX and Prosigna rely heavily on the expression of estrogen-related genes for diagnosing breast cancer, changes in expression of these genes across the menstrual cycle may affect the diagnosis of breast cancer by these tests.
Despite the known role of estrogen and progesterone on the function of the breast and on breast cancer risk, the effect of menstrual cycling on breast tumors remains unknown. In support of the possibility that menstrual cycle critically affects the gene-expression profile of breast cancers, a recent in vitro study has suggested that the combination of estrogen and progesterone results in the switching from a Luminal A to Basal-like intrinsic subtype in breast cancer cells, and increases the Oncotype DX Recurrence Score (43) compared to estrogen treatment alone. Tests that utilize gene expression profiling in breast cancer classification were developed and validated from studies predominantly in postmenopausal women, and there is a scarcity of research on how applicable these biomarkers are to premenopausal women, and the extent to which this impacts on treatment response. It is important to understand how hormonal fluctuations affect predictive and prognostic biomarkers, to provide premenopausal women with the optimal treatment for their individual cancer.
Conclusion
Breast cancer clinics are increasingly adopting gene expression profiling to subtype tumors and identify the best therapies. However, despite their availability to young women, such tests were largely developed and validated in postmenopausal women – patients in whom fluctuations in estrogen and progesterone associated with the menstrual cycle are absent. Yet, these hormones are highly likely to affect breast cancer gene expression in premenopausal women – and the diagnosis and treatment trajectories that stem from its measurement – could fundamentally depend on a patient’s menstrual cycle stage at the time of tissue sampling. Leading diagnostic tests harness intrinsic subtyping of breast cancers, but whether these tests are accurate for premenopausal women remains a startlingly open question. Quite simply, young women may be at risk of receiving inaccurate subtype diagnoses; with ramifications spanning inaccurate prognoses, suboptimal and unnecessary treatments, and reduced survival.
Author Contributions
All authors contributed intellectually to the development and writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by the Hospital Research Foundation (THRF), the Australian National Breast Cancer Foundation (NBCF), and TQEH Haem/Onc Scheme A. WI is an NBCF/THRF Fellow.
References
3. Brennan M, French J, Houssami N, Kirk J, Boyages J. Breast cancer in young women. Aust Fam Physician (2005) 34(10):851–5.
4. Fata JE, Chaudhary V, Khokha R. Cellular turnover in the mammary gland is correlated with systemic levels of progesterone and not 17beta-estradiol during the estrous cycle. Biol Reprod (2001) 65(3):680–8. doi:10.1095/biolreprod65.3.680
5. Ramakrishnan R, Khan SA, Badve S. Morphological changes in breast tissue with menstrual cycle. Mod Pathol (2002) 15(12):1348–56. doi:10.1097/01.MP.0000039566.20817.46
6. Chavez-MacGregor M, van Gils CH, van der Schouw YT, Monninkhof E, van Noord PA, Peeters PH. Lifetime cumulative number of menstrual cycles and serum sex hormone levels in postmenopausal women. Breast Cancer Res Treat (2008) 108(1):101–12. doi:10.1007/s10549-007-9574-z
7. Collaborative Group on Hormonal Factors in Breast Cancer. Menarche, menopause, and breast cancer risk: individual participant meta-analysis, including 118 964 women with breast cancer from 117 epidemiological studies. Lancet Oncol (2012) 13(11):1141–51. doi:10.1016/S1470-2045(12)70425-4
8. Cheang MC, Voduc KD, Tu D, Jiang S, Leung S, Chia SK, et al. Responsiveness of intrinsic subtypes to adjuvant anthracycline substitution in the NCIC.CTG MA.5 randomized trial. Clin Cancer Res (2012) 18(8):2402–12. doi:10.1158/1078-0432.CCR-11-2956
9. Chia SK, Bramwell VH, Tu D, Shepherd LE, Jiang S, Vickery T, et al. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin Cancer Res (2012) 18(16):4465–72. doi:10.1158/1078-0432.CCR-12-0286
10. Andres AC, Strange R. Apoptosis in the estrous and menstrual cycles. J Mammary Gland Biol Neoplasia (1999) 4(2):221–8. doi:10.1023/A:1018737510695
11. Need EF, Atashgaran V, Ingman WV, Dasari P. Hormonal regulation of the immune microenvironment in the mammary gland. J Mammary Gland Biol Neoplasia (2014) 19(2):229–39. doi:10.1007/s10911-014-9324-x
12. Ferguson JE, Schor AM, Howell A, Ferguson MW. Changes in the extracellular matrix of the normal human breast during the menstrual cycle. Cell Tissue Res (1992) 268(1):167–77. doi:10.1007/BF00338066
13. Navarrete MA, Maier CM, Falzoni R, Quadros LG, Lima GR, Baracat EC, et al. Assessment of the proliferative, apoptotic and cellular renovation indices of the human mammary epithelium during the follicular and luteal phases of the menstrual cycle. Breast Cancer Res (2005) 7(3):R306–13. doi:10.1186/bcr994
14. Vogel PM, Georgiade NG, Fetter BF, Vogel FS, McCarty KS Jr. The correlation of histologic changes in the human breast with the menstrual cycle. Am J Pathol (1981) 104(1):23–34.
15. Giovannelli P, Di Donato M, Giraldi T, Migliaccio A, Castoria G, Auricchio F. Targeting rapid action of sex-steroid receptors in breast and prostate cancers. Front Biosci (Elite Ed) (2012) 4:453–61. doi:10.2741/e390
16. Ferguson DJ, Anderson TJ. Morphological evaluation of cell turnover in relation to the menstrual cycle in the “resting” human breast. Br J Cancer (1981) 44(2):177–81. doi:10.1038/bjc.1981.168
17. Watts CK, Sweeney KJ, Warlters A, Musgrove EA, Sutherland RL. Antiestrogen regulation of cell cycle progression and cyclin D1 gene expression in MCF-7 human breast cancer cells. Breast Cancer Res Treat (1994) 31(1):95–105. doi:10.1007/BF00689680
18. Dubik D, Dembinski TC, Shiu RP. Stimulation of c-myc oncogene expression associated with estrogen-induced proliferation of human breast cancer cells. Cancer Res (1987) 47(24 Pt 1):6517–21.
19. Dubik D, Shiu RP. Transcriptional regulation of c-myc oncogene expression by estrogen in hormone-responsive human breast cancer cells. J Biol Chem (1988) 263(25):12705–8.
20. Planas-Silva MD, Weinberg RA. Estrogen-dependent cyclin E-cdk2 activation through p21 redistribution. Mol Cell Biol (1997) 17(7):4059–69. doi:10.1128/MCB.17.7.4059
21. Foster JS, Henley DC, Bukovsky A, Seth P, Wimalasena J. Multifaceted regulation of cell cycle progression by estrogen: regulation of Cdk inhibitors and Cdc25A independent of cyclin D1-Cdk4 function. Mol Cell Biol (2001) 21(3):794–810. doi:10.1128/MCB.21.3.794-810.2001
22. Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li YQ, et al. Estrogen receptor alpha mediates proliferation of breast cancer MCF-7 cells via a p21/PCNA/E2F1-dependent pathway. FEBS J (2014) 281(3):927–42. doi:10.1111/febs.12658
23. Kandouz M, Lombet A, Perrot JY, Jacob D, Carvajal S, Kazem A, et al. Proapoptotic effects of antiestrogens, progestins and androgen in breast cancer cells. J Steroid Biochem Mol Biol (1999) 69(1–6):463–71. doi:10.1016/S0960-0760(99)00069-2
24. Martin LA, Dowsett M. BCL-2: a new therapeutic target in estrogen receptor-positive breast cancer? Cancer Cell (2013) 24(1):7–9. doi:10.1016/j.ccr.2013.06.006
25. Musgrove EA, Hamilton JA, Lee CS, Sweeney KJ, Watts CK, Sutherland RL. Growth factor, steroid, and steroid antagonist regulation of cyclin gene expression associated with changes in T-47D human breast cancer cell cycle progression. Mol Cell Biol (1993) 13(6):3577–87. doi:10.1128/MCB.13.6.3577
26. Musgrove EA, Lee CS, Sutherland RL. Progestins both stimulate and inhibit breast cancer cell cycle progression while increasing expression of transforming growth factor alpha, epidermal growth factor receptor, c-fos, and c-myc genes. Mol Cell Biol (1991) 11(10):5032–43. doi:10.1128/MCB.11.10.5032
27. Wong MS, Murphy LC. Differential regulation of c-myc by progestins and antiestrogens in T-47D human breast cancer cells. J Steroid Biochem Mol Biol (1991) 39(1):39–44. doi:10.1016/0960-0760(91)90010-3
28. Musgrove EA, Swarbrick A, Lee CS, Cornish AL, Sutherland RL. Mechanisms of cyclin-dependent kinase inactivation by progestins. Mol Cell Biol (1998) 18(4):1812–25. doi:10.1128/MCB.18.4.1812
29. Hurd C, Khattree N, Alban P, Nag K, Jhanwar SC, Dinda S, et al. Hormonal regulation of the p53 tumor suppressor protein in T47D human breast carcinoma cell line. J Biol Chem (1995) 270(48):28507–10. doi:10.1074/jbc.270.48.28507
30. Vazquez F, Rodriguez-Manzaneque JC, Lydon JP, Edwards DP, O’Malley BW, Iruela-Arispe ML. Progesterone regulates proliferation of endothelial cells. J Biol Chem (1999) 274(4):2185–92. doi:10.1074/jbc.274.4.2185
31. Selam B, Kayisli UA, Mulayim N, Arici A. Regulation of Fas ligand expression by estradiol and progesterone in human endometrium. Biol Reprod (2001) 65(4):979–85. doi:10.1095/biolreprod65.4.979
32. Beleut M, Rajaram RD, Caikovski M, Ayyanan A, Germano D, Choi Y, et al. Two distinct mechanisms underlie progesterone-induced proliferation in the mammary gland. Proc Natl Acad Sci U S A (2010) 107(7):2989–94. doi:10.1073/pnas.0915148107
33. Mukherjee A, Soyal SM, Li J, Ying Y, He B, DeMayo FJ, et al. Targeting RANKL to a specific subset of murine mammary epithelial cells induces ordered branching morphogenesis and alveologenesis in the absence of progesterone receptor expression. FASEB J (2010) 24(11):4408–19. doi:10.1096/fj.10-157982
34. Fata JE, Kong YY, Li J, Sasaki T, Irie-Sasaki J, Moorehead RA, et al. The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell (2000) 103(1):41–50. doi:10.1016/S0092-8674(00)00103-3
35. Fernandez-Valdivia R, Mukherjee A, Ying Y, Li J, Paquet M, DeMayo FJ, et al. The RANKL signaling axis is sufficient to elicit ductal side-branching and alveologenesis in the mammary gland of the virgin mouse. Dev Biol (2009) 328(1):127–39. doi:10.1016/j.ydbio.2009.01.019
36. Gonzalez-Suarez E, Branstetter D, Armstrong A, Dinh H, Blumberg H, Dougall WC. RANK overexpression in transgenic mice with mouse mammary tumor virus promoter-controlled RANK increases proliferation and impairs alveolar differentiation in the mammary epithelia and disrupts lumen formation in cultured epithelial acini. Mol Cell Biol (2007) 27(4):1442–54. doi:10.1128/MCB.01298-06
37. Brisken C, Heineman A, Chavarria T, Elenbaas B, Tan J, Dey SK, et al. Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev (2000) 14(6):650–4. doi:10.1101/gad.14.6.650
38. Clarke RB, Howell A, Potten CS, Anderson E. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res (1997) 57(22):4987–91.
39. Graham JD, Clarke CL. Physiological action of progesterone in target tissues. Endocr Rev (1997) 18(4):502–19. doi:10.1210/edrv.18.4.0308
40. Murphy LC, Murphy LJ, Dubik D, Bell GI, Shiu RP. Epidermal growth factor gene expression in human breast cancer cells: regulation of expression by progestins. Cancer Res (1988) 48(16):4555–60.
41. Carvajal A, Espinoza N, Kato S, Pinto M, Sadarangani A, Monso C, et al. Progesterone pre-treatment potentiates EGF pathway signaling in the breast cancer cell line ZR-75. Breast Cancer Res Treat (2005) 94(2):171–83. doi:10.1007/s10549-005-7726-6
42. Murphy LJ, Sutherland RL, Stead B, Murphy LC, Lazarus L. Progestin regulation of epidermal growth factor receptor in human mammary carcinoma cells. Cancer Res (1986) 46(2):728–34.
43. Need EF, Selth LA, Trotta AP, Leach DA, Giorgio L, O’Loughlin MA, et al. The unique transcriptional response produced by concurrent estrogen and progesterone treatment in breast cancer cells results in upregulation of growth factor pathways and switching from a Luminal A to a Basal-like subtype. BMC Cancer (2015) 15:791. doi:10.1186/s12885-015-1819-3
44. Troyer KL, Lee DC. Regulation of mouse mammary gland development and tumorigenesis by the ERBB signaling network. J Mammary Gland Biol Neoplasia (2001) 6(1):7–21. doi:10.1023/A:1009560330359
45. Tanos T, Rojo L, Echeverria P, Brisken C. ER and PR signaling nodes during mammary gland development. Breast Cancer Res (2012) 14(4):210. doi:10.1186/bcr3166
46. Dickson RB, Huff KK, Spencer EM, Lippman ME. Induction of epidermal growth factor-related polypeptides by 17 beta-estradiol in MCF-7 human breast cancer cells. Endocrinology (1986) 118(1):138–42. doi:10.1210/endo-118-1-138
47. Bates SE, Davidson NE, Valverius EM, Freter CE, Dickson RB, Tam JP, et al. Expression of transforming growth factor alpha and its messenger ribonucleic acid in human breast cancer: its regulation by estrogen and its possible functional significance. Mol Endocrinol (1988) 2(6):543–55. doi:10.1210/mend-2-6-543
48. Ciarloni L, Mallepell S, Brisken C. Amphiregulin is an essential mediator of estrogen receptor alpha function in mammary gland development. Proc Natl Acad Sci U S A (2007) 104(13):5455–60. doi:10.1073/pnas.0611647104
49. Eigeliene N, Elo T, Linhala M, Hurme S, Erkkola R, Harkonen P. Androgens inhibit the stimulatory action of 17beta-estradiol on normal human breast tissue in explant cultures. J Clin Endocrinol Metab (2012) 97(7):E1116–27. doi:10.1210/jc.2011-3228
50. Kariagina A, Xie J, Leipprandt JR, Haslam SZ. Amphiregulin mediates estrogen, progesterone, and EGFR signaling in the normal rat mammary gland and in hormone-dependent rat mammary cancers. Horm Cancer (2010) 1(5):229–44. doi:10.1007/s12672-010-0048-0
51. Applanat MP, Buteau-Lozano H, Herve MA, Corpet A. Vascular endothelial growth factor is a target gene for estrogen receptor and contributes to breast cancer progression. Adv Exp Med Biol (2008) 617:437–44. doi:10.1007/978-0-387-69080-3_42
52. Dabrosin C, Margetts PJ, Gauldie J. Estradiol increases extracellular levels of vascular endothelial growth factor in vivo in murine mammary cancer. Int J Cancer (2003) 107(4):535–40. doi:10.1002/ijc.11398
53. Nicholson RI, McClelland RA, Finlay P, Eaton CL, Gullick WJ, Dixon AR, et al. Relationship between EGF-R, c-erbB-2 protein expression and Ki67 immunostaining in breast cancer and hormone sensitivity. Eur J Cancer (1993) 29A(7):1018–23. doi:10.1016/S0959-8049(05)80215-1
54. Nicholson RI, McClelland RA, Gee JM, Manning DL, Cannon P, Robertson JF, et al. Epidermal growth factor receptor expression in breast cancer: association with response to endocrine therapy. Breast Cancer Res Treat (1994) 29(1):117–25. doi:10.1007/BF00666187
55. Rimawi MF, Shetty PB, Weiss HL, Schiff R, Osborne CK, Chamness GC, et al. Epidermal growth factor receptor expression in breast cancer association with biologic phenotype and clinical outcomes. Cancer (2010) 116(5):1234–42. doi:10.1002/cncr.24816
56. Changavi AA, Shashikala A, Ramji AS. Epidermal growth factor receptor expression in triple negative and nontriple negative breast carcinomas. J Lab Physicians (2015) 7(2):79–83. doi:10.4103/0974-2727.163129
57. Early Breast Cancer Trialists’ Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet (2005) 365(9472):1687–717. doi:10.1016/S0140-6736(05)66544-0
58. Early Breast Cancer Trialists’ Collaborative Group, Peto R, Davies C, Godwin J, Gray R, Pan HC, et al. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet (2012) 379(9814):432–44. doi:10.1016/S0140-6736(11)61625-5
59. Fisher B, Dignam J, Wolmark N, DeCillis A, Emir B, Wickerham DL, et al. Tamoxifen and chemotherapy for lymph node-negative, estrogen receptor-positive breast cancer. J Natl Cancer Inst (1997) 89(22):1673–82. doi:10.1093/jnci/89.22.1673
60. Griggs JJ, Hawley ST, Graff JJ, Hamilton AS, Jagsi R, Janz NK, et al. Factors associated with receipt of breast cancer adjuvant chemotherapy in a diverse population-based sample. J Clin Oncol (2012) 30(25):3058–64. doi:10.1200/JCO.2012.41.9564
61. Oestreicher N, Ramsey SD, Linden HM, McCune JS, van’t Veer LJ, Burke W, et al. Gene expression profiling and breast cancer care: what are the potential benefits and policy implications? Genet Med (2005) 7(6):380–9. doi:10.1097/01.GIM.0000170776.31248.75
62. Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature (2000) 406(6797):747–52. doi:10.1038/35021093
63. Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A (2001) 98(19):10869–74. doi:10.1073/pnas.191367098
64. Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A (2003) 100(14):8418–23. doi:10.1073/pnas.0932692100
65. Yersal O, Barutca S. Biological subtypes of breast cancer: prognostic and therapeutic implications. World J Clin Oncol (2014) 5(3):412–24. doi:10.5306/wjco.v5.i3.412
66. Eroles P, Bosch A, Perez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev (2012) 38(6):698–707. doi:10.1016/j.ctrv.2011.11.005
67. Prat A, Fan C, Fernandez A, Hoadley KA, Martinello R, Vidal M, et al. Response and survival of breast cancer intrinsic subtypes following multi-agent neoadjuvant chemotherapy. BMC Med (2015) 13:303. doi:10.1186/s12916-015-0540-z
68. Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol (2012) 9(1):16–32. doi:10.1038/nrclinonc.2011.177
69. Heitz F, Harter P, Lueck HJ, Fissler-Eckhoff A, Lorenz-Salehi F, Scheil-Bertram S, et al. Triple-negative and HER2-overexpressing breast cancers exhibit an elevated risk and an earlier occurrence of cerebral metastases. Eur J Cancer (2009) 45(16):2792–8. doi:10.1016/j.ejca.2009.06.027
70. Bastien RR, Rodriguez-Lescure A, Ebbert MT, Prat A, Munarriz B, Rowe L, et al. PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC Med Genomics (2012) 5:44. doi:10.1186/1755-8794-5-44
71. Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol (2009) 27(8):1160–7. doi:10.1200/JCO.2008.18.1370
72. Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res (2010) 12(5):R68. doi:10.1186/bcr2635
73. Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, et al. Tailoring therapies – improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol (2015) 26(8):1533–46. doi:10.1093/annonc/mdv221
74. Gnant M, Filipits M, Greil R, Stoeger H, Rudas M, Bago-Horvath Z, et al. Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: using the PAM50 Risk of Recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Ann Oncol (2014) 25(2):339–45. doi:10.1093/annonc/mdt494
75. van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AAM, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med (2002) 347(25):1999–2009. doi:10.1056/NEJMoa021967
76. Sabatier R, Finetti P, Guille A, Adelaide J, Chaffanet M, Viens P, et al. Claudin-low breast cancers: clinical, pathological, molecular and prognostic characterization. Mol Cancer (2014) 13:228. doi:10.1186/1476-4598-13-228
77. Hugh J, Hanson J, Cheang MC, Nielsen TO, Perou CM, Dumontet C, et al. Breast cancer subtypes and response to docetaxel in node-positive breast cancer: use of an immunohistochemical definition in the BCIRG 001 trial. J Clin Oncol (2009) 27(8):1168–76. doi:10.1200/JCO.2008.18.1024
78. Carey LA. Through a glass darkly: advances in understanding breast cancer biology, 2000–2010. Clin Breast Cancer (2010) 10(3):188–95. doi:10.3816/CBC.2010.n.026
79. Creighton CJ. The molecular profile of luminal B breast cancer. Biologics (2012) 6:289–97. doi:10.2147/BTT.S29923
80. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature (2012) 490(7418):61–70. doi:10.1038/nature11412
81. Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res (2010) 16(21):5222–32. doi:10.1158/1078-0432.CCR-10-1282
82. Wallden B, Storhoff J, Nielsen T, Dowidar N, Schaper C, Ferree S, et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics (2015) 8:54. doi:10.1186/s12920-015-0129-6
83. Badve SS, Baehner FL, Gray RP, Childs BH, Maddala T, Liu ML, et al. Estrogen- and progesterone-receptor status in ECOG 2197: comparison of immunohistochemistry by local and central laboratories and quantitative reverse transcription polymerase chain reaction by central laboratory. J Clin Oncol (2008) 26(15):2473–81. doi:10.1200/JCO.2007.13.6424
84. Kraus JA, Dabbs DJ, Beriwal S, Bhargava R. Semi-quantitative immunohistochemical assay versus oncotype DX((R)) qRT-PCR assay for estrogen and progesterone receptors: an independent quality assurance study. Mod Pathol (2012) 25(6):869–76. doi:10.1038/modpathol.2011.219
85. Park MM, Ebel JJ, Zhao W, Zynger DL. ER and PR immunohistochemistry and HER2 FISH versus oncotype DX: implications for breast cancer treatment. Breast J (2014) 20(1):37–45. doi:10.1111/tbj.12223
86. Dabbs DJ, Klein ME, Mohsin SK, Tubbs RR, Shuai Y, Bhargava R. High false-negative rate of HER2 quantitative reverse transcription polymerase chain reaction of the Oncotype DX test: an independent quality assurance study. J Clin Oncol (2011) 29(32):4279–85. doi:10.1200/JCO.2011.34.7963
87. Dvorak L, Dolan M, Fink J, Varghese L, Henriksen J, Gulbahce HE. Correlation between HER2 determined by fluorescence in situ hybridization and reverse transcription-polymerase chain reaction of the oncotype DX test. Appl Immunohistochem Mol Morphol (2013) 21(3):196–9. doi:10.1097/PAI.0b013e3182632ff5
88. Baehner FL, Achacoso N, Maddala T, Shak S, Quesenberry CP Jr, Goldstein LC, et al. Human epidermal growth factor receptor 2 assessment in a case-control study: comparison of fluorescence in situ hybridization and quantitative reverse transcription polymerase chain reaction performed by central laboratories. J Clin Oncol (2010) 28(28):4300–6. doi:10.1200/JCO.2009.24.8211
89. Martin M, Prat A, Rodriguez-Lescure A, Caballero R, Ebbert MT, Munarriz B, et al. PAM50 proliferation score as a predictor of weekly paclitaxel benefit in breast cancer. Breast Cancer Res Treat (2013) 138(2):457–66. doi:10.1007/s10549-013-2416-2
90. Liu S, Chapman JA, Burnell MJ, Levine MN, Pritchard KI, Whelan TJ, et al. Prognostic and predictive investigation of PAM50 intrinsic subtypes in the NCIC CTG MA.21 phase III chemotherapy trial. Breast Cancer Res Treat (2015) 149(2):439–48. doi:10.1007/s10549-014-3259-1
91. Nielsen T, Wallden B, Schaper C, Ferree S, Liu S, Gao D, et al. Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer (2014) 14:177. doi:10.1186/1471-2407-14-177
92. Sestak I, Cuzick J, Dowsett M, Lopez-Knowles E, Filipits M, Dubsky P, et al. Prediction of late distant recurrence after 5 years of endocrine treatment: a combined analysis of patients from the Austrian breast and colorectal cancer study group 8 and arimidex, tamoxifen alone or in combination randomized trials using the PAM50 risk of recurrence score. J Clin Oncol (2015) 33(8):916–22. doi:10.1200/JCO.2014.55.6894
93. Dowsett M, Sestak I, Lopez-Knowles E, Sidhu K, Dunbier AK, Cowens JW, et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol (2013) 31(22):2783–90. doi:10.1200/JCO.2012.46.1558
94. Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med (2004) 351(27):2817–26. doi:10.1056/NEJMoa041588
95. de Boer RH, Baker C, Speakman D, Chao CY, Yoshizawa C, Mann GB. The impact of a genomic assay (Oncotype DX) on adjuvant treatment recommendations in early breast cancer. Med J Aust (2013) 199(3):205–8. doi:10.5694/mja12.11334
96. Mamounas EP, Tang G, Fisher B, Paik S, Shak S, Costantino JP, et al. Association between the 21-gene recurrence score assay and risk of locoregional recurrence in node-negative, estrogen receptor-positive breast cancer: results from NSABP B-14 and NSABP B-20. J Clin Oncol (2010) 28(10):1677–83. doi:10.1200/JCO.2009.23.7610
97. Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol (2006) 24(23):3726–34. doi:10.1200/JCO.2005.04.7985
98. Tang G, Shak S, Paik S, Anderson SJ, Costantino JP, Geyer CE Jr, et al. Comparison of the prognostic and predictive utilities of the 21-gene Recurrence Score assay and Adjuvant! for women with node-negative, ER-positive breast cancer: results from NSABP B-14 and NSABP B-20. Breast Cancer Res Treat (2011) 127(1):133–42. doi:10.1007/s10549-010-1331-z
99. Esteva FJ, Sahin AA, Cristofanilli M, Coombes K, Lee SJ, Baker J, et al. Prognostic role of a multigene reverse transcriptase-PCR assay in patients with node-negative breast cancer not receiving adjuvant systemic therapy. Clin Cancer Res (2005) 11(9):3315–9. doi:10.1158/1078-0432.CCR-04-1707
100. Gianni L, Zambetti M, Clark K, Baker J, Cronin M, Wu J, et al. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J Clin Oncol (2005) 23(29):7265–77. doi:10.1200/JCO.2005.02.0818
101. Habel LA, Shak S, Jacobs MK, Capra A, Alexander C, Pho M, et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res (2006) 8(3):R25. doi:10.1186/bcr1412
102. Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol (2010) 11(1):55–65. doi:10.1016/S1470-2045(09)70314-6
103. Goldstein LJ, Gray R, Badve S, Childs BH, Yoshizawa C, Rowley S, et al. Prognostic utility of the 21-gene assay in hormone receptor-positive operable breast cancer compared with classical clinicopathologic features. J Clin Oncol (2008) 26(25):4063–71. doi:10.1200/JCO.2007.14.4501
104. Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Prospective validation of a 21-gene expression assay in breast cancer. N Engl J Med (2015) 373(21):2005–14. doi:10.1056/NEJMoa1510764
105. Filipits M, Rudas M, Jakesz R, Dubsky P, Fitzal F, Singer CF, et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin Cancer Res (2011) 17(18):6012–20. doi:10.1158/1078-0432.CCR-11-0926
106. Martin M, Brase JC, Calvo L, Krappmann K, Ruiz-Borrego M, Fisch K, et al. Clinical validation of the EndoPredict test in node-positive, chemotherapy-treated ER+/HER2- breast cancer patients: results from the GEICAM 9906 trial. Breast Cancer Res (2014) 16(2):R38. doi:10.1186/bcr3642
107. Muller BM, Brase JC, Haufe F, Weber KE, Budzies J, Petry C, et al. Comparison of the RNA-based EndoPredict multigene test between core biopsies and corresponding surgical breast cancer sections. J Clin Pathol (2012) 65(7):660–2. doi:10.1136/jclinpath-2012-200716
108. Dubsky P, Filipits M, Jakesz R, Rudas M, Singer CF, Greil R, et al. EndoPredict improves the prognostic classification derived from common clinical guidelines in ER-positive, HER2-negative early breast cancer. Ann Oncol (2013) 24(3):640–7. doi:10.1093/annonc/mds334
109. Dubsky P, Brase JC, Jakesz R, Rudas M, Singer CF, Greil R, et al. The EndoPredict score provides prognostic information on late distant metastases in ER+/HER2- breast cancer patients. Br J Cancer (2013) 109(12):2959–64. doi:10.1038/bjc.2013.671
110. Muller BM, Keil E, Lehmann A, Winzer KJ, Richter-Ehrenstein C, Prinzler J, et al. The EndoPredict gene-expression assay in clinical practice – performance and impact on clinical decisions. PLoS One (2013) 8(6):e68252. doi:10.1371/journal.pone.0068252
111. Buus R, Sestak I, Kronenwett R, Denkert C, Dubsky P, Krappmann K, et al. Comparison of EndoPredict and EPclin with oncotype dx recurrence score for prediction of risk of distant recurrence after endocrine therapy. J Natl Cancer Inst (2016) 108(11):djw149. doi:10.1093/jnci/djw149
112. Martin M, Brase JC, Ruiz A, Prat A, Kronenwett R, Calvo L, et al. Prognostic ability of EndoPredict compared to research-based versions of the PAM50 risk of recurrence (ROR) scores in node-positive, estrogen receptor-positive, and HER2-negative breast cancer. A GEICAM/9906 sub-study. Breast Cancer Res Treat (2016) 156(1):81–9. doi:10.1007/s10549-016-3725-z
113. van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature (2002) 415(6871):530–6. doi:10.1038/415530a
114. Bueno-de-Mesquita JM, Linn SC, Keijzer R, Wesseling J, Nuyten DSA, van Krimpen C, et al. Validation of 70-gene prognosis signature in node-negative breast cancer. Breast Cancer Res Treat (2009) 117(3):483–95. doi:10.1007/s10549-008-0191-2
115. Buyse M, Loi S, van’t Veer L, Viale G, Delorenzi M, Glas AM, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst (2006) 98(17):1183–92. doi:10.1093/jnci/djj329
116. Straver ME, Glas AM, Hannemann J, Wesseling J, van de Vijver MJ, Rutgers EJ, et al. The 70-gene signature as a response predictor for neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat (2010) 119(3):551–8. doi:10.1007/s10549-009-0333-1
117. Knauer M, Mook S, Rutgers EJ, Bender RA, Hauptmann M, van de Vijver MJ, et al. The predictive value of the 70-gene signature for adjuvant chemotherapy in early breast cancer. Breast Cancer Res Treat (2010) 120(3):655–61. doi:10.1007/s10549-010-0814-2
118. Bueno-de-Mesquita JM, van Harten WH, Retel VP, van’t Veer LJ, van Dam FS, Karsenberg K, et al. Use of 70-gene signature to predict prognosis of patients with node-negative breast cancer: a prospective community-based feasibility study (RASTER). Lancet Oncol (2007) 8(12):1079–87. doi:10.1016/S1470-2045(07)70346-7
119. Drukker CA, Bueno-de-Mesquita JM, Retel VP, van Harten WH, van Tinteren H, Wesseling J, et al. A prospective evaluation of a breast cancer prognosis signature in the observational RASTER study. Int J Cancer (2013) 133(4):929–36. doi:10.1002/ijc.28082
120. Wittner BS, Sgroi DC, Ryan PD, Bruinsma TJ, Glas AM, Male A, et al. Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin Cancer Res (2008) 14(10):2988–93. doi:10.1158/1078-0432.CCR-07-4723
121. Mook S, Schmidt MK, Viale G, Pruneri G, Eekhout I, Floore A, et al. The 70-gene prognosis-signature predicts disease outcome in breast cancer patients with 1-3 positive lymph nodes in an independent validation study. Breast Cancer Res Treat (2009) 116(2):295–302. doi:10.1007/s10549-008-0130-2
122. Mook S, Schmidt MK, Weigelt B, Kreike B, Eekhout I, van de Vijver MJ, et al. The 70-gene prognosis signature predicts early metastasis in breast cancer patients between 55 and 70 years of age. Ann Oncol (2010) 21(4):717–22. doi:10.1093/annonc/mdp388
123. Drukker CA, van Tinteren H, Schmidt MK, Rutgers EJ, Bernards R, van de Vijver MJ, et al. Long-term impact of the 70-gene signature on breast cancer outcome. Breast Cancer Res Treat (2014) 143(3):587–92. doi:10.1007/s10549-013-2831-4
124. Cardoso F, van’t Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med (2016) 375(8):717–29. doi:10.1056/NEJMoa1602253
125. Zhang Y, Schnabel CA, Schroeder BE, Jerevall PL, Jankowitz RC, Fornander T, et al. Breast cancer index identifies early-stage estrogen receptor-positive breast cancer patients at risk for early- and late-distant recurrence. Clin Cancer Res (2013) 19(15):4196–205. doi:10.1158/1078-0432.CCR-13-0804
126. Sgroi DC, Sestak I, Cuzick J, Zhang Y, Schnabel CA, Schroeder B, et al. Prediction of late distant recurrence in patients with oestrogen-receptor-positive breast cancer: a prospective comparison of the breast-cancer index (BCI) assay, 21-gene recurrence score, and IHC4 in the TransATAC study population. Lancet Oncol (2013) 14(11):1067–76. doi:10.1016/S1470-2045(13)70387-5
127. Sgroi DC, Chapman JA, Badovinac-Crnjevic T, Zarella E, Binns S, Zhang Y, et al. Assessment of the prognostic and predictive utility of the Breast Cancer Index (BCI): an NCIC CTG MA.14 study. Breast Cancer Res (2016) 18(1):1. doi:10.1186/s13058-015-0660-6
128. Jerevall PL, Ma XJ, Li H, Salunga R, Kesty NC, Erlander MG, et al. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br J Cancer (2011) 104(11):1762–9. doi:10.1038/bjc.2011.145
129. Jerevall PL, Brommesson S, Strand C, Gruvberger-Saal S, Malmstrom P, Nordenskjold B, et al. Exploring the two-gene ratio in breast cancer – independent roles for HOXB13 and IL17BR in prediction of clinical outcome. Breast Cancer Res Treat (2008) 107(2):225–34. doi:10.1007/s10549-007-9541-8
130. Ma XJ, Salunga R, Dahiya S, Wang W, Carney E, Durbecq V, et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin Cancer Res (2008) 14(9):2601–8. doi:10.1158/1078-0432.CCR-07-5026
131. Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell (2004) 5(6):607–16. doi:10.1016/j.ccr.2004.05.015
132. Jankowitz RC, Cooper K, Erlander MG, Ma XJ, Kesty NC, Li H, et al. Prognostic utility of the breast cancer index and comparison to Adjuvant! Online in a clinical case series of early breast cancer. Breast Cancer Res (2011) 13(5):R98. doi:10.1186/bcr3038
133. Mathieu MC, Mazouni C, Kesty NC, Zhang Y, Scott V, Passeron J, et al. Breast Cancer Index predicts pathological complete response and eligibility for breast conserving surgery in breast cancer patients treated with neoadjuvant chemotherapy. Ann Oncol (2012) 23(8):2046–52. doi:10.1093/annonc/mdr550
134. Habel LA, Sakoda LC, Achacoso N, Ma XJ, Erlander MG, Sgroi DC, et al. HOXB13:IL17BR and molecular grade index and risk of breast cancer death among patients with lymph node-negative invasive disease. Breast Cancer Res (2013) 15(2):R24. doi:10.1186/bcr3402
135. Sanft T, Aktas B, Schroeder B, Bossuyt V, DiGiovanna M, Abu-Khalaf M, et al. Prospective assessment of the decision-making impact of the Breast Cancer Index in recommending extended adjuvant endocrine therapy for patients with early-stage ER-positive breast cancer. Breast Cancer Res Treat (2015) 154(3):533–41. doi:10.1007/s10549-015-3631-9
136. Ricketts D, Turnbull L, Ryall G, Bakhshi R, Rawson NS, Gazet JC, et al. Estrogen and progesterone receptors in the normal female breast. Cancer Res (1991) 51(7):1817–22.
137. Soderqvist G, von Schoultz B, Tani E, Skoog L. Estrogen and progesterone receptor content in breast epithelial cells from healthy women during the menstrual cycle. Am J Obstet Gynecol (1993) 168(3 Pt 1):874–9. doi:10.1016/S0002-9378(12)90837-6
138. Atalay C, Kanlioz M, Altinok M. Menstrual cycle and hormone receptor status in breast cancer patients. Neoplasma (2002) 49(4):278.
139. Pujol P, Daures JP, Brouillet JP, Chang S, Rouanet P, Bringer J, et al. A prospective prognostic study of the hormonal milieu at the time of surgery in premenopausal breast carcinoma. Cancer (2001) 91(10):1854–61. doi:10.1002/1097-0142(20010515)91:10<1854::AID-CNCR1206>3.3.CO;2-P
140. Dekker TJ, Smit VT, Hooijer GK, Van de Vijver MJ, Mesker WE, Tollenaar RA, et al. Reliability of core needle biopsy for determining ER and HER2 status in breast cancer. Ann Oncol (2013) 24(4):931–7. doi:10.1093/annonc/mds599
141. Jeselsohn RM, Werner L, Regan MM, Fatima A, Gilmore L, Collins LC, et al. Digital quantification of gene expression in sequential breast cancer biopsies reveals activation of an immune response. PLoS One (2013) 8(5):e64225. doi:10.1371/journal.pone.0064225
142. Riis ML, Luders T, Markert EK, Haakensen VD, Nesbakken AJ, Kristensen VN, et al. Molecular profiles of pre- and postoperative breast cancer tumours reveal differentially expressed genes. ISRN Oncol (2012) 2012:450267. doi:10.5402/2012/450267
143. Lopez-Knowles E, Gao Q, Cheang MC, Morden J, Parker J, Martin LA, et al. Heterogeneity in global gene expression profiles between biopsy specimens taken peri-surgically from primary ER-positive breast carcinomas. Breast Cancer Res (2016) 18(1):39. doi:10.1186/s13058-016-0696-2
144. Hosoda M, Yamamoto M, Nakano K, Hatanaka KC, Takakuwa E, Hatanaka Y, et al. Differential expression of progesterone receptor, FOXA1, GATA3, and p53 between pre- and postmenopausal women with estrogen receptor-positive breast cancer. Breast Cancer Res Treat (2014) 144(2):249–61. doi:10.1007/s10549-014-2867-0
145. Haynes BP, Viale G, Galimberti V, Rotmensz N, Gibelli B, A’Hern R, et al. Expression of key oestrogen-regulated genes differs substantially across the menstrual cycle in oestrogen receptor-positive primary breast cancer. Breast Cancer Res Treat (2013) 138(1):157–65. doi:10.1007/s10549-013-2426-0
Keywords: premenopausal breast cancer, intrinsic subtyping, menstrual cycle, gene expression, hormones
Citation: Bernhardt SM, Dasari P, Walsh D, Townsend AR, Price TJ and Ingman WV (2016) Hormonal Modulation of Breast Cancer Gene Expression: Implications for Intrinsic Subtyping in Premenopausal Women. Front. Oncol. 6:241. doi: 10.3389/fonc.2016.00241
Received: 30 August 2016; Accepted: 27 October 2016;
Published: 14 November 2016
Edited by:
José - Bines, Instituto Nacional de Câncer, BrazilReviewed by:
Tomas Reinert, Instituto do Câncer Mãe de Deus e Instituto DeVita, BrazilPaul N. Mainwaring, ICON Cancer Care, Australia
Copyright: © 2016 Bernhardt, Dasari, Walsh, Townsend, Price and Ingman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wendy V. Ingman, d2VuZHkuaW5nbWFuQGFkZWxhaWRlLmVkdS5hdQ==
 Sarah M. Bernhardt
Sarah M. Bernhardt Pallave Dasari1,2
Pallave Dasari1,2 Timothy J. Price
Timothy J. Price Wendy V. Ingman
Wendy V. Ingman